Abstract
OBJECTIVE:
To identify genes implicated in sudden sensorineural hearing loss (SSNHL) and localize their expression in the cochlea to further explore potential pathogenic mechanisms and therapeutic targets.
STUDY DESIGN:
Systematic literature review and bioinformatics analysis
DATA SOURCES:
The following sources were searched from inception through July 2, 2020: PubMed-NCBI, MEDLINE, Embase, CINAHL, Cochrane Library, ClinicalTrials.gov, OpenGrey, GreyNet, GreyLiterature Report, and European Union Clinical Trials Registry. PubMed-NCBI and MEDLINE were additionally searched for human temporal bone histopathologic studies related to SSNHL.
METHODS:
Literature review of candidate SSNHL genes was conducted according to PRISMA guidelines. Existing temporal bone studies from SSNHL patients were analyzed to identify the most commonly affected inner ear structures. Previously published single-cell and single-nucleus RNA-Seq datasets of the adult mouse stria vascularis, as well as postnatal day 7 and 15 mouse cochlear hair cells and supporting cells, were utilized for localization of the SSNHL-related genes curated through literature review.
CONCLUSIONS:
We report 92 unique single nucleotide polymorphisms (SNPs) in 76 different genes that have been investigated in relation to SSNHL in the literature. We demonstrate that a subset of these genes are expressed by cell types in the adult mouse stria vascularis and organ of Corti, consistent with findings from temporal bone studies in human subjects with SSNHL. We highlight several potential genetic targets relevant to current and possible future SSNHL treatments.
Keywords: sudden sensorineural hearing loss, single nucleotide polymorphism, RNA-Seq, cochlea, temporal bone
INTRODUCTION
Sudden sensorineural hearing loss (SSNHL) is most commonly defined as a hearing loss of at least 30 decibels that spans a minimum of three contiguous audiometric frequencies and rapidly develops within a 72-hour period.1 The estimated incidence of SSNHL in the U.S. has been reported to be between 5 to 27 per 100,000 people per year 2,3 with an average of over 66,000 new cases per year.3 While some SSNHL cases can be attributed to an identifiable etiology, as high as 71% of cases remain idiopathic.4 Multiple studies have analyzed structures of the inner ear in patients with SSNHL, and have reported abnormalities in various cell types.5–11 However, the exact pathogenic mechanisms underlying SSNHL are not yet well characterized.
A growing body of research has focused on identifying factors implicated in the disease process, including genetic susceptibility. Over 65 potential genetic targets have been investigated in relationship to SSNHL to date12, however, the expression of these candidate genes in cochlear cell types remains largely undefined. This study aimed to synthesize the available literature on genes and single nucleotide polymorphisms (SNPs) implicated in SSNHL. We reviewed the literature on human temporal bone histopathology studies to identify specific inner ear structures that have been most implicated in SSNHL to guide our gene localization. We then aimed to localize expression of the potential candidate SSNHL genes utilizing recently published single-cell and single-nucleus RNA-sequencing datasets from the adult mouse stria vascularis, as well as other published single-cell datasets from the cochlea, in order to provide greater insight into the mechanisms and potential anatomic locations of SSNHL pathogenesis.
METHODS
Literature Review
Systematic Review of SSNHL-Implicated Genes
A systematic review of genes implicated in SSNHL was conducted in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) reporting guidelines.13 The following databases and grey literature sources were searched from inception through July 2, 2020: PubMed-NCBI, MEDLINE, Embase, CINAHL, Cochrane Library, ClinicalTrials.gov, OpenGrey, GreyNet, GreyLiterature Report, and European Union Clinical Trials Registry. No language restriction was employed. The following search terms were utilized: sudden hearing loss AND polymorphism (text words), sudden hearing loss AND genes (text words), sudden deafness (text words), and hearing loss, sudden/ge [Genetics] (MeSH term). Article titles and abstracts were screened for eligibility before full-text articles were obtained and assessed for inclusion. Additional relevant articles were identified through reference lists of included studies or through manual searches, and were used to contextualize our findings. Data from included studies was extracted and compiled in a standardized electronic data collection sheet. The primary outcome of interest was the gene(s) or polymorphism(s) investigated in relation to SSNHL.
The strength of clinical data was graded independently by two authors according to the following rating scheme for individual studies, modified from the Oxford Centre for Evidence-Based Medicine levels of evidence: level 1, randomized clinical trials or systematic reviews with meta-analysis; level 2, controlled trials without randomization or prospective comparative cohort trials; level 3, case-control studies or retrospective cohort studies; level 4, case series with or without intervention or cross-sectional studies; level 5, opinion of respected authorities or case reports. The Newcastle Ottawa Scale was used to assess risk of bias for case-control studies, where studies receiving a score of greater than equal or to 7 were considered high quality and a low risk of bias.14 The Joanna Briggs Institute Critical Appraisal Tools for analytical cross-sectional studies and case reports were utilized to assess risk of bias for each respective study type.15,16
Curation of SSNHL Gene List
The results of our literature search were compiled to create a list of SSNHL-implicated genes. We detailed which genes were supported by the literature to have a significant association with SSNHL versus those that were not found to have a significant association (Supplemental Digital Content, Tables 1–2). Genes were deemed to have a “significant association” if studies reported the gene to have statistically significant differential expression in SSNHL patients compared to controls. Genes deemed to have “no significant association” were reported in studies but were not found to have statistically significant differential expression between SSNHL patients and controls. The final curated list of SSNHL-implicated genes included all genes regardless of association and was utilized for cross-referencing with RNA-sequencing datasets as further described below.
Temporal Bone Histopathology Literature Review and Analysis
A supplemental literature review of temporal bone histopathology studies relating to SSNHL cases was also performed through a similar search method as described above. The MEDLINE and Pubmed-NCBI databases were searched from inception through January 2, 2021. The following search terms were utilized: sudden hearing loss AND histopathology (text words), sudden hearing loss AND temporal bone (text words). The processes for identifying eligible studies and data collection were conducted as described above.
Temporal bone histopathology analyses were performed by combining results from published studies evaluating temporal bone samples of individuals with SSNHL.5–10 The data consisted of 71 ears (68 patients) with SSNHL and were standardized into a table by coding either “1” or “0” for the presence of abnormality (1=abnormal) in the following inner ear structures: hair cells, supporting cells, tectorial membrane, stria vascularis, spiral ganglion cells, and spiral limbus (Supplemental Digital Content, Table 3). Organ of Corti abnormalities were defined as loss of hair cells and/or supporting cells compared to control subjects. Tectorial membrane and stria vascularis abnormalities were defined as the presence of degeneration or atrophy of these structures compared to controls. Spiral ganglion cells were analyzed by the method described by Otte et al.17 The raw spiral ganglion cell counts from the Khetarpal et al.8 data were determined to be abnormal if the measured number of spiral ganglion cells was less than the mean minus one standard deviation (n=9; mean = 23,815 cells; standard deviation = 3797) of measured spiral ganglion cells in healthy, control ears from Ungar et al.11 The finalized data was compiled into a single spreadsheet and the number of abnormal cell type findings was divided by the total number of analyzed subjects for each ear structure to give a percentage of subjects with abnormal findings. Subjects from studies that did not report findings for a particular cell type were excluded from the final counts and analysis for that cell type.
Bioinformatics
Data and Software Availability
Our curated list of SSNHL-implicated genes was cross-referenced against the following previously published datasets. Single cell and single nucleus RNA-Seq datasets of postnatal day 30 (P30) mouse stria vascularis18 were utilized (GEO Accession ID: GSE136196) (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE136196) and are available through the gene Expression Analysis Resource (gEAR) Portal, a website for visualization and comparative analysis of multi-omic data, with an emphasis on hearing research (https://umgear.org//index.html?layout_id=b50cae7a). Single cell RNA-Seq dataset from the P7 developing mouse cochlea including IHCs, OHCs, and supporting cells including both inner phalangeal, pillar, and Deiters cells 19 was utilized (GEO Accession ID: GSE137299) and are available through gEAR (https://umgear.org//index.html?layout_id=f7baf4ea). Finally, previously published single cell RNA-Seq datasets of postnatal day 15 (P15) inner hair cells (IHCs), outer hair cells (OHCs), and Deiters cells20 were utilized (GEO Accession ID: GSE114157) (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE114157) and are available through the Molecular Otolaryngology and Renal Research Laboratories (MORL) (https://morlscrnaseq.org/). Previously published datasets from the P25–27 adult mouse spiral ganglion neurons21 and from the adult human cochlea22 were also utilized for additional gene localization analysis, included in the supplemental materials (Supplemental Digital Content, Figures 2–3).
Data Visualization
P30 SV scRNA-Seq & snRNA-Seq.
Previously published P30 SV scRNA-Seq and snRNA-Seq data were preprocessed by Scanpy (v1.5.1) with criteria as previously described.18,23
P7 IHCs, OHCs, and supporting cells.
Previously published normalized dataset from Kolla and colleagues 19 was processed by Scanpy (v1.5.1). “Rik” and “Gm-” genes were filtered in all downstream analysis. Cell clustering and annotation was performed using modularity-based clustering with Leiden algorithm (resolution=0.8) implemented in Scanpy. Heatmaps were plotted by Seaborn (v0.10.1).
P15 IHCs, OHCs, and Deiters cells.
Data expression matrix from previously published P15 IHC, OHC, and Deiters cell single cell RNA-Seq dataset was normalized by Scanpy (v1.5.1) using the function pp.normalize_total with parameter “exclude_highly_expressed” set as True. Normalized dataset was then logarithmized by the function pp.log1p. Heatmaps were plotted by Seaborn (v0.10.1).
Information on the data visualization from the P25–27 spiral ganglion neurons and the human cochlea datasets are included as supplements (Supplemental Digital Content, Figures 2–3).
RESULTS
Systematic Literature Review of SSNHL-Implicated Genes
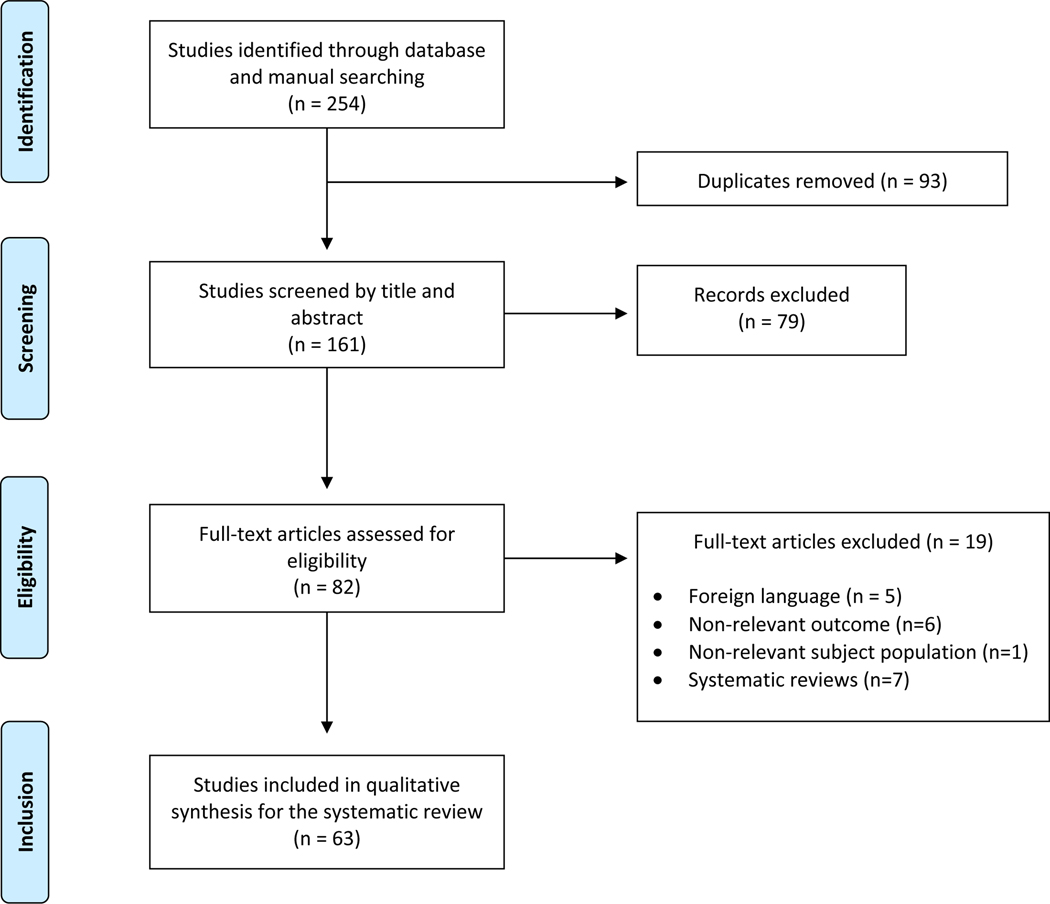
A PRISMA-based systematic literature review was conducted to curate a list of SSNHL-implicated genes (see Fig. 1 for complete search strategy). Database searching initially identified 254 articles of potential relevance. After removal of duplicate texts, 161 articles remained and were screened for eligibility. Exclusion criteria removed studies that did not relate to SSNHL or did not include genetic analysis (n=7), full-text articles in a foreign language (n=5), and other systematic reviews (n=7). In total, 63 studies fully satisfied the inclusion criteria and were included in the final review. The majority of studies (87%) were rated to be of clinical strength level 3, with the remaining few rated to be level 2 or level 4, in accordance with the modified Oxford Centre for Evidence-Based Medicine grading scale described above. When assessing risk of bias, all included case-control studies received a score of 7 or greater on the Newcastle Ottawa Scale, indicating high quality and low risk of bias (Supplemental Digital Content, Table 4A). The single analytical cross-sectional study and the four case reports included in the review were evaluated according to the Joanna Briggs Institute Critical Appraisal methodology and met all criteria within each assessment tool. (Supplemental Digital Content, Table 4B).
Figure 1. PRISMA Flow Diagram for Systematic Literature Review of SSNHL-Implicated Genes.
Flow diagram illustrates the literature review and study selection process conducted according to PRISMA reporting guidelines for published studies investigating genes related to SSNHL. A total of 63 studies satisfied inclusion criteria and were included in the final review.
The 63 included studies involved 6,165 subjects with SSNHL (52.1% male, 47.9% female, mean age of 50.1f ± 12.16). Additional demographics and characteristics of included subjects are detailed in Table 1. Not all studies consistently reported on features related to past medical history and associated symptoms of SSNHL; the table values only reflect data from studies that did report on those measures. Of the 63 studies, 25.4% recorded a history of hypertension and diabetes mellitus, 20.6% reported on dyslipidemia, and 15.9% reported on smoking status. With regard to SSNHL-associated symptoms, tinnitus, vertigo, and hearing recovery were tracked in 31.7%, 36.5%, and 50.8% of studies, respectively. Many studies employed varied methodologies to categorize levels of hearing recovery. The full list of included studies along with study design, population, subject number, and investigated genes is detailed in Supplemental Digital Content, Table 5.
Table 1.
Subject Demographics of Included SSNHL and Temporal Bone Studies
| N (Average %) | # Studies Reporting Characteristic | |
|---|---|---|
|
| ||
| SSNHL Study Demographics | ||
| Total # subjects | 6,165 | 63 |
| Male | 3,213 (52.1%) | 63 |
| Female | 2,952 (47.9%) | 63 |
| Mean Age (years) | 50.1 ± 12.16 | 63 |
| Unilateral SSNHL | 6,145 (99.7%) | 63 |
| Bilateral SSNHL | 20 (0.3%) | 63 |
| Tinnitus | 1252 (69.1%) | 20 |
| Vertigo | 572 (34.4%) | 23 |
| Hearing recovery | 1254 (54.9%) | 32 |
| DM | 236 (15.2%) | 16 |
| Hypertension | 365 (22.9%) | 16 |
| Dyslipidemia | 276 (22.3%) | 13 |
| Smoking | 325 (28.5%) | 10 |
| Temporal Bone Study Demographics | ||
| Total # subjects | 68 | 6 |
| Male | 42 (61.8%) | 6 |
| Female | 26 (38.2%) | 6 |
| Mean age of SSNHL onset (years) | 48.3 ± 22.1 | 6 |
| Mean age of death (years) | 60.1 ± 17.3 | 6 |
| SSNHL laterality, right ear | 38 (55.9%) | 6 |
| SSNHL laterality, left ear | 27 (39.7%) | 6 |
| SSNHL laterality, bilateral | 3 (4.4%) | 6 |
Literature Review of Temporal Bone Studies in SSNHL
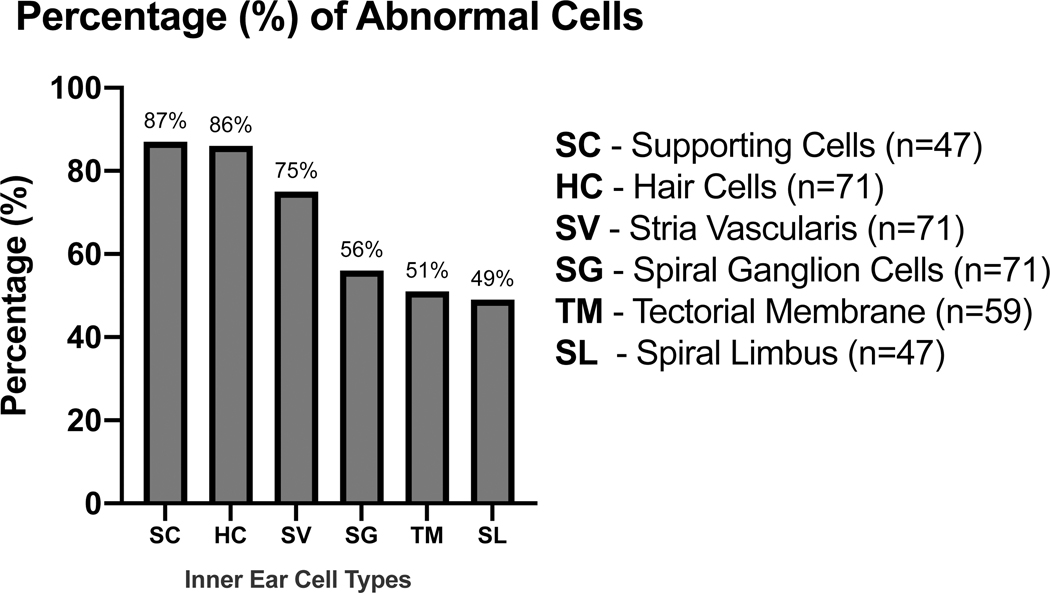
Six temporal bone studies were identified for inclusion that reported histopathological findings in patients with SSNHL. These studies report 71 ears (68 subjects) identified, with 38% female (n = 26), average age of onset 48.3 years (standard deviation = 22.1 years), and average age of death 60.1 years (n = 56 subjects, standard deviation = 17.3 years) (Table 1). The most common structural abnormalities were seen in the supporting cells (87%), hair cells (86%), and stria vascularis (75%), followed by spiral ganglion cells (56%), tectorial membrane (51%), and spiral limbus (49%) (Fig. 2).
Figure 2. Distribution of Inner Ear Cell Type Abnormalities Identified in Human Temporal Bone Studies.
Published temporal bone studies reporting histopathological findings from patients with SSNHL were analyzed to determine the distribution of inner ear cell types found to have structural abnormalities. Graph displays inner ear cell types along the horizontal axis and percentage of abnormal cells on the vertical axis. Cell types include supporting cells (SC), hair cells (HC), stria vascularis (SV), spiral ganglion (SG), tectorial membrane (TM), and spiral limbus (S). Percentage of abnormal cells was calculated by dividing the number of abnormal samples by the total number of analyzed subjects for each cell type. Subjects from studies that did not report findings for particular cell types were excluded from the analysis. Cell type abnormality was defined by criteria that varied by study and included metrics such as significantly decreased cell count and/or presence of structural degeneration or atrophy in SSNHL patient samples compared to controls. Cell type-specific definitions for abnormality are provided in the methods.
Candidate Genes Implicated in SSNHL
We present a list of 92 unique SNPs in 76 different genes that have been investigated in relation to SSNHL. 47 genes were found by at least one study to have a significant association with SSNHL, while the remaining 29 were never found to have a significant association. For 9 of the 47 genes of significance, there was conflicting evidence in the literature where at least one additional study did not find a significant relation to SSNHL for the same gene. Notably, 15 of the 76 total genes had multiple SNPs investigated across different studies, and the relationship to SSNHL was found to differ between some of the distinct SNPs. We provide documentation of each investigated gene and SNP along with their respective supporting and non-supporting studies (Supplemental Digital Content, Tables 1–2).
Gene Target Association with SSNHL Susceptibility
The majority of genetic targets found to have a significant association with SSNHL were suggested to play a role in disease susceptibility. Specifically, 23 genes had at least one SNP that was correlated with an increased risk of SSNHL. Increased risk was determined by a significantly higher SNP frequency observed in affected SSNHL subjects compared to controls. In contrast, the following seven genes had at least one SNP correlated with a decreased risk: APOE, DNMT1, FCRL3, GRHL2, GPX3, HSPA1A, SERPINE124–30 (Supplemental Digital Content, Table 1). Decreased risk was inferred by the observation of a significantly lower SNP frequency in SSNHL subjects compared to controls, or by a significantly higher SNP frequency in controls compared to SSNHL subjects. Interestingly, there were two genes, DNMT1 and FCRL3, for which specific genotypes in different SNPs of each gene were found to have opposite associations with SSNHL risk.24,26
Localization of Candidate SSNHL Genes to Regions and Cell Types in the Cochlea with Single Cell Resolution.
The cochlea is composed of a diverse collection of intricately arranged cell types that function together to enable hearing. The advent of publicly available single cell RNA-sequencing datasets from the mammalian cochlea provides the opportunity to characterize candidate gene expression in silico at the single cell level. Our list of SSNHL-implicated genes was cross-referenced against previously published single cell and single nucleus RNA-seq datasets from the postnatal day 30 (P30) mouse stria vascularis (SV).18 Heatmaps demonstrating SSNHL-implicated gene expression amongst SV cell types are shown in both the single cell (Fig. 3A) and single nucleus (Fig. 3B). For each heatmap, gene expression is displayed along the vertical axis while SV cell types are displayed along the horizontal axis. Expression is in normalized transcript counts with an increasingly dark purple color indicating a higher number of transcripts in a given cell. Genes that demonstrate enriched expression in the SV include Sod1, Gpx3,Mif, Gjb2, Nr3c1, Clock, Gpx1, Sod2, and Nr3c2.
Figure 3A. Expression of SSNHL-investigated genes in the adult mouse stria vascularis utilizing a single cell RNA-Seq adult mouse stria vascularis dataset (Korrapati, Taukulis et al., 2019).
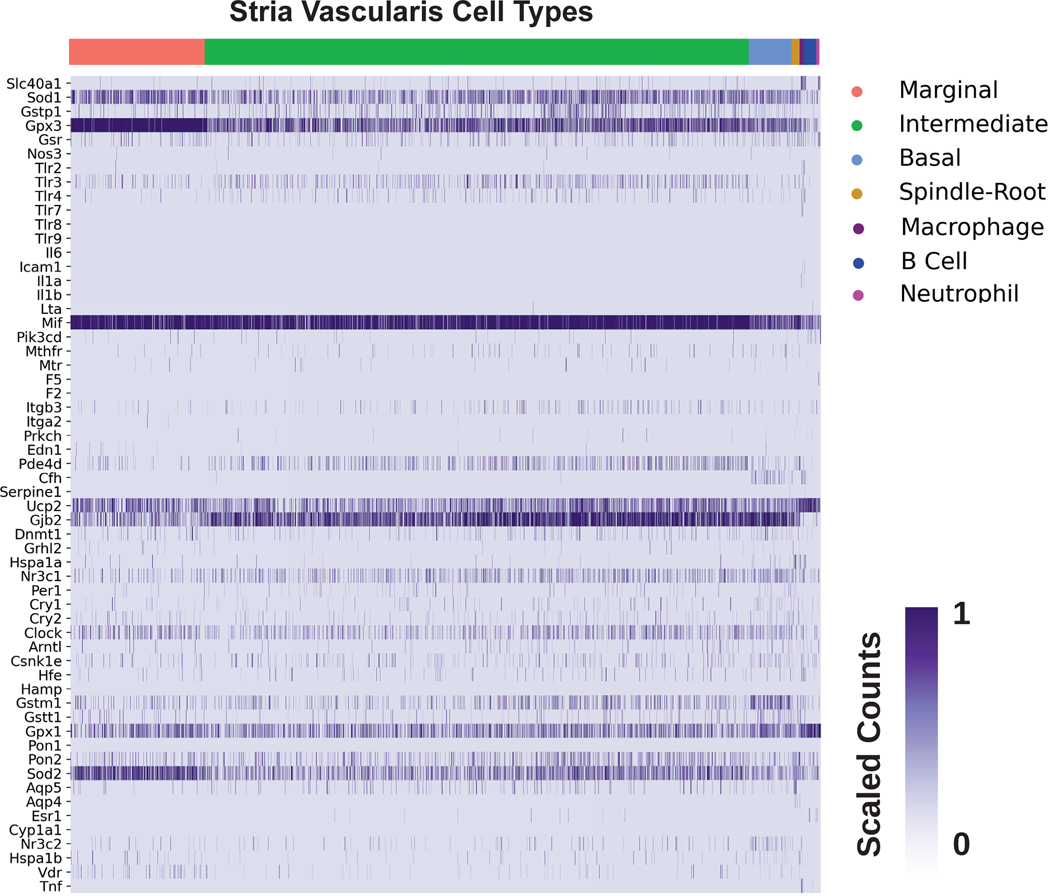
Heatmap displays cells along the horizontal axis with cell types grouped by color and SSNHL-investigated genes along the vertical axis. The darker the bar the more highly expressed the gene is in a given cell. Cell types include SV cell types (marginal cells in pink, intermediate cells in green, basal cells in light blue), cells at the boundary of the SV (spindle-root cells in gold, fibrocytes in light green), as well as immune cells (macrophages in purple, B cells in blue, and neutrophils in magenta). Spindle and root cell profiles were not distinguishable from each other in this dataset.
Figure 3B. Expression of SSNHL-investigated genes in the adult mouse stria vascularis utilizing a single nucleus RNA-Seq adult mouse stria vascularis dataset (Korrapati, Taukulis et al., 2019).
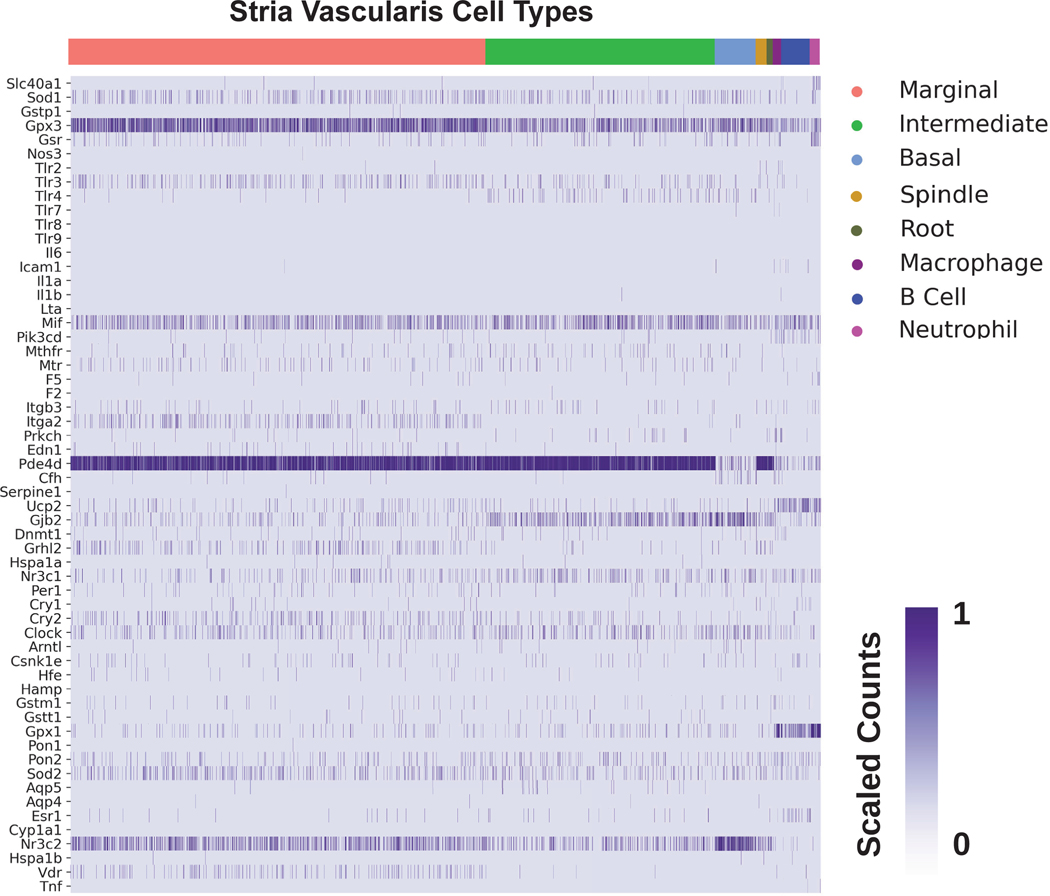
Heatmap displays cells along the horizontal axis with cell types grouped by color and SSNHL-investigated genes along the vertical axis. The darker the bar the more highly expressed the gene is in a given cell. Cell types include SV cell types (marginal cells in pink, intermediate cells in green, basal cells in light blue), cells at the boundary of the SV (spindle cells in gold, root cells in dark green, fibrocytes in light green), as well as immune cells (macrophages in purple, B cells in blue, and neutrophils in magenta).
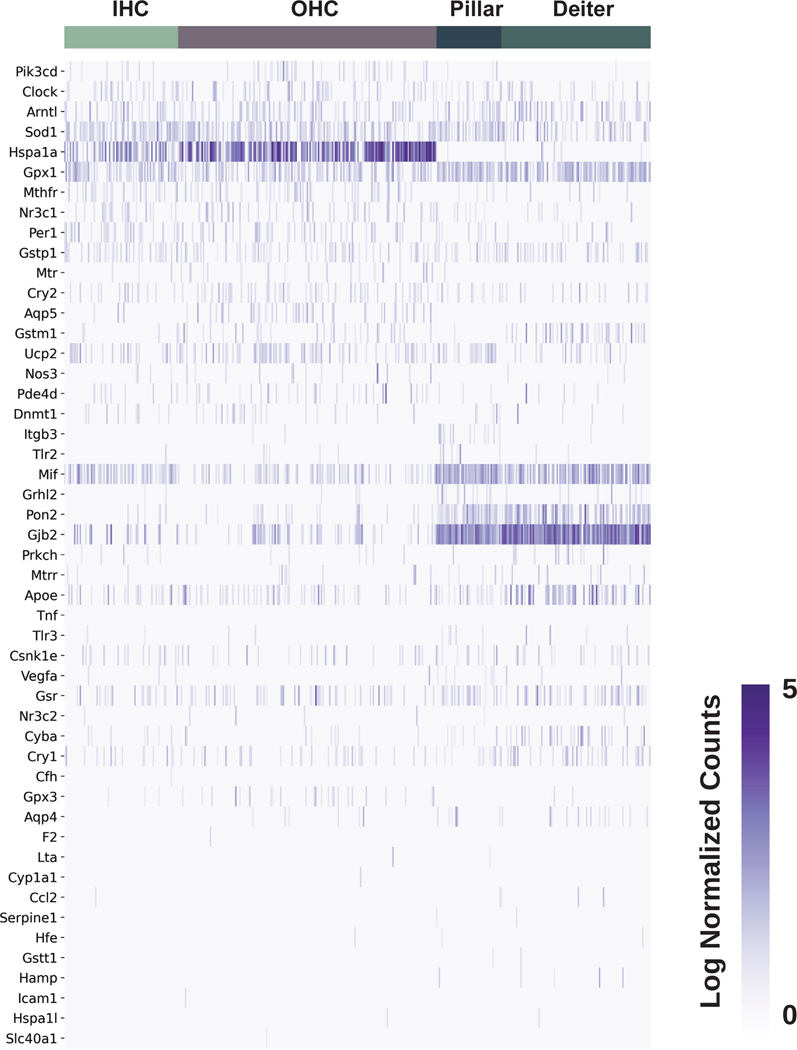
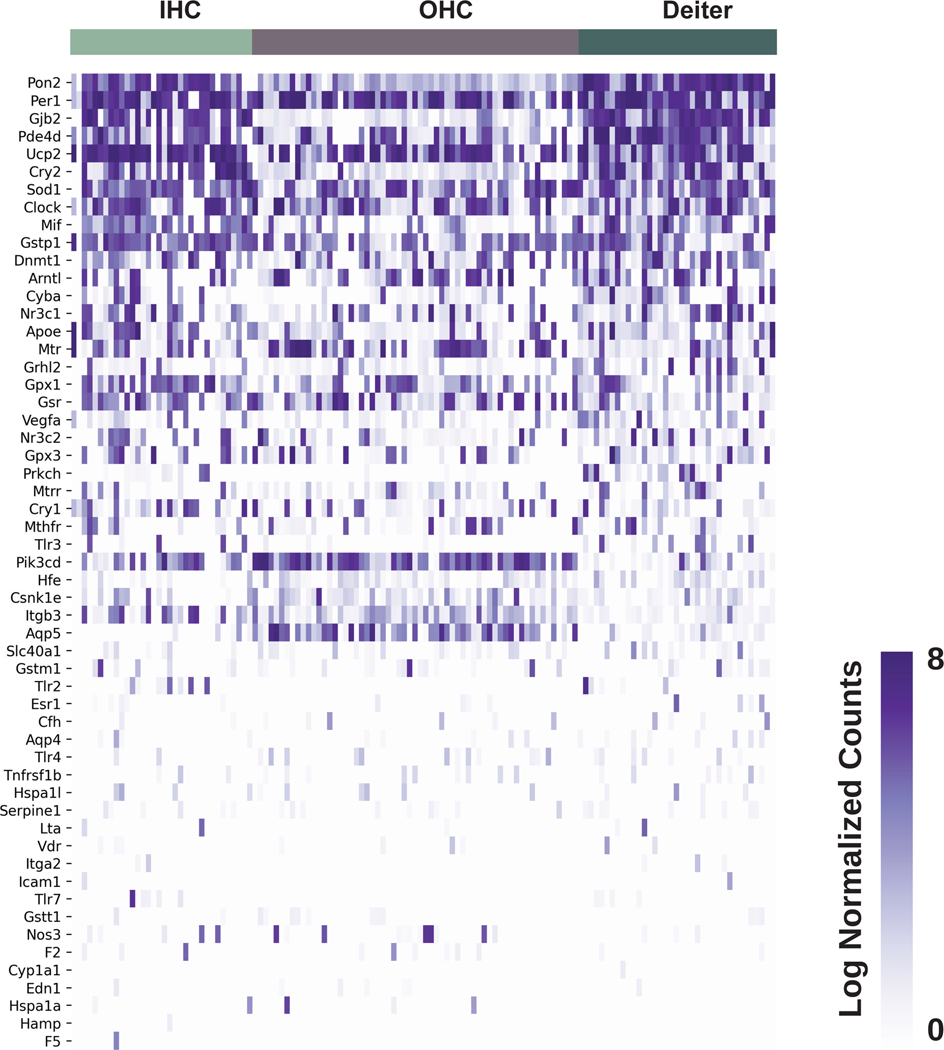
By cross-referencing SSNHL-implicated genes against a recently published P7 mouse organ of Corti scRNA-Seq dataset,19 heatmaps depicting inner and outer hair cells, pillar cells, and Deiters cells demonstrate enrichment for Sod1, Hspa1a, Gpx1, and Mif. Transcripts for Apoe, Gjb2, and Pon2 are enriched in pillar and Deiters cells (Fig. 4). Finally, in examining a previously published P15 scRNA-Seq dataset of inner and outer hair cells as well as Deiter cells,20 heatmaps demonstrate expression of Sod1, Gpx1,Mif, Gjb2, Nr3c1, Nr3c2, and Gpx3 transcripts (Fig. 5).
Figure 4. Expression of SSNHL-investigated genes in P7 mouse organ of Corti utilizing a single cell RNA-Seq P7 mouse organ of Corti dataset (Kolla et al., 2020).
Heatmap displays cells along the horizontal axis with cell types grouped by color and SSNHL-investigated genes along the vertical axis. The darker the bar the more highly expressed the gene is in a given cell. Cell types in the organ of Corti analyzed for this heatmap include inner hair cells (IHC) in pastel green, outer hair cells (OHC) in gray, pillar cells in midnight blue, and Deiters cells in forest green.
Figure 5. Expression of SSNHL-investigated genes in P15 mouse organ of Corti utilizing a single cell RNA-Seq P15 mouse organ of Corti dataset (Ranum et al., 2019).
Heatmap displays cells along the horizontal axis with cell types grouped by color and SSNHL-investigated genes along the vertical axis. The darker the bar the more highly expressed the gene is in a given cell. Cell types in the organ of Corti available for analysis include inner hair cells (IHC) in blue, outer hair cells (OHC) in gold, and Deiters cells in green.
Violin plots further demonstrate the differential expression of glutathione peroxidase isoforms amongst cochlear cell types (Supplemental Digital Content, Fig. 1). For each violin plot, cell types within a tissue (organ of Corti or SV) are displayed along the horizontal axis and expression level in normalized counts is displayed along the vertical axis. The wider the violin, the more cells at a given level of expression. Of note, in the SV datasets, Gpx3 appears to be preferentially expressed amongst SV marginal, intermediate, and basal cells over Gpx1 (Supplemental Digital Content, Fig. 1A–B) compared to the relative enrichment of Gpx1 transcripts over Gpx3 transcripts amongst cells in the Organ of Corti (Supplemental Digital Content, Fig. 1C–D).
Gene localization results utilizing the P25–27 spiral ganglion neurons and human cochlea datasets are available in the supplemental materials (Supplemental Digital Content, Figures 2–3).
DISCUSSION
This study provides a novel, comprehensive report of 92 SNPs in 76 distinct genes that have been investigated as candidate SSNHL genetic targets. Our review of temporal bone histopathology studies in SSNHL cases demonstrates that the organ of Corti (supporting cells and hair cells) and the stria vascularis (SV) are the most commonly affected structures. Lastly, our analysis localizes expression of SSNHL-implicated genes to the SV and organ of Corti based upon single-cell and single-nucleus RNA sequencing datasets. This data demonstrates that genes implicated in SSNHL, some of which were previously identified through peripheral blood assays, are expressed by relevant cell types and tissues in the cochlea. This localization establishes a rationale for examining the role of these genes in mediating SSNHL pathogenesis.
Human Temporal Bone Studies Identify Inner Ear Structures Implicated in SSNHL Pathogenesis
Our review of published human temporal bone studies summarizes available data regarding the location of cochlear pathology in SSNHL. These studies identify multiple abnormalities in inner ear structures including the supporting cells, hair cells, stria vascularis (SV), spiral ganglion cells, tectorial membrane, and spiral limbus. Importantly, in addition to hair cells and supporting cells, these studies reveal that the SV is the next most commonly involved structure after the organ of Corti. While much attention has been focused on hair cell loss, these analyses suggest additional attention to the role of the SV in SSNHL may be warranted. With this in mind, leveraging knowledge from published single cell RNA-sequencing datasets offers the opportunity to begin to understand the role that the SV may play in SSNHL by identifying involved cell types and possibly by identifying critical regionally specific cell signaling pathways.
Localization of SSNHL-Implicated Genes to Inner Ear Cell Types
Cross-referencing of SSNHL-implicated genes against single cell RNA-sequencing datasets suggests that many genes related to hearing loss susceptibility are expressed by critical cell types in the inner ear. Several genes that demonstrate prominent expression in the cochlea include superoxide dismutase 1 (Sod1) and 2 (Sod2), as well as macrophage migration inhibitory factor (Mif) (Fig. 3–6). Superoxide dismutase 1 (Sod1) and 2 (Sod2) encode a copper- and zinc-binding catalase and a manganese-binding catalase, respectively, that are responsible for the destruction of free superoxide radicals and have been implicated in SSNHL.31,32 Sod1 has previously been shown to be expressed in the SV and to a lesser extent the organ of Corti in mice and rats, with evidence of increased expression in the setting of oxidative stress.33,34 Sod1 knockout mice have been shown to be more susceptible to noise-induced permanent threshold shifts compared to control wildtype mice.35 Overexpression of both Sod1and Sod2 in mouse models has been implicated in mitigation of aminoglycoside ototoxicity.36–38 Our finding that Sod1 and Sod2 are prominently expressed in the cochlea further complements the existing literature that implicates Sod1 and Sod2 in hearing loss pathogenesis.
Macrophage migration inhibitory factor (Mif) was another gene found to be prominently expressed in the cochlea — again consistent with previous studies that have indicated a role for Mif in hearing loss.39 Mif has been shown to be expressed in the SV, spiral ligament, spiral limbus, organ of Corti, and Reissner’s membrane,40 and its expression appears to be increased in the setting of experimentally-induced inflammation with LPS treatment in mice.41 Mif knockout mice appear to be more susceptible to age-related hearing loss than wildtype mice.40 Some evidence furthermore suggests that SSNHL patients demonstrate higher levels of circulating MIF when compared to normal-hearing patients, and that higher circulating MIF levels and possibly MIF-related SNPs may impact steroid-responsiveness of SSNHL.39,42 Together, these observations suggest that genes implicated in hearing los susceptibility are involved at the tissue and cellular level in the cochlea in SSNHL.
Implication of Gene Expression Localization for Understanding SSNHL Therapeutic Targets
Our analysis localizes several genes relevant to SSNHL treatment — such as Gpx1, Gpx3, Nr3c1, and Nr3c2 — to cochlear cell types, contributing to our understanding of both current and potential future therapeutic targets. Glutathione peroxidases, Gpx1 and Gpx3, have been shown to play a role in otoprotection from oxidative damage with loss of glutathione peroxidases (GPx) associated with increased hearing loss susceptibility due to noise, aminoglycosides, and cisplatin 43–45 as well as SSNHL.28,32 Furthermore, increased expression of GPx has been associated with protection of hair cells in the presence of cisplatin,37,46 and administration of a GPx mimetic, ebselen, appears to result in some degree of otoprotection after both cisplatin- and noise-induced hearing loss.45,47–49 Interestingly, ebselen (SPI-1005) is currently being investigated as a treatment for Meniere’s disease (https://www.clinicaltrials.gov/ct2/show/NCT03325790), a disease that can often present with sudden changes in hearing. Our analysis suggests that glutathione peroxidase isoforms (Gpx1 and Gpx3) are differentially expressed amongst cochlear cell types with preferential expression of Gpx3 in the SV, with notably prominent expression in SV marginal cells (Supplemental Digital Content, Fig. 1). Whether drugs targeting GPx and potentially its different isoforms in the cochlea might be effective in the treatment of SSNHL remains to be evaluated, and presents an avenue for future research.
Nr3c1and Nr3c2 encode glucocorticoid and mineralocorticoid receptors, respectively. Previous studies have localized glucocorticoid receptors to inner ear cell types in both mice and humans.50,51 Consistent with these observations, we demonstrate that Nr3c1 and Nr3c2 expression is localized to both the organ of Corti and the SV. Since steroids are largely the only approved pharmacologic treatment option for patients with SSNHL1, understanding the localization of these receptors to cochlear cell types is relevant to understanding the effect of steroids in the inner ear. Steroids are thought to interact with both glucocorticoid and mineralocorticoid receptors.52–57 Trune and colleagues have suggested that, in the setting of autoimmune inner ear disease, the effects of steroids mediated by mineralocorticoid action may partially account for the steroid-responsiveness of hearing loss in these patients.52,58 With this assertion in mind, it is interesting that certain SNPs of NR3C1, which may alter glucocorticoid receptor expression, have been associated with poor glucocorticoid response in SSNHL.59 Given these observations, it is critical to further understand cell types and tissues that appear to be affected in humans with SSNHL in order to better appreciate the mechanisms underlying clinical responses to therapeutics.
Limitations
There are a few limitations that are important to acknowledge when discussing our findings. First, associations between SSNHL and many of the candidate genes were based only on differential gene expression or detection of gene-related SNPs in peripheral blood samples rather than local inner ear tissue samples. Furthermore, many of the candidate genes were selected for investigation based upon proposed pathways involved in SSNHL pathogenesis such as inflammation, oxidative stress, and thrombosis, regardless of existing supporting evidence. All of the studies included in the systematic literature review were observational, and thus a causal relationship between the identified candidate gene targets and SSNHL cannot be fully evaluated. We additionally recognize that differential gene expression via RNA-sequencing does not directly establish functional significance of the localized genes. Nonetheless, by synthesizing the available literature and localizing expression of the investigated genes to relevant regions and cell types in the mammalian cochlea, we make necessary initial steps towards characterizing the genetic and mechanistic underpinnings of SSNHL.
CONCLUSIONS
In summary, we present a novel report of the 96 potential genetic targets for SSNHL that have been investigated in the literature to date; 47 genes for which the association reached significance, and 23 genes of which were correlated with an increased risk of SSNHL. We localize these genes implicated in SSNHL to cochlear structures consistent with human temporal bone studies and to specific cochlear cell types, including those in the organ of Corti and in the SV. Importantly, we identify and localize gene expression relevant to current treatments for SSNHL, contributing to our understanding of the fundamental role of these genes in the etiology of SSNHL and presenting avenues for further study.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Intramural Research Program of the NIH, NIDCD to M.H. (DC000088). The authors would like to acknowledge Wade Chien, MD and Nyall London, MD, PhD, who provided helpful feedback and review of this paper.
Footnotes
FINANCIAL DISCLOSURES: The authors have no financial disclosures or sponsors.
DECLARATIONS OF INTEREST: None.
REFERENCES
- 1.Chandrasekhar SS, Tsai Do BS, Schwartz SR, et al. Clinical practice guideline: Sudden hearing loss (update) executive summary. Otolaryngol Head Neck Surg. 2019;161(2):195–210. doi: 10.1177/0194599819859883 [doi]. [DOI] [PubMed] [Google Scholar]
- 2.Byl FM Jr., Sudden hearing loss: Eight years’ experience and suggested prognostic table. Laryngoscope. 1984;94(5 Pt 1):647–661. [PubMed] [Google Scholar]
- 3.Alexander TH, Harris JP. Incidence of sudden sensorineural hearing loss. Otol Neurotol. 2013;34(9):1586–1589. doi: 10.1097/MAO.0000000000000222 [doi]. [DOI] [PubMed] [Google Scholar]
- 4.Chau JK, Lin JR, Atashband S, Irvine RA, Westerberg BD. Systematic review of the evidence for the etiology of adult sudden sensorineural hearing loss. Laryngoscope. 2010;120(5):1011–1021. doi: 10.1002/lary.20873 [doi]. [DOI] [PubMed] [Google Scholar]
- 5.Merchant SN, Adams JC, Nadol JB Jr., Pathology and pathophysiology of idiopathic sudden sensorineural hearing loss. Otol Neurotol. 2005;26(2):151–160. doi: 00129492–200503000-00004 [pii]. [DOI] [PubMed] [Google Scholar]
- 6.Schuknecht HF, Donovan ED. The pathology of idiopathic sudden sensorineural hearing loss. Arch Otorhinolaryngol. 1986;243(1):1–15. doi: 10.1007/BF00457899 [doi]. [DOI] [PubMed] [Google Scholar]
- 7.Yoon TH, Paparella MM, Schachern PA, Alleva M. Histopathology of sudden hearing loss. Laryngoscope. 1990;100(7):707–715. doi: 10.1288/00005537-199007000-00006 [doi]. [DOI] [PubMed] [Google Scholar]
- 8.Khetarpal U, Nadol JB, Glynn RJ. Idiopathic sudden sensorineural hearing loss and postnatal viral labyrinthitis: A statistical comparison of temporal bone findings. Ann Otol Rhinol Laryngol. 1990;99(12):969–976. doi: 10.1177/000348949009901207 [doi]. [DOI] [PubMed] [Google Scholar]
- 9.Vasama JP, Linthicum FH. Idiopathic sudden sensorineural hearing loss: Temporal bone histopathologic study. Ann Otol Rhinol Laryngol. 2000;109(6):527–532. doi: 10.1177/000348940010900601 [doi]. [DOI] [PubMed] [Google Scholar]
- 10.Bahmad F Jr., Temporal bone histopathology: Idiopathic sudden hearing loss. Braz J Otorhinolaryngol. 2008;74(1):159. doi: S1808–8694(15)30770–9 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ungar OJ, Handzel O, Santos F. Rate of spiral ganglion cell loss in idiopathic sudden sensorineural hearing loss. Otol Neurotol. 2018;39(10):e944–e949. doi: 10.1097/MAO.0000000000001992 [doi]. [DOI] [PubMed] [Google Scholar]
- 12.Cao Z, Gao J, Huang S, et al. Genetic polymorphisms and susceptibility to sudden sensorineural hearing loss: A systematic review. Audiol Neurootol. 2019;24(1):8–19. doi: 10.1159/000497032 [doi]. [DOI] [PubMed] [Google Scholar]
- 13.Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. Int J Surg. 2010;8(5):336–341. doi: 10.1016/j.ijsu.2010.02.007 [doi]. [DOI] [PubMed] [Google Scholar]
- 14.Wells GA, Shea B, O’Connell D, Peterson J, Welch V, Losos M, Tugwell P. The Newcastle-Ottawa Scale (NOS) for assessing the quality if nonrandomized studies in meta-analyses, 2012. Available from: http://wwwohrica/programs/clinical_epidemiology/oxfordasp. [Google Scholar]
- 15.Moola S, Munn Z, Tufanaru C, Aromataris E, Sfetcu R, Currie M, Qureshi R, Mattis P, Lisy K, Mu P-FE, Munn Z (Editors). Joanna Briggs Institute Reviewer’s Manual. The Joanna Briggs Institute, 2017. Available from https://reviewersmanual.joannabriggs.org/ [DOI] [PubMed] [Google Scholar]
- 16.Moola S, Munn Z, Tufanaru C, Aromataris E, Sears K, Sfetcu R, Currie M, Qureshi R, Mattis P, Lisy K, Mu P-F. Chapter 7: Systematic reviews of etiology and risk. In: Aromataris E, Munn Z (Editors). Joanna Briggs Institute Reviewer’s Manual. The Joanna Briggs Institute, 2017. Available from https://reviewersmanual.joannabriggs.org/ [Google Scholar]
- 17.Otte J, Schunknecht HF, Kerr AG. Ganglion cell populations in normal and pathological human cochleae. implications for cochlear implantation. Laryngoscope. 1978;88(8 Pt 1):1231–1246. doi: 10.1288/00005537-197808000-00004 [doi]. [DOI] [PubMed] [Google Scholar]
- 18.Korrapati S, Taukulis I, Olszewski R, et al. Single cell and single nucleus RNA-seq reveal cellular heterogeneity and homeostatic regulatory networks in adult mouse stria vascularis. Front Mol Neurosci. 2019;12:316. doi: 10.3389/fnmol.2019.00316 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolla L, Kelly MC, Mann ZF, et al. Characterization of the development of the mouse cochlear epithelium at the single cell level. Nat Commun. 2020;11(1):2389-y. doi: 10.1038/s41467-020-16113-y [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ranum PT, Goodwin AT, Yoshimura H, et al. Insights into the biology of hearing and deafness revealed by single-cell RNA sequencing. Cell Rep. 2019;26(11):3160–3171.e3. doi: S2211–1247(19)30232–3 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shrestha BR, Chia C, Wu L, Kujawa SG, Liberman MC, Goodrich LV. Sensory Neuron Diversity in the Inner Ear Is Shaped by Activity. Cell. 2018;174(5):1229–1246.e17. doi: 10.1016/j.cell.2018.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schrauwen I, Hasin-Brumshtein Y, Corneveaux JJ, et al. A comprehensive catalogue of the coding and non-coding transcripts of the human inner ear. Hear Res. 2016;333:266–274. doi: 10.1016/j.heares.2015.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu S, Olszewski R, Nelson L, Gallego-Martinez A, Lopez-Escamez JA, Hoa M. Identification of potential meniere’s disease targets in the adult stria vascularis. Front Neurol. 2021;12:630561. doi: 10.3389/fneur.2021.630561 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seker Yildiz K, Durmus K, Donmez G, Arslan S, Altuntas EE. Studying the association between sudden hearing loss and DNA N-methyltransferase 1 (DNMT1) genetic polymorphism. J Int Adv Otol. 2017;13(3):313–317. doi: 10.5152/iao.2017.2723 [doi]. [DOI] [PubMed] [Google Scholar]
- 25.Hamidi AK, Yazdani N, Seyedjavadi KH, et al. MTHFR AND ApoE genetic variants association with sudden sensorineural hearing loss. Am J Otolaryngol. 2019;40(2):260–264. doi: S0196–0709(18)30926–8 [pii]. [DOI] [PubMed] [Google Scholar]
- 26.Liu H, Gu Z, Kang HY, et al. FCRL3 gene polymorphisms confer risk for sudden sensorineural hearing loss in a chinese han population. Gene. 2015;570(1):89–94. doi: 10.1016/j.gene.2015.06.005 [doi]. [DOI] [PubMed] [Google Scholar]
- 27.Lin X, Teng Y, Lan J, He B, Sun H, Xu F. GRHL2 genetic polymorphisms may confer a protective effect against sudden sensorineural hearing loss. Mol Med Rep. 2016;13(3):2857–2863. doi: 10.3892/mmr.2016.4871 [doi]. [DOI] [PubMed] [Google Scholar]
- 28.Chien CY, Huang TY, Tai SY, et al. Glutathione peroxidase 3 gene polymorphisms and the risk of sudden sensorineural hearing loss. Kaohsiung J Med Sci. 2017;33(7):359–364. doi: S1607–551X(16)30427–2 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chien CY, Chang NC, Tai SY, Wang LF, Wu MT, Ho KY. Heat shock protein 70 gene polymorphisms in sudden sensorineural hearing loss. Audiol Neurootol. 2012;17(6):381–385. doi: 10.1159/000341815 [doi]. [DOI] [PubMed] [Google Scholar]
- 30.Cho SH, Chen H, Kim IS, et al. Association of the 4 g/5 g polymorphism of plasminogen activator inhibitor-1 gene with sudden sensorineural hearing loss. A case control study. BMC Ear Nose Throat Disord. 2012;12:5–5. doi: 10.1186/1472-6815-12-5 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitoh R, Nishio SY, Ogawa K, et al. SOD1 gene polymorphisms in sudden sensorineural hearing loss. Acta Otolaryngol. 2016;136(5):465–469. doi: 10.3109/00016489.2015.1116047 [doi]. [DOI] [PubMed] [Google Scholar]
- 32.Teranishi M, Uchida Y, Nishio N, et al. Polymorphisms in genes involved in oxidative stress response in patients with sudden sensorineural hearing loss and meniere’s disease in a japanese population. DNA Cell Biol. 2012;31(10):1555–1562. doi: 10.1089/dna.2012.1631 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McFadden SL, Ding D, Salvi R. Anatomical, metabolic and genetic aspects of age-related hearing loss in mice. Audiology. 2001;40(6):313–321. [PubMed] [Google Scholar]
- 34.Lopez IA, Acuna D, Beltran-Parrazal L, Espinosa-Jeffrey A, Edmond J. Oxidative stress and the deleterious consequences to the rat cochlea after prenatal chronic mild exposure to carbon monoxide in air. Neuroscience. 2008;151(3):854–867. doi: S0306–4522(07)01351–6 [pii]. [DOI] [PubMed] [Google Scholar]
- 35.Ohlemiller KK, McFadden SL, Ding DL, et al. Targeted deletion of the cytosolic cu/zn-superoxide dismutase gene (Sod1) increases susceptibility to noise-induced hearing loss. Audiol Neurootol. 1999;4(5):237–246. doi: 13847 [pii]. [DOI] [PubMed] [Google Scholar]
- 36.Sha SH, Zajic G, Epstein CJ, Schacht J. Overexpression of copper/zinc-superoxide dismutase protects from kanamycin-induced hearing loss. Audiol Neurootol. 2001;6(3):117–123. doi: 46818 [pii]. [DOI] [PubMed] [Google Scholar]
- 37.Kawamoto K, Sha SH, Minoda R, et al. Antioxidant gene therapy can protect hearing and hair cells from ototoxicity. Mol Ther. 2004;9(2):173–181. doi: 10.1016/j.ymthe.2003.11.020 [doi]. [DOI] [PubMed] [Google Scholar]
- 38.Zhong Z, Fu X, Li H, et al. Citicoline protects auditory hair cells against neomycin-induced damage. Front Cell Dev Biol. 2020;8:712. doi: 10.3389/fcell.2020.00712 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yazdani N, Kakavand Hamidi A, Ghazavi H, et al. Association between macrophage migration inhibitory factor gene variation and response to glucocorticoid treatment in sudden sensorineural hearing loss. Audiol Neurootol. 2015;20(6):376–382. doi: 10.1159/000438741 [doi]. [DOI] [PubMed] [Google Scholar]
- 40.Kariya S, Okano M, Maeda Y, et al. Role of macrophage migration inhibitory factor in age-related hearing loss. Neuroscience. 2014;279:132–138. doi: 10.1016/j.neuroscience.2014.08.042 [doi]. [DOI] [PubMed] [Google Scholar]
- 41.Ishihara H, Kariya S, Okano M, Zhao P, Maeda Y, Nishizaki K. Expression of macrophage migration inhibitory factor and CD74 in the inner ear and middle ear in lipopolysaccharide-induced otitis media. Acta Otolaryngol. 2016;136(10):1011–1016. doi: 10.1080/00016489.2016.1179786 [doi]. [DOI] [PubMed] [Google Scholar]
- 42.Zhu WY, Jin X, Ma YC, Liu ZB. Correlations of MIF polymorphism and serum levels of MIF with glucocorticoid sensitivity of sudden sensorineural hearing loss. J Int Med Res. 2020;48(4):300060519893870. doi: 10.1177/0300060519893870 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohlemiller KK, McFadden SL, Ding DL, Lear PM, Ho YS. Targeted mutation of the gene for cellular glutathione peroxidase (Gpx1) increases noise-induced hearing loss in mice. J Assoc Res Otolaryngol. 2000;1(3):243–254. doi: 10.1007/s101620010043 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klemens JJ, Meech RP, Hughes LF, Somani S, Campbell KC. Antioxidant enzyme levels inversely covary with hearing loss after amikacin treatment. J Am Acad Audiol. 2003;14(3):134–143. [PubMed] [Google Scholar]
- 45.Rybak LP, Husain K, Morris C, Whitworth C, Somani S. Effect of protective agents against cisplatin ototoxicity. Am J Otol. 2000;21(4):513–520. [PubMed] [Google Scholar]
- 46.Gozeler MS, Ekinci Akdemir FN, Yildirim S, Sahin A, Eser G, Askin S. Levosimendan ameliorates cisplatin-induced ototoxicity: Rat model. Int J Pediatr Otorhinolaryngol. 2019;122:70–75. doi: S0165–5876(19)30166–1 [pii]. [DOI] [PubMed] [Google Scholar]
- 47.Lynch ED, Gu R, Pierce C, Kil J. Reduction of acute cisplatin ototoxicity and nephrotoxicity in rats by oral administration of allopurinol and ebselen. Hear Res. 2005;201(1–2):81–89. doi: S0378–5955(04)00249–7 [pii]. [DOI] [PubMed] [Google Scholar]
- 48.Kim SJ, Park C, Han AL, et al. Ebselen attenuates cisplatin-induced ROS generation through Nrf2 activation in auditory cells. Hear Res. 2009;251(1–2):70–82. doi: 10.1016/j.heares.2009.03.003 [doi]. [DOI] [PubMed] [Google Scholar]
- 49.Kil J, Pierce C, Tran H, Gu R, Lynch ED. Ebselen treatment reduces noise induced hearing loss via the mimicry and induction of glutathione peroxidase. Hear Res. 2007;226(1–2):44–51. doi: S0378–5955(06)00217–6 [pii]. [DOI] [PubMed] [Google Scholar]
- 50.Shimazaki T, Ichimiya I, Suzuki M, Mogi G. Localization of glucocorticoid receptors in the murine inner ear. Ann Otol Rhinol Laryngol. 2002;111(12 Pt 1):1133–1138. doi: 10.1177/000348940211101213 [doi]. [DOI] [PubMed] [Google Scholar]
- 51.Kumagami H, Terakado M, Takahashi H. Distribution of glucocorticoid receptors and 11beta-hydroxysteroid dehydrogenase isoforms in the human inner ear. Otol Neurotol. 2013;34(1):151–157. doi: 10.1097/MAO.0b013e31826a55ad [doi]. [DOI] [PubMed] [Google Scholar]
- 52.Trune DR, Kempton JB. Blocking the glucocorticoid receptor with RU-486 does not prevent glucocorticoid control of autoimmune mouse hearing loss. Audiol Neurootol. 2009;14(6):423–431. doi: 10.1159/000241899 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Claire M, Faraj H, Grassy G, Aumelas A, Rondot A, Auzou G. Synthesis of new 11 beta-substituted spirolactone derivatives. relationship with affinity for mineralocorticoid and glucocorticoid receptors. J Med Chem. 1993;36(16):2404–2407. doi: 10.1021/jm00068a018 [doi]. [DOI] [PubMed] [Google Scholar]
- 54.Munck A, Mendel DB, Smith LI, Orti E. Glucocorticoid receptors and actions. Am Rev Respir Dis. 1990;141(2 Pt 2):2. [PubMed] [Google Scholar]
- 55.Lee JH, Marcus DC. Nongenomic effects of corticosteroids on ion transport by stria vascularis. Audiol Neurootol. 2002;7(2):100–106. doi: 57657 [pii]. [DOI] [PubMed] [Google Scholar]
- 56.Rupprecht R, Reul JM, van Steensel B, et al. Pharmacological and functional characterization of human mineralocorticoid and glucocorticoid receptor ligands. Eur J Pharmacol. 1993;247(2):145–154. doi: 10.1016/0922-4106(93)90072-h [doi]. [DOI] [PubMed] [Google Scholar]
- 57.Pondugula SR, Sanneman JD, Wangemann P, Milhaud PG, Marcus DC. Glucocorticoids stimulate cation absorption by semicircular canal duct epithelium via epithelial sodium channel. Am J Physiol Renal Physiol. 2004;286(6):1127. doi: 10.1152/ajprenal.00387.2003 [doi]. [DOI] [PubMed] [Google Scholar]
- 58.Trune DR, Kempton JB, Gross ND. Mineralocorticoid receptor mediates glucocorticoid treatment effects in the autoimmune mouse ear. Hear Res. 2006;212(1–2):22–32. doi: S0378–5955(05)00305–9 [pii]. [DOI] [PubMed] [Google Scholar]
- 59.Kitoh R, Nishio SY, Usami SI. Prognostic impact of gene polymorphisms in patients with idiopathic sudden sensorineural hearing loss. Acta Otolaryngol. 2017;137(sup565):S24–S29. doi: 10.1080/00016489.2017.1296971 [doi]. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Our curated list of SSNHL-implicated genes was cross-referenced against the following previously published datasets. Single cell and single nucleus RNA-Seq datasets of postnatal day 30 (P30) mouse stria vascularis18 were utilized (GEO Accession ID: GSE136196) (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE136196) and are available through the gene Expression Analysis Resource (gEAR) Portal, a website for visualization and comparative analysis of multi-omic data, with an emphasis on hearing research (https://umgear.org//index.html?layout_id=b50cae7a). Single cell RNA-Seq dataset from the P7 developing mouse cochlea including IHCs, OHCs, and supporting cells including both inner phalangeal, pillar, and Deiters cells 19 was utilized (GEO Accession ID: GSE137299) and are available through gEAR (https://umgear.org//index.html?layout_id=f7baf4ea). Finally, previously published single cell RNA-Seq datasets of postnatal day 15 (P15) inner hair cells (IHCs), outer hair cells (OHCs), and Deiters cells20 were utilized (GEO Accession ID: GSE114157) (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE114157) and are available through the Molecular Otolaryngology and Renal Research Laboratories (MORL) (https://morlscrnaseq.org/). Previously published datasets from the P25–27 adult mouse spiral ganglion neurons21 and from the adult human cochlea22 were also utilized for additional gene localization analysis, included in the supplemental materials (Supplemental Digital Content, Figures 2–3).