Summary
Upregulated signal flow through RAS and the mitogen-associated protein kinase (MAPK) cascade is the unifying mechanistic theme of the RASopathies, a family of disorders affecting development and growth. Pathogenic variants in more than 20 genes have been causally linked to RASopathies, the majority having a dominant role in promoting enhanced signaling. Here, we report that SPRED2 loss of function is causally linked to a recessive phenotype evocative of Noonan syndrome. Homozygosity for three different variants—c.187C>T (p.Arg63∗), c.299T>C (p.Leu100Pro), and c.1142_1143delTT (p.Leu381Hisfs∗95)—were identified in four subjects from three families. All variants severely affected protein stability, causing accelerated degradation, and variably perturbed SPRED2 functional behavior. When overexpressed in cells, all variants were unable to negatively modulate EGF-promoted RAF1, MEK, and ERK phosphorylation, and time-course experiments in primary fibroblasts (p.Leu100Pro and p.Leu381Hisfs∗95) documented an increased and prolonged activation of the MAPK cascade in response to EGF stimulation. Morpholino-mediated knockdown of spred2a and spred2b in zebrafish induced defects in convergence and extension cell movements indicating upregulated RAS-MAPK signaling, which were rescued by expressing wild-type SPRED2 but not the SPRED2Leu381Hisfs∗95 protein. The clinical phenotype of the four affected individuals included developmental delay, intellectual disability, cardiac defects, short stature, skeletal anomalies, and a typical facial gestalt as major features, without the occurrence of the distinctive skin signs characterizing Legius syndrome. These features, in part, characterize the phenotype of Spred2−/− mice. Our findings identify the second recessive form of Noonan syndrome and document pleiotropic consequences of SPRED2 loss of function in development.
Keywords: RASopathies, RAS, MAPK, Noonan syndrome, neurofibromin, neurofibromatosis, SPRED, SPRED1, SPRED2, Legius syndrome
Introduction
Dysregulated signaling through the RAS/mitogen-activated protein kinase (MAPK) cascade has profound consequences on cell behavior and development. Impaired function of the three members of the RAS subfamily or each tier of the MAPK backbone (i.e., RAF, MEK, and ERK proteins) dramatically affects cell homeostasis1,2 and perturbs developmental programs.3, 4, 5, 6, 7 On the other hand, enhanced or prolonged activation of this cascade due to activating variants in HRAS (MIM: 190020), KRAS (MIM: 190070), NRAS (MIM: 164790), BRAF (MIM: 164757), RAF1 (MIM: 164760), MAP2K1 (MIM: 176872), MAP2K2 (MIM: 601263), or MAPK1 (MIM: 176948) and an increasing number of genes encoding proteins acting as positive regulators of this signaling cascade is a major event contributing to oncogenesis and causes a family of diseases affecting development and growth collectively known as the RASopathies.8, 9, 10, 11 Equivalent consequences have been associated with loss-of-function (LoF) variants affecting negative regulators causing defective signaling switch-off.12, 13, 14, 15
In line with the shared pathogenesis and the pleiotropic consequences of aberrant RAS/MAPK signaling in development, RASopathies are multisystem disorders with significant clinical overlap. Major features include a distinctive facies, postnatally reduced growth, a wide spectrum of congenital heart defects (CHD), hypertrophic cardiomyopathy (HCM), cognitive impairment and behavioral anomalies, skeletal and skin features, as well as variable predisposition to malignancies.8,10,16,17 These disorders are generally transmitted as dominant traits, although recessive inheritance has recently been recognized in Noonan syndrome (NS [MIM: PS163950]),18, 19 the most clinically variable condition among RASopathies. While all RASopathies are rare disorders, NS and neurofibromatosis type 1 (NF1 [MIM: 162210]) are relatively common, which is likely due to the underlying genetic heterogeneity (NS), large size of the implicated gene (NF1), relatively mild impact on fitness (i.e., a significant proportion of families segregating the two disorders), and the selfish behavior of mutations in spermatogonial stem cells.8, 10,20
Pathogenic variants in more than 20 genes have been reported to cause RASopathies.21, 22, 23, 24 Among these, only a minority (i.e., CBL [MIM: 165360], LZTR1 [MIM: 600574], NF1 [MIM: 613113], and SPRED1 [MIM: 609291]) encode proteins functioning as negative modulators of the RAS-MAPK pathway. In this context, inefficient signaling switch-off operating at the level of RAS proteins has been established as the mechanism of disease underlying NF1 and Legius syndrome (LGSS [MIM: 611431]).25 NF1 is characterized by Lisch nodules on the iris, cutaneous café-au-lait macules (CALMs), axillary/inguinal freckling, skeletal defects, learning disabilities, neurofibromas, and increased risk for certain cancers (Table S1).26 The disorder is caused by heterozygous inactivating variants in NF1, which encodes neurofibromin, a multidomain protein functioning as a RAS-specific GTPase activating protein (GAP) negatively controlling RAS activity by promoting conversion of active GTP-bound RAS to its inactive, GDP-bound state.27,28 Of note, a small proportion of subjects with pathogenic NF1 variants shows clinical overlap with NS, resulting in the so-called neurofibromatosis-Noonan syndrome (MIM: 601321).29,30 Similar to NF1, LGSS is characterized by CALMs, with or without freckling; in LGSS, however, Lisch nodules and the NF1-associated neurofibromas and malignancies do not occur (Table S1).31 In affected subjects, developmental delay (DD), behavioral anomalies, and hyperactivity, as well as facial features suggestive of NS are commonly reported. The disorder is caused by heterozygous mutations in the Sprouty-related, EVH1 domain containing 1 (SPRED1) gene,32 encoding a protein inhibiting RAS function by promoting neurofibromin translocation to the plasma membrane and its binding to GTP-bound RAS.33,34 SPRED binding to neurofibromin is a required step for the ability of the GAP to suppress RAS function, and a recent study has solved the structure of the ternary complex involving SPRED1, neurofibromin, and GTP-bound KRAS, providing mechanistic insights of this regulatory circuit negatively controlling RAS-MAPK signaling.34
The three members of the SPRED family (i.e., SPRED1, SPRED2, and SPRED3) share a similar domain organization and function.25,35 These proteins are characterized by an N-terminal Enabled/VASP homology 1 (EVH1) domain, a central c-Kit related binding domain (KBD) (missing in SPRED3), and a C-terminal cysteine-rich Sprouty-related (SPR) domain. The EVH1 domain is essential for the inhibitory function of SPRED proteins exerted on RAS since it mediates binding of the protein to neurofibromin.34,36 The SPR domain, which is shared with the members of the Sprouty family, undergoes post-translational processing (i.e., palmitoylation) and is responsible for binding of SPRED proteins to the cytoplasmic leaflet of membranes.37, 38, 39 The three proteins negatively regulate signal traffic through the RAS-MAPK pathway elicited by multiple growth factors (e.g., FGF and EGF) and cytokines, with SPRED1 and SPRED2 sharing the stronger suppression activity.39 Of note, partial redundancy in Spred1 and Spred2 function has been documented.40
Despite the successful use of exome/genome sequencing in understanding the molecular bases of RASopathies during the last years, a large proportion of individuals with a clinical diagnosis of RASopathy or having an unclassified but RASopathy-related phenotype remains molecularly unsolved. By a “functional candidacy” strategy performed in the frame of the NSEuroNet consortium, using parallel sequencing of a panel of genes functionally related to the currently known RASopathy genes, we identify SPRED2 (MIM: 609292) as a gene implicated in a RASopathy clinically resembling NS. Biochemical, in vitro and in vivo functional studies indicate that the identified SPRED2 variants are inactivating and that SPRED2 LoF results in an enhanced, stimulus-dependent activation of the MAPK cascade. Remarkably and differently from LGSS, this new condition is transmitted as a recessive trait, indicating a differential requirement of SPRED2 and SPRED1 function in developmental processes.
Material and methods
Subjects
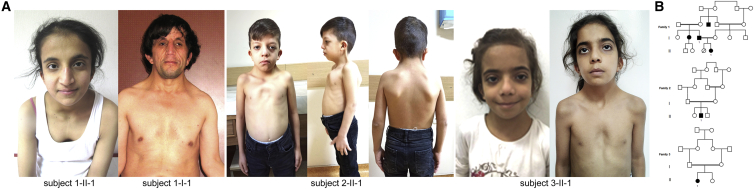
Subjects from three unrelated consanguineous families were included in the study. Clinical data, DNA samples, and skin biopsies were collected from the participating families after written informed consent was obtained. DNA samples and primary fibroblast lines were collected, stored, and used following procedures in accordance with the ethical standards of the declaration of Helsinki protocols, under research projects approved by the Review Boards of the participating institutions (Ospedale Pediatrico Bambino Gesù, Rome; Otto von Guericke University, Magdeburg; CHU Hôpital Robert Debré, Paris). Explicit permission was obtained to publish the photographs of the subjects shown in Figure 1. The additional RASopathy cohorts included in the study were referred for diagnostic genetic testing because of a suspected RASopathy.
Figure 1.
Clinical features and pedigrees of the subjects with bi-allelic inactivating SPRED2 variants
(A) Facial features of the four individuals included in this study. Note the occurrence of bitemporal narrowing, hypertelorism, down-slanting palpebral fissures, ptosis, low-set/posteriorly rotated ears with evident antitragus, wide nasal bridge, and low posterior hairline with a webbed/short neck. Typical NS chest anomalies (superior pectus carinatum and inferior pectus excavatum) are also evident. Craniofacial features resemble NS. The NS facial gestalt is particularly represented in subject 2-II-1.
(B) Pedigree charts of the three families with bi-allelic inactivating SPRED2 variants. All probands were born from first cousins. Note the high inbreeding of family 1.
Molecular analyses
Mutation scan of the known genes implicated in RASopathies and a selected panel of candidates was performed at the reference centers for RASopathy testing of the Robert Debré University Hospital (Paris, France), Magdeburg University Hospital (Magdeburg, Germany), Ospedale Pediatrico Bambino Gesù (Rome, Italy), Policlinico S. Orsola (Bologna, Italy), and Fondazione IRCCS Casa Sollievo della Sofferenza (San Giovanni Rotondo, Italy). The panels used included PTPN11 (MIM: 176876), CBL, SOS1 (MIM: 182530), SOS2 (MIM: 601247), HRAS, KRAS, NRAS, RIT1 (MIM: 609591), RRAS (MIM: 165090), RRAS2 (MIM: 600098), MRAS (MIM: 608435), NF1, SPRED1, LZTR1, SHOC2 (MIM: 602775), PPP1CB (MIM: 600590), RAF1, BRAF, MAP2K1, MAP2K2, SPRED2, and a variable additional list of candidates. Enrichment of target genes was achieved using a Nextera Rapid Capture Custom Enrichment kit (Illumina) (Magdeburg), SureSelectQXT target enrichment system (Agilent) (Paris, San Giovanni Rotondo), and multiplex PCR using an Ion AmpliSeq On-Demand panel (ThermoFisher Scientific) (Bologna). In all analyses, a minimum depth of 100× for all target regions was generally attained (>20×, Paris; >200× Magdeburg). Alignment and variant calling were performed with the MiSeq Reporter software (Illumina), followed by annotation of VCF output files using Variant Studio v.2.2 (Illumina) (Magdeburg, San Giovanni Rotondo), Ion Reporter Software (ThermoFisher Scientific) (Bologna), or BenchlabNGS Alissa Interpret v.5.2 (Agilent) (Paris). All reported variants were visualized with the Integrative Genomics Viewer v.2.3 or Alamut Visual v.2.15 software.
Exome sequencing (trio-based approach) was performed using DNA samples obtained from peripheral blood leukocytes. Target enrichment was performed using the SureSelect Clinical Research Exome v.2 and SureSelect Exome v.7 (Agilent). WES data processing, including sequence alignment to GRCh37 or GRCh38, and variant filtering and prioritization by allele frequency, predicted functional impact, and inheritance were performed as previously reported.24,41,42
Structural modeling analysis
Structural analyses used the recently solved structure of the complex in which the neurofibromin GAP-related domain (GRD) interacts with the EVH1 domain of SPRED1 and GTP-bound KRAS (PDB: 6v65) as a template,34 by using the SWISS-MODEL automated protein structure homology modeling server.43 The 3-dimensional structure was visualized using the VMD visualization software.44 The structural impact of the p.Leu100Pro change was explored using the UCSF Chimera package.45 The side-chain orientations were obtained with the Dunbrack backbone-dependent rotamer library,46 choosing the best rotamer with minimal/no steric clashes with neighboring residues.
Reagents and expression constructs
Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum (FBS), phosphate-buffered saline (PBS), glutamine, and antibiotics were obtained from Euroclone. Fugene6 transfection reagent and complete protease inhibitor cocktail tablets were purchased from Roche Diagnostics. Epidermal growth factor (EGF) was from Invitrogen. Cycloheximide (CHX), MG132, and Bafilomycin A1 were from Sigma-Aldrich. Protein G Sepharose was obtained from GE Healthcare. Trans-Blot Turbo Transfer Packs were from Bio-Rad Laboratories. ECL Western Blotting Detection reagents were purchased from Pierce Biotechnology. Alexa Fluor 488 phalloidin dye was from Molecular Probes. Vectashield anti-fade medium containing DAPI was purchased from Vector Laboratories. The following antibodies were used: mouse monoclonal IgG1 anti-Xpress and mouse monoclonal IgG2a anti-V5 (Invitrogen); rabbit polyclonal anti-GAPDH (zebrafish) (Genetex); rabbit monoclonal GM130 (Abcam); mouse monoclonal anti-neurofibromin (H12) and mouse monoclonal anti-GAPDH (Santa Cruz Biotechnology); rabbit polyclonal anti-p44/42 ERK1/2 (MAPK3/1), mouse monoclonal anti-phospho-p44/42 ERK1/2 (MAPK3/1) (Thr202/Tyr204), mouse monoclonal anti-MEK1/2, and rabbit polyclonal anti-phospho-MEK1/2 (Ser217/221) (Cell Signaling Technology); rabbit polyclonal anti-RAF1 (Abcam); mouse monoclonal anti-phospho-RAF1 (Ser338) (Millipore); horseradish peroxidase-conjugated anti-rabbit or anti-mouse (Sigma-Aldrich); goat anti-mouse IgG conjugated to Alexa Fluor 594 and Alexa Fluor 488, goat anti-mouse IgG1 and goat anti-mouse IgG2a conjugated to Alexa Fluor 647 and Alexa Fluor 594, respectively, and goat anti-rabbit conjugated to Alexa Fluor 594 (Molecular Probes).
The entire SPRED2 and LZTR1, and neurofibromin GRD coding sequences were cloned into the pcDNA6/Xpress-HisC, pcDNA6.2/V5-DEST and pcDNA6/V5-HisA eukaryotic expression vectors (Invitrogen), respectively. Mutations were introduced using primed amplification by polymerase chain reaction (site-directed mutagenesis) using the QuiKChange II Site-Directed Mutagenesis kit (Stratagene), as previously described.47 All constructs were bidirectionally Sanger-sequenced for their entire open reading frame.
Cell culture, transfection, inhibitor treatment, and EGF stimulation
COS-1, HEK293T (ATCC), and fibroblast cells were maintained in DMEM medium supplemented with 10% heat-inactivated FBS, 1% L-glutamine, and antibiotics. Sub-confluent cells were transfected using the Fugene6 transfection reagent according to the manufacturer’s instructions. Cells were treated with CHX (10 μg/mL), MG132 (50 μM), or Bafilomycin A1 (200 nM) to assess protein stability and degradation pathways. Serum-free DMEM and EGF were utilized to starve and stimulate cells, respectively.
Cell homogenate and protein assays
Cells were lysed in radio-immune precipitation assay (RIPA) buffer (pH 8.0) containing 20 mM NaF, 1 mM Na3VO4, and protease inhibitors. Lysates were kept on ice for 30 min and then centrifuged at 16,000 × g for 20 min at 4°C. Supernatants were collected and their protein concentration was determined by the bicinchoninic acid assay,48 using bovine serum albumin as a standard.
Immunoblotting
Assays were as previously reported.47 In brief, cell lysates were resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel. After electrophoresis, proteins were transferred to a nitrocellulose membrane using the Trans-Blot Turbo transfer system. Blots were blocked with 5% non-fat milk powder in PBS containing 0.1% Tween-20 for 1 h and incubated with specific antibodies overnight (pRAF1/MEK/ERK and coimmunoprecipitation assays) or for 1 h (SPRED2 level analyses). Primary and secondary antibodies were diluted in blocking solution. Immunoreactive proteins were detected by an enhanced chemiluminescence (ECL) detection kit. Densitometric analysis of protein bands was performed using NineAlliance UVITEC software (UVITEC).
RAF1, MEK, and ERK phosphorylation assays
RAF1, MEK, and ERK phosphorylation assays were performed on transfected HEK293T cells seeded in 6-well plates the day before transfection (70%–80% confluence) and on sub-confluent primary fibroblasts. Cells were serum-starved for 16 h and then stimulated with EGF (transfected cells: 30 ng/mL, assessed 5 and 10/15 min after stimulation; primary fibroblasts: 10 ng/mL, assessed 15, 30, and 60 min after stimulation) or left unstimulated.
Confocal laser scanning microscopy
For subcellular localization analyses, COS-1 cells were seeded at the density of 1.5 × 104 in 24-well cluster plates onto 12-mm coverslips. After 24 h of culture in complete medium, cells were transfected with the indicated constructs. At 24 h after transfection, cells were treated with bafilomycin A1 (200 nM, 4 h) or left untreated and fixed with 3% paraformaldehyde. For GRD-SPRED2 co-localization analysis, transfected COS-1 cells were serum-starved for 16 h and then stimulated with EGF (30 ng/mL, 5 min). Fixed cells were permeabilized with 0.5% Triton X-100 and stained with a mouse monoclonal IgG1 anti-Xpress followed by goat anti-mouse IgG1 Alexa Fluor 647 (SPRED2 in SPRED2-neurofibromin GRD co-localization assays) or goat anti-mouse IgG Alexa Fluor 594 (SPRED2 localization) and then with Alexa Fluor 488 phalloidin dye or using a mouse monoclonal IgG2a anti-V5 followed by goat anti-mouse IgG2a Alexa Fluor 594 (GRD in SPRED2-neurofibromin GRD co-localization assays) or goat anti-mouse IgG Alexa Fluor 488 (V5-tagged LZTR1) and a rabbit monoclonal GM130 followed by goat anti-rabbit IgG Alexa Fluor 594. After staining, coverslips were extensively rinsed and then mounted on the microscope slides by using Vectashield with DAPI mounting medium. Confocal analysis was performed by a Leica TCS-SP8X (Leica Microsystems) equipped with a 405 nm diode laser and a white light laser (WLL) source using excitation spectral laser lines at 488 nm, 594 nm, and 647 nm. Cells stained only with the fluorochrome-conjugated secondary antibody were used to set up acquisition parameters. Signals from different fluorescent probes were taken in sequential scanning mode. Image processing used Adobe Photoshop 7.0 software (Adobe Systems Incorporated).
Co-immunoprecipitation assays
HEK293T cells transfected with WT or variant SPRED2 constructs were serum-starved, stimulated with EGF (30 ng/mL), and lysed in IP buffer containing 25 mM Tris–HCl (pH 7.4), 1% Triton X-100, 2 mM EDTA (pH 8.0), 150 mM NaCl, and protease inhibitors. Samples were centrifuged at 10,000 × g (20 min, 4°C), supernatants were collected, and equal amounts of total proteins were immunoprecipitated using an anti-Xpress antibody cross-linked to Protein G Sepharose beads (2 h, 4°C). The beads were recovered by centrifugation and washed six times with IP buffer. Finally, the immunoprecipitated proteins were eluted with sample buffer by incubating at 95°C (5 min) and stored at −20°C until western blot (WB) analysis (WT SPRED2, 10 μL; mock, 30 μL; SPRED2Leu100Pro, 30 μL), due to the accelerated degradation of the variant. Immunoprecipitated endogenous neurofibromin was normalized to the amount of the relative immunoprecipitated SPRED2 proteins.
Zebrafish studies
Zebrafish were cultured following standard protocols.49 Fish were housed in a water circulating system (Tecniplast) under controlled conditions (light/dark 14:10 h, 28°C, 350–400 uS, pH 6.8–7.2) and fed daily with dry and live (A. salina) food. All the experimental protocols were approved by the Italian Ministry of Health (23/2019-PR).
Embryos were obtained from natural mating of NHGRI pairs.50 Selected pairs did not carry variation within the 24-nucleotide stretch targeted by the designed ATG-morpholino (MO) sequences (Figure S1). Co-injection of spred2a and spred2b ATG-MOs targeting both spred2a and spred2b (6 ng) was performed at the 1-cell stage. Co-injection of p53 ATG-MO was used to control for p53-dependent neural toxicity.51,52 Wild-type (WT) and variant (p.Leu381Hisfs∗95) SPRED2 sequences were cloned into a pCS-Dest vector (gift from Nathan Lawson, plasmid 22423, Addgene) via LRII clonase-mediated recombination (ThermoFisher). Plasmids were digested and linearized with KpnI (New England Biolabs), and mRNA was produced using mMessage mMachine SP6 transcription kit and poly(A) tailing kit (Thermo Fisher). All cloned sequences were confirmed by bidirectional Sanger sequencing.
Injection of in vitro synthesized capped mRNAs encoding SPRED2 alleles (1.5 pg) was used in the rescue experiments. Injected embryos were cultured under standard conditions at 28°C in fresh E3 medium and non-injected fish were used as controls together with fish injected with individual SPRED2 mRNAs. To assess pErk levels in morphants, embryonic tissues (12 hpf) were homogenized in lysis buffer (10 mM Tris-HCl [pH 7.4], 2 mM EDTA, 150 mM NaCl, 1% Triton X-100) supplemented with protease inhibitors. Following centrifugation (20,000 × g, 10 min, 4°C), supernatants were collected and protein concentration was determined by Bradford assay (Bio-Rad). WB analysis was performed as reported above. At the end of gastrulation (12 hpf), embryos were imaged using a Leica M205 stereomicroscope. For the phenotype penetrance analysis of zebrafish morphants, fully developed embryos without recognizable defects were classified as “normal,” embryos showing mild developmental delay within the segmentation period (10–11 hpf) as “mild,” and embryos showing developmental arrest in any phase of gastrula period (epiboly between 60%–90%) as “severe.” Only “normal” and “mild” subgroups were considered for major and minor axes measurements. The axes of the embryos and the elongation ratio (major-to-minor) were measured and calculated using Fiji.53 Data were analyzed independently by two researchers, and statistical assessments were performed using GraphPad Prism. Normality tests (Anderson-Darling, D’Agostino & Pearson, Shapiro-Wilk, and Kolmogorov-Smirnov tests) were run to assess the normal distribution of the data. Two-tailed Fisher exact test was used to assess the statistical significance of phenotypes in morphants (2×2 contingency table). For the elongation analysis, one-way ANOVA with two-stage Benjamini, Krieger, and Yekutieli procedure was run for controlling the false discovery rate of multiple comparisons; Student’s t test was used to assess the statistical significance of pErk levels in morphants.
Spred2 knock-out mouse phenotyping
Spred2−/− mice were raised by crossbreeding heterozygous Spred2+/− mice derived from the embryonic stem cell clone XB228 (BayGenomics RRID: CVCL_PG99). To avoid inbreeding effects, only mice of a mixed 129/Ola × C57BL/6 outbred background at the indicated ages were used. All procedures involving animals were in accordance with European guidelines (2010/63/EU) and were approved by the government of Lower Franconia.
For electrocardiographic (ECG) recordings, mice were anesthetized using 1.5% isoflurane and kept on a warming plate (37°C) throughout the experiment. ECGs were captured at a sampling rate of 4,000 points per second using a PowerLab 8/35 system equipped with BIOamplifiers (ADInstruments) for 10 min, and original ECG recordings were analyzed with the ECG analysis module of LabChart Pro 8.1.8 (ADInstruments).
To quantify the relative volumes of the brain ventricle system, 100-μm sections were cut from perfusion-fixed brains using a VT1000S vibratome (Leica). Individual brain slices were photographed and the background removed. In the resulting images, the ventricle areas were measured using ImageJ (https://imagej.nih.gov). Volumes were calculated using the original slice thickness of 100 μm. Relative volumes were calculated in relation to whole brain volumes. The three-dimensional brain models were compiled with ImageJ and visualized with Imaris (Oxford Instruments). To investigate hippocampus formation, 10-μm-thick brain sections were stained according to the standard Nissl staining procedure. Levels of Spred2 were assessed using a polyclonal rabbit anti-Spred2 antibody (residues 96–415).54 To assess Ras activity, hippocampus regions were dissected from mouse brains on a metal plate cooled with ice and homogenized in an assay-dependent buffer using a plastic douncer fitting into a 1.5 mL microreaction tube. Pan-Ras Activation Assay Kit (#STA-400, Cell Biolabs) was used to detect active Ras, following the manufacturer’s protocol. Fifty milligrams of tissue were lysed in 1 mL assay buffer and 1 mg total protein was used for each Ras pull-down. To investigate kidney morphology, 10 μm-thick paraffin-embedded kidney sections were stained according to the standard hematoxylin-eosin staining protocol.
Results
Bi-allelic variants in SPRED2 cause a disorder resembling Noonan syndrome
Three probands (subjects 1-II-1, 2-II-1, and 3-II-1) suspected to be affected by NS or a related RASopathy were referred for diagnostic genetic testing (Figure 1A). The pedigrees of the three consanguineous families are shown in Figure 1B. Genomic DNA obtained from peripheral blood leukocytes was scanned for variations using an “extended” panel of RASopathy genes opportunely designed to include a set of candidate genes selected in the frame of the NSEuroNet consortium. In family 1, parallel sequencing in the proband identified a previously unreported frameshift variant in SPRED2, c.1142_1143delTT (p.Leu381Hisfs∗95; GenBank: NM_181784.2) in homozygosity, affecting the C terminus of the protein, a region highly intolerant for variation (Figure S2). The variant was predicted to result in a longer open reading frame characterized by a divergent C-terminal tail lacking key cysteine residues required for proper SPRED2 posttranslational processing and plasma membrane targeting (Figure S3). Segregation analysis confirmed homozygosity in the affected father (1-I-1) and heterozygosity in the apparently healthy mother. In family 2, analysis in the proband (2-II-1) identified a homozygous missense change (c.299T>C [p.Leu100Pro]), which was private and altered an invariantly conserved residue among orthologs and paralogs (Figure S3) located within a second highly constrained region of the protein (Figure S2). The variant was inherited by his apparently healthy consanguineous parents, who were confirmed to be heterozygous for the missense change. Of note, somatic mutations of this residue and the adjacent Gly99 have previously been reported in cancer (COSMIC database), and multiple pathogenic missense changes affecting the adjacent residue Thr101 in SPRED1 (p.Thr102Met/Lys/Arg) had causally been linked to LGSS,55,56 supporting the functional and clinical relevance of the variant. In this subject, a homozygous condition for a missense variant in LZTR1 (c.1856G>A [p.Arg619His]; GenBank: NM_006767.3) was also identified. This variant had been reported in public databases (rs568213908, ExAC 0.04%) and was classified as a variant of uncertain significance by ClinVar. In silico prediction tools consistently considered the missense change as tolerated (CADD [v.1.6, hg19] = 16.59; REVEL = 0.069; M-CAP = 0.012 [tolerated]; SIFT: tolerated; PolyPhen-2: benign). Arg619 is only moderately conserved, and His at the same codon is found in multiple mammalian species, suggesting that this missense change does not adversely affect protein function. To further confirm these predictions, the LZTR1Arg619His cDNA was generated by site-directed mutagenesis, and functional characterization of the V5-tagged LZTR1 variant was assessed in terms of protein stability and subcellular localization and by evaluating its impact on pERK levels, which confirmed a behavior equivalent to the WT protein (Figure S4).57 Finally, a previously unreported homozygous SPRED2 nonsense change, c.187C>T, predicting protein truncation at the N terminus (p.Arg63∗), was identified in subject 3-II-1, who had clinical features suggestive of NS. Parallel/Sanger sequencing confirmed the heterozygosity condition in the apparently unaffected sib and parents.
The clinical data of the four affected subjects from the three families are shown in Table 1, and their detailed clinical history is provided in the supplemental note (case reports). Shared features of the four affected individuals included DD/intellectual disability (ID), speech delay, and learning difficulties. Short stature (<−2 SD) with relative macrocephaly was documented in three individuals. Hypotonia was a common feature during childhood. Cardiac involvement occurred in all individuals. Specifically, HCM was documented in subject 1-I-1, asymmetrical interventricular septal hypertrophy and pulmonary valve stenosis (PVS) requiring balloon valvuloplasty was present in subject 2-II-1, and PVS occurred in subject 3-II-1. The distinctive craniofacial appearance included bitemporal narrowing (4/4 cases), hypertelorism (3/4 cases), ptosis (2/4 cases), down-slanting palpebral fissures (3/4 cases), low-set/posteriorly rotated ears (4/4 cases), prominent nasal bridge (4/4 cases), anteverted nares (3/4), long/prominent philtrum (3/4), and low posterior hairline with a webbed/short neck (4/4 cases), which are observed as major features in RASopathies. Typical NS chest anomalies (pectus carinatum and/or excavatum) and other skeletal defects (spine) were also common findings. Cryptorchidism and bleeding diathesis occurred in single individuals. No lymphedema or skin/ectodermal anomalies were noted except for the occurrence of deep palmar creases (2/4 cases). In particular, CALMs and freckling were not observed. None of the four subjects was diagnosed with cancer.
Table 1.
Clinical features of subjects with bi-allelic pathogenic SPRED2 variants
| Case ID | 1-II-1 | 1-I-1 | 2-II-1 | 3-II-1 |
|---|---|---|---|---|
| Ethnicity | Turkish | Turkish | Turkish | Tunisian |
| Sex | female | male | male | female |
| Age at last examination | 14 years 2 mo | 39 years | 8 years | 11 years 11 mo |
| SPRED2 variant | ||||
| cDNA change (NM_181784.2) | c.1142_1143delTT | c.1142_1143delTT | c.299T>C | c.187C>T |
| AA change (NP_861449.2) | p.Leu381Hisfs∗95 | p.Leu381Hisfs∗95 | p.Leu100Pro | p.Arg63∗ |
| Inheritance | bi-allelic | bi-allelic | bi-allelic | bi-allelic |
| ExAC/gnomAD frequency | – | – | – | – |
| Growth at last examination | 14 years 2 mo | 39 years | 8 years | 11 years 11 mo |
| Height (SD) | 145 cm (−2.41) | 144 cm (−4.35) | 122.2 cm (−1.00) | 136.5 cm (−1.50) |
| Weight (SD) | 42 kg (−1.02) | 56 kg (−1.29) | 22 kg (−1.09) | 27.5 kg (−2.30) |
| Head circumference (SD) | 57 cm (+1.32) | 56 cm (−1.00) | 51 cm (+0.86) | 55 cm (+1.00) |
| Development | ||||
| Developmental delay | yes | yes (by history) | mild | no |
| Intellectual disability | mild | mild (by history) | mild | yes |
| Language delay | yes | yes | yes | no |
| Learning disorder | yes | yes (by history) | yes | attention deficit |
| Neurological features | ||||
| Hypotonia | yes | yes (by history) | during infancy | no |
| Cardiovascular features | ||||
| Congenital heart defect | yes (mild aortic insufficiency, mitral valve prolapse) | no | yes (pulmonary valve stenosis, pulmonary balloon valvuloplasty; small secundum ASD) | yes (pulmonary valve stenosis) |
| Hypertrophic cardiomyopathy | yes (focal interventricular septum hypertrophy) | yes | yes (asymmetrical hypertrophy of the interventricular septum) | no |
| Skeletal features | ||||
| Chest | pectus excavatum | pectus excavatum | pectus carinatum superiorly and pectus excavatum inferiorly; wide and short shield chest | pectus excavatum |
| Hyperlaxity | yes | no | yes | yes |
| Other skeletal anomalies | cubitus valgus | no | limited extension of elbows, cubitus valgus, winged shoulder blades, kyphosis, mild pes valgus and pes planus | kyphosis, clinodactyly, abnormal toe position |
| Facial features | ||||
| Bitemporal narrowing | yes | yes | yes | yes |
| Hypertelorism | yes | no | yes | yes |
| Low-set and/or posteriorly rotated ears | yes | yes | yes | yes |
| Prominent nasal bridge | yes | yes | yes | yes |
| Low posterior hairline | yes | yes | yes | yes |
| Short/webbed neck | yes | yes | yes | yes |
| Other dysmorphism or clinical features | helix folding anomaly, dysmorphic ear lobe | downward slanted palpebral fissures, prominent nasolabial folds, long philtrum | triangular coarse face, sparse eyebrows, sparse eyelashes, downward slanted palpebral fissures, epicanthus, nasolacrimal duct stenosis, prominent nasolabial sulci, pointed receding chin | high cranial vault, triangular and coarse face, downward slanted palpebral fissures, ptosis, prominent philtrum, large mouth, thick lips, micrognathia, high arched/narrow palate |
| Skin | ||||
| Café-au-lait spots | no | no | no | no |
| Freckling | no | no | no | no |
| Other skin/ectodermal features | nevi | no | sparse and curly hair, sparse and thin eyebrows and eyelashes, scaly and dry skin, eczematous skin, loose and thick skin, deep palmar creases | hyperhydrosis, deep palmar creases |
| Other features/sign | ||||
| Cryptorchidism | N/A | no | bilateral | N/A |
| Lymphatic involvement | no | no | no | no |
| Bleeding/easy bruising | yes | no | partial FXII deficiency (24.8% activity) | no |
| Hematological abnormalities | lymphopenia | no | no | no |
| Other | no | tiny hemangiomas | atopic skin features, nasolacrimal duct stenosis, exotropia, bone pain and myalgia | transient splenomegaly that gradually disappeared between age 2.5 and 8.5 years, sigmoid dolichocolon |
| Imaging | brain MRI: mild left cerebral hemisphere enlargement | N/A | trans-fontanelle USG: left lateral ventriculomegaly; renal USG: bilateral grade 2 medullary nephrocalcinosis |
N/A |
RASopathy-causing SPRED2 variants affect protein synthesis/stability, localization, and function
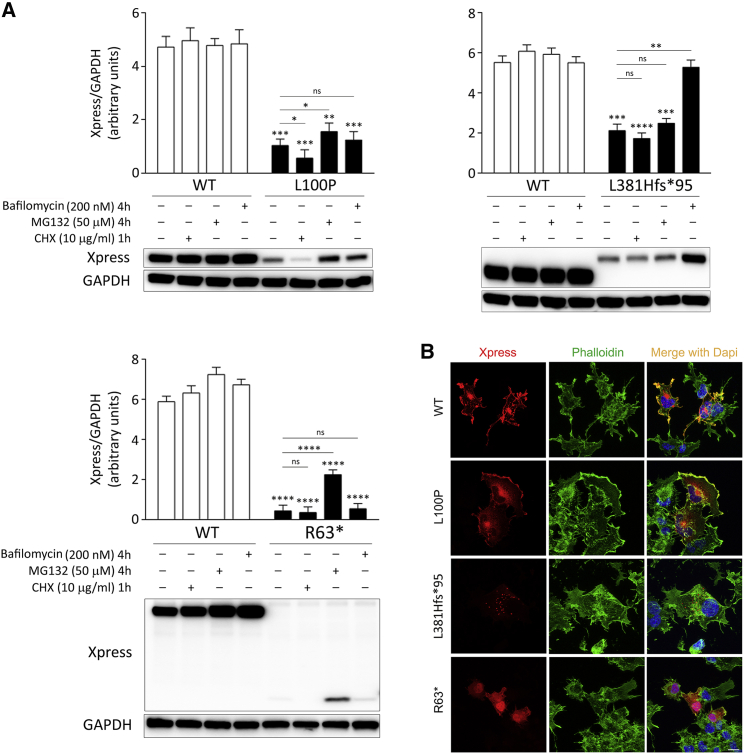
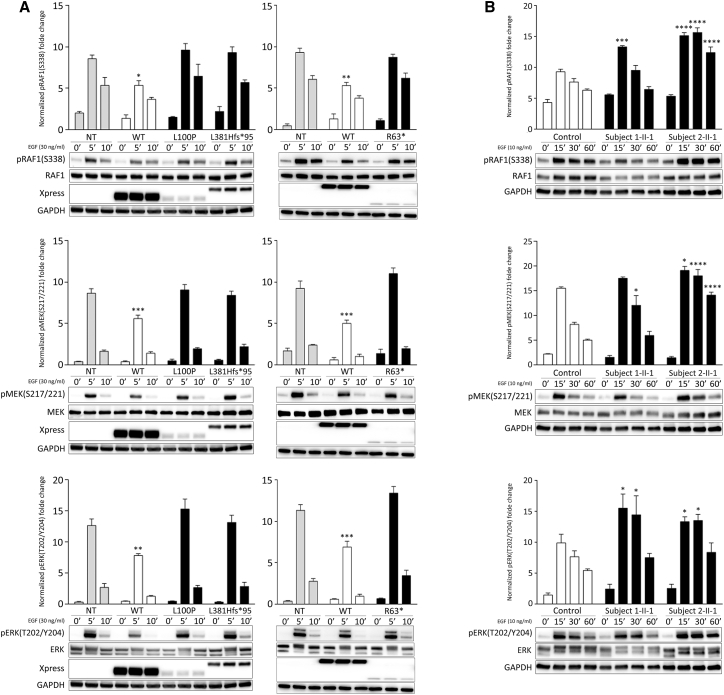
The homozygous occurrence of the three SPRED2 variants in the four affected individuals and the predicted LoF impact for two of them supported a model in which disease-associated SPRED2 variants have disruptive consequences on SPRED2 synthesis, stability, and/or function. According to a model recently developed to predict occurrence of nonsense-mediated mRNA decay (NMD) triggered by premature stop codons,58 the c.187C>T change (subject 3-II-1) was expected to cause SPRED2 mRNA degradation (NMD score > 0.6). Since biological material from subject 3-II-1 (or her parents) was not available to validate this prediction, we assume that NMD likely contributes to the LoF impact of the c.187C>T substitution, even though the synthesis of a residual truncated SPRED2 protein cannot be ruled out a priori. To validate the impact of the identified variants on SPRED2 stability, the three Xpress-tagged SPRED2 proteins were generated, and the protein level of each was assessed in transiently transfected COS-1 cells, basally and after 1 h treatment with the protein synthesis inhibitor CHX. Immunoblotting and relative quantitative analyses revealed a variably reduced level of the three proteins, indicating that the tested variants significantly impact SPRED2 stability (Figure 2A). As expected, the strongest effect was documented for the truncating variant at the N terminus. Treatment with MG132, an inhibitor of proteasomal activity, and bafilomycin, a late-stage inhibitor of autophagy and lysosomal protein degradation, variably rescued the reduced levels of SPRED2Arg63∗ and SPRED2Leu381Hisfs∗95, respectively, indicating a different route of degradation for these proteins (Figure 2A). On the other hand, blocking either of the two pathways had little effect on the accelerated degradation of SPRED2Leu100Pro, indicating that both routes actively contribute to the degradation of this variant, possibly with a more relevant role of the proteasome, as documented by the slight but statistically significant increased level of the protein in cells treated with MG132.
Figure 2.
Biochemical characterization of the RASopathy-causing SPRED2 proteins
(A) Accelerated degradation of the SPRED2Arg63∗ (R63∗), SPRED2Leu100Pro (L100P), and SPRED2Leu381Hisfs∗95 (L381Hfs∗95) proteins. WB analysis shows WT and variant Xpress-tagged SPRED2 protein levels in transfected COS-1 cells, basally and after CHX (10 μg/mL), MG132 (50 μM), or bafilomycin A1 (200 nM) treatment. As shown, accelerated degradation of SPRED2Arg63∗ mainly occurs via the proteasome, while SPRED2Leu381Hisfs∗95 is degraded primarily via autophagy/lysosomes. Both pathways are involved in the accelerated degradation of SPRED2Leu100Pro, with a more relevant role of the proteasome (slight but statistically significant increased protein level in cells treated with MG132). GAPDH was used as loading control. Representative blots (below) and mean ± SD densitometry values (above) of three independent experiments are shown. Asterisks indicate statistically significant differences (∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, ns indicates not significant; two-way ANOVA followed by Sidak’s multiple comparison test).
(B) Subcellular localization of transiently expressed Xpress-tagged WT or mutated SPRED2 proteins in COS-1 cells under steady-state conditions revealed by confocal microscopy analysis. Cells were stained using an anti-Xpress monoclonal antibody and Alexa Fluor 594 goat anti-mouse secondary antibody (red). The F-actin dye Alexa Fluor 488 phalloidin (green) was used to stain the cortical actin associated with the plasma membrane. Merged images with nuclei (DAPI staining, blue) are displayed on the right panels. Scale bar, 10 μm.
The negative modulatory role of SPRED proteins on RAS signaling requires their targeting to the plasma membrane, which is mediated by the SPR domain.33 Indeed, SPRED variants lacking the SPR domain fail to translocate to the plasma membrane and downmodulate MAPK signaling.36 By confocal laser scanning microscopy analysis of COS-1 cells transiently transfected to express an Xpress-tagged SPRED2, we confirmed the localization of the WT protein to the plasma membrane, as shown by co-localization with cortical F-actin (Figure 2B). Assessment of the subcellular localization of the three SPRED2 variants documented that, similar to the WT protein, residual SPRED2Leu100Pro showed, in part, proper targeting to the plasma membrane. In contrast, a diffuse localization in both cytoplasm and nucleus was documented for SPRED2Arg63∗, and a punctate localization pattern was observed for SPRED2Leu381Hisfs∗95, suggestive of vesicular distribution (Figure 2B). For the latter, treatment with bafilomycin resulted in an accumulation of puncta (Figure S5), which is in line with the preferential degradation of this variant via lysosomes.
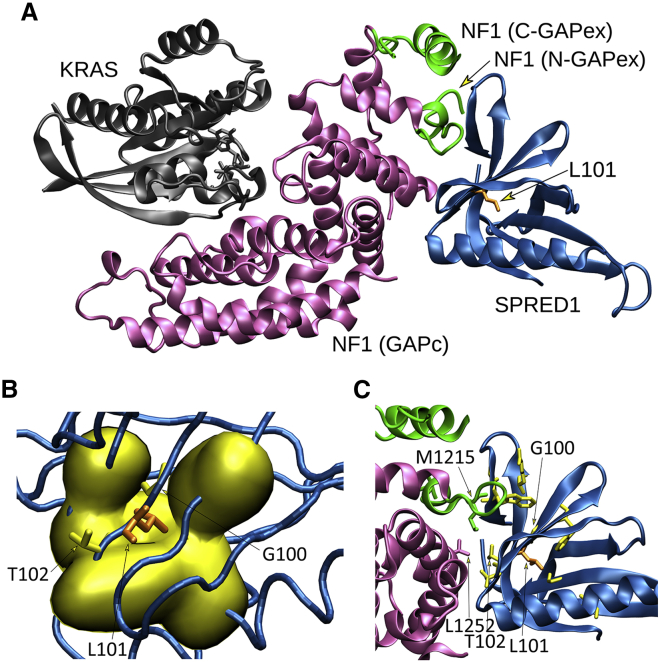
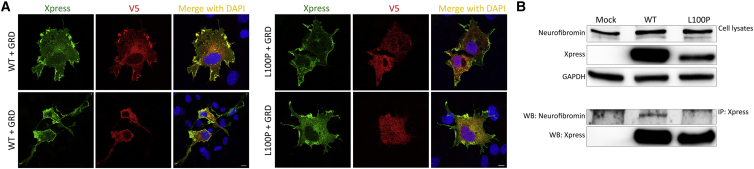
Different from the other variants, the residual SPRED2Leu100Pro protein retained a proper localization to the membrane, which is consistent with an unaffected SPR domain. Leu100 is located within the EVH1 domain, which is known to mediate protein-protein interactions. In SPRED1, this domain binds to the GRD of neurofibromin33,34 and is required for the negative modulation of RAS function. The EVH1 of SPRED2 binds to neurofibromin with comparable affinity.33,59 Remarkably, different LGSS-causing missense SPRED1 changes affecting a residue adjacent to Leu100 in SPRED2 (Thr102) and impairing binding to neurofibromin have been reported.34,60 Based on these considerations, we hypothesized a perturbation of the ability of SPRED2Leu100Pro to promote efficient translocation of neurofibromin to the plasma membrane as a contributing molecular mechanism underlying the functional dysregulation of this protein. To assess this hypothesis, we first explored the possible structural impact of the p.Leu100Pro substitution using the recently solved structure of the complex in which the neurofibromin GRD interacts with the EVH1 domain of SPRED1 and GTP-bound KRAS.34 SPRED1 and SPRED2 EVH1 domains show a high homology in sequence and all residues flanking Leu100 (Leu101 in SPRED1) are conserved. A closer view into the complex revealed that Leu101 localized in a β sheet of the EVH1 domain mediating direct interaction with the neurofibromin GRD (Figure 3A). Its lateral chain is buried in the core of the domain and contributes to the intramolecular binding network stabilizing the region (Figure 3B). Specifically, Leu101 is at the center of a hydrophobic cluster formed by Ala19, Val21, Val42, Trp92, Phe103, and Phe112, which are all conserved in SPRED2. Leu101 forms a main chain hydrogen bond with His90. The adjacent Thr102 (Thr101 in SPRED2) is an exposed residue with its lateral chain directly contributing to the intermolecular binding network with the neurofibromin GRD (Figure 3C).34,60 The Leu-to-Pro substitution is expected to result in a substantial conformational rearrangement of this region and affect the ability of the EVH1 domain to efficiently bind to the GRD of neurofibromin. These considerations suggest that besides its impact on stability and the observed accelerated degradation, the p.Leu100Pro substitution likely results in a SPRED2 protein with decreased and/or less stable binding to neurofibromin, which is in line with previously published data documenting a similar behavior of SPRED1 variants causing LGSS.34,59,60 To validate this hypothesis we evaluated the ability of SPRED2Leu100Pro to bind to neurofibromin. First, we transfected COS-1 cells with V5-tagged neurofibromin GRD alone or with Xpress-tagged WT or variant SPRED2 constructs. Following starvation and EGF stimulation, subcellular localization of SPRED2 proteins and neurofibromin GRD was analyzed by confocal microscopy. Expressed alone, neurofibromin GRD localized primarily in the cytoplasm (data not shown). When co-expressed, co-localization of WT SPRED2 and neurofibromin GRD at the plasma membrane was observed, indicating that SPRED2 facilitates plasma membrane translocation of the domain (Figure 4A). In contrast, GRD-SPRED2Leu100Pro co-localization at the plasma membrane was not observed in cells overexpressing the SPRED2 variant, indicating that this protein fails to recruit neurofibromin GRD to the plasma membrane. To confirm the defective interaction of SPRED2Leu100Pro with neurofibromin, we performed co-immunoprecipitation assays in cells overexpressing the WT and variant SPRED2 constructs in HEK293T cells, starved and stimulated with EGF. As expected, endogenous neurofibromin co-immunoprecipitated with the WT SPRED2 protein but did not with SPRED2Leu100Pro (Figure 4B). Overall, the in silico and in vitro data consistently indicate that the residual SPRED2Leu100Pro protein is able to translocate to the plasma membrane but has a defective ability to mediate neurofibromin localization at the plasma membrane.
Figure 3.
Structural impact of the p.Leu100Pro substitution in SPRED2
(A) The crystallographic complex of neurofibromin (NF1) GRD (GAPc, magenta) with the EVH1 domain of SPRED1 (blue) and GTP-bound KRAS (gray) is shown. The N- and C-terminal GAPex stretches encompassing GAPc are shown in green. The lateral chain of Leu101 (L101) (corresponding to Leu100 in SPRED2) is shown (orange).
(B) Surface visualization (Gaussian density surface) of the hydrophobic residues close to Leu101 (i.e., Ala19, Val21, Val42, Trp92, Phe103, and Phe112) is shown in yellow. Lateral chains of Leu101 and adjacent residues, Gly100 (G100) and Thr102 (T102), mutated in LGSS, are shown.
(C) Enlarged view of the neurofibromin and SPRED1 interacting surfaces. Relevant residues are indicated using the single-letter amino acid code. Lateral chains of SPRED1 residues (Arg24, Gly30, Trp31, Val44, Gly62, Thr88, Trp92, Gly100, and Thr102) mutated in LGSS are shown (yellow), together with Leu101 (orange). Of note, Thr102 interacts with both GAPc (Leu1252) and GAPex (Met1215).
Figure 4.
p.Leu100Pro results in a defective SPRED2 binding to neurofibromin
(A) Subcellular localization of transiently expressed V5-tagged GRD and Xpress-tagged WT or SPRED2Leu100Pro (L100P) proteins in COS-1 cells after stimulation with EGF (30 ng/mL, 5 min) revealed by confocal microscopy analysis. Cells were stained using anti-Xpress IgG1 and anti-V5 IgG2a monoclonal antibodies and revealed by goat anti-mouse IgG1 Alexa Fluor 647 (pseudocolored in green) and goat anti-mouse IgG2a Alexa Fluor 594 (red) secondary antibodies, respectively. Nuclei are visualized by DAPI staining (blue). Scale bar, 10 μm.
(B) Co-immunoprecipitation assays. Lysates from HEK293T cells transiently transfected to express WT or variant Xpress-tagged SPRED2 protein were serum-starved, stimulated with EGF (30 ng/mL), and immunoprecipitated with an anti-Xpress antibody and assayed by western blotting using the indicated antibodies.
RASopathy-causing SPRED2 variants fail to downmodulate MAPK signaling
SPRED2-decreased stability, impaired plasma membrane localization, and defective neurofibromin binding are expected to converge toward a flawed downmodulation of RAS-MAPK signaling. We explored the consequences of the identified SPRED2 variants on this pathway by transiently transfecting each NS-associated SPRED2 protein in HEK293T cells and assessing the phosphorylation levels of the three tiers of the MAPK cascade, under basal conditions and following EGF stimulation, in time-course experiments (Figure 5A). None of the overexpressed SPRED2 variants promoted a significant perturbation of the dynamics and strength of RAF1, MEK, and ERK phosphorylation, while expression of the WT protein was associated with a slight but significant downmodulation of signaling when compared to control cells (mock transfection). Primary fibroblasts obtained from skin biopsies from subjects 1-II-1 (p.Leu381Hisfs∗95) and 2-II-1 (p.Leu100Pro) were used in time-course experiments to assess the MAPK pathway activation in response to EGF stimulation. These assays confirmed a variably enhanced RAF1, MEK, and ERK phosphorylation in cells with bi-allelic SPRED2 variants compared to control cells (Figure 5B), indicating MAPK signaling upregulation. Notably, in both fibroblast lines, signaling was not constitutively activated, retaining dependence on EGF stimulation. These findings confirmed that the bi-allelic SPRED2 variants result in inactive proteins unable to efficiently downmodulate signal flow through the RAS-MAPK cascade, and that primary fibroblasts of subjects with impaired SPRED2 function show enhanced stimulus-dependent activation of this pathway.
Figure 5.
Impact of SPRED2 mutations on MAPK signaling
(A) Overexpression of the RASopathy-causing SPRED2Arg63∗ (R63∗), SPRED2Leu100Pro (L100P), and SPRED2Leu381Hisfs∗95 (L381Hfs∗95) proteins in HEK293T cells do not significantly perturb RAF1, MEK, and ERK phosphorylation as assessed by time-course experiments. Of note, expression of WT SPRED2 down-modulates ERK phosphorylation. HEK293T cells were transiently transfected with the indicated Xpress-tagged SPRED2 constructs, serum-starved and treated with 30 ng/mL EGF for 5 or 10 min, or left unstimulated. Equal amounts of cell lysates were resolved on 10% polyacrylamide gel. Representative blots (below) and mean ± SD densitometry values (above) of three independent experiments are shown. Asterisks indicate statistically significant differences compared with control cells (NT, mock transfection) at the corresponding time upon EGF stimulation (∗p < 0.05; ∗∗p < 0.005; ∗∗∗p < 0.001; two-way ANOVA followed by Tukey’s multiple comparison test).
(B) Primary fibroblasts from subjects 1-II-1 (p.Leu381Hisfs∗95) and 2-II-1 (p.Leu100Pro) show variably enhanced RAF1, MEK, and ERK phosphorylation levels compared to control cells. Fibroblasts were starved for 16 h and then stimulated with EGF (10 ng/mL), in time-course experiments, or left unstimulated. Equal amounts of cell lysates were resolved on 10% polyacrylamide gel. Representative blots (below) and graphs reporting mean ± SD densitometry values (above) of three independent experiments are shown. Asterisks indicate statistically significant differences in the phosphorylation levels compared to control cells at the corresponding experimental points (∗p < 0.05; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; two-way ANOVA followed by Tukey’s multiple comparison test).
In RASopathies, depending on the implicated gene/variant, dysregulated signal flow through the MAPK and/or phosphatidylinositol-3 kinase (PI3K)-AKT cascades is observed.61, 62, 63 Consistent with previous data indicating that SPRED proteins do not participate in the control of PI3K-AKT signaling, no significant difference in the extent of EGF-stimulated AKT phosphorylation was documented by time-course experiments performed in primary fibroblasts from subjects 1-II-1 and 2-II-1 (Figure S6), confirming a specific functional link between SPRED2 and the MAPK cascade, at least in the present experimental conditions.
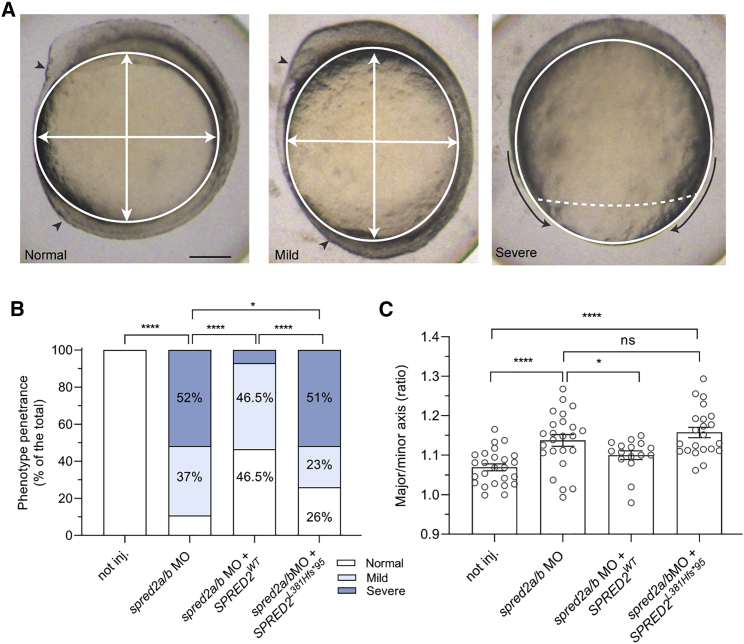
To validate the consequences of SPRED2 LoF in developmental processes reliant on MAPK signaling, we employed a MO-mediated spred2 knockdown approach in zebrafish embryos to assess any perturbation of convergence and extension movements during gastrulation, a well-established readout of upregulated RAS-MAPK signaling that can be quantitatively evaluated by determining the major-to-minor axis ratio of embryos.64, 65, 66 We first confirmed that the two annotated zebrafish spred2a and spred2b genes are homologous to human SPRED2 by sequence alignment and phylogenetic analysis (Figure S7A) and documented that both genes are expressed during zebrafish embryogenesis, including early stages of development (Figure S7B). ATG-targeting MOs for both paralogs (Figure S1) were co-injected in embryos deriving from opportunely selected zebrafish pairs (Figure S1). spred2a and spred2b gene silencing was associated with increased Erk phosphorylation levels (Figure S8) and resulted in a high incidence of embryos showing delayed gastrulation, which arrested between 60% and 90% of epiboly (classified as “severe”), or delay in elongation during the segmentation stage, with reduced formation of the head and tailbud (classified as “mild”) (Figure 6A). The severe phenotype was rescued, in part, by concomitant microinjection of WT SPRED2 mRNA, with a negligible effect of SPRED2Leu381Hisfs∗95 (Figure 6B). Assessment of the major-to-minor axis ratio at 12 hpf documented a slight but statistically significant yolk elongation in spred2 morphants, indicating enhanced MAPK signaling. Again, measuring the ratio in embryos co-injected with WT SPRED2 mRNA documented a rescue of the phenotype (Figure 6C). However, no rescue of the phenotype was observed in embryos co-injected with an equal amount of SPRED2Leu381Hisfs∗95 cDNA, confirming the LoF impact of the variant.
Figure 6.
Consequences of MO-mediated Spred2 silencing on early zebrafish embryonic development
(A) Phenotype classification (normal, mild, and severe) and quantification in embryos at around 11–12 hpf injected with antisense oligonucleotides (MO) (6 ng) against spred2a and spred2b with or without mRNA encoding WT SPRED2 (1.5 pg). Representative pictures for the different phenotypic classes are shown in the upper panel. Arrowheads indicate the head and tailbuds, black arrows show defective epiboly movements, white arrows indicate the major and minor axes. Scale bar: 125 μm.
(B) Overall number of embryos (%) exhibiting the different phenotypes (not injected, n = 24; MO, n = 19; MO + WT SPRED2, n = 19; MO + SPRED2Leu381Hisfs∗95 [SPRED2L381Hfs∗95], n = 27). Two-tailed Fisher exact test was used to compare the prevalence of “normal” and “severe” phenotypic classes (∗p < 0.05, ∗∗∗∗p < 0.0001).
(C) Quantification of the oval shape (major-to-minor axis ratio) (not injected, n = 25; MO, n = 26; MO + WT SPRED2, n = 16; MO + SPRED2Leu381Hisfs∗95 [SPRED2L381Hfs∗95], n = 22). Mean ± SEM is shown and one-way ANOVA with two-stage Benjamini, Krieger, and Yekutieli procedure was used for statistical assessment (∗p < 0.05, ∗∗∗∗p < 0.0001).
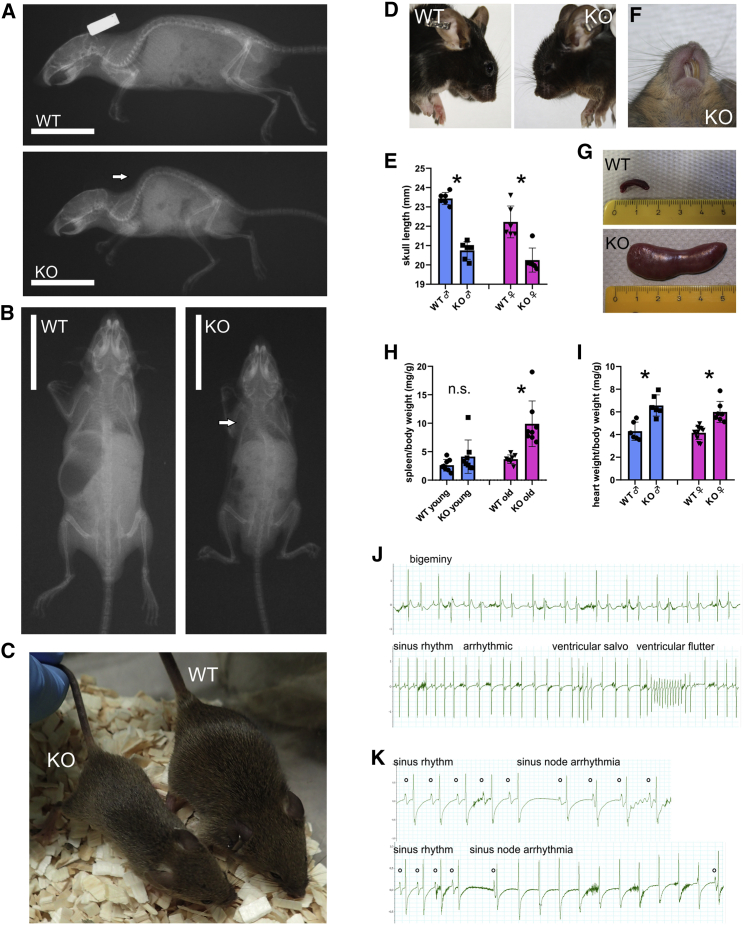
Spred2 KO mouse phenotyping
The previously generated Spred2−/− mouse model was phenotypically inspected to investigate the occurrence of the features and signs characterizing the subjects with bi-allelic inactivating SPRED2 variants.54 Compared to their age-matched and gender-matched Spred2+/+ littermates, Spred2−/− mice regularly developed kyphosis and scoliosis (Figures 7A and 7B). The skeletal defects were accompanied by growth retardation, as previously reported (Figure 7C).54 Of note, altered craniofacial development was also appreciated (Figure 7D). Specifically, mice showed a dome-shaped forehead and a shortened nose. Quantification of skull length in X-ray images revealed a gender-independent reduction of skull length in Spred2−/− mice (Figure 7E). These craniofacial features were often associated with a misalignment of the incisors (Figure 7F). Increasing with age, Spred2−/− mice developed splenomegaly (extreme example in Figure 7G), as documented by the increased spleen to body weight ratio (Figure 7H). Suggestive of cardiac hypertrophy, assessment of heart weight revealed a gender-independent increase of heart weight to body weight ratio in KO mice (Figure 7I), in line with the previously reported hypertrophic state of cardiomyocytes.67 In addition, consistent with previous observations,67 various ventricular cardiac arrhythmias were recorded in Spred2−/− mice (Figure 7J), as well supraventricular arrhythmias (Figure 7K). Notably, Spred2−/− mice showed an elevated volume of the ventricular system of the brain (Figure S9), and the dentate gyrus region of the hippocampus appeared not fully developed, which was accompanied by an enhanced Ras activity in this region (Figure S10). Furthermore, with low penetrance, mice developed dilated Bowman’s capsules, polycystic kidneys, and unilateral hydronephrosis (Figure S11). On the whole, the collected data indicated that Spred2−/− mice have craniofacial defects, reduced body size, splenomegaly, and cardiac hypertrophy with arrhythmias, similar to what is observed in the human disorder.
Figure 7.
Phenotypic assessment of Spred2 KO mice
(A) Soft X-ray images showing skeletal abnormalities in a Spred2−/− (KO) mouse compared to an age-matched WT littermate. Arrow labels kyphosis.
(B) X-ray images documenting scoliosis (arrow) in a KO mouse compared to an age-matched WT littermate.
(C) Representative photograph of male littermates at the age of 6 weeks showing growth retardation of a KO mouse (left) compared to an age-matched control animal (right).
(D) Comparison of the head shape of male littermates at the age of 6 months.
(E) Quantification of skull lengths taken from X-ray images (n = 6, each group; ∗p < 0.05).
(F) Representative photograph showing misaligned incisors in KO mice.
(G) Examples of spleens of mice at the age of 12 months showing a dramatic increase in spleen size of a KO mouse.
(H) Quantification of spleen to body weight ratios in young (6–8 weeks) and old (12 months) mice (n = 8 in each group; ∗p < 0.05).
(I) Comparison of heart weight to body weight ratios (n = 6 mice in each group; ∗p < 0.05).
(J) Examples of ventricular arrhythmias in KO mice at the age of 12 months.
(K) Examples of supraventricular arrhythmias in KO mice at the age of 12 months.
Discussion
Our findings provide evidence that loss of SPRED2 function underlies a recessive developmental disorder within the RASopathy clinical spectrum resembling NS. Complementary in silico, in vitro, and in vivo analyses documented that pathogenic SPRED2 variants can affect protein stability, causing accelerated degradation, but can also affect proper targeting of SPRED2 to the plasma membrane or impair its binding to neurofibromin. These effects converge toward an upregulation of MAPK signaling. Remarkably, the associated clinical features significantly differ from those resulting from SPRED1 haploinsufficiency occurring in LGSS.
SPRED2 is a member of a family of proteins functioning as negative regulators of signaling elicited by cell-surface receptor tyrosine kinases. Among these, four paralogs of the Sprouty subfamily (SPRY1-4) have been identified in mammals and documented to suppress the activation of the MAPK cascade induced in response to a variety of growth factors.68 Their negative modulatory function is exerted at the level of RAS activation by preventing SOS translocation to membranes and its positive modulatory role on RAS function.69 A negative regulatory role of these proteins on MAPK signaling by binding and inactivating RAF has also been suggested.70 On the other hand, the three members of the SPRED subfamily negatively control MAPK signaling by promoting binding of neurofibromin to activated, GTP-bound RAS.25 Among these, SPRED3 appears to have a lower inhibitory activity,39 possibly as a result of a reduced affinity in neurofibromin binding.59 Similarly, wide and partially overlapping expression patterns have been reported for SPRED1 and SPRED2 (Genotype-Tissue Expression Project [GTEx] portal, Human Protein Atlas), with the former being more broadly expressed during embryogenesis71 and the latter more extensively expressed in adult organs and tissues.35 In contrast, Spred3 expression appears more restricted, being limited to the brain (GTEx portal).39 These observations suggest a differential role of the three proteins in controlling MAPK signaling, which is in line with the different phenotypes characterizing the generated KO mice. Indeed, Spred1−/− mice show a lower body weight, a shortened face, spatial learning and memory deficits, and eosinophilia,72,73 while Spred2−/− mice were reported to have a markedly reduced growth and shorter long bones,54 cardiac fibrosis and arrhythmias, and a shortened lifespan.67 The different features previously documented in these models, together with those observed in the present study (see below), indicate a differential role of these proteins in controlling bone morphogenesis, hematopoiesis, cardiac function, and multiple cellular processes (present study),25,35,68 even though the finding that Spred1 and Spred2 KO mice are viable while Spred1/2 double KO mice are embryonic lethal supports partial redundancy in function.40
Similar to what is observed in affected individuals, Spred2−/− mice showed signs of cardiac arrhythmias and hypertrophy, reduced growth and weight, kyphosis, cranial anomalies, splenomegaly, and enlargement of the brain ventricle system. The observed shortened skull, the splenomegaly, and the cardiac hypertrophy in Spred2−/− mice resemble phenotypes of the Raf1L613V, KrasV14I, and Ptpn11D61G/+ mouse models of NS.74, 75, 76 However, neither the pulmonary valve stenosis documented in affected individuals was replicated in Spred2−/− mice, nor other changes in right ventricular hemodynamics were detected in mice. So far, malformation of the gyrus dentatus and kidney features were detected only in mice, possibly due to the small number of identified subjects with bi-allelic inactivating SPRED2 variants or/and young age of the affected subjects. The obsessive-compulsive disorder-like behavior observed in Spred2−/− mice is supportive for potential behavioral changes in affected individuals;77 on the other hand, no data are currently available concerning cognitive or learning aspects in these mice.
Among the members of the SPRY/SPRED family, only SPRED1 had previously been established to be implicated in a Mendelian disorder,13 while heterozygous missense variants of SPRY4 (MIM: 607984) had been reported as a contributing event in congenital hypogonadotropic hypogonadism (MIM: 615266), a condition characterized by oligogenic inheritance.78 Here, we report that SPRED2 is also causally associated with a Mendelian disorder. The four affected individuals from the three unrelated consanguineous families here reported were identified in the frame of a multicentric collaborative effort (NSEuroNet consortium). Two unrelated individuals were identified at the University Hospital of Magdeburg among 320 subjects with a tentative diagnosis of RASopathy found negative for mutations in previously known genes. One individual (3-II-1) was identified among 1,664 affected individuals screened at the Robert Debré Hospital, Paris, for diagnostic testing for NS or a related disorder. Finally, no bi-allelic occurrence of functionally relevant SPRED2 variants was identified among 40 subjects with a suspected clinical diagnosis of RASopathy, negative for pathogenic variants in known genes implicated in these disorders, who were assessed by exome sequencing (Ospedale Pediatrico Bambino Gesù, Rome) and among 471 affected individuals referred for genetic testing because of a suspected RASopathy screened at the Policlinico S. Orsola, Bologna, and Fondazione Casa Sollievo della Sofferenza, San Giovanni Rotondo (among these 57/471 were positive for pathogenic PTPN11 variants). Overall, these findings indicate that mutations in SPRED2 likely account for a relatively small proportion of affected individuals with clinical diagnosis suggestive of NS and are very uncommon in a non-consanguineous setting, though structural rearrangements (e.g., large intragenic deletions) and variants affecting regulatory and deep intronic regions were not systematically considered in our mutational scanning effort.
In contrast with the significant overlap between LGSS and NF1, the clinical features occurring in the identified subjects with bi-allelic inactivating SPRED2 variants are more closely related to NS (Table S1). LGSS, which is caused by heterozygous inactivating variants in SPRED1, is a substantially mild version of NF1.31 In LGSS, multiple CALMs freckling, DD, learning difficulties, and behavioral problems commonly occur. NS-like craniofacial features are also a relatively common finding, while Lisch nodules, neurofibromas, optic gliomas, malignant peripheral nerve sheath tumors, and NF1-specific bone lesions are essentially absent. Different from what is observed in LGSS, the phenotypes associated with disrupted SPRED2 function fit well within the clinical spectrum of NS, with short stature, a short/webbed neck with a low posterior hairline, ID, and craniofacial and thorax features resembling NS. Cardiac involvement (CHD and HCM), which is generally not observed in LGSS and is a recurrent feature of NS, was reported in three of the four subjects, and included ECG anomalies, HCM, and PVS. Of note, CALMs or axillary/inguinal freckling were not reported in these subjects. Moreover, differently from LGSS, the present condition is inherited as a recessive trait, which further supports partial redundancy. The distinct clinical presentations associated with SPRED1 haploinsufficiency and SPRED2 LoF support the idea that these proteins play different roles in development. While dedicated studies are required to dissect the role of these proteins in the modulation of RAS-MAPK signaling, multiple factors likely contribute to this picture. On one hand, the two genes are characterized by only partially overlapping expression patterns in adult tissues and during development, which are expected to contribute to the “unique” ability of the two proteins to control the amplitude and duration of activation of the MAPK cascade in response of various extracellular signals and in a specific temporal and spatial manner. On the other hand, it is also likely that the two proteins have different strengths in contributing to the tuning mechanism regulating the intensity and dynamics of MAPK signaling, and that the cellular context might be relevant for such function. Overall, both quantitative and qualitative aspects likely are relevant in explaining the diverse phenotype associated with decreased/loss of function of SPRED1 and SPRED2. While dedicated efforts are required to answer these questions, a noticeable finding of this study is the occurrence of conserved themes in the functional dysregulation associated with pathogenic SPRED1 and SPRED2 variants. Previous functional characterization of the molecular causes leading to SPRED1 haploinsufficiency in LGSS documented various aspects of the biochemical behavior, including defective protein synthesis or accelerated degradation, aberrant localization, and impaired interaction with neurofibromin, as the invariantly occurring events.32,34,59 The same key aspects were found to be affected by the bi-allelic SPRED2 variants implicated in the present recessive NS-like disorder.
To conclude, we provide evidence that inactivating bi-allelic variants in SPRED2 underlie a rare recessive clinical phenotype that is distinct from LGSS and resembles NS. These pathogenic variants affect different properties of the protein but are functionally equivalent, causing loss of SPRED2 function and upregulation of MAPK signaling. Further studies are required to more accurately understand the different modulatory function of SPRED proteins in intracellular signaling and development.
Acknowledgments
The authors thank the subjects and their families for participating in this study. This work was supported by grants from EJP-RD (NSEuroNet, to H.C., M.Z. [FZK 01GM1921A], and M.T.), AIRC (IG 21614 to M.T.), Ministero della Salute (Ricerca Corrente 2020 and 2021 to A.D.L. and M.T.; CCR-2017-23669081 and RCR-2020-23670068_001 to M.T.; RF-2018-12366931 to F.C.R., G.C., and B.D.), Ministero della Ricerca (FOE 2019 to M.T.), German Research Foundation (SCHU1600/6-1 to K.S.), and German Federal Ministry of Education and Research - BMBF (German Network for RASopathy Research “GeNeRARe” [FKZ: 01GM1902A]) to M.Z. This work has been carried out in the frame of the European Reference Network on Rare Congenital Malformations and Rare Intellectual Disability (ERN-ITHACA) activity.
Declaration of interests
The authors declare no competing interests.
Published: October 8, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.09.007.
Data and code availability
The pathogenic variants identified in this work have been submitted to ClinVar with accession numbers: c.187C>T (p.Arg63∗), SCV001809842; c.299T>C (p.Leu100Pro), SCV001805865; and c.1142_1143delTT (p.Leu381Hisfs∗95), SCV001805864.
Web resources
ExAC Browser, http://exac.broadinstitute.org/
gnomAD Browser, https://gnomad.broadinstitute.org/
GTEx Portal, https://gtexportal.org/home/
MetaDome, https://stuart.radboudumc.nl/metadome/
OMIM, https://www.omim.org/
pyBoxshade, https://github.com/mdbaron42/pyBoxshade
RCSB Protein Data Bank, http://www.rcsb.org/pdb/home/home.do
SWISS-MODEL, https://swissmodel.expasy.org/
The Human Protein Atlas, http://www.proteinatlas.org/
Supplemental Information
References
- 1.Yoon S., Seger R. The extracellular signal-regulated kinase: multiple substrates regulate diverse cellular functions. Growth Factors. 2006;24:21–44. doi: 10.1080/02699050500284218. [DOI] [PubMed] [Google Scholar]
- 2.Castellano E., Santos E. Functional specificity of ras isoforms: so similar but so different. Genes Cancer. 2011;2:216–231. doi: 10.1177/1947601911408081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson L., Greenbaum D., Cichowski K., Mercer K., Murphy E., Schmitt E., Bronson R.T., Umanoff H., Edelmann W., Kucherlapati R., Jacks T. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes Dev. 1997;11:2468–2481. doi: 10.1101/gad.11.19.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krens S.F., Spaink H.P., Snaar-Jagalska B.E. Functions of the MAPK family in vertebrate-development. FEBS Lett. 2006;580:4984–4990. doi: 10.1016/j.febslet.2006.08.025. [DOI] [PubMed] [Google Scholar]
- 5.Thomas G.M., Huganir R.L. MAPK cascade signalling and synaptic plasticity. Nat. Rev. Neurosci. 2004;5:173–183. doi: 10.1038/nrn1346. [DOI] [PubMed] [Google Scholar]
- 6.Fuentes-Mateos R., Jimeno D., Gómez C., Calzada N., Fernández-Medarde A., Santos E. Concomitant deletion of HRAS and NRAS leads to pulmonary immaturity, respiratory failure and neonatal death in mice. Cell Death Dis. 2019;10:838. doi: 10.1038/s41419-019-2075-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayashi S., Ogura Y. ERK signaling dynamics in the morphogenesis and homeostasis of Drosophila. Curr. Opin. Genet. Dev. 2020;63:9–15. doi: 10.1016/j.gde.2020.01.004. [DOI] [PubMed] [Google Scholar]
- 8.Tartaglia M., Gelb B.D. Disorders of dysregulated signal traffic through the RAS-MAPK pathway: phenotypic spectrum and molecular mechanisms. Ann. N Y Acad. Sci. 2010;1214:99–121. doi: 10.1111/j.1749-6632.2010.05790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pylayeva-Gupta Y., Grabocka E., Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat. Rev. Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rauen K.A. The RASopathies. Annu. Rev. Genomics Hum. Genet. 2013;14:355–369. doi: 10.1146/annurev-genom-091212-153523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simanshu D.K., Nissley D.V., McCormick F. RAS proteins and their regulators in human disease. Cell. 2017;170:17–33. doi: 10.1016/j.cell.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Upadhyaya M., Shen M., Cherryson A., Farnham J., Maynard J., Huson S.M., Harper P.S. Analysis of mutations at the neurofibromatosis 1 (NF1) locus. Hum. Mol. Genet. 1992;1:735–740. doi: 10.1093/hmg/1.9.735. [DOI] [PubMed] [Google Scholar]
- 13.Brems H., Chmara M., Sahbatou M., Denayer E., Taniguchi K., Kato R., Somers R., Messiaen L., De Schepper S., Fryns J.P., et al. Germline loss-of-function mutations in SPRED1 cause a neurofibromatosis 1-like phenotype. Nat. Genet. 2007;39:1120–1126. doi: 10.1038/ng2113. [DOI] [PubMed] [Google Scholar]
- 14.Martinelli S., De Luca A., Stellacci E., Rossi C., Checquolo S., Lepri F., Caputo V., Silvano M., Buscherini F., Consoli F., et al. Heterozygous germline mutations in the CBL tumor-suppressor gene cause a Noonan syndrome-like phenotype. Am. J. Hum. Genet. 2010;87:250–257. doi: 10.1016/j.ajhg.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamamoto G.L., Aguena M., Gos M., Hung C., Pilch J., Fahiminiya S., Abramowicz A., Cristian I., Buscarilli M., Naslavsky M.S., et al. Rare variants in SOS2 and LZTR1 are associated with Noonan syndrome. J. Med. Genet. 2015;52:413–421. doi: 10.1136/jmedgenet-2015-103018. [DOI] [PubMed] [Google Scholar]
- 16.Aoki Y., Niihori T., Inoue S., Matsubara Y. Recent advances in RASopathies. J. Hum. Genet. 2016;61:33–39. doi: 10.1038/jhg.2015.114. [DOI] [PubMed] [Google Scholar]
- 17.Tajan M., Paccoud R., Branka S., Edouard T., Yart A. The RASopathy family: consequences of germline activation of the RAS/MAPK pathway. Endocr. Rev. 2018;39:676–700. doi: 10.1210/er.2017-00232. [DOI] [PubMed] [Google Scholar]
- 18.Johnston J.J., van der Smagt J.J., Rosenfeld J.A., Pagnamenta A.T., Alswaid A., Baker E.H., Blair E., Borck G., Brinkmann J., Craigen W., et al. Members of the Undiagnosed Diseases Network Autosomal recessive Noonan syndrome associated with biallelic LZTR1 variants. Genet. Med. 2018;20:1175–1185. doi: 10.1038/gim.2017.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pagnamenta A.T., Kaisaki P.J., Bennett F., Burkitt-Wright E., Martin H.C., Ferla M.P., Taylor J.M., Gompertz L., Lahiri N., Tatton-Brown K., et al. DDD Study Delineation of dominant and recessive forms of LZTR1-associated Noonan syndrome. Clin. Genet. 2019;95:693–703. doi: 10.1111/cge.13533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maher G.J., Ralph H.K., Ding Z., Koelling N., Mlcochova H., Giannoulatou E., Dhami P., Paul D.S., Stricker S.H., Beck S., et al. Selfish mutations dysregulating RAS-MAPK signaling are pervasive in aged human testes. Genome Res. 2018;28:1779–1790. doi: 10.1101/gr.239186.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grant A.R., Cushman B.J., Cavé H., Dillon M.W., Gelb B.D., Gripp K.W., Lee J.A., Mason-Suares H., Rauen K.A., Tartaglia M., et al. Assessing the gene-disease association of 19 genes with the RASopathies using the ClinGen gene curation framework. Hum. Mutat. 2018;39:1485–1493. doi: 10.1002/humu.23624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niihori T., Nagai K., Fujita A., Ohashi H., Okamoto N., Okada S., Harada A., Kihara H., Arbogast T., Funayama R., et al. Germline-Activating RRAS2 Mutations Cause Noonan Syndrome. Am. J. Hum. Genet. 2019;104:1233–1240. doi: 10.1016/j.ajhg.2019.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Capri Y., Flex E., Krumbach O.H.F., Carpentieri G., Cecchetti S., Lißewski C., Rezaei Adariani S., Schanze D., Brinkmann J., Piard J., et al. Activating Mutations of RRAS2 Are a Rare Cause of Noonan Syndrome. Am. J. Hum. Genet. 2019;104:1223–1232. doi: 10.1016/j.ajhg.2019.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Motta M., Pannone L., Pantaleoni F., Bocchinfuso G., Radio F.C., Cecchetti S., Ciolfi A., Di Rocco M., Elting M.W., Brilstra E.H., et al. Enhanced MAPK1 Function Causes a Neurodevelopmental Disorder within the RASopathy Clinical Spectrum. Am. J. Hum. Genet. 2020;107:499–513. doi: 10.1016/j.ajhg.2020.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lorenzo C., McCormick F. SPRED proteins and their roles in signal transduction, development, and malignancy. Genes Dev. 2020;34:1410–1421. doi: 10.1101/gad.341222.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Legius E., Messiaen L., Wolkenstein P., Pancza P., Avery R.A., Berman Y., Blakeley J., Babovic-Vuksanovic D., Cunha K.S., Ferner R., et al. International Consensus Group on Neurofibromatosis Diagnostic Criteria (I-NF-DC) Revised diagnostic criteria for neurofibromatosis type 1 and Legius syndrome: an international consensus recommendation. Genet. Med. 2021;23:1506–1513. doi: 10.1038/s41436-021-01170-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scheffzek K., Ahmadian M.R., Wiesmüller L., Kabsch W., Stege P., Schmitz F., Wittinghofer A. Structural analysis of the GAP-related domain from neurofibromin and its implications. EMBO J. 1998;17:4313–4327. doi: 10.1093/emboj/17.15.4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bos J.L., Rehmann H., Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 29.De Luca A., Bottillo I., Sarkozy A., Carta C., Neri C., Bellacchio E., Schirinzi A., Conti E., Zampino G., Battaglia A., et al. NF1 gene mutations represent the major molecular event underlying neurofibromatosis-Noonan syndrome. Am. J. Hum. Genet. 2005;77:1092–1101. doi: 10.1086/498454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pinna V., Lanari V., Daniele P., Consoli F., Agolini E., Margiotti K., Bottillo I., Torrente I., Bruselles A., Fusilli C., et al. p.Arg1809Cys substitution in neurofibromin is associated with a distinctive NF1 phenotype without neurofibromas. Eur. J. Hum. Genet. 2015;23:1068–1071. doi: 10.1038/ejhg.2014.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brems H., Legius E. Legius syndrome, an Update. Molecular pathology of mutations in SPRED1. Keio J. Med. 2013;62:107–112. doi: 10.2302/kjm.2013-0002-re. [DOI] [PubMed] [Google Scholar]
- 32.Brems H., Pasmant E., Van Minkelen R., Wimmer K., Upadhyaya M., Legius E., Messiaen L. Review and update of SPRED1 mutations causing Legius syndrome. Hum. Mutat. 2012;33:1538–1546. doi: 10.1002/humu.22152. [DOI] [PubMed] [Google Scholar]
- 33.Stowe I.B., Mercado E.L., Stowe T.R., Bell E.L., Oses-Prieto J.A., Hernández H., Burlingame A.L., McCormick F. A shared molecular mechanism underlies the human rasopathies Legius syndrome and Neurofibromatosis-1. Genes Dev. 2012;26:1421–1426. doi: 10.1101/gad.190876.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan W., Markegard E., Dharmaiah S., Urisman A., Drew M., Esposito D., Scheffzek K., Nissley D.V., McCormick F., Simanshu D.K. Structural Insights into the SPRED1-Neurofibromin-KRAS Complex and Disruption of SPRED1-Neurofibromin Interaction by Oncogenic EGFR. Cell Rep. 2020;32:107909. doi: 10.1016/j.celrep.2020.107909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bundschu K., Walter U., Schuh K. Getting a first clue about SPRED functions. BioEssays. 2007;29:897–907. doi: 10.1002/bies.20632. [DOI] [PubMed] [Google Scholar]
- 36.King J.A., Straffon A.F., D’Abaco G.M., Poon C.L., i S.T., Smith C.M., Buchert M., Corcoran N.M., Hall N.E., Callus B.A., et al. Distinct requirements for the Sprouty domain for functional activity of Spred proteins. Biochem. J. 2005;388:445–454. doi: 10.1042/BJ20041284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wakioka T., Sasaki A., Kato R., Shouda T., Matsumoto A., Miyoshi K., Tsuneoka M., Komiya S., Baron R., Yoshimura A. Spred is a Sprouty-related suppressor of Ras signalling. Nature. 2001;412:647–651. doi: 10.1038/35088082. [DOI] [PubMed] [Google Scholar]
- 38.Lim J., Yusoff P., Wong E.S., Chandramouli S., Lao D.H., Fong C.W., Guy G.R. The cysteine-rich sprouty translocation domain targets mitogen-activated protein kinase inhibitory proteins to phosphatidylinositol 4,5-bisphosphate in plasma membranes. Mol. Cell. Biol. 2002;22:7953–7966. doi: 10.1128/MCB.22.22.7953-7966.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kato R., Nonami A., Taketomi T., Wakioka T., Kuroiwa A., Matsuda Y., Yoshimura A. Molecular cloning of mammalian Spred-3 which suppresses tyrosine kinase-mediated Erk activation. Biochem. Biophys. Res. Commun. 2003;302:767–772. doi: 10.1016/s0006-291x(03)00259-6. [DOI] [PubMed] [Google Scholar]
- 40.Taniguchi K., Kohno R., Ayada T., Kato R., Ichiyama K., Morisada T., Oike Y., Yonemitsu Y., Maehara Y., Yoshimura A. Spreds are essential for embryonic lymphangiogenesis by regulating vascular endothelial growth factor receptor 3 signaling. Mol. Cell. Biol. 2007;27:4541–4550. doi: 10.1128/MCB.01600-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Radio F.C., Pang K., Ciolfi A., Levy M.A., Hernández-García A., Pedace L., Pantaleoni F., Liu Z., de Boer E., Jackson A., et al. SPEN haploinsufficiency causes a neurodevelopmental disorder overlapping proximal 1p36 deletion syndrome with an episignature of X chromosomes in females. Am. J. Hum. Genet. 2021;108:502–516. doi: 10.1016/j.ajhg.2021.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin Y.C., Niceta M., Muto V., Vona B., Pagnamenta A.T., Maroofian R., Beetz C., van Duyvenvoorde H., Dentici M.L., Lauffer P., et al. Genomics England Research Consortium SCUBE3 loss-of-function causes a recognizable recessive developmental disorder due to defective bone morphogenetic protein signaling. Am. J. Hum. Genet. 2021;108:115–133. doi: 10.1016/j.ajhg.2020.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38, 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 45.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 46.Dunbrack R.L.J., Jr. Rotamer libraries in the 21st century. Curr. Opin. Struct. Biol. 2002;12:431–440. doi: 10.1016/s0959-440x(02)00344-5. [DOI] [PubMed] [Google Scholar]
- 47.Motta M., Chillemi G., Fodale V., Cecchetti S., Coppola S., Stipo S., Cordeddu V., Macioce P., Gelb B.D., Tartaglia M. SHOC2 subcellular shuttling requires the KEKE motif-rich region and N-terminal leucine-rich repeat domain and impacts on ERK signalling. Hum. Mol. Genet. 2016;25:3824–3835. doi: 10.1093/hmg/ddw229. [DOI] [PubMed] [Google Scholar]
- 48.Smith P.K., Krohn R.I., Hermanson G.T., Mallia A.K., Gartner F.H., Provenzano M.D., Fujimoto E.K., Goeke N.M., Olson B.J., Klenk D.C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 49.Westerfield M. Fourth Edition. Univ. of Oregon Press; Eugene: 2000. The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio) [Google Scholar]
- 50.LaFave M.C., Varshney G.K., Vemulapalli M., Mullikin J.C., Burgess S.M. A defined zebrafish line for high-throughput genetics and genomics: NHGRI-1. Genetics. 2014;198:167–170. doi: 10.1534/genetics.114.166769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bedell V.M., Westcot S.E., Ekker S.C. Lessons from morpholino-based screening in zebrafish. Brief. Funct. Genomics. 2011;10:181–188. doi: 10.1093/bfgp/elr021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robu M.E., Larson J.D., Nasevicius A., Beiraghi S., Brenner C., Farber S.A., Ekker S.C. p53 activation by knockdown technologies. PLoS Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bundschu K., Knobeloch K.P., Ullrich M., Schinke T., Amling M., Engelhardt C.M., Renné T., Walter U., Schuh K. Gene disruption of Spred-2 causes dwarfism. J. Biol. Chem. 2005;280:28572–28580. doi: 10.1074/jbc.M503640200. [DOI] [PubMed] [Google Scholar]
- 55.Messiaen L., Yao S., Brems H., Callens T., Sathienkijkanchai A., Denayer E., Spencer E., Arn P., Babovic-Vuksanovic D., Bay C., et al. Clinical and mutational spectrum of neurofibromatosis type 1-like syndrome. JAMA. 2009;302:2111–2118. doi: 10.1001/jama.2009.1663. [DOI] [PubMed] [Google Scholar]
- 56.Spencer E., Davis J., Mikhail F., Fu C., Vijzelaar R., Zackai E.H., Feret H., Meyn M.S., Shugar A., Bellus G., et al. Identification of SPRED1 deletions using RT-PCR, multiplex ligation-dependent probe amplification and quantitative PCR. Am. J. Med. Genet. A. 2011;155A:1352–1359. doi: 10.1002/ajmg.a.33894. [DOI] [PubMed] [Google Scholar]
- 57.Motta M., Fidan M., Bellacchio E., Pantaleoni F., Schneider-Heieck K., Coppola S., Borck G., Salviati L., Zenker M., Cirstea I.C., Tartaglia M. Dominant Noonan syndrome-causing LZTR1 mutations specifically affect the Kelch domain substrate-recognition surface and enhance RAS-MAPK signaling. Hum. Mol. Genet. 2019;28:1007–1022. doi: 10.1093/hmg/ddy412. [DOI] [PubMed] [Google Scholar]
- 58.Lindeboom R.G.H., Vermeulen M., Lehner B., Supek F. The impact of nonsense-mediated mRNA decay on genetic disease, gene editing and cancer immunotherapy. Nat. Genet. 2019;51:1645–1651. doi: 10.1038/s41588-019-0517-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hirata Y., Brems H., Suzuki M., Kanamori M., Okada M., Morita R., Llano-Rivas I., Ose T., Messiaen L., Legius E., Yoshimura A. Interaction between a Domain of the Negative Regulator of the Ras-ERK Pathway, SPRED1 Protein, and the GTPase-activating Protein-related Domain of Neurofibromin Is Implicated in Legius Syndrome and Neurofibromatosis Type 1. J. Biol. Chem. 2016;291:3124–3134. doi: 10.1074/jbc.M115.703710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Führer S., Ahammer L., Ausserbichler A., Scheffzek K., Dunzendorfer-Matt T., Tollinger M. NMR resonance assignments of the EVH1 domain of neurofibromin’s recruitment factor Spred1. Biomol. NMR Assign. 2017;11:305–308. doi: 10.1007/s12104-017-9768-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fragale A., Tartaglia M., Wu J., Gelb B.D. Noonan syndrome-associated SHP2/PTPN11 mutants cause EGF-dependent prolonged GAB1 binding and sustained ERK2/MAPK1 activation. Hum. Mutat. 2004;23:267–277. doi: 10.1002/humu.20005. [DOI] [PubMed] [Google Scholar]
- 62.Hanna N., Montagner A., Lee W.H., Miteva M., Vidal M., Vidaud M., Parfait B., Raynal P. Reduced phosphatase activity of SHP-2 in LEOPARD syndrome: consequences for PI3K binding on Gab1. FEBS Lett. 2006;580:2477–2482. doi: 10.1016/j.febslet.2006.03.088. [DOI] [PubMed] [Google Scholar]
- 63.Motta M., Sagi-Dain L., Krumbach O.H.F., Hahn A., Peleg A., German A., Lissewski C., Coppola S., Pantaleoni F., Kocherscheid L., et al. Activating MRAS mutations cause Noonan syndrome associated with hypertrophic cardiomyopathy. Hum. Mol. Genet. 2020;29:1772–1783. doi: 10.1093/hmg/ddz108. [DOI] [PubMed] [Google Scholar]
- 64.Anastasaki C., Estep A.L., Marais R., Rauen K.A., Patton E.E. Kinase-activating and kinase-impaired cardio-facio-cutaneous syndrome alleles have activity during zebrafish development and are sensitive to small molecule inhibitors. Hum. Mol. Genet. 2009;18:2543–2554. doi: 10.1093/hmg/ddp186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Runtuwene V., van Eekelen M., Overvoorde J., Rehmann H., Yntema H.G., Nillesen W.M., van Haeringen A., van der Burgt I., Burgering B., den Hertog J. Noonan syndrome gain-of-function mutations in NRAS cause zebrafish gastrulation defects. Dis. Model. Mech. 2011;4:393–399. doi: 10.1242/dmm.007112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jindal G.A., Goyal Y., Yamaya K., Futran A.S., Kountouridis I., Balgobin C.A., Schüpbach T., Burdine R.D., Shvartsman S.Y. In vivo severity ranking of Ras pathway mutations associated with developmental disorders. Proc. Natl. Acad. Sci. USA. 2017;114:510–515. doi: 10.1073/pnas.1615651114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ullrich M., Aßmus B., Augustin A.M., Häbich H., Abeßer M., Martin Machado J., Werner F., Erkens R., Arias-Loza A.P., Umbenhauer S., et al. SPRED2 deficiency elicits cardiac arrhythmias and premature death via impaired autophagy. J. Mol. Cell. Cardiol. 2019;129:13–26. doi: 10.1016/j.yjmcc.2019.01.023. [DOI] [PubMed] [Google Scholar]
- 68.Kawazoe T., Taniguchi K. The Sprouty/Spred family as tumor suppressors: Coming of age. Cancer Sci. 2019;110:1525–1535. doi: 10.1111/cas.13999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hanafusa H., Torii S., Yasunaga T., Nishida E. Sprouty1 and Sprouty2 provide a control mechanism for the Ras/MAPK signalling pathway. Nat. Cell Biol. 2002;4:850–858. doi: 10.1038/ncb867. [DOI] [PubMed] [Google Scholar]
- 70.Yusoff P., Lao D.H., Ong S.H., Wong E.S., Lim J., Lo T.L., Leong H.F., Fong C.W., Guy G.R. Sprouty2 inhibits the Ras/MAP kinase pathway by inhibiting the activation of Raf. J. Biol. Chem. 2002;277:3195–3201. doi: 10.1074/jbc.M108368200. [DOI] [PubMed] [Google Scholar]
- 71.Engelhardt C.M., Bundschu K., Messerschmitt M., Renné T., Walter U., Reinhard M., Schuh K. Expression and subcellular localization of Spred proteins in mouse and human tissues. Histochem. Cell Biol. 2004;122:527–538. doi: 10.1007/s00418-004-0725-6. [DOI] [PubMed] [Google Scholar]
- 72.Inoue H., Kato R., Fukuyama S., Nonami A., Taniguchi K., Matsumoto K., Nakano T., Tsuda M., Matsumura M., Kubo M., et al. Spred-1 negatively regulates allergen-induced airway eosinophilia and hyperresponsiveness. J. Exp. Med. 2005;201:73–82. doi: 10.1084/jem.20040616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Denayer E., Ahmed T., Brems H., Van Woerden G., Borgesius N.Z., Callaerts-Vegh Z., Yoshimura A., Hartmann D., Elgersma Y., D’Hooge R., et al. Spred1 is required for synaptic plasticity and hippocampus-dependent learning. J. Neurosci. 2008;28:14443–14449. doi: 10.1523/JNEUROSCI.4698-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Araki T., Mohi M.G., Ismat F.A., Bronson R.T., Williams I.R., Kutok J.L., Yang W., Pao L.I., Gilliland D.G., Epstein J.A., Neel B.G. Mouse model of Noonan syndrome reveals cell type- and gene dosage-dependent effects of Ptpn11 mutation. Nat. Med. 2004;10:849–857. doi: 10.1038/nm1084. [DOI] [PubMed] [Google Scholar]
- 75.Hernández-Porras I., Fabbiano S., Schuhmacher A.J., Aicher A., Cañamero M., Cámara J.A., Cussó L., Desco M., Heeschen C., Mulero F., et al. K-RasV14I recapitulates Noonan syndrome in mice. Proc. Natl. Acad. Sci. USA. 2014;111:16395–16400. doi: 10.1073/pnas.1418126111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu X., Simpson J., Hong J.H., Kim K.-H., Thavarajah N.K., Backx P.H., Neel B.G., Araki T. MEK-ERK pathway modulation ameliorates disease phenotypes in a mouse model of Noonan syndrome associated with the Raf1(L613V) mutation. J. Clin. Invest. 2011;121:1009–1025. doi: 10.1172/JCI44929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ullrich M., Weber M., Post A.M., Popp S., Grein J., Zechner M., Guerrero González H., Kreis A., Schmitt A.G., Üçeyler N., et al. OCD-like behavior is caused by dysfunction of thalamo-amygdala circuits and upregulated TrkB/ERK-MAPK signaling as a result of SPRED2 deficiency. Mol. Psychiatry. 2018;23:444–458. doi: 10.1038/mp.2016.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Miraoui H., Dwyer A.A., Sykiotis G.P., Plummer L., Chung W., Feng B., Beenken A., Clarke J., Pers T.H., Dworzynski P., et al. Mutations in FGF17, IL17RD, DUSP6, SPRY4, and FLRT3 are identified in individuals with congenital hypogonadotropic hypogonadism. Am. J. Hum. Genet. 2013;92:725–743. doi: 10.1016/j.ajhg.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The pathogenic variants identified in this work have been submitted to ClinVar with accession numbers: c.187C>T (p.Arg63∗), SCV001809842; c.299T>C (p.Leu100Pro), SCV001805865; and c.1142_1143delTT (p.Leu381Hisfs∗95), SCV001805864.