Abstract
Recent advances in extracellular vesicle biology have uncovered a substantial role in maintaining cell homeostasis in health and disease conditions by mediating intercellular communication, thus catching the scientific community’s attention worldwide. Extracellular microvesicles, called exosomes, functionally transfer biomolecules such as proteins and non-coding RNAs from one cell to another, influencing the local environment’s biology. Although numerous advancements have been made in treating cancer patients with immune therapy, controlling the disease remains a challenge in the clinic due to tumor-driven interference with the immune response and inability of immune cells to clear cancer cells from the body. The present review article discusses the recent findings and knowledge gaps related to the role of exosomes derived from tumors and the tumor microenvironment cells in tumor escape from immunosurveillance. Further, we highlight examples where exosomal non-coding RNAs influence immune cells’ response within the tumor microenvironment and favor tumor growth and progression. Therefore, exosomes can be used as a therapeutic target for the treatment of human cancers.
Keywords: Cancer; Exosomes; Extracellular vesicle; microRNA, LncRNA; Immune cell dysfunction; Immune evasion; Immunotherapy
1. Introduction
1.1. Extracellular vesicles (EVs):
The EVs are double-layer phospholipid membrane vesicles released by almost every cell into the extracellular environment to facilitate cell-to-cell communication [1, 2]. EVs contain signaling molecules, including DNA, RNA, proteins, lipids, and metabolites enveloped by the phospholipid layer, which enable them to regulate various cellular functions in acceptor cells [3, 4]. Besides, the loss of materials in the EVs from secreting cells facilitates the acquisition of new differentiation phenotype in acceptor cells or the clearance of undesirable proteins, DNA, or RNA, to maintain cellular homeostasis [5–7]. In the studies published so far, EVs are usually characterized by their size, origin, composition, and functions. The two main broad categories of EVs are based on their origin such as plasma membrane-derived (microvesicles and macrovesicles) or endosome-derived (exosomes). EVs that originate from outward budding and fission of plasma membrane with a diameter range from 200 to 1000 nm are traditionally considered microvesicles [8, 9]. Macrovesicles are large-sized heterogenous vesicles (1000–10,000 nm) reported in cancer cell secretions, including prostate cancer and leukemia, to facilitate cancer progression [10–12]. Macrovesicles (also known as oncosomes) secreted by primary B-cell precursor acute myeloid leukemia (AML) blasts and cell lines are internalized by bone marrow stromal cells (BMSCs), in which they induce metabolic reprogramming to favor cancer progression [10]. Di Vizio et al., reported 1000–10,000 nm sized EVs derived from cell-surface protrusions of amoeboid-shaped prostate cancer cells [11]. These were termed macrosomes and contain metalloproteinases, RNA, caveolin-1, and the GTPase ADP-ribosylation factor 6. Macrosomes are abundant in metastatic prostate cancer secretions and associate with disease progression [11]. The International Society for Extracellular Vesicles (ISEV), recommends to use operational terms for EV subtypes unless they establish specific markers unique to the subcellular origin of EVs or show EV release from a source inside cells as imaged by live microscopy [13]. As per ISEV-2018 guidelines, EVs can be differentiated based on size (if <200 nm, small EVs and >200 nm are medium or large EVs), biochemical composition, or cell of origin (podocyte EVs, hypoxic EVs oncosomes, apoptotic bodies etc.) [13]. Apoptotic bodies are well-known EVs formed by plasma membrane blebbing in the dying apoptotic cell and have a 500 nm −10 µm diameter range [14]. Apoptotic body formation facilitates the clearance of aged, damaged, or infected cells from the system to maintain homeostasis. The apoptotic bodies are phagocytosed and removed by surrounding cells without triggering an inflammatory response. Apoptotic bodies are characterized by phosphatidylserine, which translocate from the lower to the upper layer of plasma membrane during apoptosis [15, 16]. Next comes exosomes which represent a more uniform population of vesicles derived from endosomes.
1.2. Exosomes and their biogenesis:
In most studies, EVs with an endosomal origin and a size range of 30–150 nm in diameter are considered exosomes [17, 18]. Exosomes are derived from the inward budding of multivesicular bodies (MVBs), originating from endosomes [19, 20]. The MVBs are formed when a part of the endosomal membrane invaginates and buds into its lumen. MVBs contain cargo molecules enclosed in membrane-bound intraluminal vesicles (ILVs), which gives MVBs a multivesicular appearance [21]. MVBs either move to the plasma membrane or fuse with lysosomes or autophagosomes to deliver their contents [4, 22]. When MVBs fuse with the plasma membrane, they release ILVs known as exosomes into the extracellular space [23]. Studies have shown that ILV biogenesis and secretion may or may not require the endosomal sorting complex required for transport (ESCRT) machinery. ESCRT complexes, together with accessory proteins, are involved in membrane bending and budding during MVB biogenesis and ILV secretion [24, 25]. Many ESCRT machinery components have been detected in exosome vesicles isolated from different sources, suggesting their role in exosome biogenesis [24, 26–29]. A study by Baietti et al. showed that during exosome biogenesis, a heparan sulfate proteoglycan called syndecan binds to its cytoplasmic adaptors synetnin and ALIX and accumulates on endosomal membranes [30]. ALIX is known to interact and recruit several ESCRT members, including TSG101 and CHMP4, during ILVs formation and budding [24]. The ALIX/Syntenin/Syndecan complex regulates the intra-luminal budding of endosomal membrane domains and supports exosome biogenesis. The silencing of ESCRT components, including CHMP2A and VPS4A/B, reduces the formation of syndecan–syntenin exosomes and prevents exosome release [30]. While Alix/Syntenin/Syndecan complex recruits CHMP4 proteins through direct interaction with ALIX, CHMP4 recruitment involves also ESCRT machinery, which promotes endosomal membrane budding and abscission [30]. Larios et al. showed that Alix recruits ESCRT-III on late endosomes containing lysobisphosphatidic acid (LBPA) and enables the sorting and delivery of tetraspanins to exosomes [31]. Furthermore, variations in the activities of different ESCRT proteins affect the size, biogenesis, and contents of EVs, which suggests ESCRTs role in their synthesis and release [21, 32–34].
The ESCRT-independent exosome biogenesis involves lipid-raft-like microdomains containing ceramide forming sphingolipids. The ceramides have cone-shaped structures that can induce inward budding of endosomes required to produce ILVs [35]. Another ESCRT-independent mechanism involves RAB GTPases that are present on the surface of endosomal membranes during ILV formation. RAB GTPases recruit specific effector proteins required for membrane budding and ILVs formation [36]. The receptor tyrosine kinases, including epidermal growth factor receptor (EGFR), insulin like growth factor 1 receptor (IGFR), platelet-derived growth factor receptor (PDGFR)-b, MET, and fibroblast growth factor receptor 2 (FGFR2) phosphorylate and activate RAB31. The activated RAB31 engages flotillin (FLOT) proteins 1 and 2 present in caveolae/lipid raft domains of endosomal membranes to facilitate entry of these kinases into ILVs. FLOT functions have been reported in clathrin-independent endocytosis and membrane curvature and budding. Additionally, RAB31 prevents the fusion of MVEs with lysosomes by recruiting GTPase-activating protein TBC1D2B, which inactivates RAB7, a key regulator in endo-lysosomal fusion [36, 37].
1.3. Exosome interaction with recipient cells:
Exosomes interact with recipient cells, either through receptors present on recipient cells that are internalized via receptor-mediated endocytosis or through direct fusion with receptor cells’ plasma membranes, and then release their contents. Receptor-mediated endocytosis involves the formation of clathrin-coated vesicles that serve to internalize exosome cargo during the fusion process. For example, dendritic cell (DC)-derived exosomes expressing major histocompatibility complex class II (MHC-II) are internalized by activated T cells through receptor-mediated endocytosis. MHC-II on DC exosomes interacts with the leukocyte function associated antigen-1 (LFA-1) receptor on activated T cells and facilitates membrane fusion and internalization [38]. Likewise, DC exosome interaction with NK cells involves TNF superfamily ligands tumor necrosis factor (TNF), Fas ligand (FasL) and TNF related apoptosis inducing ligand (TRAIL), whose receptors are present on NK cells. The receptor-independent direct fusion of exosomes or EVs with the recipient cells occurs through various mechanisms like caveolin mediated endocytosis [39], macropinocytosis, lipid raft mediated endocytosis or phagocytosis. In caveolin-mediated endocytosis, plasma membrane invaginations called caveolae are formed at sites rich with proteins and cholesterol. Integral membrane protein caveolins and adaptor proteins (cavins) are involved in the organization and internalization of these caveolae, whereas membrane-remodeling GTPase, dynamin facilitates caveolae budding [40, 41]. Genetic silencing of caveolin-1 or dynamin inhibition significantly suppresses exosome uptake in cells [39]. Macropinocytosis does not need clathrin to form vesicles and uses membrane protrusions, termed lamellipodia or membrane ruffles made of the actin cytoskeleton to internalize extracellular materials [42]. Studies have suggested that inhibiting micropinocytosis by inhibitors of actin assembly, such as 5-(N-ethyl-N-isopropyl)amirolide (EIPA) or genistein, affects exosome uptake and internalization [43, 44]. Lipid raft mediated endocytosis is clathrin-independent internalization of EVs utilizing plasma membrane microdomains enriched in cholesterol and sphingolipids known as lipid rafts [45]. Inhibitors of cholesterol transport or sphingolipid synthesis significantly reduce exosome uptake, whereas lipid raft components flotillin and annexin A2 (AnxA2), a Ca(2+)-dependent phospholipid-binding protein that can induce lipid domain formation, promote exosome intake [46–48]. In phagocytic cells, exosomes are internalized by phagocytosis and targeted to phagolysosomes. This process requires actin polymerization and dynamin accumulation and depends upon phosphatidylinositol 3-kinase signaling [49].
The exosome EVs are essential mediators of cell-cell communication and are secreted by many cell types, including immune cells [50], adipocytes [51], nerve cells [52], stem cells [53], bone and muscle cells [54, 55], skin cells [56], endothelial cells [57] and cancer cells [58]. Exosomes have been detected in biological fluids such as blood [59], urine [60], semen [61], and breast milk [62]. Due to their essential role in cell-cell communication and transfer of cargo molecules, they are involved in the pathogenesis of many diseases, including neurological [63], cardiovascular [50], kidney [64], liver [65], and cancer [66]. Cancer cells heavily depend on EV or exosome-mediated signaling in the tumor microenvironment (TME) to facilitate communications between cancer and TME cells that are required for cancer growth. This review will focus on exosome-mediated immune suppression in the tumor microenvironment and its role in immune escape and tumor growth. In the review, we discuss the latest on the role of exosomes derived from cancer cells or from cells of the tumor microenvironment. and the roles these exosomes play in suppressing innate and adaptive immune responses, favoring cancer growth, and spreading to other organs. Finally, we discuss the potential role of exosomes in cancer immunotherapy and the identification of cancer biomarkers.
2. Cancer and Immune system
The abnormal division of cells leads to cancer growth and development. Cancer cells effectively evade the immune system, which continuously hunts for abnormal cells in the body [67]. The various means by which cancer cells bypass the immune system include weakening the immune system by the formation of immune-suppressive TME consisting of corrupt extracellular matrix (ECM) [68], inflammatory cells [69] and tumor-promoting immune cells [70]; change in tumor cell phenotype including loss or downregulation of immune recognition molecules [71]; and overexpression of immune check point molecules like programmed death ligand 1 (PDL1) or cytotoxic T-lymphocyte associated protein 4 (CTLA-4) which help them evade effector T cells or become apoptosis-resistant [72]. The cancer-derived exosomal mediated communication between cancer and TME cells plays an important role in generating an immune-suppressive pro-tumor microenvironment.
2.1. Cancer-derived exosomes and the suppression of the immune system:
Cancer exosomes help cancer cells grow, metastasize and become resistant to drug treatment [73, 74]. For example, exosomes secreted from prostate cancer cells contain PD-L1, which suppresses T cell functions and promotes tumor progression and growth. PD-L1 works by binding to its receptor programmed death (PD)-1 on T cells, which inhibits T cell proliferation and activation [75]. Exposure of tumor cells to exosomes lacking PD-L1 induces a robust immune memory response and makes these cells resistant to PD-L1 mediated immune suppression [76].
Poggio et al. injected prostate cancer cell line TRAMP-C2 in the flanks of mice, in a preclinical prostate cancer model. In the first group, one flank of mice received wild-type (WT) PD-L1 cells, whereas the other side received PD-L1 null cells, or nSMase2 null and Rab27a null, two enzyme critical in exosome biogenesis. In the second group, both flanks were injected with PD-L1 WT cells. The tumors grew more slowly and had massive infiltration of immune cells in the first group compared to the second. This supports the notion that in the first group, immune cells in the mutant side could be activated due to loss of PD-L1, generate a memory response, and migrate to the WT side via draining lymph nodes to inhibit tumor growth [76]. In the second group, where PD-L1 null or inhibition of exosome biogenesis show similar findings supports the vital role of exosomes in cancer cell communication and signaling.
Further, it was found that PD-L1 levels in the circulating exosomes were higher in patients with metastatic melanoma than healthy individuals. Incubation of PD-L1 containing exosomes with CD8 T cells inhibits their functions and facilitates melanoma progression [77]. Similar findings have been seen in head and neck squamous cell carcinoma patients where PD-L1 expression correlates with The Union for International Cancer Control (UICC) malignant tumor stage and lymph node status [78]. Exosomes of patients with active head and neck squamous cell carcinomas (HNSCC) have a high PD-L1 level compared to treated patients with no evidence of disease. Moreover, PD-L1 high exosomes isolated from HNSCC patients downregulate activation marker CD69 in effector T cells. This suggests that exosomes are effective carriers that can transfer immune suppressive molecules from cancer cells to nearby or distant immune cells.
Furthermore, the proteomic profiling of neuroblastoma (NB)-derived exosomes reveals the presence of B7-H3 and NB marker protein GD2 ganglioside (disialoganglioside) [79–82]. B7-H3 is a cell surface protein that interacts with its ligands, including checkpoint markers CTLA-4, PD-1, and CD28 on immune cells, and inhibits their further activation [83]. This helps in maintaining immune homeostasis and prevents autoimmunity. However, B7-H3 overexpression, as seen in many tumor types, can promote immune suppression and facilitate cancer progression. For example, inhibition of B7-H3 reduces tumor growth and enhances T cell and natural killer (NK) cell cytotoxic functions in NB [79, 84]. B7-H3 overexpression in medulloblastoma cells increases the production of exosomes containing B7-H3 and other pro-tumorigenic molecules like Janus kinase (JAK)-signal transducer and activator of transcription (STAT), platelet-derived growth factor (PDGF), phosphoinositide 3-kinases (PI3K), and angiogenesis-associated molecules. B7-H3 overexpressing exosomes are taken up by nearby stromal or cancer cells, suggesting that they could have a role in modulating the TME. Moreover, exosomes isolated from human papillomavirus (HPV) positive HNSCC cells are enriched in B7-H3 and CD47, whereas HPV negative exosomes contain growth-promoting MUC-1 and human leukocyte antigen (HLA)-DA [85]. CD47 is an essential immune regulatory molecule that is overexpressed in many cancer types [86]. CD47 interacts with signal receptor protein alpha (SIRPα), which is present on phagocytic cells, including macrophages and dendritic cells, inhibiting their functions [87]. This suggests that exosomes have strong potential to facilitate cancer-immune interactions that can play an important role in forming the tumor-favorable immune microenvironment.
Exosomes derived from glioblastoma stem cells (GSC) increase monocytes’ viability and polarizes them into the tumor promoting M2 macrophage phenotype. GSC-derived exosomes induce cytoskeletal changes in monocytes like enlarged cytoplasm and filopodial extensions [88]. Filopodia functions in cell migration and could plays an important role in tumor associated macrophage (TAM) recruitment during cancer progression [89, 90]. Injection of exosomes derived from gastric cancer (GC) cell lines into C57BL/6 mice induces an immunosuppressive microenvironment in the lungs. The infiltrating immune cells include high CD4+ T cells and myeloid-derived suppressor cells (MDSCs) and low CD8+ T cell and NK cell populations [91]. These exosomes also enhance lung metastasis in mice harboring mouse forestomach carcinoma cells suggesting their role in forming metastatic niches [91].
A study by Yin et al. showed the role of tumor-derived exosomes in delivering fatty acids to dendritic cells to disrupt their function[92]. Fatty acid absorption in recipient dendritic cells causes the accumulation of excessive lipids, which activates peroxisome proliferator-activated receptor (PPAR) signaling [92]. PPAR is a regulator of fatty acid metabolism and is crucial for maintaining lipid homeostasis [93]. PPAR activation induces fatty acid oxidation in the dendritic cells, which switches their metabolism from glycolysis state to oxidative phosphorylation. Metabolic reprogramming in dendritic cells makes them defective in priming CD8+ T cells and suppresses their immune regulatory functions. Inhibition of PPAR significantly enhances the anti-tumor efficacy of immunotherapy with anti-PD-L1 antibodies and improves the functionality of CD8+ T cells [92]. A schematic representation of the role of cancer cell-derived exosomes in immune evasion is given in Figure 1.
Figure 1:
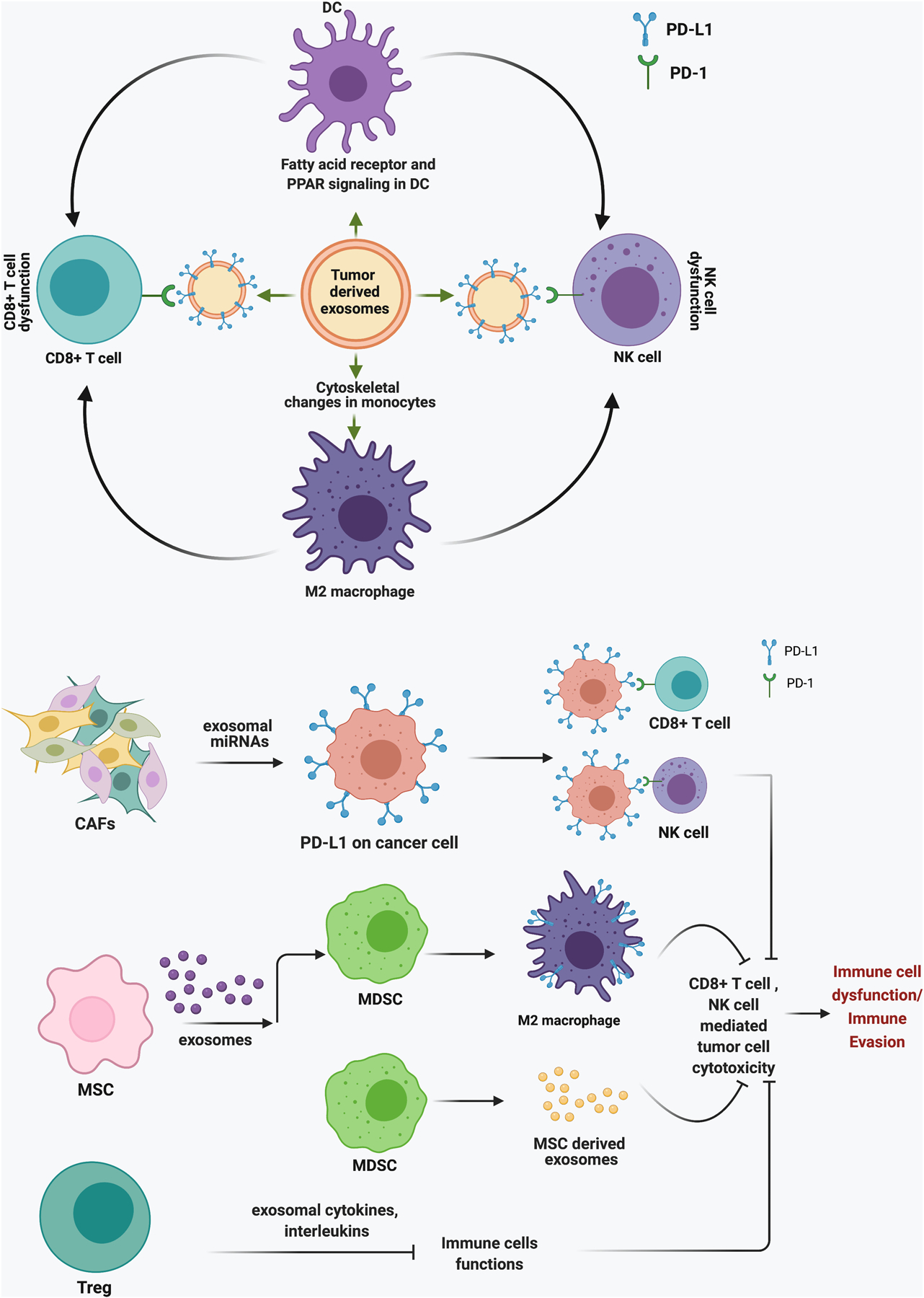
Schematic representation for the role of cancer cell-derived exosomes in promoting immune evasion through M2 microphases and dendritic cells.
2.2: Exosomes derived from the TME and their role in immune suppression:
This section discusses exosomes secreted from TME cells and their role in shaping the cancer immune microenvironment to promote cancer growth and development.
2.2.1. Immune cells derived exosomes:
2.2.1.1. Lymphocytes:
In the TME, most immune cells are either ineffective or facilitate tumor-growth by suppressing the activation of effector immune cells. A well-known example of immunosuppressive cells is regulatory T cells (Tregs), a specialized subpopulation of CD4+CD25+ T cells whose primary function is to suppress the immune response to maintain homeostasis [70]. Tregs inhibit CD8+ T cytotoxic lymphocyte (CTL) functions by suppressing their proliferation and cytokine production. The immune-inhibitory functions of Tregs are associated with an inhibited anti-tumor immune response and unfavorable prognosis in cancer patients. Exosomes isolated from CD4+CD25+ Tregs inhibit proliferation and perforin and interferon gamma (IFN-γ) expression in CD8+ CTLs [94, 95]. Inhibiting NF-kB signaling in DC by adenoviral gene transfer of the dominant-negative form of IKK2 makes the DCs capable of converting naïve T cells into regulatory T cells (dnIKK2-Treg). Naïve T cells exposed to dnIKK2-Treg-derived exosomes inhibit the activation and proliferation of other T cells. Interestingly, these cells do not express CD25 and FoxP3, common markers present on Tregs [96]. Moreover, few antigen-specific FoxP3+ and FoxP3- Tregs-derived exosomes contain the immunosuppressive cytokine IL-35 on their surface, which induces immune suppression of many non-IL-35 producing naïve T and B cells. The latter acquire and express IL-35 on their membranes, making them capable of inducing suppression in other naïve T cells. The recipient immune cells’ growth is inhibited either by IL-10 or by exhaustion due to constitutively activated IL-10 signaling via IL-35 receptors present on their surfaces [97]. Moreover, exosomes isolated from B cells of patients with relapsing-remitting multiple sclerosis induce death in oligodendrocytes [98]. In multiple sclerosis, oligodendrocytes are damaged, which causes demyelination and death of neurons.
2.2.1.2. TAMs:
Another important immune suppressor in the TME are TAMs, which are tumor-infiltrating or resident macrophages reprogrammed by cancer cells [99]. TAMs are recruited to the cancer site by chemoattractants, including cytokines and chemokines secreted by cancer or TME cells. These chemoattractants play an important role in TAM differentiation towards an immunosuppressive and tumor-promoting ‘M2-like’ phenotype. Most of the TAMs in the TME are of M2 phenotype and support tumor progression and metastasis [100, 101]. Lan et al. reported that exosomes derived from M2 macrophages positively regulate cell mobility, migration, and invasion in colorectal tumors [102]. M2-derived exosomes contain a high expression of microRNAs (miRs) miR-21–5p and miR-155–5p, which bind to the 3’-UTR (untranslated region) BRG1 gene in recipient cancer cells. BRG1 is highly expressed in colorectal cancers, but its overexpression inhibits migration and invasion in colorectal cancer cell lines [103]. miR-21–5p and miR-155–5p binding decrease BRG1 expression in cancer cells, promoting tumor growth, migration and invasion [102]. Furthermore, incubation of M2 macrophage-derived exosomes with GC cells promotes cell migration and invasion. M2 polarized TAMs are enriched in apolipoprotein E, which they deliver to GC cells. Exosomal transfer of apolipoprotein E activates phosphoinositide 3-kinase (PI3K)/Akt signaling in the recipient cancer cells and promotes their migration. A study by Wu et al. shows that M2 TAMs deliver integrin, αMβ2 (CD11b/CD18) to hepatocellular carcinoma (HCC) cells via exosomes. αMβ2 activates the MMP-9 pathway in recipient HCC cells, supporting their migration and promoting lung metastasis [104].
2.2.1.3. Myeloid-derived suppressor cells (MDSCs):
MDSCs are a heterogenous population of immune cells derived from myeloid progenitors in the bone marrow and share some properties with both neutrophils and monocytes [105, 106]. MDSCs have strong immune suppressive functions and play an important role in protecting cancer from patients’ immune system [107]. MDSCs secrete inhibitory cytokines and express high levels of reactive oxygen species (ROS), nitric oxide synthase (iNOS), and arginase 1 (ARG1), which contribute to immune suppression [108–110]. MDSC-derived exosomes are enriched with various types of proteins [111, 112], mRNAs[112], and micro RNAs (miRNAs) [112] involved in a vast array of functions, including exosome and endosome biogenesis, proteasomal pathway, angiogenesis, metabolism, chemotaxis, inflammation, and immune suppression [113]. This suggests that MDSCs exosomes could influence a variety of functions in the recipient cells. Exosomes isolated from MDSCs, infiltrating tumors or the spleen of tumor-bearing mice promote invasion and migration of cancer cells and inhibit CD8+ T cell proliferation. Furthermore, mice treated with MDSCs-derived exosomes show a reduction in the total CD8+ T cell population. These mice have increased M2-macrophage (CD11b+CD206+) infiltration in the spleen compared to the untreated control group [114]. In the TME, MDSCs differentiate into TAMs or DCs and function as a source of TAM replenishment [115]. Exosomes secreted from mesenchymal stromal cells (MSCs) differentiate monocytic MDSCs (M-MDSCs) into immunosuppressive M2 macrophages and promote epithelial to mesenchymal transition (EMT) of tumor cells. MSC-derived exosomes induce PD-L1 expression in M2 polarized macrophages and immature M-MDSC precursors, which induces apoptotic PD-1 signaling in CD8+ CTLs and protects tumor cells from antitumor immunity [116].
2.2.2. Cancer-associated fibroblasts (CAFs):
Fibroblasts are mesenchymal origin cells that produce stroma components and activate during wound healing [117]. Activated fibroblasts are large spindle-shaped cells, also known as myofibroblasts, which revert to normal phenotype after the wound healing [118]. In tumors, CAF become constitutively activated, support tumor growth, and suppress the immune system by secreting various growth factors and cytokines [119, 120]. Exosomes secreted from breast tumor isolated CAFs promote PD-L1 expression in breast cancer cell lines [121]. Upregulated PD-L1 suppresses the tumor-killing activity of T and NK cells and induces apoptosis in T cells. Mechanistically, CAF-derived exosomes induce recipient cancer cells to express elevated levels of miR-92, which directly binds and inhibits large tumor suppressor kinase 2 (LATS2). LATS2 is an upstream negative regulator of YAP1, which is the transcription coactivator of PD-L1. LATS2 inhibition promotes YAP1 nuclear translocation and increases PD-L1 transcription [121]. Moreover, CAF-derived exosomes promote growth, survival, and drug resistance in pancreatic and colorectal cancer cells [122, 123]. Exosome-mediated communication between CAFs and tumor cells provides a double-edged advantage by promoting cancer cell growth and escape from immune surveillance.
2.2.3. Bone marrow stromal cells (BMSCs):
BMSCs consist of a heterogeneous population of cells abundant within the bone marrow and can differentiate into many cell types, including bone-forming osteoblasts and osteoclasts, chondrocytes, adipocytes, fibroblasts, myocytes, macrophage, pericytes, endothelial cells, and neural cells. Due to their multipotent nature, these are also referred to as mesenchymal stem cells or bone marrow mesenchymal cells. BMSCs play a crucial role in forming a premetastatic niche by regulating cancer cells’ metastasis and homing in the bone marrow [124]. BMSC-derived exosomes suppress the expansion of effector CD8+ T cells and increase CD62L+CD44- naïve T cell population in mice with graft versus host disease (GVHD) [125]. Coculture of T or B cells with BMSCs inhibits their proliferation, suggesting an immune-suppressive function for BMSCs when in the context of a normal microenvironment [126]. In the TME, BMSC exosomes mediate the activation and expansion of MDSCs, which suppress T cell activity and promote tumor growth [127]. BMSC exosomes isolated from the bone marrow of mice with multiple myeloma promote survival and expansion of MDSCs by activating STAT-3 signaling. Moreover, BMSC exosomes increase STAT-3 expression in cancer cells, which induces EMT and promotes their metastasis [127]. In summary, the exosome release from TME is crucial for communication between cancer, immune, and non-immune cells. Exosomes directly or indirectly influence signaling in the recipient or non-recipient cells and play a decisive role in shaping tumor fate and immune response.
2.3: Exosomal non-coding RNAs and their role in immune suppression:
Molecular characterization of exosomes reveals the presence of several non-coding RNAs that may vary depending upon the origin of cells [128]. Non-coding RNAs, as their name suggests, do not code for proteins but regulate mRNAs and proteins at transcriptional or translational levels. Exosomal non-coding RNAs are key messenger molecules in the immune system regulating innate and adaptive immune responses. In immune-suppressed TME, there is a continuous flow of information through these RNAs among immune and cancer cells, which plays an essential role in orchestrating the TME. The following section will discuss the role of exosomal non-coding RNAs, in particular, miRNAs (20–25 nucleotides) and long non-coding RNAs or lncRNAs (>200 nucleotides) secreted by cancer or TME cells in immune regulation and silencing.
2.3.1: miRNAs:
miRNAs usually bind with the 3’UTR sequences of their target mRNAs which induces their degradation and represses translation. Due to their nature, exosomal miRNAs can regulate a vast majority of the recipient cells’ functions. Many studies have reported miRNA dysregulation in cancers and its use as potential biomarkers for human cancer prognosis or diagnosis [129, 130]. Studies have shown that miRNAs secreted in exosomes could influence TME and cancer progression. One example is the way in which exosomes secreted from TAMs increase the ratio of immunosuppressive CD4+ Tregs to antitumor CD4+T helper 17 cells (Treg/Th17 ratio) by transferring miR-29a-3p and miR-21–5p. The higher Treg/Th17 ratio is associated with enhanced growth and metastasis of epithelial ovarian cancer cells [131]. Another exosomal miRNA, miR-1246, released from ovarian cancer cells, negatively regulates caveolin-1 in the recipient M2-macrophages [132]. Caveolin-1 is a plasma membrane protein whose deletion in metastasis-associated macrophages increases metastasis and angiogenesis [133]. This suggests that exosomal miR-1246 dependent Caveolin-1 downregulation could influence pro-tumor TAM functions in the TME. Guo et al. found that mouse 4T1 breast cancer cell-derived exosomes contain miR-183–5p which can induce proinflammatory cytokines in the recipient macrophages [134]. miR-183–5p targets 3’UTR of the protein phosphatase 2 catalytic subunit alpha gene (PPP2CA), which encodes a negative regulator of NF-κB signaling. PPP2CA downregulation increases NF-κB subunit p65 phosphorylation and promotes its activation. Activated NF-κB signaling in macrophages is associated with the secretion of proinflammatory cytokines IL-1b, IL-6, and TNF-α [134].
Furthermore, exosomes secreted from melanoma cells can transfer miR-498 and miR-3187–3p to CD8+ T cells and inhibit their function. miR-498 binds to the 3’UTR of tumor necrosis factor alpha (TNFα) in CD8+ T cells and downregulates its expression, which inhibits its secretion from CD8+ T cells. miR-3187–3p also targets the CD45 gene PTPRC 3’UTR and inhibits CD45 mediated T-cell receptor (TCR) signaling and activation [135].
Exosomes isolated from sera of nasopharyngeal carcinoma (NPC) contain miR-24–3p, whose level correlates with a worse disease-free survival prognosis for patients. Exosomal miR-24–3p binds and inhibits FGF11 mRNA, which is associated with decreased phosphorylation of extracellular signal-regulated kinase (ERK), STAT-1, and STAT-3 in T cells. miR-24–3p recipient T cells show reduced proliferation, lower proportions of IFNγ-producing Th1 cells and IL-17-producing Th17 cells, and expansion of immunosuppressive Foxp3+ Tregs [136]. Another T-cell suppressor miRNA, hsa-miR-125b-5p, is increased in the plasma exosomes from patients with Non-Small Cell Lung Cancer (NSCLC) compared to healthy individuals. miR-125b-5p inhibits the secretion of INFγ and TNFα in γδT cells and induces apoptosis [137]. NSCLC patients who have undergone anti-PD1 immunotherapy have decreased exosomal hsa-miR-125b-5p levels in plasma [138]. This suggests that hsa-miR-125b-5p could be a promising marker in pathological conditions. Overall, miRNAs are important constituents in exosomes that can play a key role in changing the immune microenvironment and the host’s response to cancer therapy. Table.1 shows some other exosomal miRNAs and their role in immune suppression.
Table.1:
Role of exosomal non-coding RNAs in immune suppression in cancer
| Exosomal miRNAs | Secreted cells | Recipient cells | Role and mechanism in immune suppression |
|---|---|---|---|
| miR-10a and miR-21 [229] | Glioma | MDSCs | Promote expansion and function of MDSCs. miR-10a inhibits RAR Related Orphan Receptor A (Rora), which results in the activation of NF-κB signaling. miR-21 suppresses phosphatase and tensin homolog (PTEN) expression and activates PI3K/Akt pathway in MDSCs. |
| miR-9 and miR-181a [230] | Breast cancer | MDSCs | miR-9 and miR-181a target suppressor of cytokine signaling-3 (SOCS3) and protein inhibitors of activated STAT 3 (PIAS3), respectively. SOCS3 and PIAS3 inhibition activates downstream JAK/STAT signaling associated with MDSC development and their suppressive effects on T cells. |
| miR-1246 [231] | Colon cancer | macrophages | M2 macrophages after receiving miR-1246 from p53 mutant colon cancer cells promote the development of large tumors and metastasis. miR-1246 expressing TAMs have enhanced TGFβ signaling which increases Treg population in mouse tumors and promote immune suppression. |
| miR-23a-3p [232] | Hepatocellular Carcinoma (HCC) | macrophages | Endoplasmic stress in HCC cells increases miR-23a-3p in HCC-derived exosomes. miR-23a-3p inhibits PTEN in recipient macrophages and induces PDL1 expression via the Akt pathway. PDL1 expressing macrophages decrease CD8+ T-cell ratio and promote T-cell apoptosis. |
| miR-208b [233] | Colorectal cancer (CRC) | T cells | miR-208b directly binds and inhibits programmed cell death factor 4 (PDCD4) in CD4+ T regs, which promotes their expansion. An increase in the percentage of Tregs promotes CRC growth and oxaliplatin resistance. |
| miR-21 [234] | EC | monocytes | EC cells transfer miR-21 to monocytes under hypoxic conditions, which transforms monocytes into M2 macrophages. |
| miR-107 [235] | GC | MDSC | miR-107 targets 3’UTRs of DICER and tumor suppressor gene PTEN in recipient HLA-DR−CD33+MDSCs. DICER downregulation promotes MDSCs expansion, whereas PTEN inhibition upregulates the PI3Kinase pathway and promotes proliferation. |
| Exosomal lncRNAs | |||
| TUC339 [236] | HCC | THP1 monocytes | TUC339 overexpression decreases production of proinflammatory cytokines IL-1β and TNF-α, T cell activator CD86 expression and phagocytic activity in THP1 cells. |
| RPPH1 [237] | Colorectal cancer (CRC) | macrophages | RPPH1 increases the expression of M2 macrophage markers CCL17, CCL18, CXCL8, IL-10, and TGF-β. M2 polarized macrophages promote CRC proliferation and metastasis. |
| SNHG16 [238] | Breast cancer | Vδ+T cells | SNHG16 sponges miR-15–5p, which blocks its inhibitory interaction with SMAD5 mRNA. SMAD5 activation induces CD73 expression. Tregs found in BC are enriched with CD73+γδ1 T cells and exert immunosuppressive functions via the adenosine pathway. |
| CRNDE-h [239] | CRC | CD4+ T cells | CRNDE-h binds RAR-related orphan receptor gamma (RORγT) and inhibits its binding with E3 ubiquitin ligase Itch. This prevents Itch mediated RORγT ubiquitination and degradation, which increases RORγT expression. RORγT bind to IL-17 promotor and triggers CD4 + T cell differentiation into immunosuppressive IL-17 producing Th17 cells. |
2.3.2: LncRNAs:
LncRNAs regulate gene expression through several different mechanisms, including influencing chromatin regulation by recruiting or binding to regulatory factors, controlling mRNA or protein stability, and regulating RNA and protein functions through direct or indirect interactions [139]. LncRNA mediated communication in the TME plays an important role in immune escape mechanisms and can interfere with the response to immunotherapy. For example, lncRNA ENST00000560647 secreted from pancreatic cancer cells inhibits DC-mediated antigen presentation and promotes escape from immune surveillance by CD8+ T cells [140]. Exosomal lncRNAs PCAT-1 is associated with immune suppression and lymph node metastasis in chemoresistant Kras mutant lung cancer patients [141]. These patients show higher serum concentration of PCAT-1 than Kras WT patients, which is associated with disease progression [141, 142]. PCAT-1 upregulation induces immunosuppressive miRNAs (miR-182 and miR-217) that are linked with CAF infiltration, an increased number of lymph node metastatic foci, and enhanced tumor growth [138].
Another lncRNA, HIF-1α-stabilizing long noncoding RNA (HISLA), which is secreted from TAMs in EVs, promotes aerobic glycolysis in breast cancer cells and is associated with disease progression, lymph node metastasis, and a dampened response to neoadjuvant chemotherapy in breast cancer patients [143]. TAM secreted HISLA binds to prolyl hydroxyl 2 (PHD2) and inhibits its binding with HIF-1α, preventing ubiquitin-mediated HIF-1α degradation. HIF-1α stabilization promotes aerobic glycolysis and induces chemotherapeutic resistance in breast cancer cells. Also, as a feedback mechanism, lactate released from tumor cells in the TME upregulates HISLA and promotes aerobic glycolysis in TAMs [143]. Aerobic glycolysis helps sustain TAMs in the hypoxic TME and is required to maintain their tumor-promoting M2-like profile [144, 145]. Interestingly, TAM promotes tumor hypoxia by upregulating AMP-activated protein kinase (AMPK) and mitochondrial biogenesis protein, and peroxisome proliferator-activated receptor-gamma coactivator (PGC-1a), which collectively increases oxygen consumption. TAM-induced hypoxia increases glycolysis and inhibits CD4 and CD8a Tcell infiltration in tumors [146]. Moreover, bladder cancer cells cultured under hypoxic conditions release lncRNA urothelial cancer-associated 1 (UCA1) in exosomes to the surrounding microenvironment. Cellular uptake of UCA1 by surrounding cancer cells facilitates their growth, migration, and invasion [147].
NSCLC-derived exosomes contain lncRNA SOX2, which promotes M2 macrophage polarization and induces resistance to EGFR‐tyrosine kinase inhibitors (TKIs) in NSCLC cancer cells. SOX2 sponges miR-627 in THP-1 macrophages, upregulating its target genes Smad2, Smad3, and Smad4, which induce M2 polarization [148]. Likewise, lncRNA distal-less homeobox 6 antisense 1 (DLX6-AS1) released from HCC exosomes induces M2 macrophage polarization by targeting miR-15a, which upregulates its target gene C-X-C Motif Chemokine Ligand 17 (CXCL17). CXCL17 upregulation promotes M2 macrophage polarization and supports migration, invasion, and EMT in HCC cells [148]. This suggests that lncRNA exchange in the TME contributes substantially to the induction of M2 macrophage polarization, which provides immunosuppressive tumor microenvironment (TME) to support tumor growth. Furthermore, lncRNA profiling in the exosomes from healthy subjects and NSCLC patients found that lncRNA metastasis associated lung adenocarcinoma transcript 1 (MALAT1) is highly expressed in exosomes and positively correlated with tumor stage and lymphatic metastasis [149]. Exosomal MALAT1 secreted from lung cancer cells inhibits phagocytosis in dendritic cells and slows down T cell proliferation [150]. MALAT-1 is suppressed during early stages of CD4+ T cell activation and its complete deletion induces robust CD4+T cell-mediated immune response to infection [151]. MALAT-1 positively regulates proto-oncogene Maf, a transcription factor of IL-10 in Th1 cells. IL-10 is an immunosuppressive cytokine, abundantly present in the TME and associated with worse outcomes in cancer patients [151–153].
CAFs secrete lncRNA Nuclear Enriched Abundant Transcript 1 (NEAT) in exosomes, contributing to growth and proliferation by recipient endometrial carcinoma (EC) cells. NEAT is more highly expressed in CAFs than normal fibroblasts. NEAT sponges miR-26a/b in EC cells, which increases the expression of its target proteins STAT-3 and STAT-3 regulated protein YKL-40. STAT-3 and YKL-40 are tumorigenic proteins involved in the tumor progression and anti-tumor immune response [154]. Table 1 shows some other exosomal lncRNAs and their role in immune suppression. The impact of cancer cell-derived exosomal non-coding RNAs on immune evasion is presented in Figure 2.
Figure 2:
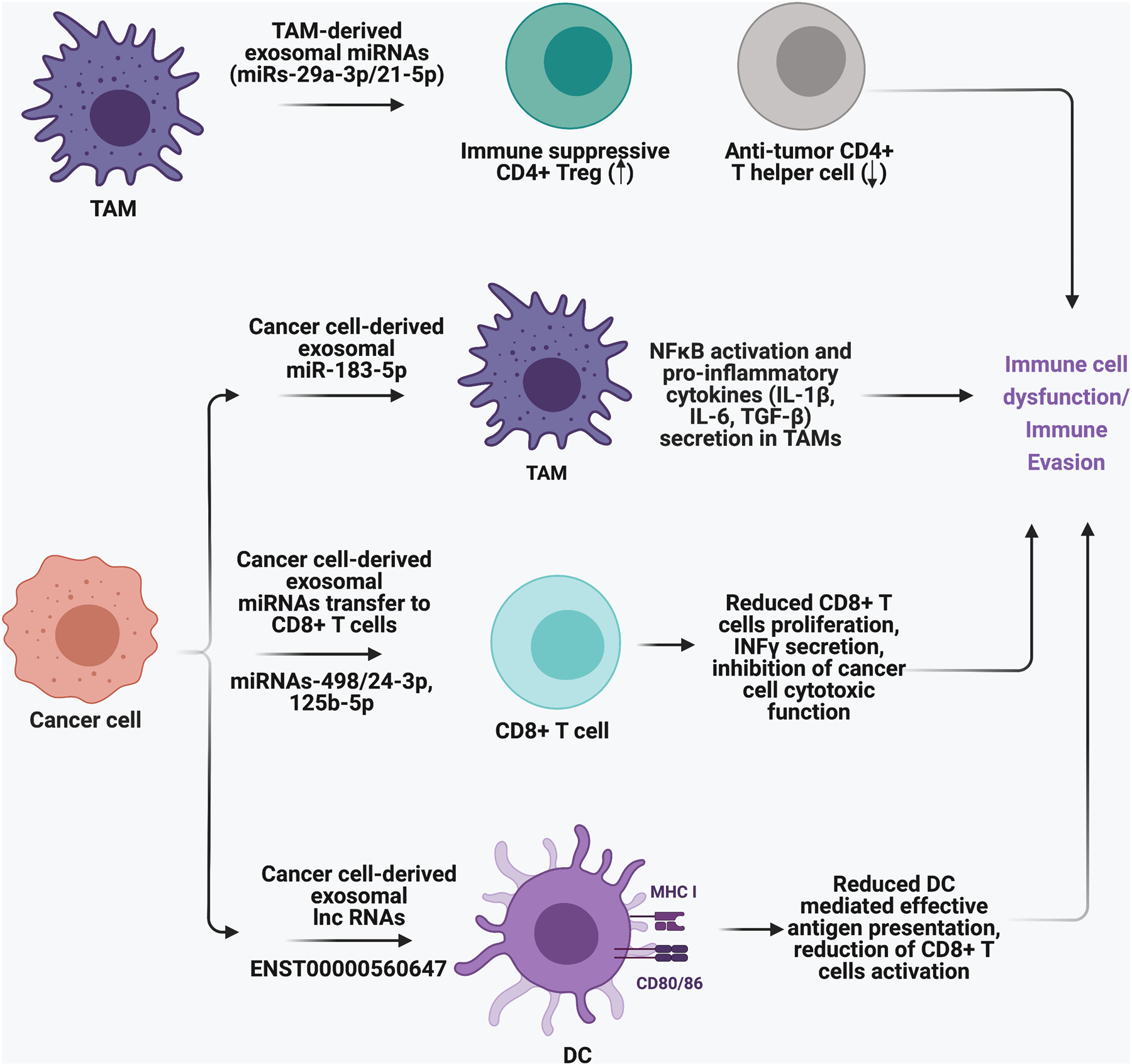
The impact of cancer cell-derived exosomal non-coding RNAs on immune evasion.
2.3.3: Ultraconserved RNAs (Uc.RNAs):
The transcribed ultraconserved regions of the genome encode a novel class of long non-coding RNA(s), called ultraconserved RNAs (Uc.RNAs) that interfere with the function of miRNAs through RNA-RNA interactions [84, 139, 155–159]. Uc.RNAs show distinct expression signatures in cancer compared to normal cells, and its deregulation is observed in many cancer types. TUC339, one of the first Uc.RNAs discovered in exosomes, is involved in regulating recipient cell functions. HCC derived exosomes contain high levels of TUC339, which promotes cell proliferation and invasion in recipient cancer cells [157, 160, 161]. Another uc.RNA, Uc.889 secreted from esophageal squamous cell carcinoma (ESCC) cells increases endothelial cell (EC) proliferation and tube formation capacity, thus promoting lymphangiogenesis. Uc.889 binds to EPH Receptor A2 (EPHA2) mRNA in the recipient EC cells and induces its degradation, which increases its target gene p38MAPK, an essential regulator of lymph angiogenesis [162]. This suggests that exosomal Uc.RNAs can potentiate tumorigenic capacities of the recipient cells in the TME. There are not many reports about the role of exosomal uc.RNAs in influencing TME, suggesting that the field is still at an early stage. However, uc.RNAs have been described to play an important role in promoting growth in many cancer types. For example, overexpression of uc.138 encoded from human transformer 2β gene (TRA2B) increases proliferation and migration in colon cancer cells and induces drug resistance [163]. Uc.63 upregulates NF-kB signaling in GC cells and facilitates GC growth and progression [164]. In addition, knockdown of Uc.63 increases cisplatin sensitivity in urothelial carcinoma (UC) cells, which is associated with low androgen receptor (AR) activity [165]. Moreover, uc.339 acts as a decoy RNA for miR-339–3p, miR-663b-3p, and miR-95–5p and mask their functions in NSCLC cells. Downregulation of miR-339–3p, miR-663b-3p, and miR-95–5p upregulates their target oncogene cyclinE2, which promotes cancer growth and proliferation [156].
2.3.4: Circular (circRNAs):
CircRNAs are a type of non-coding RNAs in which 3’ and 5’ ends of a linear RNA molecule are covalently joined to form a closed single-stranded circular loop [166]. Due to this closed-loop structure, there are no free terminal ends in the circRNAs, which protects them from degradation by ribonucleases and provides high stability [167]. Cancer cells secrete circRNAs to influence their interaction with the microenvironment components, including immune cells. HCC cells transfer circ_0074854 to macrophages via exosomes to induce M2 macrophage polarization. Circ_0074854 polarized M2 macrophages promote HCC cell growth, migration, invasion, and EMT. In addition, Circ_0074854 interacts and stabilizes RNA binding protein HuR in HCC cells, supporting their growth [168]. Another HCC-secreted circRNA, circular ubiquitin-like with PHD and ring finger domain 1 RNA (circUHRF1), induces NK cell dysfunction and is associated with poor clinical outcomes in HCC patients. CircUHRF1 binds miR-449c in the recipient NK cells and sponges its activity, upregulating its miR-449c target mRNA T cell immunoglobulin and mucin domain-containing protein3 (TIM3) [169]. TIM3 is an inhibitory receptor on NK and T cells, and its downregulation improves NK cell-mediated cytotoxicity in human cancers [170]. Moreover, CircUHRF1 overexpression significantly decreases NK-cell infiltration in HCC tumors and increases resistance to anti-PD1 therapy [169].
Furthermore, circ-CPA4 stabilizes PD-L1 in NSCLC cells, inducing CD8+ T cell inactivation and promoting tumor growth and chemoresistance. Circ-CPA4 acts as a Competing endogenous RNA (CeRNA) for let7 miRNA by masking its inhibitory binding with the 3’UTR of PDL1 mRNA. This increases exosomal levels of PD-L1, which inhibits CD8+T cell functions and promotes NSCLC growth, EMT, and cisplatin insensitivity[171]. Likewise, circRNA-002178 stabilizes PDL1 expression in lung adenocarcinoma (LUAD) cells by sponging miR-34, thus hampering the anti-tumor CD8+ T cell response. LUAD cells use exosomes to transfer circRNA-002178, also a sponge for miR-28, into CD8+ T cells. As a result, miR-28 target gene PD1 is upregulated in CD8+ T cells, making them more vulnerable to PDL1 mediated inhibition [172]. This implies that circRNA-002178 has a dual mechanism of action in inducing anti-tumor immunity by regulating PDL1 in cancer cells and PD1 in immune cells. In conclusion, the role of exosomal cirRNAs in cancer and anti-tumor immunity are beginning to come into focus; they are important players in TME communication and could be potential therapeutic targets in cancer therapy.
3. Role of exosomes in cancer immunotherapy
EVs or exosomes can act as a friend or foe in cancer treatment. Blockage of crucial exosome-mediated communication in the TME slows down tumor progression [173, 174], inhibits reprogramming of tumor microenvironment [76, 175–177], and improves drug efficacy by overcoming drug resistance [178, 179]. In contrast, exosomes secreted by activated immune cells carrying tumor-inhibitory molecules can be used to stimulate anti-tumor immune responses in cold tumors surrounded by the suppressed immune microenvironment. Exosomes are highly efficient delivery vehicles because of their biocompatibility, high specificity for the target, and low clearance [180, 181]. Exosomes can target tumor cells for delivery of various therapeutic molecules like anti-tumor cytokines IL-2 and IFN-α, agonists of costimulatory immune receptors, tumor antigens with adjuvants, tumor-suppressive or immune-stimulating miRNAs, or proteins, etc. Therefore, the use of exosomes as immunotherapeutic delivery systems could overcome significant limitations of cancer immunotherapy[182]. This section will discuss dual applications of exosomes in cancer therapeutics, as a druggable cancer target and also a promising delivery vehicle by which to target tumor cells.
3.1. Exosomes as a therapeutic target:
Inhibition of exosome secretion via knockdown of Ras-related protein (Ral) family members RalA and RalB significantly decreases the metastatic potential of breast tumor cells [183]. RalA and RalB depleted cells have altered MVB biogenesis and secrete fewer EVs than their wild-type (WT) counterparts. These EVs have low vascular permeability and do not promote pre-metastatic niche formation in the lungs of tumor-bearing mice [183]. Exosomes isolated from chemoresistant K-Ras positive lung cancer patients contain chromatin remodeling proteins involved in cancer progression, tumor growth promotion and lymph node metastasis. Co-treatment of Kras and exosome inhibitors suppresses tumor progression and decreases immunosuppressive miR-146 in lung cancer tissues which is associated with improved (Th-1)-mediated immune response [176]. Furthermore, inhibition of exosome-mediated signaling using exosome secretion inhibitor GW4869 or Rab27a knockdown inhibits breast tumor growth and sensitizes tumors to anti-PDL1 therapy in a mouse model. Interestingly, similar inhibition of tumor growth is not seen in immunodeficient nude mice under identical experimental conditions suggesting that exosome targeting induces immune-mediated cancer cell killing [184].
Biswas and colleagues have shown that exosomes secreted from MSCs promote the accumulation of immunosuppressive CD206+ M-MDSCs and CD206+ M2-macrophages in TME. MSC-derived exosomes induce PDL1 expression in these cells, which is associated with enhanced tumor growth, accumulation of immunosuppressive factors like TGF-β, high MSC infiltration and reduction in tumor-killing INFγ+CD8+ T cells. Treatment of MSCs with GW4869 inhibits the upregulation of PDL1 in macrophages and M2-macrophage polarization [116]. Moreover, studies have screened pharmacologically active compounds or approved drugs to identify inhibitors of exosome biogenesis and secretion for potential use in cancer treatment [173, 185]. One such compound is calpeptin which selectively inhibits calpains, a calcium-dependent cysteine protease involved in vesicle secretion. Calpeptin inhibits vesicle secretion in prostate cancer cells treated with docetaxel and increases drug accumulation inside cancer cells, significantly improving the therapeutic response [186]. The other EV or exosome inhibitors that have shown anticancer activity in different cancer types are Y27632 (competitive inhibitor of both Rho-associated protein kinases (ROCK) 1 and 2)[187, 188], manumycin (Ras farnesyltransferase inhibitor)[185, 189], pantethine (derivative of pantothenic acid, also known as vitamin B5)[190, 191] and tipifarnib (farnesyltransferase inhibitor) [173, 192].
3.2. Exosome use to overcome immunotherapy resistance:
Immune cells rely on immunogenic epitopes to recognize cancer cells as foreign. However, high mutational burden, low or no immunogenicity, exhaustion of immune cells, and overexpression of immune checkpoint proteins are primary reasons immunotherapy fails. Exosomes have the ability to modulate and potentiate innate and adaptive immune responses against tumor cells. For example, tumor peptide pulsed DC-derived exosomes can prime and stimulate T and NK cells against various cancers [193–196].
Phase-II clinical studies of combination therapy with DC exosomes and chemotherapy in NSCLC patients show promising improvement in immune cell response and overall survival [197, 198]. Moreover, exosomes prepared from activated DCs pulsed with ovalbumin and modified with anti-CTLA antibody induce a robust activation of effector and memory T cells and shows high anti-tumor activity compared to untreated DC exosomes [199]. In normal cells, CTLA-4 on T cells binds to APCs B7 receptors to turn off the T-cell mediated immune response; otherwise, it can lead to the development of antiproliferative or autoimmune diseases [200, 201]. In tumors, the CTLA4-B7 signaling is overexpressed, which keeps cancer cells away from the reach of T cells [202]. Further, blockade of CTLA-4 has been shown to boost T cell activation and memory in a poor immunogenic spontaneous murine tumor model. The genetic modification of the costimulatory molecule’s overexpression on leukemia cells enhances anti-leukemia immunity [203, 204].
Exosomes derived from leukemia cells modified to express two B7 costimulatory molecules (CD80, CD86) significantly upregulated the expression of B7 receptors on DCs. B7 upregulation is associated with improved DC function, enhanced CTL response, and better overall survival in leukemic mice [205]. This suggests that exosome use along with immune check point inhibitors could be a promising innovative treatment to overcome lower expression of costimulatory molecules in the cold tumors. The other resistance mechanism associated with anti–CTLA-4 therapy is the upregulation of other immune checkpoint molecules, including V-domain Ig suppressor of T cell activation (VISTA) and PDL1 [206]. Chimeric antigen receptor T (CART) cell-derived exosomes can be used to overcome PDL1 induced resistance to anti-tumor immunity. Fu and colleagues have shown that exosomes derived from CART cells carry CAR on their surface and do not express PD1, which prevents them from being suppressed by PD-L1 [207]. Therefore, compared to CART cells, CART-derived exosomes are more effective in inducing anti-tumor activity and do not show significant loss of cytolytic activity in the presence of PDL1 overexpression [207]. Due to low risk of toxicity and their high capacity to penetrate solid tumor mass compared to CART cells, exosomes might be a better option to overcome therapeutic efficiency limitations against resistant tumors.
Furthermore, loss or low expression of antigen-presenting machinery (APM) components like MHC-I or β2microglobulin on APCs or cancer cells are frequently observed in many tumor types [208–210]. Loss of antigen presentation is one of the common immune escape mechanisms, which plays an important role in immunotherapy resistance. Exosomes can be used to load and transfer APM components to APCs in the TME. Exosomes purified from DCs and directly loaded with tumor-derived MHC-I or II-restricted peptides can effectively transfer tumor peptides to APCs and elicit potent antigen-specific CD8+ T cells in vitro [211]. Besides, exosomes derived from B16F1 murine melanoma cells expressing MHC-II transactivator (encoded by gene CIITA)(B16F1-CIITA) trigger potent tumor-specific CD4+ Th1 type cell-mediated immune responses against melanoma cells. Compared to B16F1-derived exosomes, B16F1-CIITA exosomes are more effective in delaying tumor growth and inducing inflammatory cytokines, including TNF-a, chemokine receptor CCR7, and Th1-polarizing cytokine IL-12 [212]. Other approaches used to stimulate antigen presentation by exosomes include use of immunostimulatory biotinylated CpG DNA along with tumor antigens [213], introduction of synthetic double stranded analog poly(I:C), a ligand for toll-like receptor 3 (TLR3) into antigen expressing exosomes [214], co-delivery of tumor-derived exosomes with α-galactosylceramide loaded DCs [215] or anchoring exosomes with superparamagnetic magnetite colloidal nanocrystal clusters (SMCNCs), which greatly enhance tumor targeting potential and can be controlled by using magnetic fields [216].
3.3. Clinical trials using exosome therapy:
In numerous in vitro and in vivo studies, exosomes have proved to be efficient delivery vehicles, which can effectively target tumor cells and generate a robust anti-tumor immune response. Due to the success of exosomes as cancer therapeutics in preclinical models, many different clinical trials of exosome-based therapies are either in process or completed. Table. 2 summarizes some clinical trials using exosomes in immunotherapy, for prognosis prediction, and to monitor treatment response.
Table.2:
Clinical trials using exosomes in cancer immunotherapy
| Identifier | Cancer type | Exosome source | Treatment strategy |
|---|---|---|---|
| NCT01159288 | NSCLC | Tumor antigen-loaded DC-derived exosomes | Metronomic cyclophosphamide (mCTX) treatment followed by vaccinations with exosomes. Findings: mCTX inhibits immunosuppressive Treg fucntions and exosomes activate innate and adaptive immunity. |
| NCT03985696 | Non-Hodgkin B-cell lymphoma | Exosomes purified from patients and cancer cells | CD20 and PDL1 expression in exosomes are analyzed to study their prognostic significance on patient outcome and rituximab response. |
| NCT02869685 | NSCLC | Plasma | PDL1 levels in plasma exosomes are analyzed after 24h and 48h of radiotherapy. If PDL1 is upregulated, radiotherapy can be combined with immunotherapy to increase treatment efficacy. |
| NCT02977468 | Triple negative breast cancer | Serum | Testing if pembrolizumab treatment alters the expression of immune tolerance markers like PDL1 in the primary tumor, circulating lymphocytes, and serum exosomes. |
| NCT04427475 | NSCLC | Plasma | Exosomal PDL1 and mRNAs are analyzed in stage IV EGFR/ALK wild-type NSCLC patients before and after treatment with anti-PD1 (pabolizumab or nafulizumab) |
| NCT02507583 | Malignant glioma | Tumor cells | Patients’ tumor cells are taken, treating them with investigational drug insulin-like growth factor receptor-1 Antisense Oligodeoxynucleotide (IGF-1R/AS ODN) and reimplanting into patients. As the tumor cells die inside the patient, they release tumor antigen containing exosomes, which can activate immune cells. |
| NCT03854032 | Stage II-IV head and neck squamous cell cancer | Peripheral blood | Patients are treated with Nivolumab and BMS986205, an indoleamine 2, 3-dioxygenase 1 inhibitor (IDO1i). Exosomes abundance and composition before and after therapy are analyzed. |
| NCT02535247 | NK and T cell non-Hodgkin lymphoma | Peripheral blood | Patients are treated with pembrolizumab alone or combined with copanlisib. Exosomes are profiled for PDL1 expression. |
3.4. Potential use of exosomes as cancer immunotherapy biomarkers:
In recent years, immunotherapy has emerged as a frontline therapy to treat many types of cancer because of its potential to induce anti-cancer immunological memory, which has longer-lasting effects than traditional therapies, and its use as personalized medicine [217, 218]. However, despite a durable response in cancer patients, the success rate of immunotherapy is low, about 15–20% [219]. One of the potential reasons for this is the lack of appropriate candidate biomarkers that can be used to determine patients who could benefit from the particular type of immunotherapy. For example, previously treated NSCLC patients who express PD-L1 benefit from pembrolizumab, a monoclonal antibody that targets PD-1, with a tumor objective response rate of 19.4% [220]. Likewise, patients showing PD-L1 upregulation by either tumor cells or tumor-infiltrating cells in multiple cancer types are more responsive to anti-PDL1 antibody (MPDL3280A) than others [221]. This implies that PDL1 levels as a biomarker may be used to identify populations who are likely to receive clinical benefit from anti-PD-L1 immunotherapy. The other classes of biomarkers being studied in cancer immunotherapy are tumor-associated proteins like HER2 [222], serum proteins like NY-ESO-1 [223], circulating immune and tumor cells [224], mismatch repair deficiency (dMMR) [225], and tumor mutational burden (TMB) [226]. Molecular characterization of exosomes from different cancer types reveals the presence of these biomarkers, suggesting that exosomes may be used to analyze cancer-specific changes and predict response to the treatment. For instance, liquid biopsy studies of advanced tumors demonstrate that DNA analysis of exosomes can better determine TMB than circulating tumor DNA (ctDNA) [227]. The combined molecular profiling of exosomal RNA and circulating tumor ctDNA isolated from cancer patients’ plasma detects a higher number of gene mutations than ctDNA and is more accurate in identifying patients who are likely to respond to therapy. High TMB in cancer patients is associated with a higher response rate to immunotherapy because more mutations or neoantigens are present for the immune cells to respond to [227]. Furthermore, other exosomal constituents like miRNAs show differential expression in cancer patients compared to healthy individuals and differentiate patients by their likelihood to respond to therapy. Low levels of exosome-derived miR-320d, miR-320c, and miR-320b are found in NSCLC patients with EGFR/ALK mutations who responded well to anti-PD1 treatment compared to non-responders or patients with progressive disease [138]. Moreover, a plasma-based miRNA signature composed of 24 miRNAs along with PD-L1 tumor expression could predict risk levels like overall response rate (ORR), progression-free survival (PFS), and overall survival (OS) in NSCLC patients treated with immune-checkpoint inhibitors (ICI) [228].
4. Concluding remarks:
Exosomes are a vital medium of communication among cells and are actively involved in exchanging information between cancer and other TME cells. Tumor cells transform their microenvironment in ways that cause immune cells either to be ineffective and not recognize tumor growth or to show pro-tumor activity. This affects the therapeutic efficacy of cancer immunotherapy because tumor cells circumvent immune responses in many different ways. In this review, we have aimed to explore the role of exosomal constituents secreted by cancer or TME cells in shaping and making tumor-promoting microenvironments. An appreciation for exosome-mediated signaling in the TME can help in explaining the complexity and heterogeneity of tumors. There is a need to better understand the exosomal constituents and their role in forming cold tumors where immune cells are a slave to their master cancer cells. Inside tumors, there is an intricate network of multicellular communication through exosomes. Therefore, exosome heterogeneity which is defined by cell type of origin, cellular conditions, molecular content, and functional impact on recipient cells, is an essential concern in TME. Different immune and non-immune cells in the TME influence each other through exosomes in ways affecting cancer growth and immune escape. This has a significant impact on tumor response to therapy and drug resistance. Hence, there is a need to better characterize the exosome-mediated signaling network in the TM and its role in cancer-mediated immune suppression and therapy resistance.
Acknowledgments:
Dr. Challagundla’s laboratory is supported in whole or part by NIH/NCI grant CA197074; the State of Nebraska and the Pediatric Cancer Research Group, part of the Child Health Research Institute, and the Department of Biochemistry & Molecular Biology start-up grants. The authors would like to thank Matthew Sandbulte, PhD of the Child Health Research Institute at Children’s Hospital & Medical Center and the University of Nebraska Medical Center for editorial assistance. Figures were prepared using BioRender.com tool.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: The authors declare no conflict of interest.
4. References
- [1].Skotland T, Sandvig K, Llorente A, Lipids in exosomes: Current knowledge and the way forward, Prog Lipid Res, 66 (2017) 30–41. [DOI] [PubMed] [Google Scholar]
- [2].Pathania AS, Challagundla KB, Exosomal Long Non-coding RNAs: Emerging Players in the Tumor Microenvironment, Molecular Therapy Nucleic Acids, (2020). [DOI] [PMC free article] [PubMed]
- [3].Kalluri R, LeBleu VS, The biology, function, and biomedical applications of exosomes, Science, 367 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Thery C, Zitvogel L, Amigorena S, Exosomes: composition, biogenesis and function, Nat Rev Immunol, 2 (2002) 569–579. [DOI] [PubMed] [Google Scholar]
- [5].Johnstone RM, Bianchini A, Teng K, Reticulocyte maturation and exosome release: transferrin receptor containing exosomes shows multiple plasma membrane functions, Blood, 74 (1989) 1844–1851. [PubMed] [Google Scholar]
- [6].Lan Y, Jin Q, Xie H, Yan C, Ye Y, Zhao X, Chen Z, Xie Z, Exosomes Enhance Adhesion and Osteogenic Differentiation of Initial Bone Marrow Stem Cells on Titanium Surfaces, Front Cell Dev Biol, 8 (2020) 583234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Takahashi A, Okada R, Nagao K, Kawamata Y, Hanyu A, Yoshimoto S, Takasugi M, Watanabe S, Kanemaki MT, Obuse C, Hara E, Exosomes maintain cellular homeostasis by excreting harmful DNA from cells, Nat Commun, 8 (2017) 15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Muralidharan-Chari V, Clancy JW, Sedgwick A, D’Souza-Schorey C, Microvesicles: mediators of extracellular communication during cancer progression, J Cell Sci, 123 (2010) 1603–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Haraszti RA, Didiot MC, Sapp E, Leszyk J, Shaffer SA, Rockwell HE, Gao F, Narain NR, DiFiglia M, Kiebish MA, Aronin N, Khvorova A, High-resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources, J Extracell Vesicles, 5 (2016) 32570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Johnson SM, Dempsey C, Chadwick A, Harrison S, Liu J, Di Y, McGinn OJ, Fiorillo M, Sotgia F, Lisanti MP, Parihar M, Krishnan S, Saha V, Metabolic reprogramming of bone marrow stromal cells by leukemic extracellular vesicles in acute lymphoblastic leukemia, Blood, 128 (2016) 453–456. [DOI] [PubMed] [Google Scholar]
- [11].Di Vizio D, Morello M, Dudley AC, Schow PW, Adam RM, Morley S, Mulholland D, Rotinen M, Hager MH, Insabato L, Moses MA, Demichelis F, Lisanti MP, Wu H, Klagsbrun M, Bhowmick NA, Rubin MA, D’Souza-Schorey C, Freeman MR, Large oncosomes in human prostate cancer tissues and in the circulation of mice with metastatic disease, Am J Pathol, 181 (2012) 1573–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Johnson SM, Dempsey C, Parker C, Mironov A, Bradley H, Saha V, Acute lymphoblastic leukaemia cells produce large extracellular vesicles containing organelles and an active cytoskeleton, J Extracell Vesicles, 6 (2017) 1294339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Thery C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin-Smith GK, Ayre DC, Bach JM, Bachurski D, Baharvand H, Balaj L, Baldacchino S, Bauer NN, Baxter AA, Bebawy M, Beckham C, Bedina Zavec A, Benmoussa A, Berardi AC, Bergese P, Bielska E, Blenkiron C, Bobis-Wozowicz S, Boilard E, Boireau W, Bongiovanni A, Borras FE, Bosch S, Boulanger CM, Breakefield X, Breglio AM, Brennan MA, Brigstock DR, Brisson A, Broekman ML, Bromberg JF, Bryl-Gorecka P, Buch S, Buck AH, Burger D, Busatto S, Buschmann D, Bussolati B, Buzas EI, Byrd JB, Camussi G, Carter DR, Caruso S, Chamley LW, Chang YT, Chen C, Chen S, Cheng L, Chin AR, Clayton A, Clerici SP, Cocks A, Cocucci E, Coffey RJ, Cordeiro-da-Silva A, Couch Y, Coumans FA, Coyle B, Crescitelli R, Criado MF, D’Souza-Schorey C, Das S, Datta Chaudhuri A, de Candia P, De Santana EF, De Wever O, Del Portillo HA, Demaret T, Deville S, Devitt A, Dhondt B, Di Vizio D, Dieterich LC, Dolo V, Dominguez Rubio AP, Dominici M, Dourado MR, Driedonks TA, Duarte FV, Duncan HM, Eichenberger RM, Ekstrom K, El Andaloussi S, Elie-Caille C, Erdbrugger U, Falcon-Perez JM, Fatima F, Fish JE, Flores-Bellver M, Forsonits A, Frelet-Barrand A, Fricke F, Fuhrmann G, Gabrielsson S, Gamez-Valero A, Gardiner C, Gartner K, Gaudin R, Gho YS, Giebel B, Gilbert C, Gimona M, Giusti I, Goberdhan DC, Gorgens A, Gorski SM, Greening DW, Gross JC, Gualerzi A, Gupta GN, Gustafson D, Handberg A, Haraszti RA, Harrison P, Hegyesi H, Hendrix A, Hill AF, Hochberg FH, Hoffmann KF, Holder B, Holthofer H, Hosseinkhani B, Hu G, Huang Y, Huber V, Hunt S, Ibrahim AG, Ikezu T, Inal JM, Isin M, Ivanova A, Jackson HK, Jacobsen S, Jay SM, Jayachandran M, Jenster G, Jiang L, Johnson SM, Jones JC, Jong A, Jovanovic-Talisman T, Jung S, Kalluri R, Kano SI, Kaur S, Kawamura Y, Keller ET, Khamari D, Khomyakova E, Khvorova A, Kierulf P, Kim KP, Kislinger T, Klingeborn M, Klinke DJ 2nd, Kornek M, Kosanovic MM, Kovacs AF, Kramer-Albers EM, Krasemann S, Krause M, Kurochkin IV, Kusuma GD, Kuypers S, Laitinen S, Langevin SM, Languino LR, Lannigan J, Lasser C, Laurent LC, Lavieu G, Lazaro-Ibanez E, Le Lay S, Lee MS, Lee YXF, Lemos DS, Lenassi M, Leszczynska A, Li IT, Liao K, Libregts SF, Ligeti E, Lim R, Lim SK, Line A, Linnemannstons K, Llorente A, Lombard CA, Lorenowicz MJ, Lorincz AM, Lotvall J, Lovett J, Lowry MC, Loyer X, Lu Q, Lukomska B, Lunavat TR, Maas SL, Malhi H, Marcilla A, Mariani J, Mariscal J, Martens-Uzunova ES, Martin-Jaular L, Martinez MC, Martins VR, Mathieu M, Mathivanan S, Maugeri M, McGinnis LK, McVey MJ, Meckes DG Jr., Meehan KL, Mertens I, Minciacchi VR, Moller A, Moller Jorgensen M, Morales-Kastresana A, Morhayim J, Mullier F, Muraca M, Musante L, Mussack V, Muth DC, Myburgh KH, Najrana T, Nawaz M, Nazarenko I, Nejsum P, Neri C, Neri T, Nieuwland R, Nimrichter L, Nolan JP, Nolte-’t Hoen EN, Noren Hooten N, O’Driscoll L, O’Grady T, O’Loghlen A, Ochiya T, Olivier M, Ortiz A, Ortiz LA, Osteikoetxea X, Ostergaard O, Ostrowski M, Park J, Pegtel DM, Peinado H, Perut F, Pfaffl MW, Phinney DG, Pieters BC, Pink RC, Pisetsky DS, Pogge von Strandmann E, Polakovicova I, Poon IK, Powell BH, Prada I, Pulliam L, Quesenberry P, Radeghieri A, Raffai RL, Raimondo S, Rak J, Ramirez MI, Raposo G, Rayyan MS, Regev-Rudzki N, Ricklefs FL, Robbins PD, Roberts DD, Rodrigues SC, Rohde E, Rome S, Rouschop KM, Rughetti A, Russell AE, Saa P, Sahoo S, Salas-Huenuleo E, Sanchez C, Saugstad JA, Saul MJ, Schiffelers RM, Schneider R, Schoyen TH, Scott A, Shahaj E, Sharma S, Shatnyeva O, Shekari F, Shelke GV, Shetty AK, Shiba K, Siljander PR, Silva AM, Skowronek A, Snyder OL 2nd, Soares RP, Sodar BW, Soekmadji C, Sotillo J, Stahl PD, Stoorvogel W, Stott SL, Strasser EF, Swift S, Tahara H, Tewari M, Timms K, Tiwari S, Tixeira R, Tkach M, Toh WS, Tomasini R, Torrecilhas AC, Tosar JP, Toxavidis V, Urbanelli L, Vader P, van Balkom BW, van der Grein SG, Van Deun J, van Herwijnen MJ, Van Keuren-Jensen K, van Niel G, van Royen ME, van Wijnen AJ, Vasconcelos MH, Vechetti IJ Jr., Veit TD, Vella LJ, Velot E, Verweij FJ, Vestad B, Vinas JL, Visnovitz T, Vukman KV, Wahlgren J, Watson DC, Wauben MH, Weaver A, Webber JP, Weber V, Wehman AM, Weiss DJ, Welsh JA, Wendt S, Wheelock AM, Wiener Z, Witte L, Wolfram J, Xagorari A, Xander P, Xu J, Yan X, Yanez-Mo M, Yin H, Yuana Y, Zappulli V, Zarubova J, Zekas V, Zhang JY, Zhao Z, Zheng L, Zheutlin AR, Zickler AM, Zimmermann P, Zivkovic AM, Zocco D, Zuba-Surma EK, Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines, J Extracell Vesicles, 7 (2018) 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Poon IKH, Parkes MAF, Jiang L, Atkin-Smith GK, Tixeira R, Gregory CD, Ozkocak DC, Rutter SF, Caruso S, Santavanond JP, Paone S, Shi B, Hodge AL, Hulett MD, Chow JDY, Phan TK, Baxter AA, Moving beyond size and phosphatidylserine exposure: evidence for a diversity of apoptotic cell-derived extracellular vesicles in vitro, J Extracell Vesicles, 8 (2019) 1608786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wickman G, Julian L, Olson MF, How apoptotic cells aid in the removal of their own cold dead bodies, Cell Death Differ, 19 (2012) 735–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Caruso S, Poon IKH, Apoptotic Cell-Derived Extracellular Vesicles: More Than Just Debris, Front Immunol, 9 (2018) 1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zaborowski MP, Balaj L, Breakefield XO, Lai CP, Extracellular Vesicles: Composition, Biological Relevance, and Methods of Study, Bioscience, 65 (2015) 783–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Soares Martins T, Catita J, Martins Rosa I, A.B.d.C.E.S. O, Henriques AG, Exosome isolation from distinct biofluids using precipitation and column-based approaches, PLoS One, 13 (2018) e0198820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sahu R, Kaushik S, Clement CC, Cannizzo ES, Scharf B, Follenzi A, Potolicchio I, Nieves E, Cuervo AM, Santambrogio L, Microautophagy of cytosolic proteins by late endosomes, Dev Cell, 20 (2011) 131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Cullen PJ, Steinberg F, To degrade or not to degrade: mechanisms and significance of endocytic recycling, Nat Rev Mol Cell Biol, 19 (2018) 679–696. [DOI] [PubMed] [Google Scholar]
- [21].Piper RC, Katzmann DJ, Biogenesis and function of multivesicular bodies, Annu Rev Cell Dev Biol, 23 (2007) 519–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Barois N, de Saint-Vis B, Lebecque S, Geuze HJ, Kleijmeer MJ, MHC class II compartments in human dendritic cells undergo profound structural changes upon activation, Traffic, 3 (2002) 894–905. [DOI] [PubMed] [Google Scholar]
- [23].Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C, Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes), J Biol Chem, 262 (1987) 9412–9420. [PubMed] [Google Scholar]
- [24].Juan T, Furthauer M, Biogenesis and function of ESCRT-dependent extracellular vesicles, Semin Cell Dev Biol, 74 (2018) 66–77. [DOI] [PubMed] [Google Scholar]
- [25].Babst M, MVB vesicle formation: ESCRT-dependent, ESCRT-independent and everything in between, Curr Opin Cell Biol, 23 (2011) 452–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Thery C, Boussac M, Veron P, Ricciardi-Castagnoli P, Raposo G, Garin J, Amigorena S, Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles, J Immunol, 166 (2001) 7309–7318. [DOI] [PubMed] [Google Scholar]
- [27].Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, Dingli F, Loew D, Tkach M, Thery C, Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes, Proc Natl Acad Sci U S A, 113 (2016) E968–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wang Z, Hill S, Luther JM, Hachey DL, Schey KL, Proteomic analysis of urine exosomes by multidimensional protein identification technology (MudPIT), Proteomics, 12 (2012) 329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Luhtala N, Aslanian A, Yates JR 3rd, Hunter T, Secreted Glioblastoma Nanovesicles Contain Intracellular Signaling Proteins and Active Ras Incorporated in a Farnesylation-dependent Manner, J Biol Chem, 292 (2017) 611–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, Zimmermann P, David G, Syndecan-syntenin-ALIX regulates the biogenesis of exosomes, Nat Cell Biol, 14 (2012) 677–685. [DOI] [PubMed] [Google Scholar]
- [31].Larios J, Mercier V, Roux A, Gruenberg J, ALIX- and ESCRT-III-dependent sorting of tetraspanins to exosomes, J Cell Biol, 219 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Romancino DP, Paterniti G, Campos Y, De Luca A, Di Felice V, d’Azzo A, Bongiovanni A, Identification and characterization of the nano-sized vesicles released by muscle cells, FEBS Lett, 587 (2013) 1379–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Stuffers S, Sem Wegner C, Stenmark H, Brech A, Multivesicular endosome biogenesis in the absence of ESCRTs, Traffic, 10 (2009) 925–937. [DOI] [PubMed] [Google Scholar]
- [34].Hessvik NP, Llorente A, Current knowledge on exosome biogenesis and release, Cell Mol Life Sci, 75 (2018) 193–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B, Simons M, Ceramide triggers budding of exosome vesicles into multivesicular endosomes, Science, 319 (2008) 1244–1247. [DOI] [PubMed] [Google Scholar]
- [36].Wei D, Zhan W, Gao Y, Huang L, Gong R, Wang W, Zhang R, Wu Y, Gao S, Kang T, RAB31 marks and controls an ESCRT-independent exosome pathway, Cell Res, (2020). [DOI] [PMC free article] [PubMed]
- [37].Vanlandingham PA, Ceresa BP, Rab7 regulates late endocytic trafficking downstream of multivesicular body biogenesis and cargo sequestration, J Biol Chem, 284 (2009) 12110–12124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Nolte-’t Hoen EN, Buschow SI, Anderton SM, Stoorvogel W, Wauben MH, Activated T cells recruit exosomes secreted by dendritic cells via LFA-1, Blood, 113 (2009) 1977–1981. [DOI] [PubMed] [Google Scholar]
- [39].Nanbo A, Kawanishi E, Yoshida R, Yoshiyama H, Exosomes derived from Epstein-Barr virus-infected cells are internalized via caveola-dependent endocytosis and promote phenotypic modulation in target cells, J Virol, 87 (2013) 10334–10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Yao Q, Chen J, Cao H, Orth JD, McCaffery JM, Stan RV, McNiven MA, Caveolin-1 interacts directly with dynamin-2, J Mol Biol, 348 (2005) 491–501. [DOI] [PubMed] [Google Scholar]
- [41].Chidlow JH Jr., Sessa WC, Caveolae, caveolins, and cavins: complex control of cellular signalling and inflammation, Cardiovasc Res, 86 (2010) 219–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Canton J, Macropinocytosis: New Insights Into Its Underappreciated Role in Innate Immune Cell Surveillance, Front Immunol, 9 (2018) 2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Nakase I, Kobayashi NB, Takatani-Nakase T, Yoshida T, Active macropinocytosis induction by stimulation of epidermal growth factor receptor and oncogenic Ras expression potentiates cellular uptake efficacy of exosomes, Sci Rep, 5 (2015) 10300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Costa Verdera H, Gitz-Francois JJ, Schiffelers RM, Vader P, Cellular uptake of extracellular vesicles is mediated by clathrin-independent endocytosis and macropinocytosis, J Control Release, 266 (2017) 100–108. [DOI] [PubMed] [Google Scholar]
- [45].Lajoie P, Nabi IR, Regulation of raft-dependent endocytosis, J Cell Mol Med, 11 (2007) 644–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Koumangoye RB, Sakwe AM, Goodwin JS, Patel T, Ochieng J, Detachment of breast tumor cells induces rapid secretion of exosomes which subsequently mediate cellular adhesion and spreading, PLoS One, 6 (2011) e24234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Izquierdo-Useros N, Naranjo-Gomez M, Archer J, Hatch SC, Erkizia I, Blanco J, Borras FE, Puertas MC, Connor JH, Fernandez-Figueras MT, Moore L, Clotet B, Gummuluru S, Martinez-Picado J, Capture and transfer of HIV-1 particles by mature dendritic cells converges with the exosome-dissemination pathway, Blood, 113 (2009) 2732–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Emam SE, Ando H, Lila ASA, Shimizu T, Okuhira K, Ishima Y, Mahdy MA, Ghazy FS, Sagawa I, Ishida T, Liposome co-incubation with cancer cells secreted exosomes (extracellular vesicles) with different proteins expressions and different uptake pathways, Sci Rep, 8 (2018) 14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Feng D, Zhao WL, Ye YY, Bai XC, Liu RQ, Chang LF, Zhou Q, Sui SF, Cellular internalization of exosomes occurs through phagocytosis, Traffic, 11 (2010) 675–687. [DOI] [PubMed] [Google Scholar]
- [50].Wu R, Gao W, Yao K, Ge J, Roles of Exosomes Derived From Immune Cells in Cardiovascular Diseases, Front Immunol, 10 (2019) 648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Lee JE, Moon PG, Lee IK, Baek MC, Proteomic Analysis of Extracellular Vesicles Released by Adipocytes of Otsuka Long-Evans Tokushima Fatty (OLETF) Rats, Protein J, 34 (2015) 220–235. [DOI] [PubMed] [Google Scholar]
- [52].Chivet M, Javalet C, Laulagnier K, Blot B, Hemming FJ, Sadoul R, Exosomes secreted by cortical neurons upon glutamatergic synapse activation specifically interact with neurons, J Extracell Vesicles, 3 (2014) 24722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Yin K, Wang S, Zhao RC, Exosomes from mesenchymal stem/stromal cells: a new therapeutic paradigm, Biomark Res, 7 (2019) 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Lyu H, Xiao Y, Guo Q, Huang Y, Luo X, The Role of Bone-Derived Exosomes in Regulating Skeletal Metabolism and Extraosseous Diseases, Front Cell Dev Biol, 8 (2020) 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Castano C, Mirasierra M, Vallejo M, Novials A, Parrizas M, Delivery of muscle-derived exosomal miRNAs induced by HIIT improves insulin sensitivity through down-regulation of hepatic FoxO1 in mice, Proc Natl Acad Sci U S A, 117 (2020) 30335–30343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Shen Z, Sun J, Shao J, Xu J, Ultraviolet B irradiation enhances the secretion of exosomes by human primary melanocytes and changes their exosomal miRNA profile, PLoS One, 15 (2020) e0237023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Davidson SM, Riquelme JA, Zheng Y, Vicencio JM, Lavandero S, Yellon DM, Endothelial cells release cardioprotective exosomes that may contribute to ischaemic preconditioning, Sci Rep, 8 (2018) 15885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Whiteside TL, Tumor-Derived Exosomes and Their Role in Cancer Progression, Adv Clin Chem, 74 (2016) 103–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Tamkovich S, Tutanov O, Efimenko A, Grigor’eva A, Ryabchikova E, Kirushina N, Vlassov V, Tkachuk V, Laktionov P, Blood Circulating Exosomes Contain Distinguishable Fractions of Free and Cell-Surface-Associated Vesicles, Curr Mol Med, 19 (2019) 273–285. [DOI] [PubMed] [Google Scholar]
- [60].Barros ER, Carvajal CA, Urinary Exosomes and Their Cargo: Potential Biomarkers for Mineralocorticoid Arterial Hypertension?, Front Endocrinol (Lausanne), 8 (2017) 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Madison MN, Jones PH, Okeoma CM, Exosomes in human semen restrict HIV-1 transmission by vaginal cells and block intravaginal replication of LP-BM5 murine AIDS virus complex, Virology, 482 (2015) 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Reif S, Elbaum Shiff Y, Golan-Gerstl R, Milk-derived exosomes (MDEs) have a different biological effect on normal fetal colon epithelial cells compared to colon tumor cells in a miRNA-dependent manner, J Transl Med, 17 (2019) 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Liu W, Bai X, Zhang A, Huang J, Xu S, Zhang J, Role of Exosomes in Central Nervous System Diseases, Front Mol Neurosci, 12 (2019) 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Thongboonkerd V, Roles for Exosome in Various Kidney Diseases and Disorders, Front Pharmacol, 10 (2019) 1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Sung S, Kim J, Jung Y, Liver-Derived Exosomes and Their Implications in Liver Pathobiology, Int J Mol Sci, 19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].LeBleu VS, Kalluri R, Exosomes as a Multicomponent Biomarker Platform in Cancer, Trends Cancer, 6 (2020) 767–774. [DOI] [PubMed] [Google Scholar]
- [67].Vinay DS, Ryan EP, Pawelec G, Talib WH, Stagg J, Elkord E, Lichtor T, Decker WK, Whelan RL, Kumara H, Signori E, Honoki K, Georgakilas AG, Amin A, Helferich WG, Boosani CS, Guha G, Ciriolo MR, Chen S, Mohammed SI, Azmi AS, Keith WN, Bilsland A, Bhakta D, Halicka D, Fujii H, Aquilano K, Ashraf SS, Nowsheen S, Yang X, Choi BK, Kwon BS, Immune evasion in cancer: Mechanistic basis and therapeutic strategies, Semin Cancer Biol, 35 Suppl (2015) S185–S198. [DOI] [PubMed] [Google Scholar]
- [68].Winkler J, Abisoye-Ogunniyan A, Metcalf KJ, Werb Z, Concepts of extracellular matrix remodelling in tumour progression and metastasis, Nat Commun, 11 (2020) 5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Coussens LM, Werb Z, Inflammation and cancer, Nature, 420 (2002) 860–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Togashi Y, Shitara K, Nishikawa H, Regulatory T cells in cancer immunosuppression - implications for anticancer therapy, Nat Rev Clin Oncol, 16 (2019) 356–371. [DOI] [PubMed] [Google Scholar]
- [71].Keener AB, Shapeshifters in cancer: How some tumor cells change phenotype to evade therapy, Nature medicine, 22 (2016) 1194–1196. [DOI] [PubMed] [Google Scholar]
- [72].Darvin P, Toor SM, Sasidharan Nair V, Elkord E, Immune checkpoint inhibitors: recent progress and potential biomarkers, Exp Mol Med, 50 (2018) 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Azmi AS, Bao B, Sarkar FH, Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review, Cancer Metastasis Rev, 32 (2013) 623–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Milman N, Ginini L, Gil Z, Exosomes and their role in tumorigenesis and anticancer drug resistance, Drug Resist Updat, 45 (2019) 1–12. [DOI] [PubMed] [Google Scholar]
- [75].Sun C, Mezzadra R, Schumacher TN, Regulation and Function of the PD-L1 Checkpoint, Immunity, 48 (2018) 434–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Poggio M, Hu T, Pai CC, Chu B, Belair CD, Chang A, Montabana E, Lang UE, Fu Q, Fong L, Blelloch R, Suppression of Exosomal PD-L1 Induces Systemic Anti-tumor Immunity and Memory, Cell, 177 (2019) 414–427 e413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Chen G, Huang AC, Zhang W, Zhang G, Wu M, Xu W, Yu Z, Yang J, Wang B, Sun H, Xia H, Man Q, Zhong W, Antelo LF, Wu B, Xiong X, Liu X, Guan L, Li T, Liu S, Yang R, Lu Y, Dong L, McGettigan S, Somasundaram R, Radhakrishnan R, Mills G, Lu Y, Kim J, Chen YH, Dong H, Zhao Y, Karakousis GC, Mitchell TC, Schuchter LM, Herlyn M, Wherry EJ, Xu X, Guo W, Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response, Nature, 560 (2018) 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Theodoraki MN, Yerneni SS, Hoffmann TK, Gooding WE, Whiteside TL, Clinical Significance of PD-L1(+) Exosomes in Plasma of Head and Neck Cancer Patients, Clinical cancer research : an official journal of the American Association for Cancer Research, 24 (2018) 896–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Marimpietri D, Petretto A, Raffaghello L, Pezzolo A, Gagliani C, Tacchetti C, Mauri P, Melioli G, Pistoia V, Proteome profiling of neuroblastoma-derived exosomes reveal the expression of proteins potentially involved in tumor progression, PLoS One, 8 (2013) e75054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Richard H, Pokhrel A, Chava S, Pathania A, Katta SS, Challagundla KB, Exosomes: Novel Players of Therapy Resistance in Neuroblastoma, Adv Exp Med Biol, 1277 (2020) 75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Mahapatra S, Challagundla KB, Cancer, Neuroblastoma, StatPearls, Place Published, 2020.
- [82].Challagundla KB, Wise PM, Neviani P, Chava H, Murtadha M, Xu T, Kennedy R, Ivan C, Zhang X, Vannini I, Fanini F, Amadori D, Calin GA, Hadjidaniel M, Shimada H, Jong A, Seeger RC, Asgharzadeh S, Goldkorn A, Fabbri M, Exosome-mediated transfer of microRNAs within the tumor microenvironment and neuroblastoma resistance to chemotherapy, Journal of the National Cancer Institute, 107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Yi KH, Chen L, Fine tuning the immune response through B7-H3 and B7-H4, Immunol Rev, 229 (2009) 145–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Pathania AS, Prathipati P, Pandey MK, Byrareddy SN, Coulter DW, Gupta SC, Challagundla KB, The emerging role of non-coding RNAs in the epigenetic regulation of pediatric cancers, Seminars in cancer biology, (2021). [DOI] [PubMed]
- [85].Ludwig S, Marczak L, Sharma P, Abramowicz A, Gawin M, Widlak P, Whiteside TL, Pietrowska M, Proteomes of exosomes from HPV(+) or HPV(−) head and neck cancer cells: differential enrichment in immunoregulatory proteins, Oncoimmunology, 8 (2019) 1593808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Takimoto CH, Chao MP, Gibbs C, McCamish MA, Liu J, Chen JY, Majeti R, Weissman IL, The Macrophage ‘Do not eat me’ signal, CD47, is a clinically validated cancer immunotherapy target, Ann Oncol, 30 (2019) 486–489. [DOI] [PubMed] [Google Scholar]
- [87].Matozaki T, Murata Y, Okazawa H, Ohnishi H, Functions and molecular mechanisms of the CD47-SIRPalpha signalling pathway, Trends Cell Biol, 19 (2009) 72–80. [DOI] [PubMed] [Google Scholar]
- [88].Gabrusiewicz K, Li X, Wei J, Hashimoto Y, Marisetty AL, Ott M, Wang F, Hawke D, Yu J, Healy LM, Hossain A, Akers JC, Maiti SN, Yamashita S, Shimizu Y, Dunner K, Zal MA, Burks JK, Gumin J, Nwajei F, Rezavanian A, Zhou S, Rao G, Sawaya R, Fuller GN, Huse JT, Antel JP, Li S, Cooper L, Sulman EP, Chen C, Geula C, Kalluri R, Zal T, Heimberger AB, Glioblastoma stem cell-derived exosomes induce M2 macrophages and PD-L1 expression on human monocytes, Oncoimmunology, 7 (2018) e1412909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Jacquemet G, Hamidi H, Ivaska J, Filopodia in cell adhesion, 3D migration and cancer cell invasion, Curr Opin Cell Biol, 36 (2015) 23–31. [DOI] [PubMed] [Google Scholar]
- [90].Ma M, Baumgartner M, Filopodia and membrane blebs drive efficient matrix invasion of macrophages transformed by the intracellular parasite Theileria annulata, PLoS One, 8 (2013) e75577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Liu J, Wu S, Zheng X, Zheng P, Fu Y, Wu C, Lu B, Ju J, Jiang J, Immune suppressed tumor microenvironment by exosomes derived from gastric cancer cells via modulating immune functions, Sci Rep, 10 (2020) 14749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Yin X, Zeng W, Wu B, Wang L, Wang Z, Tian H, Wang L, Jiang Y, Clay R, Wei X, Qin Y, Zhang F, Zhang C, Jin L, Liang W, PPARalpha Inhibition Overcomes Tumor-Derived Exosomal Lipid-Induced Dendritic Cell Dysfunction, Cell Rep, 33 (2020) 108278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Varga T, Czimmerer Z, Nagy L, PPARs are a unique set of fatty acid regulated transcription factors controlling both lipid metabolism and inflammation, Biochim Biophys Acta, 1812 (2011) 1007–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Chen L, Huang H, Zhang W, Ding F, Fan Z, Zeng Z, Exosomes Derived From T Regulatory Cells Suppress CD8+ Cytotoxic T Lymphocyte Proliferation and Prolong Liver Allograft Survival, Med Sci Monit, 25 (2019) 4877–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Xie Y, Zhang X, Zhao T, Li W, Xiang J, Natural CD8(+)25(+) regulatory T cell-secreted exosomes capable of suppressing cytotoxic T lymphocyte-mediated immunity against B16 melanoma, Biochem Biophys Res Commun, 438 (2013) 152–155. [DOI] [PubMed] [Google Scholar]
- [96].Aiello S, Rocchetta F, Longaretti L, Faravelli S, Todeschini M, Cassis L, Pezzuto F, Tomasoni S, Azzollini N, Mister M, Mele C, Conti S, Breno M, Remuzzi G, Noris M, Benigni A, Extracellular vesicles derived from T regulatory cells suppress T cell proliferation and prolong allograft survival, Sci Rep, 7 (2017) 11518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Sullivan JA, Tomita Y, Jankowska-Gan E, Lema DA, Arvedson MP, Nair A, Bracamonte-Baran W, Zhou Y, Meyer KK, Zhong W, Sawant DV, Szymczak-Workman AL, Zhang Q, Workman CJ, Hong S, Vignali DAA, Burlingham WJ, Treg-Cell-Derived IL-35-Coated Extracellular Vesicles Promote Infectious Tolerance, Cell Rep, 30 (2020) 1039–1051 e1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Benjamins JA, Nedelkoska L, Touil H, Stemmer PM, Carruthers NJ, Jena BP, Naik AR, Bar-Or A, Lisak RP, Exosome-enriched fractions from MS B cells induce oligodendrocyte death, Neurol Neuroimmunol Neuroinflamm, 6 (2019) e550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Aras S, Zaidi MR, TAMeless traitors: macrophages in cancer progression and metastasis, Br J Cancer, 117 (2017) 1583–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Jackute J, Zemaitis M, Pranys D, Sitkauskiene B, Miliauskas S, Vaitkiene S, Sakalauskas R, Distribution of M1 and M2 macrophages in tumor islets and stroma in relation to prognosis of non-small cell lung cancer, BMC Immunol, 19 (2018) 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Jayasingam SD, Citartan M, Thang TH, Mat Zin AA, Ang KC, Ch’ng ES, Evaluating the Polarization of Tumor-Associated Macrophages Into M1 and M2 Phenotypes in Human Cancer Tissue: Technicalities and Challenges in Routine Clinical Practice, Front Oncol, 9 (2019) 1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Lan J, Sun L, Xu F, Liu L, Hu F, Song D, Hou Z, Wu W, Luo X, Wang J, Yuan X, Hu J, Wang G, M2 Macrophage-Derived Exosomes Promote Cell Migration and Invasion in Colon Cancer, Cancer Res, 79 (2019) 146–158. [DOI] [PubMed] [Google Scholar]
- [103].Pyo JS, Son BK, Oh D, Kim EK, BRG1 is correlated with poor prognosis in colorectal cancer, Hum Pathol, 73 (2018) 66–73. [DOI] [PubMed] [Google Scholar]
- [104].Wu J, Gao W, Tang Q, Yu Y, You W, Wu Z, Fan Y, Zhang L, Wu C, Han G, Zuo X, Zhang Y, Chen Z, Ding W, Li X, Lin F, Shen H, Tang J, Zhang Y, Wang X, M2 macrophage-derived exosomes facilitate hepatocarcinoma metastasis by transferring alphaM beta2 integrin to tumor cells, Hepatology, (2020). [DOI] [PMC free article] [PubMed] [Retracted]
- [105].Youn JI, Gabrilovich DI, The biology of myeloid-derived suppressor cells: the blessing and the curse of morphological and functional heterogeneity, Eur J Immunol, 40 (2010) 2969–2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Veglia F, Perego M, Gabrilovich D, Myeloid-derived suppressor cells coming of age, Nat Immunol, 19 (2018) 108–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].De Sanctis F, Solito S, Ugel S, Molon B, Bronte V, Marigo I, MDSCs in cancer: Conceiving new prognostic and therapeutic targets, Biochim Biophys Acta, 1865 (2016) 35–48. [DOI] [PubMed] [Google Scholar]
- [108].Cimen Bozkus C, Elzey BD, Crist SA, Ellies LG, Ratliff TL, Expression of Cationic Amino Acid Transporter 2 Is Required for Myeloid-Derived Suppressor Cell-Mediated Control of T Cell Immunity, J Immunol, 195 (2015) 5237–5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Ohl K, Tenbrock K, Reactive Oxygen Species as Regulators of MDSC-Mediated Immune Suppression, Front Immunol, 9 (2018) 2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Kumar V, Patel S, Tcyganov E, Gabrilovich DI, The Nature of Myeloid-Derived Suppressor Cells in the Tumor Microenvironment, Trends Immunol, 37 (2016) 208–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Gato M, Blanco-Luquin I, Zudaire M, de Morentin XM, Perez-Valderrama E, Zabaleta A, Kochan G, Escors D, Fernandez-Irigoyen J, Santamaria E, Drafting the proteome landscape of myeloid-derived suppressor cells, Proteomics, 16 (2016) 367–378. [DOI] [PubMed] [Google Scholar]
- [112].Geis-Asteggiante L, Belew AT, Clements VK, Edwards NJ, Ostrand-Rosenberg S, El-Sayed NM, Fenselau C, Differential Content of Proteins, mRNAs, and miRNAs Suggests that MDSC and Their Exosomes May Mediate Distinct Immune Suppressive Functions, J Proteome Res, 17 (2018) 486–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Fenselau C, Ostrand-Rosenberg S, Molecular cargo in myeloid-derived suppressor cells and their exosomes, Cell Immunol, 359 (2021) 104258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Rashid MH, Borin TF, Ara R, Piranlioglu R, Achyut BR, Korkaya H, Liu Y, Arbab AS, Critical immunosuppressive effect of MDSCderived exosomes in the tumor microenvironment, Oncol Rep, (2021). [DOI] [PMC free article] [PubMed]
- [115].Tcyganov E, Mastio J, Chen E, Gabrilovich DI, Plasticity of myeloid-derived suppressor cells in cancer, Curr Opin Immunol, 51 (2018) 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Biswas S, Mandal G, Roy Chowdhury S, Purohit S, Payne KK, Anadon C, Gupta A, Swanson P, Yu X, Conejo-Garcia JR, Bhattacharyya A, Exosomes Produced by Mesenchymal Stem Cells Drive Differentiation of Myeloid Cells into Immunosuppressive M2-Polarized Macrophages in Breast Cancer, J Immunol, 203 (2019) 3447–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].LeBleu VS, Neilson EG, Origin and functional heterogeneity of fibroblasts, FASEB J, 34 (2020) 3519–3536. [DOI] [PubMed] [Google Scholar]
- [118].Tomasek JJ, Gabbiani G, Hinz B, Chaponnier C, Brown RA, Myofibroblasts and mechano-regulation of connective tissue remodelling, Nat Rev Mol Cell Biol, 3 (2002) 349–363. [DOI] [PubMed] [Google Scholar]
- [119].Sahai E, Astsaturov I, Cukierman E, DeNardo DG, Egeblad M, Evans RM, Fearon D, Greten FR, Hingorani SR, Hunter T, Hynes RO, Jain RK, Janowitz T, Jorgensen C, Kimmelman AC, Kolonin MG, Maki RG, Powers RS, Pure E, Ramirez DC, Scherz-Shouval R, Sherman MH, Stewart S, Tlsty TD, Tuveson DA, Watt FM, Weaver V, Weeraratna AT, Werb Z, A framework for advancing our understanding of cancer-associated fibroblasts, Nat Rev Cancer, 20 (2020) 174–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].LeBleu VS, Kalluri R, A peek into cancer-associated fibroblasts: origins, functions and translational impact, Dis Model Mech, 11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Dou D, Ren X, Han M, Xu X, Ge X, Gu Y, Wang X, Cancer-Associated Fibroblasts-Derived Exosomes Suppress Immune Cell Function in Breast Cancer via the miR-92/PD-L1 Pathway, Front Immunol, 11 (2020) 2026. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [122].Oszvald A, Szvicsek Z, Papai M, Kelemen A, Varga Z, Tolgyes T, Dede K, Bursics A, Buzas EI, Wiener Z, Fibroblast-Derived Extracellular Vesicles Induce Colorectal Cancer Progression by Transmitting Amphiregulin, Front Cell Dev Biol, 8 (2020) 558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Richards KE, Zeleniak AE, Fishel ML, Wu J, Littlepage LE, Hill R, Cancer-associated fibroblast exosomes regulate survival and proliferation of pancreatic cancer cells, Oncogene, 36 (2017) 1770–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Goldstein RH, Reagan MR, Anderson K, Kaplan DL, Rosenblatt M, Human bone marrow-derived MSCs can home to orthotopic breast cancer tumors and promote bone metastasis, Cancer Res, 70 (2010) 10044–10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Fujii S, Miura Y, Fujishiro A, Shindo T, Shimazu Y, Hirai H, Tahara H, Takaori-Kondo A, Ichinohe T, Maekawa T, Graft-Versus-Host Disease Amelioration by Human Bone Marrow Mesenchymal Stromal/Stem Cell-Derived Extracellular Vesicles Is Associated with Peripheral Preservation of Naive T Cell Populations, Stem Cells, 36 (2018) 434–445. [DOI] [PubMed] [Google Scholar]
- [126].Khare D, Or R, Resnick I, Barkatz C, Almogi-Hazan O, Avni B, Mesenchymal Stromal Cell-Derived Exosomes Affect mRNA Expression and Function of B-Lymphocytes, Front Immunol, 9 (2018) 3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Wang J, De Veirman K, De Beule N, Maes K, De Bruyne E, Van Valckenborgh E, Vanderkerken K, Menu E, The bone marrow microenvironment enhances multiple myeloma progression by exosome-mediated activation of myeloid-derived suppressor cells, Oncotarget, 6 (2015) 43992–44004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Silva M, Melo SA, Non-coding RNAs in Exosomes: New Players in Cancer Biology, Curr Genomics, 16 (2015) 295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Blenkiron C, Miska EA, miRNAs in cancer: approaches, aetiology, diagnostics and therapy, Hum Mol Genet, 16 Spec No 1 (2007) R106–113. [DOI] [PubMed] [Google Scholar]
- [130].Condrat CE, Thompson DC, Barbu MG, Bugnar OL, Boboc A, Cretoiu D, Suciu N, Cretoiu SM, Voinea SC, miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis, Cells, 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Zhou J, Li X, Wu X, Zhang T, Zhu Q, Wang X, Wang H, Wang K, Lin Y, Wang X, Exosomes Released from Tumor-Associated Macrophages Transfer miRNAs That Induce a Treg/Th17 Cell Imbalance in Epithelial Ovarian Cancer, Cancer Immunol Res, 6 (2018) 1578–1592. [DOI] [PubMed] [Google Scholar]
- [132].Kanlikilicer P, Bayraktar R, Denizli M, Rashed MH, Ivan C, Aslan B, Mitra R, Karagoz K, Bayraktar E, Zhang X, Rodriguez-Aguayo C, El-Arabey AA, Kahraman N, Baydogan S, Ozkayar O, Gatza ML, Ozpolat B, Calin GA, Sood AK, Lopez-Berestein G, Exosomal miRNA confers chemo resistance via targeting Cav1/p-gp/M2-type macrophage axis in ovarian cancer, EBioMedicine, 38 (2018) 100–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Celus W, Di Conza G, Oliveira AI, Ehling M, Costa BM, Wenes M, Mazzone M, Loss of Caveolin-1 in Metastasis-Associated Macrophages Drives Lung Metastatic Growth through Increased Angiogenesis, Cell Rep, 21 (2017) 2842–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Guo J, Duan Z, Zhang C, Wang W, He H, Liu Y, Wu P, Wang S, Song M, Chen H, Chen C, Si Q, Xiang R, Luo Y, Mouse 4T1 Breast Cancer Cell-Derived Exosomes Induce Proinflammatory Cytokine Production in Macrophages via miR-183, J Immunol, 205 (2020) 2916–2925. [DOI] [PubMed] [Google Scholar]
- [135].Vignard V, Labbe M, Marec N, Andre-Gregoire G, Jouand N, Fonteneau JF, Labarriere N, Fradin D, MicroRNAs in Tumor Exosomes Drive Immune Escape in Melanoma, Cancer Immunol Res, 8 (2020) 255–267. [DOI] [PubMed] [Google Scholar]
- [136].Ye SB, Zhang H, Cai TT, Liu YN, Ni JJ, He J, Peng JY, Chen QY, Mo HY, Jun C, Zhang XS, Zeng YX, Li J, Exosomal miR-24–3p impedes T-cell function by targeting FGF11 and serves as a potential prognostic biomarker for nasopharyngeal carcinoma, J Pathol, 240 (2016) 329–340. [DOI] [PubMed] [Google Scholar]
- [137].Zhu Y, Zhang S, Li Z, Wang H, Li Z, Hu Y, Chen H, Zhang X, Cui L, Zhang J, He W, miR-125b-5p and miR-99a-5p downregulate human gammadelta T-cell activation and cytotoxicity, Cell Mol Immunol, 16 (2019) 112–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Peng XX, Yu R, Wu X, Wu SY, Pi C, Chen ZH, Zhang XC, Gao CY, Shao YW, Liu L, Wu YL, Zhou Q, Correlation of plasma exosomal microRNAs with the efficacy of immunotherapy in EGFR / ALK wild-type advanced non-small cell lung cancer, J Immunother Cancer, 8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Pathania AS, Challagundla KB, Exosomal Long Non-coding RNAs: Emerging Players in the Tumor Microenvironment, Mol Ther Nucleic Acids, 23 (2021) 1371–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Chen J, Wang S, Jia S, Ding G, Jiang G, Cao L, Integrated Analysis of Long Non-Coding RNA and mRNA Expression Profile in Pancreatic Cancer Derived Exosomes Treated Dendritic Cells by Microarray Analysis, J Cancer, 9 (2018) 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Domvri K, Petanidis S, Anestakis D, Porpodis K, Bai C, Zarogoulidis P, Freitag L, Hohenforst-Schmidt W, Katopodi T, Exosomal lncRNA PCAT-1 promotes Kras-associated chemoresistance via immunosuppressive miR-182/miR-217 signaling and p27/CDK6 regulation, Oncotarget, 11 (2020) 2847–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Zang Q, Xu L, Li J, Jia H, GATA6 activated long non-coding RNA PCAT1 maintains stemness of non-small cell lung cancer by mediating FRK, J BUON, 25 (2020) 2371–2381. [PubMed] [Google Scholar]
- [143].Chen F, Chen J, Yang L, Liu J, Zhang X, Zhang Y, Tu Q, Yin D, Lin D, Wong PP, Huang D, Xing Y, Zhao J, Li M, Liu Q, Su F, Su S, Song E, Extracellular vesicle-packaged HIF-1alpha-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells, Nat Cell Biol, 21 (2019) 498–510. [DOI] [PubMed] [Google Scholar]
- [144].Zheng X, Mansouri S, Krager A, Grimminger F, Seeger W, Pullamsetti SS, Wheelock CE, Savai R, Metabolism in tumour-associated macrophages: a quid pro quo with the tumour microenvironment, Eur Respir Rev, 29 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].M.d.-B. N, Duncan-Moretti J, C.d.-C. H, Saldanha-Gama R, Paula-Neto HA, G.D. G, L.S. R, Barja-Fidalgo C, Aerobic glycolysis is a metabolic requirement to maintain the M2-like polarization of tumor-associated macrophages, Biochim Biophys Acta Mol Cell Res, 1867 (2020) 118604. [DOI] [PubMed] [Google Scholar]
- [146].Jeong H, Kim S, Hong BJ, Lee CJ, Kim YE, Bok S, Oh JM, Gwak SH, Yoo MY, Lee MS, Chung SJ, Defrene J, Tessier P, Pelletier M, Jeon H, Roh TY, Kim B, Kim KH, Ju JH, Kim S, Lee YJ, Kim DW, Kim IH, Kim HJ, Park JW, Lee YS, Lee JS, Cheon GJ, Weissman IL, Chung DH, Jeon YK, Ahn GO, Tumor-Associated Macrophages Enhance Tumor Hypoxia and Aerobic Glycolysis, Cancer Res, 79 (2019) 795–806. [DOI] [PubMed] [Google Scholar]
- [147].Xue M, Chen W, Xiang A, Wang R, Chen H, Pan J, Pang H, An H, Wang X, Hou H, Li X, Hypoxic exosomes facilitate bladder tumor growth and development through transferring long non-coding RNA-UCA1, Mol Cancer, 16 (2017) 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Zhou D, Xia Z, Xie M, Gao Y, Yu Q, He B, Exosomal long non-coding RNA SOX2 overlapping transcript enhances the resistance to EGFR-TKIs in non-small cell lung cancer cell line H1975, Hum Cell, (2021). [DOI] [PubMed]
- [149].Zhang R, Xia Y, Wang Z, Zheng J, Chen Y, Li X, Wang Y, Ming H, Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer, Biochemical and biophysical research communications, 490 (2017) 406–414. [DOI] [PubMed] [Google Scholar]
- [150].Liu Y, Yin Z, Lu P, Ma Y, Luo B, Xiang L, Zhang W, He Y, Liang X, Lung Carcinoma Cells Secrete Exosomal MALAT1 to Inhibit Dendritic Cell Phagocytosis, Inflammatory Response, Costimulatory Molecule Expression and Promote Dendritic Cell Autophagy via AKT/mTOR Pathway, Onco Targets Ther, 13 (2020) 10693–10705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Hewitson JP, West KA, James KR, Rani GF, Dey N, Romano A, Brown N, Teichmann SA, Kaye PM, Lagos D, Malat1 Suppresses Immunity to Infection through Promoting Expression of Maf and IL-10 in Th Cells, J Immunol, 204 (2020) 2949–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Cao S, Liu J, Song L, Ma X, The protooncogene c-Maf is an essential transcription factor for IL-10 gene expression in macrophages, J Immunol, 174 (2005) 3484–3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Zhao S, Wu D, Wu P, Wang Z, Huang J, Serum IL-10 Predicts Worse Outcome in Cancer Patients: A Meta-Analysis, PLoS One, 10 (2015) e0139598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Fan JT, Zhou ZY, Luo YL, Luo Q, Chen SB, Zhao JC, Chen QR, Exosomal lncRNA NEAT1 from cancer-associated fibroblasts facilitates endometrial cancer progression via miR-26a/b-5p-mediated STAT3/YKL-40 signaling pathway, Neoplasia, 23 (2021) 692–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Mestdagh P, Fredlund E, Pattyn F, Rihani A, Van Maerken T, Vermeulen J, Kumps C, Menten B, De Preter K, Schramm A, Schulte J, Noguera R, Schleiermacher G, Janoueix-Lerosey I, Laureys G, Powel R, Nittner D, Marine JC, Ringner M, Speleman F, Vandesompele J, An integrative genomics screen uncovers ncRNA T-UCR functions in neuroblastoma tumours, Oncogene, 29 (2010) 3583–3592. [DOI] [PubMed] [Google Scholar]
- [156].Vannini I, Wise PM, Challagundla KB, Plousiou M, Raffini M, Bandini E, Fanini F, Paliaga G, Crawford M, Ferracin M, Ivan C, Fabris L, Davuluri RV, Guo Z, Cortez MA, Zhang X, Chen L, Zhang S, Fernandez-Cymering C, Han L, Carloni S, Salvi S, Ling H, Murtadha M, Neviani P, Gitlitz BJ, Laird-Offringa IA, Nana-Sinkam P, Negrini M, Liang H, Amadori D, Cimmino A, Calin GA, Fabbri M, Transcribed ultraconserved region 339 promotes carcinogenesis by modulating tumor suppressor microRNAs, Nature communications, 8 (2017) 1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Mudgapalli N, Shaw BP, Chava S, Challagundla KB, The Transcribed-Ultra Conserved Regions: Novel Non-Coding RNA Players in Neuroblastoma Progression, Noncoding RNA, 5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Nallasamy P, Chava S, Verma SS, Mishra S, Gorantla S, Coulter DW, Byrareddy SN, Batra SK, Gupta SC, Challagundla KB, PD-L1, inflammation, non-coding RNAs, and neuroblastoma: Immuno-oncology perspective, Seminars in cancer biology, 52 (2018) 53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Chava S, Reynolds CP, Pathania AS, Gorantla S, Poluektova LY, Coulter DW, Gupta SC, Pandey MK, Challagundla KB, miR-15a-5p, miR-15b-5p, and miR-16–5p inhibit tumor progression by directly targeting MYCN in neuroblastoma, Mol Oncol, 14 (2020) 180–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Braconi C, Valeri N, Kogure T, Gasparini P, Huang N, Nuovo GJ, Terracciano L, Croce CM, Patel T, Expression and functional role of a transcribed noncoding RNA with an ultraconserved element in hepatocellular carcinoma, Proceedings of the National Academy of Sciences of the United States of America, 108 (2011) 786–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Kogure T, Yan IK, Lin WL, Patel T, Extracellular Vesicle-Mediated Transfer of a Novel Long Noncoding RNA TUC339: A Mechanism of Intercellular Signaling in Human Hepatocellular Cancer, Genes Cancer, 4 (2013) 261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Ding Z, Yan Y, Guo YL, Wang C, Esophageal carcinoma cell-excreted exosomal uc.189 promotes lymphatic metastasis, Aging (Albany NY), 13 (2021) 13846–13858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Kuwano Y, Nishida K, Rokutan K, Overexpression of the transcribed ultraconserved region Uc.138 accelerates colon cancer progression, Sci Rep, 11 (2021) 8667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Sakamoto N, Sekino Y, Fukada K, Pham QT, Honma R, Taniyama D, Ukai S, Takashima T, Hattori T, Naka K, Tanabe K, Ohdan H, Yasui W, Uc.63+ contributes to gastric cancer progression through regulation of NF-kB signaling, Gastric Cancer, 23 (2020) 863–873. [DOI] [PubMed] [Google Scholar]
- [165].Sekino Y, Sakamoto N, Ishikawa A, Honma R, Shigematsu Y, Hayashi T, Sentani K, Oue N, Teishima J, Matsubara A, Yasui W, Transcribed ultraconserved region Uc.63+ promotes resistance to cisplatin through regulation of androgen receptor signaling in bladder cancer, Oncol Rep, 41 (2019) 3111–3118. [DOI] [PubMed] [Google Scholar]
- [166].Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK, Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures, Proc Natl Acad Sci U S A, 73 (1976) 3852–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Jeck WR, Sharpless NE, Detecting and characterizing circular RNAs, Nat Biotechnol, 32 (2014) 453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Wang Y, Gao R, Li J, Tang S, Li S, Tong Q, Li S, Downregulation of hsa_circ_0074854 Suppresses the Migration and Invasion in Hepatocellular Carcinoma via Interacting with HuR and via Suppressing Exosomes-Mediated Macrophage M2 Polarization, Int J Nanomedicine, 16 (2021) 2803–2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ, Shi GM, Cai JB, Ke AW, Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma, Molecular cancer, 19 (2020) 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Xu L, Huang Y, Tan L, Yu W, Chen D, Lu C, He J, Wu G, Liu X, Zhang Y, Increased Tim-3 expression in peripheral NK cells predicts a poorer prognosis and Tim-3 blockade improves NK cell-mediated cytotoxicity in human lung adenocarcinoma, Int Immunopharmacol, 29 (2015) 635–641. [DOI] [PubMed] [Google Scholar]
- [171].Hong W, Xue M, Jiang J, Zhang Y, Gao X, Circular RNA circ-CPA4/ let-7 miRNA/PD-L1 axis regulates cell growth, stemness, drug resistance and immune evasion in non-small cell lung cancer (NSCLC), J Exp Clin Cancer Res, 39 (2020) 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Wang J, Zhao X, Wang Y, Ren F, Sun D, Yan Y, Kong X, Bu J, Liu M, Xu S, circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma, Cell Death Dis, 11 (2020) 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Datta A, Kim H, McGee L, Johnson AE, Talwar S, Marugan J, Southall N, Hu X, Lal M, Mondal D, Ferrer M, Abdel-Mageed AB, High-throughput screening identified selective inhibitors of exosome biogenesis and secretion: A drug repurposing strategy for advanced cancer, Sci Rep, 8 (2018) 8161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Bobrie A, Krumeich S, Reyal F, Recchi C, Moita LF, Seabra MC, Ostrowski M, Thery C, Rab27a supports exosome-dependent and -independent mechanisms that modify the tumor microenvironment and can promote tumor progression, Cancer Res, 72 (2012) 4920–4930. [DOI] [PubMed] [Google Scholar]
- [175].Guarino BD, Adapala RK, Kanugula AK, Lenkey NM, Dougherty JA, Paruchuri S, Khan M, Thodeti CK, Extracellular Vesicles From Pathological Microenvironment Induce Endothelial Cell Transformation and Abnormal Angiogenesis via Modulation of TRPV4 Channels, Front Cell Dev Biol, 7 (2019) 344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Petanidis S, Domvri K, Porpodis K, Anestakis D, Freitag L, Hohenforst-Schmidt W, Tsavlis D, Zarogoulidis K, Inhibition of kras-derived exosomes downregulates immunosuppressive BACH2/GATA-3 expression via RIP-3 dependent necroptosis and miR-146/miR-210 modulation, Biomed Pharmacother, 122 (2020) 109461. [DOI] [PubMed] [Google Scholar]
- [177].Zhou C, Wei W, Ma J, Yang Y, Liang L, Zhang Y, Wang Z, Chen X, Huang L, Wang W, Wu S, Cancer-secreted exosomal miR-1468–5p promotes tumor immune escape via the immunosuppressive reprogramming of lymphatic vessels, Mol Ther, 29 (2021) 1512–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Li T, Tao Z, Zhu Y, Liu X, Wang L, Du Y, Cao J, Wang B, Zhang J, Hu X, Exosomal annexin A6 induces gemcitabine resistance by inhibiting ubiquitination and degradation of EGFR in triple-negative breast cancer, Cell Death Dis, 12 (2021) 684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Ren J, Ding L, Zhang D, Shi G, Xu Q, Shen S, Wang Y, Wang T, Hou Y, Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19, Theranostics, 8 (2018) 3932–3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Somiya M, Yoshioka Y, Ochiya T, Biocompatibility of highly purified bovine milk-derived extracellular vesicles, J Extracell Vesicles, 7 (2018) 1440132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Smith JA, Leonardi T, Huang B, Iraci N, Vega B, Pluchino S, Extracellular vesicles and their synthetic analogues in aging and age-associated brain diseases, Biogerontology, 16 (2015) 147–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Neviani P, Wise PM, Murtadha M, Liu CW, Wu CH, Jong AY, Seeger RC, Fabbri M, Natural Killer-derived exosomal miR-186 inhibits neuroblastoma growth and immune escape mechanisms, Cancer research, (2018). [DOI] [PMC free article] [PubMed]
- [183].Ghoroghi S, Mary B, Larnicol A, Asokan N, Klein A, Osmani N, Busnelli I, Delalande F, Paul N, Halary S, Gros F, Fouillen L, Haeberle AM, Royer C, Spiegelhalter C, Andre-Gregoire G, Mittelheisser V, Detappe A, Murphy K, Timpson P, Carapito R, Blot-Chabaud M, Gavard J, Carapito C, Vitale N, Lefebvre O, Goetz JG, Hyenne V, Ral GTPases promote breast cancer metastasis by controlling biogenesis and organ targeting of exosomes, Elife, 10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Yang Y, Li CW, Chan LC, Wei Y, Hsu JM, Xia W, Cha JH, Hou J, Hsu JL, Sun L, Hung MC, Exosomal PD-L1 harbors active defense function to suppress T cell killing of breast cancer cells and promote tumor growth, Cell Res, 28 (2018) 862–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Datta A, Kim H, Lal M, McGee L, Johnson A, Moustafa AA, Jones JC, Mondal D, Ferrer M, Abdel-Mageed AB, Manumycin A suppresses exosome biogenesis and secretion via targeted inhibition of Ras/Raf/ERK1/2 signaling and hnRNP H1 in castration-resistant prostate cancer cells, Cancer Lett, 408 (2017) 73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Jorfi S, Ansa-Addo EA, Kholia S, Stratton D, Valley S, Lange S, Inal J, Inhibition of microvesiculation sensitizes prostate cancer cells to chemotherapy and reduces docetaxel dose required to limit tumor growth in vivo, Sci Rep, 5 (2015) 13006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Zhu Y, Howard GA, Pittman K, Boykin C, Herring LE, Wilkerson EM, Verbanac K, Lu Q, Therapeutic Effect of Y-27632 on Tumorigenesis and Cisplatin-Induced Peripheral Sensory Loss through RhoA-NF-kappaB, Mol Cancer Res, 17 (2019) 1910–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Nam GH, Lee EJ, Kim YK, Hong Y, Choi Y, Ryu MJ, Woo J, Cho Y, Ahn DJ, Yang Y, Kwon IC, Park SY, Kim IS, Combined Rho-kinase inhibition and immunogenic cell death triggers and propagates immunity against cancer, Nat Commun, 9 (2018) 2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Singha PK, Pandeswara S, Venkatachalam MA, Saikumar P, Manumycin A inhibits triple-negative breast cancer growth through LC3-mediated cytoplasmic vacuolation death, Cell Death Dis, 4 (2013) e457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Roseblade A, Luk F, Ung A, Bebawy M, Targeting microparticle biogenesis: a novel approach to the circumvention of cancer multidrug resistance, Curr Cancer Drug Targets, 15 (2015) 205–214. [DOI] [PubMed] [Google Scholar]
- [191].Penet MF, Krishnamachary B, Wildes F, Mironchik Y, Mezzanzanica D, Podo F, de Reggi M, Gharib B, Bhujwalla ZM, Effect of Pantethine on Ovarian Tumor Progression and Choline Metabolism, Front Oncol, 6 (2016) 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Ho AL, Brana I, Haddad R, Bauman J, Bible K, Oosting S, Wong DJ, Ahn MJ, Boni V, Even C, Fayette J, Flor MJ, Harrington K, Kim SB, Licitra L, Nixon I, Saba NF, Hackenberg S, Specenier P, Worden F, Balsara B, Leoni M, Martell B, Scholz C, Gualberto A, Tipifarnib in Head and Neck Squamous Cell Carcinoma With HRAS Mutations, J Clin Oncol, 39 (2021) 1856–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Viaud S, Terme M, Flament C, Taieb J, Andre F, Novault S, Escudier B, Robert C, Caillat-Zucman S, Tursz T, Zitvogel L, Chaput N, Dendritic cell-derived exosomes promote natural killer cell activation and proliferation: a role for NKG2D ligands and IL-15Ralpha, PLoS One, 4 (2009) e4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Thery C, Duban L, Segura E, Veron P, Lantz O, Amigorena S, Indirect activation of naive CD4+ T cells by dendritic cell-derived exosomes, Nat Immunol, 3 (2002) 1156–1162. [DOI] [PubMed] [Google Scholar]
- [195].Yao Y, Chen L, Wei W, Deng X, Ma L, Hao S, Tumor cell-derived exosome-targeted dendritic cells stimulate stronger CD8+ CTL responses and antitumor immunities, Biochem Biophys Res Commun, 436 (2013) 60–65. [DOI] [PubMed] [Google Scholar]
- [196].Munich S, Sobo-Vujanovic A, Buchser WJ, Beer-Stolz D, Vujanovic NL, Dendritic cell exosomes directly kill tumor cells and activate natural killer cells via TNF superfamily ligands, Oncoimmunology, 1 (2012) 1074–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Besse B, Charrier M, Lapierre V, Dansin E, Lantz O, Planchard D, Le Chevalier T, Livartoski A, Barlesi F, Laplanche A, Ploix S, Vimond N, Peguillet I, Thery C, Lacroix L, Zoernig I, Dhodapkar K, Dhodapkar M, Viaud S, Soria JC, Reiners KS, Pogge von Strandmann E, Vely F, Rusakiewicz S, Eggermont A, Pitt JM, Zitvogel L, Chaput N, Dendritic cell-derived exosomes as maintenance immunotherapy after first line chemotherapy in NSCLC, Oncoimmunology, 5 (2016) e1071008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Markov O, Oshchepkova A, Mironova N, Immunotherapy Based on Dendritic Cell-Targeted/-Derived Extracellular Vesicles-A Novel Strategy for Enhancement of the Anti-tumor Immune Response, Front Pharmacol, 10 (2019) 1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Phung CD, Pham TT, Nguyen HT, Nguyen TT, Ou W, Jeong JH, Choi HG, Ku SK, Yong CS, Kim JO, Anti-CTLA-4 antibody-functionalized dendritic cell-derived exosomes targeting tumor-draining lymph nodes for effective induction of antitumor T-cell responses, Acta Biomater, 115 (2020) 371–382. [DOI] [PubMed] [Google Scholar]
- [200].Tivol EA, Borriello F, Schweitzer AN, Lynch WP, Bluestone JA, Sharpe AH, Loss of CTLA-4 leads to massive lymphoproliferation and fatal multiorgan tissue destruction, revealing a critical negative regulatory role of CTLA-4, Immunity, 3 (1995) 541–547. [DOI] [PubMed] [Google Scholar]
- [201].Rudd CE, Taylor A, Schneider H, CD28 and CTLA-4 coreceptor expression and signal transduction, Immunol Rev, 229 (2009) 12–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Seliger B, Marincola FM, Ferrone S, Abken H, The complex role of B7 molecules in tumor immunology, Trends Mol Med, 14 (2008) 550–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].van Elsas A, Hurwitz AA, Allison JP, Combination immunotherapy of B16 melanoma using anti-cytotoxic T lymphocyte-associated antigen 4 (CTLA-4) and granulocyte/macrophage colony-stimulating factor (GM-CSF)-producing vaccines induces rejection of subcutaneous and metastatic tumors accompanied by autoimmune depigmentation, J Exp Med, 190 (1999) 355–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Ramagopal UA, Liu W, Garrett-Thomson SC, Bonanno JB, Yan Q, Srinivasan M, Wong SC, Bell A, Mankikar S, Rangan VS, Deshpande S, Korman AJ, Almo SC, Structural basis for cancer immunotherapy by the first-in-class checkpoint inhibitor ipilimumab, Proc Natl Acad Sci U S A, 114 (2017) E4223–E4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Hu W, Huang F, Ning L, Hao J, Wan J, Hao S, Enhanced immunogenicity of leukemia-derived exosomes via transfection with lentiviral vectors encoding costimulatory molecules, Cell Oncol (Dordr), 43 (2020) 889–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Barrueto L, Caminero F, Cash L, Makris C, Lamichhane P, Deshmukh RR, Resistance to Checkpoint Inhibition in Cancer Immunotherapy, Transl Oncol, 13 (2020) 100738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Fu W, Lei C, Liu S, Cui Y, Wang C, Qian K, Li T, Shen Y, Fan X, Lin F, Ding M, Pan M, Ye X, Yang Y, Hu S, CAR exosomes derived from effector CAR-T cells have potent antitumour effects and low toxicity, Nat Commun, 10 (2019) 4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Garcia-Lora A, Algarra I, Garrido F, MHC class I antigens, immune surveillance, and tumor immune escape, J Cell Physiol, 195 (2003) 346–355. [DOI] [PubMed] [Google Scholar]
- [209].Ogretmen B, McCauley MD, Safa AR, Molecular mechanisms of loss of beta 2-microglobulin expression in drug-resistant breast cancer sublines and its involvement in drug resistance, Biochemistry, 37 (1998) 11679–11691. [DOI] [PubMed] [Google Scholar]
- [210].Wang H, Liu B, Wei J, Beta2-microglobulin(B2M) in cancer immunotherapies: Biological function, resistance and remedy, Cancer Lett, 517 (2021) 96–104. [DOI] [PubMed] [Google Scholar]
- [211].Hsu DH, Paz P, Villaflor G, Rivas A, Mehta-Damani A, Angevin E, Zitvogel L, Le Pecq JB, Exosomes as a tumor vaccine: enhancing potency through direct loading of antigenic peptides, J Immunother, 26 (2003) 440–450. [DOI] [PubMed] [Google Scholar]
- [212].Lee YS, Kim SH, Cho JA, Kim CW, Introduction of the CIITA gene into tumor cells produces exosomes with enhanced anti-tumor effects, Exp Mol Med, 43 (2011) 281–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Morishita M, Takahashi Y, Matsumoto A, Nishikawa M, Takakura Y, Exosome-based tumor antigens-adjuvant co-delivery utilizing genetically engineered tumor cell-derived exosomes with immunostimulatory CpG DNA, Biomaterials, 111 (2016) 55–65. [DOI] [PubMed] [Google Scholar]
- [214].Damo M, Wilson DS, Simeoni E, Hubbell JA, TLR-3 stimulation improves anti-tumor immunity elicited by dendritic cell exosome-based vaccines in a murine model of melanoma, Sci Rep, 5 (2015) 17622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Liu H, Chen L, Liu J, Meng H, Zhang R, Ma L, Wu L, Yu S, Shi F, Li Y, Zhang L, Wang L, Feng S, Zhang Q, Peng Y, Wu Q, Liu C, Chang X, Yang L, Uemura Y, Yu X, Liu T, Co-delivery of tumor-derived exosomes with alpha-galactosylceramide on dendritic cell-based immunotherapy for glioblastoma, Cancer Lett, 411 (2017) 182–190. [DOI] [PubMed] [Google Scholar]
- [216].Qi H, Liu C, Long L, Ren Y, Zhang S, Chang X, Qian X, Jia H, Zhao J, Sun J, Hou X, Yuan X, Kang C, Blood Exosomes Endowed with Magnetic and Targeting Properties for Cancer Therapy, ACS Nano, 10 (2016) 3323–3333. [DOI] [PubMed] [Google Scholar]
- [217].Maciejko L, Smalley M, Goldman A, Cancer Immunotherapy and Personalized Medicine: Emerging Technologies and Biomarker-Based Approaches, J Mol Biomark Diagn, 8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Waldman AD, Fritz JM, Lenardo MJ, A guide to cancer immunotherapy: from T cell basic science to clinical practice, Nat Rev Immunol, 20 (2020) 651–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].https://www.hopkinsmedicine.org/inhealth/about-us/immunotherapy-precision-medicine-action-policy-brief.html.
- [220].Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, Patnaik A, Aggarwal C, Gubens M, Horn L, Carcereny E, Ahn MJ, Felip E, Lee JS, Hellmann MD, Hamid O, Goldman JW, Soria JC, Dolled-Filhart M, Rutledge RZ, Zhang J, Lunceford JK, Rangwala R, Lubiniecki GM, Roach C, Emancipator K, Gandhi L, K.−. Investigators, Pembrolizumab for the treatment of non-small-cell lung cancer, N Engl J Med, 372 (2015) 2018–2028. [DOI] [PubMed] [Google Scholar]
- [221].Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, Sosman JA, McDermott DF, Powderly JD, Gettinger SN, Kohrt HE, Horn L, Lawrence DP, Rost S, Leabman M, Xiao Y, Mokatrin A, Koeppen H, Hegde PS, Mellman I, Chen DS, Hodi FS, Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients, Nature, 515 (2014) 563–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Carney WP, HER2 status is an important biomarker in guiding personalized HER2 therapy, Per Med, 2 (2005) 317–324. [DOI] [PubMed] [Google Scholar]
- [223].Merhi M, Raza A, Inchakalody VP, Siveen KS, Kumar D, Sahir F, Mestiri S, Hydrose S, Allahverdi N, Jalis M, Relecom A, Al Zaidan L, Hamid MSE, Mostafa M, Gul ARZ, Uddin S, Al Homsi M, Dermime S, Persistent anti-NY-ESO-1-specific T cells and expression of differential biomarkers in a patient with metastatic gastric cancer benefiting from combined radioimmunotherapy treatment: a case report, J Immunother Cancer, 8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Bonomi M, Ahmed T, Addo S, Kooshki M, Palmieri D, Levine BJ, Ruiz J, Grant S, Petty WJ, Triozzi PL, Circulating immune biomarkers as predictors of the response to pembrolizumab and weekly low dose carboplatin and paclitaxel in NSCLC and poor PS: An interim analysis, Oncol Lett, 17 (2019) 1349–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Marcus L, Lemery SJ, Keegan P, Pazdur R, FDA Approval Summary: Pembrolizumab for the Treatment of Microsatellite Instability-High Solid Tumors, Clin Cancer Res, 25 (2019) 3753–3758. [DOI] [PubMed] [Google Scholar]
- [226].Strickler JH, Hanks BA, Khasraw M, Tumor Mutational Burden as a Predictor of Immunotherapy Response: Is More Always Better?, Clin Cancer Res, 27 (2021) 1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Lazzari C, Gregorc V, Cangi MG, Giovannetti E, Bulotta A, Santarpia M, Combined exosomal RNA and circulating tumor DNA for epidermal growth factor mutation detection in non-small cell lung cancer, J Thorac Dis, 10 (2018) S4023–S4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Boeri M, Milione M, Proto C, Signorelli D, Lo Russo G, Galeone C, Verri C, Mensah M, Centonze G, Martinetti A, Sottotetti E, Pastorino U, Garassino MC, Sozzi G, Circulating miRNAs and PD-L1 Tumor Expression Are Associated with Survival in Advanced NSCLC Patients Treated with Immunotherapy: a Prospective Study, Clin Cancer Res, 25 (2019) 2166–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Guo X, Qiu W, Liu Q, Qian M, Wang S, Zhang Z, Gao X, Chen Z, Xue H, Li G, Immunosuppressive effects of hypoxia-induced glioma exosomes through myeloid-derived suppressor cells via the miR-10a/Rora and miR-21/Pten Pathways, Oncogene, 37 (2018) 4239–4259. [DOI] [PubMed] [Google Scholar]
- [230].Jiang M, Zhang W, Zhang R, Liu P, Ye Y, Yu W, Guo X, Yu J, Cancer exosome-derived miR-9 and miR-181a promote the development of early-stage MDSCs via interfering with SOCS3 and PIAS3 respectively in breast cancer, Oncogene, 39 (2020) 4681–4694. [DOI] [PubMed] [Google Scholar]
- [231].Cooks T, Pateras IS, Jenkins LM, Patel KM, Robles AI, Morris J, Forshew T, Appella E, Gorgoulis VG, Harris CC, Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246, Nat Commun, 9 (2018) 771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Liu J, Fan L, Yu H, Zhang J, He Y, Feng D, Wang F, Li X, Liu Q, Li Y, Guo Z, Gao B, Wei W, Wang H, Sun G, Endoplasmic Reticulum Stress Causes Liver Cancer Cells to Release Exosomal miR-23a-3p and Up-regulate Programmed Death Ligand 1 Expression in Macrophages, Hepatology, 70 (2019) 241–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [233].Ning T, Li J, He Y, Zhang H, Wang X, Deng T, Liu R, Li H, Bai M, Fan Q, Zhu K, Ying G, Ba Y, Exosomal miR-208b related with oxaliplatin resistance promotes Treg expansion in colorectal cancer, Mol Ther, (2021). [DOI] [PMC free article] [PubMed]
- [234].Xiao L, He Y, Peng F, Yang J, Yuan C, Endometrial Cancer Cells Promote M2-Like Macrophage Polarization by Delivering Exosomal miRNA-21 under Hypoxia Condition, J Immunol Res, 2020 (2020) 9731049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].Ren W, Zhang X, Li W, Feng Q, Feng H, Tong Y, Rong H, Wang W, Zhang D, Zhang Z, Tu S, Zhu X, Zhang Q, Exosomal miRNA-107 induces myeloid-derived suppressor cell expansion in gastric cancer, Cancer Manag Res, 11 (2019) 4023–4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].Li X, Lei Y, Wu M, Li N, Regulation of Macrophage Activation and Polarization by HCC-Derived Exosomal lncRNA TUC339, Int J Mol Sci, 19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Liang ZX, Liu HS, Wang FW, Xiong L, Zhou C, Hu T, He XW, Wu XJ, Xie D, Wu XR, Lan P, LncRNA RPPH1 promotes colorectal cancer metastasis by interacting with TUBB3 and by promoting exosomes-mediated macrophage M2 polarization, Cell Death Dis, 10 (2019) 829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].Ni C, Fang QQ, Chen WZ, Jiang JX, Jiang Z, Ye J, Zhang T, Yang L, Meng FB, Xia WJ, Zhong M, Huang J, Breast cancer-derived exosomes transmit lncRNA SNHG16 to induce CD73+gammadelta1 Treg cells, Signal Transduct Target Ther, 5 (2020) 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Sun J, Jia H, Bao X, Wu Y, Zhu T, Li R, Zhao H, Tumor exosome promotes Th17 cell differentiation by transmitting the lncRNA CRNDE-h in colorectal cancer, Cell Death Dis, 12 (2021) 123. [DOI] [PMC free article] [PubMed] [Google Scholar]


