Abstract
To determine the physiological roles of peroxisome proliferator-activated receptor β (PPARβ), null mice were constructed by targeted disruption of the ligand binding domain of the murine PPARβ gene. Homozygous PPARβ-null term fetuses were smaller than controls, and this phenotype persisted postnatally. Gonadal adipose stores were smaller, and constitutive mRNA levels of CD36 were higher, in PPARβ-null mice than in controls. In the brain, myelination of the corpus callosum was altered in PPARβ-null mice. PPARβ was not required for induction of mRNAs involved in epidermal differentiation induced by O-tetradecanoylphorbol-13-acetate (TPA). The hyperplastic response observed in the epidermis after TPA application was significantly greater in the PPARβ-null mice than in controls. Inflammation induced by TPA in the skin was lower in wild-type mice fed sulindac than in similarly treated PPARβ-null mice. These results are the first to provide in vivo evidence of significant roles for PPARβ in development, myelination of the corpus callosum, lipid metabolism, and epidermal cell proliferation.
In the past 10 years, specific roles for peroxisome proliferator-activated receptor α (PPARα) and PPARγ have emerged while information defining PPARβ-dependent processes is lacking. PPARs are members of the nuclear receptor superfamily (34). The three PPARs exhibit unique tissue distribution, are encoded by separate genes in all species examined to date, and are designated by the subtypes α, β (δ, NUC1), and γ (14, 18, 34, 47, 48). Acting as regulatory transcription factors, the PPARs heterodimerize with retinoid X receptors and modulate gene expression in target genes containing peroxisome proliferator-responsive elements (PPREs) in response to ligand activation.
The three PPARs have related but distinct activities. Activation of PPARα can occur as a result of cold shock (19), food restriction (26), dietary fatty acids (44), and treatment with the hypolipidemic fibrate class of drugs (31). Peroxisomal and mitochondrial β-oxidizing enzymes, microsomal ω-oxidizing enzymes, hepatic fatty acid binding protein, carnitine palmitoyltransferases, and a number of apolipoproteins are all regulated by PPARα ligands/activators (3, 26, 31, 38, 41, 44). These data, obtained in part from the PPARα-null mouse, provide strong in vivo evidence that PPARα regulates lipid metabolism by regulating gene expression of numerous proteins which are clinically relevant for a number of diseases including diabetes, obesity, and atherosclerosis.
Another PPAR isoform, PPARγ, is required for adipocyte differentiation and regulation of adipocyte-specific genes such as the gene for adipocyte fatty acid binding protein aP2 (47). Similar to PPARα, PPARγ is activated by specific ligands, most notably the thiazolidinedione drugs used for type 2 diabetes therapy (32). The phenotype of a PPARγ-null mouse line is embryo lethal due in part to disrupted placental function (4). Tetraploid rescue experiments to bypass the placental defect confirmed an in vivo role for the receptor in adipogenesis (4). Analysis of heterozygotes and chimeras also established a role for PPARγ in adipocyte function and glucose homeostasis (29, 45). Thus, it is clear from null mouse studies that there are distinct metabolic roles for PPARα and PPARγ.
The function of PPARβ has remained elusive. While PPARβ is ubiquitously expressed, some tissues express relatively higher levels of the mRNA including the brain, adipose tissue, and skin (2, 8). Expression of PPARβ is considerably higher in the developing neural tube and the epidermis during rat development (9). No target genes that are controlled only by PPARβ have been identified, but activators for PPARβ including fatty acids (27), bezafibrate (28), and a furan-conjugated linoleic acid metabolite (39) are reported to activate reporter gene constructs containing PPREs through PPARβ. Despite the lack of a specific PPARβ ligand to induce activation, there are several reports suggesting roles for PPARβ in adipocyte differentiation (5), brain function (51), epidermal differentiation (37), uterine implantation (33), and colon cancer (20). In large part, these studies are correlative associations; definitive proof for PPARβ function requires the use of a null mouse model. In the present study, a PPARβ-null mouse was generated and characterized to identify physiological functions dependent on PPARβ.
MATERIALS AND METHODS
Construction of the targeting vector.
Genomic clones corresponding to mouse PPARβ (mPPARβ) were obtained by screening an amplified Sv/129 genomic mouse liver library (Stratagene, La Jolla, Calif.) with a partial (nucleotides [nt] 140 to 1039, 900 bp) mPPARβ cDNA (2) obtained by reverse transcription-PCR (RT-PCR) of RNA from the 3T3 adipocyte cell line. Since there is significant homology between the other PPARs, these clones were subsequently screened with mPPARγ and mPPARα cDNA probes. The PPARβ genomic clones did not hybridize with the two cDNAs. Restriction mapping and sequencing analysis of these clones resulted in the identification of one 9.5-kb genomic clone that contained the last exon and intron of the mPPARβ gene and that was used for constructing the targeting vector. To disrupt the mPPARβ gene, the 1.14-kb phosphoribosyltransferase II gene conferring neomycin resistance (NEO; derived from plasmid pMC1NeoPolyA; Stratagene) was inserted into the XbaI site of the last exon in the same direction of transcription of the genomic clone. The targeting vector contained 1.8 kb of homologous sequence 5′, and 3.5 kb of homologous sequence 3′, of the NEO cassette. A herpes simplex virus thymidine kinase gene inserted at the 5′ end of the construct allowed negative selection.
Electroporation and selection of recombinant ES cells.
Conditions for embryonic stem (ES) cell culture and electroporation have been previously described (31). Twenty-five micrograms of XhoI-linearized targeting vector was used to electroporate Sv/129 ES cells (Genome Systems, St. Louis, Mo.). Of the 56 ES clones that were picked up after positive and negative selection, four were positive for recombination as verified by Southern analysis using both the genomic and NEO-specific probes.
Generation of chimeras.
One of the positive ES clones (JP31) was used for microinjections into recipient C57BL/6N blastocysts as previously described (31). Five chimeras with >60% agouti coat color were used to breed with C57BL/6N females, and one of these produced agouti offspring. The genotype of the F1 agouti litter was determined by Southern blot analysis of BamHI-digested tail DNA isolated from 3-week-old pups. Mice heterozygous for the disrupted PPARβ gene were mated, and homozygous PPARβ-null mice were identified by Southern blot analysis. Since F2 offspring did not exhibit Mendelian distributions of genotypes, F1 heterozygotes were bred with wild-type C57BL/6N mice to obtain F2 heterozygotes. The heterozygous F2 offspring from these matings were subsequently used to establish a colony of homozygous mice, and normal Mendelian distributions were obtained in the F3 generation. The genetic background of mice produced from this colony was on average 75% C57BL/6N, and the mice were used for all experiments unless otherwise noted.
Southern blot analysis.
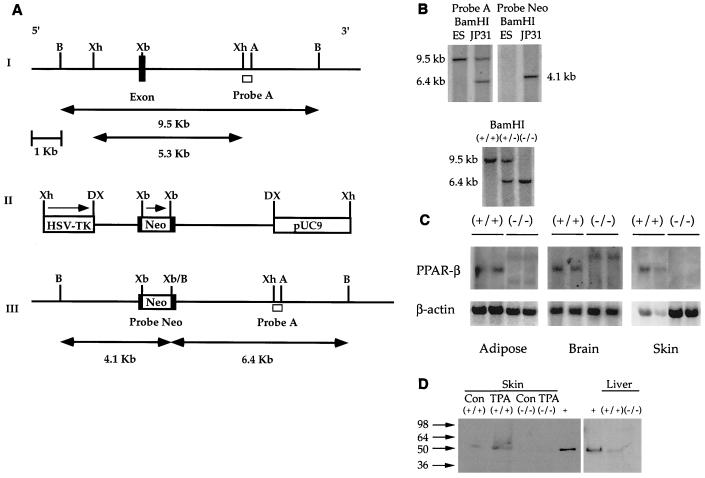
DNA was isolated from ES cells and mouse tails (30), digested with BamHI, electrophoresed, blotted to nylon membranes, and fixed as previously described (31). The blot was hybridized with 3′-flanking probe A, a 650-bp XhoI-AflII fragment. Probe A hybridizes to a 9.5-kb BamHI restriction fragment from wild-type genomes (see Fig. 1A and B). When one allele of the mPPARβ gene is replaced with the targeting vector sequence by homologous recombination, probe A hybridizes with a 6.4-kb BamHI restriction fragment (see Fig. 1A and B). An internal NEO probe was used to hybridize with DNA digested with BamHI to demonstrate single-copy insertion of the targeting vector by a homologous recombination event (Fig. 1B).
FIG. 1.
Targeted disruption of the mPPARβ gene. (A) Strategy for the mPPARβ knockout. (I) Partial map of a mouse genomic fragment containing the second-to-last and last exons encoding the mPPARβ ligand binding domain. Restriction enzymes: B, BamHI; Xh, XhoI; Xb, XbaI; A, AflII. The wild-type 9.5-kb BamHI fragment detected by probe A, a 0.65-kb XhoI-AflII fragment from the 3′ end of the mPPARβ genomic DNA, is indicated. (II) Targeting vector with a total of 5.3 kb of homologous sequence contained in the XhoI fragment of the genomic clone. The 1.14-kb NEO gene in the same orientation relative to mPPARβ transcription was inserted into the XbaI site indicated. The NEO cassette introduces a new BamHI restriction site used for genotyping by Southern blot analysis. A pMCITK expression cassette (herpes simplex virus thymidine kinase [HSV-TK]) was added at the 3′ end of the construct for negative selection. DX, disrupted XhoI site. (III) The expected homologous recombination event of mPPARβ. When one allele of the mPPARβ gene was replaced with the targeting vector sequences by homologous recombination, a 6.4-kb restriction fragment appeared when the gene was analyzed with probe A. (B) Genomic Southern blots of ES cell DNA (top) and Southern blot of tail DNA from wild-type (+/+), heterozygous (+/−), and homozygous mutant (−/−) mice (bottom). (C) Northern analysis of PPARβ mRNA in selected tissues from wild-type (+/+) and PPARβ-null (−/−) mice. (D) Western blot analysis of skin and liver from wild-type (+/+) and PPARβ-null (−/−) mice. Samples from skin of TPA-treated mice were also analyzed. +, positive control. Con, control.
Northern blot analysis.
Total RNA was isolated from gonadal adipose samples (adipose tissue from one or two mice) after disruption of cells in guanidine thiocyanate. Total RNA from corpus callosum and skin samples was isolated using Trizol reagent and the manufacturer's recommended procedures (GIBCO-BRL, Grand Island, N.Y.). Five to 10 μg of total RNA was electrophoresed on a 1.0% agarose gel containing 0.22 M formaldehyde, transferred to a nylon membrane, and baked in a vacuum oven to fix the RNA. Membranes were hybridized in ULTRAhyb hybridization buffer (Ambion, Austin, Tex.) with one of the following previously described cDNA probes: mPPARα (22), mPPARβ(δ) (2), mPPARγ (27), mouse myelin basic protein (MBP) (23), mouse proteolipid protein (PLP) (21), rat transglutaminase I (TG-I) (42), rat involucrin (12), small proline-rich (SPR) proteins SPR1A (25) and SPR2H (46), mouse cyclin B1 (10), mouse cyclin-dependent kinase-1 (CDK-1) (49), mouse CDK-4 (36), mouse proliferating cellular nuclear antigen (PCNA) (40), or mouse β-actin (31). Mouse cDNA fragments for ornithine decarboxylase (ODC), CD36, and acyl coenzyme A synthases (ACS) ACS-2 and ACS-3 were obtained by cloning as described below.
RT-PCR cloning of mouse cDNAs.
Mouse cDNA clones for CD36, ACS-2, ACS-3, and ODC were obtained by RT-PCR from 0.5 μg of total RNA isolated from adipose tissue, whole brain, or O-tetradecanoylphorbol-13-acetate (TPA)-treated skin. The PCR primers selected were based on the published cDNA sequences of mouse CD36 (13), rat ACS-2 (16), rat ACS-3 (15), and mouse ODC (24). The second-strand cDNA was amplified by subsequent PCR with designed primers for each gene. The amplified cDNA fragment for mouse CD36 was 1,102 bp, corresponding to nt 229 to 1330. The amplified cDNA fragment for mouse ACS-2 was 942 bp, corresponding to nt 61 to 1002. The mouse cDNA fragment for ACS-2 was 95% homologous with the rat sequence. The amplified cDNA fragment for mouse ACS-3 was 810 bp, corresponding to nt 56 to 865. The mouse cDNA fragment for ACS-3 was 94.2% homologous with the rat sequence. The amplified cDNA fragment for mouse ODC was 1,009 bp, corresponding to nt 855 to 1863. The identity of each clone was confirmed by sequencing. The BLASTN software, version 2.1.10 (National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, Md.), was used to show that all four of these cloned RT-PCR products (CD36, ODC, ACS-2, and ACS-3) were only significantly homologous with the respective mRNA of interest (1).
Western blot analysis.
Nuclear extracts were obtained from skin and liver samples by grinding tissue submerged in liquid nitrogen with a mortar and pestle. After centrifugation, nuclei were resuspended in a lysis buffer (20 mM HEPES, 0.4 M sodium chloride, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 1 mM sodium vanadate). The protein concentration was quantified (BCA kit; Pierce, Rockford, Ill.), and 50 μg of protein was separated on a 10% gel by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane. After being blocked overnight in Tris-buffered saline plus Tween 20 (TBST)–5% milk at 4°C, the membrane was incubated at room temperature with an anti-PPARβ antibody raised against an amino terminus peptide (Santa Cruz Biotechnology, Santa Cruz, Calif.) for 2 h. After being washed with TBST, the membrane was incubated with horseradish peroxidase-conjugated donkey anti-goat antibody, followed by a washing with TBST. Detection of PPARβ protein was performed using a chemiluminescence kit (ECL; Amersham Life Science, Cleveland, Ohio). A detergent extract of transfected COS cells expressing PPARβ was used as a positive control (kindly provided by John Woods and David Moller, Merck Pharmaceuticals).
Animal studies.
To assess body weight gain, male and female wild-type and PPARβ-null mice were weighed on postnatal day 3, week 4, week 8, week 10, and weeks 48 to 54. Mice were fed stock rodent chow and were provided water ad libitum.
The effect of 24-h food restriction on body temperature, physical activity, and adipocyte lipid metabolism was determined for male and female wild-type and PPARβ-null mice. Biotelemetry chips (Mini Mitter, Sunriver, Oreg.) were implanted into the abdomens of mice under anesthesia to monitor body temperature and motor activity (17). After surgery, mice were allowed to recover before baseline activity levels were determined. Values obtained after 1 week of monitoring showed a typical diurnal variation. After this period, food was removed from each mouse and body temperature and motor activity were measured during the next 24-h fasting period. Daily food intakes were also measured over a 12-day period using mice that were individually housed.
Two separate cohorts of animals were used to determine the effect of 24-h fasting on adipose mRNA levels. The first group of male and female wild-type and PPARβ-null mice were euthanized, and gonadal adipose tissue was weighed and snap frozen for future RNA analysis. The second group of mice were fasted for 24 h and adipose tissue was collected as before for RNA analysis.
To assess the role of PPARβ in brain, male and female wild-type and PPARβ-null mice, 12 or 36 weeks of age, were euthanized. Brains were removed and assessed for myelination as described below. To analyze RNA expression in specific regions of the brain, the corpus callosum, cerebellum, and brain stem were dissected and RNA was isolated from these samples as described above.
To assess the role of PPARβ in the epidermal response to TPA, female wild-type and PPARβ-null mice, 8 weeks of age, were shaved to remove back hair. Twenty-four hours later, either 5 μg of TPA (Sigma Chemical Co., St. Louis, Mo.) dissolved in 200 μl of acetone or 200 μl of acetone was applied to the shaved area. Eight or 48 h after TPA application, mice were euthanized and the skin was removed and snap frozen in liquid nitrogen. Total RNA was isolated as described above. Another section of skin was also removed and placed in 10% phosphate-buffered formalin for further histological analysis of epidermal cell proliferation.
To assess the effect of the nonsteroidal anti-inflammatory drug (NSAID) sulindac on TPA-induced inflammation, female wild-type and PPARβ-null mice, 8 weeks of age, were fed a diet containing 0.32 g of sulindac/kg for 10 days. Mice were then shaved to remove back hair and 24 h later were treated topically with either acetone or 5 μg of TPA dissolved in acetone. Eight hours after TPA treatment, skin sections were obtained from euthanized mice and histological analysis for inflammation and hyperplasia was performed.
Skin histology.
Tissues were fixed in 10% neutral buffered formalin (Fisher Scientific, Fair Lawn, N.J.) and embedded in paraffin, and 4- to 6-μm-thick sections were prepared. Sections were stained with hematoxylin and eosin, and the epidermis was evaluated for hyperplastic growth.
Brain histology.
Brains were removed immediately after euthanasia and frozen on dry ice. Sagittal sections (10 μm) were cut with a cryostat. Sections were stored at −70°C until use. A Luxol fast blue (LFB) solution was prepared by dissolving 0.2 g of LFB (Sigma Chemical Company) in 200 ml of 95% ethanol and adding 1 ml of 10% acetic acid. After removal from the freezer and equilibration at room temperature, brain sections were fixed in 4% paraformaldehyde for 15 min. Following two rinses in distilled water, sections were dehydrated with successive immersion in 75, 95, and 100% ethanol. Sections were immersed in LFB solution overnight at 60°C in tightly sealed staining jars. Removal of excess LFB with 95% ethanol rinses was followed by rinsing with distilled water and then immersing for 30 s in 0.05% lithium carbonate (Sigma Chemical Company). The sections were subjected to several changes of 70% ethanol until the grey and white matter were clearly distinguished. Thereafter, the sections were washed thoroughly in distilled water and counterstained with 1% methyl green (Fisher Scientific) for 5 min and rinsed with tap water. Sections were then destained with successive incubations in 80, 90, and 100% ethanol, cleared with a 5-min incubation in xylene, and mounted. Sections were examined under a Zeiss microscope.
RESULTS
PPARβ-null mice are smaller with reduced adipose stores.
Targeted disruption of the ligand binding domain of the mPPARβ gene was performed by inserting a phosphoribosyltransferase II expression cassette into the last exon of the gene (Fig. 1A and B). Successful integration of the targeting vector into the mouse genome was confirmed by Southern blot analysis (Fig. 1B). Northern blot analysis of RNA from selected tissues demonstrated successful disruption of the PPARβ gene. In the brain, an mRNA fragment ∼1 kb larger than the wild-type mRNA was detected in null mice at a substantially lower level than in wild-type mice (Fig. 1C). In adipose tissue, both the larger mRNA and a truncated form were also detected in null mice and the levels of these transcripts were substantially lower than levels of wild-type PPARβ mRNA expression (Fig. 1C). Both of these mRNA transcripts were also detected in liver mRNA from null mice (data not shown). However, despite the presence of these mRNAs, expression of the PPARβ protein was not detected in hepatic nuclear extracts from PPARβ-null mice (Fig. 1D). Neither the larger nor the truncated mRNA was detected in the skin of PPARβ-null mice (Fig. 1C). Western blot analysis of nuclear extracts from skin of wild-type mice demonstrated an increase in PPARβ protein levels as a result of TPA, while expression of the PPARβ protein was not detected in either control or TPA-treated skin samples from PPARβ-null mice (Fig. 1D).
Breeding mixed-genetic-background (C57BL/6N × Sv/129) heterozygous offspring resulted in fewer null mice than expected (Table 1). An analysis of embryos on gestation day 10 (GD10) and fetuses on GD18 revealed that the absence of PPARβ was not lethal to embryo or fetal development since relatively normal distributions of genotypes were found and the conceptus morphology appeared grossly normal (Table 2; data not shown). However, PPARβ-null fetuses on GD18 had significantly smaller crown-to-rump lengths and weighed less than controls (Table 3). F2 offspring were subsequently backcrossed one generation with C57BL/6N mice, and the heterozygous mice from these matings were used to establish homozygous wild-type and PPARβ-null colonies. The colony of PPARβ-null mice reproduced successfully, and normal Mendelian genotype distributions were found from subsequent heterozygous matings (Table 1).
TABLE 1.
Genotype of litters from matings of heterozygous PPARβ mice
| Genetic background | No. of litters | No. of mice (%) of genotype:
|
|||
|---|---|---|---|---|---|
| +/+ | +/− | −/− | Total | ||
| C57BL/6N × Sv/129 | 60 | 147 (36) | 240 (58) | 24 (6) | 411 |
| C57BL/6Na | 10 | 18 (24) | 39 (53) | 17 (23) | 74 |
Enriched as described in Materials and Methods; at least 75% C57BL/6N.
TABLE 2.
Genotypes of embryos and fetuses from heterozygous PPARβ mice on a mixed genetic background (C57BL/6N × Sv/129)
| GD | No. of litters | No. of mice (%) of genotype:
|
|||
|---|---|---|---|---|---|
| +/+ | +/− | −/− | Total | ||
| 10 | 7 | 9 (17) | 36 (68) | 8 (15) | 53 |
| 18 | 9 | 23 (36) | 25 (36) | 16 (25) | 64 |
TABLE 3.
Developmental assessment of GD18 fetuses from C57BL/6N × Sv/129 micea
| Genotype | n | Fetal wt (% control) | Crown-rump length (% control) |
|---|---|---|---|
| +/+ | 23 | 100 ± 0 | 100 ± 0 |
| +/− | 25 | 102 ± 2 | 100 ± 1 |
| −/− | 16 | 70 ± 5* | 86 ± 3* |
GD18 fetuses were weighed, crown-to-rump measurements were obtained, and tails from fetuses were used for genotyping. Data are mean percentages of wild-type controls ± standard errors of the means. ∗, P < 0.05 by one-way analysis of variance.
Postnatal development of PPARβ-null mice between 3 days and 48 weeks appeared grossly normal except that they were significantly smaller than controls (Table 4). This effect was more pronounced in female mice than in males. Contributing to the smaller body weights were smaller gonadal fat stores in the PPARβ-null mice than in the wild-type mice (Table 5). The difference in gonadal adipose stores was not found in older mice aged 48 to 54 weeks (data not shown). Food consumption normalized for body weight indicated that the male PPARβ-null mice consumed more energy than wild-type controls (Table 6). Total oxygen consumption rates corrected for body weight for wild-type males, PPARβ-null males, wild-type females, and PPARβ-null females were 10.2 ± 0.2, 11.5 ± 0.6, 11.4 ± 0.4, and 12.2 ± 0.3 ml/(g of body weight)0.75/h, respectively (n = 5 mice per group). While oxygen consumption tended to be higher in PPARβ-null mice than in controls, this was not significantly different between genotypes.
TABLE 4.
Body weights in wild-type (+/+) and PPARβ-null (−/−) mice
| Time | Mean wt ± SEM for mice of indicated sex and genotypea
|
|||
|---|---|---|---|---|
| Male
|
Female
|
|||
| +/+ | −/− | +/+ | −/− | |
| 3 days | 4.1 ± 0.1 | 3.4 ± 0.2* | 3.6 ± 0.2 | 2.9 ± 0.3* |
| 4 weeks | 15.9 ± 0.7 | 14.8 ± 0.5* | 14.8 ± 1.3 | 11.8 ± 0.4* |
| 8 weeks | 23.8 ± 1.1 | 22.3 ± 0.9* | 22.2 ± 0.5 | 18.0 ± 0.5* |
| 10 weeks | 27.6 ± 1.1 | 23.1 ± 1.4* | 24.0 ± 1.0 | 20.9 ± 1.0* |
| 48–54 weeks | 33.1 ± 0.7 | 31.3 ± 0.9 | 29.8 ± 0.9 | 25.4 ± 0.9* |
A total of 8, 5, 5, 5, and 10 mice of each genotype were weighed after 3 days, 4 weeks, 8 weeks, 10 weeks, and 48 to 54 weeks, respectively. ∗, significantly different from wild-type control (P < 0.05 by Student's t test).
TABLE 5.
Gonadal adipose stores in wild-type (+/+) and PPARβ-null (−/−) micea
| Genotype | Sex | Body wt (g) | Amt of adipose tissue (% body wt) |
|---|---|---|---|
| +/+ | Male | 27.3 ± 0.8 | 1.26 ± 0.15 |
| −/− | Male | 24.7 ± 0.8* | 1.16 ± 0.16 |
| +/+ | Female | 21.2 ± 0.6 | 1.56 ± 0.36 |
| −/− | Female | 17.2 ± 0.8* | 1.02 ± 0.09* |
Relative amount of gonadal adipose tissue is weight in grams divided by body weight in grams times 100. A total of five mice per group, 8 weeks of age, were examined. ∗, significantly different from wild-type control (P < 0.05 by one-way analysis of variance).
TABLE 6.
Food consumption in wild-type (+/+) and PPARβ-null (−/−) micea
| Genotype | Sex | Body wt (g) | Food intake (g/day) | Normalized food intake (g/day/BW0.75)b |
|---|---|---|---|---|
| +/+ | Male | 25.9 ± 0.8 | 4.25 ± 0.20 | 0.370 ± 0.011 |
| −/− | Male | 24.4 ± 1.0 | 4.83 ± 0.21 | 0.439 ± 0.023* |
| +/+ | Female | 25.1 ± 1.3 | 5.01 ± 0.16 | 0.445 ± 0.014 |
| −/− | Female | 21.1 ± 0.1* | 4.64 ± 0.14 | 0.470 ± 0.015 |
Fourteen-week-old mice (n = 5/group) were studied while housed one mouse per cage. Values are means ± standard errors of the means. ∗, significantly different from wild-type control (P < 0.05 by Student's t test).
Food intake normalized to body weight (BW).
PPARβ-null mice respond similarly to wild-type mice after fasting.
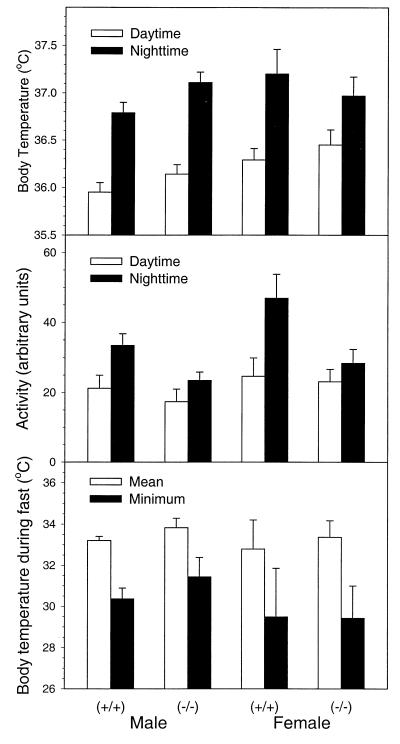
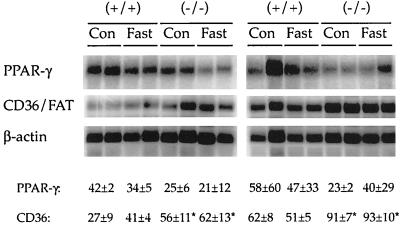
Body temperatures and basal activity levels of wild-type and PPARβ-null mice were similar and showed a normal circadian rhythm (increased body temperature and activity during the dark cycle). Fasting for 24 h caused similar decreases in body temperature in both genotypes (Fig. 2). Levels of weight loss during a 24-h fast were not different in the two genotypes and ranged from 7 to 9% of total body weight. Serum analysis revealed no consistent differences between genotypes. Typical changes in serum chemistry associated with fasting were detected in both genotypes including increased free fatty acids and β-hydroxybutyrate and decreased triglycerides, and no change in blood urea nitrogen was detected (data not shown). Since fatty acid transporters can be regulated by PPARs, levels of CD36 (also known as FAT) were quantified in adipose RNA. Constitutive expression of adipocyte CD36 mRNA was higher in PPARβ-null mice than in controls, while levels of PPARγ mRNA in adipose tissue of the PPARβ-null mice and controls were similar (Fig. 3). Fasting had no effect on either mRNA, as similar expression patterns were observed after fasting (Fig. 3).
FIG. 2.
Body temperature and activity in wild-type (+/+) and PPARβ-null (−/−) mice. Daytime (10 a.m. to 5 p.m.) and nighttime (10 p.m. to 5 a.m.) measurements were made continuously for 14-week-old mice. The body temperature during a 24-hour fast is reported both as the mean of hours 15 to 24 and the minimum during the fast. Values are means ± standard errors of the means (n = 5/group).
FIG. 3.
Effect of 24-h fasting on gonadal adipose mRNA of PPARγ and CD36/FAT in wild-type (+/+) and PPARβ-null (−/−) mice. Male and female +/+ and −/− mice (8 weeks of age) were used. For expression of CD36 and PPARγ mRNA 5 μg of total RNA was subjected to Northern analysis. Values for the respective hybridization signals normalized to β-actin are means ± standard deviations. ∗, significantly different from wild-type control (P < 0.05). Con, control.
PPARβ-null mice have altered myelination in the central nervous system.
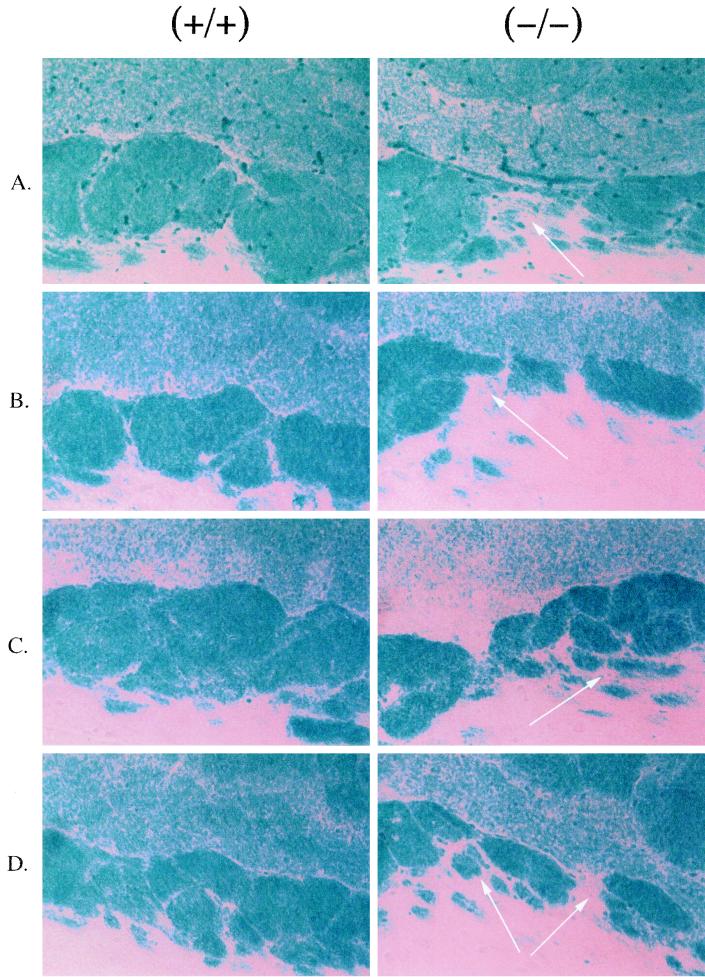
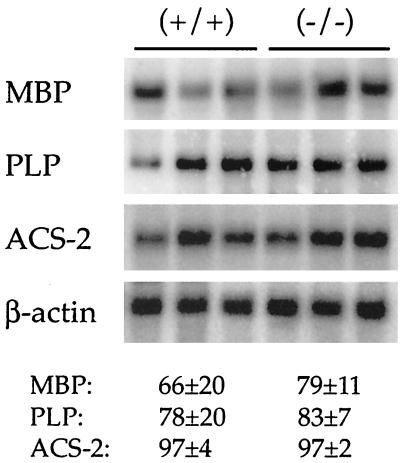
Since expression of PPARβ mRNA is reported to be high in the developing neural tubes of embryos and fetuses as well as the adult rodent brain (8, 9, 11, 51), brains from PPARβ-null mice were examined. The diameters of the brains of PPARβ-null mice were significantly smaller than those of wild-type mice, which is likely due to the relatively smaller size of the PPARβ-null mice (data not shown). Histological examination revealed alterations in the extent of myelination in the corpus callosum compared to controls (Fig. 4). This difference was found more often in female mice than in males (three of five females; two of seven males). No consistent differences in myelination of other brain regions including the cerebellum and brain stem between genotypes were found. Levels of mRNA encoding proteins that are important in the myelination process, such as MBP and PLP, were similar in the corpus callosums from both genotypes (Fig. 5). Since two ACS are expressed in the developing rodent brain and have important roles in fatty acid utilization (15, 43, 50) and since recent data suggest that PPARβ regulates ACS-2 (6), expression of the mRNAs for these enzymes were also analyzed. As shown in Fig. 5, mRNA levels for ACS-2 were similar between genotypes. The levels of RNA encoding ACS-3 were also similar between genotypes (data not shown).
FIG. 4.
Altered myelination of corpus callosum in PPARβ-null mice. Sagittal sections (10 μm) were cut and stained with LFB as described in Materials and Methods. Magnification, ×170. Arrows, regions of altered myelination. (A) Twelve-week-old females; (B) 36-week-old females; (C) 12-week-old males; (D) 36-week-old males.
FIG. 5.
Northern analysis of selected mRNAs from corpora callosa from wild-type (+/+) and PPARβ-null (−/−) mice. The corpus callosum was dissected, RNA was isolated, and 5 μg of total RNA was subjected to Northern analysis. Shown are MBP, PLP, and brain ACS-2. Values for the respective hybridization signals normalized to β-actin are means ± standard deviations.
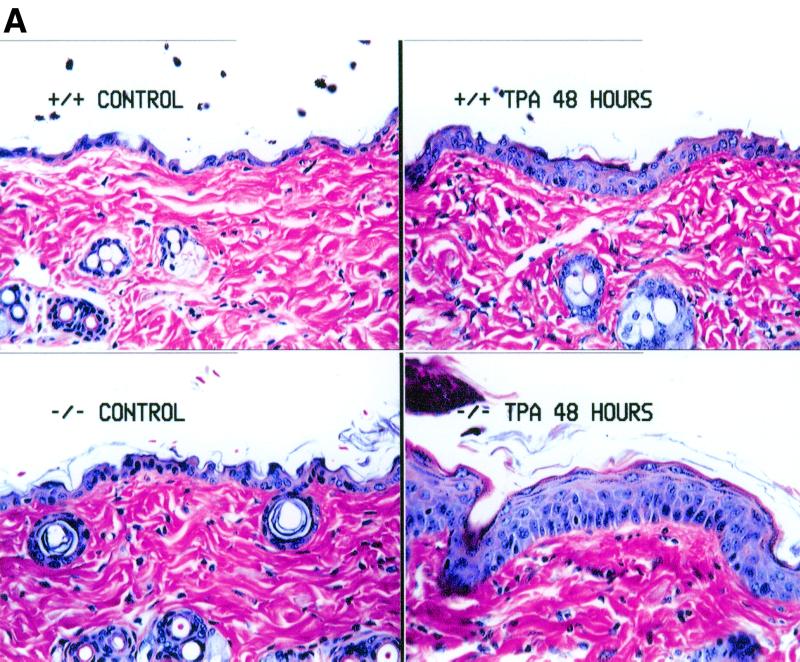
PPARβ deficiency results in accentuated TPA-induced hyperplasia.
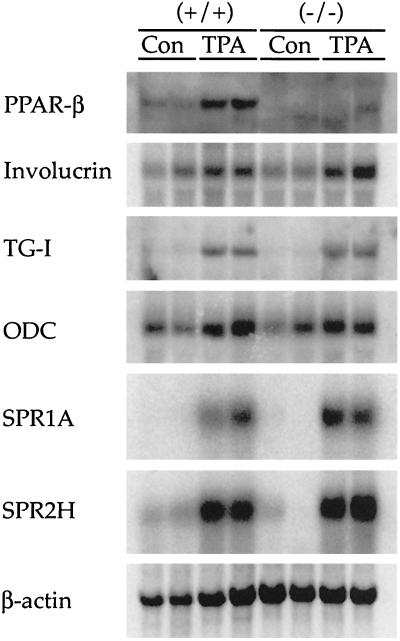
Since induction of PPARβ mRNA is coincident with increased expression of mRNAs encoding TG-I and SPR proteins in keratinocytes cultured in the presence of TPA (37), the epidermal response to TPA in PPARβ-null mice was assessed. Topical application of TPA to wild-type mice caused an increase in the mRNA encoding PPARβ, involucrin, ODC, TG-I, and SPR proteins SPR1A and SPR2H 8 h after TPA treatment (Fig. 6). However, induction of mRNAs encoding proteins associated with differentiation of the epidermis was also found in TPA-treated PPARβ-null mice despite the absence of PPARβ mRNA (Fig. 6). Expression of PPARα and PPARγ mRNA was not detectable in any of the skin RNA samples (data not shown). Interestingly, the hyperplastic response typically observed in the epidermis 48 h after TPA treatment was greater in the PPARβ-null mice than in controls (Fig. 7A), and this effect was found at both low and high doses (2.5 and 10 μg, respectively; data not shown). Associated with the enhanced hyperplastic response observed in the PPARβ-null mice were higher levels of mRNA encoding proteins involved in cell cycle regulation including CDK-1, CDK-4, cyclin B1, and PCNA (Fig. 7B).
FIG. 6.
Northern analysis of skin mRNAs in wild-type (+/+) and PPARβ-null (−/−) mice 8 h after TPA. Ten micrograms of total RNA was analyzed from representative skin from two mice. mRNAs associated with epidermal differentiation and cell proliferation were measured. Con, control.
FIG. 7.
Analysis of TPA-induced hyperplasia in skin of wild-type (+/+) and PPARβ-null (−/−) mice 48 h posttreatment. (A) Histological examination of representative skin topically treated with 5 μg of TPA 48 h posttreatment. Note the enhanced hyperplasia of the epidermis in the −/− skin compared to similarly treated +/+ skin. Magnification, ×267. (B) Northern analysis of skin RNA 48 h after TPA. Total RNA from skin was isolated and analyzed for mRNAs of genes involved in cell proliferation as described in Materials and Methods. mRNAs for the indicated proteins were measured. Values for the respective hybridization signals normalized to β-actin are means ± standard deviations. ∗, significantly different from wild-type control (P < 0.05). Con, control.
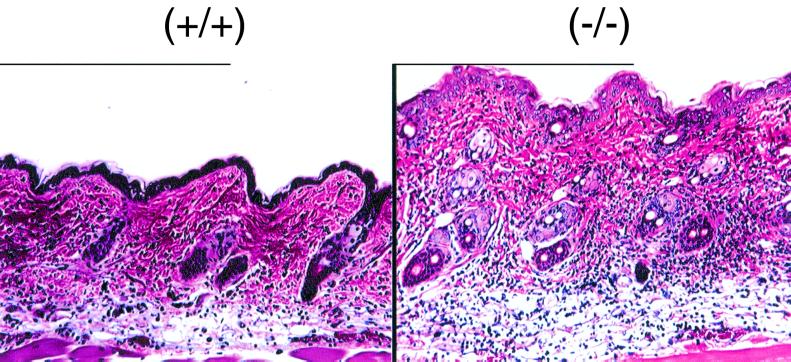
PPARβ-null mice are refractory to the anti-inflammatory drug sulindac.
Since it was recently suggested that the PPARβ may influence the effect of the NSAID sulindac (20), the effect of this drug on the inflammatory response induced by TPA was examined. Wild-type mice fed a sulindac-containing diet and then treated with TPA showed no signs of epidermal hyperplasia and mild to moderate inflammation (Fig. 8). In contrast, inflammation was more severe in TPA-treated PPARβ-null mice than in controls (Fig. 8). Further, early hyperplasia was also detected in TPA-treated PPARβ-null mice but was not observed in TPA-treated wild-type mice (Fig. 8).
FIG. 8.
TPA-induced inflammation in skin of representative wild-type (+/+) and PPARβ-null (−/−) mice fed a sulindac diet, 8 h after TPA treatment. Mice were fed 0.32 g of sulindac/kg of diet for 10 days and treated topically with 5 μg of TPA as described in Materials and Methods. Magnification, ×132. Note that there is less inflammation (blue cells) in dermis and subcutaneous tissue in the wild-type section and more infiltration of inflammatory cells in the dermis and subcutaneous tissue in the PPARβ-null section. The epidermis of the PPARβ-null mouse is approximately twice the size of the wild-type mouse epidermis.
DISCUSSION
Developmental role of PPARβ.
The phenotype of a PPARβ-null mouse line offers clues to the function of this receptor. Since the number of homozygous null offspring was less than expected from heterozygote breedings of the original mixed-genetic-background mice, PPARβ may have a role in embryonic, fetal, and/or postnatal development. However, the normal distribution of genotypes and gross morphology of PPARβ-null conceptuses on GD10 and -18 do not support the hypothesis that PPARβ is required for implantation (33). Nonetheless, these results do provide evidence that, in the absence of PPARβ, development is impaired since both the weights of GD18 fetuses and the postnatal weights of PPARβ-null mice are significantly lower than those of wild-type mice, in particular of female null mice.
The role of the PPARβ in adipocyte function.
The phenotype of the PPARβ-null mouse also indicates that the receptor is involved in adipocyte function. Indeed, overexpression of PPARβ in fibroblasts promotes induction of adipocyte differentiation (5). In the absence of PPARβ, adipose stores are smaller and constitutive expression of CD36/FAT mRNA is higher than those for wild-type mice. Thus the smaller adipose tissue may be due to alterations in fatty acid transport. However, the influence of fasting on measures of lipolysis was not different for the different genotypes, indicating that the role of PPARβ in adipose metabolism may be complex. While it is known that the CD36 gene is responsive to PPAR activators in a tissue-specific manner (35, 38), these data do not address whether PPARβ is required for inducible expression of this gene.
PPARβ and brain development.
The alteration in myelination of the corpus callosum is another unique phenotype of the PPARβ-null mouse. There are a number of possible mechanisms that could explain this effect. PPARβ may be required during postnatal development of the brain, functioning as a regulator of genes involved in this process. However, three likely candidate genes were unaffected in the PPARβ-null mouse corpus callosum including genes for MBP, PLP, and two brain-specific ACS (ACS-2 and ACS-3). For PLP, there is a reported PPRE in the promoter of the gene (7), and thus it is surprising that the level of its RNA is unaffected since PPARβ is the more predominant PPAR expressed in the brain. Reduced fatty acid utilization resulting from reduced acyl coenzyme A derivatives is also not likely to contribute to altered myelination since no difference in ACS-2 expression was found. Despite recent in vitro evidence that PPARβ regulates ACS-2 mRNA upon activation (6), these data demonstrate that constitutive expression of this gene is not influenced by the absence of PPARβ. Lastly, MBP RNA was not different for the different genotypes. Combined, these results suggest that the alteration in myelination observed in the PPARβ-null mouse corpus callosum is the result of events that are mediated by PPARβ during development but that were not detectable at the age we analyzed. Further analysis of this process is needed, but these data do provide strong evidence that PPARβ is required for brain development, possibly for regulation of genes that have not been identified. It is noteworthy that preliminary behavioral assessments of older mice using a rotorod revealed no differences between PPARβ-null and wild-type mice. While further behavioral studies are warranted, the physiological and behavioral consequences of the altered myelination remain a mystery.
The role of PPARβ in skin.
The PPARβ gene is one of the genes involved in epidermal cell proliferation and differentiation induced by TPA (37). It was hypothesized that PPARβ is required for induction of other genes involved in epidermal differentiation since PPARβ mRNA is increased coincidently with TG-I and SPR1A in vitro (37). Data provided from PPARβ-null mice demonstrate that PPARβ is not required for this effect, since mRNAs for TG-I, involucrin, ODC, SPR1A, and SPR2H were all induced to similar levels in the skin of wild-type and PPARβ-null mice treated with TPA. Since PPARβ mRNA is increased, the role of this receptor in the TPA response is of great interest and may provide a useful model to identify more-specific roles for PPARβ. The finding that the hyperplastic response to TPA is enhanced in the PPARβ-null mice suggests the possibility that this receptor is involved in cell cycle control. Support for a putative role of PPARβ in cell cycle control is provided by the recent report that colon carcinomas have elevated levels of PPARβ (20).
Interestingly, PPARβ-null mice fed the NSAID sulindac were more sensitive to the inflammatory response induced by TPA. These data indicate that the sulindac-mediated anti-inflammatory response is dependent on PPARβ. These data support recent observations that sulindac inhibits PPARβ from binding to recognition sites of unidentified target genes (20). Further support for a role for PPARβ in cell cycle control is also provided by the observation that an early hyperplastic response not found in wild-type mice was observed in TPA-treated PPARβ-null mice. The precise mechanisms for these effects are unknown, but these data clearly demonstrate that PPARβ can influence the effects of sulindac. Further studies are necessary to delineate the mechanisms underlying the PPARβ influence on cell cycle regulation in both tumor promotion and tumor formation.
The PPARβ-null mouse model.
In summary, this is the first report that provides in vivo evidence for the roles of PPARβ in development, lipid metabolism, myelination of the corpus callosum, and epidermal cell proliferation. These results support previous reports suggesting a role for this receptor in adipose tissue and brain, with a consistent theme of lipid metabolism being demonstrated for all three PPAR subtypes. In addition, these studies significantly extend our understanding of other important physiological functions that are likely regulated by PPARβ by showing that development and cell proliferation are also likely targets of this nuclear receptor.
ACKNOWLEDGMENTS
We gratefully acknowledge Karen Chandross for dissection of specific brain regions for RNA isolation, Colin Stewart for his analysis of GD10 embryos, Debra Wolgemuth for providing the mouse CDK-1, CDK-4, and cyclin B1 cDNA plasmids, Robert Rice for providing the rat TG-I and involucrin cDNA plasmids, and Tonja Kartasova for providing the mouse SPR1A and SPR2H cDNA plasmids.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amri E Z, Bonino F, Ailhaud G, Abumrad N A, Grimaldi P A. Cloning of a protein that mediates transcriptional effects of fatty acids in preadipocytes. Homology to peroxisome proliferator-activated receptors. J Biol Chem. 1995;270:2367–2371. doi: 10.1074/jbc.270.5.2367. [DOI] [PubMed] [Google Scholar]
- 3.Aoyama A, Peters J M, Iritani N, Nasu-Nakajima T, Furihata K, Hashimoto T, Gonzalez F J. Altered constitutive expression of fatty acid-metabolizing enzymes in mice lacking the peroxisome proliferator-activated receptor α (PPARα) J Biol Chem. 1998;273:5678–5684. doi: 10.1074/jbc.273.10.5678. [DOI] [PubMed] [Google Scholar]
- 4.Barak Y, Nelson M C, Ong E S, Jones Y Z, Ruiz-Lozano P, Chien K R, Koder A, Evans R E. PPARγ is required for placental, cardiac, and adipose tissue development. Mol Cell. 1999;4:585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- 5.Bastie C, Holst D, Gaillard D, Jehl-Pietri C, Grimaldi P A. Expression of peroxisome proliferator-activated receptor PPARδ promotes induction of PPARγ and adipocyte differentiation in 3T3C2 fibroblasts. J Biol Chem. 1999;274:21920–21925. doi: 10.1074/jbc.274.31.21920. [DOI] [PubMed] [Google Scholar]
- 6.Basu-Modak S, Braissant O, Escher P, Desvergne B, Honegger P, Wahli W. Peroxisome proliferator-activated receptor β regulates acyl-CoA synthetase 2 in reaggregated rat brain cell cultures. J Biol Chem. 1999;274:35881–35888. doi: 10.1074/jbc.274.50.35881. [DOI] [PubMed] [Google Scholar]
- 7.Bogazzi F, Hudson L D, Nikodem V M. A novel heterodimerization partner for thyroid hormone receptor. Peroxisome proliferator-activated receptor. J Biol Chem. 1994;269:11683–11686. [PubMed] [Google Scholar]
- 8.Braissant O, Foufelle F, Scotto C, Dauca M, Wahli W. Differential expression of peroxisome proliferator-activated receptors (PPARs): tissue distribution of PPAR-α, -β, and -γ in the adult rat. Endocrinology. 1996;137:354–366. doi: 10.1210/endo.137.1.8536636. [DOI] [PubMed] [Google Scholar]
- 9.Braissant O, Wahli W. Differential expression of peroxisome proliferator-activated receptor-α, -β, and -γ during rat embryonic development. Endocrinology. 1998;139:2748–2754. doi: 10.1210/endo.139.6.6049. [DOI] [PubMed] [Google Scholar]
- 10.Chapman D L, Wolgemuth D J. Identification of a mouse B-type cyclin which exhibits developmentally regulated expression in the germ line. Mol Reprod Dev. 1992;33:259–269. doi: 10.1002/mrd.1080330305. [DOI] [PubMed] [Google Scholar]
- 11.Cullingford T E, Bhakoo K, Peuchen S, Dolphin C T, Patel R, Clark J B. Distribution of mRNAs encoding the peroxisome proliferator-activated receptor α, β, and γ and the retinoid X receptor α, β, and γ in rat central nervous system. J Neurochem. 1998;70:1366–1375. doi: 10.1046/j.1471-4159.1998.70041366.x. [DOI] [PubMed] [Google Scholar]
- 12.Djian P, Phillips M, Easley K, Huang E, Simon M, Rice R H, Green H. The involucrin genes of the mouse and the rat: study of their shared repeats. Mol Biol Evol. 1993;10:1136–1149. doi: 10.1093/oxfordjournals.molbev.a040069. [DOI] [PubMed] [Google Scholar]
- 13.Endemann G, Stanton L W, Madden K S, Bryant C M, White R T, Protter A A. CD36 is a receptor for oxidized low density lipoprotein. J Biol Chem. 1993;268:11811–11816. [PubMed] [Google Scholar]
- 14.Fruchart J C, Duriez P, Staels B. Peroxisome proliferator-activated receptor-α activators regulate genes governing lipoprotein metabolism, vascular inflammation and atherosclerosis. Curr Opin Lipidol. 1999;10:245–257. doi: 10.1097/00041433-199906000-00007. [DOI] [PubMed] [Google Scholar]
- 15.Fujino T, Kang M J, Suzuki H, Iijima H, Yamamoto T. Molecular characterization and expression of rat acyl-CoA synthetase 3. J Biol Chem. 1996;271:16748–16752. doi: 10.1074/jbc.271.28.16748. [DOI] [PubMed] [Google Scholar]
- 16.Fujino T, Yamamoto T. Cloning and functional expression of a novel long-chain acyl-CoA synthetase expressed in brain. J Biochem (Tokyo) 1992;111:197–203. doi: 10.1093/oxfordjournals.jbchem.a123737. [DOI] [PubMed] [Google Scholar]
- 17.Gavrilova O, Leon L R, Marcus-Samuels B, Mason M M, Castle A L, Refetoff S, Vinson C, Reitman M L. Torpor in mice is induced by both leptin-dependent and -independent mechanisms. Proc Natl Acad Sci USA. 1999;96:14623–14628. doi: 10.1073/pnas.96.25.14623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gelman L, Auwerx J. Peroxisome proliferator-activated receptors: mediators of a fast food impact on gene regulation. Curr Opin Clin Nutr Metab Care. 1999;2:307–312. doi: 10.1097/00075197-199907000-00010. [DOI] [PubMed] [Google Scholar]
- 19.Guardiola-Diaz H M, Rehnmark S, Usuda N, Albrektsen T, Feltkamp D, Gustafsson J A, Alexson S E. Rat peroxisome proliferator-activated receptors and brown adipose tissue function during cold acclimatization. J Biol Chem. 1999;274:23368–23377. doi: 10.1074/jbc.274.33.23368. [DOI] [PubMed] [Google Scholar]
- 20.He T C, Chan T A, Vogelstein B, Kinzler K W. PPARδ is an APC-regulated target of nonsteroidal anti-inflammatory drugs. Cell. 1999;99:335–345. doi: 10.1016/s0092-8674(00)81664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hudson L D, Berndt J A, Puckett C, Kozak C A, Lazzarini R A. Aberrant splicing of proteolipid protein mRNA in the dysmyelinating jimpy mutant mouse. Proc Natl Acad Sci USA. 1987;84:1454–1458. doi: 10.1073/pnas.84.5.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Issemann I, Green S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature. 1990;347:645–650. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- 23.Jordan C A, Friedrich V L, Jr, Godfraind C, Cardellechio C B, Holmes K V, Dubois-Dalcq M. Expression of viral and myelin gene transcripts in a murine CNS demyelinating disease caused by a coronavirus. Glia. 1989;2:318–329. doi: 10.1002/glia.440020505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kahana C, Nathans D. Nucleotide sequence of murine ornithine decarboxylase mRNA. Proc Natl Acad Sci USA. 1985;82:1673–1677. doi: 10.1073/pnas.82.6.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kartasova T, Darwiche N, Kohno Y, Koizumi H, Osada S, Huh N, Lichti U, Steinert P M, Kuroki T. Sequence and expression patterns of mouse SPR1: correlation of expression with epithelial function. J Investig Dermatol. 1996;106:294–304. doi: 10.1111/1523-1747.ep12340741. [DOI] [PubMed] [Google Scholar]
- 26.Kersten S, Seydoux J, Peters J M, Gonzalez F J, Desvergne B, Wahli W. Peroxisome proliferator-activated receptor α mediates the adaptive response to fasting. J Clin Investig. 1999;103:1489–1498. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kliewer S A, Forman B M, Blumberg B, Ong E S, Borgmeyer U, Mangelsdorf D J, Umesono K, Evans R M. Differential expression and activation of a family of murine peroxisome proliferator-activated receptors. Proc Natl Acad Sci USA. 1994;91:7355–7359. doi: 10.1073/pnas.91.15.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krey G, Braissant O, L'Horset F, Kalkhoven E, Perroud M, Parker M G, Wahli W. Fatty acids, eicosanoids, and hypolipidemic agents identified as ligands of peroxisome proliferator-activated receptors by coactivator-dependent receptor ligand assay. Mol Endocrinol. 1997;11:779–791. doi: 10.1210/mend.11.6.0007. [DOI] [PubMed] [Google Scholar]
- 29.Kubota N, Terauchi Y, Miki H, Tamemoto H, Yamauchi T, Komeda K, Satoh S, Nakano R, Ishii C, Sugiyama T, Eto K, Tsubamoto Y, Okuno A, Murakami K, Sekihara H, Hasegawa G, Naito M, Toyoshima Y, Tanaka S, Shiota K, Kitamura T, Fujita T, Ezaki O, Aizawa S, Nagai R, Tobe K, Kimura S, Kadowaki T. PPARγ mediates high-fat diet-induced adipocyte hypertrophy and insulin resistance. Mol Cell. 1999;4:597–609. doi: 10.1016/s1097-2765(00)80210-5. [DOI] [PubMed] [Google Scholar]
- 30.Laird P W, Zijderveld A, Linders K, Rudnicki M A, Jaenisch R, Berns A. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee S S, Pineau T, Drago J, Lee E J, Owens J W, Kroetz D L, Fernandez-Salguero P M, Westphal H, Gonzalez F J. Targeted disruption of the α isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol Cell Biol. 1995;15:3012–3022. doi: 10.1128/mcb.15.6.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lehmann J M, Moore L B, Smith-Oliver T A, Wilkison W O, Willson T M, Kliewer S A. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor γ (PPARγ) J Biol Chem. 1995;270:12953–12956. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- 33.Lim H, Gupta R A, Ma W G, Paria B C, Moller D E, Morrow J D, DuBois R N, Trzaskos J M, Dey S K. Cyclo-oxygenase-2-derived prostacyclin mediates embryo implantation in the mouse via PPARδ. Genes Dev. 1999;13:1561–1574. doi: 10.1101/gad.13.12.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mangelsdorf D J, Evans R M. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 35.Martin G, Schoonjans K, Lefebvre A M, Staels B, Auwerx J. Coordinate regulation of the expression of the fatty acid transport protein and acyl-CoA synthetase genes by PPARα and PPARγ activators. J Biol Chem. 1997;272:28210–28217. doi: 10.1074/jbc.272.45.28210. [DOI] [PubMed] [Google Scholar]
- 36.Matsushime H, Ewen M E, Strom D K, Kato J Y, Hanks S K, Roussel M F, Sherr C J. Identification and properties of an atypical catalytic subunit (p34PSK-J3/cdk4) for mammalian D type G1 cyclins. Cell. 1992;71:323–334. doi: 10.1016/0092-8674(92)90360-o. [DOI] [PubMed] [Google Scholar]
- 37.Matsuura H, Adachi H, Smart R C, Xu X, Arata J, Jetten A M. Correlation between expression of peroxisome proliferator-activated receptor β and squamous differentiation in epidermal and tracheobronchial epithelial cells. Mol Cell Endocrinol. 1999;147:85–92. doi: 10.1016/s0303-7207(98)00214-7. [DOI] [PubMed] [Google Scholar]
- 38.Motojima K, Passilly P, Peters J M, Gonzalez F J, Latruffe N. Expression of putative fatty acid transporter genes are regulated by peroxisome proliferator-activated receptor α and γ activators in a tissue- and inducer-specific manner. J Biol Chem. 1998;273:16710–16714. doi: 10.1074/jbc.273.27.16710. [DOI] [PubMed] [Google Scholar]
- 39.Moya-Camarena S Y, van den Heuvel J P, Belury M A. Conjugated linoleic acid activates peroxisome proliferator-activated receptor α and β subtypes but does not induce hepatic peroxisome proliferation in Sprague-Dawley rats. Biochim Biophys Acta. 1999;1436:331–342. doi: 10.1016/s0005-2760(98)00121-0. [DOI] [PubMed] [Google Scholar]
- 40.Peters J M, Aoyama T, Cattley R C, Nobumitsu U, Hashimoto T, Gonzalez F J. Role of peroxisome proliferator-activated receptor α in altered cell cycle regulation in mouse liver. Carcinogenesis. 1998;19:1989–1994. doi: 10.1093/carcin/19.11.1989. [DOI] [PubMed] [Google Scholar]
- 41.Peters J M, Hennuyer N, Staels B, Fruchart J C, Fievet C, Gonzalez F J, Auwerx J. Alterations in lipoprotein metabolism in peroxisome proliferator-activated receptor α-deficient mice. J Biol Chem. 1997;272:27307–27312. doi: 10.1074/jbc.272.43.27307. [DOI] [PubMed] [Google Scholar]
- 42.Phillips M A, Stewart B E, Qin Q, Chakravarty R, Floyd E E, Jetten A M, Rice R H. Primary structure of keratinocyte transglutaminase. Proc Natl Acad Sci USA. 1990;87:9333–9337. doi: 10.1073/pnas.87.23.9333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reddy T S, Bazan N G. Long-chain acyl CoA synthetase in microsomes from rat brain gray matter and white matter. Neurochem Res. 1985;10:377–386. doi: 10.1007/BF00964606. [DOI] [PubMed] [Google Scholar]
- 44.Ren B, Thelen A P, Peters J M, Gonzalez F J, Jump D B. Polyunsaturated fatty acid suppression of hepatic fatty acid synthase and S14 gene expression does not require peroxisome proliferator-activated receptor α. J Biol Chem. 1997;272:26827–26832. doi: 10.1074/jbc.272.43.26827. [DOI] [PubMed] [Google Scholar]
- 45.Rosen E D, Sarraf P, Troy A E, Bradwin G, Moore K, Milstone D S, Spiegelman B M, Mortensen R M. PPARγ is required for the differentiation of adipose tissue in vivo and in vitro. Mol Cell. 1999;4:611–617. doi: 10.1016/s1097-2765(00)80211-7. [DOI] [PubMed] [Google Scholar]
- 46.Song H J, Poy G, Darwiche N, Lichti U, Kuroki T, Steinert P M, Kartasova T. Mouse Sprr2 genes: a clustered family of genes showing differential expression in epithelial tissues. Genomics. 1999;55:28–42. doi: 10.1006/geno.1998.5607. [DOI] [PubMed] [Google Scholar]
- 47.Spiegelman B M. PPAR-γ: adipogenic regulator and thiazolidinedione receptor. Diabetes. 1998;47:507–514. doi: 10.2337/diabetes.47.4.507. [DOI] [PubMed] [Google Scholar]
- 48.Spiegelman B M, Flier J S. Adipogenesis and obesity: rounding out the big picture. Cell. 1996;87:377–389. doi: 10.1016/s0092-8674(00)81359-8. [DOI] [PubMed] [Google Scholar]
- 49.Th'ng J P, Wright P S, Hamaguchi J, Lee M G, Norbury C J, Nurse P, Bradbury E M. The FT210 cell line is a mouse G2 phase mutant with a temperature-sensitive CDC2 gene product. Cell. 1990;63:313–324. doi: 10.1016/0092-8674(90)90164-a. [DOI] [PubMed] [Google Scholar]
- 50.Vaswani K K, Ledeen R W. Long-chain acyl-coenzyme A synthetase in rat brain myelin. J Neurosci Res. 1987;17:65–70. doi: 10.1002/jnr.490170110. [DOI] [PubMed] [Google Scholar]
- 51.Xing G, Zhang L, Zhang L, Heynen T, Yoshikawa T, Smith M, Weiss S, Detera-Wadleigh S. Rat PPAR δ contains a CGG triplet repeat and is prominently expressed in the thalamic nuclei. Biochem Biophys Res Commun. 1995;217:1015–1025. doi: 10.1006/bbrc.1995.2871. [DOI] [PubMed] [Google Scholar]