Abstract
Purpose:
The delineation of organs-at-risk (OARs) is fundamental to cone-beam CT (CBCT)-based adaptive radiotherapy treatment planning, but is time-consuming, labor-intensive, and subject to inter-operator variability. We investigated a deep learning-based rapid multi-organ delineation method for use in CBCT-guided adaptive pancreatic radiotherapy.
Methods and materials:
To improve the accuracy of OAR delineation, two innovative solutions have been proposed in this study. First, instead of directly segmenting organs on CBCT images, a pre-trained cycle-consistent generative adversarial network (cycleGAN) was applied to generating synthetic CT images given CBCT images. Second, an advanced deep learning model called mask-scoring regional convolutional neural network (MS R-CNN) was applied on those synthetic CT to detect the positions and shapes of multiple organs simultaneously for final segmentation. The OAR contours delineated by the proposed method were validated and compared with expert-drawn contours for geometric agreement using the Dice similarity coefficient (DSC), 95th percentile Hausdorff distance (HD95), mean surface distance (MSD), and residual mean square distance (RMS).
Results:
Across eight abdominal OARs including duodenum, large bowel, small bowel, left and right kidneys, liver, spinal cord and stomach, the geometric comparisons between automated and expert contours are as follows: 0.92 (0.89 - 0.97) mean DSC, 2.90 mm (1.63 mm - 4.19 mm) mean HD95, 0.89 mm (0.61 mm - 1.36 mm) mean MSD, and 1.43 mm (0.90 mm - 2.10 mm) mean RMS. Compared to the competing methods, our proposed method had significant improvements (p < 0.05) in all the metrics for all the eight organs. Once the model was trained, the contours of eight OARs can be obtained on the order of seconds.
Conclusion:
We demonstrated the feasibility of a synthetic CT-aided deep learning framework for automated delineation of multiple OARs on CBCT. The proposed method could be implemented in the setting of pancreatic adaptive radiotherapy to rapidly contour OARs with high accuracy.
Keywords: Cone-beam computed tomography, multi-organ segmentation, automated contouring, pancreatic cancer, adaptive radiation therapy, deep learning, mask R-CNN
1. INTRODUCTION
Pancreatic cancer is the fourth leading cause of cancer-related mortality in the United States, with a 5-year survival rate below 10% 1. Radiation therapy is often recommended for unresectable or locally advanced disease 2-4. The efficacy of radiotherapy is highly dependent upon adequate delivery of dose to the target 5-7. Highly conformal dose delivery to target regions can be achieved by modern radiotherapy techniques8-10. However, modern radiotherapy techniques are generally very sensitive to geometrical changes resulting from uncertainties in patient setup and inter-fractional changes in patient anatomy 11-13. In pancreatic radiotherapy, optimal delivery of ablative doses with minimal toxicity to adjacent organs at risk (OARs) is rather challenging due to two competing factors: the necessity for adequate margin between the planning target volume (PTV) and clinical target volume (CTV) to account for inter- and intra-fractional variation in setup position and physiologic motion (e.g. breathing motion),14-16 and the high radiation sensitivity of surrounding OARs including stomach, duodenum, bowels, kidney, liver, and spinal cord. Late fatal toxicities have been reported using dose-escalated stereotactic body radiotherapy (SBRT) 17. While SBRT using non-ablative doses may result in low toxicity, overall survival remains suboptimal 18. Respiratory-gating and image-guided radiotherapy (IGRT) have been developed to manage intra- and inter-fractional variation in position 15,19,20. Common clinical practice is to reposition the patient based on rigid registration of daily cone-beam CT (CBCT) images and the planning CT obtained at simulation.
Online adaptive planning with treatment field re-optimization based on daily CBCT imaging has been proposed to further leverage IGRT for managing inter-fraction position variations 21-23. However, clinical implementation of online adaptive radiotherapy (ART) is hampered by technical limitations resulting in prohibitively long re-planning times. To achieve practical clinical utility, online ART must be rapid, completed within the time a patient is typically held immobilized during treatment delivery. Segmentation of targets and OARs on the daily imaging is a fundamental, but time-consuming step in the replanning process. Manual delineation could highly prolong the entire adaptive re-planning process. Methods have been investigated for CBCT-based auto OAR delineation by contour propagation through rigid or deformable registration between a daily CBCT and the planning CT 24-29. However, the accuracy of contour propagation from planning CT to CBCT relies heavily upon registration quality,30 and manual editing and verification are often necessary. Those semi-automated methods to some extent speed up the OAR contouring, but the improvement is still very limited. Thus, a faster OAR delineation method is highly desired for implementation of CBCT-based plan adaptation.
Deep learning-based segmentation algorithms have achieved promising performance in CT contouring for many anatomical sites 31-38. Clinically acceptable contours of multiple organs from CT can be obtained within a minute once the model was trained as reported in very recent studies 31,32. Compared to traditional segmentation methods39,40, convolutional neural networks (CNNs) and their variations are among the most widely investigated algorithms for those automated contouring methods 31-34. CNNs extract abstract features from original images to obtain the final segmentations by an end-to-end process. However, due to the inferior image quality of CBCT compared with CT, it is difficult to obtain contours by directly applying existing CNN-based algorithms on CBCT images 37,41. Therefore, studies have to be carried out to investigate the feasible of deep learning-based segmentation on CBCT images. In this study, a two-in-one deep learning model is investigated for fully-automated CBCT-based segmentation of up to eight OARs in the setting of pancreatic radiotherapy. First, a synthetic CT (sCT) 42 is obtained from CBCT using a cycle-consistent generative adversarial network (cycleGAN)-based algorithm, which has achieved impressive success in cross-modality medical image synthesis 43,44. Synthetic image aided segmentation has been demonstrated as a promising method for improving the accuracy of segmentation in our previous studies45-47. Compared to these previous investigations, in this work, we use the strategy of image synthesis to improve the image quality of the original CBCT. We hypothesize that with the improved image quality of sCT compared to the original CBCT, the segmentation performance can be improved. In the second step, a mask scoring regional convolutional neural network (MS R-CNN) is applied to the sCT to obtain final contours of multiple organs simultaneously. Compared to our previous studies45-47, MS R-CNN firstly detects the organs’ regions-of-interest (ROIs) and then segments organs’ binary mask within these ROIs. One hundred and twenty sets of CBCT images from patients with pancreatic cancer were used to assess the performance of our proposed method.
2. METHODS AND MATERIALS
2.A. CT synthesis
Synthetic CT images were generated from daily CBCT images using cycleGAN 48, a variation of the generative adversarial network (GAN) 49. GAN and its variations have achieved impressive success in image-to-image translation due to the formulation of their characteristic adversarial loss function. The adversarial loss causes two models (a generator, which creates synthetic images from a known distribution of target images, and a discriminator, which is meant to identify counterfeits) to work against each other to improve their performance until the synthesized images are indistinguishable from the originals. Figure 1 shows the network architecture of the cycleGAN for synthesis of CT from CBCT. The generator consists of one convolutional layer with a stride size of 1×1×1, two convolutional layers with a stride size of 2×2×2, nine ResNet blocks, two deconvolution layers and two convolutional layers with a stride size of 1×1×1. The discriminator is composed of six convolutional layers with a stride size of 2×2×2, one fully connected layer and a sigmoid operation layer. In the training process, given the paired CBCT and CT, an initial mapping is learned to generate a synthetic CT image (sCT) from CBCT to fool the discriminator. In contrast, the discriminator is trained to differentiate CT from sCT. The discriminator and the generator are pitted against each other in a sequential zero-sum game, with each round of training leading to more accurate sCT generation. CycleGAN extends this framework by introducing a second generator-discriminator pair that conducts an inverse transformation from sCT back to CBCT and comparing it against the original CBCT distribution to further constrain the synthesis task and improve the quality of sCT generation.
FIG. 1.
The schematic flow diagram of the cycle-consistent generative adversarial network (cycleGAN). Syn., synthetic, Conv., convolution, DeConv., deconvolution.
2.B. Mask scoring regional convolutional neural network (MS R-CNN)
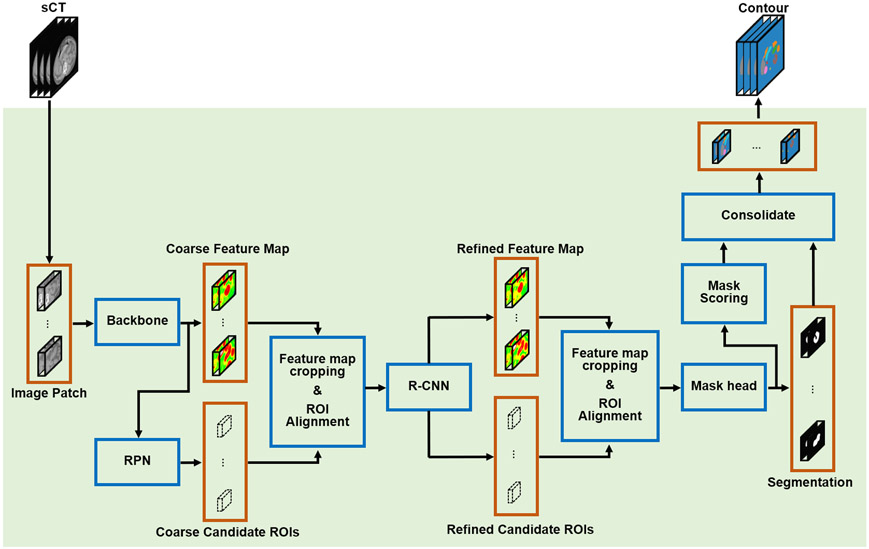
Taking sCT generated by cycleGAN as an input, multi-organ segmentations are obtained through MS R-CNN, a variation on mask R-CNN 50. Mask R-CNN for tumor and organ segmentation has been previously investigated 51-53. The mask scoring strategy aims to improve final segmentation accuracy by reducing mismatches between mask score and mask quality that were observed in analyses of the original mask R-CNN algorithm 54. The network architecture of mask scoring R-CNN is shown in Figure 2. Two rounds of feature extraction are implemented through five primary subnetworks, including a backbone, a regional proposal network (RPN), a R-CNN, a mask network, and a mask scoring network. In the first step, coarse feature maps are extracted by a backbone network taking sCT as input. An encoding-decoding feature pyramid network 55 with six hierarchical levels of resolution is adopted as the backbone network in this study (Figure S1). Each level consists of three ResNet layers and one convolution or one deconvolution layer. Hierarchical features in the decoding pathway are enhanced by lateral connections to the encoding pathways at the same resolution. The coarse feature maps obtained in the first step are then used to predict the coarse candidate regions-of-interest (ROIs) by a RPN 56 composed of two pathways (Figure S2), anchor classification and anchor regression, each with a series of convolutional operations followed by a fully-connected layer. The coarse feature maps are then cropped and realigned by coarse candidate ROIs, which are input to the R-CNN network (Figure S2) to obtain refined feature maps in the second step of feature extraction. R-CNN has an architecture similar to RPN, but in R-CNN, the input feature maps are interpolated before being fed into the network. Meanwhile, the refined candidate ROIs are simultaneously predicted by R-CNN. The refined feature maps are cropped and realigned by the refined candidate ROIs and fed into the mask network to predict ROI segmentations. The mask network is composed of four convolutional layers followed by a softmax layer (Figure S3). The final contour for each organ is obtain by consolidating these ROI segmentations according to the mask score assigned to each ROI segmentation. In conventional mask R-CNN, those mask scores are generated by the mask network along with the ROI segmentation prediction; in this study, we adopt the mask scoring strategy 54 to predict the mask scores using a discrete mask scoring network composed of five convolutional layers followed a fully-connected layer and a sigmoid layer (Figure S3).
FIG. 2.
The schematic flow diagram of the mask scoring R-CNN for segmentation. RPN, regional proposal network, R-CNN, regional convolutional neural network, sCT, synthetic CT, ROI, region-of-interest.
The mask scoring R-CNN model was trained using five types of loss functions to separately supervise the backbone, RPN, R-CNN, mask, and mask scoring subnetworks. The loss function for backbone is the binary cross entropy (BCE) which was introduced in our previous work 57. The second and third types of loss functions are identically defined for RPN and R-CNN. Take the RPN as an example, the loss LRPN can be defined as:
| (1) |
| (2) |
| (3) |
| (4) |
where, IoU is intersection over union, IROI is the ground truth ROI of an organ, is the predicted ROI, (xCi, yCi, zCi) and (, , ) are respectively the central coordinate of ground truth and predicted ROIs, wi, hi, and di are the dimensions of ground truth ROI along x, y, and z directions, and , , are that for the predicted ROI.
The fourth type of loss function is utilized to supervise the mask subnetwork, which can be defined as:
| (5) |
where, and are respectively the ground truth and predicted binary mask.
The last type of loss function is an IoU loss which is used for supervising the mask scoring subnetwork. The loss function is defined as:
| (6) |
where, IoU and respectively indicate the ground truth IoU and the predicted IoU.
2.C. Data preparation
Two separate datasets were collected for training and validation of the proposed method. The first consists of CT and CBCT images from 35 pancreatic cancer patients. This set was used to train and test the cycleGAN for sCT generation. CT images were acquired on a Siemens SOMATOM Definition AS (Siemens, Erlangen, Germany) at 120 kVp for the initial treatment planning. CBCT images were acquired immediately prior to the delivery of the first treatment fraction on a Varian TrueBeam System (Varian Medical Systems, Inc. Palo Alto, USA) at 125 kVp. CT and CBCT images for each patient were deformably registered using multi-pass algorithm integrated in Velocity AI 3.2.1 (Varian Medical Systems, Inc. Palo Alto, USA) to minimize mismatches of organ shape and location between CT and CBCT. Then CBCT images were resampled to guarantee that CBCT and CT images have identical data volume and voxel size using Velocity AI 3.2.1.
The second dataset consists of 120 sets of CBCT images and corresponding ground truth OAR contours from 40 patients with pancreatic cancer. In our clinic, CBCT images were taken in the daily patient setup, so, we selected three sets of these CBCT images from each patient. This set was used to train and test the integrated cycleGAN and mask scoring R-CNN auto-segmentation model. For each patient, three sets of CBCT images and corresponding OAR contours were collected within treatment fractions. Manual contours were initially created by radiation oncologists using the planning CT prior to delivery of the first treatment fraction. These were propagated to CBCT with deformable registration using Velocity AI 3.2.1. The propagated CBCT contours were adjusted and verified by two radiation oncologists to remove error introduced by registration and propagation. From these, consensus was reached to obtain the final multi-organ segmentations, which are regarded as ground truth. Both radiation oncologists were blinded to the patient’s clinical information.
2.D. Network training and validation
The cycleGAN was trained and tested on the first dataset. A leave-one-out cross validation approach was applied wherein, for each of 35 experiments, images belonging to each of the 35 patients were sequentially omitted during training for use as test data. The cycleGAN was trained and tested as that was detailed in our previous studies46,58. Here, the learning rate for Adam optimizer was set to 2e-4. The training was stopped after 150000 iterations. After the cycleGAN was trained and tested, it was then integrated into the segmentation model between the CBCT and the input of the mask scoring R-CNN model.
For training and validation of the integrated pre-trained cycleGAN and mask scoring R-CNN model, the second dataset was separated into two groups: a training set of 96 CBCT images and corresponding contours from 32 patients and a test set of 24 CBCT images and corresponding contours from 8 patients. The trainable parameters of the model were entirely tuned using the training set, while the testing set was utilized for quantitative statistical analysis. And there’s no overlapping between the training and test images. The model was implemented in-house in Python 3.7 and TensorFlow 1.14 on a NVIDIA Tesla V100 GPU with 32 GB of memory within the Ubuntu 18.04 LTS environment. The learnable parameters of the mask scoring R-CNN model were optimized using the Adam optimizer with a learning rate of 2e-4 in each training epoch. Training was accomplished over 400 epochs, each with a batch size of 20. Once trained, inference compute time is maintained on the order of seconds.
3. RESULTS
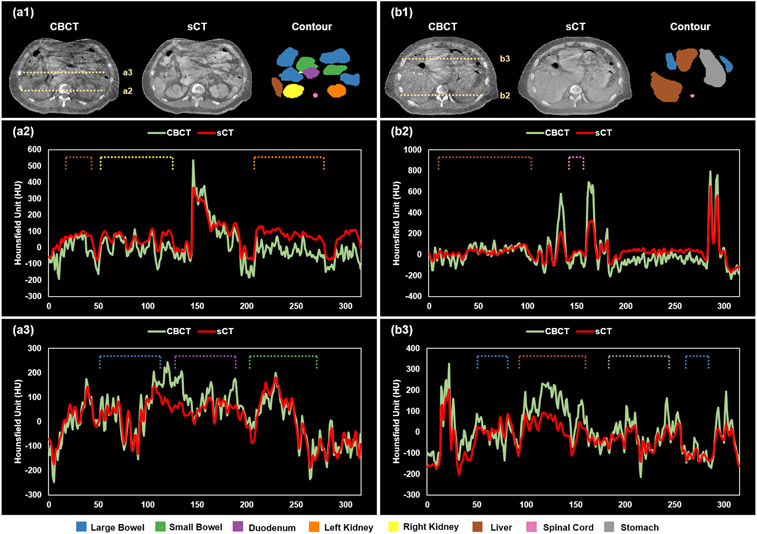
The incorporation of a cycleGAN into the segmentation model aims to improve the quality of CBCT images. As mentioned in the above section 2.D, the cycleGAN was trained with the Adam optimizer set to be 2e-4 and stopped after 150000 iterations. To closely investigate on the image quality improvement of synthetic CT versus given CBCT images, we obtained the output from cycleGAN and did one-to-one comparisons between sCT and CBCT images. Figure 3 demonstrates image quality improvement of sCT relative to CBCT. The cross-sectional profiles were respectively plotted for CBCT and sCT. Figs. 3(a2) and 3(a3) show the profiles along the dashed lines in Fig. 3(a1), while Figs. 3(b2) and 3(b3) are the profiles along the dashed lines in Fig. 3(b1). To provide better illustration, the ground truth contours for all the eight organs including large bowel, small bowel, duodenum, left/right kidney, liver, spinal cord, and stomach were also shown. In Figs. 3(a2), 3(a3), 3(b2), and 3(b3), dashed lines indicate the ground truth profile of each organ. From these images and profiles, we can see that the quality of CBCT was improved in sCT. In particular, artifacts and noise were notably mitigated within each organ, while organ boundaries remain sharp.
FIG. 3.
An illustrative example of improved image quality in synthetic CT compared to CBCT. Panels (a1) and (b1) show matching transverse slices of CBCT and synthetic CT (sCT) with corresponding ground truth organ contours. Panels (a2) and (a3) show the Hounsfield unit profiles of CBCT and sCT images along the dashed lines in (a1). Similarly, (b2) and (b3) show the profiles of CBCT and sCT along the dashed lines in (b1).
The experiments were conducted to validate the proposed mask scoring R-CNN based on the details that described in the above section 2.D. The performance of the proposed segmentation method using sCT as input to a mask scoring R-CNN (sCT MS R-CNN) was compared with MS R-CNN with CBCT as input (CBCT-only), U-Net with sCT input (sCT U-Net), and deep attention fully convolution network (DA-FCN)37 with sCT input. The U-Net is implemented in 3D convolutional layers manner59. The input size of U-Net and the proposed MS R-CNN were same, and the same training and testing data were used for both U-Net and the proposed MS R-CNN. The learning rate and number of training epochs were set based on U-Net’s best performance on validation set. Superior performance of the proposed method is demonstrated in an illustrative example in Figure 4, where, from top to bottom, rows contain CBCT images, sCT images, ground truth multi-organ contours, the predictions of the proposed, CBCT-only predictions, sCT U-Net predictions, and DA-FCN predictions. When evaluated geometrically, the proposed method achieves better performance on large bowel, small bowel, duodenum, left/right kidney, liver, spinal cord, and stomach.
FIG. 4.
An illustrative example of the benefit of our proposed method compared with the CBCT-only, sCT U-net, and DA-FCN methods. Proposed is the prediction of the proposed method, CBCT-only is the prediction of the CBCT-only method, U-net is the prediction of the sCT U-net method, and DA-FCN is the prediction of deep attention fully convolution network taking the sCT as the input.
To quantify the performance of our method, the Dice similarity coefficient (DSC), 95th percentile Hausdorff distance (HD95), mean surface distance (MSD), and residual mean square distance (RMS) 60 were calculated and compared to values obtained for the CBCT-only, sCT U-net and DA-FCN predictions. Statistical analysis was also performed using student’s paired t-test. Results are summarized in Table 1. Our method achieved higher DSC values and lower HD95, MSD, and RMS on all investigated OARs compared to CBCT-only, U-net, or DA-FCN method. In particular, mean DCS, HD95, MSD and RMS values of 0.92, 2.90 mm, 0.89 mm, and 1.43 mm were achieved by our proposed method. The statistical results listed in Table 1 show that compared to U-net method, the proposed method had significant improvements (p < 0.05) in all the metrics for all the eight organs, while significant improvements (p < 0.05) were also observed across all metrics for stomach, duodenum, small bowel, left/right kidney and spinal cord relative to the CBCT-only method.
Table 1.
Overall quantitative results achieved by the proposed, CBCT-only, U-net, and DA-FCN methods.
| Organ | Method | DSC | HD95 (mm) | MSD (mm) | RMS (mm) |
|---|---|---|---|---|---|
| Large Bowel | Ⅰ. Proposed | 0.91±0.06 | 4.19±3.94 | 1.36±1.24 | 2.10±1.83 |
| Ⅱ. CBCT-only | 0.90±0.07 | 4.54±3.43 | 1.40±1.11 | 2.27±1.62 | |
| Ⅲ. U-net | 0.89±0.04 | 5.30±2.95 | 1.70±0.91 | 2.63±1.40 | |
| IV. DA-FCN | 0.90±0.04 | 5.23±2.55 | 1.67±0.94 | 2.56±1.25 | |
| p value (Ⅰ vs. Ⅱ) | 0.20 | 0.06 | 0.45 | 0.07 | |
| p value (Ⅰ vs. Ⅲ) | <0.01 | <0.01 | <0.01 | <0.01 | |
| p value (Ⅰ vs. IV) | 0.06 | 0.01 | <0.01 | 0.04 | |
| Small Bowel | Ⅰ. Proposed | 0.92±0.05 | 2.69±1.79 | 0.82±0.57 | 1.45±0.95 |
| Ⅱ. CBCT-only | 0.90±0.04 | 3.53±1.72 | 1.01±0.48 | 1.79±0.76 | |
| Ⅲ. U-net | 0.86±0.06 | 5.13±2.04 | 1.56±0.59 | 2.58±1.00 | |
| IV. DA-FCN | 0.86±0.05 | 5.08±1.53 | 1.52±0.43 | 2.53±0.72 | |
| p value (Ⅰ vs. Ⅱ) | <0.01 | <0.01 | <0.01 | <0.01 | |
| p value (Ⅰ vs. Ⅲ) | <0.01 | <0.01 | <0.01 | <0.01 | |
| p value (Ⅰ vs. IV) | <0.01 | <0.01 | <0.01 | <0.01 | |
| Duodenum | Ⅰ. Proposed | 0.89±0.13 | 2.86±3.30 | 0.90±1.13 | 1.39±1.51 |
| Ⅱ. CBCT-only | 0.88±0.13 | 3.25±2.89 | 1.02±1.04 | 1.62±1.43 | |
| Ⅲ. U-net | 0.84±0.12 | 4.59±3.06 | 1.48±1.05 | 2.27±1.41 | |
| IV. DA-FCN | 0.84±0.09 | 4.54±2.16 | 1.47±0.80 | 2.25±1.21 | |
| p value (Ⅰ vs. Ⅱ) | <0.01 | 0.04 | <0.01 | <0.01 | |
| p value (Ⅰ vs. Ⅲ) | <0.01 | <0.01 | <0.01 | <0.01 | |
| p value (Ⅰ vs. IV) | <0.01 | <0.01 | <0.01 | <0.01 | |
| Left Kidney | Ⅰ. Proposed | 0.95±0.03 | 2.72±1.91 | 0.77±0.51 | 1.28±0.80 |
| Ⅱ. CBCT-only | 0.94±0.04 | 3.11±2.51 | 0.86±0.63 | 1.46±0.99 | |
| Ⅲ. U-net | 0.93±0.03 | 3.55±1.79 | 1.12±0.47 | 1.79±0.83 | |
| IV. DA-FCN | 0.94±0.03 | 3.47±1.60 | 1.06±0.40 | 1.75±0.74 | |
| p value (Ⅰ vs. Ⅱ) | 0.03 | 0.03 | 0.02 | 0.01 | |
| p value (Ⅰ vs. Ⅲ) | <0.01 | <0.01 | <0.01 | <0.01 | |
| p value (Ⅰ vs. IV) | <0.01 | <0.01 | <0.01 | <0.01 | |
| Right Kidney | Ⅰ. Proposed | 0.95±0.02 | 2.70±1.63 | 0.77±0.43 | 1.28±0.70 |
| Ⅱ. CBCT-only | 0.94±0.03 | 3.18±2.29 | 0.86±0.51 | 1.48±0.90 | |
| Ⅲ. U-net | 0.93±0.02 | 3.35±1.87 | 1.04±0.40 | 1.70±0.78 | |
| IV. DA-FCN | 0.94±0.02 | 3.26±1.30 | 1.00±0.35 | 1.64±0.62 | |
| p value (Ⅰ vs. Ⅱ) | 0.04 | 0.03 | 0.02 | 0.01 | |
| p value (Ⅰ vs. Ⅲ) | <0.01 | <0.01 | <0.01 | <0.01 | |
| p value (Ⅰ vs. IV) | <0.01 | <0.01 | <0.01 | <0.01 | |
| Liver | Ⅰ. Proposed | 0.97±0.03 | 3.30±3.71 | 0.95±1.23 | 1.64 ±1.62 |
| Ⅱ. CBCT-only | 0.96±0.04 | 3.57±3.51 | 0.95±0.98 | 1.86±1.63 | |
| Ⅲ. U-net | 0.95±0.03 | 4.30±2.80 | 1.22±0.81 | 2.16±1.22 | |
| IV. DA-FCN | 0.95±0.02 | 4.28±1.30 | 1.20±0.54 | 2.02±0.43 | |
| p value (Ⅰ vs. Ⅱ) | 0.18 | 0.05 | 0.89 | 0.21 | |
| p value (Ⅰ vs. Ⅲ) | <0.01 | <0.01 | <0.01 | <0.01 | |
| p value (Ⅰ vs. IV) | <0.01 | 0.02 | 0.16 | 0.16 | |
| Spinal Cord | Ⅰ. Proposed | 0.90±0.10 | 1.63±1.35 | 0.61±0.51 | 0.90±0.62 |
| Ⅱ. CBCT-only | 0.89±0.09 | 1.93±1.17 | 0.68±0.49 | 1.00±0.63 | |
| Ⅲ. U-net | 0.85±0.08 | 2.33±1.02 | 0.86±0.44 | 1.18±0.52 | |
| IV. DA-FCN | 0.86±0.07 | 2.33±0.75 | 0.84±0.38 | 1.16±0.43 | |
| p value (Ⅰ vs. Ⅱ) | 0.01 | <0.01 | <0.01 | <0.01 | |
| p value (Ⅰ vs. Ⅲ) | <0.01 | <0.01 | <0.01 | <0.01 | |
| p value (Ⅰ vs. IV) | <0.01 | <0.01 | <0.01 | <0.01 | |
| Stomach | Ⅰ. Proposed | 0.94±0.09 | 3.08±3.84 | 0.90±1.44 | 1.43±1.91 |
| Ⅱ. CBCT-only | 0.92±0.11 | 3.91±4.83 | 1.20±1.61 | 1.92±2.23 | |
| Ⅲ. U-net | 0.91±0.09 | 3.97±3.49 | 1.61±1.28 | 1.97±1.69 | |
| IV. DA-FCN | 0.92±0.03 | 3.89±2.07 | 1.24±1.11 | 1.95±1.53 | |
| p value (Ⅰ vs. Ⅱ) | <0.01 | <0.01 | <0.01 | <0.01 | |
| p value (Ⅰ vs. Ⅲ) | <0.01 | <0.01 | <0.01 | <0.01 | |
| p value (Ⅰ vs. IV) | <0.01 | <0.01 | <0.01 | <0.01 |
4. DISCUSSION
The primary clinical challenge in pancreatic radiotherapy is safely delivering ablative doses to the tumor target while sparing adjacent organs at risk, especially the duodenum 18. Online adaptive radiotherapy offers a promising solution to this problem by minimizing the impact of inter-fractional uncertainty through rapid replanning based on daily imaging 21-23. A key to the clinical implementation of rapid treatment replanning is automated segmentation of tumor targets and organs at risk, which would effectively reduce the time needed for this labor-intensive task from tens of minutes to seconds. While MR-LINAC 22,61 and CT-On-Rails15,62 technologies have been deployed in some centers, CBCT remains the most widely available modality for 3D imaging inside treatment rooms. However, CBCT imaging may only be considered a viable option for ART once the challenges of poor image quality are overcome to allow for accurate automated segmentation. The proposed method provides an encouraging solution for accelerated OAR delineation with high accuracy through deep learning. Two primary innovative solutions have been proposed within the framework. First, rather than segmenting directly on CBCT images, a cycleGAN-based algorithm has been adopted to improve the quality of CBCT images. As seen from Figure 3, the images (sCT) achieve better quality than original CBCT images for the majority of OARs. Second, an advanced network architecture has been implemented for image segmentation on those sCT images. By integrating mask R-CNN framework into the segmentation task, our method can avoid the effect of irrelevant features that are learned from the background region of CBCT. This is because the RPN and RCNN subnetworks in mask R-CNN can obtain the location of ROIs of organs. We then cropped the feature maps within these ROIs for further segmentation. Moreover, our method can diminish the misclassification issue in conventional mask R-CNN model by integrating mask scoring subnetwork into mask R-CNN framework. The results (Figure 4 and Table 1) demonstrate that our proposed method outperforms the other methods presented. The comparisons between the proposed and CBCT-only method show the benefit of quality-improvement by cycleGAN, while the improvements achieved by the proposed method over the sCT U-Net and DA-FCN methods demonstrate the advantages of the mask scoring R-CNN architecture in segmentation.
The primary limitation of this study lies in the uncertainty of image registration. The paired CT and CBCT images used in training the cycleGAN model were deformably registered using the multi-pass deformable registration algorithm integrated in the clinical software Velocity AI 3.2.1 (Varian Medical Systems, Inc. Palo Alto, USA). Registration error might affect sCT generation from CBCT. To minimize anatomic variation between CBCT and CT in this study and thereby minimize registration error, planning CT acquired during simulation and CBCT acquired the first treatment fraction were paired for training and testing the cycleGAN. A typical interval between simulation and delivery of the first treatment fraction is approximately one week in our clinic. This strategy was used in our previous study63, showing minimized effect in the sCT generation. Therefore, in this study, we assume the anatomic difference between planning CT and CBCT is minor and can be ignored. Note that CBCT-CT registration also happened in the step of obtaining the ground truth contour on each CBCT. However, we expect the registration error also had minimal effect to this study since, as discussed above, the propagated contours from CT to CBCT have been further edited and verified by experienced physicians. Certainly, manual contouring also has uncertainties. To minimize the uncertainties, in this study, the ground truth contours were obtained by manual editing on the propagated contours performed by two radiation oncologists using software (Velocity, Varian Medical Systems) to reach a consensus. Both readers were blinded to the patients’ clinical information.
5. CONCLUSIONS
In this study, we developed a deep learning-based framework for accelerated multi-organ segmentation using CBCT. Segmentation accuracy is dramatically improved using a synthetic CT intermediate. The proposed method could be incorporated into a workflow for pancreatic adaptive radiotherapy to rapidly contour OARs during treatment replanning with high accuracy. The promising initial verification results in this study demonstrate the advantages of our proposed method for automated multi-organ contouring in pancreatic radiotherapy. Our future studies will be focused on the translational investigations on routine clinical practice to further demonstrate the capability of our proposed method. The existing limitations as mentioned in the above discussion section such as the uncertainties lie in the ground truth and image registration will be carefully studied as the following steps of this work.
Supplementary Material
ACKNOWLEDGMENTS
This research is supported in part by the National Cancer Institute of the National Institutes of Health under Award Number R01CA215718 (XY), the Department of Defense (DoD) Prostate Cancer Research Program (PCRP) Award W81XWH-17-1-0438 (TL) and W81XWH-19-1-0567 (XD), and Emory Winship Cancer Institute pilot grant (XY).
Footnotes
DISCLOSURES
The authors declare no conflicts of interest.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal AJ Cacjfc. Cancer statistics, 2019. 2019;69(1):7–34. [DOI] [PubMed] [Google Scholar]
- 2.Chuong MD, Springett GM, Freilich JM, et al. Stereotactic body radiation therapy for locally advanced and borderline resectable pancreatic cancer is effective and well tolerated. 2013;86(3):516–522. [DOI] [PubMed] [Google Scholar]
- 3.Petrelli F, Comito T, Ghidini A, Torri V, Scorsetti M, Barni SJ IJoROBP. Stereotactic body radiation therapy for locally advanced pancreatic cancer: a systematic review and pooled analysis of 19 trials. 2017;97(2):313–322. [DOI] [PubMed] [Google Scholar]
- 4.Loehrer P Sr, Powell M, Cardenes H, et al. A randomized phase III study of gemcitabine in combination with radiation therapy versus gemcitabine alone in patients with localized, unresectable pancreatic cancer: E4201. 2008;26(15_suppl):4506–4506. [Google Scholar]
- 5.Cellini F, Arcelli A, Simoni N, et al. Basics and Frontiers on Pancreatic Cancer for Radiation Oncology: Target Delineation, SBRT, SIB Technique, MRgRT, Particle Therapy, Immunotherapy and Clinical Guidelines. 2020;12(7):1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunner TB, Nestle U, Grosu A-L, Partridge MJ R, Oncology. SBRT in pancreatic cancer: What is the therapeutic window? 2015;114(1):109–116. [DOI] [PubMed] [Google Scholar]
- 7.Reyngold M, Parikh P, Crane CHJ RO. Ablative radiation therapy for locally advanced pancreatic cancer: techniques and results. 2019;14(1):95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Webb S. Intensity-modulated radiation therapy. CRC Press; 2015. [Google Scholar]
- 9.Otto K Volumetric modulated arc therapy: IMRT in a single gantry arc [published online ahead of print 2008/02/26]. Med Phys. 2008;35(1):310–317. [DOI] [PubMed] [Google Scholar]
- 10.Lomax AJ, Boehringer T, Coray A, et al. Intensity modulated proton therapy: a clinical example [published online ahead of print 2001/04/25]. Med Phys. 2001;28(3):317–324. [DOI] [PubMed] [Google Scholar]
- 11.Paganetti H Range uncertainties in proton therapy and the role of Monte Carlo simulations [published online ahead of print 2012/05/11]. Phys Med Biol. 2012;57(11):R99–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arts T, Breedveld S, de Jong MA, et al. The impact of treatment accuracy on proton therapy patient selection for oropharyngeal cancer patients [published online ahead of print 2017/10/28]. Radiother Oncol. 2017;125(3):520–525. [DOI] [PubMed] [Google Scholar]
- 13.Albertini F, Bolsi A, Lomax AJ, Rutz HP, Timmerman B, Goitein G. Sensitivity of intensity modulated proton therapy plans to changes in patient weight [published online ahead of print 2008/01/18]. Radiother Oncol. 2008;86(2):187–194. [DOI] [PubMed] [Google Scholar]
- 14.Jayachandran P, Minn AY, Van Dam J, Norton JA, Koong AC, Chang DTJ IJoROBP. Interfractional uncertainty in the treatment of pancreatic cancer with radiation. 2010;76(2):603–607. [DOI] [PubMed] [Google Scholar]
- 15.Liu F, Erickson B, Peng C, Li XAJ IJoROBP. Characterization and management of interfractional anatomic changes for pancreatic cancer radiotherapy. 2012;83(3):e423–e429. [DOI] [PubMed] [Google Scholar]
- 16.Chen I, Mittauer K, Henke L, et al. Quantification of interfractional gastrointestinal tract motion for pancreatic cancer radiation therapy. 2016;96(2):E144. [Google Scholar]
- 17.De Lange S, Van Groeningen C, Meijer O, et al. Gemcitabine–radiotherapy in patients with locally advanced pancreatic cancer. 2002;38(9):1212–1217. [DOI] [PubMed] [Google Scholar]
- 18.Tchelebi LT, Zaorsky NG, Rosenberg JC, et al. Reducing the Toxicity of Radiotherapy for Pancreatic Cancer With Magnetic Resonance-guided Radiotherapy. 2020;175(1):19–23. [DOI] [PubMed] [Google Scholar]
- 19.Campbell WG, Jones BL, Schefter T, Goodman KA, Miften MJ R, Oncology. An evaluation of motion mitigation techniques for pancreatic SBRT. 2017;124(1):168–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reese AS, Lu W, Regine WF. Utilization of intensity-modulated radiation therapy and image-guided radiation therapy in pancreatic cancer: is it beneficial? Paper presented at: Seminars in Radiation Oncology 2014. [DOI] [PubMed] [Google Scholar]
- 21.Lim-Reinders S, Keller BM, Al-Ward S, Sahgal A, Kim AJ IJoROBP. Online adaptive radiation therapy. 2017;99(4):994–1003. [DOI] [PubMed] [Google Scholar]
- 22.Boldrini L, Cusumano D, Cellini F, Azario L, Mattiucci GC, Valentini VJ RO. Online adaptive magnetic resonance guided radiotherapy for pancreatic cancer: state of the art, pearls and pitfalls. 2019;14(1):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brock KK. Adaptive radiotherapy: moving into the future. Paper presented at: Seminars in radiation oncology 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woerner AJ, Choi M, Harkenrider MM, Roeske JC, Surucu MJ Ticr, treatment. Evaluation of deformable image registration-based contour propagation from planning CT to cone-beam CT. 2017;16(6):801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Hoisak JD, Li N, et al. Dosimetric benefit of adaptive re-planning in pancreatic cancer stereotactic body radiotherapy. 2015;40(4):318–324. [DOI] [PubMed] [Google Scholar]
- 26.Ziegler M, Nakamura M, Hirashima H, et al. Accumulation of the delivered treatment dose in volumetric modulated arc therapy with breath-hold for pancreatic cancer patients based on daily cone beam computed tomography images with limited field-of-view. 2019;46(7):2969–2977. [DOI] [PubMed] [Google Scholar]
- 27.Houweling AC, Crama K, Visser J, et al. Comparing the dosimetric impact of interfractional anatomical changes in photon, proton and carbon ion radiotherapy for pancreatic cancer patients. 2017;62(8):3051. [DOI] [PubMed] [Google Scholar]
- 28.Stankiewicz M, Li W, Rosewall T, et al. Patterns of practice of adaptive re-planning for anatomic variances during cone-beam CT guided radiotherapy. 2019;12:50–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakao M, Nakamura M, Mizowaki T, Matsuda TJ MIA. Statistical deformation reconstruction using multi-organ shape features for pancreatic cancer localization. 2020.101829. [DOI] [PubMed] [Google Scholar]
- 30.Wu RY, Liu AY, Williamson TD, et al. Quantifying the accuracy of deformable image registration for cone-beam computed tomography with a physical phantom [published online ahead of print 2019/09/22]. J Appl Clin Med Phys. 2019;20(10):92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang H, Chen X, Liu Y, et al. Clinically applicable deep learning framework for organs at risk delineation in ct images. Nature Machine Intelligence. 2019;1(10):480–491. [Google Scholar]
- 32.van Rooij W, Dahele M, Brandao HR, Delaney AR, Slotman BJ, Verbakel WF. Deep Learning-Based Delineation of Head and Neck Organs at Risk: Geometric and Dosimetric Evaluation. International Journal of Radiation Oncology* Biology* Physics. 2019;104(3):677–684. [DOI] [PubMed] [Google Scholar]
- 33.Zhou X, Takayama R, Wang S, Hara T, Fujita H. Deep learning of the sectional appearances of 3D CT images for anatomical structure segmentation based on an FCN voting method [published online ahead of print 2017/07/22]. Med Phys. 2017;44(10):5221–5233. [DOI] [PubMed] [Google Scholar]
- 34.Weston AD, Korfiatis P, Kline TL, et al. Automated abdominal segmentation of CT scans for body composition analysis using deep learning. Radiology. 2019;290(3):669–679. [DOI] [PubMed] [Google Scholar]
- 35.Gibson E, Giganti F, Hu Y, et al. Automatic Multi-Organ Segmentation on Abdominal CT With Dense V-Networks [published online ahead of print 2018/07/12]. IEEE Trans Med Imaging. 2018;37(8):1822–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang S, He K, Nie D, Zhou S, Gao Y, Shen D. CT male pelvic organ segmentation using fully convolutional networks with boundary sensitive representation. Med Image Anal. 2019;54:168–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong X, Lei Y, Tian S, et al. Synthetic MRI-aided multi-organ segmentation on male pelvic CT using cycle consistent deep attention network. Radiother Oncol. 2019;141:192–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dai X, Lei Y, Zhang Y, et al. Automatic Multi-Catheter Detection using Deeply Supervised Convolutional Neural Network in MRI-guided HDR Prostate Brachytherapy. Med Phys. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao Y, Corn B, Schifter D, Tannenbaum A. Multiscale 3D shape representation and segmentation with applications to hippocampal/caudate extraction from brain MRI [published online ahead of print 2011/11/29]. Med Image Anal. 2012;16(2):374–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fu Y, Lei Y, Wang T, Curran WJ, Liu T, Yang X. A review of deep learning based methods for medical image multi-organ segmentation [published online ahead of print 2021/05/17]. Phys Med. 2021;85:107–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dai X, Lei Y, Wang T, et al. Synthetic MRI-aided Head-and-Neck Organs-at-Risk Auto-Delineation for CBCT-guided Adaptive Radiotherapy. arXiv preprint arXiv:04275. 2020. [Google Scholar]
- 42.Wang YN, Liu CB, Zhang X, Deng WW. Synthetic CT Generation Based on T2 Weighted MRI of Nasopharyngeal Carcinoma (NPC) Using a Deep Convolutional Neural Network (DCNN). Front Oncol. 2019;9:1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dai X, Lei Y, Fu Y, et al. Multimodal MRI Synthesis Using Unified Generative Adversarial Networks. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang T, Lei Y, Fu Y, Curran WJ, Liu T, Yang XJ apa. Medical Imaging Synthesis using Deep Learning and its Clinical Applications: A Review. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu Y, Lei Y, Wang T, et al. Pelvic multi-organ segmentation on cone-beam CT for prostate adaptive radiotherapy. Med Phys. 2020;47(8):3415–3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Janopaul-Naylor J, Lei Y, Liu Y, et al. Synthetic CT-aided Online CBCT Multi-Organ Segmentation for CBCT-guided Adaptive Radiotherapy of Pancreatic Cancer. Int J Radiat Oncol Biol Phys. 2020;108(3):S7–S8. [Google Scholar]
- 47.Lei Y, Wang T, Tian S, et al. Male pelvic multi-organ segmentation aided by CBCT-based synthetic MRI. 2020;65(3):035013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu J-Y, Park T, Isola P, Efros AA. Unpaired image-to-image translation using cycle-consistent adversarial networks. Paper presented at: Proceedings of the IEEE international conference on computer vision 2017. [Google Scholar]
- 49.Goodfellow I, Pouget-Abadie J, Mirza M, et al. Generative adversarial nets. Paper presented at: Advances in neural information processing systems 2014. [Google Scholar]
- 50.He K, Gkioxari G, Dollár P, Girshick R. Mask r-cnn. Paper presented at: Proceedings of the IEEE international conference on computer vision 2017. [Google Scholar]
- 51.Zhang R, Cheng C, Zhao X, Li XJ Mi. Multiscale Mask R-CNN–Based Lung Tumor Detection Using PET Imaging. 2019;18:1536012119863531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chiao J-Y, Chen K-Y, Liao KY-K, Hsieh P-H, Zhang G, Huang T-CJ M. Detection and classification the breast tumors using mask R-CNN on sonograms. 2019;98(19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu M, Dong J, Dong X, Yu H, Qi L. Segmentation of lung nodule in CT images based on mask R-CNN. Paper presented at: 2018 9th International Conference on Awareness Science and Technology (iCAST) 2018. [Google Scholar]
- 54.Huang Z, Huang L, Gong Y, Huang C, Wang X. Mask scoring r-cnn. Paper presented at: Proceedings of the IEEE conference on computer vision and pattern recognition 2019. [Google Scholar]
- 55.Lin T-Y, Dollár P, Girshick R, He K, Hariharan B, Belongie S. Feature pyramid networks for object detection. Paper presented at: Proceedings of the IEEE conference on computer vision and pattern recognition 2017. [Google Scholar]
- 56.Ren S, He K, Girshick R, Sun J. Faster r-cnn: Towards real-time object detection with region proposal networks. Paper presented at: Advances in neural information processing systems 2015. [DOI] [PubMed] [Google Scholar]
- 57.Lei Y, Tian S, He X, et al. Ultrasound prostate segmentation based on multidirectional deeply supervised V-Net [published online ahead of print 2019/05/11]. Med Phys. 2019;46(7):3194–3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harms J, Lei Y, Wang T, et al. Paired cycle-GAN-based image correction for quantitative cone-beam computed tomography. Med Phys. 2019;46(9):3998–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O. 3D U-Net: learning dense volumetric segmentation from sparse annotation. Paper presented at: International conference on medical image computing and computer-assisted intervention 2016. [Google Scholar]
- 60.Dai X, Lei Y, Wang T, et al. Head-and-neck organs-at-risk auto-delineation using dual pyramid networks for CBCT-guided adaptive radiotherapy. Physics in Medicine Biology. 2021;66(4):045021. [DOI] [PubMed] [Google Scholar]
- 61.Olberg S, Green O, Cai B, et al. Optimization of treatment planning workflow and tumor coverage during daily adaptive magnetic resonance image guided radiation therapy (MR-IGRT) of pancreatic cancer. 2018;13(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Loi M, Magallon-Baro A, Suker M, et al. Pancreatic cancer treated with SBRT: Effect of anatomical interfraction variations on dose to organs at risk. 2019;134:67–73. [DOI] [PubMed] [Google Scholar]
- 63.Liu Y, Lei Y, Wang T, et al. CBCT-based synthetic CT generation using deep-attention cycleGAN for pancreatic adaptive radiotherapy. Med Phys. 2020;47(6):2472–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.