Abstract
Apoptosis-associated speck-like protein containing a caspase recruitment domain (ASC) is a key adaptor protein of inflammasomes and a proapoptotic molecule; however, its roles in signal transduction in pancreatic ductal adenocarcinoma (PDAC) cells remain unknown. Here, we clarified the role and mechanisms of action of ASC in PDAC using clinical evidence and in vitro data. ASC expression in PDAC tissues was analyzed using public tumor datasets and immunohistochemistry results of patients who underwent surgery, and PDAC prognosis was investigated using the Kaplan–Meier Plotter. ASC expression in PDAC cells was downregulated using small-interfering RNA, and gene expression was assessed by RNA sequencing. Review of the Oncomine database and immunostaining of surgically removed tissues revealed elevated ASC expression in PDAC tumors relative to non-tumor tissue, indicating poor prognosis. We observed high ASC expression in multiple PDAC cells, with ASC silencing subsequently inhibiting PDAC cell growth and altering the expression of cell cycle-related genes. Specifically, ASC silencing reduced cyclin D1 levels and stopped the cell cycle at the G1 phase but did not modulate the expression of any apoptosis-related molecules. These results show that ASC inhibited tumor progression via cell cycle modulation in PDAC cells and could be a potential therapeutic target.
Subject terms: Cell growth, Gastroenterology
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is a very aggressive disease with the poorest prognosis among human cancers1. Despite the introduction of new treatment modalities in recent years, treatment options remain limited, and chemotherapy has not been fully effective2,3. Thus, there is an urgent need to identify novel targets and mechanisms underlying PDAC progression.
Apoptosis-associated speck-like protein containing a caspase recruitment domain [ASC; also called PYD- and CARD-domain-containing protein (PYCARD) and target of methylation-induced silencing 1 (TMS1)] reportedly forms aggregates in human myelocytic leukemia HL-60 cells undergoing apoptosis4. ASC is a key adaptor protein in the formation of various inflammasomes and plays crucial roles in caspase-1 activation and the secretion of interleukin (IL)-1β and IL-18 in innate immune cells5–7. However, ASC has also been identified as a target of TMS1 and a gene that is silenced by DNA methyltransferase 1 in breast cancer. Similar to that in breast cancer, ASC expression is decreased by methylation in cancers, such as melanoma8, ovarian cancer9, prostate cancer10, colorectal cancer11, and hepatocellular carcinoma12. It is also reportedly involved in the progression of melanoma13 and oral cavity squamous cell carcinoma14. However, the roles of ASC in PDAC have not been elucidated to date. These conflicting data suggest that the role of ASC in cancer might vary depending on the pathological tumor type, stage of tumor development, and tumor microenvironment15. In this study, we identified the role and mechanisms of ASC in PDAC based on clinical evidence and in vitro data obtained using PDAC cells.
Methods
Analysis of Oncomine data
To determine the expression pattern of ASC in PDAC, we used datasets from the Oncomine database (www.oncomine.org). The ASC gene was queried in the database, and results were filtered by selecting PDAC versus Normal Analysis. Data were displayed using Box Chart. P-values for each group were calculated using the Student’s t test. Details of standardized normalization techniques and statistical calculations are provided in the Oncomine website.
Patients and pancreas specimens
PDAC specimens and adjacent normal pancreatic tissues were obtained from patients who underwent surgery at the Ehime University Hospital. All the specimens used in this study were obtained from eight samples collected between 2018 and 2019 from PDAC patients. Written informed consent was obtained from all enrolled participants, and the study protocol conformed to the ethical guidelines of the Declaration of Helsinki and was approved by the Institutional Review Board of Ehime University Hospital (approval no. 1712007).
Immunohistochemistry (IHC) of PDAC specimens
Pancreatic tissues were fixed in formalin, sections (3-µm thick) were cut from each block, and adjacent sections were stained using IHC. Paraffin-embedded samples were dewaxed and rehydrated, and antigen retrieval was performed by autoclaving for 1 min at 110 °C in citrate buffer (pH 6.0). Endogenous peroxidase activity was inactivated by incubation with methanol containing 1% hydrogen peroxidase for 20 min. The sections were then incubated in 1% blocking goat serum for 30 min to reduce nonspecific reactions. For IHC, the sections were probed overnight with the anti-ASC antibody (1:200; 10500-1-AP; Proteintech, Tokyo, Japan) at 4 °C. The tissue sections were then incubated with a peroxidase-labeled secondary antibody (Histofine Simplestain Max POR; Nichirei, Tokyo, Japan) for 1 h at room temperature and then with Simple Stain DAB solution (Nichirei). Photomicrographs were obtained using a Nikon ECLPSE 50i with a Nikon digital camera (DS-F11; Nikon, Tokyo, Japan). Staining intensity was scored with no prior knowledge of clinicopathological results, in the following manner: 0, no staining; 1, weak staining; 2, moderate staining; and 3, strong staining. Immune-reaction intensity (optical density, OD) was examined using ImageJ (NIH, Bethesda, MD, USA) and calculated as OD = log [max intensity/mean intensity (max intensity = 255].
The Kaplan–Meier Plotter
The Kaplan–Meier Plotter (https://kmplot.com/analysis/) is a web server for the discovery and validation of survival biomarkers16. We used the survival-analysis tool in the Kaplan–Meier Plotter to analyze the prognostic significance of ASC mRNA. Survival analysis was performed using the Kaplan–Meier method, and a hazard ratio (HR) with 95% confidence intervals (CIs) and a log-rank P-value were calculated. Kaplan–Meier Plotter was also used to compare the predictive value of several gene combinations in patients with low and high mutational burden.
Cancer Cell Line Encyclopedia (CCLE) analysis
ASC mRNA levels in a series of cancers were analyzed using the CCLE (https://portals.broadinstitute.org/ccle/home). According to the website, raw microarray data of PDAC cell lines were converted to a single value for each probeset using the Robust Multi-array Average algorithm and quantile normalization.
Cells and culture conditions
Four PDAC cell lines (PANC-1, BxPC-3, AsPC-1, and MIA PaCa-2), which were initially generated using samples from PDAC patients, and hTERT-HPNE cells were obtained from the American Type Culture Collection (Manassas, VA, USA). PANC-1 and MIA PaCa-2 cells were cultured in Dulbecco's modified Eagle medium (DMEM; Life Technologies, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Life Technologies) and 1% penicillin. BxPC-3 and AsPC-1 cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 10% FBS and 1% penicillin. hTERT-HPNE cells were cultured in 75% DMEM and 25% Medium M3 Base with 5% FBS, 10 ng/mL human recombinant epidermal growth factor, 5.5 mM d-glucose, and 750 ng/mL puromycin. Microphotographs were obtained after various treatments using an inverted microscope (AxioVertA1 FL; ZEISS, Oberkochen, Germany) equipped with an Axiocam MRm camera (ZEISS).
RNA extraction, cDNA synthesis, and real-time reverse transcription polymerase chain reaction (RT-PCR)
Total RNA was extracted using TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA), and the RNA was reverse transcribed using RT-PCR kits (Applied Biosystems, Tokyo, Japan). Real-time PCR was performed using a LightCycler 480 (Roche, Tokyo, Japan) with SYBR Green I (Roche). Sequences of the primers used to amplify human genes are indicated in Table S1. Glyceraldehyde 3-phosphate dehydrogenase served as an internal reference gene, and the relative change was calculated by relative quantification using the formula 2−ΔΔCt.
Western blotting
Radioimmunoprecipitation assay buffer [50 mM HEPES, KOH (pH 7.9), 150 mM NaCl, 1.5 mM MgCl2, and 1.0% (v/v) NP-40] was used to lyse cells or homogenize tumors for protein extraction. For western blotting, 20 µg of protein was loaded onto 4–12% Bis–Tris gels (Life Technologies), followed by transfer onto Immobilon-P membranes (Millipore, Bedford, MA, USA) and incubation with the relevant primary antibodies (1:1000–3000 dilutions) (Table S2). Appropriate species-specific conjugated secondary antibody kits were obtained commercially (GE Healthcare, Tokyo, Japan). Proteins were detected using the ECL prime kit or the ECL kit (GE Healthcare) and an ImageQuant LAS 4000 system (GE Healthcare).
RNA interference
Small-interfering (si)RNAs targeting ASC included the following: (1) 5′-AACUGGACCUGCAAGGACUUG-3′ (Sigma-Aldrich Japan, Tokyo, Japan)17 and (2) 5′-CCGCCGAGGAGCUCAAGAA-3′ (Hs_PYCARD_8145; Mission siRNAs; Sigma-Aldrich Japan). Control siGENOME non-targeting siRNA sequences (Thermo Scientific) included the following: 5′-UAAGGCUAUGAAGAGAUAC-3′, 5′-AUGUAUUGGCCUGUAUUAG-3′, 5′-AUGAACGUGAAUUGCUCAA-3′, and 5′-UGGUUUACAUGUCGACUAA-3′. Cell lines were transfected with 25 pM siRNA using Lipofectamine 2000 (Life Technologies), and 3 days after transfecting siRNA, mRNA and protein were extracted and analyzed.
Cell-proliferation assay
Cell viability was quantified by an MTS assay (Promega, Fitchburg, WI, USA). Cells (2 × 103) were seeded in 96-well plates and transfected with siRNA 1 day later (siRNA-transfection day was set as day 0). At each time point, cells were treated with MTS reagent and incubated for 1 h. Absorbance at 452 nm was recorded using a plate reader (Biochrom Asys Expert 96; Biochrom, Cambridge, UK).
RNA sequencing (RNA-seq) and data analyses
Total RNA was collected from PANC-1 and AsPC-1 cells at 72 h after transfection with either control siRNA, ASC siRNA (1), or ASC siRNA (2), and the integrity of isolated RNA was verified using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). RNA samples with an RNA integrity number greater than eight were normalized to 100 ng/μL before further analyses. RNA-seq libraries were prepared using the Illumina TruSeq Stranded mRNA sample prep kit setA according to manufacturer instructions (Illumina, San Diego, CA, USA) and subsequently validated to ensure an average size of ~ 330 to ~ 340 bp using a 2100 Bioanalyzer and the Agilent DNA1000 kit. Paired-end reads (75 bp) were sequenced using the MiSeq reagent kit V3 150 cycle on a MiSeq system (Illumina). Mapping to human genome data hg19 was performed using Tophat (https://ccb.jhu.edu/software/tophat/index.shtml), and the expression analysis tool Cufflinks (http://cole-trapnell-lab.github.io/cufflinks/) was used to normalize each sample before determining the differences in expression between control siRNA (n = 3), ASC siRNA (1) (n = 3), and ASC siRNA (2) (n = 3) using the tmm package in R (https://www.r-project.org/). Raw and processed data were submitted to the Gene Expression Omnibus (GSE168842) database. Pathway and process enrichment analyses were performed using all genes that after alignment and normalization were found to be downregulated with an adjusted p < 0.05 using Metascape18 and gene set enrichment analysis (GSEA)19.
Cell cycle analysis
Cells were seeded at a density of 1 × 106 cells/well in 10-cm dishes. At 72 h after siRNA transfection, a BD Cycletest TM Plus DNA kit (BD Biosciences, Franklin Lakes, NJ, USA) was used to purify the sample according to manufacturer instructions, and cell cycle distribution was evaluated by flow cytometry (Gallios flow cytometer; Beckman Coulter, Brea, CA, USA).
Apoptosis assay
PANC-1 cells were transfected with control siRNA or ASC siRNA, and a Mebstain apoptosis terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) kit II (MBL, Nagoya, Japan) was used to detect oligonucleosomal DNA breaks in PANC-1 cells 3 and 5 days after transfection.
Proteome profiling was performed using commercially available human apoptosis proteome profiler arrays (R&D Systems, Minneapolis, MN, USA) according to manufacturer instructions. Expression levels of 35 apoptosis-related proteins (effector and signaling molecules) were evaluated by densitometric analyses of the arrays using ImageJ (NIH).
Cytokine quantification in tumor cells
Tumor cells were plated onto 6-well plates in 2 mL DMEM or RPMI 1640 supplemented with 10% FBS and incubated for 24 h at 37 °C. Medium was replaced with 2 mL of fresh medium in each well, and the cells incubated for another 48 h. Supernatants were collected, and the levels of IL-1β were assessed using enzyme-linked immunosorbent assay (ELISA; human IL-1β/IL-1F2 Quantikine ELISA Kit; R&D Systems).
Statistical analysis
All statistical analyses were performed using JMP software (v.14.0; SAS Institute, Tokyo, Japan). Data are expressed as the mean and standard deviation. Differences were analyzed using Student’s t-test, the Wilcoxon test, and χ2 test. Statistical significance was defined as p < 0.05 based on a two-tailed test. Correlations between two variables were evaluated by using Pearson's correlation coefficient.
Results
ASC is highly expressed in PDAC
Examination of the Oncomine database showed that ASC mRNA was significantly upregulated in PDAC in multiple microarray datasets20–25 (Fig. 1a). A microarray meta-analysis of these six datasets revealed that ASC mRNA was significantly upregulated in PDAC (p = 0.039). Accordingly, examination of immunostaining scores for the surgically removed tissues revealed that the immunostaining intensity was predominantly stronger in PDAC tissues relative to that in tissues surrounding the tumor (Fig. 1b,c). Quantitative assessment of the immunostaining results indicated a significantly higher OD in PDAC tissue relative to that in tissues surrounding the tumor (OD: 0.409 vs. 0.216, respectively; p = 0.0181) (Fig. 1d). Furthermore, ASC expression in ducts and acinar cells was weak in tissues distant from the tumor (Fig. 1e); however, we observed strong expression of ASC in either metaplastic or normal ducts within their nearby normal tissue relative to that in acinar tissue (Fig. 1f), and ASC expression was even stronger in cancerous lesions than surrounding tissues (Fig. 1g).
Figure 1.
ASC expression according to Oncomine and clinical PDAC tissues. (a) Relative expression of ASC in normal/benign pancreas tissue (N) versus PDAC (T) according to six independent profiling datasets (surveyed using Oncomine). (b) Evaluation of ASC expression in human PDAC tissue according to IHC analysis. Data were generated from three representative PDAC tissues expressing various ASC levels and determined using the anti-ASC antibody. Scale bar: 200 μm. (c) IHC intensity score among 20 evaluated patients. (d) Immune-reaction intensity according to the OD and determined using ImageJ (n = 20). Error bars indicate the standard error of the mean based on eight independent experiments. (e–g) Expression of ASC protein in pancreatic tissue distant form the tumor (e), tissue surrounding the PDAC tissue (f), and PDAC tissue (g) according to IHC analysis. Scale bar: 100 μm.
High ASC expression predicts poor prognosis of PDAC
To explore the clinical significance of ASC in PDAC, we performed Kaplan–Meier analysis, with the results showing that patients with elevated ASC mRNA expression had shorter overall survival (OS; n = 177; HR = 1.79, 95% CI 1.17–2.75) (Fig. 2a). We then determined whether ASC mRNA expression is predictive of outcomes in a cohort of tumors with low and high mutation burden. We found that elevated ASC mRNA expression was predictive of poor prognosis in a cohort of patients harboring tumors with low mutation burden (n = 83; HR = 2.78, 95% CI 1.5–5.16; log-rank p = 0.00073) (Fig. 2b), whereas high ASC mRNA expression did not contribute to prognosis in tumors with high mutation burden (n = 84; HR = 0.7, 95% CI 0.38–1.29; log-rank p = 0.25) (Fig. 2c). Based on evaluations of tumor grade, we found that elevated ASC mRNA expression was predictive of poor prognosis in a cohort of patients with grade 1 and grade 2 tumors but not grade 3 (grade 1: n = 31; HR = 1,300,997,289; 95% CI 0–inf; log-rank p = 0.0044; grade 2: n = 94; HR = 1.88, 95% CI 1.04–3.4; log-rank p = 0.033; and grade 3: n = 48; HR = 1.89, 95% CI 0.9–3.95; log-rank p = 0.087) (Fig. 2d–f).
Figure 2.
OS evaluated between PDAC patients with high and low expression of ASC. (a) OS of all PDAC patients (n = 177) and those with (b) high TMB (n = 84) and (c) low TMB (n = 83). (d) OS of PDAC patients with histological grade 1 (n = 31), (e) grade 2 (n = 94), and (f) grade 3 (n = 48) tumors. TMB tumor mutation burden.
Downregulation of ASC expression inhibits the growth of PDAC cells
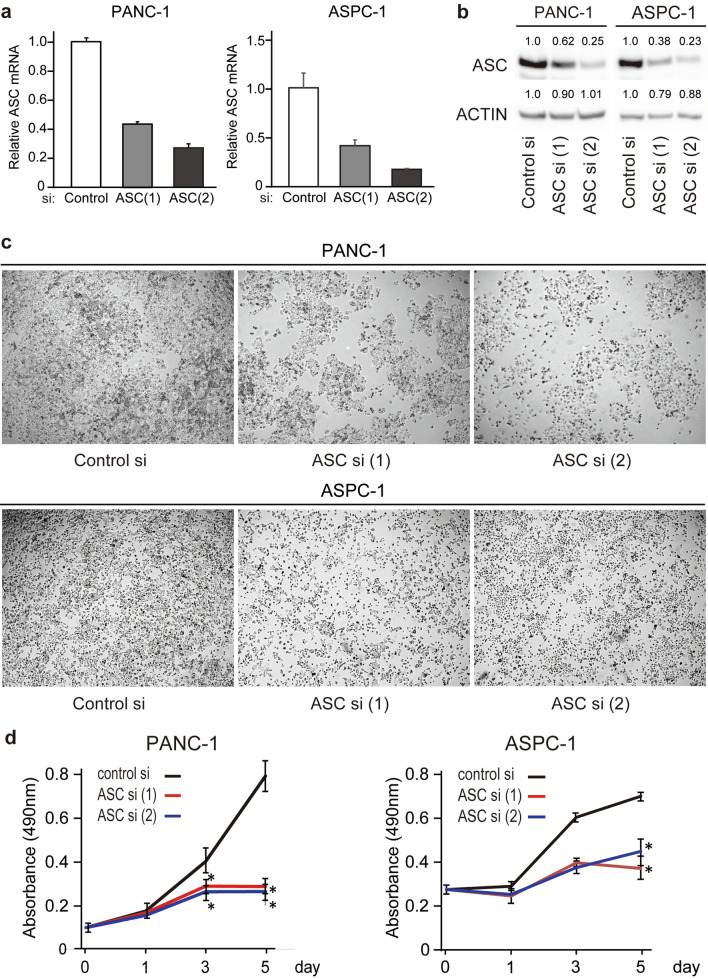
We then examined ASC expression in PDAC cell lines, finding increased expression in PANC-1, ASPC-1, and BXPC-3 cells and decreased expression in Mia-PaCa-2 cells relative to that in the pancreatic ductal epithelial cell line hTERT-HPNE, which exhibits an acinar associated with the ductal metaplasia phenotype, according to real-time RT-PCR (Fig. 3a). Additionally, we confirmed ASC expression in PDAC cell lines using the CCLE database (Fig. S1). Additionally, western blotting identified high ASC expression in PANC-1, ASPC-1, and BxPC-3 cells, low expression in hTERT-HPNE cells, and no expression in MIA PaCa-2 cells (Fig. 3b). These findings are in agreement with those of our evaluation of human tissues, with high ASC expression observed in multiple cancer cell lines relative to normal pancreatic duct epithelial cells. To investigate the role of ASC, we knocked down ASC expression using two ASC siRNAs in PANC-1 and ASPC-1 cells, both of which exhibit relatively high ASC expression. Following confirmation of successful knockdown of ASC levels by real-time RT-PCR and western blotting (Fig. 4a,b), microscopy-based observation of cells revealed inhibited proliferation (Fig. 4c). Evaluation of cell proliferation by MTS assay confirmed this observation, and that this effect increased over time (p < 0.05) (Fig. 4d). Notably, suppressed proliferation following ASC knockdown was not observed in Mia PACA-2 or hTERT-HPNE cells (Fig. S2). These results indicated that ASC is mainly involved in PDAC cell proliferation.
Figure 3.

ASC expression in human PDAC cells. Four human PDAC cell lines and one human pancreatic duct cell line were used to screen levels of ASC mRNA and protein expression using quantitative real-time RT-PCR and western blot. (a) ASC mRNA expression. Data represent the mean ± standard error of the mean of four replicates. (b) ASC protein expression. The original gel images of Western blotting are shown in Fig. S6.
Figure 4.
Changes in PDAC cells following ASC silencing. (a) ASC mRNA expression following transfection with control and ASC siRNA. Data represent the mean ± standard error of the mean of three to four replicates. (b) Western blot analysis of ASC protein expression following transfection with control and ASC siRNA. Relative densitometry data as compared with controls are given for each lane. The original gel images of Western blotting are shown in Fig. S7. (c) Photographs of wells on day 5 after siRNA transfection. (d) Viability of cells transfected with control or ASC siRNA. Data represent the mean ± standard error of the mean of three replicates. *p < 0.05, vs. control, Student’s t test.
ASC regulates the cell cycle in PDAC cells
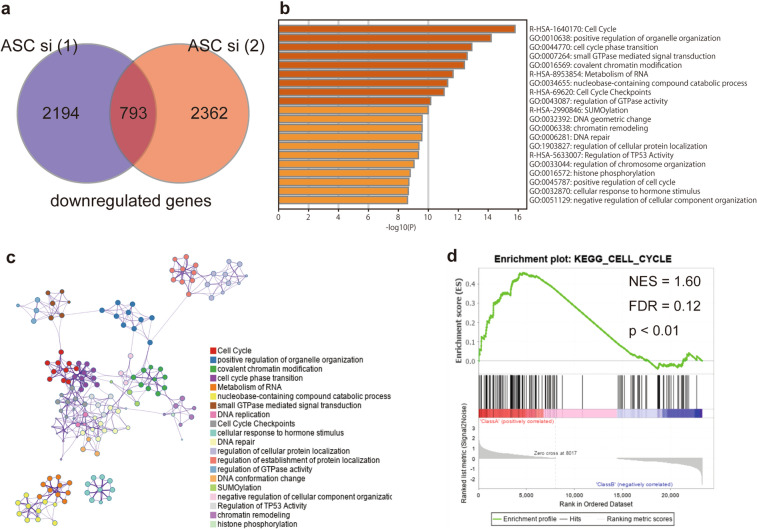
To elucidate the detailed molecular mechanism underlying ASC-mediated PDAC cell proliferation, we performed RNA-seq analysis on control PANC-1 cells and ASC-silenced PANC-1 cells. We found that the total number of RNAs in the silenced cells significantly decreased relative to the control [ASC siRNA (1) and (2): 2194 and 2364, respectively]. Additionally, we analyzed 793 RNAs that were commonly reduced in the two ASC siRNA groups using Metascape (Fig. 5a). The results showed that significantly enriched signaling pathways included the cell cycle, positive regulation of organelle organization, and cell cycle phase transition. The top 20 clusters of significantly enriched terms are shown in Fig. 5b,c. Furthermore, GSEA revealed significant relationships between the expression of cell cycle-related genes (normalized enrichment score = 1.60; and false discovery rate = 0.12; p < 0.01). (Fig. 5d). Similar data were obtained in RNA-seq experiments using AsPC-1 cells (Fig. S3). Moreover, we observed no changes in the expression of apoptosis-related genes in either the PANC-1 or the AsPC-1 cell lines [Kyoto Encyclopedia of Genes and Genomes (KEGG) apoptosis pathway26: p = 0.72, q = 0.85 in PANC-1 cells; and p = 0.42, q = 0.72 in AsPC-1 cells]. These results indicated that the reduced cell-proliferation phenotypes observed in ASC-silenced cells were induced by altered expression of cell cycle-related genes.
Figure 5.
Functional enrichment analysis using Metascape and GSEA. (a) Venn diagram showing overlapping RNAs detected via RNA-seq analysis of downregulated RNAs at 72-h post-transfection of either ASC (1) or (2) siRNA and relative to control siRNA-transfected PANC-1 cells. (b) Functional enrichment results. Heatmap showing the top 20 clusters colored according to the p-value (darker color indicates a lower p-value). (c) Network of the top 20 clusters of enriched terms. Each node indicates an enrichment term colored by cluster ID. Nodes sharing the same cluster are generally located in close proximity. Terms with a kappa similarity ≥ 0.3 are connected. (d) GSEA for the gene signature of KEGG_CELL_CYCLE. NES normalized enrichment score, FDR false discovery rate.
ASC knockdown reduces cyclin D1 (CCND1) level and stops the cell cycle at the G1 phase
We then employed flow cytometry to determine whether ASC knockdown indeed affected the cell cycle in PDAC cells, with the results showing a substantial increase in the population of cells in the G1 phase and a concomitant reduction of cells in the S phase (Fig. 6a). Based on this result, we focused on CCNs that regulate the cell cycle in the G1 phase. Real-time RT-PCR revealed that expression of the G1-phase gene CCND1 was decreased by ASC silencing in PANC-1 cells and AsSPC-1 cells (Fig. 6b,c), whereas CCND2 expression was decreased in PANC-1 and undetected in AsPC-1 cells. Additionally, CCND3 expression increased in PANC-1 cells and decreased in AsPC-1 cells, and we observed no change in CCNE1 and CCNE2 expression in PANC-1 cells but slight reductions in AsPC-1 cells. Western blotting subsequently confirmed changes in CCND1 protein levels corresponding to the observed downregulation of gene expression in both cell lines (Fig. 6d). These results suggested that ASC-silenced cells were arrested in the G1 phase due to regulated expression of cell cycle-related genes, which subsequently decreased cell proliferation.
Figure 6.
ASC silencing reduces CCND1 expression and blocks the cell cycle at the G1 phase. (a) Quantification of three experimental replicates of cell cycle phase distribution. Error bars indicate the standard error of the mean based on three independent experiments. *p < 0.05 vs. control, Student’s t test. (b, c) mRNA levels of CCNDs and CCNEs according to real-time RT-PCR following ASC silencing in PANC-1 and ASPC-1 cells. Data represent the mean ± standard error of the mean of three to four replicates. (d) CCND1 expression following ASC silencing according to western blot. Relative densitometry data as compared with controls are given for each lane. The original gel images of Western blotting are shown in Fig. S8.
Downregulation of ASC level does not modulate the expression of apoptosis-related molecules in PDAC cells
ASC is reportedly associated with apoptosis; therefore, we performed TUNEL staining and antibody arrays to observe changes in apoptosis in PDAC cells due to ASC downregulation. TUNEL staining revealed no changes in apoptosis upon ASC silencing at 3- and 5-days post-transfection with control and ASC siRNA (Figs. 7a,b; and S4). Furthermore, examination of apoptosis using antibody arrays revealed no changes in the expression of apoptosis-related proteins, including caspase-3, upon ASC downregulation (Fig. 7c,d).
Figure 7.
ASC silencing does not induce apoptosis-related changes in PDAC cells. (a) IHC staining of PANC-1 cells to detect apoptosis induction and TUNEL staining at 72-h post-transfection with control and ASC siRNA. Scale bar: 500 µm. (b) Quantitation of apoptotic cells based on TUNEL staining at 72-h post-transfection with control and ASC siRNA. Data represent the mean ± standard error of the mean of five areas. (c) Changes in the expression of apoptosis-related proteins at 72-h post-transfection with control and ASC (2) siRNA in PANC-1 cells and analyzed using human apoptosis proteome profiler arrays. Boxes show the localization of procaspase-3 and cleaved caspase-3 on the membrane. The cytokine array coordinates are as follows: positive ctrl (A1, A2); positive ctrl (A23, A24); Bad (B1, B2); Bax (B3, B4); Bcl-2 (B5, B6); Bcl-x (B7, B8); procaspase-3 (B9, B10); cleaved caspase-3 (B11, B12); catalase (B13, B14); cIAP-1 (B15, B16); cIAP-2 (B17, B18); claspin (B19, B20); clusterin (B21, B22); cytochrome c (B23, B24); TRAIL R1/DR4 (C1, C2); TRAIL R2/DR5 (C3, C4); FADD (C5, C6); Fas/TNFRSF6/CD95 (C7, C8); HIF-1α (C9, C10); HO-1/HMOX/HSP32 (C11, C12); HO-2/HMOX2 (C13, C14); HSP27 (C15, C16); HSP60 (C17, C18); HSP70 (C19, C20); HTRA2/0mi (C21, C22); livin (C23, C24); PON2 (D1, D2); p21/IP1/CDKN1A (D3, D4); p27/Kip (C5, D6); phospho-p53 (S15) (D7, D8); phospho-p53 (S46) (D9, D10); phospho-p53 (S392) (D11, D12); phospho-Rad17 (S635) (D13, D14); SMAD/Diablo (D15, D16); survivin (D17, D18); TNF RI/TNFRSF1A (D19, D20); XIAP (D21, D22); negative ctrl (D23, D24); and positive ctrl (E1, E2). (d) Quantitative analysis of procaspase-3 and cleaved caspase-3. Data represent the average of two spots.
Discussion
In recent years, increasing evidence has shown that ASC plays an important role in the interaction between PDAC cells and cells in the surrounding microenvironment (e.g., tumor-associated macrophages and cancer-associated fibroblasts)27,28; however, the role of ASC in signal transduction in PDAC cells has not been fully investigated. The present study demonstrated that downregulation of ASC induced changes in cell cycle in PDAC cells and inhibited proliferation.
A previous report identified methylated regions associated with ASC in 23% of papillary thyroid carcinomas and none in normal thyroid samples29. In other carcinomas, ASC methylation has been reported in 63.6% of prostate cancer, 40% of breast cancer cases, 32.1% of lung adenocarcinomas, 13.2% of lung squamous cell carcinomas, 38.5% of lung large-cell carcinomas, and 60% of lung small-cell carcinomas10,29,30. Although there are no studies on the frequency of ASC methylation in PDAC, a previous report indicates that > 90% of PDAC cells are IHC-positive for ASC28. In the present study, we identified ASC overexpression in human PDAC cells through bioinformatics methods. Additionally, IHC confirmed that ASC was more highly expressed in tumor tissues than in paired paracancerous tissues, as previously reported28. ASC expression is enhanced in ducts near the tumor and in metaplastic ducts and further enhanced in cancer, although rarely seen in ducts distant from the tumor. This suggests that ASC may be involved in acinar-duct metaplasia, which contributes to the development of pancreatic cancer, and might be an important factor for carcinogenesis.
Moreover, three of four PDAC cell lines exhibited ASC overexpression relative to levels observed in hTERT-HPNE cells generated from human pancreatic duct samples with acinar cells associated with the ductal metaplasia phenotype31. These results indicated that ASC expression might be suppressed by methylation in PDAC cell lines, such as MIA PACA-2; however, ASC is also considered to be highly expressed relative to levels observed in non-cancerous tissue. These results suggest that ASC methylation and expression vary according to the tissue and cancer type. Protti et al.15 consider ASC as a tumor suppressor when its expression in tumor cells is lower than that in normal epithelial cells and non-tumor adjacent tissues and tumor-promoting when its expression in tumor cells is higher than that in normal tissue. In the present study, we found that elevated ASC mRNA expression in tumor tissues was a factor associated with poor prognosis for PDAC. Tumors with low mutational burden are reportedly less responsive to current immunotherapies and require new therapeutic targets. Therefore, ASC might represent a potential therapeutic target in tumors with low mutational burden, as we found that ASC expression is a prognostic factor for PDAC. In particular, it would by clinically important as a prognostic factor in grade 2 tumors, which are the most common in PDAC32. These findings suggest that ASC might be an essential biomarker of PDAC and contribute to PDAC progression.
There has been one report on ASC using PDAC cells. The results of this report showed that increased ASC expression due to demethylation in MIA PaCa-2 cells resulted in increased sensitivity to gemcitabine and docetaxel33; however, there have been no studies reporting the detailed molecular function(s) of ASC in PDAC. Previous reports indicate that ASC plays an important role in apoptosis induction in various cancers34,35; however, in the present study, we observed no induction of apoptosis following ASC silencing in PDAC cells. Additionally, in IMR90-E1A cells, ASC silencing significantly inhibited etoposide-induced apoptosis; however, in the absence of etoposide stimulation, there was no change in the percentage of apoptotic cells relative to controls36. These findings suggest that ASC does not affect apoptosis in pancreatic cancer cells, at least in the absence of exogenous apoptotic stimuli. On the other hand, ASC is also a key adaptor molecule of inflammasomes and activates procaspase-1, which is necessary for processing IL-1β and IL1837,38. In the present study, we found that IL-1β level in the supernatant was below the measurement sensitivity in PANC-1 and AsPC-1 cells (Fig. S5). Moreover, RNA-seq showed that ASC silencing did not induce changes in inflammasome-related gene groups (data not shown). Therefore, these findings suggest that ASC does not contribute to changes in apoptosis or inflammasome formation in PDAC cells.
We observed suppression of PDAC cell proliferation following ASC silencing; however, this was not observed in MIA PACA-2 cells with poor ASC expression, suggesting that this is not an off-target effect. Additionally, because ASC-related effects were not observed in hTERT-HPNE cells, this suggests that ASC-specific effects are unique to PDAC cells. A previous report demonstrated that ASC silencing resulted in reduced cell viability, suppressed tumor growth, and arrested the cell cycle in the G1 phase in metastatic melanoma. The authors hypothesized that ASC might contribute to a positive feedback loop in cases of autoinflammation, where upregulated nuclear factor-κB expression results in transactivation of pro-IL-1β to promote autocrine IL-1 signaling in metastatic melanoma13. However, because no IL-1β was detected in the culture supernatant of PDAC cells in the present study and a previous report39, we speculated that other pathways are responsible for the changes in cell proliferation. Here, RNA-seq analysis showed that ASC silencing induced changes in the expression of cell cycle-related genes. The G0/G1 phase of the cell cycle is regulated by CCNs, including CCND and CCNE, among which we observed significant changes in only CCND1 expression. CCND1 and CCND3 are often differentially overexpressed in PDAC40, and CCND2 reportedly plays a role in the proliferation of pancreatic islet β-cells41, although its expression is infrequently detected in PDAC tissues and cells40,42. In the present study, ASC silencing induced a significant reduction in CCND1 expression in PANC-1 and AsPC-1 cells, whereas CCND3 expression increased in PANC-1 cells and decreased in AsPC-1 cells, and CCND2 was undetectable in AsPC-1 cells. Moreover, CCNE1 and CCNE2 expression did not show the same changes in PANC-1 and ASPC-1 cells. Based on these results, we speculated that CCND1 contributes to the change in cell proliferation observed following ASC silencing in PDAC cells. CCND1 expression is associated with anchorage-independent growth, tumorigenicity, angiogenesis, hypoxia response, and resistance to chemotherapeutic agents43. Furthermore, it plays an important role in G1/S-phase transition and cell proliferation44. Because ASC silencing resulted in an increase in the population of cells in the G1 phase and a concomitant reduction of cells in the S phase, we suggest that ASC regulates CCND1 expression and promotes G1/S-phase transition.
In summary, we found that downregulated ASC expression inhibited tumor progression by modulating the cell cycle in PDAC cells. These findings suggest ASC as a possible therapeutic target in PDAC; however, further studies are warranted to investigate this possibility.
Supplementary Information
Acknowledgements
We thank Mr. Kenji Tanimoto, Ms. Chie Takeichi, and Ms. Takana Fujino for their valuable technical assistance. We also thank Dr. Naohito Tokunaga and the staff of the Division of Analytical Bio-Medicine and the Division of Laboratory Animal Research, the Advanced Research Support Center (ADRES). This work was partially supported by JSPS KAKENHI (No. JP18K15819 to M.K.).
Author contributions
M.K. designed and performed the experiments, analyzed the data, and wrote the manuscript; K.S., Y.I., K.K., and T.K. provided advice and conducted the molecular experiments; T.W. and J.M. supervised the experiments and revised the manuscript; and Y.H. supervised the experiments and critically revised the manuscript. All authors reviewed the final version of the manuscript and agreed to its submission for publication.
Data availability
The datasets generated and/or analyzed during this study are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-01465-2.
References
- 1.Hidalgo M. Pancreatic cancer. N. Engl. J. Med. 2010;362:1605–1617. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- 2.Conroy T, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 2011;364:1817–1825. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- 3.Von Hoff DD, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N. Engl. J. Med. 2013;369:1691–1703. doi: 10.1056/NEJMoa1304369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Masumoto J, et al. ASC, a novel 22-kDa protein, aggregates during apoptosis of human promyelocytic leukemia HL-60 cells. J. Biol. Chem. 1999;274:33835–33838. doi: 10.1074/jbc.274.48.33835. [DOI] [PubMed] [Google Scholar]
- 5.Taniguchi S, Sagara J. Regulatory molecules involved in inflammasome formation with special reference to a key mediator protein, ASC. Semin. Immunopathol. 2007;29:231–238. doi: 10.1007/s00281-007-0082-3. [DOI] [PubMed] [Google Scholar]
- 6.Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 7.Rathinam VA, Vanaja SK, Fitzgerald KA. Regulation of inflammasome signaling. Nat. Immunol. 2012;13:333–342. doi: 10.1038/ni.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guan X, et al. ASC/TMS1, a caspase-1 activating adaptor, is downregulated by aberrant methylation in human melanoma. Int. J. Cancer. 2003;107:202–208. doi: 10.1002/ijc.11376. [DOI] [PubMed] [Google Scholar]
- 9.Akahira J, et al. Promoter methylation status and expression of TMS1 gene in human epithelial ovarian cancer. Cancer Sci. 2004;95:40–43. doi: 10.1111/j.1349-7006.2004.tb03168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Das PM, et al. Methylation mediated silencing of TMS1/ASC gene in prostate cancer. Mol. Cancer. 2006;5:28. doi: 10.1186/1476-4598-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riojas MA, et al. Methylation-induced silencing of ASC/TMS1, a pro-apoptotic gene, is a late-stage event in colorectal cancer. Cancer Biol. Ther. 2007;6:1710–1716. doi: 10.4161/cbt.6.11.4829. [DOI] [PubMed] [Google Scholar]
- 12.Zhang C, et al. Transcriptional silencing of the TMS1/ASC tumour suppressor gene by an epigenetic mechanism in hepatocellular carcinoma cells. J. Pathol. 2007;212:134–142. doi: 10.1002/path.2173. [DOI] [PubMed] [Google Scholar]
- 13.Liu W, et al. Dual role of apoptosis-associated speck-like protein containing a CARD (ASC) in tumorigenesis of human melanoma. J. Invest. Dermatol. 2013;133:518–527. doi: 10.1038/jid.2012.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu CS, et al. ASC contributes to metastasis of oral cavity squamous cell carcinoma. Oncotarget. 2016;7:50074–50085. doi: 10.18632/oncotarget.10317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Protti MP, De Monte L. Dual role of inflammasome adaptor ASC in cancer. Front. Cell Dev. Biol. 2020;8:40. doi: 10.3389/fcell.2020.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagy Á, Lánczky A, Menyhárt O, Győrffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci. Rep. 2018;8:9227–9227. doi: 10.1038/s41598-018-27521-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dowds TA, Masumoto J, Zhu L, Inohara N, Núñez G. Cryopyrin-induced interleukin 1beta secretion in monocytic cells: Enhanced activity of disease-associated mutants and requirement for ASC. J. Biol. Chem. 2004;279:21924–21928. doi: 10.1074/jbc.M401178200. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Y, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Segara D, et al. Expression of HOXB2, a retinoic acid signaling target in pancreatic cancer and pancreatic intraepithelial neoplasia. Clin. Cancer Res. 2005;11:3587–3596. doi: 10.1158/1078-0432.Ccr-04-1813. [DOI] [PubMed] [Google Scholar]
- 21.Grützmann R, et al. ADAM9 expression in pancreatic cancer is associated with tumour type and is a prognostic factor in ductal adenocarcinoma. Br. J. Cancer. 2004;90:1053–1058. doi: 10.1038/sj.bjc.6601645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishikawa M, et al. Experimental trial for diagnosis of pancreatic ductal carcinoma based on gene expression profiles of pancreatic ductal cells. Cancer Sci. 2005;96:387–393. doi: 10.1111/j.1349-7006.2005.00064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pei H, et al. FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell. 2009;16:259–266. doi: 10.1016/j.ccr.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buchholz M, et al. Transcriptome analysis of microdissected pancreatic intraepithelial neoplastic lesions. Oncogene. 2005;24:6626–6636. doi: 10.1038/sj.onc.1208804. [DOI] [PubMed] [Google Scholar]
- 25.Badea L, Herlea V, Dima SO, Dumitrascu T, Popescu I. Combined gene expression analysis of whole-tissue and microdissected pancreatic ductal adenocarcinoma identifies genes specifically overexpressed in tumor epithelia. Hepatogastroenterology. 2008;55:2016–2027. [PubMed] [Google Scholar]
- 26.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daley D, et al. NLRP3 signaling drives macrophage-induced adaptive immune suppression in pancreatic carcinoma. J. Exp. Med. 2017;214:1711–1724. doi: 10.1084/jem.20161707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brunetto E, et al. The IL-1/IL-1 receptor axis and tumor cell released inflammasome adaptor ASC are key regulators of TSLP secretion by cancer associated fibroblasts in pancreatic cancer. J. Immunother. Cancer. 2019;7:45. doi: 10.1186/s40425-019-0521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siraj AK, et al. Demethylation of TMS1 gene sensitizes thyroid cancer cells to TRAIL-induced apoptosis. J. Clin. Endocrinol. Metab. 2011;96:E215–224. doi: 10.1210/jc.2010-0790. [DOI] [PubMed] [Google Scholar]
- 30.Conway KE, et al. TMS1, a novel proapoptotic caspase recruitment domain protein, is a target of methylation-induced gene silencing in human breast cancers. Cancer Res. 2000;60:6236–6242. [PubMed] [Google Scholar]
- 31.Lee KM, Yasuda H, Hollingsworth MA, Ouellette MM. Notch 2-positive progenitors with the intrinsic ability to give rise to pancreatic ductal cells. Lab. Invest. 2005;85:1003–1012. doi: 10.1038/labinvest.3700298. [DOI] [PubMed] [Google Scholar]
- 32.Hlavsa J, et al. Tumor grade as significant prognostic factor in pancreatic cancer: Validation of a novel TNMG staging system. Neoplasma. 2018;65:637–643. doi: 10.4149/neo_2018_171012n650. [DOI] [PubMed] [Google Scholar]
- 33.Ramachandran K, Miller H, Gordian E, Rocha-Lima C, Singal R. Methylation-mediated silencing of TMS1 in pancreatic cancer and its potential contribution to chemosensitivity. Anticancer Res. 2010;30:3919–3925. [PubMed] [Google Scholar]
- 34.Manji GA, et al. PYPAF1, a PYRIN-containing Apaf1-like protein that assembles with ASC and regulates activation of NF-kappa B. J. Biol. Chem. 2002;277:11570–11575. doi: 10.1074/jbc.M112208200. [DOI] [PubMed] [Google Scholar]
- 35.Kitazawa M, et al. ASC induces apoptosis via activation of caspase-9 by enhancing gap junction-mediated intercellular communication. PLoS ONE. 2017;12:e0169340. doi: 10.1371/journal.pone.0169340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohtsuka T, et al. ASC is a Bax adaptor and regulates the p53-Bax mitochondrial apoptosis pathway. Nat. Cell Biol. 2004;6:121–128. doi: 10.1038/ncb1087. [DOI] [PubMed] [Google Scholar]
- 37.Martinon F, Burns K, Tschopp J. The inflammasome: A molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 38.Dinarello CA. Interleukin-1 beta, interleukin-18, and the interleukin-1 beta converting enzyme. Ann. N. Y. Acad. Sci. 1998;856:1–11. doi: 10.1111/j.1749-6632.1998.tb08307.x. [DOI] [PubMed] [Google Scholar]
- 39.Greco E, et al. Pancreatic cancer cells invasiveness is mainly affected by interleukin-1β not by transforming growth factor-β1. Int. J. Biol. Markers. 2005;20:235–241. doi: 10.1177/172460080502000406. [DOI] [PubMed] [Google Scholar]
- 40.Ebert MP, et al. Induction and expression of cyclin D3 in human pancreatic cancer. J. Cancer Res. Clin. Oncol. 2001;127:449–454. doi: 10.1007/s004320100235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Georgia S, Bhushan A. Beta cell replication is the primary mechanism for maintaining postnatal beta cell mass. J. Clin. Invest. 2004;114:963–968. doi: 10.1172/jci22098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsubayashi H, et al. Methylation of cyclin D2 is observed frequently in pancreatic cancer but is also an age-related phenomenon in gastrointestinal tissues. Clin. Cancer Res. 2003;9:1446–1452. [PubMed] [Google Scholar]
- 43.Joung YH, et al. Hypoxia activates the cyclin D1 promoter via the Jak2/STAT5b pathway in breast cancer cells. Exp. Mol. Med. 2005;37:353–364. doi: 10.1038/emm.2005.45. [DOI] [PubMed] [Google Scholar]
- 44.Tane S, et al. Repression of cyclin D1 expression is necessary for the maintenance of cell cycle exit in adult mammalian cardiomyocytes. J. Biol. Chem. 2014;289:18033–18044. doi: 10.1074/jbc.M113.541953. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during this study are available from the corresponding author upon reasonable request.