Abstract
Post-translational modification (PTM) of proteins is of critical importance to the regulation of many cellular processes in eukaryotic organisms. One of the most well-studied protein PTMs is methylation, wherein an enzyme catalyzes the transfer of a methyl group from a cofactor to a lysine or arginine side chain. Lysine methylation is especially abundant in the histone tails and is an important marker for denoting active or repressed genes. Given their relevance to transcriptional regulation, the study of methyltransferase function through in vitro experiments is an important stepping stone toward understanding the complex mechanisms of regulated gene expression. To date, most methyltransferase characterization strategies rely on the use of radioactive cofactors, detection of a methyl transfer byproduct, or discontinuous-type assays. Although such methods are suitable for some applications, information about multiple methylation events and kinetic intermediates is often lost. Herein, we describe the use of two-dimensional NMR to monitor mono-, di-, and trimethylation in a single reaction tube. To do so, we incorporated 13C into the donor methyl group of the enzyme cofactor S-adenosyl methionine. In this way, we may study enzymatic methylation by monitoring the appearance of distinct resonances corresponding to mono-, di-, or trimethyl lysine without the need to isotopically enrich the substrate. To demonstrate the capabilities of this method, we evaluated the activity of three lysine methyltransferases, Set7, MWRAD2 (MLL1 complex), and PRDM9, toward the histone H3 tail. We monitored mono- or multimethylation of histone H3 tail at lysine 4 through sequential short two-dimensional heteronuclear single quantum coherence experiments and fit the resulting progress curves to first-order kinetic models. In summary, NMR detection of PTMs in one-pot, real-time reaction using facile cofactor isotopic enrichment shows promise as a method toward understanding the intricate mechanisms of methyltransferases and other enzymes.
Significance
Existing methods to monitor lysine methylation kinetics rely on exogenous probes, indirect readout, and/or discontinuous detection. We present a platform for robust and reproducible detection and analysis of methyltransferase activity; we employed NMR spectroscopy using a simple 13C-enriched cofactor for the continuous and direct detection of multiple enzymatic methylation events. Isotopic enrichment of the substrate protein is not required. Using three different methyltransferases, we demonstrated the ability of this method to resolve lysine mono-, di-, and trimethylation in a single reaction. Kinetic traces may be globally fit to extract kinetic parameters for each methylation event. We anticipate that this data collection and analysis pipeline may extend to the detection of other post-translational modifications through the use of similar isotope-enriched probes.
Introduction
Dynamic post-translational modification (PTM) of histones is a critical regulator of the formation and maintenance of the chromatin structure that ultimately dictates transcriptional activation (1,2). The histone N-terminal disordered tails are subject to myriad modifications, such as methylation, acetylation, and phosphorylation, among others (3), and such modifications are sensed by downstream effector proteins. Lysine side chain methylation in the histone tails is one example of a PTM that influences chromatin architecture by gatekeeping nucleosome association with chromatin remodeling enzymes, other histone-modifying enzymes, transcription factors, and DNA repair machinery (4). The specific lysine target and number of methyl groups incorporated influence the recruitment of enzymes and transcription factors. For example, trimethylation of the histone H3 tail at lysine 4 (H3K4) is associated with transcriptional activation (5), whereas H3K9 trimethylation demarcates inactive genes (3,6). The study of histone modification by lysine methyltransferases (KMTs) has been central to understanding the interplay between PTMs and regulated gene expression (6, 7, 8); however, we now understand that methylation events are implicated in the functional regulation of nonhistone proteins as well, including transcription factors and signaling proteins (9, 10, 11, 12). Given the importance of methylation events to general and specific transcriptional control, we must employ robust and reliable methods to characterize such protein modifications.
At present, KMT activity is studied through various direct (e.g., mass spectrometry) and indirect (e.g., enzyme-coupled fluorescence) methods. Mass spectrometry (MS) is a powerful discontinuous-type method used to detect methylated products from in vitro reactions or material isolated from cells (13, 14, 15). In particular, MS-based methods offer quantitation and site-specific resolution of methylation events (16, 17, 18), but detailed and high-resolution kinetic information is inherently lost because of the discontinuous nature of MS detection. Alternatively, the incorporation of radioisotopes—usually 3H or 14C—into the methyl donor cofactor S-adenosyl methionine (SAM) permits autoradiographic detection with high sensitivity and is adaptable to high-throughput and plate formats (19, 20, 21). Radiation-based methods may also be coupled to MS to validate the number and location of methylation events (22,23). By necessity, however, these methods require the use of radiolabeled SAM, which adds a radiation safety barrier to such studies, and these studies remain discontinuous. Continuous-detection assays are advantageous because they provide real-time kinetic information and often rely on spectrophotometric detection (24), thus alleviating the need to use a radiolabeled cofactor. Many colorimetric-based detection strategies are amenable to high-throughput formats and can be used with conventional plate readers (25). Such assays rely on enzymatic steps to convert the reaction byproduct, S-adenosyl homocysteine, into a spectrophotometrically observable compound (26, 27, 28, 29). The rate of S-adenosyl homocysteine generation provides an indirect readout of KMT activity but inherently obscures multiple methylation events and site specificity information.
NMR has been leveraged toward the study of post-translational modifications broadly (30, 31, 32), in addition to specific detection of methyl-lysine. The chemical shift of the lysine backbone peak generally does not respond to methylation events, but selective 13C enrichment allows detection of methylated species by changes in the lysine CH2ε chemical shift. Importantly, mono-, di-, and trimethylated lysines yield distinct peaks, which enables kinetic analysis by tracking each methyl-state peak over time (33). To date, NMR detection of methylation has relied on selective or uniform isotope labeling of the substrate, which by necessity limits the number of potential methylation targets that may be studied through existing methods. In contrast, detection of protein phosphorylation by NMR exploits the naturally abundant spin-1/2 31P and so has been far more widely adopted. We have leveraged this advantage previously to observe continuous and real-time tracking of phosphorylation or dephosphorylation in the RNA Pol II C-terminal domain (34,35). Thus, we posited that in analogy to phosphorylation, enrichment of an enzyme cofactor needed for lysine methylation with an NMR-active nucleus may extend the utility of NMR as a tool for KMT characterization by circumventing the requirement that substrates be expressible with isotopic enrichment. Facile adaptation of PTM detection platforms to low-abundance or otherwise challenging protein systems is especially important given emerging insights into the prevalence of nonhistone protein methylation targets (9).
Here, we present a platform and analysis pipeline for continuous, real-time, and directly detected methyltransferase kinetics analysis. SAM enriched with 13C in the donor methyl group (Fig. 1 A) provides a probe used to monitor methyltransferase activity by direct detection of the covalent modification. To test this method, we characterized the methyltransferase activity toward the histone H3 tail for three out of the dozens of known KMTs (7). Set7 (also known as SetD7, Set9 (36), and Set7/9 (37)) is a monomethyltransferase with specificity for H3K4 (38, 39, 40), mixed-lineage leukemia protein-1 (MLL1) within the MWRAD2 core complex has dimethyltransferase activity toward H3K4 (41,42), and PRDM9 (also known as Meisetz (43,44)) is a meiosis-specific trimethyltransferase with activity toward H3K4 and H3K36 (22,45).
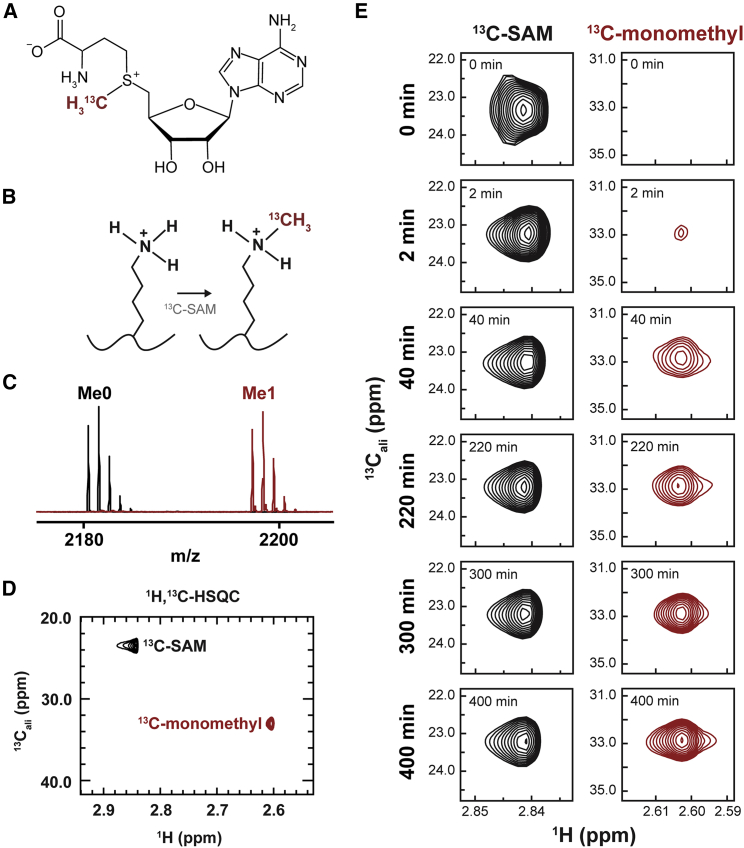
Figure 1.
13C-enriched SAM allows detection of methyl-lysine over time. (A) Structure of S-adenosyl methionine (SAM) depicting 13C enrichment on the donor methyl group. (B) Scheme describing lysine methylation by Set7 (13C-methyl group shown in red). (C) Endpoint matrix-assisted desorption/ionization-time-of-flight mass spectrum of unmodified H3 (2183 m/z, black) and 13C-methylated H3 (2197 m/z). (D) Endpoint NMR spectrum of 13C-monomethylated H3. The peak corresponding to unreacted SAM (black) is distinct from the peak corresponding to 13C-monomethylated H3 (red). (E) Sample time points of HSQC spectra showing buildup of monomethyl peak (red) and modest depletion of SAM peak (black). To see this figure in color, go online.
We find that 13C- mono-, di-, and trimethylated H3K4 each yield distinct resonances in a traditional 1H, 13C-heteronuclear single quantum coherence (HSQC) experiment; therefore, this method can report on all three distinct methyl-lysine states in a single experiment (vide infra). Further, by acquiring several short experiments in sequence and leveraging the quantitative nature of the NMR signal, it becomes possible to directly track the substrate methylation state over time to probe enzyme activity. This method requires isotopic enrichment only of the methyl donor cofactor; the user need not enrich the substrate protein or peptide, use radiolabeled materials, or SAM analogs. To our knowledge, this is the first application of NMR spectroscopy toward the facile characterization of methyltransferase activity by directly detecting on the transferred methyl group. The application of this method to nonhistone protein substrates, other methyltransferases, and even other PTM events in which the enzyme cofactors can be similarly enriched in NMR-active isotopes is expected to yield broad advances for the study of PTM as a regulatory strategy.
Materials and methods
Plasmids
The plasmid containing the Escherichia coli codon-optimized DNA sequence encoding amino acids 109–366 of the human Set7 protein (hereby referred to as Set7) was obtained from Addgene (Addgene #: 40746; Watertown, MA). The protein product of this Set7 construct within the backbone pET28a vector is Set7 residues 109–366 with a preceding Met residue as a start codon and a C-terminal noncleavable 6× His tag. The plasmid containing human DNA sequence encoding amino acids 195–415 of the PRDM9 protein (hereby referred to as PRDM9) within a modified pET28 backbone was obtained from Addgene (Addgene #: 162257). The resulting protein product of the PRDM9 construct is an N-terminal 6× His tag and a tobacco etch virus (TEV) protease recognition site followed by PRDM9 residues 195–415. For MWRAD2, human DNA sequences for each of the following MWRA complex subunits were cloned into the pST44 polycistronic expression vector (46): MLL1 SET domain (aa 3745–3969, Uniprot #: Q03164), WDR5 (aa 2–334, Uniprot #: P61964), RbBP5 (aa 1–538, Uniprot #: Q15291), and Ash2L (aa 1–534, Uniprot #: Q9UBL3-3). The WDR5 subunit harbors an N-terminal 6× His tag followed by a TEV protease recognition sequence. The plasmid containing His-(TEV)-DPY-30 (human aa 1–99, Uniprot #: Q9C005) was described previously (41). A pET19b plasmid containing the SAM synthetase gene from Methanocaldococcus jannaschii (Mj) was a gift from the laboratory of Dr. Squire J. Booker at The Pennsylvania State University (State College, PA).
Protein expression and purification
BL-21(DE3) FhuA2- E. coli cells containing the Set7-His plasmid were grown at 37°C until optical density at 600 nm (OD600) = 0.6–0.8 in Lysogeny Broth (LB) containing 50 μg/mL kanamycin, at which point the cultures were cooled on ice for 15 min. Expression was induced by the addition of 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). After incubation at 15°C for 18 h, cultures were harvested by centrifugation at 3700 × g and either stored at −80°C or lysed immediately. Rosetta BL-21(DE3) cells containing the His-(TEV)-PRDM9 plasmid were grown in LB containing 50 μg/mL kanamycin and 25 μg/mL chloramphenicol at 37°C until OD600 = ∼1.0. Expression was induced by the addition of 0.5 mM IPTG, and the cultures were incubated for 18 h at 15°C, after which point the cells were harvested by centrifugation as described above and either frozen at −80°C or lysed immediately.
Rosetta BL-21(DE3) pLysS E. coli were transformed with the polycistronic pST44 plasmid containing the MWRA subunits and plated on LB agar containing 50 μg/mL carbenicillin and 20 μg/mL chloramphenicol (46,47). Starter cultures were prepared by inoculating 50 mL of Terrific Broth II containing 50 μg/mL carbenicillin and 20 μg/mL chloramphenicol with one colony from the transformation plate. Cultures were grown overnight (∼16 h) at 30°C. 1 L of Terrific Broth II (with antibiotics) was inoculated by diluting the starter culture by 1:50. The cells grew at 37°C until OD600 = ∼1.0, at which point the cultures were chilled at 4°C for 1 h. Expression was induced by the addition of 1 mM IPTG, and the cultures were incubated at 16°C for 20–22 h. Cells were harvested by centrifugation and stored at −80°C until purification. DPY-30 was expressed using the same conditions as the MWRA complex.
Mj SAM synthetase was expressed as described previously (48) with some modifications. Briefly, BL-21(DE3) FhuA2− E. coli cells containing the Mj SAM synthetase plasmid were grown overnight at 37°C in 200 mL of LB containing 100 μg/mL ampicillin. Three liters of LB containing 100 μg/mL ampicillin were inoculated with 50 mL of turbid overnight culture and grown at 37°C until OD600 = 0.6. Protein expression was induced by adding IPTG to a final concentration of 0.5 mM. After an additional incubation at 37°C for 3 h, the cultures were harvested by centrifugation at 3700 × g, and the resulting cell pellets were either lysed immediately or stored at −80°C until purification.
Cell pellets of Set7-His or His-(TEV)-PRDM9 were resuspended in lysis buffer 1 (50 mM Tris-HCl (pH 7.5), 500 mM NaCl, 20 mM imidazole, and 2 mM β-mercaptoethanol (βME)) supplemented with 1 mM phenylmethanesulfonylfluoride fluoride and 1× EDTA-free protease inhibitor cocktail (MilliporeSigma, Burlington, MA; #: 539137). Cells were lysed by sonication on ice, and the lysate was clarified by centrifugation at 14,000 × g at 4°C for 30 min. The clarified lysate was passed through a 5-μm syringe filter and applied to Ni-NTA resin (G-Biosciences, St. Louis, MO) equilibrated with lysis buffer. Bound proteins were washed with lysis buffer 1 supplemented with 0.1% Triton X-100 followed by lysis buffer 1. His-tagged proteins were eluted from the column with elution buffer 1 (50 mM Tris-HCl (pH 7.5), 500 mM NaCl, 200 mM imidazole, and 2 mM βME).
For Set7-His, the nickel column eluate was dialyzed against 4 L 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 2 mM βME for 16 h at 4°C. The dialysate was concentrated and further purified by separation on an Superdex 200 column in 25 mM Tris-HCl (pH 7.5) (at 4°C), 200 mM NaCl, and 5 mM dithiothreitol (DTT). Fractions containing pure Set7-His by SDS-PAGE analysis were pooled and concentrated in a 3 K molecular weight cut-off (MWCO) centrifugal filter (MilliporeSigma) before being aliquoted and frozen at −80°C. In the case of the TEV-cleavable PRDM9 construct, the Ni-NTA eluate was subjected to cleavage by 1 mg TEV protease per liter of culture under dialysis against 4 L of lysis buffer 1 for 16 h at 4°C. The dialysate was applied to a second Ni-NTA column, and flowthrough fractions were concentrated in a 3 K MWCO centrifugal filter. Concentrated PRDM9 was further purified by fast protein liquid chromatography on a Superdex 100 gel filtration column in 20 mM Tris-HCl (pH 7.5) (at 4°C) and 150 mM NaCl. Fractions containing pure PRDM9 were pooled and concentrated before being aliquoted and frozen at −80°C.
Cell pellets containing MWRA complex were lysed by microfluidizer in lysis buffer 2 (50 mM Tris-HCl (pH 7.5), 300 mM NaCl, 30 mM imidazole, 2 mM DTT, and 1 μM ZnCl2) supplemented with EDTA-free protease inhibitor cocktail (Roche, Basel, Switzerland). The lysate was clarified by centrifugation at 25,000 × g at 4°C for 30 min. The supernatant was diluted 1:5 in lysis buffer 2 and applied to tandem HisTrap 5 mL nickel affinity columns (GE Healthcare, Chicago, IL). Bound MWRA complex was washed with 10 column volumes of lysis buffer 2 and then eluted with a 25 column-volume linear gradient of elution buffer 2 (composed of lysis buffer 2 plus 500 mM imidazole). Fractions containing pure MWRA complex were pooled and treated with 0.1 mg/mL TEV protease and dialyzed against lysis buffer 2. The dialysate was passed over the HisTrap columns, and fractions containing cleaved MWRA were pooled, concentrated, and further purified by gel filtration on a Superdex 200 (16/60) column (GE Healthcare) in 20 mM Tris-HCl (pH 7.5), 300 mM NaCl, 1 mM TCEP, and 1 μM ZnCl2. Fractions containing pure MWRA were pooled. DPY-30, expressed and purified as previously described (41), was added in twofold molar excess to purified MWRA. The resulting complex was purified by multiple rounds of gel filtration in 20 mM Tris-HCl (pH 7.5), 300 mM NaCl, 1 mM TCEP, and 1 μM ZnCl2. Pure MWRAD2 complex was concentrated, aliquoted, and flash frozen for storage at −80°C. Nota bene, MWRAD2 is referred to in the text as MLL1 for concision.
Cell pellets containing Mj SAM synthetase were resuspended in lysis buffer 3 (100 mM HEPES (pH 7.5), 300 mM KCl, 5% glycerol, and 2 mM TCEP) and supplemented with 0.1% (v/v) Triton X-100, 1 mg/mL lysozyme, 0.1 mg/mL DNase I, and 1 mM phenylmethanesulfonylfluoride fluoride. The cell suspension was incubated with stirring at ambient temperature for 30 min before sonication. Cells were lysed by five cycles (each spaced by at least 5 min) of the following: 1 min of total sonication time at 50% amplitude with a 1-s pulse/1-s off protocol. The lysate was heat shocked at 75°C for 1 h and then clarified by centrifugation at 14,000 × g for 1 h at 4°C. The clarified lysate was passed over a Ni-NTA column equilibrated in lysis buffer 3, and the bound protein was washed with >15 column volumes of lysis buffer 3. His-tagged proteins were eluted in 100 mM HEPES (pH 7.5), 150 mM KCl, 250 mM imidazole, 10% glycerol, and 4 mM TCEP. The elution fraction was dialyzed against 100 mM Tris (pH 8.0), 50 mM KCl, 10% glycerol, and 4 mM TCEP for 24 h at 4°C and then concentrated in an Amicon centrifugal concentrator. Concentrated Mj SAM synthetase was aliquoted and stored at −80°C.
Peptide synthesis and preparation
All solid-phase synthesized peptides used in this study were purchased from GenScript (Piscataway, NJ). H31–20 peptide contains the following residues: ARTKQTARKSTGGKAPRKQL. A mutant H3 peptide with lysines 9, 14, and 18 mutated to arginines (abbreviated “H3K4KR”) has the following sequence: ARTKQTARRSTGGRAPRRQL. The peptides were reconstituted in 50 mM Tris-HCl (pH 8.5) and 200 mM NaCl and stored as frozen aliquots.
Synthesis and purification of 13C-SAM
13C-enriched S-adenosyl methionine (SAM) was synthesized enzymatically in-house using Mj SAM synthetase. SAM synthesis reactions contained 100 μM SAM synthetase, 15 mM ATP, and 10 mM L-methionine (methyl-13C, 99%; Cambridge Isotope Laboratories, Tewksbury, MA) in 50 mM Tris (pH 8.0), 50 mM KCl, and 50 mM MgCl2. Reactions were carried out at 22°C for at least 4 h; SAM production was monitored by 1H NMR. Reactions were quenched by adding fuming hydrochloric acid to adjust the reaction pH to ∼5.0 and precipitate the SAM synthetase. Precipitated protein was removed by centrifugation at 10,000 × g for 5 min at 22°C, followed by filtration of the supernatant by 0.2-μm syringe filter. The estimated yield for enzymatic conversion of L-methionine (limiting) and ATP to S-adenosyl methionine, based on NMR resonance intensities, is 40–50%. To purify the SAM, the remaining reaction volume was diluted fivefold into 1 mM sodium acetate (pH 5.0) and passed over a CM52 cation exchange column in the same buffer. The column was washed with 10 column volumes of 1 mM sodium acetate (pH 5.0), and then the SAM was eluted with two column volumes of 40 mM sulfuric acid. The eluate was aliquoted, lyophilized, and stored at −80°C. The concentration of SAM was determined by resuspending a small amount of lyophilized SAM in 10 mM sulfuric acid and measuring the absorbance at 280 nm using ε = 15,400 M−1 cm−1. The estimated yield for enzymatic conversion of L-methionine (limiting) and ATP to S-adenosyl methionine, based on NMR resonance intensities, is 40–50%.
Methyltransferase reactions
For MS analysis, methylation was carried out in one bulk reaction: methylation was initiated by the addition of Set7 and each time point, as indicated, was stopped by diluting a portion of the reaction mixture into a solution with a final composition of 5% aqueous acetonitrile (ACN) and 0.5% formic acid, followed by storage at −80°C until analysis by MS. Set7 methylation reactions detected by NMR were carried out at 298 K (25°C) (nota bene, we observed a substantial decrease in the 13C-SAM signal at 310 K (37°C) that was likely associated with degradation at this temperature (Fig. S1 B)). Set7 methylation reactions detected by NMR spectroscopy contained 50 μM H31–20 peptide, 400 μM 13C-SAM, and 5 μM Set7 in 50 mM Tris-HCl (pH 8.5), 50 mM NaCl, 1 mM DTT, and 1% D2O. Total reaction time monitored by real-time NMR was at least 400 min, which was sufficient to generate almost entirely monomethylated H3K4. Set7 methylation reactions for the purpose of generating premethylated H3K4 for subsequent treatment with MLL1 or PRDM9 were conducted with 50 μM H3K4KR mutant peptide, 400 μM 13C-SAM, and 10 μM Set7 in 50 mM Tris-HCl (pH 8.5), 50 mM NaCl, 1 mM DTT for >18 h at 298 K (25°C). The entire reaction tube was flash frozen and stored at −80°C for up to several months. Complete monomethylation of H3K4 was validated by MS.
All MLL1 methylation reactions toward unmodified H31–20 or H3K4KR were carried out at 288 K (15°C). Reactions consisted of 50 μM H31–20 peptide, 400 μM 13C-SAM, and 1 μM MLL1 in 50 mM Tris-HCl (pH 8.5), 200 mM NaCl, 1 mM TCEP, and 1% D2O. The lower temperature and higher salt concentration in MLL1 and PRDM9 reactions, compared with the Set7 reactions, were used to better resolve the accumulation and depletion of intermediate methyl-lysine states. MLL1 reactions toward premonomethylated H3K4KR peptide were carried out by supplementing a Set7-treated H3K4KR reaction with 1 μM MLL1, 1 mM TCEP, 1% D2O, and an additional 200 μM 13C-SAM. MLL1 treatment of premethylated H3K4KR was executed at 50 mM NaCl. In all cases, reactions were initiated by the addition of the enzyme.
Preliminary attempts to methylate H31–20 with PRDM9 and 13C-SAM revealed an aberrant fourth methyl mark, evidenced by substantial peak intensity at late time points in the monomethyl trace (Fig. S2, E and F). Endpoint MS confirmed a mixed population of monomethylated and tetramethylated H3 peptide. Sequencing by tandem MS (MS/MS) (described below) indicated that, in addition to the trimethylation of H3K4, H3K9 was monomethylated by PRDM9. For the sake of simplicity, the H31–20 mutant peptide, in which every lysine except Lys4 was replaced with an arginine residue (H3K4KR), was used in subsequent experiments involving PRDM9. Trimethylation reactions were carried out with 50 μM H3K4KR mutant peptide, 400 μM 13C-SAM, and 1 μM PRDM9 in 50 mM Tris-HCl (pH 8.5), 200 mM NaCl, 1 mM DTT, and 1% D2O. PRDM9 reactions toward premonomethylated H3K4KR peptide were carried out by supplementing a Set7-treated H3K4KR reaction with 1 μM PRDM9, 1 mM TCEP, 1% D2O, and an additional 200 μM 13C-SAM. PRDM9 treatment of premonomethylated H3K4KR was executed at 50 mM NaCl. To probe PRDM9 activity toward predimethylated species, a MLL1-dimethylated H3K4KR sample was supplemented with 200 μM 13C-SAM, 1% D2O, and 1 μM PRDM9. All PRDM9 methylation reactions were carried out at 288 K (15°C) and initiated by the addition of enzyme.
Matrix-assisted laser desorption/ionization-time-of-flight and tandem time-of-flight mass spectrometry
Mass spectra were acquired on an Ultraflextreme instrument (Bruker, Billerica, MA). Before MS analysis, all samples were desalted using Pierce C-18 Spin Columns (Thermo Fisher Scientific, Waltham, MA) and dried in a speed-vac. Desiccated samples were resuspended in 30% aqueous ACN and mixed 1:1 with matrix solution. The matrix solution was composed of either 10 mg/mL 4-chloro-α-cyanocinnamic acid or 10 mg/mL α-cyano-4-hydroxycinnamic acid in 50% aqueous ACN and 2.5% formic acid. The instrument was calibrated using a homemade bovine serum albumin digest (in the appropriate corresponding matrix) in reflector positive detection mode in the 500–5000 mass/charge (m/z) range.
The extent of methylation by Set7 was assessed by comparing the relative intensities of the unmodified peptide mass to the methylated peptide mass (+15 Da per methyl group for 13C-SAM). Methylated peptides from final time points were subjected to fragmentation by MS/MS in LIFT mode. Expected peptide fragment masses were calculated using the MS-product mode in the Protein Prospector web tool (see Table S2). The theoretical peaks were assigned by hand to the experimental fragments corresponding to methylated H3 peptide. The MS/MS peaks and corresponding peptide fragments are shown in Figs. S1 D and S2, B, D and F. All mass spectra were plotted, visualized, and analyzed using in-house Python scripts.
NMR data collection
All NMR data were collected on a Bruker Avance NEO 600 MHz spectrometer equipped with a 5 mm TCI triple-resonance cryoprobe and a SampleJet autosampler. Data acquisition and initial processing (See “Real-time NMR data processing and analysis”) were carried out in Topspin 4.0.9 (Bruker). Each kinetics data set was collected via a series of (up to 110) short two-dimensional (2D) 1H, 13C-HSQC experiments. The starting parameters for all experiments were as follows: four scans (four dummy scans), 1024 1H points × 64 13C points, with sweep widths of 1.5 and 70.0 ppm, respectively. The 1H and 13C transmitter frequency offsets were set to 2.95 and 40.0 ppm, respectively. Endpoint spectra of Set7-methylated H31–20 containing sodium trimethylsilylpropanesulfonate (DSS) to test linearity with concentration were acquired with the same parameters as described above, except the 1H sweep width was widened to 9.8 ppm. For several concentrations of 13C-methylated H3K4, the intensities of the monomethyl and the trimethylsilyl group of DSS (held at 500 μM for all experiments) were measured in NMRFAM-Sparky NMR-FAM. The ratio of monomethyl and trimethylsilyl peak intensities was plotted against methylated H3K4 concentration, and the resulting data were fit by linear regression in Python (Fig. S1 E).
In each data set, the first experiment was collected on the reaction contents before adding enzyme and beginning methyl transfer. Although ultimately omitted from kinetic analysis, experiment 1 was used for sample-specific setup (tuning/matching, shim, and 90° 1H pulse length) and was set up as described above (total experiment time ∼6 min). After experiment 1, experiment 2 was set up in the same fashion, carrying over the pulse calibration from experiment 1 and all other parameters from experiment 1. The indicated enzyme was added to the reaction tube, and the sample was immediately returned to the magnet. The time course deadtime is the time between the addition of the enzyme and the beginning of experiment 2. To ensure efficient mixing, the bulk sample was removed from the NMR tube, added to a separate tube containing the enzyme, mixed thoroughly by pipetting, and then returned to the NMR tube. The only command performed after the addition of enzyme was to recover the deuterium lock. Experiments 3 through n, where n is the total number of data points desired, were set up identically to experiments 1 and 2, except that the number of dummy scans was changed from 4 to 128. Therefore, each time point was evenly spaced by ∼10 min. Immediately after termination of experiment 2, the remainder of the time course was initiated using the “multizg” command on experiment 3 and setting the number of experiments as desired.
Real-time NMR data processing and analysis
The 13C-SAM peak in experiment 1 was used to determine the phase correction for each time course. The following processing steps were applied to each multizg experiment (“time point”) sequentially, and output files were stored for further processing and analysis. First, the experiment 1 phase correction was applied to all time points. Next, the signal from buffer components was suppressed by applying a qfil baseline correction to the appropriate region of the spectrum. Then, the intensity scaling factor (NC_proc) for each time point was set to 10. Finally, one-dimensional (1D) projections of the 2D data were calculated for a row corresponding to the mono-, di-, or trimethyl peak chemical shift (Table S2). Custom Python scripts to run within Topspin 4 for this processing step are available at https://github.com/idpemery/rt-methylation.
After export of 1D projections for each time point from Topspin, an in-house Python script searched each projection text file and concatenated the intensities of the maximum of each 1D projection into a list of intensity values that correspond to each experimental time point. This step was performed individually for each methyl state, e.g., a dimethyltransferase assay would yield two files in this step, one for each mono- and dimethyl events. The user may opt to remove the first time point in this step. The time point list (in minutes) for each time course was generated based on the deadtime and experiment lengths and saved as a separate text file.
Subsequent plotting and global fitting and analysis was carried out in MATLAB (The MathWorks, Natick, MA) using the equations below (all scripts and example data can be found at https://github.com/idpemery/rt-methylation). Briefly, the integrated rate equations (see below) were coded as independent functions. Distinct for the number of methyl groups installed, the global fitting script called the appropriate function(s) into a nested objective function. To enable fitting to shared parameters across all methylation events in a single experiment, the global fitting (objective) function used all integrated rate functions as inputs. For example, a trimethyltransferase reaction requires three separate integrated rate functions (for each mono-, di-, and trimethyl signal). The objective function in the trimethyltransferase global fitting script calls all three integrated rate functions simultaneously and with shared rate constant parameters. Next, using user-inputted initial value parameters, the “time” list, and an array of scaled peak intensities (adjusted for number of spins contributing to the total signal) for each methyl-lysine peak, the objective function was optimized using lsqcurvefit. Importantly, the user must specify initial conditions that are qualitatively aligned with the faster and slower reaction rates by observing the raw data. Failure to do this causes the minimization program to incorrectly assign rate constant parameters because of a divide-by-zero error resulting from difference expressions in the denominators of various terms in the integrated rate equations (49). The resulting shared parameters were fed back into each integrated rate function to generate a nonlinear least-squares fit for each kinetic trace.
Set7 (monomethyltransferase)
Methylation Scheme:
Differential Rate Equations:
| (1) |
| (2) |
Integrated Rate Equations:
| (3) |
| (4) |
MLL1 (dimethyltransferase)
Methylation Scheme:
Differential Rate Equations:
| (5) |
| (6) |
| (7) |
Integrated Rate Equations:
| (8) |
| (9) |
| (10) |
PRDM9 (trimethyltransferase)
Methylation Scheme:
Differential Rate Equations:
| (11) |
| (12) |
| (13) |
| (14) |
Integrated Rate Equations:
| (15) |
| (16) |
| (17) |
| (18) |
Dimethyltransferase (pre-Me1 substrate)
Methylation Scheme:
Differential Rate Equations:
| (19) |
| (20) |
Integrated Rate Equations:
| (21) |
| (22) |
Trimethyltransferase (pre-Me1 substrate)
Methylation Scheme:
Differential Rate Equations:
| (23) |
| (24) |
| (25) |
Integrated Rate Equations:
| (26) |
| (27) |
| (28) |
Results and discussion
We began by studying the activity of the well-characterized enzyme Set7 toward the H3 tail. In addition to its role in transcriptional regulation through H3K4 activity (5,36), Set7 also targets numerous nonhistone substrates including p53 (10), TAF10 (50), and pancreatic factor Pdx1 (51). Wild-type Set7 reliably installs just one methyl group onto its targeted lysine side chains (11,52) and thus was a suitable prototype enzyme by which to generate 13C-methylated H3K4 for detection by NMR spectroscopy. A reference 1H, 13C-HSQC spectrum (Fig. S1 A) was generated through overnight incubation of a histone H3-derived peptide (human amino acids 1–21) with recombinant Set7 in the presence of 13C-SAM as a cofactor (Fig. 1 A). The transfer of 13C-methyl from SAM to the lysine side chain (Fig. 1 B) results in the appearance of a peak that is distinct from the peak associated with unreacted 13C-SAM (Fig. 1 C; Fig. S1 A). Importantly, the peak height is quantitatively proportional to the number of spins that contribute to the signal and, therefore, allows use to evaluate the degree of methylation for up to three methylation events. Endpoint matrix-assisted desorption/ionization-time-of-flight mass spectrometry (MALDI-TOF MS) showed nearly complete (>99%) monomethylation of the H3 peptide and tandem MS/MS confirmed that lysine 4 is the sole target (Fig. 1 C; Fig. S1 D).
Next, we sought to determine if continuous in situ monitoring of monomethylation through 1H, 13C-HSQC was suitable for kinetic analysis. Acquisition of several short (∼6 min), sequential 1H, 13C-HSQC experiments on a reaction containing H3 peptide, Set7, and 13C-SAM allowed us to observe the gradual appearance of the unique peak corresponding to 13C-monomethylated H3K4. Select time points from the continuous NMR acquisition are shown in Fig. 1 E. Importantly, this assay, which yielded almost 18 h of data with ∼10-min resolution, required less than an hour of hands-on experiment time. Thus, with the proper consideration of experimental conditions (e.g., enzyme concentration and SAM concentration), direct detection of 13C-methylated substrate accumulation through NMR is achievable. Having demonstrated the technique through monomethylation by Set7, we next turned our attention to two multimethyltransferase enzymes, which pose a greater challenge for state-of-the-art methods.
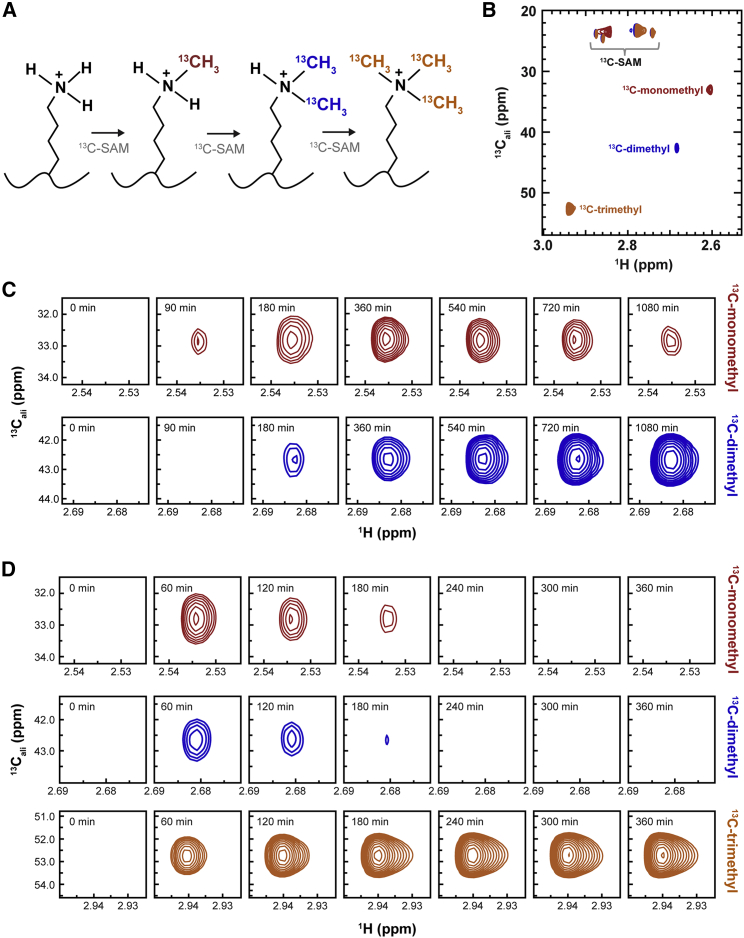
We selected the putative di- and trimethyltransferases MLL1 and PRDM9, respectively, to test whether our real-time NMR assay could simultaneously resolve multiple 13C-methyl groups on a single lysine. The MLL1 methyltransferase activity is specific to Hox genes and is associated with certain human leukemias (53, 54, 55). Importantly, absent the rest of the WRAD2 subunits, MLL1 displays only monomethyltransferase activity toward H3K4 in vitro (41). Therefore, the experiments conducted here used the entire MLL1 core complex (MWRAD2) to ensure H3K4 dimethylation; however, we note that MLL1 appears to modestly trimethylate H3K4, which is consistent with previous results (41) (Fig. S2 A). PRDM9 is a trimethyltransferase that canonically trimethylates histone H3 lysine residues to demarcate double-stranded DNA break loci associated with meiotic prophase (56,57). Although the in vivo activity of PRDM9 is targeted toward H3K4 and H3K36 (58), in vitro, including our work (Fig. S2, E and F), note activity toward H3K9 and H3K18 as well (59). To focus our study on the trimethylation of H3K4, the H3 peptide used in PRDM9 characterization contained multiple Lys-to-Arg mutations (H3K4KR; see Materials and methods).
H3 peptide was subjected to enzymatic mono-, di-, or trimethylation using 13C-SAM (Fig. 2 A) and monitored for up to 18 h through sequential acquisition of 10-min 1H, 13C-HSQC experiments. Each methyl-lysine state is observable as a distinct peak in the final time point spectrum (Fig. 2 B). By tracking each methyl-state peak over time, we observe a monotonic increase of the terminal methyl species and, for MLL1 and PRDM9, build-up and subsequent depletion of the intermediate methylation states (Fig. 2, C and D). Importantly, MLL1 and PRDM9 reaction conditions were selected to slow the kinetics to sufficiently sample the intermediate methyl states. Peak height of each methyl-lysine state was plotted against time for each enzyme (Fig. 3, A–C). The terminal methyl-state progress curves for each enzyme show that H3K4 was almost entirely methylated on the timescale of the experiment (Fig. 1 C; Figs. S1 D and S2, A–D). Direct comparison of MLL1 and PRDM9 methylated species over time from Fig. 2, C and D, respectively, implicates MLL1 as a nonprocessive enzyme and PRDM9 as a processive enzyme. In the case of MLL1, this assignment comes from the observation of the accumulation and persistence of the monomethyl H3K4 intermediate, followed by subsequent, but incomplete, depletion of the intermediate state upon dimethylation (60) (Fig. 2 C). The nonprocessivity of MLL1 was first described through single-turnover experiments (41). In contrast, the mono- and dimethyl intermediates in the PRDM9 reaction are rapidly converted to the next methyl state without substantial accumulation (Fig. 2 D), which is characteristic of a processive enzyme.
Figure 2.
Substrate methylation using 13C-SAM yields distinct peaks corresponding to mono-, di-, and tri-methyl-lysine. (A) Color-coded methylation scheme by a tri-methyltransferase enzyme. (B) 2D 1H, 13C-HSQC spectra showing methyl-lysine resonances at final experiment time points (red = Set7, blue = MLL1, and orange = PRMD9). (C) Sample time points of HSQC spectra showing the resonance intensities of the mono- and dimethyl peaks over time installed by MLL1. (D) Sample time points of PRDM9-installed mono-, di-, and trimethyl marks. To see this figure in color, go online.
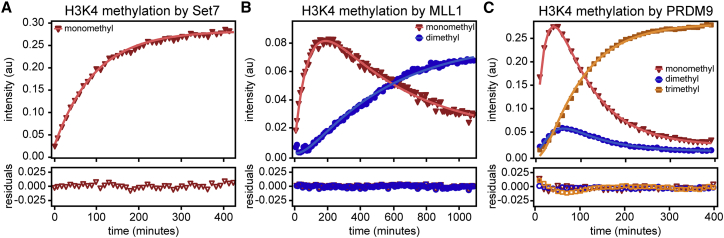
Figure 3.
Kinetic profiles and fits for mono-, di-, and trimethylation. (A) Progress curve (red triangles) and nonlinear fit (solid line) to the signal from Set7-monomethylated H3 peptide over time. (B) Progress curve tracking H3 mono- (red triangles) and di-methylation (blue circles) by MLL1. Nonlinear fits are shown by the solid lines. (C) Progress curves for mono- (red triangles), di- (blue circles), and trimethylation (orange squares) activity of PRDM9 toward H3K4. Fits are shown as solid lines. Rate constants extracted from global fits of methyltransferase data are shown in Table 1. Residuals for each fit are plotted below each progress curve panel. To see this figure in color, go online.
For each enzyme, we fit the methyl-state progress curves to first-order integrated rate equations (see Materials and methods) (Fig. 3, A–C). For Set7, each technical replicate was plotted and fitted individually, and the extracted rate constants (k1) were averaged (Table 1). For MLL1 and PRDM9, the mono- and multimethylation data were fit globally within each replicate to extract rate constants shared between all methylation events. The average of rate constants from three technical replicates are reported in Table 1. The residuals of global fitting are shown below each plot (Fig. 3, A–C), and the SD from the average of rate constants across replicates is reported in Table 1. Monomethylation of H3K4 by MLL1 is about five times faster than the following dimethylation step, which is consistent with previous reports (41,47). PRDM9 activity monomethyl and trimethyl rate constants are comparable in magnitude and suggest that mono- and trimethylation steps are about five times faster than dimethylation by PRDM9. The finding that dimethylation is the slowest step in stepwise methylation of H3K4 by PRDM9 is consistent with existing kinetic information (22). Acknowledging the use of reaction conditions that allowed for deliberate slowing of the MLL1 and PRDM9 methyl transfer, our kinetic parameters are in qualitative agreement with previous studies.
Table 1.
Rate constants extracted from global fits of kinetic data
| Set7 | MLL1 | PRDM9 | MLL1-Me1a | PRDM9-Me1 | |
|---|---|---|---|---|---|
| k1 (min−1) | 0.0151 ± 0.0062b | 0.0092 ± 0.0030 | 0.0401 ± 0.0031 | NA | NA |
| k2 (min−1) | NA | 0.0018 ± 0.0002 | 0.0121 ± 0.0031 | 0.0017 ± 0.0008 | 0.0119 ± 0.0032 |
| k3 (min−1) | NA | NA | 0.0506 ± 0.0070 | NA | 0.1982 ± 0.0560 |
NA, not applicable.
Reported rate constants represent the average of two data sets.
Reported error is the SD from the average of three technical replicates and global fits.
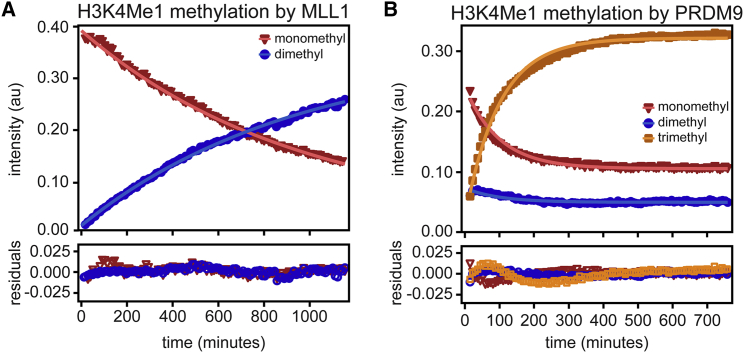
We next applied this method to monitor multimethyltransferase activity toward a monomethylated substrate (H3K4Me1) to assess the impact of a priming methyl mark on downstream methyltransferase activity. Beyond the clear biological significance of this approach, this experiment was important for comparison with methods that use radioisotopes to evaluate KMT activity, which generally rely on the use of premethylated substrate peptides to monitor di- or trimethylation activity. In this manner, the enzyme activity toward a given substrate encompasses, at minimum, a binding step before the methylation step of interest. Notably, our use of real-time NMR to observe mono-, di-, and trimethylation events in a single assay circumvents the requirement of feeding premethylated substrate to the enzyme and thus separates the binding and methylation steps.
To compare our novel method with existing methods, we repeated the MLL1 and PRDM9 activity assays using H3 peptide that was entirely K4-monomethylated by Set7 using 13C-SAM (Fig. 4, A and B). By globally fitting the monotonically decreasing monomethyl signal alongside the multimethylation events, we extracted rate constants that more closely reflect the experimental conditions of traditional discontinuous assays (Table 1). For MLL1, the dimethylation rate constant (k2) is almost the same as that for MLL1 dimethylation of unmodified H3K4. In the case of PRDM9 given the H3K4Me1 substrate, dimethylation still appears to be the rate-limiting step. To achieve near-complete trimethylation, PRDM9-H3K4Me1 experiments were conducted at 25°C and 50 mM sodium chloride, in contrast to assays using unmodified H3K4 (15°C and 200 mM sodium chloride). Interestingly, we did not observe increased overall activity; whereas the trimethylation step given H3K4Me1 was about four times faster than trimethylation of unmodified H3K4, the dimethyl rate constant was the same within error regardless of starting peptide. Discontinuous-type assays that require the use of premethylated substrates to assay di- or tri- methylation events may conflate methylation activity with an initial binding event that may or may not be relevant to an enzyme’s canonical activity. The current methodology allows total control so that the substrate presented to the enzyme can better represent the (presumed) biological substrate.
Figure 4.
Multimethyltransferase activity toward H3K4Me1. (A) Representative progress curve tracking the disappearance of H3K4Me1 and appearance of H3K4Me2 catalyzed by MLL1. Mono- and dimethyl data points are red triangles and blue circles, respectively, and their nonlinear fits are shown as solid lines. The data were globally fit to a three-state model to extract a dimethyl rate constant, k2. (B) Representative plot tracking trimethylation of H3K4Me1 by PRMD9. Monomethyl data and fit are shown in red, dimethyl data and fit are blue, and trimethyl data and fit are orange. The data were globally fit to a four-state model to extract di- and trimethyl rate constants (k2 and k3, respectively). Residuals for each fit are plotted by color beneath each panel. Rate constants from global fitting of each data set are shown in Table 1. To see this figure in color, go online.
Conclusions
Given the critical importance of PTMs to myriad cellular processes (1), the study of the enzymes responsible for such modifications is integral to a mechanistic understanding of the regulation of those processes. In particular, histone 3 lysine 4 methylation demarcates loci of active gene expression (3). Existing methods by which to study KMT activity fall primarily into three categories: radioisotope incorporation, spectrophotometric detection on a reaction byproduct, or MS detection. Although each of the aforementioned strategies have advantages and disadvantages, no existing method affords both direct detection of the covalent modification and continuous kinetic information simultaneously.
The real-time NMR detection and analysis platform that we describe here offers a continuous, one-pot assay to resolve multiple methylation events. By conducting methyltransferase reactions using 13C-SAM, investigators can directly observe up to three methyl groups on a single lysine side chain in a short 2D NMR experiment. To demonstrate the utility of this method, we characterized three KMTs with different putative specificities. Set7, MLL1, and PRDM9 were used as prototype enzymes to assess mono-, di-, and trimethyltransferase activity, respectively. Global fitting of multimethylation data to first-order rate equations yielded reproducible rate constants for each methylation step from a single overnight reaction. Further, the adaptability of this method to study unmodified and premethylated substrates in the same experimental framework permits the direct study of the biologically appropriate substrate of a given enzyme.
Another advantage of this method is that direct detection of PTMs by NMR does not require uniformly or site-specifically isotope-labeled substrate. Therefore, this real-time platform is amenable to the study of virtually any methyltransferase that uses SAM as a cofactor and can be made to perform on the minutes-hours timescale. For the observation of faster kinetic phases and/or in facilities in which robust detection using short experiments is not feasible, the end user may employ a chemical quench-flow time point collection format coupled to downstream analysis by 2D NMR. Inclusion of an NMR standard, such as DSS, will allow for quantitative comparison across discrete time points while retaining the label-free advantages of the one-pot NMR method. At present, however, assignment of targeted lysine residues is not trivial by NMR; degeneracy of chemical shifts for methylation on different lysines prohibits the unambiguous identification of modified lysines by NMR. Instead, as we have done in this study, lysine targets may be determined or validated by mutagenic or mass spectrometric methods.
Given the emerging insights into the importance of PTMs not only on histones but across the proteome (24), this method may find its greatest impact through application to nonhistone substrates. Treatment of a 13C-methylated substrate with a demethylase may also be a tractable application of the real-time NMR method presented here. Further, we envision potential applications for this detection and analysis strategy in the characterization of other PTMs if an appropriate spin 1/2 probe (cofactor) may be employed. In summary, the real-time NMR strategy that we present for the characterization of methyltransferase activity not only gives a robust framework for the study of KMTs but may ultimately extend to the study of other enzymes, substrates, and PTMs.
Author contributions
E.T.U., K.E.W.N., and S.A.S. designed the experiments. S.A.S. supervised the work. E.T.U. and K.E.W.N. performed the experiments. E.T.U. analyzed the data and wrote the manuscript. All authors revised the manuscript and provided commentary.
Acknowledgments
We acknowledge Dr. Bo Wang and Dr. Squire Booker for the gift of the 13C-SAM and assistance in generating our own. We also thank Dr. Erik Cook for some preliminary experiments.
This work was funded by National Institutes of Health (NIH) grant R01DK121509 and U.S. National Science Foundation grant MCB-1515974 to S.A.S., NIH National Research Service Award predoctoral fellowship F31DK124047 to E.T.U. (formerly G.A.U.), and NIH grant R01CA140522 to M.S.C.
Editor: Wendy Shaw.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2021.09.034.
Supporting material
References
- 1.Jenuwein T., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 2.Lawrence M., Daujat S., Schneider R. Lateral thinking: how histone modifications regulate gene expression. Trends Genet. 2016;32:42–56. doi: 10.1016/j.tig.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 3.Bannister A.J., Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Musselman C.A., Lalonde M.E., et al. Kutateladze T.G. Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 2012;19:1218–1227. doi: 10.1038/nsmb.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruthenburg A.J., Allis C.D., Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol. Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 6.Hyun K., Jeon J., et al. Kim J. Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 2017;49:e324. doi: 10.1038/emm.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greer E.L., Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murray K. The occurence of iε-N-methyl lysine in histones. Biochemistry. 1964;3:10–15. doi: 10.1021/bi00889a003. [DOI] [PubMed] [Google Scholar]
- 9.Levy D. Lysine methylation signaling of non-histone proteins in the nucleus. Cell. Mol. Life Sci. 2019;76:2873–2883. doi: 10.1007/s00018-019-03142-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chuikov S., Kurash J.K., et al. Reinberg D. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- 11.Couture J.F., Collazo E., et al. Trievel R.C. Structural basis for the methylation site specificity of SET7/9. Nat. Struct. Mol. Biol. 2006;13:140–146. doi: 10.1038/nsmb1045. [DOI] [PubMed] [Google Scholar]
- 12.Biggar K.K., Li S.S. Non-histone protein methylation as a regulator of cellular signalling and function. Nat. Rev. Mol. Cell Biol. 2015;16:5–17. doi: 10.1038/nrm3915. [DOI] [PubMed] [Google Scholar]
- 13.Bonaldi T., Regula J.T., Imhof A. The use of mass spectrometry for the analysis of histone modifications. Methods Enzymol. 2004;377:111–130. doi: 10.1016/S0076-6879(03)77006-2. [DOI] [PubMed] [Google Scholar]
- 14.Afjehi-Sadat L., Garcia B.A. Comprehending dynamic protein methylation with mass spectrometry. Curr. Opin. Chem. Biol. 2013;17:12–19. doi: 10.1016/j.cbpa.2012.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zee B.M., Levin R.S., et al. Garcia B.A. In vivo residue-specific histone methylation dynamics. J. Biol. Chem. 2010;285:3341–3350. doi: 10.1074/jbc.M109.063784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith C.M., Gafken P.R., et al. Smith D.L. Mass spectrometric quantification of acetylation at specific lysines within the amino-terminal tail of histone H4. Anal. Biochem. 2003;316:23–33. doi: 10.1016/s0003-2697(03)00032-0. [DOI] [PubMed] [Google Scholar]
- 17.Britton L.M., Gonzales-Cope M., et al. Garcia B.A. Breaking the histone code with quantitative mass spectrometry. Expert Rev. Proteomics. 2011;8:631–643. doi: 10.1586/epr.11.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao B., Jing C., et al. Gamblin S.J. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature. 2003;421:652–656. doi: 10.1038/nature01378. [DOI] [PubMed] [Google Scholar]
- 19.Gowher H., Zhang X., et al. Jeltsch A. Avidin plate assay system for enzymatic characterization of a histone lysine methyltransferase. Anal. Biochem. 2005;342:287–291. doi: 10.1016/j.ab.2005.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rathert P., Cheng X., Jeltsch A. Continuous enzymatic assay for histone lysine methyltransferases. Biotechniques. 2007;43:602–604. doi: 10.2144/000112623. 606 passim. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suh-Lailam B.B., Hevel J.M. A fast and efficient method for quantitative measurement of S-adenosyl-L-methionine-dependent methyltransferase activity with protein substrates. Anal. Biochem. 2010;398:218–224. doi: 10.1016/j.ab.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 22.Eram M.S., Bustos S.P., et al. Vedadi M. Trimethylation of histone H3 lysine 36 by human methyltransferase PRDM9 protein. J. Biol. Chem. 2014;289:12177–12188. doi: 10.1074/jbc.M113.523183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilczek C., Chitta R., et al. Shechter D. Protein arginine methyltransferase Prmt5-Mep50 methylates histones H2A and H4 and the histone chaperone nucleoplasmin in Xenopus laevis eggs. J. Biol. Chem. 2011;286:42221–42231. doi: 10.1074/jbc.M111.303677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo M. Current chemical biology approaches to interrogate protein methyltransferases. ACS Chem. Biol. 2012;7:443–463. doi: 10.1021/cb200519y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collazo E., Couture J.F., et al. Trievel R.C. A coupled fluorescent assay for histone methyltransferases. Anal. Biochem. 2005;342:86–92. doi: 10.1016/j.ab.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 26.Duchin S., Vershinin Z., et al. Aharoni A. A continuous kinetic assay for protein and DNA methyltransferase enzymatic activities. Epigenetics Chromatin. 2015;8:56. doi: 10.1186/s13072-015-0048-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burgos E.S., Walters R.O., et al. Shechter D. A simplified characterization of S-adenosyl-l-methionine-consuming enzymes with 1-Step EZ-MTase: a universal and straightforward coupled-assay for in vitro and in vivo setting. Chem. Sci. (Camb.) 2017;8:6601–6612. doi: 10.1039/c7sc02830j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dorgan K.M., Wooderchak W.L., et al. Hevel J.M. An enzyme-coupled continuous spectrophotometric assay for S-adenosylmethionine-dependent methyltransferases. Anal. Biochem. 2006;350:249–255. doi: 10.1016/j.ab.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 29.Hendricks C.L., Ross J.R., et al. Zhou Z.S. An enzyme-coupled colorimetric assay for S-adenosylmethionine-dependent methyltransferases. Anal. Biochem. 2004;326:100–105. doi: 10.1016/j.ab.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 30.Theillet F.X., Smet-Nocca C., et al. Selenko P. Cell signaling, post-translational protein modifications and NMR spectroscopy. J. Biomol. NMR. 2012;54:217–236. doi: 10.1007/s10858-012-9674-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liokatis S., Klingberg R., et al. Schwarzer D. Differentially isotope-labeled nucleosomes to study asymmetric histone modification crosstalk by time-resolved NMR spectroscopy. Angew. Chem. Int.Engl. 2016;55:8262–8265. doi: 10.1002/anie.201601938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamah A., Huvent I., et al. Smet-Nocca C. Nuclear magnetic resonance analysis of the acetylation pattern of the neuronal Tau protein. Biochemistry. 2014;53:3020–3032. doi: 10.1021/bi500006v. [DOI] [PubMed] [Google Scholar]
- 33.Theillet F.X., Liokatis S., et al. Selenko P. Site-specific mapping and time-resolved monitoring of lysine methylation by high-resolution NMR spectroscopy. J. Am. Chem. Soc. 2012;134:7616–7619. doi: 10.1021/ja301895f. [DOI] [PubMed] [Google Scholar]
- 34.Gibbs E.B., Lu F., et al. Showalter S.A. Phosphorylation induces sequence-specific conformational switches in the RNA polymerase II C-terminal domain. Nat. Commun. 2017;8:15233. doi: 10.1038/ncomms15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gibbs E.B., Cook E.C., Showalter S.A. Application of NMR to studies of intrinsically disordered proteins. Arch. Biochem. Biophys. 2017;628:57–70. doi: 10.1016/j.abb.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 36.Nishioka K., Chuikov S., et al. Reinberg D. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 2002;16:479–489. doi: 10.1101/gad.967202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson J.R., Jing C., et al. Xiao B. Crystal structure and functional analysis of the histone methyltransferase SET7/9. Cell. 2002;111:105–115. doi: 10.1016/s0092-8674(02)00964-9. [DOI] [PubMed] [Google Scholar]
- 38.Wang H., Cao R., et al. Zhang Y. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol. Cell. 2001;8:1207–1217. doi: 10.1016/s1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- 39.Kwon T., Chang J.H., et al. Cho Y. Mechanism of histone lysine methyl transfer revealed by the structure of SET7/9-AdoMet. EMBO J. 2003;22:292–303. doi: 10.1093/emboj/cdg025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo H.B., Guo H. Mechanism of histone methylation catalyzed by protein lysine methyltransferase SET7/9 and origin of product specificity. Proc. Natl. Acad. Sci. USA. 2007;104:8797–8802. doi: 10.1073/pnas.0702981104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patel A., Dharmarajan V., et al. Cosgrove M.S. On the mechanism of multiple lysine methylation by the human mixed lineage leukemia protein-1 (MLL1) core complex. J. Biol. Chem. 2009;284:24242–24256. doi: 10.1074/jbc.M109.014498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Southall S.M., Wong P.S., et al. Wilson J.R. Structural basis for the requirement of additional factors for MLL1 SET domain activity and recognition of epigenetic marks. Mol. Cell. 2009;33:181–191. doi: 10.1016/j.molcel.2008.12.029. [DOI] [PubMed] [Google Scholar]
- 43.Hayashi K., Matsui Y. Meisetz, a novel histone tri-methyltransferase, regulates meiosis-specific epigenesis. Cell Cycle. 2006;5:615–620. doi: 10.4161/cc.5.6.2572. [DOI] [PubMed] [Google Scholar]
- 44.Hayashi K., Yoshida K., Matsui Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature. 2005;438:374–378. doi: 10.1038/nature04112. [DOI] [PubMed] [Google Scholar]
- 45.Wu H., Mathioudakis N., et al. Kadlec J. Molecular basis for the regulation of the H3K4 methyltransferase activity of PRDM9. Cell Rep. 2013;5:13–20. doi: 10.1016/j.celrep.2013.08.035. [DOI] [PubMed] [Google Scholar]
- 46.Tan S., Kern R.C., Selleck W. The pST44 polycistronic expression system for producing protein complexes in Escherichia coli. Protein Expr. Purif. 2005;40:385–395. doi: 10.1016/j.pep.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 47.Namitz K.E.W., Tan S., Cosgrove M.S. Hierarchical assembly of the MLL1 core complex within a biomolecular condensate regulates H3K4 methylation. bioRxiv. 2019 doi: 10.1101/870667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iwig D.F., Booker S.J., Booker S.J. Insight into the polar reactivity of the onium chalcogen analogues of S-adenosyl-L-methionine. Biochemistry. 2004;43:13496–13509. doi: 10.1021/bi048693+. [DOI] [PubMed] [Google Scholar]
- 49.Bollinger J.M., Jr., Krebs C. Stalking intermediates in oxygen activation by iron enzymes: motivation and method. J. Inorg. Biochem. 2006;100:586–605. doi: 10.1016/j.jinorgbio.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 50.Kouskouti A., Scheer E., et al. Talianidis I. Gene-specific modulation of TAF10 function by SET9-mediated methylation. Mol. Cell. 2004;14:175–182. doi: 10.1016/s1097-2765(04)00182-0. [DOI] [PubMed] [Google Scholar]
- 51.Maganti A.V., Maier B., et al. Mirmira R.G. Transcriptional activity of the islet β cell factor Pdx1 is augmented by lysine methylation catalyzed by the methyltransferase Set7/9. J. Biol. Chem. 2015;290:9812–9822. doi: 10.1074/jbc.M114.616219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Del Rizzo P.A., Couture J.F., et al. Trievel R.C. SET7/9 catalytic mutants reveal the role of active site water molecules in lysine multiple methylation. J. Biol. Chem. 2010;285:31849–31858. doi: 10.1074/jbc.M110.114587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Milne T.A., Briggs S.D., et al. Hess J.L. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol. Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 54.Terranova R., Agherbi H., et al. Djabali M. Histone and DNA methylation defects at Hox genes in mice expressing a SET domain-truncated form of Mll. Proc. Natl. Acad. Sci. USA. 2006;103:6629–6634. doi: 10.1073/pnas.0507425103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ziemin-van der Poel S., McCabe N.R., et al. Rowley J.D. Identification of a gene, MLL, that spans the breakpoint in 11q23 translocations associated with human leukemias. Proc. Natl. Acad. Sci. USA. 1991;88:10735–10739. doi: 10.1073/pnas.88.23.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mzoughi S., Tan Y.X., et al. Guccione E. The role of PRDMs in cancer: one family, two sides. Curr. Opin. Genet. Dev. 2016;36:83–91. doi: 10.1016/j.gde.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 57.Diagouraga B., Clément J.A.J., et al. Baudat F. PRDM9 methyltransferase activity is essential for meiotic DNA double-strand break formation at its binding sites. Mol. Cell. 2018;69:853–865.e6. doi: 10.1016/j.molcel.2018.01.033. [DOI] [PubMed] [Google Scholar]
- 58.Paigen K., Petkov P.M. PRDM9 and its role in genetic recombination. Trends Genet. 2018;34:291–300. doi: 10.1016/j.tig.2017.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koh-Stenta X., Joy J., et al. Hill J. Characterization of the histone methyltransferase PRDM9 using biochemical, biophysical and chemical biology techniques. Biochem. J. 2014;461:323–334. doi: 10.1042/BJ20140374. [DOI] [PubMed] [Google Scholar]
- 60.Dirk L.M., Flynn E.M., et al. Houtz R.L. Kinetic manifestation of processivity during multiple methylations catalyzed by SET domain protein methyltransferases. Biochemistry. 2007;46:3905–3915. doi: 10.1021/bi6023644. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.