Abstract
Infections are a major cause of premature death. Fast and accurate laboratory diagnostics of infectious diseases is a key condition for the timely initiation and success of treatment. Potentially, it can reduce morbidity, as well as prevent the outbreak and spread of dangerous epidemics. The traditional methods of laboratory diagnostics of infectious diseases are quite time- and labour-consuming, require expensive equipment and trained personnel, which is crucial within limited resources. The fast biosensor-based methods that combine the diagnostic capabilities of biomedicine with modern technological advances in microelectronics, optoelectronics, and nanotechnology make an alternative.
The modern achievements in the development of label-free biosensors make them promising diagnostic tools that combine rapid detection of specific molecular markers, simplicity, ease-of-use, efficiency, accuracy, and cost-effectiveness with the tendency to the development of portable platforms. These qualities exceed the generally accepted standards of microbiological and immunological diagnostics and open up broad prospects for using these analytical systems in clinical practice directly at the site of medical care provision (point-of-care, POC concept).
A wide variety of modern biosensor designs are based on the use of diverse formats of analytical and technological strategies, identification of various regulatory and functional molecular markers associated with infectious pathogens. The solution to the existing problems in biosensing will open up great prospects for these rapidly developing diagnostic biotechnologies.
Keywords: biosensors, label-free biosensor, laboratory diagnostics, infectious diseases, sensory strategies, molecular markers
Introduction
The most terrible tragedies of mankind in recent centuries have been associated with the outbreaks and spread of pandemic infections that have claimed hundreds of millions of lives. Despite the obvious success of the global health care system, the risk of epidemics of known, new, and recurring infections remains a serious threat to the world’s population. The bacterial and viral infectious diseases with the fecal-oral mechanism of infection claim about 2 million lives annually. The recent outbreaks of Ebola, Zika, Dengue, Middle East respiratory syndrome, severe acute respiratory syndrome, and H5N1 influenza, as well as the increasing resistance of bacteria to antimicrobial drugs, increase the urgency of searching for new effective diagnostic tools aimed at early and rapid detection of pathogens [1–3].
With broadening our knowledge on the complex biochemical processes underlying the pathogenesis of infectious processes, it has become necessary to develop more sensitive and highly specific diagnostic strategies. They are based on the identification of molecular markers, profiling of microorganisms without cultivation, enrichment, and isolation of pure cultures. These methods will become ideal analytical tools for controlling pathogenic microorganisms and make a basis for identifying the relationship between molecular structures and biological processes [1, 4, 5].
Classical microbiological and immunoserological methods, as well as modern diagnostic platforms such as ELISA and chemiluminescence analysis, PCR, flow cytometry, and mass spectrometry (MALDI) adopted in recent decades, prevail when accurate verification of infectious agents is needed in centralized laboratories of medical hospitals and centers. However, these diagnostic tools require expensive equipment, long testing times, and qualified personnel, and are not always available for small hospitals, especially under limited economic resources and the decentralized infrastructure of medical facilities [4, 6, 7].
The biosensor technologies that have emerged over recent years and are actively developing serve as innovative platforms for analyzing biomarkers of the infectious process have a high potential to become affordable, fast and reliable in operation, highly specific and sensitive tools for timely and true diagnosis of bacterial and viral diseases [8, 9]. The economic feasibility and ease-of-use of these portable analytical systems are fully consistent with the modern global concept of point-of-care testing (laboratory testing at the site of treatment).
Modern diagnostic technologies based on the point-of-care concept
In the global practice of infectious disease diagnostics, the point-of-care strategy is becoming increasingly important, based on modern molecular diagnostic technologies, including laboratory testing by the medical personnel at the patient’s bedside or self-monitoring of certain laboratory parameters by patients at home [8, 9].
The diagnostic platforms-precursors of this innovative strategy include qualitative and semi-quantitative test systems for identifying specific antigens [9] and antibodies [10], as well as gene amplification products [11, 12], based on latex agglutination, immunochromatography, and variations of lateral flow immunoassay (lateral flow assay, LFA; and lateral flow immunoassay, LFIA), which have not lost their significance for the diagnosis of infectious diseases these days [10, 11, 13–16] (Figure 1).
Figure 1. Schematic representation of the lateral-flow immunoassay mechanism.
The sample containing the test antigen (analyte) is applied to the sample application pad and migrates to the conjugate. The specific reagent with the target analyte migrates to the test line, where they form a complex with antibodies (source: authors)
For example, in their recent study, Jørgensen et al. [17] successfully tested the first commercial combined test for the detection of Streptococcus pneumoniae and Legionella pneumophila urinary antigens with the LFIA method. This has increased the popularity of the universal technology that is equally effective in the sandwich analysis format for both high-molecular antigens of microorganisms and antibodies to them in biosubstrates and low-molecular analytes [16, 18–20].
Today, the test systems based on the LFIA method in both standard and multiplex formats make up the most part of the global segment of rapid laboratory diagnostics [14, 16, 18, 19]. However, despite the obvious attractiveness of LFA and LFIA methods, significant disadvantages of immunoassay hinder the expansion of practical use of these diagnostic platforms in the diagnosis of bacterial and viral infections (Table 1).
Table 1.
Advantages and disadvantages of the test-systems on the lateral-flow immunoassay platform
| Advantages | Disadvantages | References |
|---|---|---|
| Inexpensive, fast, and easy-to-use tests; long shelf-life of the test systems | Applicable only for primary screening and require confirmation of positive results by independent methods | [13, 17] |
| No special temperature conditions for storage are required | Special equipment (scanners, reflectometers, CCD cameras) and software are required to obtain quantitative results | [16, 20] |
| No special equipment is required | Technological improvement of the method increases the cost and duration of the analysis | [14, 18] |
| Qualified personnel is not required; can be operated by general practitioners or home patients | In the competitive format, the response negatively correlates with concentration | [15, 19, 21] |
| The visual result is clear and readily visible | Potential technical errors in sample application may affect accuracy and reproducibility of the result | [18, 19, 21] |
| Tests are usually sold in kits with a set of all the items necessary to perform a test | An increase in the sensitivity of tests is associated with the use of gold and silver nanoparticles or an enzyme, which limits shelf-life, increases the cost of the analysis, and breaks the one-step mode of the test | [15, 19, 21] |
| Possible increase in the sensitivity of the test systems when using plasmon resonance, surface-enhanced Raman scattering (SERS), chemiluminescent or fluorescent labels |
A test sample must be in the form of a solution Pre-dissolution of dry samples is mandatory Low analyte content in the solution requires sample concentrating |
Biosensor technologies
Thanks to the impressive advances in molecular biology and nanotechnology since the beginning of the twenty-first century, biosensors are becoming increasingly developed and used for diagnosis both in vitro and in vivo. Over the past 10–15 years, the research and development of these highly selective analytical devices has become a popular and most actively developing biotechnological trend, the most attractive alternative to lateral-flow immunoassay methods [22, 23].
With putting these fast-acting, sensitive, and inexpensive autonomous sensors into the laboratory practice, the most advanced achievements in various biotechnological fields, including healthcare are expected to appear in the near future. Biosensors have shown great potential for use in medical laboratory diagnostics, as well as a tool for immediate detection in real-time of several markers of bacterial and viral infections. The microorganisms consist of a wide range of macromolecules with electrochemically active groups that can react with free electrons of the electrode surface [22, 23]. Controlling these processes with the use of physical and chemical methods enables to detect and study infectious pathogens. In this case, changes in temperature and pH are used as additional analytical information. Due to these properties, biosensors are considered as a powerful diagnostic technology in the point-of-care strategy for detecting infectious diseases at the initial stage, monitoring the development of the pathological process, and carrying out epidemiological studies [22, 24, 25].
Moreover, due to the selective capability of the devices based on the advances in modern nanoelectronics to receive and convert biosignals, it has become possible to conduct quantitative monitoring of the infectious process [22, 24–27]. For example, by now, we have accumulated experience in using electrochemical biosensors to monitor biofilm formation in real time [22, 25, 26] and other dormant forms of bacteria [27], sepsis development [11, 12, 28, 29], spore formation [30–32].
We shall consider the main types of biosensors and the most common methods of biosensing, as well as modern achievements in designing these analytical devices. Impressive results have been obtained in the use of biosensors in various fields of biology, ecology, toxicology, parasitology, criminology, medicine, and microbiology over the recent years. However, this review will focus on the prospects for using modern analytical devices in the laboratory diagnostics of infectious diseases.
Main types of biosensors and their functioning
The study of the molecular basis of pathogenicity of microorganisms as well as the search and development of highly effective and sensitive methods for the identification of pathogenic microorganisms have always been the focus of attention for researchers. Besides, timely laboratory diagnosis is the key to successful treatment of infections, as well as prevention of the occurrence and spread of epidemics. For the diagnosis of infectious diseases, monitoring and early detection of the markers of infectious agents are essential. Thus, the development and practical use of biosensors based on modern advances in molecular biology and nanotechnologies fully comply with the current goals of global healthcare and are aimed at solving stated problems [22–24, 26].
According to the definition of IUPAC (International Union of Pure and Applied Chemistry) [33], biosensors are integrated autonomous devices that represent quantitative or semi-quantitative analytical information about the target analyte using a biological element of biorecognition (bioreceptor) located in spatial contact with the transducer. Biosensors do not require additional reagents and differ from other analytical systems in this respect. Thus, biosensors are portable analytical devices equipped with biological elements that can potentially control the biochemical parameters of physiological and pathological processes that are not accessible to modern analytical tools [25]. These are unique detectors the action of which is based on the specific interaction of biomolecules and bioreceptors and which are used to detect and identify the minimum concentrations of various analytes [23, 25, 26]. When a bioreceptor binds to a target molecule, the ligand-receptor interaction is converted into an optical, spectral, or electrochemical signal the power of which is proportional to the analyte concentration [26].
The idea of developing biosensors appeared more than half a century ago, and putting it into practice started [34, 35]. The very first sensors were designed to quantify relatively simple biochemical analytes (glucose, myoglobin, urea, cholesterol, prothrombin). The glucose meters of various modifications for monitoring patients’ blood glucose levels at home are an example of the most common modern biosensors. The glucose oxidase or glucose dehydrogenase enzymes are used as a biosensing component. They are immobilized on the surface of the electrode and break down glucose. The products of enzymatic reactions are converted into a physicochemical signal [25, 26, 36]. However, only in the recent years, thanks to the integration of nanoelectronics and biochemistry, the idea of biosensing has been widely developed. There is a wide range of biosensors that use biological materials to recognize certain biomarkers of the infectious process, which are quantified using optical, micromechanical, interferometric, and other alternative types of transducers [7, 21, 25, 26, 37].
In the modern world, a considerable growth of interest is observed in biosensor technologies, which are rightly considered to be one of the rising trends in the scientific and technical sphere [30, 38–40]. According to the experts’ forecasts, in 10–15 years the market for these analytical devices will exceed $ 70 billion [25, 26]. Most of them are focused on conducting laboratory studies of biofluids for early and accurate quantitative rapid identification of the molecular markers of myocardial infarction, diabetes, sepsis, as well as identification of the markers of parasitic and infectious diseases [25, 26, 41, 42].
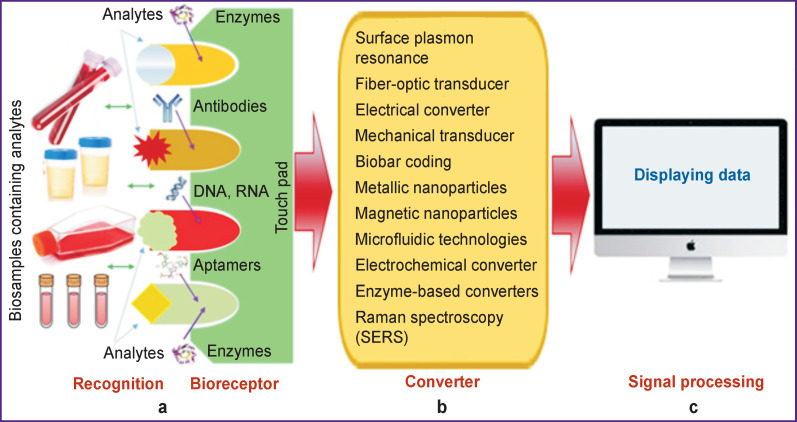
Structurally, biosensors are a complex consisting of three main functional segments:
a bioreceptor (a biosensing component) with the elements on the sensor plate for recognizing target analytes (target molecules) contained in the biosubstrates;
a transducer operating on physical and chemical principles (electrochemical, spectroscopic, or optical);
an electronic device for signal processing, recording, and displaying data in a convenient (analog or digital) form for the researcher (Figure 2).
Figure 2. Schematic diagram of the biosensor design.
The main parts are a bioreceptor with recognition elements for selective (specific) binding of target analytes contained in biosubstrates (a); converter (b); and also an electronic device for signal processing and data display (c) (source: authors)
The analysis of the literature demonstrates interest in the development and use of these analytical devices in ecology, toxicology, agriculture, biosafety, and medicine, including the clinical diagnosis of infectious diseases and septic conditions. Over the decades of the development of the biosensing technology, a large number of structurally different sensors and attempts to systematize them have been proposed [25, 26, 41–43].
Currently, the biosensor classification is based on the nature of the biochemical component used, the analytical tasks to be solved, the type of signal transducer, the intended application field, and the generated signal. The technical strategies conditioning the ways of further detection (transduction) of the signal/event determine the basic principle of differentiation of the analytical devices [43, 44].
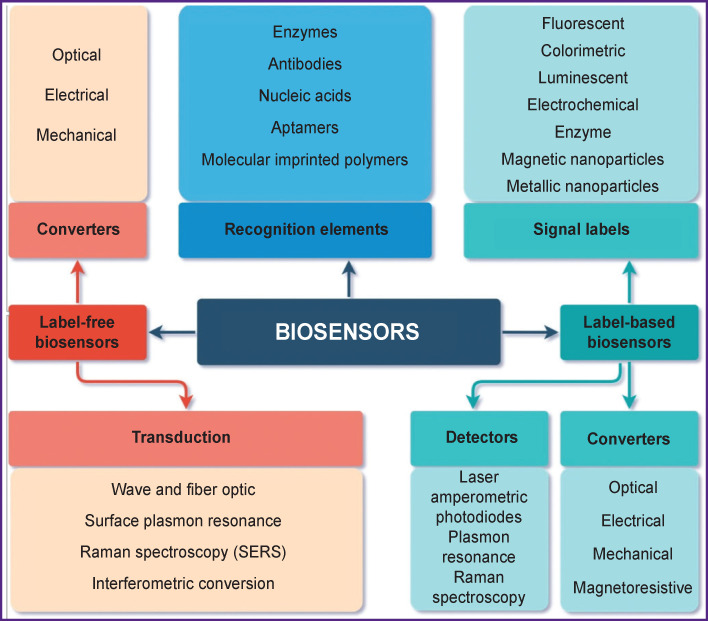
According to this principle, in one of the proposed IUPAC classifications [33], all biosensors are divided into two large groups based on technical strategies and design differences used in the development of detection methods by analytical devices: label-free and label-based ones (Figure 3).
Figure 3. Classification of biosensors based on constructive strategies of detection methods: labeled and label-free biosensors (adapted from [33]).
The recognition elements (bioreceptors) are common to all types of biosensors used in biomedical diagnostics: immunoglobulins (antibodies), enzymes (or microbial cell homogenates), nucleic acids (DNA, RNA, and PNA — peptide nucleic acids) [45–47], microbial cells (microorganisms) [5, 42, 44], and aptamers (short DNA and RNA oligonucleotides that can specifically bind to specific targets-molecules) [3, 40, 48]. These receptor biomolecules with concentrations ranging from 1 to 5 mg/mm2 are immobilized on a solid sensor substrate (matrix) by covalent binding or biotin-avidin interaction. They serve for selective binding and identification of target analytes (ligands) in biological fluids (whole blood, serum, plasma, urine, saliva, liquor, tissue and cell cultures extracts) [3, 5, 49–51].
When designing highly sensitive biosensors, the correct choice of the matrix and conditions for immobilization of bioreceptors is of key importance. When using non-covalent binding, the receptor is retained on the sensor substrate of the transducer due to electrostatic, van der Waals, or ion interactions that hold the biomolecules fairly firmly. The main advantage of this type of immobilization is that the matrix does not affect the biological properties of the receptor [25, 26]. With covalent binding to the surface of the sensor matrix of the transducer, the biomolecules are held firmly, which prevents them from leaching out of the matrix, and this is of key importance when designing a reusable biosensor [40, 42, 52, 53].
The new-generation devices more often comprise nanomaterials, the unique catalytic efficiency and adsorption properties of which are ensured by the optimal physical and chemical characteristics of the sensor substrate surface [26, 40, 54]. At the same time, neither bioreceptors nor analytes undergo conformational changes and loss of biological activity, which ultimately ensures effective interaction of the ligand with the receptor, which is transmitted as a specific equivalent amplified signal [25, 26, 54, 55].
The mechanism of transmitting a ligand-receptor interaction signal and its transduction is another important functional element of biosensors. Transmission is carried out with electrodes (gold, silver, platinum, mercury, and others with various surface modifications) and graphite pastes [25, 53, 56, 57]. This biochemical process is detected and converted into quantitatively detectable physical parameters with a certain type of physical and chemical converters that provide optical (responding to changes in physical and optical parameters), piezoelectric (quartz crystal microbalance technology), electrochemical (operating on the principle of measuring electric current) or micromechanical signals that are processed by the processor and analyzed at the output [26, 54–56] (see Figure 3).
The principle of operation of bioreceptors can also be represented as three consecutive stages: recognition of the target ligand in the biosubstrate by a specific bioelement located on the touch panel; transduction of information about the biochemical reaction into the form of an electrochemical signal; transduction of this signal into a form convenient for reading or processing by the researcher [26, 51, 55, 56].
For example, in the biosensors, where the enzymes immobilized on the sensor plates serve as a recognition element, the substrates from the biomaterial in the presence of catalysts enter a biochemical reaction with them. The resulting product is determined with the aid of an electrode that transduces the biochemical reaction into an electrochemical signal, the magnitude of which is proportional to the amount of substrate in the studied biomaterial [22, 57, 58].
In recent decades, the key task of interdisciplinary research into the design of modern biosensors (in fact, representing the first generation of bioelectronic devices) has been to improve the parameters of close interaction of biochemical and physical functional elements in order to increase their sensitivity, selectivity, and reduce the detection limits of target analytes [40, 59, 60]. These characteristics of the analytical systems are of prime importance in the diagnosis of infectious diseases.
The progress in the development of biosensor diagnostics of bacterial and viral infections has been achieved mainly due to modern improvements in the methods used for the identification of specific markers [22, 61–64].
Together with the already used analytical devices, where immunoglobulins and enzymes in the form of bacterial homogenates are applied as bioreceptors, in recent years, whole-cell microbial biosensors have been introduced, in which live natural or engineered microorganisms (for example, Escherichia coli or Staphylococcus aureus) integrated on the sensory substrate assimilate target organic compounds from biosubstrates (for example, antibodies from blood serum), they themselves acting as a sensitive mechanism [25, 61, 64]. In this case, a positive respond to the promoter of the target molecule after its transport through the cell membrane and diffusion inside the bacterial cell causes the expression of the reporter gene, which is recorded as a quantitative response using optical [7, 20, 23, 63, 64] or electrochemical signals [45, 46, 65].
The use of reporter genes to identify factors that trigger genetic response in living microorganisms was proposed in the middle of the last century [34, 35] when the functioning of the lactose operon of E. coli (lac-operon) and its relationship with the patterns of metabolism and growth of microbes were described. These fundamental studies were confirmed in subsequent years by studying the role and structure of DNA and other reporter genes, such as xylE and tfdA, which are currently used actively as a biophysical model for environmental research [26, 66, 67] (Figure 4).
Figure 4. Functional diagram of the lactose lac-operon of E. coli in microbial biosensors (source: authors).
Liu et al. [60] reported on designing a biosensor that uses synthetic antimicrobial peptides as new recognition bioreceptors. The proposed analytical device in combination with the impedance recognition method allowed rapid and quantitative identification of bacterial pathogens in biosubstrates (E. coli, S. aureus, Pseudomonas aeruginosa, and Staphylococcus epidermidis) with concentrations ranging from 102 CFU/ml. Besides, this sensor enabled the differentiation between live bacteria and dead ones.
Another group of researchers proposed a biosensor design for highly sensitive and rapid identification of S. aureus, where a bacteriophage with a detection range of 4·108 CFU/ml was used as a receptor [68].
Further improvement of microbial biosensors has been due to the emergence of regulatory biosensing associated with advances in molecular genetic technologies and the discovery of new mechanisms for detecting various extracellular and intracellular signals, as well as their subsequent optical and electrochemical transduction [45, 46, 69, 70]. The development of new technologies and advances in synthetic biology resulted in the appearance of biosensors with recombinant nucleic acid receptors and aptamers which are successfully used for diagnosing infections [5, 40, 71, 72]. These technologies provided an increase in the sensitivity of these analytical devices by allosteric regulation of the metabolic signaling pathways of microorganisms, aimed at selective detection of specific biomarkers — small molecules of microbial origin [40, 45, 69–72].
Label-free biosensors
With the development of modern technologies in infectious disease diagnostics and epidemiology, label-free biosensors have become increasingly more widespread. They enable screening of intermolecular interactions and cellular reactions, provide detailed information about the selectivity of bacterial exotoxins and the specificity of antimicrobial agents, the interaction of antigen with antibody, as well as the kinetics of the inflammatory process, immunological and serological reactions [60, 73].
Currently, there is a wide range of analytical devices for analyzing biospecific ligand-receptor interactions in label-free biosensors. In these highly sensitive and functional systems, the binding reactions of the target analyte to the bioreceptor can be studied without the use of any enzyme, radioactive, or fluorescent labels [11, 61, 74, 75]. Such biosensors do not need expensive reagents and markers, which ensures their cost-effectiveness. These analytical systems are capable of monitoring the reactions of ligand-receptor interaction that occur when target analytes bind to molecular elements immobilized on the sensor substrate (antibodies, enzymes, nucleic acids, aptamers) [61, 62, 76, 77].
This type of biosensor requires only one recognition element, which simplifies the analysis scheme, reduces its duration, and the cost of reagents. The current generation of label-free biosensors allows quantitative measurements of biomolecular reaction products in real time, which makes it possible to perform continuous data recording that enables kinetic monitoring of the parameters of the recognition process in ligand-receptor interactions [27, 78].
An important advantage of using label-free biosensors is that the target analytes are detected in their natural form, without labeling or chemical modification, which means that they can be saved for further analysis (Table 2).
Table 2.
Advantages of modern label-free biosensors over similar analytical label-based devices
| Advantages | References |
|---|---|
| Simplified pattern of analysis | [3, 46, 48, 73, 79, 80] |
| Shortened analysis duration (rapid response time) | [7, 78, 81, 82] |
| Lower analysis cost | [7, 59, 83, 84] |
| Reduced consumption of organic solvents | [30, 61, 77, 85] |
| Portability and small dimensions | [30, 40, 71, 80] |
| No qualified medical personnel required | [3, 7, 36, 61, 80, 82, 85] |
| Opportunity of quantitative measuring of biomolecules in the real-time mode | [25, 26, 77, 82, 84] |
| Detection of target analytes in natural forms, without modifications and labels | [22, 30, 71, 81, 83] |
| High sensitivity | [22, 25, 26, 40, 61, 80, 82] |
| Direct measurement of analytes | [40, 48, 61, 83] |
| Opportunity of detecting small molecules | [3, 7, 25, 26, 40, 78] |
| Opportunity of multiplexing | [59, 61, 73, 85] |
| Access to kinetic and thermodynamic parameters | [22, 26, 36, 80, 83] |
In recent decades, numerous studies have been conducted to develop new types of receptors [60, 68, 73] and recognition methods in label-free biosensors that can generate a signal directly after binding to a recognition element. In this context, there have been proposed many physical and chemical types of transducers that convert the results of bioreceptor binding of targets (for example, an increase in mass, resistivity, and surface refractive indices), which are recognized in various ways [64, 70, 73, 77, 86].
The optical, (piezo)electrical, or (micro)mechanical transducers are among the promising methods for recognizing ligand-receptor interaction signals in label-free biosensors used for the diagnosis of various infectious diseases. These biosensing methods are enhanced by surface plasmon resonance (SPR) [78], surface Raman spectrometry (SERS) [36, 38, 39], quartz crystal microbalance [86], and microcantilever sensors [78, 87, 88].
The biosensors with optical transducers, considered as the main tools for signal perception, are one of the most powerful detection and analysis tools widely used in biomedical research and practical medicine [8, 9, 72, 88, 89]. These transducers are based on measuring changes in optical properties in the presence of the analyte, such as absorption, reflectivity, radiation, or interferometric pattern, which can be detected by a photodetector. They are immune to electromagnetic interference, can perform remote sensing, and have a number of advantages, including high sensitivity, direct real-time measurement, and multiplexing (simultaneous detection of multiple analytes). The microbial biosensors that detect interactions between microorganisms and target ligands are no exception [5, 42–44, 54].
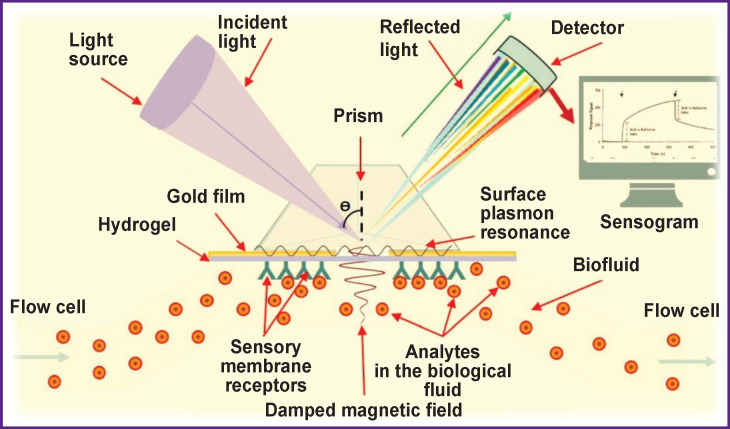
Due to a variety of detection methods using optical transducers in label-free biosensors, the authors of the review limited themselves to the devices that have proved successful in the detection of infectious disease pathogens. The cutting-edge technologies in the design of label-free optical biosensors with the focus on the diagnosis of bacterial infections are associated with the development of modern methods of transduction (fiber-optical and damped electromagnetic field systems, surface plasmon resonance, Raman spectroscopy or interferometry) and new recognition elements (molecular-imprinted polymers) [69, 78, 88, 90–92] (Figure 5).
Figure 5. Modern transduction systems in optical biosensors are based on the effect of surface plasmon resonance and damped electromagnetic field (source: authors).
Among the modern technologies used for the clinical diagnostics of bacterial and viral infections, the development of electrochemical methods of optical biosensing based on nucleic acids presents interest. The recognition elements used in these types of analytical devices include DNA, RNA, PNA, and aptamers. For example, nowadays the sensor technology of DNA hybridization based on electrochemical (impedance spectroscopy) and optical methods which recognizes the complementary target DNA chain of a pathogenic microorganism is gaining popularity, [46, 47, 71, 93, 94]. Due to the ease of use and high sensitivity, electrochemical label-free biosensors have become the most widely used ones for detecting infectious pathogens in recent years (Table 3).
Table 3.
Examples of modern designs of label-free biosensors for the detection of pathogenic bacteria and viruses
| Recognizing bioreceptor | Transduction method | Test models of pathogens (sensitivity) | References |
|---|---|---|---|
| Bacteriophage | Photoluminescence | S. aureus (4·108 CFU/ml) | [68, 92] |
| Antimicrobial peptides | Impedancemetry | E. coliS. epidermidis , S. aureus, (P. 10aeruginosa2 CFU/ml) , | [60, 73, 91] |
| Antibacterial Zn-CuO and nanoparticles graphene oxide Man/MUA-MH/Au* | Impedancemetry Electrochemical impedance spectroscopy | E. coli, S. aureus (50 CFU/ml); antibacterial effect — 100%, 30 min | [36, 79, 85, 87] |
| Thiolated G protein on gold electrodes and gold nanoparticles | Cyclic voltammetry Electrochemical impedance spectroscopy | S. typhimurium (2.16·106 CFU/ml) E. coli (50–103 CFU/ml) | [93] |
| Enzymes | Electrochemical | E. coli O157:H7 (150 CFU/ml) | [55, 57, 58] |
| Nucleic acids (DNA, RNA) | Electrochemical | S. S. typhimurium aureus (140 (48 CFU/CFU/ml) ml) | [11, 18, 71] |
| Nucleic acids (DNA, RNA) | Electrochemical | S. aureus, M. tuberculosis | [11, 45–47] |
| Aptamer on gold nanoparticles | Autofluorescence quenching | S. typhimurium (48 CFU/ml) | [3, 5, 40] |
| Monoclonal antibodies | Optical | S. Listeria enteritidis monocytogenes (80 CFU/ ml) | [14, 16, 88, 94] |
| Thiolated aptamer | Impedancemetry | Shigella dysenteriae | [8, 95] |
| Nucleic acids (DNA, RNA) | Electrochemical impedance spectroscopy | M. tuberculosis | [62, 88, 96] |
| Monoclonal antibodies | Surface plasmon resonance | Enterococcus faecalis (104–108 CFU/ml) | [86, 90] |
| Aptamer | Impedancemetry | Bacillus Bacillus cereus anthracis (104–10(spores) 6 CFU/ml) | [3, 5, 30, 40, 48] |
| Nucleic acids (DNA, RNA) | Electrochemical Cyclic impedance voltammetry spectroscopy | Salmonella spp. | [71] |
| Enzyme (graphene simulator quantum dots) | Electrochemical | (5 (Yersinia milk)–30 enterocolitica (serum) CFU/ml) | [48, 80, 97] |
| Monoclonal (long-term fiber antibodies lattices) | Surface plasmon resonance | S. aureus (224 CFU/ml, 30 min) | [69, 78, 90] |
| Monoclonal antibodies | Visualization | Salmonella enteritidis (102–108 CFU/ml) | [81] |
| Nucleic acids (DNA, aptamer) | Electrochemical | Avian influenza virus H5N1 (AIV) | [45] |
| Nucleic acids (DNA) | Electrochemical impedance | Zika virus (25.0±1.7 nmol) | [46] |
| Aptamer (rGO-TiO2) | Electrochemical | Salmonella (10enterica–108 CFU/, typhimurium ml) | [80] |
| Nucleic acids (DNA) | Piezoelectric | Clostridium specificity difficile (sensitivity — 95%) — 95%, | [85] |
| Monoclonal antibodies | Surface plasmon resonance | M. tuberculosis (102–106 CFU/ml) | [88, 96] |
| Aptamer | Fluorescent | Salmonella typhimurium (6·10 CFU/ml) | [82] |
| Monoclonal antibodies | Potentiometry | Salmonella typhimurium (106 CFU/ml) | [84] |
| Nucleic acids (DNA) | Electrochemical impedance | M. tuberculosis (102–106 CFU/ml) | [47] |
| Aptamer (RNA) | Fluorescent | S. aureus (102–106 CFU/ml) | [3] |
* Man/MUA-MH/Au — mannose/11-mercaptoundecanoic acid/6-mercapto-hexanol/gold.
The appearance of aptamer-based biosensors (aptasensors) and recombinant nucleic acids as recognition elements has resulted from the development of new technologies and advances in synthetic aptamer biology [3, 40, 48]. These types of analytical devices are very promising due to the high specificity and stability of nucleic receptors, low cost, and the potential for developing various sensor platforms [40, 48].
For example, in a recent study by Sheng et al. [3], they reported the creation of a label-free biosensor with an RNA aptamer that allows rapid quantitative detection of food pathogens. In the proposed aptasensor the RNA-aptamer acts as “antibodies against nucleic acids” of target microorganisms. The oligonucleotide nature of aptamers makes it possible to amplify or chemically synthesize a desired pool with a high frequency and in any quantity, which makes it possible to create highly specific homogeneous sensors providing accurate quantitative detection of pathogen nucleic chains [50, 51, 80, 82, 95].
The authors [3] demonstrated the effective and rapid detection of S. aureus, selected as the target pathogen for their aptosensor in food and water. The quantitative assessment of ligand-receptor interaction in the proposed biosensor variant was performed with specific fluorescent dyes for nucleic acids (Sybr Gold and Sybr Green I). It is noteworthy that rapid detection of the pathogen did not require special sample preparation (purification, enrichment).
These modern innovations make optical biosensors more versatile than other types of sensor technologies [62, 72, 74, 76]. They make it possible not only to quantify low-molecular organic molecules (less than 1000 Da), but also to conduct early diagnostics, quantitative analysis of the course of infectious diseases, as well as to carry out epidemiological monitoring [98, 99].
In optical biosensors, the use of the transduction method based on the surface plasmon resonance effect and the phenomenon of total internal light reflection is common [69, 78, 84, 90]. This effect occurs when the angle of incidence is beyond the critical angle with the change of refractive index that occurs on ligand binding to the receptor and increasing the substance mass on the touch sensor pad, and in the reverse process — on reducing the mass-dissociation of the ligand-receptor complex. In this case, an electromagnetic vanishing wave occurs, which, when damped, penetrates the medium with a lower refractive index and creates a so-called damped electromagnetic field (see Figure 5).
If gold (or silver) is sprayed on a hydrogel plate, then another phenomenon occurs associated with the presence of free electrons in this metal(s), which are exposed to the alternating electric field when illuminated. These electrons can collectively vibrate and resonate, adjusting to the frequency of incident light (these electron vibrations in precious metal nanoparticles are called plasmonic ones [61, 62, 69, 87]), and the unique properties of nanometals increase the sensitivity and selectivity of biosensors [71, 90, 95]. Thus, plasmon resonance significantly increases the damped electromagnetic field, reduces the intensity of reflected plane-polarized light, and also allows detecting target analytes in very low concentrations in various biosubstrates without preliminary sample preparation [71, 80, 81, 95].
Among the family of optical label-free biosensors, SPR is one of the most accessible, developed, and most successfully used technologies in recent years for the diagnostics of infectious diseases and septic conditions. This is due to the high sensitivity and versatility of this type of sensors, which also allow real-time detection and direct measurement of the kinetics of molecular ligand-receptor interaction [71, 80]. For example, in recent years, a number of researchers [61, 62, 78, 88, 96] have proposed SPR-based biosensors for detecting and monitoring urinal M. tuberculosis biomolecules and non-tuberculosis CFP10 and MPT64 mycobacteria, which, among numerous mycobacteria antigens, interact most strongly with related anti-CFP10 and anti-MPT64 antibodies on the immunosensor matrix.
The only drawback of optical label-free biosensors that use prismatic light refraction and SPR effects is their relatively large size, which is incompatible with mobile use in the point-of-care mode [80, 81]. Therefore, a promising alternative to this type of optical sensors is fiber-optic biosensors, which are perfectly suitable for designing miniature portable devices, have a low cost, and have successfully proved themselves for clinical diagnostics [80].
Label-free optical biosensors have been consistently used for characterization and screening of molecular interactions in clinical laboratories. For example, Golichenari et al. [61] presented a detailed overview of successful uses of the most promising optical label-free biosensors for rapid, highly effective, and accessible detection and quantification of M. tuberculosis, mycobacterial proteins, and IFN-γ cytokine as the most important markers in the early diagnosis of tuberculosis.
The majority of modern biosensors intended for the detection of infectious pathogens are based on electrochemical conversion of the signal. These sensors are based on measuring changes in the current, electrochemical potential, and impedancemetry as a means of conversion of biochemical reactions [46, 47, 85]. When designing modern electrochemical biosensors for detecting bacterial pathogens, particular attention is paid to bioelectrodes. The most widely used materials are thin polymer films, nanostructured metal oxides, self-organizing monolayers of organic molecules (SAM), and carbon nanostructures (nanotubes, fullerenes, and graphenes) [48]. For example, SAM are considered to be an ideal material for the immobilization of nucleic acids when designing biosensors for detecting bacteria [48, 80, 97].
The advantages of these devices include portability and simple measuring equipment. Together with their cost-effectiveness, high sensitivity, and large linearity detection range, the electrochemical sensors are capable to work with small sample volumes. Moreover, the result is not affected by sample turbidity, unlike the optical methods based on spectroscopic transduction [74, 100].
Conclusion
The analysis of the research results published in recent years has shown that the development of molecular biology methods with the aid of nanotechnology opens up broad prospects for designing new biosensor platforms with highly efficient, highly sensitive, and highly selective detection of molecular infectious biomarkers.
Medical biosensors as a new type of diagnostic tools are at the initial stage of their development. However, the first decades of practical application of these analytical devices in healthcare have shown their absolute attractiveness and prospects for detecting bacterial and viral pathogens. The development and implementation of biosensor technologies in clinical laboratory practice is a modern non-alternative strategy for reducing infectious diseases in the regions with a low level of healthcare, where cheap and highly effective diagnostics can play a key role in timely verification of pathogens. The development of inexpensive and affordable analytical devices for clinical diagnostics with lower detection limits for pathogens is necessary due to the importance of diagnosing infectious diseases at the preclinical stage. In this regard, successful experiments in designing and using biosensors for the detection of spore and uncultivated bacteria with an assessment of their viability pose interest [31, 32, 79, 81, 87, 101].
This review focuses on a large group of biosensors that are more available and do not need labels for the reproduced signal, but have rather complex transduction systems. However, the future is for simpler and more portable diagnostic analytical systems that do not require complex conversion platforms (surface plasmon resonance and surface Raman spectrometry) and can detect multiple pathogens simultaneously on the base of multiplex analysis. Such biosensors will be able to solve the global problem of effective control of infectious diseases.
Modern trends in the development of biosensor medical technologies are associated with the development of new materials for designing transducers and conditions for more effective ligand-receptor interaction. The prospects for expanding the practical applications of biosensor technologies are related to clinical diagnostics which meets the requirements of personalized medicine, and are equally attractive for doctors and patients, particularly when verifying pathogens of infectious diseases.
Moreover, one of the key modern trends in the development of clinical laboratory diagnostics is non-invasive testing, which does not involve blood sampling. In this regard, highly sensitive, miniature, and portable medical biosensors with their capability to continuously monitor in vivo metabolites, drugs and molecular markers of the infectious process will soon play a leading role.
Funding Statement
Research funding and conflicts of interest. The study was not funded by any sources, and there are no conflicts of interest related to this study.
References
- 1.Havelaar A.H., Kirk M.D., Torgerson P.R., Gibb H.J., Hald T., Lake R.J., Praet N., Bellinger D.C., De Silva N.R., Gargouri N., Speybroeck N., Cawthorne A., Mathers C., Stein C., Angulo F.J., Devleesschauwer B. World Health Organization Foodborne Disease Burden Epidemiology Reference Group. World Health Organization global estimates and regional comparisons of the burden of foodborne disease in 2010. PLoS Med. 2015;12(12):e1001923. doi: 10.1371/journal.pmed.1001923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matea C.T., Mocan T., Tabaran F., Pop T., Mosteanu O., Puia C., Iancu C., Mocan L. Quantum dots in imaging, drug delivery and sensor applications. Int J Nanomedicine. 2017;12:5421–5431. doi: 10.2147/ijn.S138624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sheng L., Lu Y., Deng S., Liao X., Zhang K., Ding T., Gao H., Liu D., Deng R., Li J. A transcription aptasensor: amplified, label-free and culture-independent detection of foodborne pathogens via light-up RNA aptamers. Chem Commun (Camb) 2019;55(68):10096–10099. doi: 10.1039/c9cc05036a. [DOI] [PubMed] [Google Scholar]
- 4.WHO. Fact sheets: infectious diseases. URL: https://www.who.int/topics/infectious_diseases/factsheets/ru/.
- 5.Su L., Jia W., Hou C., Lei Y. Microbial biosensors: a review. Biosens Bioelectron. 2011;26(5):1788–1799. doi: 10.1016/j.bios.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Sin M.L., Mach K.E., Wong P.K., Liao J.C. Advances and challenges in biosensor-based diagnosis of infectious diseases. Expert Rev Mol Diagn. 2014;14(2):225–244. doi: 10.1586/14737159.2014.888313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peltomaa R., Glahn-Martínez B., Benito-Peña E., Moreno-Bondi M.C. Optical biosensors for label-free detection of small molecules. Sensors (Basel) 2018;18(12):4126. doi: 10.3390/s18124126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zarei M. Infectious pathogens meet point-of-care diagnostics. Biosens Bioelectron. 2018;106:193–203. doi: 10.1016/j.bios.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 9.Kozel T.R., Burnham-Marusich A.R. Point-of-care testing for infectious diseases: past, present, and future. J Clin Microbiol. 2017;55(8):2313–2320. doi: 10.1128/jcm.00476-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim H., Chung D.R., Kang M. A new point-of-care test for the diagnosis of infectious diseases based on multiplex lateral flow immunoassays. Analyst. 2019;144(8):2460–2466. doi: 10.1039/c8an02295j. [DOI] [PubMed] [Google Scholar]
- 11.Fabri-Faja N., Calvo-Lozano O., Dey P., Terborg R.A., Estevez M.C., Belushkin A., Yesilköy F., Duempelmann L., Altug H., Pruneri V., Lechuga L.M. Early sepsis diagnosis via protein and miRNA biomarkers using a novel point-of-care photonic biosensor. Anal Chim Acta. 2019;1077:232–242. doi: 10.1016/j.aca.2019.05.038. [DOI] [PubMed] [Google Scholar]
- 12.Min J., Nothing M., Coble B., Zheng H., Park J., Im H., Weber G.F., Castro C.M., Swirski F.K., Weissleder R., Lee H. Integrated biosensor for rapid and point-of-care sepsis diagnosis. ACS Nano. 2018;12(4):3378–3384. doi: 10.1021/acsnano.7b08965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Safenkova I.V., Panferov V.G., Panferova N.A., Varitsev Y.A., Zherdev A.V., Dzantiev B.B. Alarm lateral flow immunoassay for detection of the total infection caused by the five viruses. Talanta. 2019;195:739–744. doi: 10.1016/j.talanta.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Y., Zhang Q., Meng Q., Wu F., Zhang L., Tang Y., Guan Y., An L. Quantum dots-based lateral flow immunoassay combined with image analysis for semiquantitative detection of IgE antibody to mite. Int J Nanomedicine. 2017;12:4805–4812. doi: 10.2147/IJN.S134539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boisen M.L., Oottamasathien D., Jones A.B., Millett M.M., Nelson D.S., Bornholdt Z.A., Fusco M.L., Abelson D.M., Oda S., Hartnett J.N., Rowland M.M., Heinrich M.L., Akdag M., Goba A., Momoh M., Fullah M., Baimba F., Gbakie M., Safa S., Fonnie R., Kanneh L., Cross R.W., Geisbert J.B., Geisbert T.W., Kulakosky P.C., Grant D.S., Shaffer J.G., Schieffelin J.S., Wilson R.B., Saphire E.O., Branco L.M., Garry R.F., Khan S.H., Pitts K.R. Viral Hemorrhagic Fever Consortium. Development of prototype filovirus recombinant antigen immunoassays. J Infect Dis. 2015;212(Suppl 2):S359–S367. doi: 10.1093/infdis/jiv353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nielsen K., Yu W.L., Kelly L., Bermudez R., Renteria T., Dajer A., Gutierrez E., Williams J., Algire J., de Eschaide S.T. Development of a lateral flow assay for rapid detection of bovine antibody to Anaplasma marginale. J Immunoassay Immunochem. 2008;29(1):10–18. doi: 10.1080/15321810701734693. [DOI] [PubMed] [Google Scholar]
- 17.Jørgensen C.S., Uldum S.A., Sørensen J.F., Skovsted I.C., Otte S., Elverdal P.L. Evaluation of a new lateral flow test for detection of Streptococcus pneumoniae and Legionella pneumophila urinary antigen. J Microbiol Methods. 2015;116:33–36. doi: 10.1016/j.mimet.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 18.Rohrman B.A., Leautaud V., Molyneux E., Richards-Kortum R.R. A lateral flow assay for quantitative detection of amplified HIV-1 RNA. PLoS One. 2012;7(9):e45611. doi: 10.1371/journal.pone.0045611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamphee H., Chaiprasert A., Prammananan T., Wiriyachaiporn N., Kanchanatavee A., Dharakul T. Rapid molecular detection of multidrug-resistant tuberculosis by PCR-nucleic acid lateral flow immunoassay. PLos One. 2015;10(9):e0137791. doi: 10.1371/journal.pone.0137791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pilavaki E., Demosthenous A. Optimized lateral flow immunoassay reader for the detection of infectious diseases in developing countries. Sensors (Basel) 2017;17(11):2673. doi: 10.3390/s17112673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Posthuma-Trumpie G.A., Korf J., van Amerongen A. Lateral flow (immuno)assay: its strengths, weaknesses, opportunities and threats. A literature survey. Anal Bioanal Chem. 2008;393(2):569–582. doi: 10.1007/s00216-008-2287-2. [DOI] [PubMed] [Google Scholar]
- 22.Bhalla N., Jolly P., Formisano N., Estrela P. Introduction to biosensors. Essays Biochem. 2016;60(1):1–8. doi: 10.1042/ebc20150001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ragavan K.V., Kumar S., Swaraj S., Neethirajan S. Advances in biosensors and optical assays for diagnosis and detection of malaria. Biosens Bioelectron. 2018;105:188–210. doi: 10.1016/j.bios.2018.01.037. [DOI] [PubMed] [Google Scholar]
- 24.Patel S., Nanda R., Sahoo S., Mohapatra E. Biosensors in health care: the milestones achieved in their development towards lab-on-chip-analysis. Biochem Res Int. 2016;2016:3130469. doi: 10.1155/2016/3130469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ebara M., editor. Biomaterials nanoarchitectonics. Elsevier Inc; 2016. p. 362. [DOI] [Google Scholar]
- 26.Sadasivuni K.K., Ponnamma D., Kim J., Cabibihan J.J., AlMaadeed M.A., editors. Biopolymer composites in electronics. Elsevier Inc; 2017. p. 544. [DOI] [Google Scholar]
- 27.Fan X., White I.M., Shopova S.I., Zhu H., Suter J.D., Sun Y. Sensitive optical biosensors for unlabeled targets: a review. Anal Chim Acta. 2008;620(1–2):8–26. doi: 10.1016/j.aca.2008.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar S., Tripathy S., Jyoti A., Singh S.G. Recent advances in biosensors for diagnosis and detection of sepsis: a comprehensive review. Biosens Bioelectron. 2019. pp. 205–215. 124–125. [DOI] [PubMed]
- 29.McLinden T., Sargeant J.M., Thomas M.K., Papadopoulos A., Fazil A. Component costs of foodborne illness: a scoping review. BMC Public Health. 2014;14:509. doi: 10.1186/1471-2458-14-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mazzaracchio V., Neagu D., Porchetta A., Marcoccio E., Pomponi A., Faggioni G., D’Amore N., Notargiacomo A., Pea M., Moscone D., Palleschi G., Lista F., Arduini F. A label-free impedimetric aptasensor for the detection of Bacillus anthracis spore simulant. Biosens Bioelectron. 2019;126:640–646. doi: 10.1016/j.bios.2018.11.017. [DOI] [PubMed] [Google Scholar]
- 31.Waller D.F., Hew B.E., Holdaway C., Jen M., Peckham G.D. Rapid detection of Bacillus anthracis spores using immunomagnetic separation and amperometry. Biosensors (Basel) 2016;6(4):61. doi: 10.3390/bios6040061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wynn D., Deo S., Daunert S. Engineering rugged field assays to detect hazardous chemicals using spore-based bacterial biosensors. Methods Enzymol. 2017;589:51–85. doi: 10.1016/bs.mie.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 33.Nič M., Jirát J., Košata B., Jenkins A., McNaught A., editors. Compendium of chemical terminology. Oxford: Blackwell Scientific Publications; 1997. [DOI] [Google Scholar]
- 34.Novick A., Weiner M. Enzyme induction as an all-or-none phenomenon. Proc Natl Acad Sci U S A. 1957;43(7):553–566. doi: 10.1073/pnas.43.7.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacob F., Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 36.Wu X., Xu C., Tripp R.A., Huang Y.W., Zhao Y. Detection and differentiation of foodborne pathogenic bacteria in mung bean sprouts using field deployable label-free SERS devices. Analyst. 2013;138(10):3005–3012. doi: 10.1039/c3an00186e. [DOI] [PubMed] [Google Scholar]
- 37.van der Meer J.R., Belkin S. Where microbiology meets microengineering: design and applications of reporter bacteria. Nat Rev Microbiol. 2010;8(7):511–522. doi: 10.1038/nrmicro2392. [DOI] [PubMed] [Google Scholar]
- 38.Marks H., Schechinger M., Garza J., Locke A., Coté G. Surface enhanced Raman spectroscopy (SERS) for in vitro diagnostic testing at the point of care. Nanophotonics. 2017;6(4):681–701. doi: 10.1515/nanoph-2016-0180. [DOI] [Google Scholar]
- 39.Kahraman M., Mullen E.R., Korkmaz A., Wachsmann-Hogiu S. Fundamentals and applications of SERS-based bioanalytical sensing. Nanophotonics. 2017;6(5):831–852. doi: 10.1515/nanoph-2016-0174. [DOI] [Google Scholar]
- 40.Urmann K., Reich P., Walter J.G., Beckmann D., Segal E., Scheper T. Rapid and label-free detection of protein a by aptamer-tethered porous silicon nanostructures. J Biotechnol. 2017;257:171–177. doi: 10.1016/j.jbiotec.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 41.Qiu C., Zhai H., Hou J. Biosensors design in yeast and applications in metabolic engineering. FEMS Yeast Res. 2019;19(8) doi: 10.1093/femsyr/foz082. foz082, [DOI] [PubMed] [Google Scholar]
- 42.González-Pabón M.J., Figueredo F., Martínez-Casillas D.C., Cortón E. Characterization of a new composite membrane for point of need paper-based micro-scale microbial fuel cell analytical devices. PLoS One. 2019;14(9):e0222538. doi: 10.1371/journal.pone.0222538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lim J.W., Ha D., Lee J., Lee S.K., Kim T. Review of micro/nanotechnologies for microbial biosensors. Front Bioeng Biotechnol. 2015;3:61. doi: 10.3389/fbioe.2015.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lei Y., Chen W., Mulchandani A. Microbial biosensors. Anal Chim Acta. 2006;568(1–2):200–210. doi: 10.1016/j.aca.2005.11.065. [DOI] [PubMed] [Google Scholar]
- 45.Lee T., Park S.Y., Jang H., Kim G.H., Lee Y., Park C., Mohammadniaei M., Lee M.H., Min J. Fabrication of electrochemical biosensor consisted of multi-functional DNA structure/porous au nanoparticle for avian influenza virus (H5N1) in chicken serum. Mater Sci Eng C Mater Biol Appl. 2019;99:511–519. doi: 10.1016/j.msec.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 46.Faria H.A.M., Zucolotto V. Label-free electrochemical DNA biosensor for zika virus identification. Biosens Bioelectron. 2019;131:149–155. doi: 10.1016/j.bios.2019.02.018. [DOI] [PubMed] [Google Scholar]
- 47.Teengam P., Siangproh W., Tuantranont A., Vilaivan T., Chailapakul O., Henry C.S. Electrochemical impedance-based DNA sensor using pyrrolidinyl peptide nucleic acids for tuberculosis detection. Anal Chim Acta. 2018;1044:102–109. doi: 10.1016/j.aca.2018.07.045. [DOI] [PubMed] [Google Scholar]
- 48.Zhang S., Ma L., Ma K., Xu B., Liu L., Tian W. Label-free aptamer-based biosensor for specific detection of chloramphenicol using AIE probe and graphene oxide. ACS Omega. 2018;3(10):12886–12892. doi: 10.1021/acsomega.8b01812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang F.Z., Keasling J. Biosensors and their applications in microbial metabolic engineering. Trends Microbiol. 2011;19(7):323–329. doi: 10.1016/j.tim.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 50.Dietrich J.A., McKee A.E., Keasling J.D. High-throughput metabolic engineering: advances in small-molecule screening and selection. Annu Rev Biochem. 2010;79:563–590. doi: 10.1146/annurev-biochem-062608-095938. [DOI] [PubMed] [Google Scholar]
- 51.Durrieu C., Lagarde F., Jaffrezic-Renault N. Nanotechnology assets in biosensors design for environmental monitoring. In: Brayner R., Fiévet F., Coradin T., editors. Nanomaterials: a danger or a promise? London: Springer; 2013. pp. 189–229. https://doi-org-443.webvpn.jnu.edu.cn/10.1007/978-1-4471-4213-3_7. [Google Scholar]
- 52.Yagi K. Applications of whole-cell bacterial sensors in biotechnology and environmental science. Appl Microbiol Biotechnol. 2007;73(6):1251–1258. doi: 10.1007/s00253-006-0718-6. [DOI] [PubMed] [Google Scholar]
- 53.Renella G., Giagnoni L. Light dazzles from the black box: whole-cell biosensors are ready to inform on fundamental soil biological processes. Chem Biol Technol Agric. 2016;3:8. doi: 10.1186/s40538-016-0059-3. [DOI] [Google Scholar]
- 54.Park M., Tsai S.L., Chen W. Microbial biosensors: engineered microorganisms as the sensing machinery. Sensors (Basel) 2013;13(5):5777–5795. doi: 10.3390/s130505777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roy V., Adams B.L., Bentley W.E. Developing next generation antimicrobials by intercepting AI-2 mediated quorum sensing. Enzym Microb Technol. 2011;49(2):113–123. doi: 10.1016/j.enzmictec.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 56.Mangwani N., Dash H.R., Chauhan A., Das S. Bacterial quorum sensing: functional features and potential applications in biotechnology. J Mol Microb Biotechnol. 2012;22(4):215–227. doi: 10.1159/000341847. [DOI] [PubMed] [Google Scholar]
- 57.Amine A., Arduini F., Moscone D., Palleschi G. Recent advances in biosensors based on enzyme inhibition. Biosens Bioelectron. 2016;76:180–194. doi: 10.1016/j.bios.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 58.Rocchitta G., Spanu A., Babudieri S., Latte G., Madeddu G., Galleri G., Nuvoli S., Bagella P., Demartis M.I., Fiore V., Manetti R., Serra P.A. Enzyme biosensors for biomedical applications: strategies for safeguarding analytical performances in biological fluids. Sensors (Basel) 2016;16(6):780. doi: 10.3390/s16060780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sang S., Wang Y., Feng Q., Wei Y., Ji J., Zhang W. Progress of new label-free techniques for biosensors: a review. Crit Rev Biotechnol. 2016;36(3):465–481. doi: 10.3109/07388551.2014.991270. [DOI] [PubMed] [Google Scholar]
- 60.Liu X., Marrakchi M., Xu D., Dong H., Andreescu S. Biosensors based on modularly designed synthetic peptides for recognition, detection and live/dead differentiation of pathogenic bacteria. Biosens Bioelectron. 2016;80:9–16. doi: 10.1016/j.bios.2016.01.041. [DOI] [PubMed] [Google Scholar]
- 61.Golichenari B., Velonia K., Nosrati R., Nezami A., Farokhi-Fard A., Abnous K., Behravan J., Tsatsakis A.M. Label-free nano-biosensing on the road to tuberculosis detection. Biosens Bioelectron. 2018;113:124–135. doi: 10.1016/j.bios.2018.04.059. [DOI] [PubMed] [Google Scholar]
- 62.Golichenari B., Nosrati R., Farokhi-Fard A., Faal Maleki M., Gheibi Hayat S.M., Ghazvini K., Vaziri F., Behravan J. Electrochemical-based biosensors for detection of Mycobacterium tuberculosis and tuberculosis biomarkers. Crit Rev Biotechnol. 2019;39(8):1056–1077. doi: 10.1080/07388551.2019.1668348. [DOI] [PubMed] [Google Scholar]
- 63.Mowbray S.E., Amiri A.M. A brief overview of medical fiber optic biosensors and techniques in the modification for enhanced sensing ability. Diagnostics (Basel) 2019;9(1):23. doi: 10.3390/diagnostics9010023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gharatape A., Yari Khosroushahi A. Optical biomarker-based biosensors for cancer/infectious disease medical diagnoses. Appl Immunohistochem Mol Morphol. 2019;27(4):278–286. doi: 10.1097/PAI.0000000000000586. [DOI] [PubMed] [Google Scholar]
- 65.Russell C., Ward A.C., Vezza V., Hoskisson P., Alcorn D., Steenson D.P., Corrigan D.K. Development of a needle shaped microelectrode for electrochemical detection of the sepsis biomarker interleukin-6 (IL-6) in real time. Biosens Bioelectron. 2019;126:806–814. doi: 10.1016/j.bios.2018.11.053. [DOI] [PubMed] [Google Scholar]
- 66.Abisado R.G., Benomar S., Klaus J.R., Dandekar A.A., Chandler J.R. Bacterial quorum sensing and microbial community interactions. mBio. 2018;9(5):e01749–18. doi: 10.1128/mbio.01749-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sin M.L., Mach K.E., Wong P.K., Liao J.C. Advances and challenges in biosensor-based diagnosis of infectious diseases. Expert Rev Mol Diagn. 2014;14(2):225–244. doi: 10.1586/14737159.2014.888313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bhardwaj N., Bhardwaj S.K., Mehta J., Kim K.H., Deep A. MOF-bacteriophage biosensor for highly sensitive and specific detection of Staphylococcus aureus. ACS Appl Mater Interfaces. 2017;9(39):33589–33598. doi: 10.1021/acsami.7b07818. [DOI] [PubMed] [Google Scholar]
- 69.Nasrin F., Chowdhury A.D., Takemura K., Lee J., Adegoke O., Deo V.K., Abe F., Suzuki T., Park E.Y. Single-step detection of norovirus tuning localized surface plasmon resonance-induced optical signal between gold nanoparticles and quantum dots. Biosens Bioelectron. 2018;122:16–24. doi: 10.1016/j.bios.2018.09.024. [DOI] [PubMed] [Google Scholar]
- 70.Damborský P., Švitel J., Katrlík J. Optical biosensors. Essays Biochem. 2016;60(1):91–100. doi: 10.1042/ebc20150010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nquyet N.T., Yen L.T.H., Doan V.Y., Hoang N.L., Van Thu V., Lan H., Trung T., Pham V.H., Tam P.D. A label-free and highly sensitive DNA biosensor based on the core-shell structured CeO2-NR@Ppy nanocomposite for Salmonella detection. Mater Sci Eng C Mater Biol Appl. 2019;96:790–797. doi: 10.1016/j.msec.2018.11.059. [DOI] [PubMed] [Google Scholar]
- 72.Campuzano S., Yáñez-Sedeño P., Pingarrón J.M. Molecular biosensors for electrochemical detection of infectious pathogens in liquid biopsies: current trends and challenges. Sensors (Basel) 2017;17(11):2533. doi: 10.3390/s17112533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu Y., Zhou H., Hu Z., Yu G., Yang D., Zhao J. Label and label-free based surface-enhanced Raman scattering for pathogen bacteria detection: a review. Biosens Bioelectron. 2017;94:131–140. doi: 10.1016/j.bios.2017.02.032. [DOI] [PubMed] [Google Scholar]
- 74.Barreiros dos Santos M., Agusil J.P., Prieto-Simón B., Sporer C., Teixeira V., Samitier J. Highly sensitive detection of pathogen Escherichia coli O157:H7 by electrochemical impedance spectroscopy. Biosens Bioelectron. 2013;45:174–180. doi: 10.1016/j.bios.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 75.Zhang J., Oueslati R., Cheng C., Zhao L., Chen J., Almeida R., Wu J. Rapid, highly sensitive detection of gram-negative bacteria with lipopolysaccharide based disposable aptasensor. Biosens Bioelectron. 2018;112:48–53. doi: 10.1016/j.bios.2018.04.034. [DOI] [PubMed] [Google Scholar]
- 76.Huang Y., Xu J., Liu J., Wang X., Chen B. Disease-related detection with electrochemical biosensors: a review. Sensors (Basel) 2017;17(10):2375. doi: 10.3390/s17102375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zanchetta G., Lanfranco R., Giavazzi F., Bellini T., Buscaglia М. Emerging applications of label-free optical biosensors. Nanophotonics. 2017;6(4):627–645. doi: 10.1515/nanoph-2016-0158. [DOI] [Google Scholar]
- 78.Dudak F.C., Boyaci I.H. Rapid and label-free bacteria detection by surface plasmon resonance (SPR) biosensors. Biotechnol J. 2009;4(7):1003–1011. doi: 10.1002/biot.200800316. [DOI] [PubMed] [Google Scholar]
- 79.Yang F., Chang T.L., Liu T., Wu D., Du H., Liang J., Tian F. Label-free detection of Staphylococcus aureus bacteria using long-period fiber gratings with functional polyelectrolyte coatings. Biosens Bioelectron. 2019;133:147–153. doi: 10.1016/j.bios.2019.03.024. [DOI] [PubMed] [Google Scholar]
- 80.Muniandy S., Teh S.J., Appaturi J.N., Thong K.L., Lai C.W., Ibrahim F., Leo B.F. A reduced graphene oxide-titanium dioxide nanocomposite based electrochemical aptasensor for rapid and sensitive detection of Salmonella enterica. Bioelectrochemistry. 2019;127:136–144. doi: 10.1016/j.bioelechem.2019.02.005. [DOI] [PubMed] [Google Scholar]
- 81.Wang Z., Yao X., Wang R., Ji Y., Yue T., Sun J., Li T., Wang J., Zhang D. Label-free strip sensor based on surface positively charged nitrogen-rich carbon nanoparticles for rapid detection of Salmonella enteritidis. Biosens Bioelectron. 2019;132:360–367. doi: 10.1016/j.bios.2019.02.061. [DOI] [PubMed] [Google Scholar]
- 82.Srinivasan S., Ranganathan V., DeRosa M.C., Murari B.M. Label-free aptasensors based on fluorescent screening assays for the detection of Salmonella typhimurium. Anal Biochem. 2018;559:17–23. doi: 10.1016/j.ab.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 83.Zhang Y., Tian J., Li K., Tian H., Xu W. Label-free visual biosensor based on cascade amplification for the detection of Salmonella. Anal Chim Acta. 2019;1075:144–151. doi: 10.1016/j.aca.2019.05.020. [DOI] [PubMed] [Google Scholar]
- 84.Silva N.F.D., Magalhães J.M.C.S., Barroso M.F., Oliva-Teles T., Freire C., Delerue-Matos C. In situ formation of gold nanoparticles in polymer inclusion membrane: application as platform in a label-free potentiometric immunosensor for Salmonella typhimurium detection. Talanta. 2019;194:134–142. doi: 10.1016/j.talanta.2018.10.024. [DOI] [PubMed] [Google Scholar]
- 85.Han S., Soylu M.C., Kirimli C.E., Wu W., Sen B., Joshi S.G., Emery C.L., Au G., Niu X., Hamilton R., Krevolin K., Shih W.H., Shih W.Y. Rapid, label-free genetic detection of enteropathogens in stool without genetic isolation or amplification. Biosens Bioelectron. 2019;130:73–80. doi: 10.1016/j.bios.2019.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lin P.H., Huang S.C., Chen K.P., Li B.R., Li Y.K. Effective construction of a high-capacity boronic acid layer on a quartz crystal microbalance chip for high-density antibody immobilization. Sensors (Basel) 2019;19(1):28. doi: 10.3390/s19010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wu R., Ma Y., Pan J., Lee S.H., Liu J., Zhu H., Gu R., Shea K.J., Pan G. Efficient capture, rapid killing and ultrasensitive detection of bacteria by a nano-decorated multi-functional electrode sensor. Biosens Bioelectron. 2018;101:52–59. doi: 10.1016/j.bios.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 88.Kimuda S.G., Biraro I.A., Bagaya B.S., Raynes J.G., Cose S. Characterising antibody avidity in individuals of varied Mycobacterium tuberculosis infection status using surface plasmon resonance. PLoS One. 2018;13(10):e0205102. doi: 10.1371/journal.pone.0205102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rebelo R., Barbosa A.I., Caballero D., Kwon I.K., Oliveira J.M., Kundu S.C., Reis R.L., Correlo V.M. 3D biosensors in advanced medical diagnostics of high mortality diseases. Biosens Bioelectron. 2019;130:20–39. doi: 10.1016/j.bios.2018.12.057. [DOI] [PubMed] [Google Scholar]
- 90.Erdem Ö., Saylan Y., Cihangir N., Denizli A. Molecularly imprinted nanoparticles based plasmonic sensors for real-time Enterococcus faecalis detection. Biosens Bioelectron. 2019;126:608–614. doi: 10.1016/j.bios.2018.11.030. [DOI] [PubMed] [Google Scholar]
- 91.Hoyos-Nogués M., Gil F.J., Mas-Moruno C. Antimicrobial peptides: powerful biorecognition elements to detect bacteria in biosensing technologies. Molecules. 2018;23(7):1683. doi: 10.3390/molecules23071683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liébana S., Brandão D., Alegret S., Pividori M.I. Electrocehmical immunosensors, genosensors and phagosensors for Salmonella detection. Anal Methods. 2014;6(22):8858–8873. doi: 10.1039/c4ay01373e. [DOI] [Google Scholar]
- 93.Cui F., Xu Y., Wang R., Liu H., Chen L., Zhang Q., Mu X. Label-free impedimetric glycan biosensor for quantitative evaluation interactions between pathogenic bacteria and mannose. Biosens Bioelectron. 2018;103:94–98. doi: 10.1016/j.bios.2017.11.068. [DOI] [PubMed] [Google Scholar]
- 94.Bu T., Huang Q., Yan L., Zhang W., Dou L., Huang L., Yang Q., Zhao B., Yang B., Li T., Wang J., Zhang D. Applicability of biological dye tracer in strip biosensor for ultrasensitive detection of pathogenic bacteria. Food Chem. 2019;274:816–821. doi: 10.1016/j.foodchem.2018.09.066. [DOI] [PubMed] [Google Scholar]
- 95.Zarei S.S., Soleimanian-Zad S., Ensafi A.A. An impedimetric aptasensor for Shigella dysenteriae using a gold nanoparticle-modified glassy carbon electrode. Mikrochim Acta. 2018;185(12):538. doi: 10.1007/s00604-018-3075-0. [DOI] [PubMed] [Google Scholar]
- 96.Chuensirikulchai K., Laopajon W., Phunpae P., Apiratmateekul N., Surinkaew S., Tayapiwatana C., Pata S., Kasinrerk W. Sandwich antibody-based biosensor system for identification of Mycobacterium tuberculosis complex and nontuberculous mycobacteria. J Immunoassay Immunochem. 2019;40(6):590–604. doi: 10.1080/15321819.2019.1659814. [DOI] [PubMed] [Google Scholar]
- 97.Savas S., Altintas Z. Graphene quantum dots as nanozymes for electrochemical sensing of Yersinia enterocolitica in milk and human serum. Materials (Basel) 2019;12(13):2189. doi: 10.3390/ma12132189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chang H.J., Voyvodic P.L., Zúñiga A., Bonnet J. Microbially derived biosensors for diagnosis, monitoring and epidemiology. Microb Biotechnol. 2017;10(5):1031–1035. doi: 10.1111/1751-7915.12791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kuss S., Amin H.M.A., Compton R.G. Electrochemical detection of pathogenic bacteria-recent strategies, advances and challenges. Chem Asian J. 2018;13(19):2758–2769. doi: 10.1002/asia.201800798. [DOI] [PubMed] [Google Scholar]
- 100.Singh R., Mukherjee M.D., Sumana G., Gupta R.K., Sood S., Malhotra B.D. Biosensors for pathogen detection: a smart approach towards clinical diagnosis. Sens Actuators B Chem. 2014;197:385–404. doi: 10.1016/j.snb.2014.03.005. [DOI] [Google Scholar]
- 101.Zhou Q., Son K., Liu Y., Revzin A. Biosensors for cell analysis. Annu Rev Biomed Eng. 2015;17:165–190. doi: 10.1146/annurev-bioeng-071114-040525. [DOI] [PubMed] [Google Scholar]