Abstract
Nuclear magnetic resonance (NMR) has become a universal method for biochemical and biomedical studies, including metabolomics, proteomics, and magnetic resonance imaging (MRI). By increasing the signal of selected molecules, the hyperpolarization of nuclear spin has expanded the reach of NMR and MRI even further (e.g. hyperpolarized solid‐state NMR and metabolic imaging in vivo). Parahydrogen (pH2) offers a fast and cost‐efficient way to achieve hyperpolarization, and the last decade has seen extensive advances, including the synthesis of new tracers, catalysts, and transfer methods. The portfolio of hyperpolarized molecules now includes amino acids, which are of great interest for many applications. Here, we provide an overview of the current literature and developments in the hyperpolarization of amino acids and peptides.
Keywords: amino acids, catalysis, hydrogenation, hyperpolarization, parahydrogen
Amino acids and peptides are vital elements of metabolic and proteomic regulations. Non‐invasive NMR and MRI techniques are highly suited for metabolomics diagnostics, but lack sufficient sensitivity. This Minireview summarizes the considerable advances in signal amplification through parahydrogen‐induced polarization of amino acids and peptides.
1. Introduction
NMR and MRI are among the most versatile and successful methods in medical and natural sciences. Despite their great success, however, the signal to noise ratio (SNR) of NMR and MRI remains a limiting factor for many applications, such as proteomics and metabolomics. Consequently, significant effort has been invested in increasing the signal and lowering the noise, such as by designing stronger magnets, matching probes and sample size, and using cryogenically cooled detectors.[ 1 , 2 ]
One approach to boost the signal by several orders of magnitude is the hyperpolarization of nuclear spins. Here, the population difference of the energy states of spins in a magnetic field is dramatically increased. This method leads to unprecedented signal to noise enhancements[ 3 , 4 , 5 , 6 , 7 ] and can achieve 10 % polarization or more.[ 8 , 9 ] To put these numbers into context: 10 % 13C polarization is equivalent to a signal enhancement of 10 000‐fold at 11.7 T or about 100 000‐fold at 1.5 T.
Hyperpolarization techniques have significantly advanced breakthrough applications such as real‐time metabolic imaging, [10] high‐resolution NMR, and solid‐state NMR.[ 11 , 12 ]
Among these applications, the hyperpolarization of amino acids (AAs) and peptides is particularly interesting, although not fully explored. AAs, as the building blocks of proteins, play essential roles in regulation and metabolism. As a consequence of the robustness and versatility of solid‐phase peptide synthesis (SPPS) [13] and the pharmacological importance of peptide‐based drugs, employing hyperpolarized peptides as markers in magnetic resonance imaging or biophysical studies has immense technical potential.
Despite the enormous signal enhancement provided by hyperpolarization, the SNR of hyperpolarized MRI is usually low, thereby resulting in limited spatial and chemical resolution.[ 14 , 15 , 16 ] The reason for the low SNR is relaxation, substantial dilution of the hyperpolarized agent when applied in vivo, and the fact that you can only encode the magnetization once, as it is not replenished. To achieve the desired SNR, enough and sufficiently polarized agents must reach the area of interest. Thus, one of the key challenges is to highly polarize a concentrated sample (sometimes measured in molar polarization, the product of polarization and concentration).
It has been estimated that the minimum 13C polarization for a single‐scan detection of l‐3,4‐dihydroxyphenylalanine (l‐DOPA), a precursor for several neurotransmitters in the human brain, is about 0.1 %. [17] For in vivo studies, however, the polarization and concentration of the injected agent often exceed 20 % and 100 mm, respectively.[ 18 , 19 ] l‐DOPA can readily cross the blood–brain barrier and can convert into dopamine within 15 s. [20]
Several other AAs and peptides have been polarized with dynamic nuclear polarization (DNP)16] or chemically induced dynamic nuclear polarization (CIDNP). [3] Alanine ethyl ester, alanine, aspartate, serine, and lysine were polarized to more than 20 % and found to have a pH‐dependent chemical shift suitable for pH imaging.[ 21 , 22 , 23 ] The metabolic conversion of DNP‐polarized alanine into lactate, pyruvate, and bicarbonate has been observed in the liver and the heart of healthy mice within one minute. [23] Imaging alanine transaminase activity may be a biomarker for pathologies such as cancer, inflammation, and infections. [24] The arginase‐mediated breakdown of hyperpolarized arginine to urea and ornithine has been demonstrated in myeloid‐derived suppressor cells (MDSCs) and may be used to image inflammation. [25] CIDNP‐polarized arginine has been applied for studying protein folding and to investigate the properties of AAs and protein radicals. [26] These studies demonstrate that the applications of hyperpolarized AAs are manifold, ranging from protein studies to cell cultures and living organisms.
Another hyperpolarization method is parahydrogen‐induced polarization (PHIP, Figure 1). The pioneering studies of Bowers and Weitekamp[ 27 , 28 ] as well as Eisenschmid et al. [29] have led to numerous applications of PHIP in catalysis, analytical chemistry, and biomedicine.[ 30 , 31 ] The advantages of PHIP are the fast and low‐cost production of hyperpolarized agents; the disadvantages include the sometimes limited polarization yield as well as the need for a catalyst and chemical reaction. The separation of the catalyst and hyperpolarized agent is essential but challenging; catalyst filtration [30] and the utilization of heterogeneous or immobilized catalysts have been suggested as ways to address this matter. [32]
Figure 1.
Parahydrogen‐based hyperpolarization: Schematic view of the spin states of dihydrogen (a,b), their populations at >300 K and ca. 25 K (c), as well as the basic reactions of hydrogenative (d) and non‐hydrogenative (e) hyperpolarization. The nuclear spins of dihydrogen form four different spin states: three ortho (triplet: T+, T0, T−; a), and one para state (singlet, S; b). At room temperature, all four states are approximately equally populated (c). By cooling the H2, the equilibrium is shifted toward the para state and almost pure pH2 is obtained at ca. 25 K (c).[ 33 , 59 ] To use the NMR‐invisible para order, pH2 is added either permanently to an unsaturated molecule (d,e), or brought into temporary contact through reversible exchange (f). Next, the spin order is converted spontaneously (free evolution) into observable polarization (pol) induced by RF pulses or variations of the magnetic field. The polarization may be observed on the added hydrogen atoms or transferred to other nuclei.
Several recent advances have led to PHIP increasingly becoming a method of choice for the polarization of many molecules at low cost and high throughput. The hyperpolarization of amino acids with PHIP is of particular interest for biochemistry and MR‐imaging. This is the focus of this Minireview.
2. Principles of PHIP
2.1. Background and Preparation
PHIP is based on the spin order of pH2. There are many variants of the method, but most feature two essential steps: the incorporation of spin order to a target molecule and converting that order into observable polarization (Figure 1).
pH2 is a convenient source of spin order as it is quickly produced in large quantities and has a long lifetime. Consisting of two protons (spin‐ particles), molecular dihydrogen H2 follows the Fermi–Dirac statistics. Hence, the total wave function must change sign when the protons are interchanged. The result is two spin isomers: parahydrogen (pH2) and orthohydrogen (oH2). For pH2, the nuclear spin wave function is antisymmetric, and the remaining part is symmetric; thus, the rotational quantum number is even: J=2n. oH2, on the other hand, has a symmetric nuclear spin state and antisymmetric rotational states (J=2n+1). As a result, there is one antisymmetric (singlet, para) nuclear spin state and three symmetric spin states (triplet, ortho). The energy difference between the ground rotational states J=0 and J=1 is much larger than the thermal energy at room temperature (about 120 cm−1), [33] such that the [oH2]/[pH2] ratio is 3:1. At 77 K, the ratio is already close to 1:1, while at 25 K it is 1:99.
Several instruments have been developed to aid the production of pH2.[ 34 , 35 , 36 ] As the oH2–pH2 transition is forbidden under conservation of quantum numbers, a conversion catalyst such as FeO(OH) is used. Once produced, pH2 may be conveniently stored in a bottle and used on demand.
2.2. Polarization
For hyperpolarization, pH2 is either added permanently [37] to a target molecule or brought into temporary contact. [38] This chemical reaction can be carried out in a test tube[ 37 , 38 ] or in a more dedicated automated system: a stand‐alone polarizer[ 39 , 40 , 41 , 42 , 43 , 44 ] or a reaction chamber in an MR system.[ 8 , 45 , 46 , 47 ] For high and reproducible polarizations, such systems are crucially important.
Hydrogenation at high magnetic fields is usually referred to as a PASADENA experiment.[ 28 , 29 ] In contrast, hydrogenation at low magnetic fields and subsequent transfer to high field is often called ALTADENA. [37]
pH2, however, has spin 0 and is NMR silent. The conversion of this spin order into observable magnetization may take place spontaneously, as the spins evolve, or it may be aided by radio‐frequency (RF) pulses [48] or variations of the magnetic field (field cycling).[ 40 , 49 , 50 , 51 ] Whereas detection of a 1H signal may be sufficient for in vitro analysis,[ 52 , 53 ] in vivo application usually necessitates transfer to a slow‐relaxing X‐nucleus such as 13C or 15N.[ 40 , 54 , 55 ]
2.3. Variations
For a long time, hydrogenative PHIP was severely hindered by the need for dedicated precursor molecules. This need was ameliorated by PHIP side‐arm hydrogenation (PHIP‐SAH), [56] signal amplification by reversible exchange (SABRE), [38] and recently SABRE‐RELAY [57] and PHIP‐X. [58] Most of these methods can be used to hyperpolarize AAs and peptides, as discussed in the following sections.
3. PHIP of Amino Acids
3.1. PHIP of AAs by Direct Hydrogenation
The most direct way to hyperpolarize AAs by PHIP is the hydrogenation of a suitable precursor. This approach was used to hyperpolarize γ‐aminobutyric acid (GABA) in water using trans‐aminocrotonic acid as a precursor (Figure 2). [60] Here, the water‐soluble catalyst 1,4‐bis‐[(phenyl‐3‐propane sulfonate)phosphine]butane(norbornadiene)rhodium(I) tetrafluoroborate was used for homogeneous hydrogenation [30] at pH=2.54. However, only a small signal enhancement of GABA was found (SNR of 3–10). Fast catalyst degradation at low pH values and a low hydrogenation yield were reported to be possible reasons. No hyperpolarization was found at neutral pH.
Figure 2.
PHIP of AAs by direct hydrogenation. [60] In this example, pH2 is added to trans‐aminocrotonic acid (TACA) by homogeneous hydrogenation to yield GABA. Although the hydrogenation was successful, only a low signal enhancement was found at pH 2.54, and none was observed at neutral pH.
Obtaining other precursors for AAs, however, is challenging due to keto‐enol tautomerization. Protection of the amino group of AAs or derivatives was suggested as a solution to this problem.[ 17 , 61 , 62 ] For example, 2‐acetamidoacrylate was hydrogenated with pH2 to yield N‐acetyl‐dl‐alanine methyl ester with [Rh(dppb)(COD)]BF4 (dppb=1,4‐bis(diphenylphosphino)butane, COD=1,5‐cyclooctadiene) in D2O (Figure 3). [62] It was also demonstrated that the solubility of the reagent was improved by adding 4.2–14.7 mm sodium dodecyl sulfate (SDS), [63] with 12.6 mm SDS resulting in the highest signal.
Figure 3.
PHIP of protected AA derivatives by direct hydrogenation. In this example, methyl 2‐acetamidoacrylate is hydrogenated to N‐acetyl‐dl‐alanine methyl ester. [62]
In a different study, four N‐acetyldehydroamino acids were synthesized, namely, precursors for N‐acetylphenylalanine (Figure 4), N‐acetyltryptophan (and N‐acetyl‐5‐methoxytryptophan), N‐acetyltyrosine (and (2S)‐2‐acetamido‐3‐(3,4‐dihydroxyphenyl) propanoic acid), and N‐acetylhistidine as well as their D (2H) and 13C isotopomers. [17] Hydrogenation was performed in CD3OD or D2O containing SDS and [Rh(dppb)(COD)]BF4. [17] The achieved proton polarization levels were in the range of P(1H)=0.4–1.1 %, where protonated precursors provided the highest polarization P(1H)=0.76–1.1 % for phenylalanine as well as tyrosine and its derivative. For these AA precursors, the hydrogenation rate was about ten times higher than for the others, which was tentatively attributed to faster hydrogenation of electron‐poor alkenes. [64] Deuteration of the phenylalanine precursor increased the polarization and T1 value from 0.76 % to 2.05 % and 1.6 s to 3.3 s, respectively. Using PH‐INEPT+, [33] the polarization was then transferred to the 13C‐carboxy group, thereby resulting in 1.3 % 13C polarization.
Figure 4.
PHIP of N‐acetyldehydroamino acids by direct hydrogenation.[ 17 , 61 ] 14 μm 13C‐1‐N‐acetylphenylalanine precursor was hydrogenated in 350 μm CD3OD in the presence of 0.7 μm [Rh(dppb)(COD)]BF4. The maximum polarization for the fully deuterated precursor was P(1H)=2.05 % and P(13C)=1.3 %. Other hydrogenation reactions were demonstrated for N‐acetyltyrosine, N‐acetyltryptophan, N‐acetylhistidine, [17] and the antibiotic thiostrepton. [61]
3.2. PHIP of Peptides by Side‐Chain Hydrogenation (PHIP‐Label)
In an alternative approach, non‐natural, unsaturated amino acid building blocks were designed for the direct addition of pH2. [60] PHIP‐active peptides can be prepared by direct SPPS synthesis by employing amino acid building blocks protected with fmoc (fluorenylmethoxycarbonyl) protecting groups and PHIP active groups.
To preserve the conformational freedom of the backbone, it was found advantageous to place the necessary unsaturated groups, such as, allyl or propargyl groups, into a side chain. A simple realization of this PHIP‐label approach was achieved using allylglycine and propargylglycine. Here, the use of the propargyl group is convenient because it allows two hydrogenations.
Körner et at. [65] evaluated the feasibility of this approach with propargylglycine (Pra). They studied the PHIP activity of a series of tripeptides. Specific amino acid combinations, such as Ala‐Pra‐Ala, Phe‐Pra‐Phe, or Met‐Pra‐Met yielded substantial signal enhancements, although an order of magnitude or so smaller than those achieved with neat Pra. Other combinations of sulfur‐containing amino acid residues, such as Cys‐Pra‐Cys, did not show any PHIP enhancement. They showed that this approach can also be applied to larger oligopeptides by employing two different decapeptides. Recently, Bouchard and co‐workers demonstrated the hydrogenation of propargylglycine to form allylglycine by using a ligand‐stabilized heterogeneous Pd‐NP catalyst. [66] Although these results clearly showed the high potential of this technique for biomedical applications, it has become clear that using Pra as the PHIP label is not generally suitable.
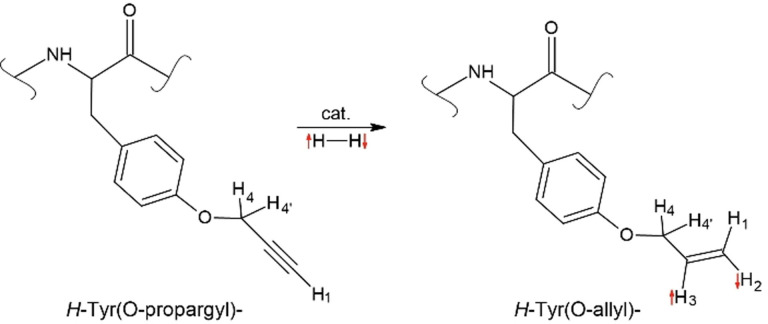
Sauer et al. [68] found that isolating the triple bond within the respective label and ensuring its accessibility to the hydrogenation catalyst were essential factors in controlling the resulting signal enhancement. In particular, the use of O‐propargyl‐l‐tyrosine yielded substantial enhancements of the 1H NMR signal in an aqueous solution (P(1H)=1.5 %). [68] They showed that this label could be introduced at any position in a peptide chain by SPPS without side‐chain protection. Such labeling was exemplified with a bioactive derivative of the sunflower trypsin inhibitor‐1 (SFTI‐1). [13] SFTI‐1, a disulfide‐bridged cyclic tetradecapeptide (Figures 5 and 6) found in nature,[ 36 , 37 , 38 , 69 , 70 , 71 ] is a very potent inhibitor of serine protease trypsin[ 71 , 72 ] and is applied for tumor diagnostics and treatment.[ 73 , 74 ]
Figure 5.
PHIP of peptides by hydrogenation of a side chain (PHIP‐label). In this example, a propargyl moiety was added to tyrosine in a peptide. The addition of pH2 results in hydrogenation of propargyltyrosine to yield a hyperpolarized allyltyrosine peptide. In the sunflower trypsin inhibitor SFTI‐1, polarizations of P(1H)=9.6 % [67] were obtained in the allyl product (right).
Figure 6.
PHIP of large peptides by direct hydrogenation of a side chain (PHIP‐label). SFTI‐Tyr(O‐propargyl) was hydrogenated in [D4]MeOH with [Rh(dppb)(COD)]BF4: A) 1D NMR (0.96 mm precursor and 4.1 mm catalyst) and B) ultrafast 2D PHIP‐TOCSY (10 s 3 bar pH2, single scan, 5 mm precursor, and 2.5 mm catalyst). Signals marked with an asterisk (*) are impurities (adapted from Sauer et al. [68] and Kiryutin et al. [67] ). For assignment of the side‐chain protons of the allyl moiety see Figure 5.
Manual ALTADENA [37] experiments with a propargyl‐functionalized tyrosine mutant of SFTI‐1 [68] revealed a 70‐fold signal enhancement of the protons in the O‐allyl‐l‐tyrosyl unit, with the inhibitory activity of the peptide also preserved.
Under PASADENA conditions, an automated PHIP setup with higher pH2 pressures [75] allowed the enhancement factors of propargyl‐SFTI‐1 to be increased to about 1200 (P(1H)=9.6 %; Figure 6). [75] The reproducibility of this setup enabled PHIP‐enhanced 2D NMR experiments, which are usually difficult because of variations in the polarization yield. To address this issue, ultrafast 2D NMR methods were suggested (Figure 6),[ 67 , 76 , 77 ] as pioneered by the Frydman group.[ 78 , 79 , 80 , 81 ] Ultrafast 2D NMR combines ideas from NMR and MRI, for example, the slice‐wise acquisition of spectra for swift biochemical analysis.
3.3. PHIP of AA Derivatives by Side‐Arm Hydrogenation (PHIP‐SAH)
One severe limitation of PHIP was the need for an unsaturated precursor that could be hydrogenated to form the desired product (e.g. an AA). Such precursors may be unavailable or unstable. A solution[ 49 , 82 ] found by Aime and co‐workers is based on the addition of an unsaturated side arm to the desired molecule (PHIP‐SAH) through an ester bond. After adding pH2 to the side arm, the polarization is transferred to the primary molecule, and the side arm is cleaved off. As an added benefit, a catalyst‐free aqueous solution of the target molecule is obtained by spontaneous separation into an organic and aqueous phase after cleavage. PHIP‐SAH led to P(13C)≈5 % for aqueous pyruvate [56] after cleavage and was used to monitor its transformation into lactate in an enzyme solution, in suspensions of cancer cell lines,[ 83 , 84 ] and in mice. [55]
PHIP‐SAH of AAs was demonstrated by Glöggler et al., [85] who esterified the carboxy groups with hydroxyethyl acrylate and then deprotected the amine group. The resulting amino acid derivatives (Figure 7) showed typical polarization on the order of P(1H)=1 % in an aqueous solution. [85]
Figure 7.
PHIP of AA derivatives through side‐arm hydrogenation (PHIP‐SAH). In this example, a protected alanine ethyl acrylate ester was hydrogenated to yield alanine ethyl propanoate in an aqueous solution. [85] Polarizations of P(1H)=0.70 % and P(13C,*)=4.4 % (for a deuterated vinyl group) were achieved at 80 °C and pH 6.5±0.5.
PHIP‐SAH was also combined with heterogeneous (HET) catalysis. [86] Here, a virtually catalyst‐free solution was obtained by the gas‐phase hydrogenation of vinyl acetate over an Rh/TiO2 powder, with subsequent condensation or dissolution of the hyperpolarized ethyl acetate and its hydrolysis. [87]
The combined HET‐PHIP‐SAH approach has been demonstrated for AAs as well (Figure 8). [88] To this end, [D3]vinyl esters of alanine and glycine (1‐13C‐labeled) were synthesized by transesterification of the corresponding N‐protected amino acids. The final unprotected amino acid esters were hydrogenated over a heterogeneous catalyst in D2O. The presence of DCl in the sample was essential to suppress the rapid decomposition of the substrates. This procedure yielded a 1H polarization of about 1 % for the resulting ethyl esters. Subsequent polarization transfer induced by RF fields provided 0.65 % and 0.8 % 13C polarization of the carbonyl carbon atom of the alanine and glycine ethyl esters, respectively. Hydrolysis of the ester bond with added NaOD resulted in a further decline in the 13C polarization to about 0.25 % and 0.29 % for alanine and glycine, respectively. Further workup of the resulting slurry, including the removal of the nanoparticulate catalyst, has not been attempted so far.
Figure 8.
PHIP of AAs by heterogeneous PHIP‐SAH. Here, unprotected amino acid esters [88] were hydrogenated over a heterogeneous catalyst (Rh nanoparticles with preadsorbed N‐acetylcysteine) by bubbling 80 % pH2 for 30 s through the slurry at 7 bar and 80 °C (0.6–0.9 mm vinylglycine and vinylalanine in D2O with 56 mm DCl; pH 2). P(1H)≈1 % was observed on the resulting ethyl esters; the subsequent RF‐induced 1H to 13C polarization transfer yielded P(13C)=0.65 % (ethylalanine) and 0.8 % (ethylglycine). The addition of a NaOD solution effected hydrolysis of the ester bond in less than 10 s and resulted in P(13C)=0.25 % (alanine) and 0.29 % (glycine).
3.4. PHIP of AAs by SABRE
In 2009, the PHIP technique termed signal amplification by reversible exchange (SABRE) was described. [38] SABRE creates hyperpolarized substrates without changing their chemical identity. SABRE releases the latent polarization of pH2 through its reversible binding to a metal complex, whereby the magnetic symmetry of its two hydrogen atoms is broken (Figure 9). Ligands within the coordination sphere of the catalyst then become hyperpolarized by spin‐order transfer through the scalar coupling network. [89] However, dissociation of a ligand or the substrate (sub, Figure 9) is required if the aim is to build‐up finite concentrations of a hyperpolarized material in solution.[ 90 , 91 ] The timescales of SABRE make it a very rapid hyperpolarization method that can create very high levels of polarization in seconds as well as continuous hyperpolarization.[ 53 , 92 , 93 ] Although its use with amino acids is still in its infancy, many other systems have been examined with great success. [94]
Figure 9.
Schematic representation of the SABRE process in which pH2 in the presence of an Ir complex hyperpolarizes the substrate.
Polarization “flows” through the scalar coupling network of the active complex when it is in an appropriate DC magnetic field [95] or on RF excitation. [96] The exact details of this spin‐order transfer have been described theoretically. Its efficiency depends on the ligand dissociation rates, the spin‐order relaxation rates, and the size of the propagating spin–spin couplings. [97] For 1H nuclei, optimal polarization transfer usually results when the sample is exposed to a static field of around 6 mT. [98] For 15N transfer, however, the larger 1H‐15N scalar couplings and differences in the involved magnetogyric ratio act to reduce the necessary field to around 200 nT. Consequently, SABRE catalysis must now take place inside a magnetic shield, [51] a refinement termed SABRE‐SHEATH (signal amplification by reversible exchange in shield enables alignment transfer to heteronuclei). This is important for studies on amino acids if 15N is to be optimally detected.[ 51 , 99 ]
Ligand dissociation from the active complex is vital for “refreshing” the latent spin order of the pH2‐derived hydride ligands during SABRE (Figure 9). This implies, for example, the involvement of short‐lived complex [Ir(H)2(η2‐H2)(IMes)(py)2]Cl when the substrate is pyridine (py; IMes=1,3‐bis(2,4,6‐trimethylphenyl)imidazol‐2‐ylidine). Density functional theory (DFT) methods have confirmed this. [91] The reversible binding of a targeting reagent to the metal center containing the pH2‐derived hydride ligands is, therefore, critical to spin‐order propagation during SABRE. Many of the iridium catalysts used for SABRE are based on N‐heterocyclic carbenes (NHCs) and exhibit the form [Ir(H)2(NHC)(sub)3]Cl. They have delivered gains to the NMR signals of 1H, 15N, 13C, 19F, 31P, and other nuclear sites in a range of molecules, including N heterocycles, nitriles, amines, and many others. [7] The substrate scope of SABRE has, therefore, been most commonly illustrated for molecules with N‐donor sites that readily ligate to the iridium center. This approach has been extended to pyridine‐tagged biocompatible molecular frameworks, including oligopeptides[ 100 , 101 ] and glycine. [102]
SABRE can involve high symmetry catalysts such as [Ir(H)2(IMes)(py)3]Cl with chemically equivalent hydride ligands. Their magnetic inequivalence is responsible for polarization flow. [91] Complexes where the hydride ligands become chemically different have also been successfully used. This situation is readily achieved when a further co‐ligand is present, since complexes of the type [Ir(H)2(IMes)(py)2(coligand)]Cl can now form when the hydride ligands lie trans to different groups.[ 103 , 104 ]
There are 21 different α‐amino acids. They can exhibit monodentate amine or carboxylate coordination in addition to forming five‐membered chelates when binding takes place through both moieties (N,O‐chelation). However, several contain additional binding sites in their side chains (e.g. the imidazole ring of histidine (His), the phenol ring of tyrosine (Tyr), and the thiol group of cysteine (Cys)). The complexation of amino acids is, therefore, complicated and will be reflected in any SABRE effects, which could occur through both types of hydride symmetry breaking processes.
The observation of direct SABRE polarization of amines [105] and carboxylates, such as acetate [106] and pyruvate, [107] via complexes such as [Ir(H)2(IMes)(NH2CH2Ph)3]Cl and [Ir(H)2(IMes)(py)2(CO2CH3)] and [Ir(H)2(IMes)(DMSO)(CO2COCH3) provides confidence that the SABRE hyperpolarization of amino acids is indeed viable through N, O‐, or N,O‐ligation modes. [108] Therefore, not surprisingly, an early study on SABRE by Appelt and co‐workers [109] suggested that all of the proteinogenic amino acids could be hyperpolarized in this way. They used [Ir(COD)(PCy3)(py)][PF6] as the SABRE catalyst precursor in [D4]MeOH. The measurements were carried out with a home‐built NMR spectrometer at proton frequencies of 166 kHz and 10 kHz. It was suggested that either [Ir(amino acid)2(H)2(PCy3)(py)][PF6] or [Ir(amino acid)(H)2(PCy3)(py)][PF6] is formed, but these complexes were not observed in solution. An enhanced 1H NMR signal was detected in single‐scan measurements of the samples with histidine, glycine, and phenylalanine. The enhanced signal was attributed to the hyperpolarized amino acid, but this work has not been developed further.
3.5. PHIP of AAs by Proton Exchange Using SABRE‐RELAY
Recently, the pool of molecules that can be hyperpolarized with pH2 was significantly extended by the introduction of SABRE‐RELAY. Here, the enhancement of a SABRE‐polarized transfer agent is passed on to another molecule through proton exchange (Figure 10). Amino acids contain exchangeable amine (NH) and carboxylate protons (COOH). When the SABRE precatalyst [IrCl(COD)(IMes)] reacts with ammonia or a suitable amine, complexes of the type [Ir(H)2(IMes)(NH3)3]Cl [105] are formed. Such complexes can hyperpolarize the exchangeable protons of these reagents (Figure 10). Once they become embedded in a second molecule, the hyperpolarization can be relayed into the core of the reagent. [57] This process has been called SABRE‐RELAY and shown to operate successfully on a range of amines, amides, carboxylic acids, alcohols, phosphates, and carbonates—again without changing their chemical identity.
Figure 10.
Schematic representation of the SABRE‐RELAY process. This catalytic cycle enables the hyperpolarization of a target substrate, such as the alcohol ROH, through the transfer of SABRE‐polarized protons from a transfer agent with labile protons, such as the illustrated amine RtNH2. Here, R is a suitable polarization receptor, e.g. CH3 or CH2‐phenyl.
The use of DNP‐hyperpolarized water as a means to enhance the sensitivity of nuclei in biomolecules such as alanine through the exchange has also been reported. [110] Water has been polarized with parahydrogen in conjunction with histidine; the observed hydride ligand signals indicate histidine binding to the SABRE catalyst. [111] The polarization of the solvent OH protons by SABRE using standard iridium‐based catalysts under slightly acidic conditions has also been reported. [112] Although this polarization is achieved by the coherent redistribution of polarization (SABRE mechanism), it is also possible that an exchange pathway involving single proton spins is feasible. [113] Consequently, there are several ways in which the successful hyperpolarization of AAs and peptides can be brought about using SABRE‐RELAY.
3.6. PHIP of AAs by Proton Exchange Using PHIP‐X
Another method that takes advantage of polarization transfer by proton exchange was introduced only recently. [58] Parahydrogen‐induced polarization relayed by proton exchange (PHIP‐X) attempts to combine the advantages of SABRE‐RELAY and PHIP, namely, the broad applicability of proton exchange and the high polarizations achieved by hydrogenative PHIP.
In PHIP‐X, pH2 is added directly to an unsaturated precursor with a labile proton (e.g. propargyl alcohol, Figure 11). The labile proton undergoes exchange and carries the polarization to the target or receiver molecule, such as water (P(1H)=0.36 %, 48 mm), ethanol (P(1H)=0.4 %, 28 mm), propargyl alcohol (P(1H)=0.41 %, 25 mm), pyruvic acid (P(1H)=0.009 %, 120 mm), or lactic acid (P(1H)=0.07 %, 29 mm). Polarization transfer from the nascent pH2 protons to the labile proton was demonstrated by variation of the magnetic field, much in the way of the classic ALTANDENA [37] experiment or SABRE‐RELAY. [57] However, other methods may also prove beneficial.
Figure 11.
Scheme of parahydrogen‐induced polarization relayed by proton exchange (PHIP‐X). First, an unsaturated transfer agent (propargyl alcohol) is hydrogenated with pH2. Then polarization is transferred to the labile OH proton and, finally, the labile proton transfers polarization to the target molecule. [58]
The 1H polarization of the transfer agent was very high, exceeding 13 %. However, each precursor can be hydrogenated and, thus, used only once. SABRE‐RELAY, instead, allows continuous re‐hyperpolarization and possibly signal accumulation or averaging.[ 53 , 92 , 93 ] PHIP‐X appears to apply to AAs in the same way as SABRE‐RELAY, since many AAs feature exchangeable protons. Difficulties arise from the fact that AAs are poorly soluble in many solvents, including those beneficial for PHIP‐X and SABRE‐RELAY. For example, water and alcohol are poor choices as solvents because they act as receiver molecules. Furthermore, the catalyst may hinder the proton exchange, and both the precursor and transfer molecule may serve as a receiver of exchanging hydrogen atoms, thereby reducing the polarization of the target molecule.
4. Conclusion and Outlook
Several recent advances have enabled the synthesis and hyperpolarization of protected AAs and derivatives. The hydrogenation of side chains (PHIP‐label) and side arms (PHIP‐SAH) has drastically enlarged the pool of available agents, now also including nonprotected AAs, peptides, and proteins. SABRE allows direct polarization of proteinogenic AAs or pyridine‐tagged AAs and oligopeptides without altering their chemical identity. SABRE also has been successfully utilized to reveal the composition of biofluids with an analyte concentration as low as 0.5 μm. [52]
The transfer of polarization by proton exchange is a new route for the polarization of AAs. For example, proton exchange of hyperpolarized water recently helped to trace RNA refolding induced by hypoxanthine. [114] We foresee that, in the same manner, pH2‐hyperpolarized water or other transfer agents such as NH3 or allyl alcohol could be utilized in analytical chemistry to probe AAs and peptides.
The hyperpolarization of AAs has never been easy with PHIP. First and foremost, suitable precursors are rare. However, this picture is about to change as new precursors are found, new protection schemes designed, and derivatives synthesized. New methods to overcome the need for unsaturated precursors altogether have been demonstrated. All these developments are reflected in the fast‐growing field of PHIP‐polarized AAs. However, more work is needed to provide clean solutions and to increase the concentration and polarizations, as already achieved by DNP. These challenges will likely be met by developing dedicated setups that allow the use of high‐pressure hydrogenation,[ 115 , 116 ] catalyst filters,[ 30 , 117 , 118 ] or heterogeneous catalysts.[ 6 , 88 ] The results achieved by DNP may prove to be very helpful to overcome these hurdles. Another issue is that only a few natural amino acids have been hyperpolarized so far—this shortcoming may be addressed by the proton‐exchange methods SABRE‐RELAY and PHIP‐X.
The hyperpolarization of AAs—fundamental building blocks of all mammalian life—is intriguing and promises many insights into biochemistry in vitro and in vivo. Whether or not hyperpolarization will develop into a widely used tool to investigate AAs is yet to be seen. Very likely, polarization levels will improve drastically as the development continues. Aside from studying metabolism, hyperpolarized AAs have already been used for angiography[ 54 , 119 ] and pH imaging. [21] It will continue to be thrilling to witness the rapid evolution of NMR and MRI with hyperpolarized amino acids.
Conflict of interest
The authors declare no conflict of interest.
Biographical Information
Andrey N. Pravdivtsev received his PhD in 2017 in Chemical Physics at Novosibirsk State University and the International Tomography Center SB RAS (in the groups of Prof. K. L. Ivanov and Prof. A. V. Yurkovskaya). In 2017–2019 he was a DAAD fellow and later postdoctoral researcher in the MRI and hyperpolarization group of Prof. J.‐B. Hövener (Kiel University). Currently, he is a BMBF Juniorverbünde fellow at Kiel University, where his research focuses on the development of PHIP and SABRE applications for in vivo MRI.

Biographical Information
Gerd Buntkowsky received his PhD in Physics at the FU Berlin in 1991. He then changed to chemistry and completed his habilitation in Physical Chemistry in 2000 and accepted a position as a full professor at the FSU Jena in 2004. Since 2009, he has been Professor of Physical Chemistry (Chair) at the Technical University Darmstadt. His research focuses on investigating the structure and dynamics of condensed matter using solid‐state NMR techniques, and the development and application of hyperpolarization techniques in solution and solid‐state NMR. He is editor in chief of Applied Magnetic Resonance.

Biographical Information
Simon B. Duckett is the Director of the Centre for Hyperpolarization in Magnetic Resonance. His group studies and develops hyperpolarization techniques with a view to translation into the clinic. In 1990, he was awarded a D.Phil. from the University of York and then undertook postdoctoral work with Prof. W. D. Jones and Prof. R. Eisenberg at the University of Rochester. He has authored over 130 publications on the parahydrogen effect.

Biographical Information
Igor V. Koptyug received his PhD in 1991 before being a postdoctoral researcher in the photochemistry group of Prof. N. J. Turro (Columbia University, New York; 1992–1995). He earned his Dr Sci. (Habilitation) in catalysis in 2003 and a title of Professor in 2006. Currently, he is the research group leader at the International Tomography Center, Siberian Branch of the Russian Academy of Sciences, Novosibirsk. His research interests include signal enhancement in NMR and applications of NMR and MRI in catalysis.

Biographical Information
Jan‐Bernd Hövener studied Physics in Münster, Heidelberg, and Nice (France). After working in New York and in Pasadena (USA), he graduated in Physics at the University of Heidelberg in the group of Prof. P. Bachert. In 2009, he joined Prof. J. K. Henning in Freiburg, and was accepted to the Academy of Excellence in 2010 and the Emmy Noether program of the DFG in 2014. In 2016, he received the venia legendi and habilitation. In 2017, he became Professor for Translational MRI at Kiel University, where he heads the Section for Biomedical Imaging and the Molecular Imaging Competence Center North (MOIN CC).

Acknowledgements
A.P. and J.‐B.H. acknowledge funding from BMBF (01ZX1915C), DFG (HO‐4602/2‐2, HO‐4602/3, GRK2154‐2019, EXC PMI2167, FOR5042, TRR287), Kiel University, and the Faculty of Medicine for supporting the MOIN CC. I.V.K. acknowledges financial support from the Russian Foundation for Basic Research (#19‐53‐12013). G.B. acknowledges funding by the DFG (Bu911/22‐2 and Bu911/29‐1). We also acknowledge Julia Kugler for help with the frontispiece. Open access funding enabled and organized by Projekt DEAL.
A. N. Pravdivtsev, G. Buntkowsky, S. B. Duckett, I. V. Koptyug, J.-B. Hövener, Angew. Chem. Int. Ed. 2021, 60, 23496.
Contributor Information
Dr. Andrey N. Pravdivtsev, Email: andrey.pravdivtsev@rad.uni-kiel.de.
Prof. Dr. Jan‐Bernd Hövener, Email: jan.hoevener@rad.uni-kiel.de.
References
- 1. Sack M., Wetterling F., Sartorius A., Ende G., Weber-Fahr W., NMR Biomed. 2014, 27, 709–715. [DOI] [PubMed] [Google Scholar]
- 2. Hirsch M. L., Kalechofsky N., Belzer A., Rosay M., Kempf J. G., J. Am. Chem. Soc. 2015, 137, 8428–8434. [DOI] [PubMed] [Google Scholar]
- 3. Kaptein R., Chem. Phys. Lett. 1968, 2, 261–267. [Google Scholar]
- 4. Ardenkjaer-Larsen J. H., J. Magn. Reson. 2016, 264, 3–12. [DOI] [PubMed] [Google Scholar]
- 5. Capozzi A., Cheng T., Boero G., Roussel C., Comment A., Nat. Commun. 2017, 8, 15757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kovtunov K. V., Pokochueva E., Salnikov O., Cousin S., Kurzbach D., Vuichoud B., Jannin S., Chekmenev E., Goodson B., Barskiy D., Koptyug I., Chem. Asian J. 2018, 13, 1857–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hövener J.-B., Pravdivtsev A. N., Kidd B., Bowers C. R., Glöggler S., Kovtunov K. V., Plaumann M., Katz-Brull R., Buckenmaier K., Jerschow A., Reineri F., Theis T., Shchepin R. V., Wagner S., Bhattacharya P., Zacharias N. M., Chekmenev E. Y., Angew. Chem. Int. Ed. 2018, 57, 11140–11162; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2018, 130, 11310–11333. [Google Scholar]
- 8. Korchak S., Mamone S., Glöggler S., ChemistryOpen 2018, 7, 672–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Radaelli A., Yoshihara H. A. I., Nonaka H., Sando S., Ardenkjaer-Larsen J. H., Gruetter R., Capozzi A., J. Phys. Chem. Lett. 2020, 11, 6873–6879. [DOI] [PubMed] [Google Scholar]
- 10. Kurhanewicz J., Vigneron D. B., Ardenkjaer-Larsen J. H., Bankson J. A., Brindle K., Cunningham C. H., Gallagher F. A., Keshari K. R., Kjaer A., Laustsen C., Mankoff D. A., Merritt M. E., Nelson S. J., Pauly J. M., Lee P., Ronen S., Tyler D. J., Rajan S. S., Spielman D. M., Wald L., Zhang X., Malloy C. R., Rizi R., Neoplasia 2019, 21, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bielytskyi P., Gräsing D., Mote K. R., Sai Sankar Gupta K. B., Vega S., Madhu P. K., Alia A., Matysik J., J. Magn. Reson. 2018, 293, 82–91. [DOI] [PubMed] [Google Scholar]
- 12. Daube D., Vogel M., Suess B., Corzilius B., Solid State Nucl. Magn. Reson. 2019, 101, 21–30. [DOI] [PubMed] [Google Scholar]
- 13. Merrifield R. B., J. Am. Chem. Soc. 1963, 85, 2149–2154. [Google Scholar]
- 14. Hurd R. E., Yen Y.-F., Chen A., Ardenkjaer-Larsen J. H., J. Magn. Reson. Imaging 2012, 36, 1314–1328. [DOI] [PubMed] [Google Scholar]
- 15. Müller C. A., Hundshammer C., Braeuer M., Skinner J. G., Berner S., Leupold J., Düwel S., Nekolla S. G., Månsson S., Hansen A. E., von Elverfeldt D., Ardenkjaer-Larsen J. H., Schilling F., Schwaiger M., Hennig J., Hövener J.-B., NMR Biomed. 2020, 33, e4291. [DOI] [PubMed] [Google Scholar]
- 16. Durst M., Koellisch U., Frank A., Rancan G., Gringeri C. V., Karas V., Wiesinger F., Menzel M. I., Schwaiger M., Haase A., Schulte R. F., NMR Biomed. 2015, 28, 715–725. [DOI] [PubMed] [Google Scholar]
- 17. Che Soon P., Xu X., Zhang B., Gruppi F., Canary J. W., Jerschow A., Chem. Commun. 2013, 49, 5304–5306. [DOI] [PubMed] [Google Scholar]
- 18. Cunningham C. H., Lau J. Y. C., Chen A. P., Geraghty B. J., Perks W. J., Roifman I., Wright G. A., Connelly K. A., Circ. Res. 2016, 119, 1177–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gallagher F. A., Woitek R., McLean M. A., Gill A. B., Garcia R. M., Provenzano E., Riemer F., Kaggie J., Chhabra A., Ursprung S., Grist J. T., Daniels C. J., Zaccagna F., Laurent M.-C., Locke M., Hilborne S., Frary A., Torheim T., Boursnell C., Schiller A., Patterson I., Slough R., Carmo B., Kane J., Biggs H., Harrison E., Deen S. S., Patterson A., Lanz T., Kingsbury Z., Ross M., Basu B., Baird R., Lomas D. J., Sala E., Wason J., Rueda O. M., Chin S.-F., Wilkinson I. B., Graves M. J., Abraham J. E., Gilbert F. J., Caldas C., Brindle K. M., Proc. Natl. Acad. Sci. USA 2020, 117, 2092–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wade L., Katzman R., Am. J. Physiol. 1975, 228, 352–359. [DOI] [PubMed] [Google Scholar]
- 21. Hundshammer C., Düwel S., Ruseckas D., Topping G., Dzien P., Müller C., Feuerecker B., Hövener J. B., Haase A., Schwaiger M., Glaser S. J., Schilling F., Sensors 2018, 18, 600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen J., Hackett E. P., Singh J., Kovács Z., Park J. M., Anal. Chem. 2020, 92, 11681–11686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jensen P. R., Karlsson M., Meier S., Duus J. Ø., Lerche M. H., Chem. Eur. J. 2009, 15, 10010–10012. [DOI] [PubMed] [Google Scholar]
- 24. Sherman K. E., Arch. Intern. Med. 1991, 151, 260–265. [PubMed] [Google Scholar]
- 25. Najac C., Chaumeil M. M., Kohanbash G., Guglielmetti C., Gordon J. W., Okada H., Ronen S. M., Sci. Rep. 2016, 6, 31397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mok K. H., Hore P. J., Methods 2004, 34, 75–87. [DOI] [PubMed] [Google Scholar]
- 27. Bowers C. R., Weitekamp D. P., Phys. Rev. Lett. 1986, 57, 2645–2648. [DOI] [PubMed] [Google Scholar]
- 28. Bowers C. R., Weitekamp D. P., J. Am. Chem. Soc. 1987, 109, 5541–5542. [Google Scholar]
- 29. Eisenschmid T. C., Kirss R. U., Deutsch P. P., Hommeltoft S. I., Eisenberg R., Bargon J., Lawler R. G., Balch A. L., J. Am. Chem. Soc. 1987, 109, 8089–8091. [Google Scholar]
- 30. Bhattacharya P., Harris K., Lin A. P., Mansson M., Norton V. A., Perman W. H., Weitekamp D. P., Ross B. D., MAGMA 2005, 18, 245–256. [DOI] [PubMed] [Google Scholar]
- 31. Bhattacharya P., Chekmenev E. Y., Perman W. H., Harris K. C., Lin A. P., Norton V. A., Tan C. T., Ross B. D., Weitekamp D. P., J. Magn. Reson. 2007, 186, 150–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kovtunov K. V., Zhivonitko V. V., Corma A., Koptyug I. V., J. Phys. Chem. Lett. 2010, 1, 1705–1708. [Google Scholar]
- 33. Green R. A., Adams R. W., Duckett S. B., Mewis R. E., Williamson D. C., Green G. G. R., Prog. Nucl. Magn. Reson. Spectrosc. 2012, 67, 1–48. [DOI] [PubMed] [Google Scholar]
- 34. Hövener J.-B., Bär S., Leupold J., Jenne K., Leibfritz D., Hennig J., Duckett S. B., von Elverfeldt D., NMR Biomed. 2013, 26, 124–131. [DOI] [PubMed] [Google Scholar]
- 35. Birchall J. R., Coffey A. M., Goodson B. M., Chekmenev E. Y., Anal. Chem. 2020, 92, 15280–15284. [DOI] [PubMed] [Google Scholar]
- 36. Du Y., Zhou R., Ferrer M.-J., Chen M., Graham J., Malphurs B., Labbe G., Huang W., Bowers C. R., J. Magn. Reson. 2020, 321, 106869. [DOI] [PubMed] [Google Scholar]
- 37. Pravica M. G., Weitekamp D. P., Chem. Phys. Lett. 1988, 145, 255–258. [Google Scholar]
- 38. Adams R. W., Aguilar J. A., Atkinson K. D., Cowley M. J., Elliott P. I. P., Duckett S. B., Green G. G. R., Khazal I. G., López-Serrano J., Williamson D. C., Science 2009, 323, 1708–1711. [DOI] [PubMed] [Google Scholar]
- 39. Kadlecek S., Vahdat V., Nakayama T., Ng D., Emami K., Rizi R., NMR Biomed. 2011, 24, 933–942. [DOI] [PubMed] [Google Scholar]
- 40. Goldman M., Jóhannesson H., Axelsson O., Karlsson M., Magn. Reson. Imaging 2005, 23, 153–157. [DOI] [PubMed] [Google Scholar]
- 41. Hövener J.-B., Chekmenev E. Y., Harris K. C., Perman W. H., Robertson L. W., Ross B. D., Bhattacharya P., Magn. Reson. Mater. Phys. 2009, 22, 111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hövener J.-B., Schwaderlapp N., Borowiak R., Lickert T., Duckett S. B., Mewis R. E., Adams R. W., Burns M. J., Highton L. A. R., Green G. G. R., Olaru A., Hennig J., von Elverfeldt D., Anal. Chem. 2014, 86, 1767–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Richardson P. M., Parrott A. J., Semenova O., Nordon A., Duckett S. B., Halse M. E., Analyst 2018, 143, 3442–3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kiryutin A. S., Pravdivtsev A. N., Ivanov K. L., Grishin Y. A., Vieth H.-M., Yurkovskaya A. V., J. Magn. Reson. 2016, 263, 79–91. [DOI] [PubMed] [Google Scholar]
- 45. Waddell K. W., Coffey A. M., Chekmenev E. Y., J. Am. Chem. Soc. 2011, 133, 97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schmidt A. B., Berner S., Schimpf W., Müller C., Lickert T., Schwaderlapp N., Knecht S., Skinner J. G., Dost A., Rovedo P., Hennig J., von Elverfeldt D., Hövener J.-B., Nat. Commun. 2017, 8, 14535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kovtunov K. V., Zhivonitko V. V., Skovpin I. V., Barskiy D. A., Salnikov O. G., Koptyug I. V., J. Phys. Chem. C 2013, 117, 22887–22893. [Google Scholar]
- 48. Bär S., Lange T., Leibfritz D., Hennig J., von Elverfeldt D., Hövener J.-B., J. Magn. Reson. 2012, 225, 25–35. [DOI] [PubMed] [Google Scholar]
- 49. Reineri F., Boi T., Aime S., Nat. Commun. 2015, 6, 5858. [DOI] [PubMed] [Google Scholar]
- 50. Pravdivtsev A. N., Ivanov K. L., Yurkovskaya A. V., Petrov P. A., Limbach H.-H., Kaptein R., Vieth H.-M., J. Magn. Reson. 2015, 261, 73–82. [DOI] [PubMed] [Google Scholar]
- 51. Theis T., Truong M. L., Coffey A. M., Shchepin R. V., Waddell K. W., Shi F., Goodson B. M., Warren W. S., Chekmenev E. Y., J. Am. Chem. Soc. 2015, 137, 1404–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Reile I., Eshuis N., Hermkens N. K. J., van Weerdenburg B. J. A., Feiters M. C., Rutjes F. P. J. T., Tessari M., Analyst 2016, 141, 4001–4005. [DOI] [PubMed] [Google Scholar]
- 53. Eshuis N., Aspers R. L. E. G., van Weerdenburg B. J. A., Feiters M. C., Rutjes F. P. J. T., Wijmenga S. S., Tessari M., Angew. Chem. Int. Ed. 2015, 54, 14527–14530; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2015, 127, 14735–14738. [Google Scholar]
- 54. Goldman M., Jóhannesson H., Axelsson O., Karlsson M., C. R. Chim. 2006, 9, 357–363. [Google Scholar]
- 55. Cavallari E., Carrera C., Sorge M., Bonne G., Muchir A., Aime S., Reineri F., Sci. Rep. 2018, 8, 8366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cavallari E., Carrera C., Aime S., Reineri F., J. Magn. Reson. 2018, 289, 12–17. [DOI] [PubMed] [Google Scholar]
- 57. Iali W., Rayner P. J., Duckett S. B., Sci. Adv. 2018, 4, eaao6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Them K., Ellermann F., Pravdivtsev A. N., Salnikov O. G., Skovpin I. V., Herges R., Hövener J.-B., arXiv 2020, https://arxiv.org/abs/2012.03626. [Google Scholar]
- 59. Ellermann F., Pravdivtsev A., Hövener J.-B., Magn. Reson. 2021, 2, 49–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Trantzschel T., Plaumann M., Bernarding J., Lego D., Ratajczyk T., Dillenberger S., Buntkowsky G., Bargon J., Bommerich U., Appl. Magn. Reson. 2013, 44, 267–278. [Google Scholar]
- 61. Gruppi F., Xu X., Zhang B., Tang J. A., Jerschow A., Canary J. W., Angew. Chem. 2012, 124, 11957–11960. [DOI] [PubMed] [Google Scholar]
- 62. Tang J. A., Gruppi F., Fleysher R., Sodickson D. K., Canary J. W., Jerschow A., Chem. Commun. 2011, 47, 958–960. [DOI] [PubMed] [Google Scholar]
- 63. Kumar A., Oehme G., Roque J. P., Schwarze M., Selke R., Angew. Chem. Int. Ed. Engl. 1994, 33, 2197–2199; [Google Scholar]; Angew. Chem. 1994, 106, 2272–2275. [Google Scholar]
- 64. Candlin J. P., Oldham A. R., Discuss. Faraday Soc. 1968, 46, 60–71. [Google Scholar]
- 65. Körner M., Sauer G., Heil A., Nasu D., Empting M., Tietze D., Voigt S., Weidler H., Gutmann T., Avrutina O., Kolmar H., Ratajczyk T., Buntkowsky G., Chem. Commun. 2013, 49, 7839–7841. [DOI] [PubMed] [Google Scholar]
- 66. McCormick J., Grunfeld A. M., Ertas Y. N., Biswas A. N., Marsh K. L., Wagner S., Glöggler S., Bouchard L.-S., Anal. Chem. 2017, 89, 7190–7194. [DOI] [PubMed] [Google Scholar]
- 67. Kiryutin A. S., Sauer G., Tietze D., Brodrecht M., Knecht S., Yurkovskaya A. V., Ivanov K. L., Avrutina O., Kolmar H., Buntkowsky G., Chem. Eur. J. 2019, 25, 4025–4030. [DOI] [PubMed] [Google Scholar]
- 68. Sauer G., Nasu D., Tietze D., Gutmann T., Englert S., Avrutina O., Kolmar H., Buntkowsky G., Angew. Chem. Int. Ed. 2014, 53, 12941–12945; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2014, 126, 13155–13159. [Google Scholar]
- 69. Korsinczky M. L. J., Schirra H. J., Rosengren K. J., West J., Condie B. A., Otvos L., Anderson M. A., Craik D. J., J. Mol. Biol. 2001, 311, 579–591. [DOI] [PubMed] [Google Scholar]
- 70. Korsinczky M. L. J., Schirra H. J., Craik D. J., Curr. Protein Pept. Sci. 2004, 5, 351–364. [DOI] [PubMed] [Google Scholar]
- 71. de Veer S. J., White A. M., Craik D., Angew. Chem. Int. Ed. 2021, 60, 8050–8071; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2021, 133, 8128–8151. [Google Scholar]
- 72. Luckett S., Garcia R. S., Barker J. J., Konarev Al. V., Shewry P. R., Clarke A. R., Brady R. L., J. Mol. Biol. 1999, 290, 525–533. [DOI] [PubMed] [Google Scholar]
- 73. Fittler H., Avrutina O., Glotzbach B., Empting M., Kolmar H., Org. Biomol. Chem. 2013, 11, 1848–1857. [DOI] [PubMed] [Google Scholar]
- 74. Fittler H., Avrutina O., Empting M., Kolmar H., J. Pept. Sci. 2014, 20, 415–420. [DOI] [PubMed] [Google Scholar]
- 75. Kiryutin A. S., Sauer G., Hadjiali S., Yurkovskaya A. V., Breitzke H., Buntkowsky G., J. Magn. Reson. 2017, 285, 26–36. [DOI] [PubMed] [Google Scholar]
- 76. Ratajczyk T., Gutmann T., Dillenberger S., Abdulhussaein S., Frydel J., Breitzke H., Bommerich U., Trantzschel T., Bernarding J., Magusin P. C. M. M., Buntkowsky G., Solid State Nucl. Magn. Reson. 2012, 43–44, 14–21. [DOI] [PubMed] [Google Scholar]
- 77. Pravdivtsev A. N., Ivanov K. L., Yurkovskaya A. V., Vieth H.-M., Sagdeev R. Z., Dokl. Phys. Chem. 2015, 465, 267–269. [Google Scholar]
- 78. Frydman L., Scherf T., Lupulescu A., Proc. Natl. Acad. Sci. USA 2002, 99, 15858–15862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Tal A., Frydman L., Prog. Nucl. Magn. Reson. Spectrosc. 2010, 57, 241–292. [DOI] [PubMed] [Google Scholar]
- 80. Giraudeau P., Frydman L., Annu. Rev. Anal. Chem. 2014, 7, 129–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.B. Gouilleux, L. Rouger, P. Giraudeau, in EMagRes, American Cancer Society, 2016, pp. 913–922.
- 82. Salnikov O. G., Chukanov N. V., Shchepin R. V., Manzanera Esteve I. V., Kovtunov K. V., Koptyug I. V., Chekmenev E. Y., J. Phys. Chem. C 2019, 123, 12827–12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Cavallari E., Carrera C., Aime S., Reineri F., ChemPhysChem 2019, 20, 318–325. [DOI] [PubMed] [Google Scholar]
- 84. Cavallari E., Carrera C., Di Matteo G., Bondar O., Aime S., Reineri F., Front. Oncol. 2020, 10, 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Glöggler S., Wagner S., Bouchard L.-S., Chem. Sci. 2015, 6, 4261–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kovtunov K. V., Salnikov O. G., Skovpin I. V., Chukanov N. V., Burueva D. B., Koptyug I. V., Pure Appl. Chem. 2020, 92, 1029–1046. [Google Scholar]
- 87. Salnikov O. G., Kovtunov K. V., Koptyug I. V., Sci. Rep. 2015, 5, 13930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kaltschnee L., Jagtap A. P., McCormick J., Wagner S., Bouchard L.-S., Utz M., Griesinger C., Glöggler S., Chem. Eur. J. 2019, 25, 11031–11035. [DOI] [PubMed] [Google Scholar]
- 89. Adams R. W., Duckett S. B., Green R. A., Williamson D. C., Green G. G. R., J. Chem. Phys. 2009, 131, 194505. [DOI] [PubMed] [Google Scholar]
- 90. Atkinson K. D., Cowley M. J., Elliott P. I. P., Duckett S. B., Green G. G. R., López-Serrano J., Whitwood A. C., J. Am. Chem. Soc. 2009, 131, 13362–13368. [DOI] [PubMed] [Google Scholar]
- 91. Cowley M. J., Adams R. W., Atkinson K. D., Cockett M. C. R., Duckett S. B., Green G. G. R., Lohman J. A. B., Kerssebaum R., Kilgour D., Mewis R. E., J. Am. Chem. Soc. 2011, 133, 6134–6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Pravdivtsev A. N., Yurkovskaya A. V., Vieth H.-M., Ivanov K. L., J. Phys. Chem. B 2015, 119, 13619–13629. [DOI] [PubMed] [Google Scholar]
- 93. Hövener J.-B., Schwaderlapp N., Lickert T., Duckett S. B., Mewis R. E., Highton L. A. R., Kenny S. M., Green G. G. R., Leibfritz D., Korvink J. G., Hennig J., von Elverfeldt D., Nat. Commun. 2013, 4, 2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Rayner P. J., Norcott P., Appleby K. M., Iali W., John R. O., Hart S. J., Whitwood A. C., Duckett S. B., Nat. Commun. 2018, 9, 4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Mewis R. E., Atkinson K. D., Cowley M. J., Duckett S. B., Green G. G. R., Green R. A., Highton L. A. R., Kilgour D., Lloyd L. S., Lohman J. A. B., Williamson D. C., Magn. Reson. Chem. 2014, 52, 358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Pravdivtsev A. N., Yurkovskaya A. V., Zimmermann H., Vieth H.-M., Ivanov K. L., RSC Adv. 2015, 5, 63615–63623. [Google Scholar]
- 97. Barskiy D. A., Pravdivtsev A. N., Ivanov K. L., Kovtunov K. V., Koptyug I. V., Phys. Chem. Chem. Phys. 2016, 18, 89–93. [DOI] [PubMed] [Google Scholar]
- 98. Pravdivtsev A. N., Yurkovskaya A. V., Vieth H.-M., Ivanov K. L., Kaptein R., ChemPhysChem 2013, 14, 3327–3331. [DOI] [PubMed] [Google Scholar]
- 99. Truong M. L., Theis T., Coffey A. M., Shchepin R. V., Waddell K. W., Shi F., Goodson B. M., Warren W. S., Chekmenev E. Y., J. Phys. Chem. C 2015, 119, 8786–8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Ratajczyk T., Gutmann T., Bernatowicz P., Buntkowsky G., Frydel J., Fedorczyk B., Chem. Eur. J. 2015, 21, 12616–12619. [DOI] [PubMed] [Google Scholar]
- 101. Ratajczyk T., Buntkowsky G., Gutmann T., Fedorczyk B., Mames A., Pietrzak M., Puzio Z., Szkudlarek P. G., ChemBioChem 2021, 22, 855–860. [DOI] [PubMed] [Google Scholar]
- 102. Chae H., Min S., Jeong H. J., Namgoong S. K., Oh S., Kim K., Jeong K., Anal. Chem. 2020, 92, 10902–10907. [DOI] [PubMed] [Google Scholar]
- 103. Fekete M., Bayfield O., Duckett S. B., Hart S., Mewis R. E., Pridmore N., Rayner P. J., Whitwood A., Inorg. Chem. 2013, 52, 13453–13461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Eshuis N., Hermkens N., van Weerdenburg B. J. A., Feiters M. C., Rutjes F. P. J. T., Wijmenga S. S., Tessari M., J. Am. Chem. Soc. 2014, 136, 2695–2698. [DOI] [PubMed] [Google Scholar]
- 105. Iali W., Rayner P. J., Alshehri A., Holmes A. J., Ruddlesden A. J., Duckett S. B., Chem. Sci. 2018, 9, 3677–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Gemeinhardt M. E., Limbach M. N., Gebhardt T. R., Eriksson C. W., Eriksson S. L., Lindale J. R., Goodson E. A., Warren W. S., Chekmenev E. Y., Goodson B. M., Angew. Chem. Int. Ed. 2020, 59, 418–423; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2020, 132, 426–431. [Google Scholar]
- 107. Iali W., Roy S. S., Tickner B. J., Ahwal F., Kennerley A. J., Duckett S. B., Angew. Chem. Int. Ed. 2019, 58, 10271–10275; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2019, 131, 10377–10381. [Google Scholar]
- 108. Vaneeckhaute E., Tyburn J.-M., Kilgour D., Kempf J. G., Taulelle F., Martens J. A., Breynaert E., J. Phys. Chem. C 2020, 124, 14541–14549. [Google Scholar]
- 109. Glöggler S., Müller R., Colell J., Emondts M., Dabrowski M., Blümich B., Appelt S., Phys. Chem. Chem. Phys. 2011, 13, 13759–13764. [DOI] [PubMed] [Google Scholar]
- 110. Harris T., Szekely O., Frydman L., J. Phys. Chem. B 2014, 118, 3281–3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Lehmkuhl S., Emondts M., Schubert L., Spannring P., Klankermayer J., Blümich B., Schleker P. P. M., ChemPhysChem 2017, 18, 2426–2429. [DOI] [PubMed] [Google Scholar]
- 112. Moreno K. X., Nasr K., Milne M., Sherry A. D., Goux W. J., J. Magn. Reson. 2015, 257, 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Emondts M., Schikowski D., Klankermayer J., Schleker P. P. M., ChemPhysChem 2018, 19, 2614–2620. [DOI] [PubMed] [Google Scholar]
- 114. Novakovic M., Olsen G. L., Pintér G., Hymon D., Fürtig B., Schwalbe H., Frydman L., Proc. Natl. Acad. Sci. USA 2020, 117, 2449–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Korchak S., Emondts M., Mamone S., Bluemich B., Glöggler S., Phys. Chem. Chem. Phys. 2019, 21, 22849–22856. [DOI] [PubMed] [Google Scholar]
- 116. Berner S., Schmidt A. B., Zimmermann M., Pravdivtsev A. N., Glöggler S., Hennig J., von Elverfeldt D., Hövener J.-B., ChemistryOpen 2019, 8, 728–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Barskiy D. A., Ke L. A., Li X., Stevenson V., Widarman N., Zhang H., Truxal A., Pines A., J. Phys. Chem. Lett. 2018, 9, 2721–2724. [DOI] [PubMed] [Google Scholar]
- 118. Kidd B. E., Gesiorski J. L., Gemeinhardt M. E., Shchepin R. V., Kovtunov K. V., Koptyug I. V., Chekmenev E. Y., Goodson B. M., J. Phys. Chem. C 2018, 122, 16848–16852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Lipsø K. W., Hansen E. S. S., Tougaard R. S., Laustsen C., Ardenkjær-Larsen J. H., Magn. Reson. Med. 2017, 78, 1131–1135. [DOI] [PubMed] [Google Scholar]