Summary
Multiple lines of evidence show that modern humans interbred with archaic Denisovans. Here, we report an account of shared demographic history between Australasians and Denisovans distinctively in Island Southeast Asia. Our analyses are based on ∼2.3 million genotypes from 118 ethnic groups of the Philippines, including 25 diverse self-identified Negrito populations, along with high-coverage genomes of Australopapuans and Ayta Magbukon Negritos. We show that Ayta Magbukon possess the highest level of Denisovan ancestry in the world—∼30%–40% greater than that of Australians and Papuans—consistent with an independent admixture event into Negritos from Denisovans. Together with the recently described Homo luzonensis, we suggest that there were multiple archaic species that inhabited the Philippines prior to the arrival of modern humans and that these archaic groups may have been genetically related. Altogether, our findings unveil a complex intertwined history of modern and archaic humans in the Asia-Pacific region, where distinct Islander Denisovan populations differentially admixed with incoming Australasians across multiple locations and at various points in time.
Keywords: archaic introgression, Denisovans, admixture, genomics, Island Southeast Asia, Philippines, Negrito, Australasian
Highlights
-
•
Comprehensive analyses of archaic ancestry among 118 Philippine ethnic groups
-
•
Ayta Magbukon display ∼30%–40% greater Denisovan ancestry than Australopapuans
-
•
The model is explained by a distinct admixture event into Negritos from Denisovans
-
•
Prior to modern humans, Islander Denisovans may have been present in the Philippines
Larena et al. reveal Philippine Ayta to possess the highest level of Denisovan ancestry in the world—∼30%–40% greater than that of Australopapuans—consistent with an independent admixture event into Negritos from Denisovans. The Philippine archipelago is thus likely inhabited by multiple archaic groups prior to the arrival of modern humans.
Introduction
Since their exit out of Africa, anatomically modern humans have overlapped and interbred with archaic hominins across time and space. Interactions with Neanderthals,1,2 Denisovans,3,4 and possibly other archaic hominins5,6 have left an indelible genetic trace in the genomes of present-day populations.7, 8, 9 All Eurasians possess uniform levels of Neanderthal ancestry, whereas Australasians uniquely have elevated levels of Denisovan ancestry relative to any other population. Denisovans were initially thought to have a simple shared demographic history with modern humans, through a single admixture event with the ancestor of Australasians (a collective term for the shared genetic ancestry between Philippine Negritos and Australopapuans).4,10,11
Later work suggested that Denisovans were likely deeply diverging structured populations who inhabited wide-ranging diverse environments, given that Denisovan ancestry can also be detected, albeit at lower levels, among East Asian, South Asian, Siberian, and Native American populations.7,12, 13, 14 Through analysis of recovered archaic tracts from population-level genomes, it was recently shown that modern humans admixed with Denisovans at least twice in the past, with Denisovan-like populations possessing varying degrees of genetic affiliation to the reference Altai Denisovan.15 Subsequent work revealed a third independent Denisovan introgression event that was exclusively found among Papuan-related populations, indicating an increasingly complex admixture history between Denisovans and Australasians.16
Although the intertwined demographic history between Denisovans and Australasians is being uncovered, our knowledge of their specific interactions in Island Southeast Asia (ISEA) remains limited. This is especially important given that some present-day populations of the Philippines, who self-identify as “Negritos” and who are genetically related to Australopapuans, display significant levels of Denisovan ancestry.4,17,18 To deepen our understanding of past Denisovan-Australasian interactions in the ISEA region, we comprehensively investigated the archaic ancestry of 1,107 individuals from 118 distinct ethnic groups of the Philippines, including 25 ethnolinguistically diverse Negrito populations (Figures 1A and S1; see STAR Methods).19
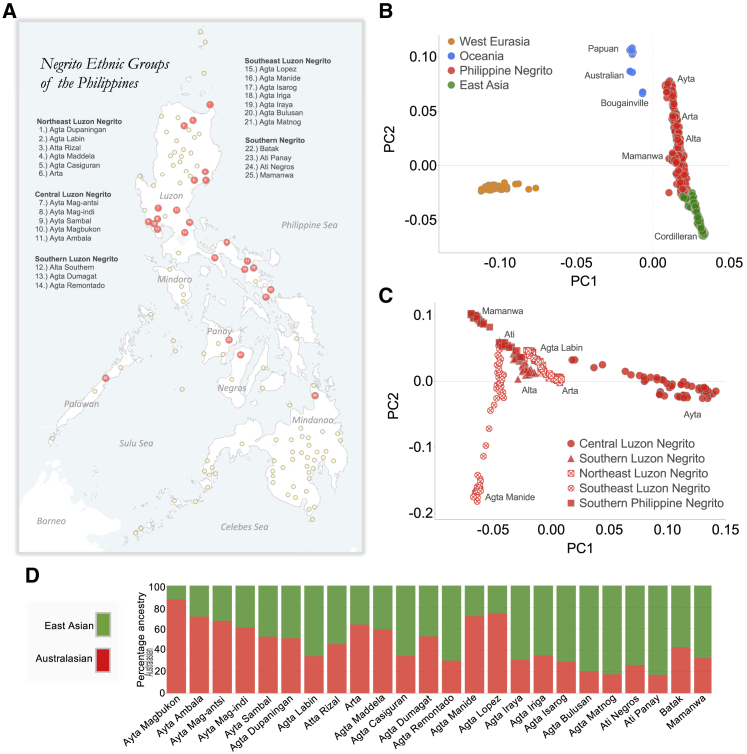
Figure 1.
Indigenous Negrito ethnic groups of the Philippines
(A) Location of the 118 Philippine ethnic groups analyzed in this study. Negrito ethnic groups are labeled with red circular markers, whereas all other non-Negrito populations are labeled with yellow circular markers. Philippine Negritos are classified based on their genetic and/or geographical clusters: Northeast Luzon Negrito; Central Luzon Negrito; Southern Luzon Negrito; Southeast Luzon Negrito; and Southern Negrito.
(B) Principal-component analysis (PCA) of West Eurasians, Australasians, and East Asians. Philippine Negritos lie between the axis defined by East Asian and Australasian-related ancestries.
(C) Subset PCA showing the five distinct population clusters of Philippine Negritos.
(D) Proportion of Australasian-related versus East-Asian-related ancestries among Philippine Negritos estimated using qpAdm.
See also Figures S1 and S2.
Results and discussion
Population structure and genetic affiliations of Philippine Negritos
Australasian-related Negritos are regarded as the earliest modern human inhabitants of Sundaland, a contiguous biogeographical region in ISEA that was largely exposed during the last glacial period (Figure S1; see STAR Methods).11,17,19 Following their divergence from Papuans ∼53 kya (95% confidence interval [CI]: 41–64 kya; see STAR Methods), Negritos migrated from Sundaland into the Philippines likely via the accessible Palawan-Mindoro or Sulu-Zamboanga corridors (Figure S1) and later diverged into the multiple and linguistically diverse Philippine Negrito groups of today (see STAR Methods). To provide an overview of the genetic affinities of present-day Philippine Negritos to other ethnic groups of the Asia-Pacific region, we performed a principal-component analysis (PCA) on a subset of populations, including Australasians, East Asians, and West Eurasians (Figure 1B). All Negritos lie between the population clusters of Australopapuans and Philippine Cordillerans, indicating admixture between the two ancestral sources: an Australasian-related and an East-Asian-related ancestry, respectively. Additionally, a subset PCA restricted to Philippine Negritos, as well as an unsupervised clustering analysis implemented in ADMIXTURE,20 reveal at least five distinct population clusters of Negritos: Central Luzon Negritos; Northeast Luzon Negritos; Southeast Luzon Negritos; Southern Luzon Negritos; and Southern Negritos (Figures 1C, S2B, and S2D; see STAR Methods).
It was previously shown that Philippine Negritos form a clade with Papuans. We then investigated further on the evidence of recent admixture between Philippine Negritos and East Asians. Using the test D(Mbuti;EastAsian,Australasian,X), we show that all Negritos exhibit significant levels of gene flow from East Asians (Figure S2E), which is attributed to recent admixture with the Austronesian-speaking migrants from the South China-Taiwan greater area.11,17 Applying a linkage disequilibrium (LD)-based admixture date inference approach in Malder,21 we estimate that the Negrito-East Asian admixture event occurred ∼2,281 years ago (95% CI: 2,083–2,523 years). We then quantified the magnitude of admixture between the two ancestral sources with the implementation of qpAdm,22 using Balangao Cordillerans as the surrogate for the least admixed East Asian source and Papuans as the surrogate for the least admixed Australasian source. Overall, Negritos exhibit varying degree of admixture with East Asians (Figures 1D, S3A, and S3B), with Central Luzon Negritos, such as the Ayta Magbukon and Ayta Ambala, presenting with the least amount of East-Asian-related ancestry, ∼10%–30%.
To identify the genetic affiliation of Australasians to archaic hominins, we performed a PCA on chimpanzee, Neanderthal, and Denisovan individuals and projected onto the plane of the first 2 PCs the present-day populations of Africans, East Asians, Philippine Negritos, and Australopapuans (Figure S2C). PC1 is defined by the chimpanzee versus archaic human axis, whereas PC2 is defined by the Denisovan versus Neanderthal axis. All present-day populations lie at the center on the plot, and an inset plot zooming in on the present-day populations demonstrates the affiliation of Australopapuans and Philippine Negritos toward the Denisovan axis. Interestingly, most Ayta Magbukon individuals lie at the edge of the Denisovan axis, even farther than that of any Papuan or Australian individuals, indicating higher levels of Denisovan ancestry in Ayta Magbukon Negritos than in Australopapuans.
Philippine Negritos exhibit the highest levels of Denisovan ancestry
Although Neanderthal ancestry is uniformly detectable in all Philippine ethnic groups (Figure S3C), at levels that are comparable to those of worldwide populations outside Africa,2,12 the degree of Denisovan ancestry is more variable (Figures 2A, 2B, and S3D; Data S1A–S1C).3,4,13,17 Denisovan ancestry is higher among Negritos, with a clear gradient that declines in proportion to the level of non-Negrito ancestry (Figure S4A). Thus, we find a strong correlation between Negrito ancestry and Denisovan ancestry (Figures 3A and S4B–S4E).
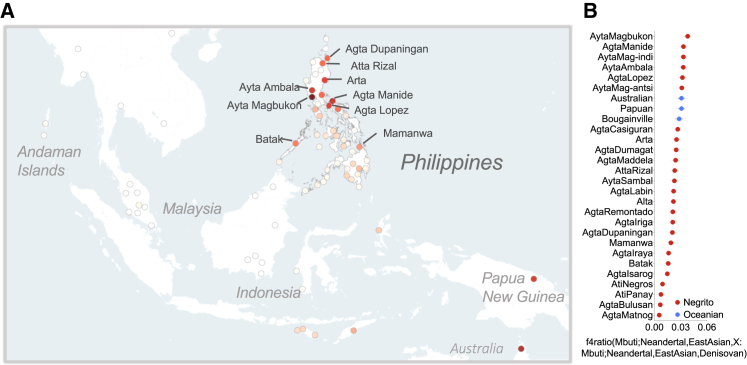
Figure 2.
Ayta Negritos exhibit the highest levels of Denisovan ancestry
(A) Map showing distribution of populations with the indicative levels of Denisovan ancestry estimated by f4-ratio (Mbuti;Neanderthal,EastAsian,X)/(Mbuti,Neanderthal,EastAsian, Denisovan) (see Data S1 for the complete list of Denisovan ancestry estimates among 118 ethnic groups of the Philippines).
(B) Asia-Pacific populations, represented by five least admixed individuals per group, with the highest levels of Denisovan ancestry estimated by f4-ratio statistics. Thick and thin error bars represent 1 and 1.96 standard error of the estimate, respectively.
See also Figures S2–S5 and Data S1.
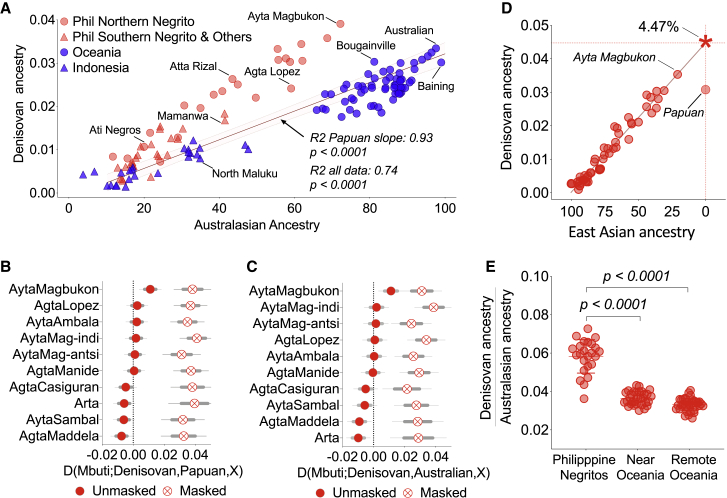
Figure 3.
Ancestral Philippine Negritos exhibit higher levels of Denisovan ancestry than Australopapuans
(A) Correlation between Denisovan ancestry and Australasian ancestry. Philippine Negritos fall outside the slope formed by populations with Papuan-related ancestry.
(B and C) Denisovan ancestry in Northern Negritos versus Papuans (B) or Australians (C), before and after accounting for East Asian admixture. Masked or unmasked labels indicate removal or non-removal of East Asian ancestry following RFMix analysis (see STAR Methods).
(D) Extrapolation on the level of Denisovan ancestry in an ancestral “unadmixed” Negrito population by plotting Denisovan ancestry and East-Asian-related ancestry.
(E) Ratio of Denisovan ancestry over Australasian-related ancestry where Philippine Negritos display significantly higher values over near Oceanians or remote Oceanians (Mann-Whitney U test). Thick and thin error bars represent 1 and 1.96 standard error of the estimate, respectively.
See also Figures S2–S5 and Data S1.
More interestingly, some Northern Negritos possess the highest point estimate of Denisovan ancestry recorded to date (Figures 2A, 2B, and S5), with Ayta Magbukon exhibiting greater Denisovan ancestry than Australopapuans based on the test D (Mbuti;Denisovan,Australian/Papuan,AytaMagbukon) (Figures 3B and 3C). The higher degree of Denisovan introgression into Ayta Magbukon can be observed despite the group’s detectable ∼10%–20% East Asian recent admixture, in contrast with the less admixed Papuan and Australian populations (Figures 3B–3D and S5A–S5D). These results are consistent when using alternative datasets, testing individuals instead of populations, accounting for ascertainment bias, or correcting for Neanderthal ancestry in these populations (Figures S5F–S5O; see STAR Methods).
The difference in the degree of Denisovan admixture is clearer when we masked out East-Asian-related admixture in Negritos (Figures 3B, 3C, and S5E; see STAR Methods). The masking step results in a distinctively higher point estimate of Denisovan ancestry, with non-overlapping confidence intervals, in most Negritos than in Papuans or Australians. Moreover, extrapolating to “no-East-Asian-admixture” on the regression line between Denisovan versus East-Asian-related ancestry among Negritos indicates a substantially higher Denisovan ancestry in the “unadmixed” ancestral Negritos (f4-ratio = 0.0447), which is 46% higher than that of Papuans (Figure 3D). Likewise, the ratio of Denisovan ancestry to Negrito or Papuan-related ancestry is significantly higher in Negritos than in Papuan-related populations (Figure 3E).
Analyses of sequence data confirm higher levels of Denisovan ancestry in Ayta Magbukon Negritos than in Papuans
We explored further our finding of high levels of Denisovan ancestry in some Philippine Negritos; generated, at high coverage (mean depth of ∼37×), whole-genome sequences of five Ayta Magbukon Negritos; and merged these data with the publicly available genome sequence datasets of archaic and modern humans (see STAR Methods).1, 2, 3,23 Following comparative analyses of high-coverage genomes of Australasians, we confirm that the Ayta Magbukon possess the highest level of Denisovan ancestry in the world (Figures 4A–4C, S6A, and S6B; Data S2A–S2C; see STAR Methods), levels that are 34%–40% higher than that of Australians or Papuans (based on the difference in Denisovan ancestry estimates using f4-ratio statistics; Figure 4B).
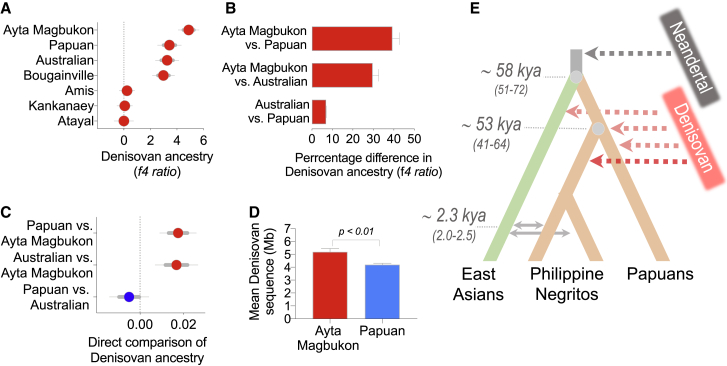
Figure 4.
Analyses of high-coverage genomes confirm higher levels of Denisovan ancestry in Ayta Magbukon relative to Australopapuans
(A) Using high-coverage genomes of Ayta Magbukon, Papuans, Australians, Bougainville Islanders, and East Asians, percentage Denisovan ancestry were estimated using the statistic f4-ratio (Mbuti;Neanderthal,Han,X)/ (Mbuti;Neanderthal,Han,Denisovan).
(B) Percentage difference in Denisovan ancestry based on f4-ratio statistics.
(C) Direct comparison of Denisovan ancestry between Australians, Papuans, and Ayta Magbukon, using the statistic D(Mbuti;Denisovan,Y1,Y2) (see Data S2C).
(D) Mean plus SEM of Denisovan sequence detected per individual using S’ approach. Inferred model of Denisovan introgression events into Philippine Negritos and Papuans is shown (see STAR Methods).
(E) Topology of East Asians and Australasians indicating the pulses of Denisovan introgression into Papuans and East Asians as previously reported in Browning et al.15 and Jacobs et al.16 and an independent pulse of Denisovan introgression into Philippine Negritos reported here. Time of divergence between East Asians and Australasians was previously reported in Malaspinas et al.24 Divergence time between Philippine Negritos and Papuans was estimated using the Two-plus-Two method (see STAR Methods; Sjödin et al.25). Mean date of admixture between Philippine Negritos and East Asians was inferred using Malder. Values between brackets represent 95% confidence interval. Thick and thin error bars represent 1 and 1.96 standard error of the estimate, respectively.
See also Figures S6 and S7 and Data S2.
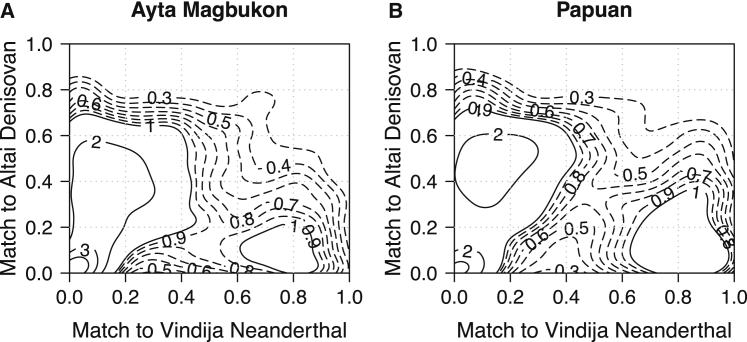
We used the S’ framework15 to identify (high confidence) archaic regions in the genomes of Ayta Magbukon and Papuans (see STAR Methods) and to exclude the potential biases in f4-ratio statistics for estimating the levels of archaic ancestry in populations.26 The average amount of Denisovan sequence per individual in Ayta Magbukon is significantly higher than that of Papuans (51.94 Mb [95% CI: 44.62.66–59.25 Mb] versus 41.96 Mb [95% CI: 39.54–44.37 Mb]; p = 3.7 × 10−3), showing that Ayta Magbukon have at least ∼24% more Denisovan ancestry than in Papuans (Figures 4D and S6C). Moreover, both Ayta Magbukon and Papuans exhibit Denisovan segments that have moderate affinity to the reference Altai Denisovan, a match rate of ∼50% (Figures 5A and 5B). This suggests that the archaic groups introgressing into the Ayta Magbukon and Papuans are likely to be distantly related to the Altai Denisovan of Siberia, consistent with previous observations.15,16
Figure 5.
Contour density plots showing the affiliation of S’-detected introgressed segments to the reference archaic genomes
Contour density plots of match rate to the reference Vindija Neanderthal and Altai Denisovan genomes, using the S’-detected introgressed segments in Ayta Magbukon Negritos (A) and Papuans (B). The numbers within the plots correspond to the height of the density for each contour line, and contour lines are shown for multiples of 1 (solid lines) and multiples of 0.1 between 0.3 and 0.9 (dashed lines). See also Figure S6.
The high levels of Denisovan ancestry in Ayta may be due to a recent Denisovan admixture event into this population. This would make the admixture tracts longer and easier to detect, which may conceivably account for the 24% excess Denisovan ancestry relative to Papuans. However, we find that this is not the case, given that the mean tract lengths are similar for the Ayta and Papuans (260 Kb [95% CI: 239–282 Kb] versus 262 Kb [95% CI: 254–271 Kb]; Mann-Whitney test; p = 0.6). Likewise, we do not find a significant difference in the cumulative distributions of Denisovan tracts in Ayta Magbukon and Papuans (Kolmogorov-Smirnov test; p = 0.70). Our findings imply that the admixture is of similar age in the two populations, which lends credence to our claim that there is more Denisovan admixture in the Ayta than in Papuans.
Tract lengths of Denisovan ancestry have previously been investigated in Papuans and in some Philippine Negrito groups with varying results. Browning et al.15 found no difference in Denisovan tract lengths attributed to the suggested two introgression events into East Asians and Papuans. Similarly, Jacobs et al.16 found no difference in mean Denisovan tract lengths in Papuans but reported a difference in the tract length distribution consistent with two different Denisovan introgressions into Papuans. Wall et al.18 found slightly longer mean tract lengths in an Aeta group (likely the Ayta Mag-antsi and Mag-indi in our dataset) than in Papuans, suggesting separate Denisovan introgression events into Aeta and Papuans. Note, however, that the mean tract lengths found by Wall et al.18 are ∼15 times shorter than those found by Jacobs et al.16 and, in our estimates, at least partly attributed to different inference approaches, making direct comparisons difficult.
Independent Denisovan introgression event into Philippine Negritos
The significantly higher level of Denisovan ancestry in Ayta Magbukon relative to Papuans highlights the possibility of an independent Denisovan introgression event in the Philippines among Negritos that is different from the Denisovan introgression event into the ancestors of Australopapuans. This observation is consistent with recent studies suggesting multiple pulses of Denisovan introgression into humans, that Denisovans were probably widespread throughout ISEA, and that Ayta Negritos were likely to have experienced a second Denisovan introgression event.12,15,16,18,27 Hence, we further examined the lines of evidence for an independent Denisovan introgression event into Negritos after the divergence between Negritos and Papuans.
First, we find that Negritos possess a Denisovan ancestry that is not associated with the Australasian-related ancestry in Australopapuans (Figure 3A; see STAR Methods). Most Negritos form a clear outlier to the slope that correlates Denisovan ancestry and Australasian-related ancestry in Oceanians and Indonesians (Figure 3A). Second, the substantially higher Denisovan ancestry in Negritos (Figures 3B, 3C, 4A–4D, and S5A–S5O), relative to Papuans, is consistent with a model of an additional Denisovan introgression event into Negritos. Alternatively, it could be caused by a dilution of Denisovan ancestry in Papuans by admixture with populations containing little Denisovan ancestry. However, neither we (Figures S7A–S7L; see STAR Methods) nor previous studies24,28 find evidence for recent gene flow from East Asians or Europeans into highland Papuans. On the contrary, Negritos do display evidence for recent admixture with Cordilleran-related East Asians (who carry very little Denisovan ancestry; Figures 1D, S3A, and S3B). Third, an explicit population topology model using qpGraph, based on a shared Denisovan introgression event into Negritos, Australians, and Papuans, was rejected (Figure 6A; see STAR Methods). This is in contrast to the models fitting a separate Denisovan introgression event into Negritos (Figures 6B and 6C; all f-stats are within 1.18 standard error).
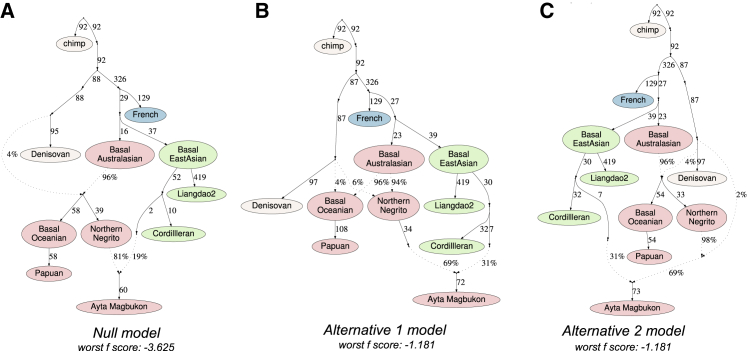
Figure 6.
Admixture graph models of Denisovan introgression into Philippine Negritos and Papuans
Admixture graphs for the null, alternative 1, and alternative 2 models indicating the number of archaic introgression events into Ayta Negritos and Papuans.
(A) Null model depicts a single Denisovan introgression event into the ancestral populations of Ayta Negritos and Papuans.
(B) Alternative 1 model depicts an additional admixture event into Ayta Negritos coming from a distinct and unsampled Denisovan population, after an admixture event in the shared ancestral population of Ayta Negritos and Papuans.
(C) Alternative 2 model depicts two completely separate Denisovan introgression events into the ancestral populations of Ayta Negritos and Papuans. Only alternative 1 and 2 models are not rejected (Z score = −1.181).
See also STAR Methods.
Simulations support two distinct Denisovan lineages introgressing independently into Ayta Negritos and Papuans
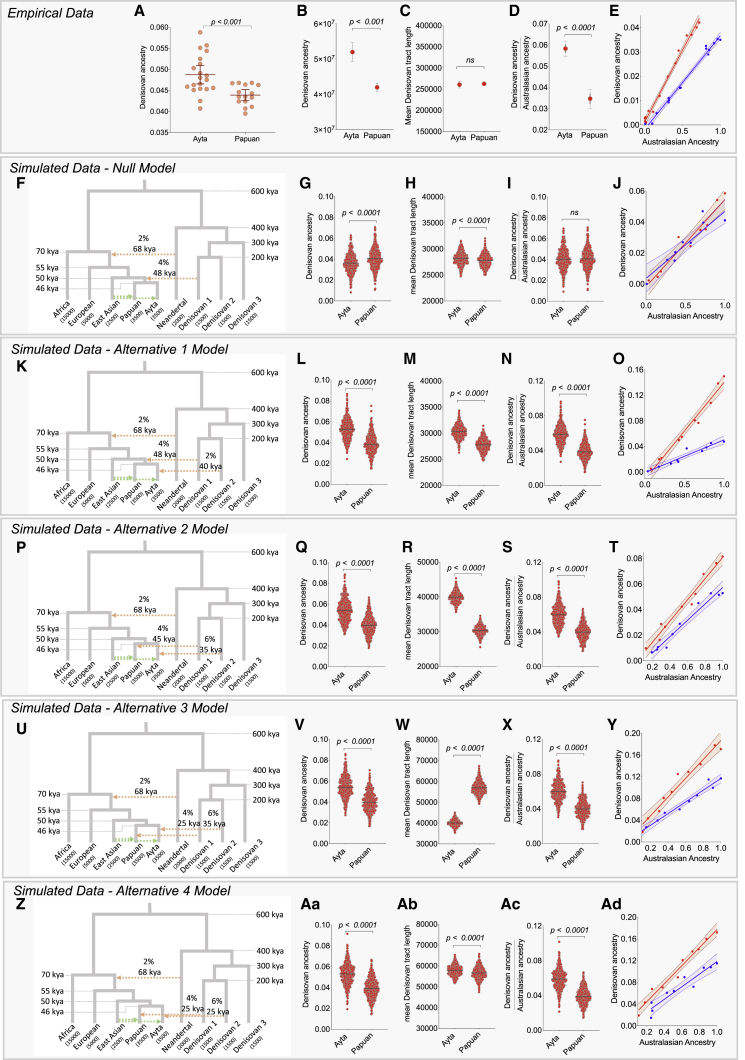
We performed simulations using the coalescent simulator msprime29 (see STAR Methods) to evaluate whether a null model of shared Denisovan introgression event into the common ancestor of Ayta Negritos and Papuans (Figure 7F) or the alternative models (Alt1, Alt2, Alt3, and Alt4) of multiple Denisovan admixture events (Figures 7K, 7P, 7U, and 7Z) produce patterns of Denisovan ancestry that are consistent with the observed data (Figures 7A–7E), where Ayta Negritos possess higher levels of Denisovan ancestry than Papuans. Alt1 model (Figure 7K) depicts an additional admixture event into Ayta Negritos coming from a distinct and unsampled Denisovan population, after an admixture event in the shared ancestral population of Ayta Negritos and Papuans. Alt2, Alt3, and Alt4 models depict completely separate Denisovan introgression events into the ancestral populations of Ayta Negritos and Papuans happening at different time periods (Alt2 and Alt3; Figures 7P and 7U) or at similar times (Alt 4; Figure 7Z).
Figure 7.
Evidence for independent Denisovan introgression event into Philippine Negritos
(A and B) Empirical evidence for higher Denisovan ancestry in Ayta relative to Papuans estimated by f4-ratio statistics on genotyped data (A) and S’ method on sequence data (B).
(C) Mean Denisovan tracts lengths in Ayta Magbukon versus Papuans.
(D) Ratio of Denisovan ancestry over Australasian ancestry in Ayta-related and Papuan-related populations.
(E) Correlation between Denisovan ancestry and Australasian ancestry, where Negritos (red line) fall outside the slope of the Papuan cline (blue line).
(F–Ad) Diagram for the null (F), alternative models 1–4 (K, P, U, and Z) indicating the number and the time of archaic introgression events into Ayta Negritos and Papuans, simulated using msprime (see STAR Methods), with the respective estimated levels of Denisovan ancestry (G, L, Q, V, and Aa), mean Denisovan tract length (H, M, R, W, and Ab), ratio of Denisovan ancestry over Australasian ancestry (I, N, S, X, and Ac), and correlation between Denisovan ancestry and Australasian ancestry (J, O, T, Y, and Ad). Australasian ancestry was estimated using qpAdm. All pairwise comparisons were tested for significance using the Mann-Whitney U test with Holm-Bonferroni correction. Error bars represent standard error of the mean.
In contrast to a null model of shared Denisovan admixture event, simulations based on a model with a separate Denisovan introgression event into Negritos (Alt1, Alt2, Alt3, and Alt4) produce patterns of Denisovan ancestry that are consistent with the observed data (Figure 7). All alternative models consistently replicate three pieces of empirical evidence: (1) higher level of Denisovan ancestry in Ayta Negritos than in Papuans; (2) higher ratio of Denisovan over Australasian ancestry in Ayta Negritos than in Papuans; and (3) Ayta Negritos falls outside the slope formed by Papuan-related groups when plotting the correlation between Denisovan ancestry and Australasian ancestry (Figure 7). Only Alt4 model, where the timing of the separate Denisovan introgression events into Ayta Negritos and Papuans occurred around the same time, produced similar levels of mean Denisovan tract length in Ayta Negritos and Papuans, a pattern consistent with the empirical data (Figures 7C and 7Ab). Consequently, only the Alt4 model produced qualitative patterns that are consistent with all of the empirical data.
Altogether, our simulations provide support for the presence of two separate Denisovan lineages that independently introgressed into the ancestors of Ayta Negritos and Papuans, likely occurring around the same time after the Negrito-Papuan divergence 53 kya (95% CI: 41–64 kya). Upon entry of the first modern human migrants into Sunda and Sahul (ancestors of Negritos and Australopapuans), these ancestral Australasian groups likely experienced admixture with deeply divergent Denisovan-related populations scattered all throughout the ISEA and the Oceania region.15,16
Despite the fact that the populations with the highest levels of Denisovan ancestry are found in the regions of ISEA and Near Oceania, no Denisovan fossils have yet been discovered in the area. This may, in part, be limited by the lack of information on the definitive phenotypic characteristics of Denisovans. Most of the available knowledge on Denisovans is based on genomic data from the Altai Denisovans of Siberia.3,10 The only other direct evidence of Denisovans outside of Siberia is from the Baishiya Karst Cave site of the Tibetan Plateau,30,31 where an ancient proteome and an ancient mitochondrial DNA analysis revealed the Xiahe hominin to be phylogenetically affiliated with the Altai Denisovan. This led to the proposition that the other previously discovered archaic humans in the region may in fact be Denisovans, such as, among others, the Xujiayao and Penghu-1 individuals (Figure S1).30,32,33 Additionally, the physical evidence for a previously undescribed hominin in Luzon 67 kya, where present-day Negritos reside,34,35 combined with the genetic evidence presented here, raises the possibility that the suggested Homo luzonensis and Denisovans were likely genetically related, either as distinct forms or possibly belonging to the same group residing on the islands.36,37 Furthermore, it is not entirely impossible that the recently identified new species of archaic hominins in the Indonesian island of Flores, the Homo floresiensis,38 may also be related to Denisovans. Hence, the presence of multiple archaic human remains in the region, together with the genomic evidence presented here and elsewhere,15,16,31,37,39 raises the possibility that the Denisovans comprised deeply structured populations with considerable genetic and phenotypic diversity, enabling them to adapt into a wide variety of environments and thus inhabit a broad geographic range across the Asia-Pacific region.36,37,40,41 This is supported by the genomic data showing that Papuans experienced a distinct Denisovan introgression event as recent as ∼25–30 kya,16,37 which is several millennia after the divergence between Ayta Negritos and Papuans ∼53 kya. Though our interpretation provides a parsimonious explanation for the overlapping presence of archaic human remains and present-day populations with high levels of Denisovan ancestry in ISEA, this is not in line with the current palaeoanthropological analyses of the available fossil data. The definitive phylogenetic relationships between Denisovans, Homo luzonensis, Homo floresiensis, and other archaic hominins in the region still remain to be determined and will likely be resolved with the availability of ancient DNA or ancient proteomic data in the future.
Conclusions
While we await the discovery and successful extraction of ancient DNA from Denisovans in challenging tropical environments of the ISEA region, a computational genetic approach appears to be the only viable alternative at this stage.7,15,42 This approach was previously shown to be successful in advancing our understanding on how archaic hominins shaped our past and our biology.7,8,40,41 Moreover, as we take full advantage of the available computational tools, we underscore the importance of including underrepresented populations in genomic investigations, such as the Philippine Negritos. Our genomic analysis on Philippine Negritos shed light on an important section of human evolutionary history: the previously unappreciated complex interactions between modern humans and Denisovans in the ISEA region.
Here, we have presented evidence for the possible presence of diverse Islander Denisovan populations, who differentially admixed with incoming Australasians across multiple locations and at various points in time. Consequently, this led to variable levels of Denisovan ancestry in the genomes of Philippine Negritos and Papuans. In ISEA, Philippine Negritos later admixed with East Asian migrants who possess little Denisovan ancestry, which subsequently diluted their archaic ancestry. Some groups though, such as the Ayta Magbukon, minimally admixed with the more recent incoming migrants. For this reason, the Ayta Magbukon retained most of their inherited archaic tracts and left them with the highest level of Denisovan ancestry in the world.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| 1,186 human samples from the Philippines |
This study and Larena et al.11 |
EGAS00001005083 |
| EGAS00001005408 | ||
| Deposited data | ||
| Philippine Genotype data | This study | EGAS00001005083 |
| Ayta Magbukon Genome data | This study | EGAS00001005407 |
| Simon’s Genome Diversity Project data | Mallick et al.23 | https://sharehost.hms.harvard.edu/genetics/reich_lab/sgdp/phased_data/PS2_multisample_public/ |
| 1000 Genomes Project data | Abecasis et al.43 | ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/ |
| Estonian Genome Diversity Project data | Pagani et al.44 | https://evolbio.ut.ee |
| Malaysian Genotype data | Aghakhanian et al.45 | N.A.; direct request from Maude Phipps |
| Indonesian Genotype data | Kusuma et al.,46,47 Cox et al.,48 and Mörseburg et al.49 | NCBI GEO GSE76645 and GSE77508; and direct requests from Murray Cox and Francois-Xavier Ricaut |
| South Asian Genotype data | Basu et al.50 | direct request from Partha Majumder |
| Human Origins (HO) Genotype data | Lazaridis et al.51 and Patterson et al.52 | https://reich.hms.harvard.edu/datasets |
| East Asian HO Genoytpe data | Wang et al.53 | https://reich.hms.harvard.edu/datasets |
| Mainland Southeast Asian HO Genoytpe data | Lipson et al.54 | https://reich.hms.harvard.edu/datasets |
| Oceanian HO Genoytpe data | Lipson et al.55 and Skoglund et al.56 | https://reich.hms.harvard.edu/datasets |
| South Asian HO Genoytpe data | Nakatsuka et al.57 | direct request from David Reich |
| Altai Neanderthal, Vindija Neanderthal, and Altai Denisovan genome data | Prüfer et al.2,14 and Meyer et al.3 | http://cdna.eva.mpg.de/neandertal/Vindija/ |
| Single nucleotide variant annotation | dbSNP | https://www.ncbi.nlm.nih.gov/projects/SNP/ |
| Software and algorithms | ||
| PLINK v1.9 | Purcell et al.58 | https://www.cog-genomics.org/plink/ |
| KING v2.2 | Manichaikul et al.59 | https://www.kingrelatedness.com/ |
| EIGENSOFT v7.2.0 | Price et al.60 and Patterson et al.61 | https://github.com/DreichLab/EIG |
| ADMIXTURE v1.3.0 | Alexander et al.20 | https://dalexander.github.io/admixture/download.html |
| CLUMPP v1.1.2 | Jakobsson and Rosenberg62 | https://rosenberglab.stanford.edu/lump.html |
| Pong v1.4 | Behr et al.63 | https://github.com/ramachandran-lab/pong |
| ADMIXTOOLS v5.0 | Patterson et al.52 | https://github.com/DreichLab/AdmixTools |
| RFMix v1.5 | Maples et al.64 | https://github.com/indraniel/rfmix |
| Malder | Loh et al.21 | https://github.com/joepickrell/malder |
| Burrows–Wheeler Aligner v0.7.17 | Li and Durbin65 | http://bio-bwa.sourceforge.net |
| Genome Analysis Toolkit v3.5 | Van der Auwera et al.,66 DePristo et al.,67 and Poplin et al.68 | https://gatk.broadinstitute.org |
| S-prime | Browning et al.15 | http://faculty.washington.edu/browning/sprime.html |
| msprime v0.6.1 | Kelleher et al.29 | https://github.com/tskit-dev/msprime/releases |
| TT-Method | Sjödin et al.25 | https://github.com/jammc313/TT-method |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Mattias Jakobsson (mattias.jakobsson@ebc.uu.se).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Human Subjects
All 1,186 samples were obtained from human subjects who are at least 18 years old. Consent was obtained from each participant, and whenever necessary, from the participant’s respective Indigenous Cultural Community Council. Community preparation and consultation, consent process, sampling, and presentation of results back to the community was implemented in partnership with the National Commission for Culture and the Arts (NCCA) and/or other local partners listed in SI Appendix 10 of related publication11. Saliva samples were collected from participants with the use of Oragen Saliva Collection Kit (DNA Genotek Inc, Ottawa, ON, Canada). Individuals were asked to report their degree of relationship to other participants, their ethnicity, as well as the ethnicity of their parents and grandparents. Only unrelated individuals (at least 3rd degree) and participants who have all of their four grandparents belonging to the same self-identified ethnic group were included in the study.
Ethical Considerations
With the aim of establishing baseline scientific data for the genetic diversity, interrelatedness, and migration history of Philippine ethnic groups, this project was implemented in partnership with the NCCA of the Philippines, pursuant to the provisions of Philippine Republic Act 7356, or the Law Creating the NCCA. In some regional areas, the project was also implemented in coordination with Local Government Units, local non-governmental and cultural organizations, local educational institutions, Indigenous Cultural Community Councils, and/or National Commission on Indigenous Peoples regional offices (see SI Appendix 10 of related publication11). In addition, the processing and analysis of samples aimed at investigating Philippine Genetic Variation was approved by the Regional Ethical Review Board in Uppsala, Sweden (Dnr 2016/103). The research project was implemented in accordance with the regulations of UNESCO International Declaration on Human Genetic Data. The ethical aspect of this research work has also been reviewed and validated by the Uppsala University Board for Investigation of Deviations from Good Research Practice, the Ethics Review Appeals Board of Sweden, and following consultation with independent bioethicists specializing in the region, the editors of Nature,37 and the editors of Proceedings of the National Academy of Sciences of the United States of America .11.
Ethnolinguistic Background of Philippine Negritos
All Philippine Negritos speak a language that is classified under the Malayo-Polynesian (MP) branch of the Austronesian language family. To date, there are ∼30 self-identified Negrito groups who speak a distinct MP language. Many of these languages are classified as severely endangered, while some, such as those of Iraya Agta, Dicamay Agta, and Villa Viciosa Agta, have already been declared extinct. In addition, the majority of these languages are classified as a first order branch within their respective subfamilies, indicating long periods of isolation among Negrito groups. This isolation likely brought about wide-ranging innovations into their languages, thereby distinguishing it from the other languages of neighboring non-Negritos.
In this study, we have included a total of 25 MP-speaking Negrito populations (Figure 1A), who following direct consultation with the respective cultural communities, are labeled in accordance with the group’s own preference to be called themselves. Most of the group names are based on the retention of a reflex of the proto-MP word ∗ʔa(R)ta, which refers to ‘Negrito person’19. The variations in their group names are alterations in the reflex of the medial consonant proto-MP ∗R, for instance the group names of Arta, Agta, Ayta, Alta, and Atta. Other group names are based on locations, such as Dupaningan and Dumagat, where both words retain the initial du as an old locative specifier. Dupaningan comes from the word dupaneng ‘other side or opposite side of the mountain’ and the locative nominalizing suffix -an. Dumagat is an endonym for the Negrito group that lives along the Umiray River, such as the Dumagat Agta of General Nakar in this study. The word Dumagat is derived from the locative specifier du ‘from’ and the place called magat ‘Magat River’, hence Dumagat would refer to the people ‘from Magat river’, indicating local historical migrations which may pertain to the origins of Dumagats from the Magat River basin.
The subsistence patterns of Philippine Negritos that we visited have increasingly shifted toward a mixed foraging and farming and/or urbanized lifestyle. Some engage in local trades and/or in seasonal contractual labor. Of all the Negrito groups we visited, only some individuals from the Agta Maddela and Agta Casiguran cultural communities retained a predominant hunting and gathering way of subsistence. This is consistent with previous observations, where in a span of four decades69, the combination of forest degradation, expansion of agricultural lands, increased accessibility brought about by rapidly expanding infrastructure development, and the inroads of modernized lifestyle have significantly affected the cultural practices of Negritos away from their traditional nomadic hunter-gathering mode of living.
Aside from the change in subsistence patterns, the religious practices of Philippine Negritos have also largely shifted in the advent of increasing development and urbanization, where Christian missionaries gradually extended their reach into the formerly inaccessible areas of Negrito cultural communities. Despite being Christianized, most Negritos still retain some traditional or animistic features in their belief system.
In this study, we grouped the Negrito populations based on their geographic location and the genetic clusters observed following PCA and Admixture analysis (see STAR Methods). The Philippine Negritos are classified into Northern Negritos of Luzon and Southern Negritos outside of Luzon. Additionally, Northern Negritos are classified into Northeast Luzon Negritos, Central Luzon Negritos, Southern Luzon Negritos, and Southeast Luzon Negritos.
Northeast Luzon Negritos
The Northeast Luzon Negritos include Negrito populations found in Cagayan, Isabela, and Quirino provinces of the Cagayan Valley region and the northeast section of Aurora province. The Northeast Luzon Negritos in this study are represented by the cultural communities of Agta Labin of Lal-lo, Cagayan; Agta Dupaningan of Palaui Island, Cagayan; Atta Rizal (also known as Agta Rizal or Atta Faire) of Rizal, Cagayan; Agta Casiguran of Casiguran, Aurora and Nagtipunan, Quirino; Agta Maddela of Maddela, Quirino; and Arta of Nagtipunan, Quirino. Northern Alta cultural communities, who found in Baler, Diteki and San Luis, Aurora, are not included in this study.
Central Luzon Negritos
The Central Luzon Negritos are the Negrito cultural communities of Bataan, Zambales, Pampanga and Tarlac provinces of the Central Luzon region. They are collectively referred to as the Ayta (or Aeta) Negritos, and speak a MP language that is classified under the Sambalic group. The five Ayta cultural communities included this study are the Ayta Ambala, Ayta Magbukon, Ayta Sambal, Ayta Mag-antsi, and Ayta Mag-indi. Ayta Ambala and Ayta Magbukon cultural communities are found in the province of Bataan. Outside of Bataan are the Ayta Sambal cultural communities in Botolan and Cabagan, Zambales; Ayta Mag-indi cultural communities in Floridablanca and Porac, Pampanga and in San Marcelino, Zambales; and Ayta Mag-antsi cultural communities in various towns of Tarlac, Zambales, and Pampanga. The Ayta group included in a recent genomic study18 likely involved the Ayta Mag-antsi and Ayta Mag-indi cultural communities of Pampanga and Tarlac provinces. To our knowledge, this is the first comprehensive genome-wide investigation of the Ayta Magbukon and Ayta Ambala ethnic groups.
Southern Luzon Negritos
The Southern Luzon Negritos include the Negrito populations residing in the Quezon and Rizal provinces of the Southern Luzon region. The Dumagat Agta of General Nakar, Quezon, Remontado Agta of Tanay, Rizal, and the Southern Alta of General Tinio, Nueva Ecija comprise the Southern Luzon Negritos represented in this study.
Southeast Luzon Negritos
The Southeast Luzon Negritos include the Negrito populations of the Bicol region and the Lopez municipality of Quezon province. In this study, the Southeast Luzon Negritos are represented by the cultural communities of Agta Lopez from Lopez, Quezon; Agta Manide from Jose Panganiban, Camarines Norte; Agta Isarog from Ocampo, Camarines Sur; Agta Iriga from Iriga City, Camarines Sur; Agta Iraya from Buhi, Camarines Sur; Agta Matnog from Matnog, Sorsogon; and Agta Bulusan from Bulusan, Sorsogon.
Phenotypically, Agta Bulusan and Agta Matnog appear more similar to the neighboring non-Negrito populations. These observations are supported by our f statistical, qpAdm, and Admixture analyses, where both groups display minimal Negrito and Denisovan ancestry. The levels of Negrito ancestry in both Agta Matnog and Agta Bulusan are similar to the levels found in neighboring non-Negrito Waray and Bicolano ethnic groups.
Southern Negritos
Southern Negritos are self-identified Negrito groups who inhabit the islands south of Luzon, such as the Batak of Palawan, the Ati of Panay and Negros Islands, and the Mamanwa of Mindanao. The Ati group in this study are represented by the cultural communities of Nagpana, Iloilo and Marikudo, Negros Occidental; while the Mamanwa and Batak are represented by the cultural communities of Lake Mainit, Surigao del Norte and Puerto Princesa City, Palawan, respectively.
Geologic Context of the Philippines and the rest of ISEA
The Philippines is an archipelago of 7,641 islands situated in the region of Island Southeast Asia (ISEA). Most of the Philippine Islands lie within the Philippine mobile plate, a complex section that is positioned between the tectonic boundaries of the Eurasian plate and the Philippine Sea plate. In contrast to the presence of dynamic seismic activity and volcanism at the plate margins of the Philippine mobile belt, the southwest section of the Philippines is more stable. This section includes the island of Palawan and those of the Sulu Archipelago. The Palawan micro-block and the Sulu micro-block form part of the northeast protrusions of the largely aseismic Sunda block.
In the course of past glacial cycles, the geography of the Philippines and the rest of ISEA has been modified by regional and global processes. Tectonic changes at the active plate margins, where the Philippine Islands lie, play some role in regional modifications. On a global scale, climate fluctuations influenced the magnitude of glaciation at the extreme latitudes, serving as the main factor determining the rise and fall of sea levels and the associated submergence or exposure of low-lying sections of landmasses70,71. The changes in sea levels, with a magnitude of up to 100-150 m, have their greatest impact on shallow continental shelves such as the large landmass of Sundaland72. These dramatic changes can influence the migration and settlement patterns of human populations, causing isolation and differentiation, bottleneck, or expansion.
The location of past shorelines and the area extent of exposed landmasses was reconstructed using the information from earth rheology parameters that are appropriate for ISEA region, ocean-floor bathymetry using the GEBCO 30 ˝ global gridded data (https://www.gebco.net/data_and_products/), and inferred models of glaciation from 140 kya to the present73. For instance, the lower sea levels throughout the last glacial period have left the shallow continental shelves of the Sunda Block largely exposed11 (Figure S1): the Malay peninsula is interconnected with the present-day major islands of Sumatra, Java, and Borneo, forming one large biogeographical region of Sundaland. Gaining access to the Philippines from Sundaland only requires minimal water crossings, as narrow as ∼3 km during the last glacial maximum ∼21 kya. From northeast Borneo, the accessible pathways are either via the Palawan-Mindoro corridor or the Sulu Archipelago-Zamboanga Peninsula corridor. Given the recent period of deglaciation and the concomitant rise in sea levels, the more tractable pathway starting ∼14.5 kya is via the southern passage of the Sibutu-Basilan Ridge.
Paleolithic Archeology of the Philippines and the Surrounds
The earliest evidence of hominin activity in the Philippines is currently located in the province of in northern Luzon74. The site was dated to ∼700 kya, and is characterized by an assemblage of in situ megafaunal fossils and lithic artifacts. A disarticulated skeleton of a local Rhinoceros species was shown to have evident cutmarks, which were attributed to anthropogenic butchery. Other archeological sites, though with limited stratigraphic information, occur throughout the Philippines; these include rockshelters, caves, and open sites containing stone artifacts (Figure S1). Likely Paleolithic sites include Arubo, Cabarruyan Island, Novaliches, Ille Cave, Bubog rockshelter, and Huluga75,76.
The only Upper Paleolithic fossilized remains of Homo sapiens that have been recovered so far in the Philippines are the osteological materials from the Tabon cave site complex of Palawan Island77. Using Uranium-series (U-series) dating, three sets of human remains were directly dated to 16.5 kya, 31 kya, and 47 kya78. More recently, a third metatarsal hominin bone was recovered from the Callao Cave of Cagayan province, and was U-series dated to 67 kya34. A more comprehensive analysis of the twelve additional hominin elements from the same site revealed a combination of ancestral and derived morphological characteristics. The combination of these traits were not found in any known species of Homo, and hence was designated as a new species of Homo luzonensis35.
Outside of the Philippines, the Deep Skull from Niah Cave, Sarawak, Borneo is the oldest fossil of an anatomically modern human in ISEA79. The Deep Skull was dated to 45–39 kya based on direct U-series dating of the cranial bone and radiocarbon dating of charcoal from nearby sediments. The majority of other Paleolithic human remains discovered in the ISEA region are classified as archaic hominins. In the island of Java, several excavations from the 1890’s to the earlier part of the 20th century put forward the early discoveries of the Java Man, Ngandong Man, Mojorkerto child, and Sangiran 2 individual; all of which were recently characterized as having features belonging to the Homo erectus species80, 81, 82, 83 and dated to as recent as 108 kya84. In 2003, a 1.1 m-tall individual was discovered at Liang Bua on the island of Flores, Indonesia38, which was also defined as a new species of archaic hominins, coined as the Homo floresiensis. The skeletal remains of Homo floresiensis are dated to 50–100 kya, while the artifacts accompanying the skeletal remains are dated to 50–190 kya85, 86, 87.
No fossilized remains of Denisovans was as of yet found in ISEA and Near Oceania. This is despite the fact that the region are inhabited by ethnic groups possessing the highest levels of Denisovan ancestry. Aside from the Altai Denisovan of Siberia, the only other direct evidence on the presence of Denisovans is the Xiahe individual from the Tibetan Plateau30,31. Our current knowledge on Denisovans is largely based on the high-coverage genomic data of Altai Denisovan from Siberia. Given the scant material that were established as definitively Denisovans, we presently carry limited information on the general physical features and phenotypic variability of Denisovans. We are not certain though whether some previously discovered archaic humans in the Asia-Pacific region may in fact be Denisovans or Denisovan-related. These fossilized remains contain some features that were either characterized as archaic or a combination of archaic and modern, which include, among others, the Dali, Maba, Xujiayao and Penghu-1 individuals30,32,33,88 (Figure S1). Moreover, it is not entirely impossible that the recently identified new species of archaic hominins in the ISEA region such as the Homo floresiensis of western Indonesia38 and Homo luzonensis of northern Philippines35, may also be Denisovans or Denisovan-related. To date, no ancient autosomal DNA material has been successfully extracted from any of these archaic fossils discovered outside of Siberia.
Method details
Sample Processing and Genotyping
In addition to the 1,090 Philippine samples genotyped and analyzed in a related study11, we generated additional data from 96 samples, which includes 75 previously unreported, self-identified Negrito individuals. DNA extraction from samples was performed using the prepIT DNA isolation kit (DNA Genotek Inc., Ottawa, ON, Canada). The DNA samples were sent for genotyping, using Infinium Omni2-5Exome-8v1-3 Bead Chip (2,612,357 SNPs), at the SNP&SEQ Technology Platform at Uppsala University. The genotyped samples had an average call rate of 99.85% and reproducibility of 99.99% (54 conflicts in 2,612,141 duplicate tests).
Filtering and Merging of Genotyped Datasets
The newly genotyped data were filtered by removing SNPs with > 10% missingness, indels, duplicates, unmapped and non-autosomal SNPs, and SNPs that were not in Hardy-Weinberg equilibrium. The dataset was then merged with the 1,090-individual Philippine dataset, and family relationships were inferred using the KING v2.2 software, where related individuals are defined by having a kinship coefficient of at least 0.0844 (3rd degree relationship). Only a single individual from each set of related pairs was retained, resulting in the exclusion of 79 individuals, and generating a final Phil_2.35M dataset with 1,107 individuals and 2,352,547 SNPs.
The Phil_2.35M dataset was merged with previously published datasets to generate four additional datasets: i) Phil_1KGP_SGDP_1.92M dataset with 1,691 individuals and 1,924,387 SNPs after merging with a worldwide panel of populations from 1000 Genomes Project43 and Simon’s Genome Diversity Project23; ii) Phil_AsiaPacific_315K dataset with 5,132 individuals and 315k SNPs after merging with the panel 1 above plus Illumina-based Malaysian45, Indonesian46, 47, 48, 49, and other Asian50 panels; iii) Phil_HO_201K dataset with 5402 individuals and 201k SNPs after merging with the Affymetrix Human Origins-based worldwide51,52, East Asian89, MSEA54, Indian57, and Oceanian55,56 panels; and iv) Phil_EGVP dataset after merging with the Estonian Genome Diversity Variation panel44.
Whole Genome Sequencing
Sample library preparation and sequencing were performed at the SNP&SEQ Technology Platform facility in Uppsala. Sequencing libraries were prepared from 100 ng DNA using the TruSeq Nano DNA sample preparation kit (Illumina Inc.), in accordance with the manufacturer’s instructions. Libraries were sequenced on the NovaSeq6000 platform, using the S4 flowcell and v1 sequencing chemistry, to obtain paired end reads of 150 bp length, and an average sequencing depth of at least 30X for each sample. The workflow for data pre-processing and variant calling was implemented following the best-practices recommendations of the Genome Analysis Toolkit (GATK) pipeline66,67,90. In addition to the five newly generated Ayta Magbukon genome sequences, we processed in parallel the publicly available SGDP genome sequences of Papuans, Australians, Bougainville Islander, Kankanaey, Amis, Atayal, Han, Japanese, French, Spanish, Mbuti, Juhoansi, Biaka, Khomani San, and Yoruba.
Reads were mapped onto the human reference genome hs37d5, using the ‘bwa mem’ algorithm of the Burrows–Wheeler Aligner v0.7.1765. Subsequently, duplicates were marked with PicardTools, and base quality scores recalibrated using the BaseRecalibrator tool of GATK v3.8. Variant calling per sample was implemented using HaplotypeCaller68 in GVCF mode, and the per-sample GVCFs were consolidated using the joint genotyping tool, GenotypeGVCFs. Variant filtering was implemented with Variant Quality Score Recalibration using the VariantRecalibrator and ApplyRecalibration tools of GATK v3.8, which generated the multi-sample VCF file containing 24,349,659 variants. The mean sequencing depth per sample for the five newly generated Ayta Magbukon genomes is 37.92x (29.12 – 56.90).
Quantification and statistical analysis
Principal Component Analysis
To determine the genetic relationships of Philippine Negritos to other populations outside of Africa, we performed a Principal component analysis (PCA) using EIGENSOFT on a subset of populations from the Phil_1KGP_SGDP_1.92M dataset with a sample size limit per population of 20 individuals and without outlier removal. The subset includes all Philippine Negritos; Cordillerans, Amis, and Atayal to represent East Asians; Australians, Papuans, and Bougainville Islanders to represent other Australasians, and all West Eurasians from the SGDP dataset (Figure 1B). PC1 is defined by the West Eurasian versus the East Asian clusters, while PC2 is defined by the Australasian versus the East Asian cluster. All Philippine Negritos form a cline between the East Asian and Australasian clusters, indicating admixture between the two sources. We then performed a subset PCA restricted to Philippine Negritos and other populations in the ISEA and Oceania region (Figure S2A). PC1 is defined by the Australopapuan versus the East Asian clusters, while PC2 is defined by the Negrito versus the Australasian cluster. Evidently, Philippine Negritos lie again on the cline between East Asians (represented by Cordillerans) and the least admixed Negrito, Ayta Magbukon, indicating variable magnitude of historical admixture between Negritos and East Asian-related populations. Moreover, Bougainville Islanders lie between the East Asian and Papuan clusters, reflecting the impact of recent admixture between the resident Papuan-related population and the migrating Austronesian-speaking populations in the past 3000 years55,56.
A subset PCA performed on Negrito ethnic groups revealed Ayta Negrito versus Alta/Agta/Arta/Atta/Dumagat Negrito clustering on PC1, and a Northern Negrito versus Southern Negrito clustering on PC2 (Figure 1C). Another subset PCA restricted to Northern Negritos revealed at least three distinct clusters of Luzon Island Negritos; the Central Luzon Negritos (Ayta Ambala, Ayta Magbukon, Ayta Mag-antsi, Ayta Mag-indi and Ayta Sambal), Southeastern Luzon Negritos (Agta Manide, Agta Lopez, Agta Iraya, Agta Iriga and Agta Isarog), and Northeast Luzon Negritos (Agta Dupaningan, Agta Labin, Atta Rizal, Agta Casiguran, Agta Maddela and Arta) (Figure S2B). The Southern Luzon Negritos (Southern Alta, Agta Dumagat and Agta Remontado) form a small cluster right next to the Northeast Luzon Negrito cluster.
Using the lsq project function of EIGENSOFT v7.160,61, a subset PCA was also performed on Africans, East Asians, Philippine Negritos, Australopapuans, Denisovan, Neanderthal, and chimpanzee to identify the genetic affiliation of modern humans to archaic hominins (Figure S2C). The present-day populations was projected onto the plane of first 2 components generated by performing PCA on chimpanzee, Neanderthal, and Denisovan individuals. Among the present-day populations, Australopapuans and Philippine Negritos lie toward the Denisovan axis; where most Ayta Magbukon individuals lie at the extreme edge, which is even farther than that of any Papuan or Australian individuals.
Admixture Analysis
To determine the overall population structure of Philippine Negritos, we performed an unsupervised clustering method implemented in ADMIXTURE20 (Figure S2D). We included in the analysis all Philippine Negrito ethnic groups, and limited the number of individuals to 10 per population. In addition, we included 15 Cordillerans and 15 Papuans to serve as the least admixed reference for East Asians and Australasians, respectively. We ran a total of 50 iterations for each of the 8 clusters using the default settings. Common modes of replicates were identified using CLUMPP62, and the major mode for each K cluster was plotted using Pong v1.463.
Assuming two clusters (K2), the populations are split into black and green components, representing the Australasian and East Asian clusters, where the Papuans represent the least admixed Australasian and the Cordillerans represent the least admixed East Asian (Figure S2D). The Philippine Negritos acquire their own cluster at K3, which is represented by the red component. The Ayta of Central Luzon represent the least admixed Philippine Negritos. From K4 to K8, the Philippine Negritos are further split into the geographic clusters, with the appearance of Central Luzon Negrito cluster at K4 ((light red component, best represented by Ayta Magbukon and Ayta Ambala), the Southern Negrito cluster at K5 (blue component, best represented by the Mamanwa), the Southeast Luzon Negrito cluster at K6 (red component, best represented by the Agta Manide and Agta Lopez), the Southern Luzon Negrito cluster at K7 (yellow component, best represented by the Alta, Agta Dumagat, and Agta Remontado), and the Northeast Luzon Negrito clusters at K8 (red component represented by the Agta Maddela, Agta Casiguran and Arta and light blue component represented by Agta Labin and Atta Rizal). Our analysis reveal at least five distinct population of Philippine Negritos, based on the high support at K = 8 where we find 47 consistent results out of 50 iterations. At K = 8, the Northeast Luzon Negritos are even further subdivided into two clusters, making the total number of distinct Negrito clusters up to six.
East Asian-related Admixture in Negritos
To determine the presence of East Asian admixture among Philippine Negritos, we implemented the test D(Mbuti;EastAsian,Papuan,X); with Balangao Cordilleran utilized as a surrogate for the least admixed East Asian source (Figure S2E). We find that all Philippine Negritos display varied levels of gene flow from East Asians, with Ayta Magbukon presenting as the least admixed. To estimate the levels of East Asian and Australasian-related ancestries in Negritos, we implemented the qpAdm22 tool of Admixtools v5.0 software package. We first prepared a new dataset with reference ancient samples, by merging Phil_1KGP_SGDP_1.92M dataset with published ancient DNA data to produce the Phil_1KGP_SGDP_Ancient_1.92M dataset, which was then haploidized and filtered to keep transversion sites only, producing the Phil_1KGP_SGDP_Ancient_Transv_317K dataset with 317,220 SNPs. The ‘left’ populations include Balangao Cordilleran of the Philippines as the surrogate for the least admixed East Asian source and Papuan as the surrogate for the least admixed Australasian source. The outgroup or ‘right’ populations include Juhoansi, Mbuti, Mota, Loschbour, Anzick1, Ust’Ishim, Sumiduouro8, and Karitiana. Overall, there is a wide variation in the magnitude of admixture with East Asians. For instance, Ayta Magbukon and Ayta Ambala presented as the least admixed Negrito populations, with an East Asian-related ancestry of only ∼10%–30% (Figure 1D). On the other hand, both Agta Bulusan and Agta Matnog have the highest levels of admixture, possessing ∼79%–85% East Asian-related ancestry. To estimate the date of admixture, we utilized MALDER91, which applies a weighted linkage disequilibrium (LD) statistic-based method and allows detection of multiple admixture events. Philippine non-Negritos, Amis, Atayal, mainland East Asians, and Australopapuans were set as putative source populations, while all Philippine Negrito groups were set as target populations. The mean date of admixture between Negritos and East Asians among all ethnic groups within the Philippines was estimated to ∼2,281 years (95% CI: 2,083 – 2,523 years).
Divergence Time between Negritos and Papuans
To estimate the divergence time between Philippine Negritos versus Papuans, we utilized the ‘TT’ method, which is based on computing sample configurations in a population divergence model25,92. This approach estimates the number of generations since a population divergence for a pair of individuals (or populations), and the method produces direct estimates in generations that are unaffected by the effective population size of the population of each of the individuals in the comparison. We utilized the publicly available genome sequence data on Papuans23 and our newly generated sequence data on Ayta Magbukon. The estimated divergence time between Ayta Magbukon Negrito and Papuans is ∼85 kya (95% CI: 76 – 95 kya). This unusually old divergence time may be attributed to the deeply diverging Denisovan ancestries found in both Papuans and Ayta Magbukon. To correct the effect of archaic introgression, we filtered out all the archaic sequences (that were identified using the S’ method) in both Ayta Magbukon and Papuans. The estimated divergence time between Ayta Magbukon Negritos and Papuans, using the filtered data, is 53 kya (95% CI: 41-64 kya). This estimated date falls shortly after the divergence between Australasians and East Asians/West Eurasians, which was previously estimated to ∼58 kya (95% CI: 51 – 72 kya)24.
D and f4-ratio statistics on genotyped data
All Philippine ethnic groups carry Neanderthal ancestry that is similar to the levels found in all worldwide populations outside of Africa (Figure S3C). This was determined using the test f4-ratio (Chimp;AltaiNeanderthal,Mbuti,X) / (Chimp;AltaiNeanderthal,Mbuti, VindijaNeanderthal). The analysis was restricted to the sites that are non-polymorphic or homozygous ancestral in the Denisovan genome (to correct for inflation of the estimate due to the presence of Denisovan ancestry in some populations). The comparable levels of Neanderthal ancestry in all populations are in line with previous findings indicating that Neanderthal introgression is likely to have been a single event that occurred when anatomically modern humans migrated out of Africa2,3. However, an alternative model of multiple episodes of Neanderthal gene flow into European and East Asians has been suggested93, which may explain the asymmetric pattern of Neanderthal allele frequencies among these populations.
We performed the test D(Mbuti;Denisovan,Han,X) or D(Mbuti;Denisovan,Dai,X) (Figures S5A and S5B) to investigate whether any X population received any gene flow from Denisovans, using East Asians as the reference population for comparison (East Asians are known to have some minimal amount of Denisovan ancestry, but which is not significant by standard D tests)12,15. Almost all ethnic groups of the Philippines exhibited significant levels of Denisovan ancestry with the highest levels found among Negritos. The results are consistent when we use the tests D(Mbuti;Denisovan,French/Norwegian,X), where we use Europeans, who are known to lack Denisovan ancestry, as the alternative reference populations for comparison (Figures S5C and S5D).
Among Negritos, Ayta Magbukon displayed the highest estimated levels of Denisovan ancestry in a worldwide panel of populations, with Ayta Magbukon exhibiting higher estimates than Papuans, Australians, or any Oceanian populations (Figures 2A, 2B, and S5A–S5D; Data S1A). These findings are consistent when we ran the D tests with different outgroups (Chimp, Juhoansi, or YRI), when we ran with Phil_HO or Phil_AsiaPacific_315K datasets, or with our Philippine dataset merged with the Estonian Genome Diversity Project dataset, or when we correct for Neanderthal ancestry in target populations with f4-ratio (Mbuti;Neanderthal,EastAsian,X:Mbuti; Neanderthal,EastAsian,Denisova) (Figures S5F–S5L; Data S1B and S1C). The findings are also consistent when we restrict the Phil_1KGP_SGDP_1.92M dataset to a panel of SNPs that are polymorphic only or ascertained for Europeans or Africans or to the H3frica SNP panel (Figures S5M–S5O). Moreover, when we test individuals instead of populations, we find that most Ayta Magbukon Negritos possess the highest level of Denisovan ancestry in the world (Figure S5L). Some Ayta Ambala, Agta Lopez and Agta Manide individuals also exhibit higher Denisovan ancestry than any other Papuan individual (Figure S5L).
RFMix-based masking of East Asian ancestry
Considering that Philippine Negritos are admixed with Cordilleran-related ancestry (Figure 1D), we implemented RFMix to produce a new dataset where we removed the Cordilleran-related alleles (and hence retain only the Negrito-related genome-regions) among Philippine Negritos. RFMix64 was implemented on a shapeit-phased94,95 subset of the Phil_1KGP_SGDP_1.92M dataset using the flag -fb 1 -e 1 -n 10 -u 1, with 15 Cordillerans as a reference for least admixed East Asian and 15 Papuans as a reference for least admixed Australasian. We find that Northern Negritos of the Philippines consistently exhibit higher Denisovan ancestry, with non-overlapping confidence intervals, relative to Australians or Papuans or to a southern Negrito, Mamanwa (Figure S5E). There is no change in the levels of Denisovan ancestry in Papuans and Australians before and after masking (the data points are overlapping), which indicates that the increase in Denisovan ancestry among Philippine Negritos is solely related to removal of confounding East Asian admixture.
We also performed the test D(Mbuti;Denisovan,Negrito,Papuan/Australian), both in the unmasked and masked versions of the dataset, with matched sample size between populations and inclusion of least admixed individuals (Figures 3B and 3C). We consistently find that Ayta Magbukon exhibit higher Denisovan ancestry relative to Papuans or Australians, and is more pronounced when we mask away East Asian-related ancestry.
Correlation analysis
A significant negative correlation is observed when we plot the proportion of East Asian ancestry versus Denisovan ancestry (R2: 0.8796, p value: < 0.0001) (Figure S4A). Likewise, a positive correlation between Negrito ancestry and Denisovan ancestry is observed when we plot the proportion of Australasian ancestry using D(Mbuti;Papuan,EastAsian,X) versus D(Mbuti;Denisovan,EastAsian,X) or D(Mbuti; Australian,EastAsian,X) versus D(Mbuti;Denisovan,EastAsian,X) (Figures S4B–S4E). Separate from the slope of Papuan-related ancestry, two additional slopes can be observed for Northern Negritos and Southern Negritos (Figures S4B and S4E). The linear clusters of Northern Negritos and Southern Negritos (together with Manobo-related populations with high Southern Negrito ancestry) form their distinct respective slopes that are also positively correlated with Denisovan ancestry.
All populations with Papuan-related ancestry (that is distinctively post the Australopapuan divergence), such as Indonesians and Oceanians, form a distinct and separate slope, separate from the slopes formed by Negritos (Figures S4B–S4E). As expected, among the Philippine ethnic groups, Sangil is a distinct outlier from the slope formed by Northern Negritos or Southern Negritos (Figures S4B–S4E). Sangil aligns more with the slope for eastern Indonesians, Bougainville islander, and Papuans. This suggests that populations with more Papuan-like ancestry exhibit a Denisovan ancestry that is distinct from the Denisovan ancestry found in Northern or Southern Negritos. Altogether, these findings, that Denisovan ancestry in Negritos is not associated with Papuan-related ancestry, indicates that the Denisovan introgression event in Negritos is independent from that of Papuans.
Sequencing Analyses of Ayta Magbukon Negritos
We have sequenced and analyzed the genomes of five Ayta Magbukon individuals and present the results in Figure 4. In order to omit potential genome-sequence-processing biases, we started from the raw sequence data (FASTQs) for all individuals (including the comparative human sequence data), and processed all the sequence data in one batch, using standard mapping and filtering criteria (see Method details section for a full description).
D and f4-ratio statistics on sequenced data
Denisovan ancestry was estimated using the test D(Outgroup;Denisovan,EastAsian,X) or the f4-ratio statistic f4(Outgroup; Neanderthal,EastAsian,X) / f4(Outgroup;Neanderthal, EastAsian,Denisovan), with Altai and Vindija individuals representing the Neanderthals, and Kankanaey Cordilleran representing East Asians. Ayta Magbukon consistently demonstrate to have the highest level of Denisovan ancestry (Figure 4A; Data S2A and S2B). To assess whether Ayta Magbukon possess higher Denisovan ancestry than highland Papuans or Australians, we implemented the test D(Outgroup;Denisovan,Papuan/Australian,AytaMagbukon), with Chimp, Juhoansi, Biaka, Khomani San, or Yoruba used as outgroups. We confirm that the Ayta Magbukon possess higher levels of Denisovan ancestry relative to that of Australians or Papuans, with D values of 0.0168 - 0.0256 and all Z score of > 3 ((Figures S6A and S6B; Data S2C). To assess the percentage difference of Denisovan ancestry in Ayta Magbukon versus Australians or Papuans, we utilized the statistic D(Mbuti,Denisovan,EastAsian,AytaMagbukon) / D(Mbuti,Denisovan,East Asian,Papuan/Australian). We find that the Ayta Magbukon possess 29%–39% more Denisovan ancestry than do Australians and Papuans (Figure 4B; Data S2A).
S’ method
For identifying archaic introgressed sequences without the use of reference archaic genomes, we utilized the recently developed S-prime (S’) method15. The method is designed to detect divergent haplotypes that are in strong LD and that are of very low frequency or absent in outgroup African population (who are known not to harbor archaic ancestry). The S’ method was demonstrated to have better power relative to other archaic reference-free methods, and is appropriate for large-scale genomic data15. For our analysis, we utilized 15 SGDP Sub-Saharan African individuals as the outgroup population (which includes the Juhoansi, Mbuti, Khomani, Yoruba, and Biaka individuals), and 5 newly sequenced Ayta Magbukon and 15 SGDP Papuan individuals as the test populations.
Following retrieval of archaic sequences using S’, we leveraged the sequences to the reference Altai Denisovan and Vindija Neanderthal genomes. First, using the masks downloaded from http://cdna.eva.mpg.de/neandertal/, we filtered out the following sites from the archaic genomes: indels, sites within tandem repeats, sites with poor mapability, depth of coverage of < 10x, or mapping quality of < 25. For each filtered site, we compared the genomes of the test individuals to the reference Denisovan and Neanderthal genomes, and report if they match or mismatch. The match rate per archaic segment was then estimated by dividing the total number of matches over the total number of sites compared. Contour densities of the Denisovan versus Neanderthal match rates were plotted using the kde2d function of the R v3.3.1 MASS package. We find that both Ayta Magbukon and Papuans display similar patterns of affinity to the reference Denisovan and Neanderthal genomes, moderate affinity or match rate of ∼50% to the Altai Denisovan and high affinity or match rate of ∼80% to the Vindija Neanderthal (Figures 5A and 5B).
Following a previous approach15, we identify the segments as putatively Denisovan with the following criteria: presence of at least 30 introgressed alleles that can be compared with the reference archaic genomes; less than 30% match rate to the Vindija/Altai Neanderthal genome, and greater than 30% match rate to the Denisovan genome. Based on this criteria, we find that the Ayta Magbukon exhibit significantly higher magnitude of Denisovan ancestry relative to that of Papuans. The average amount of Denisovan sequence detected in Ayta Magbukon and Papuans are 100.38 Mb (95% CI: 89.66 – 111.11 Mb) and 79.35 Mb (95% CI: 76.89 – 81.79 Mb), respectively (p = 3.0x10-4); a difference of ∼27%.
To classify the introgressed haplotypes into Denisovan versus Neanderthal source, we applied the criteria used in a previous work15. We classified segments as putatively Neanderthal with the following conditions: presence of at least at least 30 introgressed alleles that can be compared with the reference archaic genomes; greater than 60% match rate to the Vindija Neanderthal genome; and less than 40% match rate to the Denisovan genome. We then identified segments as putatively Denisovan with the following conditions: presence of at least at least 30 introgressed alleles that can be compared with the reference archaic genomes; greater than 40% match rate to the Denisovan genome; and less than 30% match rate to the Vindija Neanderthal genome. Any segment that indistinguishable from Neanderthal or Denisovan are categorized as ambiguous.
The summary of archaic sequences per individual is presented in Figure S6C. Using the aforementioned criteria that distinguishes between different archaic sources, Ayta Magbukon and Papuans display similar amounts of Neanderthal sequences [(45.35 Mb (95% CI: 41.71 – 48.99 Mb) versus 46.47 Mb (95%CI: 44.96 – 47.97 Mb)] and ambiguous sequences [(7.52 Mb (95% CI: 5.47 – 9.57 Mb) versus 6.92 Mb (95%CI: 6.25 – 7.60 Mb)]. Consistently, we find Ayta Magbukon to present with significantly higher levels of Denisovan ancestry relative to Papuans (Figure 4D). The average amount of Denisovan sequences detected in Ayta Magbukon and Papuans are 51.94 Mb (95% CI: 44.62.66 – 59.25 Mb) and 41.96 Mb (95% CI: 39.54 – 44.37 Mb), respectively (p = 3.7x10-3); a difference of ∼24%.
Correlation Between Denisovan and Australasian ancestry
We plotted the correlation between Denisovan and Australasian-related ancestry among Negrito and Papuan-related groups. Denisovan ancestry was estimated using f4-ratio statistics, while Australasian-related ancestry was estimated using qpAdm. Consistent with the correlation analysis presented earlier, Negritos fall outside the slope formed by the Papuan-related populations. Furthermore, an important aspect of this study is the availability of Negritos with varying levels of admixture with East Asian-related populations. This allowed us for the first time to plot and extrapolate the level of Denisovan ancestry in an ‘unadmixed’ ancestral Negrito population (D = 4.47), which demonstrably have at least 46% more Denisovan ancestry than the highlander Papuans (Figure 3D). In addition, our analysis also demonstrated for the first time that Negritos have significantly higher levels of ‘Denisovan richness’ in their Australasian ancestry than Oceanians, which we define as the ratio of Denisovan ancestry over Australasian ancestry (Figure 3E).
Altogether, the evidence presented here, that Denisovan ancestry in Negritos is not associated with Papuan-related ancestry and that Negritos have higher levels of Denisovan richness, provide support to a demographic model where Negritos receive an additional and/or an independent Denisovan introgression event.
Evaluating Recent Admixture in Philippine Negritos versus Papuans
The lower levels of Denisovan ancestry in Papuans could be explained by dilution via recent admixture with non-Papuan populations with no detectable or low levels of Denisovan ancestry. To determine the presence of gene flow from populations with little or no Denisovan ancestry into Papuans or Philippine Negritos, we implemented the test D(Outgroup;X,Papuan, Philippine Negrito); with Balangao, Han, or Dai as a surrogate for East Asian; Even and Yakut as a surrogate for Siberian or North Asian; Karitiana or Mixe as a surrogate for Native American; Irula as a surrogate for South Asian; French or Sardinian as a surrogate for European; and Luhya or Mbuti as a surrogate for African (Figures S7A–S7L). Based on the various combination of D tests listed above, we do not find any population that shares more alleles with Papuans relative to Negritos (Figures S7A–S7L). In fact, it is the Negritos that consistently displayed evidence for recent admixture, given the high levels of allele sharing between Negritos and East Asians, relative to Papuans.
Admixture Graph Analysis
We constructed admixture graphs using the qpGraph tool of the AdmixTools v5.0 software package to determine which population-history model best fits the data that accounts for the number of Denisovan introgression events into Philippine Negritos and Papuans. Using the Phil_1KGP_SGDP_Ancient_Transv_317K dataset, we incorporarted in the model the chimpanzee as an outgroup, French to represent West Eurasians, Balangao Cordilleran and Liangdao 2 to represent East Asians, and Australian, Papuan, and Ayta Magbukon to represent Australasians (Figures 6A–6C). The qpGraph tool was ran with the following parameters: outpop (NULL), blgsize (0.05), diag (0.0001), hires (YES), and lsqmode (YES). A model is rejected when the worst fitting f statistical test has a significant Z score of > | 3 |. A model with a single Denisovan introgression event shared between Philippine Negritos and Australopapuans is rejected (worst-fitting f score Z = −3.6). Models that allow independent Denisovan introgression events into Negritos and Papuans are not rejected (worst-fitting f score of Z = −1.18 for both models), providing support that Philippine Negritos likely admixed with Denisovans independently from the Denisovan introgression event that occurred in Australopapuans.
Simulation of Demographic Scenarios
The coalescent simulator msprime29 was utilized to evaluate whether a null model (Figure 7F) of single Denisovan introgression event into the common ancestor of Ayta Negritos and Papuans or alternative models (Figures 7K, 7P, 7U, and 7Z) of multiple Denisovan admixture events are consistent with our empirical data: higher levels of Denisovan ancestry in Ayta Negritos than Papuans (Figures 7A and 7B), similar Denisovan tract lengths between Ayta Negritos and Papuans (Figure 7C), higher ratio of Denisovan over Australasian ancestry in Ayta Negritos relative to Papuans (Figure 7D), and Ayta Negritos is an outlier to the Papuan-Denisovan slope formed after correlating Denisovan and Australasian ancestry (Figure 7E). For each of the models shown in Figures 7F, 7K, 7P, 7U, and 7Z, 500 replicate simulations were performed, with 10 samples of 100M sequence taken from each of 8 populations. The parameters fixed across all models are shown in Table S1, and parameters that differed between models are shown in Table S2. All scripts used to generate data are available at https://github.com/jammc313/Philippine_Denisovan_Ancestry.
The null model (Figure 7F) includes an admixture event coming from an unsampled Denisovan population that diverged from the ‘Altai’ Denisovan 300 kya, into the shared ancestral population of Papuan and Ayta (4%, 48 kya). An alternative model (Alt1) follows an identical setup (Figure 7K), plus an additional admixture event (2%) coming from a distinct and unsampled Denisovan population, (also diverged from the Altai Denisovan 300 kya), into the (ancestors of) an Ayta population (40 kya), resulting in a total of 4% + 2% = 6% Denisovan admixture in the (ancestral) Ayta population. A second alternative model (Alt2) includes Denisovan admixture events private to both Papuans (4%, 45kya) and Ayta (6%, 35 kya) (Figure 7P). Again, the sources of Denisovan admixture are distinct Denisovan populations both diverged from the ‘Altai’ Denisovan 300 kya. A third alternative model (alt3) is similar to alt2, but with the Denisovan introgression event into Papuans occurring more recently at 25 kya (Figure 7U). The last alternative model (alt4) is similar to alt2, but with the independent Denisovan introgression events into Ayta and Papuans occurring around the same at 25 kya (Figure 7Z). The parameters of all models were kept as general as possible, with population sizes, admixture dates, and admixture proportions all chosen in accordance with the general consensus from previous studies24,27,28.
For all the simulations, we plotted the levels of Denisovan ancestry based on f4-ratio (Africa;Neanderthal,EastAsian,X)/(Africa;Neanderthal,EastAsian,Denisovan), mean tract lengths of Denisovan ancestry, and the ratio of Denisovan ancestry over Australasian ancestry. All pairwise comparisons were tested for significance using the Mann-Whitney U test with Holm-Bonferroni correction. The correlation between Denisovan and Australasian ancestry were also plotted following additional simulations with incremental 10% East Asian admixture into Ayta and Papuans.
The simulated models were leveraged against the empirical data using the following four parameters: level of Denisovan ancestry, mean Denisovan tract length, ratio of Denisovan ancestry over Australasian ancestry, and correlation between Denisovan ancestry and Australasian ancestry. Except for mean Denisovan tract length, the null model produced patterns of Denisovan ancestry that are not consistent with the observed data (Figures S7F–S7J). On the other hand, all the alternative models consistently replicate three pieces of empirical evidence: i) higher level of Denisovan ancestry in Ayta Negritos than Papuans, ii) higher ratio of Denisovan over Australasian ancestry in Ayta Negritos than Papuans, and iii) Ayta Negritos falls outside the slope formed by Papuan-related groups when plotting the correlation between Denisovan ancestry and Australasian ancestry (Figures S7K–S7Ad). Among the alternative models, only Alt4 model produced patterns of mean Denisovan tract length that is approximating similar levels in Ayta Negritos and Papuans, supporting a demographic scenario where the timing of the separate Denisovan introgression event into Ayta Negritos and Papuans happened around the same time, (Figures 7C and 7Ab).
Acknowledgments
We are grateful to all of the volunteers and indigenous cultural communities who participated in this research. The project is recognized by and implemented in partnership with the National Commission for Culture and the Arts (NCCA) of the Philippines, in accordance with the provisions of Philippine Republic Act 7356 or the Law Creating the NCCA. The project was reviewed and approved by the Regional Ethical Review Board in Uppsala, Sweden (Dnr 2016/103). We are thankful to Felipe Mendoza de Leon, Jr., NCCA chairperson 2010–2016, for his support. We also thank the regional offices of the National Commission for Indigenous Peoples (NCIP), Local Government Units, and/or regional cultural organizations for facilitating fieldwork. See SI Appendix 10 of Larena et al.11 for the complete list of acknowledgments. This work was supported by the Swedish Research Council grant no. 642-2013-8019 to M.J. and 2020-04789 to M.L. and the Knut and Alice Wallenberg Foundation grant to M.J. The computations were performed at the Swedish National Infrastructure for Computing (SNIC-UPPMAX).
Author contributions
M.L., C.S., and M.J. designed research; M.L., C.E., R. Reveche, O.C., J.-Y.H., K.P.H., D.G., J.R., F.P.A., V.M., L.S.A., A. Manera, C.T., L.J., G.S., R.M.-T., M.S.L., R.D.V., A.L., R.L.S., E. Marte., P.M., A. Morales, I.J., R. Reyes, B.B., E.B., J.C.S., M.J.T.K., A.A., R.B.C.-A., E. Molanida, J.-H.L., and J.T. conducted field work, performed community preparation and validation, and provided samples; M.L., F.S.-Q., J.M., C.B., L.G., M.V., H.E., S.Y.W.H., L.R., K.L., H.M., C.S., P.E., and M.J. performed research and analysis; M.L., F.S.-Q., and J.M. analyzed genetic data; and M.L. and M.J. wrote the paper with input from all authors.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We worked to ensure ethnic or other types of diversity in the recruitment of human subjects. One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. One or more of the authors of this paper self-identifies as a member of the LGBTQ+ community. The author list of this paper includes contributors from the location where the research was conducted who participated in the data collection, design, analysis, and/or interpretation of the work.
Published: August 12, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cub.2021.07.022.
Contributor Information
Maximilian Larena, Email: maximilian.larena@ebc.uu.se.
Mattias Jakobsson, Email: mattias.jakobsson@ebc.uu.se.
Supplemental information
A) Denisovan ancestry estimation using D statistics. B) Denisovan ancestry estimation using f4-ratio statistics. C) f4-ratio statistics performed on 5 least admixed individuals per population.
A) Denisovan ancestry estimation using D statistics, and percentage difference Denisovan ancestry in Ayta Magbukun and AustraloPapuans. B) Denisovan ancestry estimation using f4-ratio statistics, and percentage difference Denisovan ancestry in Ayta Magbukon and AustraloPapuans. C) Direct comparison of Denisovan ancestry in Ayta Magbukon versus AustraloPapuans using D-statistics.
Data and code availability
All requests to access the genotype and sequence data presented in this work are managed through the Data Access Committee of the official data repositories at the European Genome-phenome Archive (https://ega-archive.org/ega/home; Database: EGAS00001005408 and EGAS00001005407). The data generated in this study can be obtained for the purposes of academic research on genetic origins and diversity following approval by the Data Access Committee. All simulation scripts used in this study are available at https://github.com/jammc313/Philippine_Denisovan_Ancestry.
References
- 1.Green R.E., Krause J., Briggs A.W., Maricic T., Stenzel U., Kircher M., Patterson N., Li H., Zhai W., Fritz M.H., et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prüfer K., de Filippo C., Grote S., Mafessoni F., Korlević P., Hajdinjak M., Vernot B., Skov L., Hsieh P., Peyrégne S., et al. A high-coverage Neandertal genome from Vindija Cave in Croatia. Science. 2017;358:655–658. doi: 10.1126/science.aao1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyer M., Kircher M., Gansauge M.T., Li H., Racimo F., Mallick S., Schraiber J.G., Jay F., Prüfer K., de Filippo C., et al. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338:222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reich D., Patterson N., Kircher M., Delfin F., Nandineni M.R., Pugach I., Ko A.M., Ko Y.C., Jinam T.A., Phipps M.E., et al. Denisova admixture and the first modern human dispersals into Southeast Asia and Oceania. Am. J. Hum. Genet. 2011;89:516–528. doi: 10.1016/j.ajhg.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mondal M., Casals F., Xu T., Dall’Olio G.M., Pybus M., Netea M.G., Comas D., Laayouni H., Li Q., Majumder P.P., et al. Genomic analysis of Andamanese provides insights into ancient human migration into Asia and adaptation. Nat. Genet. 2016;48:1066–1070. doi: 10.1038/ng.3621. [DOI] [PubMed] [Google Scholar]
- 6.Hammer M.F., Woerner A.E., Mendez F.L., Watkins J.C., Wall J.D. Genetic evidence for archaic admixture in Africa. Proc. Natl. Acad. Sci. USA. 2011;108:15123–15128. doi: 10.1073/pnas.1109300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sankararaman S., Mallick S., Patterson N., Reich D. The combined landscape of Denisovan and Neanderthal ancestry in present-day humans. Curr. Biol. 2016;26:1241–1247. doi: 10.1016/j.cub.2016.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vernot B., Tucci S., Kelso J., Schraiber J.G., Wolf A.B., Gittelman R.M., Dannemann M., Grote S., McCoy R.C., Norton H., et al. Excavating Neandertal and Denisovan DNA from the genomes of Melanesian individuals. Science. 2016;352:235–239. doi: 10.1126/science.aad9416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vernot B., Akey J.M. Resurrecting surviving Neandertal lineages from modern human genomes. Science. 2014;343:1017–1021. doi: 10.1126/science.1245938. [DOI] [PubMed] [Google Scholar]
- 10.Reich D., Green R.E., Kircher M., Krause J., Patterson N., Durand E.Y., Viola B., Briggs A.W., Stenzel U., Johnson P.L., et al. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468:1053–1060. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larena M., Sanchez-Quinto F., Sjödin P., McKenna J., Ebeo C., Reyes R., Casel O., Huang J.Y., Hagada K.P., Guilay D., et al. Multiple migrations to the Philippines during the last 50,000 years. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2026132118. e2026132118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skoglund P., Jakobsson M. Archaic human ancestry in East Asia. Proc. Natl. Acad. Sci. USA. 2011;108:18301–18306. doi: 10.1073/pnas.1108181108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qin P., Stoneking M. Denisovan ancestry in East Eurasian and Native American populations. Mol. Biol. Evol. 2015;32:2665–2674. doi: 10.1093/molbev/msv141. [DOI] [PubMed] [Google Scholar]
- 14.Prüfer K., Racimo F., Patterson N., Jay F., Sankararaman S., Sawyer S., Heinze A., Renaud G., Sudmant P.H., de Filippo C., et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Browning S.R., Browning B.L., Zhou Y., Tucci S., Akey J.M. Analysis of human sequence data reveals two pulses of archaic Denisovan admixture. Cell. 2018;173:53–61.e9. doi: 10.1016/j.cell.2018.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacobs G.S., Hudjashov G., Saag L., Kusuma P., Darusallam C.C., Lawson D.J., Mondal M., Pagani L., Ricaut F.X., Stoneking M., et al. Multiple deeply divergent Denisovan ancestries in Papuans. Cell. 2019;177:1010–1021.e32. doi: 10.1016/j.cell.2019.02.035. [DOI] [PubMed] [Google Scholar]
- 17.Jinam T.A., Phipps M.E., Aghakhanian F., Majumder P.P., Datar F., Stoneking M., Sawai H., Nishida N., Tokunaga K., Kawamura S., et al. Discerning the origins of the Negritos, first Sundaland people: deep divergence and archaic admixture. Genome Biol. Evol. 2017;9:2013–2022. doi: 10.1093/gbe/evx118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.GenomeAsia 100K Consortium The GenomeAsia 100K Project enables genetic discoveries across Asia. Nature. 2019;576:106–111. doi: 10.1038/s41586-019-1793-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reid L.A. Who are the Philippine negritos? Evidence from language. Hum. Biol. 2013;85:329–358. doi: 10.3378/027.085.0316. [DOI] [PubMed] [Google Scholar]
- 20.Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loh P.-R., Lipson M., Patterson N., Moorjani P., Pickrell J.K., Reich D., Berger B. Inferring admixture histories of human populations using linkage disequilibrium. Genetics. 2013;193:1233–1254. doi: 10.1534/genetics.112.147330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harney É., Patterson N., Reich D., Wakeley J. Assessing the performance of qpAdm: a statistical tool for studying population admixture. Genetics. 2021;217:iyaa045. doi: 10.1093/genetics/iyaa045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mallick S., Li H., Lipson M., Mathieson I., Gymrek M., Racimo F., Zhao M., Chennagiri N., Nordenfelt S., Tandon A., et al. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature. 2016;538:201–206. doi: 10.1038/nature18964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malaspinas A.S., Westaway M.C., Muller C., Sousa V.C., Lao O., Alves I., Bergström A., Athanasiadis G., Cheng J.Y., Crawford J.E., et al. A genomic history of Aboriginal Australia. Nature. 2016;538:207–214. doi: 10.1038/nature18299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sjödin P., McKenna J., Jakobsson M. Estimating divergence times from DNA sequences. Genetics. 2021;217:iyab008. doi: 10.1093/genetics/iyab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rogers A.R., Bohlender R.J. Bias in estimators of archaic admixture. Theor. Popul. Biol. 2015;100C:63–78. doi: 10.1016/j.tpb.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 27.Bergström A., McCarthy S.A., Hui R., Almarri M.A., Ayub Q., Danecek P., Chen Y., Felkel S., Hallast P., Kamm J., et al. Insights into human genetic variation and population history from 929 diverse genomes. Science. 2020;367:eaay5012. doi: 10.1126/science.aay5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bergström A., Oppenheimer S.J., Mentzer A.J., Auckland K., Robson K., Attenborough R., Alpers M.P., Koki G., Pomat W., Siba P., et al. A Neolithic expansion, but strong genetic structure, in the independent history of New Guinea. Science. 2017;357:1160–1163. doi: 10.1126/science.aan3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelleher J., Etheridge A.M., McVean G. Efficient coalescent simulation and genealogical analysis for large sample sizes. PLoS Comput. Biol. 2016;12:e1004842. doi: 10.1371/journal.pcbi.1004842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen F., Welker F., Shen C.-C., Bailey S.E., Bergmann I., Davis S., Xia H., Wang H., Fischer R., Freidline S.E., et al. A late Middle Pleistocene Denisovan mandible from the Tibetan Plateau. Nature. 2019;569:409–412. doi: 10.1038/s41586-019-1139-x. [DOI] [PubMed] [Google Scholar]
- 31.Zhang D., Xia H., Chen F., Li B., Slon V., Cheng T., Yang R., Jacobs Z., Dai Q., Massilani D., et al. Denisovan DNA in Late Pleistocene sediments from Baishiya Karst Cave on the Tibetan Plateau. Science. 2020;370:584–587. doi: 10.1126/science.abb6320. [DOI] [PubMed] [Google Scholar]
- 32.Dennell R., Martinón-Torres M., de Castro J.-M.B., Xing G. A demographic history of late Pleistocene China. Quat. Int. 2020;559:4–13. [Google Scholar]
- 33.Ao H., Liu C.-R., Roberts A.P., Zhang P., Xu X. An updated age for the Xujiayao hominin from the Nihewan Basin, North China: implications for Middle Pleistocene human evolution in East Asia. J. Hum. Evol. 2017;106:54–65. doi: 10.1016/j.jhevol.2017.01.014. [DOI] [PubMed] [Google Scholar]
- 34.Mijares A.S., Détroit F., Piper P., Grün R., Bellwood P., Aubert M., Champion G., Cuevas N., De Leon A., Dizon E. New evidence for a 67,000-year-old human presence at Callao Cave, Luzon, Philippines. J. Hum. Evol. 2010;59:123–132. doi: 10.1016/j.jhevol.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 35.Détroit F., Mijares A.S., Corny J., Daver G., Zanolli C., Dizon E., Robles E., Grün R., Piper P.J. A new species of Homo from the Late Pleistocene of the Philippines. Nature. 2019;568:181–186. doi: 10.1038/s41586-019-1067-9. [DOI] [PubMed] [Google Scholar]
- 36.Teixeira J.C., Jacobs G.S., Stringer C., Tuke J., Hudjashov G., Purnomo G.A., Sudoyo H., Cox M.P., Tobler R., Turney C.S.M., et al. Widespread Denisovan ancestry in Island Southeast Asia but no evidence of substantial super-archaic hominin admixture. Nat. Ecol. Evol. 2021;5:616–624. doi: 10.1038/s41559-021-01408-0. [DOI] [PubMed] [Google Scholar]
- 37.Choin J., Mendoza-Revilla J., Arauna L.R., Cuadros-Espinoza S., Cassar O., Larena M., Ko A.M., Harmant C., Laurent R., Verdu P., et al. Genomic insights into population history and biological adaptation in Oceania. Nature. 2021;592:583–589. doi: 10.1038/s41586-021-03236-5. [DOI] [PubMed] [Google Scholar]
- 38.Brown P., Sutikna T., Morwood M.J., Soejono R.P., Jatmiko, Saptomo E.W., Due R.A. A new small-bodied hominin from the Late Pleistocene of Flores, Indonesia. Nature. 2004;431:1055–1061. doi: 10.1038/nature02999. [DOI] [PubMed] [Google Scholar]
- 39.Massilani D., Skov L., Hajdinjak M., Gunchinsuren B., Tseveendorj D., Yi S., Lee J., Nagel S., Nickel B., Devièse T., et al. Denisovan ancestry and population history of early East Asians. Science. 2020;370:579–583. doi: 10.1126/science.abc1166. [DOI] [PubMed] [Google Scholar]
- 40.Huerta-Sánchez E., Jin X., Asan, Bianba Z., Peter B.M., Vinckenbosch N., Liang Y., Yi X., He M., Somel M., et al. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature. 2014;512:194–197. doi: 10.1038/nature13408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gittelman R.M., Schraiber J.G., Vernot B., Mikacenic C., Wurfel M.M., Akey J.M. Archaic hominin admixture facilitated adaptation to out-of-Africa environments. Curr. Biol. 2016;26:3375–3382. doi: 10.1016/j.cub.2016.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tucci S., Vohr S.H., McCoy R.C., Vernot B., Robinson M.R., Barbieri C., Nelson B.J., Fu W., Purnomo G.A., Sudoyo H., et al. Evolutionary history and adaptation of a human pygmy population of Flores Island, Indonesia. Science. 2018;361:511–516. doi: 10.1126/science.aar8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A., 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pagani L., Lawson D.J., Jagoda E., Mörseburg A., Eriksson A., Mitt M., Clemente F., Hudjashov G., DeGiorgio M., Saag L., et al. Genomic analyses inform on migration events during the peopling of Eurasia. Nature. 2016;538:238–242. doi: 10.1038/nature19792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aghakhanian F., Yunus Y., Naidu R., Jinam T., Manica A., Hoh B.P., Phipps M.E. Unravelling the genetic history of Negritos and indigenous populations of Southeast Asia. Genome Biol. Evol. 2015;7:1206–1215. doi: 10.1093/gbe/evv065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kusuma P., Brucato N., Cox M.P., Pierron D., Razafindrazaka H., Adelaar A., Sudoyo H., Letellier T., Ricaut F.X. Contrasting linguistic and genetic origins of the Asian source populations of Malagasy. Sci. Rep. 2016;6:26066. doi: 10.1038/srep26066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kusuma P., Brucato N., Cox M.P., Letellier T., Manan A., Nuraini C., Grangé P., Sudoyo H., Ricaut F.X. The last sea nomads of the Indonesian archipelago: genomic origins and dispersal. Eur. J. Hum. Genet. 2017;25:1004–1010. doi: 10.1038/ejhg.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cox M.P., Hudjashov G., Sim A., Savina O., Karafet T.M., Sudoyo H., Lansing J.S. Small traditional human communities sustain genomic diversity over microgeographic scales despite linguistic isolation. Mol. Biol. Evol. 2016;33:2273–2284. doi: 10.1093/molbev/msw099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mörseburg A., Pagani L., Ricaut F.-X., Yngvadottir B., Harney E., Castillo C., Hoogervorst T., Antao T., Kusuma P., Brucato N., et al. Multi-layered population structure in Island Southeast Asians. Eur. J. Hum. Genet. 2016;24:1605–1611. doi: 10.1038/ejhg.2016.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Basu A., Sarkar-Roy N., Majumder P.P. Genomic reconstruction of the history of extant populations of India reveals five distinct ancestral components and a complex structure. Proc. Natl. Acad. Sci. USA. 2016;113:1594–1599. doi: 10.1073/pnas.1513197113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lazaridis I., Nadel D., Rollefson G., Merrett D.C., Rohland N., Mallick S., Fernandes D., Novak M., Gamarra B., Sirak K., et al. Genomic insights into the origin of farming in the ancient Near East. Nature. 2016;536:419–424. doi: 10.1038/nature19310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Patterson N., Moorjani P., Luo Y., Mallick S., Rohland N., Zhan Y., Genschoreck T., Webster T., Reich D. Ancient admixture in human history. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang C.-C., Yeh H.-Y., Popov A.N., Zhang H.-Q., Matsumura H., Sirak K., Cheronet O., Kovalev A., Rohland N., Kim A.M., et al. Genomic insights into the formation of human populations in East Asia. Nature. 2021;591:413–419. doi: 10.1038/s41586-021-03336-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lipson M., Cheronet O., Mallick S., Rohland N., Oxenham M., Pietrusewsky M., Pryce T.O., Willis A., Matsumura H., Buckley H., et al. Ancient genomes document multiple waves of migration in Southeast Asian prehistory. Science. 2018;361:92–95. doi: 10.1126/science.aat3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lipson M., Skoglund P., Spriggs M., Valentin F., Bedford S., Shing R., Buckley H., Phillip I., Ward G.K., Mallick S., et al. Population turnover in remote Oceania shortly after initial settlement. Curr. Biol. 2018;28:1157–1165.e7. doi: 10.1016/j.cub.2018.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Skoglund P., Posth C., Sirak K., Spriggs M., Valentin F., Bedford S., Clark G.R., Reepmeyer C., Petchey F., Fernandes D., et al. Genomic insights into the peopling of the Southwest Pacific. Nature. 2016;538:510–513. doi: 10.1038/nature19844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakatsuka N., Moorjani P., Rai N., Sarkar B., Tandon A., Patterson N., Bhavani G.S., Girisha K.M., Mustak M.S., Srinivasan S., et al. The promise of discovering population-specific disease-associated genes in South Asia. Nat. Genet. 2017;49:1403–1407. doi: 10.1038/ng.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Manichaikul A., Mychaleckyj J.C., Rich S.S., Daly K., Sale M., Chen W.-M. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 61.Patterson N., Price A.L., Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jakobsson M., Rosenberg N.A. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- 63.Behr A.A., Liu K.Z., Liu-Fang G., Nakka P., Ramachandran S. pong: fast analysis and visualization of latent clusters in population genetic data. Bioinformatics. 2016;32:2817–2823. doi: 10.1093/bioinformatics/btw327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maples B.K., Gravel S., Kenny E.E., Bustamante C.D. RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am. J. Hum. Genet. 2013;93:278–288. doi: 10.1016/j.ajhg.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J., et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M., et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Poplin R., Ruano-Rubio V., DePristo M.A., Fennell T.J., Carneiro M.O., Van der Auwera G.A., Kling D.E., Gauthier L.D., Levy-Moonshine A., Roazen D., et al. Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv. 2018 doi: 10.1101/201178. [DOI] [Google Scholar]
- 69.Headland T.N. Thirty endangered languages in the Philippines. Work Papers of the Summer Institute of Linguistics, University of North Dakota Session. 2003;47:1. [Google Scholar]
- 70.Lambeck K., Chappell J. Sea level change through the last glacial cycle. Science. 2001;292:679–686. doi: 10.1126/science.1059549. [DOI] [PubMed] [Google Scholar]
- 71.Lambeck K., Yokoyama Y., Purcell T. Into and out of the last glacial maximum: sea-level change during oxygen isotope stages 3 and 2. Quat. Sci. Rev. 2002;21:343–360. [Google Scholar]
- 72.Hall R. Southeast Asia’s changing palaeogeography. Blumea. 2009;54:148–161. [Google Scholar]
- 73.Lambeck K., Rouby H., Purcell A., Sun Y., Sambridge M. Sea level and global ice volumes from the last glacial maximum to the holocene. Proc. Natl. Acad. Sci. USA. 2014;111:15296–15303. doi: 10.1073/pnas.1411762111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ingicco T., van den Bergh G.D., Jago-on C., Bahain J.-J., Chacón M.G., Amano N., Forestier H., King C., Manalo K., Nomade S., et al. Earliest known hominin activity in the Philippines by 709 thousand years ago. Nature. 2018;557:233–237. doi: 10.1038/s41586-018-0072-8. [DOI] [PubMed] [Google Scholar]
- 75.Pawlik A.F., Piper P.J. The Philippines from c. 14,000 to 4,000 cal. BP in regional context. Camb. Archaeol. J. 2019;29:1–22. [Google Scholar]
- 76.Dizon E.Z., Pawlik A.F. The lower Palaeolithic record in the Philippines. Quat. Int. 2010;223:444–450. [Google Scholar]
- 77.Détroit F., Dizon E., Falguères C., Hameau S., Ronquillo W., Sémah F. Upper Pleistocene Homo sapiens from the Tabon cave (Palawan, The Philippines): description and dating of new discoveries. C. R. Palevol. 2004;3:705–712. [Google Scholar]
- 78.Corny J., Garong A.M., Sémah F., Dizon E.Z., Bolunia M.J.L.A., Bautista R., Détroit F. Paleoanthropological significance and morphological variability of the human bones and teeth from Tabon Cave. Quat. Int. 2016;416:210–218. [Google Scholar]
- 79.Barker G., Barton H., Bird M., Daly P., Datan I., Dykes A., Farr L., Gilbertson D., Harrisson B., Hunt C., et al. The ‘human revolution’ in lowland tropical Southeast Asia: the antiquity and behavior of anatomically modern humans at Niah Cave (Sarawak, Borneo) J. Hum. Evol. 2007;52:243–261. doi: 10.1016/j.jhevol.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 80.Bartstra G.-J., Soegondho S., van der Wijk A. Ngandong man: age and artifacts. J. Hum. Evol. 1988;17:325–337. [Google Scholar]
- 81.Antón S.C. Developmental age and taxonomic affinity of the Mojokerto child, Java, Indonesia. Am. J. Phys. Anthropol. 1997;102:497–514. doi: 10.1002/(SICI)1096-8644(199704)102:4<497::AID-AJPA6>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 82.Kaifu Y., Aziz F., Indriati E., Jacob T., Kurniawan I., Baba H. Cranial morphology of Javanese Homo erectus: new evidence for continuous evolution, specialization, and terminal extinction. J. Hum. Evol. 2008;55:551–580. doi: 10.1016/j.jhevol.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 83.Choi K., Driwantoro D. Shell tool use by early members of Homo erectus in Sangiran, central Java, Indonesia: cut mark evidence. J. Archaeol. Sci. 2007;34:48–58. [Google Scholar]
- 84.Rizal Y., Westaway K.E., Zaim Y., van den Bergh G.D., Arthur Bettis 3rd E., Morwood M.J., Huffman O.F., Grün R., Joannes-Boyau R., Bailey R.M., et al. Last appearance of Homo erectus at Ngandong, Java, 117,000-108,000 years ago. Nature. 2020;577:381–385. doi: 10.1038/s41586-019-1863-2. [DOI] [PubMed] [Google Scholar]
- 85.Brumm A., van den Bergh G.D., Storey M., Kurniawan I., Alloway B.V., Setiawan R., Setiyabudi E., Grün R., Moore M.W., Yurnaldi D., et al. Age and context of the oldest known hominin fossils from Flores. Nature. 2016;534:249–253. doi: 10.1038/nature17663. [DOI] [PubMed] [Google Scholar]
- 86.van den Bergh G.D., Kaifu Y., Kurniawan I., Kono R.T., Brumm A., Setiyabudi E., Aziz F., Morwood M.J. Homo floresiensis-like fossils from the early Middle Pleistocene of Flores. Nature. 2016;534:245–248. doi: 10.1038/nature17999. [DOI] [PubMed] [Google Scholar]
- 87.Sutikna T., Tocheri M.W., Morwood M.J., Saptomo E.W., Jatmiko, Awe R.D., Wasisto S., Westaway K.E., Aubert M., Li B., et al. Revised stratigraphy and chronology for Homo floresiensis at Liang Bua in Indonesia. Nature. 2016;532:366–369. doi: 10.1038/nature17179. [DOI] [PubMed] [Google Scholar]
- 88.Kaifu Y. Archaic hominin populations in Asia before the arrival of modern humans: their phylogeny and implications for the “Southern Denisovans”. Curr. Anthropol. 2017;58:S418–S433. [Google Scholar]
- 89.Ning C., Li T., Wang K., Zhang F., Li T., Wu X., Gao S., Zhang Q., Zhang H., Hudson M.J., et al. Ancient genomes from northern China suggest links between subsistence changes and human migration. Nat. Commun. 2020;11:2700. doi: 10.1038/s41467-020-16557-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pickrell J.K., Patterson N., Loh P.R., Lipson M., Berger B., Stoneking M., Pakendorf B., Reich D. Ancient west Eurasian ancestry in southern and eastern Africa. Proc. Natl. Acad. Sci. USA. 2014;111:2632–2637. doi: 10.1073/pnas.1313787111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schlebusch C.M., Malmström H., Günther T., Sjödin P., Coutinho A., Edlund H., Munters A.R., Vicente M., Steyn M., Soodyall H., et al. Southern African ancient genomes estimate modern human divergence to 350,000 to 260,000 years ago. Science. 2017;358:652–655. doi: 10.1126/science.aao6266. [DOI] [PubMed] [Google Scholar]
- 93.Villanea F.A., Schraiber J.G. Multiple episodes of interbreeding between Neanderthal and modern humans. Nat. Ecol. Evol. 2019;3:39–44. doi: 10.1038/s41559-018-0735-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Delaneau O., Marchini J., Zagury J.F. A linear complexity phasing method for thousands of genomes. Nat. Methods. 2011;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 95.Delaneau O., Marchini J., 1000 Genomes Project Consortium Integrating sequence and array data to create an improved 1000 Genomes Project haplotype reference panel. Nat. Commun. 2014;5:3934. doi: 10.1038/ncomms4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) Denisovan ancestry estimation using D statistics. B) Denisovan ancestry estimation using f4-ratio statistics. C) f4-ratio statistics performed on 5 least admixed individuals per population.
A) Denisovan ancestry estimation using D statistics, and percentage difference Denisovan ancestry in Ayta Magbukun and AustraloPapuans. B) Denisovan ancestry estimation using f4-ratio statistics, and percentage difference Denisovan ancestry in Ayta Magbukon and AustraloPapuans. C) Direct comparison of Denisovan ancestry in Ayta Magbukon versus AustraloPapuans using D-statistics.
Data Availability Statement
All requests to access the genotype and sequence data presented in this work are managed through the Data Access Committee of the official data repositories at the European Genome-phenome Archive (https://ega-archive.org/ega/home; Database: EGAS00001005408 and EGAS00001005407). The data generated in this study can be obtained for the purposes of academic research on genetic origins and diversity following approval by the Data Access Committee. All simulation scripts used in this study are available at https://github.com/jammc313/Philippine_Denisovan_Ancestry.