Summary
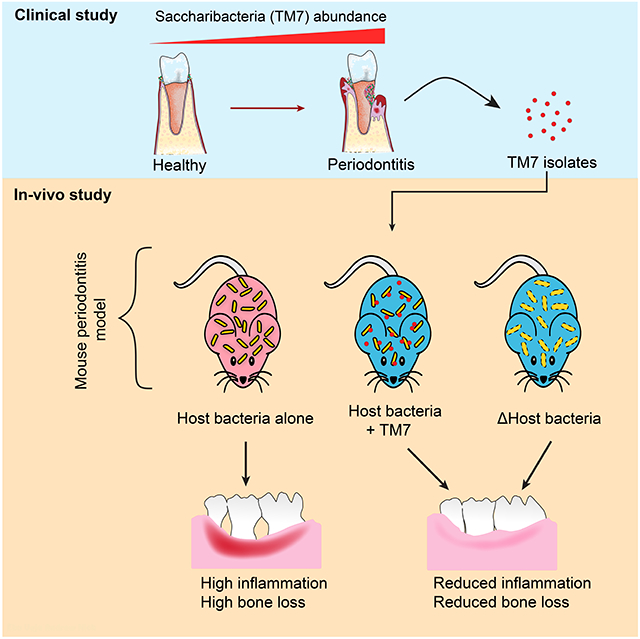
Saccharibacteria (TM7) are obligate epibionts living on the surface of their host bacteria and are strongly correlated with dysbiotic microbiomes during periodontitis and other inflammatory diseases, suggesting they are putative pathogens. However, due to the recalcitrance of TM7 cultivation, causal research to investigate their role in inflammatory diseases is lacking. Here, we isolated multiple TM7 species on their host bacteria from periodontitis patients. These TM7 species reduce inflammation and consequential bone loss by modulating host bacterial pathogenicity in a mouse ligature-induced periodontitis model. Two host bacterial functions involved in collagen binding and utilization of eukaryotic sialic acid are required for inducing bone loss and are altered by TM7 association. This TM7-mediated down-regulation of host bacterial pathogenicity is shown for multiple TM7/host bacteria pairs, suggesting that, in contrast to their suspected pathogenic role, TM7 could protect mammalian hosts from inflammatory damage induced by their host bacteria.
Graphical Abstract
eTOC Blurb
If certain bacteria are increased in disease, does that mean they are bad? Chipashvili et. al show that this may not be the case for ultrasmall Saccharibacteria. By using a mouse model for periodontitis, they determined that Saccharibacteria offer protection by reducing the pathogenicity of other bacteria in inflammatory diseases.
Introduction
Shaped by an extended history of co-evolution, the relationship between mammalian hosts and their commensal bacteria is of mutual benefit and critical for human health (Kodaman et al., 2014). However, the molecular function of the majority of these microorganisms remains elusive (Lloyd-Price et al., 2016). One such nascent and minimally characterized group of bacteria are from phylum Saccharibacteria, designated initially as candidate division TM7, which have interspecies symbiotic interactions within the mammalian microbiome (Bor et al., 2019). Currently, there is limited understanding of their impact on the eukaryotic host, including humans. Recent cultivation of human oral TM7 offers a unique opportunity to explore this unknown frontier (He et al., 2015), and in the process unveils the biology of a ubiquitous yet unique microbial group (McLean et al., 2020).
To date, TM7 are detected as commensal members of the healthy human oral, stomach, skin, and intestinal microbiome communities (Bik et al., 2006; Camanocha and Dewhirst, 2014; Gao et al., 2007; Zhu et al., 2018). In contrast to the remaining human-associated microbiota, however, TM7 have extremely distinct biology. The first cultivated TM7 member from the human oral cavity, strain TM7x (formally Nanosynbacter lyticus), was found to live on the surface of another bacteria, referred to as host bacteria, Actinomyces odontolyticus strain XH001 (He et al., 2015). Initial studies showed that TM7x is ultrasmall in size (200-500 nm) and behaves as an episymbiont that can kill its host bacteria but also form a stable relationship (Bor et al., 2018; Utter et al., 2020). In addition to TM7x, multiple oral TM7 species have been cultivated, all of which are ultrasmall and live on the surface of Actinomyces or other Actinobacteria (Bor et al., 2020; Cross et al., 2019; Murugkar et al., 2020). TM7 is one of the >73 phyla that make up a major lineage of uncultivated bacteria, termed the Candidate Phyla Radiation (Brown et al., 2015; Castelle and Banfield, 2018; Hug et al., 2016). Most recently, the first environmental TM7 species has been cultivated and it also displays an obligate symbiotic lifestyle (Batinovic et al., 2021).
Based on 16S rRNA profiling studies, TM7 flourish in an inflammatory environment. Their relative abundance is increased in vaginosis, various lung diseases, and inflammatory bowel disease (Table S1). Gastrointestinal disease and TM7 have been further correlated by multiple rodent model studies where TM7 have increased representation in colitis, irritable bowel syndrome, and colon cancer (Table S1). The human oral cavity is the most prominent anatomical site where TM7 are detected and studied (Jaffe et al., 2021; McLean et al., 2020). They are increased in various oral diseases such as periodontitis (Abusleme et al., 2013; Griffen et al., 2012), gingivitis (Al-Kamel et al., 2019; Huang et al., 2011), leukocyte adhesion deficiency I (Moutsopoulos et al., 2015), and conditions such as smoking (Duan et al., 2017; Ganesan et al., 2017). Many of the oral studies focused on periodontitis in particular (Galimanas et al., 2014; Papapanou et al., 2020; Wei et al., 2019), and in some instances, TM7 made up more than 20% of the bacterial abundance in periodontal plaque (Liu et al., 2012). Based on these studies, TM7 have been labeled as a pathogenic group of bacteria that may initiate or exacerbate the progression of periodontitis. Conversely, its known host bacteria, Actinomyces, are correlated with a healthy microbiome indicated by its decreased relative abundance but not absolute abundance in periodontal disease (Abusleme et al., 2013; Griffen et al., 2012). However, these reports are all based on culture-independent sequencing studies, lacking causation or even directionality. In fact, there is substantial evidence demonstrating that Actinobacteria, including Actinomyces, can induce inflammation and consequential bone loss in rodent as well as tissue culture models (Burckhardt et al., 1981; Jordan et al., 1965; Sato et al., 2012; Shimada et al., 2012; Socransky et al., 1970; Takada et al., 1993). Actinomyces also have been shown to induce multiple other human diseases such as Actinomycosis (Könönen and Wade, 2015). Thus, the pathogenicity of Actinomyces is far from clear, and conducting causal research to experimentally investigate the role of TM7 and their host bacteria in disease pathogenesis is warranted.
To elucidate the complex three-way interaction dynamics between TM7, their host bacteria, and the eukaryotic mucosal interface, we systematically characterized multiple clinical isolates of TM7 phylotypes in a ligature-induced periodontitis murine model. Surprisingly, TM7/host bacteria pairs induced significantly less bone loss compared to host bacteria alone. We identified two gene functions in one of the host bacteria, XH001, that are down-regulated by TM7, leading to a diminished inflammatory response and bone loss. Altogether, these results suggest that TM7 alone may not directly exacerbate inflammatory processes leading to periodontal disease but rather demonstrate a unique ability to impact the host bacterial physiology and pathogenicity.
Results
Increased relative abundance of TM7 in periodontal plaque
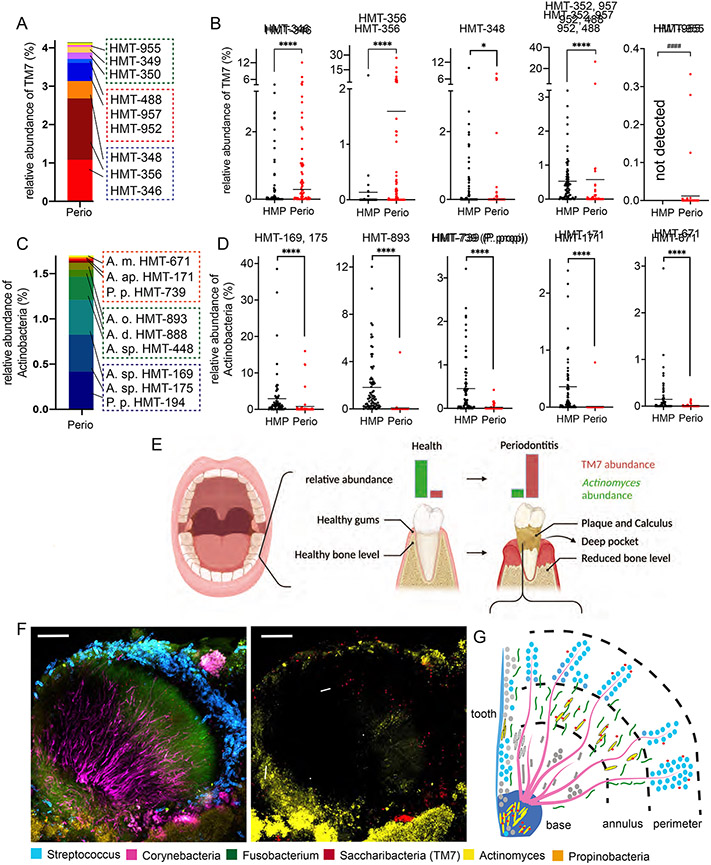
In order to identify patients with increased TM7/host bacteria, we utilized the 16S rRNA sequencing database from a previously established patient cohort with moderate to severe periodontal disease (N=32, mixed gender/race) at the Forsyth Institute Center for Clinical and Translational Research (see methods). The data were analyzed for TM7 and various Actinobacteria species belonging to known TM7 hosts Pseudoproprionibacterium and Actinomyces spp. (Figure 1A-D). The species taxonomic classification is designated by Human Microbial Taxon IDs (HMT; (Escapa et al., 2018). On average, patient subgingival plaque samples from periodontal pockets (referred to as periodontal plaque throughout the manuscript) contained ~4% TM7 and ~2% Actinobacteria species (Figure 1A, 1C), while some individual patient samples contained up to 20% of the total relative abundance as TM7 and Actinobacteria (Figure S1A, S1B). Several TM7 species HMT-346, 356, 348, and a cluster of HMT-952 related species (HMT-352, 957, 488) dominated the periodontal plaque (Figure 1A), which is consistent with previous studies (Griffen et al., 2012; McLean et al., 2020). Comparing these samples to the 77 healthy human dental plaque samples from the Human Microbiome Project (HMP) illustrated that the periodontal plaque had significantly higher TM7 relative abundance (see methods) (Figure 1B). We could not directly compare the HMT-952 related species since we could not individually resolve these species in the HMP 16S rRNA data. Actinobacteria species HMT-175, 169, and 893 dominated the periodontal plaque (Figure 1C), and their individual species relative abundances were significantly lower than in the health-associated HMP plaque samples (Figure 1D). Actinomyces spp. HMT-194, 448, and 888 were detected in only a few periodontal plaques, while showing a high relative abundance in HMP dental plaque. These results are consistent with previous observations, and based on these increased and decreased abundances of TM7 and Actinomyces, they are considered disease- and health-associated bacteria, respectively (Figure 1E) (Abusleme et al., 2013).
Figure 1.
Diversity and localization of TM7 bacteria. (A and C) Overall 16S rRNA relative abundance of TM7 (A) and Actinobacteria (C) species by their Human Microbiome Taxon (HMT). Red boxes highlight strains isolated/used in Figure 2, while blue and green boxes highlight abundant and less abundant species respectively. (B and D) Individual species comparison of panel A and C (red dots) versus 77 healthy plaque samples (black dots) from the human microbiome project (HMP). Each dot indicates an individual patient sample (n=32-77). ####, comparison between detected vs not-detected; *p < 0.05, ****p < 0.0001 by Mann-Whitney U test. See Also Figure S1. (E) Graphic summary depicting the periodontal disease progression from health to disease with TM7 (red) and Actinomyces (green). (F) FISH-Spectral composite image of periodontal plaque samples that were stained with six individual bacterial genus-specific 16S rRNA probes. Localization patterns showing all bacteria (right) or only TM7/Actinomyces (left: arrow and arrowheads respectively). Scale bars = 30 μm. (G) Depiction of gingival plaque biofilm structure from panel F.
To further determine the presence and subgingival biogeography of TM7 and Actinomyces, we conducted Fluorescence In Situ Hybridization combined with Spectral Imaging on periodontal plaque from the aforementioned patient cohort (Figure 1F) (Mark Welch et al., 2016). Beyond the three focal bacterial taxa (TM7, Actinomyces, Pseudoproprionibacterium), we also stained the three common oral bacteria (Streptococcus, Corynebacteria and Fusobacterium) that form the backbone of the classic hedgehog structure (Figure 1G). Within this structure, TM7 and Actinomyces colocalize to what is considered the annulus and perimeter region of the hedgehog, while Actinomyces can also independently localize to the base region (Figure 1F, 1G, S1C). A few TM7 localized independently from Actinomyces, possibly representing additional host bacteria. Pseudoproprionibacterium detection was minimal in these structures.
Isolation and cultivation of TM7 bacteria from the periodontal plaque
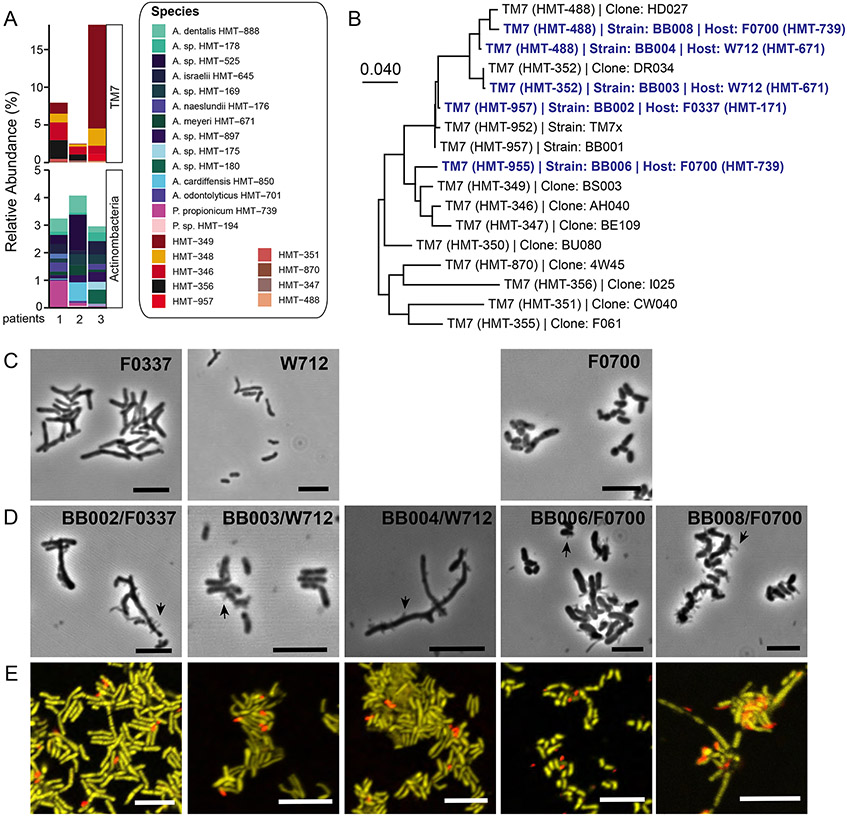
Three periodontitis patients with a high abundance of TM7 were selected and re-sampled from the 32-patient cohort. These patients harbored similar TM7 and Actinobacteria profiles as the larger cohort, although some TM7 species such as HMT-488 and HMT-957 were more pronounced (Figure 1A, 2A). With the previously identified host bacteria (Table S2) (Bor et al., 2020; Murugkar et al., 2020), we “baited” five TM7 strains (BB002, BB003, BB004, BB006, and BB008) belonging to 4 species (HMT-957, 352, 488, 955) on three host bacteria (strains: F0337, W712, and F0700) (Table S3). TM7 species HMT-957, 352, and 488 were closely related by 16S phylogeny while HMT-955 was taxonomically distant (Figure 2B). All four TM7 species and three “bait” species were readily detected in the periodontal plaque samples (Figure 1A, 1C, 2A). Furthermore, all five TM7 strains dependently grew on the surface of host bacteria (Figure 2C, 2D, 2E). Phylogenetically, three host bacteria are distant from one another (Bor et al., 2020). F0337 and W712 are Actinomyces, while F0700 is Pseudopropionibacterium and related to well-known skin microbe P. acnes. Both F0337 (unnamed Actinomyces sp. similar to A. naeslundii) and W712 (A. meyeri) have been linked to human infections previously (Könönen and Wade, 2015) while P. acnes is a well-known opportunistic pathogen (Achermann et al., 2014; McLaughlin et al., 2019).
Figure 2.
Cultivation of TM7 species from periodontitis. (A) 16S rRNA profiling of 3 patients that were sampled for the cultivation study. (B) Neighbor-joining tree for TM7 using full-length 16S rRNA sequences using MEGA X. Isolated TM7 and their host bacteria are highlighted in blue. Host bacteria alone (C) and with TM7 (D) are imaged by phase-contrast microscopy. Black arrows are showing TM7. Scale bars are 5 μm. (E) FISH composite images of coculture containing TM7 (red) and host bacteria (yellow) stained with 16S rRNA specific DNA probes. See also Figure S2. Scale bars are 5 μm.
Bulk growth analysis revealed that BB002, BB003, and BB004 slightly decreased their host bacterial growth, reminiscent of TM7x (Bor et al., 2018), while BB006 and BB008 more drastically inhibited their host bacterial growth and their final cell density (Figure S2A-F). To propagate BB006 and BB008, we had to supplement with fresh host bacteria and medium in every passage. Image analysis revealed that BB006 and BB008 tend to induce host bacterial aggregation while other strains did not (Figure S2B, S2D, S2F). This aggregation was not seen in the host bacteria alone control.
Presence of TM7 isolates decrease inflammatory bone loss
To evaluate the pathogenic nature of isolated TM7 bacteria, a murine oral infection model was employed, where the oral bacteria are reduced by antibiotic treatment followed by repeated inoculation of the host bacteria (Graves et al., 2008). We used two strains of mice (BALB/cJ and C57BL/6J), however neither showed bacterial colonization nor inflammation-mediated bone loss compared to controls (Figure S3A).
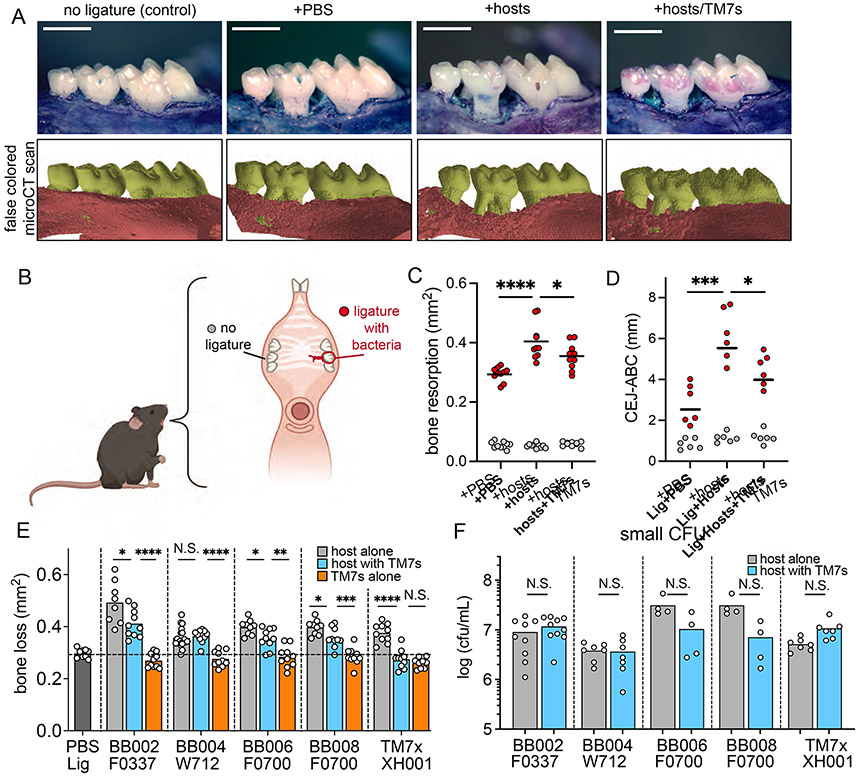
To overcome this challenge, we moved to a well-established murine ligature-induced periodontitis model to facilitate colonization of bacterial pathogens within the gingival pocket (Abe and Hajishengallis, 2013; Marchesan et al., 2018). In this model, a silk ligature soaked in phosphate buffered saline (PBS) or with an experimental bacterial group is applied to maxillary second molars of C57BL/6J mouse maxillae for four weeks (Figure 3A, 3B). Typically, the ligature alone induces inflammatory bone loss by accumulating mouse resident oral microbiota, and bone loss is usually arrested and plateaus after nine days (Marchesan et al., 2018). Addition of human pathogenic bacteria to the ligature induces further inflammation and bone loss (Lin et al., 2014). For our initial study, we applied a mixture of all three host bacteria with or without all five TM7 strains (Figure 3C). Consistent with previous studies, ligature with PBS, referred to as “baseline”, induced more inflammatory bone loss compared to the no-ligature group (Figure 3A, 3C). Comparatively, the host bacterial mixture on the ligature induced significantly more bone loss compared to baseline, revealing that these host bacteria can exacerbate periodontal destruction initiated by the ligature. Unexpectedly, the TM7/host bacteria mixture had significantly reduced inflammatory bone loss compared to the host bacteria group (Figure 3A, 3C). To further validate these findings, the maxillae were 2D imaged with a microCT scanner (Figure 3A, S3B), and the distance between the cemento-enamel junction (CEJ) and the alveolar bone crest (ABC) was measured (Figure S3C). The macroscopic and 2D microCT measurements revealed the same bone loss difference between groups (Figure 3D).
Figure 3.
Ligature-induced periodontitis supplemented with bacteria. (A) Top: Buccal view of alveolar bone loss from the mice without ligature (control), or with ligature soaked with phosphate buffer saline (PBS), mixture of 3 host bacteria, or all hosts with all TM7s. Bottom: 2D microCT scanned 3D representation of the molars. (B) Diagram of ligature placement around the second molar. (C) Cumulative 2D area measurements of bone resorption between PBS, hosts, and hosts with TM7s (red dots) from panel A. Internal control (no ligature) side of the bone measurement is shown in gray circles. (D) Cumulative distance between CEJ and ABC measurements of bone resorption from microCT images in panel A. (E) Cumulative 2D area measurements of bone resorption in mice with ligature soaked with host alone, host with TM7, or TM7 alone cultures. (F) Number of small CFUs (host bacteria) on the ligature. (C-F) Results are shown as mean (bar) ± SD. Each dot indicates an individual mouse (n = 6-10). N.S., not significant; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 by one-way ANOVA with Bonferroni post hoc test (C, D and E) or Mann-Whitney U test (F). See also Figure S3 and S4.
To determine which TM7 strains decreased inflammatory bone loss caused by their host bacteria, we tested each pair separately, barring BB003 which had inconsistent growth and inadequate cell numbers in the coculture. All host bacteria alone groups showed significantly increased inflammatory bone loss compared to baseline, while all TM7-containing coculture groups showed decreased inflammatory bone loss compared to the host bacteria alone, with the exception of BB004/host bacteria, which had bone loss similar to the host bacteria alone (Figure 3E). We also isolated each TM7 strain by separating them from the host bacteria (see methods) and applied them onto the ligature without the host bacteria, which resulted in no additional bone loss compared to the baseline (Figure 3E). The bone loss phenotype was drastic in some cases: F0337 induced significantly higher inflammatory bone loss relative to other host bacteria such that teeth alignment was disrupted frequently (Figure 3E, S3D). However, BB002 decreased this bone loss to the extent that tooth alignment was preserved. In addition to the BB00 strains from the periodontal patients, we also tested our previously characterized TM7x/XH001 pair in this model. TM7x (HMT-952) and XH001 (HMT-701) originally were isolated from healthy human saliva, and TM7x is a completely different species than the BB00 strains (Figure 2B, Table S3). Both are readily detected in the periodontal plaque samples (Figure 1A, 1C), and HMT-952 has been shown to be associated with periodontal disease. XH001 alone increased the inflammatory bone loss significantly while TM7x/XH001 induced bone loss that was indistinguishable from the baseline (Figure 3E, S3E), illustrating that even a TM7 strain from a healthy microbiome has the capability to decrease bone loss induced by its host bacteria.
TM7 do not restrict host bacterial viability in vivo
To determine the colonized bacteria inhabiting the ligature after 4 weeks of application, we analyzed the ligature microbiome through 16S rRNA sequencing (Figure S4A). At baseline, the ligature microbiome was dominated by mouse oral resident Firmicutes and Proteobacteria (~99%), and no TM7 or Actinobacteria were detected. In contrast, in ligatures soaked with TM7 and/or their host bacteria, we observed colonization by both bacteria to varying degrees in host alone and host with TM7 groups, while TM7 alone group did not colonize the ligature (Figure S4A). The remainder of the microbiome was mostly dominated by resident Fimicutes, and the diversity of these residual bacteria were not significantly different, suggesting that the impact of TM7/host bacteria on the overall composition of ligature microbiome is minimal.
In general, TM7 strains colonize the ligature relatively well in the presence of their host bacteria. In particular, F0337 alone and BB002/F0337 coculture comprised up to 80-90% of the total relative abundance of the ligature microbiome (Figure S4A), and BB002 took up over half (50-60%) of the relative abundance in BB002/F0337 group. This is consistent with the drastic inflammatory bone loss data for F0337 (Figure 3E). Furthermore, the presence of TM7 strains resulted in a lower relative abundance of host bacteria compared to host bacteria alone, except for BB004/W712. However, relative abundance does not reflect the absolute number of host bacteria. To reconcile this disparity, we plotted absolute counts of 16S rRNA reads as well as plated ligature microbes on blood agar media to recover viable bacterial colony forming units (CFUs). The read counts showed very similar colonization by the host bacteria in the presence or absence of TM7 (Figure S4B). Absolute read counts were not compared between TM7/host bacteria groups (e.g., BB002/F0337 vs. TM7x/XH001) for technical reasons (see methods). The plating resulted in a mix of two types of colonies: small and large (Figure S4C). PCR determined small and large CFUs as host bacteria (Actinobacteria) and resident bacteria (Firmicutes), respectively. TM7 bacteria do not form CFUs. Similar to the read counts, enumeration of the small CFUs showed similar number of host bacteria with or without TM7 (Figure 3F), suggesting that TM7 do not suppress their host bacterial viability on the ligature. Total CFUs (small + large colonies), however, were higher in host bacteria alone and co-culture with TM7 groups compared to baseline, but no significant difference between them (Figure S4D). This indicates that host bacteria colonize on the ligature in addition to the native bacteria rather than replacing them. TM7 alone controls showed a similar level of total CFUs compared to baseline and had no small colonies.
Inhibition of bone loss is through an attenuated inflammatory response
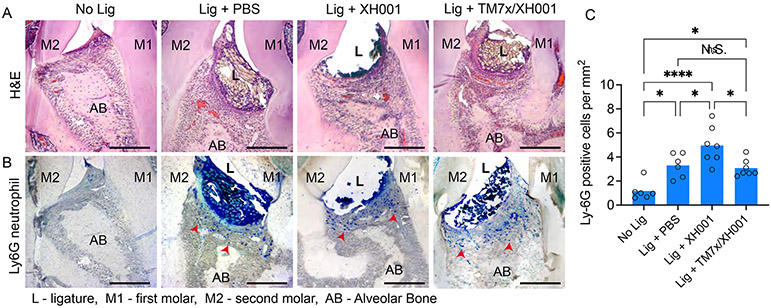
It has been shown that ligature-induced periodontitis occurs through increased gingival inflammation and cytokine response (Marchesan et al., 2018). To investigate direct innate and adaptive immune responses to these treatments in the gingival tissue surrounding the ligature, we measured the pro-inflammatory cytokines (IL-1, IL-6, IL-17, TNFα) at day 7 and 28 following treatment. We utilized the TM7x/XH001 co-culture since they showed the most drastic bone loss inhibition (Figure 3E) despite having a similar number of host bacteria (Figure 3F). In addition, TM7x/XH001 have the most well-understood biology with many experimental tools available. By day 7 of cytokine measurements, we observed a significant difference between no ligature control and ligature with PBS or XH001 (Figure S5A). The differences were smaller and not significant at day 28 (Figure S5B). Conversely, although there was a similar pattern, we did not see a significant difference between ligature with PBS control compared to ligature with XH001. This was anticipated knowing that the ligature alone is colonized by the resident microbiome and induces a robust inflammatory response, masking the host bacterial effect. To circumvent this assay parameter, we directly visualized the interproximal area between the distal root of the first molar and the mesial root of the second molar by hematoxylin and eosin (H&E), and neutrophil antibody (Ly6G) staining (Figure S5C). H&E staining clearly showed the infiltrating inflammatory cells (polynuclear structures), loss of connective tissue attachment, and alveolar bone resorption in all groups except the no-ligature group (Figure 4A). The neutrophil antibody staining illustrated clear blue neutrophil cells in the tissue aligning the ligature (Figure 4B). To show where the ligature was placed, we sectioned the histology samples with the embedded ligatures. These ligature fragments had polynucleated immune cells such as neutrophils (shown with dark blue nuclei), suggesting that immune cells are infiltrating deep into the ligature. Quantification of the neutrophils by antibody staining showed that XH001 alone group had drastically more neutrophils than both baseline and the host bacteria with TM7x (Figure 4C).
Figure 4.
Tissue immune response. (A-B) Representative histology images of no ligature control and ligature with PBS, XH001, and TM7x/XH001 stained with hematoxylin & eosin (A) or immunohistochemistry for neutrophil-expressed Ly6G (B). Red arrows indicate stained neutrophil cells, and scale bars are 100 μm. (C) Numeration of neutrophils in Ly6G-stained images. Six or more sections were analyzed from independent maxillary samples per group. Results are shown as mean (bar) ± SD. N.S., not significant; *p < 0.05, ****p < 0.0001 by one-way ANOVA followed by Bonferroni post hoc test. See also Figures S5.
TM7 decrease inflammatory bone loss by modulating their host bacterial pathogenicity
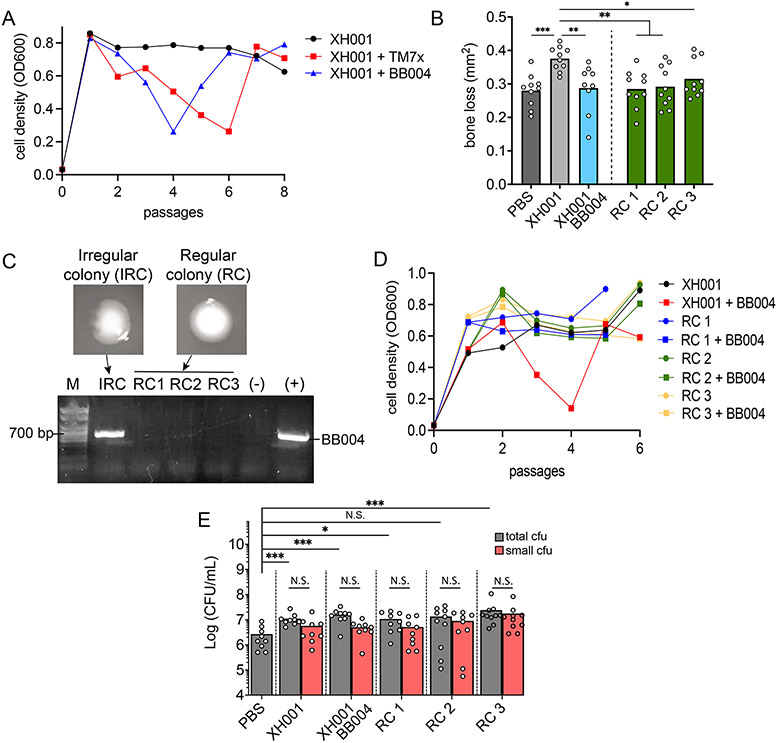
Our current evidence suggests that TM7 do not decrease the inflammatory bone loss through a change in the ligature microbiome composition nor the viable host bacterial count (Figure 3F, S4A). Alternative possibilities include TM7 directly influencing the innate immune response and/or changing its host bacterial pathogenicity. The observation that BB004/W712 and W712 had similar levels of inflammatory bone loss gave us a strategy to separate these possibilities (Figure 3E). First, we isolated BB004 away from the W712 host bacteria and infected XH001 as a new host. This process has been shown in our previous studies where TM7x can infect both W712 and XH001 (Utter et al., 2020). During infection, we observed a typical growth crash and recovery pattern of XH001 where BB004 killed their host bacteria first followed by establishment of a stable episymbiotic relationship where both parties coexist (Figure 5A). This newly established BB004/XH001 coculture resulted in decreased inflammatory bone loss similar to TM7x/XH001 coculture (Figure 5B) compared to XH001 alone control. This result is in opposition of what we observed with the BB004/W712 inflammatory bone-loss, suggesting that BB004 is influencing XH001’s pathogenicity but not W712’s. However, it is still possible that BB004 phenotype is altered, not its host bacteria.
Figure 5.
TM7 alter host bacterial pathogenicity. (A) XH001p was infected with TM7x (red squares) or BB004 (blue triangles), and the subsequent cell density (OD600) was measured across time (passages). (B) 2D alveolar bone loss area analysis from ligature experiment with PBS, XH001p, BB004/XH001 and three XH001np regular colonies (RC). (C) PCR test differences in regular versus irregular colonies (IRC) from established BB004/XH001 coculture. (D) Same infection experiment as in panel A but this time XH001np RC were re-infected with BB004. (E) Quantification of total and small CFUs from experiment in panel B. (B and E) Results are shown as mean (bar) ± SD. Each dot indicates an individual ligature (n = 8-10). N.S., not significant; *p < 0.05, **p < 0.01, ***p < 0.001 by one-way ANOVA followed by Bonferroni post hoc test (B) or Mann-Whitney U test (E).
We previously have shown that once TM7x/XH001 establish stable coculture, they form two types of colonies on a blood agar plate: regular and irregular (Bor et al., 2018). Regular colonies do not contain TM7x while irregular colonies do. The same phenomenon was observed with BB004/XH001 coculture, and we used PCR to validate both the presence and absence of BB004 in the regular and irregular colonies (Figure 5C). Subsequently, we isolated three regular colonies from the BB004/XH001 coculture, representing host bacteria that were previously exposed to BB004 but do not contain BB004, and tested their growth crash phenotype by infecting them with isolated BB004. We did not observe a growth crash similar to the initial infection of the XH001 (Figure 5D). We refer to these XH001 as nonpermissive XH001 strain (XH001np) while those that continue to show the crash phenotype are referred to as permissive (XH001p). This illustrates a fundamental phenotype difference between XH001p and XH001np due to prior exposure to BB004. Therefore, we tested whether this change was enough to decrease their ability to induce inflammatory bone loss in absence of BB004. Surprisingly, while both XH001p and XH001np colonize the ligature at similar levels (Figure 5E), all three XH001np regular colonies from the BB004/XH001 coculture induced significantly less bone loss compared to XH001p control (Figure 5B).
TM7 modify host bacterial metabolism and cell surface proteins
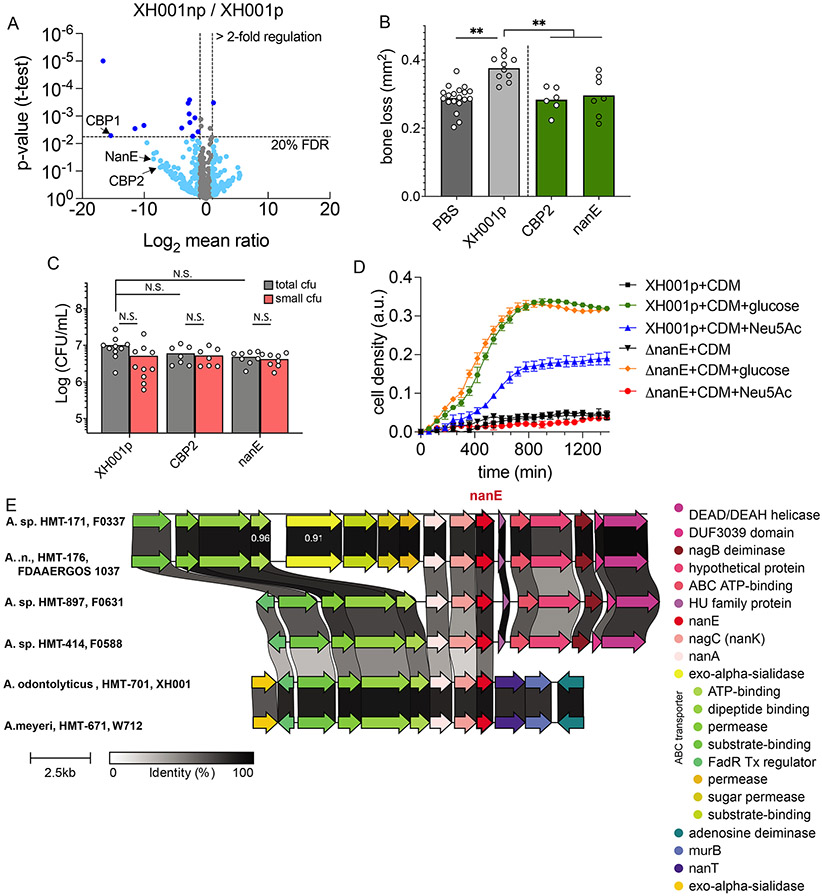
BB004 most likely induces widespread changes in its host bacteria, similar to our previous study on TM7x (He et al., 2015). To identify inflammation-associated genes that are attenuated by BB004 association, we focused on the bacterial surface proteins that are exposed to immune cells. Cell wall material was isolated from XH001p and XH001np cells. To analyze cell wall-associated proteins, the isolated material was treated with mutanolysin followed by mass spectrometry analysis (see methods). We identified multiple down-regulated proteins in XH001np compared to XH001p while few highly up-regulated genes from our analysis (Figure 6A, Table S4). Three down-regulated proteins were of interest due to their previously shown involvement in bacterial pathogenicity: significantly down-regulated collagen-binding protein 1 (CBP1: 43,822-fold, p-value = 0.005, FDR = 19.7%) and marginally down-regulated CBP2 (172-fold, p-value = 0.074, FDR = 47.6%) and N-acetylmannosamine-6-phosphatate 2-epimerase enzyme (NanE: 364-fold, p-value = 0.036, FDR = 41.9%) (Table S4). Collagen binding proteins facilitate direct binding and interaction of bacteria to organisms in the environment, both prokaryotic and eukaryotic (Arora et al., 2021) while the operon containing nanE has been shown to offer a growth advantage to pathogenic bacteria by allowing them to use sialic acid from the eukaryotic host as an energy source (Chang et al., 2004; Olson et al., 2013). Although NanE is a cytoplasmic enzyme, previous studies have shown that some dominant cytoplasmic proteins also show up in bacterial cell wall preparations (He and De Buck, 2010). XH001 genome carries a full set of genes in the nanE operon (nanA, nanK, nanE, and nanT) plus at least one sialidase enzyme, suggesting that XH001 is able to utilize sialic acid (N-acetylneuraminic acid, Neu5Ac) as an energy source (Figure S6A, S6B).
Figure 6.
Modified genes in host bacteria. (A) Comparative mass spectrophotometer analysis of cell wall proteins from XH001p and RC of XH001np shown by volcano plot. The threshold is set for genes that are increased >2-fold (x-axis) and with FDR <20% (y-axis). Dark blue dots are significantly regulated genes by these two criteria, while light blue dots are marginally significantly regulated genes that have higher than 20% FDR. (B) Top genes from panel A were deleted using homologous recombination in XH001p. These mutants were tested in our murine ligature model and the bone loss was measured in each mutant. (C) CFU quantification of panel B mutants on the ligature. (D) Bulk growth curve measurement of XH001p and ΔnanE mutant growing on minimal CDM medium supplemented with buffer, glucose or N-acetylneuraminic acid (Neu5Ac). See also Figure S6. (E) Complete genomes of Actinomyces were downloaded from NCBI RefSeq, aligned, and queried for the nanE gene (red) and its operon. Conserved synteny of nanE operon in three tested Actinomyces host bacteria (XH001, W712, F0337) and three additional taxonomically distance Actinomyces spp. (F0631, F0588, FDAAERGOS 1037). (B and C) Results are shown as mean (bar) ± SD. Each dot indicates an individual ligature (n = 8-10). N.S., not significant; **p < 0.01 by one-way ANOVA followed by Bonferroni post hoc test.
To investigate the hypothesized genetic basis for pathogenesis, we targeted cbp1, cbp2, and nanE loci for deletion in XH001p strain, utilizing a recently developed genetic system (Bedree et al., 2018). We were able to delete cbp2 and nanE genes but not cbp1 due to its gene duplication found in the XH001 genome. This makes it extremely difficult to delete in the GC-rich Actinomyces bacteria. Pathogenicity of Δcbp2 and ΔnanE mutants were tested in our murine ligature model in the absence of BB004. Interestingly, both mutants did not induce bone loss to the extent of the XH001p parent strain (Figure 6B), illustrating that these genes are important for XH001’s pathogenicity. We further investigated their ability to colonize the ligature in the gingiva and found that both mutants had normal colonization ability (Figure 6C). This is reflective of our BB004/XH001 coculture, which also colonizes the ligature normally. The functional ability of ΔnanE mutant to break down Neu5Ac as an energy source was tested next, where wild type XH001p grew normally on minimal medium supplemented with Neu5Ac but ΔnanE mutant had completely compromised function (Figure 6D). Insertion of selection marker in nanE gene could result in disruption of the downstream gene (nanT) in the operon (Figure S6A). Therefore, we tested the transcript level of nanT gene in XH001p and ΔnanE mutant, which showed no significant difference (Figure S6C).
To investigate the broader patterns of these genes in hosts, we searched the genomes of related organisms for CBP2 and nanE. A previous homology assessment of 23 Actinomyces genomes predicted only one CBP2 homolog in Actinomyces spp. ICM39 (Utter et al., 2020), and a broader HMMER search (https://www.ebi.ac.uk/Tools/hmmer/) of the XH001 CBP2 sequence against reference proteomes only identified high-confidence Actinomyces homologs in an unassembled whole-genome shotgun dataset. Both NCBI Conserved Domain Database and PFAM domain searches revealed a clear bacterial Ig domain, but these domains were broadly shared among diverse cell wall genes, precluding further phylogenetic investigation. However, comparison of the genetic context of nanE revealed striking operon-level conservation within Actinomyces hosts (Figure 6E). All studied Actinomyces strains encoded a nanE gene immediately downstream of nanA and nagC (nanK), which are required for breakdown of Neu5Ac, although no nanE gene was identified for P. propionicum. Thus, nanE and the broader sialic acid degradation pathway appears important to symbiotic Actinomyces for their survival and growth due to its high conservation.
Discussion
The relative abundance of TM7 has been found to increase in many diseases and based on these studies, TM7 are thought to help initiate or exacerbate pathogenesis. Within the human oral cavity, TM7 are associated with periodontal disease and grouped into the notoriously pathogenic “red complex” (Abusleme et al., 2013). At the opposite end of the spectrum, their host Actinobacteria have decreased relative abundance in periodontitis, and they are grouped into the health-associated “green complex”. Our study provides strong evidence that oral Actinobacteria are potential pathobionts with their pathogenicity suppressed by TM7 in a mouse model. This, combined with previous reports that Actinomyces can cause human diseases (Könönen and Wade, 2015), supports that Actinomyces may have the potential to be human pathogen. TM7 on the other hand are likely “inflammophilic” (inflammation loving) bacteria (Hajishengallis, 2014) that thrive in inflammatory nutrition-rich conditions. Although not all TM7 species have been tested at this time, the data directly challenge the current notion that TM7 are overtly pathogenic bacteria and reverses the assumption that high relative abundance induces more inflammation.
Periodontal disease associated TM7 are quite diverse. Species such as HMT-346, 356, and 952 populate in larger quantities while HMT-350, 349, and 955 show smaller numbers. Both abundant and less abundant species had the ability to decrease inflammatory bone loss induced by their host bacteria. TM7 do not seem to change the composition and diversity of ligature microbiome, nor do they affect the number of viable host bacteria on the ligature. In contrast, TM7 decrease their host bacterial growth in bulk in vitro measurements. This could be explained by the fact that in vitro and in vivo conditions are drastically different and the non-infected host bacteria can out-grow TM7 under in vivo condition (Bor et al., 2018). Addition of host bacteria to the ligature does increase the total bacterial load on the ligature, suggesting an important putative mechanism for increased inflammation. However, whenever there are TM7 present, the bone loss decreased despite having similar number of host bacteria, arguing that host bacterial pathogenicity plays a larger role rather than total bacterial load. This was further validated by TM7-free XH001np strains that had a completely altered phenotype (no growth crash) due to previous BB004 exposure. The transition between XH001p to XH001np could be the result of a genetic and/or phenotypic as shown by our previous study (Bor et al., 2018). Furthermore, the phenotypic change is relatively stable, and could be due to various characterized bacterial mechanisms such as phase variation or epigenetics. In this study, we further discovered altered protein expression profile between XH001p and XH001np, although additional study is needed to determine how TM7x modify these genes.
Collagen is the most abundant protein in the human body and an integral part of the extracellular matrix. Many gram-positive human oral pathogens use collagen-binding surface proteins to interact and colonize the gingival tissue, as well as interact with neighboring eukaryotic or prokaryotic cells (Arora et al., 2021). Consistent with this idea, deletion of CBP2 in our experiment decreased XH001p’s pathogenicity in the absence of TM7 bacteria. We initially hypothesized that these mutants would show a disadvantage in colonizing the inflammatory environment due to impaired collagen binding. However, Δcbp2 mutants still colonized the ligature akin to XH001p, which is consistent with our TM7/host bacteria colonization, suggesting that pre-soaking the ligature allows Δcbp2 mutants to adhere to the ligature regardless of their ability to bind tissue matrix. The ΔnanE mutant in our study also had impaired pathogenicity. Previous studies have demonstrated that human pathogens with intact nanE operons can survive/colonize better in mouse models and cause increased disease severity due to their ability to metabolize sialic acid (Chang et al., 2004; Olson et al., 2013). Despite diminished sialic acid metabolism in the ΔnanE mutant, our CFU quantification demonstrated XH001p-level colonization of the ligature. This phenomenon could be from sufficient non-sialic acid carbohydrates available from the destruction of periodontal tissue by the ligature. Or, Actinomyces spp. could use the sialic acid degradation pathway not for energy metabolism but for biosynthesis or modification of cell envelope components, as metabolic modeling predicts Actinomyces hosts provide GlcNAc-modified envelope components to TM7 (Bernstein et al., 2019). Furthermore, sialic acid function and modification on eukaryotic cells are essential for regulating immune cell responses such as macrophages (Varki, 2017). Whichever the case, further genetic analysis is warranted to determine if these genes or their homologs are crucial for other TM7 host bacteria.
One important remaining question is why do TM7 increase in abundance in periodontal plaque if they do not contribute to the disease? The simplest answer could be that they are inflammophilic bacteria that can efficiently catabolize and utilize nutrients from the inflammatory destruction of tissues without necessarily mounting an inflammatory response (Hajishengallis, 2014). The inflammatory destruction combined with the availability of the host bacteria in the natural environment can result in the bloom of TM7 bacteria, similar to what we observed under laboratory conditions where single host bacteria can be infected by 50 or more TM7 cells (Bor et al., 2018). This is consistent with some of the patient samples containing an extremely high number of TM7 bacteria and the high TM7 colonization on the ligature in our results. To directly test this, however, longitudinal studies needed to follow the progression of periodontal disease ab initio.
Mammals likely have had multiple independent acquisitions of TM7 across time, resulting in large genomic diversity, and that Neanderthal dental calculus contain abundant TM7 species, alluding to its evolutionarily long and symbiotic existence with mammals (McLean et al., 2020). However, symbiosis exists on a parasite-mutualist continuum (Drew et al., 2021). Our study shows that despite TM7 initial killing of the host bacteria, once the stable relationship is developed, the host bacteria could benefit by reducing antigenicity, likely transitioning from pathogen to commensal. This mutualism may ensure long-term survival for the TM7/host bacteria within the eukaryotic interface. However, during dysbiosis and inflammation, this dynamic is ablated and TM7 proliferate while host bacteria decrease in numbers. Beyond the human oral cavity, TM7 increase within many disease states, yet it is unclear exactly what role they play at this time. Thus, we suggest TM7 ameliorate host bacteria-induced inflammation through reducing their virulence-related functions and this may be a general mechanism for the majority of TM7 bacteria.
Limitations of the Study
Due to ethical concerns, the causative research on periodontal pathogens cannot be done on humans, and currently, we rely on animal models. One of the key experimental models for studying periodontal diseases is the murine ligature-induced periodontitis model (Abe and Hajishengallis, 2013). Ligatures are thought to cause minimal trauma while the native mouse oral bacteria colonizing the ligature drive the majority of the damage (Marchesan et al., 2018). This, combined with distinct mouse dental anatomy, therefore, does not ultimately reflect the natural etiology of human periodontitis associated with microbiome dysbiosis.
In our experiments, we applied TM7/host bacteria on the ligature. This has been done previously with both mice/human pathogens and commensals (Jiao et al., 2013; Lin et al., 2014; Marchesan et al., 2018), showing that pathogens induce additional bone loss compared to ligature alone control while commensals do not. We show that TM7/host bacteria colonize the ligature even after 4 weeks, and their interaction in vivo is fully supported by the fact that TM7 cannot colonize the ligature on their own. As with most studies using this model, we acknowledge that TM7/host bacteria could be having complex interactions with the endogenous murine oral bacteria that we cannot account for. Future studies will consider using germ free mice.
STAR Method
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to the lead contact and corresponding author, Batbileg Bor (bbor@forsyth.org).
Material Availability
All unique reagents and bacterial strains generated in this study are available from the Lead Contact with a complete Material Transfer Agreement.
Data and code availability
The raw sequencing data and original code generated by this study were deposited on Mendeley at http://dx.doi.org/10.17632/vp68zv9wj9.1, with the exception of initial 32 patient 16S rRNA sequencing data. These data were collected as a confidential medical record at Forsyth Center for Clinical and Translational Research, and we cannot release the raw data publicly due to ethical prohibition. Individuals who are interested in these data can contact the Lead Contact. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [1] partner repository with the dataset identifier PXD026999 and 10.6019/PXD026999.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Bacterial strains and culture conditions
Host bacterial strains, their sources, and growth conditions are listed in Table S2 while coculture conditions are listed in Table S3. Before each experiment, cells from frozen stock were recovered and passaged twice in brain heart infusion (BHI) broth to ensure homogeneity. Most oral Actinomyces spp. are facultative anaerobes, but some are anaerobic while others are aerobic. We used microaerophilic conditions due to our previous finding that the majority of the host bacteria grow under this condition (Bor et al., 2020).
Subjects and sampling
All patients were enrolled at the Center for Clinical and Translational Research under Forsyth IRB approved protocols and all subjects provided informed consent prior sampling. Patients with Stage III/IV periodontitis, 18 years and older were recruited for the 32 patient cohort data that we used under the IRB protocol number IRB #16-07. Out of 32 patient cohort, 3 patients with a high abundance of TM7 were enrolled in the current study (IRB# 18-06) to provide additional oral samples for TM7 bacterial isolation and imaging. Gender balance and racial diversity were sought. Sub-gingival plaque samples were collected using Gracey curettes from 6 mm pocket depth and quickly suspended in 1 mL BHI medium. The samples were processed within the same day (see below).
Mice strains and handling
Mice were all acquired from Jackson Laboratories and were all 9-week-old males at the start of the experiment. C57BL/6J and BALB/cJ were the two species of mice used for the oral infection model. Only C57BL/6J were used for the ligature-induced periodontitis model. All animal experiments were carried out in accordance with the NIH Guide for the Care and Use of Laboratory Animals, and were approved by the local animal care committee at the Forsyth Institute. C57BL/6J mice are more commonly used due to their readily available genetic variants while BALB/cJ strain exhibits more susceptibility to inflammatory bone loss (Baker et al., 2000).
Murine oral infection model
Bacteria were all cultured in BHI medium under 2% oxygen microaerophilic conditions at 37°C, and passaged every day (24 hours) at least two times before being used for the experiment. Mice were given water with antibiotics (ANI Pharmaceuticals) 1mg/mL sulfamethoxazole and 200 ug/mL trimethoprim in order to reduce the original oral flora for 4 days. Mice were then given regular sterile water for 24 hours. Bacteria with cell density of 15 in PBS was mixed with 3% carboxymethylcellulose (CMC) solution 1:1 in order to make a solution (109 cells/mL) that can be easily applied to the mouse oral cavity. Mice were then anesthetized using isoflurane, and oral infection was performed using a blunt-ended needle. About 50uL of solution was applied to each side of the oral cavity, making sure to cover the gingival areas around the molars. Oral infection was performed once a day for the first two days of the experiment. After this initial infection, oral infection was performed every other day for two weeks (7 times). The control group was given a mixture of PBS and CMC instead of bacteria. The experiment was terminated after 42 days from the initial oral infection. Mice were euthanized using CO2.
Murine ligature-induced periodontitis model
Ligature was applied to the maxillary second molar of the mice similar to previous studies (Abe and Hajishengallis, 2013). A silk ligature corresponding to 6-0 thickness (Teleflex 6-0) was soaked in a CMC solution with 109 bacterial cells/mL. For the controls CMC with PBS was used instead of bacteria. Mice were anesthetized by intraperitoneal injection using 80 mg/kg Ketamine and 16 mg/kg Xylazine before tying the ligature using a surgeon’s knot. The mice were allowed to recover overnight and treated with CMC solution containing PBS or bacteria for three alternating days following the ligature application. The trial was terminated 28 days from the initial ligature placement. The mice were euthanized using CO2.
METHOD DETAILS
16S rRNA profiling/analysis of periodontal patient samples
Three periodontal patient plaque samples were resuspended by vortexing. 0.5 mL of the sample was pelleted and resuspended in 150 μL of phosphate buffer saline (PBS). Genomic DNA (gDNA) was isolated by MasterPure gram-positive DNA purification kit (Epicenter) according to the manufactory’s protocol with a slight modification of bead beating step. Briefly, 150 μL of PBS bacteria was mixed with 150μL of Tris-EDTA (TE) buffer and transferred to a stock tube containing glass beads (Sigma G8772). The final concentration of lysozyme was added to the final concentration of 2 mg/mL, and incubated for 1 hour at 37°C. Subsequently, the cells were disrupted by bead beater for 3 x 30 s each at 6 m/s speed with 1 minute short pauses at 4°C. g DNA was isolated using the manufacturer’s protocol here on. Isolated gDNA was sent to Zymo sequencing core for sequencing of the V1-V3 region of the 16S rRNA gene using 250bp pair-end sequencing chemistry.
16S rRNA gene sequences were analyzed to produce exact amplicon sequence variants (ASVs) with DADA2 (Callahan et al., 2016) following the developer’s recommendations. A detailed, reproducible workflow, and the ASV abundances and representative sequences are stored on Mendeley (http://dx.doi.org/10.17632/vp68zv9wj9.1). Briefly, sequences were trimmed to 240nt and filtered reads to have no more than 2 expected errors and truncated tails after a base with a quality score of 2. Forward and reverse reads were denoised individually and merged with a custom merging approach as the 240+240nt read length had enough overlap to merge sequences from taxa with shorter V1V3 amplicons (e.g., TM7) but insufficient for those producing longer amplicons (e.g., Actinomyces spp.). Therefore, our merging strategy first merged forward and reverse reads for which sufficient overlap existed, and then it concatenated the remaining read pairs with N’s in between. Chimaeras were removed using removeBimeraDenovo with the ‘consensus’ method. Representative ASV sequences were then exported and annotated using a custom BLAST-based pipeline to identify the closest (highest percent identity of the longest alignment length) representative taxa in the eHOMD 16S rRNA reference database. Note that an ASV’s best match could be to multiple eHOMD references. For the HMP amplicon data, ASV (oligotype) counts, and representative data were downloaded from the supplemental data of Eren et al. (2014) (Eren et al., 2014). HMP oligotype reference sequences were put through the same custom BLAST annotation system to update the names to the latest eHOMD.
Isolation and culturing of TM7 bacteria
Periodontitis-associated TM7 bacteria were cultured by the previously described host bacteria “Bait” method (Bor et al., 2020). Briefly, ultrasmall TM7 bacteria were separated from other bacteria in clinical samples (0.5 mL) by filtration through 0.45 μm syringe filters with a membrane (PVDF; Millipore). The filtered cells were collected and added directly to broth cultures of several potential host “Bait” species (Table S2) that were diluted to an optical density of 0.037 (OD600). Resultant cocultures were passaged every 24 hours for at least 5 passages. The presence of TM7 cells in the final cultures was confirmed by microscopic examination and PCR by TM7-specific primers.
FISH, spectral imaging, and analysis of dental plaques
Subgingival periodontal plaque was collected from the above three patients with periodontitis, each of whom had given informed consent (IRB #18-06). Volunteers refrained from oral hygiene for 12 hrs and food intake for 2 hrs before sample collection. The whole arch subgingival plaque was carefully collected using Gracey curettes. Samples were directly resuspended and fixed in 4% Formaldehyde in PBS for 3 hrs on ice, then rinsed with 2 changes of 1x PBS. Whole-mount FISH was performed with minor adjustments according to a previous study (Mark Welch et al., 2016). Briefly, 30 μL sample in PBS was applied gently to each Gold Seal UltraStick slide (Thermo Fisher, MA, USA) with a wide pipette tip to preserve as much spatial structure as possible. Slides were then incubated in 10 mM TE buffer (pH 7.4) with lysozyme (2 mg/ml, L-6876, Sigma-Aldrich) at 37 °C for 10 min, followed by rinsing with PBS. FISH was carried out by applying the hybridization solution (900mM NaCl, 20mM Tris, pH 7.5, 0.01% SDS, 20% (vol/vol) formamide, each probe at a final concentration of 5 ng/μl) to slides and incubated for 3 h at 46 °C. Slides were then washed in wash buffer (215 mM NaCl, 20 mM Tris, pH 7.5, 5 mM EDTA) at 48 °C for 15 min, then further washed in two changes of 0.1x SSC (15 mM NaCl, 150 μM sodium citrate) at 37 °C for 10 min each. Slides were air-dried and mounted in ProLong Gold Antifade Solution (ThermoFisher) using #1.5 cover glass and allowed to cure in the dark overnight. Spectral imaging was performed using a Zeiss LSM 780 Confocal Microscope with a 40x 1.4 N.A. Plan-Apochromat objective. Each field of view was scanned using Lambda Mode and excited by the 405-, 488-, 561- and 633- nm laser lines simultaneously to acquire the spectral dataset with 8.9 nm spectral resolution over 32 detector elements. Linear unmixing was performed using the Zeiss ZEN software and reference spectra files for each fluorophore, which were acquired with the same FISH and imaging protocols using cultured cells labeled with single fluorophores. Unmixed images were processed and assembled with false colors using Fiji (Schindelin et al., 2012). The oligonucleotide sequences can be found in the key resources table. Confocal microscopy was performed at the Forsyth Institute Advanced Microscopy Core Facility (RRID:SCR_021121).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Purified anti-mouse Ly-6G antibody | BioLegend | Cat# 127601 |
| AP goat anti-rat IgG (Mouse Absorbed) polymer kit, Alkaline Phosphatase | Vector | Cat# MP-5444 |
| Bacterial strains | ||
| See details in supplemental tables S2, S3 | ||
| Biological samples | ||
| Murine oral ligature | from this study | N/A |
| Murine gingival tissue | from this study | N/A |
| Murine maxilla with associated molar teeth | from this study | N/A |
| Critical commercial assays | ||
| DAB substrate Kit, Peroxidase (HRP) | Vector | Ref# SK-4100 |
| MasterPure Gram Positive DNA purification kit | Epicentre | Cat# MGP04100 |
| Blue Substrate Kit, Alkaline Phosphatase | Vector | Ref# SK-5300 |
| ImmPRESS-AP Goat Anti-Rat IgG (Mouse Adsorbed) Polymer Kit, AP | Vector | Ref# MP-5444 |
| Immunocal Formic Acid Bone Decalcifier | StatLab | Cat# 1414-32 |
| PureLink RNA mini kit | Invitrogen | Cat# 12183018A |
| Oligo(dT)12-18 primer | Invitrogen | Cat# 18418012 |
| dNTP mix | Invitrogen | Cat# 18427013 |
| Superscript II Reverse Transcriptase | Invitrogen | Cat# 18064014 |
| TaqMan Fast Advanced Mix | Applied Biosystems | Cat# 4444556 |
| TaqMan Gene Expression Assa (FAM) | Applied Biosystems | Cat# 4453320 |
| PowerUp SYBR Green Master Mix | Applied Biosystems | Cat# A25743 |
| Oligonucleotide sequences | This study | See Table S5 |
| Deposited data | ||
| 16S rRNA raw amplicon data from periodontal patient plaque | Mendeley Dataset | http://dx.doi.org/10.17632/vp68zv9wj9.1–Perio_3 |
| 16S rRNA raw amplicon data from mouse ligatures | Mendeley Dataset | 10.17632/vp68zv9wj9.1–Ligature_microbiome |
| Reproducible code: | Mendeley Dataset | 10.17632/vp68zv9wj9.1–narrative_methods |
| Experimental models: Organisms/strains | ||
| C57BL/6J | Jackson Laboratories | Stock #000664 |
| BALBc/J | Jackson Laboratories | Stock #000651 |
| Software and algorithms | ||
| Forsyth Institute eHOMD pipeline | Forsyth Institute | https://microbiome.forsyth.org/ |
| FIJI | NIH Image | https://imagej.net/Fiji |
| Other | ||
| TM7 Probe 5’-Cy5-CCTACGCAACTCTTTACGCC | IDT and Biomers | From (Bor et al., 2020) |
| Actinomyces specific Probe 5’-ATTO425-ATCCAGCTACCGTCAACC | Biomers | From (Gmür and Lüthi-Schaller, 2007) |
| Propionibacteria Probe 5’-Cy3-TCAGGAAAGGTCCAGAGA | Biomers | This study |
| Fusobacterium Probe 5’-AlexaFluor488-GGCTTCCCCATCGGCATT | Biomers | From (Valm et al., 2011) |
| Streptococcus Probe 5’-ATTO532-TAGCCGTCCCTTTCTGGT | Biomers | From (Paster et al., 1998) |
| Corynebacterium Probe 5’-ATTO620-AGTTATGCCCGTATCGCCTG | Biomers | From (Mark Welch et al., 2016) |
| Next generation sequencing core | Zymo | https://www.zymoresearch.com/pages/microbiome-analysis-services |
Host bacterial growth monitoring with and without TM7 (oCelloScope)
Monoculture of host bacteria and coculture of host bacteria with TM7 were recovered from frozen stocks and passaged twice in culture medium to ensure homogeneity. Five replicate samples (250 μL) of each culture were loaded into 96-well plates with starting optical density (OD600) of 0.05. The analogous image-based cell density measurement (TANormalized) was acquired using OcelloScope (BioSense Solutions) every 60 minutes for the indicated amount of time (Figure S2). The growth curve is graphed by plotting time versus arbitrary unit (a.u.) of the OcelloScope measurement which is equivalent to cell density measurement at 600 nm. Representative phase-contrast images for various time points were also displayed from the growth curve.
Bone resorption analysis (area, CEJ-ABC)
Resorption Area:
Maxillae were dissected at 28 days after ligature treatment. Gingival tissue was collected for cytokine analysis, and the remaining tissue was removed using beetle colonies for two weeks. The de-fleshed maxillae were then bleached by washing in 5% hydrogen peroxide for 8 hours, and then stained by 1% Toluidine Blue O (Sigma) for 5-10 seconds. The specimens were photographed using a digital stereomicroscopy on a custom-made stage-holder with maxillary teeth at a certain angle to enhance visualization. The area from the cementoenamel junction to the alveolar bone crest of the second molar was measured using FIJI software by drawing an area of interest (Schindelin et al., 2012).
Micro-Computer Tomography Imaging and Analysis:
Maxillae with soft tissues removed were scanned in a μCT-40 (Scanco, Brüttisellen, Switzerland) at 70kVp, 114μA, 8W, 8μm voxel size, and 300 milliseconds integration time. To reduce noise, samples were scanned in ddH2O. The microCT files were converted into DICOM image stacks using SCANCO medical evaluation (Version 1.1.19) and subsequently imported into Imaris Microscopy Image Analysis Software. Samples were oriented in a standardized manner and a 100 μm thick cross-section was taken in a coronal plane through the distal root of the maxillary right and left second molars for subsequent assessment of bone loss in Fiji version 2.0.0 (Schindelin et al., 2012). Buccal and palatal bone loss was measured as the distance between the cementoenamel junction (CEJ) and alveolar bone crest (ABC).
Ligature microbiome analysis and quantification
Ligature from the mouse 2nd molar was removed and placed in 150 μL of sterile PBS buffer. Cells from the ligature were resuspended by vortexing the sample. 10 μL of the cells were diluted 101-4 and plated on blood BHI agar and incubated for 3 days in a microaerophilic chamber before counting the small and large colonies (Figure S4C). The remaining 140 μL cells were mixed with 150 μL of TE buffer and the gDNA was isolated using MasterPure DNA isolation kit (see above) supplemented with bead beating. Isolated DNA was sent for 16S rRNA sequencing to Forsyth Institute or Zymo sequencing core (http://dx.doi.org/10.17632/vp68zv9wj9.1).
Relative abundance and absolute read counts were extracted from Forsyth or Zymo sequencing core analysis output table. For absolute reads, we only compared within TM7/host bacteria groups (e.g., XH001 vs. TM7x/XH001) and not between groups (e.g., TM7x/XH001 vs. BB002/F0337). This is due to the fact that these groups were sequenced separately from different preparations, and on different sequencing platforms, resulting in a large difference between the total read counts.
Cytokine quantification from the tissue surrounding the ligature
Ligature soaked with control or bacteria were applied for 7 or 28 days. At those times, ligature was removed, and tissue surrounding the ligature area was harvested according to previous protocol (Marchesan et al., 2018). Total RNA was isolated and purified from the gingival tissues by PureLink RNA Mini Kit (Invitrogen). cDNA was then synthesized by using the RNA in with oligo primer (Invitrogen) and reverse transcription enzymes (Invitrogen) according to the manufacturer’s instructions. The expression levels of inflammatory genes such as interleukin 1 (IL-1), interleukin 6 (IL-6), interleukin 17 (IL-17), and tumor necrosis factor-alpha (TNFα) were detected by qPCR using the Thermo Fisher Scientific QuantStudio 3 Real-Time PCR System, with the TaqMan Fast Advanced Master Mix (Applied Biosystems). The detection was conducted in triplicates, and the expression levels were normalized by β-actin mRNA level. Relative fold change in gene expression was determined by using the 2−ΔΔCT method as described in (Livak and Schmittgen, 2001).
Histology and immunohistochemistry quantification
The maxillae were dissected at 11 days after experimental periodontitis treatment and were subsequently fixed with 10% buffered formalin. For tissue staining, the maxillae were then decalcified with Immunocal (StatLab) for 72 hours, and then processed through alcohol and xylene. Tissues were then embedded into paraffin wax. Sections were sliced at 5 μm thickness using a microtome (Leica HMT55). Serial sections were then stained with H&E and immunohistochemistry. For immunohistochemistry staining, tissues were de-waxed and then blocking was done for 20 min at room temperature in 3% normal goat serum, the slices were incubated overnight using the primary antibody of rat-anti-mouse Ly6G (BioLegend) at 1/500 dilution. Then, goat-anti-rat secondary antibodies (Vector) were used to further stain the slices on the following day.
Ly-6G positive cells were quantified at 200X magnification with the region of interest being the gingival connective tissue between the root surface of the first and second molars, from the epithelial border down to the alveolar bone crest (Xiao et al., 2017). The periodontal ligament was excluded from quantitative analysis. A minimum of 6 hemimaxillae were examined per group. The immunopositive cells were counted by a blinded observer per unit area with ImageJ software, and data are expressed as the number per mm2.
Growth crash experiment
Bacterial cultures were grown for two passages in BHI and aliquoted into fresh tubes (x3) with each tube containing a final OD600 of 0.037. The cells were pelleted by centrifuging at 13,000 x g for 5 minutes and resuspended in 300 μL of fresh BHI. To each tube of host bacteria, 4μL of isolated TM7 was added. For the controls, we added PBS alone. The mixed cells were incubated for 10 minutes in room temperature and mixed with an additional 2.2 mL of fresh BHI, bringing the final volume to 2.5 mL. This culture was incubated for 24 hours in a microaerophilic chamber at 37°C and passaged into fresh media at a final concentration of OD~0.1 (final volume 4mL). We repeated the passaging thereafter every 24 hours by transferring the culture in fresh medium to a final volume of 4 mL with OD~0.1. By performing passaging, we tried to mimic continuous culture as much as possible and ensure that nutrient is not a limiting factor for host bacterial growth. During every passage, the optical (cell) density was monitored at 600 nm using a Spectronic Genesys 5 spectrophotometer.
Cell surface protein profiling by mass spectrometry
Isolation and washing of bacterial cell walls.
Total cell wall proteins of Actinomyces were isolated using a modified protocol by (Yu et al., 2012) that was adapted for nanoLC-MS/MS instead of 2D-gel analysis. XH001p and XH001np cultures (5 biological replicates each) were grown for 24 hrs in 100 mL of BHI, harvested at OD600 0.5-0.8 by centrifugation (7,000 x g for 5 min at 4 °C). Each pellet was washed twice with 1.0 mL 4 °C cold PBS, centrifuged and re-suspended in 750 μL ice-cold PBS. Cell suspensions were mixed with equal volumes of 4% (w/v) SDS in PBS and boiled for 15 min in a water bath. Boiling of the cells in SDS dissolved membranes and released cytoplasmic and membrane proteins into solution including well-described surface protein markers such as S-layer proteins (Yu et al., 2012). To release remaining cytosolic proteins repeated freeze-thaw cycles (3 x for 10 min at −80 °C) were used to crack cells open. Soluble protein fractions were recovered by centrifugation at 18,000 x g for 20 min in RT followed by three wash steps with 1 mL of 2 % (w/v) SDS. To eliminate SDS from samples, cell wall pellets were transferred into clean tubes and washed twice with 1.0 mL TE buffer (25 mM Tris-HCl, pH 7.5) containing 10% isopropanol and centrifuging samples for 5 min at 18,000 x g. An additional wash step with 1.0 mL TE containing 1 M NaCl was employed to released proteins attached to the cell wall fraction through ionic interactions. Two additional wash steps with 1.0 mL TE followed to eliminate NaCl from specimens (18,000 x g for 5 min at RT). The last two wash fractions were subjected to SDS-PAGE analysis to test for residual soluble proteins.
Digestion of cell wall with mutanolysin.
10,000 U of mutanolysin (Sigma-Aldrich, Burlington, MA, USA) were dissolved in 1 mL of 50% Glycerol/50% 25mM Tris-HCl, 50 mM NaCl, pH 7.5 and 50 μl aliquots prepared and kept at −20°C until further use. To release cell wall associated proteins, each purified Actinomyces cell wall was resuspended to a final concentration of 20 μg/mL in 750 μL TE containing 100 U of mutanolysin and incubated for 3-4 h at 37 °C. Dissolved cell wall proteins were separated from insoluble materials by centrifugation at 18,000 x g for 15 min at 4 °C.
Protein sample preparation.
Cell wall protein extracts were prepared for mass spectrometric analysis by using a modified version of the filter-aided sample preparation (FASP) protocol described by (Wiśniewski et al., 2009). Specifically, 500 μL of cell wall extracts were ultrafiltrated in Vivaspin filter units (molecular weight cut-off: 10,000 Da; Sartorius, Göttingen, Germany) for 60 min at 14,000 x g at 4 °C. Retentates were washed with 400 μL UA buffer (8 M urea, 50 mM Tris pH 8.0, 10 mM DTT), 200 μL UA buffer, 100 μL 50 mM iodoacetamide in 50 mM ammonium bicarbonate (ABC) buffer after 15 min incubation, 400 μL 50 mM and 200 μl ABC 50 mM buffer. Subsequently, retentates were reconstituted in 100 μL 50 mM ABC buffer and transferred into new microcentrifuge tubes. Total protein content was quantified by using the Micro BCA Assay (Pierce, Dallas, TX, USA). Equivalents of 80 μg of protein were diluted in ABC buffer to a final volume of 80 μL. Protein samples were digested overnight at 37°C with 10 μL trypsin (0.1 μg/μL; Promega, Madison, WI, USA) in ABC buffer with an additional bolus of 10 μL trypsin added in the morning. Samples were incubated for an additional 2 hrs at 37 °C to cleave the remaining undigested proteins. Protein digests were transferred into new Vivaspin filter units and centrifuged for 30 min at 12,000 × g. To release peptides trapped in the filter units 100 μL ABC buffer was added and centrifuged again for 30 min at 12,000 × g. Filtrates from respective samples were combined and acidified to a final concentration of 0.5 % (v/v) trifluoroacetic acid (TFA). Acidified digests were subjected to C18 solid-phase extraction using 100 μL C18 Tips (Pierce, Dallas, TX, USA) according to specifications provided by the manufacturer. Purified peptides were released from the C18 resin with 50% (v/v) acetonitrile containing 0.1 % (v/v) formic acid, evaporated to dryness in a Vacufuge plus (Eppendorf, Hamburg, Germany) and resuspended in 25 μL 0.1 % (v/v) formic acid. Insoluble compounds were removed by centrifugation prior to analysis by nanoflow-liquid chromatography-tandem mass spectrometry (nanoLC-MS/MS).
LC-MS/MS Analysis.
LC-MS/MS analyses of tryptically digested cell wall protein samples were performed on an Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) interfaced with an Easy-nLC 1000 system coupled online with an EASY-Spray source. The LC-MS/MS system was operated using Xcalibur software (Thermo Fisher Scientific). For the analyses, 5 μL of tryptic digest extracts were injected and trapped on an Acclaim PepMap 100 (100 μm x 2 cm; Thermo Fisher Scientific) trap column and separated on a PepMap RSLC C18 (ES901, 50 μm x 15 mm, 100 A particle size; Thermo Fisher Scientific) analytical column. The flow rate was set to 300 nL/min, the column temperature to 45°C, and the ion source temperature to 275°C with an electrical potential of 1800 V in positive ionization mode. Peptides were chromatographically separated with a 2-32% 150 min linear acetonitrile/water gradient in 0.1% formic acid. Mass spectra were acquired in data-dependent acquisition mode with high resolution MS1 acquisition (Resolution (R) = 60,000; automatic gain control (AGC) target setting 3.0 x 105 ions, maximal injection time of 250 ms) and MS2 (R = 15,000, AGC target 5 x 104 ions, maximal injection time of 150 ms). The 20 most intense precursor ions with a minimum intensity of 5 .0 × 104 ions were selected (1.6 Da isolation window) for fragmentation in the HCD cell (collision energy set to 30). The dynamic exclusion window was set to 40s.
Protein identification, quantitation, and statistical analysis.
The raw data was processed and searched with the PEAKS Studio 8.5 software package (Bioinformatics Solutions Inc., Waterloo, Ontario, Canada) using the Actinomyces odontolytica UniProt database (UniProt release July 2020, 1925 entries). Raw data were processed using the following specifications: merge scans (retention time window: 10 ppm, precursor m/z tolerance: 10 ppm, precursor mass and charge states z = 1-10). Other data pre-processing (centroiding, deisotoping, and deconvolution) was executed automatically. The following search parameters were used: precursor mass error tolerance 10 ppm; fragment ion mass error tolerance 0.05 Da; trypsin enzyme specificity with cleavage prior to proline permitted; fixed modifications: carbamidomethylation (C); variable modifications: phosphorylation (STY), oxidation (M), pyro-glu (Q), deamidation (NQ); one non-specific cleavage specificity on one terminus, maximal two missed cleavages, maximal two variable posttranslational modifications per peptide. The false discovery rate for peptide identifications was controlled using the decoy-fusion method. A 1% peptide FDR was applied at the peptide spectrum match level and only protein hits above the significance threshold of 20 (−10lgP) were retained for further analysis. For the relative quantitation of proteins, the peak areas of the top 3 most abundant unique peptides were employed. Raw protein peak areas were exported as a CSV table and analyzed by using Microsoft Excel. All protein abundance differences with log2 mean ratio ≤ or ≥ 1 and FDR ≤ 20 were considered significant.
Generating XH001p mutants
XH001p Δcbp2 and ΔnanE strains created in this study were constructed and verified as previously described (Bedree et al., 2018) with modifications: the chromosomal gene deletion constructs contained ~1.0 kb up/downstream homology fragments to augment the double-crossover homologous recombination event that generated a 3.0 kb construct. For nanE, the deletion constructs were designed in such a way that the selection marker was inserted in the opposite transcriptional direction than the nanE gene and its operon. This helps avoid disruption of the expression of downstream genes due to the lack of transcriptional termination in the selection marker. However, even with this orientation, the selection maker could still affect the downstream gene nanT due to transcriptional interference (Figure S6A). To test this, we measured the relative expression of nanT in the ΔnanE mutant by RT-qPCR (described below) and did not find a significant difference compared to XH001p (Figure S6C). Primers 1-6 (cbp2) and 7-12 (nanE) were used to generate the individual fragments via fusion PCR (Shevchuk et al., 2004) as previously depicted (Bedree et al., 2018). Additionally, 2 μg of the gene deletion constructs (DNA) for nanE and cbp2, native genes designated as APY09_01955 and APY09_04670, respectively, were used for transformation. Mid-log exponential phase cells (0.5 and 0.7 OD600) were harvested for electrocompetent cell preparation to increase transformation efficiency. BHI agar and media (Difco Laboratories, Detroit, Michigan) were supplemented with 250 μg/ml of Kanamycin sulfate (Fischer Bioreagents, Hampton, NH, United States) for Δcbp2 and ΔnanE mutant selection during transformation. gDNA was isolated as previously described (Truett et al., 2000) for template gDNA to verify Δcbp2 and ΔnanE allelic replacement of the native genes with the Kanamycin resistance cassette. Primers 13/14 and 15/16 were used to generate 700-750 bp amplicons confirmed by DNA sequencing (Psomagen, Inc. Boston, Cambridge, MA, United States). To evaluate the ablation of cbp2 mRNA transcripts via cDNA, primers 17-22 were used to amplify (~800-850 bp amplicons) three separate and equidistant loci within CBP due to large gene length (7.5kb). Primers 23/24 were used to amplify (~650 bp amplicon) the full length nanE gene. All PCR reactions were carried out using Q5® High-Fidelity DNA Polymerase (New England Biolabs, Ipswich, MA) following the manufacturer’s protocol for PCR cycling parameters. An annealing temperature of 65°C was used for all PCR reactions, except primers 17-22 used annealing temperature of 59°C. All confirmed Δcbp2 and ΔnanE lacked mRNA transcripts. All primers used in this study are listed in Supplementary Table S5.
The complementation experiments for the two deleted genes were not conducted for two reasons: currently we do not have a well-developed expression system in Actinomyces odontolyticus strain XH001, and even if we develop the additional genetic tools, it will still be difficult to select and maintain for a plasmid with antibiotics once the mutants are in the mouse ligature/tooth itself. Treating the mouse with antibiotics for 28 days to select for the plasmid would impact the mouse physiology and the mouse oral/gut microbiome composition. Therefore, we reasoned analyzing the mRNA level and testing the mutant’s phenotype is sufficient (Figure 6D).
Quantitative Real-Time PCR gene expression analysis for nanT
Isolation of mRNA, construction of cDNA, as well as quantitative real-time PCR was performed as previously described (Bor et al., 2016). PowerUp SYBR Green Master Mix (Applied Biosystems) was used to perform the qPCR with the Thermo Fisher Scientific QuantStudio 3 Real-Time PCR System. The final RT-qPCR mixture (10μL) contained 1x SYBR Green Master Mix, 1μg cDNA, and 750nM of the appropriate forward and reverse RT-qPCR primers (Table S5) designed for the XH001 16S and nanT genes. Three biological replicates of XH001p and nanE mutant were used to determine the variation in the experiment (depicted by standard deviation). The detection of the nanT gene transcript was conducted in triplicates, and the expression levels were normalized by 16S level. Relative fold change in gene expression was determined by using the 2−ΔΔCT method as described in (Figure S6C) (Livak and Schmittgen, 2001).
Gene neighborhood analysis for nanE
Gene neighborhoods were generated with Clinker (Gilchrist and Chooi, 2021) on 6 complete genomes downloaded from NCBI (F0337, FDAARGOS 1037, F0631, F0588, XH001, and W712). Alignments were performed using default parameters and colored manually in Inkscape.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis of the 16S rRNA profiling is described in the Sequencing Data Analysis section above. Analysis for all other experiments was performed by using GraphPad Prism 9.0 software from GraphPad Software Inc. (La Jolla, CA, USA). In our graphs, we presented all individual points in the bar graphs to illustrate the spread of the data and the bar graph, and the error bars are expressed as the mean ± SD. The difference between the two groups was shown by the Mann-Whitney U test while multiple group comparisons were performed by one-way ANOVA, with Bonferroni’s post hoc test to identify differences between specific groups. A p-value of < 0.05 was considered to be statistically significant. Our initial ligature experiments were done on individual animals where ligature was tied to one side while the other side was not treated with a ligature (internal control). However, once we established that the bone loss was robust, to minimize the number of animals, we applied two ligatures to each mouse on both second molars. Therefore, initial experiments used individual animals as the unit of measurement while later experiments used five mice per group for up to ten ligatures per group measurements.
Supplementary Material
Table S4. Differentially expressed cell wall genes from the volcano plot. Related to Figure 6.
Highlights.
Saccharibacteria (TM7) is an epibiont that lives on the surface of Actinobacteria
Multiple TM7 isolates from periodontitis patients decrease inflammatory bone loss
TM7 reduce bone loss by modulating the pathogenicity of their host Actinobacteria
TM7 could serve a protective role for mammalian hosts in inflammatory diseases
Acknowledgments
We thank Xiaozhe Han and Yufeng Wang for ligature training; Alpdogan Kantarci, Thomas Van Dyke and Ning Yu for productive discussion on immunology; Floyd Dewhirst and Mircea Podar for host bacterial strains. We were supported by The National Institute of Dental and Craniofacial Research of the National Institutes of Health under Awards 1R01DE023810, 1R01DE020102, 1R01DE026186 (to X.H., and J.S.M); F31DE026057 (to J.K.B.); and 1K99DE027719-01 (to B.B.). Additional support was provided to D.R.U by the National Science Foundation Graduate Research Fellowship Program under Grant DGE1745303 (to D.R.U). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or NSF.
Footnotes
Declaration of Interest
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abe T, and Hajishengallis G (2013). Optimization of the ligature-induced periodontitis model in mice. Journal of Immunological Methods 394, 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abusleme L, Dupuy AK, Dutzan N, Silva N, Burleson JA, Strausbaugh LD, Gamonal J, and Diaz PI (2013). The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. The ISME Journal 7, 1016–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achermann Y, Goldstein EJC, Coenye T, and Shirtliff ME (2014). Propionibacterium acnes: from Commensal to Opportunistic Biofilm-Associated Implant Pathogen. Clinical Microbiology Reviews 27, 419–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Kamel A, Baraniya D, Al-Hajj WA, Halboub E, Abdulrab S, Chen T, and Al-Hebshi NN (2019). Subgingival microbiome of experimental gingivitis: shifts associated with the use of chlorhexidine and N-acetyl cysteine mouthwashes. J Oral Microbiol 11, 1608141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora S, Gordon J, and Hook M (2021). Collagen Binding Proteins of Gram-Positive Pathogens. Front Microbiol 12, 628798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker PJ, Dixon M, and Roopenian DC (2000). Genetic Control of Susceptibility toPorphyromonas gingivalis-Induced Alveolar Bone Loss in Mice. Infect. Immun 68, 5864–5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batinovic S, Rose JJA, Ratcliffe J, Seviour RJ, and Petrovski S (2021). Cocultivation of an ultrasmall environmental parasitic bacterium with lytic ability against bacteria associated with wastewater foams. Nat Microbiol 6, 703–711. [DOI] [PubMed] [Google Scholar]
- Bedree JK, Bor B, Cen L, Edlund A, Lux R, McLean JS, Shi W, and He X (2018). Quorum Sensing Modulates the Epibiotic-Parasitic Relationship Between Actinomyces odontolyticus and Its Saccharibacteria epibiont, a Nanosynbacter lyticus Strain, TM7x. Front. Microbiol 9, 2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein DB, Dewhirst FE, and Segrè D (2019). Metabolic network percolation quantifies biosynthetic capabilities across the human oral microbiome. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, Perez-Perez G, Blaser MJ, and Relman DA (2006). Molecular analysis of the bacterial microbiota in the human stomach. Proc. Natl. Acad. Sci. U.S.A 103, 732–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bor B, Poweleit N, Bois JS, Cen L, Bedree JK, Zhou ZH, Gunsalus RP, Lux R, McLean JS, He X, et al. (2016). Phenotypic and Physiological Characterization of the Epibiotic Interaction Between TM7x and Its Basibiont Actinomyces. Microb Ecol 71, 243–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bor B, McLean JS, Foster KR, Cen L, To TT, Serrato-Guillen A, Dewhirst FE, Shi W, and He X (2018). Rapid evolution of decreased host susceptibility drives a stable relationship between ultrasmall parasite TM7x and its bacterial host. Proc. Natl. Acad. Sci. U.S.A 115, 12277–12282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bor B, Bedree JK, Shi W, McLean JS, and He X (2019). Saccharibacteria (TM7) in the Human Oral Microbiome. J Dent Res 98, 500–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bor B, Collins AJ, Murugkar PP, Balasubramanian S, To TT, Hendrickson EL, Bedree JK, Bidlack FB, Johnston CD, Shi W, et al. (2020). Insights Obtained by Culturing Saccharibacteria With Their Bacterial Hosts. J Dent Res 002203452090579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CT, Hug LA, Thomas BC, Sharon I, Castelle CJ, Singh A, Wilkins MJ, Wrighton KC, Williams KH, and Banfield JF (2015). Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 523, 208–211. [DOI] [PubMed] [Google Scholar]
- Burckhardt JJ, Gaegauf-Zollinger R, Schmid R, and Guggenheim B (1981). Alveolar bone loss in rats after immunization with Actinomyces viscosus. Infect Immun 31, 971–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, and Holmes SP (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13, 581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camanocha A, and Dewhirst FE (2014). Host-associated bacterial taxa from Chlorobi, Chloroflexi, GN02, Synergistetes, SR1, TM7, and WPS-2 Phyla/candidate divisions. Journal of Oral Microbiology 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelle CJ, and Banfield JF (2018). Major New Microbial Groups Expand Diversity and Alter our Understanding of the Tree of Life. Cell 172, 1181–1197. [DOI] [PubMed] [Google Scholar]
- Chang D-E, Smalley DJ, Tucker DL, Leatham MP, Norris WE, Stevenson SJ, Anderson AB, Grissom JE, Laux DC, Cohen PS, et al. (2004). Carbon nutrition of Escherichia coli in the mouse intestine. Proc Natl Acad Sci U S A 101, 7427–7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipashvili Otto; Utter Daniel R.; Bedree Joseph K.; Ma Yansong; Schulte Fabian; Mascarin Gabrielle; Alayyoubi Yasmin; Chouhan Deepak; Hardt Markus; Bidlack Felicitas; Hasturk Hatice; He Xuesong; McLean Jeffrey S.; Bor Batbileg (2021), “Ultrasmall episymbiotic Saccharibacteria suppresses gingival inflammation and bone loss through host bacterial modulation”, Mendeley Data, V1, doi: 10.17632/vp68zv9wj9.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross KL, Campbell JH, Balachandran M, Campbell AG, Cooper SJ, Griffen A, Heaton M, Joshi S, Klingeman D, Leys E, et al. (2019). Targeted isolation and cultivation of uncultivated bacteria by reverse genomics. Nat. Biotechnol 37, 1314–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew GC, Stevens EJ, and King KC (2021). Microbial evolution and transitions along the parasite–mutualist continuum. Nat Rev Microbiol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Wu T, Xu X, Chen D, Mo A, Lei Y, Cheng L, Man Y, Zhou X, Wang Y, et al. (2017). Smoking May Lead to Marginal Bone Loss Around Non-Submerged Implants During Bone Healing by Altering Salivary Microbiome: A Prospective Study. J Periodontol 88, 1297–1308. [DOI] [PubMed] [Google Scholar]
- Eren AM, Borisy GG, Huse SM, and Mark Welch JL (2014). Oligotyping analysis of the human oral microbiome. Proc Natl Acad Sci U S A 111, E2875–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escapa IF, Chen T, Huang Y, Gajare P, Dewhirst FE, and Lemon KP (2018). New Insights into Human Nostril Microbiome from the Expanded Human Oral Microbiome Database (eHOMD): a Resource for the Microbiome of the Human Aerodigestive Tract. MSystems 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galimanas V, Hall MW, Singh N, Lynch MDJ, Goldberg M, Tenenbaum H, Cvitkovitch DG, Neufeld JD, and Senadheera DB (2014). Bacterial community composition of chronic periodontitis and novel oral sampling sites for detecting disease indicators. Microbiome 2, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan SM, Joshi V, Fellows M, Dabdoub SM, Nagaraja HN, O’Donnell B, Deshpande NR, and Kumar PS (2017). A tale of two risks: smoking, diabetes and the subgingival microbiome. ISME J 11, 2075–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Tseng C, Pei Z, and Blaser MJ (2007). Molecular analysis of human forearm superficial skin bacterial biota. Proc. Natl. Acad. Sci. U.S.A 104, 2927–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist CLM, and Chooi Y-H (2021). clinker & clustermap.js: automatic generation of gene cluster comparison figures. Bioinformatics btab007. [DOI] [PubMed] [Google Scholar]
- Gmür R, and Lüthi-Schaller H (2007). A combined immunofluorescence and fluorescent in situ hybridization assay for single cell analyses of dental plaque microorganisms. Journal of Microbiological Methods 69, 402–405. [DOI] [PubMed] [Google Scholar]
- Graves DT, Fine D, Teng Y-TA, Van Dyke TE, and Hajishengallis G (2008). The use of rodent models to investigate host-bacteria interactions related to periodontal diseases. J Clin Periodontol 35, 89–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, Podar M, and Leys EJ (2012). Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. The ISME Journal 6, 1176–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G (2014). The inflammophilic character of the periodontitis-associated microbiota. Mol Oral Microbiol 29, 248–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Osaka T, Tawaratsumida K, Yamazaki T, Tada H, Chen GY, Tsuneda S, Núñez G, and Inohara N (2010). Transitions in Oral and Intestinal Microflora Composition and Innate Immune Receptor-Dependent Stimulation during Mouse Development. IAI 78, 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, and De Buck J (2010). Cell wall proteome analysis of Mycobacterium smegmatis strain MC2 155. BMC Microbiol 10, 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, McLean JS, Edlund A, Yooseph S, Hall AP, Liu S-Y, Dorrestein PC, Esquenazi E, Hunter RC, Cheng G, et al. (2015). Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc. Natl. Acad. Sci. U.S.A 112, 244–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Yang F, Zeng X, Chen J, Li R, Wen T, Li C, Wei W, Liu J, Chen L, et al. (2011). Preliminary characterization of the oral microbiota of Chinese adults with and without gingivitis. BMC Oral Health 11, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, Butterfield CN, Hernsdorf AW, Amano Y, Ise K, et al. (2016). A new view of the tree of life. Nature Microbiology 1, 16048. [DOI] [PubMed] [Google Scholar]
- Jaffe AL, He C, Keren R, Valentin-Alvarado LE, Munk P, Bouma-Gregson K, Farag IF, Amano Y, Sachdeva R, West PT, et al. (2021). Patterns of gene content and co-occurrence constrain the evolutionary path toward animal association in CPR bacteria (Microbiology). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Darzi Y, Tawaratsumida K, Marchesan JT, Hasegawa M, Moon H, Chen GY, Núñez G, Giannobile WV, Raes J, et al. (2013). Induction of Bone Loss by Pathobiont-Mediated Nod1 Signaling in the Oral Cavity. Cell Host & Microbe 13, 595–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan HV, Fitzgerald RJ, and Stanley HR (1965). Plaque formation and periodontal pathology in gnotobiotic rats infected with an oral actinomycete. Am J Pathol 47, 1157–1167. [PMC free article] [PubMed] [Google Scholar]
- Kodaman N, Pazos A, Schneider BG, Piazuelo MB, Mera R, Sobota RS, Sicinschi LA, Shaffer CL, Romero-Gallo J, de Sablet T, et al. (2014). Human and Helicobacter pylori coevolution shapes the risk of gastric disease. Proceedings of the National Academy of Sciences 111, 1455–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Könönen E, and Wade WG (2015). Actinomyces and related organisms in human infections. Clin Microbiol Rev 28, 419–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Bi L, Yu X, Kawai T, Taubman MA, Shen B, and Han X (2014). Porphyromonas gingivalis Exacerbates Ligature-Induced, RANKL-Dependent Alveolar Bone Resorption via Differential Regulation of Toll-Like Receptor 2 (TLR2) and TLR4. Infect. Immun 82, 4127–4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Faller LL, Klitgord N, Mazumdar V, Ghodsi M, Sommer DD, Gibbons TR, Treangen TJ, Chang Y-C, Li S, et al. (2012). Deep Sequencing of the Oral Microbiome Reveals Signatures of Periodontal Disease. PLoS ONE 7, e37919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, and Schmittgen TD (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2–ΔΔCT Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lloyd-Price J, Abu-Ali G, and Huttenhower C (2016). The healthy human microbiome. Genome Med 8, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesan J, Girnary MS, Jing L, Miao MZ, Zhang S, Sun L, Morelli T, Schoenfisch MH, Inohara N, Offenbacher S, et al. (2018). An experimental murine model to study periodontitis. Nat Protoc 13, 2247–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE, and Borisy GG (2016). Biogeography of a human oral microbiome at the micron scale. Proc. Natl. Acad. Sci. U.S.A 113, E791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin J, Watterson S, Layton AM, Bjourson AJ, Barnard E, and McDowell A (2019). Propionibacterium acnes and Acne Vulgaris: New Insights from the Integration of Population Genetic, Multi-Omic, Biochemical and Host-Microbe Studies. Microorganisms 7, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean JS, Bor B, Kerns KA, Liu Q, To TT, Solden L, Hendrickson EL, Wrighton K, Shi W, and He X (2020). Acquisition and Adaptation of Ultra-small Parasitic Reduced Genome Bacteria to Mammalian Hosts. Cell Rep 32, 107939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutsopoulos NM, Chalmers NI, Barb JJ, Abusleme L, Greenwell-Wild T, Dutzan N, Paster BJ, Munson PJ, Fine DH, Uzel G, et al. (2015). Subgingival microbial communities in Leukocyte Adhesion Deficiency and their relationship with local immunopathology. PLoS Pathog 11, e1004698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugkar PP, Collins AJ, Chen T, and Dewhirst FE (2020). Isolation and cultivation of candidate phyla radiation Saccharibacteria (TM7) bacteria in coculture with bacterial hosts. Journal of Oral Microbiology 12, 1814666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson ME, King JM, Yahr TL, and Horswill AR (2013). Sialic acid catabolism in Staphylococcus aureus. J Bacteriol 195, 1779–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papapanou PN, Park H, Cheng B, Kokaras A, Paster B, Burkett S, Watson CW-M, Annavajhala MK, Uhlemann A-C, and Noble JM (2020). Subgingival microbiome and clinical periodontal status in an elderly cohort: The WHICAP ancillary study of oral health. J Periodontol 91 Suppl 1, S56–S67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster BJ, Bartoszyk IM, and Dewhirst FE (1998). Identification of oral streptococci using PCR-based, reverse-capture, checkerboard hybridization. Methods in Cell Science 20, 223–231. [Google Scholar]
- Sato T, Watanabe K, Kumada H, Toyama T, Tani-Ishii N, and Hamada N (2012). Peptidoglycan of Actinomyces naeslundii induces inflammatory cytokine production and stimulates osteoclastogenesis in alveolar bone resorption. Arch. Oral Biol 57, 1522–1528. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada E, Kataoka H, Miyazawa Y, Yamamoto M, and Igarashi T (2012). Lipoproteins of Actinomyces viscosus induce inflammatory responses through TLR2 in human gingival epithelial cells and macrophages. Microbes Infect 14, 916–921. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Hubersak C, and Propas D (1970). Induction of periodontal destruction in gnotobiotic rats by a human oral strain of Actinomyces naeslundii. Arch Oral Biol 15, 993–995. [DOI] [PubMed] [Google Scholar]
- Takada H, Kimura S, and Hamada S (1993). Induction of inflammatory cytokines by a soluble moiety prepared from an enzyme lysate of Actinomyces viscosus cell walls. J Med Microbiol 38, 395–400. [DOI] [PubMed] [Google Scholar]
- Truett GE, Heeger P, Mynatt RL, Truett AA, Walker JA, and Warman ML (2000). Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques 29, 52, 54. [DOI] [PubMed] [Google Scholar]
- Utter DR, He X, Cavanaugh CM, McLean JS, and Bor B (2020). The saccharibacterium TM7x elicits differential responses across its host range. ISME J 14, 3054–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valm AM, Welch JLM, Rieken CW, Hasegawa Y, Sogin ML, Oldenbourg R, Dewhirst FE, and Borisy GG (2011). Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proceedings of the National Academy of Sciences 108, 4152–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A (2017). Are humans prone to autoimmunity? Implications from evolutionary changes in hominin sialic acid biology. J Autoimmun 83, 134–142. [DOI] [PubMed] [Google Scholar]
- Wei Y, Shi M, Zhen M, Wang C, Hu W, Nie Y, and Wu X (2019). Comparison of Subgingival and Buccal Mucosa Microbiome in Chronic and Aggressive Periodontitis: A Pilot Study. Front Cell Infect Microbiol 9, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewski JR, Zougman A, Nagaraj N, and Mann M (2009). Universal sample preparation method for proteome analysis. Nat Methods 6, 359–362. [DOI] [PubMed] [Google Scholar]
- Xiao E, Mattos M, Vieira GHA, Chen S, Corrêa JD, Wu Y, Albiero ML, Bittinger K, and Graves DT (2017). Diabetes Enhances IL-17 Expression and Alters the Oral Microbiome to Increase Its Pathogenicity. Cell Host & Microbe 22, 120–128.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Xu X, Peng Y, Luo Y, and Yang K (2012). Cell wall proteome of Clostridium thermocellum and detection of glycoproteins. Microbiological Research 167, 364–371. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Dupont CL, Jones MB, Pham KM, Jiang Z-D, DuPont HL, and Highlander SK (2018). Visualization-assisted binning of metagenome assemblies reveals potential new pathogenic profiles in idiopathic travelers’ diarrhea. Microbiome 6, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S4. Differentially expressed cell wall genes from the volcano plot. Related to Figure 6.
Data Availability Statement
The raw sequencing data and original code generated by this study were deposited on Mendeley at http://dx.doi.org/10.17632/vp68zv9wj9.1, with the exception of initial 32 patient 16S rRNA sequencing data. These data were collected as a confidential medical record at Forsyth Center for Clinical and Translational Research, and we cannot release the raw data publicly due to ethical prohibition. Individuals who are interested in these data can contact the Lead Contact. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [1] partner repository with the dataset identifier PXD026999 and 10.6019/PXD026999.