Abstract
Satellite myoblasts serve as stem cells in postnatal skeletal muscle, but the genes responsible for choosing between growth versus differentiation are largely undefined. We have used a novel genetic approach to identify genes encoding proteins whose dominant negative inhibition is capable of interrupting the in vitro differentiation of C2C12 murine satellite myoblasts. The screen is based on fusion of a library of cDNA fragments with the lysosomal protease cathepsin B (CB), such that the fusion protein intracellularly diverts interacting factors to the lysosome. Among other gene fragments selected in this screen, including those of known and novel sequence, is the retinoblastoma protein (RB) pocket domain. This unique dominant negative form of RB allows us to genetically determine if MyoD and RB associate in vivo. The dominant negative CB-RB fusion produces a cellular phenotype indistinguishable from recessive loss of function RB mutations. The fact that the dominant negative RB inhibits myogenic differentiation in the presence of nonlimiting concentrations of either RB or MyoD suggests that these two proteins do not directly interact. We further show that the dominant negative RB inhibits E2F1 but cannot inhibit a forced E2F1-RB dimer. Therefore, E2F1 is a potential mediator of the dominant negative inhibition of MyoD by CB-RB during satellite cell differentiation. We propose this approach to be generally suited to the investigation of gene function, even when little is known about the pathway being studied.
Satellite cells are a lineage derived from somites that reside under the basement membrane of the myofiber and are responsible for replenishing skeletal muscle during growth in the postnatal period and in response to exercise and injury in the adult animal (24). As muscles hypertrophy, the satellite cells divide and fuse in order to increase the complement of myonuclei in myofibers. Transplant studies in chick embryos indicate that satellite cells are unable to take part in muscle embryogenesis, suggesting that they serve a specialized function as a presumptive skeletal muscle stem cell (3). Satellite cells are required to both proliferate and terminally differentiate, but invariably the proportion of satellite cells in a muscle remains constant, independent of both the age of the animal and the size of the muscle, and returns to this fixed value at the conclusion of muscle regeneration following injury (23).
Understanding the molecular mechanisms responsible for the switch between proliferation and differentiation in satellite cells could be a key to understanding skeletal muscle regeneration in response to disease and trauma. However, relatively little is known about the mechanisms at work to ensure maintenance of the satellite cell population. Most of this knowledge comes from embryological studies of mesoderm differentiation in somites or the developing limb bud that have been extrapolated to tissue culture models of satellite myoblast differentiation. As with embryonic myogenesis, expression of MyoD or the related basic helix-loop-helix (bHLH) myogenic transcription factors is required for the in vitro differentiation of C2C12 and other satellite myoblast cell lines (17). The myogenic bHLH factors heterodimerize with E protein partners to activate transcription through E box enhancer motifs. The MEF2 family of MADS box transcription factors act as coregulators of transcription through interaction with the basic region of myogenic bHLH proteins. The bHLH proteins somehow lead to activation of the retinoblastoma protein (RB), which in turn titrates E2F factors to effect cell cycle exit upon terminal differentiation. These transcriptional complexes then induce a cascade of gene activation and repression events in which as many as 20,000 genes are differentially regulated (4, 14).
To identify genetic pathways responsible for controlling decisions between growth and differentiation in skeletal muscle satellite cells, we have adopted a novel approach to dominant negative mutation that requires no a priori knowledge of the pathway under study. The strategy (15) is based on the demonstration that the lysosomal localization signal in the protease preprocathepsin B (CB) acts in cis dominance to other subcellular localization signals. By fusing CB to a fragment of a gene encoding a subunit of a multimeric complex, the CB fusion protein can dominantly inhibit the function of associating proteins through diversion of the interacting complex from its usual subcellular localization to the hydrolytic environment of the lysosome.
Here we have constructed a library composed of skeletal muscle cDNA fragments fused downstream to CB. This library conceivably presents all of the cell's expressed genes in a dominant negative form. The library was stably transected into cultured satellite myoblasts in which the majority of cells normally differentiate into irreversibly growth-arrested myotubes upon serum starvation. By selection from the population of transfected myoblasts those clones capable of cell cycle reentry and continued growth upon serum repletion, the library becomes enriched for cDNA fragments whose dominant negative inhibition can abrogate terminal differentiation. Using this approach, we have identified several cDNA fragments, some corresponding to previously known genes and others novel, with apparent roles in switching from differentiation to growth in satellite myoblasts. We initially focus our investigations on the properties of unique dominant negative RB pocket domain fragments. Heretofore, all available mutants of RB have been recessive in nature. The ability of a dominant negative RB to inhibit myogenesis in the apparent presence of nonlimiting concentrations of functional wild-type RB and MyoD implies that the two proteins do not have a significant in vivo interaction. We discuss this result with respect to its implications for cell cycle regulation in satellite myoblast differentiation.
MATERIALS AND METHODS
Cells and cultivation.
C2C12, a subclone of the C2 mouse myoblast cell line, and NIH 3T3, a mouse fibroblast cell line, were obtained from the American Type Culture Collection. Polyclonal populations of C2C12 cells retrovirally tagged with β-galactosidase have been previously described (4). Growth medium (GM) for C2C12 cells was Dulbecco's modified Eagle's medium (DMEM) supplemented with 15% fetal bovine serum; NIH 3T3 cells were cultured in DMEM supplemented with 10% fetal calf serum. Penicillin-streptomycin (1%) was added to the media. Cell differentiation was induced by placing cells in a differentiation medium (DM) (DMEM containing 2% heat-inactivated horse serum) for 3 to 5 days.
Plasmids and CB fusion cDNA library.
pCS2+, pCS2+myc6, pCS2+CB-myc6, pCS2+myc6-MyoD, pEMSVscribe, and the expression vector for mouse MyoD cDNA (pEMSV-MyoD) have been described previously (15). pCS2+CB-myc6-(neo) and pCS2+myc6-(neo) were made by ligating a BamHI-NheI Klenow filled-in fragment derived from plasmid pCD2 into the StuI sites of pCS2+CB-myc6 and pCS2+myc6, respectively. This fragment contains the coding region of the neomycin resistance (neo) gene under control of the simian virus 40 enhancer/promoter for selecting stably transfected cell lines using the antibiotic G418. E2F1 and E2F1-RB fusion plasmids were provided by William Sellers (Harvard University). The CB fusion library was constructed by ligating PCR-amplified inserts derived from an adult human skeletal muscle cDNA library (Clontech HL3000s) into the EcoRI and XbaI sites of pCS2+CB-myc6-(neo). The vector contained a hexameric Myc epitope tag (myc6) for staining with a mouse hybridoma antibody (9E10) against Myc to distinguish transfected cells. pCS2+CB-myc6-RBf1-(neo) and pCS2+CB-myc6-RBf2-(neo) were recovered by plasmid rescue experiments from selected undifferentiated cell lines. pCS2+myc6-RBf1-(neo) and pCS2+myc6-RBf2-(neo) were constructed by ligating the EcoRI-XbaI fragments of pCS2+CB-myc6-RBf1-(neo) and pCS2+CB-myc6-RBf2-(neo) into the EcoRI and XbaI sites of pCS2+myc6-(neo), respectively. All plasmid DNAs were purified on Qiagen columns.
Transient and stable transfection.
Cells (5 × 105) were seeded onto 60-mm-diameter plates in GM and transiently transfected about 8 h later by CaPO4 precipitation using HEPES-buffered saline (15) with 10 μg of DNA (unless stated otherwise). CaPO4-DNA precipitate remained on the cells for about 17 h before feeding with fresh GM. Cell extracts and β-galactosidase assay were performed as before (15). To establish stable transfectants, C2C12 cells were transfected with 20 μg of plasmid DNA containing a neo gene; 17 h later, transfectants were selected by placing cells in GM containing G418 (500 μg/ml; Gibco). G418-resistant cell lines were isolated approximately 14 days later and propagated. Immunofluorescence staining was subsequently performed on the transfected cells. To select undifferentiated cell lines, 17 h after transfection, cells were transferred to 15-cm-diameter plates and placed in GM containing G418 (500 μg/ml) for approximately 2 weeks. The G418-resistant cells were induced to differentiate by 3 to 5 days of incubation in DM and then were trypsinized and placed in GM until they reached 60 to 80% confluency. This process was repeated three times. The undifferentiated cells were further screened by staining the myc6 epitope with the anti-Myc monoclonal antibody 9E10. Cell lines expressing myc6 were subsequently used for isolating genes by plasmid rescue experiment.
Colony formation assay.
C2C12 cells were transfected with either the pCS2+(neo) parental plasmid or the various CB-RB fusion constructs. G418 selection began 24 h after transfection. Approximately 2 weeks later, when macroscopic colonies became detectable, the cells were washed with phosphate-buffered saline (PBS), fixed in 50% methanol–50% acetone for 5 min, and stained with 0.4% crystal violet–20% ethanol for 15 min; then colonies were counted. For the cell growth speed assay, stably transfected cells were plated at clonal density. The number of cells per clone was determined approximately 1 week later.
Plasmid rescue.
Stably transfected undifferentiated cell lines were cultured in GM and harvested, and genomic DNA was isolated using the Qiagen tissue kit according to the manufacturer's instructions. The genomic DNA was digested to completion with EcoRV, sites for which are not present in the vector used for stable transfection. The restriction fragments were diluted to a final concentration of 1 μg/ml in ligation buffer (50 mM Tris-HCl [pH 7.6], 10 mM MgCl2, 1 mM ATP, 1 mM dithiothreitol) and circularized by adding 10 U of T4 DNA ligase per ml at 16°C for about 16 h. DNA was ethanol precipitated then resuspended in Tris-EDTA buffer. The plasmids were recovered into the Escherichia coli DH5α′ by electrotransformation. Following 17 h of incubation at 37°C on agar plates containing appropriate antibiotics, plasmids were extracted by sodium dodecyl sulfate-alkaline lysis, and the recovered inserts were sequenced by dideoxy-chain termination methods. Two RB fragments were recovered. RBf1 starts at residue 369 and ends at 896; RBf2 includes residues 390 to 815. Both of these fragments contain most of the entire pocket domain and a portion of C-terminal region of RB. CB-myc6-RBf1ΔC has a carboxyl-terminal deletion between residues 703 and 896.
Immunofluorescent staining.
After transfection or inducing differentiation, cells were washed three times with PBS, fixed for 3 min with 50% methanol–50% acetone, and incubated for 60 min at room temperature with the appropriate primary antibody at the indicated dilution in PBS: 1:3 of anti-Myc epitope mouse hybridoma supernatant (9E10) (28), 1:200 of rabbit polyclonal anti-MyoD (27) (a gift from L. Snider), 1:10 of mouse monoclonal anti-skeletal myosin (MF-20), and 1:200 of rabbit polyclonal anti-skeletal myosin (Sigma). Secondary detection was carried out by incubation for 30 min at room temperature with 1:200 dilution of fluorescein- or rhodamine-conjugated, non-cross-reactive, goat anti-mouse or anti-rabbit antibodies, respectively (Jackson ImmunoResearch Laboratories). Nuclei were counterstained by incubation for 3 min using DAPI (4′,6-diamidino-2-phenylindole; 0.5 μg/ml). Epifluorescence and phase-contrast photomicroscopy were performed with digitally captured images composited with Adobe Photoshop software (to adjust size, brightness, and contrast).
BrdU incorporation.
Cells were grown in GM until they reached about 50% confluency, induced to differentiate for 3 days, and then stimulated with DMEM supplemented with 20% fetal bovine serum containing 10 μM 5-bromo-2′-deoxyuridine BrdU (Boehringer Mannheim) for 36 h. Cells were washed three times with PBS, fixed for 10 min in ice-cold 70% ethanol–3.7% formaldehyde–5% glacial acetic acid, and then washed three times more with PBS. After myosin heavy chain (MHC) was detected using rabbit polyclonal anti-skeletal myosin and secondary rhodamine-conjugated goat anti-rabbit antibody, the immunocomplexes were fixed for 10 min with 2% paraformaldehyde at room temperature, and then BrdU incorporation was detected using a BrdU labeling and detection kit as instructed by the manufacturer (Boehringer Mannheim).
RESULTS
The strategy of dominant negative screening.
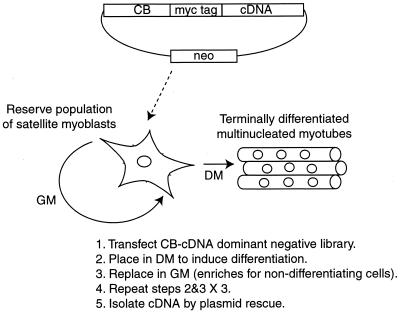
These investigations were conducted with mouse C2C12 satellite myoblasts, which can be induced by serum starvation to form differentiated multinucleated myotubes that permanently exit from the cell cycle. To isolate genes that participate in muscle cell differentiation using the dominant negative system, we created a library of human skeletal muscle cDNAs fused downstream to mouse CB. The library was stably transfected into C2C12 cells, and clones that no longer permanently growth arrest upon serum starvation were amplified by allowing for a period of regrowth with serum repletion (Fig. 1). Specifically, terminally differentiated cells lose the ability to reattach to the plates when replacing them to GM following trypsinization and should, in any event, fail to divide even if they are passaged. There is always a small number of C2C12 cells constituting the reserve population of satellite myoblasts that down-regulate MyoD and Myf-5 and physiologically avoid entry into the differentiation program even under serum-starved conditions (35). Therefore, we performed growth selection with three alternating cycles of serum starvation to remove the small population of cells that failed to differentiate.
FIG. 1.
Strategy for isolating genes that contribute to muscle cell differentiation.
We initially performed control experiments to determine if this approach was capable of enriching for nondifferentiating subclones. Two sets of C2C12 cells were prepared. The first population was stably transfected with the CB-cDNA fusions. Upon initial G418 selection, there were about 2,000 colonies that were then subsequently maintained as a polyclonal population. The second population was tagged by stable integration of a β-galactosidase marker gene and is therefore capable of staining blue with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). To test whether the library had any influence on the relative growth or differentiation of C2C12 cells, we mixed the β-galactosidase-tagged population with the population transfected with the library and grew them for three passages either in serum-containing GM or in alternating cycles of serum-depleted DM and GM. To determine the relative composition of the combined population at the conclusion of the experiment, the cells were plated at clonal density, stained blue with X-Gal to detect β-galactosidase activity, and scored for the ratio of blue to white colonies. This allows us to test whether the frequency of spontaneously nondifferentiating clones exceeds that induced in cells transfected with the library. If the library does have an effect on inhibiting the differentiation of the cells, then it is expected that after cycling between GM and DM, the final distribution of the combined population will shift to a much greater proportion of white cells. The results are shown in Table 1. On the first line is a control experiment of plating only the cells transfected with the library, which lack β-galactosidase activity. As expected, the cells remained white after three passages in either GM alone or in GM alternating with DM. On the last line is the opposite control in which only the β-galactosidase-tagged cells were plated. Again as expected, the cells remained blue after three passages in either set of conditions. The cells were then mixed at two different ratios. On the second line of Table 1, 104 blue cells (not transfected with the library) were mixed with an equal number of white cells (transfected with the library). After three passages in GM alone, the clonal composition of the population remained with a nearly equal proportion of the two starting cell types. However, following three passages alternating between GM and DM, the proportion of cells not expressing β-galactosidase, and therefore derived by transfection with the CB fusion library, was enriched (from 46 to 97%). We repeated this experiment but this time starting with a ratio of blue to white cells of 100:1. Again, the clonal composition of the population of cells was nearly unchanged following three passages in GM alone, but white clones transfected with the library were greatly enriched following selection in alternating cycles of GM and DM (accounting for 74% of the final population). Finally, we performed the converse experiment (data not shown) in which we chose a nondifferentiating β-galactosidase-tagged C2C12 clone (previously isolated through gene-trapping experiments [4]) and mixed it at a ratio of 1:1,000 with untransfected C2C12 cells. After three cycles of alternating GM and DM conditions, the nondifferentiating clone represented about 10% of the clonal composition of the population of the cells but remained at about 0.08% of the population when passaged only in GM. We conclude both that nondifferentiating clones can be specifically enriched against the physiologically nondifferentiating population of reserve satellite C2C12 myoblasts and that expression of the transfected CB-cDNA fusion library generates nondifferentiating clones at a frequency significantly greater than background.
TABLE 1.
Enrichment for nondifferentiating clones by alternate rounds of selection between growth and differentiation conditions
| Blue:whitea ratios upon initial seeding (% white) | Blue:white clones after 3 passages in:
|
|
|---|---|---|
| GM (% white) | DM alternating with GM (% white) | |
| All white (100) | 0:191 (100) | 0:58 (100) |
| 1:1 (50) | 142:119 (46) | 2:64 (97) |
| 99:1 (1) | 312:1 (0.3) | 36:101 (74) |
| All blue (0) | 263:0 (0) | 76:0 (0) |
Blue versus white distinction is made with reference to staining for β-galactosidase activity with X-Gal.
Recovery of cDNAs that inhibit differentiation.
The CB-cDNA library, inserted into a vector containing the neo gene, was stably transfected into C2C12 myoblasts, and approximately 5,000 to 10,000 clones were selected in GM with G418. The G418-resistant transfectants were screened by immunofluorescent staining for expression of the Myc epitope-tagged CB fusion genes, and it was found that most of the selected clones (>90%) were positive for the Myc epitope in a pattern consistent with lysosomal distribution (data not shown). Nevertheless, only one-half of the cDNA fragments are expected in the sense orientation; only one-third of those will be in the correct reading frame; fewer still of the cDNAs containing residual 5′ untranslated regions will have open reading frames (ORFs) through this segment. We anticipate then that no more than one-sixth of the clones in the library will be functional. However, some cells may be transfected with multiple copies of different plasmids from the fusion library. It is also possible that some cDNAs can negate differentiation through expression as antisense messages, independently of fusion with CB. In fact, selection of antisense messages from libraries is another approach that has been used to identify genes participating in phenotypic pathways involving growth and differentiation in other types of cells (as reviewed in reference 15). It is therefore somewhat difficult to estimate the functional complexity of the transfected library.
Following initial selection for transfected myoblasts, the population of G418-resistant cells was maintained polyclonally and cycled in alternate plating between differentiation and growth media. After the second round of selection between the two conditions, the cells generally did not appear to morphologically differentiate into multinucleated myotubes, as judged by visible inspection (data not shown). A third round of selection between differentiation and growth media was performed to ensure further selection against physiologically nondifferentiating reserve satellite myoblasts. The polyclonal population of cells that remained after three cycles of selection for nondifferentiation were then used as a source of DNA for the isolation of the cDNA fragments by plasmid rescue in E. coli.
Fifteen clones were initially sequenced following plasmid rescue. The recovered cDNAs are listed in Table 2. Both the pocket domain of RB and antisense orientations of myosin light chain (MLC) were recovered twice. Among other previously known sequences are the tumor suppressor gene product Bin1, previously implicated in C2C12 cell differentiation (30), the alpha subunit of the G stimulatory signal conduction protein (Gsα), and myoglobin. Three novel ORFs were also recovered. One of these, CAGH32, corresponds to a cDNA previously isolated from a brain library selected for sequences with long CAG triplet repeat tracts; however, the CAG repeat-encoding portion does not appear in the recovered fragment. Although all of the predicted recovered clones contained a predicted ORF of at least 20 residues, it is possible that some of these sequences, such as myoglobin or MHC, were incidentally recovered as a consequence of their abundance in the library among the background population of physiologically nondifferentiating cells. This is less likely to be the case for RB, a ubiquitous protein with known myogenic function in which different and overlapping sequences were recovered as two independent events. Our first effort was therefore to determine whether these candidates truly are capable of dominant negative inhibition of satellite myogenesis. We chose to initially study the CB-RB fusion.
TABLE 2.
cDNAs encoding known proteins recovered from screening dominant negative library
| Gene product | GenBank accession no. | Nucleotide position | Protein position | Orientation | No. of times recovered |
|---|---|---|---|---|---|
| RB | M28419 | 1243–2826 | 369–896 | Sense | 2 |
| 1306–2583 | 390–815 | ||||
| Bin1 | AF068914 | 712–1420 | 218–440 | Sense | 1 |
| Gsα | X56009 | 608–1123 | 203–374 | Sense | 1 |
| Ryanodine receptor | J05200 | 3196–4941 | 1031–1612 | Sense | 1 |
| Nebulin | U35637 | 7–1788 | 3–596 | Sense | 1 |
| Enolase | NM-001976 | 10–948 | 1–316a | Sense | 1 |
| Telethonin | NM-003673 | 17–382 | 3–124 | Sense | 1 |
| Myoglobin | NM-005368 | 116–458 | 16–130 | Sense | 1 |
| MHC | M25133 | 3198–3363 | 6–60 | Sense | 1 |
| MLC | S69022 | 43–159 | NA | Antisense | 2 |
| 41–163 |
Includes ORF in 5′ untranslated region.
The pocket domain of RB inhibits muscle cell differentiation when fused downstream of CB.
We isolated two different cDNA fragments of the RB gene pocket domain fused downstream of CB-myc6, CB-myc6-RBf1 and CB-myc6-RBf2, apparently independently selected from two different clones in the library. The pocket is required for the binding of viral transactivating proteins such as the adenovirus E1A protein, simian virus 40 large T (tumor) antigen, and human papillomavirus E7 protein (9, 10, 11). This segment of the protein sequence is also sufficient for the binding of certain cellular transcription factors such as E2F, MyoD, and c-Myc (5, 7, 8, 12). Our isolation of the pocket domain of RB suggests that this region of the protein sequence may also allow for the inhibition of skeletal muscle cell differentiation by the dominant negative system.
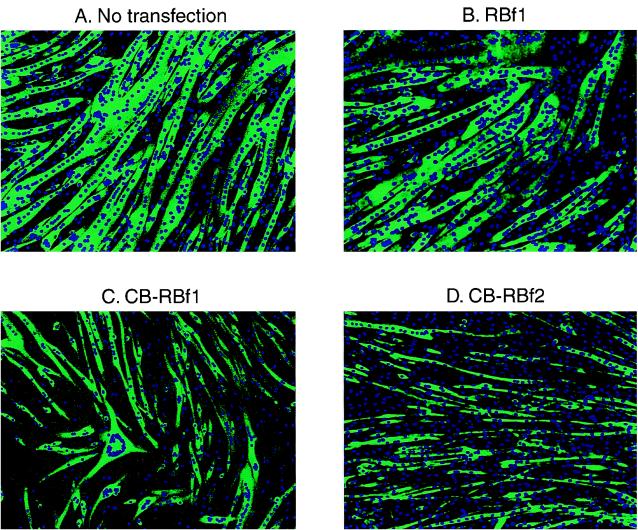
To confirm the inhibitory effects of the CB-RB fusion proteins on the differentiation of skeletal muscle cell, we reconstructed the CB-RB fusion obtained by plasmid rescue and regenerated polyclonal populations of stably transfected C2C12 cells. Immunofluorescent staining with an anti-MHC antibody reveals that cells stably transfected with control RBf1 (Fig. 2B) or RBf2 (data not shown) differentiated as well as did the wild-type cells (Fig. 2A) or cells transfected with vector alone (data not shown). Cells that were stably transfected with CB-RB fusion plasmids also entered the differentiation pathway, as evidenced by the induction of myogenin (by immunofluorescent staining [data not shown]), and phenotypically differentiated by the confirmation of the staining for the differentiation marker MHC (Fig. 2C and D). But compared to these controls, myotubes resulting from transfection with CB-RBf1 (Fig. 2C) or CB-RBf2 (Fig. 2D) were thinner, smaller, and contained fewer nuclei. There also appeared to be a greater fraction of MHC-negative mononuclear cells. Table 3 lists the average number of nuclei apparent in myotubes transfected with the different constructs. Myotubes overexpressing myc6-RBf1 or myc6-RBf2 fusion protein contain similar numbers of nuclei as those transfected with the control vectors expressing myc6 or CB-myc6. In contrast, the overexpression of CB-myc6-RBf1 or CB-myc6-RBf2 reduced the number of nuclei in each myotube to approximately one-fourth of the level for those transfected with the control vectors. Although RB loss has been shown to specifically lead to loss of late markers (18) of the differentiation program, immunofluorescent staining of the cells in the presence or absence of CB-RB (not shown) did not reveal any significant differences in the intensity of expression of an early marker (myogenin) compared to a late marker (MHC).
FIG. 2.
Immunofluorescence photomicrographs of stably transfected C2C12 cells (after inducing differentiation) doubly stained with DAPI for nuclear DNA and with an antibody to MHC (detected with a secondary fluorescein-conjugated antibody). (A) Untransfected C2C12 cells; (B) cells after transfection with pCS2+RBf1; (C) cells after transfection with pCS2+CB-myc6-RBf1; (D) cells after transfection with pCS2+CB-myc6-RBf2.
TABLE 3.
CB-RB reduces number of nuclei in myotubes
| Stably transfected plasmid | No. of nuclei/MHC- positive myotubea |
|---|---|
| pCS2+myc6 | 35 ± 5 |
| pCS2+CB-myc6 | 34 ± 4 |
| pCS2+myc6-RBf1 | 36 ± 5 |
| pCS2+myc6-RBf2 | 39 ± 3 |
| pCS2+CB-myc6-RBf1 | 8 ± 1 |
| pCS2+CB-myc6-RBf2 | 9 ± 2 |
Mean number of nuclei per myotube in 25 randomly picked MHC-positive myotubes. The standard deviation is indicated.
We sought further confirmation of the inhibitory function of the CB-RB fusion. We investigated whether overexpression of the CB-RB fusion proteins inhibits the ability of myogenic bHLH factors to induce skeletal muscle differentiation in fibroblasts. We transiently cotransfected NIH 3T3 cells with MyoD and the CB-RB fusions, induced the NIH 3T3 cells to differentiate by serum starvation, and assayed for differentiation by double immunofluorescence staining with MyoD and MHC antibodies. Table 4 indicates the total number of MyoD- and MHC-positive cells in three different microscopic fields. Cells transfected with CB-myc6-RB, myc6-RB, or vector alone showed no positive staining for MHC. When MyoD was transfected by itself, all NIH 3T3 cells in which MyoD expression was detectable were positive for MHC expression. Cotransfection of MyoD with myc6-RBf1 or myc6-RBf2 acted similarly to transfection of MyoD alone. In contrast, the proportion of MHC-positive cells was significantly reduced when MyoD was cotransfected with CB-myc6-RBf1 or CB-myc6-RBf2. Similar results were obtained by cotransfecting Myf-5 with CB-myc6-RBf1 or CB-myc6-RBf2 (data not shown). These data indicate that overexpression of CB-RB fusion proteins is also capable of inhibiting the myogenic factor-induced differentiation of fibroblasts.
TABLE 4.
CB-RB inhibits MyoD-induced NIH 3T3 cell differentiation
| Plasmid cotransfecteda | No. of MyoD-positive cells | No. of MHC-positive cells | Inhibition of differentiationb (%) |
|---|---|---|---|
| None | 18 | 18 | 0 |
| pCS2+myc6 | 20 | 19 | 5 |
| pCS2+CB-myc6 | 19 | 16 | 16 |
| pCS2+myc6-RBf1 | 20 | 19 | 5 |
| pCS2+myc6-RBf2 | 21 | 21 | 0 |
| pCS2+CB-myc6-RBf1 | 17 | 6 | 65 |
| pCS2+CB-myc6-RBf2 | 19 | 7 | 63 |
Plasmids were cotransfected along with pEMSV-MyoD.
Calculated by the formula (number of MyoD-positive cells − number of MHC-positive cells)/number of MyoD-positive cells. Cells were counted from a total of three representative fields.
Myotubes overexpressing CB-RB fusion reenter the cell cycle.
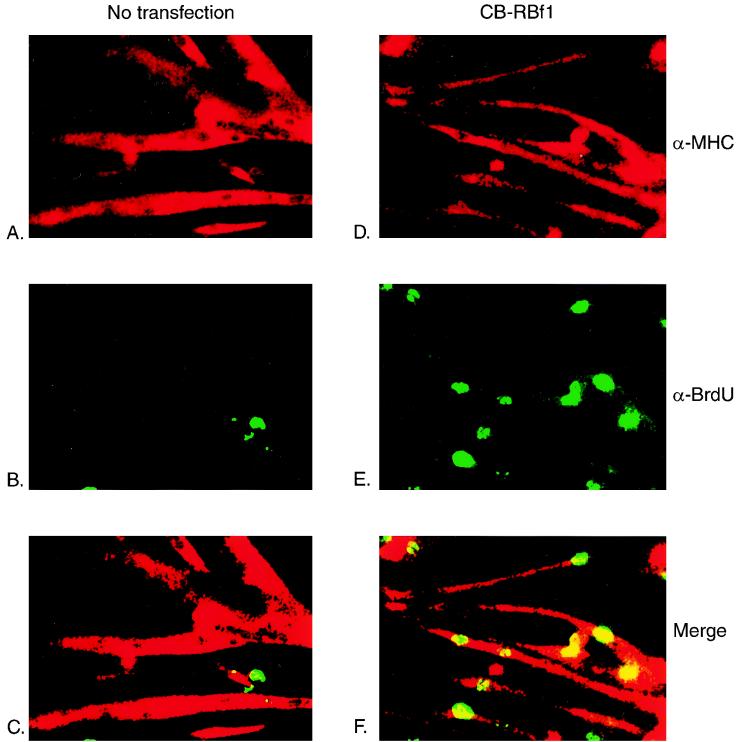
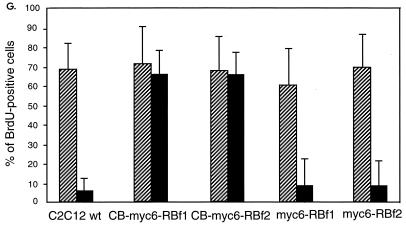
Analysis of myoblasts derived from RB-deficient embryos produced by targeted homologous recombination has revealed that these cells do not permanently withdraw from the cell cycle after differentiating into multinucleated myotubes (18, 22). To investigate whether the overexpression of CB-myc6-RB fusion proteins enables cells to reenter the cell cycle, we determined if myotubes expressing these constructs could incorporate the thymidine analog BrdU, which is taken as a marker of S-phase activity, as do RB−/− myoblasts. After inducing differentiation, the cells were stimulated with high-serum medium containing BrdU for 36 h and subsequently fixed and stained for the detection of BrdU uptake. Double immunofluorescent staining confirms that terminally differentiated MHC-positive C2C12 myotubes (Fig. 3A) fail to reenter the cell cycle after serum stimulation, as evidenced by absence of BrdU incorporation (Fig. 3B and C). In contrast, many of the MHC-positive myotubes that were stably transfected with CB-myc6-RBf1 (Fig. 3D) or CB-myc6-RBf2 (data not shown) do incorporate BrdU (Fig. 3E and F). Figure 3G summarizes the efficiencies of BrdU uptake of proliferating and serum-starved cells in response to serum stimulation. Myotubes that were stably transfected with control myc6-RBf1 or myc6-RBf2 appeared similarly to wild-type C2C12 cells, but up to about 70% of myotubes transfected with CB-myc6-RBf1 or CB-myc6-RBf2 incorporate BrdU. These data indicate that myotubes overexpressing CB-RB fusion proteins are able to reenter the cell cycle.
FIG. 3.
Myotubes stably transfected with CB-RB fusion plasmids are able to reenter the cell cycle. C2C12 cells that were either not transfected (A to C) or transfected with a vector expressing CB-myc6-RBf1 (D to F) were induced to differentiate in DM and then cultured in GM containing 10 μM BrdU for 36 h. Cells were subsequently fixed, permeabilized, and immunostained with antibodies for MHC (secondarily detected with rhodamine-conjugated antibody) and BrdU (secondarily detected with fluorescein-conjugated antibody). Note the specific uptake of BrdU in the nuclei of CB-myc6-RBf1 stably transfected myotubes. (G) Quantification of BrdU-positive nuclei. Approximately 150 to 250 cells were counted for each column. The diagonal striped and filled bars represent the populations of BrdU-positive cells while proliferating in GM and following 3 days of serum starvation in DM, respectively. wt, wild type.
The inhibition of satellite cell differentiation by CB-RB cannot be rescued with overexpression of MyoD.
It has been reported that RB and MyoD directly bind to each other through a region that involves the pocket and bHLH domains, respectively (5), and that this interaction is required for both the permanent withdrawal of muscle cells from the cell cycle and the activation of myogenic differentiation. The isolation of the RB pocket domain using the dominant negative system thus indicates a possibility that the CB-RB fusion protein might inhibit both myogenesis and permanent cell cycle withdrawal through a direct interaction with MyoD. If so, overexpression of MyoD might relieve the inhibition of muscle cell differentiation resulting from the overexpression of CB-RB fusion proteins. To test this possibility, we transiently transfected myc6-MyoD expression plasmid into a CB-RBf1 or CB-RBf2 stably transfected cell line and, 1 day later, induced the cells to differentiate. Double immunofluorescent staining with an anti-Myc antibody and an anti-MHC antibody showed that the overexpression of myc6-MyoD did not normalize the morphologic appearance of the myotubes, as they remained small, thin, and with few nuclei (Table 5). In previous experiments with dominant negative CB fusions employing lacZ and MyoD (15) we were able to detect redistribution of targeted interacting factors to the lysosome. In the experiments here we found no evidence from immunofluorescent staining of a direct interaction between the CB-RB fusion and MyoD, in that the staining for MyoD remained exclusively nuclear (data not shown). These two observations suggest that the inhibition of muscle cell differentiation by the CB-RB fusion proteins is unlikely to be the direct consequence of interaction with MyoD.
TABLE 5.
MyoD overexpression cannot rescue inhibition of differentiation by CB-RB
| Plasmid used to stably transfect C2C12 cells | Plasmid transiently transfected | No. of nuclei pera:
|
|
|---|---|---|---|
| myc6-negative myotube | myc6-positive myotube | ||
| pCS2+CB-RBf1-(neo) | pCS2+myc6-MyoD | 8 | 10 |
| pCS2+myc6 | 9 | 8 | |
| pCS2+CB-RBf2-(neo) | pCS2+myc6-MyoD | 7 | 7 |
| pCS2+myc6 | 10 | 9 | |
Nuclei were scored by indirect immunofluorescent staining with the anti-Myc epitope monoclonal antibody 9E10. Numbers represent the mean of 25 MHC-positive myotubes. Comparable results were obtained in at least two independent experiments.
The pocket B domain of RB is essential for MyoD binding both in vitro and in vivo (5). To investigate if CB-RB fusion proteins inhibit muscle cell differentiation through the same region, we deleted a part of the pocket B region from CB-myc6-RBf1 (CB-myc6-RBf1ΔC) and tested its effect on C2C12 cells differentiation. Immunofluorescent staining of cells stably transfected with CB-myc6-RBf1ΔC indicates that they are just as impaired in differentiative capacity as those transfected with CB-myc6-RBf1 or CB-myc6-RBf2 (data not shown). This result indicates both that direct interaction between MyoD and RB is unlikely to be required for satellite myogenesis and that the inhibitory effect results from inhibition of third-party factor(s) interfacing exclusively with the pocket A domain of RB. This conclusion is bolstered by our inability to complement the defective differentiation phenotype by stable transfection of RB (not shown).
CB-RB increases cell growth in colony formation assays.
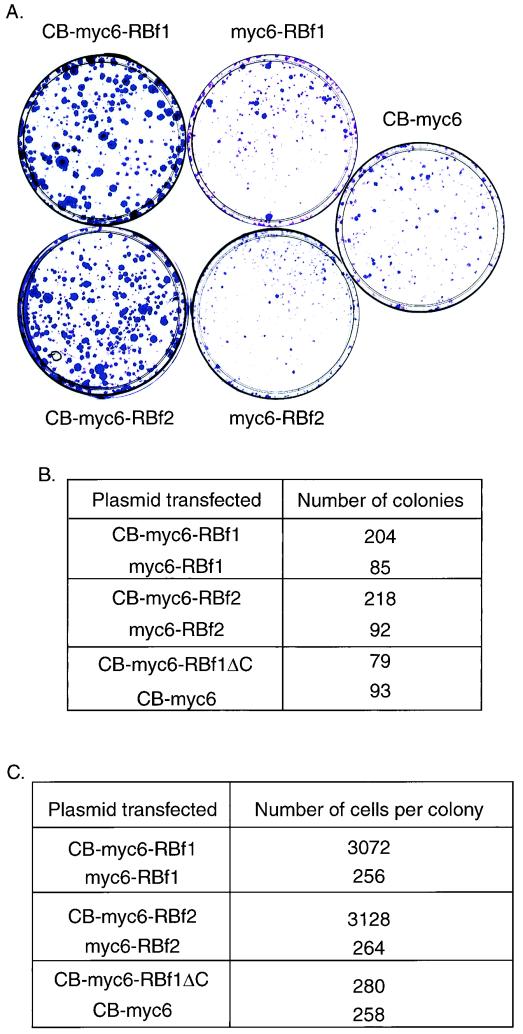
When the stably transfected cell lines were selected with G418, we noticed that a greater number of G418-resistant colonies were formed on the plates transfected with CB-RBf1 or CB-RBf2 fusion constructs than on those transfected with control constructs. To investigate whether this is due to increased cellular growth resulting from expression of the CB-RB fusion, we performed a colony formation assay, a method previously established for documenting disruption of pathways involving tumor suppressor genes (1), including the RB gene (20). Figure 4A shows representative plates, and Fig. 4B shows colony counts from this experiment. When transfected with CB-myc6-RBf1 or CB-myc6-RBf2 fusion constructs, C2C12 cells revealed a large increase in colony-forming efficiency, with colonies that appeared larger, compared to transfection with the control RBf2 or CB-myc6 constructs. Overexpression of the pocket B deletion mutant of CB-myc6-RBf1 (CB-myc6-RBf1ΔC) had no effect on colony formation efficiency.
FIG. 4.
The CB-RB fusion protein increases cell growth. (A) C2C12 cells were transfected with 10 μg of the indicated plasmids and grown in the presence of G418 for 2 weeks. G418-resistant colonies were then stained with crystal violet. (B) Number of colonies counted per plate in an experiment similar to that shown in panel A. (C) Number of cells per clone. The indicated stably transfected cells were plated at clonal density and grown in the presence of G418 for 1 week. Then cells were stained as for panel A, and the mean number of cells per colony was counted.
To rule out the possibility that the observed differences in clonal number did not trivially result from transfection efficiency, we also performed a cell growth speed assay. We split stably transfected cell lines, plated them at clonal density, and then grew them under normal selection conditions with G418. Cells stably transfected with CB-myc6-RBf1 or CB-myc6-RBf2 formed colonies comprised of a greater number of cells compared to those resulting from transfection with the control expression vector. Cells stably transfected with CB-myc6-RBf1ΔC formed colonies similar in size to those transfected with the empty expression vector. Figure 4C represents the average cell number of each colony on different plates and demonstrates that cell lines transfected with CB-myc6-RBf1 or CB-myc6-RBf2 grow more quickly. We conclude that the CB-RB fusion protein disrupts an activity of RB related to regulation of cell growth, but because the pocket B deletion mutant had no effect on cell growth but retains the capacity to inhibit C2C12 cell differentiation, fusion with CB may disrupt two unique functions of RB that are spatially separated in the primary sequence.
CB-RB inhibits transcription activation by MyoD.
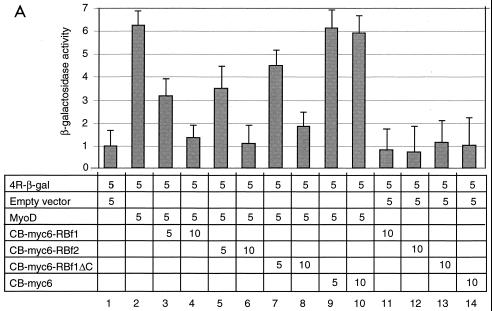
To address whether the inhibition of muscle cell differentiation by overexpression of the CB-RB fusion protein is achieved by inhibiting muscle gene expression, we determined if the CB-RB fusion could inhibit transactivation by MyoD. We used a minimal 4R enhancer containing four concatenated MyoD binding sites (4R-β-gal) as a reporter (32) to transiently transfect NIH 3T3 cells. Cotransfection of MyoD increased the reporter activity by about sixfold compared to control cotransfection with an empty expression vector (Fig. 5A, columns 1 and 2). Addition of CB-myc6-RBf1 or CB-myc6-RBf2 resulted in inhibition of reporter activity by MyoD in a concentration-dependent manner (columns 3 to 6). The pocket B deletion mutation of CB-myc6-RBf1 also proved inhibitory (columns 7 and 8), albeit somewhat less so; the finding that the pocket B deletion mutant retains activity agrees with the finding that this region is not necessary to inhibit myogenic differentiation. Addition of the control expression vector CB-myc6 had no effect on reporter activity (columns 9 and 10), nor did the various constructs in the absence of MyoD (columns 11 through 14). It has been shown that RB is specifically needed to activate the transcriptional potential of MEF2 (19). We therefore also monitored the effects of CB-RB on MyoD activation of the muscle creatine kinase (MCK) reporter, which requires MEF2 activity (Fig. 5B). The actions of CB-RB are similar on this promoter. Taken together, these data indicate that the CB-RB fusion proteins inhibit muscle cell differentiation by inhibiting the transactivation function of MyoD.
FIG. 5.
The CB-RB fusion inhibits MyoD transcriptional activation of 4R-β-gal (A) and MCK (B) reporters. NIH 3T3 cells were transiently cotransfected with the indicated quantities (in micrograms) of MyoD, plasmids expressing either CB-myc6-RBf1, CB-myc6-RBf2, CB-myc6-RBf1ΔC, or CB-myc6 (as a control), and either the 4R-β-gal or MCK-β-gal reporter. Induction is normalized in each experiment by parallel cotransfection with the reporter (5 μg) and the empty vector (5 μg) to which the β-galactosidase activity is arbitrarily assigned a value of 1 (column 1). The reported β-galactosidase activity is thus expressed as a ratio of the activity for the conditions in lane 1. Error bars represent standard deviation of triplicate experiments.
CB-RB inhibits E2F1 but not a forced E2F1-RB dimer.
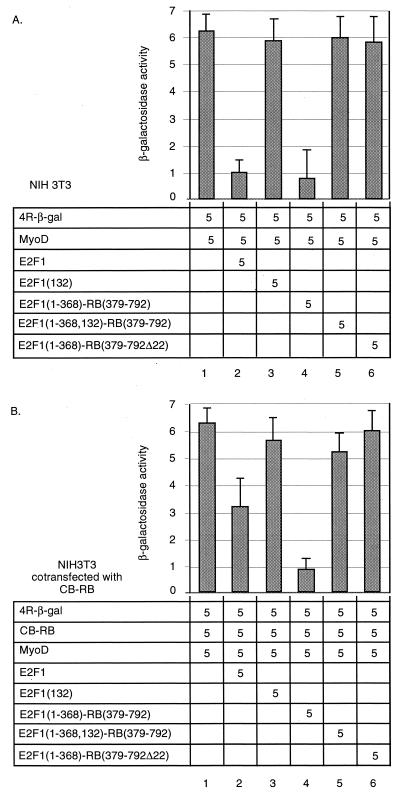
In the simplest model of myogenesis and cell cycle control, RB is antagonized by binding with E2F1. E2F1 is, accordingly, generally inhibitory to MyoD-induced activation of a variety of muscle promoters (29). We therefore determined if E2F1 can be inhibited by CB-RB. E2F1 was cotransfected into NIH 3T3 cells with MyoD and the 4R promoter reporter (Fig. 6A). As expected, E2F1 inhibits transactivation of the 4R promoter by MyoD (columns 2 versus 1), whereas E2F(132), which contains a point mutation at amino acid 132 that disrupts DNA binding by E2F1, does not (column 3). Similarly, a chimera in which the E2F1 transactivation domain was replaced with the RB pocket domain, E2F1(1-368)-RB(379-792)—previously shown to function as a transcriptional repressor on a variety of promoters (25)—also functions to repress transactivation of the 4R reporter in this assay (column 4). An E2F1-RB fusion construct containing the point mutation at amino acid 132 in the E2F1 DNA binding domain, E2F1(1-368, 132)-RB(379-792) (column 5) or, conversely, a construct in which the E2F1 transactivation domain was replaced with a tumor-derived RB mutant which deleted exon 22, E2F1(1-368)-RB(379-792Δex22) (column 6), each failed to repress the 4R reporter. When this assay was repeated with the additional cotransfection of CB-RB (Fig. 6B), E2F1 failed to fully repress 4R promoter activation (column 2), whereas the chimeric E2F1(1-368, 132)-RB(379-792) retains this activity (column 4). We therefore conclude that normally, E2F1 inhibits MyoD-dependent activation of an E-box reporter. In the presence of the dominant negative CB-RB, however, E2F1 is an ineffective inhibitor of MyoD-dependent activation of E boxes, but the forced E2F1-RB dimer remains as an effective inhibitor, presumably because it is resistant to titration of E2F1 from the RB complex.
FIG. 6.
CB-RB can inhibit E2F1 activity. NIH 3T3 cells (A) or NIH 3T3 cells transiently cotransfected with CB-RB (B) were transiently transfected with the indicated quantity (in micrograms) of vectors encoding the 4R-β-gal reporter and various constructs, and reporter activity was determined as for Fig. 5. Error bars represent standard deviation of triplicate experiments.
Two additional tests of E2F1 mediation were also imposed. Stable transfection of E2F1 did not rescue the defective myogenic phenotype of C2C12 cells that were stably transfected with CB-RB (not shown). Further, we were unable to detect any redistribution of E2F1 from the nucleus to the lysosome upon coexpression with CB-RB (not shown). These observations indicate that E2F1 may help to mediate interactions between MyoD and RB in this system, but that the result may be complicated and dependent, as well, upon additional, unidentified factors.
DISCUSSION
To investigate genetic pathways responsible for decisions between differentiation and growth in satellite myoblasts, we have used a novel screen based on dominant negative mutation. As with other genetic approaches, its value derives from the absence of a requirement of prior molecular knowledge of the pathway under study. Although several new potential candidate genes have emerged, we chose to initially validate this strategy by investigating the properties of RB, since it is known from prior phenomenological investigations to participate in myogenesis.
The RB gene is the archetypal tumor suppressor gene. To our knowledge, no dominant-acting mutations have either been identified in cancer or engineered in laboratory investigations of its function. Because of the central importance of RB in cell cycle control, it has presumably evolved a sequence somewhat resistant to constitutive activation via mutagenic mechanisms likely to be encountered in tumors. Instead, it follows the “two hit” paradigm in which inactivation of both alleles is required for loss of activity. All RB mutants to date therefore behave recessively, and all cellular studies utilizing mutant RB proteins in myogenesis have been conducted in RB−/− backgrounds resulting from cell lines derived from either knockout mice or from tumors lacking RB function. The unique availability here of a dominant-acting RB not only validates the potential power of this approach but also allows us to address questions in myogenesis that have previously been unanswerable through a genetic strategy.
Specifically, there have been ambiguous data on whether or not a direct physical association between MyoD and RB occurs during myogenesis in vivo. It was demonstrated by affinity chromatography (5) that MyoD and RB associate in vitro and that the interaction requires an intact bHLH domain in MyoD and pocket domain of RB; in vivo MyoD-RB interactions were demonstrated by coimmunoprecipitation experiments. However, supershift electrophoretic mobility assays with antibodies to MyoD or RB failed to detect complexes on MyoD (E box) binding sites (6), and there was no difference in electrophoretic mobility between myogenic bHLH complexes formed in extracts from RB deficient compared to RB wild-type cells (18). Genetic experiments have incompletely resolved the issue. Myoblasts obtained from RB knockout mice can still differentiate into myotubes (albeit with attenuated expression of late differentiation markers such as MHC) but demonstrate an absence of permanent cell cycle withdrawal (18). C2C12 cells stably transfected with antisense RB behave similarly (13). The conclusion drawn from these experiments is that morphological differentiation and cell cycle arrest are disassociable activities of RB and that the latter might be independent of MyoD. This is confirmed by studies performed in RB−/− osteosarcoma cells that show that mutation in the RB pocket domain has independent effects on the ability of RB to induce G1/S growth arrest or alternatively to cooperate in transcriptional activation with MyoD (26). Nevertheless, with both knockouts and antisense, potential interactions between MyoD and RB are voided by the absence of RB in the cell. Our results here indicate that cells expressing the dominant negative RB have a phenotype similar to the antisense RB C2C12 cells or the RB knockout myoblasts. The unique feature of our approach is that both RB and MyoD are still expressed at normal levels in the cell and are therefore free to associate, should that be important. However, we conclude that a direct association between MyoD and RB is not of significance, because we are unable to rescue the phenotype with overexpression of MyoD or RB, detect in vivo interaction between the RB pocket domain and MyoD (by examining for cytoplasmic redistribution of MyoD with CB-RB), or prove a requirement for the pocket B domain for the dominant negative effect on either morphological differentiation or transcriptional activation (though it does appear important to colony formation efficiency). The most parsimonious interpretation for all of these observations is that in vivo interactions between RB and MyoD are conducted indirectly through a third party that is inhibited by the dominant negative RB. We conclude that the prior observations of direct physical association of RB and MyoD may not be physiologically relevant.
The question then becomes, what factors are targeted by the CB-RB fusion in order to explain the mechanism of inhibition of satellite cell differentiation? One possibility is the E2F family of transcription factors, which physically associate through the RB pocket domain and play a central role in the regulation of cell cycle progression (21, 31). It has been proposed that RB serves simply to sequester E2F and prevent it from driving entry into S phase by repressing differentiation-specific promoters. Accumulating evidence, however, suggests that the function of E2F is rather more complex. The RB-E2F transcriptional complex, rather than merely serving to sequester E2F, can actively repress transcription (25), and E2F knockout mice paradoxically display atrophy of some tissues while developing tumors of other cell types (34). Based on the observations that E2F loses activity as a transcriptional repressor in satellite myoblasts expressing the CB-RB fusion, whereas a forced E2F-RB heterodimer retains activity, we suggest that E2F1 is a potential mediator of MyoD and RB interactions. On the other hand, E2F1 appears predominantly as a repressor of myogenesis in our assays, so some other factors that promote myogenesis must also be inactivated by the CB-RB fusion.
Another previously known protein that we have isolated in this screen is Bin1, a Myc-interacting protein found in both the nucleus and cytoplasm that is upregulated with C2C12 differentiation (30). Antisense expression of Bin1 inhibits C12C12 differentiation, whereas overexpression of Bin1 promotes differentiation (30). In the screen performed here, overexpression of sense Bin1, when fused with CB, inhibits differentiation, as is the case for RB, thereby providing additional evidence that the novel genes identified in this screen are also likely to participate in satellite myogenesis. Further study of Bin1 in the function of satellite cell differentiation is thus warranted.
Yet another protein identified in this screen is Gsα. The stimulatory G protein is a key regulator of adenylate cyclase in a signaling cascade leading to the production of cyclic AMP (cAMP). cAMP transduces mitogenic signals by binding to the regulatory subunits of the cAMP-dependent protein kinase A. It has previously been shown that elevated levels of the intracellular signaling molecule cAMP and overexpression of protein kinase A inhibit myogenic differentiation (16, 33). We have yet to explore the mechanism through which Gsα affects myogenic differentiation in our system at the molecular level, but it presumably does so through diverting some factor(s) required for mitogenic signal conduction. We believe that the approach to dominant negative mutation taken here offers the possibility of identifying novel genes with important roles in the differentiation of satellite muscle cells and that this strategy may prove suitable for investigating gene function in other developmental models.
ACKNOWLEDGMENTS
We thank William Sellers for his gift of the E2F1-RB constructs.
This study was supported by Doris Duke Charitable Research Foundation grant T98006.
REFERENCES
- 1.Baker S J, Markowitz S, Fearon E R, Willson J K V, Vogelstein B. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science. 1990;249:912–915. doi: 10.1126/science.2144057. [DOI] [PubMed] [Google Scholar]
- 2.Cavanaugh A H, Hempel W M, Taylor L J, Rogalsky V, Todorov G, Rothblum L I. Activity of RNA polymerase I transcription factor UBF blocked by Rb gene product. Nature. 1995;9:177–180. doi: 10.1038/374177a0. [DOI] [PubMed] [Google Scholar]
- 3.Chevallier A, Pauto M P, Harris A J, Kieny M. On the non-equivalence of skeletal muscle satellite cells and embryonic myoblasts. Arch Anat Microsc. 1987;75:161–166. [PubMed] [Google Scholar]
- 4.Gogos J A, Thompson R, Lowry W, Sloane B F, Weintraub H, Horwitz M. Gene trapping in differentiating cell lines: regulation of the lysosomal protease cathepsin B in skeletal myoblast growth and fusion. J Cell Biol. 1996;134:837–847. doi: 10.1083/jcb.134.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gu W, Schneider J W, Condorelli G, Kaushal S, Mahdavi V, Nadal-Ginard B. Interaction of myogenic factors and the retinoblastoma protein mediates muscle cell commitment and differentiation. Cell. 1993;72:309–342. doi: 10.1016/0092-8674(93)90110-c. [DOI] [PubMed] [Google Scholar]
- 6.Halevy O, Novitch B G, Spicer D B, Skapek S X, Rhee J, Hannon G J, Beach D, Lassar A B. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science. 1995;267:1018–1021. doi: 10.1126/science.7863327. [DOI] [PubMed] [Google Scholar]
- 7.Helin K, Lees J A, Vidal M, Dason N, Harlow E, Fattaey A. A cDNA encoding a pRB-binding protein with properties of the transcription factor E2F. Cell. 1992;70:337–350. doi: 10.1016/0092-8674(92)90107-n. [DOI] [PubMed] [Google Scholar]
- 8.Hiebert S W, Chellappan S P, Horowitz J M, Nevins J R. The interaction of RB with E2F coincides with an inhibition of the transcriptional activity of E2F. Genes Dev. 1992;6:177–185. doi: 10.1101/gad.6.2.177. [DOI] [PubMed] [Google Scholar]
- 9.Hu Q, Dyson N, Harlow E. The regions of the retinoblastoma protein needed for binding to adenovirus E1A or SV40 large T antigen are common sites for mutations. EMBO J. 1990;9:1147–1155. doi: 10.1002/j.1460-2075.1990.tb08221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang S, Wang N, Tseng B Y, Lee W, Lee E H. Two distinct and frequently mutated regions of retinoblastoma protein are required for binding to SV40 T antigen. EMBO J. 1990;9:1815–1822. doi: 10.1002/j.1460-2075.1990.tb08306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaelin W G, Ewen M E, Livingston D M. Definition of the minimal simian virus 40 large T antigen- and adenovirus E1A-binding domain in the retinoblastoma gene product. Mol Cell Biol. 1990;10:3761–3769. doi: 10.1128/mcb.10.7.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaelin W G, Pallas D C, DeCaprio J A, Kaye F J, Livingston D M. Identification of cellular proteins that can interact specifically with the T/A-binding region of the retinoblastoma gene product. Cell. 1991;64:521–532. doi: 10.1016/0092-8674(91)90236-r. [DOI] [PubMed] [Google Scholar]
- 13.Kobyashi M, Yamauchi Y, Tanaka A. Stable expression of antisense Rb-1 RNA inhibits terminal differentiation of mouse myoblast C2 cells. Exp Cell Res. 1998;239:40–49. doi: 10.1006/excr.1997.3880. [DOI] [PubMed] [Google Scholar]
- 14.Leibovitch M P, Leibovitch S A, Harel J, Kruh J. Changes in the frequency and diversity of messenger RNA populations in the course of myogenic differentiation. Eur J Biochem. 1979;97:321–326. doi: 10.1111/j.1432-1033.1979.tb13117.x. [DOI] [PubMed] [Google Scholar]
- 15.Li F-Q, Coonrod A, Horwitz M. Preferential MyoD homodimer formation demonstrated by a general method of dominant negative mutation employing fusion with a lysosomal protease. J Cell Biol. 1996;135:1043–1057. doi: 10.1083/jcb.135.4.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li L, Heller H R, Czech M, Olson E N. Cyclic AMP-dependent protein kinase inhibits the activity of myogenic helix-loop-helix proteins. Mol Cell Biol. 1992;12:4478–4485. doi: 10.1128/mcb.12.10.4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Molkentin J D, Black B L, Martin J F, Olson E N. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- 18.Novitch B G, Mulligan G J, Jacks T, Lassar A B. Skeletal muscle cells lacking the retinoblastoma protein display defects in muscle gene express and accumulate in S and G2 phases of the cell cycle. J Cell Biol. 1996;135:441–456. doi: 10.1083/jcb.135.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Novitch B G, Spicer D B, Kim P S, Cheung W L, Lassar A B. pRb is required for MEF2-dependent gene expression as well as cell-cycle arrest during skeletal muscle differentiation. Curr Biol. 1999;9:449–459. doi: 10.1016/s0960-9822(99)80210-3. [DOI] [PubMed] [Google Scholar]
- 20.Qin X-Q, Chittenden T, Livingston D M, Kaelin W G. Identification of a growth suppression domain within the retinoblastoma gene product. Genes Dev. 1992;6:953–964. doi: 10.1101/gad.6.6.953. [DOI] [PubMed] [Google Scholar]
- 21.Sanchez A, Dynlacht B. Transcriptional control of the cell cycle. Curr Opin Cell Biol. 1996;8:318–324. doi: 10.1016/s0955-0674(96)80004-4. [DOI] [PubMed] [Google Scholar]
- 22.Schneider J W, Gu W, Zhu L, Mahdavi V, Nadal-Ginard B. Reversal of terminal differentiation mediated by p107 in Rb−/− muscle cells. Science. 1994;264:1467–1471. doi: 10.1126/science.8197461. [DOI] [PubMed] [Google Scholar]
- 23.Schultz E. Satellite cell proliferative compartments in growing skeletal muscle. Dev Biol. 1996;175:84–94. doi: 10.1006/dbio.1996.0097. [DOI] [PubMed] [Google Scholar]
- 24.Schultz E, McCormick K M. Skeletal muscle satellite cells. Rev Physiol Biochem Pharmacol. 1995;123:213–257. doi: 10.1007/BFb0030904. [DOI] [PubMed] [Google Scholar]
- 25.Sellers W R, Rodgers J W, Kaelin W G. A potent transrepression domain in the retinoblastoma protein induces a cell cycle arrest when bound to E2F sites. Proc Natl Acad Sci USA. 1995;92:11544–11548. doi: 10.1073/pnas.92.25.11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sellers W R, Novitch B G, Miyake S, Heith A, Otterson G A, Kaye F J, Lassar A B, Kaelin W G. Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor cell growth. Genes Dev. 1998;12:95–106. doi: 10.1101/gad.12.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tapscott S J, Davis R, Thayer M J, Cheng P F, Weintraub H, Lassar A B. MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science. 1988;242:405–411. doi: 10.1126/science.3175662. [DOI] [PubMed] [Google Scholar]
- 28.Turner D L, Weintraub H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- 29.Wang J, Huang Q, Tang W, Nadal-Ginard B. E2F1 inhibition of transcription activation by myogenic basic helix-loop-helix regulators. J Cell Biochem. 1996;62:405–410. doi: 10.1002/(SICI)1097-4644(199609)62:3%3C405::AID-JCB10%3E3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 30.Wechsler-Reya R J, Elliott K J, Pendergast G C. A role for the putative tumor suppressor Bin1 in muscle cell differentiation. Mol Cell Biol. 1998;18:566–575. doi: 10.1128/mcb.18.1.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 32.Weintraub H, Davis R, Lockshon D, Lassar A. MyoD binds cooperatively to two sites in a target enhancer sequence: occupancy of two sites is required for activation. Proc Natl Acad Sci USA. 1990;87:5623–5627. doi: 10.1073/pnas.87.15.5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winter B, Braun T, Arnold H H. cAMP-dependent protein kinase represses myogenic differentiation and the activity of the muscle-specific helix-loop-helix transcription factors Myf-5 and MyoD. J Biol Chem. 1993;268:9869–9878. [PubMed] [Google Scholar]
- 34.Yamasaki L, Jacks T, Bronson R, Goillot E, Harlow E, Dyson N J. Tumor induction and tissue atrophy in mice lacking E2F-1. Cell. 1996;85:537–548. doi: 10.1016/s0092-8674(00)81254-4. [DOI] [PubMed] [Google Scholar]
- 35.Yoshida N, Yoshida S, Koishi K, Masuda Y, Nabeshima Y. Cell heterogeneity upon myogenic differentiation: down-regulation of MyoD and Myf-5 generates ‘reserve cells.’. J Cell Sci. 1998;111:769–779. doi: 10.1242/jcs.111.6.769. [DOI] [PubMed] [Google Scholar]
- 36.Zacksenhaus E Z, Jiang Z, Chung D, Marth J D, Phillips R A, Gallie B L. pRB controls proliferation, differentiation, and death of skeletal muscle cells and other lineages during embryogenesis. Genes Dev. 1996;10:3051–3064. doi: 10.1101/gad.10.23.3051. [DOI] [PubMed] [Google Scholar]