Abstract
Maternal anxiety during pregnancy is associated with adverse fetal, neonatal, and child outcomes, but biological mechanisms remain unclear. Altered fetal DNA methylation (DNAm) has been proposed as a potential underlying mechanism. In the current study, we performed a meta-analysis to examine the associations between maternal anxiety, measured prospectively during pregnancy, and genome-wide DNAm from umbilical cord blood. Sixteen non-overlapping cohorts from 12 independent longitudinal studies of the Pregnancy And Childhood Epigenetics Consortium participated, resulting in a combined dataset of 7 243 mother-child dyads. We examined prenatal anxiety in relation to genome-wide DNAm and differentially methylated regions. We observed no association between the general symptoms of anxiety during pregnancy or pregnancy-related anxiety, and DNAm at any of the CpG sites, after multiple-testing correction. Further, we identify no differentially methylated regions associated with maternal anxiety. At the cohort-level, of the 21 associations observed in individual cohorts, none replicated consistently in the other cohorts. In conclusion, contrary to some previous studies proposing cord blood DNAm as a promising potential mechanism explaining the link between maternal anxiety during pregnancy and adverse outcomes in offspring, we found no consistent evidence for any robust associations between maternal anxiety and DNAm in cord blood. Larger studies and analysis of DNAm in other tissues may be needed to establish subtle or subgroup-specific associations between maternal anxiety and the fetal epigenome.
INTRODUCTION
Anxiety symptoms during pregnancy are common, and an estimated 15% of pregnant women fulfill the diagnostic criteria of any anxiety disorder.1 Perinatal anxiety may not only affect the mother, but could also have long-lasting implications for the offspring. Children of mothers with high levels of prenatal anxiety are more likely to be preterm and small-for-gestational-age, and to have emotional and behavioral problems that may persist beyond childhood.2–4
The biological pathways underlying maternal mental health during pregnancy and offspring development are poorly understood. One proposed pathway is fetal programming through epigenetic modifications: prenatal stressors may alter fetal DNA methylation (DNAm), which may affect downstream gene expression, neuroendocrine functioning, and behavioral development.5 Several animal studies have supported this hypothesis.5 However, human evidence of associations between maternal anxiety during pregnancy and offspring DNAm is mixed and mostly based on small samples.6 A recent epigenome-wide association study (EWAS) found that high levels of prenatal anxiety predicted differential DNAm at 13 CpG sites in cord blood (N=445), but these findings did not replicate in another sample (N=969).14
We present a meta-analysis of epigenome-wide associations between prenatal general- and pregnancy-related anxiety and cord blood DNAm, from 12 independent longitudinal studies (total N=7,243).
MATERIALS AND METHODS
Participating studies
Sixteen cohorts from 12 independent longitudinal studies, in the Pregnancy And Childhood Epigenetics Consortium (PACE),15 participated in this meta-analysis. These were the 1) Avon Longitudinal Study of Parents and Children (ALSPAC) (United Kingdom),16 2) Born in Bradford (BiB) (United Kingdom),17 subdivided into South Asian (BiB-1) and European (BiB-2) cohorts, 3) Drakenstein Child Health Study (DCHS) (South Africa),18 comprising the DCHS-450k and DCHS-EPIC cohorts, differentiated by the choice of DNAm array (Illumina 450k and EPIC, respectively), 4) EDEN Mother-Child Cohort (EDEN) (France),19 5) Generation R Study (Generation R) (Netherlands),20 6) Genome-Wide Population-based Association Study of Extremely Overweight Young Adults (GOYA) (Denmark; weight distribution of the subsample used for these analyses corresponds to the general population),21 7) INfancia y Medio Ambiente (INMA) (Spain),22 8) Norwegian Mother, Father and Child Cohort Study (MoBa) (Norway),23 comprising the MoBa-1 and the MoBa-2 cohorts, two independent samples from the MoBa study, 9) Prediction and Prevention of Preeclampsia and Intrauterine Growth Restriction study (PREDO) (Finland),24 comprising the PREDO-450k and the PREDO-EPIC cohort, differentiated by Illumina 450k vs EPIC DNAm array, respectively, 10) Programming Research in Obesity, Growth, Environment and Social Stressors study (PROGRESS) (Mexico),25 11) PRogramming of Intergenerational Stress Mechanisms study (PRISM) (USA),26 and 12) Project Viva (Viva) (USA).27
See Supplementary methods and PACE profile15 for details.
Measures
Exposure: Maternal anxiety during pregnancy
Anxiety can be assessed using general self-report instruments or instruments specifically designed for pregnant women, which may be more predictive of perinatal outcomes and include pregnancy-related items such as childbirth-related worries.28 We examined the two separately: we refer to anxiety symptoms measured using general instruments as general anxiety (not to be confused with generalized anxiety disorder), and pregnancy-related anxiety symptoms measured using pregnancy-specific instruments as pregnancy-related anxiety. In one cohort (INMA), mothers reported, during prenatal interviews, if a physician had diagnosed them with any anxiety disorder: we included INMA in the primary meta-analysis of general anxiety (Table 1). Instruments are described in Supplementary methods.
Table 1.
Measurement of maternal anxiety across the participating studies
| ALSPAC | BiB-1 | BiB-2 | DCHS −450k | DCHS-EPIC | EDEN | Generation R | GOYA | INMA | MoBa-1 | MoBa-2 | PREDO-450k | PREDO-EPIC | PRISM | PROGRESS | Viva | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total number of mother-child dyads | 659 | 384 | 375 | 104 | 149 | 151 | 1292 | 488 | 336 | 1049 | 636 | 581 | 120 | 155 | 313 | 451 |
| Maternal general anxiety during pregnancy | ||||||||||||||||
| Measure name | CCEI | GHQ-28 | GHQ-28 | SRQ-20 | SRQ-20 | STAI-ST | BSI | SCL-92 | Diagnosis | SCL-8 | SCL-8 | STAI-S | STAI-S | STAI-T | STAI-T | none |
| Number of mother-child dyads | 659 | 384 | 375 | 104 | 149 | 151 | 1186 | 488 | 336 | 1049 | 636 | 581 | 120 | 155 | 313 | 0 |
| Number of assessments during pregnancy* | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 14 | 14 | 1 | 1 | - |
| Timing of assessment, gestational weeks | 18–32 | 26–28 | 26–28 | 28–32 | 28–32 | 20–24 | 18–25 | ~30 | 12 | 30 | 30 | 12–38 | 12–38 | 7–39 | 18–35 | - |
| Mean (standard deviation) | 4.7 (3.2) | 7.0 (4.7) | 6.7 (4.4) | 5.7 (4.1) | 5.3 (3.9) | 9.4 (8.0) | 0.2 (0.3) | 1.0 (1.2) | - | 4.8 (1.4) | 4.8 (1.4) | 33.9 (7.8) | 34.4 (8.3) | 16.6 (4.8) | 18.9 (5.1) | - |
| Range | 0 to 16 | 0 to 20 | 0 to 20 | 0 to 18 | 0 to 17 | 0 to 34 | 0 to 1.1 | 0 to 6 | - | 4 to 16 | 4 to 16 | 20 to 58 | 20 to 60 | 10 to 32 | 10 to 33 | - |
| High general anxiety, N (%) | 88 (13%) | 59 (15%) | 41 (11%) | 26 (25%) | 28 (19%) | 23 (15%) | 170 (14%) | 147 (30%) | 73 (22%) | 198 (19%) | 61 (10%) | 85 (15%) | 20 (17%) | 25 (16%) | 50 (16%) | - |
| Maternal pregnancy-related anxiety during pregnancy | ||||||||||||||||
| Measure name | none | none | none | none | none | none | POQ | none | none | none | none | none | none | PAS-1 | PAS-2 | PRAS |
| Number of mother-child dyads | 0 | 0 | 0 | 0 | 0 | 0 | 1197 | 0 | 0 | 0 | 0 | 0 | 0 | 128 | 313 | 451 |
| Number of assessments during pregnancy* | - | - | - | - | - | - | 1 | - | - | - | - | - | - | 1 | 1 | 1 |
| Timing of assessment, gestational weeks | - | - | - | - | - | - | <18 | - | - | - | - | - | - | 7–39 | 7–39 | 10 |
| Mean (standard deviation) | - | - | - | - | - | - | 0.7 (0.3) | - | - | - | - | - | - | 5.1 (4.2) | 18.8 (5.5) | 0.7 (1.3) |
| Range | - | - | - | - | - | - | 0 to 1.7 | - | - | - | - | - | - | 0 to 21 | 7 to 28 | 0 to 7 |
| High pregnancy-related anxiety, N (%) | - | - | - | - | - | - | 208 (17%) | - | - | - | - | - | - | 14 (11%) | 47 (15%) | 85 (19%) |
Abbreviations:
BSI: Brief Symptom Inventory, 6-item anxiety subscale;
CCEI: Crown-Crisp Experiental Index, 8-item anxiety subscale;
GHQ-28: General Health Questionnaire (28-item version), 7-item anxiety subscale;
N: number of mothers in the high anxiety group;
PAS-1: Pregnancy Anxiety Scale, 7-item abbreviated version, each item scored from 0 to 3;
PAS-2: Pregnancy Anxiety Scale, 7 item abbreviated version, each item scored from 1 to 4;
POQ: Pregnancy Outcome Questionnaire, 13-item version;
PRAS: Pregnancy-Related Anxiety Scale, 7-item version;
SCL-8: Symptom Checklist (8-item version), 5-item anxiety subscale;
SCL-92: Symptom Checklist (92-item version), 3-item anxiety subscale;
SRQ-20: Self-Report Questionnaire, 20-item anxiety and mood scale;
STAI-ST: State-Trait Anxiety Inventory, 40-item version including the 20 Trait and 20 State Anxiety items, each item scored from 0–3;
STAI-S: State-Trait Anxiety Inventory, 20-item State-Anxiety subscale, each item scored from 1–4;
STAI-T: State-Trait Anxiety Inventory, 10-item abbreviated version of the Trait-Anxiety subscale, each item scored from 1–4
Number of assessments refers to the number of times each mother filled out the anxiety questionnaire (or was interviewed) during pregnancy
Each cohort dichotomized questionnaire scores, contrasting the highest-scoring 15% of mothers (“high-anxiety” group), against other mothers.1,4 Scores were neither transformed nor standardized. In sensitivity analyses, we used continuous questionnaire scores (winsorized at +/− 3SD): higher scores reflect more symptoms. Cohorts with repeated measures of anxiety during pregnancy calculated mean scores (Table 1).
Outcome: DNAm measurements
Newborn umbilical cord blood DNAm was assessed with the Illumina® HumanMethylation450 (450k) or HumanMethylationEPIC (EPIC) BeadChip assay at Illumina or cohort-specific laboratories. Cohorts performed sample processing, quality control and normalization as described in Supplementary methods.
We used normalized, untransformed beta values, ranging from 0 (completely unmethylated) to 1 (completely methylated), after trimming extreme outliers (3×interquartile range from the quartile limit).
We excluded probes mapped to X/Y chromosomes, polymorphic CpGs (overlapping with known single-nucleotide-polymorphisms [SNPs]),29 control or cross-reactive probes (targeting repetitive sequences/co-hybridizing to alternate sequences).30,31
Function-related information was derived from GeneCards and GWAS Catalog (https://www.genecards.org; https://www.ebi.ac.uk/gwas, accessed August 17th, 2020).
Covariates
Based on theoretical expectations,3 we adjusted for child sex, maternal age, socioeconomic status (SES, maternal education/family income), cell counts, ethnicity/ancestry (if applicable) and technical covariates (e.g. batch) (Supplementary methods). We used reference-based projection/quadratic programming to estimate cell type proportions:32,33 EDEN used the Houseman reference panel,32 PRISM a combined panel,33,34 and others the Bakulski panel.33
In a sensitivity analysis, we added the maternal smoking covariate. This was a separate model because causal associations between smoking and anxiety could be bidirectional and smoking could mediate some effects on DNAm (Supplementary methods).
Statistical analyses
Cohort-specific EWAS
Cohort-level EWASes were performed according to a predefined analysis plan. We used robust multiple linear regression (rlm; MASS R-package) to control for potential heteroscedasticity and influential DNAm outliers. We included participants with complete information. Cohorts excluded known chromosomal abnormalities, multiple births, and one random sibling per sibling pair. Models are described in Supplementary methods.
Primary meta-analysis
We examined associations between maternal general anxiety during pregnancy and DNAm at 364,659 CpG sites across 15 cohorts (ALSPAC, BiB-1, BiB-2, DCHS-450k, DCHS-EPIC, EDEN, Generation R, GOYA, INMA, MoBa-1, MoBa-2, PREDO-450k, PREDO-EPIC, PRISM, PROGRESS). We combined 450k and EPIC data and only included sites that are available on the 450k. We performed a sample-size-weighted meta-analysis using R 3.6.1 (https://www.r-project.org/) and METAL (version released 2018–08-28),35 choosing the sample-size-weighted approach because of variation across anxiety scales in different cohorts. Probes were annotated using meffil (hg19/b37).29
To assess epigenome-wide statistical inflation, we calculated cohort- and meta-analysis-level genomic inflation factor lambdas (λ) and examined quantile-quantile plots.
Cohort-level results were meta-analyzed at Erasmus MC; a shadow meta-analysis was conducted independently at the University of Helsinki.
Secondary meta-analyses
We performed two secondary meta-analyses. First, we examined associations between pregnancy-related anxiety and DNAm (Generation R, PRISM, PROGRESS, Viva); second, associations between general anxiety and DNAm at the 439,294 EPIC-assay-specific CpGs (BiB-1, BiB-2, DCHS-EPIC, PROGRESS).
Multiple-testing correction
All reported p-values are two-sided. For multiple-testing correction, we used Bonferroni-correction based on the number of probes (primary meta-analysis cut-off p<1.37×10−7). Additionally, we examined our results using the Benjamini Hochberg False-Discovery-Rate method (FDR).36 When we observed no statistically significant associations, we used a suggestive p-value threshold (p<5×10−5) to select CpGs for follow-up analyses.
Sensitivity analyses
We repeated the primary and secondary meta-analyses twice: first, adding smoking into covariates, and second, modelling anxiety as continuous. To address potential across-study heterogeneity, we examined I2 statistics, and repeated the primary meta-analysis only including cohorts with 1) 450k or 2) EPIC array, 3) predominantly European-ancestry cohorts (ALSPAC, BiB-2, EDEN, Generation R, GOYA, INMA, MoBa, PREDO), and 4) cohorts using the Bakulski panel for cell type correction.33
We identified the 30 probes with the largest effect estimates for general anxiety in the three largest cohorts (Generation R, MoBa-1, ALSPAC; 10 probes/cohort), and examined whether effects were consistent across cohorts. This was repeated with pregnancy-related anxiety (largest cohorts: Generation R, Viva, PROGRESS).
DMR analysis
To identify differentially methylated regions (DMRs), we used dmrff:37 it addresses the correlation structure of DNAm and can be used to identify DMRs at the meta-analytic level. Ten cohorts (ALSPAC, EDEN, GOYA, Generation R, INMA, MoBa-1, PREDO-450k, PREDO-EPIC, PRISM, PROGRESS) calculated correlations between each CpG and the next 20 CpGs downstream. We used data on CpGs covered by 450k. We identified candidate regions: sets of nearby CpG sites (≤500bp between consecutive sites) that associated with anxiety (nominal p-values<0.05 in cohort-level EWAS, effect estimates in the same direction), and, using an extension of inverse-variance weighted meta-analysis, calculated the dmrff statistics. We then meta-analyzed those statistics by selecting sub-regions with the most extreme meta-analysis statistic. P-values for candidate regions were Bonferroni-corrected.
Blood-brain comparisons
To examine correlation between blood and brain DNAm levels at CpG sites with meta-analytical p<5×10–5, we used two tools. The first by Hannon et al38 reports correlations between DNAm in blood vs prefrontal cortex, entorhinal cortex, superior temporal gyrus, and cerebellum. Secondly, BECon shows correlations between DNAm in blood vs parietal, anterior prefrontal, and ventral temporal cortex (Brodmann 7, 10, 20, respectively).39
Code and data availability and ethical statements
All studies acquired approval from local ethics committees and informed consent from participants. Site-level meta-analytical results are publicly available (https://doi.org/10.5281/zenodo.4147817). For access to cohort-level data, requests can be sent directly to individual studies. Analytical codes can be requested from authors.
RESULTS
Study characteristics
Table 2 describes our 7,243 mother-child dyads. Supplementary Table 1 shows characteristics stratified by low-vs-high anxiety.
Table 2.
Characteristics across participating studies
| ALSPAC, n (%) | BiB-1, n (%) | BiB-2, n (%) | DCHS −450k, n (%) | DCHS-EPIC, n (%) | EDEN, n (%) | Generation R, n (%) | GOYA, n (%) | INMA, n (%) | MoBa-1, n (%) | MoBa-2, n (%) | PREDO-450k, n (%) | PREDO-EPIC, n (%) | PRISM, n (%) | PROGRESS, n (%) | Viva, n (%) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of mother-child dyads | 659 | 384 | 375 | 104 | 149 | 151 | 1186 | 488 | 336 | 1049 | 636 | 581 | 120 | 155 | 313 | 451 |
| Methylation array | 450k | EPIC | EPIC | 450k | EPIC | 450k | 450k | 450k | 450k | 450k | 450k | 450k | EPIC | 450k | EPIC | 450k |
| Child sex, female | 343 (52%) | 178 (46%) | 181 (48%) | 43 (41%) | 67 (45%) | 61 (40%) | 597 (50%) | 238 (49%) | 164 (49%) | 489 (47%) | 283 (44%) | 299 (51%) | 60 (50%) | 71 (46%) | 144 (46%) | 215 (48%) |
| Maternal characteristics | ||||||||||||||||
| Age at delivery, years, mean (SD) | 29.9 (4.4) | 28.4 (5.2) | 27.2 (6.2) | 26.7 (5.7) | 27.6 (5.8) | 30.4 (4.9) | 32.2 (4.1) | 29.9 (4.0) | 30.3 (4.0) | 29.9 (4.3) | 30.0 (4.4) | 33.5 (5.5) | 32.3 (4.8) | 31.2 (5.5) | 28.1 (5.4) | 32.7 (5.2) |
| Tobacco exposure during pregancy | ||||||||||||||||
| no smoking | 568 (86%) | 371 (97%) | 246 (66%) | 77 (74%) | 125 (84%) | 121 (80%) | 923 (78%) | 383 (78%) | 237 (71%) | 741 (71%) | 475 (75%) | 556 (96%) | 111 (93%) | 142 (92%) | 222 (71%)* | 397 (88%) |
| smoking during pregnancy | 91 (14%) | 13 (3.4%) | 129 (34%) | 27 (26%) | 24 (16%) | 30 (20%) | 263 (22%) | 105 (22%) | 99 (29%) | 308 (29%) | 161 (25%) | 25 (4%) | 9 (8%) | 13 (8%) | 91 (29%)* | 54 (12%) |
| quit smoking in early pregnancy | 21 (3%) | n/a | n/a | n/a | n/a | 12 (8%) | 107 (9%) | 39 (8%) | n/a | 156 (15%) | 97 (15%) | 22 (4%) | 5 (4%) | n/a | n/a | n/a |
| continued smoking in pregnancy | 70 (11%) | n/a | n/a | n/a | n/a | 18 (12%) | 156 (13%) | 66 (14%) | n/a | 152 (14%) | 64 (10%) | 3 (1%) | 4 (3%) | n/a | n/a | n/a |
| Socioeconomic status ** | ||||||||||||||||
| Household income | ||||||||||||||||
| high, n (%) | n/a | n/a | n/a | 19 (18%) | 22 (15%) | 31 (21%) | n/a | n/a | 73 (22%) | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| medium-level, n (%) | n/a | n/a | n/a | 40 (38%) | 67 (45%) | 104 (69%) | n/a | n/a | 104 (31%) | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| low, n (%) | n/a | n/a | n/a | 45 (43%) | 60 (40%) | 16 (11%) | n/a | n/a | 159 (47%) | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| Maternal education | ||||||||||||||||
| upper tertiary, n (%) | 345 (52%) | 161 (42%) | 152 (41%) | n/a | n/a | n/a | 467 (39%) | 279 (57%) | n/a | 467 (45%) | 258 (41%) | 194 (33%) | 45 (38%) | 125 (81%) | 70 (22%) | 299 (66%) |
| lower tertiary, n (%) | n/a | n/a | n/a | 321 (27%) | n/a | 336 (32%) | 212 (33%) | 138 (24%) | 29 (24%) | |||||||
| secondary, n (%) | 263 (40%) | 223 (58%) | 223 (59%) | n/a | n/a | n/a | 380 (32%) | 167 (34%) | n/a | 77 (7%) | 46 (7%) | 231 (40%) | 40 (33%) | 30 (19%) | 110 (35%) | 152 (34%) |
| primary, n (%) | 51 (8%) | n/a | n/a | n/a | 18 (2%) | 42 (9%) | n/a | 169 (16%) | 120 (19%) | 18 (3%) | 6 (5%) | 133 (42%) | ||||
| Psychiatric medication, n (%) *** | 21 (3%) | n/a | n/a | n/a | n/a | 6 (4%) | 26 (2%) | n/a | 11 (3%) | 11 (1%) | 8 (1%) | 16 (3%) | 5 (4%) | 11 (7%) | n/a | 14 (3%) |
describes exposure to cigarette smoke at home during pregnancy instead of maternal smoking, see Supplementary Methods for details
different categorizations were used as deemed suitable per cohort: please see Supplementary Methods for details
includes medications during pregnancy such as antidepressants and anxiolytes: please see Supplementary Methods for details
General anxiety during pregnancy and DNAm
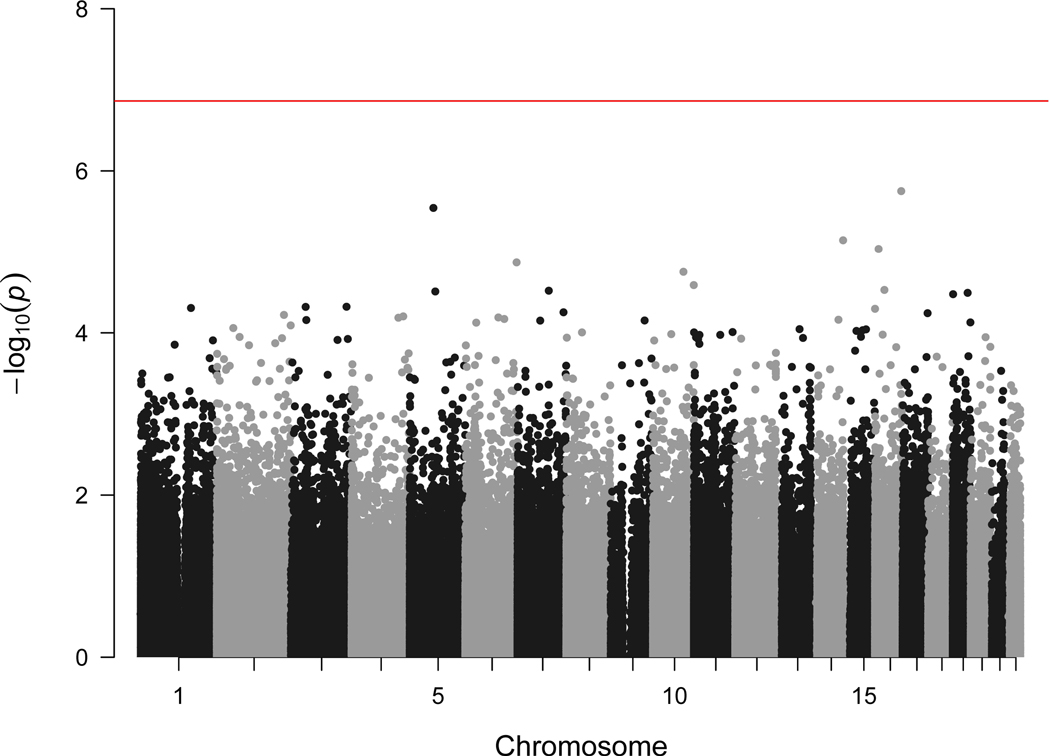
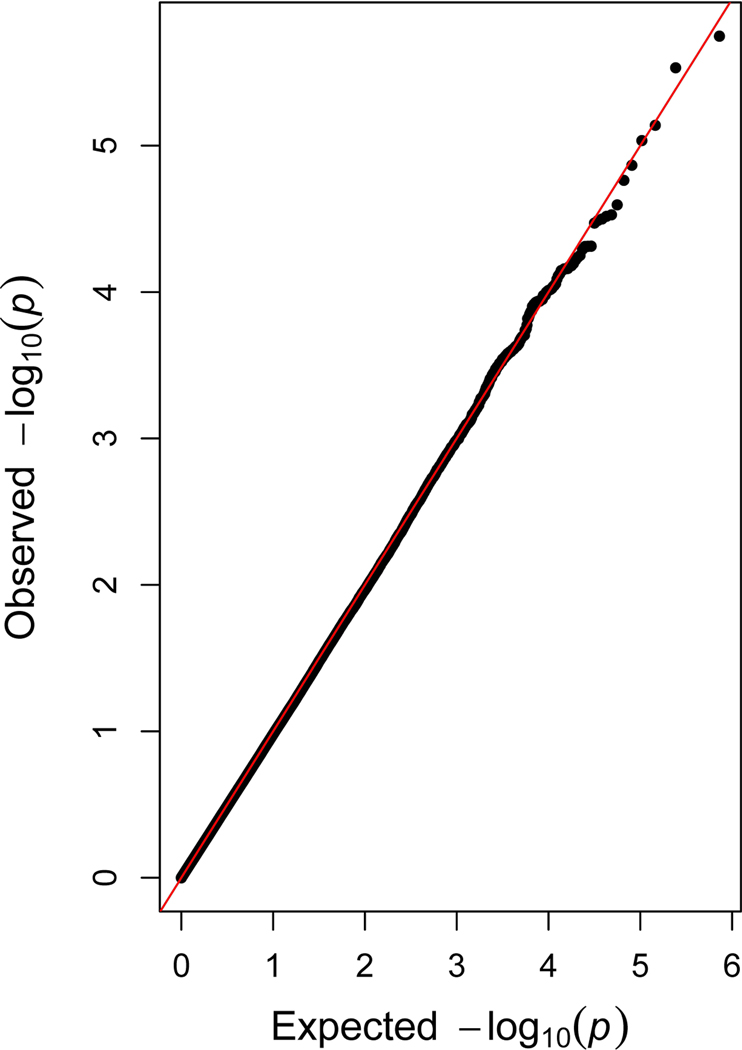
We observed no associations between maternal general anxiety during pregnancy and DNAm of 364,659 CpG sites in cord blood, among 6,686 mother-child dyads, when using Bonferroni correction (p<1.37×10−7) (Figure 1A). We found no evidence of genomic inflation (λ=0.96, quantile-quantile plot in Figure 1B).
Figure 1.
Manhattan and quantile-quantile plot showing meta-analytic associations between general anxiety during pregnancy and cord blood DNA methylation (within 15 cohorts, maximum N=6 686 mother-child dyads)
Panel A. Manhattan plot (red line indicates the Bonferroni threshold)
Panel B. Quantile-quantile plot (λ=0.962)
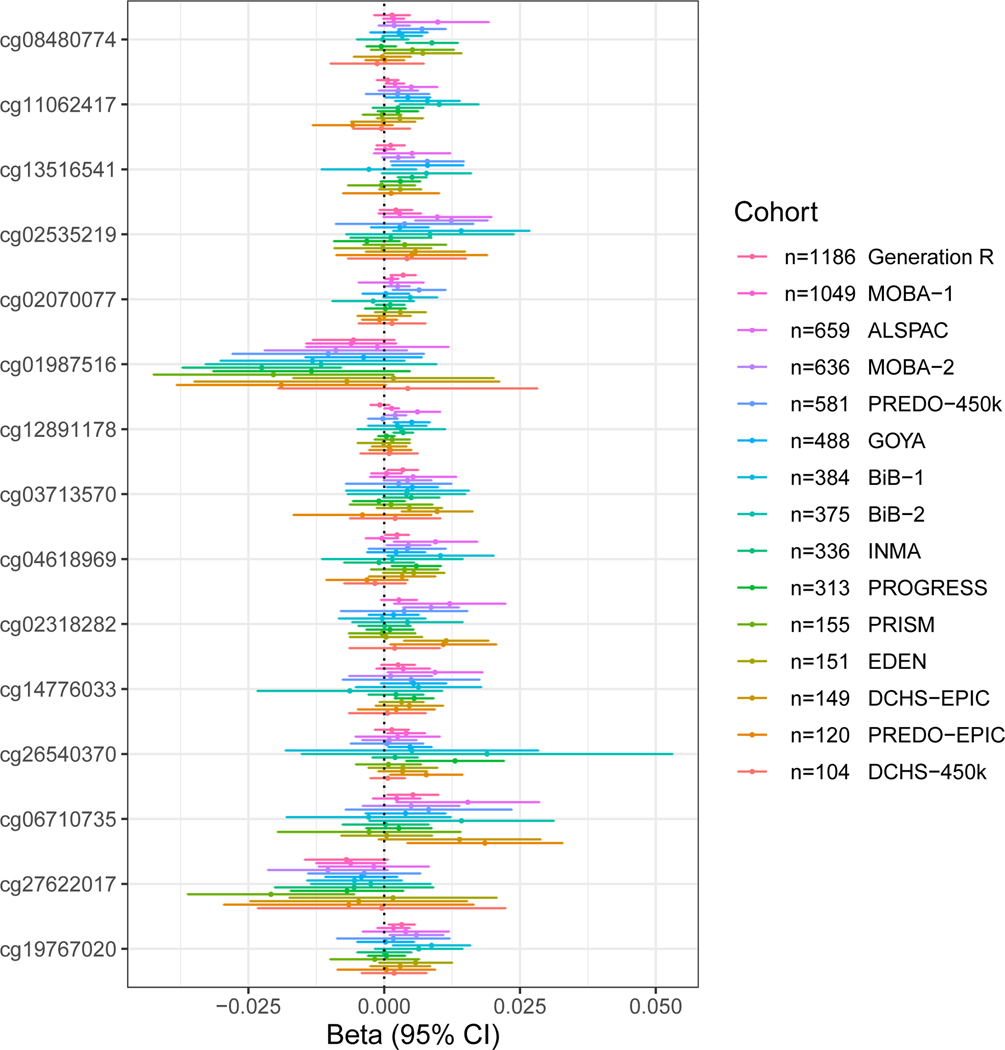
In Table 3, we list the 15 CpG sites that were most strongly associated (p<5×10−5) with maternal general anxiety during pregnancy. Figure 2 shows a regression coefficient forest-plot at these 15 CpG sites across cohorts: effect direction was not consistent across cohorts for any of these sites.
Table 3.
General maternal anxiety during pregnancy and cord blood DNAm: associations with p-value below 5e-5.
| CpG site | Chr | Coordinate | P-value | Direction | N of Cohorts | N of Individuals | I2 | Heterogeneity p-value | Illumina annotated gene |
|---|---|---|---|---|---|---|---|---|---|
| cg08480774 | 16 | 85676306 | 1.79E-06 | ++−−−+++++++++− | 15 | 6653 | 23.7 | 0.19 | KIAA0182 |
| cg11062417 | 5 | 76578232 | 2.94E-06 | +++−−+++++++−−+ | 15 | 6676 | 6.1 | 0.38 | PDE8B |
| cg13516541 | 14 | 105239354 | 7.25E-06 | +−+??++++++++−+ | 13 | 6395 | 34.8 | 0.10 | AKT1 |
| cg02535219 | 16 | 11876908 | 9.23E-06 | +++++−++++++++− | 15 | 6675 | 0 | 0.55 | ZC3H7A |
| cg02070077 | 6 | 168199753 | 1.36E-05 | ++−+−+++++++−?+ | 14 | 6501 | 0 | 0.61 | - |
| cg01987516 | 10 | 101282726 | 1.73E-05 | −−−+−+−−−−−−−−− | 15 | 6669 | 0 | 0.68 | - |
| cg12891178 | 10 | 135085127 | 2.54E-05 | +++++−−++++−+++ | 15 | 6596 | 37.5 | 0.07 | ADAM8 |
| cg03713570 | 16 | 30789484 | 2.97E-05 | ++++++++++++−+− | 15 | 6662 | 0 | 0.45 | - |
| cg04618969 | 7 | 101848538 | 3.04E-05 | +++−++++−−++−++ | 15 | 6675 | 15.9 | 0.28 | CUX1 |
| cg02318282 | 5 | 82789386 | 3.17E-05 | +−++++?++++++−+ | 14 | 5481 | 18.1 | 0.26 | VCAN |
| cg14776033 | 19 | 49240808 | 3.24E-05 | ++−++++++++++?+ | 14 | 6515 | 0 | 0.60 | RASIP1 |
| cg26540370 | 19 | 1374742 | 3.38E-05 | +++++++++++++++ | 15 | 6668 | 0 | 0.53 | MUM1 |
| cg06710735 | 3 | 48264567 | 4.86E-05 | +−+?+++++++++−+ | 14 | 6577 | 0 | 0.60 | CAMP |
| cg27622017 | 3 | 181447418 | 4.87E-05 | −−−−−+−−−−−−−−− | 15 | 6679 | 0 | 0.93 | SOX2OT |
| cg19767020 | 1 | 164582668 | 4.88E-05 | +++++++++++++−+ | 15 | 6655 | 0 | 0.77 | PBX1 |
Abbreviations: Chr: chromosome; Direction: direction of effect per cohort, in which + signifies a positive effect estimate, - signifies a negative effect estimate, and ? refers to a lack of effect estimate in the particular study due to lack of data on the methylation of the specific site (cohorts are listed in alphabetical order: ALSPAC, BiB-1, BiB-2, DCHS-450k, DCHS-EPIC, EDEN, Generation R, GOYA, MoBa-1, MoBa-2, PREDO-450k, PREDO-EPIC, PRISM, PROGRESS); I2: this heterogeneity statistic describes the variation attributable to heterogeneity across studies (high I2 values suggest high heterogeneity); N of cohorts: number of cohorts included in analysis; N of Individuals: number of mother-child dyads included in the analysis; p-value: meta-analytic p-value for association between DNA methylation at the CpG site in question and anxiety)
Figure 2.
Forest plot showing associations between general anxiety during pregnancy and cord blood DNA methylation for the most significant associations (p<5×10−5), across all cohorts with available data, in order of sample size.
Secondary meta-analyses
Pregnancy-related anxiety
We found no association between pregnancy-related anxiety and DNAm among the 2,089 mother-child dyads in four cohorts with available data (Figure SF1, λ=0.92). Figure SF2 shows the 8 CpG sites that were most strongly associated (p<5×10−5) with pregnancy-related anxiety and the variation in estimates across cohorts. At two of these sites (cg18769357, cg03985478), all four cohorts reported the association in the same direction: for cg18769357, these associations were only nominally significant (p<0.05) in Generation R and PRISM; for cg03985478, they were only nominally significant in Generation R. Only one site, cg02624770, had a nominally significant association with pregnancy-related anxiety across the three largest cohorts (N>200): the direction was negative suggesting hypomethylation. None of the sites most strongly associated (p<5×10−5) with pregnancy-related anxiety overlapped with the sites most strongly associated (p<5×10−5) with general anxiety (Supplementary Table 2).
EPIC-only sites
We found no association between general anxiety during pregnancy and DNAm at the 439,294 CpG sites that are included only on the EPIC array among 1,221 mother-child dyads from four cohorts (Figure SF3, λ=1.02). Figure SF4 shows all 40 EPIC-only sites that were most strongly (p<5×10−5) associated with general anxiety. We identified 26 sites where general anxiety during pregnancy associated with hypermethylation across all four cohorts, and 3 sites where it associated with hypomethylation. (Figure SF4). Five sites (cg02306798, cg20639127, cg20817245, cg21202789, cg22566694) had a nominally significant (p<0.05) association in the same direction (all positive) with general anxiety across the three largest cohorts (N>200) (Figure SF4).
FDR
We additionally applied the FDR instead of Bonferroni method to correct for multiple testing. We identified no associations passing this more lenient threshold in the primary, nor the secondary meta-analyses.
Sensitivity analyses
We examined results after cohorts additionally adjusted for maternal smoking during pregnancy (all cohorts), or when treating the exposure as continuous (available in 14 cohorts, maximum N=6,350) and identified no associations between general or pregnancy-related anxiety and DNAm at any of the 450k or EPIC-assay sites (corrected for multiple testing).
We repeated the primary meta-analysis in cohorts with 1) 450k (maximum N=5,345), 2) EPIC (maximum N=1,341), 3) predominantly European participants (maximum N=5,581), and 4) Bakulski panel33 (N=6,380). Again, no associations were observed.
Figure SF5 illustrates associations between DNAm and general (panel A) and pregnancy-related (B) anxiety, for 30 probes with the largest effect estimates in the largest cohorts. We found no consistent associations across cohorts at these sites.
DMR analysis
We identified no DMRs associated with general/pregnancy-related anxiety (after Bonferroni correction) (Supplementary Table 3).
Blood-brain comparisons
Among CpGs closest to statistical significance, none showed consistent, wide-spread correlations between blood and brain DNAm.
Figure SF6 shows that two of the 15 sites most strongly associated (p<5×10–5) with general anxiety (cg13516541, cg01987516) exhibited a relatively concordant DNAm pattern (r>0.41, p<0.001) across blood vs the prefrontal cortex and superior temporal gyrus.38 Two of the 8 sites most strongly associated with pregnancy-related anxiety (cg01093369, cg03985478) showed high correlation between blood vs brain DNAm (r>0.44, p<0.001).38
Figure SF7 shows that among the sites most strongly related to general anxiety, one (cg14776033) showed moderate negative correlations between blood and the brain regions using BECon (r=−0.51…−0.31).39 One site among those most strongly associated with pregnancy-related anxiety (cg14200609) showed moderate negative correlations between blood and all brain regions (r=−0.53…−0.26).
Sites of interest: look-up
Supplementary Table 4 shows meta-analytical results for CpGs associated with maternal anxiety/depression in a previous EWAS,40 and CpGs annotated to genes of interest for perinatal mental health epigenetics (NR3C1, NR3C2, FKBP5, HSD11B2, OXTR).e.g. 6,13,41 None were consistently associated with maternal anxiety in our meta-analysis (non-corrected p-values>0.001, inconsistent direction of effects).
Cohort-level results
Supplementary Table 5 shows lambdas and Bonferroni-corrected cohort-level hits. At genome-wide level, we observed 12 significant cohort-level associations between maternal general anxiety and DNAm. Five of these sites map to genes associated with neurodevelopmental/mental health-related phenotypes (cg06113534/DSCC1, cg06861562/C5orf43, cg15475427/BMP8A, cg19614454/REPS1, cg12670525/PARP6), however none of the 12 cohort-level hits overlapped across two or more cohorts, nor was the effect direction consistent across all cohorts for any of these sites.
We found 9 cohort-level associations between pregnancy-related anxiety and DNAm (Supplementary Table 5). At only one of these sites, cg19660243, effect estimates were in the same direction (positive) in all cohorts. Two sites mapped to genes previously linked to neurodevelopmental/mental health phenotypes (cg11920519/MAP1LC3A, cg15401317/NARS2), but showed no consistent pattern of findings across cohorts.
We advise a very cautious interpretation of these 21 cohort-level associations. They are likely false positives, given the lack of consistency across cohorts.
DISCUSSION
We found no robust evidence of associations between cord blood DNAm and either general symptoms of anxiety during pregnancy or pregnancy-related anxiety across 16 non-overlapping cohorts within 12 independent longitudinal studies. These results were consistently supported by a comprehensive set of additional analyses.
We dichotomized exposures to address different questionnaire score distributions, harmonize questionnaire-based with diagnosis-based measures, and define a subgroup of interest across a continuum of symptom severity. The 15% cut-off was validated against diagnostic interviews in our largest cohort,4 and is in line with estimated global prevalence of prenatal anxiety disorders (~15%),1 and previous studies in non-clinical populations.42,43 Nonetheless, by modelling anxiety with a dichotomous approach, we inevitably lose information and power. Thus, we also examined anxiety symptoms as continuous scores (excluding INMA, for which only a binary diagnosis was available). Consistent with our main results, these analyses revealed no associations between anxiety and DNAm.
DNAm at nearby sites is correlated, and there is disagreement over the optimal method of genome-wide correction. We confirmed our findings using FDR and Bonferroni methods. No meta-analytical associations would survive correction for multiple testing even if we used a more lenient 450k-array P-value cut-off (2.4×10–7).44 Removal of invariable probes could reduce the number of tests. However, invariable probes differ per tissue and we are not aware of a cord-blood-specific reference list. Our results would not change if we filtered out across-tissue invariable probes (n=42,315).45 We show the meta-analytic results for sites closest to the Bonferroni threshold, to facilitate evaluating across-cohort consistency. To further address non-independence between CpGs, we searched for DMRs and found none. Finally, we uploaded the meta-analytical results for all probes in a public repository, to enable others to examine specific sites or apply future filtering methods.
We combined data from different countries and different arrays. Overall DNAm patterns are, however, highly correlated between the 450k and EPIC arrays.46 Sensitivity analyses did not suggest that array differences would represent a source of substantial bias. Similarly, we ran sensitivity analyses including only the predominantly European cohorts to address potential issues arising from ethnic variation. The results were unchanged.
These findings are in line with some,7,14 but not all previous studies.8–10,12,13 One epigenome-wide study used Viva (N=445) as a discovery and Generation R (N=969) as a replication sample (both studies included in this meta-analysis),14 while others relied on smaller samples from individual cohorts (N=45–576).
A possible interpretation of our results is that maternal symptoms of anxiety may not substantially affect circulating fetal DNAm, and the associations between prenatal anxiety and child phenotypes that have been observed in several studies2,3 are explained by other mechanistic pathways. These could include, for example, the postnatal extension of maternal anxiety and subsequent differences in parenting and the home environment, increased risk of perinatal complications/interventions, or confounding by genetic and environmental risk factors which could affect both anxiety and child outcomes. Anxiety may also impact methylation in specific target tissues only without reflection of these changes in blood.
Alternative interpretations may be considered in light of our study limitations. First, maternal anxiety may associate with cord blood DNAm, but the effects at individual sites could be so subtle that using the current methods, they could not be detected. With >7,000 mother-child dyads, our study is substantially larger than previous studies,11,14 and meta-analyses of similar/smaller samples have shown robust associations between cord blood DNAm and other prenatal exposures such as maternal smoking,47 however we may have insufficient power to detect all meaningful change in DNAm. DNAm variation is site-specific, and while some evidence has suggested associations between cord blood DNAm and child neurodevelopmental outcomes,48 more information about the magnitude of associations between cord blood DNAm and relevant child phenotypes is needed before deciding what constitutes a meaningful change in DNAm. In particular, analyses of pregnancy-related anxiety were based on a smaller sample and could be underpowered. Although we observed no meta-analytic association between pregnancy-related anxiety and cord blood DNAm, some CpGs exhibited nominal-level associations with pregnancy-related anxiety in the largest cohorts. Similarly, we report some nominal associations between general anxiety and DNAm at sites included only on the EPIC array across the largest cohorts. These could be of interest for future studies.
Second, there may be subgroup-specific associations between maternal anxiety and DNAm that merit future research. Effects specific to most severe or certain types of anxiety could go unnoticed in our population-based cohorts due to rarity/selective attrition. We lacked power to examine trait-anxiety as a separate construct49 or effects mediated by medication. Furthermore, measurement error and variation across cohorts in the timing and instrument used to measure anxiety likely contributed to reduced power. However, questionnaires were chosen per cohort to reliably measure anxiety within the specific population, and it is unlikely that instrument differences would hide robust, wide-spread associations between anxiety and DNAm. Most scales were previously validated in perinatal settings.e.g. 4,28,50–52 For example, GHQ, BSI, and STAI were validated against perinatal diagnostic interviews,4,50 and CCEI correlates highly with STAI-S/STAI-T during pregnancy (r=.70/r=.76, respectively)52 (Supplementary Methods contains more psychometric information). Further, we examined pregnancy-related anxiety separately as recommended.28 Nonetheless, some symptom- or disorder-specific effects may have gone unnoticed. Further, we encourage research into potential sex differences. We included sex as a covariate but did not examine sex-specificity, to limit the number of tests to those with the most power.
Our choice to focus on anxiety alone may be considered another limitation. For example, we did not examine comorbidity between anxiety and depression, which is common and could increase the risk for poor perinatal outcomes compared to anxiety alone.53 We encourage research on related prenatal exposures, including depression, stressful events, and substance use disorder, for which the existing small epigenetic studies have shown mixed results. e.g. 14,54
Finally, emerging cell-type correction methods could prove useful. However, we used a validated approach to address cell composition, and 14 of 16 cohorts used the same reference panel.33 A study assessing prenatal phthalate exposure and cord blood DNAm found only small differences when three different cell-type reference panels were used.55 Also, no significant results emerged in analyses only including cohorts with the Bakulski panel.33 Different reference panels were, thus, unlikely to strongly influence our results.
Our study has several strengths. Firstly, the collaboration across studies allowed us to examine the association between maternal anxiety and cord blood DNAm among >7,000 mother-child dyads. Secondly, we examined pregnancy-related anxiety, which may be more closely associated with perinatal outcomes, compared to general anxiety symptoms.28 Thirdly, combining data from several cohorts with the EPIC array, we examined DNAm across hundreds of thousands of sites that have not been examined before, in particular increasing coverage of regulatory regions. Our null findings do not definitely refute a link between prenatal anxiety and epigenetic programming. Future studies of other tissues of interest, such as the brain and the placenta, and less well-understood forms of epigenetic alterations, such as histone modifications, could prove relevant. Also, studies with larger samples and higher resolution could elucidate subtle effects.
Conclusions:
This meta-analysis suggests little evidence of a relation between maternal anxiety during pregnancy and differential DNAm in cord blood. While this is a considerably large study on maternal mental health and offspring DNAm, even larger studies with validated clinical diagnoses instead of self-reported symptoms may be needed to establish subtle or subgroup-specific associations between maternal anxiety and the fetal epigenome. Other tissues, epigenetic mechanisms, and aspects of perinatal mental health warrant exploration, as we begin to disentangle the many biological mechanisms that could explain how perinatal maternal mental health affects the next generation.
Supplementary Material
ACKNOWLEDGEMENTS
Acknowledgements for each of the participating studies are listed in the Funding and Acknowledgements supplement.
CONFLICT OF INTEREST
One author (Dan J Stein) received research grants and/or consultancy honoraria from Lundbeck and Sun; the other authors confirm they have no financial relationships with commercial interests to disclose. Funding for each of the participating studies is listed in the Funding and Acknowledgements supplement. There was no editorial direction or censorship from the sponsors.
REFERENCES
- 1.Dennis CL, Falah-Hassani K, Shiri R. Prevalence of antenatal and postnatal anxiety: Systematic review and meta-analysis. Br J Psychiatry. 2017;210(5):315–323. [DOI] [PubMed] [Google Scholar]
- 2.Grigoriadis S, Graves L, Peer M, Mamisashavili L, Tomlinson G, Vigod SN, et al. Maternal anxiety during pregnancy and the association with adverse perinatal outcomes: Systematic review and meta-analysis. J Clin Psychiatry. 2018;79(5): 17r12011. [DOI] [PubMed] [Google Scholar]
- 3.Stein A, Pearson RM, Goodman SH, Rapa E, Rahman A, McCallum M, et al. Effects of perinatal mental disorders on the fetus and child. Lancet. 2014;384:1800–1819. [DOI] [PubMed] [Google Scholar]
- 4.Henrichs J, Schenk JJ, Roza SJ, van den Berg MP, Schmidt HG, Steegers EAP, et al. Maternal psychological distress and fetal growth trajectories: The Generation R study. Psychol Med. 2010;40(4):633–643. [DOI] [PubMed] [Google Scholar]
- 5.Cao-Lei L, Rooij SR De, King S, Matthews SG, Metz GAS, Roseboom TJ, et al. Prenatal stress and epigenetics. Neurosci Biobehav Rev. In press. [DOI] [PubMed] [Google Scholar]
- 6.Ryan J, Mansell T, Fransquet P, Saffery R. Does maternal mental well-being in pregnancy impact the early human epigenome? Epigenomics. 2017;9(3):313–322. [DOI] [PubMed] [Google Scholar]
- 7.Mansell T, Vuillermin P, Ponsonby AL, Collier F, Saffery R, Ryan J. Maternal mental well-being during pregnancy and glucocorticoid receptor gene promoter methylation in the neonate. Dev Psychopathol. 2016;28(4):1421–1430. [DOI] [PubMed] [Google Scholar]
- 8.Mansell T, Novakovic B, Meyer B, Rzehak P, Vuillermin P, Ponsonby A-L, et al. The effects of maternal anxiety during pregnancy on IGF2 / H19 methylation in cord blood. Transl Psychiatry. 2016;6:e765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J, Li Q, Rialdi A, Mystal E, Ly J, Finik J, et al. Influences of maternal stress during pregnancy on the epigenome: Comparison of placenta and umbilical cord blood. J Depress Anxiety. 2014;3(2):152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vangeel EB, Izzi B, Hompes T, Vansteelandt K, Lambrechts D, Freson K, et al. DNA methylation in imprinted genes IGF2 and GNASXL is associated with prenatal maternal stress. Genes, Brain Behav. 2015;14(8):573–582. [DOI] [PubMed] [Google Scholar]
- 11.Vangeel EB, Pishva E, Hompes T, van den Hove D, Lambrechts D, Allegaert K, et al. Newborn genome-wide DNA methylation in association with pregnancy anxiety reveals a potential role for GABBR1. Clin Epigenetics. 2017;9:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S, Devlin AM. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics. 2008;3(2):97–106. [DOI] [PubMed] [Google Scholar]
- 13.Hompes T, Izzi B, Gellens E, Morreels M, Fieuws S, Pexters A, et al. Investigating the influence of maternal cortisol and emotional state during pregnancy on the DNA methylation status of the glucocorticoid receptor gene (NR3C1) promoter region in cord blood. J Psychiatr Res. 2013;47(7):880–891. [DOI] [PubMed] [Google Scholar]
- 14.Cardenas A, Faleschini S, Cortes Hidalgo A, Rifas-Shiman SL, Baccarelli AA, DeMeo DL, et al. Prenatal maternal antidepressants, anxiety, and depression and offspring DNA methylation: Epigenome-wide associations at birth and persistence into early childhood. Clin Epigenetics. 2019;11(1):56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felix JF, Joubert BR, Baccarelli AA, Sharp GC, Almqvist C, Annesi-Maesano I, et al. Cohort profile: Pregnancy and childhood epigenetics (PACE) consortium. Int J Epidemiol. 2018;47(1):22–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraser A, Macdonald-Wallis C, Tilling K, Boyd A, Golding J, Davey Smith G, et al. Cohort profile: The Avon longitudinal study of parents and children: ALSPAC mothers cohort. Int J Epidemiol. 2013;42(1):97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wright J, Small N, Raynor P, Tuffnell D, Bhopal R, Cameron N, et al. Cohort profile: The Born in Bradford multi-ethnic family cohort study. Int J Epidemiol. 2013;42(4):978–991. [DOI] [PubMed] [Google Scholar]
- 18.Stein DJ, Koen N, Donald KA, Adnams CM, Koopowitz S, Lund C, et al. Investigating the psychosocial determinants of child health in Africa: The Drakenstein Child Health Study. J Neurosci Methods. 2015;252:27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heude B, Forhan A, Slama R, Douhaud L, Bedel S, Saurel-Cubizolles M-J, et al. Cohort Profile: The EDEN mother-child cohort on the prenatal and early postnatal determinants of child health and development. Int J Epidemiol. 2016;45(2):353–363. [DOI] [PubMed] [Google Scholar]
- 20.Kooijman MN, Kruithof CJ, van Duijn CM, Duijts L, Franco OH, van IJzendoorn MH, et al. The Generation R Study: Design and cohort update 2017. Eur J Epidemiol. 2016;31(12):1243–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paternoster L, Evans DM, Aagaard Nohr E, Holst C, Gaborieau V, Brennan P, et al. Genome-wide population-based association study of extremely overweight young adults - the GOYA study. PLoS One. 2011;6(9):e24303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guxens M, Ballester F, Espada M, Fernández MF, Grimalt JO, Ibarluzea J, et al. Cohort profile: The INMA-INfancia y Medio Ambiente-(environment and childhood) project. Int J Epidemiol. 2012;41(4):930–940. [DOI] [PubMed] [Google Scholar]
- 23.Magnus P, Birke C, Vejrup K, Haugan A, Alsaker E, Daltveit AK, et al. Cohort profile update: The Norwegian Mother and Child Cohort Study (MoBa). Int J Epidemiol. 2016;45(2):382–388. [DOI] [PubMed] [Google Scholar]
- 24.Girchenko P, Lahti M, Tuovinen S, Savolainen K, Lahti J, Binder EB, et al. Prediction and Prevention of Preeclampsia and Intrauterine Growth Restriction (PREDO) study. Int J Epidemiol. 2017;46(5):1380–1381. [DOI] [PubMed] [Google Scholar]
- 25.Tamayo y Ortiz M, Téllez-Rojo MM, Trejo-Valdivia B, Schnaas L, Osorio-Valencia E, Coull B, et al. Maternal stress modifies the effect of exposure to lead during pregnancy and 24-month old children’s neurodevelopment. Environ Int. 2017;98:191–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brunst KJ, Enlow MB, Kannan S, Carroll KN, Coull BA, Wright RJ. Effects of prenatal social stress and maternal dietary fatty acid ratio on infant temperament: Does race matter? Epidemiol. 2014;4(4):1000167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oken E, Baccarelli AA, Gold DR, Kleinman KP, Litonjua AA, De Meo D, et al. Cohort profile: Project Viva. Int J Epidemiol. 2015;44:37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brunton RJ, Dryer R, Saliba A, Kohlhoff J. Pregnancy anxiety: A systematic review of current scales. J Affect Disord. 2015;176:24–34. [DOI] [PubMed] [Google Scholar]
- 29.Min JL, Hemani G, Davey Smith G, Relton C, Suderman M. Meffil: Efficient normalization and analysis of very large DNA methylation datasets. Bioinformatics. 2018;34(23):3983–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen YA, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8(2):203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCartney DL, Walker RM, Morris SW, McIntosh AM, Porteous DJ, Evans KL. Identification of polymorphic and off-target probe binding sites on the Illumina Infinium MethylationEPIC BeadChip. Genomics Data. 2016;9:22–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bakulski KM, Feinberg JI, Andrews SV, Yang J, Brown S, McKenney SL, et al. DNA methylation of cord blood cell types: Applications for mixed cell birth studies. Epigenetics. 2016;11(5):354–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Goede OM, Razzaghian HR, Price EM, Jones MJ, Kobor MS, Robinson WP, et al. Nucleated red blood cells impact DNA methylation and expression analyses of cord blood hematopoietic cells. Clin Epigenetics. 2015;7:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willer CJ, Li Y, Abecasis GR. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57(1):289–300. [Google Scholar]
- 37.Suderman M, Staley JR, French R, Arathimos R, Simpkin A, Tilling K. Dmrff: Identifying Differentially Methylated Regions efficiently with power and control. BioRxiv. Preprint. doi: 10.1101/508556 [DOI] [Google Scholar]
- 38.Hannon E, Lunnon K, Schalkwyk L, Mill J. Interindividual methylomic variation across blood, cortex, and cerebellum: Implications for epigenetic studies of neurological and neuropsychiatric phenotypes. Epigenetics. 2015;10(11):1024–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edgar RD, Jones MJ, Meaney MJ, Turecki G, Kobor MS. BECon: A tool for interpreting DNA methylation findings from blood in the context of brain. Transl Psychiatry. 2017;7:e1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Non AL, Binder AM, Kubzansky LD, Michels KB. Genome-wide DNA methylation in neonates exposed to maternal depression, anxiety, or SSRI medication during pregnancy. Epigenetics. 2014;9(7):964–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Conradt E, Lester BM, Appleton AA, Amstrong DA, Marsit CJ. The roles of DNA methylation of NR3C1 and 11β-HSD2 and exposure to maternal mood disorder in utero on newborn neurobehavior. 2013;8(12):1321–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heron J, O’Connor TG, Evans J, Golding J, Glover V, ALSPAC Study Team. The course of anxiety and depression through pregnancy and the postpartum in a community sample. J Affect Disord. 2004;80(1):65–73. [DOI] [PubMed] [Google Scholar]
- 43.Glover V, O’Connor TG, Heron J, Golding J, ALSPAC Study Team. Antenatal maternal anxiety is linked with atypical handedness in the child. Early Hum Dev. 2004;79(2):107–118. [DOI] [PubMed] [Google Scholar]
- 44.Saffari A, Silver MJ, Zavattari P, Moi L, Columbano A, Meaburn EL, et al. Estimation of a significance threshold for epigenome-wide association studies. Genet Epidemiol. 2018;42(1):20–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edgar RD, Jones MJ, Robinson WP, Kobor MS. An empirically driven data reduction method on the human 450K methylation array to remove tissue specific non-variable CpGs. Clin Epigenetics. 2017;9:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Solomon O, MacIsaac J, Quach H, Tindula G, Kobor MS, Huen K, et al. Comparison of DNA methylation measured by Illumina 450K and EPIC BeadChips in blood of newborns and 14-year-old children. Epigenetics. 2018;13(6):655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Joubert BR, Felix JF, Yousefi P, Bakulski KM, Just AC, Breton C, et al. DNA methylation in newborns and maternal smoking in pregnancy: Genome-wide consortium meta-analysis. Am J Hum Genet. 2016;98(4):680–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hodyl NA, Roberts CT, Bianco-Miotto T. Cord blood DNA methylation biomarkers for predicting neurodevelopmental outcomes. Genes (Basel). 2016;7(12):117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spielberger CD. State-Trait Anxiety Inventory for Adults. Manual, Instrument, and Scoring Guide. Palo Alto, CA: Consulting Psychologists Press; 1983. [Google Scholar]
- 50.Meades R, Ayers S. Anxiety measures validated in perinatal populations: A systematic review. J Affect Disord. 2011;133(1–2):1–15. [DOI] [PubMed] [Google Scholar]
- 51.Evans K, Spiby H, Morrell CJ. A psychometric systematic review of self-report instruments to identify anxiety in pregnancy. J Adv Nurs. 2015;71(9):1986–2001. [DOI] [PubMed] [Google Scholar]
- 52.O’Connor TG, Ben-Shlomo Y, Heron J, Golding J, Adams D, Glover V. Prenatal anxiety predicts individual differences in cortisol in pre-adolescent children. Biol Psychiatry. 2005;58(3):211–217. [DOI] [PubMed] [Google Scholar]
- 53.Field T, Diego M, Hernandez-Reif M, Figueiredo B, Deeds O, Ascencio A, et al. Comorbid depression and anxiety effects on pregnancy and neonatal outcome. Infant Behav Dev. 2010;33:23–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rijlaarsdam J, Pappa I, Walton E, Bakermans-Kranenburg MJ, Mileva-Seitz VR, Rippe RCA, et al. An epigenome-wide association meta-analysis of prenatal maternal stress in neonates: A model approach for replication. Epigenetics. 2016;11(2):140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Solomon O, Yousefi P, Huen K, Gunier R, Escudero-Fung M, Barcellos LF, et al. Prenatal phthalate exposure and altered patterns of DNA methylation in cord blood. Env Mol Mutagen. 2017;58(6):398–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.