Abstract
Exploring reliable and highly-sensitive SARS-CoV-2 antibody diagnosis by point-of-care (POC) manner, holds great public health significance for extensive COVID-19 screening and controlling. Unfortunately, the currently applied gold based lateral flow immunoassay (GLFIA) may expose both false-negative and false-positive interpretations owing to the sensitivity and specificity limitations, which may cause significant risk and waste of public resources for large population screening. To simultaneously overcome the drawbacks of GLFIA, a novel fluorescent LFIA based on signal amplification and dual-antigen sandwich structure was established with largely improved sensitivity and specificity. The compact three-dimensional incorporation of hydrophobic quantum dots within dendritic affinity templates and multilayer surface derivation guaranteed a high and robust fluorescence of single label, which lowered the false negative rate of GLFIA prominently. A dual-antigen sandwich structure using labeled/immobilized SARS-CoV-2 spike receptor binding domain antigen for capturing total human SARS-CoV-2 antibody was developed, instead of general indirect antibody capturing approach, to reduce the false positive rate of GLFIA. Over 300 cases of COVID-19 negative and 97 cases of COVID-19 positive samples, the current assay revealed a 100% sensitivity and 100% specificity confirmed by both polymerase chain reaction (PCR) and chemiluminescence immunoassay (CLIA), compared with the considerable misinterpretation cases by currently applied GLFIA. The quantitative results verified by receiver operating characteristic curve and other statistical analysis indicated a well-distinguished positive/negative sample groups. The proposed strategy is highly sensitive towards low concentrated SARS-CoV-2 antibody serums and highly specific towards serums from COVID-19 negative persons and patients infected by other viruses.
Keywords: Quantum dots assembly, Signal amplification, Dual-antigen sandwich immunoassay, Point-of-care testing, Misinterpretation
1. Introduction
COVID-19, which is caused by human pathogenic virus-severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has been declared as a pandemic by the World Health Organization on March 11, 2020, and seriously threatened the global public health for more than one year (Zhou et al., 2020), and also resulted in massive economic and social damages. Due to high transmissivity and asymptomatic carriage rate of SARS-CoV-2, well-control of the pandemic is an unprecedented challenge. Although different SARS-CoV-2 vaccines have been developed and gradually put into use, their effectiveness still need to be verified (Alturki et al., 2020; Moore and Klasse, 2020). So far, the most effective way to prevent the spread of pandemic is to detect SARS-CoV-2 infection in asymptomatic and presymptomatic individuals in the mass population. Therefore, lateral flow immunoassay (LFIA) as a promising point-of-care (POC) method (Gong et al., 2017; Hu et al., 2019; Huang et al., 2013; Wang et al., 2019), is exerting a key role in the epidemic prevention and control because of the characteristics of rapidity, portability and affordability (Hu et al., 2017; Huang et al., 2016; Wang et al., 2020a; Wu et al., 2016).
The colloidal gold lateral flow immunoassay (GLFIA) is the most commonly applied LFIA, which can be read visually without the requirement of sophisticated facilities and healthcare professionals (Li et al., 2020b; Liu et al., 2020a). The major drawback of this method refers to the low sensitivity with limited precise quantification ability and detection sensitivity. Therefore, a certain false negative cases would be mistakenly considered as non-infectious, resulting in a huge risk of further transmission in the community (Chen et al., 2020; Huang et al., 2013; Koller et al., 2021). Compared with colorimetric analysis, the fluorometric LFIA provides improved signal contrast and lower background interference (Huang et al., 2020; Wang et al., 2021). Quantum dots (QDs) as an emerging class of fluorescent label exhibit high single-particle brightness, none photo-bleaching and versatile nanostructure engineering for integration and functionalization (Zhou et al., 2015). Up to date, a series of synthetic strategies have been developed for water-dispersible QDs structures. The micellization is a widely adopted and effective approach for phase-transfer and assembling of hydrophobic QDs (Guo et al., 2019; Huang et al., 2012; Zhou et al., 2011). To achieve a better control over the dimension and monodispersity of the QDs composites, the surface assembling of QDs by silica colloids (Huang et al., 2014; Lee et al., 2010; Lu et al., 2011) and near surface encapsulating of QDs by polystyrene latex were developed (Han et al., 2001; Hu et al., 2016; Zhang et al., 2016). Yet, the above templated synthetic approaches could not utilize the inner space of a template to achieve high units packing density and high single-colloid brightness. Compared with traditional mesoporous silica, the dendritic silica colloid with central radial pore structure is a promising nano-carrier for bio-macromolecules and nanoparticles with three dimensional incorporation manner (Yang et al., 2016; Yue et al., 2015). The integration of dendritic silica templates with high-quality hydrophobic quantum dots would effectively enhance the single-label brightness for sensitive SARS-CoV-2 antibody detection.
Moreover, the current SARS-CoV-2 antibody immunoassays (including GLFIA) are predominantly based on the indirect format, which utilizes immobilized antigen and anti-human IgG/IgM antibody for sandwiching the analyte antibody (Liu et al., 2020b). Nevertheless, in a large population screening, there are still false positive cases in an indirect detection (Feng et al., 2020; Shaw et al., 2020). In addition to the target antibodies, there are extremely high concentrated non-targeted primary antibodies in serum matrix, some of which with strong hydrophobic binding ability would adhere to the nano-label surface or membrane surface. The immobilized or labeled secondary antibodies may react with any primary antibodies including the non-specifically absorbed ones to produce high background or false positive results (Boukli et al., 2020; Ge et al., 2012). In addition, rheumatoid factors (RF) in the samples, for similar reasons, will also cause a certain proportion of false positive results (Wang et al., 2020b).
Considering the two major drawbacks for the commonly used GLFIA, namely the simultaneous high false-negative rate and high false-positive rate, the novel strategy combining signal amplification and modified immuno-structure is necessary and promising for lowering the misinterpretation rate of SARS-CoV-2 antibody POC testing. Herein, a compact three-dimensional QDs assemblies consisting of dendritic silica templates, hydrophobic QDs, silica-encapsulating layers and hydrophilic polymer layers were fabricated, which served as an efficient fluorescent label to overcome the relative low-sensitivity of the commercial GLFIA. On the other hand, the dual-antigen sandwich format utilizing both labeled antigen and capturing antigen to sandwich the target antibody can effectively exclude the interference of primary antibodies in general indirect-model, thus providing higher specificity against SARS-CoV-2 antibody (Qin et al., 2020). Noticeably, the dual-antigen sandwich can give an advantage of detecting total rather than class-specific antibodies (Sotnikov et al., 2021). The total screening of all types of antibodies including IgA, IgM and IgG is favorable for improving the detection rate of SARS-COV-2 and is more consistent and useful in patients with atypical symptoms or in presymptomatic cases (Ma et al., 2020). Using both immobilized and labeled SARS-CoV-2 spike receptor binding domain (RBD), the total SARS-CoV-2 antibody was detected in real serums by POC manner with significantly improved sensitivity and specificity.
2. Materials and methods
2.1. Ethics statement
All of the human serum samples from 97 COVID-19 patients and 300 non-COVID-19 persons (detected by Reverse Transcription-Polymerase Chain Reaction, RT-PCR) were provided by Xinyu People's hosipital in Jiangxi, China. The study was approved by the Ethics Committee of Xinyu People's hosipital with case number of RMYY20201009001.
2.2. Materials and reagents
Details of the materials and reagents are presented in the supporting information.
2.3. Synthesis and surface modification of SQS nanospheres
Details of the synthesis and surface modification are presented in the supporting information.
2.4. Preparation of SARS-CoV-2 spike RBD-Conjugated iSQS
The iSQS was generated by the reaction of the carboxyl group of SQS-PAA nanospheres with the amino group of RBD to form amide bond. The synthesis step was based on the previous protocol by adopting different nanospheres and antigen (Huang et al., 2021).
2.5. Fabrication of SQS-LFIA strips
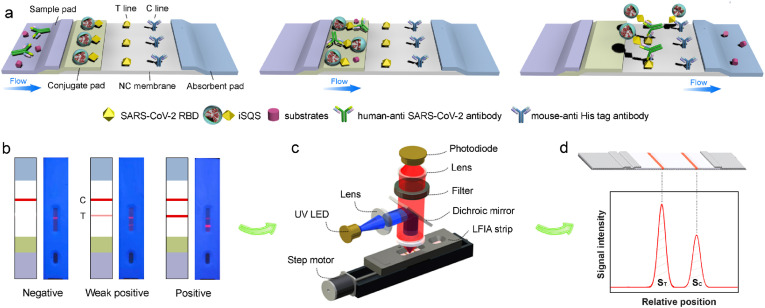
The LFIA strip for SARS-CoV-2 total antibody detection was shown in Fig. 4a. Prior to assembly, the conjugate pad was first treated with optimized blocking buffers and dried at 37 °C. The iSQS reporter, RBD and mouse anti-His antibody were dispersed on the strip according to the previous protocol (Huang et al., 2020). The details are shown in supporting information.
Fig. 4.
(a) Schematic illustration of the working principle of dual-antigen sandwich type SQS-LFIA for SARS-CoV-2 antibody detection. (b) Representative visual results of the strips: negative, weak positive and strong positive samples. (c) Setup of the portable optical scanner for LFIA strips using fluorescent detection. (d) Representative scanning curves of fluorescent signals on the test line and control line of an LFIA strip.
2.6. Analysis of clinical serum samples via SQS-LFIA strip
All of the samples including human serums (10 μL) were added into 90 μL of running buffer, and the mixture were added onto the sample pad of a SQS-LFIA strip. The RBD-conjugated labels were captured at the test line and control line as the liquid migrated from the sample pad toward the absorbent pad. Each test was repeated three times and the corresponding fluorescence intensities were collected after 15 min.
2.7. Characterization
The transmission electron microscopy (TEM) and scanning electron microscopy (SEM) images were acquired by FEI-F20 and FEI NanoSEM450 electron microscope, respectively. The Fourier transform infrared (FTIR) spectra were acquired by Nicolet 6700 spectrometer. The dynamic light scattering (DLS) and zeta potential were recorded by a Malvern Zetasizer NanoZS Instrument. The UV-vis absorption and fluorescence spectra were performed with Shimadzu UV-2550 spectrophotometer and Hitachi F-2700 fluorometer, respectively. The fluorescence quantum yield (QY) of QDs and their derivate nanostructures were determined according to a previous procedure (Grabolle et al., 2009) using rhodamine 6G as a standard. The fluorescent signals of SQS-LFIA strips were recorded by an optical scanner (TND09-M-Y, Shenzhen TND Technology Co., Ltd.).
2.8. Data analysis
The specificity and sensitivity of 300 negative samples and 97 positive samples were calculated according to the following formulas:
| Specificity (%) = 100 × [True negative/(True Negative + False Positive)] |
| Sensitivity (%) = 100 × [True Positive/(True Positive + False Negative)] |
Nonparametric 2-sided Mann Whitney U test and receiver operating characteristic (ROC) curve were calculated by SPSS software.
3. Results and discussion
3.1. Fabrication of three dimensional QDs assembly labels
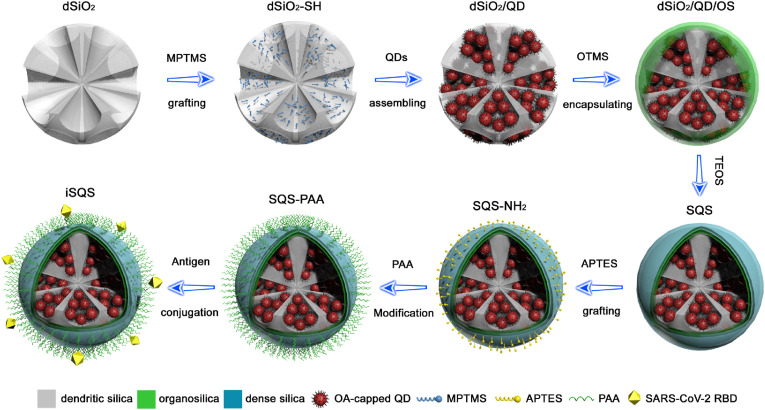
Highly fluorescent hydrophobic QDs were densely incorporated into mercapto-group grafted dendritic silica templates (dSiO2) via strong thiol-metal coordination to form the dSiO2/QD assemblies, as illustrated in Fig. 1 . The organosilica (OS) and dense silica layers encapsulation was performed for the phase transfer of the hydrophobic assemblies and the formation of dSiO2/QD/SiO2 (SQS) spheres, respectively. After a sequential grafting of amino-groups and carboxyl-groups by silanization and polymer conjugation, the SARS-CoV-2 spike RBD was covalently coupled with the polymer surface to form the immuno-SQS (iSQS) labels for LFIA.
Fig. 1.
Schematic illustration for the synthetic route and structure of the SQS based nanolabel.
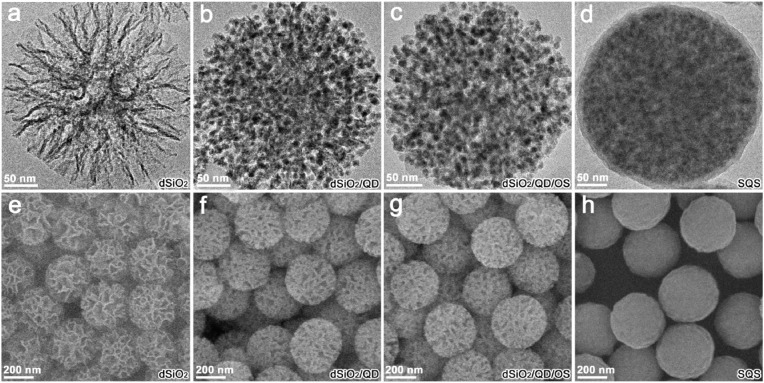
The TEM images indicate a compact assembling of oleic acid capped CdSe/CdS/ZnS QDs (Fig. S1) within the dendritic pore channels of dSiO2 templates (Fig. 2 a–b). The encapsulation of the OS layer from the hydrolysis and condensation of OTMS maintained the high loading capacity of the QDs by a gentle hydrophobic interaction (Fig. 2c). And the vitreophilic surface favored the homogeneous deposition of silica shell to form the SQS (Fig. 2d). The SEM images confirm the gradual shrinking of the dendritic pores by QDs incorporating and silica matrix padding, revealing a smooth spherical morphology of the final labels (Fig. 2e–h). The gradual surface modifications of the nanostructures were verified by FTIR spectra as shown in Fig. S2. The dendritic silica templates reveal characteristic bands of Si−O−Si stretching vibrations (1052 and 794 cm−1) and stretching/bending vibrations of O−H from silanol groups (3370 and 1630 cm−1). The hydrophobic dSiO2/QD assemblies show a combination feature of the Si−O−Si stretching vibrations with the emerging C−H stretching/bending vibrations (2925, 2850, 1460 cm−1) and C=O stretching vibration (1550 cm−1), which were attributed to the silica scaffold and alkyl chain/carboxylate group from oleic acid ligands of QDs. The organosilica layer formation and silica shell growth caused enhanced characteristic absorptions at 3370 and 1630 cm−1, which were assigned to the stretching/bending vibrations of O−H from silanol groups.
Fig. 2.
(a–d) TEM images of dSiO2 template (a), dSiO2/QD assembly (b), dSiO2/QD/OS composite (c), and SQS nanosphere (d). (e–h) SEM images of dSiO2 templates (e), dSiO2/QD assemblies (f), dSiO2/QD/OS composites (g), and SQS nanospheres (h).
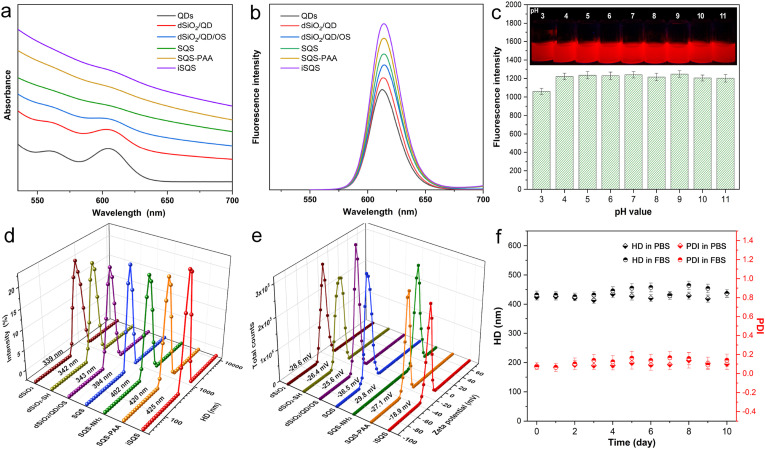
The UV-vis absorption spectra indicated a preserving of the exciton absorption peak of QDs during the processing and a combination of the absorption feature of QDs and light scattering of silica matrix of the final labels (Fig. 3 a). The fluorescent emission characters including emission peak wavelength and full width at half maximum of the QDs were well retained during the synthetic steps (Fig. 3b and Table S1), owing to the preserving of QDs’ hydrophobic ligands by organic-phase assembling and alkylsilane encapsulation. The final quantum yield (QY) of the QDs from iSQS was calculated as 44% which was 58% preservation of the original QY in chloroform. Meanwhile, the SQS-PAA fluorescent labels under a variety of pH values (3∼11) and after long term storage in aqueous solution all exhibited a robust fluorescence emission which is quite favorable for practical diagnosis (Fig. 3c, Fig. S3).
Fig. 3.
UV-vis absorption spectra (a) and fluorescence spectra (b) of QDs and composite nanospheres at different synthetic stages. (c) Fluorescence intensities at the maximum emission wavelength of SQS-PAA aqueous dispersions with different pH values. (d–e) HD (d) and ZP (e) distributions of the nanostructures at different synthetic stages. (f) HD and PDI variations of SQS-PAA dispersed in PBS and FBS over storage time.
The dSiO2 templates and their derivate nanostructures all reveal a well-defined single hydrodynamic diameter (HD) distribution peak, with a gradual increasing of the HD as shown in Fig. 3d. After the incorporation of hydrophobic QDs and OS layer encapsulation, the HD of the assemblies maintained unchanged, possibly owing to the implanting of nanoparticles in the inner space of dendritic silica. While the dense silica growth caused an obvious increase of the HD which is consistant with the TEM observation. The SQS revealed a zeta potential (ZP) of -36.5 mV possibly owing to the surface silanol-groups and nonporous nature of SQS (Fig. 3e). The amino-groups grafting by silanization using (3-aminopropyl) triethoxysilane (APTES) caused minor change in HD but a positive ZP of 29.8 mV. After the conjugation of polyacrylic acid (PAA) layer, the whole assembly exhibited an enlarged HD and a reverse charge of -27.1 mV, which were originated from the warping structure of the polymer chains and the abundant carboxyl-groups from PAA, respectively. The consecutive grafting of amino groups and carboxyl groups also resulted in FTIR absorption bands of N−H bending vibration (1530 cm−1) and C=O stretching vibration (1680 cm−1), respectively (Fig. S2), suggesting the introduction of the functional groups. After incubation in PBS and FBS for days, the HD and polydispersity index (PDI) of SQS-PAA remained nearly invariable (Fig. 3f), indicating an excellent dispersion state for biological labeling. The covalently bonding of SQS-PAA with SARS-CoV-2 RBD resulted in a slight increase of the HD and a prominent decline of the ZP to -18.9 mV, which is probably due to the surface conjugated protein and blocking agents (Huang et al., 2021).
3.2. Preparation, optimization and properties of SQS-LFIA
The conjugation of RBD with SQS was confirmed by fluorescence microscopy (Fig. S4), where the FITC labeled anti-His tag antibodies could specifically bind with RBD and stain the iSQS with green fluorescence. Fig. 4 illustrates the principle of SARS-CoV-2 antibody detection based on the formation of dual-antigen immunosandwich format. After the dropping of serum sample containing SARS-CoV-2 antibodies, the iSQS labels on conjugate pad are rehydrated, and specifically bind with antibodies. The immunocomplexs are then captured by the immobilized antigen (RBD) on the T line, with the free iSQS labels flowing over the C line and captured by the mouse anti-His tag antibodies (Fig. 4a). The SARS-CoV-2 antibody levels correspond to fluorescence band intensities of T line which could be distinguished by naked eye (Fig. 4b). The quantitative analysis is achieved by a portable fluorescence scanner (Fig. 4c) using the fluorescence intensity on T line (ST) (Fig. 4d).
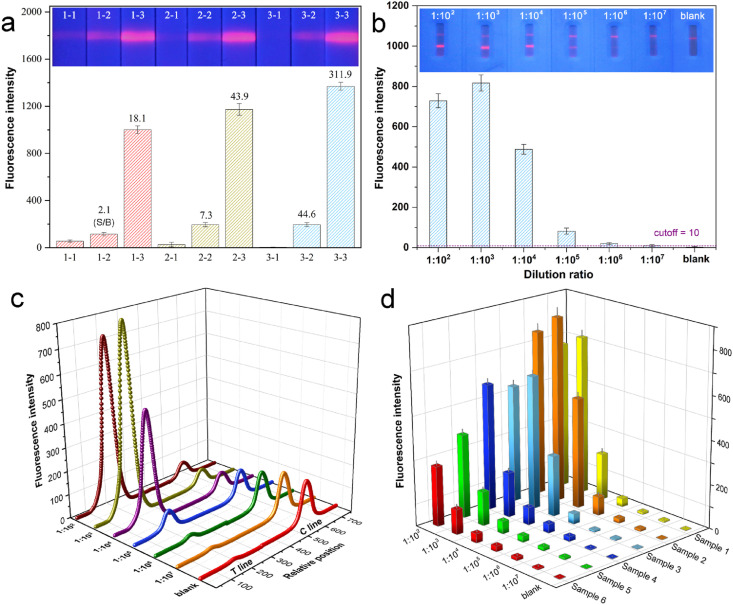
According to the relationship of the fluorescence intensity against immunoassay time (Fig. S5), the fluorescent signal on T line increased gradually and reached a plateau after 15 min of sample addition. To eliminate the non-specific binding of optical labels to prevent false positive results and improve the sensitivity, the blocking of NC membrane was carefully optimized. As depicted in Fig. 5 a, by the consecutive introduction of high concentrated salt and sodium caseinate, the ST values for blank sample decreased remarkably and approached 0 which was completely invisible to the naked eye. This was probably attributed to the screening of the interactions of charged species with NC membrane by salt (Güven et al., 2014), and the blocking of microporous surface of NC membrane by high-molecular protein (Huber et al., 2009). Noticeably, for both weak and strong positive samples, the relevant ST signals rose gradually after the optimizations. The ST ratios of positive sample to blank (S/B) increased dramatically by 21.2 and 17.2 times for weak and strong positive samples after the final optimization, which greatly promoted the sensitivity of the LFIA against real serum samples. Both of the conjugation mass ratio of SQS:RBD and the concentration of RBD on T-line were subsequently optimized. As the mass ratio of SQS:RBD varied from 30:1 to 10:1, both the ST signals and the S/B values increased continuously (Fig. S6). Considering the upper limit of instrument signal range, 10:1 was adopted for further research. Moreover, three concentrations of RBD on T-line were selected for comparison. Fig. S7 indicates that when the RBD concentration changed from 0.5 mg/mL to 1.0 mg/mL, the signals and S/B were significantly improved. The increase was not obvious from 1.0 mg/mL to 1.5 mg/mL, so 1.0 mg/mL was chosen for the follow-up study. The repeated determination of both weak positive and strong positive serum samples indicated CV values of 7.09% and 5.16% (Table S2), respectively, showing a satisfactory reproducibility of the SQS-LFIA for serum samples. The long term stability of the strips was also evaluated as shown in Fig. S8. Both the ST fluorescence intensities of the strips under 4 °C and 25 °C storage conditions (sealed in aluminum foil bag with desiccant) almost remained stable with minor decreasing over 60 days, which facilitates the storage and long term usage.
Fig. 5.
(a) Fluorescence images and corresponding intensities of SQS-LFIA with various blocking buffer treatments (1−, 2−, 3−) for blank sample (−1), medium positive (−2) and strong positive (−3) SARS-CoV-2 antibody samples. Value above each histogram represent the ratio of the response positive signal to blank signal (S/B). (b) Fluorescence images and corresponding intensities of SQS-LFIA for a serum sample of SARS-CoV-2 positive patient with different dilution ratios. (c) Scanning curves of SQS-LFIA strips by the portable optical scanner corresponding to the results of (b). (d) Fluorescence intensities of six different serum samples from SARS-CoV-2 positive patients with different dilutions ratios.
The detection performance of the SQS-LFIA for the main types of antibodies against SARS-CoV-2 was investigated (Fig. S9). The fluorescence intensities of T lines increased gradually with the rose of specific antibody concentrations (SARS-CoV-2 IgG, SARS-CoV-2 IgM or SARS-CoV-2 IgA). By the linear correlations of the signals to the logarithms of antibodies concentrations, the limit of detection (LOD) for SARS-CoV-2 IgG, SARS-CoV-2 IgM and SARS-CoV-2 IgA antibodies were calculated as 0.10 ng/mL, 0.04 ng/mL and 0.06 ng/mL, with the corresponding limit of quantitation (LOQ) calculated as 0.33 ng/mL, 0.13 ng/mL and 0.20 ng/mL, according to previous method (Singh et al., 2020). The response of SQS-LFIA for each antibody type verified the capability of the platform for sensitive POC screening of total SARS-CoV-2 antibodies.
The detection performance of the assay for continuously diluted serums of SARS-CoV-2 positive patients (dilution ratio from 1:102 to 1:107) was evaluated (Fig. 5b). The serum specimens from 300 SARS-CoV-2 negative persons were first examined by SQS-LFIA. The cutoff value calculated by the fluorescence signals was 10, which was expressed as the average signal intensity of the negative samples plus 3 times the standard deviation (Liu et al., 2021). The fluorescence images (Fig. 5b) revealed a stepwise decrease of ST with the increase of dilution ratio, with the fluorescence band distinguishable to naked eye even at a dilution ratio of 1:106. The fluorescent scanning curves by the optical scanner (Fig. 5c) indicated a gradual decline of ST and an increase of SC, with the exception of a hook effect at 1:102 dilution ratio. The quantitative data (Fig. 5b) confirmed that the ST signals all exceeded the cutoff value until a dilution ratio of 1:106 was reached, verifying the reliability of the SQS-LFIA for SARS-CoV-2 antibody with low concentrations. The feasibility of the LFIA for clinical samples was further examined by testing the serums collected from 6 SARS-CoV-2 positive patients (Fig. 5d). The serum samples almost exhibited a concentration gradient dependent signal response, with a gradual dropping of the ST values by the dilution, except for those at very high concentration (dilution ratio of 1:102), probably by the analyte saturation effect. The result confirmed the applicability of the SQS-LFIA for quantification for a wide range of clinical samples. Encouragingly, all the 6 serums with dilution ratios lower than 1:106 exhibited ST values above the cutoff value, which meant that the low-concentrated antibodies from serum samples could be recognized by the quantitative analysis.
3.3. Clinical performances and practicability of the SQS-LFIA
The GLFIA has become the most widely adopted and massively consumed commercial kit for POC testing of SARS-CoV-2 antibodies. Inevitably, the commercial GLFIA may expose false-positive and false-negative results owing to the indirect immuno-structure and colorimetric labels. For the serum specimens containing 300 negative samples (N1–N300) and 97 positive samples (P301–P397), the commercial GLFIA kit revealed 5 false-positive results from negative samples and 6 false-negative results from positive samples (results provided by Xinyu people's hospital). The misinterpretation probability against SARS-CoV-2 infected patients and healthy persons by GLFIA reached 6.2% and 1.7%, respectively, which is consistent with previous literatures using colloidal gold strips (Li et al., 2020a; Liu et al., 2020a). And the high misdiagnosis probability (especially high false negative rate) in relatively large population screening would cause a significant risk of public health hazards as well as a great waste of labor and financial resources for retesting and verification (Koller et al., 2021).
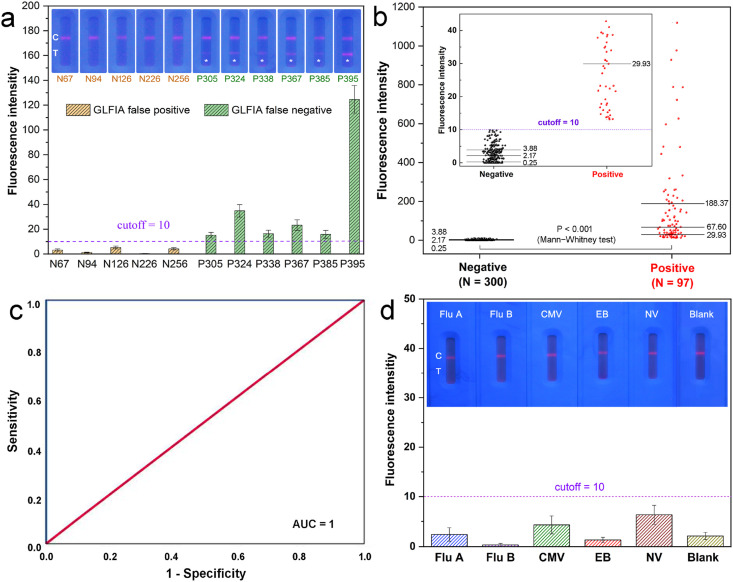
The 11 mismatched clinical cases by GLFIA against RT-PCR method were examined by SQS-LFIA. As displayed in Fig. 6 a, the SQS-LFIA quantitative results of the five false positive samples by GLFIA (N67, N94, N126, N226, N256) were all lower than the cutoff value of SQS-LFIA. On the contrary, although the fluorescence signals of GLFIA false negative samples (P305, P324, P338, P367, P385) determined by SQS-LFIA were relatively low, all the ST values exceeded the cutoff value of 10. Encouragingly, the fluorescent bands could be distinguished by naked eye under a UV lamp from the negative samples, revealing an improved sensitivity against GLFIA. The commercial chemiluminescence immunoassay (CLIA) kit was further utilized to verify the existence of SARS-CoV-2 antibodies. As shown in Table S3, all the misdiagnosed samples by GLFIA could be properly identified by SQS-LFIA which is coincident with both RT-PCR and CLIA results.
Fig. 6.
(a) Fluorescence images and corresponding intensities of SQS-LFIA for the 11 mismatched cases by GLFIA against RT-PCR. (b) Box-diagram for the quantitative detection of SARS-CoV-2 antibody in 397 clinical samples by SQS-LFIA. The medians (midlines), the third/first quartiles (upper and lower lines) and the sample numbers (N) of each group are labeled in the diagram. The inset shows an enlarged box-diagram with low fluorescence intensity range (lower than 45). (c) ROC analysis to assess the detection capability of SQS-LFIA. (d) Fluorescence images and corresponding intensities of SQS-LFIA for Flu A, Flu B, CMV, EB and NV antibodies positive serum samples (tested by ELISA kits with signal/cutoff ratios of 5.5, 7.2, 15.0, 9.6 and 7.7, respectively, and signal/cutoff >1.0 means positive result).
We further examined the total 397 serum specimens by SQS-LFIA and the corresponding fluorescent intensities were recorded in the box-diagram (Fig. 6b). The ST values of all the 300 negative samples were below the cutoff value, with a median of 2.17. On the contrary, the ST values of 97 positive samples were all above the cutoff value, with a median of 67.6 which was 31.6 times larger than that of negative samples. From the enlarged box-diagram, the top part of the negative samples and the bottom part of the positive samples could be clearly distinguished. The significance of difference between the two groups was also confirmed by non-parametric test (Mann-Whitney test) with p < 0.001. The receiver operating characteristic (ROC) curve was adopted to validate the diagnostic performance of the current method (Fig. 6c), which indicates an integrated area under curve (AUC) value of 1, illustrating an excellent identification of the positive and negative sample groups. Impressively, over the total 397 clinical samples, the detected results by SQS-LFIA were completely coincident with the clinically used molecular and immunologic diagnostic methodologies (Table S4), showing a sensitivity and specificity of both 100%. Compared with the commercial GLFIA (with a sensitivity of 93.8% and a specificity of 98.3%), the SQS-LFIA simultaneously improved the detection sensitivity and specificity, while maintaining the POC manner for field-based and non-professional usages.
Table S5 illustrates the sensitivity and specificity of reported combination detection of SARS-CoV-2 antibodies in clinical serum samples. As indicated, most of the reported colloidal gold based LFIA and the CLIA methods using indirect immunostructure showed a relatively insufficient sensitivity (82.3%∼95.85%) accompanied by certain false-positive results. And the combination detection of IgA & IgM & IgG by the developed SQS-LFIA reached a 100% sensitivity/specificity in a relatively large quantity of clinical samples (300 negatives plus 97 positives). The detection specificity of the SQS-LFIA against respiratory virus and other common virus was investigated (Fig. 6d). For the clinical serum samples containing Influenza A (Flu A), Influenza B (Flu B), Cytomegalovirus (CMV), Epstein-Barr virus (EB), Norwalk virus (NV) antibodies (verified by commercial ELISA kits with signal/cutoff ratios of 5.5, 7.2, 15.0, 9.6 and 7.7, respectively), both the naked eye inspection and fluorescence quantification indicated negative results for SARS-CoV-2 antibodies, demonstrating a satisfactory specificity of the SQS-LFIA for clinical POC testing.
To evaluate the practicability of the SQS-LFIA for potential commercialization, the costs of the main constituents from the immunoassay strip were listed in Table S6. The total cost for a single test of SARS-CoV-2 antibody is estimated as $0.2, which is satisfactory considering the general price of commercial GLFIA for SARS-CoV-2 antibody (selling price around $1.0–1.5 per test). Moreover, compared with the price of PCR (selling price around $3.0–4.0 per test), the affordability of the current assay is quite competitive. The low cost and excellent clinical performance indicate the commercial potential of the developed sensor and provide a good tool for SARS-CoV-2 antibody detection. Meanwhile, considering the complexity of fluorescence-based LFIA over traditional GLFIA, the large-scale fabrication of QDs based nanolabels and high-performance hand-held reading devices should be the aims of next stage to further improve the practicability of this platform.
4. Conclusion
We have explored a remarkably sensitive and specific SQS-LFIA platform for detecting SARS-CoV-2 total antibody, which was realized by compact QDs assembly reporters and dual-antigen sandwich immuno-structure. Compared with commercial GLFIA, the amplification of fluorescence signals was achieved by compact 3D assembling of QDs within dendritic templates to reduce the false-negative probability. While the dual-antigen immuno-sandwich format was utilized instead of the general indirect manner to resist the false-positive probability, and to detect the total antibody of IgA & IgM & IgG that may be produced in different periods after immunization. Both the naked eye inspection and quantification results of SQS-LFIA could properly identify all the misinterpreted clinical samples by commercial GLFIA. Over the 300 negative and 97 positive specimens, the detection results were exactly coincident with both commercial RT-PCR and CLIA results, showing a sensitivity and specificity of 100%. The SQS-LFIA combines the superiority of high sensitivity and specificity with the speediness and portability of POC testing platform, holding great significance for on-site and accurate serological screening and monitoring of COVID-19.
Funding
This work was supported by the National Natural Science Foundation of China (U1909214, 21875218 and 51872261), the Zhejiang Provincial Public Technology Research Project (LGF20B010001) and the Zhejiang Provincial Natural Science Foundation (LY21B050001, LR19E020002).
CRediT author statement
Jianghua Jia: Methodology, Writing - original draft. Lijiao Ao: Methodology, Formal analysis. Yongxin Luo: Resource (clinical samples and diagnosis). Tao Liao: Investigation, Formal analysis. Liang Huang: Supervision, Project administration, Writing - review & editing. Dinglv Zhuo: Methodology. Chenxing Jiang: Investigation, Methodology. Jing Wang: Supervision, Writing - review & editing. Jun Hu: Supervision, Project administration, Resources.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113810.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Alturki S.O., Connors J., Cusimano G., Kutzler M.A., Izmirly A.M., Haddad E.K. Front. Immunol. 2020;11:1880. doi: 10.3389/fimmu.2020.01880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukli N., Le Mene M., Schnuriger A., Cuervo N.S., Laroche C., Morand-Joubert L., Gozlan J. J. Clin. Microbiol. 2020;58(11) doi: 10.1128/JCM.01352-20. e01352-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Zhang Z., Zhai X., Li Y., Lin L., Zhao H., Bian L., Li P., Yu L., Wu Y., Lin G. Anal. Chem. 2020;92(10):7226–7231. doi: 10.1021/acs.analchem.0c00784. [DOI] [PubMed] [Google Scholar]
- Feng M., Chen J., Xun J., Dai R., Zhao W., Lu H., Xu J., Chen L., Sui G., Cheng X. ACS Sens. 2020;5(8):2331–2337. doi: 10.1021/acssensors.0c00927. [DOI] [PubMed] [Google Scholar]
- Ge M., Luo W., Jiang D., Li R., Zhao W., Chen G., Yang X., Yu X. Clin. Vaccine Immunol. 2012;19(9):1480–1486. doi: 10.1128/CVI.00234-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong X., Cai J., Zhang B., Zhao Q., Piao J., Peng W., Gao W., Zhou D., Zhao M., Chang J. J. Mater. Chem. B. 2017;5(26):5079–5091. doi: 10.1039/c7tb01049d. [DOI] [PubMed] [Google Scholar]
- Grabolle M., Spieles M., Lesnyak V., Gaponik N., Eychmüller A., Resch-Genger U. Anal. Chem. 2009;81(15):6285–6294. [Google Scholar]
- Guo L., Shao Y., Duan H., Ma W., Leng Y., Huang X., Xiong Y. Anal. Chem. 2019;91(7):4727–4734. doi: 10.1021/acs.analchem.9b00223. [DOI] [PubMed] [Google Scholar]
- Güven E., Duus K., Lydolph M.C., Jørgensen C.S., Laursen I., Houen G. J. Immunol. Methods. 2014;403(1):26–36. doi: 10.1016/j.jim.2013.11.014. [DOI] [PubMed] [Google Scholar]
- Han M., Gao X., Su J.Z., Nie S. Nat. Biotechnol. 2001;19(7):631–635. doi: 10.1038/90228. [DOI] [PubMed] [Google Scholar]
- Hu J., Zhang Z.L., Wen C.Y., Tang M., Wu L.L., Liu C., Zhu L., Pang D.W. Anal. Chem. 2016;88(12):6577–6584. doi: 10.1021/acs.analchem.6b01427. [DOI] [PubMed] [Google Scholar]
- Hu J., Jiang Y.Z., Wu L.L., Wu Z., Bi Y., Wong G., Qiu X., Chen J., Pang D.W., Zhang Z.L. Anal. Chem. 2017;89(24):13105–13111. doi: 10.1021/acs.analchem.7b02222. [DOI] [PubMed] [Google Scholar]
- Hu J., Jiang Y.Z., Tang M., Wu L.L., Xie H.Y., Zhang Z.L., Pang D.W. Anal. Chem. 2019;91(1):1178–1184. doi: 10.1021/acs.analchem.8b05154. [DOI] [PubMed] [Google Scholar]
- Huang L., Luo Z., Han H. Chem. Commun. 2012;48(49):6145–6147. doi: 10.1039/c2cc32084c. [DOI] [PubMed] [Google Scholar]
- Huang L., Wu Q., Wang J., Foda M., Liu J., Cai K., Han H. Chem. Commun. 2014;50(22):2896–2899. doi: 10.1039/c3cc48405j. [DOI] [PubMed] [Google Scholar]
- Huang L., Jin J., Ao L., Jiang C., Hu J. Appl. Mater. Interfaces. 2020;12(52):58149–58160. doi: 10.1021/acsami.0c18667. [DOI] [PubMed] [Google Scholar]
- Huang L., Zhang Y., Liao T., Xu K., Jiang C., Zhuo D., Wang Y., Wen H., Wang J., Ao L., Hu J. Small. 2021;17(25) doi: 10.1002/smll.202100862. [DOI] [PubMed] [Google Scholar]
- Huang X., Aguilar Z.P., Li H., Lai W., Wei H., Xu H., Xiong Y. Anal. Chem. 2013;85(10):5120–5128. doi: 10.1021/ac400502v. [DOI] [PubMed] [Google Scholar]
- Huang X., Aguilar Z.P., Xu H., Lai W., Xiong Y. Biosens. Bioelectron. 2016;75:166–180. doi: 10.1016/j.bios.2015.08.032. [DOI] [PubMed] [Google Scholar]
- Huber D., Rudolf J., Ansari P., Galler B., Führer M., Hasenhindl C., Baumgartner S. Anal. Bioanal. Chem. 2009;394(2):539–548. doi: 10.1007/s00216-009-2698-8. [DOI] [PubMed] [Google Scholar]
- Koller G., Morrell A.P., Galão R.P., Pickering S., MacMahon E., Johnson J., Ignatyev K., Neil S.J.D., Elsharkawy S., Fleck R., Machado P.M.P., Addison O. ACS Appl. Mater. Interfaces. 2021;13(22):25694–25700. doi: 10.1021/acsami.1c04283. [DOI] [PubMed] [Google Scholar]
- Lee B., Kim Y., Lee S., Kim Y.S., Wang D., Cho J. Angew. Chem. Int. Ed. 2010;49(2):359–363. doi: 10.1002/anie.200905596. [DOI] [PubMed] [Google Scholar]
- Li H., Liu Z., He Y., Qi Y., Chen J., Ma Y., Liu F., Lai K., Zhang Y., Jiang L., Wang X., Ge J. Clin. Transl. Med. 2020;10(2):e90. doi: 10.1002/ctm2.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S., Sun R., Wang Y., Hu B., Chen W., Zhang Y., Wang J., Huang B., Lin Y., Yang J., Cai W., Wang X., Cheng J., Chen Z., Sun K., Pan W., Zhan Z., Chen L., Ye F. J. Med. Virol. 2020;92(9):1518–1524. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Mao B., Martinez V., Chen X., Li Y., He L., Chen S., Guo X., Shen X., Bao X., Shen H., Lenna S., Qian P., Wu L., Li C. RSC Adv. 2020;10(47):28041–28048. doi: 10.1039/d0ra04107f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Dai E., Xiao R., Zhou Z., Zhang M., Bai Z., Shao Y., Qi K., Tu J., Wang C., Wang S. Sens. Actuators B Chem. 2021;329:129196. doi: 10.1016/j.snb.2020.129196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Liu L., Kou G., Zheng Y., Ding Y., Ni W., Wang Q., Tan L., Wu W., Tang S., Xiong Z., Zheng S. J. Clin. Microbiol. 2020;58(6) doi: 10.1128/JCM.00461-20. https://pubmed.ncbi.nlm.nih.gov/32229605/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Gao C., Zhang Q., Chi M., Howe J.Y., Yin Y. Nano Lett. 2011;11(8):3404–3412. doi: 10.1021/nl201820r. [DOI] [PubMed] [Google Scholar]
- Ma H., Zeng W., He H., Zhao D., Jiang D., Zhou P., Cheng L., Li Y., Ma X., Jin T. Cell. Mol. Immunol. 2020;17(7):773–775. doi: 10.1038/s41423-020-0474-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J.P., Klasse P.J. J. Virol. 2020;94(17) doi: 10.1128/JVI.01083-20. e01083-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y.J., Sha R.C., Feng Y.C., Huang Y.C. J. Clin. Lab. Anal. 2020;34(11) doi: 10.1002/jcla.23481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw A.M., Hyde C., Merrick B., James-Pemberton P., Squires B.K., Olkhov R.V., Batra R., Patel A., Bisnauthsing K., Nebbia G., MacMahon E., Douthwaite S., Malim M., Neil S., Martinez Nunez R., Doores K., Mark T.K.I., Signell A.W., Betancor G., Wilson H.D., Galão R.P., Pickering S., Edgeworth J.D. Analyst. 2020;145(16):5638–5646. doi: 10.1039/d0an01066a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N., Huang L., Wang D.B., Shao N., Zhang X.E. Anal. Chem. 2020;92(15):10569–10577. doi: 10.1021/acs.analchem.0c01464. [DOI] [PubMed] [Google Scholar]
- Sotnikov D.V., Zherdev A.V., Dzantiev B.B. Sensors. 2021;21(1):39–54. doi: 10.3390/s21010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Wang C., Wang X., Wang K., Zhu Y., Rong Z., Wang W., Xiao R., Wang S. ACS Appl. Mater. Interfaces. 2019;11(21):19495–19505. doi: 10.1021/acsami.9b03920. [DOI] [PubMed] [Google Scholar]
- Wang C., Yang X., Gu B., Liu H., Zhou Z., Shi L., Cheng X., Wang S. Anal. Chem. 2020;92(23):15542–15549. doi: 10.1021/acs.analchem.0c03484. [DOI] [PubMed] [Google Scholar]
- Wang J., Jiang C., Jin J., Huang L., Yu W., Su B., Hu J. Angew. Chem. Int. Ed. 2021;60(23):13042–13049. doi: 10.1002/anie.202103458. [DOI] [PubMed] [Google Scholar]
- Wang Q., Du Q., Guo B., Mu D., Lu X., Ma Q., Guo Y., Fang L., Zhang B., Zhang G., Guo X. J. Clin. Microbiol. 2020;58(6) doi: 10.1128/JCM.00375-20. e00375-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Yuan H., Zhou C., Mao M., Liu Q., Shen H., Cen Y., Qin Z., Ma L., Song L.L. Biosens. Bioelectron. 2016;77:464–470. doi: 10.1016/j.bios.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Bernardi S., Song H., Zhang J., Yu M., Reid J.C., Strounina E., Searles D.J., Yu C. Chem. Mater. 2016;28(3):704–707. [Google Scholar]
- Yue Q., Li J., Luo W., Zhang Y., Elzatahry A.A., Wang X., Wang C., Li W., Cheng X., Alghamdi A., Abdullah A.M., Deng Y., Zhao D. J. Am. Chem. Soc. 2015;137(41):13282–13289. doi: 10.1021/jacs.5b05619. [DOI] [PubMed] [Google Scholar]
- Zhang D., Jiang Y., Yang H., Zhu Y., Zhang S., Zhu Y., Wei D., Lin Y., Wang P., Fu Q., Xu H., Gu H. Adv. Funct. Mater. 2016;26(34):6146–6157. [Google Scholar]
- Zhou C., Yuan H., Shen H., Guo Y., Li X., Liu D., Xu L., Ma L., Li L.S. J. Mater. Chem. 2011;21(20):7393–7400. [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Yang Y., Zhang C. Chem. Rev. 2015;115(21):11669–11717. doi: 10.1021/acs.chemrev.5b00049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.