Summary
Congenital disorders of glycosylation (CDGs) form a group of rare diseases characterized by hypoglycosylation. We here report the identification of 16 individuals from nine families who have either inherited or de novo heterozygous missense variants in STT3A, leading to an autosomal-dominant CDG. STT3A encodes the catalytic subunit of the STT3A-containing oligosaccharyltransferase (OST) complex, essential for protein N-glycosylation. Affected individuals presented with variable skeletal anomalies, short stature, macrocephaly, and dysmorphic features; half had intellectual disability. Additional features included increased muscle tone and muscle cramps. Modeling of the variants in the 3D structure of the OST complex indicated that all variants are located in the catalytic site of STT3A, suggesting a direct mechanistic link to the transfer of oligosaccharides onto nascent glycoproteins. Indeed, expression of STT3A at mRNA and steady-state protein level in fibroblasts was normal, while glycosylation was abnormal. In S. cerevisiae, expression of STT3 containing variants homologous to those in affected individuals induced defective glycosylation of carboxypeptidase Y in a wild-type yeast strain and expression of the same mutants in the STT3 hypomorphic stt3-7 yeast strain worsened the already observed glycosylation defect. These data support a dominant pathomechanism underlying the glycosylation defect. Recessive mutations in STT3A have previously been described to lead to a CDG. We present here a dominant form of STT3A-CDG that, because of the presence of abnormal transferrin glycoforms, is unusual among dominant type I CDGs.
Keywords: congenital disorders of glycosylation, oligosaccharyltransferase complex, glycosylation, dominant inheritance
Introduction
Congenital disorders of glycosylation (CDGs) are a group of rare disorders that disrupt protein or lipid glycosylation and lead to a wide range of often multisystemic clinical presentations. Over 140 genetically distinct CDGs have been reported.1, 2, 3, 4 This number is expected to increase further because at least 2% of the human genome encodes proteins involved in glycosylation.5 CDGs that affect the biosynthesis of the lipid-linked oligosaccharide (LLO) in the ER and/or the transfer of the N-glycan to recipient precursor glycoproteins are termed type I CDGs.6
N-glycan transfer to a recipient protein is performed by the oligosaccharyltransferase (OST) complex. More specifically, the LLO is transferred from a lipid dolichol carrier to N-X-S/T sequons of nascent proteins. The OST is a multimeric protein complex containing either STT3A (OST-A complex) or STT3B (OST-B complex) as catalytic subunits and a range of accessory proteins.7,8 In humans, the OST-A and OST-B complexes both consist of six subunits (ribophorin-I, ribophorin-II, DDOST, TMEM258, DAD1, and OST4). The OST-A also contains the KRTCAP2 and OSTC subunits and the OST-B multimer either MAGT1 or TUSC3.7,9 In humans, the STT3A- and STT3B-containing OST complexes have slightly different functions and kinetic properties. The OST-A catalyzes the co-translational N-linked glycosylation of the majority of proteins upon entry into the ER, whereas the OST-B performs the posttranslational glycosylation of sites that are missed by OST-A, such as those located toward the C terminus of glycoproteins.10
In recent years, pathogenic variants in several OST subunits have been shown to result in autosomal-recessive or X-linked type I CDGs. These are OST48-CDG (MIM: 614507),11 TUSC3-CDG (MIM: 611093),12 MAGT1-CDG (MIM: 301031),13 STT3A-CDG (MIM: 615596), and STT3B-CDG (MIM: 615597).14 For STT3A-CDG, two missense variants in STT3A (GenBank: NM_001278503.1, c.1877T>C [p.Val626Ala] and c.1079A>C [p.Tyr360Ser]) have been reported in a total of nine individuals from four families in which only homozygous individuals were affected. In these individuals, STT3A-CDG led to a severe, mostly neurological phenotype consisting of developmental delay (DD), intellectual disability (ID), microcephaly, hypotonia, and seizures.14,15 Biochemically, affected individuals had abnormal serum transferrin glycosylation consistent with a type I CDG. Clotting abnormalities caused by factor VIII and vWF underglycosylation have also been reported in one case.16
Herein, we describe 16 individuals from nine families with a total of six dominantly inherited or de novo heterozygous missense variants in STT3A. Individuals were identified by a type 1 transferrin isoelectrofocusing pattern and showed a broad spectrum of clinical features, including facial dysmorphism, skeletal anomalies, variable DD, ID, speech delay, muscle cramps, and early onset severe osteoarthritis. In all cases, the heterozygous missense substitutions identified (GenBank: NM_001278503.1; c.137A>G [p.His46Arg]; c.479G>A [p.Arg160Gln]; c.985C>T [p.Arg329Cys]; c.1213C>T [p.Arg405Cys]; c.1214G>A [p.Arg405His]; c.1589A>C [p.Tyr530Ser]; c.1637C>T [p.Thr546Ile]) are located at or surrounding the catalytic site of STT3A. Together with biochemical studies in fibroblasts and yeast, our data indicate STT3A-CDG as the only described dominant type I CDG with abnormal N-glycosylation measurable by the detection of serum transferrin glycoforms. Therefore, STT3A-CDG presents with two forms of inheritance, autosomal recessive14,15 and inherited or de novo autosomal dominant as we describe in the present manuscript.
Material and methods
Ethics statement
Written informed consent (verbal assent when appropriate) was obtained from parents of legal guardians of all participants, and assent was obtained from the participants. Clinical, biochemical, genetic, and demographic data were collected in affected individuals as part of the CDG natural history study (IRB: 19-005187). Fibroblast cultures were collected as part of the clinical care. Residual, de-identified material was used in this study (including protein expression, mRNA expression, functional measurements) under ethics agreements from the Mayo Clinic, Rochester, MN, United States (IRB: 16-004682); University Hospital Leuven, Belgium (study number: S58358); and Radboud University Medical Center, the Netherlands (2019-5591; 2020-6588).
In case of research on human material, this was approved by the ethical committee of the University Hospital Leuven (study number: S58358; individuals 1.I.1, 1.II.1, 4.II.1, 5.II.1, 6.II.1), Radboud University Medical Center (study numbers: 2019-5591 and 2020-6588; individuals 1.I.1, 1.II.1, 1.II.2, 3.II.1, 9.I.1, 9.II.1), or an IRB-approved protocol (IRB: 16-004682; individual 2.I.1). This study was performed in accordance with ethical principles for medical research outlined in the Declaration of Helsinki. Consent was obtained with ethical approval in all cases. Individuals were gathered through the Euroglycanet and FCDGC networks following identification of heterozygous variants in STT3A by exome sequencing (ES) or genome sequencing (GS).
Cell lines
CRISPR/Cas9-engineered HEK293 knockout cell lines for either STT3A or STT3B were previously described.17 Primary fibroblasts from affected individuals and controls were grown from skin biopsies. Cell lines were cultured in DMEM/F12 (Life Technologies) supplemented with 10% FBS (Clone III, HyClones) and antibiotics streptomycin (100 μg/mL), penicillin (100 U/mL), and amphotericin (0.5 μg/mL) at 37°C under 5% CO2.
CDG glycosylation studies
Transferrin N-glycosylation was analyzed by isoelectric focusing, immunoblot, HPLC, mass spectrometry,18 or high-resolution quadrupole time of flight (QTOF) mass spectrometry.19 Analysis of lipid-linked and protein-linked oligosaccharide analysis was performed as described.20
Next-generation sequencing
All individuals were investigated via ES and/or GS as part of a cohort of individuals with biomarkers indicative of type I CDG. This was performed at several centers, and filtering and variant curation were prioritized on the basis of the analysis of known or predicted type I CDG-associated genes. In all individuals, no other pathogenic or probably pathogenic candidate variants were identified.
Genetic findings were confirmed by Sanger sequencing of genomic DNA surrounding each respective variant. When DNA from family members was available, inheritance patterns were also confirmed by Sanger sequencing of genomic DNA.
RNA extraction, real-time quantitative PCR (qPCR), and Sanger sequencing of cDNA
We isolated total RNA from cell lines with the RNeasy Mini kit (QIAGEN) and performed DNase treatment (Roche Diagnostics) to remove contaminating genomic DNA. Subsequently, reverse transcription was performed on 2 μg of purified total RNA with the First-Strand cDNA synthesis kit (GE Healthcare).
Transcribed cDNA was used for qPCRs for STT3A (GenBank: NM_001278503.1) and HPRT1 (GenBank: NM_000194), which was used as an endogenous control for normalization. PCR primers were designed via the NCBI primer-BLAST software and synthesized by Integrated DNA Technologies. PCR primer sequences can be found in Table S1. qPCRs were performed with the 2 × LightCycler 480 SYBR Green I Master. Data were analyzed with the LightCycler 480 Software (both Roche Applied Science).
Sanger sequencing of cDNA was performed by PCR amplification of the areas surrounding each respective variant, and primers used can be found in Table S1.
Immunoblotting
Protein lysates from affected and age-matched control fibroblasts were prepared by the addition of RIPA buffer (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS, and 1× cOmplete protease inhibitor cocktail [Sigma Aldrich]) at 4°C, mechanical lysis and incubation for 30 min followed by centrifugation at 4°C (15,000 rcf, 10 min). Supernatant was taken for analysis and the total protein concentration was calculated with the Pierce BCA protein assay kit (Thermo Fisher Scientific). Ten micrograms of protein lysate were analyzed by SDS/PAGE and immunoblotted onto a nitrocellulose membrane (Thermo Fisher Scientific) with the indicated antibodies. Signal detection was performed by autoradiography with ImageQuant LAS 4000 (GE Healthcare), and quantification was performed with the ImageJ software. All antibodies used for immunoblotting can be found in Table S2. S. cerevisiae protein lysates were prepared identically, with the exception that lysis was performed by the addition of 0.5 mm glass beads (Sigma Aldrich) followed by 4 × 1 min vortex cycles, alternating with 1 min on ice.
Yeast culture and preparation
Yeast strains were purchased from Euroscarf (Germany) and are reported in Table S3. Plasmids containing mutant and wild-type S. cerevisiae STT3 cDNA were based on the pESC-URA cloning vector and purchased from GenScript, NL. Briefly, STT3 cDNA was inserted downstream of the GAL10 inducible promoter and upstream of a FLAG-tag (DYKDDDDK). Complete sequences are available on request.
For transformation, yeast strains were first precultured overnight in YPED medium (2% glucose). Wild-type yeast was cultured at 30°C. The temperature-sensitive stt3-7 strain was cultured at 22°C. We then diluted cultures to 0.3 OD600 and incubated them for 4–6 h to ensure log phase conditions before their undergoing lithium-acetate transformation via a commercial Yeast Transformation Kit (Sigma Aldrich). We plated transformed yeast strains onto SD medium (2% glucose) lacking uracil (-URA) to allow selection of transformed colonies. Galactose induction of transformed yeast was performed by culturing colonies in SD medium (-URA) containing 2% galactose overnight before harvesting lysates for immunoblotting (see above).
Statistical analysis
All analyses were carried out via GraphPad Prism version 9.20 for Mac (GraphPad Software, La Jolla, CA). Statistical analysis was performed via one-way ANOVA followed by Tukey’s multiple comparisons test.
Results
Clinical description
We report on 16 affected individuals from nine families (eight females and eight males, age range 3–55 years) with variable dysmorphic features, short stature, musculoskeletal, and joint involvement. Ten individuals had variable degrees of developmental delay and/or intellectual disability (Table 1; Figures 1A–1H; Figure 2A).
Table 1.
Summary of genetic, laboratory, and clinical data for individuals with autosomal-dominant STT3A-CDG
| Family 1 | Family 2 | Family 3 | Family 4 | Family 5 | Family 6 | Family 7 | Family 8 | Family 9 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Individual | 1.I.1 (mother) | 1.II.1 | 1.II.3 | 2.I.1 (father) | 2.II.1 | 3.II.1 | 4.II.1 | 5.II.1 | 6.II.1 | 7.II.1 | 8.I.1 (father) | 8.II.1 | 8.II.2 | 8.II.3 | 9.I.1 (mother) | 9.II.1 |
| Sex | F | M | M | M | F | F | F | F | F | M | M | M | M | F | F | M |
| Current age | 55 years | 30 years | 27 years | 42 years | 3 years | 15 years | 24 years | 18 years | 12 years | 27 years | 39 years | 11 years | 7 years | 3 years | 30 years | 3 years |
| Consanguinity | – | – | – | – | – | no | no | No | no | no | no | no | no | no | no | no |
| Ancestry | European | European | European | European | European | European | Turkish | Anglo-Australian | European | European | ||||||
| Genetic and biochemical data | ||||||||||||||||
| Variant cDNA (GenBank: NM_001278503.1) | c.1637C>T | c.1589A>C | c.137A>G | c.479G>A | c.1213C>T | c.1213C>T | c.1213C>T | c.1214G>A | c.1214G>A | c.1214G>A | c.1214G>A | c.985C>T | c.985C>T | |||
| Variant protein | p.(Thr546Ile) | p.(Tyr530Ser) | p.(His46Arg) | p.(Arg160Gln) | p.(Arg405Cys) | p.(Arg405Cys) | p.(Arg405Cys) | p.(Arg405His) | p.(Arg405His) | p.(Arg405His) | p.(Arg405His) | p.(Arg329Cys) | p.(Arg329Cys) | |||
| Mode of inheritance | inherited | inherited | de novo | de novo | no parental DNA | de novo | no maternal DNA | no parental DNA | inherited | inherited | inherited | no parental DNA | inherited | |||
| Age at CDG diagnosis | 45 years | 20 years | 17 years | 39 years | 3 months | 10 years | 22 years | 8 years | 7 months | 14 years | 28 years | 9 months | 7 months | 3 weeks | 30 years | 3 years |
| Under-glycosylation | ∼25% (IEF) | ∼25% (IEF) | 38% (QTOF) | ∼12% (MS) | ∼18% (IEF) | 8% (HPLC) | WB profile CDG-1 | WB profile CDG-1 | ∼21% (CE) | 56% | 18% (HPLC) | 15%–18% (HPLC) | 13%–15% (HPLC) | 13% (HPLC) | 8.5% | 11.1% |
| Clinical symptoms and signs according to age | ||||||||||||||||
| Prenatal | ||||||||||||||||
| Intrauterine growth restriction | – | – | – | – | – | – | + | – | + | – | – | – | – | – | – | – |
| Pediatric age | ||||||||||||||||
| Dysmorphic features | ||||||||||||||||
| Head circumference | >95th pc | normal | 95th pc | >95th pc | 75th pc | normal | normal | normal | 3rd pc | 25th pc | >98th pc | ≫99th pc | ≫99th pc | >99th pc | normal | normal |
| Long face | – | – | – | – | – | – | – | – | + | – | – | + | + | – | – | – |
| High anterior hairline | + | – | + | + | + | – | – | – | + | + | – | + | + | + | – | – |
| Short palpebral fissures | – | + | – | + | – | – | – | – | + | + | – | + | – | + | – | – |
| Wide nasal bridge | + | – | – | – | – | – | – | – | + | + | – | + | + | + | – | – |
| Long/protruding ears | – | – | + | – | – | – | – | – | + | – | – | – | – | – | – | – |
| Thin upper lip vermilion | + | – | – | + | + | + | – | – | + | – | – | + | + | + | – | – |
| Prognathism | + | + | + | + | – | – | – | – | + | – | – | – | – | – | – | – |
| Inverted nipples | – | – | – | – | – | + | – | – | + | – | – | + | + | – | – | – |
| Abnormal fat-distribution | – | – | – | – | + | + | – | – | + | – | - | - | - | - | - | |
| Neurological | ||||||||||||||||
| Motor developmental delay | +(mild) | – | + (mild) | – | – | −4 | +(mild) | +(mild) | +(severe) | – | not known | +(mild) | + moderate | +(mild-mod.) | not known | not known |
| Speech delay | – | – | + | – | – | + (mild) | – | + | +(nonverbal) | – | not known | +moderate | +(nonverbal) | +(mild-mod.) | not known | not known |
| Learning problems | + | – | + | – | – | + | + | + | + | + | + | + | + (autism) | + | – | not known |
| Increased muscle tone | + | + | + | – | – | – | – | – | + (spastic diplegia) | + | N/A | – | –(hypotonia) | –(hypotonia) | – | – |
| Behavior abnormalities | – | – | – | – | – | – | – | – | aggressivity | – | aggressivity | – | – | – | not known | not known |
| Strabismus | – | – | – | – | – | – | – | – | +(retin. dystrophy) | – | – | – | + | – | not known | not known |
| Brain MRI | N/A | N/A | N/A | N/A | N/A | persistent cavum septum pellucidum | N/A | N/A | arachnoid cyst, large posterior fossa | N/A | N/A | N/A | perinatal subdural hematoma | N/A | N/A | N/A |
| Other | – | – | – | – | – | obesity | – | – | failure to thrive | – | – | constipation | – | – | jaundice | jaundice |
| Adolescence/adulthood | ||||||||||||||||
| Musculoskeletal involvement | ||||||||||||||||
| Short stature | −3 SD | −2 SD | −3 SD | −2 SD | – | – | −3 SD | −2 SD | −2 SD | <−2 SD | – | – | – | – | – | – |
| Skeletal abnormalities | + (meta-physeal dysplasia) | + | + (short arms) | – | – | – | + (epi-metaphyseal dysplasia) | + | + (spondylo-metaphyseal dysplasia brachydactyly) | + (scoliosis, dysplastic L5 vertebra) | – | + (delayed closure of large anterior fontanelle) | + (delayed closure of large anterior fontanelle) | + (large anterior fontanelle) | – | – |
| Osteoarthritis | + | + | + | + | – | – | – | – | – | – | – | – | – | – | not known | – |
| Muscle cramps | + | + | + | + | – | – | + | – | + | – | – | – | – | – | + | – |
| Muscle hypertrophy | – | – | + | – | – | – | + | N/A | + (biceps /quadriceps) | + (biceps /quadriceps) | – | – | – | – | – | – |
| Intellectual development | ||||||||||||||||
| Intellectual disability | + (mild) | – | + (mild) | – | N/A | + (mild) | + (moderate) | + | + (severe) | – | + (mild) | + (moderate) | + (severe) | N/A | – | N/A |
| Other | – | – | – | – | – | eczema, hyper-trichosis | normal pregnancy | – | mastoid granulomatosis | easy bruising | delayed puberty dysarthria | delayed puberty | – | – | spherocytosis | spherocytosis |
| Laboratory abnormalities | ||||||||||||||||
| Increased PT/APTT | – | – | – | – | – | – | – | – | + | N/A | – | – | +/− | – | – | – |
| Decreased coagulation factors/inhibitors | – | – | – | + (ferritin) | + (protein C) | – | + (FXI, VIII, IX) | – | + (FXI, VIII) + (protein C) | – | N/A | – | – | N/A | – | – |
| Increased serum transaminases | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Altered hormone levels | – | – | – | – | – | increased dehydroepiandrosterone | – | – | decreased prolactin and elevated TSH | decreased GH (on GH therapy) | – | – | – | – | – | – |
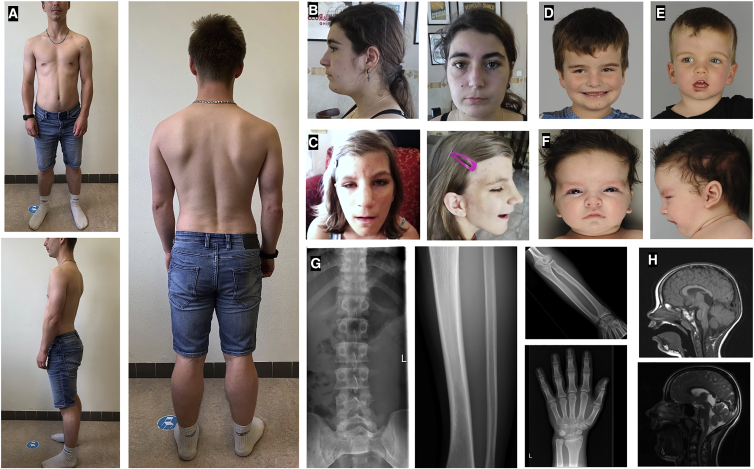
Figure 1.
Clinical images of affected individuals
(A) Individual 1.II.2; inverted nipples, long arms, shorter lower extremities, significant lordosis. Scoliosis and winged scapulae. Pseudohypertrophy of the calves.
(B) Individual 3.II.1; wide nasal base, low hairline, narrow nasal tip. Highly arched eyebrows.
(C) Individual 6.II.1; wide nasal base, bitemporal narrowing, low hairline, prominent nose, narrow nasal tip. Low-set ears, deeply set eyes, highly arched eyebrows.
(D and E) Individuals 8.II.1 and 8.II.3; macrocephaly, high anterior hairline, low-set ears, midface retrusion in both male siblings, downslanted palpebral fissures, short nose, and tented upper lip vermilion in younger brother.
(F) Individual 8.II.2; macrocephaly, high anterior hairline, narrow forehead, midface retrusion, short nose, low-set and posteriorly rotated ears, micrognathia, and down-turned corners of the mouth
(G) Individual 1.II.2; skeletal survey at age 16 years. Sclerotizing lesions of the sacroiliacal region, scoliosis. Bowing of the tibia and irregular shape of the ulna. Larger 2nd and 3rd metacarpals.
(H) Individual 6.II.1; brain MRI at ages 2 (T1) and 12 years (T2). Supratentorial, interhemispheric, non-progressive arachnoid cyst in the pyneal region having the same signal intensity as CSF. Slightly enlarged posterior fossa.
Figure 2.
Heterozygous variants in STT3A lead to variable clinical features, including facial dysmorphia and skeletal abnormalities
(A) Selected clinical features in individuals with dominant STT3A-CDG; N/A, not applicable. Features present in only single individuals are not shown. For a further summary of clinical features, see Table 1.
(B) Pedigrees of affected families showing presence of the variant in each family
All children were born at term. Two had intrauterine growth retardation. Six individuals had macrocephaly and one had progressive microcephaly. Dysmorphic features were subtle in most individuals and included high anterior hairline, short palpebral fissures, thin upper lip, broad base of the nose, and prognathism. Protruding ears were noted in four affected individuals (Figures 1A–1H). Three children had abnormal fat distribution and fat pads around the hips and four had inverted nipples. One child had hypertrichosis.
The children were generally healthy up until young adult age. There was a mild to moderate motor developmental delay in six individuals who were able to stand and walk around the age of 2 to 3 years. One child (6.II.1) developed severe motor delay with spasticity and ataxia. This individual was later diagnosed with retinal dystrophy, spastic diplegia, and an enlarged posterior fossa with an arachnoid cyst. This child, and one other, had strabismus. Increased muscle tone was noted in five individuals, while two had hypotonia. Speech delay was documented in seven children, and two of them developed no speech. One (3.II.1) was obese at the age of 10 years with a BMI of 29.1 (99th percentile). Gastrointestinal abnormalities were not significant. No cardiac, renal, or pulmonary symptoms were noted. Two children had spherocytosis.
A short stature was present in eight individuals and skeletal abnormalities in ten. Out of the ten individuals with skeletal abnormalities, three had mild metaphyseal flaring (in one affected individual [4.II.1] in combination with epiphyseal abnormalities), two had vertebral abnormalities, and one (1.II.3) had tibial and ulnar epiphyseal deformity and scoliosis on X-ray (Figures 1A–1H). Three children had a (late closing) large anterior fontanel. Four young adults had early signs of arthritis from the age of 30 years, necessitating bilateral hip replacement in one of the individuals (2.I.1) at the age of 40 years. Muscle cramps were present in seven individuals. Intellectual disability (ID) was diagnosed in nine individuals (one adult had a history of subtle learning difficulties but no ID).
Laboratory abnormalities were uncommon. Transaminase levels were normal. Decreased coagulation factors were detected in two individuals. Decreased testosterone levels were present in one child, and increased thyroid-stimulating hormone (TSH) level in another child, without clinical symptoms of hypothyroidism. Delayed puberty was detected in two cases. One child received growth hormone therapy.
Glycobiological findings
All affected individuals underwent metabolic testing, including screening for CDGs. In four out of nine families, CDG screening was abnormal in two generations, suggestive of dominant inheritance. All individuals showed a type 1 transferrin glycosylation profile. High-resolution mass spectrometry of intact transferrin indicated an increase of monoglycosylated transferrin in all individuals analyzed and a minor increase of nonglycosylated transferrin in some (Figure 3 and Table 1; Figure S1B). These results point to a biochemical defect in the synthesis of the LLO or its transfer to nascent proteins in the endoplasmic reticulum.
Figure 3.
Mass spectrometry analysis confirms N-glycosylation defect consistent with a type I CDG
Analysis of lipid-linked oligosaccharides and protein-linked oligosaccharides in fibroblasts from individuals 4.II.1, 5.II.1, and 6.II.1 showed that both these analytes were the same in affected individuals and control individuals (Figure S2). This suggests that the defect causing a type I CDG was not causing the accumulation of truncated LLO species, such as is seen in some CDG-I disorders upstream of the OST.20
Identification of heterozygous STT3A variants in all affected individuals
All individuals included in the study were investigated via ES or GS. No potentially pathogenic homozygous candidate variants were identified in any of the known or suspected CDG-associated genes. because affected individuals were present in multiple generations (families 1 and 2, five individuals in total), filtering was expanded to heterozygous variants in known type I CDG-associated genes, resulting in the identification of heterozygous missense variants in STT3A (Table 1; Figure 2B). On further analysis of other unsolved CDG-I cases, an additional 11 individuals were identified with potentially damaging heterozygous variants in STT3A. Only one of the variants (family 8) had been identified in population databases such as gnomAD:21 c.1214G>A (p.Arg405His), which has an allele frequency of 2/282,294. All the variants are conserved among species and predicted to be deleterious by in silico algorithms such as REVEL22 and CADD.23 No other candidate variants were identified in known CDG-associated genes.
The heterozygous missense variants segregated with the inheritance patterns observed in our cohort. The variants in family 1 (p.Thr546Ile), family 2 (p.Tyr530Ser), family 8 (p.Arg405His), and family 9 (p.Arg329Cys) were only present in affected individuals. In other affected individuals from which parental material was available, the variants were de novo (family 3 [p.His46Arg], family 4 [p.Arg160Gln], and family 6 [p.Arg405Cys]). The parents of probands in these cases were accordingly unaffected.
Effect of STT3A variants on RNA and protein expression and stability
In view of the recessive inheritance of known CDGs, we first embarked to exclude potentially undetected intronic variants affecting the expression of STT3A. RT-cDNA from individuals 1.I.1, 1.II.2, 4.II.1, 5.II.1, and 6.II.1 was produced from mRNA extracted from cultured primary dermal fibroblasts. qPCR analysis showed that expression in all individuals was similar to controls (Figure S3). In addition, Sanger sequencing at the location of the respective STT3A variants showed that the reference and mutant alleles were expressed at similar levels in all individuals (Figure 4A). The same analysis was also performed for individuals 3.II.1 and 7.II.1 In these cases, both alleles were also shown to be expressed equally (data not shown).
Figure 4.
Characterization of OST stability and glycosylation defects in fibroblasts from affected individuals
(A) Sequencing analysis of total mRNA from fibroblasts showed that both variant and wild-type alleles are expressed at similar levels in individuals 1.I.1, 1.II.2, 4.II.1, 5.II.1, and 6.II.1.
(B) Immunoblot analysis of STT3A and KRTCAP2 in whole cell protein lysates collected from fibroblasts of individuals 1.I.1, 1.II.2, 2.I.1 (STT3A only), 4.II.1, 5.II.1, and 6.II.1 shows levels of STT3A and KRTCAP2 are normal in affected individuals but reduced in previously identified AR STT3A-CDG individuals. Error bars indicate SEM of three repeated immunoblot analyses from three separate whole cell lysates from affected fibroblasts or controls. Average of controls is the mean of three immunoblot analyses from three whole cell lysates collected from three separate control fibroblast lines. STT3A levels of individual 2.I.1 were measured with a different antibody (invitrogen, see Table S2), resulting in altered immunoblot appearance. Representative immunoblot is shown.
(C) Analyses of fibroblasts from individual 2.I.1 via antibodies that recognize glycosylated epitopes show reduced signals for ICAM1 and LAMP1. Immunoblot analysis of fibroblasts from individuals 1.I.1, 1.II.2, 4.II.1, 5.II.1, and 6.II.1 show underglycosylation of LAMP1 indicated by increased electrophoretic mobility, similar to that seen in a STT3A−/− HEK293 cell line. No underglycosylation of LAMP1 was visualized in previously identified AR STT3A-CDG individuals
Heterozygous carriers of variants known to cause autosomal-recessive STT3A-CDG (c.1877T>C [p.Val626Ala] and c.1079A>C [p.Tyr360Ser]) are asymptomatic. However, these variants cause protein instability and lead to reduced steady state levels of STT3A.14,16 We therefore sought to elucidate whether the variants in individuals in our cohort cause haploinsufficiency of STT3A and whether this could lead to a CDG.
Having proven in fibroblasts that STT3A RNA is expressed at similar levels as controls, and that both the WT and mutant alleles are expressed similarly, we also investigated STT3A steady state levels in cultured fibroblasts from individuals 1.I.1, 1.II.2, 2.I.1, 4.II.1, 5.II.1, and 6.II.1 by immunoblotting and found them to be normal (Figure 4B). STT3A was undetectable in an STT3A−/− HEK293 cell line and reduced by 74% and 53% in two fibroblast lines from individuals with autosomal-recessive STT3A-CDG, confirming previous findings.14,16 In addition, the protein steady state levels of KRTCAP2 were also measured from the same fibroblasts. The OST-A subunit KRTCAP2 requires STT3A for stability within the OST-A and in the absence of STT3A, KRTCAP2 levels are greatly reduced.9 Indeed, in autosomal-recessive STT3A-CDG fibroblasts, KRTCAP2 levels were lowered by 61% and 68% (Figure 4B). In fibroblasts from individuals in our cohort, KRTCAP2 levels were found to be at least 80% those found in control fibroblasts. These data indicate that the intact OST-A complex is present in fibroblasts from individuals in our cohort and are present at the same or similar levels to those in controls. Together with results showing that expression of both alleles at RNA level was equal, this implies that both wild-type and mutant STT3A are present in the intact OST.
Abnormal glycosylation in affected fibroblasts
In order to confirm that the glycosylation abnormalities in fibroblasts were specific to OST-A (STT3A) dysfunction, we performed immunoblotting of LAMP1 from fibroblast lines derived from individuals 1.I.1, 1.II.2, 4.II.1, 5.II.1, and 6.II.1. LAMP1 is a heavily glycosylated lysosomal membrane protein that is underglycosylated in STT3A−/− HEK293 cells but not in STT3B−/− HEK293 cells. In affected individuals, all showed an intermediate LAMP1 hypoglycosylation pattern indicative of a partial deficiency of OST-A activity. The same pattern was not identified in previously described autosomal-recessive STT3A-CDG cases, indicating an alternate pathomechanism (Figure 4C). Similarly, immunoblotting for LAMP1 in fibroblasts of individual 2.I.1 showed a 58% reduced expression via an antibody that recognizes a glycosylated region of the protein. In the same individual, a similar reduction (49%) was seen in the levels of ICAM1, another heavily glycosylated membrane protein used as a marker of hypoglycosylation.
Molecular modeling of the mutations in STT3A and the OST complex
Molecular modeling of STT3A within the OST complex predicts that each of the substituted amino acids are localized at or near the catalytic site and are thus important for the transfer of the N-glycan to recipient glycoproteins (Figure 5A). This is supported by cryo-electron microscopy of the human OST-A complex,7 as well as recent work in yeast and bacteria showing the deleterious effect of active site STT3 substitutions.24,25 All variants identified in our cohort are in conserved regions of at least six amino acids in all eukaryotes (Figure 5B). Indeed, several of these residues (Arg329, Arg405, and Tyr530) have previously been experimentally determined as essential for OST function in a yeast model. Arg405 is known to stabilize dolichol-pyrophosphate (DolPP) during the nucleophilic substitution reaction while LLO transfer occurs, and Tyr530 forms a hydrogen bind with the reducing-end GlcNAc moiety.7,24 Indeed, multiple mutations induced at the yeast STT3 active site (p.Asp47Ala, p.Asp166Ala, p.Glu168Gln, p.Glu350Ala, p.Arg404Ala, and p.Lys586Ala) have been shown to affect metal ion, LLO, or protein substrate binding without affecting stability of the OST complex.24
Figure 5.
Molecular modeling and conservation of substituted amino acid residues in individuals with dominant STT3A-CDG
(A) Molecular modeling of STT3A active site showing the location of heterozygous missense variants identified in affected individuals. The 3D model (PDB: 6S7O) is derived from the cryo-EM structure of the OST-A collected by Ramirez et al.7 and visualized via PyMOL 2.4.2.
(B) Sequence alignment of the loci of heterozygous missense variants identified in affected individuals. Sequences shown are H. sapiens STT3A (UniProtKB: P46977) M. musculus stt3a (UniProtKB: P46978), H. sapiens STT3B (UniProtKB: Q8TCJ2), and S. cerevisiae STT3 (UniProtKB: P39007). Alignment performed with blastp (web resources).
S. cerevesiae modeling of substitutions homologous to STT3A heterozygous variants found in humans produces a dominant negative effect on N-glycosylation
The conservation in eukaryotes of the amino acid positions of the variants in our cohort allowed their study in yeast. WT (BY4741) S. cerevesiae were transformed with a galactose-inducible plasmid vector containing cDNA of either WT S. cerevesiae STT3 (transcript ID: YGL022W) or STT3 containing mutations corresponding to the conserved loci found in our cohort (Table S4). WT and mutant STT3 were also transformed into the previously studied stt3-7 strain containing the p.Ser552Pro substitution in STT3.26 The p.Ser552Pro variant leads to deficient oligosaccharyltransferase activity, measurable by immunoblotting as underglycosylation of carboxypeptidase Y (CPY), a marker of yeast N-glycosylation defects with four N-linked glycosylation sites (Figure 6A).26, 27, 28, 29
Figure 6.
S. cerevisiae as a model of dominant STT3A-CDG
(A) Schematic of the S. cerevisiae carboxypeptidase Y (CPY) protein showing the locations of four glycosylation sites found underglycosylated in the stt3-7 mutant S. cerevisiae strain.
(B) Immunoblot analysis of whole cell protein extracts from WT (BY4741) S. cerevisiae expressing STT3 containing missense substitutions found in affected individuals show an induced underglycosylation of CPY. Expression of WT and p.Val616Ala STT3 had no effect on CPY glycosylation. Culture conditions and plasmids used for transformation are indicated. Variants indicated correspond to those altered from the WT S. cerevisiae STT3 (Table S4). Average number of glycans and percentage underglycosylation were calculated from three replicate immunoblots; representative blot is shown. Error bars represent SEM.
(C) Immunoblot analysis of whole cell protein extracts from stt3-7 (STT3 p.Ser552Pro; BY4741) S. cerevisiae expressing STT3 containing missense substitutions found in affected individuals show increased underglycosylation of STT3. Culture conditions and plasmids used for transformation are indicated. Variants indicated correspond to those altered from the WT S. cerevisiae STT3 (Table S4). Average number of glycans and percentage underglycosylation were calculated from three replicate immunoblots; representative blot is shown. Error bars represent SEM.
Expression in WT yeast of STT3 containing substitutions at each locus corresponding to those found in our cohort induced clear underglycosylation of CPY ranging from 27%–40% of total CPY, whereas <3% is underglycosylated in WT yeast (Figure 6B). This indicates a dominant negative effect on N-linked glycosylation. Furthermore, expression of STT3 containing a variant homologous to that known to cause autosomal-recessive STT3A-CDG (human, p.Val626Ala; yeast, p.Val616Ala) had no effect on the glycosylation of CPY, supporting evidence that the pathomechanism of this variant is due to instability of STT3A containing p.Val626Ala.
In stt3-7 S. cerevesiae, expression of WT STT3 was able to partially rescue underglycosylation of CPY (2% galactose stt3-7 CPY = 80% underglycosylation; 2% galactose stt3-7 + WT STT3 CPY = 35% underglycosylation), whereas expression of the variants of the affected individuals in our cohort were unable to do so (Figure 6C). In fact, all but one of these variants (p.Tyr521Ser) worsened the observed N-glycosylation defect to 91%–94% underglycosylation. As well as providing further evidence of a dominant negative effect on N-glycosylation, this indicates that these variants are more detrimental to OST activity than p.Ser552Pro, previously found to cause a 98% reduction in oligosaccharyltransferase activity.26
Discussion
We report a dominant CDG caused by variants in a subunit of the OST complex caused by de novo and autosomal-dominantly inherited heterozygous missense variants close to the active site of STT3A. Because STT3A is the essential and catalytic subunit of the OST-A, this is likely to disrupt transfer of N-glycans onto recipient glycoproteins, causing a type I CDG. Most CDGs are autosomal-recessive disorders. However, recently several types caused by heterozygous variants have been identified, or dominant forms of existing recessive CDG. Several of these are closely linked to the biosynthesis or transfer of N-glycans to nascent glycoproteins (i.e., ALG8-CDG [MIM: 617874],30 ALG9-CDG [MIM: 263210],31 DHDDS-CDG [MIM: 617836],32 and NUS1-CDG [MIM: 617831]33). Others have less well-defined mechanisms leading to the dysregulation of trafficking or homeostasis in the ER and Golgi. These include SLC37A4-CDG (MIM: 619525)34 and dominant COG4-CDG (Saul-Wilson syndrome; [MIM: 618150]).35 Until now, only autosomal-recessive variants have been identified as leading to STT3A-CDG (MIM: 615596).
A combination of factors provides conclusive evidence for pathogenicity of the heterozygous STT3A variants identified in our cohort. First, the clustering of multiple variants at highly conserved amino acids around the catalytic site of STT3A, several of them de novo, in individuals with biomarkers indicative of type I CDG and a coherent shared phenotype. All individuals in this study have undergone ES or GS without the identification of further candidate variants in genes known or suspected to have a link to CDGs. Second, glycosylation abnormalities are present in the plasma and fibroblasts of affected individuals in this study. Underglycosylation of LAMP1 measured by immunoblotting resembles that seen in STT3A−/− HEK293 cells. Third, in a yeast model, expression of STT3 containing substitutions corresponding to those found in the affected individuals induced a glycosylation defect measured by underglycosylation of CPY, even when present alongside endogenous WT STT3 in an otherwise healthy yeast strain. This demonstrates a dominant negative effect of these mutant forms of STT3 upon N-glycosylation.
Clinically, autosomal-dominant STT3A-CDG is characterized by a variable phenotype consisting of intellectual disability, dysmorphic features, short stature, skeletal dysplasia, and muscle cramps. This expands the phenotypic spectrum of STT3A-CDG. The presence of muscle hypertrophy and muscle cramps have so far not been reported in other N-glycosylation defects. We detected these features in half of our cohort and mostly in adults. Another suggestive feature for the syndrome is the presence of osteoarthritis. Autosomal-recessive STT3A-CDG has been reported in association with failure to thrive, microcephaly, developmental delay, optic atrophy, and seizures, symptoms that were not common in the autosomal-dominant form. Unfortunately, because of the broad spectrum of clinical features, we could not delineate a genotype-phenotype correlation of the dominant versus recessive forms of STT3A-CDG.
Out of the twelve OST subunits, a CDG has been reported as caused by variants in only five of them. This is in contrast to steps upstream of the OST complex within the N-glycan and dolichol biosynthesis pathways. For example, pathogenic recessive variants have been identified at almost every known step of the Dolichol-P-P-GlcNAc2Man9Glc3 biosynthesis pathway.1,36 This could be due to the critical importance of the OST complex for glycan transfer. The previous missense variants identified in STT3A (leading to p.Val626Ala and p.Tyr360Ser) are both far from the catalytic site and pathogenicity is thought to be due to instability causing reduced protein levels rather than complete loss of the catalytic subunit.14 We have shown that in cases of dominant STT3A-CDG, the mutant protein is stable but most likely inactive. Probably, homozygous variants that fully ablate the activity of the OST complex are lethal in utero and only those that cause a partial perturbation of OST activity are compatible with life.
Mechanistically, it is important that all variants identified in this cohort are located at the catalytic region of STT3A. Some variants are predicted to alter the shape of the active site in a manner that could alter catalytic activity (c.137G>A [p.His46Arg], c.1589A>C [p.Tyr530Ser], and c.1637C>T [p.Thr546Ile]), whereas others are predicted to lead to the substitution of amino acid residues directly in contact with the LLO (c.479G>A [p.Arg160Gln], c.985C>T [p.Arg329Cys], c.1213C>T [p.Arg405Cys], and c.1214G>A [p.Arg405His]). Steady-state protein levels of STT3A were normal in all affected subjects, and cDNA showed similar expression of both the WT and mutant alleles. This indicates that the mutant forms of STT3A are conserved in an intact but catalytically dysfunctional OST-A.
One inherited variant identified in the cohort (c.1214G>A [p.Arg405His]; family 8) was identified in the gnomAD21 population database with an allele frequency of 2/292,294. One of these individuals is Swedish and the other is Estonian, and one of the two was 45–50 years old when genomic data was generated. Unfortunately, further detail on these two individuals is not available. However, it is possible that mildly affected individuals are present in gnomAD.21 In our cohort, several adults with no neurological phenotype or only mild learning disabilities are present.
Most enzyme deficiencies are recessive because of the active allele’s ability to compensate for the defective allele in cases of heterozygosity. However, as for the specific dominant mechanism in cases of dominant STT3A-CDG, it is possible that the abnormal OST-A complex successfully receives peptides through the ER translocon but is unable to catalyze the attachment of N-glycans, simply leading to underglycosylated proteins’ being released into the ER lumen. Because this translocation into the ER lumen is unidirectional, there is no way for the glycosylation of these proteins to be “rescued” (although partial rescue could be performed by the STT3B-containing OST-B17). In this way, the dominant effect could be caused by an increase of underglycosylated proteins in the ER lumen and further downstream in the Golgi and as secreted proteins that are dysfunctional. This offers an explanation as to why in cases of autosomal-recessive STT3A-CDG, heterozygous carriers are unaffected because the WT allele is able to perform adequate glycosylation to compensate for the reduced stability of the protein derived from the mutant allele.
As for the dominant form of STT3A-CDG described here, there are other possibilities as to the mechanism. For example, a change in active site conformation could exclude the nascent glycoprotein from entering the catalytic site altogether, thus “blocking” translocation. This explanation would however be difficult to reconcile with a dominant mechanism. Perhaps a more likely possibility is derived from the fact that the OST complex in both yeast and humans is known to hydrolyse the LLO and produce free oligosaccharides in the absence of a recipient asparagine for N-glycosylation. Indeed, upon destabilization of the OST complex caused by dominant C-terminal truncating mutations in TREX1, increased LLO hydrolysis and subsequent free glycan release occurs.37 Perhaps several of the variants in our cohort lead to increased LLO hydrolysis, inducing an N-glycosylation defect through this means.
The reported cases of recessive STT3A-CDG are characterized by a relatively severe disorder with mostly central nervous system involvement. Dominant STT3A-CDG appears to cause a more broad phenotype that includes muscle cramps and skeletal abnormalities and also causes underglycosylation of LAMP1 measured by immunoblotting; in recessive individuals, LAMP1 is normally glycosylated (Figure 4C).
It is therefore interesting to consider whether the pathomechanisms of the two forms of STT3A-CDG could differ and how this could explain the alternate biochemistry and phenotypes. It is possible that this difference lies in the fact that in the cases of dominant STT3A-CDG studied here, levels of OST complex proteins (including STT3A) in fibroblasts were normal, whereas in the two recessive individuals studied, levels appeared reduced. This would indicate that in the case of dominant STT3A-CDG, glycoproteins pass through the OST complex (and the translocon) but enter the ER lumen hypoglycosylated because of the pathogenic substitutions around the STT3A active site. Or, as mentioned above, the mechanism is in fact linked to hydrolysis of the LLO by mutant STT3A. If there is simply inadequate amounts of active OST to perform N-glycosylation in autosomal-recessive STT3A-CDG, this could be important. Quite simply, different glycoproteins could be underglycosylated in each of the two forms of the disorder, leading to alternate phenotypes and biochemistry.
However, when considering these differences, it should also be noted that recessive STT3A-CDG has only been identified in two families, which had a total of two different pathogenic variants. It is possible that the identification of additional affected individuals will expand the phenotypic spectrum.
To conclude, we describe a dominant form of STT3A-CDG caused by heterozygous missense variants at or surrounding the active site of STT3A that lead to a stable and intact OST complex but ablate oligosaccharyltransferase activity. Affected individuals have consistent biomarkers indicative of an N-linked glycosylation defect and a broad clinical presentation including variable intellectual disability, facial dysmorphisms, and musculoskeletal abnormalities. Further biochemical studies will reveal the precise dominant effect of the variants identified at the active site of STT3A.
Acknowledgments
The authors would like to thank the families that have participated in this study. Our thanks to François Foulquier and Geoffroy Bettignies for their valuable technical and intellectual input. Thank you to Reid Gilmore for supplying us with an antibody for detection of STT3A via immunoblotting. This research was supported by ERA-Net for Research on Rare Diseases (ERA-NET Cofund action; FWO GOI2918N; EUROGLYCAN-omics) (G.M., D.L., C.T.), a Marie Curie Individual Fellowship (H2020-MSCA-IF-2019; project ID: 894669) (M.P.W.), the Jaeken Theunissen CDG Fund, R01DK99551, and The Rocket Fund (H.H.F. and B.G.N.). Additional funding was supplied by 1U54NS115198-01 from the National Institute of Neurological Diseases and Stroke (NINDS), the National Center for Advancing Translational Sciences (NCATS), and the Rare Disorders Consortium Disease Network (RDCRN). This research was supported, in part, by the Instituto de Salud Carlos III (PI19/01310) and the Centro de Investigación Biomédica en Red de Enfermedades Raras (CIBERER). This study was also supported by the Agència de Gestió d’Ajuts Universitaris i de Recerca (AGAUR, 2017:SGR 1428) and the CERCA Programme/Generalitat de Catalunya.
Declaration of interests
The authors declare no competing interests.
Published: October 14, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.09.012.
Contributor Information
Matthew P. Wilson, Email: matthew.wilson@kuleuven.be.
Dirk J. Lefeber, Email: dirk.lefeber@radboudumc.nl.
Data and code availability
This study did not generate datasets or code.
Web resources
blastp, https://blast.ncbi.nlm.nih.gov/
Genome Aggregation Database (gnomAD), https://gnomad.broadinstitute.org/
Online Mendelian Inheritance in Man (OMIM), https://www.omim.org/
Supplemental information
References
- 1.Ondruskova N., Cechova A., Hansikova H., Honzik T., Jaeken J. Congenital disorders of glycosylation: Still “hot” in 2020. Biochim. Biophys. Acta, Gen. Subj. 2021;1865:129751. doi: 10.1016/j.bbagen.2020.129751. [DOI] [PubMed] [Google Scholar]
- 2.Jaeken J., Péanne R. What is new in CDG? J. Inherit. Metab. Dis. 2017;40:569–586. doi: 10.1007/s10545-017-0050-6. [DOI] [PubMed] [Google Scholar]
- 3.Freeze H.H. Improving biochemical markers for disorders of N-glycosylation. Ann. Transl. Med. 2019;7(Suppl 6):S176. doi: 10.21037/atm.2019.07.79. S176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ng B.G., Freeze H.H. Perspectives on Glycosylation and Its Congenital Disorders. Trends Genet. 2018;34:466–476. doi: 10.1016/j.tig.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freeze H.H., Chong J.X., Bamshad M.J., Ng B.G. Solving glycosylation disorders: fundamental approaches reveal complicated pathways. Am. J. Hum. Genet. 2014;94:161–175. doi: 10.1016/j.ajhg.2013.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaeken J., Hennet T., Matthijs G., Freeze H.H. CDG nomenclature: Time for a change! Biochim. Biophys. Acta - Mol. Basis Dis. 2009;1792:825–826. doi: 10.1016/j.bbadis.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramírez A.S., Kowal J., Locher K.P. Cryo-electron microscopy structures of human oligosaccharyltransferase complexes OST-A and OST-B. Science. 2019;366:1372–1375. doi: 10.1126/science.aaz3505. [DOI] [PubMed] [Google Scholar]
- 8.Lu H., Fermaintt C.S., Cherepanova N.A., Gilmore R., Yan N., Lehrman M.A. Mammalian STT3A/B oligosaccharyltransferases segregate N-glycosylation at the translocon from lipid-linked oligosaccharide hydrolysis. Proc. Natl. Acad. Sci. USA. 2018;115:9557–9562. doi: 10.1073/pnas.1806034115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shrimal S., Cherepanova N.A., Gilmore R. DC2 and KCP2 mediate the interaction between the oligosaccharyltransferase and the ER translocon. J. Cell Biol. 2017;216:3625–3638. doi: 10.1083/jcb.201702159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cherepanova N.A., Venev S.V., Leszyk J.D., Shaffer S.A., Gilmore R. Quantitative glycoproteomics reveals new classes of STT3A- and STT3B-dependent N-glycosylation sites. J. Cell Biol. 2019;218:2782–2796. doi: 10.1083/jcb.201904004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones M.A., Ng B.G., Bhide S., Chin E., Rhodenizer D., He P., Losfeld M.E., He M., Raymond K., Berry G., et al. DDOST mutations identified by whole-exome sequencing are implicated in congenital disorders of glycosylation. Am. J. Hum. Genet. 2012;90:363–368. doi: 10.1016/j.ajhg.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al-Amri A., Saegh A.A., Al-Mamari W., El-Asrag M.E., Ivorra J.L., Cardno A.G., Inglehearn C.F., Clapcote S.J., Ali M. Homozygous single base deletion in TUSC3 causes intellectual disability with developmental delay in an Omani family. Am. J. Med. Genet. A. 2016;170:1826–1831. doi: 10.1002/ajmg.a.37690. [DOI] [PubMed] [Google Scholar]
- 13.Blommaert E., Péanne R., Cherepanova N.A., Rymen D., Staels F., Jaeken J., Race V., Keldermans L., Souche E., Corveleyn A., et al. Mutations in MAGT1 lead to a glycosylation disorder with a variable phenotype. Proc. Natl. Acad. Sci. USA. 2019;116:9865–9870. doi: 10.1073/pnas.1817815116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shrimal S., Ng B.G., Losfeld M.E., Gilmore R., Freeze H.H. Mutations in STT3A and STT3B cause two congenital disorders of glycosylation. Hum. Mol. Genet. 2013;22:4638–4645. doi: 10.1093/hmg/ddt312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh A., Urquhart J., Daly S., Ferguson A., Scotcher D., Morris A.A.M., Clayton-Smith J. Phenotypic Heterogeneity in a Congenital Disorder of Glycosylation Caused by Mutations in STT3A. J. Child Neurol. 2017;32:560–565. doi: 10.1177/0883073817696816. [DOI] [PubMed] [Google Scholar]
- 16.Chang I.J., Byers H.M., Ng B.G., Merritt J.L., 2nd, Gilmore R., Shrimal S., Wei W., Zhang Y., Blair A.B., Freeze H.H., et al. Factor VIII and vWF deficiency in STT3A-CDG. J. Inherit. Metab. Dis. 2019;42:325–332. doi: 10.1002/jimd.12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cherepanova N.A., Gilmore R. Mammalian cells lacking either the cotranslational or posttranslocational oligosaccharyltransferase complex display substrate-dependent defects in asparagine linked glycosylation. Sci. Rep. 2016;6:20946. doi: 10.1038/srep20946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carchon H.A., Chevigné R., Falmagne J.B., Jaeken J. Diagnosis of congenital disorders of glycosylation by capillary zone electrophoresis of serum transferrin. Clin. Chem. 2004;50:101–111. doi: 10.1373/clinchem.2003.021568. [DOI] [PubMed] [Google Scholar]
- 19.van Scherpenzeel M., Steenbergen G., Morava E., Wevers R.A., Lefeber D.J. High-resolution mass spectrometry glycoprofiling of intact transferrin for diagnosis and subtype identification in the congenital disorders of glycosylation. Transl. Res. 2015;166:639–649.e1. doi: 10.1016/j.trsl.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 20.Thiel C., Rind N., Popovici D., Hoffmann G.F., Hanson K., Conway R.L., Adamski C.R., Butler E., Scanlon R., Lambert M., et al. Improved diagnostics lead to identification of three new patients with congenital disorder of glycosylation-Ip. Hum. Mutat. 2012;33:485–487. doi: 10.1002/humu.22019. [DOI] [PubMed] [Google Scholar]
- 21.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ioannidis N.M., Rothstein J.H., Pejaver V., Middha S., McDonnell S.K., Baheti S., Musolf A., Li Q., Holzinger E., Karyadi D., et al. REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am. J. Hum. Genet. 2016;99:877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rentzsch P., Schubach M., Shendure J., Kircher M. CADD-Splice-improving genome-wide variant effect prediction using deep learning-derived splice scores. Genome Med. 2021;13:31. doi: 10.1186/s13073-021-00835-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wild R., Kowal J., Eyring J., Ngwa E.M., Aebi M., Locher K.P. Structure of the yeast oligosaccharyltransferase complex gives insight into eukaryotic N-glycosylation. Science. 2018;359:545–550. doi: 10.1126/science.aar5140. [DOI] [PubMed] [Google Scholar]
- 25.Yamasaki T., Kohda D. Uncoupling the hydrolysis of lipid-linked oligosaccharide from the oligosaccharyl transfer reaction by point mutations in yeast oligosaccharyltransferase. J. Biol. Chem. 2020;295:16072–16085. doi: 10.1074/jbc.RA120.015013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spirig U., Glavas M., Bodmer D., Reiss G., Burda P., Lippuner V., te Heesen S., Aebi M. The STT3 protein is a component of the yeast oligosaccharyltransferase complex. Mol. Gen. Genet. 1997;256:628–637. doi: 10.1007/s004380050611. [DOI] [PubMed] [Google Scholar]
- 27.Körner C., Knauer R., Stephani U., Marquardt T., Lehle L., von Figura K. Carbohydrate deficient glycoprotein syndrome type IV: deficiency of dolichyl-P-Man:Man(5)GlcNAc(2)-PP-dolichyl mannosyltransferase. EMBO J. 1999;18:6816–6822. doi: 10.1093/emboj/18.23.6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imbach T., Burda P., Kuhnert P., Wevers R.A., Aebi M., Berger E.G., Hennet T. A mutation in the human ortholog of the Saccharomyces cerevisiae ALG6 gene causes carbohydrate-deficient glycoprotein syndrome type-Ic. Proc. Natl. Acad. Sci. USA. 1999;96:6982–6987. doi: 10.1073/pnas.96.12.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cantagrel V., Lefeber D.J., Ng B.G., Guan Z., Silhavy J.L., Bielas S.L., Lehle L., Hombauer H., Adamowicz M., Swiezewska E., et al. SRD5A3 is required for converting polyprenol to dolichol and is mutated in a congenital glycosylation disorder. Cell. 2010;142:203–217. doi: 10.1016/j.cell.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Besse W., Dong K., Choi J., Punia S., Fedeles S.V., Choi M., Gallagher A.R., Huang E.B., Gulati A., Knight J., et al. Isolated polycystic liver disease genes define effectors of polycystin-1 function. J. Clin. Invest. 2017;127:1772–1785. doi: 10.1172/JCI90129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Besse W., Chang A.R., Luo J.Z., Triffo W.J., Moore B.S., Gulati A., Hartzel D.N., Mane S., Torres V.E., Somlo S., Mirshahi T., Regeneron Genetics Center ALg9 mutation carriers develop kidney and liver cysts. J. Am. Soc. Nephrol. 2019;30:2091–2102. doi: 10.1681/ASN.2019030298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamdan F.F., Myers C.T., Cossette P., Lemay P., Spiegelman D., Laporte A.D., Nassif C., Diallo O., Monlong J., Cadieux-Dion M., et al. Deciphering Developmental Disorders Study High Rate of Recurrent De Novo Mutations in Developmental and Epileptic Encephalopathies. Am. J. Hum. Genet. 2017;101:664–685. doi: 10.1016/j.ajhg.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu S.H., Wang T., Wiggins K., Louie R.J., Merino E.F., Skinner C., Cassera M.B., Meagher K., Goldberg P., Rismanchi N., et al. Lysosomal cholesterol accumulation contributes to the movement phenotypes associated with NUS1 haploinsufficiency. Genet. Med. 2021;23:1305–1314. doi: 10.1038/s41436-021-01137-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ng B.G., Sosicka P., Fenaille F., Harroche A., Vuillaumier-Barrot S., Porterfield M., Xia Z.-J., Wagner S., Bamshad M.J., Vergnes-Boiteux M.-C., et al. University of Washington Center for Mendelian Genomics (UW-CMG) A mutation in SLC37A4 causes a dominantly inherited congenital disorder of glycosylation characterized by liver dysfunction. Am. J. Hum. Genet. 2021;108:1040–1052. doi: 10.1016/j.ajhg.2021.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferreira C.R., Xia Z.J., Clément A., Parry D.A., Davids M., Taylan F., Sharma P., Turgeon C.T., Blanco-Sánchez B., Ng B.G., et al. Undiagnosed Diseases Network. Scottish Genome Partnership A Recurrent De Novo Heterozygous COG4 Substitution Leads to Saul-Wilson Syndrome, Disrupted Vesicular Trafficking, and Altered Proteoglycan Glycosylation. Am. J. Hum. Genet. 2018;103:553–567. doi: 10.1016/j.ajhg.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Varki A., Cummings R.D., Esko J.D., Stanley P., Hart G.W., Aebi M., Darvill A.G., Kinoshita T., Packer N.H., Prestegard J.H., et al. Cold Spring Harbor Laboratory Press; 2017. Essentials of Glycobiology. [PubMed] [Google Scholar]
- 37.Hasan M., Fermaintt C.S., Gao N., Sakai T., Miyazaki T., Jiang S., Li Q.Z., Atkinson J.P., Morse H.C., 3rd, Lehrman M.A., Yan N. Cytosolic Nuclease TREX1 Regulates Oligosaccharyltransferase Activity Independent of Nuclease Activity to Suppress Immune Activation. Immunity. 2015;43:463–474. doi: 10.1016/j.immuni.2015.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate datasets or code.