Abstract
A large proportion of heritability for prostate cancer risk remains unknown. Transcriptome-wide association study combined with validation comparing overall levels will help to identify candidate genes potentially playing a role in prostate cancer development. Using data from the Genotype-Tissue Expression Project, we built genetic models to predict normal prostate tissue gene expression using the statistical framework PrediXcan, a modified version of the unified test for molecular signatures, and Joint-Tissue Imputation. We applied these prediction models to the genetic data of 79,194 prostate cancer cases and 61,112 controls to investigate the associations of genetically determined gene expression with prostate cancer risk. Focusing on associated genes, we compared their expression in prostate tumor versus normal prostate tissue, compared methylation of CpG sites located at these loci in prostate tumor versus normal tissue, and assessed the correlations between the differentiated genes’ expression and the methylation of corresponding CpG sites, by analyzing The Cancer Genome Atlas (TCGA) data. We identified 573 genes showing an association with prostate cancer risk at a false discovery rate (FDR) ≤ 0.05, including 451 novel genes and 122 previously reported genes. Of the 573 genes, 152 showed differential expression in prostate tumor versus normal tissue samples. At loci of 57 genes, 151 CpG sites showed differential methylation in prostate tumor versus normal tissue samples. Of these, 20 CpG sites were correlated with expression of 11 corresponding genes. In this TWAS, we identified novel candidate susceptibility genes for prostate cancer risk, providing new insights into prostate cancer genetics and biology.
Keywords: transcriptome-wide association study, genetic factors, prostate cancer, gene expression
Introduction
Prostate cancer remains the most frequently diagnosed malignancy in men (1). It is critical to better elucidate its etiology which is currently poorly understood. Age, ethnicity, and family history are the few established risk factors for prostate cancer (2, 3). It has been estimated that the heritability of prostate cancer is approximately 58% (4). To date, genome-wide association studies (GWAS) have identified over 200 genetic loci harboring risk variants of prostate cancer, but overall, these variants explain less than half of the familial risk (5, 6). Studies leveraging expression quantitative trait loci (eQTL) analyses have implicated that specific GWAS-identified risk variants could regulate the expression of target genes which might play important roles in prostate carcinogenesis (7-9). However, the target genes responsible for a majority of GWAS-identified association signals remain unknown.
Transcriptome-wide association study (TWAS) is a design to uncover disease susceptibility genes by imputing gene expression levels into GWAS datasets, which can significantly improve the power to identify gene-disease associations (10, 11). This design has been proven to be useful for identifying multiple new candidate susceptibility genes across human malignancies (12-18). For prostate cancer, three TWAS studies have been published, in which significant associations for 316 genes have been identified (12,15,18). Mancuso et al. assessed genetically predicted expression in 45 tissue types beyond the prostate (15). Focusing on prostate, the authors evaluated normal prostate tissue (the Genotype-Tissue Expression dataset (GTEx), n=87), prostate tumor tissue, and tumor adjacent normal prostate tissue (The Cancer Genome Atlas (TCGA)). Emami et al. leveraged prediction models built using a large Mayo Clinic dataset comprising genetic data and gene expression data of fresh frozen normal prostate tissue obtained from patients with either radical prostatectomy or cystoprostatectomy (N=471) (18). Wu et al. developed prostate tissue prediction models using gene expression data of normal prostate tissue (GTEx, n=73) (12). The authors also built cross-tissue models aiming to increase the statistical power for genes with genetic regulatory mechanisms that are shared across different tissues.
It has been well established that tumor growth can influence gene expression in surrounding tissues, and thus the expression of some genes could be substantially different in tumor-adjacent normal tissue compared with that in normal tissue from subjects without cancer. Therefore, ideally, to study prostate cancer susceptibility genes, gene expression prediction models derived from normal prostate tissue from healthy subjects without cancer should be used. Recently, the last version (v8) of the GTEx project has been released (19). In this dataset, 221 subjects, primarily of European ancestry, have both genotyping and normal prostate tissue transcriptome data available. Leveraging this large reference dataset for normal prostate tissue from subjects without cancer, we applied several state-of-the-art modeling strategies, including the modified UTMOST (unified test for molecular signatures) (20), the newly developed Joint-Tissue Imputation (JTI) method (21), as well as PrediXcan (10), to develop comprehensive normal prostate tissue gene expression genetic prediction models. We conducted a comprehensive prostate cancer TWAS to identify novel susceptibility gene candidates for this common malignancy. Focusing on associated genes, we further evaluated their expression in prostate tumor and tumor adjacent normal prostate tissue samples in The Cancer Genome Atlas (TCGA).
Materials and Methods
Transcriptome and genome data from the GTEx project (version 8)
To develop genetic imputation models for genes expressed in normal prostate tissue, we used transcriptome and genome data from postmortem/organ procurement cases for the GTEx project (v8). Details of the GTEx v8 dataset have been described elsewhere (https://gtexportal.org/home/documentationPage). Detailed information on RNA sequencing experiments, whole genome sequencing (WGS) and quality control (QC) of the transcriptome and genome data have been described elsewhere (22, 23).
Building normal prostate tissue gene expression prediction models
The PrediXcan, modified UTMOST, and JTI frameworks were used to build three separate sets of normal prostate tissue expression genetic prediction models. The detailed information for model development has been described elsewhere (21). Briefly, the residuals of the normalized gene expression levels (19) were used after regressing out covariates, including sex (not applied to the single-tissue approach PrediXcan), platform, first five principal components (PCs), and probabilistic estimation of expression residuals (PEER) factors. SNPs within 1 Mb upstream and downstream of the gene body were considered as predictor variables in the models. LD-pruning (r2=0.9) was performed before model training to reduce the computational burden, and no significant difference in prediction quality from applying LD pruning (10).
For PrediXcan model training (24), the elastic net was applied. Five-fold cross validation was performed to generate the prediction models and to evaluate their prediction performance.
For modified UTMOST (20), the effect sizes were estimated by minimizing the loss function with a LASSO penalty for within-tissue effects, and a group-LASSO penalty for cross-tissue effects in the joint-tissue prediction model. The group penalty term could share the information from feature (SNP) selection across all the tissues. λ1 and λ2 were tuned to optimize the problem for the within-tissue and cross-tissue penalization, respectively. Five-fold cross-validation was performed for hyperparameter tuning (25). Notably, we modified the original script of UTMOST by using uniform hyper-parameters across different folds to make the hyper-parameters directly comparable (17, 26). We confirmed that the modified UTMOST gave an unbiased estimate of prediction performance using empirical datasets (21). Details of the modification can be found at https://github.com/gamazonlab/MR-JTI/blob/master/README.md. JTI estimates the gene expression profile similarity and the regulatory profile similarity (here, generated from the DNase I hypersensitivity sites in the promoter region) for each tissue-tissue pair (21). The two similarity measures were combined using hyper-parameters, which were tuned using five-fold cross validation. For all the prediction models, genes with a good prediction quality from five-fold cross-validation (r > 0.1 and P < 0.05 for the correlation between the observed and the predicted expression) were defined as imputable genes and were used for downstream analyses.
Associations between genetically determined gene expression in prostate tissue and prostate cancer risk
We investigated the associations of genetically determined gene expression in prostate tissue with prostate cancer risk using the GWAS summary statistics generated from 79,194 cases and 61,112 controls of European ancestry in the PRACTICAL consortium. The detailed information of this meta-analysis has been described elsewhere (27). Briefly, a total of 46,939 cases and 27,910 controls were genotyped using OncoArray including 570,000 SNPs (http://epi.grants.cancer.gov/oncoarray/). The SNP data were imputed using the 1000 Genomes Project (1KGP; 2014 June release) data as reference. Data from seven previous GWAS or high-density SNP panels imputed to 1KGP, including UK stage 1 and UK stage 2, BPC3, NCI PEGASUS, iCOGS, CaPS 1, and CaPS 2, were also included. An inverse variance fixed-effect approach was used to meta-analyze logistic regression summary statistics.
Using S-PrediXcan (28, 29), the associations of genetically determined gene expression with prostate cancer risk were estimated based on prediction weights, GWAS summary statistics, and a SNP-correlation (LD) matrix (11, 14). For a majority of the tested genes, most of the corresponding predicting SNPs were used for the association analyses (e.g., ≥80% predicting SNPs used for 99.2% of the tested genes). A Benjamini-Hochberg false discovery rate (FDR) of < 0.05 was used to adjust for multiple comparisons.
Comparison of expression of associated genes and DNA methylation levels of CpG sites at loci of associated genes in prostate tumor samples versus tumor adjacent normal prostate samples in TCGA
To further assess whether TWAS identified associated genes show differential expression in prostate tumor versus tumor adjacent normal prostate tissue samples, we compared the directly measured expression of these genes in TCGA data. The detailed information for the study dataset, data QC, processing, and analyses have been described elsewhere (30). In brief, gene expression data of 468 prostatic tumor samples and 51 tumor-adjacent normal prostate tissue samples were analyzed (30). For genes showing differential expression in tumor versus normal prostate tissue samples with directions of effect consistent with those in TWAS, we further evaluated whether there was additional evidence from differential DNA methylation levels of CpG sites at the same loci. This will help to prioritize promising candidates for future functional analysis based on findings from this work. For this analysis, we analyzed data from 469 prostatic tumor tissue samples and 50 histologically normal prostatic tissue samples in TCGA, as described elsewhere (31). Focusing on CpG sites demonstrating differential methylation in tumor versus normal samples, we further evaluated correlations of their methylation with the expression of nearby genes in 34 histologically normal prostate tissue samples. Depending on the distribution, either a Pearson or a Spearman correlation test was conducted. An FDR corrected significance threshold at <0.05 was used in each of the validation analyses.
Functional enrichment analyses using Ingenuity Pathway Analysis (IPA)
We performed functional enrichment analysis for the genes identified to be associated with prostate cancer risk. Using the IPA software, we estimated top associated diseases and bio-functions, canonical pathways, and top-level networks (32).
Results
Prostate tissue gene expression prediction model building
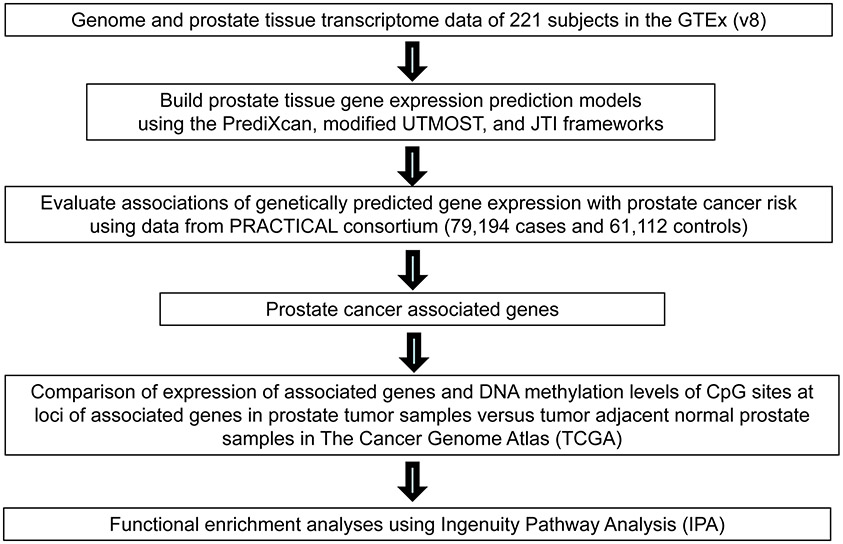
The overall study design is presented in Figure 1. Using PrediXcan, a modified UTMOST, and JTI framework, we built three separate sets of prediction models for 11,536 genes with a model performance r (correlation between genetically determined gene expression and measured expression) > 0.1 and P < 0.05. Detailed information regarding the number of prediction models built according to different performance thresholds and gene types is shown in Supplementary Table 1.
Figure 1.
Study design flow chart.
Associations of predicted gene expression in prostate tissue with prostate cancer risk
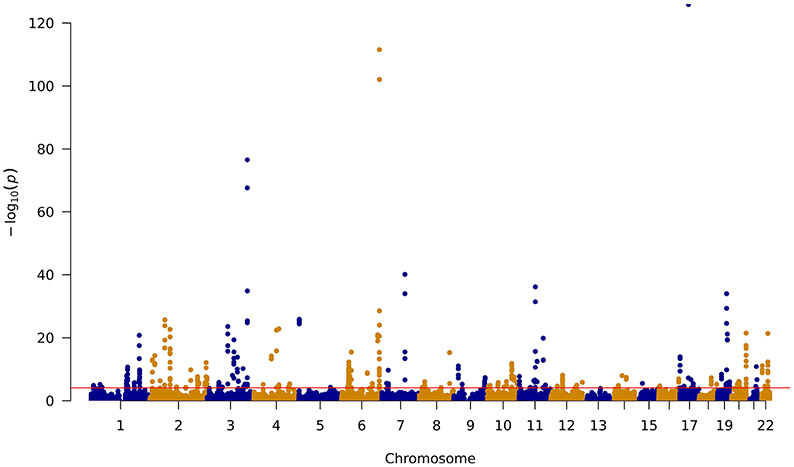
By analyzing 24,238 prediction models for 11,536 genes, we identified 573 genes associated with prostate cancer risk at P ≤ 2.01×10−3, a false discovery rate (FDR)-corrected significance level (Table 1 and Table 2; Supplementary Table 2-5; Figure 2). Of these, 140 genes were associated with prostate cancer risk at a Bonferroni-corrected significance level (P ≤ 2.06×10−6).
Table 1.
Eighteen previous reported candidate target genes of GWAS-identified prostate cancer risk variants based on fine-mapping or eQTL
| Gene | Region | Classification | R2a | OR (95% CI) | P-value | P-value after FDRa | Closest risk SNPsb | Model | Reference (PMID) |
|---|---|---|---|---|---|---|---|---|---|
| SV2A | 1q21.2 | protein_coding | 0.02 | 0.9 (0.86-0.95) | 4.39×10−5 | 2.60×10−3 | rs56391074 | PrediXcan | 26611117 |
| RN7SL832P | 2p25.1 | lncRNA | 0.03 | 0.73 (0.67-0.79) | 9.99×10−14 | 3.19×10−11 | rs9287719 | Modified UTMOST | 27526323 |
| 0.08 | 0.9 (0.86-0.94) | 8.94×10−7 | 8.81×10−5 | rs9287719 | JTI | ||||
| EFHD1 | 2q37.1 | protein_coding | 0.12 | 0.89 (0.84-0.93) | 3.79×10−6 | 3.23×10−4 | rs59308963 | PrediXcan | 27409348 |
| 0.13 | 0.9 (0.86-0.94) | 9.29×10−6 | 6.84×10−4 | rs59308963 | Modified UTMOST | ||||
| 0.14 | 0.9 (0.86-0.94) | 2.46×10−6 | 2.22×10−4 | rs59308963 | JTI | ||||
| HDLBP | 2q37.3 | protein_coding | 0.03 | 1.09 (1.04-1.14) | 4.30×10−4 | 1.57×10−2 | rs111770284 | JTI | 24907074 |
| HLA-DQA2 | 6p21.32 | protein_coding | 0.62 | 1.04 (1.03-1.06) | 1.33×10−6 | 1.27×10−4 | rs9296068 | PrediXcan | 26611117 |
| 0.63 | 1.04 (1.02-1.06) | 2.71×10−5 | 1.72×10−3 | rs9296068 | Modified UTMOST | ||||
| 0.64 | 1.04 (1.02-1.05) | 8.36×10−6 | 6.22×10−4 | rs9296068 | JTI | ||||
| HLA-DRB1 | 6p21.32 | protein_coding | 0.17 | 0.9 (0.87-0.94) | 4.19×10−7 | 4.62×10−5 | rs3129859 | PrediXcan | 26611117 |
| NOTCH4 | 6p21.32 | protein_coding | 0.13 | 1.06 (1.03-1.1) | 5.28×10−4 | 1.84×10−2 | rs3096702 | PrediXcan | 26611117 |
| 0.2 | 1.06 (1.02-1.09) | 6.86×10−4 | 2.25×10−2 | rs3096702 | Modified UTMOST | ||||
| 0.24 | 1.05 (1.02-1.08) | 9.94×10−4 | 2.97×10−2 | rs3096702 | JTI | ||||
| HLA-C | 6p21.33 | protein_coding | 0.22 | 0.96 (0.94-0.98) | 4.50×10−4 | 1.63×10−2 | rs2596546 | PrediXcan | 26611117 |
| MICA | 6p21.33 | protein_coding | 0.51 | 1.03 (1.01-1.05) | 1.21×10−3 | 3.45×10−2 | rs2596546 | JTI | 26611117 |
| HOTTIP | 7p15.2 | lncRNA | 0.05 | 0.77 (0.71-0.83) | 2.19×10−10 | 4.28×10−8 | rs200362064 | JTI | 26611117 |
| RAB14 | 9q33.2 | protein_coding | 0.04 | 1.26 (1.1-1.43) | 5.60×10−4 | 1.93×10−2 | rs1571801 | Modified UTMOST | 25371445 |
| PSMB7 | 9q33.3 | protein_coding | 0.02 | 0.89 (0.84-0.95) | 1.01×10−4 | 5.04×10−3 | rs1571801 | JTI | 27409348 |
| AS3MT | 10q24.32 | protein_coding | 0.43 | 1.04 (1.02-1.06) | 6.84×10−4 | 2.25×10−2 | rs34032774 | PrediXcan | 26611117, 26162851, 24907074 |
| 0.46 | 1.04 (1.02-1.06) | 4.46×10−4 | 1.61×10−2 | rs34032774 | Modified UTMOST | ||||
| 0.48 | 1.03 (1.01-1.05) | 4.08×10−4 | 1.52×10−2 | rs34032774 | JTI | ||||
| NT5C2 | 10q24.32-33 | protein_coding | 0.02 | 1.24 (1.16-1.33) | 8.67×10−11 | 1.84×10−8 | rs34032774 | Modified UTMOST | 26611117 |
| C1QL4 | 12q13.12 | protein_coding | 0.25 | 1.09 (1.06-1.13) | 6.21×10−8 | 8.50×10−6 | rs10875943 | PrediXcan | 24907074 |
| 0.26 | 1.1 (1.06-1.14) | 5.66×10−7 | 6.05×10−5 | rs10875943 | Modified UTMOST | ||||
| 0.25 | 1.09 (1.05-1.12) | 1.17×10−7 | 1.48×10−5 | rs10875943 | JTI | ||||
| C14orf39 | 14q23.1 | protein_coding | 0.06 | 0.87 (0.8-0.94) | 9.71×10−4 | 2.92×10−2 | rs7153648 | PrediXcan | 26611117 |
| 0.11 | 0.9 (0.84-0.96) | 1.48×10−3 | 3.99×10−2 | rs7153648 | JTI | ||||
| SYNJ2BP | 14q24.2 | protein_coding | 0.07 | 1.25 (1.16-1.36) | 3.82×10−8 | 5.54×10−6 | rs7141529 | Modified UTMOST | 26611117 |
| 0.08 | 1.2 (1.12-1.28) | 1.65×10−7 | 2.05×10−5 | rs7141529 | JTI | ||||
| ZNF652 | 17q21.32 | protein_coding | 0.04 | 1.36 (1.16-1.6) | 1.89×10−4 | 8.26×10−3 | rs11650494 | JTI | 26162851, 26025378, 24907074 |
R2: prediction performance (R2) derived using GTEx data. P value: derived from association analyses of 79,194 cases and 61,112 controls; associations with FDR-corrected P value ≤ 0.05 considered significant
Risk SNPs identified in previous GWAS or fine-mapping studies. The risk SNP closest to the gene is presented.
lncRNA: long noncoding RNAs.
Table 2.
The validation study to test for 11 genes transcriptomic and methylomic expression level associations with prostate cancer at a FDR-corrected significance level
| Associations in TWAS | Genes showing differential expression in tumor vs normal prostate samples |
CpG sites showing significant differential methylation levels between prostate tumor and tumor adjacent normal samples |
Biologically significant differential methylation levels between prostate tumor and tumor adjacent normal samples |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene Name | region | Classification | R2a | OR (95%CI) | P-value | P-value after FDRa | model | FC b | P-value after FDR b | Probe ID | FC b | P-value after FDR b | P-value after FDR b | P-valueb | r b | |
| ADAM15 | 1q22 | protein_coding | 0.02 | 1.35 (1.23-1.48) | 1.59×10−10 | 3.23×10−8 | JTI | 1.36 | 4.51×10−8 | cg13069100 | 1.48 | 5.35×10−22 | 3.31×10−5 | 1.75×10−6 | 0.72 | Pearson |
| 0.05 | 1.23 (1.16-1.3) | 1.94×10−11 | 4.57×10−9 | PrediXcan | ||||||||||||
| LDAH | 2p24.1 | protein_coding | 0.05 | 0.8 (0.75-0.85) | 3.45×10−12 | 9.09×10−10 | JTI | −1.73 | 2.84×10−11 | cg20248458 | 2.00 | 6.36×10−13 | 0.02 | 1.65×10−3 | −0.52 | Pearson |
| 0.13 | 0.9 (0.88-0.93) | 1.25×10−12 | 3.48×10−10 | PrediXcan | ||||||||||||
| 0.13 | 0.87 (0.83-0.9) | 4.71×10−15 | 1.78×10−12 | Modified UTMOST | ||||||||||||
| AMT | 3p21.31 | protein_coding | 0.33 | 0.95 (0.92-0.97) | 5.42×10−5 | 3.10×10−3 | Modified UTMOST | −1.87 | 3.42×10−12 | cg02251567 | 1.70 | 4.13×10−20 | 1.65×10−7 | 1.10×10−9 | −0.83 | Pearson |
| cg05924489 | 2.17 | 3.73×10−20 | 5.23×10−7 | 1.38×10−8 | −0.80 | Pearson | ||||||||||
| 0.35 | 0.96 (0.94-0.98) | 5.24×10−5 | 3.02×10−3 | PrediXcan | cg14184400 | 1.66 | 9.00×10−19 | 5.23×10−7 | 3.51×10−7 | −0.80 | Pearson | |||||
| cg15246085 | 1.74 | 2.65×10−20 | 5.23×10−7 | 2.40×10−7 | −0.80 | Pearson | ||||||||||
| 0.32 | 0.95 (0.93-0.97) | 1.81×10−5 | 1.22×10−3 | JTI | cg17877071 | 1.56 | 3.62×10−19 | 8.34×10−7 | 2.91×10−7 | −0.79 | Pearson | |||||
| cg20191453 | 1.71 | 1.23×10−17 | 8.34×10−7 | 3.31×10−8 | −0.79 | Pearson | ||||||||||
| cg21926782 | 1.45 | 4.80×10−17 | 1.07×10−6 | 3.51×10−7 | −0.78 | Pearson | ||||||||||
| UBA7 | 3p21.31 | protein_coding | 0.07 | 0.84 (0.76-0.94) | 1.62×10−3 | 0.04 | Modified UTMOST | −1.26 | 6.88×10−5 | cg13343687 | 1.59 | 3.16×10−20 | 0.04 | 3.76×10−3 | −0.48 | Pearson |
| SRGAP3 | 3p25.3 | protein_coding | 0.03 | 0.88 (0.81-0.95) | 1.05×10−3 | 0.03 | Modified UTMOST | −1.15 | 0.04 | cg12669543 | 1.72 | 2.20×10−17 | 0.02 | 2.42×10−3 | −0.50 | Pearson |
| 0.04 | 0.88 (0.81-0.95) | 1.04×10−3 | 0.03 | PrediXcan | ||||||||||||
| ATF6B | 6p21.33 | protein_coding | 0.12 | 1.07 (1.03-1.12) | 5.92×10−4 | 0.02 | Modified UTMOST | 1.07 | 0.03 | cg09149894 | −1.15 | 9.65×10−4 | 0.03 | 3.42×10−3 | −0.49 | Pearson |
| 0.13 | 1.07 (1.04-1.1) | 3.60×10−5 | 2.19×10−3 | PrediXcan | ||||||||||||
| 0.03 | 1.17 (1.09-1.26) | 2.67×10−5 | 1.71×10−3 | JTI | ||||||||||||
| VARS2 | 6p21.33 | protein_coding | 0.44 | 1.05 (1.03-1.07) | 6.49×10−7 | 6.78×10−5 | JTI | 1.42 | 3.01×10−10 | cg00933603 | −1.08 | 0.02 | 6.34×10−5 | 3.78×10−6 | −0.70 | Pearson |
| 0.43 | 1.06 (1.03-1.08) | 2.99×10−7 | 3.39×10−5 | Modified UTMOST | ||||||||||||
| 0.47 | 1.05 (1.03-1.07) | 1.05×10−7 | 1.35×10−5 | PrediXcan | ||||||||||||
| SMYD4 | 17p13.3 | protein_coding | 0.12 | 0.92 (0.88-0.96) | 4.34×10−4 | 0.02 | JTI | −1.38 | 6.71×10−9 | cg16616918 | 1.46 | 5.91×10−14 | 9.36×10−5 | 6.20×10−6 | −0.69 | Pearson |
R2: prediction performance (R2) derived using GTEx data. P value: derived from association analyses of 79,194 cases and 61,112 controls; associations with FDR-corrected P value < 0.05 considered significant.
P value: derived from 34 prostatic tumor tissue samples and paired normal prostatic tissue samples.
Figure 2.
Manhattan plot of association results obtained from the prostate cancer transcriptome-wide association study. The red line represents P = 2.01 × 10−3 (FDR-corrected P value ≤ 0.05). Each dot represents the genetically predicted expression of one specific gene by prostate tissue prediction models: the x axis represents the genomic position of the corresponding gene, and the y axis represents the negative logarithm of the association P value.
We identified 249 associated genes at least 500kb away from any GWAS identified prostate cancer risk variants (9, 33-38) (Supplementary Table 2). Of these, an association between lower predicted expression and increased prostate cancer risk was detected for 138 genes, and an association between higher predicted expression and increased prostate cancer risk was identified for 111 genes. Among them, 37 genes were suggested by all three genetic prediction models (PrediXcan, modified UTMOST, and JTI). We also identified 194 novel genes at known prostate cancer susceptibility loci (9, 32-37). Among them, 37 genes were suggested by all three genetic models (Supplementary Table 3). Furthermore, we observed significant associations for 104 genes that had been previously reported as candidate prostate cancer susceptibility genes in published TWAS. Reassuringly, for a majority of them (69 genes), the directions of the associations were consistent in published TWAS vs the current study (Supplementary Table 4). For 19 of the genes, both positive and inverse associations were reported in previous TWAS (Supplementary Table 5). For the remaining 16 genes, the reported associations in previous TWAS had different directions compared with those in the current study (Supplementary Table 6). For 15 of these 16 genes, their associations in the previous TWAS were based on prediction models of either non-prostate tissues or prostate tumor tissue (Supplementary Table 6). We also identified an additional 18 genes that had been previously reported as candidate target genes of known prostate cancer risk variants identified through functional studies and/or eQTL analyses (Table 1). Overall, we were able to identify 451 novel candidate susceptibility genes for prostate cancer and confirmed 122 genes known to potentially play a role in prostate cancer susceptibility.
Measured expression of associated genes and DNA methylation levels of CpG sites at loci of associated genes in prostate tumor samples versus tumor adjacent normal prostate tissue samples
Of the 573 associated genes identified in this TWAS, 152 genes showed a differential expression in prostate tumor and tumor adjacent normal tissue (FDR<0.05) with directions consistent with those identified in TWAS (Supplementary Table 7). At these loci, 2,353 CpG sites showed differential methylation levels between prostate tumor and tumor adjacent normal samples (FDR < 0.05) (Supplementary Table 8). Among them, 151 CpG sites were further found to be biologically significant in that the average methylation beta value needed to be >0.5 in one of the tested groups (tumor or normal samples) and be <0.5 in the other group (Supplementary Table 8). Of those, the methylation of 14 CpG sites was significantly correlated with the expression of corresponding genes in normal samples. Finally, after aggregating results from the above analyses, we identified 8 genes showing consistent effect directions that were supported by these complementary analyses (Table 2).
Pathway enrichment analyses
The results of IPA (32) suggested potential enrichment of cancer-related functions for the TWAS identified genes (Supplementary Table 9). The top canonical pathways included Antigen Presentation Pathway (P = 1.08 × 10−9), Th1 Pathway (P = 6.36 × 10−5), T Cell Exhaustion Signaling Pathway (P = 8.72 × 10−5), Allograft Rejection Signaling (P = 1.54 × 10−4), and OX40 Signaling Pathway (P = 1.96 × 10−4).
Discussion
Leveraging the largest available reference dataset of normal prostate tissue transcriptome and the state-of-the-art modified UTMOST, JTI, and PrediXcan modeling strategies, we performed a comprehensive TWAS to evaluate the relationship between genetically determined gene expression levels in prostate tissue and prostate cancer risk throughout the human genome. We identified 573 genes with genetically determined expression to be associated with prostate cancer risk (FDR ≤ 0.05), including 451 novel genes that have not been reported in published TWAS. The present study provides substantial new information to improve the understanding of genetics and etiology for prostate cancer.
Previously, we developed a gene-level association analysis approach named PrediXcan which applies elastic net to develop gene expression genetic prediction models (39). Owing to the fact that approaches such as PrediXcan do not take into account the similarity of genetic regulation for genes across different human tissues, analysis becomes challenging when the effective number of relevant tissue samples is small (40, 41). To overcome this potential limitation in our study, we also leveraged two other modelling strategies, modified UTMOST and JTI. UTMOST is a powerful method to jointly analyze data from multiple genetically-correlated tissues which has obvious advantages compared with many other methods (23). The gene expression imputation accuracy was shown to be improved by 38.6% across tissues for the original UTMOST method compared with PrediXcan. We further modified the model training approach to obtain a reliable estimate of the imputation performance. JTI, a method recently developed by us, borrows information from the other tissues in a tissue-dependent manner. i.e., weighing up more relevant tissues and weighing down less relevant tissues by integrating high-throughput functional genomic data (ENCODE (42) and Roadmap (43)) to improve prediction quality. For highly tissue-specific genes, JTI automatically reduces to single-tissue PrediXcan by a grid-search based hyper-parameter tuning. By evaluating prediction performance in independent datasets, JTI demonstrated higher statistical power than PrediXcan and modified UTMOST for many genes. Overall, by leveraging these three strategies with complementary strengths, it is expected that we could have high statistical power to fully identify gene-prostate cancer associations. Reassuringly, for a majority of genes that showed a significant association, their associations based on the other tested models (when available) also demonstrated consistent directions and nominal significance (P<0.05). The results were shown in Supplementary Table 8.
It is reassuring that for previous TWAS identified genes that also showed a significant association in the current study, for a large proportion (66%) of them the association directions were consistent. Only 15% of those genes showed inconsistent directions of associations, of which associations of most such genes in prior studies were based on non-prostate or prostate tumor but not normal prostate tissue prediction models. This supports the validity of the current study, though further studies of these genes are needed to validate the direction of association.
Of the TWAS identified genes, 152 showed differential expression between prostate tumor and tumor adjacent normal tissue samples, with consistent directions of effect in the TCGA dataset. Meanwhile, 151 CpG sites at loci of such genes showed differential methylation levels in prostate tumor versus tumor adjacent normal tissue samples. Of them, 20 CpG sites were further correlated with expression of 11 corresponding nearby genes in tumor adjacent normal prostate tissue samples. Such additional functional analyses leveraging measured gene expression and DNA methylation data provided a list of promising genes for further characterization. For several of our identified genes, there is already some evidence from published literature supporting their potential roles in human tumorigenesis. For example, one of the identified genes, the human A Disintegrin and Metalloproteinase 15 (ADAM15) is a multi-domain disintegrin protease (44). Activated ADAM15 was reported as a key modulator of cell-cell and cell-matrix interactions, and to be involved in the proteolysis of cytokines, growth factors and adhesion molecules (45, 46). Previous research supported that the expression of ADAM15 mRNA and its protein levels were increased in prostate cancer compared with normal prostate and its protein level was increased significantly during metastatic progression (44). There was accumulating evidence supporting that gene ADAM15 and its protein might play important roles in prostate cancer biology. Tissue microarrays (TMAs) analysis revealed that ADAM15 protein was overexpressed in prostate cancer specimens compared with benign prostate tissue specimens and its expression was also increased significantly during metastatic progression (45, 47). Abdo J Najy et al. found that ADAM15 played an important role in prostate tumor cell interaction with vascular endothelium and the metastatic progression of PC-3 prostate cancer cells (33). Our results also supported higher mRNA expression of ADAM15 in prostate tumors versus tumor adjacent normal tissues. Another gene that we identified was glutathione S-transferase pi 1, which is an isozyme encoded by the GSTP1 gene (11q13.2) that plays an important regulatory role in detoxification, anti-oxidative damage, and the occurrence of cancers (48). The over-expression of GSTP1 inhibits the viability and motility of prostate cancer in vitro and in vivo through targeting Myelocytomatosis Viral Oncogene Homolog (MYC) and inactivating MEK/ERK1/2 pathway (49). Another study found that the deletion of GSTP1 might lead to the accumulation of oxidative DNA base damage and promote the survival of prostate cancer cells under long-term oxidative stress (50). Zhang L et al. found that functional inactivation of GSTP1 could increase the susceptibility to oxidative stress and enhance the risk of developing prostate carcinoma (51). Kamińska K et al. observed that hypermethylation of GSTP1 could result in down-regulation of its gene expression as compared to wild type fibroblasts in prostate cancer cell lines (52). In a systematic review and meta-analysis, Zhou et al. observed that GSTP1 promoter methylation was higher in prostate cancer patients than in controls (53). Another study found that GSTP1 methylation was stable over time in negative prostate biopsies, and could predict missed cancer with high specificity (54). Patel, PG et al. developed a three-gene biomarker signature (GAS6/GSTP1/HAPLN3) to discriminate benign and malignant prostate tissue with low false positive and negative rates (below 7%) (55). In the current study, higher expression or hypomethylation of GSTP1 tended to be associated with a decreased risk of prostate cancer. Another gene, LDAH, has been reported to promote cholesterol ester turnover in macrophages, and have an effect on the development of prostate cancer (56). Currall BB et al. validated using both in vitro and in vivo models that loss of LDAH resulted in an increased risk of prostate cancer (57).
In this study, we leveraged complementary, state-of-the-art modelling strategies to develop comprehensive normal prostate tissue genetic prediction models, which brings increased statistical power for detecting gene expression-prostate cancer risk associations. In the main TWAS analysis, for the overall tests, we conducted FDR correction to reduce type 1 error rate. The associations detected by more than one modeling approach may represent more credible ones. However, several potential limitations also need to be acknowledged. Although in the current design we used one possible design by comparing the genes’ overall expression in tumor vs tumor adjacent normal prostate tissue samples, there is a possibility that such evidence alone may yield false positive or false negative findings. Similarly, the incorporation of methylation comparison was aimed to prioritize promising genes for further functional characterization, but such a design may produce false positive or false negative findings as well. Thus, further functional studies will be needed to investigate whether these genes could play a causal role in prostate tumorigenesis. In addition, our validation analysis using TCGA data was based on a comparison between tumor versus tumor-adjacent normal tissues. Since the molecular profiles of tumor adjacent normal tissues may have been affected by the tumor and were not the same as those of completely normal tissue, future studies using completely normal tissues are warranted to further verify our findings.
In conclusion, in this large-scale TWAS of prostate cancer, we identified 573 genes with genetically determined expression in prostate tissue to be associated with prostate cancer risk, including 451 novel genes. We provided additional evidence from both measured gene expression and DNA methylation supporting potential roles of 11 genes. We believe that such analyses can help us further understand the associations identified in TWAS. Further investigation of these genes will provide new insights into the biology and genetics of prostate cancer.
Supplementary Material
Novelty & Impact Statements.
A large proportion of heritability for prostate cancer risk remains unknown. In a transcriptome-wide association study (TWAS), the authors identified 573 candidate genes for prostate cancer risk, including 451 novel genes and 122 previously reported genes. 152 of the genes and 151 CpG sites at loci of such 152 genes showed differential expression/methylation levels in prostate tumor versus tumor adjacent normal tissue samples. These findings provide new insights into prostate cancer genetics and biology.
Acknowledgments
We thank The PRACTICAL, CRUK, BPC3, CAPS, PEGASUS consortia for making the prostate cancer GWAS summary statistics publicly available. The authors also would like to thank all of the individuals for their participation in the parent PRACTICAL studies and all the researchers, clinicians, technicians and administrative staff for their contribution to the studies. This study is supported by the University of Hawaii Cancer Center. Nancy J Cox is supported by U01HG009086. E.R.G. is supported by NIH/NHGRI R35HG010718, NIH/NHGRI R01HG011138, NIH/NIA AG068026, and NIH/NIGMS R01GM140287. Duo Liu is partially supported by the Harbin Medical University Cancer Hospital. Yanfa Sun is partially supported by the Department of Education of Fujian Province, P R China. The Prostate cancer genome-wide association analyses are supported by the Canadian Institutes of Health Research, European Commission’s Seventh Framework Programme grant agreement n° 223175 (HEALTH-F2-2009-223175), Cancer Research UK Grants C5047/A7357, C1287/A10118, C1287/A16563, C5047/A3354, C5047/A10692, C16913/A6135, and The National Institute of Health (NIH) Cancer Post-Cancer GWAS initiative grant: No. 1 U19 CA 148537-01 (the GAME-ON initiative). We would also like to thank the following for funding support: The Institute of Cancer Research and The Everyman Campaign, The Prostate Cancer Research Foundation, Prostate Research Campaign UK (now PCUK), The Orchid Cancer Appeal, Rosetrees Trust, The National Cancer Research Network UK, The National Cancer Research Institute (NCRI) UK. We are grateful for support of NIHR funding to the NIHR Biomedical Research Centre at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust. The Prostate Cancer Program of Cancer Council Victoria also acknowledge grant support from The National Health and Medical Research Council, Australia (126402, 209057, 251533, 396414, 450104, 504700, 504702, 504715, 623204, 940394, 614296), VicHealth, Cancer Council Victoria, The Prostate Cancer Foundation of Australia, The Whitten Foundation, PricewaterhouseCoopers, and Tattersall’s. EAO, DMK, and EMK acknowledge the Intramural Program of the National Human Genome Research Institute for their support. Genotyping of the OncoArray was funded by the US National Institutes of Health (NIH) [U19 CA 148537 for ELucidating Loci Involved in Prostate cancer SuscEptibility (ELLIPSE) project and X01HG007492 to the Center for Inherited Disease Research (CIDR) under contract number HHSN268201200008I] and by Cancer Research UK grant A8197/A16565. Additional analytic support was provided by NIH NCI U01 CA188392 (PI: Schumacher). Funding for the iCOGS infrastructure came from: the European Community’s Seventh Framework Programme under grant agreement n° 223175 (HEALTH-F2-2009-223175) (COGS), Cancer Research UK (C1287/A10118, C1287/A 10710, C12292/A11174, C1281/A12014, C5047/A8384, C5047/A15007, C5047/A10692, C8197/A16565), the National Institutes of Health (CA128978, CA128813) and Post-Cancer GWAS initiative (1U19 CA148537, 1U19 CA148065 and 1U19 CA148112 – the GAME-ON initiative), the Department of Defense (W81XWH-10-1-0341), the Canadian Institutes of Health Research (CIHR) for the CIHR Team in Familial Risks of Breast Cancer, Komen Foundation for the Cure, the Breast Cancer Research Foundation, and the Ovarian Cancer Research Fund. The BPC3 was supported by the U.S. National Institutes of Health, National Cancer Institute (cooperative agreements U01-CA98233 to D.J.H., U01-CA98710 to S.M.G., U01-CA98216 toE.R., and U01-CA98758 to B.E.H., and Intramural Research Program of NIH/National Cancer Institute, Division of Cancer Epidemiology and Genetics). CAPS GWAS study was supported by the Swedish Cancer Foundation (grant no 09-0677, 11-484, 12-823), the Cancer Risk Prediction Center (CRisP; www.crispcenter.org), a Linneus Centre (Contract ID 70867902) financed by the Swedish Research Council, Swedish Research Council (grant no K2010-70X-20430-04-3, 2014-2269). PEGASUS was supported by the Intramural Research Program, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health.
Abbreviations
- ADAM15
A Disintegrin and Metalloproteinase 15
- eQTL
expression quantitative trait loci
- FDR
false discovery rate
- GTEx
Genotype-Tissue Expression dataset
- GWAS
genome-wide association studies
- IPA
Ingenuity Pathway Analysis
- MYC
Myelocytomatosis Viral Oncogene Homolog
- PC
sprincipal components
- PEER
probabilistic estimation of expression residuals
- QC
quality control
- TCGA
The Cancer Genome Atlas
- TMAs
Tissue microarrays
- TWAS
transcriptome-wide association study
- UTMOST
unified test for molecular signatures
- WGS
whole genome sequencing
Footnotes
Conflict of Interest
JBN is a partner of Genomix Inc. Genomix Inc. had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. E.R.G. receives an honorarium from the journal Circulation Research of the American Heart Association, as a member of the Editorial Board. There is no conflict of interest for other authors.
Data Availability Statement
The summary statistics of prostate cancer GWAS studies in PRACTICAL consortium are available at http://practical.icr.ac.uk/blog/?page_id=8164. The GTEx transcriptome and genome data are publically available via dbGaP (www.ncbi.nlm.nih.gov/gap; dbGaP Study Accession: phs000424.v8.p2). Further information and the full association results from our main analyses and are available upon request.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA: A Cancer Journal for Clinicians 2020;70:7–30 [DOI] [PubMed] [Google Scholar]
- 2.Tsodikov A, Gulati R, Heijnsdijk EA, Pinsky PF, Moss SM, Qiu S, et al. Reconciling the effects of screening on prostate cancer mortality in the ERSPC and PLCO trials. Annals of internal medicine 2017;167:449–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.American Cancer Society CDoPH, California Cancer Registry. California Cancer Facts & Figures 2017. 2017
- 4.Hjelmborg JB, Scheike T, Holst K, Skytthe A, Penney KL, Graff RE, et al. The heritability of prostate cancer in the Nordic Twin Study of Cancer. Cancer Epidemiology and Prevention Biomarkers 2014;23:2303–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benafif S, Kote-Jarai Z, Eeles RA; PRACTICAL Consortium. A Review of Prostate Cancer Genome-Wide Association Studies (GWAS). Cancer Epidemiol Biomarkers Prev. 2018; 27(8): 845–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takata R, Takahashi A, Fujita M, Momozawa Y, Saunders EJ, Yamada H, Maejima K, Nakano K, Nishida Y, Hishida A, Matsuo K, Wakai K, Yamaji T, Sawada N, Iwasaki M, Tsugane S, Sasaki M, Shimizu A, Tanno K, Minegishi N, Suzuki K, Matsuda K, Kubo M, Inazawa J, Egawa S, Haiman CA, Ogawa O, Obara W, Kamatani Y, Akamatsu S, Nakagawa H. 12 new susceptibility loci for prostate cancer identified by genome-wide association study in Japanese population. Nat Commun. 2019; 10(1): 4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gusev A, Shi H, Kichaev G, Pomerantz M, Li F, Long HW, et al. Atlas of prostate cancer heritability in European and African-American men pinpoints tissue-specific regulation. Nature communications 2016;7:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Penney KL, Sinnott JA, Tyekucheva S, Gerke T, Shui IM, Kraft P, et al. Association of prostate cancer risk variants with gene expression in normal and tumor tissue. Cancer Epidemiology and Prevention Biomarkers 2015;24:255–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thibodeau SN, French A, McDonnell S, Cheville J, Middha S, Tillmans L, et al. Identification of candidate genes for prostate cancer-risk SNPs utilizing a normal prostate tissue eQTL data set. Nature communications 2015;6:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gamazon ER, Wheeler HE, Shah KP, Mozaffari SV, Aquino-Michaels K, Carroll RJ, et al. A gene-based association method for mapping traits using reference transcriptome data. Nature genetics 2015;47:1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gusev A, Ko A, Shi H, Bhatia G, Chung W, Penninx BW, et al. Integrative approaches for large-scale transcriptome-wide association studies. Nature genetics 2016;48:245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu L, Wang J, Cai Q, Cavazos TB, Emami NC, Long J, et al. Identification of novel susceptibility loci and genes for prostate cancer risk: a transcriptome-wide association study in over 140,000 European descendants. Cancer research 2019;79:3192–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu L, Shi W, Long J, Guo X, Michailidou K, Beesley J, et al. A transcriptome-wide association study of 229,000 women identifies new candidate susceptibility genes for breast cancer. Nature genetics 2018;50:968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu Y, Beeghly-Fadiel A, Wu L, Guo X, Li B, Schildkraut JM, et al. A transcriptome-wide association study among 97,898 women to identify candidate susceptibility genes for epithelial ovarian cancer risk. Cancer research 2018;78:5419–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mancuso N, Gayther S, Gusev A, Zheng W, Penney KL, Kote-Jarai Z, et al. Large-scale transcriptome-wide association study identifies new prostate cancer risk regions. Nature communications 2018;9:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang T, Choi J, Kovacs MA, Shi J, Xu M, Goldstein AM, et al. Cell-type–specific eQTL of primary melanocytes facilitates identification of melanoma susceptibility genes. Genome research 2018;28:1621–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu D, Zhou D, Sun Y, Zhu J, Ghoneim D, Wu C, Yao Q, Gamazon ER, Cox NJ, Wu L. A Transcriptome-Wide Association Study Identifies Candidate Susceptibility Genes for Pancreatic Cancer Risk. Cancer Res. 2020; 80(20):4346–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emami NC, Kachuri L, Meyers TJ, Das R, Hoffman JD, Hoffmann TJ, et al. Association of imputed prostate cancer transcriptome with disease risk reveals novel mechanisms. Nature communications 2019;10:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369(6509):1318–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu Y, Li M, Lu Q, Weng H, Wang J, Zekavat SM, et al. A statistical framework for cross-tissue transcriptome-wide association analysis. Nature genetics 2019;51:568–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou D, Jiang Y, Zhong X, Cox NJ, Liu C, Gamazon ER. A unified framework for joint-tissue transcriptome-wide association and Mendelian randomization analysis. Nat Genet. 2020. doi: 10.1038/s41588-020-0706-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Consortium G. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 2015;348:648–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Consortium G. Genetic effects on gene expression across human tissues. Nature 2017;550:204–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barbeira AN, et al. Widespread dose-dependent effects of RNA expression and splicing on complex diseases and traits. BioRxiv, 2019; 814350 [Google Scholar]
- 25.Liu D, Zhou D, Sun Y, Zhu J, Ghoneim D, Wu C, Yao Q, Gamazon ER, Cox NJ, Wu L. A Transcriptome-Wide Association Study Identifies Candidate Susceptibility Genes for Pancreatic Cancer Risk. Cancer Res. 2020; 80(20): 4346–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun Yanfa, Zhou Dan, Rahman Md Rezanur, Zhu Jingjing, Ghoneim Dalia, Cox Nancy J, Beach Thomas G, Wu Chong, Gamazon Eric R, Wu Lang. A transcriptome-wide association study identifies novel blood-based gene biomarker candidates for Alzheimer's disease risk. Hum Mol Genet. 2021;ddab229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schumacher FR, Al Olama AA, Berndt SI, Benlloch S, Ahmed M, Saunders EJ, et al. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nature genetics 2018;50:928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barbeira AN, Dickinson SP, Bonazzola R, Zheng J, Wheeler HE, Torres JM, et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nature communications 2018;9:1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nature genetics 2016;48:1279–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nikas JB, Mitanis NT, Nikas EG. Whole Exome and Transcriptome RNA-Sequencing Model for the Diagnosis of Prostate Cancer. ACS Omega 2020;5:481–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nikas JB, Nikas EG. Genome-wide DNA methylation model for the diagnosis of prostate cancer. ACS Omega 2019;4:14895–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krämer A, Green J, Pollard J Jr, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 2014;30:523–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Najy Abdo J, Day Kathleen C, Day Mark L. ADAM15 supports prostate cancer metastasis by modulating tumor cell-endothelial cell interaction. Cancer Res. 2008; 68(4): 1092–9 [DOI] [PubMed] [Google Scholar]
- 34.Iczkowski KA. Genomic Pathology and Cancer Biomarkers: Prostate Cancer. Critical Reviews™ in Oncogenesis 2017;22 [DOI] [PubMed] [Google Scholar]
- 35.Santric V, Djokic M, Suvakov S, Pljesa-Ercegovac M, Nikitovic M, Radic T, et al. GSTP1 rs1138272 Polymorphism Affects Prostate Cancer Risk. Medicina 2020;56:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Yuan Y, Chen Y, Wang Z, Li F, Zhao Q. Association between GSTP1 Ile105Val polymorphism and urinary system cancer risk: evidence from 51 studies. OncoTargets and therapy 2016;9:3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Z, Jiang D, Wang C, Garzotto M, Kopp R, Wilmot B, et al. Polymorphisms in oxidative stress pathway genes and prostate cancer risk. Cancer Causes & Control 2019;30:1365–75 [DOI] [PubMed] [Google Scholar]
- 38.Conti DV, Darst BF, Moss LC, Saunders EJ, Sheng X, Chou A, et al. Trans-ancestry genome-wide association meta-analysis of prostate cancer identifies new susceptibility loci and informs genetic risk prediction. Nat Genet 2021;53:65–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hendriks RJ, Dijkstra S, Smit FP, Vandersmissen J, Van de Voorde H, Mulders PF, et al. Epigenetic markers in circulating cell-free DNA as prognostic markers for survival of castration-resistant prostate cancer patients. The Prostate 2018;78:336–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu Y, Lu Q, Liu W, Zhang Y, Li M, Zhao H. Joint modeling of genetically correlated diseases and functional annotations increases accuracy of polygenic risk prediction. PLoS genetics 2017;13:e1006836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duong D, Gai L, Snir S, Kang EY, Han B, Sul JH, et al. Applying meta-analysis to genotype-tissue expression data from multiple tissues to identify eQTLs and increase the number of eGenes. Bioinformatics 2017;33:i67–i74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.ENCODE Project Consortium. The ENCODE (ENCyclopedia of DNA elements) project. Science, 2004; 306, 636–640 [DOI] [PubMed] [Google Scholar]
- 43.Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, et al. Integrative analysis of 111 reference human epigenomes. Nature, 2015; 518(7539): 317–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lucas N and Day ML (2009). The role of the disintegrin metalloproteinase ADAM15 in prostate cancer progression. J Cell Biochem 106(6), 967–974 [DOI] [PubMed] [Google Scholar]
- 45.Edwards DR, Handsley MM, and Pennington CJ (2008). The ADAM metalloproteinases. Mol Aspects Med 29(5), 258–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.White JM (2003). ADAMs: modulators of cell-cell and cell-matrix interactions. Curr Opin Cell Biol 15(5), 598–606 [DOI] [PubMed] [Google Scholar]
- 47.Kuefer R, Day KC, Kleer CG, Sabel MS, Hofer MD, Varambally S, Zorn CS, Chinnaiyan AM, Rubin MA, Day ML. ADAM15 disintegrin is associated with aggressive prostate and breast cancer disease. Neoplasia. 2006; 8(4):319–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cui Jian, Li Guoqing, Yin Jie, Li Linwei, Tan Yue, Wei Haoran, et al. GSTP1 and cancer: Expression, methylation, polymorphisms and signaling (Review). Int J Oncol 2020; 56(4): 867–878 [DOI] [PubMed] [Google Scholar]
- 49.Wang X-x, Jia H-t, Yang H, Luo M-h, Sun T. Overexpression of glutathione S-transferase P1 inhibits the viability and motility of prostate cancer via targeting MYC and inactivating the MEK/ERK1/2 pathways. Oncology Research Featuring Preclinical and Clinical Cancer Therapeutics 2020 [DOI] [PubMed] [Google Scholar]
- 50.Mian OY, Khattab MH, Hedayati M, Coulter J, Abubaker-Sharif B, Schwaninger JM, et al. GSTP1 Loss results in accumulation of oxidative DNA base damage and promotes prostate cancer cell survival following exposure to protracted oxidative stress. The Prostate 2016;76:199–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang L, Meng X, Pan C, Qu F, Gan W, Xiang Z, et al. piR-31470 epigenetically suppresses the expression of glutathione S-transferase pi 1 in prostate cancer via DNA methylation. Cellular Signalling 2020;67:109501. [DOI] [PubMed] [Google Scholar]
- 52.Kamińska K, Białkowska A, Kowalewski J, Huang S, Lewandowska MA. Differential gene methylation patterns in cancerous and non-cancerous cells. Oncology reports 2019;42:43–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou X, Jiao D, Dou M, Chen J, Li Z, Li Y, et al. Association of glutathione-S-transferase p1 gene promoter methylation and the incidence of prostate cancer: a systematic review and meta-analysis. Journal of cancer research and clinical oncology 2019;145:1939–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fiano V, Zugna D, Grasso C, Trevisan M, Delsedime L, Molinaro L, et al. DNA methylation in repeat negative prostate biopsies as a marker of missed prostate cancer. Clinical epigenetics 2019;11:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patel PG, Wessel T, Kawashima A, Okello JB, Jamaspishvili T, Guérard KP, et al. A three-gene DNA methylation biomarker accurately classifies early stage prostate cancer. The Prostate 2019;79:1705–14 [DOI] [PubMed] [Google Scholar]
- 56.Kory N, Grond S, Kamat SS, Li Z, Krahmer N, Chitraju C, et al. Mice lacking lipid droplet-associated hydrolase, a gene linked to human prostate cancer, have normal cholesterol ester metabolism. Journal of lipid research 2017;58:226–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Currall BB, Chen M, Sallari RC, Cotter M, Wong KE, Robertson NG, et al. Loss of LDAH associated with prostate cancer and hearing loss. Human molecular genetics 2018;27:4194–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The summary statistics of prostate cancer GWAS studies in PRACTICAL consortium are available at http://practical.icr.ac.uk/blog/?page_id=8164. The GTEx transcriptome and genome data are publically available via dbGaP (www.ncbi.nlm.nih.gov/gap; dbGaP Study Accession: phs000424.v8.p2). Further information and the full association results from our main analyses and are available upon request.