Abstract
Liver-specific gene expression is controlled by a heterogeneous group of hepatocyte-enriched transcription factors. One of these, the winged helix transcription factor hepatocyte nuclear factor 3β (HNF3β or Foxa2) is essential for multiple stages of embryonic development. Recently, HNF3β has been shown to be an important regulator of other hepatocyte-enriched transcription factors as well as the expression of liver-specific structural genes. We have addressed the role of HNF3β in maintenance of the hepatocyte phenotype by inactivation of HNF3β in the liver. Remarkably, adult mice lacking HNF3β expression specifically in hepatocytes are viable, with histologically normal livers and normal liver function. Moreover, analysis of >8,000 mRNAs by array hybridization revealed that lack of HNF3β affects the expression of only very few genes. Based on earlier work it appears that HNF3β plays a critical role in early liver development; however, our studies demonstrate that HNF3β is not required for maintenance of the adult hepatocyte or for normal liver function. This is the first example of such functional dichotomy for a tissue specification transcription factor.
Hepatic gene expression is controlled by a diverse set of transcription factors, none of which is expressed exclusively in the liver. Among these are the hepatocyte nuclear factor 3 α, β, and γ proteins (HNF3α, HNF3β, and HNF3γ, respectively), which were first discovered by their ability to bind to the promoters of the liver-specific genes encoding α1-antitrypsin and transthyretin (8, 20, 21). The HNF3 genes are closely related to the Drosophila melanogaster gene forkhead, which is essential for the proper formation of the foregut and hindgut (32). This fact together with the observation that the mouse HNF3 genes are expressed early during the formation of definite endoderm led to the hypothesis that the HNF3 proteins function in mammalian liver and gut development (3, 23, 27).
Recently, the nomenclature of the winged helix/forkhead transcription factor gene family has been revised, and according to this new nomenclature the genetic loci encoding HNF3α, HNF3β, and HNF3γ are now known as Foxa1, Foxa2, and Foxa3, respectively, in mouse (Fox refers to forkhead box) (17).
Analysis of the crystal structure of the DNA binding domain of HNF3γ showed a striking similarity to the winged helix domain of linker histones (7). The HNF3 proteins can bind to and reposition nucleosomes in the albumin gene which correlates with an active albumin enhancer (6, 22, 29). As the HNF3 proteins can alter chromatin and are the earliest known factors expressed in the prehepatic endoderm, the HNF3 proteins were proposed to act as genetic potentiators of the hepatic differentiation program (34).
During formation of the definite endoderm, HNF3β is activated first, followed by HNF3α, and finally HNF3γ (3, 23, 27). HNF3β is required for the development of the node and for visceral endoderm formation because these structures are missing or abnormal in embryos homozygous for a targeted null mutation in the HNF3β gene (2, 10, 33). However, the functions of HNF3β in the differentiation of the hepatic primordium or in hepatic metabolism could not be addressed using the HNF3β null allele, as even embryos obtained from tetraploid embryonic stem (ES) cell aggregations lacked foregut and midgut endoderm (10).
In contrast to the HNF3β−/− mice, embryos deficient for HNF3α or HNF3γ develop normally to term and show no obvious morphological liver phenotype (15, 16, 28). HNF3α mutant mice had unchanged expression of liver-specific HNF3 target genes involved in metabolism and glucose homeostasis. However, the transcription of several HNF3 targets was reduced by 50 to 70% in the HNF3γ mutants. Due to the similarity of the HNF3 proteins it seems likely that these transcription factors can at least partially compensate for each other during liver development, when all three genes are expressed concurrently.
Recently, the regulatory function of HNF3β has been studied in visceral endoderm derived from HNF3β−/− embryoid bodies in vitro (12). In this model, the lack of HNF3β resulted in a reduction of the mRNA levels for the transcription factors HNF1α and HNF4α and a complete elimination of the HNF3α transcript, suggesting that HNF3β regulates a transcription factor network required for differentiation and metabolism (12). To test the role of HNF3β in regulating the hepatic transcriptional program in vivo, we have generated mice lacking HNF3β specifically in hepatocytes using the Cre-loxP recombination system. Here we describe the liver-specific gene expression profile and metabolic regulation in these hepatocyte-specific HNF3β knockout mice.
MATERIALS AND METHODS
Generation of HNF3βloxP/loxP Alb.Cre mice.
Lambda phage clones containing the murine HNF3β gene had been isolated from a mouse ES cell (strain 129) library previously (2). The targeting vector is depicted in Fig. 1A and contains approximately 5 kb of homology regions. This targeting construct was electroporated into E14/1 mouse ES cells (18). Stably transfected cells were isolated after selection in G418 (350 μg/ml; Gibco), and 340 clones were screened for the desired homologous recombination event. A 0.8-kb XhoI/BglII fragment (3′ probe in Fig. 1A) was used as an external probe for Southern blot analysis of DNA digested with BglII (data not shown). The neo-tk cassette was then removed from correctly targeted clones by partial Cre-mediated recombination. Two clones were expanded and transfected transiently with 2 μg of Cre expression vector (pIC-Cre [13]). Two days posttransfection, cells were treated with 2 μM gancyclovir to select for the cells that had lost the neo-tk cassette. Ninety-six individual gancyclovir-resistant clones were analyzed by Southern blotting (data not shown). Approximately 20% of the clones were identified to have undergone partial recombination to generate the HNF3βloxP allele, thereby removing the neo-tk cassette and one loxP site but leaving both exon 3 and its flanking loxP sites intact. Two of the HNF3βloxP ES cell clones were injected into C57BL/6J mouse blastocysts. Blastocysts were transferred to pseudopregnant NMRI females, and chimeric offspring were identified by the presence of agouti hair. Chimeric males were mated to C57BL/6 females to obtain ES cell-derived offspring that were analyzed by Southern blotting of tail DNA to identify the HNF3βloxP/+ heterozygote mice. Germ line chimeras were crossed to CD1 outbred mice, as this is the strain of mice used previously for the analysis of the HNF3β null allele (2). Heterozygotes were mated inter se to generate homozygous HNF3βloxP/loxP mice. The derivation of the Alb.Cre transgenic line has been described previously (25), and mice were kept on a CD1 background.
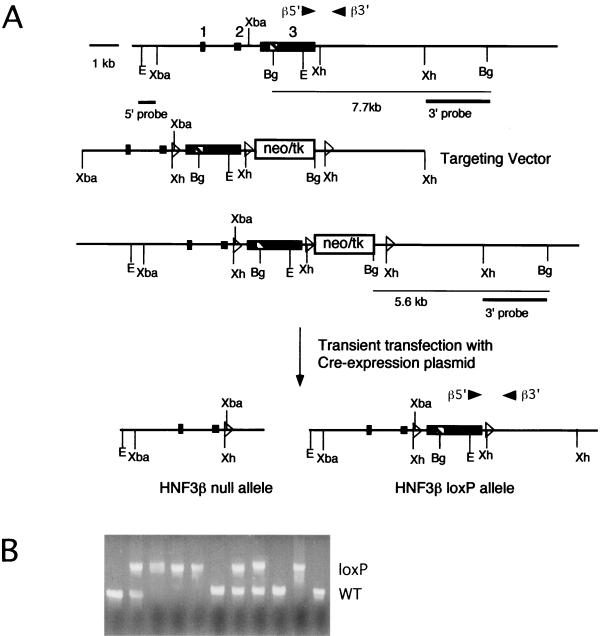
FIG. 1.
Cre-loxP-mediated targeting of the HNF3β gene and generation of homozygous HNF3βloxP/loxP mice. (A) Targeting vector for the HNF3βloxP allele. Primers β5′ and β3′ were used in PCR genotype analysis. (First line) Gene structure of the endogenous HNF3β locus. Exons are indicated as boxes, the striped box represents the winged helix domain, open triangles represent loxP sites, and arrow heads represent primer positions. (Second line) Targeting vector which introduces a cassette containing the neomycin-herpes simplex virus-thymidine kinase selection cassette (neo-tk) flanked by loxP sites downstream of exon 3. An additional loxP site was introduced in the intron upstream of exon 3. (Third line) Gene structure of homologous recombinants. The 3′ probe was used for the Southern blot analysis (data not shown). (Fourth line) Cre-mediated deletion results in either the HNF3β null allele (deletion of exon 3) or the HNF3βloxP allele (exon 3 flanked by loxP sites). Abbreviations: Bg, BglII; E, EcoRI; Xh, XhoI; Xba, XbaI. (B) HNF3βloxP/loxP mice are viable and healthy. Shown are the results of PCR genotype analysis of a litter from mating HNF3βloxP/+ heterozygotes inter se. The HNF3βloxP allele segregates in expected Mendelian frequency. WT, wild type.
Genotype analysis.
The genotypes of all offspring were analyzed using DNA isolated either from the yolk sac of embryos or the tails of 4-week-old mice. The 5′ and 3′ primers for the Alb.Cre transgene (PCR product of 232 bp) were 5′-GCGGCATGGTGCAAGTTGAAT-3′ and 5′-CGTTCACCGGCATCAACGTTT-3′, respectively. The 5′ and 3′ primers for the HNF3β gene whose positions are depicted in Fig. 1a were 5′-CCCCTGAGTTGGCGGTGGT-3′ and 5′-TTGCTCACGGAAGAGTAGCC-3′, respectively, which produce an ∼290-bp amplified fragment for the wild-type HNF3β allele and an ∼450-bp amplified fragment for the HNF3βloxP allele. PCRs were carried out for 32 cycles (95°C for 45 s, 60°C for 45 s; 72°C for 90 s) in a buffer containing 1.5 mM MgCl2.
Immunohistochemistry.
Adult and fetal tissue samples were fixed in 4% paraformaldehyde overnight at 4°C, embedded in paraffin, cut to 6-μm-thick sections and applied to Probe-on Plus slides (Fisher Scientific). Deparaffinized and rehydrated slides were subjected to microwave antigen retrieval by boiling for 6 min in a 10 mM citric acid buffer, pH 6.0, and allowed to cool for 10 min at room temperature (RT). Slides were washed in phosphate-buffered saline (PBS), then blocked with avidin and biotin blocking agent (Vector) for 15 min each at RT, and then blocked with protein blocking reagent (Immunotech) for 20 min at RT. The primary antibody, a 1:10,000 dilution of polyclonal rabbit anti-HNF-3β, kindly provided by Thomas M. Jessell (Columbia University, New York, N.Y.), was diluted in PBS containing 0.1% bovine serum albumin (BSA) and 0.2% Triton X-100 (PBT) and incubated with the sections overnight at 4°C. Slides were washed in PBS, then incubated with goat anti-rabbit biotinylated secondary antibody (Biomeda Biostain Rabbit IgG [AP] kit) diluted in PBT for 30 min at 37°C, washed in PBS, and incubated with alkaline phosphatase-conjugated ABC reagent (Biomeda) diluted in PBT for 30 min at 37°C. Slides were washed in PBS and then washed in a solution containing 100 mM NaCl, 50 mM MgCl2, 100 mM Tris (pH 9.5), and 0.1% Tween 20. Signal was developed in Nitro Blue Tetrazolium (0.35 mg/ml; Boehringer Mannheim), BCIP (5-bromo-4-chloro-3-indolylphosphate (0.18 mg/ml; Boehringer Mannheim) for 8 min at RT. Slides were washed in water, counterstained in a 2% neutral red solution for 20 min, dehydrated, mounted, and viewed using Nomarski optics on a Nikon X4Z microscope.
RNA analysis and liver function tests.
Northern blot, RNase protection, and reverse transcription-PCR analysis were performed as described previously (12, 16). The primers used for reverse-transcription-PCR are available upon request. Microarray hybridizations were carried out by Incyte Pharmaceuticals, Inc. (Palo Alto, Calif.). For liver function tests, mice were fasted for 9 h before sacrifice by carbon dioxide asphyxiation. Whole venous blood obtained from the inferior vena cava was mixed with an anticoagulant consisting of trasylol, EDTA, and leupeptin and centrifuged for 5 min at 14,000 × g to obtain plasma. Plasma parameters were determined by Ani Lytics, Inc. (Gaithersburg, Md.).
Preparation of nuclear extracts from liver.
Livers from 16-week-old adult mice were extracted, rinsed in cold PBS, minced, and dounced in homogenization buffer (2 M sucrose, 10 mM HEPES (pH 7.6), 25 mM KCl, 1 mM EDTA, 0.15 mM spermine, 0.5 mM spermidine, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 0.5 mM dithiothreitol [DTT], and 10% glycerol) until most of the cells were disrupted but nuclei were still intact. The liver homogenates were then layered on top of homogenization buffer in an ultracentrifuge tube and centrifuged at 27,000 rpm (90,000 × g) for 55 min at 4°C. The nuclear pellet was resuspended in 0.1 ml of nuclear storage buffer (20 mM Tris [pH 7.9], 75 mM NaCl, 0.5 mM EDTA, 0.125 mM PMSF, 0.85 mM DTT, and 50% glycerol). A 0.01-ml aliquot of the nuclear pellet was used to normalize nuclear extract concentration as follows: 0.2 ml of 1% sodium dodecyl sulfate was added to the nuclear pellet aliquot, vortexed, and centrifuged at 15,000 × g for 3 min. Absorbance at 260 nm was assayed to estimate the amount of nucleic acid in the nuclear extract. The remainder of the nuclear extract was centrifuged at 3,000 rpm for 15 min at 4°C. The nuclei were resuspended in nuclear storage buffer at 25 μg/ml. Nine volumes of 1.1× NUN solution (27.5 mM HEPES [pH 7.6], 1.1 M urea, 0.33 M NaCl, 1.1 mM DTT, and 1.1% NP-40) was added, vortexed briefly, incubated on ice for 15 min, and centrifuged at 14,000 rpm at 4°C. Glycerol was added to the supernatant containing the nuclear extract to a 10% final volume and the solution was dialyzed against a 1,000× volume of Gorski dialysis buffer (25 mM HEPES [pH 7.9], 40 mM KCl, 0.1 mM EDTA, 0.5 mM PMSF, 1 mM DTT, and 10% glycerol) for 2 h at 4°C. Samples were subsequently centrifuged at 14,000 rpm for 1 min at 4°C, aliquoted, and stored at −80°C. Additionally, all buffers contained the following protease inhibitors: 0.1 mM benzamidine; 1 μg of antipain, leupeptin, and soybean trypsin inhibitor per ml; and 2 μg of aprotinin and bestatin per ml.
Electrophoretic mobility shift assays (EMSAs).
The binding reaction contained 2.0 μg of nuclear extract in a 15-μl solution of 10 mM Tris (pH 7.5), 50 mM NaCl, 1 mM EDTA, 1 mM 2-mercaptoethanol, and 1% Ficoll, with 200 ng of poly(dI-dC) and 2 μg of bovine serum albumin. The mixtures were incubated at RT for 10 min, and then an 80-fold molar excess of competitor oligonucleotide (as indicated) and 0.1 ng (5 × 104 to 10 × 104 cpm) of oligonucleotide probe were added to the reaction mixture. Incubation was continued for an additional 20 min prior to electrophoresis on 8% polyacrylamide gels in 1× Tris-borate-EDTA, and gels were dried and exposed to a PhosphorImager cassette. Oligonucleotide probes were labeled with [32P]dCTP by filling in the ends with Klenow polymerase I. The sequences of the double-stranded oligonucleotides used as probes or competitors have been described previously (9).
Glucose tolerance test.
Overnight-fasted 8- to 15-week-old mice were injected intraperitoneally with 5 mg of glucose per g of body weight. Blood samples were obtained from the tail vein, and glucose levels were measured immediately before and 30, 60, and 120 min after injection using a Glucometer Elite device (Bayer, Inc.).
RESULTS
Generation of HNF3βloxP mice.
We have employed conditional gene inactivation using Cre-loxP-mediated recombination to study the role of HNF3β in the liver, as embryos homozygous for a HNF3β null allele die before hepatic differentiation begins (13, 25, 31). To achieve this, we constructed a targeting vector that introduced two loxP sites flanking exon 3 of the HNF3β gene, which contains the bulk of the protein-coding sequence, including the DNA binding domain (14), and a third loxP site downstream of a selection cassette consisting of the neomycin resistance and herpes simplex virus thymidine kinase genes (neo-tk) (Fig. 1A). We chose exon 3 because deletion of this exon in the germ line resulted in complete inactivation of the HNF3β gene (2, 33). After electroporation of mouse ES cells, screening of 340 neomycin (G418)-resistant colonies yielded three correctly targeted clones (data not shown). As the presence of the neo-tk cassette might interfere with the promoter activity and/or mRNA processing of the HNF3β gene, this neo-tk cassette was subsequently deleted in two of the targeted clones by transient transfection with a Cre expression vector. As depicted in Fig. 1A, this can result in either an HNF3βnull allele or the desired HNF3βloxP allele. Southern blot analysis revealed approximately 20% of the 96 gangcyclovir-resistant clones tested to be HNF3βloxP ES cell clones (data not shown). Germ line chimeras and mice heterozygous for the HNF3βloxP allele were obtained for two independent ES cell lines injected. Both lines gave identical results in the tissue-specific gene ablation experiments.
To assess whether introduction of the two loxP sites in the second intron and 3′ untranslated region of the HNF3β gene affects gene function, heterozygous HNF3βloxP mice were bred inter se, and the resulting offspring were genotyped by PCR analysis. Of 27 offspring genotyped at 4 weeks of age, seven were HNF3βloxP/loxP, in close agreement with the expected Mendelian frequency (Fig. 1b). HNF3βloxP/loxP mice are fertile, healthy, and have normal growth curves (data not shown). Thus, insertion of the two loxP sites into the HNF3β locus does not appear to interfere with HNF3β function in this genetic background.
Ablation of the HNF3β gene in the liver.
In order to investigate liver development and function in hepatocyte-specific HNF3β knockout mice, we utilized a transgenic mouse with Cre under the control of the albumin promoter (23). HNF3βloxP heterozygous mice carrying this Alb.Cre transgene (HNF3βloxP/+; Alb.Cre) were bred with HNF3βloxP homozygous mice, and embryos were collected from various stages of gestation. As shown in Fig. 2A through F, no morphological abnormalities were observed in the livers of HNF3βloxP/loxP; Alb.Cre embryos compared to their wild-type control littermates. Furthermore, adult livers that lack HNF3β have a normal morphology (compare Fig. 2G and H; also data not shown). HNF3βloxP/loxP; Alb.Cre mice were born in the expected Mendelian distribution, and no significant differences in body weights were observed between these mice and their wild-type control littermates (data not shown). Liver-specific HNF3β knockout mice were fertile and produced normal-sized litters.
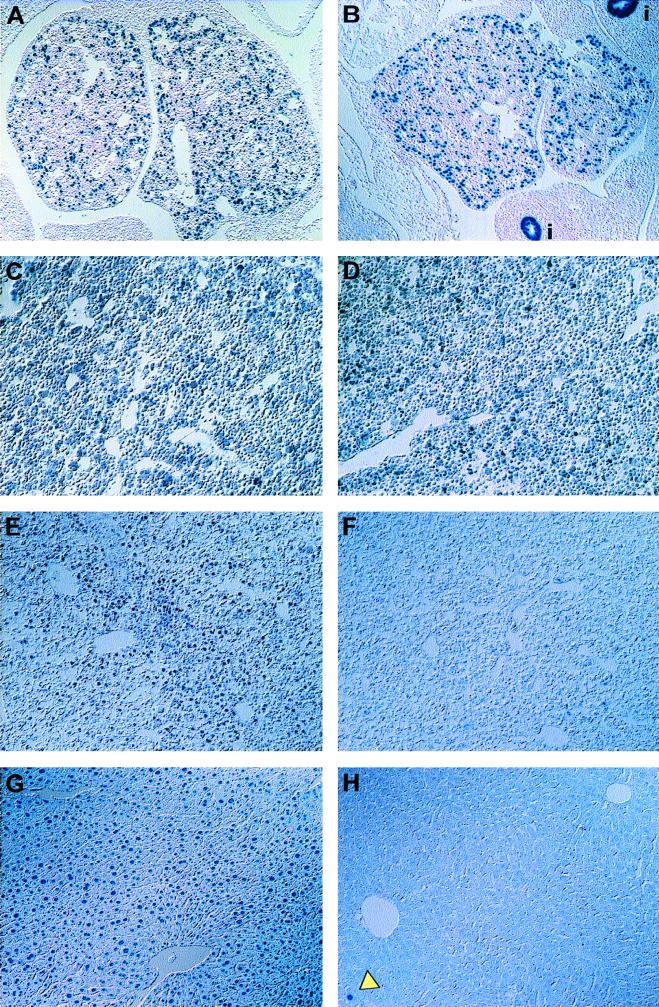
FIG. 2.
The HNF3βloxP allele is excised by the Alb.Cre transgene. (A to G) Immunohistochemical analysis of liver sections from 11.5 (A and B), 14.5 (C and D), and 18.5 (E and F) d.p.c. embryos and 10-week-old adult (G and H) mice stained with anti-HNF3β antibody. HNF3βloxP/loxP; Alb.Cre mice express HNF3β by 14.5 d.p.c. (B and D), but HNF-3β protein is reduced in 18.5-d.p.c. fetal livers and absent from adult livers (F and H). The arrowhead in panel H shows a rare HNF3β immunoreactive nucleus. Images were obtained using Nomarski optics of livers from wild-type control mice (A, C, E, and G) and mutant HNF3βloxP/loxP; Alb.Cre mice (B, D, G, and H). Images are shown at ×90 (A, B, G, and H) or ×180 (D to F) magnification.
In light of the normal development of hepatic HNF3β knockout mice, we investigated whether we had indeed deleted the HNF3β gene in the liver. Therefore, steady-state HNF3β mRNA levels were analyzed in total liver RNA obtained from adult HNF3βloxP/loxP; Alb.Cre or control mice using quantitative RNase protection analysis. HNF3β message was undetectable in the livers of 10-week-old HNF3βloxP/loxP; Alb.Cre mice (Fig. 3A), indicating that the Cre recombinase had efficiently excised the loxP-flanked HNF3β gene in hepatocytes.
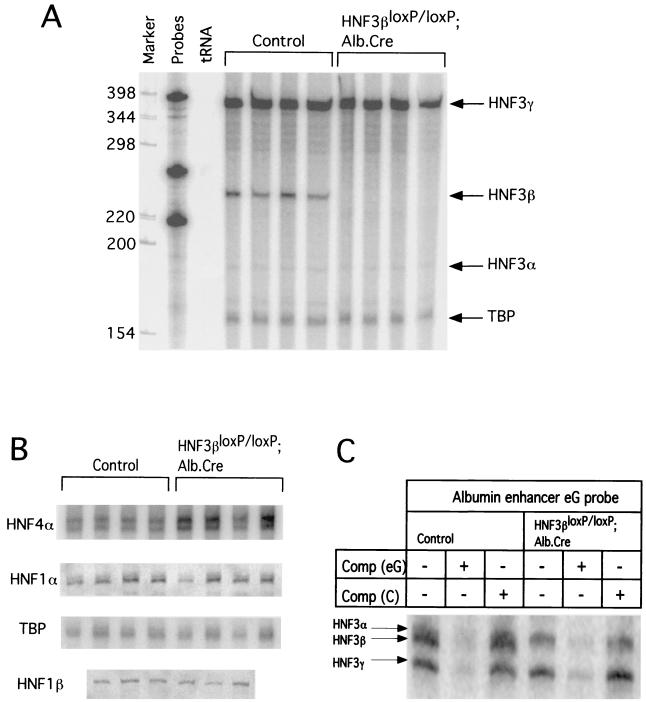
FIG. 3.
Expression of hepatic transcription factors in adult livers from HNF3βloxP/loxP; Alb.Cre. mice. (A) Total RNA (30 μg) from livers of wild type control or HNF3βloxP/loxP; Alb.Cre mice was analyzed by RNase protection assay for mRNA levels of all HNF3 genes. HNF3β mRNA is no longer expressed in livers of HNF3βloxP/loxP; Alb.Cre mice, whereas HNF3α and HNF3γ steady-state mRNA levels are unchanged in HNF3βloxP/loxP; Alb.Cre mice compared to controls. (B) Total RNA (30 μg) from livers of wild-type control or HNF3βloxP/loxP; Alb.Cre mice was analyzed by RNase protection assay for mRNA levels of other liver-enriched HNF genes. HNF1α and HNF1β steady-state mRNA concentrations are unaltered whereas HNF4α mRNA levels are slightly increased in HNF3βloxP/loxP; Alb.Cre mice compared to controls when quantitated using PhosphorImager analysis (data not shown). TATA box binding protein (TBP) served as the loading control. (C) Nuclear binding activities of HNF3α and HNF3γ are not increased in HNF3βloxP/loxP; Alb.Cre hepatocytes compared to controls when analyzed using an EMSA. A radioactive oligonucleotide probe for the albumin enhancer eG site was incubated with 2 μg of nuclear extract isolated from wild-type control or HNF3βloxP/loxP; Alb.Cre liver. An 80-fold molar excess of nonradioactive competitor oligonucleotide (Comp) was added as indicated: −, no indicated competitor added; eG, HNF3 binding site from albumin enhancer; C, non-specific competitor containing CCAAT site from albumin promoter. Specific HNF3 binding complexes are indicated. HNF3α and -β complexes from liver extracts migrate very closely together, as shown previously (9).
Next, we wanted to determine the stage of fetal development at which excision occurs. We analyzed the disappearance of HNF3β protein in livers obtained from HNF3βloxP/loxP; Alb.Cre mice and their control littermates during various developmental stages using immunohistochemical staining with an antibody specific to HNF3β. Although the endogenous albumin promoter is active during the onset of liver development (approximately 9.5 days postcoitum [d.p.c.] in the mouse [4]), HNF3β inactivation was not evident in livers from midgestation HNF3βloxP/loxP; Alb.Cre embryos (compare Fig. 2A and B and Fig. 2C and D). By 18.5 d.p.c., however, the vast majority of hepatocytes from such embryos had lost expression of HNF3β (compare Fig. 2E and F). HNF3β deletion is restricted to the liver, as HNF3β is still expressed in other organ systems where HNF3β is normally expressed (data not shown). Finally, HNF3β protein is missing in over 99.9% of hepatocytes in 10-week-old mice (compare Fig. 2G and H), consistent with the absence of HNF3β mRNA shown in Fig. 3A. Thus, we conclude that HNF3β is not required for the maintenance of hepatocytes in late fetal and postnatal development.
Ablation of HNF3β does not lead to dramatic changes in the hepatic transcriptional program.
The expression of many genes required for yolk sac and liver metabolism, including apolipoproteins, aldolase B, and albumin, was dramatically decreased or completely absent in embryoid bodies derived from ES cells lacking HNF3β (12). In addition, expression of HNF3α was absent and that of HNF1α and HNF4α was reduced dramatically. As visceral endoderm differentiated in vitro expresses many of the same genes as hepatocytes in vivo, the notion was put forward that HNF3β directs a regulatory network of transcription factors and their targets in the metabolism of yolk sac and liver. However, as shown in Fig. 4A expression levels of mRNAs encoding apolipoproteins, albumin, and gluconeogenic enzymes are not significantly reduced in adult livers of HNF3βloxP; Alb.Cre mice, despite the fact that these livers are devoid of HNF3β mRNA and protein.
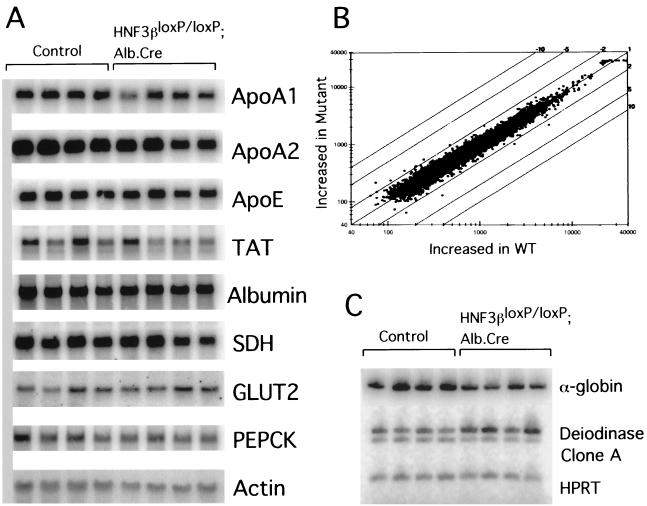
FIG. 4.
Analysis of steady-state mRNA levels and microarray expression profile of potential HNF3 targets in liver. (A) Total RNA (10 μg) from livers of wild-type control (Control) or HNF3βloxP/loxP; Alb.Cre mice was separated on denaturing agarose gels, blotted onto nylon membrane, and hybridized to the probes indicated (Apo, apolipoprotein; SDH, serine dehydrogenase; PEPCK, phosphoenolpyruvate carboxykinase; TAT, tyrosine amino transferase; GLUT2, glucose transporter 2). β-Actin (Actin) served as a loading control. PhosphorImager analysis did not reveal significant differences between the mRNA levels between the control and HNF3βloxP/loxP; Alb.Cre mice (data not shown). (B) Scatter plot analysis representing expression profiles between livers from wild-type control and HNF3βloxP/loxP; Alb.Cre mice using an oligonucleotide microarray representing 8,700 mouse cDNA and EST transcripts. (C) Reverse transcription-PCR analysis of total RNA from livers of wild-type control (Control) or HNF3βloxP/loxP; Alb.Cre mice using [α-32P]dATP. Steady-state mRNA levels of α-globin (accession no. AA109900) is decreased twofold, whereas deiodinase (accession no. AA212899) is induced twofold in livers of HNF3βloxP/loxP; Alb.Cre mice compared to controls. Signals were quantified using PhosphorImager analysis (data not shown). Clone A (accession no. AA267590) EST mRNA levels were not significantly different between wild-type and mutant livers, although the microarray hybridization had indicated a threefold difference.
In several knockouts of transcription factors belonging to gene families, targeted mutation of one gene led to an upregulation of other family members, demonstrating the existence of regulatory networks between these transcription factors (15, 26). As mentioned above, HNF3β also regulates a network of transcription factors in visceral endoderm differentiated from embryoid bodies in vitro that includes HNF1α, HNF4α, and HNF3α (12). Therefore, we analyzed the steady-state mRNA levels of these genes by quantitative RNase protection assay. Consistent with the findings by Duncan et al. (12), mRNA levels of HNF3γ were unchanged in the livers from mutant animals (Fig. 3A). However, in contrast to the situation in embryoid bodies, there was no decrease in the transcript levels of HNF3α or HNF1α but a small (60%) increase in HNF4α relative mRNA levels in liver (Fig. 3A and B). Furthermore, the relative mRNA levels for HNF1β and other liver-enriched transcription factors are unaffected (Fig. 3B and data not shown).
To address the possibility that the remaining HNF3 factors could compensate for lack of HNF3β function at a posttranscriptional level via increased protein stabilization or nuclear localization, we assayed nuclear HNF3 binding activity of hepatocytes that lack HNF3β and compared these to wild-type controls using an electrophoretic mobility shift assay with the albumin enhancer eG site, which contains an HNF3 binding site, as a probe. As demonstrated in Fig. 3C, there is no upregulation of HNF3α or HNF3γ nuclear binding activity in hepatocyte nuclei that lack HNF3β protein.
To identify other potential liver-specific targets of HNF3β, we used mRNA from livers of 10-week-old HNF3βloxP/loxP; Alb.Cre mice and wild-type control littermates and screened for altered expression profiles using cDNA and expressed sequence tag (EST) microarray hybridization. As shown in the scatter plot in Fig. 4B, the overall transcriptional program is strikingly similar in livers containing or lacking HNF3β. Out of 8,700 mouse transcripts analyzed, three genes were reduced approximately two- to threefold in livers that lack HNF3β compared to control livers (Fig. 4B), whereas one gene was induced by more than twofold. To confirm whether differences revealed by the microarray hybridization were real and not just an artifact of the assay, the transcript abundance of three of these differentially expressed genes was analyzed by quantitative reverse transcription-PCR in livers of HNF3βloxP/loxP; Alb.Cre mice and control mice (Fig. 4C). After first determining the number of PCR cycles required for exponential amplification for each primer pair (data not shown), we compared mRNA levels in livers from four wild-type control and four mutant animals. As shown in Fig. 4C, deiodinase mRNA levels are induced two-fold in livers that lack HNF3β, in close agreement with the result from the array hybridization. Likewise, α-globin mRNA levels were reduced by approximately twofold, as had been indicated by the microarray data. These results confirm the usefulness of microarray hybridization for obtaining accurate comparisons of the global transcriptional program between wild-type and mutant animals. These very limited changes in the overall gene expression pattern between HNF3β wild-type and mutant livers also argue that HNF3β is not required for the maintenance of the global hepatic transcriptional program.
Physiological consequence of hepatocyte-specific HNF3β gene inactivation.
To assess whether loss of HNF3β in the liver affects liver metabolism of the HNF3βloxP; Alb.Cre mice, we measured various serum parameters that are indicative of hepatic metabolism. There were no significant differences in these parameters between HNF3βloxP; Alb.Cre mice and wild-type control littermates, as summarized in Table 1.
TABLE 1.
Circulating intermediates in plasma in HNF3βloxP/loxP; Alb.Cre and control micea
| Intermediate | Concn in plasma in mice
|
Reference range (concn) | |
|---|---|---|---|
| Control | HNF3βloxP/loxP; Alb.Cre | ||
| Creatine phosphokinase (U/liter) | 2,241 ± 1,445 | 2,114 ± 574 | 0 – 800 |
| Lactate dehydrogenase (U/liter) | 581 ± 120 | 580 ± 86 | 260 – 680 |
| AST-SGOTb (U/liter) | 150 ± 36 | 139 ± 21 | 72 – 288 |
| ALT-SGPTc (U/liter) | 60 ± 29 | 32 ± 4 | 24 – 140 |
| Gamma glutamyl transferase (U/liter) | 0 ± 0 | 0.83 ± 0.50 | 0 – 2 |
| Amylase (U/liter) | 2,199 ± 160 | 2,778 ± 303 | 602 – 2,311 |
| Bilirubin, total (mg/dl) | 0.67 ± 0.45 | 0.27 ± 0.05 | 0.0 – 0.9 |
| Bilirubin, direct (mg/dl) | 0.13 ± 0.09 | 0.07 ± 0.03 | 0.0 – 0.2 |
| Uric acid (mg/dl) | 4.4 ± 1.9 | 1.9 ± 0.2 | 2.2 – 4.6 |
| Blood urea nitrogen (mg/dl) | 21.3 ± 1.4 | 19.8 ± 1.5 | 9.0 – 28.0 |
| Creatinine (mg/dl) | 0.38 ± 0.03 | 0.33 ± 0.03 | 0.2 – 0.7 |
| Albumin (g/dl) | 2.9 ± 0.5 | 2.3 ± 0.4 | 2.6 – 4.6 |
| Protein, total (g/dl) | 4.7 ± 0.2 | 3.9 ± 0.4 | 4.0 – 6.2 |
Metabolic parameters were experimentally determined or analyzed by Ani Lytics, Inc., as described under Materials and Methods. Values are represented as means ± standard errors of the means. Comparisons were made among mice of similar age and sex (n = 6 animals for each group).
Aspartate aminotransferase-serum glutamic oxalacetic transaminase.
Alanine aminotransferase-serum glutamic pyruvic transaminase.
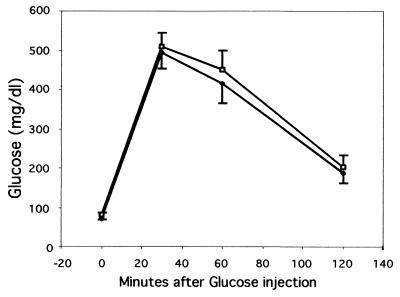
Many liver-specific genes that encode glycolytic or gluconeogenic enzymes have an HNF3 binding site located in their cis-regulatory element. Embryoid bodies derived from ES cells lacking HNF3β have decreased or absent expression of the genes encoding the glycolytic enzymes l-type pyruvate kinase and aldolase B (12). As HNF3β potentially regulates genes required for normal glucose homeostasis in vivo, we performed glucose tolerance tests on adult HNF3βloxP; Alb.Cre mice and wild-type control littermates to examine whether they respond normally to a glucose challenge. Mice that lack hepatic HNF3β clear excess blood glucose at a rate similar to that of their wild-type control littermates (Fig. 5). Therefore, hepatic HNF3β appears not essential for normal glucose homeostasis in vivo.
FIG. 5.
Mice lacking hepatic HNF3β have normal glucose homeostasis. Glucose challenge of age-matched HNF3βloxP/loxP; Alb.Cre (□) and wild-type control (⧫) mice. Blood glucose levels are shown at indicated time points after intraperitoneal administration of glucose. Values are means ± standard errors of the means (error bars) of seven (control) or nine (HNF3βloxP/loxP; Alb.Cre) animals. There was no significant difference between experimental and control groups as determined by Students's t test for each time point.
DISCUSSION
We have employed conditional gene ablation of HNF3β using the Cre-loxP recombination system to generate a mouse model that lacks HNF3β specifically in the liver. While hepatocyte-specific inactivation of HNF3β was greater than 99.9% efficient in the adult liver, inactivation of HNF3β gene expression was not evident until the end of fetal development, although the albumin gene is activated on day 9 of gestation (4). A similar temporal discrepancy between Cre expression and gene inactivation has been observed when the Pdx1 gene was ablated in β-cells of the pancreas using the Cre-loxP system (1). Several factors could account for this delay in gene inactivation. First, plasmid transgene expression levels depend on the choice of the promoter as well as position effects after insertion of the transgene into the genome. Secondly, before HNF3β (or PDX1) protein expression is extinguished, Cre must first be expressed by the tissue-specific promoter and transported to the nucleus where it must recombine both loxP-flanked alleles. Finally, depending on the half-life of the mRNA and protein remaining after Cre-mediated recombination, hours or days might be required until expression is completely lost. Therefore, using the HNF3βloxP/loxP; Alb.Cre mice we were unable to determine whether HNF3β has a role in initiating the hepatic differentiation program during early liver development, as HNF3β was still present at this time. A Cre transgene expressed specifically in the prehepatic endoderm may be used in the future to investigate whether HNF3β is required for organogenesis of the liver from foregut endoderm.
Analyses of the promoters of liver-specific genes have identified binding sites for multiple transcription factors, leading to the hypothesis that cooperation between several liver-enriched transcription factors regulates hepatic gene expression (reviewed in reference 5). This model would predict that loss of one transcription factor alone would have minor consequences for the global liver gene expression profile. In contrast to this model, the idea has been put forth that a transcriptional hierarchy among the liver-enriched transcription factors exists to maintain the hepatic transcriptional program (11, 12, 19, 30). Visceral endoderm expresses many genes encoding metabolic enzymes also active in the liver. For this reason, the contribution of HNF3β to their regulation was investigated in visceral endoderm derived from embryoid bodies lacking HNF3β (12). Under these in vitro conditions, loss of HNF3β resulted in a dramatic alteration in mRNA abundance of the genes encoding hepatocyte-enriched transcription factors (HNF3α, HNF1α, and HNF4α) and their target genes that are involved in normal hepatic metabolism and differentiation. In contrast, mice lacking HNF3β in the liver exhibited only minor changes in the transcriptional program.
Several explanations could account for the mild hepatic phenotype in HNF3βloxP/loxP; Alb.Cre mice. First, compensatory binding by HNF3α and HNF3γ proteins could explain the largely unchanged hepatic gene expression profile. It has been shown, for instance, that HNF3α can bind to and transactivate the same cis-regulatory elements as HNF3β in a native chromosomal context (12). However, our evidence clearly shows the lack of upregulation of the remaining HNF3 genes at either the transcriptional (Fig. 3A) or posttranscriptional level (Fig. 3C) in livers from HNF3βloxP/loxP; Alb.Cre mice, in contrast to what had been observed in livers from HNF3γ mutant animals (15). Alternatively, hepatic gene expression may be redundantly regulated by a combination of liver-enriched and ubiquitous transcription factors, any one of which might play only a minor role in its regulation. This notion is supported by gene targeting of HNF1α, in which many of the previously known HNF1α targets identified in vitro were not altered in the mutant animals (24).
In summary, HNF3β gene targeting studies have identified several steps in mammalian development that are critically dependent on HNF3β. Regarding endoderm development, these include the formation of a functional node, from which definite endoderm is derived, the development of foregut and midgut (but not hindgut) endoderm, and the differentiation of functionally competent visceral endoderm (2, 10, 33). While the molecular targets of HNF3β in the development of the node and definitive endoderm are largely unknown, it appears that the activation of HNF4α and HNF1α and their target genes are dependent on HNF3β in visceral endoderm, at least in vitro. However, our studies have shown that gene regulation in visceral endoderm and liver are not necessarily parallel, as HNF3β is not required for the maintenance of the network of HNF transcription factors or for the global transcriptional program in hepatocytes.
ACKNOWLEDGMENTS
N.J.S. and S.-L.A. contributed equally to this work.
We are grateful to M. Birnbaum, L. Greenbaum, and M. A. Lazar for critical reading of the manuscript; T. Jessell for providing the HNF3β antibody; K. Zaret, P. Bossard, and L. Greenbaum for providing assistance with the preparation of nuclear extracts and the EMSA; R. Ahima for assistance with the glucose tolerance test; and G. Schütz for his support during the initial phase of the project.
Our studies were facilitated by the Center for Molecular Studies in Digestive and Liver Disease at the University of Pennsylvania (P30 DK50306). This work was supported by the NIDDK (RO1 DK55342 to KHK and RO1s DK42502 and DK42612 to MAM), the Association pour la Recherche sur le Cancer, the Institut National de la Santé et de la Recherche Médicale, the Centre National de la Recherche Scientifique, and the Centre Hospitalier Universitaire Régional (grant to S.L.A.). N.J.S. was supported through an NIH pre-doctoral training grant (5-T32-GM08216).
REFERENCES
- 1.Ahlgren U, Jonsson J, Jonsson L, Simu K, Edlund H. beta-cell-specific inactivation of the mouse Ipf1/Pdx1 gene results in loss of the beta-cell phenotype and maturity onset diabetes. Genes Dev. 1998;12:1763–1768. doi: 10.1101/gad.12.12.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ang S L, Rossant J. HNF-3 beta is essential for node and notochord formation in mouse development. Cell. 1994;78:561–574. doi: 10.1016/0092-8674(94)90522-3. [DOI] [PubMed] [Google Scholar]
- 3.Ang S L, Wierda A, Wong D, Stevens K A, Cascio S, Rossant J, Zaret K S. The formation and maintenance of the definitive endoderm lineage in the mouse: involvement of HNF3/forkhead proteins. Development. 1993;119:1301–1315. doi: 10.1242/dev.119.4.1301. [DOI] [PubMed] [Google Scholar]
- 4.Cascio S, Zaret K S. Hepatocyte differentiation initiates during endodermal-mesenchymal interactions prior to liver formation. Development. 1991;113:217–225. doi: 10.1242/dev.113.1.217. [DOI] [PubMed] [Google Scholar]
- 5.Cereghini S. Liver-enriched transcription factors and hepatocyte differentiation. FASEB J. 1996;10:267–282. [PubMed] [Google Scholar]
- 6.Cirillo L A, McPherson C E, Bossard P, Stevens K, Cherian S, Shim E Y, Clark K L, Burley S K, Zaret K S. Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. EMBO J. 1998;17:244–254. doi: 10.1093/emboj/17.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark K L, Halay E D, Lai E, Burley S K. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 8.Costa R H, Grayson D R, Darnell J J. Multiple hepatocyte-enriched nuclear factors function in the regulation of transthyretin and alpha 1-antitrypsin genes. Mol Cell Biol. 1989;9:1415–1425. doi: 10.1128/mcb.9.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DiPersio C M, Jackson D A, Zaret K S. The extracellular matrix coordinately modulates liver transcription factors and hepatocyte morphology. Mol Cell Biol. 1991;11:4405–4414. doi: 10.1128/mcb.11.9.4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dufort D, Schwartz L, Harpal K, Rossant J. The transcription factor HNF3beta is required in visceral endoderm for normal primitive streak morphogenesis. Development. 1998;125:3015–3025. doi: 10.1242/dev.125.16.3015. [DOI] [PubMed] [Google Scholar]
- 11.Duncan S A, Nagy A, Chan W. Murine gastrulation requires HNF-4 regulated gene expression in the visceral endoderm: tetraploid rescue of Hnf-4(−/−) embryos. Development. 1997;124:279–287. doi: 10.1242/dev.124.2.279. [DOI] [PubMed] [Google Scholar]
- 12.Duncan S A, Navas M A, Dufort D, Rossant J, Stoffel M. Regulation of a transcription factor network required for differentiation and metabolism. Science. 1998;281:692–695. doi: 10.1126/science.281.5377.692. [DOI] [PubMed] [Google Scholar]
- 13.Gu H, Marth J D, Orban P C, Mossmann H, Rajewsky K. Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- 14.Kaestner K H, Hiemisch H, Luckow B, Schütz G. The HNF-3 gene family of transcription factors in mice: gene structure, cDNA sequence, and mRNA distribution. Genomics. 1994;20:377–385. doi: 10.1006/geno.1994.1191. [DOI] [PubMed] [Google Scholar]
- 15.Kaestner K H, Hiemisch H, Schütz G. Targeted disruption of the gene encoding hepatocyte nuclear factor 3γ results in reduced transcription of hepatocyte-specific genes. Mol Cell Biol. 1998;18:4245–4251. doi: 10.1128/mcb.18.7.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaestner K H, Katz J, Liu Y, Drucker D J, Schütz G. Inactivation of the winged helix transcription factor HNF3alpha affects glucose homeostasis and islet glucagon gene expression in vivo. Genes Dev. 1999;13:495–504. doi: 10.1101/gad.13.4.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaestner K H, Knochel W, Martinez D E. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000;14:142–146. [PubMed] [Google Scholar]
- 18.Kühn R, Rajewski K, Müller W. Generation and analysis of interleukin-4 deficient mice. Science. 1991;254:707–710. doi: 10.1126/science.1948049. [DOI] [PubMed] [Google Scholar]
- 19.Kuo C J, Conley P B, Chen L, Sladek F M, Darnell J E, Jr, Crabtree G R. A transcriptional hierarchy involved in mammalian cell-type specification. Nature. 1992;355:457–461. doi: 10.1038/355457a0. [DOI] [PubMed] [Google Scholar]
- 20.Lai E, Prezioso V R, Smith E, Litvin O, Costa R H, Darnell J E., Jr HNF-3A, a hepatocyte-enriched transcription factor of novel structure is regulated transcriptionally. Genes Dev. 1990;4:1427–1436. doi: 10.1101/gad.4.8.1427. [DOI] [PubMed] [Google Scholar]
- 21.Lai E, Prezioso V R, Tao W F, Chen W S, Darnell J E., Jr Hepatocyte nuclear factor 3 alpha belongs to a gene family in mammals that is homologous to the Drosophila homeotic gene fork head. Genes Dev. 1991;5:416–427. doi: 10.1101/gad.5.3.416. [DOI] [PubMed] [Google Scholar]
- 22.McPherson C E, Shim E Y, Friedman D S, Zaret K S. An active tissue-specific enhancer and bound transcription factors existing in a precisely positioned nucleosomal array. Cell. 1993;75:387–398. doi: 10.1016/0092-8674(93)80079-t. [DOI] [PubMed] [Google Scholar]
- 23.Monaghan A P, Kaestner K H, Grau E, Schütz G. Postimplantation expression patterns indicate a role for the mouse forkhead/HNF-3 alpha, beta and gamma genes in determination of the definitive endoderm, chordamesoderm and neuroectoderm. Development. 1993;119:567–578. doi: 10.1242/dev.119.3.567. [DOI] [PubMed] [Google Scholar]
- 24.Pontoglio M, Barra J, Hadchouel M, Doyen A, Kress C, Bach J P, Babinet C, Yaniv M. Hepatocyte nuclear factor 1 inactivation results in hepatic dysfunction, phenylketonuria, and renal Fanconi syndrome. Cell. 1996;84:575–585. doi: 10.1016/s0092-8674(00)81033-8. [DOI] [PubMed] [Google Scholar]
- 25.Postic C, Shiota M, Niswender K D, Jetton T L, Chen Y, Moates J M, Shelton K D, Lindner J, Cherrington A D, Magnuson M A. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem. 1999;274:305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- 26.Rudnicki M A, Braun T, Hinuma S, Jaenisch R. Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell. 1992;71:383–390. doi: 10.1016/0092-8674(92)90508-a. [DOI] [PubMed] [Google Scholar]
- 27.Sasaki H, Hogan B L. Differential expression of multiple fork head related genes during gastrulation and axial pattern formation in the mouse embryo. Development. 1993;118:47–59. doi: 10.1242/dev.118.1.47. [DOI] [PubMed] [Google Scholar]
- 28.Shih D Q, Navas M A, Kuwajima S, Duncan S A, Stoffel M. Impaired glucose homeostasis and neonatal mortality in hepatocyte nuclear factor 3alpha-deficient mice. Proc Natl Acad Sci USA. 1999;96:10152–10157. doi: 10.1073/pnas.96.18.10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shim E Y, Woodcock C, Zaret K S. Nucleosome positioning by the winged helix transcription factor HNF3. Genes Dev. 1998;12:5–10. doi: 10.1101/gad.12.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stoffel M, Duncan S A. The maturity-onset diabetes of the young (MODY1) transcription factor HNF4alpha regulates expression of genes required for glucose transport and metabolism. Proc Natl Acad Sci USA. 1997;94:13209–13214. doi: 10.1073/pnas.94.24.13209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsien J Z, Chen D F, Gerber D, Tom C, Mercer E H, Anderson D J, Mayford M, Kandel E R, Tonegawa S. Subregion- and cell type-restricted gene knockout in mouse brain. Cell. 1996;87:1317–1326. doi: 10.1016/s0092-8674(00)81826-7. [DOI] [PubMed] [Google Scholar]
- 32.Weigel D, Jurgens G, Kuttner F, Seifert E, Jäckle H. The homeotic gene fork head encodes a nuclear protein and is expressed in the terminal regions of the Drosophila embryo. Cell. 1989;57:645–658. doi: 10.1016/0092-8674(89)90133-5. [DOI] [PubMed] [Google Scholar]
- 33.Weinstein D C, Ruiz i Altaba A, Chen W S, Hoodless P, Prezioso V R, Jessell T M, Darnell J E., Jr The winged-helix transcription factor HNF-3 beta is required for notochord development in the mouse embryo. Cell. 1994;78:575–588. doi: 10.1016/0092-8674(94)90523-1. [DOI] [PubMed] [Google Scholar]
- 34.Zaret K S. Molecular genetics of early liver development. Annu Rev Physiol. 1996;58:231–251. doi: 10.1146/annurev.ph.58.030196.001311. [DOI] [PubMed] [Google Scholar]