Abstract
Tat stimulates human immunodeficiency virus type 1 (HIV-1) transcriptional elongation by recruitment of carboxyl-terminal domain (CTD) kinases to the HIV-1 promoter. Using an immobilized DNA template assay, we have analyzed the effect of Tat on kinase activity during the initiation and elongation phases of HIV-1 transcription. Our results demonstrate that cyclin-dependent kinase 7 (CDK7) (TFIIH) and CDK9 (P-TEFb) both associate with the HIV-1 preinitiation complex. Hyperphosphorylation of the RNA polymerase II (RNAP II) CTD in the HIV-1 preinitiation complex, in the absence of Tat, takes place at CTD serine 2 and serine 5. Analysis of preinitiation complexes formed in immunodepleted extracts suggests that CDK9 phosphorylates serine 2, while CDK7 phosphorylates serine 5. Remarkably, in the presence of Tat, the substrate specificity of CDK9 is altered, such that the kinase phosphorylates both serine 2 and serine 5. Tat-induced CTD phosphorylation by CDK9 is strongly inhibited by low concentrations of 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole, an inhibitor of transcription elongation by RNAP II. Analysis of stalled transcription elongation complexes demonstrates that CDK7 is released from the transcription complex between positions +14 and +36, prior to the synthesis of transactivation response (TAR) RNA. In contrast, CDK9 stays associated with the complex through +79. Analysis of CTD phosphorylation indicates a biphasic modification pattern, one in the preinitiation complex and the other between +36 and +79. The second phase of CTD phosphorylation is Tat-dependent and TAR-dependent. These studies suggest that the ability of Tat to increase transcriptional elongation may be due to its ability to modify the substrate specificity of the CDK9 complex.
Human immunodeficiency virus type 1 (HIV-1) encodes a transactivator protein, Tat, which stimulates transcription elongation through interaction with the transactivation response (TAR) RNA element located at the 5′ end of nascent transcripts (12, 28, 68, 75). In view of the observations that hyperphosphorylation of the carboxyl-terminal domain (CTD) of the large subunit of RNA polymerase II (RNAP II) correlates with the formation of processive elongation complexes (11) and that Tat transactivation requires the CTD (6, 44, 46, 79), it has been proposed that a critical step in Tat transactivation is mediated through a cellular kinase(s) (68, 80). Two cyclin-dependent kinase (CDK)–cyclin pairs, present in two distinct transcription factor complexes, have been implicated as Tat cofactors which could phosphorylate the CTD (54, 68). TFIIH, a general transcription factor which contains nine polypeptides (ERCC3/XPB, ERCC2/XPD, p62, p54, p44, CDK7 [MO15], cyclin H, MAT1, and p34) (13, 24), possesses CTD kinase activity (14, 37). The kinase activity of TFIIH resides in the CDK7 subunit (15, 58, 61, 62). In association with cyclin H and Mat1, CDK7 forms the CDK-activating kinase (CAK) complex that phosphorylates CDKs involved in the regulation of the cell cycle (19, 42, 43, 53, 67). The association of CAK with core TFIIH switches its substrate specificity from CDKs to the CTD of RNAP II (57, 81). Interestingly, the yeast homologue of CDK7, Kin28, is found only in a complex with TFIIH and is devoid of CAK activity (8).
The second CTD kinase, TAK (Tat-associated kinase), was first reported by Herrmann and Rice (22). It was later found that the human positive transcription elongation factor complex called P-TEFb, first identified in and purified from Drosophila extracts, is actually equivalent to TAK (41, 85). P-TEFb most likely functions by phosphorylating RNAP II CTD and preventing polymerase arrest (40). Cloning and sequence analysis of the small subunit of the Drosophila P-TEFb complex revealed its extensive sequence identity (72%) to a previously identified cdc2-related human kinase termed PITALRE (now referred to as CDK9) (18, 85). Importantly, immunodepletion of CDK9 from HeLa nuclear extract eliminated basal transcription elongation and Tat transactivation (39, 84, 85). Further, the addition of the affinity-purified human P-TEFb complex completely restored these two processes (84).
The interaction between Tat and P-TEFb is mediated through human cyclin T1 (51, 76). The function of the Tat–P-TEFb complex is mediated through the high-affinity, loop-specific binding of the Tat–P-TEFb complex to the TAR RNA structure, and the formation of the tripartite complex between Tat, cyclin T1, and TAR depends on the 5′ bulge and central loop in TAR (2, 16, 17, 26, 32, 54, 68, 76). The P-TEFb-associated CDK9 kinase then induces phosphorylation of RNAP II CTD and perhaps of other proteins present in the transcription complex, leading to a transition from nonprocessive to processive transcription. More recent observations indicate that a critical cysteine residue (C261) which is not conserved in the murine cyclin T1 protein (Y261) is important for the formation of the tripartite complex between Tat, cyclin T1, and TAR (2, 16, 17, 26, 32). Interestingly, a reciprocal exchange of a cysteine to a tyrosine at position 261 (C261↔Y261) between human cyclin T1 (hT1) and the murine cyclin T1 (mT1) renders hT1 inactive and mT1 active for human Tat transactivation (2, 16, 17, 26, 32). Thus, the ability of Tat to recruit cyclin T1-CDK9 to TAR not only stimulates HIV-1 transcriptional elongation but also governs the species specificity of HIV-1 Tat transactivation. It was originally proposed that both TFIIH and P-TEFb may act sequentially and in a concerted manner, promoting hyperphosphorylation of RNAP II CTD and increasing polymerase processivity (27, 84). At present, however, the role of TFIIH in HIV-1 transcription is controversial (5).
The mammalian CTD consists of 52 repeats of heptapeptide Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7 and is phosphorylated mostly at serine residues during transcription. However, the exact mechanism of CTD phosphorylation remains elusive (11, 71). Our results indicate that CDK7 (TFIIH) and CDK9 (P-TEFb) both associate with the HIV-1 preinitiation complex (PIC) and function to hyperphosphorylate the CTD of RNAP II. In basal transcription, CDK7 and CDK9 facilitate transcription activity: CDK7 phosphorylates CTD serine 5, and CDK9 phosphorylates CTD serine 2. In the presence of Tat, CDK7 is not required for HIV-1 transcription. Remarkably, Tat modifies the substrate specificity of PIC CDK9, allowing CDK9 to phosphorylate serine 2 and serine 5. The substrate specificity of CDK9 was confirmed using recombinant P-TEFb. In the absence of Tat, P-TEFb phosphorylates the CTD at serine 2. In the presence of Tat, P-TEFb phosphorylates the CTD at positions 2 and 5. Phosphorylation at positions 2 and 5 is sensitive to low concentrations of 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB), an inhibitor of transcription elongation by RNAP II. These observations provide significant insight into the complex process of Tat transactivation and provide the first experimental evidence that Tat modifies the substrate specificity of P-TEFb.
MATERIALS AND METHODS
Antibodies.
Anti-CTD RNAP II monoclonal antibodies 8WG16, H5 (phosphoserine 2), H14 (phosphoserine 5) (49, 69), and anti-Tat monoclonal antibody are products of BAbCO. Anti-CDK9 (PITALRE) antibody was purchased from Biodesign Company. Anti-CDK7, anti-cyclin H, anti-Mat1, anti-p62 (subunits of TFIIH), and anti-CDK8 antibodies are products of Santa Cruz Biotechnology.
Biotinylation of template DNAs.
The wild type, TATA box mutant, and TAR mutant TM26 (4) HIV-1 long terminal repeat (LTR) templates (nucleotides [nt] −110 to +168) were amplified by PCR with the forward primer 5′ biotinylated-TAT GGA TTT ACA AGG GAC TTT C-3′ and the reverse primer 5′-GAT CCG ATT ACT AAA AGG G-3′. The primers were synthesized and biotinylated by Lofstrand.
Expression and purification of recombinant P-TEFb proteins.
The production of recombinant P-TEFb proteins was carried out as described by Peng et al. (51).
Immunodepletion of CDK8, CDK7, and/or CDK9 from HeLa nuclear extract.
HeLa nuclear extract (100 μl) in 0.8 M KCl buffer D (20 mM HEPES [pH 7.9], 15% glycerol, 800 mM KCl, 10 mM MgCl2, 0.2 mM EDTA [pH 8.0], 0.1% NP-40, and 1 mM dithiothreitol [DTT]) was incubated with 20 μl of protein A-Sepharose beads to which anti-CDK8, anti-CDK7 and/or anti-CDK9 had been prebound (10 μg of immunoglobulin G). Antigen-antibody complexes were removed by centrifugation. After the procedure was repeated twice, depleted nuclear extract was dialyzed against 0.1 M KCl buffer D and assayed by Western blot analysis.
Purification of PICs.
Reaction mixtures (30 μl) contained 15 μl of HeLa nuclear extract, 1.0 μg of biotinylated templates and 1.0 μg of poly(dI-dC) in the absence or presence of Tat protein (100 ng). The in vitro transcription (IVT) buffer contained 50 mM KCl, 6.25 mM MgCl2, 20 mM HEPES (pH 7.9), 2 mM DTT, 0.5 mM EDTA (pH 8.0), 10 μM ZnSO4, 10 mM creatine phosphate, 100 μg of creatine kinase/ml, and 8.5% glycerol (1× IVT buffer). After a 30-min incubation at 30°C, streptavidin-coated magnetic beads (Dynabeads; Dynal) preequilibrated in binding buffer (20 mM HEPES [pH 7.9], 80 mM KCl, 10 mM MgCl2, 2 mM DTT, 10 μM ZnSO4, 100 μg of bovine serum albumin/ml, 0.05% NP-40, and 10% glycerol) were then added to the reactions, and the mixtures were further incubated for 30 min at 30°C. The immobilized templates were then harvested using a magnetic stand, and the PICs were washed extensively with 1× IVT buffer. In vitro transcription and Western blot analysis could then be performed using the purified PICs assembled on the immobilized templates.
Western blot analysis of the purified PICs.
The purified PICs assembled on the immobilized templates were heated for 10 min at 100°C in sodium dodecyl sulfate (SDS)-loading buffer. The released proteins were fractionated by electrophoresis on SDS–4 to 20% polyacrylamide gels and then transblotted onto polyvinylidene fluoride (PVDF) membranes (Millipore). CDK9 (a subunit of P-TEFb), CDK7, cyclin H, Mat1, and p62 (subunits of TFIIH), CDK8, and Tat were detected with specific antibodies as indicated above.
In vitro transcription with the purified PICs.
In vitro transcription reactions (100 μl) were set up by resuspending the purified PICs in 100 μl of 1× IVT buffer, 50 μM ATP, 50 μM CTP, 50 μM GTP, 10 μCi of [α-32P]UTP, and 10 units of RNasin (Promega). The transcription reactions were allowed to take place for 60 min at 30°C. The radiolabeled transcripts were fractionated by electrophoresis on 6% denaturing polyacrylamide gels and detected by PhosphorImager.
Kinase reactions in the purified PICs and immunoprecipitation of the phosphorylated RNAP II.
Kinase reactions were performed by mixing the purified PICs with 10 μCi of [γ-32P]ATP in 100 μl of 1× IVT buffer. After an incubation of 10 min at 30°C, the PICs were separated from the supernatants and washed extensively. Five hundred microliters of RIPA buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) was then added into the tubes containing PICs immobilized by streptavidin-coated beads, and the mixtures were incubated for 120 min at 4°C with rocking. The supernatants were saved, and the phosphorylated RNAP II was immunoprecipitated by monoclonal antibody 8WG16.
Kinase reactions in the purified PICs and Western blot analysis of the phosphorylated RNAP II.
Kinase reactions were done by mixing the purified PICs with 50 μM ATP in 100 μl of 1× IVT buffer. After an incubation of 10 min at 30°C, the PICs were separated from the supernatants and heated for 10 min at 100°C in SDS-loading buffer. The released proteins were fractionated on SDS–4% polyacrylamide gels and then transblotted onto PVDF membranes. RNAP II was detected with anti-CTD monoclonal antibody 8WG16, H5, or H14.
DRB sensitivity assay of distinct RNAP II CTD phosphorylation sites.
Biotinylated HIV-1 LTR templates were incubated with CDK7-depleted extract, and the PICs were then purified with streptavidin-coated magnetic beads. For the inhibition assays, the purified PICs were incubated with ATP in the presence of different concentrations of DRB and washed extensively. The PICs were then heated 10 min at 100°C in SDS-loading buffer. The released proteins were fractionated on SDS–4% polyacrylamide gels and then transblotted onto PVDF membranes. Phosphorylated RNAP II was detected with anti-CTD monoclonal antibody H5 or H14. Alternatively, the kinase reaction buffer contained [γ-32P]ATP, and phosphorylated RNAP II was immunoprecipitated by anti-CTD monoclonal antibody H5 or H14. The activity was determined by direct quantitation using the Molecular Dynamics ImageQuant system.
Stepwise transcription.
The purified PICs were incubated with 50 μM ATP for 10 min and then washed extensively with 1× IVT buffer. The PICs were walked to position U14 by incubation with 50 μM CTP, GTP, and UTP for 5 min at 30°C and then washed extensively with 1× IVT buffer. The transcriptional elongation complexes (TECs) stalled at U14 were walked stepwise along the DNA by repeated incubation with different sets of three nucleoside triphosphates (NTPs) and then washed extensively with 1× IVT buffer to remove the unincorporated NTPs. When indicated, RNA transcripts were labeled with [α-32P]UTP and analyzed on 15% denaturing polyacrylamide gels. To analyze the components of TECs, the TECs were elongated with cold NTPs. The TECs that were stalled at different stages were analyzed by Western blot. To detect phosphorylation of RNAP II CTD during transcription, the phosphorylated CTD was labeled with [γ-32P]ATP during stepwise transcription.
CTD kinase assay.
The CTD kinase assays were performed by mixing 100 ng of glutathione S-transferase (GST)–CTD, 100 ng of P-TEFb, 10 μM ATP, and 10 μCi of [γ-32P]ATP in the absence or presence of Tat and incubating for 60 min at 23°C. The total reaction volume was 30 μl, and the final conditions were 50 mM Tris-HCl (pH 7.5), 5 mM DTT, 5 mM MnCl2, 4 mM MgCl2, and 10 μM ZnSO4. The phosphorylated GST-CTD was then immunoprecipitated with anti-CTD monoclonal antibody H5 or H14 and fractionated by electrophoresis on SDS–8% polyacrylamide gels. The labeled products were detected by PhosphorImager.
RESULTS
Tat stimulates the formation of transcriptionally active PICs on the biotinylated HIV-1 LTR templates.
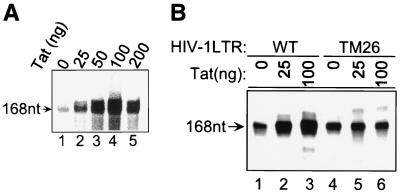
We have used an immobilized template assay to isolate basal and Tat PICs and study their transcription and CTD kinase activities. We first demonstrated that Tat specifically activates transcription. HIV-1 LTR promoter templates were 5′ end labeled with biotin at position −110 as described in Materials and Methods. The biotinylated templates were incubated with HeLa nuclear extract in the absence or presence of Tat. PICs were subsequently isolated using streptavidin-coated magnetic beads, and in vitro transcription assays were performed. In the absence of Tat, a low level of basal HIV-1 transcription was observed (Fig. 1A, lane 1). The addition of increasing amounts of Tat protein to the preincubation mixture significantly increased transcription from the HIV-1 promoter (Fig. 1A, lanes 2 to 5). Optimum Tat transactivation was observed when approximately 100 ng of Tat was added to the reaction mixture (Fig. 1A, lane 4). The addition of the adenosine analogue DRB to the transcription reaction mixture inhibited Tat transactivation (data not shown).
FIG. 1.
Tat-stimulated transcription from HIV-1 LTR. In vitro transcription reactions were performed with the purified PICs, and the transcripts were labeled with [α-32P]UTP. The runoff transcripts are 168 nt, as indicated. (A) Tat stimulated transcription from the wild-type HIV-1 LTR. (B) Tat was not able to activate transcription from the TAR mutant (TM26) HIV-1 LTR. A comparison of the transcription activities of wild-type HIV-1 LTR (lanes 1 to 3) and TM26 (lanes 4 to 6) is shown.
To demonstrate the specificity of the in vitro transcription system, we utilized a template which contains a base substitution in the TAR RNA bulge. This mutation knocks out the ability of Tat to bind TAR RNA, inhibiting Tat transactivation in vitro and in vivo (4). Consistent with previous reports, the TAR RNA mutation (TM26) inhibited the ability of Tat to transactivate the template (Fig. 1B, lanes 4 to 6). The TM26 mutation did not significantly affect the level of basal transcription. To further demonstrate the specificity of the Tat transactivation, we utilized Tat mutants with single amino acid substitutions at lysine 41 or cysteine 22. Consistent with previous results, the mutants failed to activate transcription (data not shown).
CDK7 and CDK9 associate with HIV-1 PICs.
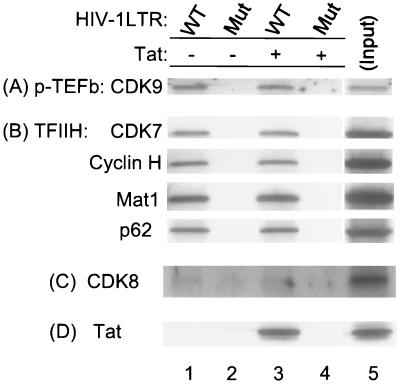
We next analyzed the relative protein composition of PICs formed in the absence or present of Tat, especially with respect to CTD kinases. Biotinylated templates were incubated with HeLa nuclear extract in the absence or presence of Tat as described in Materials and Methods. Parallel binding reactions were carried out with a TATA box mutant HIV-1 LTR template as a negative control for nonspecific binding of protein to the templates. Figure 2 shows the Western blot analysis for P-TEFb (CDK9), TFIIH (CDK7, cyclin H, Mat1, and p62 subunits), CDK8, and Tat. The results of these assays demonstrate that both CDK7 and CDK9 are present in the HIV-1 PICs. Interestingly, the amount of either kinase is equal in the absence or presence of Tat with the wild-type HIV-1 LTR template (Fig. 2A and B, lanes 1 and 3). The appearance of proteins bound to the templates is specific. Parallel assays performed with a TATA box mutant HIV-1 LTR, which is transcriptionally inactive, failed to precipitate proteins associated with either the P-TEFb complex or the TFIIH complex (Fig. 2, lanes 2 and 4). In contrast to the results obtained with CDK7 and CDK9, we find that CDK8 is not present in the HIV-1 PIC in the absence or presence of Tat (Fig. 2C, lanes 1 and 3).
FIG. 2.
CDK7 and CDK9, but not CDK8, are components of the HIV-1 PIC. Association reactions (30 μl) were performed with 15 μl of HeLa nuclear extract, 1.0 μg of biotinylated HIV-1 LTR templates, and 1.0 μg of poly(dI-dC) in the absence (lanes 1 and 2) or presence (lanes 3 and 4) of Tat. PICs were purified with streptavidin-coated magnetic beads. Western blot analysis of the purified HIV-1 PICs was then done with anti-CDK9, anti-CDK7, anti-cyclin H, anti-Mat1, anti-p62, anti-CDK8, and anti-Tat antibodies. An HIV-1 LTR TATA box mutant (Mut) was used as a parallel control (lanes 2 and 4). WT, wild type.
Basal and Tat-activated transcription differ in their requirements for CDK7 and CDK9.
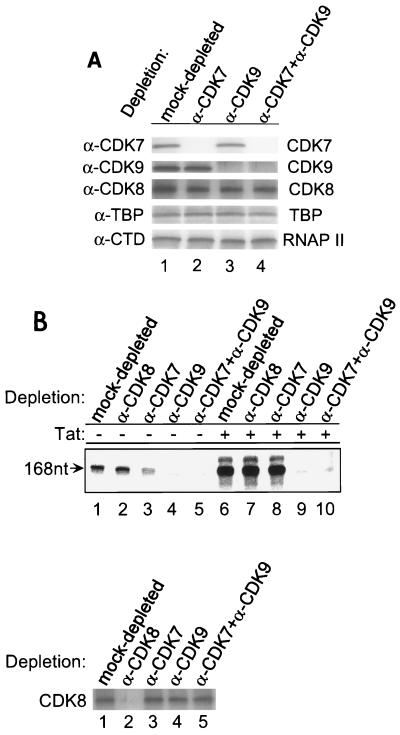
To determine the relative importance of CDK7 and CDK9 in HIV-1 transcription, HeLa nuclear extracts were treated with anti-CDK7 and/or anti-CDK9 antibodies and subsequently used for in vitro transcription assays. Western blot analysis of the antibody-treated extracts demonstrated that CDK7 and CDK9 had been specifically depleted (Fig. 3A, panels 1 and 2). CDK7 was detected in the mock-depleted control (panel 1, lane 1) and CDK9-depleted (lane 3) extracts, but not in the CDK7-depleted extracts (lanes 2 and 4). Similarly, CDK9 was detected in the mock-depleted (panel 2, lane 1) and CDK7-depleted (lane 2) extracts, but not in the CDK9-depleted extracts (lanes 3 and 4). Western blot analysis of the same extracts with anti-CDK8, anti-TBP, or anti-RNAP II antibody demonstrated the specificity of the clearing, since no change in the level of CDK8, TBP, or RNAP II was observed in the immunodepleted extracts (Fig. 3A, panels 3, 4, and 5).
FIG. 3.
The effects of the CTD kinase activities of CDK7 and CDK9 on HIV-1 transcription. (A) Western blot analysis of mock-depleted, CDK7-depleted, CDK9-depleted, or CDK7- and CDK9-depleted extracts with anti-CDK7, anti-CDK9, anti-CDK8, anti-TBP, or anti-CTD of RNAP II antibody. Panels 3, 4, and 5 demonstrate that depletions did not change the level of CDK8, other general transcription factors, or RNAP II. (B) The effects of the CTD kinase activities of CDK7 and CDK9 on HIV-1 transcription. Biotinylated HIV-1 LTR templates were incubated with mock-depleted, CDK8-depleted, CDK7-depleted, CDK9-depleted, and CDK7- and CDK9-depleted extracts in the absence (lanes 1 to 5) or presence (lanes 6 to 10) of Tat, and PICs were then purified with streptavidin-coated magnetic beads. In vitro transcription was done with the purified PICs, and transcripts were labeled with [α-32P]UTP and fractionated on 6% denaturing polyacrylamide gel containing 7 M urea in 1× TBE buffer (top). The Western blot analysis of kinase-depleted extracts was done with anti-CDK8 antibody (bottom).
Biotinylated HIV-1 LTR templates were incubated with the mock-depleted, CDK8-depleted, CDK7-depleted, CDK9-depleted, and CDK7- and CDK9-depleted extracts in the absence or presence of Tat, and PICs were purified with streptavidin-coated magnetic beads. First, in vitro transcription reactions were performed with the purified PICs, and the result is shown in Fig. 3B (top). In the absence of Tat, the depletion of CDK7 or CDK9 decreased HIV-1 transcription approximately 3- and 10-fold, respectively (lanes 1 to 4). In the presence of Tat, no quantitative decrease in transcription was observed in the absence of CDK7 (lanes 6 and 8). In contrast, CDK9 depletion decreased Tat transactivation by more than 16-fold to essentially background levels (lanes 6 and 9). When both CDK7 and CDK9 were depleted from the extract, the basal transcription and Tat transactivation were eliminated (lanes 5 and 10).
It has been reported that CDK8 has in vitro CTD phosphorylation activity. To investigate whether CDK8 functions in HIV-1 transcription, in vitro transcription was also performed with the CDK8-depleted extract (Fig. 3B, bottom). The results of this experiment demonstrate that CDK8 depletion does not affect either basal transcription or Tat transactivation (Fig. 3B, top, lanes 1, 2, 6, and 7). Consistent with these results, the experiments presented above and in Fig. 2C demonstrate that CDK8 is barely detected in HIV-1 PICs (Fig. 2, compare with CDK7 and CDK9). It is also important to point out that CDK7 and/or CDK9 depletions did not affect the level of CDK8 in extracts (Fig. 3B, bottom). These results suggest that CDK8 does not play a critical role in HIV-1 transcription.
CDK7 and CDK9 phosphorylate the CTD in the HIV-1 PICs.
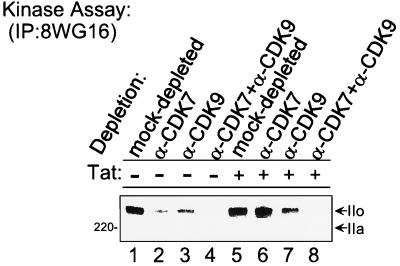
We next analyzed the effect of CDK7 and CDK9 on phosphorylation of RNAP II in the HIV-1 PICs. Consistent with the recent publication by Isel and Karn (25), the presence or absence of Tat does not affect the extent of CTD phosphorylation in the PICs. Equal CTD phosphorylation and conversion of RNAP II from form IIa to form IIo were observed in both the basal and Tat PICs (Fig. 4, lanes 1 and 5). The migration position of RNAP II form IIa was determined by Western blot analysis of HeLa nuclear extracts.
FIG. 4.
Phosphorylation of RNAP II CTD in HIV-1 PICs. Biotinylated HIV-1 LTR templates were incubated with mock-depleted, CDK7-depleted, CDK9-depleted, or CDK7- and CDK9-depleted extracts in the absence (lanes 1 to 4) or presence (lanes 5 to 8) of Tat, and PICs were then purified with streptavidin-coated magnetic beads. Kinase reactions were performed with the purified PICs, and phosphorylated RNAP II was labeled with [γ-32P]ATP and immunoprecipitated (IP) with anti-CTD monoclonal antibody 8WG16.
Hyperphosphorylation of the CTD in the basal transcription complex was decreased approximately twofold by depletion of either CDK7 or CDK9 (Fig. 4, lanes 1 to 3). In the presence of Tat, no detectable quantitative difference in hyperphosphorylation of the RNAP II CTD was observed in PICs from extract from which CDK7 was immunodepleted (compare lane 6 with lane 5). However, immunodepletion of CDK9 decreased the hyperphosphorylation of the RNAP II CTD by approximately twofold (lane 7). Depletion of both CDK7 and CDK9 from the HeLa extract decreased hyperphosphorylation of the CTD in the PICs 10-fold (lanes 4 and 8). This result demonstrates that both the CDK7 and CDK9 kinases phosphorylate the RNAP II CTD in the HIV-1 transcription complex. It is important to point out that the extent of CTD phosphorylation may not strictly correlate with transcription, in part because it is difficult to determine the number of active PICs under different experimental conditions.
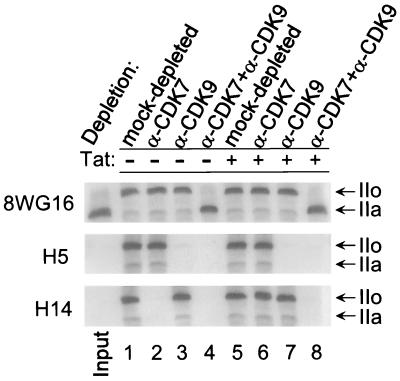
CDK9 and CDK7, respectively, phosphorylate serine 2 and serine 5 of the RNAP II CTD in HIV-1 PICs.
The RNAP II CTD is composed of multiple repeats of the heptapeptide sequence Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7 (11, 45, 71). Patturajan et al. (49) have described monoclonal antibodies which recognize different phosphoamino acid epitopes on the CTD. To investigate the specificity of CTD phosphorylation, we have compared the phosphorylation pattern of the RNAP II CTD in HIV-1 PICs with antibodies specific for phosphorylated serine 2 (H5 antibody) and serine 5 (H14 antibody). Monoclonal antibody 8WG16 was used to detect both the unphosphorylated IIa and phosphorylated IIo forms of RNAP II. Incubation of the PICs with ATP resulted in the conversion of RNAP II from the IIa to the IIo form (Fig. 5, 8WG16 antibody, input versus lanes 1 and 5). With parallel assays run with extracts with either CDK7 or CDK9 depleted, demonstrated in the basal HIV-1 transcription initiation complex, serine 2 was phosphorylated by CDK9 (H5 antibody, lanes 2 and 3), while serine 5 was phosphorylated by CDK7 (H14 antibody, lanes 2 and 3). Depletion of both CDK7 and CDK9 from the extracts abolished the phosphorylation of the CTD in the HIV-1 PIC (Fig. 5, lane 4).
FIG. 5.
CDK9 and CDK7 phosphorylated serine 2 and serine 5, respectively, of the RNAP II CTD in HIV-1 PICs. Biotinylated HIV-1 LTR templates were incubated with mock-depleted, CDK7-depleted, CDK9-depleted, or CDK7- and CDK9-depleted extracts in the absence (lanes 1 to 4) or presence (lanes 5 to 8) of Tat. PICs were then purified with streptavidin-coated magnetic beads. The PICs were then incubated with 50 μM ATP for 10 min in order to have RNAP II CTD phosphorylated and washed extensively. Western blot analysis of the PICs was done with anti-CTD monoclonal antibodies 8WG16, H5, or H14.
FIG. 8.
The recruitment of the Tat–P-TEFb complex by TAR binding during elongation. Biotinylated wild-type (WT) or TAR mutant (TM26) HIV-1 LTR templates were incubated with CDK9-depleted extract, and PICs were purified with streptavidin-coated magnetic beads. The transcription complexes were walked stepwise along the templates to +79 (as described in Materials and Methods). The runoff transcription was then performed by incubating the TECs stalled at +79 with ATP, CTP, GTP, and [α-32P]UTP. P-TEFb and Tat were added at different sites, as indicated.
The analysis of kinase activity in HIV-1 PICs formed in the presence of Tat provided evidence that the substrate specificity of CDK9 is altered in the Tat complexes. As with the results obtained with the basal HIV-1 PICs, CDK9 was required to phosphorylate serine 2 (Fig. 5, middle, lanes 6 and 7). Remarkably, the phosphorylation of serine 5 was maintained in the HIV-1 Tat PICs assembled from CDK7-depleted extracts (Fig. 5, bottom, compare lane 2 with lane 6). Since the serine 5 kinase activity was lost when CDK9 was also depleted from the extract (Fig. 5, bottom, lane 8), this result suggests that CDK9 phosphorylates serine 5 in the HIV-1 Tat transcription complex. In the absence of Tat, CDK9 phosphorylates only serine 2. It should be stressed that these studies, which clearly show the specificity of CDK9 kinase activity in the absence or presence of Tat, are done in the context of the transcription complex and the authentic substrate, the CTD of RNAP II.
CTD phosphorylation by Tat-modified CDK9 is sensitive to DRB.
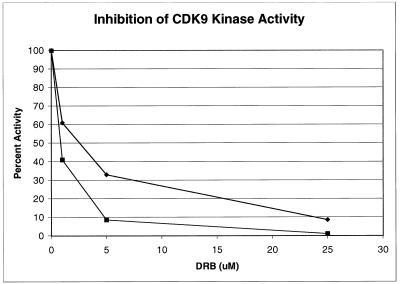
Tat transactivation has been shown to be preferentially sensitive to the adenosine analogue DRB, which inhibits RNAP II elongation. We next, therefore, tested the sensitivity of serine 2 and serine 5 phosphorylation to DRB. CDK7-depleted extract specific for CDK9 serine 2 and serine 5 kinase activities was used in the inhibition assay. The results of this experiment demonstrate that serine 2 and serine 5 phosphorylation by Tat-induced CDK9 is sensitive to low concentrations of DRB. At a concentration of 5 μM DRB, serine 2 phosphorylation and serine 5 phosphorylation were inhibited by approximately 70 or 90%, respectively (Fig. 6).
FIG. 6.
CTD phosphorylation by Tat-modified CDK9 is sensitive to DRB. Biotinylated HIV-1 LTR templates were incubated with CDK7-depleted extract in the presence of Tat, and PICs were then purified with streptavidin-coated magnetic beads. The inhibition assays were performed by incubating the purified PICs with ATP in the presence of different concentrations of DRB. Western blot analyses of the complexes were done with anti-CTD monoclonal antibody H5 or H14, and the activities were determined by direct quantitation using the Molecular Dynamics ImageQuant. The top curve (♦) indicates the inhibition of serine 2 phosphorylation, while the bottom curve (▪) indicates the inhibition of serine 5 phosphorylation.
CDK7 is released between positions +14 and +36, while CDK9 remains stably associated with the HIV-1 TECs.
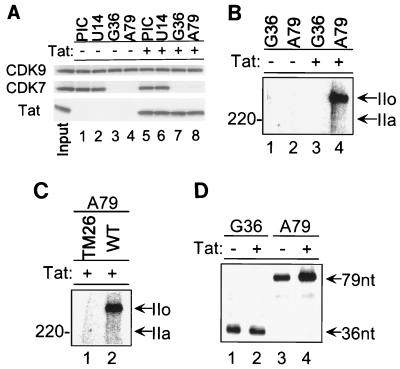
Since CDK7 and Tat-induced CDK9 phosphorylate serine 5, the ability of Tat to modify the substrate specificity of the CDK9 complex would most likely become important when CDK7 was released from the transcription complex. Interestingly, on other polymerase II promoters, TFIIH is released once the polymerase has traveled approximately 30 nt (83). To see whether the same phenomenon was observed with the HIV-1 transcription complex, we used the immobilized template assay and isolated TECs stalled at +14, +36, or +79 (see Materials and Methods). The protein composition of transcription complexes was subsequently analyzed by Western blotting. The results of this experiment demonstrate that CDK7 is released from the template between +14 and +36 (Fig. 7A). The absence or presence of Tat does not affect the stability of TFIIH (CDK7) with the template. In contrast, CDK9 is stably associated with the TECs through position +79 (Fig. 7A). The amount of CDK9 associated with the complex is not modified by the presence or absence of Tat.
FIG. 7.
Stepwise walking of RNAP II elongation complexes and TAR-dependent rephosphorylation of RNAP II CTD during elongation. The purified PICs were incubated with 50 μM ATP for 10 min and then washed extensively. The PICs were walked to position U14 by incubation with 50 μM CTP, GTP, and UTP for 5 min at 30°C and then washed extensively. The TECs stalled at U14 were walked stepwise along the DNA by repeated incubation with different sets of three NTPs and then washed extensively to remove the unincorporated NTPs. (A) Western blot analysis of PICs and stalled TECs. (B and C) TAR-dependent rephosphorylation of RNAP II CTD during elongation. (D) Transcription facilitated by TAR-dependent rephosphorylation of RNAP II CTD during elongation.
A second phase of RNAP II CTD phosphorylation occurs between +36 and +79.
The above-described results suggest that Tat-induced phosphorylation of serine 5 by CDK9 might be important after transcription has reached the +36 position, at which time CDK7 has been released from the complex. To analyze phosphorylation of the RNAP II CTD during transcription elongation, PICs were first incubated with cold ATP. The transcription complexes were then stalled at various positions downstream of the RNA initiation site by sequential incubation with different combinations of three NTPs. Kinase reactions were subsequently performed in the presence of [γ-32P]ATP. RNAP II was immunoprecipitated from the reactions and analyzed by SDS gel electrophoresis. The results of this study provided two very important observations. First, in the transcription complex assembled in the absence of Tat, there was no subsequent phosphorylation of RNAP II once the elongation complex had reached +36 to +79 (Fig. 7B, lanes 1 and 2). Second, in the presence of Tat there was an additional phase of CTD phosphorylation that occurred between nt +36 and +79 (Fig. 7B, lanes 3 and 4). This phosphorylation must be due to CDK9 activity, since CDK7 had been released from the template.
Of importance, the secondary phase of CTD phosphorylation also correlates with the synthesis of TAR RNA structure that has the binding site for Tat. The TM26 HIV-1 LTR carries base substitutions in the bulge of TAR structure which have been shown to inhibit Tat interaction with the RNA (4). When RNA was synthesized from this template, no postinitiation phosphorylation of RNAP II was detected (Fig. 7C).
Finally, the results presented in Fig. 7D suggest that the second phase of RNAP II CTD phosphorylation correlates with Tat transactivation. HIV-1 transcription PICs were isolated and incubated in the presence of sequential nucleotide mixtures to stall complexes at different stages of transcription. [α-32P]UTP was added to the transcription reactions to label the nascent RNA. Tat did not have any detectable effect on transcription through the first 36 bases (lanes 1 and 2). In contrast, when the complexes were allowed to synthesize the TAR RNA enhancer and proceed to nt +79, an increase in the level of transcription was observed in the presence of Tat (Fig. 7D, lanes 3 and 4) which represents the initial stages of Tat transactivation. The rather modest increase in [α-32P]UTP incorporation is due to the fact that we are only analyzing transcription of 43 bases of RNA. Importantly, the sizes of the 36- and 79-base RNA transcripts confirm that the transcription complexes are indeed stalled at the indicated sites on the template DNA.
The recruitment of the Tat–P-TEFb complex by TAR stimulates HIV-1 transcription elongation.
To investigate whether the Tat–P-TEFb complex can be recruited after initiation to rescue transcription from the stalled transcription complexes, the experiment was designed as indicated in the legend to Fig. 8. Biotinylated wild-type or TAR mutant (TM26) HIV-1 LTR templates were incubated with CDK9-depleted extract, and the PICs were purified with streptavidin-coated magnetic beads. The transcription complexes were then walked stepwise along the templates to +79 (as described in Materials and Methods). The runoff transcription was performed by incubating the TECs stalled at +79 with ATP, CTP, GTP, and [α-32P]UTP. P-TEFb and Tat were added at different sites as indicated in Fig. 8. Interestingly, the results presented here imply that P-TEFb can indeed be recruited during elongation as a complex with Tat (lanes 10 to 12) and that the recruitment of the Tat–P-TEFb complex was TAR dependent (lanes 4 to 12). It should be noted that the level of transcription facilitated by the recruitment of Tat–P-TEFb through TAR binding during elongation was significantly lower than that observed when Tat and P-TEFb were present during the assembly of the PICs (lanes 12 and 15). These results suggest that while Tat–P-TEFb may enter at a later point, perhaps the most efficient entry is during the PIC formation, so that efficient conversion to the elongation complex is achieved.
Tat directly modifies the substrate specificity of CDK9 in the recombinant P-TEFb complex.
The above-described experiments suggest that Tat modifies the substrate specificity of the CDK9-containing P-TEFb complex. To demonstrate this point directly, CTD kinase assays were performed with recombinant P-TEFb (Fig. 9A) (51). Following the kinase reaction, equal aliquots of the same kinase assay were immunoprecipitated with phosphoserine 2 (H5)- or phosphoserine 5 (H14)-specific antibody. The results presented in Fig. 9B demonstrate several important points about the functional interaction between Tat and CDK9. First, in the absence of Tat, CDK9 phosphorylates the CTD at serine 2 but not at serine 5 (Fig. 9B, lane 1). In the presence of Tat, a slight increase in the level of serine 2 phosphorylation was observed. The addition of 200 ng of Tat increased serine 2 phosphorylation approximately twofold (top panel, lane 8). The addition of a Tat mutant containing an amino acid substitution at cysteine 22 failed to increase CDK9 activity (Fig. 9B, top, compare lane 1 with lanes 2 to 5).
FIG. 9.
Tat directly modified the substrate specificity of P-TEFb in a CTD kinase assay. (A) A silver-stained SDS-polyacrylamide gel electrophoresis of recombinant P-TEFb fractions. (B) CTD kinase assay. The assays were performed by mixing 100 ng of GST-CTD, 100 ng of P-TEFb, 10 μM ATP, and 10 μCi of [γ-32P]ATP in the absence or presence of Tat and incubating for 60 min at 23°C. The total reaction mixture volume was 30 μl, and the final conditions were 50 mM Tris-HCl (pH 7.5), 5 mM DTT, 5 mM MnCl2, 4 mM MgCl2, and 10 μM ZnSO4. The phosphorylated GST-CTD was then immunoprecipitated (IP) with anti-CTD monoclonal antibody H5 (top) or H14 (bottom) and fractionated by electrophoresis on SDS–8% polyacrylamide gels. Numbers at left represent molecular masses in kilodaltons. The labeled products were detected by PhosphorImager. Numbers at the top show the amounts of Tat (GST-Tat 72) or a Tat mutant (GST-Tat72Cys22) that were added, expressed in nanograms. Lane M, molecular mass marker. (C) DRB sensitivity assay of serine 2 phosphorylation (lanes 1 to 4) and serine 5 phosphorylation (lanes 5 to 8). Micromolar concentrations of DRB are expressed. Numbers at left, molecular masses in kilodaltons.
Remarkably, in the presence of Tat, CDK9 was also able to phosphorylate serine 5 (bottom, compare lane 1 with lanes 6 to 9). Importantly, the Tat mutant Cys22 failed to activate serine 5 phosphorylation (bottom, lanes 2 to 4). The fold increase in Tat-induced serine 5 phosphorylation is difficult to calculate, since there is no serine 5 phosphorylation in the absence of Tat. However, the level of serine 5 phosphorylation was equivalent to the level of serine 2 phosphorylation. In results similar to those presented in Fig. 6, the addition of low concentrations of DRB to the kinase reactions inhibited serine 2 and serine 5 phosphorylation by Tat-induced CDK9 (Fig. 9C). These results provide direct evidence that Tat modifies the substrate specificity of the CDK9 enzyme and that the induction of kinase activity observed in the presence of Tat is primarily due to phosphorylation at serine 5. It is important to point out that we cannot eliminate the possibility that Tat simply markedly enhances the activity of P-TEFb kinase. The activation must, however, be selective toward serine 5 phosphorylation.
DISCUSSION
The RNAP II CTD heptapeptide Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7 contains three serine residues at positions 2, 5, and 7. The CTD is heavily phosphorylated in vivo, and substitution of nonphosphorylatable amino acids at position 2 or 5 of the Saccharomyces cerevisiae CTD is lethal (77, 82). Phosphorylation of the CTD is temporally linked to the transition between transcription initiation and elongation. Human CTD kinases specific for serine 5, serines 2 and 5, or serines 2 and 7 have been characterized (11, 71). It has been reported that the TFIIH kinase phosphorylates serine 5 of the CTD (58, 71). The analyses of TFIIH kinase activity in HIV-1 transcription complexes presented in this manuscript are consistent with these findings. The substrate specificity of the P-TEFb complex has been more difficult to distinguish. Ramanathan et al. have recently reported that serine 5 is essential for phosphorylation by the Tat-TAK complex (Tat–P-TEFb complex) (55). Using an in vitro kinase assay, the investigators demonstrated that phosphorylation of the CTD peptide was abolished when a mutation was introduced at serine 5. Mutation of serines 2 and 7 did not affect the activity of the Tat-TAK complex. The authors did not include reactions which contained P-TEFb alone in the absence of Tat. Thus, it was not obvious that the serine 5 phosphorylation observed in their assay is the Tat-modified function of P-TEFb. Moreover, the specificity of the P-TEFb kinase alone cannot be distinguished. Patturajan et al. have recently reported that the deletion of yeast CTDK-I, a CDK kinase closely related to P-TEFb, eliminates the increase in CTD serine 2 phosphorylation during a response to nutrient depletion (48). In our studies, we provide clear evidence that recombinant P-TEFb phosphorylates the CTD at serine 2. In the presence of Tat, the substrate specificity of P-TEFb is altered such that it phosphorylates serine 2 and serine 5. It is important to point out that the change in substrate specificity is reproduced in the natural setting of the kinase, the transcription complex, and the natural substrate, the RNAP II CTD.
A unique feature of Tat transactivation of HIV-1 transcription has been observed, which is that it is preferentially inhibited by DRB, an adenosine analogue that targets RNAP II-mediated elongation. P-TEFb has been distinguished from other transcription factors by its sensitivity to very low doses of DRB. In this regard, the results presented in this report demonstrate that serine 2 phosphorylation and serine 5 phosphorylation by Tat-activated P-TEFb are sensitive to DRB and that at a concentration of 5 μM DRB, serine 2 and serine 5 phosphorylation was inhibited by approximately 70 or 90%, respectively.
It will be of interest to determine whether the initial phosphorylation at serine 2 and serine 5 within the PICs is important for Tat transactivation. Okamoto et al. (44) have shown that the CTD is not required for basal transcription and for the formation of short, attenuated transcripts. In contrast, transcriptional activation by Tat in vivo and in vitro requires the CTD. It is possible that the critical step of CTD phosphorylation takes place in the elongation complex, after CDK7 has been released, TAR RNA has been synthesized, and Tat–P-TEFb has been recruited to the complex. TAR likely acts to recruit the Tat–P-TEFb complex to the elongation complex in this activation process. It will also be of interest to determine whether phosphorylation of serine 2, serine 5, or serines 2 and 5 is required for Tat transactivation at this stage.
In a very elegant analysis of the fate of transcription factors during the transition from initiation to elongation, Zawel et al. have demonstrated that TFIID remains promoter bound, wherease TFIIB, TFIIE, TFIIF, and TFIIH are released rapidly (83). TFIIH release occurs after the complex reaches +30 to +50. Consistent with these studies, our analysis indicates that TFIIH is released between +14 and +36 (Fig. 7A). Previous observations suggest that Tat interacts with a target cellular protein (as a cofactor of Tat) and that the interaction of Tat with its cellular cofactor is a prerequisite for TAR binding (1, 38). Recent studies strongly imply that the Tat cofactor is a cellular protein kinase termed TAK which interacts with the activation domain of Tat and phosphorylates CTD (21, 22, 79). It has been found that a human positive-acting transcription elongation factor complex called P-TEFb is actually equivalent to TAK (85). Several observations further suggest that the recruitment of the Tat–P-TEFb complex by TAR binding to elongating complexes is the critical step in Tat transactivation (22, 29, 40, 41). Similar to the results obtained in this study, recent observations indicate that the entry of the Tat–P-TEFb complex into the transcription complex comes at the time of PIC assembly (47, 52). Our results presented here are consistent with the observations that Tat–P-TEFb associates with HIV-1 PICs. Importantly, in contrast to the TFIIH-CDK7 complex, which is released from the complex between +14 and +36, P-TEFb remains stably associated with the TECs. The results presented in this study further suggest that the ability of Tat to alter the substrate specificity of CDK9 would allow the continued hyperphosphorylation of the RNAP II CTD at serine 2 and serine 5 in the Tat transcription elongation complex. As a general transcription elongation factor, it will also be important to determine how P-TEFb acts on other promoters and whether other activators affect the activity of P-TEFb.
Genetic data from several groups show that TAR can be functionally replaced by heterologous RNA structures. The subsequent recruitment of Tat to these RNA targets by fusion of Tat to an RNA binding domain can clearly fully activate HIV-1 LTR-dependent transcription (59, 70). The results suggest that TAR acts only as an interface. Further, the recruitment of P-TEFb to an HIV-1 LTR containing a heterologous promoter-proximal target by fusion of cyclin T1 to an RNA binding domain is both necessary and sufficient for full activation of transcription from HIV-1 LTR in vivo. The authors of the latter study suggest that Tat does not activate the P-TEFb in any specific way but rather serves as an interface between the RNA and the enzyme complex (3). The results presented in the latter study demonstrate that Tat, in fact, does modify the activity of the P-TEFb-associated CDK9 kinase, altering the specificity of CTD phosphorylation to allow CDK9 to phosphorylate serine 5.
Multiple kinases appear to be involved in phosphorylating the CTD in vivo (7, 35, 50). In yeast, at least three distinct complexes have been described, Kin28-CCL1 (8, 15, 72, 73), SRB10-SRB11 (36), and CTDK1 (a possible yeast homologue of P-TEFb) (48, 63). In higher eukaryotes, there are three homologues, CDK7-cyclin H (14, 37, 58, 60–62), CDK8-cyclin C (34, 56, 65), and P-TEFb (40, 41). The observations demonstrate that CDK8-cyclin C (SRB10-SRB11, the yeast homologue) associates with the RNAP II holoenzyme (34, 36, 56). However, only a small portion (less than 10% in mammals and 6% in yeast) of total RNAP II was found to be associated in a holoenzyme form with CDK8 (SRB10) in vivo (30, 34). It has been reported that SRB10-SRB11 (CDK8-cyclin C) is a negative regulator of transcription (20, 65) and that SRB10 is not a general repressor of protein-coding genes (36, 66). CTD phosphorylation by SRB10-SRB11 kinase prevents the assembly of the PIC and thereby represses the transcription of specific genes (20), including those involved in cell type specificity (74), meiosis (64, 66), sugar utilization (31, 36), and stress response (9). Our results presented in this report suggest that CDK8 may not function in HIV-1 transcription and Tat transactivation.
Finally, it is of interest to consider the possibility that phosphorylation of the RNAP II CTD may have a direct effect on capping of the pre-mRNAs. Capping is targeted to nascent RNAs through binding of the guanyltransferase to the phosphorylated CTD. Guanyltransferase binds CTD peptides containing phosphate groups at either serine 2 or serine 5. Interestingly, it has recently been reported that binding of guanyltransferase to CTDs containing a phosphorylated serine 5 specifically stimulates enzymatic activity by enhancing the affinity for GTP and increasing the yield of enzyme-GMP intermediate (23). A CTD containing phosphorylated serine 2 has no effect on enzymatic activity. It will be of importance to determine if the Tat-directed P-TEFb phosphorylation at serine 5 contributes to the capping of the viral pre-mRNA. Several studies have previously shown that Tat enhances the translation of mRNAs synthesized from the HIV-1 LTR (10, 33, 78). As capping is known to markedly increase the efficiency of translation of mRNAs, it will be interesting to determine whether Tat enhances recruitment of capping enzymes to the hyperphosphorylated CTD.
REFERENCES
- 1.Alonso A, Derse D, Peterlin B M. Human chromosome 12 is required for optimal interactions between Tat and TAR of human immunodeficiency virus type 1 in rodent cells. J Virol. 1992;66:4617–4621. doi: 10.1128/jvi.66.7.4617-4621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bieniasz P D, Grdina T A, Bogerd H P, Cullen B R. Recruitment of a protein complex containing Tat and cyclin T1 to TAR governs the species specificity of HIV-1 Tat. EMBO J. 1998;17:7056–7065. doi: 10.1093/emboj/17.23.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bieniasz P D, Grdina T A, Bogerd H P, Cullen B R. Recruitment of cyclin T1/P-TEFb to an HIV type 1 long terminal repeat promoter proximal RNA target is both necessary and sufficient for full activation of transcription. Proc Natl Acad Sci USA. 1999;96:7791–7796. doi: 10.1073/pnas.96.14.7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boris-Lawrie K A, Brady J N, Kumar A. Sequences within the R region of the long terminal repeat activate basal transcription from the HIV-1 promoter. Gene Expr. 1992;2:215–230. [PMC free article] [PubMed] [Google Scholar]
- 5.Chen D, Zhou Q. Tat activates human immunodeficiency virus type 1 transcriptional elongation independent of TFIIH kinase. Mol Cell Biol. 1999;19:2863–2871. doi: 10.1128/mcb.19.4.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chun R F, Jeang K T. Requirements for RNA polymerase II carboxyl-terminal domain for activated transcription of human retroviruses human T-cell lymphotropic virus I and HIV-1. J Biol Chem. 1996;271:27888–27894. doi: 10.1074/jbc.271.44.27888. [DOI] [PubMed] [Google Scholar]
- 7.Cisek L J, Corden J L. Phosphorylation of RNA polymerase by the murine homologue of the cell-cycle control protein cdc2. Nature. 1989;339:679–684. doi: 10.1038/339679a0. [DOI] [PubMed] [Google Scholar]
- 8.Cismowski M J, Laff G M, Solomon M J, Reed S I. KIN28 encodes a C-terminal domain kinase that controls mRNA transcription in Saccharomyces cerevisiae but lacks cyclin-dependent kinase-activating kinase (CAK) activity. Mol Cell Biol. 1995;15:2983–2992. doi: 10.1128/mcb.15.6.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper K F, Mallory M J, Smith J B, Strich R. Stress and developmental regulation of the yeast C-type cyclin Ume3p (Srb11p/Ssn8p) EMBO J. 1997;16:4665–4675. doi: 10.1093/emboj/16.15.4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cullen B R. Trans-activation of human immunodeficiency virus occurs via a bimodal mechanism. Cell. 1986;46:973–982. doi: 10.1016/0092-8674(86)90696-3. [DOI] [PubMed] [Google Scholar]
- 11.Dahmus M E. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- 12.Dingwall C, Ernberg I, Gait M J, Green S M, Heaphy S, Karn J, Lowe A D, Singh M, Skinner M A, Valerio R. Human immunodeficiency virus 1 Tat protein binds trans-activation-responsive region (TAR) RNA in vitro. Proc Natl Acad Sci USA. 1989;86:6925–6929. doi: 10.1073/pnas.86.18.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drapkin R, Reinberg D. The multifunctional TFIIH complex and transcriptional control. Trends Biochem Sci. 1994;19:504–508. doi: 10.1016/0968-0004(94)90139-2. [DOI] [PubMed] [Google Scholar]
- 14.Feaver W J, Gileadi O, Li Y, Kornberg R D. CTD kinase associated with yeast RNA polymerase II initiation factor b. Cell. 1991;67:1223–1230. doi: 10.1016/0092-8674(91)90298-d. [DOI] [PubMed] [Google Scholar]
- 15.Feaver W J, Svejstrup J Q, Henry N L, Kornberg R D. Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell. 1994;79:1103–1109. doi: 10.1016/0092-8674(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 16.Fujinaga K, Taube R, Wimmer J, Cujec T P, Peterlin B M. Interactions between human cyclin T, Tat, and the transactivation response element (TAR) are disrupted by a cysteine to tyrosine substitution found in mouse cyclin T. Proc Natl Acad Sci USA. 1999;96:1285–1290. doi: 10.1073/pnas.96.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garber M E, Wei P, KewalRamani V N, Mayall T P, Herrmann C H, Rice A P, Littman D R, Jones K A. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 1998;12:3512–3527. doi: 10.1101/gad.12.22.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grana X, De Luca A, Sang N, Fu Y, Claudio P P, Rosenblatt J, Morgan D O, Giordano A. PITALRE, a nuclear CDC2-related protein kinase that phosphorylates the retinoblastoma protein in vitro. Proc Natl Acad Sci USA. 1994;91:3834–3838. doi: 10.1073/pnas.91.9.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harper J W, Elledge S J. The role of Cdk7 in CAK function, a retro-retrospective. Genes Dev. 1998;12:285–289. doi: 10.1101/gad.12.3.285. [DOI] [PubMed] [Google Scholar]
- 20.Hengartner C J, Myer V E, Liao S M, Wilson C J, Koh S S, Young R A. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol Cell. 1998;2:43–53. doi: 10.1016/s1097-2765(00)80112-4. [DOI] [PubMed] [Google Scholar]
- 21.Herrmann C H, Rice A P. Specific interaction of the human immunodeficiency virus Tat proteins with a cellular protein kinase. Virology. 1993;197:601–608. doi: 10.1006/viro.1993.1634. [DOI] [PubMed] [Google Scholar]
- 22.Herrmann C H, Rice A P. Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxyl-terminal domain of the large subunit of RNA polymerase II: candidate for a Tat cofactor. J Virol. 1995;69:1612–1620. doi: 10.1128/jvi.69.3.1612-1620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho C K, Shuman S. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol Cell. 1999;3:405–411. doi: 10.1016/s1097-2765(00)80468-2. [DOI] [PubMed] [Google Scholar]
- 24.Joeijmakers J H, Egly J M, Vermeulen W. TFIIH: a key component in multiple DNA transactions. Curr Opin Genet Dev. 1996;6:26–33. doi: 10.1016/s0959-437x(96)90006-4. [DOI] [PubMed] [Google Scholar]
- 25.Isel C, Karn J. Direct evidence that HIV-1 tat stimulates RNA polymerase II carboxyl-terminal domain hyperphosphorylation during transcriptional elongation. J Mol Biol. 1999;290:929–941. doi: 10.1006/jmbi.1999.2933. [DOI] [PubMed] [Google Scholar]
- 26.Ivanov D, Kwak Y T, Nee E, Guo J, Garcia-Martinez L F, Gaynor R B. Cyclin T1 domains involved in complex formation with Tat and TAR RNA are critical for tat-activation. J Mol Biol. 1999;288:41–56. doi: 10.1006/jmbi.1999.2663. [DOI] [PubMed] [Google Scholar]
- 27.Jones K A. Taking a new TAK on tat transactivation. Genes Dev. 1997;11:2593–2599. doi: 10.1101/gad.11.20.2593. [DOI] [PubMed] [Google Scholar]
- 28.Kao S Y, Calman A F, Luciw P A, Peterlin B M. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987;330:489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- 29.Keen N J, Churcher M J, Karn J. Transfer of Tat and release of TAR RNA during the activation of the human immunodeficiency virus type-1 transcription elongation complex. EMBO J. 1997;16:5260–5272. doi: 10.1093/emboj/16.17.5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koleske A J, Young R A. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 31.Kuchin S, Yeghiayan P, Carlson M. Cyclin-dependent protein kinase and cyclin homologs SSN3 and SSN8 contribute to transcriptional control in yeast. Proc Natl Acad Sci USA. 1995;92:4006–4010. doi: 10.1073/pnas.92.9.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwak Y T, Ivanov D, Guo J, Nee E, Gaynor R B. Role of the human and murine cyclin T proteins in regulating HIV-1 tat-activation. J Mol Biol. 1999;288:57–69. doi: 10.1006/jmbi.1999.2664. [DOI] [PubMed] [Google Scholar]
- 33.Laspia M F, Rice A P, Mathews M B. Synergy between HIV-1 Tat and adenovirus E1A is principally due to stabilization of transcriptional elongation. Genes Dev. 1990;4:2397–2408. doi: 10.1101/gad.4.12b.2397. [DOI] [PubMed] [Google Scholar]
- 34.Leclerc V, Tassan J P, O'Farrell P H, Nigg E A, Leopold P. Drosophila Cdk8, a kinase partner of cyclin C that interacts with the large subunit of RNA polymerase II. Mol Biol Cell. 1996;7:505–513. doi: 10.1091/mbc.7.4.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee J M, Greenleaf A L. CTD kinase large subunit is encoded by CTK1, a gene required for normal growth of Saccharomyces cerevisiae. Gene Expr. 1991;1:149–167. [PMC free article] [PubMed] [Google Scholar]
- 36.Liao S M, Zhang J, Jeffery D A, Koleske A J, Thompson C M, Chao D M, Viljoen M, van Vuuren H J, Young R A. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature. 1995;374:193–196. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- 37.Lu H, Zawel L, Fisher L, Egly J M, Reinberg D. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature. 1992;358:641–645. doi: 10.1038/358641a0. [DOI] [PubMed] [Google Scholar]
- 38.Madore S J, Cullen B R. Genetic analysis of the cofactor requirement for human immunodeficiency virus type 1 Tat function. J Virol. 1993;67:3703–3711. doi: 10.1128/jvi.67.7.3703-3711.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mancebo H S, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, Peng J, Blau C, Hazuda D, Price D, Flores O. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 1997;11:2633–2644. doi: 10.1101/gad.11.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marshall N F, Peng J, Xie Z, Price D H. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 41.Marshall N F, Price D H. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem. 1995;270:12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- 42.Morgan D O. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 43.Morgan D O, Fisher R P, Espinoza F H, Farrell A, Nourse J, Chamberlin H, Jin P. Control of eukaryotic cell cycle progression by phosphorylation of cyclin-dependent kinases. Cancer J Sci Am. 1998;4(Suppl. 1):S77–S83. [PubMed] [Google Scholar]
- 44.Okamoto H, Sheline C T, Corden J L, Jones K A, Peterlin B M. Trans-activation by human immunodeficiency virus Tat protein requires the C-terminal domain of RNA polymerase II. Proc Natl Acad Sci USA. 1996;93:11575–11579. doi: 10.1073/pnas.93.21.11575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 46.Parada C A, Roeder R G. Enhanced processivity of RNA polymerase II triggered by Tat-induced phosphorylation of its carboxy-terminal domain. Nature. 1996;384:375–378. doi: 10.1038/384375a0. [DOI] [PubMed] [Google Scholar]
- 47.Parada C A, Roeder R G. A novel RNA polymerase II-containing complex potentiates Tat-enhanced HIV-1 transcription. EMBO J. 1999;18:3688–3701. doi: 10.1093/emboj/18.13.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patturajan M, Conrad N K, Bregman D B, Corden J L. Yeast carboxyl-terminal domain kinase I positively and negatively regulates RNA polymerase II carboxyl-terminal domain phosphorylation. J Biol Chem. 1999;274:27823–27828. doi: 10.1074/jbc.274.39.27823. [DOI] [PubMed] [Google Scholar]
- 49.Patturajan M, Schulte R J, Sefton B M, Berezney R, Vincent M, Bensaude O, Warren S L, Corden J L. Growth-related changes in phosphorylation of yeast RNA polymerase II. J Biol Chem. 1998;273:4689–4694. doi: 10.1074/jbc.273.8.4689. [DOI] [PubMed] [Google Scholar]
- 50.Payne J M, Dahmus M E. Partial purification and characterization of two distinct protein kinases that differentially phosphorylate the carboxyl-terminal domain of RNA polymerase subunit IIa. J Biol Chem. 1993;268:80–87. [PubMed] [Google Scholar]
- 51.Peng J, Zhu Y, Milton J T, Price D H. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 1998;12:755–762. doi: 10.1101/gad.12.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ping Y H, Rana T M. Tat-associated kinase (P-TEFb): a component of transcription preinitiation and elongation complexes. J Biol Chem. 1999;274:7399–7404. doi: 10.1074/jbc.274.11.7399. [DOI] [PubMed] [Google Scholar]
- 53.Poon R Y, Yamashita K, Howell M, Ershler M A, Belyavsky A, Hunt T. Cell cycle regulation of the p34cdc2/p33cdk2-activating kinase p40MO15. J Cell Sci. 1994;107:2789–2799. doi: 10.1242/jcs.107.10.2789. [DOI] [PubMed] [Google Scholar]
- 54.Price D H. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol Cell Biol. 2000;20:2629–2634. doi: 10.1128/mcb.20.8.2629-2634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramanathan Y, Reza S M, Young T M, Mathews M B, Pe'ery T. Human and rodent transcription elongation factor P-TEFb: interactions with human immunodeficiency virus type 1 Tat and carboxy-terminal domain substrate. J Virol. 1999;73:5448–5458. doi: 10.1128/jvi.73.7.5448-5458.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rickert P, Seghezzi W, Shanahan F, Cho H, Lees E. Cyclin C/CDK8 is a novel CTD kinase associated with RNA polymerase II. Oncogene. 1996;12:2631–2640. [PubMed] [Google Scholar]
- 57.Rossignol M, Kolb-Cheynel I, Egly J M. Substrate specificity of the cdk-activating kinase (CAK) is altered upon association with TFIIH. EMBO J. 1997;16:1628–1637. doi: 10.1093/emboj/16.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roy R, Adamczewski J P, Seroz T, Vermeulen W, Tassan J P, Schaeffer L, Nigg E A, Hoeijmakers J H, Egly J M. The MO15 cell cycle kinase is associated with the TFIIH transcription-DNA repair factor. Cell. 1994;79:1093–1101. doi: 10.1016/0092-8674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 59.Selby M J, Peterlin B M. Trans-activation by HIV-1 Tat via a heterologous RNA binding protein. Cell. 1990;62:769–776. doi: 10.1016/0092-8674(90)90121-t. [DOI] [PubMed] [Google Scholar]
- 60.Serizawa H, Conaway R C, Conaway J W. A carboxyl-terminal-domain kinase associated with RNA polymerase II transcription factor delta from rat liver. Proc Natl Acad Sci USA. 1992;89:7476–7480. doi: 10.1073/pnas.89.16.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Serizawa H, Makela T P, Conaway J W, Conaway R C, Weinberg R A, Young R A. Association of Cdk-activating kinase subunits with transcription factor TFIIH. Nature. 1995;374:280–282. doi: 10.1038/374280a0. [DOI] [PubMed] [Google Scholar]
- 62.Shiekhattar R, Mermelstein F, Fisher R P, Drapkin R, Dynlacht B, Wessling H C, Morgan D O, Reinberg D. Cdk-activating kinase complex is a component of human transcription factor TFIIH. Nature. 1995;374:283–287. doi: 10.1038/374283a0. [DOI] [PubMed] [Google Scholar]
- 63.Sterner D E, Lee J M, Hardin S E, Greenleaf A L. The yeast carboxyl-terminal repeat domain kinase CTDK-I is a divergent cyclin–cyclin-dependent kinase complex. Mol Cell Biol. 1995;15:5716–5724. doi: 10.1128/mcb.15.10.5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Strich R, Slater M R, Esposito R E. Identification of negative regulatory genes that govern the expression of early meiotic genes in yeast. Proc Natl Acad Sci USA. 1989;86:10018–10022. doi: 10.1073/pnas.86.24.10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun X, Zhang Y, Cho H, Rickert P, Lees E, Lane W, Reinberg D. NAT, a human complex containing Srb polypeptides that functions as a negative regulator of activated transcription. Mol Cell. 1998;2:213–222. doi: 10.1016/s1097-2765(00)80131-8. [DOI] [PubMed] [Google Scholar]
- 66.Surosky R T, Strich R, Esposito R E. The yeast UME5 gene regulates the stability of meiotic mRNAs in response to glucose. Mol Cell Biol. 1994;14:3446–3458. doi: 10.1128/mcb.14.5.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tassan J P, Schultz S J, Bartek J, Nigg E A. Cell cycle analysis of the activity, subcellular localization, and subunit composition of human CAK (CDK-activating kinase) J Cell Biol. 1994;127:467–478. doi: 10.1083/jcb.127.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Taube R, Fujinaga K, Wimmer J, Barboric M, Peterlin B M. Tat transactivation: a model for the regulation of eukaryotic transcriptional elongation. Virology. 1999;264:245–253. doi: 10.1006/viro.1999.9944. [DOI] [PubMed] [Google Scholar]
- 69.Thompson N E, Steinberg T H, Aronson D B, Burgess R R. Inhibition of in vivo and in vitro transcription by monoclonal antibodies prepared against wheat germ RNA polymerase II that react with the heptapeptide repeat of eukaryotic RNA polymerase II. J Biol Chem. 1989;264:11511–11520. [PubMed] [Google Scholar]
- 70.Tiley L S, Madore S J, Malim M H, Cullen B R. The VP16 transcription activation domain is functional when targeted to a promoter-proximal RNA sequence. Genes Dev. 1992;6:2077–2087. doi: 10.1101/gad.6.11.2077. [DOI] [PubMed] [Google Scholar]
- 71.Trigon S, Serizawa H, Conaway J W, Conaway R C, Jackson S P, Morange M. Characterization of the residues phosphorylated in vitro by different C-terminal domain kinases. J Biol Chem. 1998;273:6769–6775. doi: 10.1074/jbc.273.12.6769. [DOI] [PubMed] [Google Scholar]
- 72.Valay J G, Simon M, Dubois M F, Bensaude O, Facca C, Faye G. The KIN28 gene is required both for RNA polymerase II mediated transcription and phosphorylation of the Rpb1p CTD. J Mol Biol. 1995;249:535–544. doi: 10.1006/jmbi.1995.0316. [DOI] [PubMed] [Google Scholar]
- 73.Valay J G, Simon M, Faye G. The kin28 protein kinase is associated with a cyclin in Saccharomyces cerevisiae. J Mol Biol. 1993;234:307–310. doi: 10.1006/jmbi.1993.1587. [DOI] [PubMed] [Google Scholar]
- 74.Wahi M, Johnson A D. Identification of genes required for alpha 2 repression in Saccharomyces cerevisiae. Genetics. 1995;140:79–90. doi: 10.1093/genetics/140.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weeks K M, Ampe C, Schultz S C, Steitz T A, Crothers D M. Fragments of the HIV-1 Tat protein specifically bind TAR RNA. Science. 1990;249:1281–1285. doi: 10.1126/science.2205002. [DOI] [PubMed] [Google Scholar]
- 76.Wei P, Garber M E, Fang S M, Fischer W H, Jones K A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 77.West M L, Corden J L. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics. 1995;140:1223–1233. doi: 10.1093/genetics/140.4.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wright C M, Felber B K, Paskalis H, Pavlakis G N. Expression and characterization of the transactivator of HTLV-III/LAV virus. Science. 1986;234:988–992. doi: 10.1126/science.3490693. [DOI] [PubMed] [Google Scholar]
- 79.Yang X, Herrmann C H, Rice A P. The human immunodeficiency virus Tat proteins specifically associate with TAK in vivo and require the carboxyl-terminal domain of RNA polymerase II for function. J Virol. 1996;70:4576–4584. doi: 10.1128/jvi.70.7.4576-4584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yankulov K, Bentley D. Transcriptional control: Tat cofactors and transcriptional elongation. Curr Biol. 1998;8:R447–R449. doi: 10.1016/s0960-9822(98)70289-1. [DOI] [PubMed] [Google Scholar]
- 81.Yankulov K Y, Bentley D L. Regulation of CDK7 substrate specificity by MAT1 and TFIIH. EMBO J. 1997;16:1638–1646. doi: 10.1093/emboj/16.7.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yuryev A, Corden J L. Suppression analysis reveals a functional difference between the serines in positions two and five in the consensus sequence of the C-terminal domain of yeast RNA polymerase II. Genetics. 1996;143:661–671. doi: 10.1093/genetics/143.2.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zawel L, Kumar K P, Reinberg D. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 1995;9:1479–1490. doi: 10.1101/gad.9.12.1479. [DOI] [PubMed] [Google Scholar]
- 84.Zhou Q, Chen D, Pierstorff E, Luo K. Transcription elongation factor P-TEFb mediates Tat activation of HIV-1 transcription at multiple stages. EMBO J. 1998;17:3681–3691. doi: 10.1093/emboj/17.13.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhu Y, Pe'ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews M B, Price D H. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]