Abstract
Snail/Slug family proteins have been identified in diverse species of both vertebrates and invertebrates. The proteins contain four to six zinc fingers and function as DNA-binding transcriptional regulators. Various members of the family have been demonstrated to regulate cell movement, neural cell fate, left-right asymmetry, cell cycle, and apoptosis. However, the molecular mechanisms of how these regulators function and the target genes involved are largely unknown. In this report, we demonstrate that human Slug (hSlug) is a repressor and modulates both activator-dependent and basal transcription. The repression depends on the C-terminal DNA-binding zinc fingers and on a separable repression domain located in the N terminus. This domain may recruit histone deacetylases to modify the chromatin and effect repression. Protein localization study demonstrates that hSlug is present in discrete foci in the nucleus. This subnuclear pattern does not colocalize with the PML foci or the coiled bodies. Instead, the hSlug foci overlap extensively with areas of the SC-35 staining, some of which have been suggested to be sites of active splicing or transcription. These results lead us to postulate that hSlug localizes to target promoters, where activation occurs, to repress basal and activator-mediated transcription.
The transcriptional regulator Snail is the prototype of a family of zinc finger proteins that participate in various developmental and physiological processes. The snail mutant was first identified in a large-scale screen for genes involved in Drosophila embryonic patterning (45). Embryos that are homozygous for loss-of-function mutations of snail exhibit defects in gastrulation, mesoderm formation, and germ band retraction (16, 45, 54). snail is expressed in the ventral cells of the blastoderm-stage embryo (1, 33, 35). By directly binding to the target promoters, Snail represses neuroectodermal genes such as rhomboid and single-minded in the mesodermal territory to prevent the mixing of cell fates (6, 29, 32, 33, 37). Snail may also regulate other target genes that are important for ventral cell invagination (8, 23, 28).
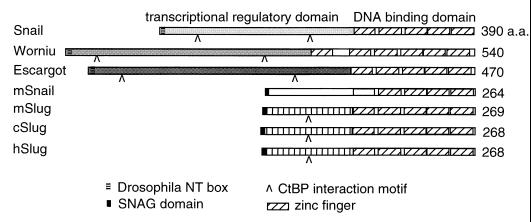
A number of genes that encode proteins with extensive homology to Snail in the zinc finger domain have been identified in various species (2, 11, 12, 19, 30, 31, 34, 38, 43, 44, 48, 50, 51, 53, 55, 58–60, 63, 65). snail and related genes in Drosophila, including escargot, worniu, and scratch, have been shown to be critical, in some cases redundantly, for wing imaginal cell development or neural cell fate determination (2, 14, 15, 22, 48). The urochordate Ciona genome has a snail homolog that is expressed in the dorsal neuroectoderm and functions as a repressor for the brachyury gene (12, 13). Similarly, a cephalochordate snail is expressed in paraxial mesoderm and lateral neural plate (34). Such expression of the protochordate snail genes is reminiscent of the embryonic patterns of vertebrate snail homologs, including those from the frog, chicken, zebra fish, and mouse (7, 19, 31, 43, 44, 50, 51, 53, 55, 58, 59). The vertebrate homologs can be further divided into the Snail and Slug subgroups (53). While both subgroups contain similar zinc finger domains in the C termini, members of the Slug subgroups are also particularly highly conserved among themselves throughout the N termini (Fig. 1).
FIG. 1.
Structural relationship of Snail family proteins. The three proteins shown on the top, Snail, Worniu, and Escargot, are from Drosophila. mSnail is from the mouse, and the three Slug proteins are from the mouse, chicken, and human, respectively. The other members of the family, including two zebra fish Snails, frog Snail and Slug, protochordate Snails, chicken Snail, human Snail, and Drosophila Scratch, are not shown here. The N termini of all three Drosophila proteins are highly divergent among themselves; these regions are also highly divergent among vertebrate and invertebrate proteins. The Drosophila N-terminal (NT) box and the vertebrate SNAG domain are different motifs, but both contain highly basic amino acid residues. The C-terminal binding protein (CtBP) interaction motif has the sequence related to P-DLS-K/R (41, 42, 47). The DBD contains four to six highly conserved zinc fingers.
The expression patterns of Snail and Slug proteins suggest conserved functions in cell migration during embryonic development. Functional studies demonstrate that chick and frog embryos incubated with Slug antisense oligonucleotides exhibit defects in early development (7, 44). These defects include epithelial-mesenchymal transition of the mesodermal cells, migration of neural crest cells, and formation of neural tubes. In contrast, null mutations of a Slug homolog in the mouse (mSlug or Slugh) do not exhibit obvious defects in the embryo (31), suggesting that the function of Slug in the mouse may be substituted by other Snail family proteins (7, 31). In addition to the possible roles in controlling cell migration, Snail-related proteins also participate in determining left-right asymmetry (30) and in regulating apoptosis (26, 39). Such diversity of cellular processes that Snail proteins are involved in underscores the importance of these evolutionarily conserved proteins. Some Snail family proteins have been demonstrated to function as transcriptional regulators, but very limited number of in vivo target genes have been identified (29, 32). Furthermore, the molecular mechanism of how this family of proteins mediate various cellular processes, which when perturbed lead to the observed phenotypes, is largely unknown.
We demonstrate here that the human Slug (hSlug) is a transcriptional repressor. Both activator-dependent and basal transcription are repressed by hSlug. The repression depends on DNA binding, but the DNA-binding zinc fingers are necessary but not sufficient to mediate repression. Deletion analysis reveals that the N terminus of hSlug, when linked to a heterologous DNA-binding domain (DBD), can mediate repression. While this 129-amino-acid (a.a.) N terminus possesses multiple regulatory motifs, the first 32 aa are responsible for most of the repressor activity. The repression is alleviated by tricostatin A (TSA), suggesting a possible involvement of histone deacetylases (HDACs) and chromatin modification. Immunofluorescence staining reveals a punctated pattern of hSlug localization in interphase nuclei. This subnuclear distribution colocalizes with SC-35 staining but not with PML foci or coiled bodies. Together, these results suggest that rather than bringing the target genes into separate chromatin domains to achieve silencing, hSlug modulates target promoters locally where both activation and repression occur.
MATERIALS AND METHODS
cDNA isolation and Northern analysis.
A sequence search was performed using the sequence of Drosophila snail to match expression sequences from the human EST (expressed sequence tag) database. The full-length Slug cDNA was subsequently identified from a melanocyte cDNA library (Soares melanocyte 2NbHM; constructed by Bento Soares and M. Fatima Bonaldo, National Institutes of Health) and confirmed by sequencing. Comparison with the genomic sequence available from the database (accession no. AF042001) (11) and Northern assay showed that it is likely a full-length cDNA. Northern analysis was performed using blots that contained approximately 2 μg of mRNA from various adult human tissues (Clontech product no. 7760-1 and 7759-1). The blots were hybridized with random prime-labeled full-length hSlug cDNA. Hybridization was carried out for 20 h at 42°C in a buffer that contained 50% formamide. The blot was then washed sequentially with buffers that contained 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate (SDS) and 0.5× SSC–0.1% SDS. The final wash was carried out at 42°C. The rat 18S rRNA probe was used to perform subsequent hybridization on the same blots.
Electrophoretic mobility shift assays.
The double-stranded Snail-binding site (SBS) was annealed from synthesized oligonucleotides with the sequences 5′-TGAGGTAGCAGGTGCACG-3′and 5′-TGAGCGTGCACCTGCTAC-3′. The underlined sequence is the core recognition sequence for Snail family proteins. The SBS mutant oligonucleotides change this core sequence to 5′-GTTACT-3′ and 5′-AGTAAC-3′, respectively. The annealed SBS was end labeled with T4 polynucleotide kinase in the presence of [γ-32P]ATP and then filled in the overhangs with Klenow polymerase in the presence of deoxynucleotides. The end-labeled DNA fragment was incubated with Escherichia coli extract programmed to express hSlug. The wild-type and zinc finger-deleted hSlug coding sequences were placed into the pAR3040 vector. The plasmids were transformed into E. coli BL21, and protein production was induced with isopropyl-β-d-thiogalactopyranoside. The end-labeled DNA and the protein extracts were allowed to incubate in a buffer as described previously (27) for 10 min at room temperature. The mixture was then analyzed in a 4% acrylamide gel, followed by autoradiography.
Plasmid construction and transfection assays.
The hSlug cDNA sequence from −85 to +984 (where +1 is the translation start site) was cloned into the pCDNA3 vector for transfection. The reporter plasmid contains a basal promoter placed upstream of the luciferase (luc) gene coding sequence. The hSlug response sequence contains three tandem repeats of the SBS (the sequence of the top strand is 5′-AGCTTAGCAGGTGCACGATATCAGCAGGTGCACCATATGAGCAGGTGCAA-3′). The SBS fragment was cloned into the HindIII site of the reporter vector containing either the 7× AP1 recognition sequence or 4× Gal4 recognition sequence. The hSlug deletion series were constructed by PCR amplification of specific regions and then fusion of various DNA fragments with the Gal4 DBD in the pCDNA3 vector. The primers for generating the deletions and fusions are P1 (5′-CGGGGTACCCCTGGCCCGCCGCGATGC-3′), P2 (5′-CGCGGATCCCTGAAACTTTTCAGCTTC-3′), P3 (5′-CGCGGATCCATGAAGCTACTGTCTTCT-3′ [for Gal4 DBD]), P4 (5′-CTAGTCTAGATCAGAATTCCGGCGATACAGT-3′ [for Gal4 DBD]), P5 (5′-CGGGGTACCATGTTTCAGTGCAATTTATGC-3′), P6 (5′-GTGGGAATTCCATATGTCAGTGTGCTACACAGCA-3′), P7 (5′-CGCGGATCCGGTGTCAGATGGAGGAGG-3′), P8 (5′-CGCGGATCCAGCAGCGGTAGTCCACAC-3′), P9 (5′-CGCGGATCCATACGGGGAAATAATCAC-3′), P10 (5′-CGCGGATCCGCTGTAGTTTGGCTTTTT-3′), P11 (5′-CGGGGTACCCGCCAGACCCGCTG GCAAGATGCTCTATGAGAGTTACTCC-3′), P12 (5′-CGGGGTACCCGCCAGACCCGCTGGCAAGATGGCATACAGCCCCATCACT-3′), and P13 (5′-GTGATTATTTCCCCGTATCCATTCCACGCCCAGCTA-3′). For generating mutants M1 to M15 by PCR, the primer combinations given in parentheses were used: M1 (P1 and P2), M2 (P5 and P6), M3 (P1 and P2), M4 (P1 and P7), M5 (P1 and P8), M6 (P1 and P9), M7 (P1 and P10), M8 (P11 and P2), M9 (P11 and P7), M10 (P11 and P8), M11 (P12 and P2), M12 (P12 and P7), M13 (P12 and P8), M14 (P1, P13, and P2), M15 (P1, P13, and P7). The fragments of M1 and M2 were cloned directly into the pCDNA3 vector, while the products of M3 to M15 were cloned into pCDNA3 vector along with the Gal4 DBD PCR fragment amplified by primers P3 and P4 to generate the fusions.
Antibody production and immunofluorescence staining.
Full-length hSlug protein was expressed in E. coli BL21 and purified by SDS-polyacrylamide gel electrophoresis. The purified proteins were used to immunize two guinea pigs (Pocono Rabbit Farms, Canadensis, Pa.). The antibodies were affinity purified by the filter binding method and then used for immunofluorescence staining. HeLa cells or 293T cells were grown on coverslips, fixed in methanol, and rehydrated with 1× phosphate-buffered saline (PBS). The cells were then blocked in PBT (1× PBS, 2% bovine serum albumin, 0.5% Triton X-100) for 20 min, with one change of the buffer. Cells were incubated with antibodies in the same buffer for 1 h, washed 10 times each with 100 μl of PBT, and then incubated with fluorochrome-conjugated secondary antibodies for 20 min in the same buffer. The cells were washed a few times with PBT, once with 1× PBS, and once with deionized water and then mounted in glycerol-antifade (1 mg of p-phenylenediamine [Sigma] per ml, 9 ml of glycerol, 1 ml of 10× PBS) medium. Confocal microscopy was carried out using a Leica TCS NT microscope. The affinity-purified hSlug antibodies were used at 1:5 dilution, and the PML (36) and coilin (67) antibodies were used at 1:100 dilution; both antibodies were raised in rabbits. The SC-35 monoclonal antibodies (Sigma) were used at 1:1,000 dilution. The hemagglutinin (HA) monoclonal antibody was used at 1:100 dilution.
RESULTS
Structure and expression of hSlug.
We searched for genes in the human EST database that have homology with Drosophila snail. Subsequently, we isolated a full-length cDNA that has homology to snail in the zinc finger-encoding region and to chicken Slug throughout the coding region. This hSlug sequence has since been reported by other laboratories (11, 26). The zinc finger domains of the Slug proteins are homologous (about 70% identity) to those of all other members of the Snail family. However, the N termini of the Slug subgroup, while 95% identical among themselves, are more divergent from other Snail proteins (Fig. 1) (31, 38, 44). Another interesting feature is that the N-terminal 7 aa are conserved among vertebrate Snail family proteins and Gfi-1 proteins. These amino acids constitute part of the SNAG (Snail/Gfi-1) domain, which has been shown in the Gfi-1 proto-oncoprotein to be essential for mediating transcriptional repression and nuclear localization (18). In addition, many Snail family proteins contain one to two short stretches that are similar to the P-DLS-R/K sequence, which are potential C-terminal binding protein corepressor interaction motifs (41, 42, 47).
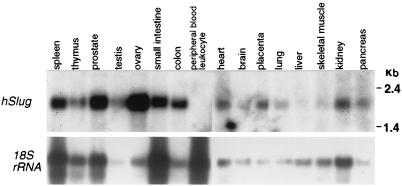
To gain insight into the possible functions of hSlug, we examined the expression in various adult human tissues. Northern analysis revealed a prominent band of approximately 2.2 kb (Fig. 2), a size similar to that of the cDNA obtained. The hybridization signals were detected in all tissues tested except peripheral blood leukocytes, similar to findings presented in a recent report (26). The level of expression was higher in the ovary than in other tissues tested. Thus, the hSlug transcript is expressed in most adult human tissues.
FIG. 2.
mRNA expression of hSlug in adult tissues. The hSlug full-length cDNA was used to prepare a radioactive probe, which was then used to hybridize with mRNA from various human tissues. The analysis reveals a single mRNA species of approximately 2.2 kb that hybridized with the probe. Expression is relatively high in the ovary and almost undetectable in peripheral blood leukocytes. All other tissues tested have detectable and variable levels of expression. The lower panel shows hybridization of the same blots after stripping using the rat 18S rRNA probe, which cross-hybridized with the homologous human transcripts.
hSlug binds to the consensus sequence that interacts with Snail family proteins.
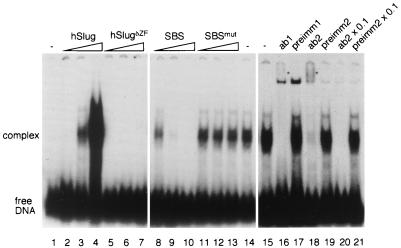
The DNA-binding ability of hSlug was examined by electrophoretic mobility shift assays. Since there is no known target gene in humans, we designed an oligonucleotide (SBS) that contains the consensus core CAGGTG sequence, to which Snail, Escargot, and mSnail can bind (14, 29, 37, 40). The bacterial extract containing hSlug exhibited a prominent DNA-binding activity that interacted with this oligonucleotide (Fig. 3). This activity was present only in extract that was programmed to express hSlug (lanes 2 to 4), not the control extract (lane 1). Furthermore, the DNA-binding activity was absent if the zinc finger domain was deleted (lanes 5 to 7). These results demonstrate that hSlug can interact with DNA and that the binding requires the zinc finger domain.
FIG. 3.
hSlug is a sequence-specific DNA-binding protein. Bacterial extracts that contained full-length or zinc finger domain-truncated hSlug proteins were incubated with a double-stranded oligonucleotide probe that contained the consensus SBS. The mixture was then analyzed on an acrylamide gel. A prominent protein-DNA complex was detected that has slower mobility than the free DNA (lanes 2 to 4, increasing amount of extract). This complex was not seen in extract that contained the zinc finger domain-deleted hSlug (lanes 5 to 7). The competition assay (lane 8 to 14) demonstrates that the wild-type, unlabeled oligonucleotide (SBS) is an efficient competitor, while the oligonucleotide that contained mutations in the recognition core (SBSmut) could not compete. One microliter each of two antisera (ab1 and ab2) raised against full-length hSlug protein was added in similar assays (lane 15 to 21). The antisera abolished complex formation, while the preimmune sera (preimm1 and 2) did not. Weak bands (*) with slower mobility were also formed when the antisera were added to the mixture, indicating the formation of complexes that contained the antibodies for hSlug.
A competition assay was carried out to test the specificity of hSlug toward the target sequence. Increasing amounts of the same, unlabeled SBS competed efficiently with the binding (Fig. 3, lanes 8 to 10), while a similar oligonucleotide that had the core consensus mutated could not compete (lanes 11 to 13). The results demonstrate that hSlug is a sequence-specific DNA-binding protein and can recognize the same target site as other members of the family. A recent study (26) using random selection also revealed that hSlug can interact with sequences containing the core consensus that we used here.
To further demonstrate that the DNA-binding activity in the bacterial extract contained the hSlug protein, we performed antibody interaction assays. Antibodies were raised using nonfusion, full-length proteins which were purified through gel electrophoresis separation and subsequent elution. Antisera obtained from two immunized animals contained activities that blocked the formation of the DNA-protein complex in the mobility shift assay (Fig. 3, lanes 16 and 18). The disappearance of the major complex was concomitant with the appearance of a weak supershift complex. The preimmune sera did not exhibit this activity (lanes 17 and 19). These results further support the conclusion that hSlug is the protein in the extracts responsible for the specific DNA-binding activity.
Repression of basal and activated transcription by hSlug.
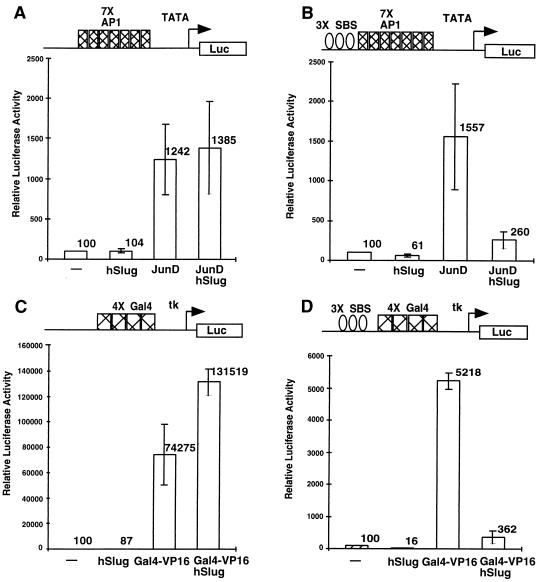
To test whether hSlug can function similar to Drosophila Snail as a transcriptional regulator, we performed transfection assays using the target DNA motif tested in the previous series of experiments. Human 293T embryonic kidney cells were cotransfected with various combinations of plasmids that contained different protein coding sequences under the control of the cytomegalovirus promoter and different target motifs placed upstream of a minimal promoter-driven luc reporter gene. The low level of activity exhibited by the luc reporter was considered the basal transcriptional activity. The addition of JunD or Gal4-VP16 activator resulted in much higher luciferase activity, which was dependent on the presence of correct targets (Fig. 4A and C), because in the absence of an AP1 or Gal4 binding motif no activation occurred (data not shown). Cotransfection of an hSlug expression plasmid into this system did not result in a significant change of reporter activity (Fig. 4A and C), demonstrating that expression of hSlug alone cannot modulate the transcriptional activities of irrelevant target genes. In contrast, when SBS were placed upstream of the activator sites, the reporter activities were much lower in the presence of cotransfected hSlug (Fig. 4B and D). The results demonstrate that hSlug, upon binding to the targets, can repress activator-dependent transcription.
FIG. 4.
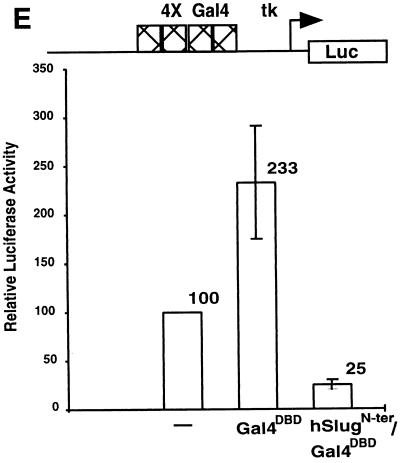
Repression of basal and activated transcription by hSlug. Human 293T cells were transfected with different combinations of expression and reporter plasmids. The specific DNA-binding sites on the promoter of the reporter plasmids used are illustrated at the top of each panel. Luciferase reporter activities were measured 48 h after transfection. In the presence of appropriate binding sites, JunD (A) and Gal4-VP16 (C) increased the reporter activity substantially, indicating activation of transcription. The addition of hSlug did not repress transcription if the binding sites for hSlug were not present (A and C). In the presence of the SBS, hSlug repressed the activated transcription by both JunD and Gal4-VP16 (B and D). Furthermore, the basal transcription was also repressed significantly by hSlug (B and D). Panel E shows that a fusion construct, the hSlug N terminus fused with the Gal4 DBD, repressed basal transcription. The reporter contained the Gal4 binding sites and was modestly activated by the GAL4 DBD alone. Therefore, occupation of the binding sites by the hSlug fusion reduces the activity to a level much lower than the basal level, suggesting active repression.
Careful examination of the reporter gene activity revealed that basal transcription was also repressed by hSlug (Fig. 4B and D). This repression of basal activity was dependent on the correct target site, since reporter plasmids that did not contain SBS were not repressed by hSlug expression (Fig. 4A and C). Although the repression of basal transcription on the 7× AP1 recognition sequence-containing reporter was not as efficient, better repression could be obtained with increasing amounts of transfected hSlug (data not shown). It may be that there are endogenous activators that interact with these AP1 sites and a higher concentration of hSlug is required to overcome this activity.
Alternatively, the apparent repression of basal transcription could be due to the competition by hSlug of an endogenous protein that activates through interaction with the SBS. To further investigate this alternative possibility, we constructed a fusion protein that contained the N terminus of hSlug and the heterologous Gal4 DBD. Cotransfection of the Gal4 DBD alone led to a modest increase in reporter activity, showing that the occupation of the target promoter by a DNA-binding protein does not automatically result in repression. The cotransfection of hSlug and the Gal4 DBD, in contrast, led to a 4 fold decrease in reporter activity compared with the reporter plasmid itself, or a 10-fold decrease compared with the reporter in the presence of Gal4 DBD (Fig. 4E). The result confirms that hSlug can repress transcription from the basal promoter.
hSlug-dependent repression is mediated through a separable N-terminal domain.
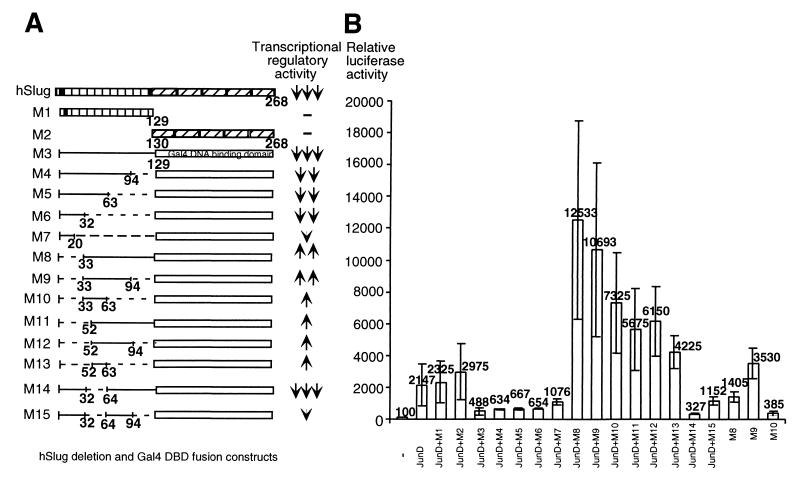
Systematic analysis of the structural requirement of hSlug reveals that a repression domain is present in the N terminus of the protein. We first examined the C-terminal DBD or the N-terminal half of hSlug and found that neither was sufficient for repressing JunD-activated transcription (Fig. 5, M1 and M2). Therefore, it is likely that the DBD brings the protein to the target promoters and the N-terminal regulatory domain mediates the repression. To test this hypothesis, we designed a construct that had the N terminus of hSlug fused with the heterologous Gal4 DBD. Cotransfection assays demonstrated that this fusion protein (construct M3) repressed the activated transcription by JunD in a Gal4 binding-site-dependent manner (Fig. 5 and data not shown). As shown in Fig. 4, this fusion protein also repressed basal transcription.
FIG. 5.
The N terminus of hSlug contains both repression and activation modules. A transfection assay using the M1 and M2 constructs and SBS-containing reporter demonstrates that neither the N or C terminus of hSlug changes the reporter activity. Various portions of the N terminus of hSlug were then fused in frame with the Gal4 DBD (A). The constructs were cotransfected with a luciferase reporter that contained both four Gal4 binding sites (for hSlug-Gal4 repression) and seven AP1 binding sites (for JunD activation) (B). The N-terminal hSlug-Gal4 DBD fusion (M3) repressed activation efficiently, demonstrating the presence of the repression domain in the N terminus. Serial deletion shows that the first 32 aa contain the most potent repression domain. This domain is dominant over the central activation domain (aa 33 to 94). The region from aa 95 to 129 contains a helper domain for repression (compare M3 and M4 with M14 and M15), since the construct M14 presents the best repressor activity. However, this helper domain is not sufficient to override the activation by the central domain (M8). Full-length hSlug, M1, and M2 were transfected with a different reporter that contained the SBS target.
The N-terminal domain was further analyzed by deleting various portions of the protein. Sequential deletion from the C terminus (constructs M4 to M6) caused only minor loss of the repressor function. Thus, the first 32 aa of the protein contains the major activity. These result are consistent with the implication of the SNAG domain in repression. However, unlike Gfi-1, the first 20 aa of hSlug is not sufficient for maximum repression. Half of the activity was lost when a smaller construct (M7) was analyzed. When the first 32 aa were deleted from the otherwise full-length N-terminal domain, the repressor activity disappeared (construct M8). Therefore, the 32-aa segment is necessary and no other region in the N terminus can replace its function.
Interestingly, deletion of the N-terminal 32 aa caused a dramatic change in regulatory activity, such that the fusion protein became a potent activator. This protein could function as an activator with or without the addition of JunD. The core activation domain was localized between aa 33 and 63 (M10), while it best activated when extended to aa 94 (M9). When the core central domain from aa 33 to 63 was deleted (M14 and M15), the activation was lost. The M14 construct, which contains all of the N terminus except the central domain actually, functioned as the best repressor. Therefore, the central domain may antagonize the repression function in the context of the full-length protein. The presence of such an activation domain implies that hSlug can be an activator in vivo or that it is an artifact of deletion manipulation (see Discussion).
The structure-function analysis, thus, demonstrates that the N terminus of hSlug contains a tripartite transcriptional regulatory domain. The core repressor domain is located in the first 32 aa of the protein. The middle 30 aa can activate transcription, but the activation potential is masked by the core repressor domain. The last 30 aa of the N terminus functions as a helper domain for repression. These different functional modules may contribute to the in vivo regulatory activities of hSlug.
hSlug-mediated repression is alleviated by an HDAC inhibitor.
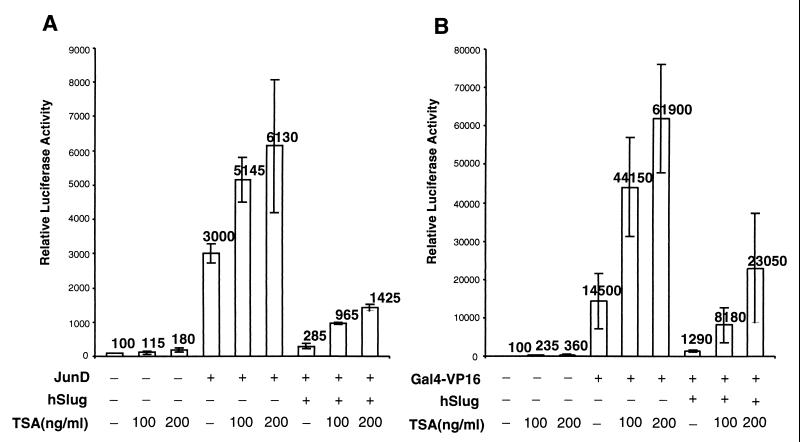
Many repressors recruit corepressor proteins to form the functional units which can modify chromatin (3, 9, 10, 66). Chromatin modification proteins include HDACs, which remove acetyl groups from the histones and lead to compacting of chromatin (4, 46). Since hSlug can repress activator-mediated and basal transcription, we tested whether hSlug-mediated repression may involve HDAC. Transfection assays were carried out in the presence of increasing amounts of the HDAC-specific inhibitor trichostatin A (TSA). As controls, the addition of TSA up to 200 ng/ml led to a modest (1.8- or 3.6-fold) increase of basal reporter activity; TSA also led to some (2- or 5-fold) increase of transcription in the presence of the activators (Fig. 6). While hSlug reduces the activator-mediated transcription approximately 10-fold, the addition of TSA led to a significant reversal of reporter activity. For JunD, TSA at 200 ng/ml increased transcription twofold, while the same concentration released the repression of hSlug fivefold (Fig. 6A). For Gal4-VP16, TSA alone increased transcription 5-fold, but the inhibitor released the repression of hSlug 18-fold (Fig. 6B). These results indicate that hSlug represses transcription possibly through the help of HDACs.
FIG. 6.
hSlug repression is affected by an HDAC inhibitor. The cultured cells were cotransfected with expression plasmids as indicated. The reporter plasmids contain the corresponding binding sites for both protein expression plasmids. The HDAC inhibitor TSA was added 24 h after transfection; the cells were harvested 24 h later. The presence of dimethyl sulfoxide solvent (−TSA) did not result in any relief of repression. Basal transcription and activated transcription could be elevated to some extent by the addition of TSA, demonstrating some nonspecific increase of transcription. However, the addition of TSA caused more significant relief of the hSlug-mediated repression (5-fold versus 2-fold in panel A; 18-fold versus 5-fold in panel B). These suggest that HDACs may mediate part of the repressor function of hSlug.
Colocalization of hSlug with transcription/splicing foci.
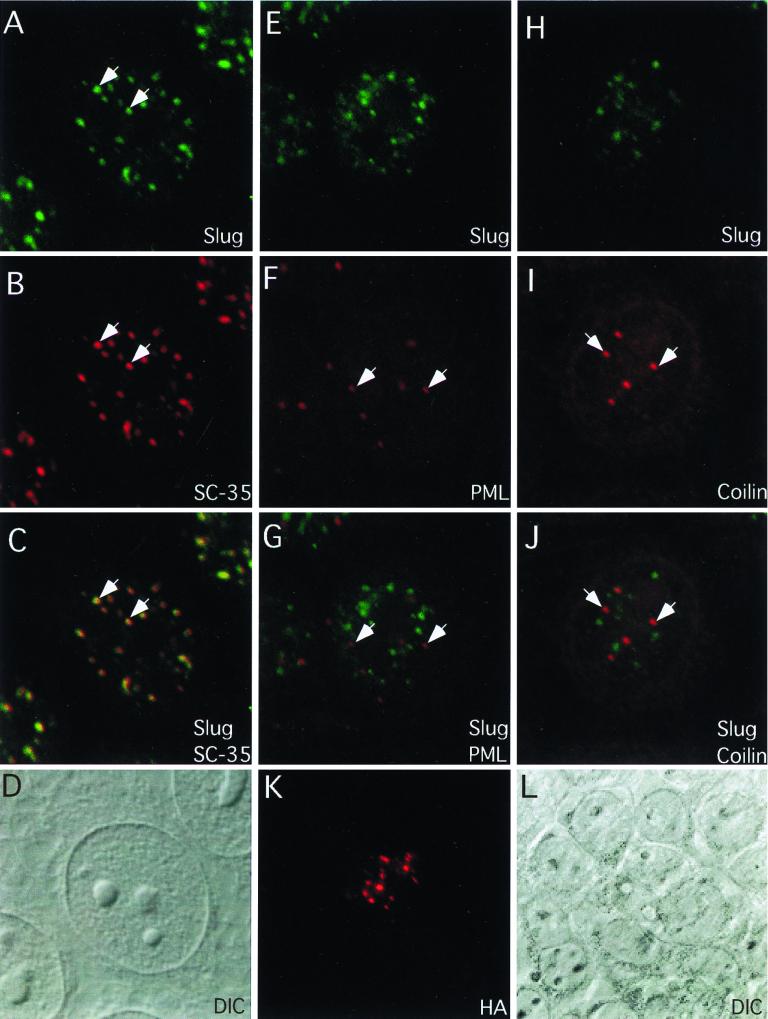
Immunofluorescence staining using anti-hSlug antibodies reveals that the proteins are present in discrete foci in interphase nuclei (Fig. 7). The hSlug antibodies were raised against bacterially expressed, gel-purified, full-length, nonfusion hSlug protein. Staining with affinity-purified antibodies revealed that more than 50 foci of hSlug could be detected in the nuclei of HeLa cells (Fig. 7A, E, and H) and 293T cells (data not shown). The staining was not observed with preimmune sera, and sera obtained from two independent animals showed identical pattern. Furthermore, the antisera showed the same staining pattern both before and after affinity purification. Also, staining was abolished when the antisera were first incubated with purified hSlug protein (data not shown). These same two antisera can inhibit in vitro hSlug DNA binding, as shown in Fig. 3. We further tested whether transfected hSlug exhibited specific nuclear staining. Full-length hSlug was fused with the HA tag, and the transfected protein was visualized by immunofluorescence staining using monoclonal anti-HA antibody. Cells that expressed the fusion protein frequently showed punctate HA staining (Fig. 7K, compare the differential interference contrast [DIC] image of surrounding cells in Fig. 7J). Thus, the subnuclear staining revealed by the anti-hSlug antibodies likely represents the distribution of endogenous hSlug protein.
FIG. 7.
Colocalization of hSlug with SC-35 domains. The antibodies raised against hSlug were affinity purified and used for immunofluorescence staining. The staining for hSlug is predominantly nuclear and punctate in HeLa cells (A to J) as well as in other cell types tested (data not shown). Furthermore, the transfected hSlug-HA fusion protein in 293T cells (K and L) also exhibited punctate nuclear staining. Double stainings were performed together with the anti-SC-35 (A to C), anti-PML (E to G), and anti-coilin (H to J) antibodies. There are fewer foci for PML, and the pattern does not overlap with that of hSlug (PML staining is indicated by arrows in panels F and G). Similar double staining with SC-35 shows that the two patterns overlap extensively (two examples are indicated by the arrows in panels A to C). The coiled bodies also contain splicing factors, but the double staining with coilin reveals that hSlug and coilin do not colocalize (coilin staining is indicated by arrows in panels I and J). All panels are images obtained from confocal microscopy.
The intensity and size vary among the hSlug foci, and these nuclear foci are not located in the nucleoli (compare the DIC image in Fig. 7D with the immunofluorescence staining in Fig. 7A). We then tested whether hSlug colocalized with other known nuclear structures. The PML protein involved in acute promyelocytic leukemia has been found in discrete nuclear foci, which colocalize with the SP100 and the HP1 heterochromatin protein foci (52, 61, 62). The double staining with PML showed that the hSlug staining did not overlap with the PML foci (Fig. 7E to G). The hSlug staining, on the other hand, is reminiscent of the punctate pattern of splicing factor SC-35 nuclear staining (25, 56, 57, 64). SC-35 foci represents active splicing sites as well as nascent transcription regions (25, 56, 57, 64). Indeed, double staining revealed that hSlug and SC-35 overlapped extensively (Fig. 7A to C).
Another well-studied nuclear structure is the coiled body, which has been suggested to be the site of spliceosome assembly (5). We tested whether hSlug might be associated with these sites by double staining of hSlug together with the monoclonal antibody R228, which recognized coilin, a resident protein of the coiled bodies. The results showed that there was no overlap of the two patterns (Fig. 7H to J). Therefore, it is more likely that hSlug is associated with active transcription or active-splicing regions, not the regions that assemble the splicing complexes.
DISCUSSION
We have demonstrated that the hSlug protein can function as a transcriptional repressor, and the repression depends on the N-terminal half, which is separable from the DNA-binding zinc fingers. The repression has a dominant effect on neighboring activator-mediated and basal transcription. hSlug appears to be colocalized with SC-35 foci in the nucleus. Such foci have been shown to be sites of active splicing and transcription. Thus, hSlug may repress gene expression by locating itself to the target sites where active transcription occurs.
The analysis of Drosophila Snail provided much information regarding the molecular function of this protein family. Snail binds directly to at least three target promoters and represses gene expression in the early embryo (6, 8, 23, 28, 29, 32, 33, 35, 37). The repression domain resides in the N terminus (17). However, this N terminus of Snail is highly divergent and is approximately twice the length of hSlug (Fig. 1). Therefore, it was not clear whether hSlug could function as a repressor, or whether the two proteins use any conserved motif to repress transcription. The results presented in this paper demonstrate that hSlug not only binds to similar target sequences but also represses transcription through the N terminus. The first 32 aa of the N terminus constitute the major repressor activity, and the region contains a partial SNAG domain. It has also been shown that mSnail functions as a repressor in cultured cells and that its SNAG domain is essential (40). However, we show here that the SNAG domain (20 aa) of hSlug is not sufficient. The most potent repression domain requires the first 32 aa, which are highly conserved among vertebrate Snail and Slug proteins. How this domain mediates repression requires further investigation.
A mechanism of repression is to heterochromatinize the target region to ensure long-range silencing for a long period of time (10, 24). Our results suggest that hSlug, although it can silence neighboring genes efficiently, does not seem to bring the target genes to heterochromatin domains. The staining of hSlug does not colocalize with the PML pattern, which has been shown to overlap with that of SP100 and heterochromatin protein HP1 (52, 61, 62). Instead, hSlug colocalizes with the splicing and transcription regions characterized by SC-35 staining (25, 56, 57, 64). Since hSlug repression is to some extent sensitive to TSA, we postulate that the repressor may recruit HDACs to modify local chromatin as part of the mechanism to inhibit transcription.
Interestingly, structure-function analysis reveals that hSlug may contain an activation domain in the N terminus (Fig. 5). Perhaps the Snail family proteins can function as activators or repressors at different target promoters (23), depending on parameters such as neighboring cofactors and binding sequences. Some other repressor proteins also contain both activation and repression modules (20, 21). Whether they are artifacts of protein dissection or represent in vivo function remains unclear. This can be verified only after more direct target genes are characterized.
Another issue that awaits investigation is whether the repression and subnuclear localization of mSlug can be linked to the biological functions and to the phenotypes observed in different organisms. mSlug has been demonstrated in prolymphocytes to possess antiapoptotic activity (26). Interestingly, the Caenorhabditis elegans protein Ces-1, a Snail family zinc finger protein most related to Scratch, was identified as an antiapoptotic molecule (39). It has been proposed based on genetic analysis that Ces-1 may be a repressor (39). The idea that the antiapoptotic activity of hSlug depends on gene repression can now be tested based on the results presented in this report.
Antisense experiments in chick and frog embryos, as well as in rat bladder carcinoma NBT-II cells, showed that the other vertebrate Slug homologs may participate in controlling cell movements during embryogenesis (7, 44, 49, 51). Gene knockout experiments in the mouse, however, demonstrate that null mutations of mSlug do not lead to any morphological phenotype (31). One possible explanation is the redundant function provided by other Snail-related proteins, as shown in frog embryos that the antisense-induced phenotype can be rescued by either Slug or Snail (7). Molecular genetic experiments in Drosophila also demonstrate possible redundant functions among different members of Snail family (2, 15). In addition to regulating cell movement, the cSnail can regulate left-right asymmetry (30), and Drosophila Snail family proteins have essential functions during nervous system development (2, 48). Details of how these proteins regulate the various biological processes are not known. However, at least in the case of Drosophila Snail, repression of the known target genes, though essential for mesoderm specification, is not sufficient to explain the gastrulation phenotype (23). Whether hSlug and other family members repress the same or different sets of target genes and how such regulation leads to the correct decision in various developmental and physiological processes remain to be determined.
ACKNOWLEDGMENTS
We thank Jeffrey Nickerson for much help with confocal microscopy, Jeanne Lawrence for providing some SC-35 antibodies, Yvonne Yannoni for the R228 antibodies, Peter Newburger for the 18S rRNA plasmid, and Settara C. Chandrasekharappa for support.
The work was funded by a research grant from the March of Dimes Birth Defects Foundation, a pilot grant from the Our Danny Cancer Fund of the University of Massachusetts Cancer Center, and a Scholar Award from the Leukemia Society of America (to Y. T. Ip) and by NIH grant R01DK52542 (to J. D. Chen).
REFERENCES
- 1.Alberga A, Boulay J-L, Kempe E, Dennefeld C, Haenlin M. The snail gene required for mesoderm formation is expressed dynamically in derivatives of all three germ layers. Development. 1991;111:983–992. doi: 10.1242/dev.111.4.983. [DOI] [PubMed] [Google Scholar]
- 2.Ashraf S, Hu X, Roote J, Ip Y T. The mesoderm determinant Snail collaborates with related zinc finger proteins to control Drosophila neurogenesis. EMBO J. 1999;18:6426–6438. doi: 10.1093/emboj/18.22.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashraf S I, Ip Y T. Transcriptional control: repression by local chromatin modification. Curr Biol. 1998;8:R683–686. doi: 10.1016/s0960-9822(98)70435-x. [DOI] [PubMed] [Google Scholar]
- 4.Ayer D E. Histone deacetylases: transcriptional repression with SINers and NuRDs. Trends Cell Biol. 1999;9:193–198. doi: 10.1016/s0962-8924(99)01536-6. [DOI] [PubMed] [Google Scholar]
- 5.Bohmann K, Ferreira J, Santama N, Weis K, Lamond A I. Molecular analysis of the coiled body. J Cell Sci Suppl. 1995;19:107–113. doi: 10.1242/jcs.1995.supplement_19.16. [DOI] [PubMed] [Google Scholar]
- 6.Boulay J L, Dennefeld C, Alberga A. The Drosophila developmental gene snail encodes a protein with nucleic acid binding fingers. Nature. 1987;330:395–398. doi: 10.1038/330395a0. [DOI] [PubMed] [Google Scholar]
- 7.Carl T F, Dufton C, Hanken J, Klymkowsky M W. Inhibition of neural crest migration in Xenopus using antisense Slug RNA. Dev Biol. 1999;213:101–115. doi: 10.1006/dbio.1999.9320. [DOI] [PubMed] [Google Scholar]
- 8.Casal J, Leptin M. Identification of novel genes in Drosophila reveals the complex regulation of early gene activity in the mesoderm. Proc Natl Acad Sci USA. 1996;93:10327–10332. doi: 10.1073/pnas.93.19.10327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J D, Li H. Coactivation and corepression in transcriptional regulation by steroid/nuclear hormone receptors. Crit Rev Eukaryot Gene Expr. 1998;8:169–190. doi: 10.1615/critreveukargeneexpr.v8.i2.40. [DOI] [PubMed] [Google Scholar]
- 10.Cockell M, Gasser S M. Nuclear compartments and gene regulation. Curr Opin Genet Dev. 1999;9:199–205. doi: 10.1016/S0959-437X(99)80030-6. [DOI] [PubMed] [Google Scholar]
- 11.Cohen M E, Yin M, Paznekas W A, Schertzer M, Wood S, Jabs E W. Human SLUG gene organization, expression, and chromosome map location on 8q. Genomics. 1998;51:468–71. doi: 10.1006/geno.1998.5367. [DOI] [PubMed] [Google Scholar]
- 12.Corbo J C, Erives A, Di Gregorio A, Chang A, Levine M. Dorsoventral patterning of the vertebrate neural tube is conserved in a protochordate. Development. 1997;124:2335–2344. doi: 10.1242/dev.124.12.2335. [DOI] [PubMed] [Google Scholar]
- 13.Fujiwara S, Corbo J C, Levine M. The snail repressor establishes a muscle/notochord boundary in the Ciona embryo. Development. 1998;125:2511–2520. doi: 10.1242/dev.125.13.2511. [DOI] [PubMed] [Google Scholar]
- 14.Fuse N, Hirose S, Hayashi S. Diploidy of Drosophila imaginal cells is maintained by a transcriptional repressor encoded by escargot. Genes Dev. 1994;8:2270–2281. doi: 10.1101/gad.8.19.2270. [DOI] [PubMed] [Google Scholar]
- 15.Fuse N, Hirose S, Hayashi S. Determination of wing cell fate by the escargot and snail genes in Drosophila. Development. 1996;122:1059–1067. doi: 10.1242/dev.122.4.1059. [DOI] [PubMed] [Google Scholar]
- 16.Grau Y, Carteret C, Simpson P. Mutations and chromosomal rearrangements affecting the expression of snail, a gene involved in embryonic patterning in Drosophila melanogaster. Genetics. 1984;108:347–360. doi: 10.1093/genetics/108.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gray S, Levine M. Short-range transcriptional repressors mediate both quenching and direct repression within complex loci in Drosophila. Genes Dev. 1996;10:700–710. doi: 10.1101/gad.10.6.700. [DOI] [PubMed] [Google Scholar]
- 18.Grimes H L, Chan T O, Zweidler-McKay P A, Tong B, Tsichlis P N. The Gfi-1 proto-oncoprotein contains a novel transcriptional repressor domain, SNAG, and inhibits G1 arrest induced by interleukin-2 withdrawal. Mol Cell Biol. 1996;16:6263–6272. doi: 10.1128/mcb.16.11.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammerschmidt M, Nusslein-Volhard C. The expression of a zebrafish gene homologous to Drosophila snail suggests a conserved function in invertebrate and vertebrate gastrulation. Development. 1993;119:1107–1118. doi: 10.1242/dev.119.4.1107. [DOI] [PubMed] [Google Scholar]
- 20.Han K, Manley J L. Functional domains of the Drosophila Engrailed protein. EMBO J. 1993;12:2723–2733. doi: 10.1002/j.1460-2075.1993.tb05934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han K, Manley J L. Transcriptional repression by the Drosophila even-skipped protein: definition of a minimal repression domain. Genes Dev. 1993;7:491–503. doi: 10.1101/gad.7.3.491. [DOI] [PubMed] [Google Scholar]
- 22.Hayashi S, Hirose S, Metcalfe T, Shirras A D. Control of imaginal cell development by the escargot gene of Drosophila. Development. 1993;118:105–115. doi: 10.1242/dev.118.1.105. [DOI] [PubMed] [Google Scholar]
- 23.Hemavathy K, Meng X, Ip Y T. Differential regulation of gastrulation and neuroectodermal gene expression by Snail in the Drosophila embryo. Development. 1997;124:3683–3691. doi: 10.1242/dev.124.19.3683. [DOI] [PubMed] [Google Scholar]
- 24.Hennig W. Heterochromatin. Chromosoma. 1999;108:1–9. doi: 10.1007/s004120050346. [DOI] [PubMed] [Google Scholar]
- 25.Huang S, Spector D L. Nascent pre-mRNA transcripts are associated with nuclear regions enriched in splicing factors. Genes Dev. 1991;5:2288–2302. doi: 10.1101/gad.5.12a.2288. [DOI] [PubMed] [Google Scholar]
- 26.Inukai T, Inoue A, Kurosawa H, Goi K, Shinjyo T, Ozawa K, Mao M, Inaba T, Look A T. Slug, a ces-1 related zinc finger transcription factor gene with antiapoptotic activity, is a downstream target of the E2A-HLF oncoprotein. Mol Cell. 1999;4:343–352. doi: 10.1016/s1097-2765(00)80336-6. [DOI] [PubMed] [Google Scholar]
- 27.Ip Y T, Kraut R, Rushlow C A, Levine M. The dorsal morphogen is a sequence-specific DNA-binding protein that interacts with a long-range repression element in Drosophila. Cell. 1991;64:439–446. doi: 10.1016/0092-8674(91)90651-e. [DOI] [PubMed] [Google Scholar]
- 28.Ip Y T, Maggert K, Levine M. Uncoupling gastrulation and mesoderm differentiation in the Drosophila embryo. EMBO J. 1994;13:5826–5834. doi: 10.1002/j.1460-2075.1994.tb06926.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ip Y T, Park R E, Kosman D, Bier E, Levine M. The dorsal gradient morphogen regulates stripes of rhomboid expression in the presumptive neuroectoderm of the Drosophila embryo. Genes Dev. 1992;6:1728–1739. doi: 10.1101/gad.6.9.1728. [DOI] [PubMed] [Google Scholar]
- 30.Issac A, Sargent M G, Cooke J. Control of vertebrate left-right asymmetry by a snail-related zinc finger gene. Science. 1997;275:1301–1304. doi: 10.1126/science.275.5304.1301. [DOI] [PubMed] [Google Scholar]
- 31.Jiang R, Lan Y, Norton C R, Sundberg J P, Gridley T. The Slug gene is not essential for mesoderm or neural crest development in mice. Dev Biol. 1998;198:277–285. [PubMed] [Google Scholar]
- 32.Kasai Y, Nambu J R, Lieberman P M, Crews S T. Dorsal-ventral patterning in Drosophila: DNA binding of Snail protein to the single-minded gene. Proc Natl Acad Sci USA. 1992;89:3414–3418. doi: 10.1073/pnas.89.8.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kosman D, Ip Y T, Levine M, Arora K. Establishment of the mesoderm-neuroectoderm boundary in the Drosophila embryo. Science. 1991;254:118–122. doi: 10.1126/science.1925551. [DOI] [PubMed] [Google Scholar]
- 34.Langeland J A, Tomas J M, Jackman W R J, Kimmel C B. An amphioxus snail gene: expression in paraxial mesoderm and neural plate suggests a conserved role in patterning the chordate embryo. Dev Genes Evol. 1998;208:569–577. doi: 10.1007/s004270050216. [DOI] [PubMed] [Google Scholar]
- 35.Leptin M. Twist and snail as positive and negative regulators during Drosophila mesoderm development. Genes Dev. 1991;5:1568–1576. doi: 10.1101/gad.5.9.1568. [DOI] [PubMed] [Google Scholar]
- 36.Li H, Leo C, Zhu J, Wu X, O'Neil J, Park E J, Chen J D. Sequestration and inhibition of Daxx-mediated transcriptional repression by PML. Mol Cell Biol. 2000;20:1784–1796. doi: 10.1128/mcb.20.5.1784-1796.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mauhin V, Lutz Y, Dennefeld C, Alberga A. Definition of the DNA-binding site repertoire for the Drosophila transcription factor. Snail. Nucleic Acids Res. 1993;21:3951–3957. doi: 10.1093/nar/21.17.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mayor R, Morgan R, Sargent M G. Induction of prospective neural crest of Xenopus. Development. 1995;121:767–777. doi: 10.1242/dev.121.3.767. [DOI] [PubMed] [Google Scholar]
- 39.Metzstein M M, Horvitz H R. The C. elegans cell death specification gene ces-1 encodes a Snail family zinc finger protein. Mol Cell. 1999;4:309–319. doi: 10.1016/s1097-2765(00)80333-0. [DOI] [PubMed] [Google Scholar]
- 40.Nakayama H, Scott I C, Cross J C. The transition to endoreduplication in trophoblast giant cells is regulated by the mSNA zinc finger transcription factor. Dev Biol. 1998;199:150–163. doi: 10.1006/dbio.1998.8914. [DOI] [PubMed] [Google Scholar]
- 41.Nibu Y, Zhang H, Bajor E, Barolo S, Small S, Levine M. dCtBP mediates transcriptional repression by Knirps, Kruppel and Snail in the Drosophila embryo. EMBO J. 1998;17:7009–7020. doi: 10.1093/emboj/17.23.7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nibu Y, Zhang H, Levine M. Interaction of short-range repressors with Drosophila CtBP in the embryo. Science. 1998;280:101–104. doi: 10.1126/science.280.5360.101. [DOI] [PubMed] [Google Scholar]
- 43.Nieto M A, Bennett M F, Sargent M G, Wilkinson D G. Cloning and developmental expression of Sna, a murine homologue of the Drosophila snail gene. Development. 1992;116:227–237. doi: 10.1242/dev.116.1.227. [DOI] [PubMed] [Google Scholar]
- 44.Nieto M A, Sargent M G, Wilkinson D G, Cooke J. Control of cell behavior during vertebrate development by slug, a zinc finger gene. Science. 1994;264:835–839. doi: 10.1126/science.7513443. [DOI] [PubMed] [Google Scholar]
- 45.Nusslein-Volhard C, Wieschaus E, Kluding H. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. I. Zygotic loci on the second chromosome. Wilhelm Roux's Arch Dev Biol. 1984;193:267–282. doi: 10.1007/BF00848156. [DOI] [PubMed] [Google Scholar]
- 46.Pazin M J, Kadonaga J T. What's up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 47.Poortinga G, Monoru W, Parkhurst S. Drosophila CtBP: a Hairy-interacting protein required for embryonic segmentation and Hairy-mediated transcriptional repression. EMBO J. 1998;17:2067–2078. doi: 10.1093/emboj/17.7.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roark M, Sturtevant M A, Emery J, Vaessin H, Grell E, Bier E. Scratch, a pan-neural gene encoding a zinc finger protein related to snail, promotes neuronal development. Genes Dev. 1995;9:2384–2398. doi: 10.1101/gad.9.19.2384. [DOI] [PubMed] [Google Scholar]
- 49.Romano L A, Runyan R B. Slug is a mediator of epithelia-mesenchymal cell transformation in the developing chicken heart. Dev Biol. 1999;212:243–254. doi: 10.1006/dbio.1999.9339. [DOI] [PubMed] [Google Scholar]
- 50.Sargent M G, Bennett M F. Identification in Xenopus of a structural homologoue of the Drosophila gene Snail. Development. 1990;109:967–973. doi: 10.1242/dev.109.4.967. [DOI] [PubMed] [Google Scholar]
- 51.Savagner P, Yamada K M, Thiery J P. The zinc-finger protein Slug causes desmosome dissociation, an initial and necessary step for growth factor-induced epithelial-mesenchymal transition. J Cell Biol. 1997;137:1403–1419. doi: 10.1083/jcb.137.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seeler J S, Marchio A, Sitterlin D, Transy C, Dejean A. Interaction of SP100 with HP1 proteins: a link between the promyelocytic leukemia-associated nuclear bodies and the chromatin compartment. Proc Natl Acad Sci USA. 1998;95:7316–7321. doi: 10.1073/pnas.95.13.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sefton M, Sanchez S, Nieto M A. Conserved and divergent roles for members of the Snail family of transcription factors in the chick and mouse embryos. Development. 1998;125:3111–3121. doi: 10.1242/dev.125.16.3111. [DOI] [PubMed] [Google Scholar]
- 54.Simpson P. Maternal-zygotic gene interactions during the formation of the dorsoventral pattern in Drosophila embryos. Genetics. 1983;105:615–632. doi: 10.1093/genetics/105.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith D E, Del Amo F F, Gridley T. Isolation of Sna, a mouse gene homologous to the Drosophila genes snail and escargot: its expression pattern suggests multiple roles during postimplantation development. Development. 1992;116:1033–1039. doi: 10.1242/dev.116.4.1033. [DOI] [PubMed] [Google Scholar]
- 56.Smith K P, Moen P T, Wydner K L, Coleman J R, Lawrence J B. Processing of endogenous pre-mRNAs in association with SC-35 domains is gene specific. J Cell Biol. 1999;144:617–629. doi: 10.1083/jcb.144.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spector D L, Fu X D, Maniatis T. Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO J. 1991;10:3467–3481. doi: 10.1002/j.1460-2075.1991.tb04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thisse C, Thisse B, Postlethwait J H. Expression of snail2, a second member of the zebrafish snail family, in cephalic mesendoderm and presumptive neural crest of wild-type and spadetail mutant embryos. Dev Biol. 1995;172:86–99. doi: 10.1006/dbio.1995.0007. [DOI] [PubMed] [Google Scholar]
- 59.Thisse C, Thisse B, Schilling T F, Postlethwait J H. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development. 1993;119:1203–1215. doi: 10.1242/dev.119.4.1203. [DOI] [PubMed] [Google Scholar]
- 60.Twigg S R F, Wilkie A O M. Characterisation of the human snail (SNAI1) gene and exclusion as an major disease gene in craniosynostosis. Hum Genet. 1999;105:320–326. doi: 10.1007/s004399900143. [DOI] [PubMed] [Google Scholar]
- 61.Vallian S, Chin K V, Chang K S. The promyelocytic leukemia protein interacts with Sp1 and inhibits its transactivation of the epidermal growth factor receptor promoter. Mol Cell Biol. 1998;18:7147–7156. doi: 10.1128/mcb.18.12.7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vallian S, Gaken J A, Trayner I D, Gingold E B, Kouzarides T, Chang K S, Farzaneh F. Transcriptional repression by the promyelocytic leukemia protein, PML. Exp Cell Res. 1997;237:371–382. doi: 10.1006/excr.1997.3801. [DOI] [PubMed] [Google Scholar]
- 63.Wada S, Saiga H. Cloning and embryonic expression of Hrsna, a snail family gene of the ascidian Halocynthia roretzi: implication in the origins of mechanisms for mesoderm specification and body axis formation in chordates. Dev Growth Differ. 1999;41:9–18. doi: 10.1046/j.1440-169x.1999.00408.x. [DOI] [PubMed] [Google Scholar]
- 64.Wansink D G, Schul W, van der Kraan I, van Steensel B, van Driel R, de Jong L. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J Cell Biol. 1993;122:283–293. doi: 10.1083/jcb.122.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Whiteley M, Noguchi P D, Sensabaugh S M, Odenwald W F, Kassis J A. The Drosophila gene escargot encodes a zinc finger motif found in snail-related genes. Mech Dev. 1992;36:117–127. doi: 10.1016/0925-4773(92)90063-p. [DOI] [PubMed] [Google Scholar]
- 66.Wolffe A P, Hayes J J. Chromatin disruption and modification. Nucleic Acids Res. 1999;27:711–720. doi: 10.1093/nar/27.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yannoni Y M, White K. Association of the neuron-specific RNA binding domain-containing protein ELAV with the coiled body in Drosophila neurons. Chromosoma. 1997;105:332–341. doi: 10.1007/BF02529748. [DOI] [PubMed] [Google Scholar]