Abstract
Background:
Autophagy, a highly conserved homeostatic mechanism, is essential for cell survival. The decline of autophagy function has been implicated in various diseases as well as aging. Although mitochondria play a key role in the autophagy process, whether mitochondrial-derived peptides are involved in this process has not been explored.
Methods:
We developed a high through put screening method to identify potential autophagy inducers among mitochondrial-derived peptides. We used three different cell lines, mice, c.elegans, and a human cohort to validate the observation.
Results:
Humanin, a mitochondrial-derived peptide, increases autophagy and maintains autophagy flux in several cell types. Humanin administration increases the expression of autophagy-related genes and lowers accumulation of harmful misfolded proteins in mice skeletal muscle, suggesting that humanin-induced autophagy potentially contributes to the improved skeletal function. Moreover, autophagy is a critical role in humanin-induced lifespan extension in C. elegans.
Conclusions:
Humanin is an autophagy inducer.
General Significance:
This paper presents a significant, novel discovery regarding the role of the mitochondrial derived peptide humanin in autophagy regulation and as a possible therapeutic target for autophagy in various age-related diseases.
Keywords: Mitochondrial-derived peptides, Humanin, Autophagy, Lifespan
Introduction
Mitochondria are multifaceted organelles. They produce ATP, maintain calcium homeostasis, and are recognized as a center of retrograde signaling. Mitochondria release calcium, reactive oxygen species (ROS), nicotinamide adenine dinucleotide (NAD+), and mitochondrial derived peptides (MDPs) and have various cellular effects including activation of nuclear genes and activation of signaling pathways [1, 2]. Mitochondria also regulate autophagy through several different mechanisms. Autophagy is essential for cell survival as it breaks down misfolded proteins, damaged organelles, and toxins [3]. The autophagic process starts with forming an autophagosome, a double membrane-bound vacuole that encloses the cytoplasmic materials in need of degradation. Mature autophagosomes fuse with the lysosome, becoming an autolysosome, where the sequestered cell contents are degraded and recycled for cellular maintenance [3]. These processes tightly regulated by multiple autophagic pathways. One of the way the mitochondria regulate autophagy is by the production of ROS, which regulates several autophagic pathways, such as ATG4–ATG8/LC3, Beclin-1, p53, PTEN, PI3K–Akt–mTOR, and MAPK signaling, especially in cancer cells [4]. When mitochondrial respiration is impaired, it directly inhibits autophagic flux and autophagy gene induction, as well as the recruitment of an important autophagy-related protein complex, Atg1-Atg13 kinase, to the pre-autophagosome [5]. This shows that properly functioning mitochondria are vital for autophagy processes. Mitochondria modulate lysosome function as well—high levels of NAD+ correlate with properly functioning lysosomes [6]. Furthermore, mitochondrial impairment disrupts endolysosomal trafficking pathways [6]. During the autophagy process, the autophagosome containing damaged proteins and organelles fuses with the cell’s endolysosomal system [7]. Lysosomes are an essential component of the autophagy process; thus, mitochondria’s regulation of lysosomes translates to another mechanism of mitochondrial regulation of autophagy.
Emerging studies demonstrate that mtDNA harbor small open reading frames (sORFs) that encode mitochondrial-derived peptides (MDPs) [8]. Humanin was the first discovered MDP and has since been implicated in multiple biological processes such as cytoprotection, apoptosis, and metabolism [1, 9-11]. This MDP is 24 amino acids in length and is encoded from the 16s ribosomal RNA coding region of the mtDNA. In terms of metabolism, humanin improves insulin sensitivity, decreases body weight gain, and decreases visceral fat levels [12, 13]. Humanin has a positive impact on bioenergetics as evident through increased basal oxygen consumption rate, increased respiration capacity, and increased ATP production in retinal pigment epithelial cells [14, 15].
In addition to its aforementioned impact on biological processes, humanin is also implicated in aging. Decreases in humanin levels have been observe in many different organisms including mice, monkeys, and humans [16], implying that insufficient humanin levels could be associated with the onset of certain age-related illnesses [16-18]. Interestingly, the naked mole rat, a model of negligible senescence [19],only shows a slight decrease in humanin levels over its exceptionally long 30 year lifespan, supporting the idea that humanin is related to biological aging [16]. Humanin also affects and is affected by known anti-aging pathways such as IGF-I [20]. Humanin reduces oxidative stress, thereby combatting mitochondrial dysfunction and diminishing levels of ROS [21]. It is speculated that exogenous treatment of humanin and other MDPs could help combat degenerative aging symptoms associated with low humanin levels and high oxidative stress. This involves diseases such as neurodegenerative disease, type 2 diabetes, cardiovascular disease, memory loss, stroke, inflammation, and amyotrophic lateral sclerosis (ALS)[17, 22-27].
The m.2706A>G polymorphism in humanin is associated with accelerated cognitive aging and a decrease in circulating humanin levels [28]. Six additional small humanin-like peptides (SHLPs 1-6) are encoded from the same 16S rRNA region as humanin [29]. Some, particularly SHLP2, have similar cytoprotective effects as humanin [29, 30]. MOTS-c, which is derived from a sORF within the mitochondrial 12S rRNA, regulates insulin sensitivity and metabolic homeostasis [31]. MOTS-c acts as a retrograde signaling molecule and translocates into the nucleus and modulates metabolic stress response [32]. Although mitochondria play important roles in the autophagy processes, whether or not mitochondrial-derived peptides are involved in modulating the autophagy process have not been elucidated.
Here, we utilized a high-throughput screening tools to investigate which MDPs induce autophagy. We found that humanin increases autophagy in various cells and skeletal muscle from old mice. Additionally, we find that autophagy plays a critical role in humanin-induced lifespan extension in C. elegans.
Materials and Methods
Reagents and Antibodies
HNG (a potent analogue of humanin with a glycine substitution, S14G), SHLPs 2 and 6 (small humanin-like peptide 2 and 6), HNG-F6A were synthesized by Genscript (Piscataway, NJ, USA). Peptides were initially dissolved in Milli-Q water. Choloroquine (Sigma, St. Louis, MO, USA) and 3-Methyladenine (3-MA; Sigma) were used for inhibiting lysosomal degradation in culture. Humanin siRNA and control siRNA were ordered from Dharmacon. Humanin siRNA sequence is 5’GCUCAUAAGGAAAGGUUAAUU3’. Control siRNA was purchased from Dharmacon: ON-TARGETplus Non-targeting Control Pool (cat#. D-001810-10-05). The following antibodies were used in this study: anti-GAPDH antibody (Cat. #5174S), anti-LC3 (Cat. #3868), anti-ATG5 antibody (Cat. #12994), anti-ATG7 antibody (Cat. #8558), anti-ATG3 (Cat. #3415), anti-Beclin-1(Cat. #3495), anti-rabbit IgG, HRP-linked antibody (Cat. #7074), and anti-mouse IgG, HRP-linked antibody (Cat. #7076). These antibodies are supplied by Cell Signaling Technology (Danvers, MA, USA). Anti-p62 (Cat. #GP62-C, Progen Biotechnik, Heidelberg, Germany) and anti-K63 ubiquitin (Cat. #05-1308, Millipore) were also used.
Cell culture and treatment
SH-SY5Y cells and B16 cells were cultured in Dulbecco's Modified Eagle Medium: Nutrient Mixture F-12 (DMEM:F-12, ThermoFisher Scientific, Waltham, MA, USA ) and in high glucose Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies, Waltham, MA, USA), respectively, supplemented with 10% fetal bovine serum (FBS; Omega Scientific, Tarzana, CA, USA) at 37°C in 5% CO2. HEK293 cells stably expressing mRFP-GFP-LC3 were established and characterized. The stable cell lines were cultured in DMEM supplemented with 10% FBS and 500 μg/ml G-418 (ThermoFisher Scientific) at 37°C in 5% CO2. siRNAs were transfected into HEK293 cells using RNAiMAX (ThermoFisher Scientific).
High Throughput Autophagy Screening Assay
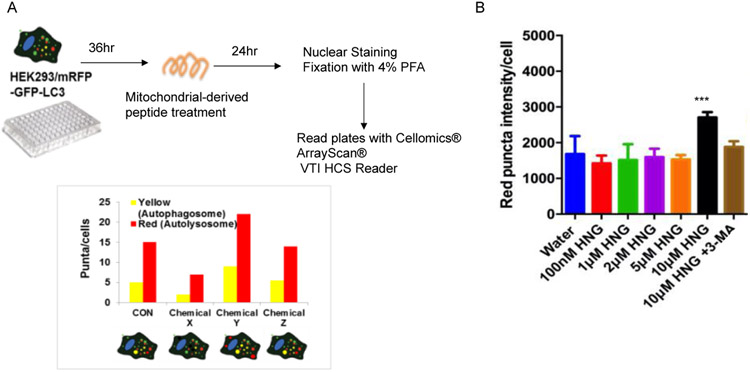
HEK293 cells stably expressing mRFP-GFP-LC3 (ptfLC3 plasmid)[33] were seeded in 96 well plate. Mitochondrial-derived peptides were treated for 24 hrs. Then, cells were fixed with 4% paraformaldehyde for 10min at RT. Nuclei were stained for 5 minutes at room temperature in PBS containing Hoechst 33258 (2 mg/ml; Invitrogen). Cells were imaged in Cellomics® ArrayScan® VTI HCS Reader (ThermoFisher Scientifc). Yellow (mRFP+ GFP) and red (mRFP only) intensity per cells were measured to quantify the autophagosomes and autolysosomes, respectively.
Western blot analysis
Cells were lysed with RIPA Lysis and Extraction Buffer (ThermoFisher Scientific) plus the Halt protease & phosphatase inhibitor cocktail (ThermoFisher Scientific). The lysates were incubated on ice for 10min then homogenized using a sonicator, and the supernatant was collected by centrifugation at 15,000 x g for 15min at 4°C. For tissue, dissected skeletal muscles were lysed with RIPA buffer and homogenized using a tissue homogenizer followed by sonication. The supernatant was collected by centrifugation at 15,000 x g for 15min at 4°C. Protein content in the cellular lysates was quantified using the Pierce™ BCA Protein Assay Kit (ThermoFisher Scientific). Predetermined amounts of proteins (10-30μg) were separated on 8-16% SDS-PAGE gels and blotted onto PVDF membranes (Biorad, Hercules, CA, USA). Membranes were incubated with primary antibody at 4°C overnight according to the manufacturer’s instructions. After several washes with Tris-buffered saline containing 0.1% Tween-20, membranes were incubated at room temperature for 1hr with the appropriate HRP-conjugated secondary antibody. Clarity™ Western ECL substrate (Biorad) was used for detecting specific bands. Membranes were imaged on a Bio-Rad ChemiDoc XRS+ imager. If necessary, relative intensities of the bands in each condition were measured using Image J, a free software program provided by National Institute of Health (Bethesda, Maryland, USA).
Imaging cells
HEK293 cells stably expressing mRFP-GFP-LC3 cells cultured on coverslips were fixed with 4% paraformaldehyde for 10 min at room temperature. After fixation, nuclei were stained for 5 minutes at room temperature in PBS containing Hoechst 33258 (2 mg/ml; Invitrogen). Coverslips were mounted with ProLong Gold antifade reagent (Invitrogen) and observed under an LSM780 confocal microscope (Carl Zeiss, Germany).
RNA extraction and qRT-PCR
Total RNA was isolated from skeletal muscle using RNeasy Fibrous Tissue Mini Kit (QIAGEN, Redwood City, CA, USA). The protocol was performed according to manufacturer’s instructions and RNA was measured at 260 nm with the Nanodrop system (ThermoFisher Scientific, Wilmington, DE, USA). RNA purity and quality were evaluated by 260/280 and 260/230 ratios. Quantified RNA was reverse transcribed with Superscript IV (ThurmoFisher Scientific). Quantitative real-time PCR (CFX system, Biorad) was used for the detection of ATG7 and ATG8 genes using SsoAdvanced Universal SYBR Green Supermix (Biorad) and following the protocol provided by the manufacturer. GAPDH were used as reference gene. The primers used for amplification are: ATG7 Forward primer (5’TCCGTTGAAGTCCTCTGCTT3’), and Reverse primer (5’ CCACTGAGGTTCACCATCCT3’); ATG8 Forward primer (5’CGGAGCTTTGAACAAAGAGTG3’) and, Reverse primer (5’TCTCTCACTCTCGTACACTTC3’); GAPDH Forward primer (5’ ACCACAGTCCATGCCATCAC3’), and Reverse primer (5’ TCCACCACCCTGTTGCTGTA3–).
Animals
C57BL/6N mice were obtained from the NIA aged mouse colony starting at 18 months of age. The mice were given twice-weekly injections of HNG at 4-mg/kg/BW or water/vehicle IP for 14 months, as described previously [28]. Survival was monitored daily and body weight and daily food intake measured weekly. All experiments with mice were performed in accordance with the appropriate guidelines and regulations and approved by the University of Southern California Institutional Animal Care and Use Committee (IACUC) under protocol #20787.
Rotarod Performance Test
At 24 months of age, the mice were placed on Rotarod performance test. During a training phase, mice were introduced to walking on the rotating rod one day before being tested. The accelerating speed (from 4 to 40 rpm within 300s) of Rotarod performance tests were performed three times a day for two days. The time until the animal falls off the rotating rod was recorded by electronically connected software.
C. elegans
Worms were synchronized via hypochlorite treatment and grown at 15°C until adulthood. They were then transferred to NGM plates supplemented with 10μM FUDR, 1mM IPTG, 100 μg/mL ampicillin, and seeded with HT115 E. coli that were transfected with the appropriate RNAi construct. They were maintained at 20°C for the remainder of their life and checked for death approximately every other day. Lifespans were analyzed using a log-rank test on the Kaplan-Meier curves.
Co-expression network analysis
The humanin transcript was correlated to all gene transcripts using data from the Mayo Clinic brain Bank series. Briefly, counts from RNASeq data were produced by SNAPR and normalized using edgeR-based implementation of the trimmed mean of M-values (TMM), which calculated counts per million (CPM). These normalized counts were assigned to the gene levels. Since the humanin ORF was not annotated, we re-analyzed available BAM files and counted transcripts, using the summarizeOverlaps method as part of the Bioconductor package in R, that mapped back to the humanin ORF against a custom mitochondrial ORFome database. We then extracted normalized humanin transcript counts and conducted a Pearson correlation for the humanin transcript against normalized counts of all genes, correcting for multiple hypothesis testing using a False Discovery Rate threshold of 0.05. Because of the experimental effects of humanin and autophagy in vitro and in vivo, we conducted a gene enrichment analysis targeted for autophagy-related genes. We created a GO-annotated autophagy gene set list, extracted significant coexpressed autophagy-related genes, and analyzed this curated autophagy-related gene set list against a universe background gene list of all significant co-expressed genes using the clusterProfiler R package, which revealed enriched GO terms relating to all pathways involved in autophagy.
Statistical analysis
Data are presented as mean ± S.E.M. Significant differences were determined by Student’s t-tests, oneway ANOVA followed by Tukey’s post hoc test using GraphPad Prism 8 software. Values of *<0.05, **<0.01, ***<0.001 were considered statistically significant. Statistics for c.elegans.
Results
High throughput screening revealed that humanin increases autophagy.
Various experimental tools to measure the autophagy processes have been developed [34]. Here, we used HEK293 cells stably expressing mRFP-GFP-LC3 to examine autophagy induction by various mitochondrial-derived peptides. Because mRFP-GFP-LC3 captures both autophagosomes and autolysosomes signals from cells, it can be used to examine the autophagy induction and autophagic flux [35]. LC3 proteins incorporated into the autophagosome’s inner and outer membranes were also carried to the autolysosome [35]. The low pH inside the lysosome quenches the fluorescent signal of GFP, whereas RFP exhibits more stable fluorescence in acidic compartments [35]. Therefore, autophagosomes and autolysosomes are labeled with yellow (mRFP+GFP) and red (mRFP only) signals, respectively [35]. We applied this principal to develop a 96-well-based imaging assay to quantify the autophagosomes and autolysosomes (Figure 1A). We treated HEK293 cells stably expressing mRFP-GFP-LC3 with several doses of different MDPs to quantify the autophagosomes and autolysosomes. Both autophagy induction and autophagic flux are important for maintaining proper protein quality control. We focused on the autolysosomes to screen the MDPs because increased autolysosomes can represent that the increased autophagy completed its function to degrade abnormal cargos. HNG, a potent analog of humanin, increased the autolysosomes in HEK293 cells (Figure 1B). On the other hand, the other three MDPs, including SHLP2, SHLP4, and SHLP6 peptides, did not change the number of autolysosomes (Supplemental figure 1). We focused on HNG, a potent analog of humanin, which was previously proposed as a dietary restriction mimetic peptide by reducing IGF-1 levels and activating AMPK [20], and has cytoprotective roles in various age-related diseases [17, 22-27]. Moreover, humanin previously has been shown to activate chaperone-mediated autophagy by interacting with hsp90, and increases substrate binding and translocation into lysosomes [36]. Co-treatment of HNG with 3- Methyladenine (3-MA) has an inhibitory effect on class III Phosphatidylinositol 3-kinases (PI-3K) activity [34], which is known to be essential for the induction of autophagy, diminishing HNG-mediated autolysosome increase (Figure 1B). This result suggests that HNG increases autophagy by an upstream effector of PI3K Class III complex formation such as ULK1 complex, AMPK, and mTOR. Previously, humanin was cloned as a IGFBP3 binding protein and humanin’s 6th amino acid, phenylalanine, is important for its interaction to IGFBP3 [10]. The humanin F6A, which replace 6th amino acid phenylalanine with alanine, did not increase the autolysosomes (Supplemental figure 1), suggesting the IGFBP3 interaction play important roles in humanin’s autophagy regulation.
Figure 1.
High throughput screening of which mitochondrial-derived peptides induce autophagy. (A) Schematic diagram of the screening. The graph shows examples of screening results. For instance, chemical x inhibits autophagy, chemical y induces autophagy, and chemical z does not alter autophagy in cells. (B) The red puncta (autolysosome) intensity per cell (n=8).
Humanin induces autophagy in several cell lines.
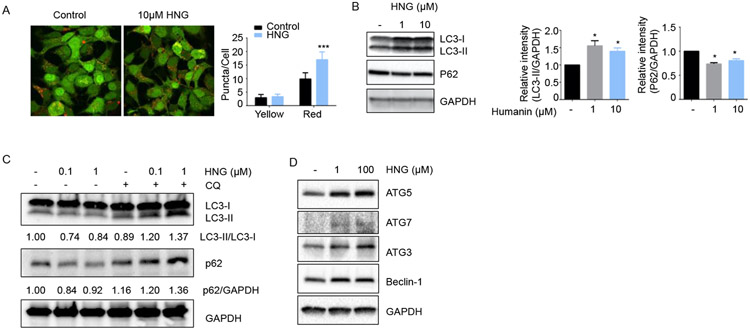
To validate the observation from the high-throughput screening, HEK293 cells stably expressing mRFP-GFP-LC3 cells were treated with 10μM HNG and imaged with a confocal microscopy (Figure 2A). Because LC3-II is localized at the autophagosomal vesicles, the punctated staining pattern of LC3-II forms are widely used to monitor autophagy. The number of yellow (autophagosomes) and red (autolysosomes) puncta per cells were quantified. HNG increase the number of red puncta, whereas the number of yellow puncta remained the same, suggesting that HNG induces the autophagy process in the cells while maintaining autophagic flux. To understand whether HNG-induced autophagy is a cell type-specific phenomenon, we used B16 melanoma cells and SH-SY5Y neuroblastoma cells to examine the HNG-induced autophagy. We measured autophagy levels in B16 cells on a biochemical level and performed immunoblot analysis using an antibody that recognizes LC3 and p62. A cytosolic form of LC3 (LC3-I) is conjugated to phosphatidylethanolamine to form LC3-phosphatidylethanolamine conjugate (LC3-II), which is recruited to autophagosomal membranes. Due to the localization of LC3-II at the autophagosomal vesicles, the formation of LC3-II is used to monitor autophagy. p62/SQSTM1 is also widely used to monitor autophagy as it is localized at the autophagic compartments and subsequently degraded through autophagy. Immunoblot analysis demonstrated that both 1 and 10 μM HNG treatment decreased levels of p62/SQSTM1, supporting the notion that HNG induces autophagy while autophagic degradation process is intact in B16 cells (Figure 2B). Both 1 and 10 μM HNG treatment induced accumulation of the endogenous the LC3-II form (Figure 2B). HNG increases both LC3-I and LC3-II in B16 cells suggesting HNG potentially increases the expression of autophagy-related genes including LC3. In SH-SY5Y cells, 0.1 and 1 μM HNG increases autophagy (Figure 2C). Using a lysomotropic agent, chloroquine (CQ), which inhibits protein degradation by raising intralysosomal pH, we specifically investigated whether the HNG-induced accumulation of LC3-II is due to enhanced synthesis of autophagosomes or reduced degradation of autolysosomes. When we compared and quantitated the levels of LC3-II in HNG-treated cells in the presence or absence of chloroquine, we found that co-treatment with a lysomotropic agent led to further increase in HNG-induced accumulation of LC3-II (Figure 2C). p62/SQSTM1 levels were lower in HNG-treated cells compared to control cells and the p62/SQSTM1 levels were elevated with co-treatment with CQ. These results validated that accumulation of LC3-II was most likely caused by an enhanced synthesis of autophagosomes. Thus, humanin induces autophagy while maintaining autophagy flux. The autophagy process requires multiple steps including initiation, elongation, and maturation. We examined whether HNG treatment increases the autophagy proteins involved in the initiation complex (Beclin-1) and LC3 lipidation during elongation (ATG7, 5, and 3). We found that both 1 and 100 μM HNG increased the expression of autophagy proteins involved in the initiation and elongation (Figure 2D).
Figure 2.
Humanin induces autophagy in several cell lines. (A) HEK293 cells stably expressing mRFP-GFP-LC3 were treated with HNG for 24hr. For quantification of either yellow (mRFP+GFP) or red (mRFP only) puncta, 80 cells were analyzed by Image J. (B) B16 melanoma cells were treated with HNG for 24hr. The expression levels of LC3 and p62/SQSTM1 were measured by western blot (n=3). (C-D) SH-SY5Y neuroblastoma cells were treated with HNG for 24hr. The expression levels of LC3, p62/SQSTM1 (n=3), and autophagy-related genes (n=2) were measured by western blot.
*<0.05,***<0.001.
Humanin increases autophagy in skeletal muscle from aged-mice
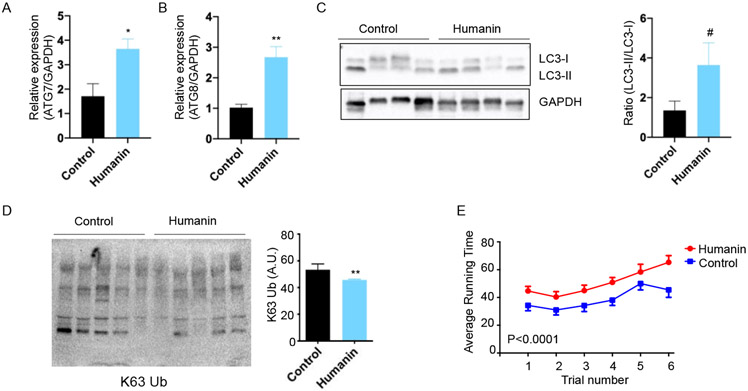
We previously published a study where we administrated HNG in 18-month-old female C57Bl/6N mice for 14 months and investigated the effects on lifespan and healthspan [28]. In the study, we determined that HNG increase the healthspan in mice by improving cognitive function and reducing inflammation [28]. In a rotarod performance test, HNG-treated mice maintained their balance longer than the control group [28]. AMPK is one of key modulators of autophagy in the skeletal muscles [37]. Previously, humanin was shown to increase the AMPK pathway in primary hepatocytes, macrophages, and myocardial cell [17, 38, 39]. HNG treatment in the old mice also increases the AMPK pathways and increases mitochondrial biogenesis in skeletal muscles [40]. Since autophagy declines with age and is implicated in multiple age-related diseases and since autophagy induction improves muscle function [41, 42], we hypothesized that HNG-treated old mice maintained their autophagy process better compared to the control group, thus improving protein and mitochondrial quality. We examined autophagy-related gene expression in skeletal muscle from the mice treated with HNG. HNG-treated mice showed elevated ATG7 and ATG8 expression compared to controls in skeletal muscle (Figures 3A and 3B). To examine whether HNG can activate autophagy in other tissues, we examined the expression of autophagy-related genes. The expression of autophagy-related genes in the experimental group was the same as the control in the liver (Supplemental figure 2). HNG treatment also induced accumulation of endogenous LC3-II in skeletal muscle (Figure 3C). Autophagy and the ubiquitin proteasome system are the major degradation systems in mammalian cells that allow for the control of protein quality and the recycling of cellular contents [43]. Their mode of action and their requirements for substrate recognition are different. Although ubiquitin is a common signal for both the autophagy and the ubiquitin-proteasome system, the ubiquitin chain type could determine the pathway of choice for protein degradation [43]. K48-linked ubiquitination is a proteasome signal, whereas K63-linked ubiquitination directs proteins towards autophagosomal degradation [43]. We thus examined whether humanin administration reduces abnormal protein accumulation, which is designated for autophagy and proteasome. The accumulation of K63-linked ubiquitinated proteins were lower in HNG-treated mice compared to control groups (Figures 3D). As shown previously, HNG-treated mice exhibit better health outcomes [28]. At 28 months of age, mice were evaluated balance and motor skills using Rotarod performance test. During the accelerating speed, the control mice maintained balance for 36 seconds and improved with time. The HNG-treated group had the same learning curve but had better performance (Figure 3E). Taken together, the finding that humanin maintain the autophagy process and reduce the burden of the abnormal protein accumulation in skeletal muscle during aging may be one of the mechanisms how humanin treated animals maintain better motor function during aging.
Figure 3.
Humanin increases autophagy in mouse skeletal muscle and improves muscle function in aged mice. (A-B) Autophagy-related protein levels were measured by qRT-PCR (n=5/group). (C) Western blot showing the LC3 expression and quantification of the LC3-II accumulation (n=4/group). (D) The western blot and quantification of K63-mediated ubiquitinated proteins (n=5/group). (E) Rotarod performance test (n=10/group). # <0.1, *<0.05, **<0.01.
Autophagy is essential for humanin-induced lifespan extension in C. elegans.
Autophagy plays crucial roles for cellular homeostasis. Dysregulation of autophagy potentially leads to the development of physiological alterations. Autophagy activity decreases with age, leading to the accumulation of damaged molecules and organelles during aging [41, 44]. Dysfunction of the autophagy process exacerbates age-related diseases such as neurodegenerative diseases and cancer [41, 44].
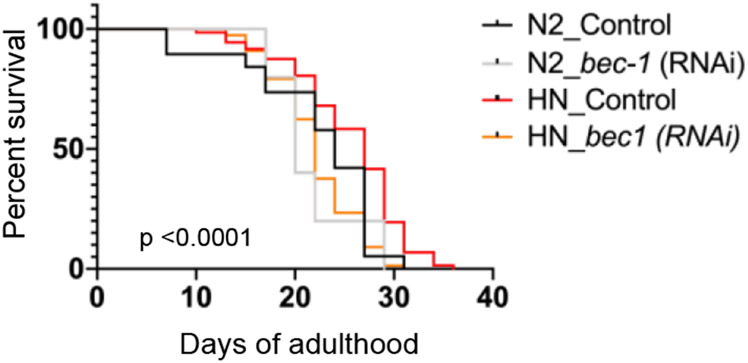
Likewise, the maintenance of proper autophagy activity contributes to extended lifespan [45-47]. Genetic studies suggested that autophagy has an essential role in the regulation of animal lifespan [48]. Basal autophagy activity is elevated in many longevity paradigms and the activity is required for lifespan extension [45-47]. The genes involved in autophagy and lysosome function are elevated in long-lived animals [49, 50]. Pharmacological treatments have been shown to extend lifespan through activation of autophagy, suggesting autophagy could be a potential target to modulate animal lifespan [42, 51, 52]. Autophagy, an evolutionary conserved lysosomal degradation pathway, interacts with various longevity signals in the regulation of C. elegans lifespan [50]. Previously we have shown that humanin transgenic worms have extended lifespan compared to control worms [16]. We hypothesized that autophagy processes potentially explain the humanin-induced lifespan extension. To address the question, we used RNAi to inhibit autophagy in humanin-transgenic worms. We knocked down bec-1, the C. elegans ortholog of Atg6/Vps30/Beclin1 and a key regulator of the autophagic machinery, using RNAi. As seen in Figure 4, HN transgenic worm increased mean lifespan from (24 days in control to 27 days in the transgenic). Depleting bec-1 function by RNAi eliminated the lifespan extension seen in humanin-transgenic worms (mean lifespan of 22 days), indicating that autophagy plays an important role in humanin-induced lifespan extension (Figure 4, n=90/group).
Figure 4.
Autophagy is essential for humanin-induced life span extension in worms.
Survival curves N2 (control) and HN (humanin transgenic) animals fed either control bacteria or bacteria expressing bec-1 dsRNA during adulthood. Mean lifespan was 24d for N2 control, 20 d for N2 bec-1 RNAi, 27d for HN control, and 22d for HN bec-1 RNAi , p <0.0001 (HN bec-1 RNAi vs HN control), p<0.01 (HN control vs N2 control), Log-rank (Mantel-Cox) test. This experiment was performed twice.
Humanin co-expresses with autophagy-annotated genes in the human brain temporal cortex.
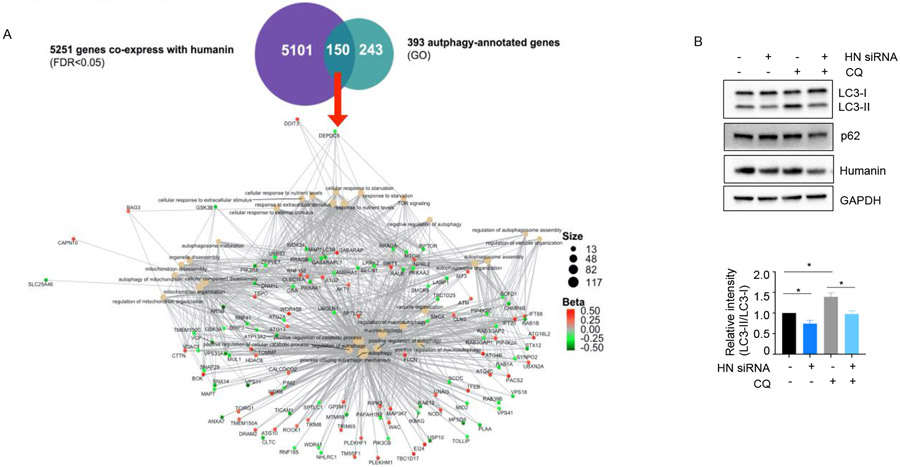
We utilized publicly available RNA-seq data to perform a co-expression network analysis. The data was derived from 78 brain temporal cortical samples without evidence of Alzheimer’s disease pathology [53]. First, 5251 genes that co-express with humanin gene expression were identified. Second, to understand the network of humanin and autophagy-related genes, we utilized 393 GO autophagy-annotated genes and merged with genes which co-express with humanin. Further enrichment analysis (i.e., analyzing the autophagy-related gene set against a background of all humanin co-expressed genes) showed that 150 gene among 393 autophagy-annotated genes were co-expressed with humanin (Figure 5A). These results suggested that humanin might regulate autophagy pathways in human brain. To understand whether endogenous humanin is involved in basal autophagy, we treated HEK293 cells with siRNA targeting endogenous humanin. Humanin siRNA reduces the LC3-II levels compared to control siRNA (Figure 5B). This suggests that endogenous humanin also play important roles in basal autophagy.
Figure 5.
Humanin expression is associated with autophagy pathway (A) Autophagy-annotated genes co-express with humanin in the human brain temporal cortex. 150 genes that co-express with humanin are involved in autophagy-related networks. (B) The expression levels of LC3, p62/SQSTM1 were measured in HEK293 cells transiently transfected with humanin siRNA (n=3).
Discussion
In this study, we examined whether mitochondrial-derived peptides induce autophagy processes in high-throughput screening. We, then, utilized several systems including cell culture, mice, C. elegans, and human brain RNA-seq to validate the targets identified in high-throughput screening. We showed that humanin, the first identified MDP, induces autophagy in several cell types and skeletal muscle from old mice. In humanin-transgenic worms, autophagy plays a critical role in lifespan extension. Furthermore, humanin co-expressed with autophagy-related genes in human brains, further evidencing humanin’s effect on autophagy pathways.
Humanin increases autophagy by activating AMPK upstream of PI3K Class III complex formation, since 3-MA blocks humanin-induced autophagy. However, we could not rule out the possibility of the involvement of other pathways activated by humanin. Humanin has been cloned as a IGFBP3 binding protein [10]. HNG increases autophagy while HNG-F6A -- an alanine replacement mutant which does not bind to IGFBP3 -- does not have the same function. Previous work showed that IGFBP-3 stimulates autophagy in breast cancer cells by binding to glucose-regulated protein 78 (GRP78) [54]. Therefore, humanin can increase autophagy via both AMPK and its binding to IGFBP-3. Further studies deciphering the detail mechanism on the interaction between humanin, AMPK, IGFBP-3 will provide us better understanding on the mechanisms of humanin in autophagy induction.
Autophagy activates to maintain muscle integrity by degrading protein aggregates and dysfunctional mitochondria in skeletal muscle [37, 47]. Preventing the accumulation of damaged mitochondria is important as it may induce oxidative damage, skeletal muscle apoptosis, and thereby lead to muscle diseases [47, 55]. In fact, several skeletal muscle diseases show dysregulation of autophagy; excessive autophagy triggers excessive muscle catabolism, which in turn leads to atrophy, whereas reduced autophagic flux exhibits muscle degeneration and dysfunctional mitochondria [47, 56, 57]. Additionally, it has been found that autophagy declines throughout the normal aging process in invertebrates and higher organisms, positively correlating with common signs of aging such as progressive decline of muscle mass, strength, and quality [57]. Autophagic removal of damaged mitochondria appears to be one of the cellular mechanisms to prevent loss of muscle mass and quality by attenuating mitochondria-induced apoptosis in a healthy cell [55-57].
Due to its cytoprotective nature, there has been growing attention on the therapeutic modulation of autophagy [42, 51, 52]. Autophagy should be stimulated when the goal is to increase normal cellular functions, but it should be inhibited when the goal is to treat cancer [58]. Having full control of the regulation of autophagy would be crucial to develop a cure for various diseases, such as skeletal muscle dysfunction, cancer, cardiovascular diseases, and neurodegeneration [3]. Age-related muscle dysfunction and myopathy is one of the leading causes of permanent disability and mortality in elders, but there is no known treatment for it [59]. Many are studying pathways and potential pharmacological treatments that modulate autophagy to consequently improve skeletal muscle health and function [42, 51, 52]. Previously, we showed humanin administration can improve skeletal muscle function in the aged mice and improves other healthspan markers [28]. Here, we showed humanin administration increased autophagy and decreased abnormal protein accumulation in skeletal muscle. These results suggest that humanin-induced autophagy potentially contributes to the improved skeletal muscle function. There is great interest in the therapeutic potential of MDP analogs in aging-related diseases and metabolism disorders, and a MOTS-c analogue entered clinical trial for NASH. A recent study showed high-intensity interval exercise increases plasma and muscle humanin expression [60]. Further studies examining whether humanin treatment indeed improves muscle mass, strength, and exercise capacity in aged mice or other mice models of muscle wasting in an autophagy dependent manner would be important to develop new therapeutic targets for muscle wasting.
In summary, our studies showed that humanin-induced autophagy plays important roles in skeletal muscle function during aging and lifespan extension in humanin transgenic worms. These results suggest that humanin is an autophagy inducer and has therapeutic potential for targeting autophagy in various age-related diseases including sarcopenia.
Supplementary Material
Supplemental figure 1. High throughput screening of mitochondrial-derived peptides. Four MDPs, including HNG F6A, SHLP2, SHLP4, and SHLP6, were treated in the screening. Red puncta (autolysosome) intensity per cells were quantified (n=8).
Supplemental figure 2. Humanin did not alter autophagy-related gene expression in mouse liver. Autophagy-related gene (ATG7 and ATG8) expression was measured by qRT-PCR (n=6/group).
Highlights.
While the first identified MDP called humanin has been linked to many beneficial effects in various age-relative diseases, this is the first report of humanin being associated with macroautophagy.
Humanin administration induces autophagy in three different cell lines, while humanin siRNA treatment reduces the basal autophagy levels.
Humanin administration in old mice increases autophagy-related gene, decreases abnormal protein accumulation, and improves exercise capacity.
Humanin is co-expressed with autophagy-related genes in human brains.
Acknowledgements
The High Throughput Autophagy Screening Assay was performed using Cellomics® ArrayScan® VTI HCS Reader in the Choi Family Therapeutic Screening Facility at the Eli and Edythe Broad CIRM Center for Regenerative Medicine and Stem Cell Research at USC. ptfLC3 (Mammalian expression of rat LC3 fused to mRFP and EGFP) was a gift from Tamotsu Yoshimori (Addgene plasmid # 21074 ; http://n2t.net/addgene:21074 ; RRID:Addgene_21074). The bec-1 RNAi clone was obtained from Dr. Malene Hansen, Sanford Burnham Prebys medical discovery institute. This work was supported by an Ellison/AFAR Postdoctoral Fellowship in Aging Research Program to SJK and by 3P01AG055369, 5R01AG068405, RF1AG061834, and R01AG069698 Award to PC.
Footnotes
Competing interests
Pinchas Cohen is a consultant and stockholder of CohBar Inc.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Kim SJ, Miller B, Kumagai H, Silverstein AR, Flores M, Yen K, Mitochondrial-derived peptides in aging and age-related diseases, Geroscience, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hunt RJ, Bateman JM, Mitochondrial retrograde signaling in the nervous system, FEBS letters, 592 (2018) 663–678. [DOI] [PubMed] [Google Scholar]
- [3].Leidal AM, Levine B, Debnath J, Autophagy and the cell biology of age-related disease, Nature cell biology, 20 (2018) 1338–1348. [DOI] [PubMed] [Google Scholar]
- [4].Li Z.-y., Yang Y, Ming M, Liu B, Mitochondrial ROS generation for regulation of autophagic pathways in cancer, Biochemical and biophysical research communications, 414 (2011) 5–8. [DOI] [PubMed] [Google Scholar]
- [5].Graef M, Nunnari J, Mitochondria regulate autophagy by conserved signalling pathways: Mitochondria regulate autophagy, The EMBO journal, 30 (2011) 2101–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Baixauli F, Acin-Perez R, Villarroya-Beltri C, Mazzeo C, Nunez-Andrade N, Gabande-Rodriguez E, Ledesma MD, Blazquez A, Martin MA, Falcon-Perez JM, Redondo JM, Enriquez JA, Mittelbrunn M, Mitochondrial Respiration Controls Lysosomal Function during Inflammatory T Cell Responses, Cell Metab, 22 (2015) 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Gomez-Suaga P, Paillusson S, Stoica R, Noble W, Hanger DP, Miller CCJ, The ER-Mitochondria Tethering Complex VAPB-PTPIP51 Regulates Autophagy, Current biology, 27 (2017) 371–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Miller B, Kim SJ, Kumagai H, Mehta HH, Xiang W, Liu J, Yen K, Cohen P, Peptides derived from small mitochondrial open reading frames: Genomic, biological, and therapeutic implications, Exp Cell Res, 393 (2020) 112056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hashimoto Y, Niikura T, Ito Y, Sudo H, Hata M, Arakawa E, Abe Y, Kita Y, Nishimoto I, Detailed characterization of neuroprotection by a rescue factor humanin against various Alzheimer's disease-relevant insults, J Neurosci, 21 (2001) 9235–9245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ikonen M, Liu B, Hashimoto Y, Ma L, Lee KW, Niikura T, Nishimoto I, Cohen P, Interaction between the Alzheimer's survival peptide humanin and insulin-like growth factor-binding protein 3 regulates cell survival and apoptosis, Proc Natl Acad Sci U S A, 100 (2003) 13042–13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Guo B, Zhai D, Cabezas E, Welsh K, Nouraini S, Satterthwait AC, Reed JC, Humanin peptide suppresses apoptosis by interfering with Bax activation, Nature, 423 (2003) 456–461. [DOI] [PubMed] [Google Scholar]
- [12].Kim SJ, Xiao J, Wan J, Cohen P, Yen K, Mitochondrially derived peptides as novel regulators of metabolism, J Physiol, 595 (2017) 6613–6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Muzumdar RH, Huffman DM, Atzmon G, Buettner C, Cobb LJ, Fishman S, Budagov T, Cui L, Einstein FH, Poduval A, Hwang D, Barzilai N, Cohen P, Humanin: a novel central regulator of peripheral insulin action, PLoS One, 4 (2009) e6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sreekumar PG, Ishikawa K, Spee C, Mehta HH, Wan J, Yen K, Cohen P, Kannan R, Hinton DR, The Mitochondrial-Derived Peptide Humanin Protects RPE Cells From Oxidative Stress, Senescence, and Mitochondrial Dysfunction, Investigative ophthalmology & visual science, 57 (2016) 1238–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Qin Q, Jin J, He F, Zheng Y, Li T, Zhang Y, He J, Humanin promotes mitochondrial biogenesis in pancreatic MIN6 β-cells, Biochemical and biophysical research communications, 497 (2018) 292–297. [DOI] [PubMed] [Google Scholar]
- [16].Yen K, Mehta HH, Kim S-J, Lue Y, Hoang J, Guerrero N, Port J, Bi Q, Navarrete G, Brandhorst S, Lewis KN, Wan J, Swerdloff R, Mattison JA, Buffenstein R, Breton CV, Wang C, Longo V, Atzmon G, Wallace D, Barzilai N, Cohen P, The mitochondrial derived peptide humanin is a regulator of lifespan and healthspan, Aging (Albany, NY.), 12 (2020) 11185–11199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Muzumdar RH, Huffman DM, Calvert JW, Jha S, Weinberg Y, Cui L, Nemkal A, Atzmon G, Klein L, Gundewar S, Yong Ji S, Lavu M, Predmore BL, Lefer DJ, Acute Humanin Therapy Attenuates Myocardial Ischemia and Reperfusion Injury in Mice, Arteriosclerosis, thrombosis, and vascular biology, 30 (2010) 1940–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Thummasorn S, Apaijai N, Kerdphoo S, Shinlapawittayatorn K, Chattipakom SC, Chattipakorn N, Humanin exerts cardioprotection against cardiac ischemia/reperfusion injury through attenuation of mitochondrial dysfunction, Cardiovascular therapeutics, 34 (2016) 404–414. [DOI] [PubMed] [Google Scholar]
- [19].Ruby JG, Smith M, Buffenstein R, Naked Mole-Rat mortality rates defy gompertzian laws by not increasing with age, Elife, 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lee C, Wan J, Miyazaki B, Fang Y, Guevara-Aguirre J, Yen K, Longo V, Bartke A, Cohen P, IGF-I regulates the age-dependent signaling peptide humanin, Aging Cell, 13 (2014) 958–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yen K, Lee C, Mehta H, Cohen P, The emerging role of the mitochondrial-derived peptide humanin in stress resistance, J Mol Endocrinol, 50 (2013) R11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kuliawat R, Klein L, Gong Z, Nicoletta-Gentile M, Nemkal A, Cui L, Bastie C, Su K, Huffman D, Surana M, Barzilai N, Fleischer N, Muzumdar R, Potent humanin analog increases glucose-stimulated insulin secretion through enhanced metabolism in the β cell, The FASEB journal, 27 (2013) 4890–4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhang W, Zhang W, Li Z, Hao J, Zhang Z, Liu L, Mao N, Miao J, Zhang L, S14G-humanin improves cognitive deficits and reduces amyloid pathology in the middle-aged APPswe/PS1dE9 mice, Pharmacol Biochem Behav, 100 (2012) 361–369. [DOI] [PubMed] [Google Scholar]
- [24].Mamiya T, Ukai M, [Gly(14)]-Humanin improved the learning and memory impairment induced by scopolamine in vivo, Br J Pharmacol, 134 (2001) 1597–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Matsuoka M, Hashimoto Y, Aiso S, Nishimoto I, Humanin and colivelin: neuronal-death-suppressing peptides for Alzheimer's disease and amyotrophic lateral sclerosis, CNS Drug Rev, 12 (2006) 113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhao S-T, Huang X.-t., Zhang C, Ke Y, Humanin Protects Cortical Neurons from Ischemia and Reperfusion Injury by the Increased Activity of Superoxide Dismutase, Neurochemical research, 37 (2012) 153–160. [DOI] [PubMed] [Google Scholar]
- [27].Zhao J, Zeng Y, Wang Y, Shi J, Zhao W, Wu B, Du H, Humanin protects cortical neurons from calyculin A-induced neurotoxicities by increasing PP2A activity and SOD, International journal of neuroscience, ahead-of-print 1-9. [DOI] [PubMed] [Google Scholar]
- [28].Yen K, Wan J, Mehta HH, Miller B, Christensen A, Levine ME, Salomon MP, Brandhorst S, Xiao J, Kim S-J, Navarrete G, Campo D, Harry GJ, Longo V, Pike CJ, Mack WJ, Hodis HN, Crimmins EM, Cohen P, Humanin Prevents Age-Related Cognitive Decline in Mice and is Associated with Improved Cognitive Age in Humans, Scientific reports, 8 (2018) 14212–14210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Cobb LJ, Lee C, Xiao J, Yen K, Wong RG, Nakamura HK, Mehta HH, Gao Q, Ashur C, Huffman DM, Wan J, Muzumdar R, Barzilai N, Cohen P, Naturally occurring mitochondrial-derived peptides are age-dependent regulators of apoptosis, insulin sensitivity, and inflammatory markers, Aging (Albany NY), 8 (2016) 796–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Nashine S, Cohen P, Nesburn AB, Kuppermann BD, Kenney MC, Characterizing the protective effects of SHLP2, a mitochondrial-derived peptide, in macular degeneration, Sci Rep, 8 (2018) 15175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zempo H, Kim S-J, Fuku N, Nishida Y, Higaki Y, Wan J, Yen K, Miller B, Vicinanza R, Miyamoto-Mikami E, Kumagai H, Naito H, Xiao J, Mehta HH, Lee C, Hara M, Patel YM, Setiawan VW, Moore TM, Hevener AL, Sutoh Y, Shimizu A, Kojima K, Kinoshita K, Arai Y, Hirose N, Maeda S, Tanaka K, Cohen P, A pro-diabetogenic mtDNA polymorphism in the mitochondrial-derived peptide, MOTS-c, Aging (Albany, NY.), 13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kim KH, Son JM, Benayoun BA, Lee C, The Mitochondrial-Encoded Peptide MOTS-c Translocates to the Nucleus to Regulate Nuclear Gene Expression in Response to Metabolic Stress, Cell metabolism, 28 (2018) 516–524.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kimura S, Noda T, Yoshimori T, Dissection of the Autophagosome Maturation Process by a Novel Reporter Protein, Tandem Fluorescent-Tagged LC3, Autophagy, 3 (2007) 452–460. [DOI] [PubMed] [Google Scholar]
- [34].Abdelfatah S, Abdellatif M, Abel S, Ahmed ZM, Alves S, Araki Y, Arguelles S, Armstrong-James D, Arnauné-Pelloquin L, Azzopardi M, Bae D-H, Bai Y, Baptista MS, Belleudi F, Belló Pérez M, Beltran S, Bezbradica JS, Boban M, Burgoyne JR, Campbell EM, Caniggia I, Carle GF, Carter AB, Cavaliere F, Cheung CHA, Coll NS, Corkery DP, Dewson G, Di Fazio P, Dikic I, Dong B, Dou J, Dowaidar M, Eichinger L, Escalante R, Feng W, Flo TH, Florio T, García-Del Portillo F, Ginet V, Giulivi C, González-Rodríguez P, Ho EA, Inoki K, Ippolito G, Jin L, Jo E-K, Kaganovich D, Kain R, Kaminska J, Kanamori H, Kho W, Ktistakis NT, Langer R, Lei Y, Li M, Lin Q, López-Pérez Ó, Lu H, Luna-Dulcey L, Luo Z, Macian F, Maejima Y, Maiti P, Manshian BB, Marconi AM, Masyuk TV, Merkley SD, Milan E, Miyazaki T, Mizushima N, Moura AF, Mukhopadhyay S, Ni H-M, O'Donovan TR, Ogawa M, Ojha R, Pan W, Roberts HC, Romero LC, Roth L, Schröder B, Selsby JT, Shojaei S, Singh SB, Soria LR, Subbian S, Sudhandiran G, Tanida I, Towns R, Ulasov IV, Varano G, Vijayan V, Weaver TE, Wildenberg ME, Zhang DD, Zhang H, Zhang Y, Zhu Y, Zientara-Rytter K, Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition) 1, Autophagy, 17 (2021) 1–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Mizushima N, Yoshimori T, Levine B, Methods in Mammalian Autophagy Research, Cell (Cambridge), 140 (2010) 313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Gong Z, Tasset I, Diaz A, Anguiano J, Tas E, Cui L, Kuliawat R, Liu H, Kühn B, Cuervo AM, Muzumdar R, Humanin is an endogenous activator of chaperone-mediated autophagy, The Journal of cell biology, 217 (2018) 635–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Sanchez AMJ, Csibi A, Raibon A, Cornille K, Gay S, Bernardi H, Candau R, AMPK promotes skeletal muscle autophagy through activation of forkhead FoxO3a and interaction with Ulk1, Journal of cellular biochemistry, 113 (2012) 695–710. [DOI] [PubMed] [Google Scholar]
- [38].Kang N, Kim KW, Shin DM, Humanin suppresses receptor activator of nuclear factor-κB ligand-induced osteoclast differentiation via AMP-activated protein kinase activation, The Korean journal of physiology & pharmacology, 23 (2019) 411–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kwon C, Sun JL, Jeong JH, Jung TW, Humanin attenuates palmitate-induced hepatic lipid accumulation and insulin resistance via AMPK-mediated suppression of the mTOR pathway, Biochemical and biophysical research communications, 526 (2020) 539–545. [DOI] [PubMed] [Google Scholar]
- [40].Xiao J, The regulation, roles, and mechanism of action of mitochondrial-derived-peptides (MDPs) in aging, University of Southern California Dissertations and Theses, (2018). [Google Scholar]
- [41].Rubinsztein David C., Mariño G, Kroemer G, Autophagy and Aging, Cell (Cambridge), 146 (2011) 682–695. [DOI] [PubMed] [Google Scholar]
- [42].Ryu D, Mouchiroud L, Andreux PA, Katsyuba E, Moullan N, Nicolet-Dit-Félix AA, Williams EG, Jha P, Fo Sasso G, Huzard D, Aebischer P, Sandi C, Rinsch C, Auwerx J, Urolithin A induces mitophagy and prolongs lifespan in C. elegans and increases muscle function in rodents, Nature medicine, 22 (2016) 879–888. [DOI] [PubMed] [Google Scholar]
- [43].Kocaturk NM, Gozuacik D, Crosstalk Between Mammalian Autophagy and the Ubiquitin-Proteasome System, Frontiers in cell and developmental biology, 6 (2018) 128–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Cuervo AM, Macian F, Autophagy and the immune function in aging, Current opinion in immunology, 29 (2014) 97–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Demontis F, Perrimon N, FOXO/4E-BP Signaling in Drosophila Muscles Regulates Organism-wide Proteostasis during Aging, Cell (Cambridge), 143 (2010) 813–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Tavernarakis N, Madeo F, Kroemer G, Can autophagy promote longevity?, Nature cell biology, 12 (2010) 842–846. [DOI] [PubMed] [Google Scholar]
- [47].Madeo F, Zimmermann A, Maiuri MC, Kroemer G, Essential role for autophagy in life span extension, The Journal of clinical investigation, 125 (2015) 85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Nakamura S, Yoshimori T, Autophagy and Longevity, Molecules and cells, 41 (2018) 65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C, A role for autophagy in the extension of lifespan by dietary restriction in C. elegans, PLoS genetics, 4 (2008) e24–e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Alicia M, Zsolt T, Matthew S, Eeva-Fiisa E, David HH, Beth L, Autophagy Genes Are Essential for Dauer Development and Life-Span Extension in C. elegans, Science (American Association for the Advancement of Science), 301 (2003) 1387–1391. [DOI] [PubMed] [Google Scholar]
- [51].Tavemarakis N, Kroemer G, Herker E, Madeo F, Sinner F, Magnes C, Laun P, Knauer H, Ring J, Schraml E, Carmona-Gutierrez D, Fróhlich K-U, Fahrenkrog B, Eisenberg T, Schroeder S, Heeren G, Breitenbach M, Fussi H, Weiskopf D, Hartl R, Criollo A, Megalou E, Deszcz L, Grubeck-Loebenstein B, Büttner S, Minois N, Ruckenstuhl C, Schauer A, Antonacci L, Induction of autophagy by spermidine promotes longevity, Nature Cell Biology, 11 (2009) 1305–1314. [DOI] [PubMed] [Google Scholar]
- [52].Bjedov I, Toivonen JM, Kerr F, Slack C, Jacobson J, Foley A, Partridge L, Mechanisms of Life Span Extension by Rapamycin in the Fruit Fly Drosophila melanogaster, Cell metabolism, 11 (2010) 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Allen M, Carrasquillo MM, Funk C, Heavner BD, Zou F, Younkin CS, Burgess JD, Chai H-S, Crook J, Eddy JA, Li H, Logsdon B, Peters MA, Dang KK, Wang X, Serie D, Wang C, Nguyen T, Lincoln S, Malphrus K, Bisceglio G, Li M, Golde TE, Mangravite LM, Asmann Y, Price ND, Petersen RC, Graff-Radford NR, Dickson DW, Younkin SG, Ertekin-Taner N, Human whole genome genotype and transcriptome data for Alzheimer's and other neurodegenerative diseases, Scientific data, 3 (2016) 160089–160089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Grkovic S, O'Reilly VC, Han S, Hong M, Baxter RC, Firth SM, IGFBP-3 binds GRP78, stimulates autophagy and promotes the survival of breast cancer cells exposed to adverse microenvironments, Oncogene, 32 (2013) 2412–2420. [DOI] [PubMed] [Google Scholar]
- [55].Amie D, Christiaan L, Apoptosis in skeletal muscle with aging, American Journal of Physiology - Regulatory, Integrative and Comparative Physiology, 282 (2002) 519–527. [DOI] [PubMed] [Google Scholar]
- [56].Zhang Y, Wang C, Zhou J, Sun A, Hueckstaedt LK, Ge J, Ren J, Complex inhibition of autophagy by mitochondrial aldehyde dehydrogenase shortens lifespan and exacerbates cardiac aging, Biochimica et biophysica acta. Molecular basis of disease, 1863 (2017) 1919–1932. [DOI] [PubMed] [Google Scholar]
- [57].Masiero E, Agatea L, Mammucari C, Blaauw B, Loro E, Komatsu M, Metzger D, Reggiani C, Schiaffino S, Sandri M, Autophagy Is Required to Maintain Muscle Mass, Cell metabolism, 10 (2009) 507–515. [DOI] [PubMed] [Google Scholar]
- [58].Chen N, Karantza V, Autophagy as a therapeutic target in cancer, Cancer biology & therapy, 11 (2011) 157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].McCormick R, Vasilaki A, Age-related changes in skeletal muscle: changes to life-style as a therapy, Biogerontology (Dordrecht), 19 (2018) 519–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Woodhead JST, D'Souza RF, Hedges CP, Wan J, Berridge MV, Cameron-Smith D, Cohen P, Hickey AJR, Mitchell CJ, Merry TL, High-intensity interval exercise increases humanin, a mitochondrial encoded peptide, in the plasma and muscle of men, Journal of applied physiology (1985), 128 (2020) 1346–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figure 1. High throughput screening of mitochondrial-derived peptides. Four MDPs, including HNG F6A, SHLP2, SHLP4, and SHLP6, were treated in the screening. Red puncta (autolysosome) intensity per cells were quantified (n=8).
Supplemental figure 2. Humanin did not alter autophagy-related gene expression in mouse liver. Autophagy-related gene (ATG7 and ATG8) expression was measured by qRT-PCR (n=6/group).