Summary
An open question in environmental ecology regards the mechanisms triggered by root chemistry to drive the assembly and functionality of a beneficial microbiome to rapidly adapt to stress conditions. This phenomenon, originally described in plant defence against pathogens and predators, is encompassed in the ‘cry‐for‐help’ hypothesis. Evidence suggests that this mechanism may be part of the adaptation strategy to ensure the holobiont fitness in polluted environments. Polychlorinated biphenyls (PCBs) were considered as model pollutants due to their toxicity, recalcitrance and poor phyto‐extraction potential, which lead to a plethora of phytotoxic effects and rise environmental safety concerns. Plants have inefficient detoxification processes to catabolize PCBs, even leading to by‐products with a higher toxicity. We propose that the ‘cry‐for‐help’ mechanism could drive the exudation‐mediated recruitment and sustainment of the microbial services for PCBs removal, exerted by an array of anaerobic and aerobic microbial degrading populations working in a complex metabolic network. Through this synergistic interaction, the holobiont copes with the soil contamination, releasing the plant from the pollutant stress by the ecological services provided by the boosted metabolism of PCBs microbial degraders. Improving knowledge of root chemistry under PCBs stress is, therefore, advocated to design rhizoremediation strategies based on plant microbiome engineering.
The ‘cry‐for‐help’ hypothesis is poorly investigated in polluted environments
An impressive body of evidence proves that the root chemistry, that includes the metabolites exudated by the plant, their breakdown compounds and the soil microbe‐degraded sub‐products, plays a crucial role in shaping the structure and functionality of the root‐associated microbiome (Olanrewaju et al., 2019). The extent and nature of exudation varies with the age of the plant (Pausch and Kuzyakov, 2018), with the plant genetics (Cordovez et al., 2021), pathogen or symbiotic interaction or exposure to abiotic stresses, such as drought, high salinity, flooding, extreme temperatures or nutrient starvation (Vives‐Peris et al., 2020). The root chemistry drives the wellness of the holobiont, the meta‐organism resulting from the proficient interactions between the plant and the associated microbiome (Vandenkoornhuyse et al., 2015). In this win–win relationship, primary and secondary metabolites, volatile organic compounds, cells debris and by‐products of senescent root cells, accounting for ~10% of the total photosynthetically fixed carbon (Pausch and Kuzyakov, 2018), are allocated in the flux of root exudation to recruit, feed and influence the metabolism of specific microbial taxa (Liu et al., 2020). Recruited soil microbes establish in the rhizo/endosphere and assist the plant homeostasis by encoding for functionalities that extend the plant genome (Berendsen et al., 2012).
Despite the accumulating evidence, the molecular mechanisms that rule over the causal connection between (i) the plant phenotypes relevant for fitness, like development, growth, and health and (ii) the microbiome structure and functionality are still unknown.
In recent years, an experimental framework related mainly to pests‐ (Cotton et al., 2019; Liu et al., 2021) and herbivore‐attacked plants (Hu et al., 2018) has been validated to describe the proficient interplay between plant and microbiomes and that is encompassed in the ‘cry‐for‐help’ hypothesis (Rolfe et al., 2019). According to this theory, under phytopathogen or herbivore‐induced stress, the plant exudation pattern is shifted to release specific chemicals that favour the recruitment of beneficial microbes or of antagonists able to hinder the growth of pathogens (Rasmann and Turlings, 2016; Carrión et al., 2019; Rolfe et al., 2019). For example, Fusarium oxysporum‐infected cucumber roots were showed to increase tryptophan and decrease raffinose exudation, enhancing the colonization ability of the beneficial strain Bacillus amyloliquefaciens and counteracting the pathogen proliferation. On the other side, B. amyloliquefaciens releases tryptophan‐dependent auxin, in a positive feedback loop to support phytohormone homeostasis (Liu et al., 2017). The ‘cry‐for‐help’ hypothesis also explains the development of disease‐suppressive properties in infested soils (Wilkinson et al., 2019) and the maintenance of this legacy to subsequent plant generations growing in the same soil (Kong et al., 2019). For instance, through a metatranscriptomic approach targeting the sugar beet rhizosphere communities growing in a Rhizoctonia‐suppressive soil, bacteria affiliated to various families, (including Oxalobacteraceae, Sphingobacteriaceae, Burkholderiaceae, Alcaligenaceae, Cystobacteraceae, Sphingomonadaceae, Cytophagaceae, Comamonadaceae and Verrucomicrobia) were demonstrated to upregulate stress‐related genes to enhance survival strategies to cope with the pathogen (Chapelle et al., 2016; Expósito et al., 2017). Similarly, entomopathogenic nematodes are attracted by (E)‐ß‐caryophyllene, exudated by maize roots to hinder the damages caused by the insect Diabrotica vergifera (Rasmann et al., 2005).
In response to pathogen and pest attack, the plant microbiome is often enriched of bacteria that potentially can enhance plant defences or directly counteract the pathogen proliferation. Members of the genera Chitinophaga, Chryseobacterium, Flavobacterium, Microbacterium, Pseudomonas, Sphingomonas, Stenotrophomonas and Xanthomonas have been recently identified among the beneficial bacteria (Liu et al., 2020).
Beyond the ‘adapt or migrate’ strategy, plants could employ the ‘cry‐for‐help’ approach to benefit from microbial associations in contaminated environments, in case the pollutants are highly recalcitrant and toxic, poorly biodegradable and phyto‐extractable. Often plants fail to achieve full metabolism of persistent organic compounds, resulting in slow removal and incomplete degradation because they lack the catabolic enzymes necessary for their complete mineralization (Eapen et al., 2007; Schwitzguébel, 2017). As a consequence, xenobiotics commonly induce molecular injuries that disrupt biochemical, physiological and signalling processes and unbalance plant growth and survival (Ramel et al., 2012). The impact of xenobiotics exposure on rhizodeposition is underrated and the knowledge is sparse, limited to heavy metals like cadmium (Bali et al., 2020) and aluminium (Saha et al., 2020). The root system plunging in polluted soil, by changing the rhizodeposit fingerprint, resorts to the ‘cry‐for‐help’ to recruit microbes for their plant‐growth promoting activities, with the potential to release the plant from stress (Vergani et al., 2017a). Furthermore, the eventual enrichment of bacteria possessing the enzymatic machinery able to degrade the pollutant can decrease its local concentration, enhancing the detoxification in the root/rhizosphere compartments. This beneficial association is the keystone for rhizoremediation strategies, consisting in pollutant removal by microorganisms in the root zone (Balloi et al., 2010; Vergani et al., 2017b).
Classified within the most deleterious persistent organic compounds (POPs) for human, animal and ecosystem health (Stockholm Convention on POPs, 2015; Hens and Hens, 2017), polychlorinated biphenyls (PCBs) are a wide class of 209 congeners containing biphenyl with one up to 10 chlorine atoms. Although their production was banned worldwide in 1979 (Passatore et al., 2014), PCBs still represent a threat for their teratogenic (Berghuis and Roze, 2019), carcinogenic (Magoni et al., 2019; Guo et al., 2020), mutagenic potential (Murati et al., 2020), together with their recalcitrance in the environment (Simhadri et al., 2020), persistence and biomagnification in the food web (Amutova et al., 2021). For land reclamation, PCB‐polluted soils should be excavated for off‐site treatment by solvent extraction, thermal alkaline dechlorination, incineration or landfilling (Campanella et al., 2002). While these techniques are expensive and, in many cases, almost infeasible due to the general large extension of the contamination (Van Aken et al., 2010), rhizoremediation offers a sustainable, potentially efficient and cost‐effective technology for PCBs removal from soil (Vergani et al., 2017b).
In this review, the role of root exudation as plant driver of the ‘cry‐for‐help’ strategy in PCBs contaminated soil is discussed, showing its central role in recruiting and sustaining the microbial services to reduce the contaminant load, thus restoring the holobiont health.
Plant fitness falters in PCBs contaminated soil
The routes of PCBs uptake and translocation in plant tissues are largely unknown: as foreign molecules not naturally present in the environment, they presumably enter plant tissues by hijacking the pathways required for the uptake of soil nutrients (Greenwood et al., 2011).
PCBs mainly absorb on root epidermal surfaces and then diffuse in less amount within the root tissues (Fig. 1), as observed in alfalfa plants exposed to PCB 28 (Teng et al., 2017). The root capacity to absorb extremely hydrophobic compounds like PCBs depends on the thickness of waxes on the root epidermis (Zhang et al., 2009). Pectins, as well, may mediate PCBs accumulation in the root tissues: the Arabidopsis mutant quasimodo1, affected in a galacturonosyltransferase enzyme, showed a reduced amount of PCB 18 in the root system (Bao et al., 2013). Although pectins are well documented in chelating heavy metals (Shao et al., 2021), their role in PCBs phyto‐extraction remains elusive.
Fig 1.
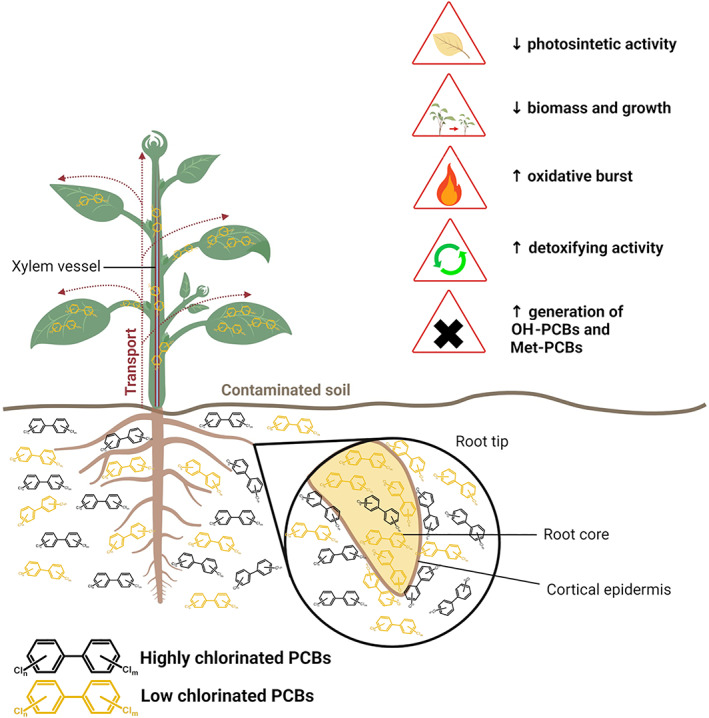
Polychlorinated biphenyls (PCBs) distribution in plant tissues and phytotoxic effects. Several studies reported a differential distribution of low and highly chlorinated PCBs in plant tissues. The external cortical epidermis of the root can interact and be enwrapped by PCBs with a different degree of chlorination, but low chlorinated PCBs are favoured in entrance in the root internal core and then can be translocated through the xylem sup in the plant aerial compartments, both stem, shoot and leaves. Plants are unable to fully mineralize PCBs and the exposition to these toxic pollutants result in a plethora of phytotoxic effects, affecting plant growth and health.
Several lines of evidence suggest that lower chlorinated PCBs are preferentially uptaken and accumulated by the roots than higher chlorinated PCBs (Asai et al., 2002; Luo et al., 2020; Fig. 1). Once collected in the root tissue, PCBs can be then translocated at some extent in the epigeal compartment (stem and leaves), as observed in pumpkin plants cultivated in a soil polluted by the commercial PCBs mixture Aroclor (Whitfield Åslund et al., 2008). Similar indications were observed in poplar plants grown in a hydroponic medium artificially contaminated by PCBs where the higher chlorinated compounds were compartmentalized in the root and the lower chlorinated ones were mobilized in the stem (Liu and Schnoor, 2009). The translocation seems to occur through the xylem sup, as documented in pumpkin plants (Greenwood et al., 2011) (Fig. 1). PCBs concentration decreases in the sup along the shoot as the distance from the roots increases, with low chlorinated compounds that are more easily mobilized toward the plant aerial compartments (Whitfield Åslund et al., 2007; Greenwood et al., 2011). PCBs translocation seems to be influenced not only by the PCB congener and the chlorine substitution pattern, but also by the plant species (Asai et al., 2002; Iwabuchi et al., 2020).
Controversial conclusions were drawn for PCBs‐induced phytotoxicity, which has been estimated on a wide array of heterogeneous experimental settings, including in vitro assay or field trial with different plant species at various developmental stages and different PCB congener's mixtures (Zeeb et al., 2006; Wang et al., 2017). Thus, variable plant responses to PCBs exposition were recorded. Some studies reported a phytotoxic effect given by PCBs on plant growth only at high concentrations (Jin et al., 2011; Wang et al., 2017), and low to moderate negative effect for lower levels of PCBs contamination (Zeeb et al., 2006; Deavers et al., 2010;Subramanian et al., 2018; Urbaniak et al., 2020). On the contrary, in some cases PCB were described to induce plant growth stimulation (Subramanian et al., 2018; Urbaniak et al., 2020), claiming for a hormesis effect (Calabrese, 2014).
A possible explanation to the observed variability in PCBs toxic effect is linked to the above‐mentioned factors affecting PCB uptake by the roots and translocation, such as the plant species and PCB hydrophobicity and bioavailability, strongly dependent on the congener and substitution patterns (Van Aken et al., 2010).
Xenobiotics responses in higher plants are still cryptic, although it is ascertained that POPs could disrupt molecular and biochemical processes, thus affecting signalling circuits (Ramel et al., 2012). Transcriptomics study of Arabidopsis plants exposed to 2,5‐dichlorobiphenyl (Jin et al., 2011) and tetrachlorobiphenyl (Subramanian et al., 2018) showed an enrichment in gene functions related to the metabolism of toxic substances and to response to oxidative stress, oomycetes and bacteria, confirming, as observed for other stresses, the convergence of biotic and abiotic signalling responses (Fujita et al., 2006).
Phenotypically, PCBs exposure showed to cause fall in germination rate (Meier et al., 1997; Subramanian et al., 2018), reduction of the plant biomass (Kučerová et al., 2000), decrease in the chlorophyll content with leaf bleaching effect (Bao et al., 2013) and burst of oxidative stress (Ahammed et al., 2013) (Fig. 1).
To cope with xenobiotics, plants adopt a three‐steps detoxifying strategy, summarized in the green liver model (Sandermann, 1994), that include: (i) activation of POPs by oxidation to produce hydroxylated molecules with a higher solubility; (ii) conjugation to generate adducts with glutathione to decrease the POPs toxicity and (iii) sequestration of the conjugates in the vacuole or incorporation into plant cell wall (Coleman et al., 1997).
Unfortunately, this detoxification mechanism is inefficient and has a limited impact on plant release from PCBs‐induced stress (Schäffner et al., 2002). Indeed, many evidence suggest that plant catabolism of PCBs is limited to tetrachlorinated and lower congeners (Kučerová et al., 2000; Van Aken et al., 2010), and, moreover, the substitution patterns of these molecules can dramatically affect their recalcitrance (Rezek et al., 2007; Van Aken et al., 2010).
Furthermore, the initial metabolism of PCBs in plants involves the activity of monooxygenases of the cytochrome P‐450 family and peroxidases that generate hydroxylated (OH‐) and methoxylated (MeO‐) PCBs as observed in rice, maize, wheat and Arabidopsis, having an enhanced toxicity than the parental molecule (Sun et al., 2016; Subramanian et al., 2018; Sun et al., 2018; Lin et al., 2020), posing a further stress for plant physiology and an environmental risk for human and ecosystem health (Wu et al., 2014). Exposure to OH‐ and MeO‐PCBs in rice triggers an enhanced energy demand, promoting the catabolism of pyruvate, the TCA cycle (tricarboxylic acid), the transfer of acetyl groups into mitochondria and finally resulting in a higher biomass reduction than the parental 2,3,4,5‐tetrachlorobiphenyl treatment (Lin et al., 2020). The plant displays the most efficient detoxification effect, glutathione‐mediated, only after exposure to the parental congener, while a vast array of less effective antioxidant mechanisms are activated by OH‐ and MeO‐PCBs, explaining, therefore, their higher phytotoxicity (Lin et al., 2020).
Recently, in the soil of a historically polluted site in Italy (Di Guardo et al., 2017), sulfonated‐ and hydroxy‐sulfonated‐PCBs were detected (Bagnati et al., 2019). The synthesis of these sulfonated compounds, maybe through the action of the glutathione S‐transferase enzyme (Blanchette and Singh, 2003), has been hypothesized to be part of the detoxifying strategy to enhance the PCBs mobility and bioaccessibility (Bagnati et al., 2019) as already demonstrated for other molecules potentially harmful for the plant (Chen et al., 2015). Plant and environmental fate of such modified PCBs remains still to be elucidated.
Knowledge about plant metabolism of xenobiotics is still fragmentary, although it was described that plants contribute to de‐adsorb PCBs from soil particles and mobilize them, up to 62% of the initial PCB contamination (Chekol et al., 2004). During growth and development, plants influence in the soil surrounding root the abundance and composition of dissolved and particulate organic carbon (DOC and POC respectively) that are the main drivers for the chemical movement of hydrophobic compounds in soil (Terzaghi et al., 2020). The increased PCB bioavailability could further compromise plant growth. Therefore, the selection of plant species effective for PCB phytoremediation like Medicago sativa (alfalfa), Lespedeza cuneate (Chinese bushclover), Lathyrus sylvestris (everlasting pea), Phalaris arundinacea (reed canary grass), Cucurbitaceae (cucurbits), Sparganium (bur‐reed), Salix alaxensis (Alaska willow) and Picea glauca (white spruce; Chekol et al., 2004; Ficko et al., 2010; Slater et al., 2012; Terzaghi et al., 2020), have been suggested as crucial for land reclamation of PCB‐polluted areas. The increase of PCB bioavailability in the rhizosphere makes these highly recalcitrant molecules more accessible also for microbes featuring the enzymatic arsenal to degrade them (Vergani et al., 2017b), generating propitiatory conditions for the ‘cry‐for‐help’ strategy. Although it can be supposed that PCB‐ induced effect on plant physiology and stress response could affect rhizodeposition, the changes in the root chemistry have still to be mechanistically demonstrated.
PCBs‐degradative potential of soil microbiomes
Despite PCBs have been introduced in the environment by human activity in relatively recent times, bacterial communities inhabiting polluted soils are well known for their potential to degrade this class of molecules, via both dechlorination and cleavage of the biphenyl ring. These capacities are probably derived from the cycling of natural organohalogens (Atashgahi et al., 2018; Temme et al., 2019) and the degradation of plant‐derived aromatic organic compounds (Fuchs et al., 2011) as a consequence of structural similarity.
PCB dechlorination has been confirmed only as anaerobic reductive activity among members of the phylum Chloroflexi, including Dehalococcoides, Dehalogenimonas and Dehalobium, either by organohalide‐respiration or by co‐metabolism (Xiang et al., 2020). There are evidences that also bacteria classified as Clostridia (Firmicutes) and Geobacteraceae (δ‐Proteobacteria) are involved in this activity (Wang and He, 2013; Praveckova et al., 2016).
The most studied PCB‐dechlorinating bacteria are strains belonging to the only described species of the genus Dehalococcoides, D. mccartyi. This species is an obligate organohalide‐respiring bacteria (OHRB), strictly anaerobic, with a small‐sized genome yet encoding for several reductive dehalogenase enzymes (RDases) active towards a broad range of chlorinated compounds that represent its sole source of energy (Taş et al., 2010; Bedard, 2014). Dehalococcoides presence and activity was reported in different environments such as polluted soils, sediments and groundwater (Taş et al., 2010), however, RDases responsible for PCB dechlorination were described for the first time in 2014 (Wang et al., 2014). Combining cultural techniques with metagenomic and metatranscriptomics, the authors identified in three new isolates of D. mccartyi different reductive dechlorinases, PcbA1, PcbA4 and PcbA5, catalysing the meta and para‐dechlorination of several PCB congeners of the mixture Aroclor 1260 (Bedard, 2014).
The role of biotic and abiotic factors influencing the activity of OHRB in soils is still poorly understood; however, the interactions with other bacteria populations, the soil type and water content seem to be crucial. A pivotal role is likely played by other bacteria providing electron donors, carbon sources and the essential corrinoid cofactors for RDases (Men et al., 2012). It has been proposed that D. mccartyi forms a core community with Methanosarcina and Desulfovibrio populations, which have been observed to support PCB dechlorination mediating acetate and hydrogen sources (Wang et al., 2019). The structure and activity of organohalide‐degrading communities are also connected to soil edaphic factors and the presence of anaerobic niches allowing the metabolism of strictly anaerobic OHRB. In a recent study, the soil water content was identified as the main parameter positively related to PCB dechlorination, increasing the cell‐to‐contaminant availability (Shen et al., 2021). Both the presence of oxygen‐depleted niches, due to root respiration activity, and the increase in water retention due to the release of exopolysaccharides, indicates the plant rhizosphere as a potential hot spot for PCB reductive dechlorination.
As most of the knowledge about PCB dechlorination deals with OHRB, far less is known about the possible role of other bacteria and associated non‐reductive enzymes. According to recent metagenomic studies, both respiratory and non‐respiratory processes seem to be widespread in terrestrial ecosystems and involved in chlorinated‐natural organic matter (Cl‐NOM) cycling (Weigold et al., 2016; Temme et al., 2019). RDases and hydrolytic or oxidative dehalogenase enzymes were retrieved in many environments, including sediments from urban lakes and in enrichment cultures from a PCB‐polluted soil, and the abundance of these enzymes increased in the presence of Cl‐NOM (Temme et al., 2019). These findings suggest that both respiring and non‐respiring dehalogenating microbial communities may be important for the dechlorination of PCB in contaminated soils, though this hypothesis requires to be further investigated.
The possibility of aerobic degradation of PCB is tightly related to the number and position of chlorine substitutions. Indeed, highly chlorinated congeners are not accessible to aromatic ring‐degrading enzymes and therefore their metabolism is dependent on the removal of chlorine by dehalogenating microorganisms (Borja et al., 2005). PCB aerobic catabolism by soil bacteria has been extensively reviewed (Abraham et al., 2002; Field and Sierra‐Alvarez, 2008; Furukawa and Fujihara, 2008). The process is known to occur mainly through the enzymes of the biphenyl pathway, encoded by the bph operon that is shared among different bacterial phyla, probably as a result of the ancient evolution of the degradation of biphenyl‐related molecules of plant origin such as the lignin complex (Fuchs et al., 2011). These genes are often found on mobile genetic elements and therefore may be spread through horizontal gene transfer. Recent studies showed that bph clusters of β‐ and γ‐proteobacteria were located as integrative conjugative elements both on plasmids or on the chromosome and could be transferred among bacterial cells (Suenaga et al., 2017; Hirose et al., 2019).
The aerobic biphenyl degradation pathways potentially lead to the conversion of the biphenyl ring into benzoate (upper pathway) and to further degradation into the TCA cycle (lower pathway). The key enzyme responsible for initiating PCB degradation is biphenyl dioxygenase (BphA), a multi‐component Rieske non‐heme iron oxygenase encoded by bphA1A2A3A4. Differences in bphA1 sequence, encoding for BphA α‐subunit, are determinant for substrate specificity and consequently biodegradation capability of different PCB congeners, as found among the most studied PCB‐degrading strains such as Pseudomonas furukawaii KF707, Paraburkholderia xenovorans LB400 and Rhodococcus jostii RHA1 (Vergani et al., 2017a). Other bph operons of bacteria belonging to different species have been sequenced, presenting some differences in their genetic organization and regulation, as well as different enzymes specificity towards ortho‐, meta‐ or para‐ substituted congeners. However, the characterization of the degradation pathways confirmed that their function is highly conserved (Gómez‐Gil et al., 2007; Ridl et al., 2018; Garrido‐Sanz et al., 2020).
Although bph pathways associated to diverse strains are incomplete, causing the accumulation of intermediates that can be toxic for other bacteria and organisms, it is important to note that the biodegradation processes by soil bacterial communities likely occur through a complex metabolic network, rather than a linear series of reactions, and involve the interaction of different populations of degraders active at different catabolic steps (Leewis et al., 2016; Duarte et al., 2017). A few studies coupling stable isotope probing (SIP) with 16S rRNA gene sequencing showed that biodegradation potential in PCBs polluted soils is widespread among different species, with a prevalence of Actinobacteria and Proteobacteria, suggesting that active populations vary depending on site and environmental conditions (Uhlik et al., 2013; Leewis et al., 2016; Jiang et al., 2018). Also, aromatic ring‐hydroxylating dioxygenases metagenomic studies highlighted a high degree of sequence diversity, indicating that the catabolic potential hosted by microbial communities inhabiting contaminated soils remains widely unexplored (Aguirre De Cárcer et al., 2007; Iwai et al., 2010; Standfuß‐Gabisch et al., 2012). In a recent study, the metabolic pathways involved in biphenyl biodegradation by a bacterial consortium isolated from PCBs polluted soil were identified via metagenomic analysis, revealing three different pathways converting biphenyl into benzoate and five pathways degrading benzoate to the TCA cycle intermediates (Garrido‐Sanz et al., 2018).
This approach also allowed to reveal which bacterial populations carried out specific reactions in the network, showing that the bphABCD operons of Rhodococcus strains were responsible of all the three upper pathways. The lower pathways were mainly initiated by Pseudomonas and Bordetella via benzoate 1,2‐dioxygenase (benABC), by Variovorax and Achromobacter that were involved in benzoate ligation with acetyl‐CoA (benzoate CoA‐ligase, blcA), while 4‐hydroxybenzoate 3‐monooxygenase (PobA) were associated to Pseudomonas, Bordetella, Achromobacter, Ralstonia and Rhodococcus. A more complex bacterial community was then involved in the subsequent catabolism of intermediates.
Besides bacteria, PCB degradation has been focused mainly on wood‐decay fungi, which have been studied due to their ligninolytic activity. In fact, as xylophagous organisms, they produce extracellular non‐specific laccases and peroxidases enzymes that efficiently attack the biphenyl ring of several PCBs congeners. Intracellular enzymes such as cytochrome P‐450 and dehydrogenases are also involved in the detoxification process (Čvančarová et al., 2012; Stella et al., 2017). Instead, the degrading activity of soil‐dwelling fungi is poorly studied. Some species of filamentous fungi belonging to the genus Penicillium, Aspergillus, Scedosporium, Doratomyces, Myceliophthora, Phoma and Thermoascus were isolated from PCBs polluted soils, and their degradation potential was characterized in a few studies (Tigini et al., 2009; Mouhamadou et al., 2013; Germain et al., 2021). PCB depletion is thought to occur mainly through the activity of laccases, however the biodegradation pathways of these microorganisms need further investigation.
Plant–microbes dialogue mediated by the root exudates: sources of carbon, co‐metabolites or signal molecules for PCB degrading bacteria
Prolonged exposure to xenobiotics in historically polluted areas promotes a Darwinian selection of holobionts that adapt as a whole to thrive in presence of the contaminant (Osmanovic et al., 2018). The ‘cry‐for‐help’ contributes to the structure of the holobiont, driving the ecosystem services provided by the microbial functionalities involved in organic pollutants degradation (Uhlik et al., 2013) (Fig. 2). Furthermore, plants adopt a variety of strategies to facilitate the microbial viability and metabolism in an adverse environment, challenged by the high organic contaminant concentration (Di Guardo et al., 2017). Such adverse conditions could lead to the entry of bacterial cells in a dormant phase, like for example the so‐called ‘viable but non culturable’ (VBNC) state, from which they can resuscitate under proper environmental and nutritional stimuli (Murugan and Vasudevan, 2018). PCBs pose a threat to bacterial cells viability, as they may accumulate in the cytoplasmatic membrane and disrupt its functionality (Chávez et al., 2006). Notably, PCB toxicity, as for plants, even for bacteria is mostly degradation‐dependent, with catabolic products such as dihydrodiols, dihydroxybiphenyls and catechols affecting cell growth and viability much more than the parental compounds due to their hydrophilic substituents, which increase the impact on the cell membrane structure and function (Cámara et al., 2004). To cope with these effects, the strong degrader Paraburkholderia xenovorans LB400 has been shown to activate an efficient detoxification response that is induced only upon PCB degradation (Parnell et al., 2006). Besides the activation of such detoxification pathways, bacterial tolerance and stress response to PCBs and related metabolites is variable and includes also structural adaptive changes of the cell membrane and the expression of nonspecific stress shock proteins (Kim and Masunaga, 2005; Chávez et al., 2006).
Fig 2.
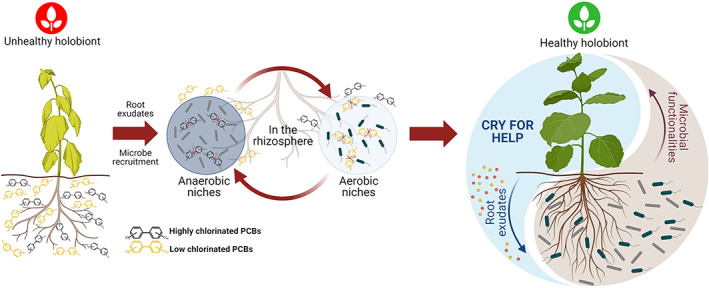
The complex network interaction between the plant ‘cry‐for‐help’ strategy and the ecological services provided by the degrader microbiome sustain the holobiont fitness in polychlorinated biphenyls (PCBs) contaminated soils. The PCB‐induced phytotoxic effects negatively impact the growth and health of plants growing in polluted soils, which modify their root chemistry in a ‘cry‐for‐help’ strategy to recruit, feed and sustain PCB‐degrading microbes in the rhizosphere. Plant primary and secondary metabolites are valuable candidates to support the microbial aerobic co‐metabolism and anaerobic dechlorination of PCBs. Different arrays of microbial populations are involved in PCBs degradation, acting in a metabolic network and in different aerobic and anaerobic micro‐niches that establish in the rhizosphere also as consequence of microbial growth and PCB metabolism, favouring further clean‐up of the pollutant. The ‘cry‐for‐help’ approach and the microbial functionalities encoded by the recruited microbiome contribute to foster this healthy system, restoring the holobiont fitness.
Among the vast array of mechanisms that plants adopt to set off the ‘cry‐for‐help’, the release of primary metabolites like sugars, amino acids and organic acids can support the growth of specific microbial taxa (Macek et al., 2000). The release of a specific profile of root exudates had been considered responsible of the decline in PCBs concentration in Salix caprea and Armoracia rusticana vegetated soil correlated positively with the population size of culturable PCB degraders enumerated in the rhizosphere of the two plant species (Ionescu et al., 2009). Moreover, the root‐associated soil was enriched with bacteria showing a broad PCB congeners catabolism, belonging to the Pseudomonas, Burkholderia and Sphingobacterium genera (Ionescu et al., 2009).
Besides being used as nutrients, plant primary metabolites can be deployed as electron donors to support aerobic co‐metabolism or anaerobic dechlorination of PCBs (Leigh et al., 2006). As a side effect, the enhanced aerobic metabolism consumes oxygen, favouring the formation of anaerobic niches for dehalogenation of high‐chlorinated PCBs (Chaudhry et al., 2005). This will presumably generate a positive feedback mechanism. The decrease of PCB concentration in the rhizosphere would remove the inhibitory effect of these molecules on plant growth, favouring root development and penetration in soil. As a consequence, oxygenation of soil particles would increase, driving the aerobic oxygenases‐mediated degradation of PCBs and generating a cascade effect of oxygen consumption with important implications for the further anaerobic depletion processes (Fig. 2).
Besides primary metabolites, plants can contribute in accelerating microbial PCB degradation by releasing secondary metabolites that act as inducers or co‐metabolites and trigger the expression of the microbial enzymatic degradation machinery (Musilova et al., 2016). These root exudate compounds include phenolic molecules like flavonoids, terpenoids, steroids and alkaloids that often share a high structure similarity with PCBs (Jha et al., 2015). Flavonoids, in particular, encompass a huge variety of molecules that influence plant development, response to stress and interaction between plants, microbes and animals (Mathesius, 2018) and have been implicated in the plant‐mediated induction of PCB‐degrader mechanisms in soil microorganisms (Toussaint et al., 2012; Pham et al., 2015; Subramanian et al., 2018). An altered exudation profile of flavonoids in Arabidopsis mutants affected the colonization and consequently the PCB degradation ability of Pseudomonas putida PLM2 (Narasimhan et al., 2003). Indeed, flavone and isoflavone were demonstrated to act as inducers of the biphenyl degradative pathway in Rhodococcus erythropolis, providing the energy necessary for the contaminant metabolism (Toussaint et al., 2012; Pham et al., 2015).
Terpenes, composed of two or more isoprene units, are responsible for the fragrances of fruits and essential oils and act as chemoattractants for soil microbes (Huang and Osbourn, 2019). The PCB degrader Pseudomonas stutzeri, exposed to the terpenes limonene and carvone, showed differential degradation ability towards the PCB mixture Delor 103, that was dependent on the concentration of the inducer (Tandlich et al., 2001).
The role of primary and secondary metabolites can also overlap by acting as attractants to stimulate chemotaxis toward the rhizosphere, as demonstrated for the strain Rhodococcus erythropolis U23A that showed motility toward phenolic compounds exudated by Arabidopsis roots (Toussaint et al., 2012). Furthermore, variation in the concentration profile of exudated nutrients would affect the residency in the rhizosphere of microbial taxa characterized by diverse life‐history strategies (i.e. copiotrophs vs. oligotrophs). This effect was recently documented by a metabolomic approach on the exudates released by Avena barbata. During plant development, besides the fast‐growing taxa that rapidly consumed the labile exudates, there were a high proportion of slow‐growing strains adapted to specialized metabolic niches (Zhalnina et al., 2018).
Rhizodeposition can also occur through decomposition of deposited litter and lysis of sloughed‐off root cells (Dennis et al., 2010), presumably containing a diverse array of metabolites. Past (Hernandez et al., 1997; Musilova et al., 2016) and recent studies (Terzaghi et al., 2019; Terzaghi et al., 2021) pinpoint for the effectiveness of the combination of compost supply and plant exudation in increasing cell viability and the microbial activity of PCB‐degrading microbiome, while decreasing the load of higher and lower‐chlorinated PCBs (Hayat et al., 2019; Di Lenola et al., 2020).
Through the mixture of organic compounds released by roots, litter decomposition and sludged root cells, plants could contribute to resuscitate microbes from the dormant VBNC state, in which they can enter as an adaptive response limiting their growth and division (Murugan and Vasudevan, 2018). Plants contribute, moreover, to increase PCB mobility and bioavailability for microbial degradation through the release of molecules that can act as surfactants (Campanella et al., 2002) or affecting the composition of particulate organic carbon in soil (Terzaghi et al., 2020).
Recent evidence suggests that the ‘cry‐for‐help’ is not a merely unidirectional strategy employed by the plant to maximize its fitness through the services provided by beneficial microbes. It implies multiple interactions and a molecular dialogue of the holobiont components, the plant and its microbiome, aimed to preserve its functionality under adverse environmental conditions (Fig. 2). Once established in the rhizosphere, after recruitment and feeding through root exudation, microbes can manipulate rhizodeposition to ensure and consolidate their metabolic niche (Korenblum et al., 2020). For example, the release of coumarins, like scopoletin, by Arabidopsis roots under iron starvation, showed to improve the colonization by the beneficial strain P. simiae that, in turn, stimulated the further exudation of scopoletin to counteract the growth of fungal and bacterial competitors that are sensitive to the antimicrobial activity of coumarins (Stringlis et al., 2018).
The chemistry and mechanisms of communication between the plant host and PCB‐degrading soil microorganisms open new challenges for ecological investigation.
Concluding remarks
Today, an arsenal of new methodologies is available to unveil the complex crosstalk between plants and the associated microbiome (Park and Ryu, 2021). The use of Arabidopsis mutant lines (Huang and Osbourn, 2019; Voges et al., 2019) and the ‐omics approaches, including culturomics for setting up microbial synthetic communities (Ziegler et al., 2013; de Souza et al., 2020) and metabolomics to identify the plant exudate compounds (Jaini et al., 2017; Pétriacq et al., 2017; Kawasaki et al., 2018; Dietz et al., 2020) potentially will allow to decipher the messages involved in the ‘cry‐for‐help’ dialogue adopted by plants as response strategy to cope with different environmental stresses (Liu et al., 2020). As recently highlighted by Stringlis and colleagues (2018), who investigated the plant response to iron deficit condition, the microbiome is not merely a receiving component of this dual system. On the contrary, the ability of certain microbes to promote the synthesis and root release of specific compounds ultimately influences the composition of the microbiome itself. The bacteria‐mediated induction of specific plant genes recalls the fine talk taking place in the establishment of the legume‐Rhizobium symbiotic relationship, where the bacterium, after rhizoplane colonization, causes morphological changes in the root epidermis related to the expression of the plant early‐noduling genes and, from the other side, in response to the flavonoids exudated by roots initiate the synthesis of the Nod Factors which in turn act as transcriptional factors for the plant (Geurts and Bisseling, 2002).
A co‐adaptative strategy is established between recruited microorganisms and plants exposed to a particular stress, as observed for drought (Williams and de Vries, 2020), metal toxicity (Timm et al., 2018), plant predation (Adaikpoh et al., 2020) and nutrient limited growth conditions (Ham et al., 2018) to support plant growth under these specific abiotic stressors (Liu et al., 2020). The coumarin scopoletin breaks this paradigm, showing that a molecule exudated under iron starvation is also involved in counteracting pathogen proliferation and favouring the recruitment of beneficial microbes (Stringlis et al., 2019). Identifying similarities and peculiarities in the root exudation profile responding to different abiotic stresses will contribute to decipher the mechanistic aspects of microbial communities assembly upon the plant ‘cry‐for‐help’ strategy, and the study of root exudation in contaminated soils will highlight the plant‐microorganisms interplay in pollutant degradation.
Improving the knowledge on plant secondary metabolites and root exudates effect in response to PCBs polluted soil paves the way for microbiome manipulation to gain ecological services like rhizoremediation. Though most of the ‘cry‐for‐help’ pioneering studies have been realized using the model plant Arabidopsis thaliana, future research could be addressed on plant species that demonstrated to induce a decrease in PCBs concentration by biostimulating the microbiome of historically and highly polluted soils (e.g., Festuca arundinacea, Medicago sativa, Cucurbita pepo ssp. pepo, Terzaghi et al., 2019). A fine metabolomic characterization of the exudation pattern of these plant species challenged in PCBs contaminated soil, coupled with metagenomic studies aimed at identifying the soil microbiome response in terms of functional traits related to biodegradation, will thus represent a milestone to steer the recruitment of PCB‐degrading microbial populations and design effective rhizoremediation strategy based on microbiome engineering.
Acknowledgements
This project has received funding from the European Union's Horizon 2020 research and innovation programme under the Marie Sklodowska‐Curie grant agreement No 841317 to ER and SB for the project “SENSE”.
References
- Abraham, W.‐R. , Nogales, B. , Golyshin, P.N. , Pieper, D.H. , and Timmis, K.N. (2002) Polychlorinated biphenyl‐degrading microbial communities in soils and sediments. Curr Opin Microbiol 5: 246–253. [DOI] [PubMed] [Google Scholar]
- Adaikpoh, B.I. , Akbar, S. , Albataineh, H. , Misra, S.K. , Sharp, J.S. , and Stevens, D.C. (2020) Myxobacterial response to methyljasmonate exposure indicates contribution to plant recruitment of micropredators. Front Microbiol 11: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre De Cárcer, D. , Martín, M. , Karlson, U. , and Rivilla, R. (2007) Changes in bacterial populations and in biphenyl dioxygenase gene diversity in a polychlorinated biphenyl‐polluted soil after introduction of willow trees for rhizoremediation. Appl Environ Microbiol 73: 6224–6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahammed, G.J. , Ruan, Y. , Zhou, J. , Xia, X. , Shi, K. , Zhou, Y. , and Yu, J. (2013) Brassinosteroid alleviates polychlorinated biphenyls‐induced oxidative stress by enhancing antioxidant enzymes activity in tomato. Chemosphere 90: 2645–2653. [DOI] [PubMed] [Google Scholar]
- Amutova, F. , Delannoy, M. , Baubekova, A. , Konuspayeva, G. , and Jurjanz, S. (2021) Transfer of persistent organic pollutants in food of animal origin – meta‐analysis of published data. Chemosphere 262: 128351. [DOI] [PubMed] [Google Scholar]
- Asai, K. , Takagi, K. , Shimokawa, M. , Sue, T. , and Hibi, A. (2002) Phytoaccumulation of coplanar PCBs by Arabidopsis thaliana . Environ Pollut 120: 509–511. [DOI] [PubMed] [Google Scholar]
- Atashgahi, S. , Häggblom, M.M. , and Smidt, H. (2018) Organohalide respiration in pristine environments: implications for the natural halogen cycle. Environ Microbiol 20: 934–948. [DOI] [PubMed] [Google Scholar]
- Bagnati, R. , Terzaghi, E. , Passoni, A. , Davoli, E. , Fattore, E. , Maspero, A. , et al. (2019) Identification of sulfonated and hydroxy‐sulfonated polychlorinated biphenyl (PCB) metabolites in soil: new classes of intermediate products of PCB degradation? Environ Sci Technol 53: 10601–10611. [DOI] [PubMed] [Google Scholar]
- Bali, A.S. , Sidhu, G.P.S. , and Kumar, V. (2020) Root exudates ameliorate cadmium tolerance in plants: a review. Environ Chem Lett 18: 1243–1275. [Google Scholar]
- Balloi, A. , Rolli, E. , Marasco, R. , Mapelli, F. , Tamagnini, I. , Cappitelli, F. , et al. (2010) The role of microorganisms in bioremediation and phytoremediation of polluted and stressed soils. Agrochimica LIV: 16–19. [Google Scholar]
- Bao, L. , Gao, C. , Li, M. , Chen, Y. , Lin, W. , Yang, Y. , et al. (2013) Biomonitoring of non‐dioxin‐like polychlorinated biphenyls in transgenic Arabidopsis using the mammalian pregnane X receptor system: a role of pectin in pollutant uptake. PLoS One 8: e79428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedard, D.L. (2014) PCB dechlorinases revealed at last. Proc Natl Acad Sci U S A 111: 11919–11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen, R.L. , Pieterse, C.M.J. , and Bakker, P.A.H.M. (2012) The rhizosphere microbiome and plant health. Trends Plant Sci 17: 478–486. [DOI] [PubMed] [Google Scholar]
- Berghuis, S.A. , and Roze, E. (2019) Prenatal exposure to PCBs and neurological and sexual/pubertal development from birth to adolescence. Curr Probl Pediatr Adolesc Health Care 49: 133–159. [DOI] [PubMed] [Google Scholar]
- Blanchette, B.N. , and Singh, B.R. (2003) An enzyme based dechlorination of a polychlorinated biphenyl (PCB) mixture, Aroclor 1248, using glutathione S‐transferases from the northern quahog Mercenaria mercenaria . J Protein Chem 22: 377–386. [DOI] [PubMed] [Google Scholar]
- Borja, J. , Taleon, D.M. , Auresenia, J. , and Gallardo, S. (2005) Polychlorinated biphenyls and their biodegradation. Process Biochem 40: 1999–2013. [Google Scholar]
- Calabrese, E.J. (2014) Hormesis: a fundamental concept in biology. Microb Cell 1: 145–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cámara, B. , Herrera, C. , Gonzalez, M. , Couve, E. , Hofer, B. , and Seeger, M. (2004) From PCBs to highly toxic metabolites by the biphenyl pathway. Environ Microbiol 6: 842–850. [DOI] [PubMed] [Google Scholar]
- Campanella, B.E. , Bock, C. , and Schröder, P. (2002) Phytoremediation to increase the degradation of PCBs and PCDD/Fs: potential and limitations. Environ Sci Pollut Res 9: 73–85. [DOI] [PubMed] [Google Scholar]
- Carrión, V.J. , Perez‐Jaramillo, J. , Cordovez, V. , Tracanna, V. , De Hollander, M. , Ruiz‐Buck, D. , et al. (2019) Pathogen‐induced activation of disease‐suppressive functions in the endophytic root microbiome. Science (80‐ ) 366: 606–612. [DOI] [PubMed] [Google Scholar]
- Chapelle, E. , Mendes, R. , Bakker, P.A.H. , and Raaijmakers, J.M. (2016) Fungal invasion of the rhizosphere microbiome. ISME J 10: 265–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry, Q. , Blom‐Zandstra, M. , Gupta, S. , and Joner, E.J. (2005) Utilising the synergy between plants and rhizosphere microorganisms to enhance breakdown of organic pollutants in the environment. Environ Sci Pollut Res 12: 34–48. [DOI] [PubMed] [Google Scholar]
- Chávez, F.P. , Gordillo, F. , and Jerez, C.A. (2006) Adaptive responses and cellular behaviour of biphenyl‐degrading bacteria toward polychlorinated biphenyls. Biotechnol Adv 24: 309–320. [DOI] [PubMed] [Google Scholar]
- Chekol, T. , Vough, L.R. , and Chaney, R.L. (2004) Phytoremediation of polychlorinated biphenyl‐contaminated soils: the rhizosphere effect. Environ Int 30: 799–804. [DOI] [PubMed] [Google Scholar]
- Chen, J. , Gao, L. , Baek, D. , Liu, C. , Ruan, Y. , and Shi, H. (2015) Detoxification function of the Arabidopsis sulphotransferase AtSOT12 by sulphonation of xenobiotics. Plant Cell Environ 38: 1673–1682. [DOI] [PubMed] [Google Scholar]
- Coleman, J.O.D. , Blake‐Kalff, M.M.A. , and Davies, T.G.E. (1997) Detoxification of xenobiotics by plants: chemical modification and vacuolar compartmentation. Trends Plant Sci 2: 144–151. [Google Scholar]
- Cordovez, V. , Rotoni, C. , Dini‐Andreote, F. , Oyserman, B. , Carrión, V.J. , and Raaijmakers, J.M. (2021) Successive plant growth amplifies genotype‐specific assembly of the tomato rhizosphere microbiome. Science of The Total Environment 772. 10.1016/j.scitotenv.2020.144825. [DOI] [PubMed] [Google Scholar]
- Cotton, T.E.A. , Pétriacq, P. , Cameron, D.D. , Al Meselmani, M. , Schwarzenbacher, R. , Rolfe, S.A. , and Ton, J. (2019) Metabolic regulation of the maize rhizobiome by benzoxazinoids. ISME J 13: 1647–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Čvančarová, M. , Křesinová, Z. , Filipová, A. , Covino, S. , and Cajthaml, T. (2012) Biodegradation of PCBs by ligninolytic fungi and characterization of the degradation products. Chemosphere 88: 1317–1323. [DOI] [PubMed] [Google Scholar]
- de Souza, R.S.C. , Armanhi, J.S.L. , and Arruda, P. (2020) From microbiome to traits: designing synthetic microbial communities for improved crop resiliency. Front Plant Sci 11: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deavers, K. , Macek, T. , Karlson, U.G. , and Trapp, S. (2010) Removal of 4‐chlorobenzoic acid from spiked hydroponic solution by willow trees (Salix viminalis). Environ Sci Pollut Res 17: 1355–1361. [DOI] [PubMed] [Google Scholar]
- Dennis, P.G. , Miller, A.J. , and Hirsch, P.R. (2010) Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol Ecol 72: 313–327. [DOI] [PubMed] [Google Scholar]
- Di Guardo, A. , Terzaghi, E. , Raspa, G. , Borin, S. , Mapelli, F. , Chouaia, B. , et al. (2017) Differentiating current and past PCB and PCDD/F sources: the role of a large contaminated soil site in an industrialized city area. Environ Pollut 223: 367–375. [DOI] [PubMed] [Google Scholar]
- Di Lenola, M. , Barra Caracciolo, A. , Ancona, V. , Laudicina, V.A. , Garbini, G.L. , Mascolo, G. , and Grenni, P. (2020) Combined effects of compost and Medicago sativa in recovery a PCB contaminated soil. Water 12: 860. [Google Scholar]
- Dietz, S. , Herz, K. , Gorzolka, K. , Jandt, U. , Bruelheide, H. , and Scheel, D. (2020) Root exudate composition of grass and forb species in natural grasslands. Sci Rep 10: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte, M. , Nielsen, A. , Camarinha‐Silva, A. , Vilchez‐Vargas, R. , Bruls, T. , Wos‐Oxley, M.L. , et al. (2017) Functional soil metagenomics: elucidation of polycyclic aromatic hydrocarbon degradation potential following 12 years of in situ bioremediation. Environ Microbiol 19: 2992–3011. [DOI] [PubMed] [Google Scholar]
- Eapen, S. , Singh, S. , and D'Souza, S.F. (2007) Advances in development of transgenic plants for remediation of xenobiotic pollutants. Biotechnol Adv 25: 442–451. [DOI] [PubMed] [Google Scholar]
- Expósito, R.G. , de Bruijn, I. , Postma, J. , and Raaijmakers, J.M. (2017) Current insights into the role of Rhizosphere bacteria in disease suppressive soils. Front Microbiol 8: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficko, S.A. , Rutter, A. , and Zeeb, B.A. (2010) Potential for phytoextraction of PCBs from contaminated soils using weeds. Sci Total Environ 408: 3469–3476. [DOI] [PubMed] [Google Scholar]
- Field, J.A. , and Sierra‐Alvarez, R. (2008) Microbial transformation and degradation of polychlorinated biphenyls. Environ Pollut 155: 1–12. [DOI] [PubMed] [Google Scholar]
- Fuchs, G. , Boll, M. , and Heider, J. (2011) Microbial degradation of aromatic compounds ‐ from one strategy to four. Nat Rev Microbiol 9: 803–816. [DOI] [PubMed] [Google Scholar]
- Fujita, M. , Fujita, Y. , Noutoshi, Y. , Takahashi, F. , Narusaka, Y. , Yamaguchi‐shinozaki, K. , and Shinozaki, K. (2006) Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol 9: 436–442. [DOI] [PubMed] [Google Scholar]
- Furukawa, K. , and Fujihara, H. (2008) Microbial degradation of polychlorinated biphenyls: biochemical and molecular features. J Biosci Bioeng 105: 433–449. [DOI] [PubMed] [Google Scholar]
- Garrido‐Sanz, D. , Manzano, J. , Martín, M. , Redondo‐Nieto, M. , and Rivilla, R. (2018) Metagenomic analysis of a biphenyl‐degrading soil bacterial consortium reveals the metabolic roles of specific populations. Front Microbiol 9: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido‐Sanz, D. , Sansegundo‐Lobato, P. , Redondo‐Nieto, M. , Suman, J. , Cajthaml, T. , Blanco‐Romero, E. , et al. (2020) Analysis of the biodegradative and adaptive potential of the novel polychlorinated biphenyl degrader rhodococcus sp. Way2 revealed by its complete genome sequence. Microb Genomics 6: 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain, J. , Raveton, M. , Binet, M.N. , and Mouhamadou, B. (2021) Screening and metabolic potential of fungal strains isolated from contaminated soil and sediment in the polychlorinated biphenyl degradation. Ecotoxicol Environ Saf 208: 111703. [DOI] [PubMed] [Google Scholar]
- Geurts, R. , and Bisseling, T. (2002) Rhizobium nod factor perception and signalling. Plant Cell 14: 239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez‐Gil, L. , Kumar, P. , Barriault, D. , Bolin, J.T. , Sylvestre, M. , and Eltis, L.D. (2007) Characterization of biphenyl dioxygenase of Pandoraea pnomenusa B‐356 as a potent polychlorinated biphenyl‐degrading enzyme. J Bacteriol 189: 5705–5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood, S.J. , Rutter, A. , and Zeeb, B.A. (2011) The absorption and translocation of polychlorinated biphenyl congeners by Cucurbita pepo ssp pepo . Environ Sci Technol 45: 6511–6516. [DOI] [PubMed] [Google Scholar]
- Guo, J.Y. , Wang, M.Z. , Wang, M.S. , Sun, T. , Wei, F.H. , Yu, X.T. , et al. (2020) The undervalued effects of polychlorinated biphenyl exposure on breast cancer. Clin Breast Cancer 20: 12–18. [DOI] [PubMed] [Google Scholar]
- Ham, B.K. , Chen, J. , Yan, Y. , and Lucas, W.J. (2018) Insights into plant phosphate sensing and signaling. Curr Opin Biotechnol 49: 1–9. [DOI] [PubMed] [Google Scholar]
- Hayat, A. , Hussain, I. , Soja, G. , Iqbal, M. , and Shahid, N. (2019) Organic and chemical amendments positively modulate the bacterial proliferation for effective rhizoremediation of PCBs‐contaminated soil. Ecol Eng 138: 412–419. [Google Scholar]
- Hens, B. , and Hens, L. (2017) Persistent threats by persistent pollutants: chemical nature, concerns and future policy regarding PCBs—what are we heading for? Toxics 6: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez, B.S. , Koh, S.C. , Chial, M. , and Focht, D.D. (1997) Terpene‐utilizing isolates and their relevance to enhanced biotransformation of polychlorinated biphenyls in soil. Biodegradation 8: 153–158. [Google Scholar]
- Hirose, J. , Fujihara, H. , Watanabe, T. , Kimura, N. , Suenaga, H. , Futagami, T. , et al. (2019) Biphenyl/PCB degrading bph genes of ten bacterial strains isolated from biphenyl‐contaminated soil in Kitakyushu, Japan: comparative and dynamic features as integrative conjugative elements (ICEs). Genes (Basel) 10: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, L. , Robert, C.A.M. , Cadot, S. , Zhang, X. , Ye, M. , Li, B. , et al. (2018) Root exudate metabolites drive plant‐soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat Commun 9: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, A.C. , and Osbourn, A. (2019) Plant terpenes that mediate below‐ground interactions: prospects for bioengineering terpenoids for plant protection. Pest Manag Sci 75: 2368–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionescu, M. , Beranova, K. , Dudkova, V. , Kochankova, L. , Demnerova, K. , Macek, T. , and Mackova, M. (2009) Isolation and characterization of different plant associated bacteria and their potential to degrade polychlorinated biphenyls. Int Biodeterior Biodegrad 63: 667–672. [Google Scholar]
- Iwabuchi, A. , Katte, N. , Suwa, M. , Goto, J. , and Inui, H. (2020) Factors regulating the di ff erential uptake of persistent organic pollutants in cucurbits and non‐cucurbits. J Plant Physiol 245: 153094. [DOI] [PubMed] [Google Scholar]
- Iwai, S. , Chai, B. , Sul, W.J. , Cole, J.R. , Hashsham, S.A. , and Tiedje, J.M. (2010) Gene‐targeted‐metagenomics reveals extensive diversity of aromatic dioxygenase genes in the environment. ISME J 4: 279–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaini, R. , Wang, P. , Dudareva, N. , Chapple, C. , and Morgan, J.A. (2017) Targeted metabolomics of the phenylpropanoid pathway in Arabidopsis thaliana using reversed phase liquid chromatography coupled with tandem mass spectrometry. Phytochem Anal 28: 267–276. [DOI] [PubMed] [Google Scholar]
- Jha, P. , Panwar, J. , and Jha, P.N. (2015) Secondary plant metabolites and root exudates: guiding tools for polychlorinated biphenyl biodegradation. Int J Environ Sci Technol 12: 789–802. [Google Scholar]
- Jiang, L. , Luo, C. , Zhang, D. , Song, M. , Sun, Y. , and Zhang, G. (2018) Biphenyl‐metabolizing microbial community and a functional operon revealed in E‐waste‐contaminated soil. Environ Sci Technol 52: 8558–8567. [DOI] [PubMed] [Google Scholar]
- Jin, X.‐F. , Shuai, J.‐J. , Peng, R.‐H. , Zhu, B. , Fu, X.‐Y. , Tian, Y.‐S. , et al. (2011) Identification of candidate genes involved in responses of Arabidopsis to polychlorinated biphenyls based on microarray analysis. Plant Growth Regul 65: 127–135. [Google Scholar]
- Kawasaki, A. , Okada, S. , Zhang, C. , Delhaize, E. , Mathesius, U. , Richardson, A.E. , et al. (2018) A sterile hydroponic system for characterising root exudates from specific root types and whole‐root systems of large crop plants. Plant Methods 14: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K.‐S. , and Masunaga, S. (2005) Behavior and source characteristic of PCBS in urban ambient air of Yokohama, Japan. Environ Pollut 138: 290–298. [DOI] [PubMed] [Google Scholar]
- Kong, H.G. , Song, G.C. , and Ryu, C.M. (2019) Inheritance of seed and rhizosphere microbial communities through plant–soil feedback and soil memory. Environ Microbiol Rep 11: 479–486. [DOI] [PubMed] [Google Scholar]
- Korenblum, E. , Dong, Y. , Szymanski, J. , Panda, S. , Jozwiak, A. , and Massalha, H. (2020) Rhizosphere microbiome mediates systemic root metabolite exudation by root‐to‐root signaling. Proc Natl Acad Sci 117: 3874–3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kučerová, P. , Macková, M. , Chromá, L. , Burkhard, J. , Tříska, J. , Demnerová, K. , and Macek, T. (2000) Metabolism of polychlorinated biphenyls by Solanum nigrum hairy root clone SNC‐9O and analysis of transformation products. Plant and Soil 225: 109–115. [Google Scholar]
- Leewis, M.C. , Uhlik, O. , and Leigh, M.B. (2016) Synergistic processing of biphenyl and benzoate: carbon flow through the bacterial community in polychlorinated‐biphenyl‐contaminated soil. Sci Rep 6: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh, M.B. , Prouzová, P. , Macková, M. , Macek, T. , Nagle, D.P. , and Fletcher, J.S. (2006) Polychlorinated biphenyl (PCB)‐degrading bacteria associated with trees in a PCB‐contaminated site. Appl Environ Microbiol 72: 2331–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, F. , Sun, J. , Liu, N. , and Zhu, L. (2020) Phytotoxicity and metabolic responses induced by tetrachlorobiphenyl and its hydroxylated and methoxylated derivatives in rice (Oryza sative L.). Environ Int 139: 105695. [DOI] [PubMed] [Google Scholar]
- Liu, H. , Brettell, L.E. , Qiu, Z. , and Singh, B.K. (2020) Microbiome‐mediated stress resistance in plants. Trends Plant Sci 25: 733–743. [DOI] [PubMed] [Google Scholar]
- Liu, H. , Li, J. , Carvalhais, L.C. , Percy, C.D. , Prakash Verma, J. , Schenk, P.M. , and Singh, B.K. (2021) Evidence for the plant recruitment of beneficial microbes to suppress soil‐borne pathogens. New Phytol 229: 2873–2885. [DOI] [PubMed] [Google Scholar]
- Liu, J. , and Schnoor, J.L. (2009) Uptake and translocation of lesser‐chlorinated polychlorinated biphenyls (PCBs) in whole hybrid poplar plants after hydroponic exposure. Chemosphere 73: 319–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Chen, L. , Wu, G. , Feng, H. , Zhang, G. , Shen, Q. , and Zhang, R. (2017) Identification of root‐secreted compounds involved in the communication between cucumber, the beneficial Bacillus amyloliquefaciens, and the soil‐borne pathogen Fusarium oxysporum . Mol Plant Microbe Interact 30: 53–62. [DOI] [PubMed] [Google Scholar]
- Luo, C. , Hu, B. , Wang, S. , Wang, Y. , Zhao, Z. , Wang, Y. , et al. (2020) Distribution and chiral signatures of polychlorinated biphenyls (PCBs) in soils and vegetables around an e‐waste recycling site. J Agric Food Chem 68: 10542–10549. [DOI] [PubMed] [Google Scholar]
- Macek, T. , Macková, M. , and Káš, J. (2000) Exploitation of plants for the removal of organics in environmental remediation. Biotechnol Adv 18: 23–34. [DOI] [PubMed] [Google Scholar]
- Magoni, M. , Donato, F. , Apostoli, P. , Rossi, G. , Comba, P. , Fazzo, L. , et al. (2019) Serum levels of polychlorinated biphenyls and risk of non‐Hodgkin lymphoma: a hospital‐based case‐control study. Chemosphere 235: 969–975. [DOI] [PubMed] [Google Scholar]
- Mathesius, U. (2018) Flavonoid functions in plants and their interactions with other organisms. Plan Theory 7: 7–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier, J.R. , Chang, L.W. , Jacobs, S. , Torsella, J. , Meckes, M.C. , and Smith, M.K. (1997) Use of plant and earthworm bioassays to evaluate remediation of soil from a site contaminated with polychlorinated biphenyls. Environ Toxicol Chem 16: 928–938. [Google Scholar]
- Men, Y. , Feil, H. , Verberkmoes, N.C. , Shah, M.B. , Johnson, D.R. , Lee, P.K.H. , et al. (2012) Sustainable syntrophic growth of Dehalococcoides ethenogenes strain 195 with desulfovibrio vulgaris hildenborough and methanobacterium congolense: global transcriptomic and proteomic analyses. ISME J 6: 410–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouhamadou, B. , Faure, M. , Sage, L. , Marçais, J. , Souard, F. , and Geremia, R.A. (2013) Potential of autochthonous fungal strains isolated from contaminated soils for degradation of polychlorinated biphenyls. Fungal Biol 117: 268–274. [DOI] [PubMed] [Google Scholar]
- Murati, T. , Miletić, M. , Pleadin, J. , Šimić, B. , and Kmetič, I. (2020) Cell membrane‐related toxic responses and disruption of intercellular communication in PCB mechanisms of toxicity: a review. J Appl Toxicol 40: 1592–1601. [DOI] [PubMed] [Google Scholar]
- Murugan, K. , and Vasudevan, N. (2018) Intracellular toxicity exerted by PCBs and role of VBNC bacterial strains in biodegradation. Ecotoxicol Environ Saf 157: 40–60. [DOI] [PubMed] [Google Scholar]
- Musilova, L. , Ridl, J. , Polivkova, M. , Macek, T. , and Uhlik, O. (2016) Effects of secondary plant metabolites on microbial populations: changes in community structure and metabolic activity in contaminated environments. Int J Mol Sci 17: 1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan, K. , Basheer, C. , Bajic, V.B. , and Swarup, S. (2003) Enhancement of plant‐microbe interactions using a rhizosphere metabolomics‐driven approach and its application in the removal of polychlorinated biphenyls. Plant Physiol 132: 146–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olanrewaju, O.S. , Ayangbenro, A.S. , Glick, B.R. , and Babalola, O.O. (2019) Plant health: feedback effect of root exudates‐rhizobiome interactions. Appl Microbiol Biotechnol 103: 1155–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmanovic, D. , Kessler, D.A. , Rabin, Y. , and Soen, Y. (2018) Darwinian selection of host and bacteria supports emergence of Lamarckian‐like adaptation of the system as a whole. Biol Direct 13: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, Y.S. , and Ryu, C.M. (2021) Understanding Plant Social Networking System: Avoiding Deleterious Microbiota but Calling Beneficials. International Journal of Molecular Sciences 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnell, J.J. , Park, J. , Denef, V. , Tsoi, T. , Hashsham, S. , Quensen, J. , and Tiedje, J.M. (2006) Coping with polychlorinated biphenyl (PCB) toxicity: physiological and genome‐wide responses of Burkholderia xenovorans LB400 to PCB‐mediated stress. Appl Environ Microbiol 72: 6607–6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passatore, L. , Rossetti, S. , Juwarkar, A.A. , and Massacci, A. (2014) Phytoremediation and bioremediation of polychlorinated biphenyls (PCBs): state of knowledge and research perspectives. J Hazard Mater 278: 189–202. [DOI] [PubMed] [Google Scholar]
- Pausch, J. , and Kuzyakov, Y. (2018) Carbon input by roots into the soil: quantification of rhizodeposition from root to ecosystem scale. Glob Chang Biol 24: 1–12. [DOI] [PubMed] [Google Scholar]
- Pétriacq, P. , Williams, A. , Cotton, A. , McFarlane, A.E. , Rolfe, S.A. , and Ton, J. (2017) Metabolite profiling of non‐sterile rhizosphere soil. Plant J 92: 147–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham, T.T.M. , Rodriguez, N.J.P. , Hijri, M. , and Sylvestre, M. (2015) Optimizing polychlorinated biphenyl degradation by flavonoid‐induced cells of the rhizobacterium Rhodococcus erythropolis U23A. PLoS One 10: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praveckova, M. , Brennerova, M.V. , Holliger, C. , De Alencastro, F. , and Rossi, P. (2016) Indirect evidence link PCB dehalogenation with geobacteraceae in anaerobic sediment‐free microcosms. Front Microbiol 7: 933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel, F. , Sulmon, C. , Serra, A.‐A. , Gouesbet, G. , and Couée, I. (2012) Xenobiotic sensing and signalling in higher plants. J Exp Bot 63: 3999–4014. [DOI] [PubMed] [Google Scholar]
- Rasmann, S. , Köllner, T.G. , Degenhardt, J. , Hiltpold, I. , Toepfer, S. , Kuhlmann, U. , et al. (2005) Recruitment of entomopathogenic nematodes by insect‐damaged maize roots. Nature 434: 732–737. [DOI] [PubMed] [Google Scholar]
- Rasmann, S. , and Turlings, T.C.J. (2016) Root signals that mediate mutualistic interactions in the rhizosphere. Curr Opin Plant Biol 32: 62–68. [DOI] [PubMed] [Google Scholar]
- Rezek, J. , Macek, T. , Mackova, M. , and Triska, J. (2007) Plant metabolites of polychlorinated biphenyls in hairy root culture of black nightshade Solanum nigrum SNC‐9O. Chemosphere 69: 1221–1227. [DOI] [PubMed] [Google Scholar]
- Ridl, J. , Suman, J. , Fraraccio, S. , Hradilova, M. , Strejcek, M. , Cajthaml, T. , et al. (2018) Complete genome sequence of pseudomonas alcaliphila JAB1 (=DSM 26533), a versatile degrader of organic pollutants. Stand Genomic Sci 13: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfe, S.A. , Griffiths, J. , and Ton, J. (2019) Crying out for help with root exudates: adaptive mechanisms by which stressed plants assemble health‐promoting soil microbiomes. Curr Opin Microbiol 49: 73–82. [DOI] [PubMed] [Google Scholar]
- Saha, B. , Swain, D. , Borgohain, P. , Rout, G.R. , Koyama, H. , and Panda, S.K. (2020) Enhanced exudation of malate in the rhizosphere due to AtALMT1 overexpression in blackgram (Vigna mungo L.) confers increased aluminium tolerance. Plant Biol 22: 701–708. [DOI] [PubMed] [Google Scholar]
- Sandermann, H. (1994) Higher plant metabolism of xenobiotics: the “green liver” concept. Pharmacogenetics 4: 225–241. [DOI] [PubMed] [Google Scholar]
- Schäffner, A. , Messner, B. , Langebartels, C. , and Sandermann, H. (2002) Genes and enzymes for in‐planta phytoremediation of air, water and soil. Acta Biotechnol 22: 141–151. [Google Scholar]
- Schwitzguébel, J.P. (2017) Phytoremediation of soils contaminated by organic compounds: hype, hope and facts. J Soil Sediment 17: 1492–1502. [Google Scholar]
- Shao, Z. , Lu, J. , Ding, J. , Fan, F. , Sun, X. , Li, P. , et al. (2021) Novel green chitosan‐pectin gel beads for the removal of Cu (II), Cd (II), Hg (II) and Pb (II) from aqueous solution. Int J Biol Macromol 176: 217–225. [DOI] [PubMed] [Google Scholar]
- Shen, R. , Yu, L. , Xu, P. , Liang, Z. , Lu, Q. , Liang, D. , et al. (2021) Water content as a primary parameter determines microbial reductive dechlorination activities in soil. Chemosphere 267: 129152. [DOI] [PubMed] [Google Scholar]
- Simhadri, J.J. , Loffredo, C.A. , Trnovec, T. , Murinova, L.P. , Nunlee‐Bland, G. , Koppe, J.G. , et al. (2020) Biomarkers of metabolic disorders and neurobehavioral diseases in a PCB‐exposed population: what we learned and the implications for future research. Environ Res 191: 110211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater, H. , Gouin, T. , and Leigh, M.B. (2012) Assessing the potential for rhizoremediation of PCB contaminated soils in northern regions using native species. Chemosphere 84: 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standfuß‐Gabisch, C. , Al‐Halbouni, D. , and Hofer, B. (2012) Characterization of biphenyl dioxygenase sequences and activities encoded by the metagenomes of highly polychlorobiphenyl‐contaminated soils. Appl Environ Microbiol 78: 2706–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stella, T. , Covino, S. , Čvančarová, M. , Filipová, A. , Petruccioli, M. , D'Annibale, A. , and Cajthaml, T. (2017) Bioremediation of long‐term PCB‐contaminated soil by white‐rot fungi. J Hazard Mater 324: 701–710. [DOI] [PubMed] [Google Scholar]
- Stockholm Convention on POPs (2015) URL http://chm.pops.int/TheConvention/ThePOPs/The1InitialPOPs/tabid/296/Default.aspx.
- Stringlis, I.A. , De Jonge, R. , and Pieterse, C.M.J. (2019) The age of Coumarins in plant‐microbe interactions. Plant Cell Physiol 60: 1405–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringlis, I.A. , Yu, K. , Feussner, K. , De Jonge, R. , Van Bentum, S. , Van Verk, M.C. , et al. (2018) MYB72‐dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc Natl Acad Sci U S A 115: E5213–E5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, S. , Schnoor, J.L. , and Van Aken, B. (2018) Effects of polychlorinated biphenyls (PCBs) and their hydroxylated metabolites (OH‐PCBs) on Arabidopsis thaliana . Environ Sci Technol 51: 7263–7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suenaga, H. , Fujihara, H. , Kimura, N. , Hirose, J. , Watanabe, T. , Futagami, T. , et al. (2017) Insights into the genomic plasticity of pseudomonas putida KF715, a strain with unique biphenyl‐utilizing activity and genome instability properties. Environ Microbiol Rep 9: 589–598. [DOI] [PubMed] [Google Scholar]
- Sun, J. , Pan, L. , Chen, J. , Li, K. , and Zhu, L. (2018) Uptake, translocation, and metabolism of hydroxylated and methoxylated polychlorinated biphenyls in maize, wheat, and rice. Environ Sci Pollut Res 25: 12–17. [DOI] [PubMed] [Google Scholar]
- Sun, J. , Pan, L. , Su, Z. , Zhan, Y. , and Zhu, L. (2016) Interconversion between methoxylated and hydroxylated polychlorinated biphenyls in rice plants: an important but overlooked metabolic pathway. Environ Sci Technol 50: 3668–3675. [DOI] [PubMed] [Google Scholar]
- Tandlich, R. , Brežná, B. , and Dercová, K. (2001) The effect of terpenes on the biodegradation of polychlorinated biphenyls by Pseudomonas stutzeri . Chemosphere 44: 1547–1555. [DOI] [PubMed] [Google Scholar]
- Taş, N. , Van Eekert, M.H.A. , De Vos, W.M. , and Smidt, H. (2010) The little bacteria that can – diversity, genomics and ecophysiology of “Dehalococcoides” spp. in contaminated environments. J Microbial Biotechnol 3: 389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temme, H.R. , Carlson, A. , and Novak, P.J. (2019) Presence, diversity, and enrichment of respiratory reductive dehalogenase and non‐respiratory hydrolytic and oxidative dehalogenase genes in terrestrial environments. Front Microbiol 10: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng, Y. , Sun, X. , Zhu, L. , Christie, P. , and Luo, Y. (2017) Polychlorinated biphenyls in alfalfa: accumulation, sorption and speciation in different plant parts. Int J Phytoremediation 19: 732–738. [DOI] [PubMed] [Google Scholar]
- Terzaghi, E. , Alberti, E. , Raspa, G. , Zanardini, E. , Morosini, C. , Anelli, S. , et al. (2021) A new dataset of PCB half‐lives in soil: effect of plant species and organic carbon addition on biodegradation rates in a weathered contaminated soil. Sci Total Environ 750: 141411. [DOI] [PubMed] [Google Scholar]
- Terzaghi, E. , Vergani, L. , Mapelli, F. , Borin, S. , Raspa, G. , Zanardini, E. , et al. (2019) Rhizoremediation of weathered PCBs in a heavily contaminated agricultural soil: results of a biostimulation trial in semi field conditions. Sci Total Environ 686: 484–496. [DOI] [PubMed] [Google Scholar]
- Terzaghi, E. , Vergani, L. , Mapelli, F. , Borin, S. , Raspa, G. , Zanardini, E. , et al. (2020) New data set of polychlorinated Dibenzo‐p‐dioxin and Dibenzofuran half‐lives: natural attenuation and rhizoremediation using several common plant species in a weathered contaminated soil. Environ Sci Technol 54: 10000–10011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tigini, V. , Prigione, V. , Di Toro, S. , Fava, F. , and Varese, G.C. (2009) Isolation and characterisation of polychlorinated biphenyl (PCB) degrading fungi from a historically contaminated soil. Microb Cell Fact 8: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timm, C.M. , Carter, K.R. , Carrell, A.A. , Jun, S.‐R. , Jawdy, S.S. , Vélez, J.M. , et al. (2018) Abiotic stresses shift belowground populus‐associated bacteria toward a core stress microbiome. mSystems 3: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint, J.P. , Pham, T.T.M. , Barriault, D. , and Sylvestre, M. (2012) Plant exudates promote PCB degradation by a rhodococcal rhizobacteria. Appl Microbiol Biotechnol 95: 1589–1603. [DOI] [PubMed] [Google Scholar]
- Uhlik, O. , Musilova, L. , Ridl, J. , Hroudova, M. , Vlcek, C. , Koubek, J. , et al. (2013) Plant secondary metabolite‐induced shifts in bacterial community structure and degradative ability in contaminated soil. Appl Microbiol Biotechnol 97: 9245–9256. [DOI] [PubMed] [Google Scholar]
- Urbaniak, M. , Lee, S. , Takazawa, M. , Mierzejewska, E. , Baran, A. , and Kannan, K. (2020) Effects of soil amendment with PCB‐contaminated sediment on the growth of two cucurbit species. Environ Sci Pollut Res 27: 8872–8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aken, B. , Correa, P.A. , and Schnoor, J.L. (2010) Phytoremediation of polychlorinated biphenyls: new trends and promises. Environ Sci Technol 44: 2767–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenkoornhuyse, P. , Quaiser, A. , Duhamel, M. , Le Van, A. , and Dufresne, A. (2015) The importance of the microbiome of the plant holobiont. New Phytol 206: 1196–1206. [DOI] [PubMed] [Google Scholar]
- Vergani, L. , Mapelli, F. , Marasco, R. , Crotti, E. , Fusi, M. , Di Guardo, A. , et al. (2017a) Bacteria associated to plants naturally selected in a historical PCB polluted soil show potential to sustain natural attenuation. Front Microbiol 8: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergani, L. , Mapelli, F. , Zanardini, E. , Terzaghi, E. , Di Guardo, A. , Morosini, C. , et al. (2017b) Phyto‐rhizoremediation of polychlorinated biphenyl contaminated soils: an outlook on plant‐microbe beneficial interactions. Sci Total Environ 575: 1395–1406. [DOI] [PubMed] [Google Scholar]
- Vives‐Peris, V. , de Ollas, C. , Gómez‐Cadenas, A. , and Pérez‐Clemente, R.M. (2020) Root exudates: from plant to rhizosphere and beyond. Plant Cell Rep 39: 3–17. [DOI] [PubMed] [Google Scholar]
- Voges, M.J.E.E.E. , Bai, Y. , Schulze‐Lefert, P. , and Sattely, E.S. (2019) Plant‐derived coumarins shape the composition of an Arabidopsis synthetic root microbiome. Proc Natl Acad Sci U S A 116: 12558–12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. , Chen, C. , Zhao, S. , and He, J. (2019) Microbial synergistic interactions for reductive dechlorination of polychlorinated biphenyls. Sci Total Environ 666: 368–376. [DOI] [PubMed] [Google Scholar]
- Wang, S. , Chng, K.R. , Wilm, A. , Zhao, S. , Yang, K.L. , Nagarajan, N. , and He, J. (2014) Genomic characterization of three unique Dehalococcoides that respire on persistent polychlorinated biphenyls. Proc Natl Acad Sci U S A 111: 12103–12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. , and He, J. (2013) Dechlorination of commercial PCBs and other multiple halogenated compounds by a sediment‐free culture containing dehalococcoides and dehalobacter. Environ Sci Technol 47: 10526–10534. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Teng, Y. , Zhang, N. , Christie, P. , Li, Z. , Luo, Y. , and Wang, J. (2017) Rhizobial symbiosis alleviates polychlorinated biphenyls‐induced systematic oxidative stress via brassinosteroids signaling in alfalfa. Sci Total Environ 592: 68–77. [DOI] [PubMed] [Google Scholar]
- Weigold, P. , El‐Hadidi, M. , Ruecker, A. , Huson, D.H. , Scholten, T. , Jochmann, M. , et al. (2016) A metagenomic‐based survey of microbial (de)halogenation potential in a German forest soil. Sci Rep 6: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield Åslund, M.L. , Rutter, A. , Reimer, K.J. , and Zeeb, B.A. (2008) The effects of repeated planting, planting density, and specific transfer pathways on PCB uptake by Cucurbita pepo grown in field conditions. Sci Total Environ 405: 14–25. [DOI] [PubMed] [Google Scholar]
- Whitfield Åslund, M.L. , Zeeb, B.A. , Rutter, A. , and Reimer, K.J. (2007) In situ phytoextraction of polychlorinated biphenyl—(PCB) contaminated soil. Sci Total Environ 374: 1–12. [DOI] [PubMed] [Google Scholar]
- Wilkinson, S.W. , Mageroslashy, M.H. , Lopez Sanchez, A. , Smith, L.M. , Furci, L. , Cotton, T.E.A. , et al. (2019) Surviving in a hostile world: plant strategies to resist pests and diseases. Annu Rev Phytopathol 57: 505–529. [DOI] [PubMed] [Google Scholar]
- Williams, A. , and de Vries, F.T. (2020) Plant root exudation under drought: implications for ecosystem functioning. New Phytol 225: 1899–1905. [DOI] [PubMed] [Google Scholar]
- Wu, X. , Kammerer, A. , and Lehmler, H. (2014) Microsomal oxidation of 2,2′,3,3′,6,6′‐Hexachlorobiphenyl (PCB 136) results in species‐dependent chiral signatures of the hydroxylated metabolites. Environ Sci Technol 48: 2436–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, Y. , Xing, Z. , Liu, J. , Qin, W. , and Huang, X. (2020) Recent advances in the biodegradation of polychlorinated biphenyls. World J Microbiol Biotechnol 36: 1–10. [DOI] [PubMed] [Google Scholar]
- Zeeb, B.A. , Amphlett, J.S. , Rutter, A. , and Reimer, K. (2006) Potential for phytoremediation of polychlorinated biphenyl‐(PCB)‐contaminated soil. Int J Phytoremediation 8: 199–221. [DOI] [PubMed] [Google Scholar]
- Zhalnina, K. , Louie, K.B. , Hao, Z. , Mansoori, N. , Nunes, U. , Shi, S. , et al. (2018) Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat Microbiol 3: 470–480. [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Chen, J. , Ni, Y. , Zhang, Q. , and Zhao, L. (2009) Uptake by roots and translocation to shoots of polychlorinated dibenzo‐p‐dioxins and dibenzofurans in typical crop plants. Chemosphere 76: 740–746. [DOI] [PubMed] [Google Scholar]
- Ziegler, M. , Engel, M. , Welzl, G. , and Schloter, M. (2013) Development of a simple root model to study the effects of single exudates on the development of bacterial community structure. J Microbiol Methods 94: 30–36. [DOI] [PubMed] [Google Scholar]


