Abstract
Adhesive hydrogels have been developed for wound healing applications. However, their adhesive performance is impaired dramatically due to their high swelling on wet tissues. To tackle this challenge, we fabricated a new type of non‐swelling protein adhesive for underwater and in vivo applications. In this soft material, the electrostatic complexation between supercharged polypeptides with oppositely charged surfactants containing 3,4‐dihydroxylphenylalanine or azobenzene moieties plays an important role for the formation of ultra‐strong adhesive coacervates. Remarkably, the adhesion capability is superior to commercial cyanoacrylate when tested in ambient conditions. Moreover, the adhesion is stronger than other reported protein‐based adhesives in underwater environment. The ex vivo and in vivo experiments demonstrate the persistent adhesive performance and outstanding behaviors for wound sealing and healing.
Keywords: bioadhesives, coacervates, supercharged polypeptides, supramolecular interactions, wound healing
Non‐swelling protein‐based coacervate adhesives for in vivo applications were fabricated by employing electrostatic complexation of supercharged polypeptides (SUPs) and oppositely charged surfactants. The adhesives exhibited superior performance compared to other reported protein‐based adhesives. The outstanding adhesion performance, good biocompatibility, and biodegradability of SUP adhesives facilitate fast wound healing and skin regeneration.
Introduction
Tissue adhesives and hemostatic agents have attracted tremendous attention as potential alternatives for tissue engineering and wound healing. [1] However, those adhesives are often associated with UV‐induced crosslinking or chemical reactions, which can lead to secondary damage to traumatized tissues. Furthermore, during the wound healing process, the very slow degradation of the adhesives prevents the migration of motile cells and inhibits the remodeling of extracellular matrix microenvironment, thus impeding wound healing. In addition to these materials, adhesive hydrogels have been investigated extensively.[ 2 , 3 , 4 , 5 , 6 , 7 ] For example, an adhesive hydrogel consisting of Ag‐Lignin NPs, acrylic acid (AA) and pectin exhibited good adhesive properties and high toughness, which can be used to repair skin defects. [8] However, adhesive hydrogels often suffer from swelling when applied on wet tissues, which then weakens their mechanical strength dramatically. [9] This shortcoming leads to the breakage of adhesive hydrogels during the process of wound healing. In addition, the swelling of the adhesive hydrogels can lead to tissue compression and severe complications in the environment of the wound. Therefore, there is an urgent need to develop alternatives that exhibit persistent adhesive performance for surgical applications.
The continuous need for novel adhesives has turned researchers’ attention to natural systems, for example, mussel secretions. In these materials, the interplay of multiple supramolecular interactions including electrostatic‐, π‐π‐, hydrogen‐ and van der Waals bonds is responsible for their superb adhesive performance. In this context, catechol‐based adhesives exhibit interesting adhesion properties to various surfaces due to the diverse covalent and noncovalent interactions of catechol binding moieties.[ 10 , 11 , 12 , 13 , 14 , 15 , 16 ] Other approaches are focusing on biomimetic small molecule systems, which exhibit weak adhesive behaviors.[ 17 , 18 , 19 ] Their biomedical applications are thus limited. In addition, a systematic study of the adhesion performance of biomacromolecular adhesives controlled by multiple supramolecular interactions has rarely been investigated.
In this work, a new class of bioengineered protein coacervates with robust adhesive performance was developed. The electrostatic complexation between supercharged polypeptides (SUPs) and biomimetic synthetic surfactants leads to liquid‐liquid phase separation and the formation of non‐swelling adhesive materials. Moreover, the synergy of catechol chemistry and electrostatic interactions allows to engineer extraordinary adhesion performance of the resulting bioglues with adhesion properties surpassing the ones of other reported systems. In vivo experiments suggest that this protein‐based adhesive coacervate is very well suited for the acceleration of wound sealing and skin regeneration.
Results and Discussion
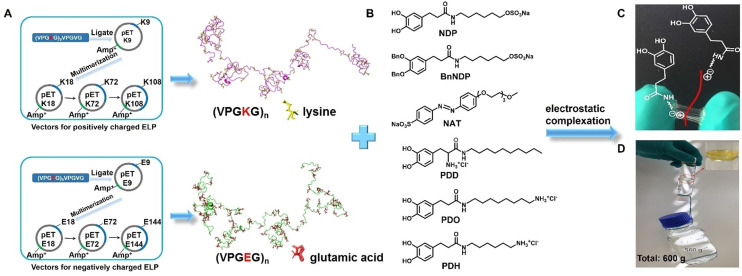
The SUPs were derived from elastin‐like polypeptides, consisting dominantly of repetitive pentapeptide sequences with the primary structure (VPGXG) n , in which the fourth position X is occupied by lysine (K) or glutamic acid (E). These SUPs were produced by recombinant DNA technology and expressed in E. Coli.[ 20 , 21 , 22 ] A series of SUPs were produced with different chain lengths and charges, including K18, K72, K108, E36, E72, and E144. The letter code denotes positive (K) or negative (E) variants while the digit indicates the number of charged amino acid residues (Figures 1 A and S1–S2, Table S1). Meanwhile, a series of surfactants containing 3,4‐dihydroxylphenylalanine (DOPA) or azobenzene (Azo) moieties were synthesized (Figure 1 B). The synthetic routes are shown in Schemes S1–S4. Starting from l‐DOPA or 3‐(3,4‐dihydroxyphenyl) propanoic acid, the corresponding negatively charged DOPA‐surfactant (NDP) and positively charged DOPA‐surfactants with different tail lengths including decyl‐ (PDD), octyl‐ (PDO), and hexyl‐ (PDH) chains were readily synthesized through stepwise protection, amination, and deprotection. Additionally, the negatively charged Azo‐based surfactant with a triethylene glycol unit (NAT) was synthesized in a one‐step procedure from 4‐hydroxyazobenzene‐4′‐sulfonic acid sodium salt hydrate and 1‐(2‐bromoethoxy)‐2‐(2‐methoxyethoxy)ethane. All surfactants were characterized by nuclear magnetic resonance spectroscopy (NMR) and mass spectrometry (MS).
Figure 1.
Preparation and characterization of the SUP‐surfactant coacervate glues. A) Schematic for the expression of supercharged polypeptides (SUPs) (K72, K108, and E36, E72, E144) and (B) chemical structures of different DOPA‐based surfactants (NDP, BnNDP, PDD, PDO, and PDH) and azo‐based surfactant (NAT). The SUP glues were prepared via electrostatic complexation of the SUP and the respective surfactants. C) Photograph of SUP‐NDP adhesive. An elastic, sticky thread can be stretched between two fingers with freshly prepared protein‐based coacervates. Here, K108‐NDP was used as a representative example. D) Photograph showing two smooth glass bottles adhered together by the SUP glue and bearing a load of 600 g. The inset is a zoom‐in of the contact area of the K108‐NAT glue connecting the two smooth glass bottoms of the bottles.
The SUP‐surfactant coacervates were prepared by electrostatic complexation of SUPs and the corresponding surfactants.[ 23 , 24 ] Typically, SUPs and surfactants were mixed in an aqueous solution with the initial molar ratio of lysine or glutamic acid to surfactant at 1:1. As a result, the solution became turbid and after centrifugation a viscous SUP‐surfactant complex was obtained. This liquid‐liquid phase separation behavior leads to the formation of protein‐surfactant coacervates.[ 25 , 26 ] The water content was ≈30 % (w/w) in the SUP‐surfactant complexes as determined by thermogravimetric analysis (TGA) (Figure S3). After lyophilizing for ≈30 min, ≈14 % of water remained in the coacervate (Figure S4), resulting in the formation of the protein‐based adhesive (Figure 1 C). To determine the composition of the SUP‐surfactant complexes quantitatively, the K18‐NDP complex was characterized by 1H NMR spectroscopy (Figure S5). The analysis revealed the stoichiometry of K18 to NDP surfactant to be 1:17 (i.e., ca. 0.95 NDP surfactant molecules per lysine of the SUP), indicating that ≈5 % of lysine moieties were not complexed with the surfactant molecules. Therefore, we hypothesized that cation‐π interactions between these free lysine residues and the DOPA phenyl groups may be present in the system, as observed in other polypeptide systems.[ 13 , 27 , 28 , 29 , 30 ] Furthermore, Figure 1 D demonstrates the robust adhesion performance of the SUP adhesives allowing to glue two glass bottles, bearing a load of 600 g.
Subsequently, the adhesion performance of SUP glues was evaluated by uniaxial tensile testing. A typical adhesion test setup is shown in Figure S6. All the tensile testing was performed under ambient conditions with different substrates, including steel, polyethylene (PE), and polyvinyl chloride (PVC). It should be noted that the SUP glues and commercial cyanoacrylate‐based glue (super glue) were tested in parallel for comparison.
The charge ratio between lysine and NDP surfactant (Figures S7–S8) (1:0.5, 1:1, 1:2, and 1:5) was investigated firstly to determine the optimal molecular composition regarding the adhesion performance. It was found that there's a substantial increase in the breaking strength as the molar ratio of lysine/surfactant is increased until a maximum value of 6.3 MPa at a 1:1 charge ratio was reached. Afterwards, the breaking strength decreased when the ratio was further increased from 1:1 to 1:5. In particular, the fracture strength decreased by about one order of magnitude when fabricating the SUP‐NDP complex in a 1:5 lysine/surfactant stoichiometry. Under this condition, the 1H NMR analysis of the glue showed that the stoichiometry of 4.9 NDP surfactant molecules to one lysine of the SUP was found in the SUP‐NDP complex (Figure S9). In this case, cationic lysine residues are neutralized by too many sulfates of NDP, leading to the difficulty in the formation of cation‐π interaction. This behavior strongly suggests that cation‐π interactions are important for the interfacial adhesion. Therefore, this supramolecular interaction, resulting from the recombinant positively charged protein and aromatic surfactants, contribute significantly to the overall adhesion performance.
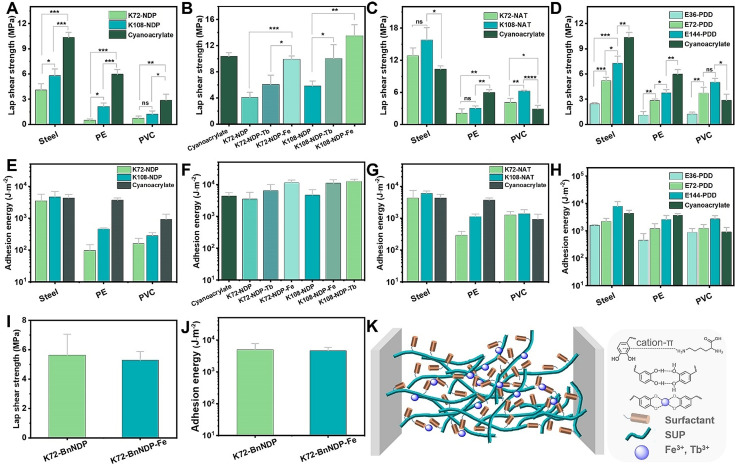
The adhesion performance of the K‐NDP glue was then further evaluated on different substrates (steel, PE, and PVC) under dry conditions. The K‐NDP glue exhibited stronger adhesion performance on a metal substrate (steel) than non‐metal substrates (PE and PVC) (Figures 2 A and S10). Taking K108‐NDP as an example, a fracture strength of 6.3 MPa on steel was observed. In contrast, the fracture strength of K108‐NDP is only 2.1 MPa on PE and 1.2 MPa on PVC, respectively. The adhesion energies of K‐NDP on different substrates are depicted in Figure 2 E. This behavior might be caused by the strong interactions between lysine residues from the SUPs and catechol moieties of the surfactants and the metal surface. Moreover, the adhesion performance of K‐NDP glues became significantly stronger when increasing the molar mass of the SUP (Figure 2).
Figure 2.
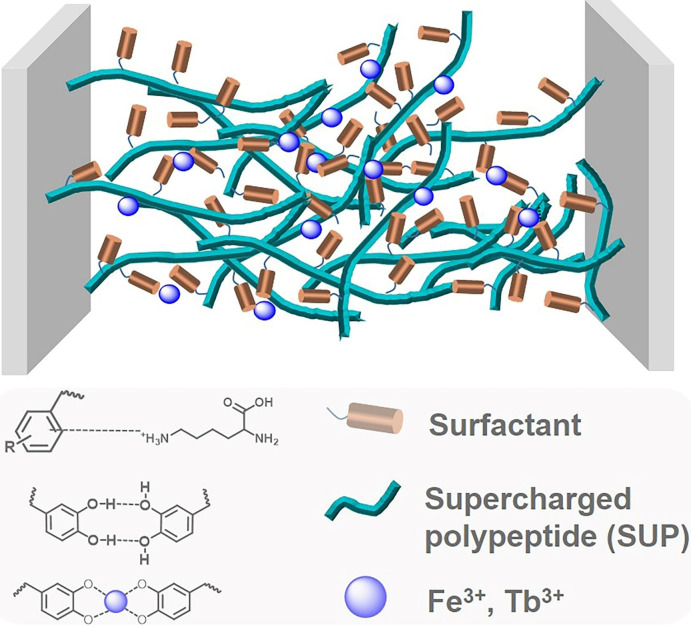
Adhesion investigation of the SUP coacervate glues. A) Lap shear strength for the SUP‐NDP adhesives on three different substrates (Steel, PE, and PVC). K72‐NDP and K108‐NDP were chosen as representative examples and cyanoacrylate was used as a control. P‐values were calculated using the student's t‐test. ns, not significant difference; *p=0.013 <0.05, *** p=0.0003 and 0.0006 <0.001 (in the group of steel), **p=0.0017 and 0.0023 <0.01 (in the group of PE), * p=0.049 <0.05, ** p=0.0077 <0.01 (in the group of PVC). B) The effect of different metal ions (Fe3+, Tb3+) on the bonding strength of SUP‐NDP adhesives on steel (K72‐NDP and K108‐NDP). *p=0.012 <0.05, ***p=0.00039 <0.001 (in the group of K72), * p=0.031 <0.05, ** p=0.0019 <0.01 (in the group of K108). C) Lap shear strength for SUP‐NAT glues (K72‐NAT and K108‐NAT) on three different substrates (Steel, PE, and PVC). *p=0.016 <0.05 (in the group of steel), **p=0.002 <0.01 (in the group of PE), *p=0.0015 <0.05, ** p=0.009 <0.01, **** p=0.00008 <0.0001 (in the group of PVC). D) Lap shear strength for SUP‐PDD glues (E36‐PDD, E72‐PDD, and E144‐PDD) on three different substrates (Steel, PE, and PVC. *p=0.018 <0.05, ** p=0.0066 <0.01, *** p=0.00041 and 0.00061 <0.001 (in the group of steel), * p=0.022 <0.05, **p=0.0032, 0.0014, and 0.0042 <0.01 (in the group of PE), * p=0.013 <0.05, ** p=0.0049 <0.01, ***p=0.00028 <0.001 (in the group of PVC). E–H) Adhesion energy of SUP adhesives under different conditions. I–J) Lap shear strength and adhesion energy of SUP‐BnNDP on steel before/after Fe3+ ion treatment. K) Schematic for the adhesion mechanism of SUP‐surfactant adhesives. Besides electrostatic interactions, van der Waals forces, hydrophobic interactions, and hydrogen bonds both inside the complex and on the interface of samples and substrates, cation–π, and metal coordination bonding between catechol units of surfactant and metal ions are important to enhance the adhesion effect of SUP‐surfactant complexes. The brown cylinder and cyan fiber represent surfactant and SUP, respectively. The deep blue sphere represents metal ions, including Fe3+ and Tb3+.
Next, the metal coordination effect on the adhesion property of K‐NDP glue was investigated (Figure 2 B). To mimic the ratio between Fe3+ ions and DOPA from mussel adhesive, a molar ratio of metal ions to NDP of 1:3 was chosen. [31] Upon adding Fe3+ ions in aqueous solution to the K‐NDP coacervate, the color of the complex turned black immediately, indicating the formation of catecholate complexes. The absorbance peak at ≈545 nm in the UV‐vis spectrum (Figure S12) confirmed the formation of bis Fe‐catecholate. [32] As a result, the bonding strength of SUP glue increased another 2.5 times in a dry environment. It should be mentioned that the adhesive performance of K108‐NDP‐Fe glue (13.5±1.6 MPa) is even superior to cyanoacrylate‐based glue (10.4±0.6 MPa) on steel. [33] This behavior indicates that the Fe3+‐catechol coordination bonds are of great importance for the overall performance of SUP‐glue. In addition to Fe3+ ions, Tb3+ ions were also used to modulate the adhesion behavior of SUP‐glues (Figures 2 B and S13). Similarly, the adhesion performances of K72‐NDP and K108‐NDP increased to 6.1±1.4 MPa and 10.0±2.1 MPa after treatment with Tb3+ ions, respectively. Similarly, high values for the adhesion energies were obtained for the addition of metal ions (Figure 2 F). To the best of our knowledge, the adhesion performance of K108‐NDP mediated by metal ions were higher than any other reported protein‐based and DOPA‐mimicking adhesive (Table S2).[ 34 , 35 ]
To further prove the importance of metal ions in the SUP coacervate glues, benzyl protection groups were installed at the catechol units in the NDP surfactant (BnNDP). The bulk adhesion strength of K‐BnNDP glue on steel was measured under the same conditions as K‐NDP. It turned out that the breaking strength of K72‐BnNDP glue was 5.7±1.4 MPa, which is comparable to the Fe3+‐mediated K72‐BnNDPglue (5.3±0.6 MPa) (Figures 2 I and S14). This indicates that the metal coordination force was absent in the K‐BnNDP system. Comparison with the breaking strength of K72‐NDP on steel (4.1±0.7 MPa, Figure 2 B), the results confirmed the synergistic effect of catechol moieties and metal ions (Fe3+ ions and Tb3+ ions) for the robust adhesion performance (Figure 2 J). It is important to note that the breaking strength of K72‐BnNDP was higher than that of K72‐NDP adhesive, suggesting additional contributions from the aromatic rings of BnNDP to the adhesion performance.
In addition to the DOPA‐inspired NDP system, the scope of surfactants was further broadened to investigate further structure property relationships of the glue. As a typical aromatic rich photo‐switchable molecule, azobenzene was selected to study the effect of an increased π‐system on the adhesive properties. The negatively charged azo‐based surfactant (NAT) was investigated under similar conditions as the NDP‐based glues (Figures 2 C,G and S15). To our surprise, the K‐NAT glues exhibited even better adhesion performance on all different substrates. In particular, the bonding strength of ≈16 MPa of K108‐NAT was higher than any other reported protein‐based adhesive and even cyanoacrylate super glue (Table S2). [34] We speculate that the robust adhesion performance of K‐NAT glues might be related to the presence of strong π‐π stacking and cation‐π interactions.[ 13 , 29 , 30 , 36 ] We also investigated photo‐modulation of the adhesion performance by switching the azobenzene moiety from trans‐ to cis configuration. However, it turned out that the light irradiation with different wavelength didn't change the adhesion performance of the K‐NAT adhesive.
Furthermore, a family of positively charged DOPA surfactants including PDD, PDO, and PDH was used to fabricate SUP glues by the complexation with anionic SUPs (E type). As shown in Figures 2 D and 2 H, the E‐PDD glues exhibited enhanced adhesion performance with increasing the SUP chain length (Figures S16–S18). The higher molecular weight of SUP would lead to the stronger supramolecular network formation of the glue system, thus enhancing the adhesion properties. Based on these results, it can be concluded that the nature of SUP does not greatly affect the adhesion properties of the glue system. Important contributions to the strong adhesion behavior of SUP glues can be ascribed to the multiple supramolecular interactions in the system, especially the cation‐π, π‐π, and metal coordination interactions (Figure 2 K).
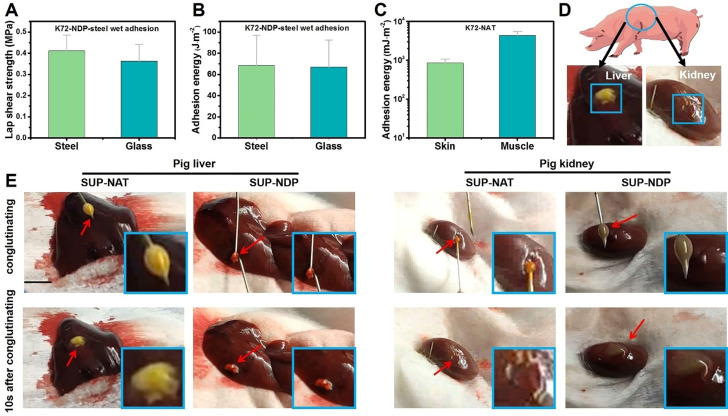
Aside from dry surfaces, wet adhesion for SUP glues was investigated. The substrates were glued together and cured in air for 30 min and then put into water bath overnight before lap shear tensile test. The SUP glue exhibited high underwater adhesion strength of ≈0.4±0.07 MPa and ≈0.36±0.08 MPa on steel and glass, respectively (Figures 3 A,B and S19), which are higher than underwater protein‐based systems reported to date.[ 37 , 38 ] Furthermore, the non‐covalent adhesive behavior endows the SUP glues with additional attractive features. Firstly, SUP glues are cleanable and recyclable. The SUP glues on surfaces can be removed under sufficiently high external shear due to the non‐covalent interactions between SUP and surfaces (Figure S20). Additionally, it was found that the regenerated SUP‐NDP glue exhibited only little less adhesion bonding strength as the original sample both in dry and wet conditions (Figures S21–22).
Figure 3.
Quantitative evaluation of the SUP glues in ex vivo and in vivo applications. A–B) The underwater adhesion test and adhesion energy of SUP adhesive on steel and glass, respectively. C) The ex vivo application of SUP glue on porcine skin and muscle. Three parallel experiments were performed for each group of specimens. D) Schematic illustration of the in vivo hemostasis model, that is, use of SUP glue on pig liver and kidney. The blue box indicates the pasting position on pig organs. E) The bleeding wound was ceased in 10 s after SUP glue was applied. The scale bar is 0.5 cm.
To explore the biocompatibility of SUP glues, we co‐cultured SUP‐surfactant complexes with A549 cancer cells and two normal cell lines including L929 cells (mouse fibroblast) and 293t cells (embryonic kidney, epithelial) in a concentration regime from 5–100 μg mL−1. After 24 h of culturing in the presence of the SUP complexes in DMEM medium, the cell viability did not drop below 80 % even at a concentration of 100 μg mL−1, confirming the non‐toxic nature of the SUP glues (Figures S23–24).
Motivated by the extraordinary performance and non‐toxic nature, ex vivo and in vivo applications of SUP glues including wound healing and adhesion on wet tissues (porcine skin and muscle) were investigated. Due to the excellent performance of SUP adhesives, two pieces of muscle and porcine skin were glued together. As shown in Figure S25, the corresponding force‐displacement curves showed that the peak force is ≈180 mN, F/w equals ≈26 N m−1, and the adhesion energy is 4393±1144 mJ m−2 on muscle, which is comparable to covalently cross‐linked adhesives on soft tissues reported earlier. [39] Adhesion performance of SUP glue on porcine skin was further evaluated exhibiting a peak force of ≈150 mN, F/w equals ≈20 N m−1, and adhesion energy is 857±218 mJ m−2 (Figure 3 C). Such behaviors might be attributed to the non‐covalent and possible covalent interactions between adhesives and tissue surfaces. Subsequently, the hemostatic properties of SUP glue were investigated in vivo. It was found that SUP glue exhibited tissue adhering and hemostatic features on bleeding models of pig liver and kidney inhibiting blood leakage within 10 s (Figures 3 D,E). At the same time, there was no swelling observed in the SUP‐surfactant coacervate glues, indicating their superior tissue adhesion behavior compared to hydrogel systems (Figure S26).[ 2 , 3 , 40 , 41 ] Regarding the control experiment employing commercial cyanoacrylate, this glue solidified quickly and the bleeding continued when the wound was treated with this material indicating that it is not suitable for hemostasis on tissues and organs (Figure S27). All the experiments above showed that the SUP glues are suitable for hemostatic application.
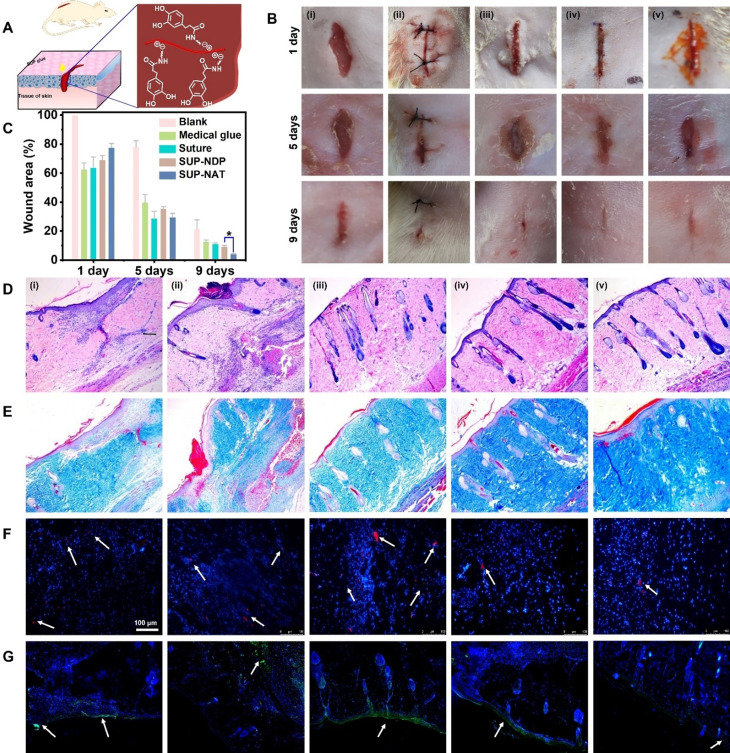
To further explore the potential of SUP glues for in vivo applications, wound sealing and healing experiments were conducted on rat skin with linear openings of 1 cm in length (Figure 4 A). The coacervates were used directly without further freeze‐drying for in vivo applications. It should be noted that the K72‐surfactant adhesives were chosen as a representative sample for in vivo tests due to the higher expression yield of K72 compared to K108. The K72‐NDP, K72‐NAT, suture closure, and commercially available cyanoacrylate‐based adhesive COMPONT® were selected as the experimental groups, and an untreated blank defect was used as a control group (Figure 4 B). The quantitative analysis of the healing progress over 9 days showed that the group treated with SUP glues outperformed the other groups (Figure 4 C) indicating the suitability of SUP glues for skin regeneration and application for topical wound healing. In stark contrast to suture closure and commercial chemical adhesives, the intrinsic biodegradability and the fact that the glue relies solely on multiple supramolecular interactions probably renders the SUP glue very well suited to accelerate the healing and regeneration of skin defects.
Figure 4.
Wound sealing and healing investigated in a rat model treated with SUP glues. A) Schematic representation of SUP glue application in vivo for healing of linear wounds. K‐NDP and K‐NAT glues were used for the experiments. B) Photographs of the wounds after 1, 5, and 9 d on rat skin. Different treatments were used for in vivo wound healing experiments. (i) blank (no treatment), (ii) suture closure, (iii) commercial medical adhesive COMPONT®, (iv) K72‐NDP glue, and (v) K72‐NAT glue. SUP glue facilitates wound healing and tissue regeneration in a 9‐day wound‐healing experiment. The scale bar is 10 mm. C) Quantitative analysis of SUP glue treatment over time monitoring the wound closure area. Three trials were performed for each measurement. Statistics were evaluated by t‐test. (* p=0.022 <0.05). D) H&E staining to investigate tissue regeneration. E) Masson's trichrome staining indicating collagen deposited within the defects. F–G) Immunofluorescence analysis of IL‐6 and TNF‐α. Red immunofluorescent staining and green immunofluorescent staining as an indicator of the levels of IL‐6 and TNF‐α (marked by white arrows), respectively. The scale bar is 100 μm.
The histological analyses, including Hematoxylin and Eosin (H&E) staining as well as Masson's trichrome staining further verified these results. The H&E staining demonstrated that wounds treated with K72‐NDP glue and K72‐NAT glue formed healthy epithelial tissue and new blood vessels, outperforming the recovery of the control groups (Figure 4 D). In addition, Masson's trichrome staining showed more collagen content with an intense blue color in the SUP glue treated groups compared to the other groups (Figure 4 E). It is well known that wound inflammation is one of the major causes of death among injured patients. Therefore, immunofluorescence analysis was used to assess the efficiency of the SUP glue in preventing wound inflammation. Two typical proinflammatory cytokines, interleukin‐6 (IL‐6) and tumor necrosis factor‐α (TNF‐α) in the wound site were chosen as an evaluation criterion in this study. As shown in Figure 4 F, a high level of IL‐6 (red fluorescence) was detected in groups of suture closure, commercial medical glue, and the control, indicating a severe inflammatory response in the traumatized area. Moreover, intensive green fluorescence was detected in these control groups, suggesting high levels of secreted TNF‐α (Figure 4 G). Encouragingly, only minor green fluorescence could be detected for SUP glue treated groups, which indicated low signs of inflammation compared with the other samples. These results suggested that the biocompatible SUP glues exhibit superior anti‐inflammation properties. Therefore, SUP glues are a potential candidate as a wound repair material, providing ideal wound healing ability while avoiding inflammation.
Conclusion
In summary, a new type of genetically engineered protein adhesive involving phase‐separated coacervates was developed. Mediated exclusively by multiple supramolecular interactions especially electrostatic forces, cation‐π interactions, and metal coordination, the proteinaceous adhesives exhibit ultra‐high adhesion performance on various substrates, soft tissues, and inner organs, which renders them superior to other reported bioinspired protein‐based adhesives. In stark contrast to adhesive hydrogels, our SUP glues do not swell and exhibit persistent adhesion performance on wounds. Notably, the biocompatibility and biodegradability of SUP‐based adhesives endows them with the appealing features to remodel the wound microenvironment and to promote rapid wound repair.
The simple fabrication strategy, allowing to electrostatically bond different surfactants equipped with various functionalities to a charged polypeptide chain, represents a novel paradigm to fabricate new bioinspired glues without large synthetic chemical efforts or lengthy cloning protocols or expression optimization. Besides, the substitution of arginine for lysine may further modulate the adhesive's performance due to stronger cation‐π and electrostatic interactions.[ 42 , 43 ] Hence, this electrostatic mediated two component glue will pave the way for promising applications in the field of surgery and soft tissue regeneration. In addition, the fusion of antimicrobial peptides or growth factors in this coacervate‐based adhesive represents further means to increase the functionality of the glue.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Acknowledgements
This research was supported by the National Key R&D Program of China (2020YFA0908900 and 2018YFA0902600), K. C. Wong Education Foundation (GJTD‐2018‐09), the National Natural Science Foundation of China (Grant No. 21877104, 21834007, and 22020102003), and the European Research Council (ERC Advanced Grant SUPRABIOTICS, Grant No. 694610 awarded to AH). J. S. is grateful for financial support from the China Scholarship Council (grant number 201507720025).
J. Sun, L. Xiao, B. Li, K. Zhao, Z. Wang, Y. Zhou, C. Ma, J. Li, H. Zhang, A. Herrmann, K. Liu, Angew. Chem. Int. Ed. 2021, 60, 23687.
Contributor Information
Prof. Andreas Herrmann, Email: herrmann@dwi.rwth-aachen.de.
Prof. Kai Liu, Email: kailiu@tsinghua.edu.cn.
References
- 1. Li J., Celiz A. D., Yang J., Yang Q., Wamala I., Whyte W., Seo B. R., Vasilyev N. V., Vlassak J. J., Suo Z., Mooney D. J., Science 2017, 357, 378–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen X., Yuk H., Wu J., Nabzdyk C. S., Zhao X., Proc. Natl. Acad. Sci. USA 2020, 117, 15497–15503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blacklow S. O., Li J., Freedman B. R., Zeidi M., Chen C., Mooney D. J., Sci. Adv. 2019, 5, eaaw3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ou X., Xue B., Lao Y., Wutthinitikornkit Y., Tian R., Zou A., Yang L., Wang W., Cao Y., Li J., Sci. Adv. 2020, 6, eabb7620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang W., Wang R., Sun Z. M., Zhu X., Zhao Q., Zhang T., Cholewinski A., Yang F., Zhao B., Pinnaratip R., Forooshani P. K., Lee B. P., Chem. Soc. Rev. 2020, 49, 433–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu Y., Meng H., Qian Z., Fan N., Choi W., Zhao F., Lee B. P., Angew. Chem. Int. Ed. 2017, 56, 4224–4228; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2017, 129, 4288–4292. [Google Scholar]
- 7. Pan F., Ye S., Wang R., She W., Liu J., Sun Z., Zhang W., Mater. Horiz. 2020, 7, 2063–2070. [Google Scholar]
- 8. Gan D., Xing W., Jiang L., Fang J., Zhao C., Ren F., Fang L., Wang K., Lu X., Nat. Commun. 2019, 10, 1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ghobril C., Grinstaff M. W., Chem. Soc. Rev. 2015, 44, 1820–1835. [DOI] [PubMed] [Google Scholar]
- 10. Shao H., Stewart R. J., Adv. Mater. 2010, 22, 729–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Seo S., Das S., Zalicki P. J., Mirsha R., Eisenbach C. D., Israelachvili J. N., Waite J. H., Ahn B. K., J. Am. Chem. Soc. 2015, 137, 9214–9217. [DOI] [PubMed] [Google Scholar]
- 12. Zhao Q., Lee D. W., Ahn B. K., Seo S., Kaufman Y., Israelachvili J. N., Waite J. H., Nat. Mater. 2016, 15, 407–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fan H., Wang J., Tao Z., Huang J., Rao P., Kurokawa T., Gong J. P., Nat. Commun. 2019, 10, 5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tiu B. D. B., Delparastan P., Ney M. R., Gerst M., Messersmith P. B., Angew. Chem. Int. Ed. 2020, 59, 16616–16624; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2020, 132, 16759–16767. [Google Scholar]
- 15. Hofman A. H., van Hees I. A., Yang J., Kamperman M., Adv. Mater. 2018, 30, 1704640. [DOI] [PubMed] [Google Scholar]
- 16. Sun J., Su J., Ma C., Göstl R., Herrmann A., Liu K., Zhang H., Adv. Mater. 2020, 32, 1906360. [DOI] [PubMed] [Google Scholar]
- 17. Maier G. P., Rapp M. V., Waite J. H., Israelachvili J. N., Science 2015, 349, 628–632. [DOI] [PubMed] [Google Scholar]
- 18. Rapp M. V., Maier G. P., Dobbs H. A., Higdon N. J., Waite J. H., Butler A., Israelachvili J. N., J. Am. Chem. Soc. 2016, 138, 9013–9016. [DOI] [PubMed] [Google Scholar]
- 19. Ahn B. K., J. Am. Chem. Soc. 2017, 139, 10166–10171. [DOI] [PubMed] [Google Scholar]
- 20. Sun J., Ma C., Maity S., Wang F., Zhou Y., Portale G., Göstl R., Roos W. H., Zhang H., Liu K., Herrmann A., Angew. Chem. Int. Ed. 2021, 60, 3222–3228; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2021, 133, 3259–3265. [Google Scholar]
- 21. Ma C., Li B., Shao B., Wu B., Chen D., Su J., Zhang H., Liu K., Angew. Chem. Int. Ed. 2020, 59, 21481–21487; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2020, 132, 21665–21671. [Google Scholar]
- 22. Ma C., Su J., Sun Y., Feng Y., Shen N., Li B., Liang Y., Yang X., Wu H., Zhang H., Herrmann A., Tanzi R. E., Liu K., Zhang C., Angew. Chem. Int. Ed. 2019, 58, 18703–18709; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2019, 131, 18876–18882. [Google Scholar]
- 23. Wang S., Bo L., Zhang H., Chen J., Sun X., Xu J., Ren T., Zhang Y., Ma C., Guo W., Liu K., Angew. Chem. Int. Ed. 2021, 60, 11252–11256; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2021, 133, 11352–11356. [Google Scholar]
- 24. Shao B., Wan S., Yang C., Shen J., Li Y., You H., Chen D., Fan C., Liu K., Zhang H., Angew. Chem. Int. Ed. 2020, 59, 18213–18217; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2020, 132, 18370–18374. [Google Scholar]
- 25. Dompé M., Cedano-Serrano F. J., Heckert O., van den Heuvel N., van der Gucht J., Tran Y., Hourdet D., Creton C., Kamperman M., Adv. Mater. 2019, 31, 1808179. [DOI] [PubMed] [Google Scholar]
- 26. Peng Q., Chen J., Zeng Z., Wang T., Xiang L., Peng X., Liu J., Zeng H., Small 2020, 16, 2004132. [DOI] [PubMed] [Google Scholar]
- 27. Gebbie M. A., Wei W., Schrader A. M., Cristiani T. R., Dobbs H. A., Idso M., Chmelka B. F., Waite J. H., Israelachvili J. N., Nat. Chem. 2017, 9, 473–479. [DOI] [PubMed] [Google Scholar]
- 28. Kim S., Yoo H. Y., Huang J., Lee Y., Park S., Park Y., Jin S., Jung Y. M., Zeng H., Hwang D. S., Jho Y., ACS Nano 2017, 11, 6764–6772. [DOI] [PubMed] [Google Scholar]
- 29. Li Y., Cheng J., Delparastan P., Wang H., Sigg S. J., DeFrates K. G., Cao Y., Messersmith P. B., Nat. Commun. 2020, 11, 3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Valois E., Mirshafian R., Waite J. H., Waite J. H., Sci. Adv. 2020, 6, eaaz6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sun J., Li B., Wang F., Feng J., Ma C., Liu K., Zhang H., CCS Chem. 2020, 2, 1669–1677. [Google Scholar]
- 32. Saiz-Poseu J., Mancebo-Aracil J., Nador F., Busqué F., Ruiz-Molina D., Angew. Chem. Int. Ed. 2019, 58, 696–714; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2019, 131, 706–725. [Google Scholar]
- 33. Bhagat V., Becker M. L., Biomacromolecules 2017, 18, 3009–3039. [DOI] [PubMed] [Google Scholar]
- 34. Burke K. A., Roberts D. C., Kaplan D. L., Biomacromolecules 2016, 17, 237–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xiao L., Wang Z., Sun Y., Li B., Wu H., Ma C., Petrovskii V. S., Gu X., Chen D., Potemkin I. I., Herrmann A., Zhang H., Liu K., Angew. Chem. Int. Ed. 2021, 60, 12082-12089; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2021, 133, 12189–12196. [Google Scholar]
- 36. He H., Zhao K., Xiao L., Zhang Y., Cheng Y., Wan S., Chen S., Zhang L., Zhou X., Liu K., Zhang H., Angew. Chem. Int. Ed. 2019, 58, 18286–18289; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2019, 131, 18454–18457. [Google Scholar]
- 37. Brennan M. J., Kilbride B. F., Wilker J. J., Liu J. C., Biomaterials 2017, 124, 116–125. [DOI] [PubMed] [Google Scholar]
- 38. Jones J. P., Sima M., O'Hara R. G., Stewart R. J., Adv. Healthcare Mater. 2016, 5, 795–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rose S., Prevoteau A., Elzière P., Hourdet D., Marcellan A., Leibler L., Nature 2014, 505, 382–385. [DOI] [PubMed] [Google Scholar]
- 40. Yang J., Bai R., Chen B., Suo Z., Adv. Funct. Mater. 2020, 30, 1901693. [Google Scholar]
- 41. Barrett D. G., Bushnell G. G., Messersmith P. B., Adv. Healthcare Mater. 2013, 2, 745–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sokalingam S., Raghunathan G., Soundrarajan N., Lee S. G., PLoS One 2012, 7, e40410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kartha K. K., Allampally N. K., Politi A. T., Prabhu D. D., Ouchi H., Albuquerque R. Q., Yagai S., Fernández G., Chem. Sci. 2019, 10, 752–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information