Abstract
Background
As an immune regulator expressed on the surface of activated T cells, programmed cell death 1 (PDCD1) plays an important role in psoriasis. However, whether PDCD1 genetic polymorphism is associated with psoriasis has yet to be explored.
Objective
To study the association between polymorphisms of the immune‐related gene PDCD1 and psoriasis susceptibility in the Chinese population, to illustrate the genetic mechanism of psoriasis and provide new research ideas for the diagnosis and treatment of psoriasis (PS).
Methods
Overall, 128 psoriasis patients and 88 healthy controls were included in this study. Using polymerase chain reaction (PCR)‐Sanger sequencing analysis, six PDCD1 single nucleotide polymorphisms (SNPs) were sequenced: PD1.1, PD1.3, PD1.4, PD1.5, PD1.6, and PD1.9.
Results
Among the six tested SNPs, PD1.6 showed a significant association with psoriasis in genotype and allele frequency distribution. The G allele of PD1.6 increased the risk of psoriasis (P = 0.03). In contrast, the other five SNPs failed to show association with psoriasis. Further analysis within the patient group showed that the frequency of the PD1.6 G allele was relatively high in severe psoriasis, but the difference was nonsignificant.
Conclusion
PDCD1 gene polymorphism is associated with psoriasis. The population carrying PD1.6 allele G are at a higher risk of developing psoriasis, though the severity of psoriasis does not correlate with PD1.6 polymorphism.
Introduction
Psoriasis is a common inflammatory skin disease often characterized by obvious erythema and overlying scales, accompanied by a qualitative itching sensation. This disease seriously affects patients' daily life, emotions, and work and is listed by the World Health Organization (WHO) as one of the 20 most intractable diseases. The incidence of psoriasis has increased over the last few decades. In China, for example, the incidence of psoriasis has increased from 0.12% in 1984 to 0.72% in recent years. 1 The incidence of this disease is higher in urban areas than in rural areas and generally lower in the south than in the north. 2 To date, the etiology and pathogenesis of psoriasis are still uncertain.
Psoriasis is generally regarded as a disorder of the immune system with genetic tendency. Multiple immune‐related gene polymorphisms, such as human leukocyte antigen‐Cw6 (HLA‐Cw6), which encodes a major histocompatibility complex class I allele, have been described to be associated with psoriasis, mainly relating to guttate‐type psoriasis and greater body surface area (BSA) involvement. Patients with positive HLA‐Cw6 also reported worsening of psoriasis during and after throat infection. 3 Several genetic associations with psoriasis have been found in critical genes for T‐cell development and polarization. One of these genes is runt‐related transcription factor 3 (RUNX3), which encodes a transcription factor in the runt‐domain containing family. RUNX3 is important for CD8+ T‐cell development, promotion of Th1 polarization, and possibly Th17 polarization. 4 The genetic basis of psoriasis is complex and includes many different genes. Thus far, genes involved in antigen presentation, T‐cell receptor development and polarization, and the nuclear factor κβ (NF‐κβ) pathway have been identified. 4
The human programmed cell death 1 (PDCD1) gene (Gene ID: 5133) is located at chromosome 2q37.3, with a total length of 9,011 bp and six exons. The total length of the messenger RNA (mRNA, NM_005018.3) is 2,097 bp. The protein precursor of PDCD1 (NP_005009) consists of 288 amino acid residues, while its mature protein contains 268 amino acid residues. The PDCD1 gene encodes a protein (PD‐1) with an immunoreceptor tyrosine‐based inhibitory motif (ITIM), which could inhibit autoreactive T cells. 5 It has been described to play an important role in the inflammatory response of psoriasis by acting as an immune regulator expressed on the surface of activated T cells. 6 De Bock et al. described a case of a 65‐year‐old woman with psoriasis exacerbation who was treated with nivolumab (anti‐PD‐1) for stage IV melanoma. They also reviewed the literature of approximately 34 other cases with an exacerbation of psoriasis during treatment with anti‐PD‐1 or PD‐L1 therapy. 7 Bartosińska et al. reported that the absolute numbers and percentages of CD4+PD1+ and CD8+PD1+ T cells were significantly decreased in psoriatic patients compared with the control group. 8 Because PD‐1 could inhibit autoreactive T cells, research should address the unresolved topic of whether genetic polymorphisms of PDCD1 are associated with psoriasis.
Although previous studies demonstrated the role of PDCD1 in psoriasis, whether PDCD‐1 gene variations influence the risk of psoriasis remains unknown. The present study investigated six functional single nucleotide polymorphisms (SNPs) that have been linked to autoimmune diseases, such as systemic lupus erythematosus, 9 , 10 rheumatoid arthritis, 11 , 12 , 13 and multiple sclerosis. 14 The locations of the SNPs in the PDCD1 gene were as follows: PD1.1 in the promoter (rs36084323, NM_005018.2:c.‐606G>A), PD1.3 in intron 4 (rs11568821, NM_005018.2:c.627+189G>A), PD1.4 in intron 4 (rs6705653, NM_005018.2: c.628‐110A>G), PD1.9 in exon 5 (rs2227982, NP_005009.2:p. Ala215 Val), PD1.5 in exon 5 (rs2227981, NP_005009.2:p.Ala268=), and PD1.6 in the 3′‐untranslated region (rs10204525, NM_005018.2:c.*889G>A).
Materials and methods
Subjects
All psoriasis patients were from the dermatology ward and outpatient department of Yueyang Hospital of Integrated Traditional Chinese and Western Medicine affiliated with Shanghai University of Traditional Chinese Medicine, China. The diagnosis of psoriasis in all patients was confirmed based on clinical manifestations and histopathological results (Fig. 1). A total of 128 patients with a complete history and an average age of 53.9 years (47 women and 81 men) were recruited for the present study from January 2017 to January 2020. There were 109 cases of psoriasis vulgaris, two cases of pustular psoriasis, seven cases of arthropathic psoriasis, and 10 cases of erythrodermic psoriasis. The severity of the skin lesions was assessed by means of the Psoriasis Area Severity Index (PASI) and BSA. For the control group, 88 normal healthy volunteers with an average age of 52.4 years (43 women and 45 men) were recruited from the Medical Center of Yueyang Hospital of Integrated Traditional Chinese and Western Medicine affiliated with Shanghai University of Traditional Chinese Medicine, China, during the same period. The study was approved by the Clinic IRB of Yueyang Hospital of Integrated Traditional Chinese and Western Medicine, Shanghai University of Traditional Chinese Medicine (Approval#:2017‐016). All healthy controls were confirmed to have no skin diseases or autoimmune diseases. Informed consent was obtained from all patients and healthy controls. Information regarding sex, age, PASI score, and BSA score is listed in Table 1.
Figure 1.
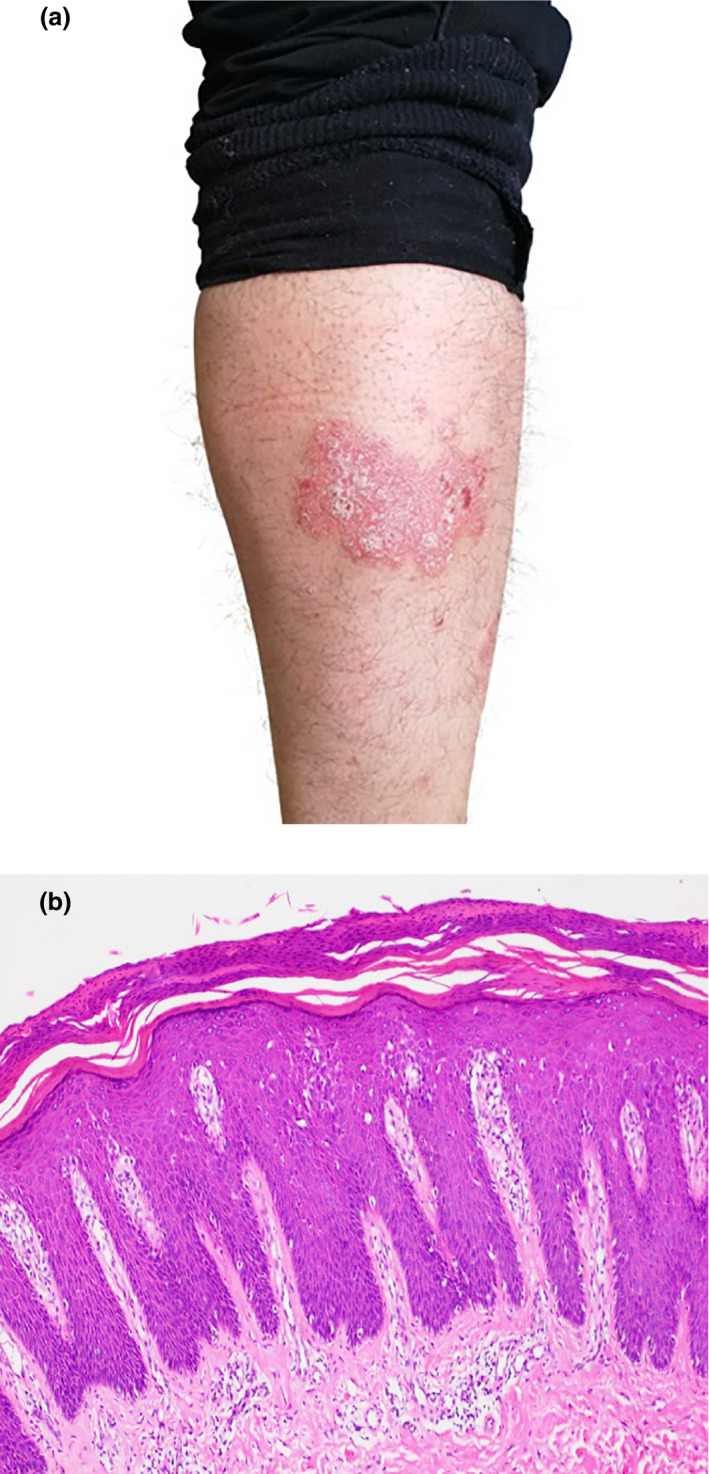
(a) A representative photo of skin lesions in a male patient with psoriasis. (b) A representative histopathological image of psoriasis (Hematoxylin and eosin, ×100)
Table 1.
Clinical information of psoriasis patients and healthy controls
| Group | Psoriasis | Control | Severe psoriasis | Mild‐moderate psoriasis | |
|---|---|---|---|---|---|
| Number | 128 | 88 | 77 | 51 | |
| Gender | |||||
| Female | 47 (36.72%) | 43 (48.86%) | 16 (20.78%) | 31 (60.78%) ## | |
| Male | 81 (63.28%) | 45 (51.14%) | 61 (79.22%) | 20 (39.22%) ## | |
| Age | |||||
| Min–max | 21–87 | 25–83 | 21–83 | 22–87 | |
|
|
53.9 ± 16.4 | 52.4 ± 15.9 | 55.71 ± 15.90 | 51.20 ± 16.80 | |
| PASI score | |||||
| Min–max | 1.5–51.6 | 12–51.6 | 1.5–9.9 | ||
|
|
15.81 ± 9.95 | 21.68 ± 8.61 | 6.96 ± 2.37 ## | ||
| BSA score | |||||
| Min–max | 2–70 | 12–70 | 2–9.6 | ||
|
|
22.69 ± 17.79 | 33.55 ± 14.98 | 6.28 ± 2.43 ## | ||
| Psoriasis subtype | Psoriasis vulgaris | Pustular psoriasis | Arthropathic psoriasis | Erythrodermic psoriasis | |
| Number | 109 (85.16%) | 2 (1.56%) | 7 (5.47%) | 10 (7.81%) | |
P < 0.01 vs. severe psoriasis.
For the analysis of PDCD1 SNPs and psoriasis severity, 128 patients with psoriasis were further divided into the severe psoriasis group (77 patients) (PASI > 10, BSA > 10%) and the mild–moderate psoriasis group (51 patients) (PASI < 10, BSA < 10%), based on PASI score and BSA score. Gender and age information is listed in Table 1.
Genotyping
A peripheral venous blood sample of 1 ml was drawn from each subject by standard venipuncture and collected into EDTA‐treated blood collection tubes. Then, genomic DNA was extracted using a commercial kit (CW2087, CoWin Biosciences, Peking, China).
The six SNPs of the PDCD1 gene, PD1.1/1.3/1.4/1.5/1.6/1.9/, were determined using polymerase chain reaction (PCR)‐based Sanger sequencing analysis. Four PCR primer pairs were designed and synthesized to amplify genomic DNA fragments containing target SNPs, as listed in Table 2. The PCR products were then purified and sequenced with an ABI 3730 DNA Analyzer (Applied Biosystems, Foster City, CA, USA) following the manufacturer's instructions.
Table 2.
Primers for PCR amplification of PDCD1 SNPs
| PCR primer pair number | Target SNPs | Upstream primer (5′‐>3′) | Downstream primer (5′‐>3′) | PCR product size (bp) |
|---|---|---|---|---|
| 1 | PD1.1 (rs36084323) | AAGCCATTTCACAAACGCCG | CATTCTGTCGGAGCCTCTGG | 408 |
| 2 | PD1.3 (rs11568821) | GCCTGCAGGACTCACATTCT | CTGTGTGATCTGGGGACACC | 422 |
| 3 |
PD1.4 (rs6705653) PD1.5 (rs2227981) PD1.9 (rs2227982) |
GGTGTCCCCAGATCACACAG | TTCTCCTGAGGAAATGCGCT | 478 |
| 4 | PD1.6 (rs10204525) | ATCCTACGGTCCCAAGGTCA | GCAGTGTGTGGATGTGAGGA | 268 |
PCR, polymerase chain reaction; SNPs, single nucleotide polymorphisms.
Statistical analyses
All statistical analyses were performed using SAS 9.0 statistical software (SAS Institute Inc., Cary, NC, USA). For comparison between the two groups (patients vs. controls and severe vs. mild–moderate psoriasis), the quantitative data (age, PASI score, and BSA score) were expressed as the mean ± standard deviation (SD) and compared using Welch's two‐sample t test, and the genotype and allele distributions of SNPs were compared by Fisher's exact test. The deviation from Hardy–Weinberg equilibrium (HWE) was examined in the control group by Fisher's exact test. The case–control association analysis for the genotypes and alleles of SNP was performed with the logistic regression model, and the odds ratios (ORs) and 95% confidence intervals (95% CIs) were calculated. Two‐tailed P < 0.05 was considered statistically significant.
Results
Six SNPs in the PDCD1 gene were successfully genotyped in 128 psoriasis patients and 88 healthy controls, with representative sequencing diagrams attached in Figure S1. The genotype and allele distributions of these SNPs are shown in Table 3. For all six SNPs, the genotype distributions in the control population conformed to HWE. Among the six SNPs, the genotype and allele distributions of PD1.6 (rs10204525) significantly differed between psoriasis patients and healthy controls (P < 0.05), while genotypic distributions of the other five SNPs showed no difference.
Table 3.
The six SNPs of the PDCD‐1 gene investigated in psoriasis cases (n = 128) and healthy controls (n = 88)
| SNP | dbSNP ID | Frequency (%) | HWE | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | P | Allele | P | P | ||||||
| PD1.1 | rs36084323 | AA | AG | GG | A | G | ||||
| Controls | 21.59 | 56.82 | 21.59 | 50.00 | 50.00 | 0.26 | ||||
| Cases | 27.34 | 51.56 | 21.10 | 0.62 | 53.13 | 46.87 | 0.56 | |||
| PD1.3 | rs11568821 | AA | AG | GG | A | G | ||||
| Controls | 0.00 | 0.00 | 100.00 | 0.00 | 100.00 | 1.00 | ||||
| Cases | 0.00 | 0.78 | 99.22 | 1.00 | 0.39 | 99.61 | 1.00 | |||
| PD1.4 | rs6705653 | AA | AG | GG | A | G | ||||
| Controls | 7.96 | 46.59 | 45.45 | 31.25 | 68.75 | 0.55 | ||||
| Cases | 7.03 | 39.06 | 53.91 | 0.50 | 26.56 | 73.44 | 0.33 | |||
| PD1.5 | rs2227981 | CC | TC | TT | C | T | ||||
| Controls | 45.45 | 46.59 | 7.96 | 68.75 | 31.25 | 0.55 | ||||
| Cases | 53.91 | 39.06 | 7.03 | 0.50 | 73.44 | 26.56 | 0.33 | |||
| PD1.6 | rs10204525 | AA | AG | GG | A | G | ||||
| Controls | 63.64 | 30.68 | 5.68 | 78.98 | 21.02 | 0.65 | ||||
| Cases | 46.09 | 46.88 | 7.03 | 0.034 | 69.53 | 30.47 | 0.035 | |||
| PD1.9 | rs2227982 | CC | CT | TT | C | T | ||||
| Controls | 20.45 | 57.96 | 21.59 | 49.43 | 50.57 | 0.18 | ||||
| Cases | 21.88 | 50.00 | 28.12 | 0.47 | 46.88 | 53.12 | 0.62 | |||
HWE, Hardy–Weinberg equilibrium; SNPs, single nucleotide polymorphisms. P values less than 0.05 are shown in bold font.
The case–control association analysis for PD1.6 showed a significant difference in codominant, dominant, and overdominant models (Table 4). Compared with the AA genotype in PD1.6, the heterozygous AG genotype was associated with a significantly increased risk of psoriasis (OR = 2.11, 95% CI 1.18–3.78, P = 0.012), as was the combination of the AG and GG genotypes (OR = 2.05, 95% CI 1.17–3.57, P = 0.012). The GG genotype in PD1.6 had a low frequency in the population. The odds ratio for the GG genotype was 1.71 (95% CI 0.54–5.41), which was increased but without statistical significance (P = 0.362), likely due to the small numbers of cases and controls with the GG genotype.
Table 4.
Case‐control association analysis of PD1.6 by using logistic regression
| Model | Genotype | Controls (%) | Cases (%) | OR (95% CI) | P value |
|---|---|---|---|---|---|
| Codominant | AA | 63.64 | 46.09 | 1 | |
| AG | 30.68 | 46.88 | 2.11 (1.18–3.78) | 0.012 | |
| GG | 5.68 | 7.03 | 1.71 (0.54–5.41) | 0.362 | |
| Dominant | AA | 63.64 | 46.09 | 1 | |
| AG+GG | 36.36 | 53.91 | 2.05 (1.17–3.57) | 0.012 | |
| Recessive | AA+AG | 94.32 | 92.97 | 1 | |
| GG | 5.68 | 7.03 | 1.26 (0.41–3.88) | 0.693 | |
| Overdominant | AA+GG | 69.32 | 53.12 | 1 | |
| AG | 30.68 | 46.88 | 1.99 (1.13–3.53) | 0.018 |
95% CI, 95% confidence interval; OR, odds ratio. P values less than 0.05 are shown in bold font.
In accordance with the genotype analysis results, the allele analysis for the six SNPs showed significant differences in allele frequencies of PD1.6, but not those of the other five SNPs, between psoriasis patients and healthy controls (Table 3). Compared with cases with the A allele, the G allele of PD1.6 carried an odds ratio of 1.65 (95% CI 1.05–2.58, P = 0.03).
In addition, we compared polymorphisms of PD1.6 (rs10204525) in 77 patients with severe psoriasis (PASI > 10, BSA > 10%) and 51 patients with mild‐moderate psoriasis (PASI < 10, BSA < 10%). Compared with the mild–moderate psoriasis group, the frequencies of genotypes AG and GG in PD1.6 were relatively high in the severe psoriasis group. However, no significant difference was detected (P = 0.118). The odds ratio for allele G in severe psoriasis was accordingly increased but without statistical significance (OR = 1.76, 95% CI 1.00–3.09, P = 0.051) (Table 5).
Table 5.
PD1.6 polymorphism in patients with severe or mild–moderate psoriasis
| Psoriasis patients | Mild–moderate (n = 51) | Severe (n = 77) | P value |
|---|---|---|---|
| PASI score () | 6.96 ± 2.37 | 21.68 ± 8.61 | <0.0001 |
| BSA score () | 6.28 ± 2.43 | 33.55 ± 14.98 | <0.0001 |
| Genotype (%) | |||
| AA | 56.86 | 38.96 | |
| AG | 39.22 | 51.95 | |
| GG | 3.92 | 9.09 | 0.118 |
| Allele (%) | |||
| A | 76.47 | 64.94 | |
| G | 23.53 | 35.06 | 0.051 |
BSA, body surface area; PASI, Psoriasis Area and Severity Index.
Discussion
Psoriasis is a chronic, recurrent inflammatory skin disease associated with immune function. The activation of the IL‐23/T17 cytokine axis is a well‐defined mechanism for the development of psoriasis. 15 The binding of the PD‐1 receptor and its ligands may inhibit the IL‐23/T17 axis and proliferation of T lymphocytes. 16 Accumulating data have suggested a role of PD1 in the pathogenesis of psoriasis. However, the genetic polymorphism of PDCD1 in psoriasis has not been addressed. The present study tested six potentially functional SNPs of the PDCD1 gene in a Chinese population and revealed that the G allele in PD1.6 increased the susceptibility to psoriasis.
The PD1 protein serves as a regulatory receptor. 17 Through binding to its ligands PD‐L1 and PD‐L2, PD1 transmits negative synergistic stimulation signals and inhibits the proliferation and activation of lymphocytes. 17 PD‐L1 is constitutively expressed in T cells, B cells, dendritic cells, macrophages, mesenchymal stem cells, bone marrow‐derived mast cells, and a range of nonhematopoietic cells. 18 PD‐L2 expression is restricted to activated dendritic cells, macrophages, and bone marrow‐derived mast cells, as well as part of the resting peritoneal B1 cells. 18 PD‐1 binding with its ligands can inhibit the T‐cell receptor (TCR) signaling pathway mediated by Src homology phosphatase 2 (SHP2) tyrosine phosphatase, which is the key to stopping the immune response. 19 In recent years, studies on PDCD1 have focused on autoimmune diseases such as tumors 20 and rheumatoid arthritis. 11 , 12 , 13
Bartosińska et al. 21 found that the percentage of CD4+‐PD‐1+ and CD8+‐PD‐1+ T cells in psoriatic patients both with and without arthritis was significantly lower than that in the control group. Impairment of the negative costimulation from PD‐1 may be another common characteristic of psoriasis both with and without arthritis. Kim et al. 22 reported that PD‐1 is overexpressed in IL‐17A‐producing T cells in imiquimod‐treated mice, as well as in patients with psoriasis. Moreover, recombinant PD‐L1‐Fc alleviates psoriatic inflammation in imiquimod‐treated mice. Considering the function of PD‐1 in psoriasis, we examined whether PD‐1 gene variations differ between psoriasis patients and the healthy population.
The present study investigated six potentially functional SNPs in the PDCD1 gene, which have frequently been studied for autoimmune diseases. For instance, the AA genotype of PD1.1, an SNP in the promoter of PDCD1, was described to be associated with a decreased risk for developing rheumatoid arthritis in the Chinese population. 12 , 13 PD1.3, an intronic SNP located in an enhancer‐like structure, showed association with the development of systemic lupus erythematosus in Europeans, 9 , 10 with the risk A allele conferring decreased transcriptional activity of the PDCD1 gene. 10
The genotype and allele frequency data of PDCD‐1 SNPs obtained by this study are in accordance with previous studies in the Chinese population. 12 , 23 For example, in our research, the frequency of the PD1.1 AA genotype was 21.59% for controls, close to 24.9% that reported in residents of Hong Kong, China. 12 In our study, the C allele frequency in PD1.5 was 68.75% for controls, close to the value of 71% reported in the Han Chinese population. 23
It is worth noting that the PD1.3 A allele showed an extremely low frequency in this study. Except for a single case of AG, only the GG genotype was observed for all the controls and patients recruited in this study. PD1.3 is located in an enhancer‐like structure in intron 4 of the PDCD‐1 gene, which is enriched in binding sites for transcription factors. In the European population, a higher frequency of PD1.3 allele A was reported (5–12%) and was found to be associated with systemic lupus erythematosus, 9 , 10 rheumatoid arthritis, 11 and multiple sclerosis. 14 In contrast, the rare polymorphism of PD1.3 in the Chinese population may indicate minor functional significance.
For PD1.6, we herein found that the G allele was related to an increased risk of psoriasis. The frequency of the PD1.6 GG genotype was low in controls (5.68%), which showed no significant increase in patients (7.03%). Nevertheless, the frequency of the PD1.6 AG genotype was significantly higher in patients (46.88%) than in controls (30.68%). The overall frequency of the G allele significantly increased from 21.02% in controls to 30.47% in patients. In previous studies of a healthy Chinese population, the frequency of PD1.6 allele G ranged between 18.4 and 45.5%, and the frequency of the PD1.6 GG genotype ranged between 1.88 and 18.8%. 24 , 25 Meanwhile, in our healthy controls, the frequency of PD1.6 G allele was 21.02%, and the frequency of the PD1.6 GG genotype was 5.68%. Our data on the PD1.6 G allele and GG genotype frequencies are located within the above range.
Regarding the functional significance of PD1.6 polymorphisms, previous studies have demonstrated its association with cancer and hepatitis B virus (HBV) and hepatitis C virus (HCV) infection. The PD1.6 AA genotype was found to be related to an increased risk of esophageal squamous cell carcinoma (ESSC) in the northern Chinese population. 26 The AA genotype of PD1.6 was reported to be protective against HBV infection in Moroccan patients. 27 In contrast, the AA genotype of PD1.6 significantly increased the risk of persistent HCV infection in the Chinese Han population aged ≤56 years, while the G allele of PD1.6 was shown to clear HCV spontaneously and play protective roles in females infected with HCV. 28 To our knowledge, the present study is the first to reveal that the PDCD1 polymorphism is associated with the risk of psoriasis.
The level of PDCD‐1 gene expression is suggested to be linked to PD1.6 polymorphism. Mediterranean HBV‐infected patients carrying GG and GA genotypes for PD1.6 displayed high PD‐1 mRNA expression. 27 Differently, a significant higher percentage of PD1 protein expression on T cells was observed for PD1.6 AA genotype as compared to GG genotype in the Chinese Han population. 28 Despite the discrepancy, regulation of PDCD1 gene expression by PD1.6 polymorphism is likely to happen. Because PD1.6 is located in the 3′‐untranslated region (3′‐UTR), PD1.6 is likely to interact with microRNAs (miRNAs), a class of small naturally occurring molecules characterized by regulating gene expression via specific binding to the 3′‐UTR of target mRNAs. As a matter of fact, miRNA‐4717 has been demonstrated to interact with PD1.6. In cells transfected with vectors containing different PD1.6 alleles, miRNA‐4717 affected luciferase activity in an allele‐specific manner. 29 In lymphocytes from chronic HBV patients with PD1.6 genotype GG, miR‐4717 mimics significantly decreased PDCD‐1 expression, suggesting that PD1.6 alleles serve as a substrate of miR‐4717 to regulate PDCD1 expression. 29
In conclusion, the present study demonstrated that the polymorphism of the immune‐related gene PDCD1 is associated with psoriasis susceptibility, supporting the immunogenic characteristics of psoriasis. The G allele of PD1.6 increases the risk of psoriasis, which may contribute to the genetic basis of psoriasis in the Chinese population.
Supporting information
Figure S1. Representative Sanger sequencing traces of PDCD‐1 SNPs.
Acknowledgments
We would like to thank everyone who helped us in this project, especially the staff in the Department of Dermatology, Yueyang Hospital. We also thank the patients and control participants for their assistance. This work was supported by the National Natural Science Foundation of China (81673977) and Shanghai Pujiang Program.
Shengyuan Hua and Bin Fan contributed equally to the article.
Conflict of interest: None.
Funding source: None.
Contributor Information
Jie Chen, Email: 0000002623@shutcm.edu.cn, Email: dercj366@163.com.
Xiao Miao, Email: 0000002623@shutcm.edu.cn, Email: dercj366@163.com.
References
- 1. Juan W. Pathogenesis and clinical diagnosis of psoriasis. Heilongjiang medicine. 2005; 29: 344–346. [in Chinese]. [Google Scholar]
- 2. National Psoriasis Epidemic Investigation Team . Distribution of psoriasis in China: a nation‐wide screening in 1984. Chin J Dermatol 1986; 19: 253–261. [in Chinese]. [Google Scholar]
- 3. Lee EB, Wu KK, Lee MP, et al. Psoriasis risk factors and triggers. Cutis 2018; 102: 18–20. [PubMed] [Google Scholar]
- 4. Harden JL, Krueger JG, Bowcock AM. The immunogenetics of psoriasis: a comprehensive review. J Autoimmun 2015; 64: 66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nickoloff BJ, Nestle FO. Recent insights into the immunopathogenesis of psoriasis provide new therapeutic opportunities. J Clin Invest 2004; 113: 1664–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hua SY, Chen J, Li B, et al. Research progress on the correlation between psoriasis and PD‐1. Chin J Dermatol Venereol 2018; 32: 588–591. [in Chinese]. [Google Scholar]
- 7. De Bock M, Hulstaert E, Kruse V, et al. Psoriasis vulgaris exacerbation during treatment with PD‐1 checkpoint inhibitor: case report and literature review. Case Rep Dermatol 2018; 10: 190–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bartosińska J, Zakrzewska E, Raczkiewicz D, et al. Suppressed programmed death 1 expression on CD4+ and CD8+ T cells in psoriatic patients. Mediators Inflamm 2017; 2017: 5385102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Prokunina L, Castillejo‐López C, Öberg F, et al. A regulatory polymorphism in PDCD1 is associated with susceptibility to systemic lupus erythematosus in humans. Nat Genet 2002; 32: 666–669. [DOI] [PubMed] [Google Scholar]
- 10. Bertsias GK, Nakou M, Choulaki C, et al. Genetic, immunologic, and immunohistochemical analysis of the programmed death 1/programmed death ligand 1 pathway in human systemic lupus erythematosus. Arthritis Rheum 2009; 60: 207–218. [DOI] [PubMed] [Google Scholar]
- 11. Prokunina L, Padyukov L, Bennet A, et al. Association of the PD‐1.3A allele of the PDCD1 gene in patients with rheumatoid arthritis negative for rheumatoid factor and the shared epitope. Arthritis Rheum 2004; 50: 1770–1773. [DOI] [PubMed] [Google Scholar]
- 12. Kong EK, Prokunina‐Olsson L, Wong WH, et al. A new haplotype of PDCD1 is associated with rheumatoid arthritis in Hong Kong Chinese. Arthritis Rheum 2005; 52: 1058–1062. [DOI] [PubMed] [Google Scholar]
- 13. Liu C, Jiang J, Gao L, et al. A promoter region polymorphism in PDCD‐1 gene is associated with risk of rheumatoid arthritis in the han Chinese population of Southeastern China. Int J Genomics 2014; 2014: 247637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kroner A, Mehling M, Hemmer B, et al. A PD‐1 polymorphism is associated with disease progression in multiple sclerosis. Ann Neurol 2005; 58: 50–57. [DOI] [PubMed] [Google Scholar]
- 15. Hawkes JE, Chan TC, Krueger JG. Psoriasis pathogenesis and the development of novel targeted immune therapies. J Allergy Clin Immunol 2017; 140: 645–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. De Bock M, Hulstaert E, Kruse V, et al. Psoriasis vulgaris exacerbation during treatment with a PD‐1 checkpoint inhibitor: case report and literature review. Case Rep Dermatol 2018; 10: 190–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fife BT, Pauken KE. The role of the PD‐1 pathway in autoimmunity and peripheral tolerance. Ann N Y Acad Sci 2011; 1217: 45–59. [DOI] [PubMed] [Google Scholar]
- 18. Keir ME, Butte MJ, Freeman GJ, et al. PD‐1 and its ligands in tolerance and immunity. Annu Rev Immunol 2008; 26: 677–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Francisco LM, Sage PT, Sharpe AH. The PD‐1 pathway in tolerance and autoimmunity. Immunol Rev 2010; 236: 219–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sunshine J, Taube JM. PD‐1/PD‐L1 inhibitors. Curr Opin Pharmacol 2015; 23: 32–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bartosińska J, Zakrzewska E, Purkot J, et al. Decreased blood CD4+PD‐1+ and CD8+PD‐1+ T cells in psoriatic patients with and without arthritis. Adv Dermatol Alergol 2018; 35: 344–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim JH, Choi YJ, Lee BH, et al. Programmed cell death ligand 1 alleviates psoriatic inflammation by suppressing IL‐17A production from programmed cell death 1‐high T cells. J Allergy Clin Immunol 2016; 137: 1466–1476 e1463. [DOI] [PubMed] [Google Scholar]
- 23. Liu X, Hu LH, Li YR, et al. Programmed cell death 1 gene polymorphisms is associated with ankylosing spondylitis in Chinese Han population. Rheumatol Int 2011; 31: 209–213. [DOI] [PubMed] [Google Scholar]
- 24. Kong LJ, Wang BT, Yang XH, et al. Association of two single nucleotide polymorphisms within the exon‐5 of programmed cell death‐1 gene and the development of systemic lupus erythematosus in Southern Chinese Han people. Fudan Univ J Med Sci 2008; 35: 1–6. [in Chinese]. [Google Scholar]
- 25. Wu Z, Miao M, Qiu Y, et al. Association between polymorphisms in PDCD1 gene and aplastic anemia in Chinese Han population. Leuk Lymphoma 2013; 54: 2251–2254. [DOI] [PubMed] [Google Scholar]
- 26. Zhou RM, Li Y, Wang N, et al. Association of programmed death‐1 polymorphisms with the risk and prognosis of esophageal squamous cell carcinoma. Cancer Genet 2016; 209: 365–375. [DOI] [PubMed] [Google Scholar]
- 27. Chihab H, Jadid FZ, Foka P, et al. Programmed cell death‐1 3'‐untranslated region polymorphism is associated with spontaneous clearance of hepatitis B virus infection. J Med Virol 2018; 90: 1730–1738. [DOI] [PubMed] [Google Scholar]
- 28. Xiao W, Zhang QI, Deng XZ, et al. Genetic variations of IL‐28B and PD‐1 are in association with the susceptibility and outcomes of HCV infection in Southeast China. Infect Genet Evol 2015; 32: 89–96. [DOI] [PubMed] [Google Scholar]
- 29. Zhang G, Li N, Li Z, et al. microRNA‐4717 differentially interacts with its polymorphic target in the PD1 3' untranslated region: a mechanism for regulating PD‐1 expression and function in HBV‐associated liver diseases. Oncotarget 2015; 6: 18933–18944. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Representative Sanger sequencing traces of PDCD‐1 SNPs.


