Abstract
We report the rapid detection of SARS-CoV-2 in infected patients (mid-turbinate swabs and exhaled breath aerosol samples) in concentrations as low as 60 copies/mL of the virus in seconds by electrical transduction of the SARS-CoV-2 S1 spike protein antigen via SARS-CoV-2 S1 spike protein antibodies immobilized on bilayer quasi-freestanding epitaxial graphene without gate or signal amplification. The sensor demonstrates the spike protein antigen detection in a concentration as low as 1 ag/mL. The heterostructure of the SARS-CoV-2 antibody/graphene-based sensor is developed through a simple and low-cost fabrication technique. Furthermore, sensors integrated into a portable testing unit distinguished B.1.1.7 variant positive samples from infected patients (mid-turbinate swabs and saliva samples, 4000–8000 copies/mL) with a response time of as fast as 0.6 s. The sensor is reusable, allowing for reimmobilization of the crosslinker and antibodies on the biosensor after desorption of biomarkers by NaCl solution or heat treatment above 40 °C.
Keywords: 2D material, Graphene, SiC, SARS-CoV-2, COVID-19, Heterostructure, Biosensor
1. Introduction
SARS-CoV-2, the virus responsible for COVID-19, can be highly infectious in recently infected individuals and asymptomatic carriers, leading to the global spread of COVID-19 (Arons et al., 2020; Gandhi et al., 2020). Remdesivir, the first treatment for COVID-19, has shown a prominent effect on inhibiting the early life cycle of coronavirus replication (Wang et al., 2020; Pizzorno et al., 2020), and monoclonal antibodies, which target the spike protein of SARS-CoV-2, is also an effective treatment for preventing death in high-risk COVID-19 patients (Jahanshahlu and Rezaei, 2020; Ledford, 2021). Vaccines, including BNT162b2, mRNA-1273, JNJ-78436735, AZD1222, and NVX-CoV2373, have been shown to prevent infection or reduce the symptoms of COVID-19 (Dagan et al., 2021; Polack et al., 2020). However, the reduced effectiveness of the vaccines and monoclonal antibodies in preventing severe illness from variants such as B.1.1.7, B.1.351, B.1.429, B.1.526, B.1.617, and P.1 remain a concern (Callaway and Ledford, 2021; Mahase, 2021). In addition, the global rollout of the vaccines will take time. The key to prevent spread is early detection before developing symptoms. Hence, ultra-fast high sensitive diagnostic methods are needed to prevent the further spread of COVID-19. Current diagnostic methods of COVID-19 are based on the antibody, antigen, and genetic material of the novel virus (Cassaniti et al., 2020; Hoffman et al., 2020; Tang et al., 2020; Porte et al., 2020; Scohy et al., 2020; Sheridan, 1038; Broughton et al., 2020). While antibody tests, which measure IgG and IgM in serum, plasma, and saliva by immunoassays, can be performed rapidly, they do not reflect the current viral infection status. They can also only diagnose those previously infected with the virus, at least 10–14 days following infection (Cassaniti et al., 2020; Hoffman et al., 2020). Real-time reverse transcription-polymerase chain reaction (RT-PCR), the golden standard for COVID-19 diagnostics, detects infection through the genetic makeup of SARS-CoV-2 (Tang et al., 2020). However, due to its sophisticated and time-consuming experimental settings, rapid and on-site diagnosis is not feasible with, for example, the Abbott ID NOW not meeting this specification. Antigen tests that directly detect the viral proteins can also be performed rapidly. However, they are less sensitive and can only detect infections with higher viral loads (>10,000 genomic copies/mL on average) compared to RT-PCR (250–1000 copies/mL). (Porte et al., 2020; Scohy et al., 2020; Sheridan, 1038).
Several groups have reported quick and sensitive ways to diagnose COVID-19 by adopting advanced techniques, such as CRISPR (Broughton et al., 2020) and SENSR (Woo et al., 2020), to detect the virus' genetic materials fluorescence within 5–10 min. Although there are several quick and, in some cases, sensitive methods to diagnose COVID-19 by detecting the virus' genetic material, sophisticated instrumentation is often required to achieve the results. In contrast, biosensors (Vermisoglou et al., 2020; Jung et al., 2010; Syu et al., 2018) have been investigated to develop early detection of pathogens because they have several advantages, including high sensitivity, fast measurement, and the use of a small amount of analytes. Solid-state biosensors are a good candidate because they operate through the material's electrical resistance, which changes proportionally to the detected molecules. For example, ZnO (Shanmugam et al., 2017), ZrO2-rGO (Kumar et al., 2016), In2O3 nanoribbon (Liu et al., 2016), Si nanowire (Yen et al., 2014; Chen et al., 2015), CuS-GO (Li et al., 2015), carbon nanohorns (Yang et al., 2014), GO (Navakul et al., 2017; Kim et al., 2013; Liu et al., 2011), rGO (Joshi et al., 2020; Maity et al., 2018; Majd and Salimi, 2018; Wu et al., 2013), and graphene (Xiang et al., 2018; Islam et al., 2019; Afsahi et al., 2018; Seo et al., 2020) typically serve as the active materials for biosensors to detect several viruses (e.g., influenza, SARS-CoV-2, ZIKA) and cancers (e.g., lung, liver, colorectal) due to their low cost, biocompatibility, good mechanical strength, and high sensitivity. Although many of these materials are promising and have yielded high sensitivity biosensors, they either suffer from false positives due to low selectivity or complex and expensive fabrication processes.
On the other hand, graphene, a single atomic layer of carbon atoms with high surface area, has shown exceptionally high sensitivity, less than 1 part per billion, with high electrical conductivity and carrier mobility (∼100,000 cm2/V theoretically) (Choi et al., 2020; Chatterjee et al., 2015; Schedin et al., 2007) which makes it very attractive for developing highly sensitive and conformable biosensors. Unfortunately, graphene by itself suffers from poor selectivity (Choi et al., 2020; Chatterjee et al., 2015) i.e,., it causes false positives, so developing a biosensor utilizing simply graphene is challenging. However, the low selectivity of graphene-based biosensors can be overcome by surface functionalization with biomarkers targeting viruses or pathogens (Maity et al., 2018; Majd and Salimi, 2018; Islam et al., 2019; Afsahi et al., 2018; Seo et al., 2020), making it possible to design novel heterostructures that interact directly with the target molecules while maintaining their attractive features. One biomarker that can be targeted by immobilization on graphene is the SARS-CoV-2 S1 spike protein antibody to detect the S1 spike protein antigens of the virus. When an antigen (or viral particle) is introduced and mixed with a polyclonal antibody, multivalent interactions lead to large, stable, highly avid interactions as the antigen can be bound by several antibodies, targeting a different epitope (Lipman et al., 2005). This bonding is reversible by its nature as non-covalent bonds. The non-covalent interactions that form the basis of antigen-antibody binding include hydrogen bonds, ionic bonds, hydrophobic interactions, and van der Waals interactions (Davies and Cohen, 1996). Antigens combine highly specifically with the corresponding fragment antigen-binding portion, constructed from heavy and light chain hypervariable regions in the antibody structure (Dreyer and Bennett, 1965). Seo et al. (2020) developed a field-effect transistor (FET) based biosensor using this biomarker gated by the SARS-CoV-2 virus, which was coupled to the graphene transistor channels (Width: 100 μm, Length: 100 μm). The biosensor resulted in a limit of detection (LOD) of 2.42 × 102 copies/mL for clinical samples, detecting the SARS-CoV-2 spike protein in concentrations as low as 1 fg/mL with 3–6% changes. However, the above FET example used thermal chemical vapor deposition (CVD) graphene on Cu foil which required wet etching transfer methods to prepare the biosensors, yielding the degraded quality of materials with impurities and defects, resulting in a detrimental effect on sensor performance and reproducibility. Moreover, transfer methods are expensive, complex, with time-consuming fabrication processes.
In this work, we combined the high sensitivity of quasi-freestanding (QFS) bilayer epitaxial graphene (EG) with the high selectivity of SARS-CoV-2 S1 spike antibodies to develop an ultra-high sensitivity biosensor for rapid detection of COVID-19. QFS EG on silicon carbide (SiC) enables a bottom-up direct immobilization of SARS-CoV-2 S1 spike protein antibody by immunoassay technique that can support large area (up to wafer size range) and yield the highest quality materials without any complex transfer methods. In addition, the QFS EG has several advantages, such as improved thickness uniformity, reduced phonon-carrier scattering, and higher mobility by hydrogen intercalation compared to conventional EG. (Daniels et al., 2017) The QFS EG demonstrates superior biosensing performance, with detection of spike protein antigen as low as 1 ag/mL and 60 copies/mL in mid-turbinate swabs and aerosols from patients with COVID-19 in as fast as 0.6 s, and an exceptional signal-to-noise ratio of 49.1 dB (average). This sensor allows for ultra-fast and ultra-sensitive diagnosis of COVID-19 in a portable and easy-to-use form with simple device fabrication, without the need of gate or signal amplification as necessary for other graphene-based biosensors (Maity et al., 2018; Majd and Salimi, 2018; Islam et al., 2019; Afsahi et al., 2018; Seo et al., 2020). Furthermore, a graphene/biomarker heterostructure-based biosensor can be easily reused multiple times after desorbing in NaCl solution (1 M) or above 40 °C and biomarkers immobilized on the biosensor again, which will make the ultimate biosensor very cost-effective and productive.
2. Material and methods
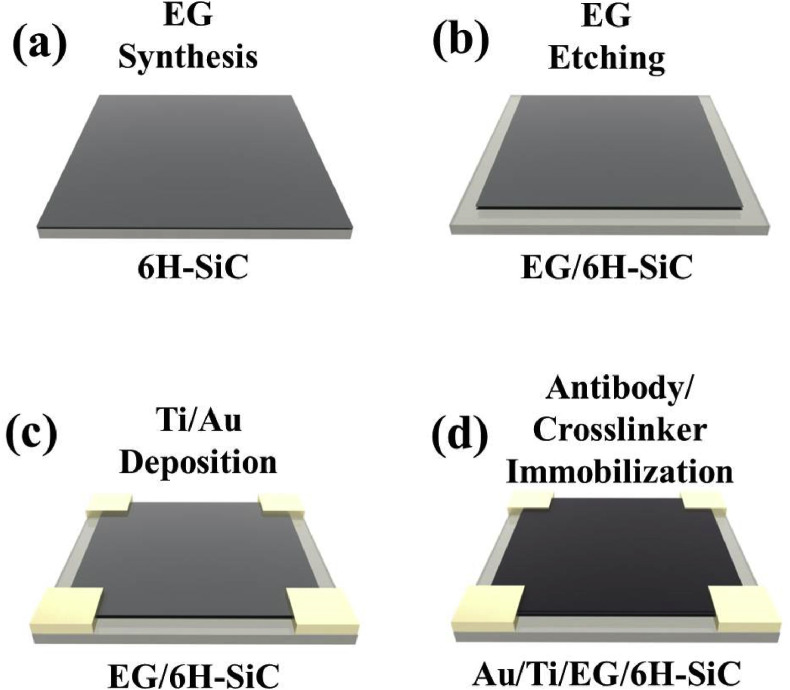
Fabrication of the biosensor for rapid detection of COVID-19 was performed using separately optimized immobilization conditions of SARS-CoV-2 spike S1 protein antibody (SinoBiological, Catalog Number: 40,592-T62) with crosslinker (poly-L-lysine, Sigma-Aldrich) and hydrogen intercalated QFS EG on silicon carbide (SiC); the latter was developed in our previous research46. The sensor was fabricated following the steps shown in Fig. 1 .
Fig. 1.
Schematic diagram of the COVID-19 sensor fabrication process. (a) Synthesis of quasi-freestanding bilayer epitaxial graphene (EG) on silicon carbide (SiC). (b) Etching of the edge side on EG using CF4 plasma with a simple mask. (c) Deposition of Ti/Au metal stack as electrodes using an e-beam evaporator. (d) Immobilization of crosslinker and SARS-CoV-2 S1 spike protein on EG. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.1. Fabrication of SARS-CoV-2 antibody/EG heterostructure based biosensors
High-quality QFS EG is synthesized through Si sublimation and subsequent hydrogen intercalation on a 4-inch diameter semi-insulating (0001) ∼0.1° off-axis 6H–SiC using an Aixtron/Epigress VP508 horizontal hot-wall reactor. After dicing it (8 mm × 8 mm), the EG/SiC samples were cleaned with acetone and isopropanol (IPA), rinsed with deionized (DI) water, and dried with pure N2 (Fig. 1(a)). And then, the mesa was etched on the EG samples by CF4 plasma (Trion IRE) with a simple mask (7 mm × 7 mm) to isolate the EG on SiC (Fig. 1(b)). The formation of the mesa was followed by the deposition of the four contact electrodes (2 mm × 2 mm), Ti/Au (30 nm/220 nm), by electron beam evaporation (Angstron Ebeam Evaporator PLC Driven B) on EG (Fig. 1(c)). After that, the crosslinker and the SARS-CoV-2 spike S1 protein antibodies were then immobilized on EG to prepare the biosensors. The crosslinker, 0.1% of poly-L -lysine (Sigma-Aldrich, P8920), was diluted in DI water to 0.01%, was immobilized on the EG for 1 h at room temperature, and then rinsed three times with phosphate-buffered saline (PBS) (1 × ). The crosslinker allows the strong adhesion with EG via the hydrophobic interaction between its butyl chains and graphene surface for further functionalization with bioactive molecules (Wang et al., 2013; Shan et al., 2009). In addition, several groups have reported effective ways to prepare the surface-functionalized graphene-based biosensors with biomarkers targeting viruses, making it possible to design antibody/graphene heterostructures (Islam et al., 2019; Afsahi et al., 2018; Seo et al., 2020). Finally, the SARS-CoV-2 spike S1 protein antibodies (1 mg/mL) were immobilized onto the crosslinker by adsorption, due to hydrophobic interaction between antibody and crosslinker (similar to immunoassay coating process) for 12 h at 4 °C after diluting the SARS-CoV-2 spike S1 protein antibody (1:1000 in ELISA coating buffer (1 × ), BioLegend, Catalog Number: 421,701).
2.2. Measurement and characterization
The structural, optical, and electrical properties of the SARS-CoV-2 antibody/EG heterostructure were investigated by atomic force microscope (AFM), X-ray photoelectron spectroscopy (XPS), and Raman spectroscopy to ensure EG substrate integrity and antibody/crosslinker bonding on EG. The fabricated sensors were exposed to SARS-CoV-2 spike S1 protein antigen (SinoBiological, Catalog Number: 40591-V08H) by dropping diluted protein (1 ag/mL to 1 μg/mL) onto the surface at room temperature. Clinical samples described below (Section 2.3) were tested in a biosafety cabinet, and cross-reactivity with four seasonal human coronaviruses was also examined. To measure the electrical response of the sensor, the sensor was attached to a chip carrier (Chelsea Technology Inc, 1.5 cm × 1.5 cm), and their Ti/Au metal contact pads were connected to the chip carrier terminals by an Au wire bonder. And then, the electrical response of the sensors was measured using the Gamry 3000 Source/Measure Unit. The input current of 10 mA was applied to the sensor directly and maintained during the measurement. The detected output electrical response was normalized as ΔV/V o, where ΔV is the change in voltage and V o is the original voltage.
2.3. Clinical samples of SARS-CoV-2 and seasonal coronaviruses
Clinical samples, including seasonal coronaviruses and SARS-CoV-2, were collected from volunteers who participated in the Prometheus@UMD (Zhu et al., 2020) and the StopCOVID@UMD studies, respectively. Both studies were approved by the Internal Review Board of the University of Maryland, College Park (No. 1004100 and 1556127, respectively). Written consent was obtained from all study participants. Mid-turbinate swabs were eluted in 1 mL of Universal Transport Media (BD). Saliva samples were stored as is or diluted with 1 × Phosphate-buffered saline containing 0.1% bovine serum albumin (PBS/BSA) when the volume was less than 1 mL. Exhaled breath aerosol condensates were collected using the Gesundheit-II (Yan et al., 2018) and concentrated to 1 mL using Centricon Plus-70 columns with 100 kDa molecular weight cutoff (Millipore Sigma). All clinical samples were aliquoted and stored at −80 °C until further analyses. The presence of seasonal coronaviruses was confirmed using TaqMan Array Cards (Thermo Fisher Scientific) that screen for common respiratory pathogens, including 229E, HKU1, NL63, and OC43 strains of human coronaviruses. Samples collected from study participants exposed to SARS-CoV-2 were screened by single-well real-time RT-PCR assays, which were carried out using the TaqPath COVID-19 Combo Kit (Thermo Fisher Scientific). SARS-CoV-2 positive samples were further quantified based on standard curves established from serial dilutions of quantified inactivated virus (BEI Resources) using the same reagents.
3. Results and discussion
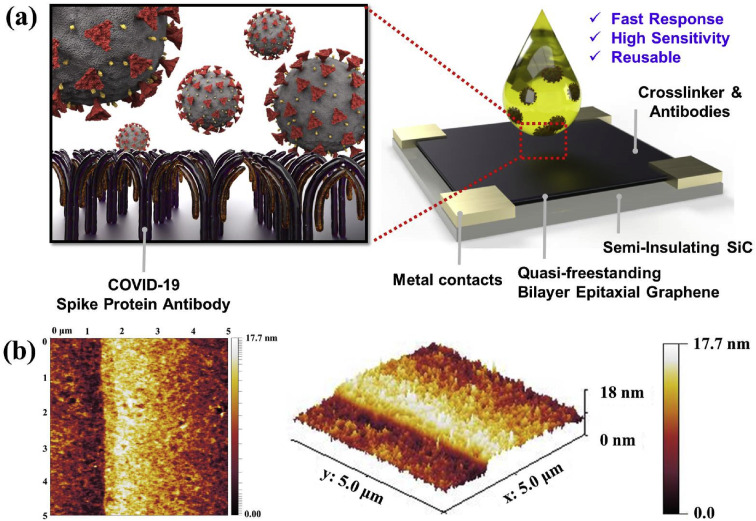
Fig. 2(a) present a schematic illustration of the COVID-19 sensor. The sensor consists of four layers: the semi-insulating SiC substrate, EG, crosslinker, SARS-CoV-2 S1 spike protein antibodies, and Ti/Au contact electrodes. The high quality and uniformity of EG enable the bottom-up direct immobilization of both crosslinker and SARS-CoV-2 S1 spike protein antibodies on EG without any complex transfer methods. EG supports large area synthesis, i.e., up to wafer size range, for commercial scale-up. Details on the EG's synthesis and properties are explained extensively in our previous work (Daniels et al., 2017). Fig. 2(b) shows the AFM images of SARS-CoV-2 S1 spike protein antibody/crosslinker prepared on EG/SiC. The immobilized antibody/crosslinker is uniform and dense on EG, indicating the root-mean-square (RMS) roughness of 1.9 nm. The RMS roughness of the crosslinker and EG are 0.8 nm and 0.7 nm, respectively (Fig. S1).
Fig. 2.
(A) shows schematic illustrations of the sensor design, and (b) shows the atomic force microscopy images of the surface on the antibody/crosslinker prepared on EG/SiC.
The bonding of both the EG and its heterostructure with the SARS-CoV-2 S1 spike protein antibodies was confirmed using XPS and Raman spectroscopy, as shown in Figs. S2 and S3. The most critical parameters in these analyses are the changes in the C1s, O1s, and N1s regions from the XPS results and the G and D peaks from the Raman results, which prove the functionalization of the graphene and the addition of other atomic media. The detailed comparison analyses are discussed in the supplementary information (Figs. S2 and S3). An appearance of new peaks in all regions (i.e., C1s, O1s, and N1s) from the XPS and a red-shifted G peak and an enhanced D peak of EG from the Raman indicate that the SARS-CoV-2 S1 spike protein antibodies/crosslinker has bonded to the EG surface successfully.
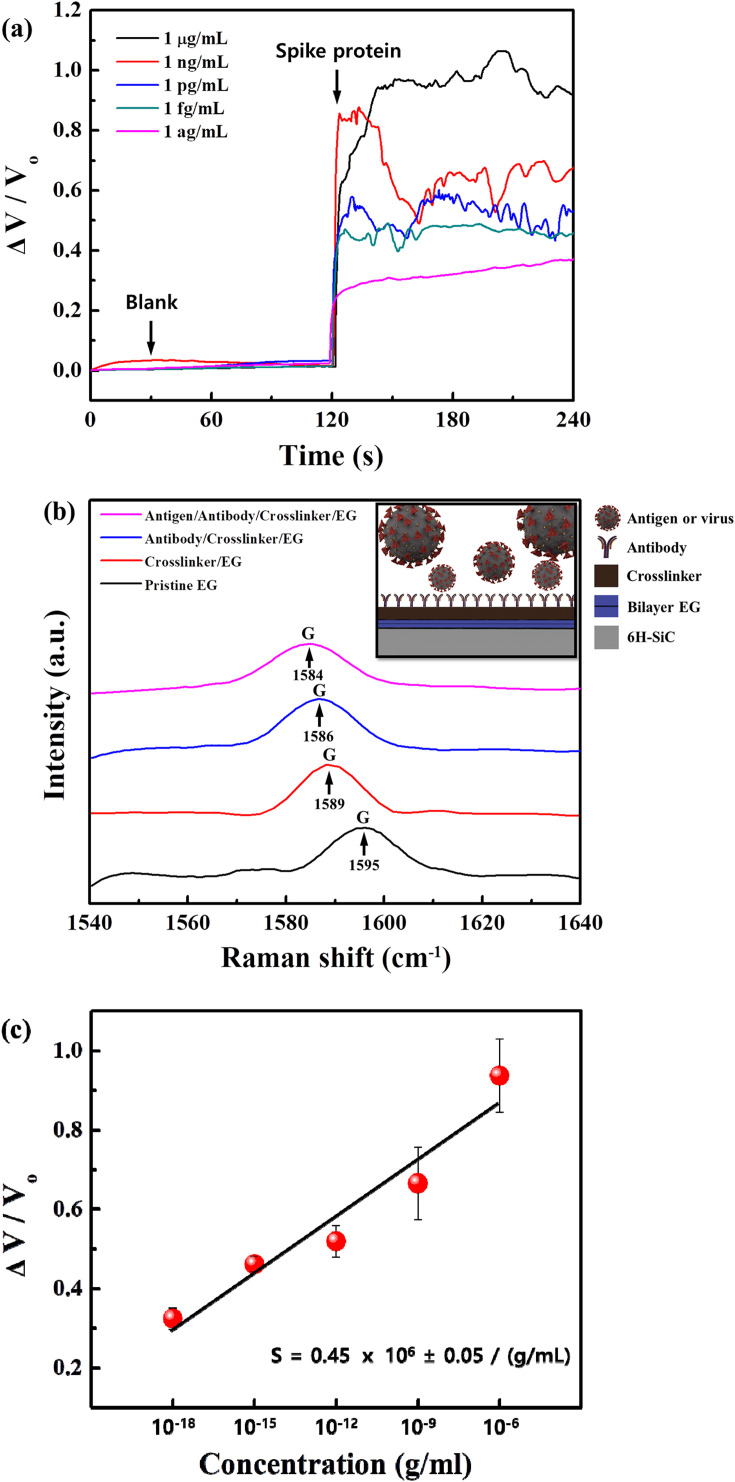
To investigate the performance of the COVID-19 sensor, we measured the sensor's response to varying concentrations of the SARS-CoV-2 S1 spike protein antigen from 1 ag/mL to 1 μg/mL as shown in Fig. 3 (a). The spike protein solutions were prepared in ELISA assay diluent (1 × ) by serial dilution, and 1 μL was dropped onto the sensor for the test. No significant changes were observed from 1 μL of pure ELISA assay diluent (i.e., blank), which served as a baseline before the spike protein testing and a means to distinguish sensor response from the ELISA assay diluent. The sensor responded to as low as 1 ag/mL of SARS-CoV-2 S1 spike protein antigen in ELISA assay diluent, which is not only significantly lower than the LOD of conventional immunoassays (∼ng/mL) but also one of the best LOD values reported on the biosensors (refer to Table S2). A pristine EG-based sensor was also tested to distinguish the sensor performance from the EG-related response and showed a response of 6.2% with 100 ng/mL (Fig. S4). To confirm the performance of the COVID-19 sensor, we tested it with 1 ag/mL of SARS-CoV-2 S1 spike protein antigen from another manufacturer (R&D Systems, Catalog Number: 10569-CV), as shown in Fig. S5. We find that the sensor clearly distinguished between a blank and 1 ag/mL protein and yielded an average response of 31%, similar to the protein concentration of 1 ag/mL from SinoBiological with a response of 32.4%. Altogether, this indicates that this COVID-19 sensor is ultra-high sensitive and selective for the SARS-CoV-2 S1 spike protein antigen. The proposed COVID-19 sensor uses polyclonal antibodies for the heterostructure with EG, which can be powerful tools for detecting an antigen with higher signal levels than monoclonal antibodies because polyclonal antibodies can recognize different epitopes on the same protein antigen. The detection limit of the proposed sensor, as shown in Fig. 3, is similar to the molecular weight of a single spike protein. This LOD is possible as the proposed sensor, with polyclonal antibodies, can detect several epitopes from degraded or fragmented protein. Fig. S6 shows the SDS-PAGE results of SARS-CoV-2 spike S1 protein antigens, and potential degradation of the protein was expected based on the bands identified under 70 kDa (predicted molecular mass is about 75 kDa by R&D Systems and SinoBiological), which might explain an extremely low LOD of the proposed sensor. Another factor that may contribute to the ultra-high sensitivity could be that the detection of the voltage change is far more sensitive than traditional methods of detecting antigen-antibody binding, even very weak antigen-antibody binding that would be washed away in traditional assays would be picked up by this sensor, leveraging the superior advantages of EG.
Fig. 3.
Detection performance of SARS-CoV-2 S1 spike protein antigen. (a) Real-time variation in COVID-19 sensor with the different concentrations ranging from 1 ag/mL to 1 μg/mL. (b) The Raman spectra show a shift in the graphene G peak position with the crosslinker, SARS-CoV-2 spike S1 protein antibody, and antigen (1 ag/mL) on the sensor. (c) The fractional change in output (ΔV/Vo) with varying SARS-CoV-2 S1 spike protein concentrations over the range of 1 ag/mL – 1 μg/mL (semi-log scale). A least-square fit line to the experimental data points is also shown, which yields an average sensitivity of 0.45 × 106 ± 0.05 / (g/mL) from the slope. Error bars indicate standard deviation based on measurements of the sensors.
However, simple charge transfer from antibody-antigen binding to the underlying QFS EG cannot fully explain the response observed with these sensors, at least within the attogram regime. The complex interplay of polarization fields at the QFS EG surface explains the significant voltage response of the sensor to 1 ag/ml SARS-CoV-2 S1 spike proteins. The origin of the p-type doping (1013 cm−2) of QFS EG on 6H–SiC is caused by a net positive polarization charge at the 6H–SiC surface. The hydrogen passivated SiC binds free electrons to the bottom surface of QFS EG, leaving behind an excess of holes (Mammadov et al., 2014). Similarly, the protonated amine groups from poly-L-lysine (PLL) that form the cation- bonds with the top surface of the QFS EG have a net positive polarization that opposes the substrate's polarization field, reducing this induced charge. This is consistent with the Raman data, which found a significant charge depletion on the graphene surface after functionalization with PLL, as indicated by a large G-peak redshift (6 cm−1) as shown in Fig. 3(b). The additional layers of SARS-CoV-2 antibody and subsequent antigen provide additional fields. As an antibody-antigen reaction occurs, electrons are transferred to the PLL, which deprotonates the amine groups in the polymer. The resulting polarization field from the PLL is reduced, but this is insufficient to explain the magnitude of the change in response as it should not significantly affect the field. However, it does produce inhomogeneity in the field, which can generate strain in the underlying graphene sheet. It has been found in the literature that small compressive strains (∼0.05%) can lead to large reductions in carrier concentration in graphene, on the order of ½ the total magnitude (Giannazzo et al., 2019; Schmidt et al., 2011). This inhomogeneity would produce compressive strain, as it removes the positive charge interacting with a positive QFS EG layer. Therefore, this strain would reduce the local carrier concentration and induce strain on other nearby C–C bonds propagating its effect. This inhomogeneity is also mobile, as cation-π bonds allow for charge transfer (Sharma et al., 2015), indicating that the non-protonated amine can move to reach the lowest energy configuration. This could delocalize this strain effect, leading to the significant resistance changes seen in our device, given the large-scale cation-π system of the PLL-QFS EG. Compared to CVD graphene (Grolltex, Inc.) with grain sizes> 80 μm, QFS EG is single-crystal across an entire wafer of SiC. On CVD graphene, we observed by Raman only a 2 cm−1 redshift in the G-peak after PLL functionalization and no response after antibody-antigen binding with exposure to 1 ag/mL of the SARS-CoV-2 S1 spike protein antigen as shown in Fig. S12. This explains why CVD graphene biosensors found in the literature require a gate or signal amplification to operate (Maity et al., 2018; Majd and Salimi, 2018; Islam et al., 2019; Afsahi et al., 2018; Seo et al., 2020). For the QFS EG, we observed a 6 cm−1 redshift in the G-peak after PLL functionalization, a 3 cm−1 redshift after antibody immobilization, and an additional 2 cm−1 redshift after antibody-antigen binding. After binding, we attribute the G-peak shift observed to strain induced on the QFS EG, enabling the transduction of very few antibody-antigen bindings as well as operation without a gate or amplification.
The sensitivity (S) of a sensor is defined as the slope of the output (ΔV/V o, where ΔV is the change in voltage and V o is the original voltage) to the input quantity (i.e., concentration ΔC) and can be determined from the slope of the least-squares line fitted to the ΔV/V o vs ΔC graph in Fig. 3(c) (Kim et al., 2019). The sensor response corresponding to each spike protein antigen concentration (ΔC) is plotted on a semi-log scale, and we find that the sensor response increases monotonically and linearly with increased spike protein antigen (1 ag/mL to 1 μg/mL), with the sensor sensitivity calculated as 0.45 × 106 ± 0.05 / (g/mL). The response time can also be estimated from the magnified time axis plot in Fig. S7 to be 1–32 s as defined by 90% of the low-to-high response (Xin et al., 2019). In addition, the signal-to-noise ratio (SNR) can be calculated, quantifying the performance of a sensor in response to a particular exposure. The SNR expressed in dB can be calculated by the equation
| SNRdB = 20log10((ΔV/Vsignal)/(ΔV/Vnoise)), | (1) |
where ΔV/V signal is the fractional change in voltage due to the exposed spike protein antigen concentration, and ΔV/V noise is the standard deviation of the voltage fluctuation at that concentration. The calculated average SNR ranging between 1 ag/mL and 1 μg/mL from Eq. (1) is 49.1 dB. The sensor's performance in terms of sensitivity, SNR, and response time, including the fabrication simplicity in an array over a large area, originates from the novel architecture of the direct synthesis of the antibody/EG heterostructure, which enables itself to adsorb specific target spike protein antigen. The EG acts as a channel to transport the carriers quickly, leveraging its high conductivity.
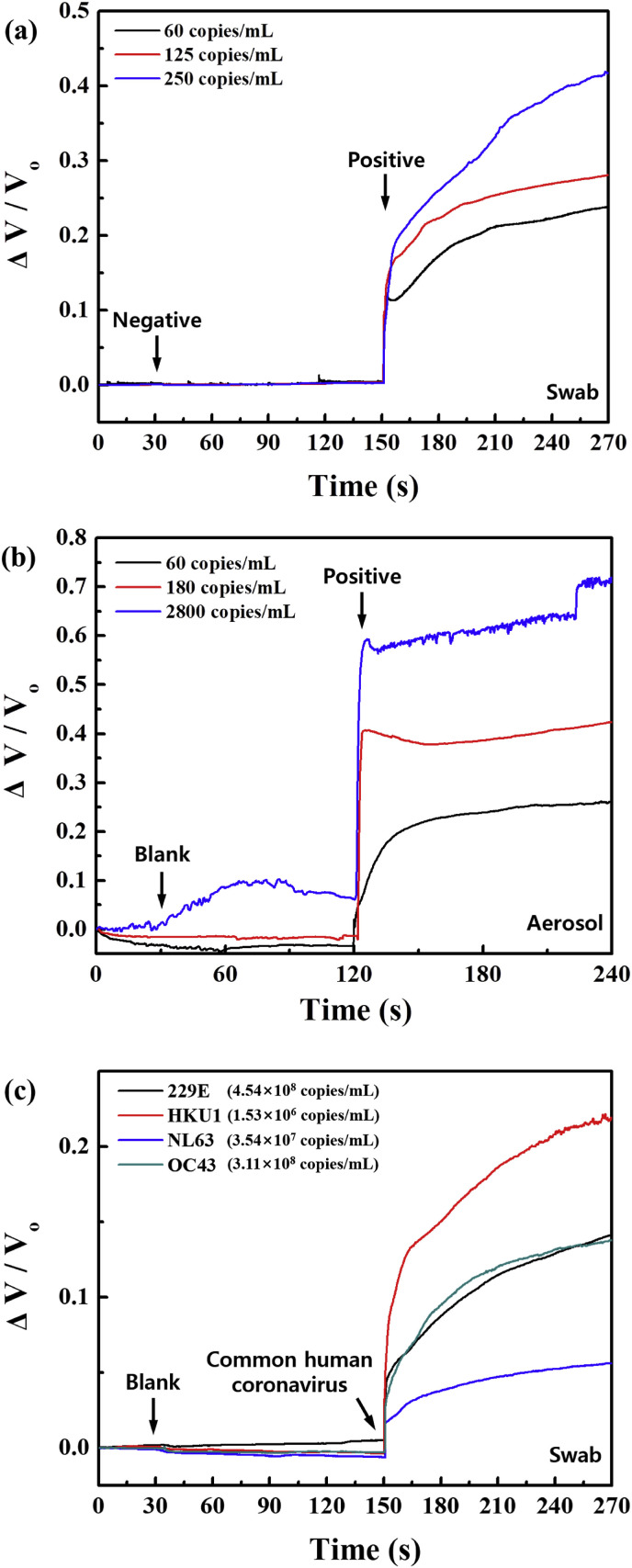
For a practical diagnosis application, we tested the COVID-19 sensor with SARS-CoV-2 virus from infected patient samples such as mid-turbinate swabs, saliva, exhaled breath aerosol samples, and common human coronaviruses. Detailed information on patient samples is described in Table S1. The patient samples were used after dissolving in 1% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) (1 × ) to minimize the nonspecific binding on the sensor surface as a blocking agent approach, and 1 μL was dropped onto the sensor for the test. The BSA is the most common economic blocking agent and is generally used to improve sensitivity and maximize SNR by reducing nonspecific background noise (Frutiger et al., 2021). To distinguish the related response from the 1% BSA in PBS (1 × ), 1 μL of pure 1% BSA in PBS (1 × ) (i.e., blank) was also tested as the baseline before the patient samples testing, which results in no significant changes. Fig. 4 (a) shows the response of the COVID-19 sensor as it is subjected to negative samples (from a patient that was tested negative by RT-PCR) and different concentrations of positive samples (swab) such as 60, 125, and 250 copies/mL of SARS-CoV-2 virus. We find that the sensor distinguishes between negative and positive samples very clearly, while the response increases monotonically with an increase in the concentration of positive samples. In general, the RT-PCR, which is the most effective standard for the COVID-19 testing currently, requires at least 250 copies/mL of swab samples for reliable results. Our sensor responds to positive samples diluted as low as 60 copies/mL and had remarkably high SNR of 64 dB ranging between 60 and 250 copies/mL, making it a strong candidate for a practical portable diagnosis application. Although 60 copies/mL roughly translates to 0.06 virus particles in a 1 μL drop on the sensor, virus particles in these clinical samples were likely disrupted by freezing and thawing. Furthermore, each virus particle contains numerous spike proteins, and infected cells, which may express spike proteins depending on the stage of infection, could also present in these samples (Sender et al., 2024). We also tested the detection performance of our sensor using saliva, and the sensor clearly distinguished between negative and positive samples (response time: 1.1 s, SNR: 39.2 dB) as shown in Fig. S8. In Table S2, we compare the best reported results for the significant performance on SARS-CoV-2 detection. Fig. 4(b) further demonstrates the sensor's efficacy in distinguishing positive exhaled breath aerosol samples based on the three different patients with the various concentrations of SARS-CoV-2 viruses such as 60, 180, and 2800 copies/mL. Notably, the sensor responded to positive exhaled breath aerosol samples very well and had high SNR of 44.1 dB. The demonstration of the exhaled breath aerosol samples using this sensor once again highlights its strong potential for a practical portable diagnosis application, especially a breathalyzer test that would allow for rapid screening of COVID-19 in populations such as airports, schools, and factories. To investigate the cross-reactivity of common coronaviruses on our COVID-19 sensor, we tested the sensor with seasonal coronaviruses (common human coronaviruses) such as 229E, HKU1, NL63, and OC43 (∼108 copies/mL), as shown in Fig. 4(c). We find that the sensor detects all four common human coronaviruses, yielding average responses of 4.5% (NL63), 10% (229E), 11% (OC43), and 18% (HKU1). This is likely due to the cross-reaction between the anti-SARS-CoV-2 S1 antibody and spike proteins of the seasonal coronaviruses. Even when exposed to a high concentration of seasonal coronaviruses (∼108 copies/mL), the sensor's responses were still lower than those when exposed to clinical samples with a significantly lower amount of SARS-CoV-2. Therefore, we believe this sensor has great potential as a diagnostic tool.
Fig. 4.
Demonstration of a practical diagnosis application of the proposed COVID-19 sensor in real-time by testing clinical samples such as (a) mid-turbinate swabs (patient #1) and (b) exhaled breath aerosol samples (patient #2, 3, and 4). (c) Real-time response of common human coronaviruses (229E, HKU1, NL63, and OC43) using swabs on the COVID-19 sensor.
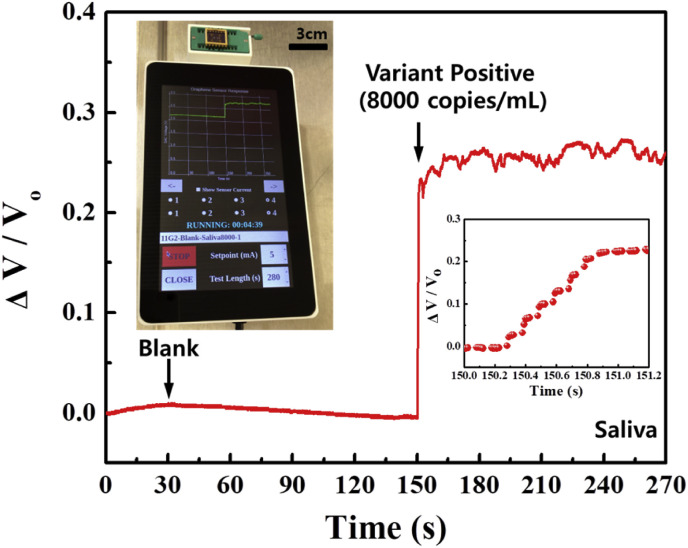
A practical application of the portable sensor was demonstrated by using it to detect the SARS-CoV-2 B.1.1.7 variant. The portable unit includes four main components: a high frequency (125 kHz) and sensitive galvanostat, a microprocessor module, a touchscreen interface, and a cloud storage link (Fig. S9). The actual sensor was physically attached to the portable unit, and positive variant samples of the swab and saliva were dropped onto the sensor for the test. Fig. 5 shows the picture of the portable unit, including the proposed sensor (inset) and the sensor responses as it is subjected to the variant saliva sample (the result of the swab sample is shown in Fig. S10 and Supplementary Video 1
Fig. 5.
Demonstration of a practical diagnosis application of the proposed portable COVID-19 sensor testing positive human sample (patient #6: saliva, 8000 copies/mL of B.1.1.7 variant) in real-time. Note that saliva samples were dissolved in 1% bovine serum albumin (BSA) in phosphate-buffered saline PBS) (1 × ), and 1 μL was dropped onto the sensor for the test. The sensor clearly distinguished variant positive sample and showed a signal-to-noise ratio of 67.57 dB and a response time of 0.6 s.
The following is the supplementary data related to this article:
4. Conclusions
An ultra-high sensitive biosensor for rapid detection of COVID-19 based on the heterostructure of SARS-CoV-2 antibody and quasi-freestanding bilayer epitaxial graphene on a SiC substrate, utilizing a simple fabrication process, has been demonstrated. The sensor exhibited a linear response to SARS-CoV-2 S1 spike protein from 1 ag/mL to 1 μg/mL, with detection in positive patient samples including B.1.1.7 variant such as the mid-turbinate swab and exhaled breath aerosol (LOD: 60 copies/mL). The portable sensor demonstrated an exceptional SNR of 67.57 dB and a fast response time of 0.6 s. This technique is still based on antigen-antibody reactions, which is the same principle as the immunoassay technique, but QFS EG allows the reaction to be transduced as an electrical signal output rather than enzymatic colorimetric enhanced signal output, without gate or signal amplification. The lower detection limit provides a quicker and more robust reaction. Further trajectory analysis with larger cohorts is needed to demonstrate its capacity for early diagnosis. With this method, we hope to provide a novel diagnostic method that allows for on-site diagnosis before going to a screening clinic, prior to developing symptoms, as well as massive screening for COVID-19 as a portable breathalyzer in airports, schools, and factories.
CRediT authorship contribution statement
Soaram Kim: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Data curation, Writing – original draft, Writing – review & editing, Visualization, Supervision, Project administration. Heeju Ryu: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing – original draft, Writing – review & editing, Visualization, Supervision. Sheldon Tai: Validation, Formal analysis, Resources, Writing – review & editing. Michael Pedowitz: Validation, Formal analysis, Investigation, Writing – original draft, Writing – review & editing, Visualization. John Robertson Rzasa: Software, Resources, Writing – review & editing. Daniel J. Pennachio: Resources. Jenifer R. Hajzus: Resources. Donald K. Milton: Resources, Writing – review & editing. Rachael Myers-Ward: Resources, Writing – review & editing. Kevin M. Daniels: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Data curation, Writing – original draft, Writing – review & editing, Visualization, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We acknowledge the financial support for this work from the University of Maryland UM Ventures MDDF fund. Clinical samples were collected as a part of Prometheus-UMD sponsored by the Defense Advanced Research projects Agency (DARPA) BTO under the auspices of Col. Matthew Hepburn through agreement N66001-18-2-4015. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position or policy of the funding agency, and no official endorsement should be inferred. Sample collection and PCR testing were also supported by the National Institute of Allergy and Infectious Diseases Centers of Excellence for Influenza Research and Surveillance (CEIRS), Grant Number HHSN272201400008C, the Centers for Disease Control and Prevention, Contract Number 200-2020-09528, and by a grant from the Bill & Melinda Gates Foundation. HR is supported by the National Research Foundation of Korea (NRF-2020R1A6A3A03037852). Work at the US Naval Research Laboratory was supported by the Office of Naval Research. This research was performed while DJP held an NRC Research Associateship award at NRL. JRH acknowledges the American Society for Engineering Education Naval Research Laboratory Postdoctoral Fellow program. These funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We express gratitude to Dr. D. Kurt Gaskill for his invaluable insight throughout this work. We wish to extend our special thanks to Daniel Lewis for the wafer dicing in this work.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113803.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Afsahi S., Lerner M.B., Goldstein J.M., Lee J., Tang X., Bagarozzi D.A., Jr., Pan D., Locascio L., Walker A., Barron F., Goldsmith B.R. Biosens. Bioelectron. 2018;100:85–88. doi: 10.1016/j.bios.2017.08.051. [DOI] [PubMed] [Google Scholar]
- Arons M.M., Hatfield K.M., Reddy S.C., Kimball A., James A., Jacobs J.R., Taylor J., Spicer K., Bardossy A.C., Oakley L.P., Tanwar S., Dyal J.W., et al. N. Engl. J. Med. 2020;382:2081–2090. doi: 10.1056/NEJMoa2008457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Stereithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.-Y., Huevara H., Wadford D.A., Chen J.S., Chiu C.Y. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E., Ledford H. Nature. 2021;590:15–16. doi: 10.1038/d41586-021-00241-6. [DOI] [PubMed] [Google Scholar]
- Cassaniti I., Novazzi F., Giardina F., Salinaro F., Sachs M., Perlini S., Bruno R., Mojoli F., Baldanti F. J. Med. Virol. 2020;92:1724–1727. doi: 10.1002/jmv.25800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee G.S., Chatterjee S., Ray A.K., Chakraborty A.K. Sensor. Actuator. B Chem. 2015;221:1170–1181. [Google Scholar]
- Chen H.-C., Chen Y.-T., Tsai R.-Y., Chen M.-C., Chen S.-., Xiao M.-C., Chen C.-L., Hua M.-Y. Biosens. Bioelectron. 2015;66:198–207. doi: 10.1016/j.bios.2014.11.019. [DOI] [PubMed] [Google Scholar]
- Choi H.J., Lee J., Byeon M., Hong E.T., Park H., Lee Y.C. ACS Appl. Nano Mater. 2020;3:2257–2265. [Google Scholar]
- Dagan N., Barda N., Kepten E., Miron O., Perchik S., Katz M.A., Hernan M.A., Lipsitch M., Reis B., Balicer R.D. N. Engl. J. Med. 2021;384:1412–1423. doi: 10.1056/NEJMoa2101765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels K.M., Jadidi M.M., Sushkov A.B., Nath A., Boyd A.K., Sridhara K., Drew H.D., Murphy T.E., Myers-Ward R.L., Gaskill D.K. 2D Mater. 2017;4 [Google Scholar]
- Davies D.R., Cohen G.H. Proc. Natl. Acad. Sci. U.S.A. 1996;93:7–12. doi: 10.1073/pnas.93.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyer W.J., Bennett J.C. Proc. Natl. Acad. Sci. U.S.A. 1965;54:864–869. doi: 10.1073/pnas.54.3.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frutiger A., Tanno A., Hwu S., Tiefenauer R.F., Voros J., Nakatsuka N. Chem. Rev. 2021;121:8095–8160. doi: 10.1021/acs.chemrev.1c00044. [DOI] [PubMed] [Google Scholar]
- Gandhi M., Yokoe D.S., Havlir D.V. N. Engl. J. Med. 2020;382:2158–2160. doi: 10.1056/NEJMe2009758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannazzo F., Shtepliuk I., Ivanov I.G., Iakimov T., Kakanakova-Georgieva A., Schiliro E., Fiorenza P., Yakimova R. Nanotechnology. 2019;30:284003. doi: 10.1088/1361-6528/ab134e. [DOI] [PubMed] [Google Scholar]
- Hoffman T., Nissen K., Krambrich J., Ronnberg B., Akaberi D., Esmaeilzadeh M., Salaneck E., Lindahl J., Lundkvist A. Infect. Ecol. Epidemiol. 2020;10:1754538. doi: 10.1080/20008686.2020.1754538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S., Shukla S., Bajpai V.K., Han Y.-K., Huh Y.S., Kumar A., Ghosh A., Handhi S. Biosens. Bioelectron. 2019;126:792–799. doi: 10.1016/j.bios.2018.11.041. [DOI] [PubMed] [Google Scholar]
- Jahanshahlu L., Rezaei N. Biomed. Pharmacother. 2020;129:110337. doi: 10.1016/j.biopha.2020.110337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi S.R., Sharma A., Kim G.-H., Jang J. Mater. Sci. Eng. C. 2020;108:110465. doi: 10.1016/j.msec.2019.110465. [DOI] [PubMed] [Google Scholar]
- Jung J.H., Cheon D.S., Liu F., Lee K.B., Seo T.S. Angew. Chem. Int. Ed. 2010;49:5708–5711. doi: 10.1002/anie.201001428. [DOI] [PubMed] [Google Scholar]
- Kim S., Ryoo S.-R., Na H.-K., Kim Y.-K., Choi B.-S., Lee Y., Kim D.-E., Min D.-H. Chem. Commun. 2013;49:8241–8243. doi: 10.1039/c3cc43368d. [DOI] [PubMed] [Google Scholar]
- Kim S., Dong Y., Hossain M.M., Gorman S., Towfeeq I., Gajula D., Childress A., Rao A.M., Koley G. ACS Appl. Mater. Interfaces. 2019;11:16006–16017. doi: 10.1021/acsami.9b01964. [DOI] [PubMed] [Google Scholar]
- Kumar S., Sharma J.G., Maji S., Malhotra B.D. Biosens. Bioelectron. 2016;78:497–504. doi: 10.1016/j.bios.2015.11.084. [DOI] [PubMed] [Google Scholar]
- Ledford H. Nature. 2021;591:513–514. doi: 10.1038/d41586-021-00650-7. [DOI] [PubMed] [Google Scholar]
- Li L., Zhang L., Yu J., Ge S., Song X. Biosens. Bioelectron. 2015;71:108–114. doi: 10.1016/j.bios.2015.04.032. [DOI] [PubMed] [Google Scholar]
- Lipman N.S., Jackson L.R., Trudel L.J., Weis-Garcia F. ILAR J. 2005;46:258–268. doi: 10.1093/ilar.46.3.258. [DOI] [PubMed] [Google Scholar]
- Liu F., Choi K.S., Park T.J., Lee S.Y., Seo T.S. BioChip. J. 2011;5:123–128. [Google Scholar]
- Liu Q., Aroonyadet N., Song Y., Wang X., Cao X., Liu Y., Cong S., Wu F., Thompson M.E., Zhou C. ACS Nano. 2016;10:10117–10125. doi: 10.1021/acsnano.6b05171. [DOI] [PubMed] [Google Scholar]
- Mahase E. BMJ. 2021;372:n597. doi: 10.1136/bmj.n597. [DOI] [PubMed] [Google Scholar]
- Maity A., Sui X., Jin B., Pu H., Bottum K.J., Huang X., Chang J., Zhou G., Lu G., Chen J. Anal. Chem. 2018;90:14230–14238. doi: 10.1021/acs.analchem.8b03226. [DOI] [PubMed] [Google Scholar]
- Majd S.M., Salimi A. Anal. Chim. Acta. 2018;1000:273–282. doi: 10.1016/j.aca.2017.11.008. [DOI] [PubMed] [Google Scholar]
- Mammadov S., Ristein J., Koch R.J., Ostler M., Raidel C., Wanke M., Vasiliauskas R., Yakimova R., Seyller T. 2D Mater. 2014;1 [Google Scholar]
- Navakul K., Warakulwit C., Yenchitsomanus P.T., Panya A., Lieberzeit P.A., Sangma C. Nanomed-Nanotechnol. 2017;13:549–557. doi: 10.1016/j.nano.2016.08.009. [DOI] [PubMed] [Google Scholar]
- Pizzorno A., Padey B., Julien T., Trouillet-Assant S., Traversier A., Errazuriz-Cerda E., Fouret J., Dubois J., Gaymard A., Lescure F.-X., Duliere V., Brun P., Constant S., Poissy J., Lina B., Yazdanpanah Y., Terrier O., Rosa-Calatrava M. Cell Rep. Med. 2020;1:100059. doi: 10.1016/j.xcrm.2020.100059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Marc G.P., Moreira E.D., Zerbini C., Bailey R., Swanson K.A., Roychoudhury S., Koury K., Li P., Kalina W.V., Cooper D., Frenck R.W., Hammitt L.L., Tureci O., Nell H., Schaefer A., Unal S., Tresnan D.B., Mather S., Dormitzer P.R., Sahin U., Jansen K.U., Gruber W.C. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porte L., Legarraga P., Vollrath V., Aguilera X., Munita J.M., Araos R., Pizarro G., Vial P., Iruretagoyena M., Dittrich S., Weitzel T. Int. J. Infect. Dis. 2020;99:328–333. doi: 10.1016/j.ijid.2020.05.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schedin F., Geim A.K., Morozov S.V., Hill E.W., Blake P., Katsnelson M.I., Novoselov K.S. Nat. Mater. 2007;6:652–655. doi: 10.1038/nmat1967. [DOI] [PubMed] [Google Scholar]
- Schmidt D.A., Ohta T., Beechem T.E. Phys. Rev. B. 2011;84:235422. [Google Scholar]
- Scohy A., Anatharajah A., Bodeus M., Kabamba-Mukadi B., Verroken A., Rodriguez-Villalobos H. J. Clin. Virol. 2020;129:104455. doi: 10.1016/j.jcv.2020.104455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sender, R., Bar-On, Y.M., Gleizer, S., Bernshtein, B., Flamholz, A., Phillips, R., & Milo, R., Proc. Natl. Acad. Sci. Unit. States Am. 118, e2024815118. [DOI] [PMC free article] [PubMed]
- Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B., Lee C.-S., Jun S., Park D., Kim H.G., Kim S.-J., Lee J.-O., Kim B.T., Park E.C., Kim S.I. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- Shan C., Yang H., Han D., Zhang Q., Ivaska A., Niu L. Langmuir. 2009;25:12030–12033. doi: 10.1021/la903265p. [DOI] [PubMed] [Google Scholar]
- Shanmugam N.R., Muthukumar S., Chaudhry S., Anguiano J., Prasad S. Biosens. Bioelectron. 2017;89:764–772. doi: 10.1016/j.bios.2016.10.046. [DOI] [PubMed] [Google Scholar]
- Sharma B., Srivastava H.K., Gayatri G., Sastry G.N. J. Comput. Chem. 2015;36:529–538. doi: 10.1002/jcc.23827. [DOI] [PubMed] [Google Scholar]
- Sheridan, C. Nat. Biotechnol. Nov. 5. doi: 10.1038/d41587-020-00021-z. Epub ahead of print. PMID: 33154578.
- Syu Y.-C., Hsu W.-E., Lin C.-T. ECS J. Solid State Sci. Technol. 2018;7:Q3196–Q3207. [Google Scholar]
- Tang Y.-W., Schmitz J.E., Persing D.H., Stratton C.W. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00512-20. e0051-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermisoglou E., Panacek D., Jayaramulu K., Pykal M., Frebort I., Kolar M., Hajduch M., Zboril R., Otyepka M. Biosens. Bioelectron. 2020;166:112436. doi: 10.1016/j.bios.2020.112436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zhao Y., Ma F.-X., Wang K., Wang F.-B., Xia & X.-H. J. Mater. Chem. B. 2013;1:1406–1413. doi: 10.1039/c2tb00454b. [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., Fu S., Gao L., Cheng Z., Lu Q., Hu Y., Luo G., Wang K., Lu Yang, Li H., Wang S., Ruan S., Yang C., Mei C., Wang Y., Ding D., Wu F., Tang X., Ye X., Ye Y., Liu B., Yang J., Yin W., Wang A., Fan G., Zhou F., Liu Z., Gu X., Xu J., Shang L., Zhang Y., Cao L., Guo T., Wan Y., Qin H., Jaing Y., Jaki T., Hayden F.G., Horby P.W., Cao B., Wang C. Lancet. 2020;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo C.H., Jang S., Shin G., Jung G.Y., Lee J.W. Bat. Biomed. Eng. 2020;4:1168–1179. doi: 10.1038/s41551-020-00617-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Xue P., Kang Y., Hui K.M. Anal. Chem. 2013;85:8661–8668. doi: 10.1021/ac401445a. [DOI] [PubMed] [Google Scholar]
- Xiang Q., Huang J., Huang H., Mao W., Ye Z. RSC Adv. 2018;8:1820–1825. doi: 10.1039/c7ra11945c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin X., Zhang Y., Guan X., Cao J., Li X., Long X., Tan X. ACS Appl. Mater. Interfaces. 2019;11:9438–9447. doi: 10.1021/acsami.8b20984. [DOI] [PubMed] [Google Scholar]
- Yan J., Grantham M., Pantelic J., Bueno de Mesquita J.P., Albert B., Liu F., Ehrman S., Milton D.K., Consortium, EMIT Proc. Natl. Acad. Sci. U.S.A. 2018;115:1081–1086. doi: 10.1073/pnas.1716561115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Han J., Zhuo Y., Yang Z., Chai Y., Yuan R. Biosens. Bioelectron. 2014;55:360–365. doi: 10.1016/j.bios.2013.12.040. [DOI] [PubMed] [Google Scholar]
- Yen P.-W., Huang C.-W., Huang Y.-J., Chen M.-C., Liao H.-H., Lu S.-S., Lin C.-Y. Biosens. Bioelectron. 2014;61:112–118. [Google Scholar]
- Zhu S., Jenkins S., Addo K., Heidarinejad M., Romo S., Layne A., Ehizibolo J., Dalgo D., Mattise N.W., Hong F., Adenaiye O.O., Bueno de Mesquita J.P., Albert B.J., Washington-Lewis R., German J., Tai S., Youssefi S., Milton D.K., Srebric J. Environ. Int. 2020;137:105537. doi: 10.1016/j.envint.2020.105537. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.