Abstract
CRAM is a cysteine-rich acidic transmembrane protein, highly expressed in the procyclic form of Trypanosoma brucei. Cell surface expression of CRAM is restricted to the flagellar pocket of trypanosomes, the only place where receptor mediated endocytosis takes place in the parasite. CRAM can function as a receptor and was hypothesized to be a lipoprotein receptor of trypanosomes. We study mechanisms involved in the presentation and routing of CRAM to the flagellar pocket of insect- and bloodstream-form trypanosomes. By deletional mutagenesis, we found that deleting up to four amino acids from the C terminus of CRAM did not affect the localization of CRAM at the flagellar pocket. Shortening the CRAM protein by 8 and 19 amino acids from the C terminus resulted in the distribution of the CRAM protein in the endoplasmic reticulum (ER) (the CRAM protein is no longer uniquely sequestered at the flagellar pocket). This result indicates that the truncation of the CRAM C terminus affected the transport efficiency of CRAM from the ER to the flagellar pocket. However, when CRAM was truncated between 29 and 40 amino acids from the C terminus, CRAM was not only distributed in the ER but also located to the flagellar pocket and spread to the cell surface and the flagellum. Replacing the CRAM transmembrane domain with the invariant surface glycoprotein 65-derived transmembrane region did not affect the flagellar pocket location of CRAM. These results indicate that the CRAM cytoplasmic extension may exhibit two functional domains: one domain near the C terminus is important for efficient export of CRAM from the ER, while the second domain is of importance for confining CRAM to the flagellar pocket membrane.
African trypanosomes are protozoan parasites, causing sleeping sickness in humans and nagana in cattle, both of which are diseases endemic in large areas of tropical Africa. Trypanosomiasis is not only a health threat; it also affects the economic viability of these areas. Trypanosoma brucei has an intricate life cycle alternating between a mammalian host and an insect vector, the tsetse fly. In the bloodstream of the mammal, the parasite is covered by a thick coat protein, the variant cell surface glycoprotein (VSG). By undergoing antigenic variation of the VSG coat, bloodstream-form trypanosomes escape host immune attack (for reviews, see references 6, 14, 38, and 53). When the fly takes a blood meal, T. brucei is transferred into the midgut of the tsetse fly and differentiates into the procyclic (or insect) form. As a result, its VSG coat is replaced by a different coat protein (the procyclic acidic repetitive protein [PARP] or procyclin [33, 39]). Both the VSG and PARP coat proteins are anchored to the lipid bilayer via a covalently attached lipid-glycosylphosphatidyl inositol (GPI) moiety (32). In addition to the surface coat protein, several invariant surface glycoproteins (ISGs) of unknown function are distributed over the surface of bloodstream-form trypanosomes and are shielded by the VSG (21, 59, 60). These are ISG65 and ISG75, described by Ziegelbauer et al. (59), and ISG70 and ISG64, described by Jackson et al. (21).
T. brucei, an extracellular organism, is dependent on host-derived nutrients for its growth and development. Accumulated evidence demonstrates that cell surface receptors for nutrient uptake, such as the low-density-lipoprotein (LDL) receptor and the transferrin receptor, are located only at the flagellar pocket of trypanosomes (12, 13, 16, 17, 28, 43, 45, 49). The flagellar pocket is a deep invagination of the surface plasma membrane, where the flagellum extends from the cell (1, 23, 54–56). Because the hemidesmosome zone between plasma and flagellar membranes closes the pocket, the flagellar pocket forms a secluded extracellular surface domain; this domain is not completely covered by the VSG coat protein, and the microtubule network does not extend beneath the membrane. In trypanosomatids, endocytosis and exocytosis occur exclusively at the flagellar pocket. All vesicular trafficking between the cytoplasm and cell surface in these highly polarized parasites is restricted to the flagellar pocket (8, 37). Materials endocytosed through the pocket are subsequently delivered to an endosome or lysosomal compartment. On the way to the pocket, it appears that membrane-bound proteins, once synthesized, travel from the endoplasmic reticulum (ER) to Golgi/trans-Golgi network and then into the flagellar pocket membrane. From the pocket, surface coat proteins and some ISGs can rapidly spread over the entire cell surface, while receptors for the uptake of macromolecules are retained in the flagellar pocket. We are investigating how trypanosomes selectively retain receptor molecules at the flagellar pocket, an issue that has not been systematically analyzed and resolved.
The flagellar pocket of trypanosomes, representing ∼0.43% of the pellicle membrane (surface area, ∼1 to 2 μm2). Accordingly, the bloodstream form of T. brucei can internalize a surface area equivalent to that of the flagellar pocket membrane every 1 to 2 min (12, 13). This internalization rate is considerably higher than that reported for mammalian cells and may be attributed to the specialized configuration of the pocket. Our studies focus on the structural organization of the flagellar pocket and the mechanisms involved in protein transport and sequestration to the flagellar pocket. Thus far, two receptor proteins located at the flagellar pocket of T. brucei have been well characterized: (i) the bloodstream-form transferrin receptor complex, which is a GPI-anchored protein (7, 28, 43, 49); and (ii) a cysteine-rich repetitive acidic transmembrane (CRAM), which may be a lipoprotein receptor in trypanosomes (24, 30, 58). Recently, Nolan et al. reported a new bloodstream form, ISG100, which is an integral membrane glycoprotein also localized at the flagellar pocket of bloodstream-form trypanosomes (34). The function of the ISG100 is not clear.
CRAM is abundantly expressed in procyclic-form trypanosomes and expressed at a low level in bloodstream-form trypanosomes (24). The CRAM protein has a predicted molecular mass of ∼130 kDa (945 amino acids) consisting of, from N terminus to C terminus, a putative N-terminal signal peptide followed by the extracellular extension of a large domain of a 12-amino-acid cysteine rich repeat (66 repeats) followed by a short unique peptide, a hydrophobic transmembrane domain, and a hydrophilic cytoplasmic extension of 41 amino acids (Fig. 1A) (24). The extracellular cysteine-rich repeat of CRAM shares high-level homology with the cysteine-rich repeat in the complement C9 protein (48). This complement-like repeat is also present in the binding domain of the LDL receptor, the LDL receptor-related protein, and the very-low-density lipoprotein receptor. Based on the structural similarity of CRAM with mammalian lipoprotein receptors, we hypothesized that CRAM might function as a lipoprotein receptor in trypanosomes. Since the CRAM protein is present only in the flagellar pocket membrane and in endocytic vesicles, targeting signals and sorting systems must be involved in determining its subcellular fate. We studied mechanisms involved in the presentation and routing of the CRAM protein to the flagellar pocket membrane by determining the amino acid sequences in CRAM that are required for residence at the flagellar pocket of trypanosomes. This study is a prerequisite to our understanding of the structure of the specialized configuration of the flagellar pocket and the unusual properties involved in the uptake of macromolecules in trypanosomes. The data obtained now facilitate a detailed molecular analysis of proteins involved in trafficking via the flagellar pocket.
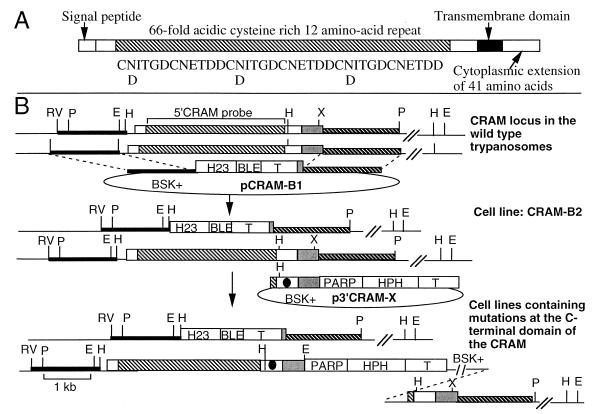
FIG. 1.
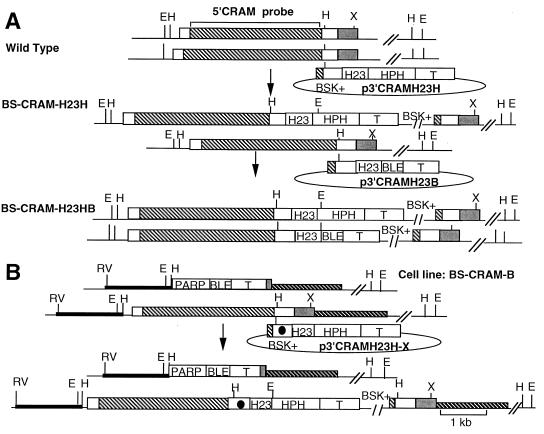
Physical maps of the CRAM locus in wild-type trypanosomes and CRAM mutant cell lines. (A) Schematic diagram of the structure of CRAM. The amino acid sequence of three contiguous cysteine-rich 12-amino-acid repeats is listed underneath the repeat domain (the shaded box region). (B) Top, structure of the CRAM locus in wild-type trypanosomes and plasmid pCRAM-B1. The large boxed region represents the CRAM gene. The open white boxes at the 5′ and 3′ ends of the CRAM gene represent the unique N- and C-terminal peptide regions, respectively; the shaded box represents the reptitive peptide region; the gray box represents the 3′ UTR of the CRAM gene. pCRAM-B1, containing the ble gene flanked by the hsp70 intergenic region promoter (H23 [27]) and the βα-tubulin intergenic region (51), was used for gene replacement. In pCRAM-B1, the 5′ targeting sequence containing the HindIII/EcoRV fragment derived from the 5′ flanking region of the CRAM locus (black bar) and the 3′ targeting sequence encodes the XhoI/PstI fragment of the 3′ flanking region of the CRAM locus (hatched bar). Middle, structure of the CRAM locus in CRAM-B2 cell line and p3′CRAM-X plasmids. One allele of the CRAM gene in the CRAM-B2 cell line was deleted and replaced by the H23-ble gene. The p3′CRAM-X plasmids, containing the hph gene flanked by the PARP promoter and the βα-tubulin intergenic region, were used for gene integration. The sequence spanning the 3′ end of the CRAM gene was used as a targeting sequence. The black dot indicates the mutation carried in the 3′ coding region of each of mutated CRAM genes. The HindIII site was used to linearize the plasmid for integration of the plasmid into the 3′ coding region of the CRAM gene. Bottom, structure of the CRAM locus of cell lines containing a mutated CRAM gene. Abbreviations: PARP, the PARP gene promoter and its 3′ splice site (41); T, intergenic region of βα-tubulin genes (51); BSK+, the plasmid vector Bluescript SK+; HPH, the hph gene; BLE, the ble gene (purchased from CAYLA Inc.); double slashes, undetermined distance; H, HindIII; E, EcoRI; P, PstI; RV, EcoRV; X, XhoI. The 5′ CRAM probe used in hybridizations is indicated at the top.
MATERIALS AND METHODS
Trypanosome strains.
The procyclic form of T. brucei stock 427-60, originally obtained from R. Brun, was maintained in SDM-79 medium at 25°C (9). The culture-adapted bloodstream form of variant 118 clone 1 was maintained in HMI-9 medium at 37°C (20). For all the experiments, early- to mid-log-phase trypanosomes (the procyclic form at a cell density of 5 × 106 to 8 × 106 cells/ml; the bloodstream form at a cell density of 1 × 106 to 2 × 106 cells/ml) were used.
Description of primary antibodies.
Anti-CRAM and anti-Tb-29 were as previously described (24, 26). Anti-Bip was obtained from J. D. Bangs (2). Monoclonal antibodies (MAbs) to p67 and GLP-1 are described elsewhere (22, 29). Rabbit- and rat-derived anti-TrpE were raised against the bacterial TrpE protein (M. G.-S. Lee, unpublished data). The anti-procyclic antibody was generated using ground procyclic trypanosome powder (Lee, unpublished).
Description of plasmid constructs and primers.
PCR-based mutagenesis was used to alter the DNA fragments encoding the various C-terminal extensions of CRAM. Primers used for PCR-based mutagenesis were p40A-30mer (CGCATATGACAACCGCGTTCTGGCGTTACC), p29A-18mer (AGATGCATCAGATTAGCA), p19S-21mer (GTCATATGCGACAACTAGGTT), p12S-36mer (GATGCATCTTTTGCCGTCCCCGTAACTTAAGCCTCA), p11S-30mer (TGATGCATCTTAGGCCTTCCCCGTAACTCA), p13A-29mer (CGGATCCTTTACGGTCACGCTTCATCTGA), and p14A-30mer (CGGATCCTTTACGGTCTCGATTAATCTGAG), which resulted in mutations described in cell lines CRAM-40, CRAM-29, CRAM-19, CRAM-14, CRAM-8, CRAM-4, andCRAM-2, respectively. To perform genetic studies of CRAM, we isolated genomic clones encoding the 5-kb region located upstream of the CRAM gene and the 2-kb region located downstream of the CRAM gene (Lee, unpublished). Plasmid pCRAM-B1 (Fig. 1), used for inactivation of one CRAM allele by gene replacement, consisted, from 5′ to 3′, of the 5′ targeting sequence derived from the EcoRV/HindIII fragment of the 5′ flanking region of the CRAM locus, a hsp70 intergenic region promoter (H23 [27]) driving the phleomycin resistance (ble) gene, followed by an αβ-tubulin intergenic region and the 3′ targeting sequence encoded the XhoI/PstI fragment of the 3′ flanking region of the CRAM locus (Fig. 1). A series of p3′CRAM-X plasmids was used to modify the C-terminal coding region of CRAM through integration events via homologous recombination. These p3′CRAM-X plasmids contain the targeting DNA fragment which extends from the last three repeats of the CRAM extracellular repeat domain to the end of the 3′ untranslated region (UTR) (24) linked to the PARP promoter driving the hygromycin resistance (hph) gene (25). The targeting DNA fragment of p3′CRAM-X either encoded the wild-type CRAM C-terminal sequence (p3′CRAM-0) or contained point mutations which encoded truncated CRAM C terminus: p3′CRAM-2, -4, -8, -14, -19, -29, and -40 (numbers indicated deleted amino acids from the C terminus of CRAM [Fig. 2A]). In p3′CRAM-40, p3′CRAM-29, and p3′CRAM-19, only a single nucleotide change was introduced (Fig. 2A). In p3′CRAM-14, p3′CRAM-8, p3′CRAM-4, and p3′CRAM-2, in addition to converting the codon at amino acids -14, -8, -4, and -2, respectively, to termination codons, additional mutations were introduced to the 3′ adjacent nucleotides, which create a diagnostic restriction enzyme site in each clone (StuI, AseI, AfluII, and BamHI in p3′CRAM-12, p3′CRAM-8, p3′CRAM-4, and p3′CRAM-2, respectively). These additional restriction enzyme site polymorphisms facilitated the identification of correct transformed cell lines (Fig. 2A). Three additional plasmids contain a large segment replacement in the CRAM C terminus: (i) p3′CRAM-XTM contains the ISG65 transmembrane domain replacing the CRAM transmembrane domain; (ii) p3′CRAM-XCD contains the ISG65 cytoplasmic domain replacing the CRAM cytoplasmic domain; and (iii) p3′CRAM-XTM.CD contains both the ISG65 transmembrane domain and cytoplasmic domain replacing those of CRAM (Fig. 3B shows the amino acid sequences). The transmembrane domain and cytoplasmic extension of ISG65 were derived from the cDNA clone pSW14 (a gift from P. Overath) (59). p3′CRAMH23H and p3′CRAMH23B are insertion plasmids for activation of the CRAM gene expression in bloodstream-form trypanosomes and contain, from 5′ to 3′, the 3′ coding region of CRAM, the hsp70 intergenic region (H23) including the 3′ UTR of the hsp70 gene, the selectable marker (hph for p3′CRAMH23H and ble for p3′CRAMH23B [see Fig. 5]), and the βα-tubulin intergenic region. p3′CRAMH23H-X plasmids including p3′CRAMH23H-4, -19, -29, and -40 were used to introduce mutations at the CRAM C terminus of bloodstream-form trypanosomes. All of the p3′CRAM-X and p3′CRAMH23H-X plasmids were linearized at the HindIII site located in the center of the targeting sequence and were electroporated into trypanosomes.
FIG. 2.
Sequences of the C-terminal extension of CRAM in different cell lines. (A) DNA nucleotide and amino acid sequences of the transmembrane and cytoplasmic domains of CRAM in wild-type trypanosomes and cell lines containing truncated CRAM cytoplasmic domain. Dotted lines indicate sequences of 100% homology. Changes of nucleotide sequences in different cell lines are indicated by underlines, and the diagnostic restriction enzyme sites resulting from mutations are indicated by typed-out sequences. (B) Comparison of the amino acid sequences of the transmembrane domain and the cytoplasmic domain of CRAM in the wild-type trypanosome and cell lines CRAM-XCD, CRAM-XTM.CD, and CRAM-XTM. The transmembrane domain is underlined. Sequences derived from the ISG65 gene are indicated by boldface.
FIG. 3.
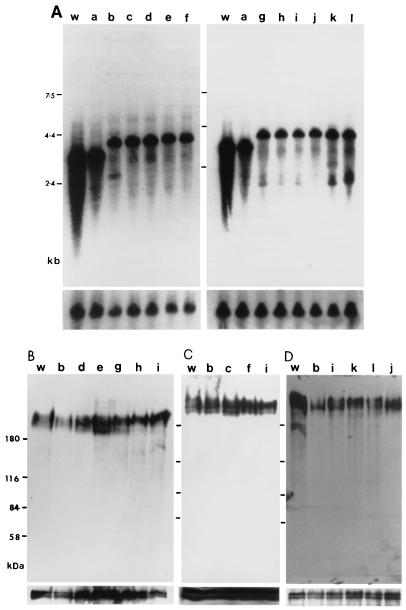
Expression of CRAM in different procyclic cell lines. (A) Northern blot analysis of the steady-state mRNA level of CRAM. Equal amounts (∼20 μg) of total RNA from the wild-type trypanosome (w), CRAM-B2 (a), CRAM-0 (b), CRAM-2 (c), CRAM-4 (d), CRAM-8 (e), CRAM-14 (f), CRAM-19 (g), CRAM-29 (h), CRAM-40 (i), CRAM-XTM (j), CRAM-XCD (k), and CRAM-XTM.CD (l) cell lines were separated in 1% formaldehyde agarose gels. The blot was hybridized to a 5′ CRAM probe (Fig. 1). The final posthybridization wash was performed in 0.1× SSC–0.1% SDS at 65°C. The bottom panels represent hybridization with the β-tubulin gene probe, indicating that approximately equal amounts of RNA were loaded in all lanes. (B to D) CRAM protein expression. Total protein lysates (∼2 × 107 trypanosomes) derived from wild-type trypanosomes (w), CRAM-B2 (a), CRAM-0 (b), CRAM-2 (c), CRAM-4 (d), CRAM-8 (e), CRAM-14 (f), CRAM-19 (g), CRAM-29 (h), CRAM-40 (i), CRAM-XTM (j), CRAM-XCD (k), and CRAM-XTM.CD (l) cell lines were size separated in 6% polyacrylamide gels and electrophoretically transferred to nitrocellulose filters. The blots were probed with anti-CRAM antibody (24). The same blots were later probed with anti-Tb-29 (B and D, bottom) (26) or the anti-procyclic antibody identifying two proteins of ∼43 to 45 kDa (C, bottom) (the anti-procyclic antibody was raised against the total procyclic proteins [Lee, unpublished]) to demonstrate equal loading in all lanes.
FIG. 5.
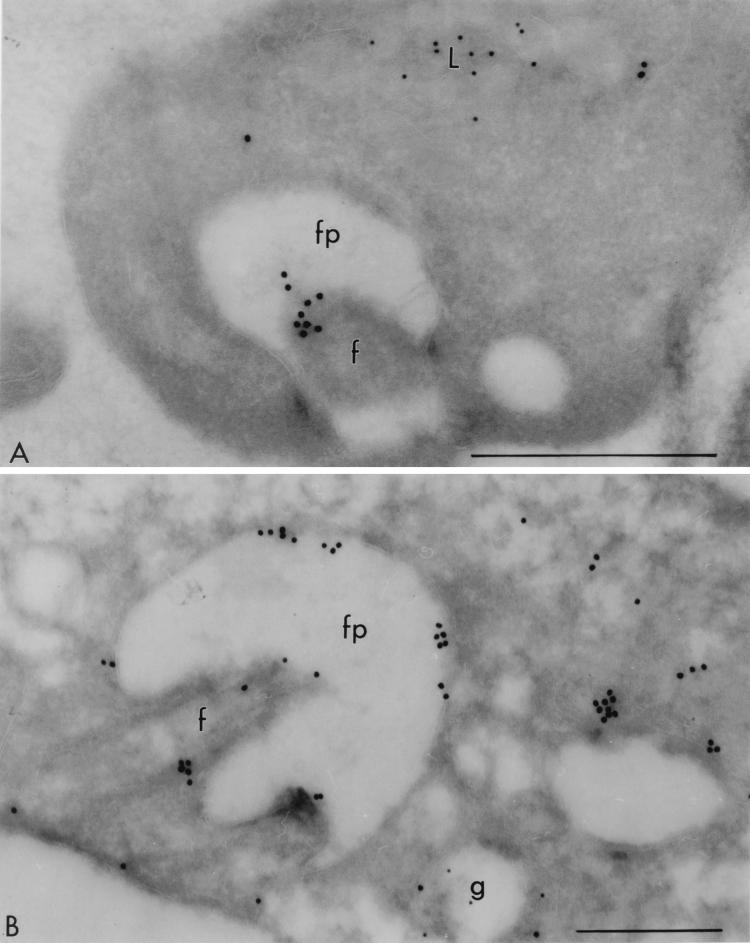
Immuno-EM localization of the CRAM protein in the CRAM-40 cell line. Panel A was probed with rabbit anti-CRAM (18 nm) and mouse MAb Ali I-218 (12 nm) directed against p67, a lysosomal glycoprotein characterized by Kelley et al. (22). The label for CRAM is observed on the flagellum. Panel B was probed with rabbit anti-CRAM (18 nm) and mouse MAb against GLP-1 (12 nm), a Golgi-associated transmembrane protein (29). An intense labeling with anti-CRAM appears on the surface of the flagellar pocket, and some label for CRAM is seen to be on the surface. fp, flagellar pocket; f, flagellum; g, Golgi; L, lysosome. Scale bars, 0.5 μm.
The trpE-CRAM fusion construct contained the PARP promoter and its 3′ splicing site driving the expression of the trpE-CRAM fusion gene which was linked to the PARP-hph transcription unit for selection of transformed cell lines. In the trpE-CRAM fusion gene, the N-terminal CRAM sequence spans from the entire 5′ UTR of CRAM to the first cysteine-rich 12-amino-acid repeat; the C-terminal CRAM-derived sequence encodes the 3′-end unique peptide region including the transmembrane domain and the cytoplasmic domain and the CRAM 3′ UTR. The plasmid was linearized at the MluI site in the βα-tubulin intergenic region located downstream of the hph gene. The plasmid was integrated into the tubulin intergenic region in transformed cell lines.
DNA transformation.
Linearized plasmid DNA (10 or 20 μg) was electroporated into trypanosomes using a BTX electroporator as previously described (25, 41). For procyclic trypanosomes, 48 h after electroporation, phleomycin (3 μg/ml) and/or hygromycin B (40 μg/ml) were added to select stably transformed trypanosomes. The individually transformed procyclic forms were cloned by limiting dilution cloning with the addition of wild-type trypanosomes (106 cells/ml). For bloodstream-form trypanosomes, 16 h after electroporation, phleomycin (1 μg/ml) and/or hygromycin B (1 μg/ml) were added to select stably transformed trypanosomes.
DNA isolation and Southern genomic blot analysis.
Nuclear DNA was isolated from trypanosomes as described elsewhere (52). Following digestion with restriction endonucleases, DNA was separated on a 0.8% agarose gel and transferred onto nitrocellulose filters. Filters were hybridized with 32P-labeled 5′ CRAM probes. Posthybridizational washes were performed to a final condition of 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate (SDS) at 65°C. All of the clonal trypanosome cell lines were first analyzed by Southern blotting analysis to confirm that the correct integration event occurred in each cell line. Then the C-terminal extension of the CRAM allele in every selected mutant cell line described in the text was isolated by PCR amplifications, and their nucleotide sequences were analyzed to confirm the presence of the correct mutations.
RNA isolation and Northern blot analysis.
All RNA samples were isolated by GuanSCN lysis (10) and purified by centrifugation through CsCl cushions. RNA samples were separated in 1% formaldehyde agarose gels and transferred to nitrocellulose filters. Northern blots were hybridized with 32P-labeled probes. Following hybridization, filters were washed to a final stringency of 0.1× SSC–0.1% SDS at 65°C.
Western blot analysis.
Total cell lysates of 2 × 107 trypanosomes were size separated in 6% polyacrylamide gels and electrophoretically transferred to nitrocellulose filters. The nitrocellulose filters were first blocked with 5% nonfat milk in TBS (50 mM Tris [pH 7.5]-buffered saline). Antibody reactions were performed in TBS with 0.2% Tween 20. Following the first antibody reaction and washing, filters were treated with horseradish peroxidase-labeled goat anti-rabbit immunoglobulin G (IgG; Sigma Chemical Co.). Following the second antibody reaction and washing, the filter was reacted with an enhanced chemiluminescence detection system (Amersham Life Science).
Nucleotide sequence analysis.
The DNA fragment spanning the mutated region of CRAM C-terminal domain in each transformed cell line was PCR amplified with a sense primer specific for the CRAM C terminus (CRAM7.1S [GTGGGGCTGTCGGCAGTTTTATTT] or CRAM6.1S [AATGCAAAGGGGAAAGGATCGAGC]) and an antisense oligomer specific for the hph gene (Hph-3 [CAGAAACTTCTCGACAGACGTCG]) or an antisense primer specific for the PARP promoter sequence (PARP3A [CGACTCACCAATAAAACGAGCCGAC]) located downstream of the CRAM C-terminal domain in transformed cell lines. Either the nucleotide sequences of amplified DNAs were determined directly or DNA was first subcloned into M13, after which their nucleotide sequences were analyzed.
Immunofluorescence microscopy.
For total cell staining, trypanosomes were harvested by centrifugation for 10 min at 400 × g and washed once with phosphate-buffered saline (PBS). Next, cells were suspended in 3.7% formaldehyde (or 4% paraformaldehyde) in PBS (pH 7.4) for 10 min in ice and neutralized with 0.1 M glycine for 10 min. Following fixation, cells were spun down and washed with PBS. Then cells were resuspended in PBS, dotted on slides, and fixed in cold methanol and cold acetone for 5 min each. After rehydration in PBS for 5 min, slides were incubated with the first antibody in the presence of 3% bovine serum albumin and 0.05% Tween 20 in PBS for 1 h. Following the first antibody reaction, slides were washed three times with PBS and then reacted with fluorescein isothiocyanate (FITC)- or rhodamine-conjugated goat-derived anti-rabbit or anti-rat IgG (Cappel) for 1 h. After the last wash, 4′,6-diamino-2-phenylindole (DAPI) was applied in water at a concentration of 0.1 μg/ml for 1 min at room temperature, after which the slides were briefly rinsed with water. The cells were mounted under a coverslip with a mounting medium containing an inhibitor that retards photobleaching (Kirkegaard & Perry Laboratories Inc.). Cells were viewed and photographed under a Leica fluorescence microscope using a 100× objective. Some of the images were directly captured by a charge-coupled device (CCD) camera and analyzed by the MetaMorph program (Universal Imaging Co.). Confocal image analysis was performed with a Molecular Dynamics confocal laser scanning microscope and ImageSpace software.
For cell surface staining, fixed and nonpermeabilized trypanosomes were used. Trypanosomes that were fixed only in 3.7% formaldehyde (or 4% paraformaldehyde) for 5 min in ice were dotted onto slides and then reacted with the first antibody in the presence of 3% bovine serum albumin. For live cell staining, 107 trypanosomes were harvested, washed, and incubated with the first antibody in 1 ml at 4°C for 1 h. Following the first antibody reaction, trypanosomes were washed twice with cold medium and once with cold PBS and spun down at 4°C. Next, cells were fixed with 3.7% formaldehyde (or 4% paraformaldehyde) as described above. After fixation, cells were dotted onto slides and reacted with the second antibody as described.
Immunoelectron microscopy (immuno-EM).
Procyclic- and bloodstream-form trypanosomes were washed in PBS and then fixed in 4% paraformaldehyde in piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) buffer (200 mM PIPES, 0.5 mM MgCl2 [pH 7.0]). Fixed cells were embedded in gelatin, and infiltrated into buffered polyvinylpyrrolidone (20%)-sucrose (2.3 M), as described by Russell et al. (42). The blocks were trimmed, frozen, and sectioned in the presence of 5% goat serum and 5% fetal calf serum in PIPES buffer. For double labeling experiments, primary antibody incubation was followed by use of gold-conjugated secondary antibodies as indicated in the figure legends. Finally, grids were washed and embedded in polyvinyl alcohol-uranyl acetate.
RESULTS
Construction of trypanosome cell lines expressing different versions of truncated CRAM or CRAM-ISG fusion genes.
The genome of T. brucei contains two alleles of the functional CRAM gene. We first deleted one allele of the CRAM gene and then performed mutagenesis on the remaining allele. This will prevent the wild-type CRAM allele from interfering with the analysis of the subcellular localization of the mutated CRAM in resulting cell lines. Since culture-adapted procyclic CRAM null mutants survive and exhibit no obvious phenotypic changes under normal culture conditions (58), we do not anticipate that mutating CRAM will lead to abnormal cell growth. The top part of Fig. 1B represents structures of the two polymorphic alleles of the CRAM gene in wild-type trypanosomes. Due to the presence of different numbers of 12-amino-acid repeats, the two alleles of CRAM slightly differ in size (Fig. 1B). The CRAM-B2 cell line contained only one functional allele of CRAM; its second CRAM allele was replaced by the ble gene via homologous recombination using the construct pCRAM-B1 (Fig. 1B; see Materials and Methods). The CRAM-B2 cell line was chosen for further mutagenesis of its remaining CRAM allele. We designed a series of integration constructs referred to as p3′CRAM-X plasmids that allow introduction of mutations at the CRAM C-terminal region (Fig. 1B; see Materials and Methods). Two types of mutant cell lines were established.
Type 1 cell lines contained point mutations that create a termination codon at different positions in the CRAM C-terminus resulting in C-terminally truncated CRAM proteins. Cell line CRAM-0 is the control cell line, which was transformed with plasmid p3′CRAM-0 encoding a wild-type CRAM C terminus. Cell lines CRAM-40, CRAM-29, CRAM-19, CRAM-12, CRAM-8, CRAM-4, and CRAM-2 express CRAM shortened by 40, 29, 19, 14, 8, 4, and 2 amino acids, respectively, from the C-terminal end (Fig. 2A).
Type 2 cell lines contained a large domain exchange at the CRAM C terminus to compare the role of the transmembrane and cytoplasmic domains of CRAM and ISG65 in the flagellar pocket localization of proteins. ISG65 is a transmembrane protein homogeneously expressed on the cell surface of bloodstream-form trypanosomes (59) (see Materials and Methods for details of the construction of cell lines and plasmids). Three cell lines were established: (i) CRAM-XTM, containing the ISG65 transmembrane domain replacing the CRAM transmembrane domain; (ii) CRAM-XCD, containing the ISG65 cytoplasmic domain replacing the CRAM cytoplasmic domain; and (iii) CRAM-XTM.CD, containing both the transmembrane and cytoplasmic domains of ISG replacing those of CRAM. The amino acid sequences spanning the CRAM C-terminal domain of these three cell lines are described in Fig. 2B.
Clonal transformants were first characterized by Southern blotting analysis. Cell lines exhibiting banding patterns expected for the correct integration event at the CRAM locus were selected for further nucleotide sequence analysis to confirm that the correct mutation was generated in each cell line. All cell lines analyzed exhibited a normal growth efficiency. We found that in each batch of the transformants, only <50% of the transformants contained the correct mutation, while the other >50% of the transformants contained the wild-type CRAM C terminus. Since the mutations created are located less than 290 bp away from the double-stranded break generated (the HindIII site), it is possible that the mismatch correction repair system has corrected the mutations during the recombinational event, as described for yeast (50).
Expression of the mutated CRAM genes in transformed trypanosome cell lines.
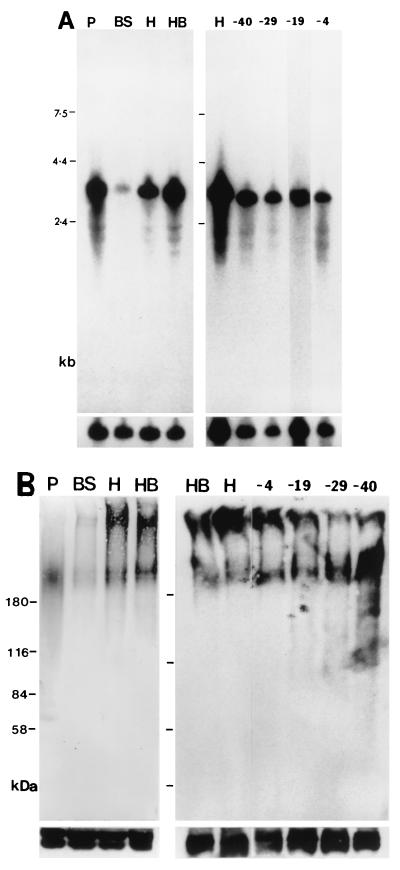
The level of mutated CRAM expression in each transformed cell line was first compared to that of wild-type trypanosomes and the CRAM-B2 cell line (containing only one CRAM allele) by Northern blot analysis (Fig. 3A). In wild-type trypanosomes, two closely migrating CRAM mRNAs of ∼3.2 kb were detected (24). The CRAM-B2 cell line expressed only the larger CRAM mRNA. The mutated CRAM mRNAs in all transformed cell lines, generated with p3′CRAM-X plasmids, are ∼0.8 kb longer than the wild-type CRAM mRNA. The CRAM mRNA expression levels among these cell lines are about equal and equivalent to ∼85% of that observed in the CRAM-B2 cell line (the calculation was based on the intensity of the autoradiograms and adjusted for the amount of RNA loaded, as determined from the amount of β-tubulin RNA). The bottom panels of Fig. 3A represent the hybridization with the β-tubulin probe indicating the relative amount of RNA loaded in each lane. Further hybridization analysis with probes encoding the PARP promoter and the hph gene indicated that the 3′ end of the CRAM mRNA in transformed cell lines expressing different versions of CRAM terminated at the downstream PARP promoter region resulting in the observed 0.8-kb-longer mRNAs (data not shown).
Relative amounts of the CRAM protein produced in different cell lines were compared by Western blot analysis. The CRAM protein in wild-type procyclics was detected as a broad band with a electrophoretic mobility at ∼200 kDa, using anti-CRAM antibodies (anti-P1-55 [24]) (Fig. 3B; we believe CRAM protein to be glycosylated). The CRAM proteins in the CRAM-B2 cell line (containing only one functional CRAM gene) and in cell lines containing different versions of truncated CRAM or CRAM-ISG65 fusion genes are expressed at similar levels and exhibit a similar banding pattern as that detected in the wild-type procyclic cell extract. We have not been able to detect a significant difference between the levels of the CRAM protein in transformed cell lines and in wild-type procyclics. However, given the heterogeneous size distribution of the CRAM protein, possibly resulting from glycosylation, it is not excluded that the Western blot may fail to give an accurate protein quantitation. A smaller size of the CRAM protein was visualized in the wild-type procyclic trypanosome in Fig. 3D. This smaller size of CRAM most likely resulted from a small amount of degraded product or nonglycosylated forms of CRAM. To ensure that similar amounts of protein were loaded in all lanes, these Western blots were further reacted with antibodies recognizing either the Tb-29 proteins (26) (Fig. 3B and D, bottom) or two constitutively expressed proteins of 43 to 45 kDa (Lee, unpublished) (Fig. 3C, bottom). As demonstrated, these control proteins were present in similar amounts in all lanes. These results indicate that the expression level of mutated CRAM proteins and the CRAM-ISG65 fusion proteins in each transformed cell line is similar to the wild-type CRAM in procyclic trypanosomes.
Subcellular localization of mutated CRAM proteins and CRAM-ISG fusion proteins in transformed procyclic cell lines.
The effects of mutations in the CRAM cytoplasmic and transmembrane domains on the fate of the CRAM or CRAM fusion proteins were examined by subcellular localization of CRAM in each type of cell line, using indirect immunofluorescence analysis. We first examined the overall distribution of CRAM in each cell line, using formaldehyde- or paraformaldehyde-fixed and permeabilized cells. Subsequently, the amount of CRAM that spread onto the cell surface in each cell line was examined by staining live trypanosomes with anti-CRAM antibody at 4°C and by surface staining of fixed and nonpermeabilized cells. The following immunofluorescence data are not quantitative, and the relative amount of the CRAM protein in each cell line was measured by Western blot analysis as described above. For each type of CRAM mutant, at least five individually transformed cell lines were examined. The results are summarized in Table 1. Like the wild-type trypanosome, the CRAM protein in CRAM-0, CRAM-2, and CRAM-4 cell lines is exclusively located at the flagellar pocket, indicating that deleting the last four amino acids of CRAM had no effect on the subcellular localization of CRAM at the flagellar pocket. A representative image from the CRAM-4 cell line (Fig. 4A, a) represents the superimposition of the fluorescence staining with anti-CRAM antibody (green) and the DNA specific dye DAPI (blue). The large and small blue dots locate the position of nucleus and kinetoplast, respectively. The green staining locates CRAM concentrated at the area of the flagellar pocket which is immediately adjacent to the kinetoplast.
TABLE 1.
Comparison of subcellular localization of CRAM in cell lines
| Cell line | Subcellular localization of CRAMa
|
|
|---|---|---|
| Total cell staining | Surface staining | |
| Wild type | FP | (FP) |
| CRAM-0 | FP | (FP) |
| CRAM-2 | FP | (FP) |
| CRAM-4 | FP | (FP) |
| CRAM-8 | ER | UD |
| CRAM-14 | ER | UD |
| CRAM-19 | ER | UD |
| CRAM-29 | ER, FP, F | F, (FP, S) |
| CRAM-40 | ER, FP, F | F, (FP, S) |
| CRAM-XTM | FP | (FP) |
| CRAM-XTM.CD | ER | UD |
| CRAM-XCD | ER | UD |
Total cell staining represents the result of staining with fixed and permeabilized trypanosomes; surface staining represents the result of staining with live trypanosomes. FP, flagellar pocket; F, flagellum; ER, endoplasmic reticulum; S, cell surface; UD, under detection level. Parentheses indicate that staining was very weak and/or could be observed only in a subpopulation of trypanosomes on each slide.
FIG. 4.
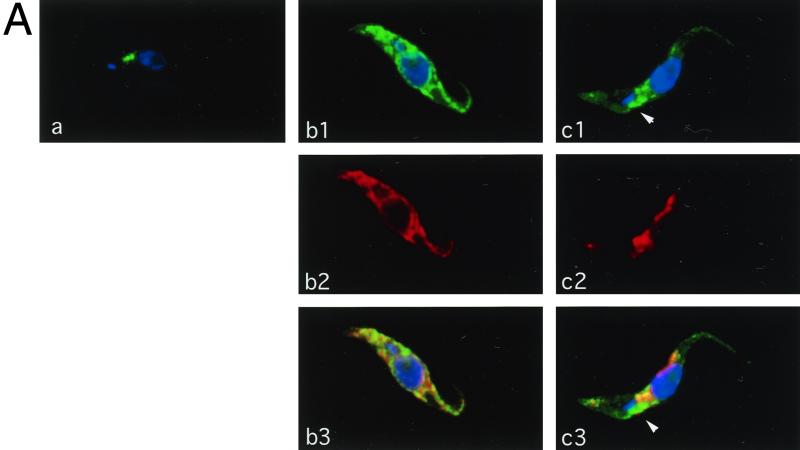
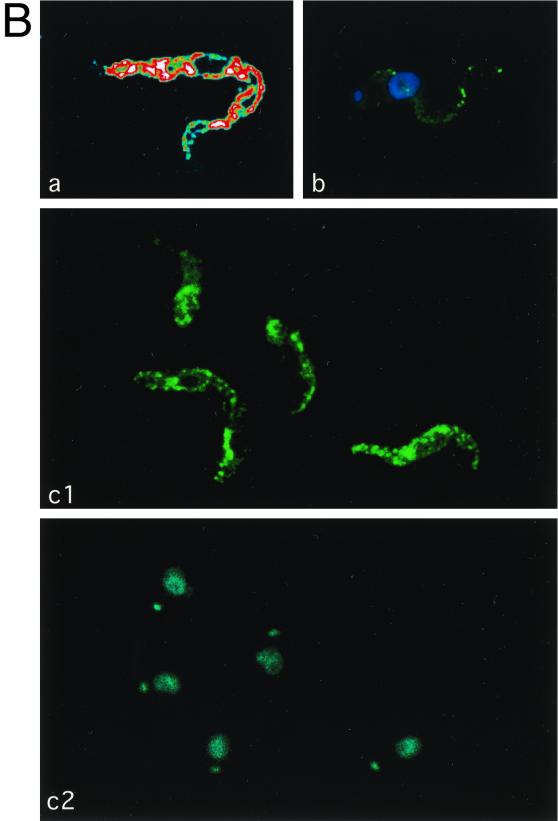
Subcellular localization of CRAM in different cell lines. (A) Staining of fixed and permeabilized trypanosomes. Slides containing fixed and permeabilized trypanosomes (a, CRAM-4 cell line; b1 to b3, CRAM-19 cell line; c1 to c3, CRAM-40 cell line) were first incubated with rat derived anti-CRAM and rabbit-derived anti-Bip antibodies, followed by reaction with FITC-conjugated goat anti-rat IgG and rhodamine-conjugated goat anti-rabbit IgG. Then cells were stained with DAPI at a concentration of 0.1 μg/ml for 1 min. The images were captured using a CCD camera and analyzed by the MetaMorph program (Universal Imaging Co.). The images are presented in pseudocolors: green for FITC labeled CRAM; red for rhodamine labeled Bip; blue for DAPI staining, which identifies the nucleus and the kinetoplast. (a, b1, and c1) Superimposed CRAM image and DAPI staining; (b2 and c2) Bip images; (b3 and c3) merging of matched CRAM images, Bip images, and DAPI staining. The white arrows indicate the area which is strongly stained with anti-CRAM but not with anti-Bip in the cell line CRAM-40. (B) CRAM distribution in the CRAM-40 cell line. (a) Confocal image of total cellular CRAM protein in a fixed and permeabilized CRAM-40 trypanosome. Different colors, arranged in the order blue-green-yellow-red-white, indicate the relative intensity of fluorescence (white, highest intensity; blue, lowest intensity). (b) Live trypanosome staining of the CRAM-40 cell line. The CRAM image and DAPI staining are superimposed. (c) Cell surface staining of fixed, nonpermeabilized CRAM-40 cells (c1, CRAM image; c2, DAPI staining). After incubation first with rabbit-derived anti-CRAM antibody and then with FITC-conjugated goat anti-rabbit IgG, the cells were stained with DAPI.
In cell lines CRAM-8 (data not shown), CRAM-14 (data not shown), and CRAM-19 (Fig. 4A, b1), the CRAM protein is no longer concentrated at the single area of the flagellar pocket but is spread throughout the cell, excluding the nucleus and kinetoplast. Frequently, a reticulum or tubule structure can be detected and in many cells, distinct perinuclear staining was apparent. This pattern is similar to the ER, a tubular network extending throughout the cell. When the cells were double stained with anti-CRAM antibody and the anti-Bip antibody (an ER-specific marker; a gift from J. D. Bangs) (2), it was obvious that the majority of CRAM in cell lines CRAM-8, CRAM-14, and CRAM-19 was colocalized with Bip at the ER. One set of representative images from CRAM-19 is shown in Fig. 4A, b1 (costaining pattern with anti-CRAM and DAPI), b2 (staining pattern with anti-Bip), and b3 (superimposition of all images). This result suggested that shortening the CRAM protein by 8 to 19 amino acids from the C terminus may have affected the efficiency of transporting CRAM from the ER to the flagellar pocket. Cell surface staining could not be observed with these cell lines.
In cell lines CRAM-29 (data not shown) and CRAM-40 (Fig. 4A, c1 to c3; Fig. 4B), the CRAM protein is still distributed throughout the ER and also spread onto the flagellum. Additionally, CRAM is highly concentrated at the area immediately adjacent to the kinetoplast—the flagellar pocket in each individual cell.
The distribution of CRAM in the CRAM-40 cell line was further confirmed by immuno-EM (Fig. 5). A significant proportion of the gold particles were found on the surface of flagellum (Fig. 5A) and at the flagellar pocket (Fig. 5B). We also found some gold particles distributed on the surface (data not shown). This result indicated that deleting >29 amino acids from the CRAM C terminus may have restored to a certain extent the ability of exporting CRAM from the ER to the flagellar pocket and resulted in some CRAM protein escape to the cell surface of flagellum (Fig. 4B, b).
We further determined to what extent the CRAM protein may have spread onto the cell surface due to the loss of different lengths of the cytoplasmic extension by comparison of surface staining of live cell at 4°C and surface staining of fixed and nonpermeabilized trypanosomes at room temperature. Following live cell staining at 4°C, significant cell surface and flagellum staining was observed only in CRAM-29 (data not shown) and CRAM-40 cell lines (Fig. 4B, b). In these cells, very light staining was found on the entire cell surface, while a strong and punctuated staining was found on the flagellum. Using live cell staining at 4°C on cell lines CRAM-0, CRAM-2, CRAM-4, and CRAM-XTM and wild-type procyclic trypanosomes, we observed that only a portion of trypanosomes had very weak staining at the flagellar pocket, which is consistent with the previous observation by fluorescence-activated cell sorting analysis (note that the labeling efficiency at the flagellar pocket of live trypanosomes at 4°C is very low [reference 24 and data not shown]). The amount of cell surface-localized CRAM in cell line CRAM-40 was further evaluated by surface staining of fixed and nonpermeabilized trypanosomes with anti-CRAM (this method gives a better labeling efficiency) (Fig. 4B, c1 and c2). The strong staining at the flagellar pocket and a strong punctuated pattern nonhomogeneously spread over the edge of the cell were revealed (Fig. 4B, c1). We did not observe significant cell surface staining in cell lines CRAM-8, CRAM-14, CRAM-19, CRAM-XTM.CD and CRAM-XCD. Based on the subcellular localization of CRAM, we hypothesized that the CRAM protein in cell lines CRAM-29 and CRAM-40 may have lost sequences required for proper routing and retention at the flagellar pocket.
In the CRAM-XTM cell line, the CRAM protein is located at the flagellar pocket as in wild-type trypanosomes, indicating that replacing the transmembrane domain did not affect the localization of the protein at the flagellar pocket (data not shown). In the CRAM-XTM.CD and CRAM-XCD cell lines, the majority of CRAM protein is still accumulated at the ER. Thus, replacing the CRAM cytoplasmic domain with that of ISG65 did not restore the ability to export CRAM from the ER (data not shown). Currently, we do not know whether mutated CRAM protein may be secreted and released into the culture media.
Expression of CRAM in the bloodstream form of T. brucei.
In bloodstream-form trypanosomes, the GPI-anchored transferrin receptor binding complex is localized at the flagellar pocket. The mechanism of anchoring the transferrin receptor complex at the flagellar pocket is unclear. It is possible that other sorting systems may operate in each life cycle stage of the parasite. We therefore compared the fate of the CRAM protein in procyclic-form and bloodstream-form trypanosomes. CRAM is expressed at a 10-fold-lower level in bloodstream-form than in procyclic-form trypanosomes (24). Because of the low-level expression of CRAM in the bloodstream form, it has been difficult to assess the cellular location of CRAM in bloodstream-form trypanosomes. By replacing the 3′ UTR of the CRAM gene with that of the hsp70 gene via homologous recombination (for details, see Materials and Methods and Fig. 6A), CRAM expression in bloodstream-form trypanosomes was up-regulated. Two bloodstream-form CRAM overexpressors, cell lines BS-CRAMH23H and BS-CRAMH23HB, were established. The BS-CRAMH23H cell line contains one allele of the activated CRAM gene; in cell line BS-CRAMH23HB, both alleles of CRAM are activated (Fig. 6A).
FIG. 6.
Physical maps. (A) The CRAM locus in wild-type trypanosomes and bloodstream-form CRAM overexpressors. Top, structure of the CRAM locus in wild-type bloodstream-form trypanosomes and plasmid p3′CRAMH23H; middle, structure of the CRAM locus in cell line BS-CRAMH23H and plasmid p3′CRAMH23B; bottom, structure of the CRAM locus in cell line BS-CRAMH23HB. (B) Construction of bloodstream-form cell lines containing mutated CRAM. Top, structure of the CRAM locus in cell line BS-CRAM-B that contained only one allele of CRAM and plasmid p3′CRAMH23H-X for introducing mutation into the C-terminal domain of the CRAM; bottom, structure of the CRAM locus in bloodstream-form cell lines expressing mutated CRAM. Symbols and abbreviations are as described for Fig. 1.
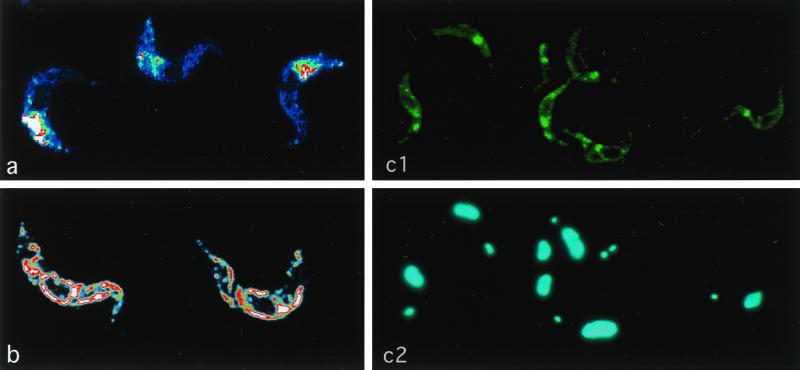
We measured the CRAM mRNA level in bloodstream-form CRAM overexpressors (Fig. 7A). In wild-type procyclics, two closely comigrating CRAM mRNAs of relatively high abundance were measured (Fig. 7A, lane P). A very low level of the CRAM mRNA was detected in the wild-type bloodstream form (Fig. 7A, lane BS). In cell line BS-CRAMH23H, the CRAM mRNA of a larger size was activated to a level equivalent to that in the procyclic form. In cell line BS-CRAMH23HB, both sizes of the CRAM mRNA were activated to a level equivalent to those in wild-type procyclics (Fig. 7A, lanes H and HB, respectively). The relative amount of the CRAM protein produced in each cell line was compared by Western blot analysis (Fig. 7B). CRAM protein was detected as a broad band of ∼200 kDa with anti-CRAM antibodies in the wild-type procyclic (Fig. 7B, lane P). Surprisingly, two sizes of the CRAM protein were detected in bloodstream form CRAM overexpressors; one is equivalent to that in the procyclic form, and the other one is much larger (Fig. 7B, lanes H and HB). The overall amount of CRAM proteins in bloodstream-form CRAM overexpressors is roughly similar to that in the procyclic form. A very low level of CRAM proteins was visualized in the cell extract derived from wild-type bloodstream form (Fig. 7B, lane BS). We do not understand the structural differences between the two sizes of CRAM proteins in cell lines BS-CRAMH23H and BS-CRAMH23HB. However, all of the CRAM protein accumulated in a single area close to the flagellar pocket of bloodstream-form CRAM overexpressors, as demonstrated by the immunofluorescence analysis of fixed and permeabilized trypanosomes (Fig. 8a; confocal images of BS-CRAMH23HB trypanosomes stained with anti-CRAM antibody). A similar result was observed in the BS-CRAMH23H cell line (data not shown). The localization of CRAM in the BS-CRAMH23HB cell line was further examined at high resolution by immuno-EM (Fig. 9A, 9B). CRAM predominantly located at the flagellar pocket and the Golgi in the BS-CRAMH23HB cell line. This result indicates that a similar sorting system may operate protein trafficking to the flagellar pocket in both the bloodstream-form and procyclic-form trypanosomes. Western blot analysis of the purified Golgi fraction showed that the relative abundance of the two sizes of CRAM protein were equally present in the Golgi fraction and the total cell lysate (data not shown). This result indicated that the Golgi did not preferentially retain a specific size class of CRAM in bloodstream-form trypanosomes.
FIG. 7.
Expression of CRAM or truncated CRAM in different bloodstream-form-transformed cell lines. (A) Northern blot analysis. Total RNAs from wild-type procyclic trypanosomes (P), wild-type bloodstream-form trypanosomes (BS), and BS-CRAMH23H (H), BS-CRAMH23HB (HB), BS-CRAM-40 (-40), BS-CRAM-29 (-29), BS-CRAM-19 (-19), and BS-CRAM-4 (-4) cell lines were separated in 1% formaldehyde agarose gels. The blot was hybridized to a 5′ CRAM probe (Fig. 1). The final posthybridization wash was performed in 0.1× SSC–0.1% SDS at 65°C. The bottom panels represent hybridization with the β-tubulin gene probe indicating the relative amount of RNA loaded in each lane. (B) Western blot analysis. Total protein lysates (∼2 × 107 trypanosomes), derived from the wild-type procyclic trypanosomes (P), the wild-type bloodstream-form trypanosomes (BS), and BS-CRAMH23H (H), BS-CRAMH23HB (HB), BS-CRAM-4 (-4), BS-CRAM-19 (-19), BS-CRAM-29 (-29), and BS-CRAM-40 (-40) cell lines were size separated in 6% polyacrylamide gels and electrophoretically transferred to nitrocellulose filters. The blots were probed with anti-CRAM antibody (24). The same blots were later probed with anti-Tb-29 (right) (26) or the anti-procyclic antibody identifying two proteins of ∼43 to 45 kDa (left) (Lee, unpublished) to demonstrate equal loading of protein.
FIG. 8.
Subcellular localization of different versions of CRAM in bloodstream-form cell lines. Trypanosomes were first incubated with rabbit-derived anti-CRAM antibody followed by reaction with FITC-conjugated goat anti-rabbit IgG. Then cells were stained with DAPI at a concentration of 0.1 μg/ml for 1 min. The images were analyzed by a confocal microscope. Different colors, arranged in the order blue-green-yellow-red-white, indicate the relative intensity of fluorescence (white, highest intensity; blue, lowest intensity). (a) Confocal image of the CRAM protein in the BS-CRAMH23HB cell line; (b) confocal image of total cellular CRAM protein in the BS-CRAM-40 cell line; (c1 and c2) surface staining of fixed, nonpermeabilized BS-CRAM-40 cells (c1, CRAM image; c2, DAPI staining).
FIG. 9.
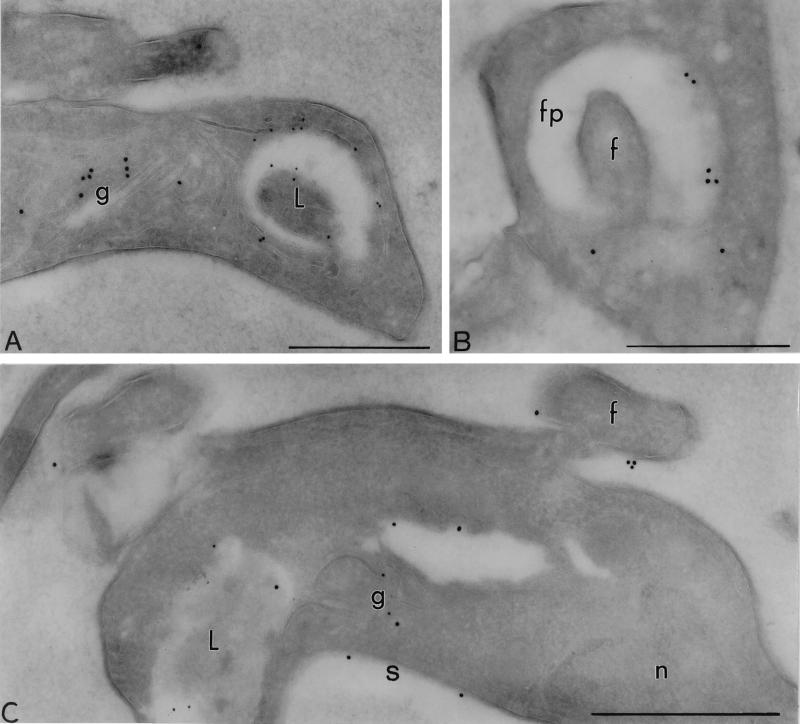
Immuno-EM localization of the CRAM protein in bloodstream-form CRAM overexpressor and BS-CRAM-40 cell line. (A and B) BS-CRAMH23HB probed with rabbit anti-CRAM (18 nm) and mouse MAb Ali I-218 (12 nm) directed against p67, a lysosomal glycoprotein (22). The label for CRAM is observed at the flagellar pocket and the Golgi in the BS-CRAMH23HB cell line. (C) The BS-CRAM-40 cell line probed with rabbit anti-CRAM (18 nm) and mouse MAb Ali I-218 against p67 (12 nm), showing that a significant amount of the CRAM protein was spread onto the surface in the BS-CRAM-40 cell line. fp, flagellar pocket; f, flagellum; g, Golgi; L, lysosome; n, nucleus; s, surface. Scale bars, 0.5 μm.
The fate of truncated CRAM proteins in bloodstream-form trypanosomes.
We subsequently addressed the importance of the C-terminal extension of CRAM in the presentation of the CRAM protein at the flagellar pocket in bloodstream-form trypanosomes. Following strategies described for the procyclic form, mutations were introduced into the CRAM gene in a previously established bloodstream-form cell line, BS-CRAM-B, that encodes only the smaller of the two CRAM alleles (the larger CRAM allele was deleted [Fig. 6B] [58]). Bloodstream-form cell lines BS-CRAM-40, BS-CRAM-29, BS-CRAM-19, and BS-CRAM-4, were established and expressed CRAM truncated by 40, 29, 19, and 4 amino acids, respectively, at its C terminus.
The expression of truncated CRAM proteins in transformed bloodstream-form cell lines was similar to those in bloodstream-form CRAM overexpressors as examined by Northern and Western blot analyses (Fig. 7). The subcellular localization of truncated CRAM protein in these bloodstream-form cell lines was examined by immunofluorescence assay. We found that (i) the CRAM protein remained at a single area close to the flagellar pocket in BS-CRAM-4 (data not shown); (ii) in BS-CRAM-19 cell line, the CRAM protein was mainly restricted inside the ER (data not shown); and (iii) in cell lines BS-CRAM-29 (data not shown) and BS-CRAM-40 (Fig. 8b and c), the CRAM protein was found at the ER, the flagellar pocket, and cell surface. Figure 8b demonstrates the Confocal image of the total CRAM distribution in cell line BS-CRAM-40. We were not able to observe a significant signal by live cell staining of the BS-CRAM-40 cell line with anti-CRAM and anti-ISG65 antibodies (as a control). However, a significant amount of surface staining was observed using fixed and nonpermeabilized trypanosomes: Fig. 8c shows that anti-CRAM stains the flagellar pocket and the outer cell surface of BS-CRAM-40 cells. The presence of CRAM on the surface of flagellum and some area of the cell surface in BS-CRAM-40 was further confirmed by immuno-EM analysis (Fig. 9C). It is possible that like the ISGs, the CRAM protein may be hidden underneath the VSG coat in the BS-CRAM-40 cell line. In summary, removing the putative sorting signal changed the fate of the CRAM protein in bloodstream-form trypanosomes. This result indicates that a similar protein sorting system may exist in both procyclic-form and bloodstream-form trypanosomes.
Can the N-terminal signal peptide of CRAM direct and confine proteins to the flagellar pocket?
To address whether the N-terminal signal peptide may play a role in confining protein to the flagellar pocket, we constructed a 5′CRAM-ISG65 fusion gene in which the N-terminal region (of 109 amino acids) of the ISG65 was replaced by that of CRAM (54 amino acids from the CRAM N terminus). When the 5′CRAM-ISG65 fusion gene was introduced into procyclic trypanosomes, we observed that the ISG65 fusion protein did not concentrate at the flagellar pocket but rather spread all over the cell (data not shown). This result indicated that the N-terminal signal peptide of CRAM is not sufficient to restrict proteins at the flagellar pocket.
Validation of the function of the cytoplasmic domain of CRAM on a transmembrane fusion protein.
We further address whether the cytoplasmic domain of CRAM is able to retain another transmembrane protein at the flagellar pocket, thus evaluating the role of the putative sorting signals on a different protein. A fusion gene containing the bacterial trpE gene flanked at its N terminus by the CRAM signal peptide and at the C terminus by the CRAM transmembrane domain and cytoplasmic extension was constructed and introduced into the βα-tubulin intergenic region (Fig. 10A). The TrpE-CRAM fusion protein was expressed in the transformed cell line at the predicted size of ∼85 kDa (data not shown). The fate of the TrpE-CRAM fusion proteins in stably transformed cell lines was analyzed by immuno-EM (Fig. 10B). The result demonstrated that the TrpE-CRAM fusion protein can indeed be colocalized with the endogenous CRAM at the flagellar pocket (Fig. 10B). However, we also found a significant amount of the fusion protein restricted inside the ER, indicating that this protein is not efficiently transported (data not shown). The result was similar for cell lines expressing an ISG65-CRAM fusion protein in which the signal peptide and the cytoplasmic domain of the ISG65 were replaced by those derived from CRAM (data not shown).
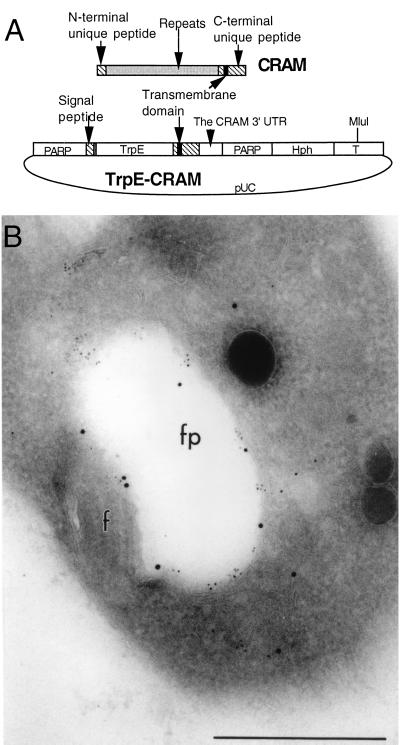
FIG. 10.
Localization of the TrpE-CRAM fusion protein at the flagellar pocket in transformed procyclic trypanosomes. (A) Diagram for the structure of the trpE-CRAM fusion gene (see Materials and Methods for the detailed description of the plasmid). The construct was linearized at the MluI site located at the center of tubulin intergenic region (T) and then electroporated into trypanosomes. (B) Immuno-EM localization of the TrpE-CRAM fusion protein in a transformed procyclic cell line. The cell line was doubled labeled with rabbit-derived anti-TrpE (15 nm of gold) (Lee, unpublished) and rat-derived anti-CRAM (5 nm of gold). Both antibodies have labeled the lumenal face of the flagellar pocket (fp). Scale bar, 0.5 μm.
DISCUSSION
Using mutagenesis, we dissected each domain of the CRAM protein and investigated its role in the targeting and retention of CRAM at the flagellar pocket of trypanosomes. Based on domain swapping experiments, we demonstrate that different N-terminal signal peptides and transmembrane domains have no influence on protein localization at the flagellar pocket. Changing the amino acid sequence of the CRAM cytoplasmic extension changes the fate of the CRAM protein in trypanosomes. Based on deletional mutation analysis, it appears that multiple functional domains may exist in the CRAM cytoplasmic extension. (i) The last four amino acids of CRAM are not required for the transport and routing of the protein at the pocket. (ii) Deletion up to amino acids −8 to −19 from the C terminus resulted in the CRAM retained in the ER, indicating that these amino acids are essential for protein export from the ER. We tentatively refer to this domain as a transport signal. (iii) Deletion of amino acids up to position −29 and −40 from the C terminus resulted in some CRAM protein being spread onto the cell surface and the surface of flagellum, though a significant amount of the protein is still retained in the ER. This result suggested that the amino acid sequences from −20 to −40 from the CRAM C terminus may be important for holding the CRAM protein at the flagellar pocket. This region is referred to as a putative flagellar pocket retention signal.
In all eukaryotes, membrane-bound proteins predominantly originate in the ER and then are sorted and transported by vesicles to their final destinations. Studies on the machinery of vesicle transport in mammalian cells and yeast have established some general rules that individual proteins are dependent on the intrinsic signals that dictate their ability to enter or not enter a given vesicle shuttle (for reviews, see references 18, 35, 40, and 44). These signals, called sorting signals, are discrete peptide domains of 4 to 25 residues or conformationally determined epitopes. A given protein can have multiple sorting signals, each identifying the fate of the protein at consecutive stages. A sorting signal which specifies movement is termed a transport signal; the one which specifies lack of movement is termed a retention signal. The transport signals can facilitate the assembly of targeted molecules into transport vesicles by interacting with secretory vesicle coat proteins (for examples, COPI and COPII in mammalian cells and yeast) and thus facilitate selective export of targeted molecules from the ER. Additional mechanisms also control export from the ER. Recent studies have indicated that after translocation across the membrane of the ER, newly synthesized proteins must be properly folded, assembled, and/or modified in order to be competent for transport from the ER to their final destination. On the other hand, misfolded, incompletely folded, and partially assembled proteins are transport incompetent, retained, and eventually degraded. This sorting process, called quality control, prevents proteins departing from the ER until fully folded (18). Thus the differences in the efficiency of folding/assembly may also attribute to the variability observed in the efficiency of protein export.
Based on knowledge obtained from study of other eukaryotes, we postulate that putative transport signals may exist in the amino acids spanning −5 to −19 (or further upstream) of the CRAM C terminus. With the loss of these signals, the CRAM protein was probably either not properly folded and/or unable to be selected by transport vesicles and was thus retained in the ER. We are currently performing experiments to distinguish these possibilities. In cell lines CRAM-29 and CRAM-40, a significant amount of CRAM protein is again targeted to the flagellar pocket. These truncated CRAM proteins presumably are able to enter a budding vesicle without acquiring the putative transport signals. We postulate that this event may occur due to the prevailing concentration of CRAM in the ER of cell lines CRAM-29 and CRAM-40, allowing transport by mechanisms similar to the process called bulk flow (36, 57). Comparing the amino acid sequence of the CRAM C terminus to the data bank, we found no obvious functional motifs similar to those involved in protein sorting and transport in higher eukaryotes. Interestingly, a 5-of-11 amino acid homology was found in the −15 to −4 amino acid sequence of the CRAM C terminus and the domain responsible for internalization in human LDL receptor (residues 801 to 812) and LDL receptor-related protein (residues 4482 to 4493) (24).
Unlike PARP, VSG, and some ISGs that are homogeneously distributed on the entire cell surface of trypanosomes, the truncated CRAM protein in cell lines CRAM-29 and CRAM-40 does not distribute uniformly onto the entire cell surface but covers predominantly the surface of the flagellum in a punctuated pattern in procyclic trypanosomes. We currently do not understand how the truncated CRAM protein selectively distributes to the flagellum in these cell lines. Nevertheless, our results indicate that sequences immediately downstream of the transmembrane domain (the putative retention signal) of CRAM are important for localization of CRAM to the flagellar pocket membrane. We hypothesize that the putative flagellar pocket retention signal may be recognized by proteins associated with the flagellar pocket, thus resulting in the retention of CRAM at the pocket membrane. Our current data do not exclude the possibility that the extracellular repeated peptide domain and its downstream adjacent unique peptide of the CRAM protein may play important roles in the correct routing of the protein to the flagellar pocket.
Our studies address unanswered questions related to vesicular trafficking in trypanosomes. Very little information is available on protein sorting and transport in trypanosomes (4, 8, 11, 37). Vesicle coat protein homologs have not yet been characterized. A recent report described the GPI-dependent secretory transport in T. brucei in which a GPI-minus VSG is secreted from transformed procyclic trypanosomes with a much reduced efficiency and a large amount of the GPI-minus VSG accumulated in the ER (3). The authors hypothesized that efficient transport of GPI-anchored proteins in procyclic trypanosomes may be mediated in a positive manner by the presence of a GPI moiety and that the delayed forward transport was not due to misfolding or misassembling in the absence of a GPI moiety (31). Hill et al. demonstrated that a structural motif in the T-lymphocyte triggering factor may be involved in the targeting of this factor to the anterior or cytoplasmic face of the flagellar pocket via interaction with trypanosome cytoskeleton (19). Several mechanisms have been documented for localization proteins to the flagellar membrane or the flagellum (5, 15, 46, 47). Studies of glucose transporters in Leishmania enriettii demonstrated that a short stretch of amino acid at the N-terminal domain of the isoform 1 glucose transporter is essential for targeting to the flagellar membrane (46, 47). Godsel and Engman showed that myristoylation and palmitoylation at the N terminus of the flagellar calcium-binding protein of Trypanosoma cruzi are required for its flagellar localization mediated through association with the flagellar plasma membrane (15). The study of flagellar morphogenesis by Gull's laboratory identified two sequence domains required for targeting and assembly of the paraflagellar rod proteins into the flagellum of trypanosomes (5). Protein sorting signals identified in the CRAM cytoplasmic domain do not resemble to those involved in targeting proteins to the flagella of trypanosomes.
The study of the fate of CRAM proteins in bloodstream-form trypanosomes indicated that similar sorting systems exist in both stages of the parasite, though unique properties during transporting proteins from the Golgi to the pocket may still operate in different life cycle stages of the parasite. In bloodstream-form trypanosomes, the GPI-anchored transferrin receptor complex is localized at the flagellar pocket. When this receptor complex was expressed in procyclic trypanosomes, it did not exclusively localize to the flagellar pocket but was expressed throughout the cell surface of the trypanosome (28). The mechanism involving anchoring of the transferrin receptor complex at the flagellar pocket of the bloodstream form is not clear, though mechanisms similar to those proposed for CRAM of the procyclic form may operate. In this event, a third protein functioning as a routing protein may be involved in holding the GPI-anchored transferrin receptor onto the flagellar pocket of bloodstream-form trypanosomes. Alternatively, other sorting mechanisms requiring novel sorting signals may also be present.
In summary, our systematic dissection of sequences involved in the localization of CRAM at the flagellar pocket of trypanosomes has identified putative sorting signals at the CRAM cytoplasmic extension. We will now use these putative sorting signals as bait to further unravel the machinery of secretory trafficking in trypanosomes.
ACKNOWLEDGMENTS
We thank P. Borst, J. de Diego, M. Muranjan, J. Raper, and L. H. T. Van der Ploeg for critical reading of the manuscript. We thank J. D. Bangs for providing anti-Bip antibody and P. Overath for providing the cDNA clone of ISG65.
This work was supported by NIH grant AI31117 to M.G.-S.L., who is a Burroughs Wellcome Fund New Investigator in Molecular Parasitology.
REFERENCES
- 1.Balber A E. The pellicle and the membrane of the flagellum, flagellar adhesion zone and flagellar pocket: functionally discrete domains of the bloodstream form of African trypanosomes. Crit Rev Immunol. 1990;10:177–201. [PubMed] [Google Scholar]
- 2.Bangs J D, Uyetake L, Brichman M J, Balber A E, Boothroyd J C. Molecular cloning and cellular localization of a Bip homologue in Trypanosoma brucei. J Cell Sci. 1993;105:1101–1113. doi: 10.1242/jcs.105.4.1101. [DOI] [PubMed] [Google Scholar]
- 3.Bangs J D, Ransom D M, McDowell M A, Brouch E M. Expression of bloodstream variant surface glycoproteins in procyclic stage Trypanosoma brucei: role of GPI anchors in secretion. EMBO J. 1997;16:4285–4294. doi: 10.1093/emboj/16.14.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bangs J D. Surface coats and secretory trafficking in African trypanosomes. Curr Opin Microbiol. 1998;4:448–454. doi: 10.1016/s1369-5274(98)80064-7. [DOI] [PubMed] [Google Scholar]
- 5.Bastin P, MacRae T H, Francis S B, Matthews K R, Gull K. Flagellar morphogenesis: protein targeting and assembly in the paraflagellar rod of trypanosomes. Mol Cell Biol. 1999;19:8191–8200. doi: 10.1128/mcb.19.12.8191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borst P. Molecular genetics of antigenic variation. Immunoparasitol Today. 1991;March:A29–A33. doi: 10.1016/S0167-5699(05)80009-X. [DOI] [PubMed] [Google Scholar]
- 7.Borst P. Transferrin receptor, antigenic variation and the prospect of a trypanosome vaccine. Trends Genet. 1991;7:307–309. doi: 10.1016/0168-9525(91)90406-g. [DOI] [PubMed] [Google Scholar]
- 8.Borst P, Fairlamb A H. Surface receptors and transporters of Trypanosoma brucei. Annu Rev Microbiol. 1998;52:745–778. doi: 10.1146/annurev.micro.52.1.745. [DOI] [PubMed] [Google Scholar]
- 9.Brun R, Schonenberger M. Cultivation and in vitro cloning of procyclic culture form Trypanosoma brucei in a semidefined medium. Acta Trop. 1979;36:289–292. [PubMed] [Google Scholar]
- 10.Chirgwin J M, Przybyla A E, MacDonald R J, Rutter W J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 11.Clayton C E, Hausler T, Blattner J. Protein trafficking in Kinetoplastid protozoa. Microbiol Rev. 1995;59:325–344. doi: 10.1128/mr.59.3.325-344.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coppens I, Opperdoes F R, Courtoy P J, Baudhuin P. Receptor mediated endocytosis in the bloodstream form of Trypanosoma brucei. J Protozool. 1987;34:465–473. doi: 10.1111/j.1550-7408.1987.tb03216.x. [DOI] [PubMed] [Google Scholar]
- 13.Coppens I, Baudhuin P, Opperdoes F R, Courtoy P J. Receptors for the host low density lipoproteins on the hemoflagellate Trypanosoma brucei: purification and involvement in the growth of the parasite. Proc Natl Acad Sci USA. 1988;85:6753–6757. doi: 10.1073/pnas.85.18.6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cross G A M. Cellular and genetic aspects of antigenic variation in trypanosomes. Annu Rev Immunol. 1990;8:83–110. doi: 10.1146/annurev.iy.08.040190.000503. [DOI] [PubMed] [Google Scholar]
- 15.Godsel L M, Engman D M. Flagellar protein localization mediated by a calcium-myristoyl/palmitoyl switch mechanism. EMBO J. 1999;18:2057–2065. doi: 10.1093/emboj/18.8.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grab D J, Wells C W, Shaw M K, Webster P, Russo D C W. Endocytosed transferrin in African trypanosomes is delivered to lysosomes and may not be recycled. Eur J Cell Biol. 1992;59:398–404. [PubMed] [Google Scholar]
- 17.Grab D J, Shaw M K, Wells C W, Verjee Y, Russo D C W, Webster P, Naessens J, Fish W R. The transferrin receptor in African trypanosomes: identification, partial characterization and subcellular localization. Eur J Cell Biol. 1993;62:114–126. [PubMed] [Google Scholar]
- 18.Hammond G, Helenius A. Quality control in the secretory pathway. Curr Opin Cell Biol. 1995;7:523–529. doi: 10.1016/0955-0674(95)80009-3. [DOI] [PubMed] [Google Scholar]
- 19.Hill K L, Hutchings N R, Russell D G, Donelson J E. A novel protein targeting domain directs proteins to the anterior cytoplasmic face of the flagellar pocket in African trypanosomes. J Cell Sci. 1999;112:3091–3101. doi: 10.1242/jcs.112.18.3091. [DOI] [PubMed] [Google Scholar]
- 20.Huirumi H, Huirumi M. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J Parasitol. 1989;75:985–989. [PubMed] [Google Scholar]
- 21.Jackson D G, Windle H J, Voorheis H P. The identification, purification, and characterization of two invariant surface glycoproteins located beneath the surface coat barrier of bloodstream forms of Trypanosoma brucei. J Biol Chem. 1993;268:8085–8095. [PubMed] [Google Scholar]
- 22.Kelley R J, Alexander D, Cowan C, Balber A E, Bangs J D. Molecular cloning of p67, a lysosomal membrane glycoprotein from Trypanosoma brucei. Mol Biochem Parasitol. 1999;98:17–28. doi: 10.1016/s0166-6851(98)00155-8. [DOI] [PubMed] [Google Scholar]
- 23.Langreth S G, Balber A E. Protein uptake and digestion in bloodstream and culture forms of T. brucei. J Protozool. 1975;22:40–53. doi: 10.1111/j.1550-7408.1975.tb00943.x. [DOI] [PubMed] [Google Scholar]
- 24.Lee G-S M, Bihain B E, Deckelbaum R J, Russell D G, Van der Ploeg L H T. Characterization of a cDNA encoding a cysteine-rich cell surface protein located in the flagellar pocket of the protozoan Trypanosoma brucei. Mol Cell Biol. 1990;10:4506–4517. doi: 10.1128/mcb.10.9.4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee G-S M, Van der Ploeg L H T. The hygromycin B-resistance-encoding gene as a selectable marker for stable transformation of Trypanosoma brucei. Gene. 1991;105:255–257. doi: 10.1016/0378-1119(91)90159-9. [DOI] [PubMed] [Google Scholar]
- 26.Lee G-S M, Russell D, D'Alesandro P A, Van der Ploeg L H T. Identification of membrane-associated proteins in Trypanosoma brucei, encoding an internal, EARLRAEE amino acid repeat. J Biol Chem. 1994;269:8408–8415. [PubMed] [Google Scholar]
- 27.Lee G-S M. An RNA polymerase II promoter in the hsp70 locus of Trypanosoma brucei. Mol Cell Biol. 1996;16:1220–1230. doi: 10.1128/mcb.16.3.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ligtenberg M J L, Bitter W, Kieft R, Steverding D, Janssen H, Calafat J, Borst P. Reconstitution of a surface transferrin binding complex in insect form Trypanosoma brucei. EMBO. 1994;13:2565–2573. doi: 10.1002/j.1460-2075.1994.tb06546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lingnau A, Zufferey R, Lingau M, Russell D G. Characterization of tGLP-1, a Golgi and lysosome-associated, transmembrane glycoprotein of African trypanosomes. J Cell Sci. 1999;112:3061–3070. doi: 10.1242/jcs.112.18.3061. [DOI] [PubMed] [Google Scholar]
- 30.Liu J, Qiao X, Du D, Lee M G-S. Receptor mediated endocytosis in the procyclic form of Trypanosoma brucei. J Biol Chem. 2000;275:12032–12040. doi: 10.1074/jbc.275.16.12032. [DOI] [PubMed] [Google Scholar]
- 31.McDowell M A, Ransom D M, Bangs J D. Glycosylphosphatidylinositol-dependent secretory transport in Trypanosoma brucei. Biochem J. 1998;335:681–689. doi: 10.1042/bj3350681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mehlert A, Zitzmann N, Richardson J M, Treumann A, Ferguson M J. The glycosylation of the variant surface glycoproteins and procyclic acidic repetitive proteins of Trypanosoma brucei. Mol Biochem Parasitol. 1998;91:145–152. doi: 10.1016/s0166-6851(97)00187-4. [DOI] [PubMed] [Google Scholar]
- 33.Mowatt M R, Clayton C E. Developmental regulation of a novel repetitive protein of Trypanosoma brucei. Mol Cell Biol. 1987;7:2838–2844. doi: 10.1128/mcb.7.8.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nolan D P, Jackson D G, Windle H J, Pays A, Geuskens M, Michel A, Voorheis H P, Pays E. Characterization of a noval stage-specific, invariant surface protein in Trypanosoma brucei containing an internal, serine-rich, repetitive motif. J Biol Chem. 1997;272:29212–29221. doi: 10.1074/jbc.272.46.29212. [DOI] [PubMed] [Google Scholar]
- 35.Pfeffer S R. Transport-vesicle targeting: tethers before SNAREs. Nat Cell Biol. 1999;1:17–22. doi: 10.1038/8967. [DOI] [PubMed] [Google Scholar]
- 36.Orci L, Glick B S, Rothman J E. A new type of coated vesicular carrier that appears not to contain Clathrin: its possible role in protein transport within the Golgi stack. Cell. 1986;46:171–184. doi: 10.1016/0092-8674(86)90734-8. [DOI] [PubMed] [Google Scholar]
- 37.Overath P, Stierhof Y D, Wiese M. Endocytosis and secretion in trypanosomatid parasites—tumultuous traffic in a pocket. Trends Cell Biol. 1997;7:27–33. doi: 10.1016/S0962-8924(97)10046-0. [DOI] [PubMed] [Google Scholar]
- 38.Pays E. Genome organization and control of gene expression in Trypanosomatids. Symp Soc Gen Microbiol. 1992;50:127–160. [Google Scholar]
- 39.Roditi I, Schwarz H, Pearson T W, Beecroft R P, Liu M K, Richardson J P, Buhring H J, Pleiss J, Bulow R, Williams R O, Overath P. Procyclin expression and loss of the variant surface glycoprotein during differentiation of Trypanosoma brucei. J Cell Biol. 1989;108:737–746. doi: 10.1083/jcb.108.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rothman J E, Wieland F T. Protein sorting by transport vesicles. Science. 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- 41.Rudenko G, Le Blancq S, Smith J, Lee M G-S, Rattray A, Van der Ploeg L H T. Procyclic acidic repetitive protein (PARP) genes located in an unusually small α-amanitin-resistant transcription unit: PARP promoter activity assayed by transient DNA transfection of Trypanosoma brucei. Mol Cell Biol. 1990;10:3492–3504. doi: 10.1128/mcb.10.7.3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Russell D G, Xu S, Chakraborty P. Intracellular trafficking and the parasitophorous vacuole of Leishmania mexicana-infected macrophages. J Cell Sci. 1992;103:1193–1210. doi: 10.1242/jcs.103.4.1193. [DOI] [PubMed] [Google Scholar]
- 43.Salmon D, Geuskens M, Hanocq F, Hanocq-Quertier J, Nolan D, Ruben L, Pays E. A novel heterodimeric transferrin receptor encoded by a pair of VSG expression site-associated genes in T. brucei. Cell. 1994;78:75–86. doi: 10.1016/0092-8674(94)90574-6. [DOI] [PubMed] [Google Scholar]
- 44.Schekman R, Orci L. Coat proteins and vesicle budding. Science. 1996;271:1526–1533. doi: 10.1126/science.271.5255.1526. [DOI] [PubMed] [Google Scholar]
- 45.Schell D, Ever R, Pries D, Ziegelbauer K, Kiefer H, Lottspeich F, Cornelissen A W C A, Overath P. A transferrin-binding protein of Trypanosoma brucei is encoded by one of the genes in the variant surface glycoprotein gene expression site. EMBO J. 1991;10:1061–1066. doi: 10.1002/j.1460-2075.1991.tb08045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Snapp E L, Landfear S M. Cytoskeleton association is important for differential targeting of glucose transporter isoforms in Leishmania. J Cell Biol. 1997;139:1775–1783. doi: 10.1083/jcb.139.7.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Snapp E L, Landfear S M. Characterization of a targeting motif for a flagellar membrane protein in Leishmania enriettii. J Biol Chem. 1999;274:29543–29548. doi: 10.1074/jbc.274.41.29543. [DOI] [PubMed] [Google Scholar]
- 48.Stanley K K, Kocher H P, Luzio J P, Jackson P, Tschopp J. The sequence and topology of human complement component C9. EMBO J. 1985;4:375–382. doi: 10.1002/j.1460-2075.1985.tb03639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steverding D, Chaudhri M, Stierhof Y D, Overath P, Ligtenberg M, Borst P. ESAG 6 and 7 products of Trypanosoma brucei form a transferrin binding protein complex. J Cell Biol. 1994;64:78–87. [PubMed] [Google Scholar]
- 50.Szostak J W, Orr-Weaver T L, Rothstein R J, Stahl F W. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 51.Thomashow L S, Milhausen M, Rutter W J, Agabian N. Tubulin genes are tandemly linked and clustered in the genome of Trypanosoma brucei. Cell. 1983;32:35–43. doi: 10.1016/0092-8674(83)90494-4. [DOI] [PubMed] [Google Scholar]
- 52.Van der Ploeg L H T, Bernards A, Rijswijk F A M, Borst P. Characterization of the DNA duplication-transposition that controls the expression of two genes for variant surface glycoproteins in Trypanosoma brucei. Nucleic Acids Res. 1982;10:593–609. doi: 10.1093/nar/10.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van der Ploeg L H T. Antigenic variation in African trypanosomes: genetic recombination and transcriptional control of VSG genes. In: Hames B D, Glover D, editors. Frontiers in molecular biology. Oxford, England: IRL Press; 1990. pp. 51–97. [Google Scholar]
- 54.Vickerman K. On the surface coat and flagellar adhesion in trypanosomes. J Cell Sci. 1969;5:163–193. doi: 10.1242/jcs.5.1.163. [DOI] [PubMed] [Google Scholar]
- 55.Vickerman K. The fine structure of Trypanosoma congolense in its bloodstream phase. J Protozool. 1969;16:54–69. doi: 10.1111/j.1550-7408.1969.tb02233.x. [DOI] [PubMed] [Google Scholar]
- 56.Webster P, Russell D G. The flagellar pocket of Trypanosomatids. Parasitol Today. 1993;9:201–206. doi: 10.1016/0169-4758(93)90008-4. [DOI] [PubMed] [Google Scholar]
- 57.Wieland F T, Gleasson M L, Serafini T A, Rothman J E. The rate of bulk flow from the endoplasmic reticulum to the cell surface. Cell. 1987;50:289–300. doi: 10.1016/0092-8674(87)90224-8. [DOI] [PubMed] [Google Scholar]
- 58.Zheng B, Yao H, Lee M G-S. Inactivation of the gene encoding the flagellar pocket protein CRAM in African trypanosomes. Mol Biochem Parasitol. 1999;100:235–242. doi: 10.1016/s0166-6851(99)00048-1. [DOI] [PubMed] [Google Scholar]
- 59.Ziegelbauer K, Multhaup G, Overath P. Molecular characterization of two invariant surface glycoproteins specific for the bloodstream stage of Trypanosoma brucei. J Biol Chem. 1992;267:10797–10803. [PubMed] [Google Scholar]
- 60.Ziegelbauer K, Overath P. Organization of two invariant surface glycoproteins in the surface coat of Trypanosoma brucei. Infect Immun. 1993;61:4540–4545. doi: 10.1128/iai.61.11.4540-4545.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]