Abstract
Background
Severe coronavirus disease 2019 (COVID-19) is characterized by impaired type I interferon activity and a state of hyperinflammation leading to acute respiratory distress syndrome. The complement system has recently emerged as a key player in triggering and maintaining the inflammatory state, but the role of this molecular cascade in severe COVID-19 is still poorly characterized.
Objective
We aimed at assessing the contribution of complement pathways at both the protein and transcriptomic levels.
Methods
To this end, we systematically assessed the RNA levels of 28 complement genes in the circulating whole blood of patients with COVID-19 and healthy controls, including genes of the alternative pathway, for which data remain scarce.
Results
We found differential expression of genes involved in the complement system, yet with various expression patterns: whereas patients displaying moderate disease had elevated expression of classical pathway genes, severe disease was associated with increased lectin and alternative pathway activation, which correlated with inflammation and coagulopathy markers. Additionally, properdin, a pivotal positive regulator of the alternative pathway, showed high RNA expression but was found at low protein concentrations in patients with a severe and critical disease, suggesting its deposition at the sites of complement activation. Notably, low properdin levels were significantly associated with the use of mechanical ventilation (area under the curve = 0.82; P = .002).
Conclusion
This study sheds light on the role of the alternative pathway in severe COVID-19 and provides additional rationale for the testing of drugs inhibiting the alternative pathway of the complement system.
Key words: Complement system, alternative pathway, COVID-19, SARS-CoV-2, immunology, hemostasis
Abbreviations used: COVID-19, Coronavirus disease 2019; SARS-CoV-2, Severe acute respiratory syndrome coronavirus-2
Introduction
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19), has to date caused more than 4 million deaths worldwide.1 Although the majority of patients remain asymptomatic or show mild-to-moderate symptoms, approximately 5% of patients display severe disease characterized by acute respiratory distress syndrome, which can result in multiorgan failure and death.2 , 3 We previously demonstrated that patients with severe and critical disease display an imbalanced immune response with impaired type I interferon activity coupled with excessive inflammation.4 Glucocorticoids have been shown to reduce COVID-19 mortality, yet complementary therapies could more specifically target certain members of the immune response.5 In this context, the complement system has emerged as an attractive candidate.
The complement system is a key player in innate immunity at the interface with the adaptive immune system.6 Activation of the complement cascade leads to the cleavage of C3 and the deposition of C3b on activating surfaces, triggering phagocytosis or cleavage of C5 into C5a and C5b, and subsequent formation of the membrane attack complex C5b9, resulting in perturbation of the cell membrane. Additionally, C3a and C5a are anaphylatoxins able to recruit and activate leukocytes, thereby bridging the gap between innate and adaptive immunity and promoting inflammation. The complement cascade can be activated through 3 different pathways, all converging to the cleavage of C3: (1) the classical pathway detects bound antibodies or other acute phase proteins via C1q; (2) the lectin pathway recognizes carbohydrate structures in pathogens and damaged membranes; and (3) the alternative pathway is in a constant state of activation unless complement inhibitory proteins are presented, a process known as tickover, and can amplify C3b formation.
Complement activation has been associated with disease severity in bacterial and viral pneumonia, respiratory distress syndrome, and multiorgan failure.7 As for SARS-CoV-2, the complement system was one of the most highly induced intracellular pathway in infected lung epithelial cells, driven by the transcription of C1r, C1s, factor B, and C3.8 Additionally, multiple products of the complement system, including sC5b9, C5a, C3bc, C3bBbP, and C4d, were found in sera of patients with COVID-19.9 Accordingly, patients with COVID-19 with severe disease displayed elevated plasma concentrations of C5a, C3a, and sC5b9,9, 10, 11, 12, 13 and genetic defects in complement regulatory genes such as CD55 and factor H were associated with disease severity.14 Additionally, anti-C5aR1 antibodies inhibited lung injury in human C5aR1 knock-in mice, indicating that targeting complement could reduce disease severity.10
What links the complement pathway to COVID-19 severity is still poorly understood, but one hypothesis lies in its association with coagulopathy.15 Severe COVID-19 has been shown to trigger thrombosis,16 , 17 and markers of coagulation have been associated with critical disease. Beyond identified connections between inflammation and coagulopathy, evidence suggests a cross talk between the complement and coagulation cascades.18 Although studies have shown association of complement activation with severe COVID-19, an integrative approach assessing the contribution of complement pathways at both protein and transcriptomic levels is lacking. To address this issue, we analyzed RNA and protein levels of components of the 3 complement pathways in patients with COVID-19 and in healthy controls.
Results and discussion
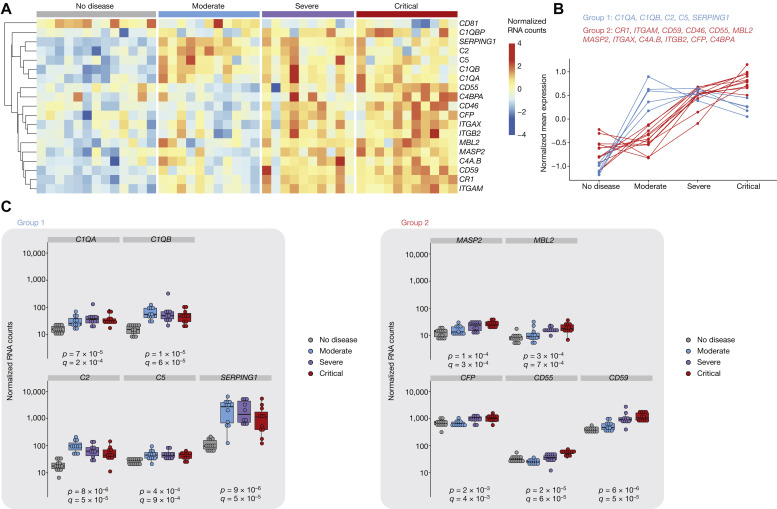
We previously analyzed whole-blood transcriptomic data from 32 patients with COVID-19 with various disease severity and 13 healthy controls.4 The main characteristics of these patients are described in Table I . To uncover the role of complement in disease severity, we determined the RNA levels of 28 complement genes with expression above the lower limit of quantification (see Fig E1 in the Online Repository at www.jacionline.org). Of those, 19 were differentially expressed depending on disease stage (Fig 1 , A). Hierarchical clustering identified 2 main gene groups (showing high intragroup correlation and including 17 of the 19 genes) displaying distinct patterns of expression: group 1 contained genes whose expressions peaked in moderate disease, whereas the group 2 genes showed increased expression in patients with severe disease and to a greater extent in patients with critical disease, whereas patients with moderate disease had expression levels that were comparable to those in the healthy controls (Fig 1, B). Group 1 included genes belonging to the classical pathway (C1QA and C1QB) and both the classical and lectin pathways (C2 and SERPING, coding for C1 inhibitor), as well as the terminal phase (C5) (Fig 1, C). In contrast, group 2 contained genes belonging to the lectin pathway (MBL2, MASP2, and C4 [the latter also belonging to the classical pathway]) and the alternative pathway (C3; its stabilizer CFP, coding for properdin; the C3 receptors ITGAM and ITGAX; and C3 regulators CR1, CD46, CD55, and CD59) (Fig 1, C).
Table I.
Patient characteristics
| Characteristics | Healthy controls |
All patients |
Disease severity |
P value | ||
|---|---|---|---|---|---|---|
| (n = 13) | (n = 32) | Moderate |
Severe |
Critical |
||
| (n = 11) | (n = 10) | (n = 11) | ||||
| Age (y) | 59.2 (45.2-60.0) | 55.6 (51.2-64.8) | 55.9 (44.7-65.1) | 53.5 (48.02-61) | 60.2 (54.8-71.65) | .23 |
| Male sex, no. (%) | 10 (77) | 24 (75) | 8 (72.7) | 9 (90) | 7 (63.6) | .37 |
| Interval from first symptoms to admission (d), median (IQR) | — | 10 (9-11) | 9 (9-11) | 10.5 (10-12) | 9 (8-11) | .06 |
| Coexisting disorder, no. (%) | ||||||
| Any | 0 (0) | 14 (44) | 2 (18) | 3 (30) | 9 (82) | .006 |
| COPD | 0 (0) | 1 (3) | 0 (0) | 0 (0) | 1 (9) | .37 |
| Diabetes | 0 (0) | 5 (16) | 0 (0) | 1 (10) | 4 (36) | .053 |
| Hypertension | 0 (0) | 10 (31) | 2 (18) | 2 (20) | 6 (55) | .12 |
| Cardiovascular disease | 0 (0) | 3 (9) | 0 (0) | 0 (0) | 3 (27) | .042 |
| Cancer or hemopathy | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | — |
| Chronic renal disease | 0 (0) | 1 (3) | 0 (0) | 0 (0) | 1 (9) | .37 |
| Overweight | 0 (0) | 2 (6) | 1 (9) | 1 (10) | 0 (0) | .5 |
| Fever on admission | ||||||
| Temperature (°C), median (IQR) | NA | 38.9 (38.5-39.4) | 38.8 (38.2-39.5) | 38.6 (38.5-39.7) | 39.0 (38.5-39.4) | — |
| Symptoms on admission, no. (%) | ||||||
| Fever | NA | 32 (100) | 11 (100) | 10 (100) | 11 (100) | — |
| Dyspnea | NA | 32 (100) | 11 (100) | 10 (100) | 11 (100) | — |
| Cough | NA | 31 (97) | 10 (91) | 10 (100) | 11 (100) | .37 |
| Fatigue | NA | 31 (97) | 10 (91) | 10 (100) | 11 (100) | .37 |
| Myalgia | NA | 20 (63) | 8 (73) | 8 (10) | 4 (36) | .082 |
| Diarrhea | NA | 11 (34) | 4 (36) | 5 (50) | 1 (9) | .091 |
| Oxygen requirement (L/min) | NA | — | 1.5 (1-3) | 5 (4-6) | MV | — |
| Clinical outcomes, no. (%) | ||||||
| Thrombotic events | NA | 3 (9) | 0 (0) | 1 (10) | 2 (18) | .34 |
| MV | NA | — | 0 (0) | 5 (50) | — | — |
| Death | NA | 5 (15.6) | 0 (0) | 0 (0) | 5 (45.5) | <.001 |
| Laboratory findings on admission, median (IQR) | ||||||
| Leukocytes (× 109/L) | NA | 6.7 (4.31-8.82) | 4.71 (3.78-5.68) | 7.78 (6.46-8.43) | 9.38 (5.48-10.49) | .038 |
| Neutrophils (× 109/L) | NA | 5.08 (3.12-7.37) | 3.25 (2.07-3.44) | 5.81 (4.74-6.36) | 7.69 (4.32-9.13) | .022 |
| Lymphocytes (× 109/L) | NA | 0.84 (0.56-1.13) | 1.00 (0.84-1.40) | 0.88 (0.57-1.12) | 0.65 (0.45-0.84) | .031 |
| Monocytes (× 109/L) | NA | 0.41 (0.23-0.52) | 0.40 (0.26-0.52) | 0.42 (0.27-0.51) | 0.33 (0.12-1.05) | .95 |
| Platelets (× 109/L) | NA | 249 (159-298) | 166 (112-251) | 229 (170-282) | 313 (199-352) | .007 |
| CRP (mg/L) | 0.7 (0.0-0.8) | 118 (55-242) | 30 (14-76) | 169 (136-249) | 159 (109-308) | <.001 |
| Lactate dehydrogenase (U/L) | 169 (155-224) | 424 (346-574) | 262 (196-454) | 411 (396-623) | 504 (426-614) | .10 |
Boldface indicates statistical significance (uncorrected P value < .05). Significance between severity groups was determined by using the Kruskal-Wallis test (for continuous variables), Fisher exact test, or χ2 test of independence where applicable (for categorical data). Percentages may not total 100 because of rounding.
COPD, Chronic obstructive pulmonary disease; CRP, C-reactive protein; IQR, interquartile range; MV, mechanical ventilation; NA, not assessed.
Fig E1.
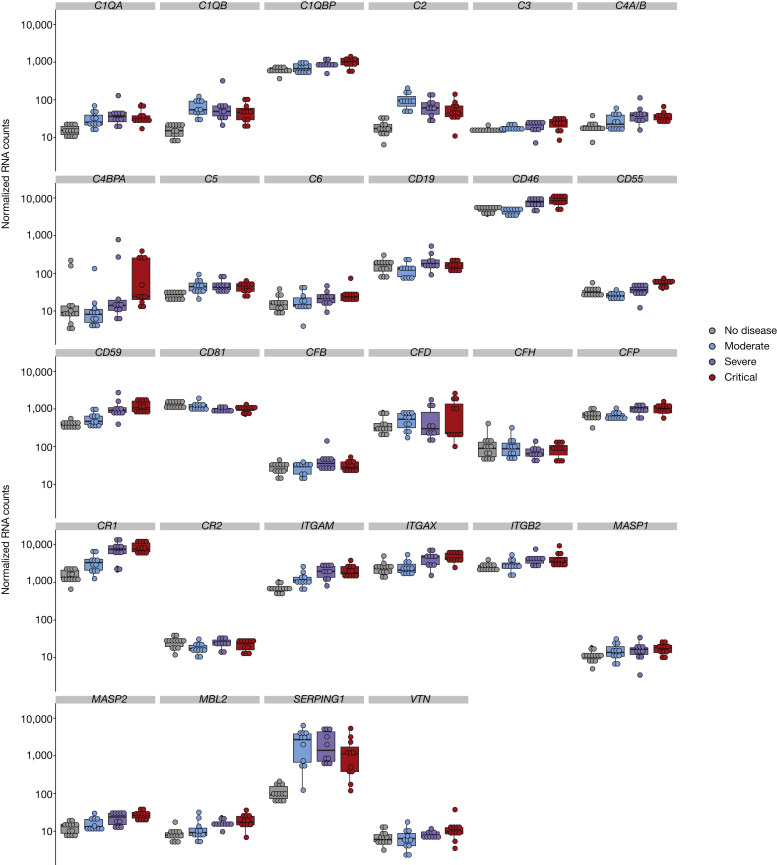
RNA expression of complement genes in whole blood. Individual dot plots showing normalized counts for each patient and all 28 complement genes with at least 1 measurement above the lower limit of quantification. Groups: healthy controls (n = 13); patients with moderate disease (n = 11); patients with severe disease (n = 10); and patients with critical disease (n = 11).
Fig 1.
Complement genes show distinct patterns of RNA expression in whole blood. A, Heatmap representation of RNA levels of the complement genes with differential expression in at least 1 severity group, measured by the NanoString nCounter technology (NanoString, Seattle, Wash). B, The 2 main gene groups, as determined by hierarchical clustering, and their expression pattern (determined as their mean expression across all individuals from a severity group). C, Individual dot plots showing normalized counts for each patient and gene of interest. Significance was determined by the Kruskal-Wallis test (comparing the 4 groups); P denotes uncorrected P values, whereas Q corresponds to false discovery rate (corrected P value). Groups: healthy controls (n = 13), patients with moderate disease (n = 11), patients with severe disease (n = 10), and patients with critical disease (n = 11).
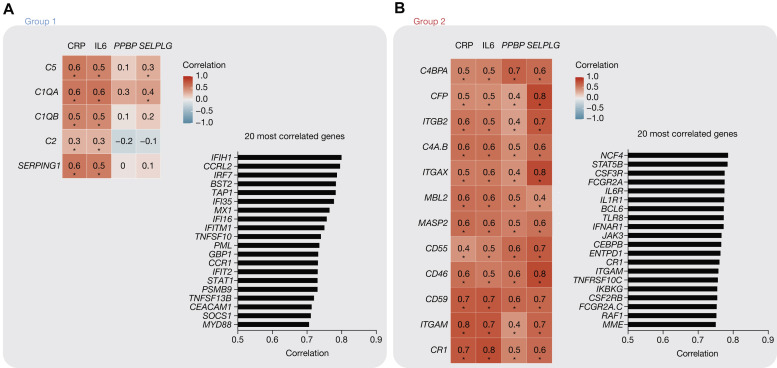
We next studied the correlation between complement gene expression and circulating C-reactive protein and IL-6 proteins on the one hand and PPBP (encoding for platelet chemokine CXCL7) and SELPLG (encoding for PSGL-1) gene expression on the other hand, which are 2 markers of coagulopathy that we previously described as predictive of intubation and death19 (Fig 2 , A and B 20). Additionally, we determined the 20 genes (among the 574-gene nCounter panel) showing highest correlation with group 1 or group 2 genes, respectively (bar graphs). Genes from group 1 were moderately correlated with markers of inflammation, whereas they showed no correlation with coagulopathy markers. They were most correlated with genes of the antiviral response (eg, IFIH1, encoding for MDA5; IRF7, an IFN regulator factor; and BST2 encoding for tetherin), which is consistent with the IFN response being a marker of moderate, but not severe, disease4 (Fig 2, A). Genes from group 2 showed moderate-to-high correlation with both inflammation and coagulopathy, and they were most correlated to genes of inflammation (Fig 2, B).
Fig 2.
Correlations of complement RNA expression with inflammation and coagulopathy markers. A, Correlation matrix (with coefficients determined by Spearman correlation [left]), and the 20 genes with highest mean correlation with group 1 genes, among the 574-gene data set. B, As in (A), for group 2 genes. Asterisks denote significant results (corrected P < .05), determined by the multiple testing procedure for correlation data developed by Cai and Liu.20
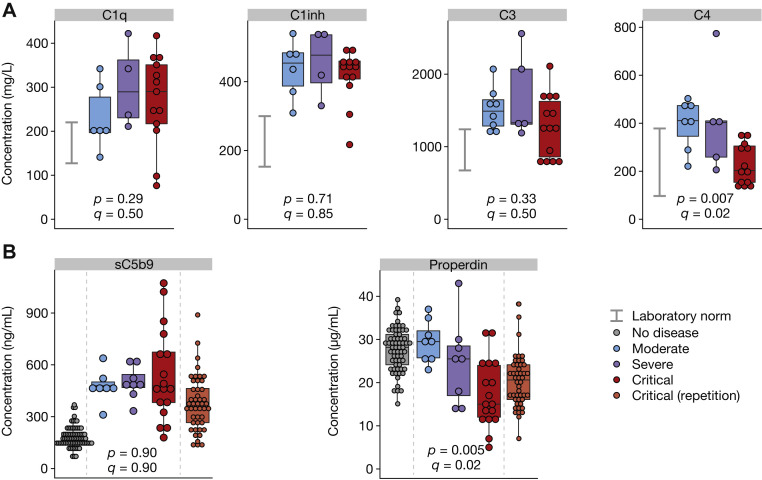
To more precisely characterize systemic complement activation in COVID-19, we measured complement protein levels from 33 patients with COVID-19 with available sera (including from 16 patients with both RNA and protein data) (Fig 3 ). Levels of C1q, C1 inhibitor, and sC5b9 were upregulated in all patients with COVID-19, regardless of severity grade, similarly to what was observed at the transcriptional level. C3 protein levels were upregulated in patients with moderate and severe disease, whereas some critical patients showed lower levels of circulating C3. The lack of a significant decrease in C3 protein may be explained by higher C3 RNA levels and subsequent protein synthesis in critical patients, and as a result, C3 consumption is counterbalanced by C3 production. C4 levels remained within normal laboratory ranges. We found that the levels of properdin decreased significantly with disease severity, whereas its RNA levels were inversely significantly increased (Fig 1, C, group 2, CFP), suggesting the deposition of properdin to complement activating surface and triggering of the alternative pathway. We confirmed this finding by using plasma samples of an independent COVID-19 intensive care unit cohort13 (Fig 3, B).
Fig 3.
Complement protein levels in circulating blood. A, Protein concentrations of C1q, C1 inhibitor (C1inh), C3, and C4 in the same samples, measured by ELISA. B, Serum concentration of sC5b9 and properdin in the same samples, as well as samples from an independent cohort of patients with critical COVID-19 and healthy controls. Significance was determined by the Kruskal-Wallis test (comparing the 3 groups); P denotes uncorrected P values, whereas Q corresponds to false discovery rate (corrected P value). Groups: healthy controls (n = 73), patients with moderate disease (n = 5-8), patients with severe disease (n = 4-8), patients with critical (n = 13-17) disease, and patients with critical disease (repetition) (n = 46). Laboratory norms are indicated with a gray bar.
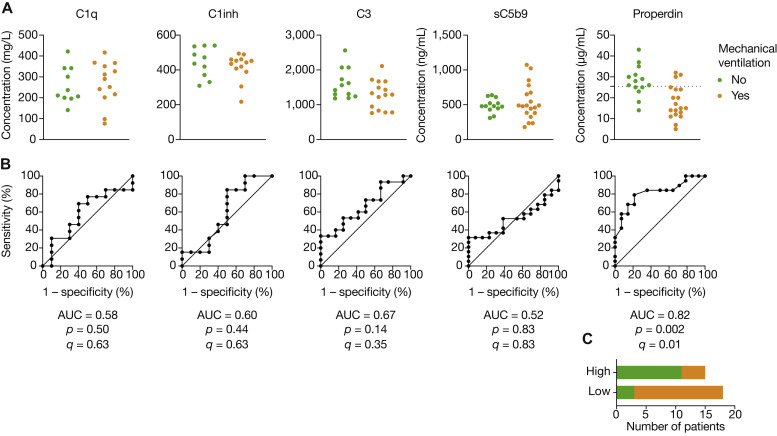
Finally, we analyzed the association of complement component with use of mechanical ventilation. Receiver operating characteristic curve analysis demonstrated that decreased properdin levels were associated with use of mechanical ventilation (Fig 4 [area under the curve = 0.82; P = .002; Q = .01]), with an 83% positive predictive value and a 73% negative predictive value with use of an optimal threshold of 25.5 μg/mL.
Fig 4.
Low properdin levels are associated with use of mechanical ventilation. A, Protein levels of patients at admission depending on (immediate or later) use of mechanical ventilation. Dashed lines represent the optimal threshold obtained by maximization of Youden index for the significant area under the curve (AUC) (properdin). B, Associated receiver operating characteristic curves, with AUC and P values; Q corresponds to false discovery rate (corrected P value). C, Bar graphs indicating the number of patients in each group depending on normalized RNA levels of a single gene with respect to the optimal threshold (properdin only). Groups: no intubation (n = 10-14) and intubation (n = 13-19).
This work constitutes the first study to our knowledge to simultaneously analyze the 3 complement pathways in patients with COVID-19 at both the RNA and protein levels, unraveling properdin consumption as a feature of severe COVID-19. Lower properdin concentration in patients with severe and critical COVID-19 cannot be explained by congenital properdin deficiency, a rare X-linked disorder associated with vulnerability for Nessieria meningitidis–driven meningitis,21 because these patients showed increased properdin RNA levels. Reduced properdin levels were found in various disease conditions, such as C3 glomerulopathy and lupus nephritis, and it was correlated with C5 convertase dysregulation in C3 glomerulopathy.22 , 23 Complement alternative pathway activation in the context of severe COVID-19 may explain the excess levels of soluble C5a and sC5b9 previously described.9 , 10 We confirmed increased sC5b9 levels in patients with COVID-19, although sC5B19 did not discriminate severity grades as potently as properdin did. Accordingly, Ma et al found only a weak association between sC5b9 levels and disease severity.12
This study has limitations: it is a monocentric, cross-sectional analysis with limited sample size, and longitudinal data will be needed to better characterize the sequential activation of complement during COVID-19. Additionally, most complement components in circulation are synthesized by the liver; therefore, whole-blood RNA quantification may offer a limited view of the complement transcriptional regulation. However, several complement proteins are produced by a wide variety of cell types24 and others are produced mainly by leukocytes, such as C1q and properdin, with the latter playing a key role in our data. An additional limitation is the substantial overlap in properdin levels between severity groups, suggesting that the impact of properdin may be driven by a subgroup of patients. Although receiver operating characteristic analysis highlighted interesting positive and negative predictive values, future work specifically designed to evaluate the predictive potential of properdin is warranted. It will be important to assess the target population and better evaluate the contributions of potential confounding factors, such as comorbidities. Moreover, we do not provide mechanistic evidence of properdin consumption at local sites of infection, and studies that include analysis of pathologic tissue samples are warranted to address this issue. Lastly, despite in vitro evidence that SARS-CoV-2 spike protein can inhibit factor H and prevent the decay of the alternative pathway C3 convertase,25 other mechanisms may be at play, such as inhibition or activation of other regulatory proteins, or nonspecific inflammation-mediated hyperactivation, and this phenomenon could be observed in other infectious conditions with excess inflammation. Further work in other disease models will help delineate the specificity of this observation and dissect its underlying mechanism.
In summary, we have shown that severe COVID-19 is characterized by increased activation of the complement alternative pathway, which was correlated with inflammation and coagulopathy markers. Specific targeting of the alternative pathway rather than the classical pathway may prove useful to control disease severity without hampering essential antiviral responses.
Key messages.
-
•
The classical pathway is activated in all patients with COVID-19, whereas hyperactivation of the lectin and alternative pathways is associated with disease severity.
-
•
Properdin RNA expression is increased in patients with severe disease, yet its protein levels are decreased, suggesting its deposition on activating surfaces.
-
•
Low properdin levels are associated with use of mechanical ventilation.
Acknowledgments
We would like to acknowledge all of the nurses, technicians, and physicians involved in all departments who were involved in management of the patients with COVID-19 in Assistance publique – Hôpitaux de Paris – Centre – Université de Paris hospitals for their help in taking care of the patients and including them in the study.
Footnotes
Supported by the Fonds IMMUNOV, for Innovation in Immunopathology; the Institut national de la santé et de la recherche médicale (Inserm); a government grant managed by the Agence national de la recherche as part of the Investment for the Future program (ANR-10-IAHU-01); and by grants from the Agence national de la recherche (ANR-flash COVID-19 “AIROCovid” and “CoVarImm”). We also acknowledge funding from the Institut Pasteur for COVID-19 Research and an Institut Imagine MD-PhD Fellowship Program award supported by the Fondation Bettencourt Schueller (to J.H.).
Disclosure of potential conflict of interest: The authors declare that they have no relevant conflicts of interest.
Methods
Patient characteristics
Recruitment was conducted between March 19, 2020, and April 3, 2020, in Cochin Hospital (Paris, France). The inclusion criteria for patients with COVID-19 were as follows: age 18 to 80 years, COVID-19 diagnosis according to the World Health Organization interim guidance, and positive RT-PCR result for SARS-CoV-2 on a respiratory sample (nasopharyngeal swab or invasive respiratory sample). Disease severity was classified at the time of admission by using the adaptation of the sixth revised trial version of the Novel Coronavirus Pneumonia Diagnosis and Treatment Guidance. Mild-to-moderate (referred to in this article as moderate) disease was defined as either mild clinical symptoms (fever, myalgia, fatigue, diarrhea) and no sign of pneumonia on thoracic computed tomography scan, or dyspnea and radiologic findings of pneumonia on thoracic computed tomography scan, with a requirement of no more than 3 L/min of oxygen that remained stable for at least the following 24 hours. Severe disease was defined as respiratory distress with a requirement of more than 3 L/min of oxygen and no other organ failure, stable for at least the following 24 hours. Critical cases were defined as respiratory failure requiring mechanical ventilation, shock, and/or other organ failure requiring an intensive care unit.
Blood sampling was performed after a median of 10 days (interquartile range = 9-11 days) after onset of first symptoms, before the initiation of any antiviral or anti-inflammatory treatment and before use of mechanical ventilation. The time interval from first symptoms to admission to the hospital (which for most patients coincided with blood sampling) are specified in Table I (see the print text). No longitudinal sampling was performed, but patients were followed up for at least 30 days. For a more detailed clinical and immunologic characterization of the cohort, refer to Hadjad et al.E1 Clinical, epidemiologic, demographic, laboratory, treatment, and outcome data were all extracted from electronic medical records by using a standardized data collection form.
To repeat the protein findings, we included plasma samples from a previously described cohort of patients in a COVID-19 intensive care unit.E2
Gene expression analysis
Detailed methods were previously reported.E1 Briefly, we analyzed 100 ng (5 μL) of total RNA from each sample by using the NanoString Human Immunology kit v2, following the manufacturer’s instructions. Raw RNA counts were adjusted by using 5 housekeeping genes selected from the 15 candidate control genes provided by NanoString, using the geNorm method. Normalized counts were log10-transformed for all subsequent analyses. A total of 35 genes belonging to the complement system were identified, of which 28 were expressed above the lower limit of quantification in at least 1 sample. Among those, 19 were differentially expressed in 1 group, as determined by an uncorrected P value less than .05 of 1-way ANOVA. A heatmap displaying these 19 genes (see Fig 1, A in the print text) was obtained by using pheatmap (package pheatmap), with the data centered to 0 and scaled to unit variance for each gene. Hierarchic clustering of these 19 genes was performed by using hclust with the default distance matrix; it revealed 2 groups with lower intragroup variance when the number of clusters was set to 4, leaving out 2 genes with different patterns (CD81 and C1QBP).
The genes with the highest correlation to a given group (see Fig 2 [bar graphs] in the print text) were determined by using as a starting gene set all of those genes among the 574 genes of the nCounter kit with at least 1 value above the lower limit of quantification, with the exception of those belonging to the complement gene group of interest. For each gene of the starting set, the mean Spearman correlation coefficient with respect to genes of the group of interest was computed. Genes were ordered by decreasing coefficients, and the first 20 genes are represented.
Protein concentrations
Plasma concentrations of C1q, C1 inhibitor, C3, and C4 were measured by nephelometry (Siemens), and sC5b9 levels were assessed by using Microvue Complement sC5b-9 Plus kit (Quidel). Levels of properdin were determined by using a homemade ELISA (sheep anti-human properdin, the Binding Site, PC116X).
Statistical analyses
Continuous variables were compared among groups by using the Kruskal-Wallis test, whereas categoric variables were compared by using the Fisher exact test or the χ2 test of independence, where applicable. No exact a priori sample size was determined owing to lack of prior knowledge of the expected effect size in such an exploratory study. However, we calculated that a sample size of 10 in each group would allow detection of a Cohen effect size of 1.5 (ie, a difference in means of 1.5 times the SD) with a power of 90% and a 5% significance level. Correlation coefficients were determined by using Spearman method to detect monotonic relationships. Receiver operating characteristic area under the curve P values were calculated by using the default method for GraphPad Prism (GraphPad Software, San Diego, Calif).E3 Specifically, following Hanley computationE4 and assuming the null hypothesis, the SE equals sqrt(0.25 + (n1 + n2 – 2)/(12 n1 n2), where n1 and n2 are the number of patients in each group. The z ratio is then equal to (area under the curve = 0.5)/SE, which follows a 2-tailed normal distribution, from which the P value is derived. Optimal thresholds were obtained by maximization of Youden index. All tests were 2 sided, and a P value less than .05 was considered statistically significant. Correction for multiple testing was obtained by using false discovery rate algorithms (Cai and Liu's procedureE5 in the case of correlation data [adapted to the high level of dependencies] and Benjamini-Hochberg procedure for all other instances [shown as q values]). Analyses were performed by using R, version 3.4.3 (CRAN), and Prism, version 8 (GraphPad Software).
References
- 1.WHO coronavirus disease (COVID-19) dashboard. World Health Organization. Available at: https://covid19.who.int. Accessed September 1, 2021.
- 2.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smadja D.M., Mentzer S.J., Fontenay M., Laffan M.A., Ackermann M., Helms J., et al. COVID-19 is a systemic vascular hemopathy: insight for mechanistic and clinical aspects. Angiogenesis. 2021;24:755–788. doi: 10.1007/s10456-021-09805-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.RECOVERY Collaborative Group. Horby P., Lim W.S., Emberson J.R., Mafham M., Bell J.L., et al. Dexamethasone in hospitalized patients with Covid-19 - preliminary report. N Engl J Med. 2021;384:693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reis E.S., Mastellos D.C., Hajishengallis G., Lambris J.D. New insights into the immune functions of complement. Nat Rev Immunol. 2019;19:503–516. doi: 10.1038/s41577-019-0168-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rittirsch D., Flierl M.A., Nadeau B.A., Day D.E., Huber-Lang M., Mackay C.R., et al. Functional roles for C5a receptors in sepsis. Nat. 2008;14:551–557. doi: 10.1038/nm1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan B., Freiwald T., Chauss D., Wang L., West E., Mirabelli C., et al. SARS-CoV-2 drives JAK1/2-dependent local complement hyperactivation. Sci Immunol. 2021;6 doi: 10.1126/sciimmunol.abg0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holter J.C., Pischke S.E., de Boer E., Lind A., Jenum S., Holten A.R., et al. Systemic complement activation is associated with respiratory failure in COVID-19 hospitalized patients. Proc Natl Acad Sci U S A. 2020;117:25018–25025. doi: 10.1073/pnas.2010540117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carvelli J., Demaria O., Vély F., Batista L., Chouaki Benmansour N., Fares J., et al. Association of COVID-19 inflammation with activation of the C5a-C5aR1 axis. Nature. 2020;588:146–150. doi: 10.1038/s41586-020-2600-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao T., Hu M., Zhang X., Li H., Zhu L., Liu H., et al. Highly pathogenic coronavirus N protein aggravates lung injury by MASP-2-mediated complement over-activation. https://www.medrxiv.org/content/10.1101/2020.03.29.20041962v2.full.pdf Available at: [DOI] [PMC free article] [PubMed]
- 12.Ma L., Sahu S.K., Cano M., Kuppuswamy V., Bajwa J., McPhatter J., et al. Increased complement activation is a distinctive feature of severe SARS-CoV-2 infection. Sci Immunol. 2021;6 doi: 10.1126/sciimmunol.abh2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peffault de Latour R., Bergeron A., Lengline E., Dupont T., Marchal A., Galicier L., et al. Complement C5 inhibition in patients with COVID-19 - a promising target? Haematologica. 2020;105:2847–2850. doi: 10.3324/haematol.2020.260117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramlall V., Thangaraj P.M., Meydan C., Foox J., Butler D., Kim J., et al. Immune complement and coagulation dysfunction in adverse outcomes of SARS-CoV-2 infection. Nat Med. 2020;26:1609–1615. doi: 10.1038/s41591-020-1021-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lo M.W., Kemper C., Woodruff T.M. COVID-19: Complement, coagulation, and collateral damage. J Immunol. 2020;205:1488–1495. doi: 10.4049/jimmunol.2000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Middeldorp S., Coppens M., van Haaps T.F., Foppen M., Vlaar A.P., Müller M.C.A., et al. Incidence of venous thromboembolism in hospitalized patients with COVID-19. J Thromb Haemost. 2020;18:1995–2002. doi: 10.1111/jth.14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oxley T.J., Mocco J., Majidi S., Kellner C.P., Shoirah H., Singh I.P., et al. Large-vessel stroke as a presenting feature of Covid-19 in the young. N Engl J Med. 2020;382:e60. doi: 10.1056/NEJMc2009787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ward P.A. The dark side of C5a in sepsis. Nat Rev Immunol. 2004;4:133–142. doi: 10.1038/nri1269. [DOI] [PubMed] [Google Scholar]
- 19.Yatim N., Boussier J., Chocron R., Hadjadj J., Philippe A., Gendron N., et al. Platelet activation in critically ill COVID-19 patients. Ann Intensive Care. 2021;11 doi: 10.1186/s13613-021-00899-1. 113-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai T.T., Liu W. Large-scale multiple testing of correlations. J Am Stat Assoc. 2016;111:229–240. doi: 10.1080/01621459.2014.999157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fijen C.A., van den Bogaard R., Schipper M., Mannens M., Schlesinger M., Nordin F.G., et al. Properdin deficiency: molecular basis and disease association. Mol Immunol. 1999;36:863–867. doi: 10.1016/s0161-5890(99)00107-8. [DOI] [PubMed] [Google Scholar]
- 22.Corvillo F., Bravo García-Morato M., Nozal P., Garrido S., Tortajada A., Rodríguez de Córdoba S., et al. Serum properdin consumption as a biomarker of C5 convertase dysregulation in C3 glomerulopathy. Clin Exp Immunol. 2016;184:118–125. doi: 10.1111/cei.12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y., Nester C.M., Martin B., Skjoedt M.-O., Meyer N.C., Shao D., et al. Defining the complement biomarker profile of C3 glomerulopathy. Clin J Am Soc Nephrol. 2014;9:1876–1882. doi: 10.2215/CJN.01820214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morgan B.P., Gasque P. Extrahepatic complement biosynthesis: where, when and why? Clin Exp Immunol. 1997;107:1–7. doi: 10.1046/j.1365-2249.1997.d01-890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu J., Yuan X., Chen H., Chaturvedi S., Braunstein E.M., Brodsky R.A. Direct activation of the alternative complement pathway by SARS-CoV-2 spike proteins is blocked by factor D inhibition. Blood. 2020;136:2080–2089. doi: 10.1182/blood.2020008248. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peffault de Latour R., Bergeron A., Lengline E., Dupont T., Marchal A., Galicier L., et al. Complement C5 inhibition in patients with COVID-19 - a promising target? Haematologica. 2020;105:2847–2850. doi: 10.3324/haematol.2020.260117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GraphPad Calculation details for ROC curves. https://www.graphpad.com/guides/prism/latest/statistics/stat_calcualtion_details_for_roc_cu.htm Available at:
- Hanley J.A., McNeil B.J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- Cai T.T., Liu W. Large-scale multiple testing of correlations. J Am Stat Assoc. 2016;111:229–240. doi: 10.1080/01621459.2014.999157. [DOI] [PMC free article] [PubMed] [Google Scholar]