Summary
Mean residual life (MRL) function defines the remaining life expectancy of a subject who has survived to a time point and is an important alternative to the hazard function for characterizing the distribution of a time-to-event variable. Existing MRL models primarily focus on studying the association between risk factors and disease risks using linear model specifications in multiplicative or additive scale. When risk factors have complex correlation structures, nonlinear effects, or interactions, the pre-fixed linearity assumption may be insufficient to capture the relationship. Single-index modeling framework offers flexibility in reducing dimensionality and modeling nonlinear effects. In this paper, we propose a class of partially linear single-index generalized MRL models, the regression component of which consists of both a semiparametric single-index part and a linear regression part. Regression spline technique is employed to approximate the nonparametric single-index function, and parameters are estimated using an iterative algorithm. Double-robust estimators are also proposed to protect against the misspecification of censoring distribution or MRL models. A further contribution of this paper is a nonparametric test proposed to formally evaluate the linearity of the single-index function. Asymptotic properties of the estimators are established, and the finite-sample performance is evaluated through extensive numerical simulations. The proposed models and inference approaches are demonstrated by a New York University Langone Health (NYULH) COVID-19 dataset.
Keywords: Counting process, Double robustness, Semiparametric regression, Spline, Survival analysis
1 |. INTRODUCTION
The mean residual life (MRL) function defines the remaining life expectancy given a subject has survived to a specific time point. For a time-to-event variable T with finite expectation, the MRL at time t is m(t) = E(T − t|T > t), and m(t|X) = E(T − t|T > t, X) as the conditional MRL function given covariates X. In contrast to the hazard function that characterizes the instantaneous risk, the remaining life expectancy has appealing interpretations and lessens communication barriers with patients in practice. Furthermore, the MRL regression model is a desired complement to Cox proportional hazards (PH) model as it directly characterizes the covariate effects on the remaining survival time.
Although the one-to-one correspondence exists among the MRL function and the hazard function, there is no straightforward relationship between the covariate effects on the MRL model and the ones on the hazard model. Under the special case of survival distribution belonging to the Hall-Wellner family, Oakes and Dasu1 showed the existence of a model that satisfies both the proportional MRL and the PH assumptions. Under general settings or with model specifications beyond two-sample comparison, a constant hazard ratio does not translate to a constant effect on the MRL or any monotonic transformation of the MRL; vice versa, neither a constant effect elongating the MRL additively or proportionally translates to any simple effect on the hazard function. Therefore, the choice between modeling the hazard or the MRL depends on the interest of scientific investigation.
During the coronavirus disease 2019 (COVID-19) pandemic, after a COVID-19 patient is hospitalized, it is of importance to estimate the remaining time to recovery (length of hospital stay) given that the patient has been hospitalized for t days, for which modeling the MRL directly addresses the question. This quantity is of interest not only for individual patients to comprehend their likelihood and speed of recovery but also for hospital management to make arrangements and adjustments to the needs of hospital beds, clinical staff, and many other clinical resources.2,3 Furthermore, to evaluate COVID-19 disease severity and progression in hospitalized patients, many biomarkers have been studied and found to play important roles in understanding the disease mechanisms and guide treatment strategies.4 Petrilli et al5 conducted a prospective cohort study in confirmed COVID-19 patients hospitalized in four acute care hospitals of the New York University Langone Health (NYULH) and showed that biomarkers of D-dimer, ferritin, and C-reactive protein (CRP) at admission were positively associated with the risk of developing critical illness that was defined as a composite endpoint of intensive care unit (ICU) admission, mechanical ventilation, and mortality. Although many researches have investigated the association of each biomarker with the COVID-19 progression,6 their relative importance and joint effects remain unclear. Motivated by our application to study the time to recovery of COVID-19 hospitalized patients, we are interested in and thus focus on modeling the MRL function in this paper. Specifically, we aim to evaluate the relative importance and overall effects of multiple biomarkers on the time to recovery, allowing potential interactions and nonlinear effects and adjusting for additional covariates.
Sun and Zhang7 proposed a class of generalized MRL (GMRL) models,
| (1) |
which allows a flexible pre-specified link function g(·) and includes multiplicative MRL models1,8,9 and additive MRL models10,11 as special cases. When β = 0, g{m0(t)} denotes the baseline MRL function. Most of the existing MRL models, however, depend on the linearity assumption of the covariate effects on the MRL function. To assess nonlinear covariate effects, Yang and Zhou12 proposed a class of semiparametric varying-coefficient MRL models that allow coefficients to vary with the level of a given variable. When this specific variable is time, the model returns to the semiparametric time-varying coefficients MRL model as a special case.13 However, when the primary interest is to explore the joint effects rather than individual covariate effect, such as the multiple biomarkers in the NYULH COVID-19 study, the linear specification of additive linear effects as in Model (1) may be insufficient. There is a growing need to develop new methodologies that can naturally accommodate flexible forms of nonlinear effects of the covariates and delineate their contributions to the joint effects. Further, flexible MRL models that could handle high-dimensional data are greatly desired.
The single-index technique14,15,16 provides an effective solution to reduce dimensionality and offers flexibility in capturing the nonlinear joint effects of multiple covariates. The relative scale and direction of the single-index coefficients reflect the contribution of each covariate to the overall effects. In the context of time-to-event data analysis, the single-index modeling technique has been incorporated into the Cox PH model,17,18 and further extended to the partially linear single-index (PLSI) PH model by allowing both linear and nonlinear components.19,20 To the best of our knowledge, there has been no research that offers both the flexibility in modeling covariates’ effects from the PLSI construct and straightforward interpretation from the MRL models. Therefore, we propose a class of PLSI generalized MRL (PLSI GMRL) models in this paper and develop estimating equations using the inverse probability of censoring weighting (IPCW) technique to account for censoring. Furthermore, we propose an extension to double-robust estimators to protect against potential model misspecifications. In addition, this paper makes an important contribution by providing a nonparametric testing procedure to evaluate the linearity of the single-index function. Failing to reject this null hypothesis suggests the linear effects of all covariates in the model, and thus we can return to the estimation procedures and inference from the generalized MRL models.7
The rest of this article is organized as follows. In Section 2, we present the proposed model and details of interpretation, estimation, inference, as well as double-robust extensions. Our proposed test statistic for the linearity of the single-index function and its nonparametric testing procedure are also presented. Moreover, we provide an extension to the restricted MRL models when it is of interest to restrict the remaining life expectancy within a time range. Section 3 presents extensive numerical simulations. In Section 4, we demonstrate our method via the application to the NYULH COVID-19 de-identified electronic health record (EHR) data. A model diagnosis method is also presented. Discussion and concluding remarks are included in Section 5. Regularity conditions, additional simulation and data application results, and technical proofs are provided in Supporting Information online.
2 |. METHODS
2.1 |. Notation
Let Y and C be the survival time and censoring time, respectively. Then T = min(Y, C) denotes the observed time subject to censoring, and δ = I(Y ≤ C) is the censoring indicator. Assume the support of C is longer than that of the survival time T to ensure an estimable MRL function. Covariates are classified into two groups a priori: X represents the pre-specified covariates to be modeled in linear form and Z represents the covariates to be included in the single-index function. We have i.i.d. copies of {Ti, δi, Xi, Zi, i = 1, …, n} and assume Y and C are conditionally independent given covariates X and Z.
The proposed PLSI GMRL model is specified as
| (2) |
where g(·) and m0(t) are similarly defined as in Model (1), α is a q-dimensional vector of coefficients characterizing the effects of X in the linear component, ψ(·) is an unknown single-index function that represents the joint effects of covariates Z, and β’s are the corresponding p-dimensional single-index coefficients representing the relative importance and direction of Z. When ψ(·) is monotone, the effects of Z can be interpreted qualitatively using the sign of its coefficient β. Specifically, if ψ(·) is monotone increasing, a positive β suggests an increased mean residual time at a larger value of the covariate, and vice versa for a negative sign. The relative importance of each covariate in ψ(·) on the MRL function can be evaluated by examining the magnitude of |β|.
Note that the absolute direction and scale of β are not identifiable since any scale and/or constant shift can be absorbed by ψ(·), so the identifiability constraint is assumed by constraining the Euclidean norm ∥β∥ = 1 with the first component to be positive. In addition, we impose the sum-to-zero constraint21 as for the identifiability of ψ(·) because the constant shift can be absorbed by m0(t). One can also let instead. Both constraints on ψ(·) solve the issue of identifiability, but the latter one will have the and its corresponding 95% confidence intervals always pass through the origin (0, 0) with centered single-index covariates, preventing any statistical inference at point 0. To ensure the MRL function is properly defined, g(·) needs to be strictly increasing, twice continuously differentiable, and m(t|X, Z) = g{m0(t) + αTX + ψ(βTZ)} is a proper MRL function for all possible values of X and Z.
Similar to Huang and Liu18 and Sun et al,19 we use spline basis functions to approximate the derivative of the unknown single-index function ψ(·),
where Bj(u),j = 1, …, k, are the B-spline basis functions, k denotes the degrees of freedom, B(u) = {B1(u), …, Bk(u)}T, and γ = {γ1, …, γk}T. We choose B-splines because of their numerical stability,18,20 but other basis functions, such as truncated power basis and P-splines, can also be adopted. The choice of approximating ψ′(·) rather than ψ(·) itself is due to two considerations: first, ψ′(·) is more frequently referred to in subsequent computation and inference steps; second, one usually can achieve better numerical stability with computing integration than differentiation. Consequently, we have , where , are the integrals of the B-spline basis functions and . Hence, Model (2) can be rewritten as
| (3) |
Throughout the article, we use quadratic B-splines in the basis expansion of ψ′(·), and ψ(·) is a cubic spline.
2.2 |. Estimation equations
For the simplicity of notation, we define V = (X, ZT)T, θ = (αT, βT, γT)T, and . We employ the technique of the inverse probability of censoring weighting (IPCW) to handle censoring in our proposed estimation and inference procedures. We first assume that C is independent of covariates V. Let G(t) be the survival function of C, and construct a stochastic process as
which is a mean-zero process at the true values of θ and m0(·). Given a pre-specified g(·) and fixed coefficients θ, m0(t) and can be estimated at each fixed t ∈ {t : 0 ≤ t ≤ τ} by solving the following estimating equation,
| (4) |
where is the Kaplan-Meier estimator of G(t). Here we assume 0 < τ = inf{t : Pr(T ≥ t) = 0} < ∞ on τ to circumvent the technical difficulty on the tail behavior of limiting distribution. For a theoretical investigation, we can adopt Ying’s approach22 on asymptotic properties beyond τ to our method. Given , we then propose estimating equations for θ as follows:
| (5) |
where and is an increasing weight function on [0, τ]. Note that other forms of H(t) can be easily adopted as long as the function is increasing and converges almost surely to a deterministic bounded function. We denote the final estimators of θ as and .
2.3 |. Implementation
In practice, instead of directly calculating the complex Jacobian matrix J(θ) = ∂U(θ)/∂θ, we consider an iterative procedure to estimate (αT, γT)T and β based on their corresponding estimating equations Uα(θ),Uγ(θ), and Uβ(θ), each of which is numerically more stable and simpler. The Jacobian matrix of each equation can be easily derived by taking the derivative with respect to the parameters of interest. The Newton-Raphson method is used to solve for these parameters iteratively, and the algorithm is depicted as follows.
Step 1: Assign initial value of βini. The initial value can be obtained from standard MRL models using R package “GMRL” with a pre-specified g(·) link function (e.g. multiplicative MRL model or additive MRL model) by assuming linear coefficients for all covariates.
Step 2: Given βini and a fixed degrees of freedom k, the covariates consist of Xi and . We then estimate α and γ simultaneously using “GMRL” package and denote the estimators by αini and γini.
Step 3: Given αini, γini and βini, update using Equation (4). Then, we fix αini and γini and update βini based on Uβ in Equation (5). Ensure ∥βini∥ = 1 with the first component to be positive.
Step 4: Repeat step 2–3 until the convergence criterion is met.
Given a fixed degrees of freedom k, we recommend that knots be placed at equally spaced sample quantiles of the single index βTZ, and obtain the corresponding estimator from equation U θ(k) = 0. Similar to the idea of Ma and Wei,23 we select the optimal k through minimizing the quadratic score function with a penalty term on the degrees of freedom as U (θ(k))TU(θ(k)) + log(n)(k + d + p + q − 2), where q and p denote, respectively, the number of covariates in linear form and inside of the single index, and d is the degree of the spline functions.
2.4 |. Inference
To satisfy the constraints ∥β∥ = 1 and β1 > 0, we reparameterize β = β(σ) = {(1 − ∥σ∥2)1/2, σ1, …, σp−1}T with σ = (σ1, …, σp−1)T. The solution to Equation (5) can be denoted as and . Note that the parametrizaiton of is purely used to develop asymptotic theory of below. Next, we define additional notations: , ,
, where g′(x) = dg(x)/dx.
Theorem 1. Under the regularity conditions (C1) to (C4) stated in Web Appendix C, we have
The estimators and exist and are consistent.
- in distribution. The variance matrix components A and can be consistently estimated by and . Specifically,
where , , υ⊗2 denotes υT for a column vector υ, Ip−1+k+q is an identity matrix of (p − 1 + k + q) dimension, 01×k and 01×q are zero vectors.
Because is analytically intractable to compute directly, we adopt a bootstrap resampling approach24 to compute the empirical standard errors (SE) of the estimators. For a fixed single index u, the 95% confidence interval for ψ(u) is given by , where and , or can be obtained as 2.5% and 97.5% sample quantiles of the estimated single-index function based on bootstrap samples.
2.5 |. Double-robust estimators
Our proposed Equation (5) uses the IPCW method and requires the modeling of censoring distribution. When censoring time is independent with covariates, the nonparametric Kaplan-Meier (KM) estimator performs well. We can easily extend to handle the covariate-dependent censoring by incorporating covariates into estimating censoring distribution, for example, using a Cox PH model. It is well known that a mis-specified censoring model may lead to biased estimators, and thus we further propose a double-robust extension, which would remain consistent when either the model for censoring distribution G(t|V) or a working model for the complete data distribution F(t|V) is correctly specified.
Define and , where and F(t|V) is the survival function of T given V. As shown in Rubin and van der Laan,25 YF,G(θ; T, δ, V) has double robustness property that E[YF,G(θ; T, δ, V)|V] = 0 when either F or G is correctly specified. Thus, we have the estimating equations for double-robust estimators as
where is an estimator for F(t|V) and can be estimated using the proposed PLSI GMRL Model (3), e.g.,
Note that and for should not depend on the censoring distribution assumption, and thus we consider obtaining the estimates from the quasi-partial score (QPS) approach proposed by Chen and Cheng.10,26 The double-robust (DR) estimators denoted as would be consistent if either the PLSI GMRL Model (3) or the censoring mechanism assumption is correct. We follow the similar iterative estimation procedures introduced in Section 2.3.
Remark: The estimating equations for the QPS approach for GMRL models are constructed based on zero-mean martingales.10,26 An advantage of the QPS approach is that it does not require any specification of the censoring distribution. In fact, the QPS approach can also be considered for the estimation and inference of the PLSI GMRL models. After approximating the single-index component using finite-dimensional B-splines, the implementation are straightforward following procedures from Chen and Cheng.26 However, when the censoring distribution is well modeled or random, the QPS method is often less efficient than the IPCW method.7,27 In our simulation studies shown below, we also compared with the QPS estimators and demonstrated that the proposed DR estimators were more efficient compared to the QPS estimators.
2.6 |. Testing linearity of single-index function
If the single-index function can be reasonably assumed to be linear for a practical application, the generalized MRL models7 would suffice to fit the data. We propose a nonparametric resampling approach28 to test the linearity of the single-index function ψ(u) by examining whether its derivative is a constant, e.g., H0 : ψ′(u) = c for a non-zero constant c. Based on the estimation steps described above, ψ′(u) is estimated by . If there is a pre-specified c of interest, e.g., c = 1 for an identity function, we can directly test the hypothesis ψ′(u) = c. For an unknown c, we estimate its value by , where a and b denote the range of interest. Thus, given a estimator , we have , where is the weak limit of . We consider the Kolmogorov and Cramer-von Mises-type test statistics as and , respectively. Take T1n as an example, we propose the nonparametric testing procedure as follows. For each resampling step b = 1, …, B:
Obtain the resampled data
Estimate ψ′(u) as based on the b-th set of resampled data
Compute the statistic of test of interest
We reject the null hypothesis if T1n is larger than , which is the empirical (1 − α)-quantile of the simulated sample , and α ∈ (0, 1) is the pre-specified nominal size.
2.7 |. Restricted MRL
Sometimes, the scientific interest in the remaining life expectancy is restricted within a finite interval. For example, in the COVID-19 application, the recovery time varied due to patients’ characteristics and disease severity, but patients who stayed in hospital longer than 30 days or even 60 days could have a very different disease course from the acute patients. Therefore, we consider a 30-day restricted MRL in the data application. The restricted MRL function for a pre-specified time τ > 0 is mτ(t) = E(Tτ − t|Tτ > 0), where 0 < t < τ, Tτ = min(T, τ), and mτ(τ) = 0. Regression models for restricted MRL have been studied under the right-censored as well as the left-truncated data.29,30,31 To incorporate the restricted MRL into our proposed PLSI GMRL model framework, we consider Equation (5) by replacing Ti with min(Ti, τ) and can follow similar estimating procedures.
3 |. SIMULATION STUDIES
3.1 |. Independent censoring
We conducted extensive simulations to evaluate the finite-sample performance of our proposed models and estimation procedures. We considered both multiplicative and additive MRL model settings, corresponding to g(t) = exp(t) and g(t) = t, respectively. The true single-index function ψ(·) was set to be linear, sine curve, and quadratic functions as follows:
S1. Linear: g{m0(t) + αTX + βTZ};
S2. Sine curve: g{m0(t) + αTX + sin(5βTZ)/2};
S3. Quadratic: g{m0(t) + αTX + 2(βTZ)2}.
The baseline MRL function was set to be m0(t) = g−1{(D1t + D2)I(D1t + D2 ≥ 0)}, where D1 > −1 and D2 > 0, and from the Hall-Wellner family. We set D1 = −0.5 and D2 = 0.5. True parameters were set as α* = (0.1, −0.1)T and β* = (0.5, 0.5, −0.5, −0.5)T. When g(t) = exp(t), covariates X = (X1, X2)T ~ U[0, 0.5] and Z = (Z1, Z2, Z3, Z4)T ~ U[0, 0.6] independently for all settings. When g(t) = t, the single-index covariates Z ~ U[0, 1] for all settings. To ensure a properly defined MRL function, we set linear covariates X ~ U[0.55, 0.9] in linear case, X ~ U[1, 1.4] in sine curve case, and X ~ U[0, 1] in quadratic case. Also, an independent censoring time C ~ exp(λ), which controlled a fixed censoring rate 10% or 20%. Throughout the simulations, we set the sample size to be 2000 or 4000. Under each simulation setting, 500 datasets were generated, and 500 bootstraps were conducted to obtain the empirical SEs of the estimators.
We used two equally spaced knots in our simulations and found that the model performance was not sensitive to the number of knots in a reasonable range (e.g., one to five knots) under our settings. The angle between the true parameter β* and its estimator , which was defined as with ⟨a, b⟩ denoting the inner product of two vectors a and b, was reported to evaluate the estimated single-index coefficients. A large value of ω indicated a large bias in the estimator .
Table 1 presents the simulation results under independent censoring with a 10% censoring rate. To better compare the single-index coefficients from the PLSI GMRL model with the GMRL model,7 we normalized estimators under the GMRL model with and β1 > 0. In the case of a linear single-index function (S1), both models performed reasonably well. The biases of were small, SDs of the estimates were close to the empirical SEs, and the coverage probabilities (CPs) of the 95% confidence intervals (CIs) were close to the nominal level. For a fixed censoring rate, both and SDs of the estimates decreased with increasing sample sizes. Compared with estimators from the multiplicative MRL model, the efficient loss of from the PLSI GMRL model was small (Table A.1).
TABLE 1.
Simulation results with independent censoring.
| α1 | α2 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Bias | SD | SE | CP | Bias | SD | SE | CP | Mean | SD | |
| g(t) = exp(t) | |||||||||||
| S1. Linear case | |||||||||||
| GMRL | 2000 | 0.001 | 0.082 | 0.088 | 95.8 | 0.006 | 0.086 | 0.088 | 95.0 | 6.90 | 3.07 |
| PLSI GMRL | 2000 | 0.001 | 0.082 | 0.088 | 96.0 | 0.006 | 0.086 | 0.089 | 95.2 | 7.41 | 3.38 |
| GMRL | 4000 | 0.001 | 0.063 | 0.062 | 94.2 | 0.001 | 0.062 | 0.062 | 95.6 | 4.74 | 1.93 |
| PLSI GMRL | 4000 | 0.001 | 0.063 | 0.062 | 94.4 | 0.001 | 0.062 | 0.062 | 96.0 | 4.87 | 1.96 |
| S2. Sine curve case | |||||||||||
| GMRL | 2000 | −0.001 | 0.084 | 0.080 | 93.4 | 0.003 | 0.078 | 0.081 | 96.4 | 3.63 | 1.53 |
| PLSI GMRL | 2000 | 0.003 | 0.083 | 0.079 | 93.2 | 0.003 | 0.076 | 0.079 | 93.2 | 3.21 | 1.30 |
| GMRL | 4000 | 0.003 | 0.056 | 0.056 | 94.8 | −0.005 | 0.058 | 0.057 | 94.4 | 2.60 | 1.12 |
| PLSI GMRL | 4000 | 0.003 | 0.055 | 0.055 | 95.4 | −0.005 | 0.057 | 0.056 | 93.8 | 2.31 | 0.98 |
| S3. Quadratic case | |||||||||||
| GMRL | 2000 | −0.005 | 0.088 | 0.087 | 95.8 | −0.005 | 0.087 | 0.087 | 95.0 | 75.67 | 31.17 |
| PLSI GMRL | 2000 | −0.004 | 0.088 | 0.086 | 95.2 | −0.008 | 0.087 | 0.086 | 94.8 | 11.43 | 5.20 |
| GMRL | 4000 | −0.001 | 0.062 | 0.061 | 93.2 | −0.004 | 0.063 | 0.061 | 94.6 | 72.02 | 30.85 |
| PLSI GMRL | 4000 | 0.001 | 0.061 | 0.060 | 94.2 | −0.005 | 0.062 | 0.061 | 93.6 | 8.01 | 3.58 |
| g(t) = t | |||||||||||
| S1. Linear case | |||||||||||
| GMRL | 2000 | 0.002 | 0.107 | 0.103 | 93.3 | −0.010 | 0.108 | 0.103 | 94.1 | 9.77 | 4.15 |
| PLSI GMRL | 2000 | 0.002 | 0.107 | 0.103 | 92.7 | −0.011 | 0.108 | 0.103 | 93.7 | 10.76 | 4.99 |
| GMRL | 4000 | −0.002 | 0.076 | 0.074 | 94.2 | 0.003 | 0.074 | 0.074 | 94.8 | 6.82 | 2.94 |
| PLSI GMRL | 4000 | −0.002 | 0.076 | 0.074 | 94.2 | 0.002 | 0.074 | 0.074 | 94.6 | 7.14 | 3.21 |
| S2. Sine curve case | |||||||||||
| GMRL | 2000 | 0.001 | 0.259 | 0.242 | 93.4 | 0.016 | 0.257 | 0.241 | 92.8 | 6.64 | 2.76 |
| PLSI GMRL | 2000 | 0.001 | 0.258 | 0.241 | 93.8 | 0.016 | 0.255 | 0.239 | 93.4 | 6.45 | 3.05 |
| GMRL | 4000 | 0.001 | 0.174 | 0.176 | 95.2 | 0.001 | 0.174 | 0.176 | 94.8 | 4.57 | 1.90 |
| PLSI GMRL | 4000 | −0.001 | 0.174 | 0.175 | 95.4 | 0.002 | 0.173 | 0.175 | 94.6 | 4.15 | 1.80 |
| S3. Quadratic case | |||||||||||
| GMRL | 2000 | −0.006 | 0.053 | 0.052 | 93.8 | 0.001 | 0.054 | 0.052 | 94.2 | 66.37 | 35.71 |
| PLSI GMRL | 2000 | −0.002 | 0.050 | 0.050 | 94.0 | 0.002 | 0.052 | 0.050 | 94.4 | 9.35 | 4.83 |
| GMRL | 4000 | −0.003 | 0.039 | 0.037 | 93.0 | −0.002 | 0.038 | 0.037 | 94.0 | 65.54 | 34.92 |
| PLSI GMRL | 4000 | 0.001 | 0.037 | 0.035 | 92.8 | 0.001 | 0.037 | 0.035 | 93.8 | 6.19 | 3.00 |
GMRL: generalized mean residual life model; PLSI GMRL: partially linear single-index generalized mean residual life model; SD: sample standard deviation; SE: mean of estimated standard error; CP: empirical coverage probability of 95% confidence interval; was calculated by arccos ; Censoring rate was 10%.
When g(t) = exp(t) and under the nonlinear single-index scenarios (S2-S3), the proposed PLSI GMRL model outperformed the multiplicative MRL model in terms of efficiency and bias. When the true ψ(·) was a sine curve (S2), we observed low biases from both models because the single index function still satisfied a monotone linear trend over the majority of the data. However, the estimators under the PLSI GMRL model were more efficient compared to those under the multiplicative MRL model (Table A.1). When the true ψ(·) was quadratic (S3), we observed large biases from the multiplicative MRL model as expected, and the performance of the PLSI GMRL model remained well. When g(t) = t, the proposed PLSI GMRL model clearly outperformed the additive MRL model when the true ψ(·) was nonlinear (S2 and S3) in terms of biases, coverage probabilities, and efficiency. The estimated single-index coefficients were summarized in Table A.2.
Figure 1 visualizes the mean of the estimated single-index function and 95% CIs using the 2.5% and 97.5% sample quantiles of the estimated single-index function from 500 simulations. The estimated single-index functions approximated the true functions very well for both g(t) = exp(t) and g(t) = t. Simulation results with 20% censored data were also provided in Table A.3. Additional simulations were conducted under other baseline MRL functions, for instance, D1 = −1/3 and D2 = 1, mimicking an exponential distribution resulting from the right skewness for survival time when all covariates equal 0. The results were summarized in Table A.4.
FIGURE 1.
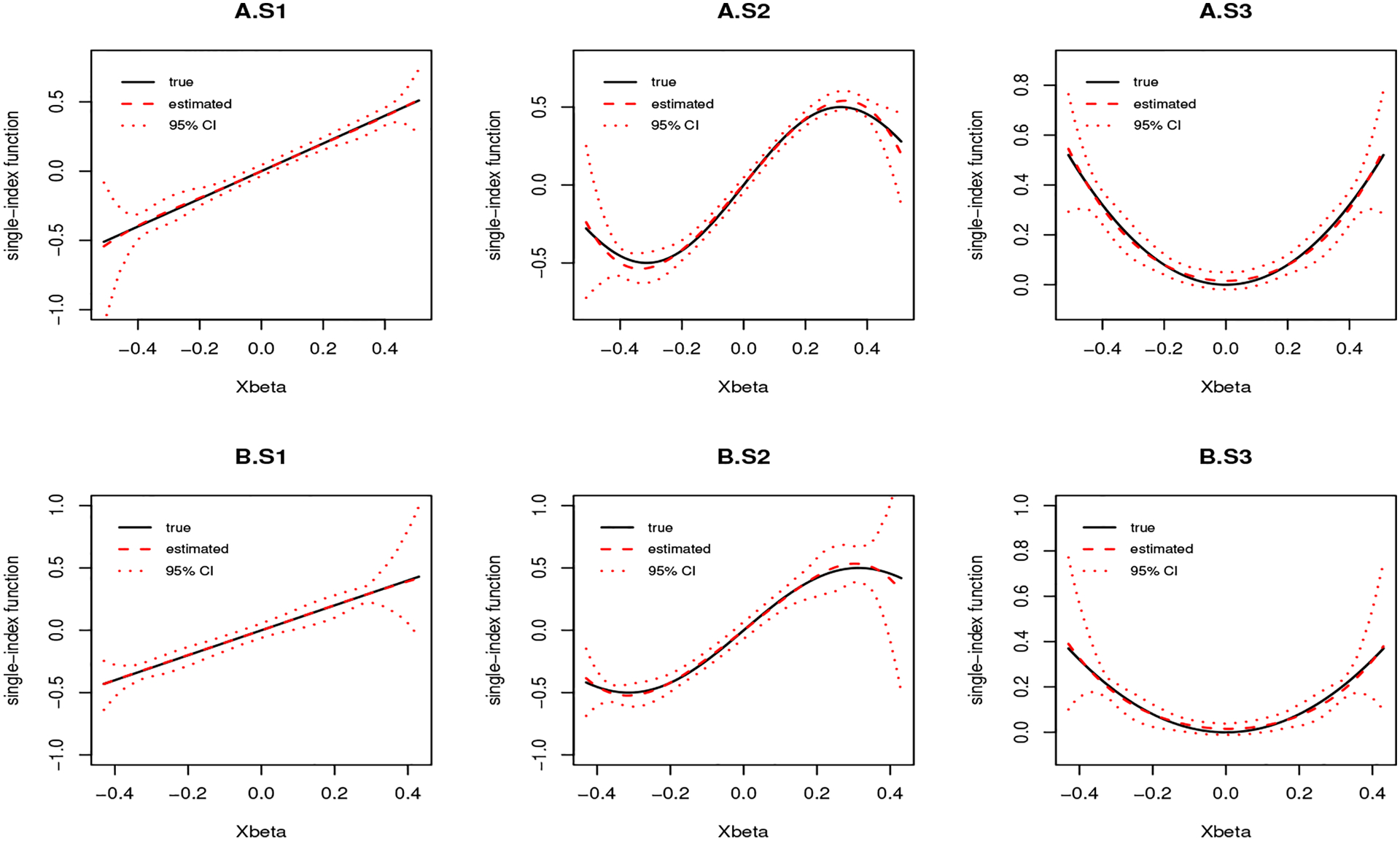
The mean of estimated single-index function with 95% confidence intervals when (A) g(t) = exp(t) and (B) g(t) = t with sample size 4000 and censoring rate 10%: (S1) linear single-index function; (S2) sine curve single-index function; (S3) quadratic single-index function.
3.2 |. Covariate-dependent censoring
In the case of covariate-dependent censoring, we evaluated the robustness of DR estimator and compared its performance to regular IPCW estimator and QPS estimator when g(t) = exp(t). Censoring time was generated from a Cox model λ(t|V) = λ0(t)exp(aTX + bTZ), where a and b were vectors with each element equal to 2 and λ0(t) = 1. All other settings remained the same as in previous simulations. Specifically, two IPCW estimators were considered here: with a mis-specified censoring model using Kaplan-Meier (KM) estimate and with a correctly specified censoring distribution using a Cox PH model. For DR estimator , we used KM estimator to approximate censoring distribution and assumed a PLSI GMRL model as the working model for . As shown in Table 2, due to the mis-specified censoring distribution, showed large biases in and , while IPCW2 estimator performed well since the censoring distribution was correctly specified. The DR estimator successfully remained robust to the misspecification, having lower biased with nominal CPs, smaller angle between and β*, and smaller SDs comparing to IPCW1. Although DR estimator required a slightly longer computation time due to modeling both censoring distribution and complete data distribution, it maintained relatively high efficiency compared with IPCW2 when censoring distribution was correctly specified. Moreover, DR estimators were more efficient in all scenarios comparing to QPS estimators.
TABLE 2.
Simulation results under g(t) = exp(t) with covariate-dependent censoring.
| α1 | α2 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Bias | SD | SE | CP | Bias | SD | SE | CP | Mean | SD | |
| S1. Linear case | |||||||||||
| IPCW1 | 2000 | −0.096 | 0.099 | 0.097 | 85.8 | −0.090 | 0.092 | 0.098 | 84.6 | 13.53 | 4.90 |
| IPCW2 | 2000 | 0.003 | 0.090 | 0.089 | 93.8 | −0.004 | 0.093 | 0.089 | 94.0 | 7.19 | 3.22 |
| DR | 2000 | 0.004 | 0.091 | 0.091 | 93.7 | 0.006 | 0.090 | 0.091 | 94.6 | 7.36 | 3.26 |
| QPS | 2000 | 0.004 | 0.102 | 0.091 | 93.8 | 0.004 | 0.093 | 0.091 | 94.7 | 7.94 | 3.56 |
| IPCW1 | 4000 | −0.096 | 0.073 | 0.072 | 72.8 | −0.095 | 0.072 | 0.073 | 69.2 | 12.44 | 3.45 |
| IPCW2 | 4000 | 0.002 | 0.064 | 0.063 | 95.6 | 0.001 | 0.065 | 0.062 | 93.6 | 4.78 | 2.08 |
| DR | 4000 | 0.003 | 0.066 | 0.066 | 95.4 | 0.004 | 0.067 | 0.066 | 94.1 | 4.96 | 2.14 |
| QPS | 4000 | 0.002 | 0.073 | 0.074 | 94.4 | 0.001 | 0.070 | 0.072 | 96.3 | 5.38 | 2.30 |
| S2. Sine curve case | |||||||||||
| IPCW1 | 2000 | −0.085 | 0.085 | 0.087 | 83.8 | −0.081 | 0.090 | 0.087 | 83.4 | 5.88 | 2.01 |
| IPCW2 | 2000 | −0.006 | 0.076 | 0.079 | 95.6 | −0.002 | 0.078 | 0.079 | 95.4 | 3.42 | 1.37 |
| DR | 2000 | −0.002 | 0.082 | 0.083 | 93.5 | 0.004 | 0.083 | 0.083 | 94.3 | 3.37 | 1.44 |
| QPS | 2000 | −0.009 | 0.096 | 0.099 | 95.2 | −0.001 | 0.096 | 0.099 | 96.2 | 3.82 | 1.67 |
| IPCW1 | 4000 | −0.081 | 0.065 | 0.067 | 74.2 | −0.081 | 0.066 | 0.068 | 75.3 | 5.50 | 1.56 |
| IPCW2 | 4000 | 0.001 | 0.056 | 0.056 | 95.8 | 0.001 | 0.057 | 0.056 | 94.2 | 2.29 | 0.97 |
| DR | 4000 | 0.002 | 0.062 | 0.062 | 94.7 | 0.002 | 0.062 | 0.063 | 93.8 | 2.31 | 0.98 |
| QPS | 4000 | 0.003 | 0.072 | 0.072 | 95.6 | −0.001 | 0.069 | 0.070 | 94.8 | 2.54 | 1.12 |
| S3. Quadratic case | |||||||||||
| IPCW1 | 2000 | −0.087 | 0.100 | 0.097 | 87.2 | −0.082 | 0.093 | 0.095 | 88.3 | 12.02 | 5.89 |
| IPCW2 | 2000 | −0.005 | 0.083 | 0.087 | 95.8 | 0.001 | 0.087 | 0.087 | 94.2 | 11.62 | 5.23 |
| DR | 2000 | 0.002 | 0.090 | 0.091 | 96.4 | 0.004 | 0.089 | 0.089 | 94.7 | 11.54 | 5.63 |
| QPS | 2000 | −0.002 | 0.096 | 0.095 | 93.7 | 0.002 | 0.091 | 0.092 | 95.6 | 13.07 | 7.07 |
| IPCW1 | 4000 | −0.082 | 0.069 | 0.069 | 75.3 | −0.081 | 0.070 | 0.071 | 76.2 | 8.92 | 3.85 |
| IPCW2 | 4000 | 0.004 | 0.062 | 0.061 | 93.4 | −0.002 | 0.062 | 0.061 | 94.0 | 8.13 | 3.62 |
| DR | 4000 | 0.004 | 0.064 | 0.064 | 94.3 | 0.005 | 0.064 | 0.065 | 95.5 | 8.40 | 3.72 |
| QPS | 4000 | 0.001 | 0.070 | 0.070 | 95.7 | 0.001 | 0.067 | 0.070 | 94.6 | 9.21 | 4.30 |
SD: sample standard deviation; SE: mean of estimated standard error; CP: empirical coverage probability of 95% confidence interval; IPCW1: inverse-probability-of-censoring weighting estimator with a mis-specified censoring distribution; IPCW2: inverse-probability-of-censoring weighting estimator with a correctly specified censoring distribution; DR: double-robust estimator; QPS: quasi-partial score estimator; was calculated by arccos ; Censoring rate was 10%.
3.3 |. Type-I error and power of the proposed test
In addition, we conducted simulations to investigate the empirical performance of our proposed nonparametric test. Assuming g(t) = exp(t) and under the null hypothesis that ψ(·) is linear (S1), we examined the type-I error rate when censoring rate was 10% and 20% with a sample size N = 500,1000, and 2000. To evaluate the power of the proposed test, we chose the nonlinear single-index function as a sine curve (S2). We used the Kolmogorov test statistic T1n throughout the simulations, and the critical values were determined by 1000 resamplings. As shown in Table 3, we observed that the empirical type-I error rate was slightly conservative, but it approached the nominal level with increasing sample size. The power showed that the test performed well to reject the null hypothesis when the true single-index function was nonlinear, and its power increased with the sample size. Overall, the proposed nonparametric test can be a useful tool in the single-index modeling framework to assess the linearity of the single-index function.
TABLE 3.
Simulation results of the nonparametric test for single-index function.
| Linear case | Since curve case | ||
|---|---|---|---|
| N | Censoring rate | Type-I error | Power |
| 500 | 10% | 0.026 | 0.194 |
| 20% | 0.032 | 0.182 | |
| 1000 | 10% | 0.029 | 0.530 |
| 20% | 0.036 | 0.422 | |
| 2000 | 10% | 0.043 | 0.892 |
| 20% | 0.040 | 0.800 |
Type-I error rate at 0.05 level.
4 |. APPLICATION
4.1 |. Time-to-recovery in Hospitalized COVID-19 Patients using NYULH EHR Data
We demonstrate the proposed models and estimation procedures through the NYULH COVID-19 EHR data. The database contained de-identified patient information regarding basic demographics, social history, medical history, medication use, lab results, and hospital encounter records since January 1st, 2020. Our analysis used data up to July 14th, 2020. Lab-confirmed COVID-19 patients who have been hospitalized within three days since diagnosis were included. Our primary interest focused on the time to recovery within 30 days since hospitalization. For patients discharged to home, acute rehabilitation facilities, and other skilled nursing facilities, the time to recovery was defined as the days from hospital admission to discharge. Similar to the Remdesivir trial for COVID-19,32 time to recovery for patients who died in hospital or discharged to hospice was set as infinity. Time to recovery for the patients who died or were hospitalized longer than 30 days were censored at 30-day. Patients who were transferred to other intermediate care facilities, discharged to long-term care hospitals, or left against medical advice were treated as censored. We considered similar covariates as in Petrilli et al5 and set age, sex, race, BMI, temperature, blood oxygen (SpO2), and medical history be linear covariates. The biomarkers of procalcitonin, CRP, troponin, D-dimer, and ferritin at hospital admission were included in the single-index component, and each biomarker has been log-transformed due to skewness and standardized with a mean of 0 and a standard deviation of 1. Correlations of these biomarkers are shown in Figure B.1. A total of 2599 patients were included in the analysis and the censoring rate was 28.4%, where over 96% of the censoring was the administrative censoring at 30-day due to patient expiration.
In this application, since majority of the censoring was due to the administrative censoring at 30-day, we directly used the KM estimator for censoring distribution estimation. In general practice, we recommend modeling the censoring mechanism first using a Cox PH model and then reduced to the nonparametric KM estimator assuming the independent censoring distribution if appropriate. The pre-specified link function g(·) provides flexibility in assessing covariate effects on the different scales of the MRL function. For illustration, we considered the candidates as either g(t) = exp(t) or g(t) = t. To assess which model fits the data better, we introduce a procedure based on the standardized score process for model diagnosis (see the remark section below). Given estimated parameters , the standardized score process over time domain t can be obtained by , where is the Jacobian matrix of . In the application, we graphically assessed the model fitting by visualizing in Figure B.2, and observed that the estimated value of the model with g(t) = exp(t) was much smaller than that of g(t) = t, indicating that g(t) = exp(t) fitted the data better. Moreover, we considered as a numerical measure for overall fit of the model, which yielded values 445.7 and 692.8 for PLSI GMRL under g(t) = exp(t) and g(t) = t, respectively, and we thus considered g(t) = exp(t) in subsequent analysis and discussion.
The empirical SEs of the estimators were obtained by 1000 bootstrapping. In addition, we assessed the linearity of the single-index function using the proposed nonparametric test and observed a p-value less than 0.001, indicating the necessity of the PLSI GMRL model to account for possible nonlinear effects. The number of knots was determined by the criteria introduced in Section 2.3, and the estimators were stable with respect to the number of knots between 2 to 7. To compare with the GMRL model,7 we standardized the estimated coefficients of biomarkers with a Euclidean norm equal to one and the first component to be positive.
Table 4 summarizes the estimated coefficients and Figure 2 visualizes the estimated single-index function. Due to the data sparsity at the tails, lower and upper 1% data were excluded from the plot. The monotone increasing trend of indicated that the expected remaining recovery time increases with the single-index value. We observed that the increasing trend flattened at the right tail, which indicated that once patients at a severe status indicated by a high level of the combination of biomarkers, one-unit change in single-index value will not change much of the MRL anymore. While for patients with less severe status, one-unit change in single-index value could significantly impact the expected remaining recovery time within the next 30 days. These observations agreed with the testing result on the linearity of the single-index function. The PLSI GMRL model indicated that biomarker levels of procalcitonin, troponin, and D-dimer had significant prolonging effects on the remaining time to recovery but not with ferritin or CRP. In addition, the estimated effect for each biomarker separately was also provided while keeping all other biomarkers at median values (Figure 2 (B) to (F)).
TABLE 4.
Model results for NYULH COVID-19 data application.
| PLSI GMRL† | GMRL† | |||||
|---|---|---|---|---|---|---|
| Estimate | 95% CI** | Estimate | 95% CI** | |||
| Baseline information (exponential of coefficient) | ||||||
| Age | 1.12* | 1.09 | 1.15 | 1.12* | 1.09 | 1.15 |
| Sex: Male | 1.06* | 1.01 | 1.12 | 1.05* | 1.01 | 1.12 |
| Race: White | 1.02 | 0.97 | 1.07 | 1.02 | 0.97 | 1.07 |
| logBMI | 1.06 | 0.94 | 1.20 | 1.06 | 0.94 | 1.20 |
| SpO2 | 0.91* | 0.89 | 0.94 | 0.91* | 0.89 | 0.94 |
| Temperature | 0.98 | 0.96 | 1.01 | 0.99 | 0.96 | 1.01 |
| Smoking: current/former | 1.00 | 0.94 | 1.06 | 1.00 | 0.95 | 1.06 |
| History of cardiac disease | 1.06* | 1.02 | 1.12 | 1.06* | 1.01 | 1.11 |
| History of pulmonary disease | 1.02 | 0.94 | 1.09 | 1.02 | 0.94 | 1.09 |
| History of diabetes | 1.03 | 0.98 | 1.08 | 1.04 | 0.98 | 1.08 |
| History of malignancy | 1.11* | 1.02 | 1.21 | 1.11* | 1.01 | 1.21 |
| Single-index component (coefficient) | ||||||
| Procalcitonin‡ | 0.91* | 0.43 | 0.98 | 0.75* | 0.51 | 0.89 |
| Troponin‡ | 0.27* | 0.05 | 0.86 | 0.34* | 0.11 | 0.56 |
| D-dimer‡ | 0.32* | 0.01 | 0.57 | 0.40* | 0.16 | 0.60 |
| Ferritin‡ | −0.01 | −0.37 | 0.13 | −0.02 | −0.29 | 0.16 |
| CRP‡ | 0.06 | −0.16 | 0.39 | 0.40* | 0.02 | 0.67 |
Estimated coefficient with p-value less than 0.05.
95% confidence intervals were estimated by 1000 simulations.
Pre-specified link function g(t) = exp(t) for both generalizedmean residual life model (GMRL) and partially linear single-index generalized mean residual life model (PLSI GMRL).
Biomarkers in single-index component were log-transformed and standardized.
FIGURE 2.
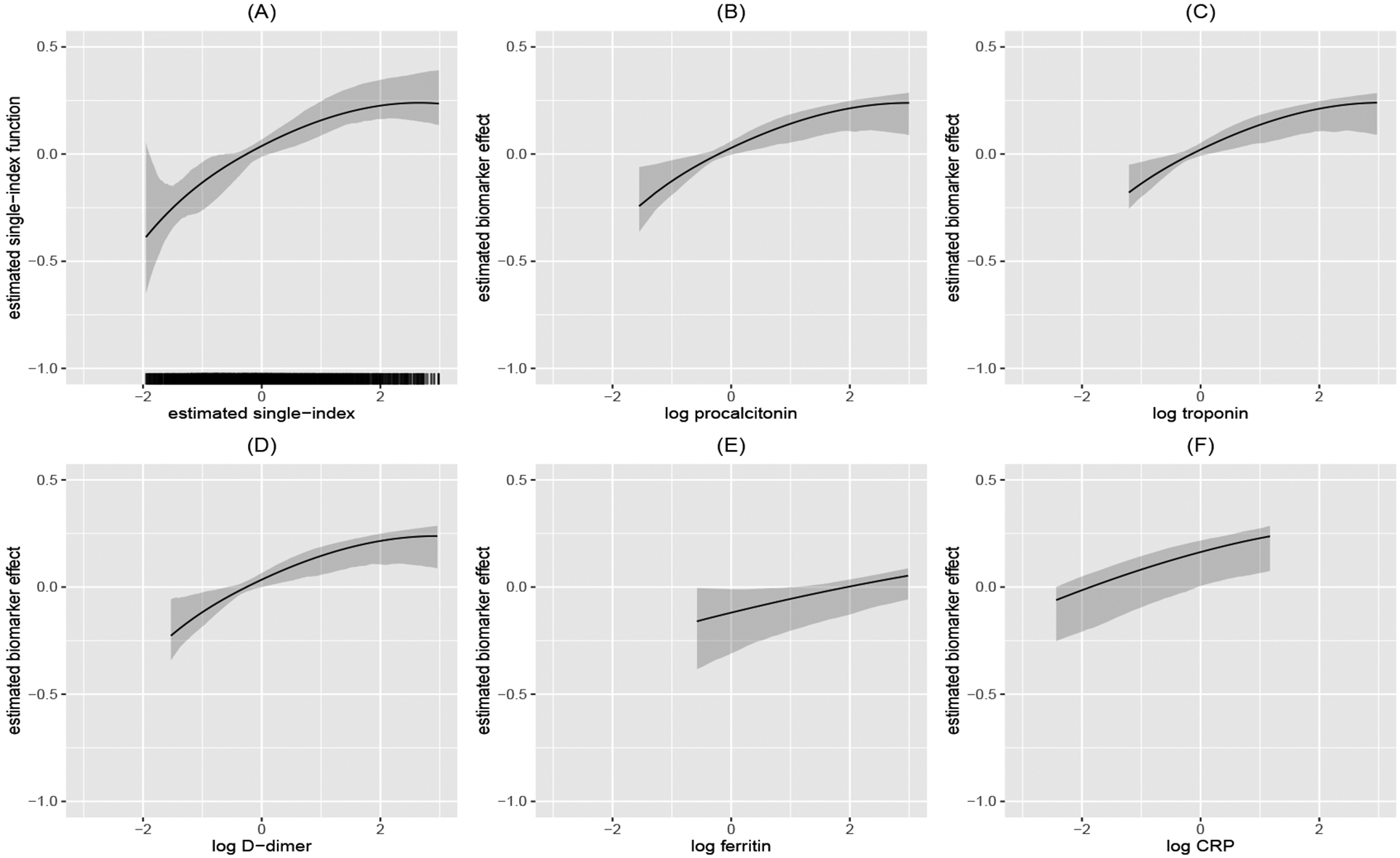
Estimated single-index function with 95% confidence intervals when g(t) = exp(t). All biomarkers were log-transformed and standardized. (A) Estimated single-index function; (B) Procalcitonin with other biomarkers fixed at median values; (C) Troponin with other biomarkers fixed at median values; (D) D-dimer with other biomarkers fixed at median values; (E) Ferritin with other biomarkers fixed at median values; (F) CRP with other biomarkers fixed at median values.
The interpretation of covariates in linear form under the PLSI GMRL model is straightforward. We observed that higher age, male, history of cardiac disease or malignancy had significant lengthening effects on the expected remaining recovery time, while a higher level of SpO2 had a significant shortening effect on the expected remaining recovery time. Keeping all other covariates fixed, the expected remaining recovery time within the next 30 days was increased by 11% for COVID-19 patients with a history of malignancy. Moreover, the estimated MRL model can enable us to predict the expected recovery time of a COVID-19 patient within 30 days since hospitalization based on baseline information at admission. As illustrated in Figure B.3, two patients were considered with different profiles and all log-transformed biomarkers in the single-index component were kept at median values except for procalcitonin. Patient A represented a 35-year-old COVID-19 confirmed Caucasian female without any smoking history or disease history, having 25 kg/m2 BMI, 90% SpO2, and 38.5 Celsius temperature at admission. Patient B had a similar profile but with a history of cardiac disease, diabetes, and malignancy. The two curves show that expected recovery time increased with a higher procalcitonin level for both patients, and patient B had a longer expected recovery time than patient A due to underlying disease history.
Remark: We further conducted simulations to evaluate the empirical performance of the numerical measures for model diagnosis. Let the sample size be N = 5000, censoring rate be 10%, true parameters maintain the same values in Section 3. The number of knots was set to be the same for comparison purpose. Specifically, when the true model was PLSI GMRL with g(t) = exp(t), the proportion of numerical measure indicating the correct model was 99.8%, 99.8%, and 81.2% for linear, sine curve, and quadratic single-index function, respectively. When the true model is PLSI GMRL with g(t) = t, the proportion of numerical measure indicating the correct model was 97.8%, 100.0%, and 89.1% for linear, sine curve, and quadratic single-index function, respectively. The empirical performance was dependent on sample size, censoring rate, and effect size.
5 |. DISCUSSION
This paper studied inference procedures for the PLSI GMRL models that allow both linear and nonlinear covariate effects. This approach reduces dimensionality and provides an estimated single-index function for summarizing the joint effects of multiple covariates. We studied the asymptotic properties of the estimators under the assumption of fixed knots due to their practicality. Furthermore, the proposed nonparametric test can be used to determine whether a nonlinear single-index function is required and is a new contribution to the single-index modeling literature. When the null hypothesis of a linear single-index function is not rejected, we can gain efficiency and straightforward interpretation by assuming a parametric form for all covariates. The proposed models incorporating single-index techniques expand the analytical toolbox to analyze the MRL for time-to-event variables, especially when multiple correlated covariates exist. Indeed, not only in survival models, the single-index technique as a powerful tool is widely considered when modeling other types of outcomes as well.16,33,34,35 With the advantage of straightforward interpretation, MRL models that directly represent an event’s residual time can be useful in many areas. Particularly during an outbreak of infectious diseases, a validated MRL model on length of hospitalization can help arrange clinical resources and facilitates planning.
Practically, MRL modeling is worthy of consideration in survival analysis under low censoring scenarios, of which the impact of a misspecified censoring model is often limited using the IPCW technique. As an alternative, the quasi-partial score (QPS) approach26,9 can also be considered for PLSI GMRL estimation and avoid modeling of censoring mechanism. In the research of GMRL modeling under subcohort sampling designs, Jin et al27 compared the performance of IPCW and QPS estimators under various censoring rates using full-cohort data. Both approaches were consistent under low censoring settings (10% or 30% censoring rate), and IPCW estimators were more efficient than QPS estimators. While for high censoring settings (80% censoring rate), GMRL models can be estimable with long-term follow-ups, and QPS estimators outperformed IPCW estimators in terms of bias and efficiency. To ensure consistency, we further studied the IPCW double-robust estimator, which requires either the model for censoring distribution or the model for complete data distribution being correctly specified. Through numerical investigations, we observed that the double-robust estimators alleviate the impact of a violation of independent censoring or a mis-specified model for covariate-dependent censoring mechanism. In addition to sample size and censoring rate, the performance of the proposed models and inference procedures would also depend on effect sizes of the covariates and signal strength of the single-index function. For a smaller sample with a moderate censoring rate (30% to 50%), the proposed method works well when the signal strength of the single-index function is relatively large.
The proposed PLSI GMRL models could also be extended to handle time-dependent covariates or time-varying coefficients, which would allow modeling repeated biomarkers and offer additional flexibility and accuracy. Moreover, the current selection of variables into the single-index function is based on the research of interest and set a prior. A formal procedure that could guide the selection of covariates into the single-index component could be a future research topic and helpful for researchers to better understand the covariate effects. For example, the approach to identifying linear versus nonlinear components by Zhang et al36 could be used first to delineate the covariate structure and separate out the nonlinear components to be included into the single-index function. In addition, when the included number of single-index variables is large, combining the PLSL GMRL models with penalties on the single-index coefficients can be further explored. It is often of interest to identify those variables contributing most to the joint effects. In this case, the PLSI GMRL model with penalization can help identify these variables and simplify the modeling and interpretation.
Supplementary Material
Table A.1 Simulation results of estimated single-index coefficients under g(t) = exp(t) with independent censoring.
Table A.2 Simulation results of estimated single-index coefficients under g(t) = t with independent censoring.
Table A.3 Simulation results under independent censoring with a 20% censoring rate.
Table A.4 Simulation results under independent censoring with a baseline MRL function
Figure B.1 Correlation matrix of Pearson correlation coefficients of five log-transformed biomarkers.
Figure B.2 Standardized score process for model diagnosis.
Figure B.3 Predicted recovery time within 30 days since hospitalization based on different procalcitonin levels for patients with different baseline disease history.
ACKNOWLEDGMENTS
The authors would like to thank the Editor, the Associated Editor, and two reviewers for their constructive and insightful suggestions that greatly improved the paper. Research reported in this manuscript was partially supported by the NIEHS of the National Institutes of Health under award number R01ES032808.
Footnotes
SUPPORTING INFORMATION
The simulation R code is available from the authors upon request. The following supporting information is available as part of the online article:
Regularity conditions and all technical proofs regarding Theorem 1 and double robust property discussed in Section 2.5.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from New York University Langone Health. Restrictions apply to the availability of these data.
References
- 1.Oakes D, Dasu T. A note on residual life. Biometrika 1990; 77(2): 409–410. [Google Scholar]
- 2.Emanuel EJ, Persad G, Upshur R, et al. Fair Allocation of Scarce Medical Resources in the Time of Covid-19. New England Journal of Medicine 2020; 382(21): 2049–2055. [DOI] [PubMed] [Google Scholar]
- 3.White DB, Lo B. A Framework for Rationing Ventilators and Critical Care Beds During the COVID-19 Pandemic. JAMA 2020; 323(18): 1773–1774. [DOI] [PubMed] [Google Scholar]
- 4.Ponti G, Maccaferri M, Ruini C, Tomasi A, Ozben T. Biomarkers associated with COVID-19 disease progression. Critical Reviews in Clinical Laboratory Sciences 2020: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petrilli CM, Jones SA, Yang J, et al. Factors associated with hospital admission and critical illness among 5279 people with coronavirus disease 2019 in New York City: prospective cohort study. BMJ 2020; 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ayanian S, Reyes J, Lynn L, Teufel K. The association between biomarkers and clinical outcomes in novel coronavirus pneumonia in a US cohort. Biomarkers in Medicine 2020; 14(12): 1091–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun L, Zhang Z. A Class of Transformed Mean Residual Life Models With Censored Survival Data. J Am Stat Assoc 2009; 104(486): 803–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maguluri G, Zhang CH. Estimation in the Mean Residual Life Regression Model. Journal of the Royal Statistical Society: Series B (Methodological) 1994: 14. [Google Scholar]
- 9.Chen YQ, Jewell NP, Lei X, Cheng SC. Semiparametric Estimation of Proportional Mean Residual Life Model in Presence of Censoring. Biometrics 2005; 61(1): 170–178. [DOI] [PubMed] [Google Scholar]
- 10.Chen YQ, Cheng S. Linear Life Expectancy Regression with Censored Data. Biometrika 2006; 93(2): 303–313. [Google Scholar]
- 11.Chen YQ. Additive Expectancy Regression. Journal of the American Statistical Association 2007; 102(477): 153–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang G, Zhou Y. Semiparametric varying-coefficient study of mean residual life models. Journal of Multivariate Analysis 2014; 128: 226–238. [Google Scholar]
- 13.Sun L, Song X, Zhang Z. Mean residual life models with time-dependent coefficients under right censoring. Biometrika 2012; 99(1): 185–197. [Google Scholar]
- 14.Stoker TM. Consistent Estimation of Scaled Coefficients. Econometrica 1986; 54(6): 1461–1481. [Google Scholar]
- 15.Hardle W, Stoker TM. Investigating Smooth Multiple Regression by the Method of Average Derivatives. Journal of the American Statistical Association 1989; 84(408): 986–995. [Google Scholar]
- 16.Ichimura H. Semiparametric least squares (SLS) and weighted SLS estimation of single-index models. Journal of Econometrics 1993; 58(1): 71–120. [Google Scholar]
- 17.Wang W. PROPORTIONAL HAZARDS REGRESSION MODELS WITH UNKNOWN LINK FUNCTION AND TIME-DEPENDENT COVARIATES. Statistica Sinica 2004; 14(3): 885–905. [Google Scholar]
- 18.Huang JZ, Liu L. Polynomial Spline Estimation and Inference of Proportional Hazards Regression Models with Flexible Relative Risk Form. Biometrics 2006; 62(3): 793–802. [DOI] [PubMed] [Google Scholar]
- 19.Sun J, Kopciuk KA, Lu X. Polynomial spline estimation of partially linear single-index proportional hazards regression models. Computational Statistics & Data Analysis 2008; 53(1): 176–188. [Google Scholar]
- 20.Shang S, Liu M, Zeleniuch-Jacquotte A, et al. Partially Linear Single Index Cox Regression Model in Nested Case-Control Studies. Comput Stat Data Anal 2013; 67: 199–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wood SN. Generalized additive models: An introduction with R. 2006.
- 22.Ying Z. A Large Sample Study of Rank Estimation for Censored Regression Data. The Annals of Statistics 1993; 21(1). [Google Scholar]
- 23.Ma Y, Wei Y. Analysis on censored quantile residual life model via spline smoothing. STAT SINICA 2012; 22(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Efron B. Bootstrap Methods: Another Look at the Jackknife. The Annals of Statistics 1979; 7(1): 1–26. [Google Scholar]
- 25.Rubin D, Laan v. dMJ. A doubly robust censoring unbiased transformation. Int J Biostat 2007; 3(1): Article 4. [DOI] [PubMed] [Google Scholar]
- 26.Chen YQ, Cheng S. Semiparametric regression analysis of mean residual life with censored survival data. Biometrika 2005; 92(1): 19–29. [Google Scholar]
- 27.Jin P, Zeleniuch-Jacquotte A, Liu M. Generalized mean residual life models for case-cohort and nested case-control studies. Lifetime Data Anal 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galvao AF, Wang L. Uniformly Semiparametric Efficient Estimation of Treatment Effects With a Continuous Treatment. Journal of the American Statistical Association 2015; 110(512): 1528–1542. [Google Scholar]
- 29.Mansourvar Z, Martinussen T, Scheike TH. Semiparametric regression for restricted mean residual life under right censoring. Journal of Applied Statistics 2015; 42(12): 2597–2613. [Google Scholar]
- 30.Mansourvar Z, Martinussen T, Scheike TH. An Additive-Multiplicative Restricted Mean Residual Life Model. Scandinavian Journal of Statistics 2016; 43(2): 487–504. [Google Scholar]
- 31.Cortese G, Holmboe SA, Scheike TH. Regression models for the restricted residual mean life for right-censored and left-truncated data. Statistics in Medicine 2017; 36(11): 1803–1822. [DOI] [PubMed] [Google Scholar]
- 32.Beigel JH, Tomashek KM, Dodd LE, et al. Remdesivir for the Treatment of Covid-19 — Preliminary Report. New England Journal of Medicine 2020; 0(0). [DOI] [PubMed] [Google Scholar]
- 33.Chaudhuri P. Global nonparametric estimation of conditional quantile functions and their derivatives. Journal of Multivariate Analysis 1991; 39(2): 246–269. [Google Scholar]
- 34.Carroll RJ, Fan J, Gijbels I, Wand MP. Generalized Partially Linear Single-Index Models. Journal of the American Statistical Association 1997; 92(438): 477–489. [Google Scholar]
- 35.Wang Y, Wu Y, Jacobson MH, et al. A family of partial-linear single-index models for analyzing complex environmental exposures with continuous, categorical, time-to-event, and longitudinal health outcomes. Environmental Health 2020;19(1): 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang HH, Cheng G, Liu Y. Linear or Nonlinear? Automatic Structure Discovery for Partially Linear Models. Journal of the American Statistical Association 2011; 106(495): 1099–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table A.1 Simulation results of estimated single-index coefficients under g(t) = exp(t) with independent censoring.
Table A.2 Simulation results of estimated single-index coefficients under g(t) = t with independent censoring.
Table A.3 Simulation results under independent censoring with a 20% censoring rate.
Table A.4 Simulation results under independent censoring with a baseline MRL function
Figure B.1 Correlation matrix of Pearson correlation coefficients of five log-transformed biomarkers.
Figure B.2 Standardized score process for model diagnosis.
Figure B.3 Predicted recovery time within 30 days since hospitalization based on different procalcitonin levels for patients with different baseline disease history.
Data Availability Statement
The data that support the findings of this study are available from New York University Langone Health. Restrictions apply to the availability of these data.


