Summary
The secretomes of filamentous fungi contain a diversity of small secreted cysteine‐rich proteins (SSCPs) that have a variety of properties ranging from toxicity to surface activity. Some SSCPs are recognized by other organisms as indicators of fungal presence, but their function in fungi is not fully understood. We detected a new family of fungal surface‐active SSCPs (saSSCPs), here named hyphosphere proteins (HFSs). An evolutionary analysis of the HFSs in Pezizomycotina revealed a unique pattern of eight single cysteine residues (C‐CXXXC‐C‐C‐C‐C‐C) and a long evolutionary history of multiple gene duplications and ancient interfungal lateral gene transfers, suggesting their functional significance for fungi with different lifestyles. Interestingly, recombinantly produced saSSCPs from three families (HFSs, hydrophobins and cerato‐platanins) showed convergent surface‐modulating activity on glass and on poly(ethylene‐terephthalate), transforming their surfaces to a moderately hydrophilic state, which significantly favoured subsequent hyphal attachment. The addition of purified saSSCPs to the tomato rhizosphere had mixed effects on hyphal attachment to roots, while all tested saSSCPs had an adverse effect on plant growth in vitro. We propose that the exceptionally high diversity of saSSCPs in Trichoderma and other fungi evolved to efficiently condition various surfaces in the hyphosphere to a fungal‐beneficial state.
Introduction
Recent studies have shown that besides enzymes and small molecules, the secretomes of filamentous and dimorphic fungi can frequently contain a diversity of small secreted cysteine‐rich proteins (SSCPs) unique for fungi (Kubicek et al., 2011, 2019; Wang et al., 2019; Feldman et al., 2020; Gao et al., 2020; Huber et al., 2020). This diverse group comprises a few distinct protein families, such as hydrophobins (HFBs) and cerato‐platanins (CPs), although most SSCPs remain unclassified. Individual fungal genomes can contain from a few dozens of such proteins in yeasts (Wang et al., 2019; Sun et al., 2020) up to several hundred in mushrooms (Zajc et al., 2013; Krizsán et al., 2019). The SSCPs are characterized by their small size (<300 amino acids, a.a.) (Pellegrin et al., 2015; Kubicek et al., 2019), the presence of a relatively short signal peptide (~20 a.a.) (Chen et al., 2013; Kim et al., 2016; Feldman et al., 2020) and cysteine enrichment. The latter feature is crucial for their properties and structure but is frequently ambiguous. For example, Kubicek et al. (2019) proposed a minimum threshold of 5% cysteine residues for SSCPs, referencing an average 1.5% content in all fungal proteins. Other authors rely on the presence of an evolutionarily conserved cysteine pattern in the primary structure, which can consist of only four residues in the CPs (Pazzagli et al., 1999; Chen et al., 2013; Gaderer et al., 2014) and up to eight in the HFBs (Wösten, 2001; Aimanianda et al., 2009; Bayry et al., 2012).
Probably the most interesting feature of SSCPs is their abundant secretion in liquid medium (up to 20%–30% of the total secretome) (Pellegrin et al., 2015; Feldman et al., 2020; Gao et al., 2020) and the unusually high polymorphism of their primary structures (Gaderer et al., 2014; Krizsán et al., 2019). Some SSCPs have a long evolutionary history of gene duplications (GDs) and lateral gene transfers (LGTs), making them frequent members of the orphomes (the genome fraction comprising orthologue‐less genes) of individual fungal taxa (Przylucka et al., 2017a; Feldman et al., 2019; Gao et al., 2020). Paradoxically, despite the abundance of SSCPs in fungal secretomes, functions have been proposed for only a few, while the role of the majority of them remains unknown (Aimanianda and Latge, 2010; Feldman et al., 2017; Wang et al., 2019; Gao et al., 2020).
Obviously, the diversity of SSCP structures corresponds to their functional versatility. Indeed, SSCPs are involved in all fungal life cycle stages and their environmental interactions (Grunbacher et al., 2014; Pellegrin et al., 2015; Feldman et al., 2017; Wang et al., 2019; Gao et al., 2020). Many are essential for specific stages of fungal reproduction and dispersal (Lugones et al., 1999; Lugones et al., 2004; Grunbacher et al., 2014; Cai et al., 2020) and for biotic interactions with plants (Frías et al., 2011; Guzmán‐Guzmán et al., 2017), animals (Andersson et al., 2013; Wang et al., 2019) and bacteria (Kombrink et al., 2019). SSCPs modulate fungal attachment and substrate colonization (Viterbo and Chet, 2006; Rosado et al., 2007; Quarantin et al., 2019), the stress response (Khalesi et al., 2016; Przylucka et al., 2017a) and general fungal fitness (Cai et al., 2020). However, numerous functional genetic studies based on the characterization of gene‐deletion mutants have demonstrated that they play only a minor or accessory role in such processes (Jeong et al., 2007; Frías et al., 2011; Frischmann et al., 2013), suggesting that the main forces driving and shaping SSCP diversity and evolution in fungi have yet to be revealed.
The large cluster of SSCP investigations reflects that other organisms use them as an indicator of fungal presence, probably because they are produced in large quantities. Therefore, SSCPs are frequently studied as effectors and elicitors, i.e. from the perspective of the host plants or animals in which the SSCPs either cause toxicity or elicit an immune response (Pellegrin et al., 2015; Dagvadorj et al., 2017; Darwiche et al., 2017; Fang et al., 2019; Li et al., 2019; Wang et al., 2019; Yu et al., 2020). These studies reveal the ecological role of individual SSCPs, but they do not sufficiently explain their function given that orthologous SSCPs are also present in saprotrophic fungi (Feldman et al., 2020; Gao et al., 2020).
The exceptionally high diversity of fungal SSCPs and their ubiquitous presence in biotrophic and saprotrophic fungi suggest their functions are related to aspects of the fungal life cycle other than host interactions (Garrigues et al., 2018; Hajdu et al., 2019; Holzknecht et al., 2020). The SSCP CPL1 in the human pathogenic yeast Cryptococcus neoformans (Tremellales, Basidiomycota) is important for its adhesion to and invasive growth into substrates (Sun et al., 2020). Similarly, at least two SSCP families in filamentous fungi (HFBs and CPs) are amphiphilic, having profound or even superior surface activity (Wösten, 2001; Gaderer et al., 2014) and the ability to reduce the force of water tension (van der Vegt et al., 1996). HFBs are particularly small (~100 a.a.) and have a conserved pattern of eight cysteine residues C‐CC‐C‐C‐CC‐C that efficiently stabilizes their tertiary structure by disulfide bonds (Linder et al., 2005). CPs are slightly larger (~150 a.a.) and have only four single cysteines. Recent physiochemical investigations and expression profile studies have shown that HFBs and CPs are probably functionally complementary (Gaderer et al., 2014). Proteins from both of these families can spontaneously assemble in interfaces, mediate the hydrophobicity of surfaces and reduce water tension (Linder et al., 2005; Gaderer et al., 2014). These properties attract research attention to HFBs for the development of multiple industrial or pharmacological applications, and to CPs for their ability to trigger plant immune responses and their role in plant pathogenicity.
Fungi from the fungivorous genus Trichoderma as well as a few other hypocrealean taxa are considered suitable models for the functional characterization of HFBs and CPs, because the respective gene families are expanded in their genomes (Kubicek et al., 2019; Gao et al., 2020). In contrast to the majority of Ascomycota fungi, which have a few HFBs and usually only one or two CPs (Chen et al., 2013), Trichoderma spp. can have seven (e.g. T. reesei and T. citrinoviride) to 15 (e.g. T. harzianum and T. gamsii) HFB‐encoding genes (Kubicek et al., 2019). All whole‐genome sequenced Trichoderma strains (September 2020) have at least three CPs, and a few species have four (Gao et al., 2020). Moreover, Trichoderma spp. are ecologically versatile, and several environmentally opportunistic species can parasitize other fungi, colonize dead wood and other cellulosic substrates, grow in soil, establish in rhizosphere, become endophytes, kill water moulds (Oomycota), control plant‐parasitic nematodes and attack immunocompromised humans (Druzhinina et al., 2011). Thus, the expansion of surface‐active SSCPs (saSSCPs) such as the HFBs and CPs in Trichoderma genomes may be linked to the diversity of the biotic and abiotic interactions of these fungi.
This study continues the evolutionary and functional characterization of saSSCPs in Trichoderma (Kubicek et al., 2008; Gao et al., 2020). We detected and characterized a new family of saSSCPs, named here as hyphosphere proteins (HFSs) and the two new Trichoderma HFBs, here named HFB11 and HFB12. The application of recombinant HFB2, HFB4, HFB12, HFS1 and EPL1 proteins (produced by Pichia pastoris) to Solanum lycopersicum (tomato) roots resulted in divergent effects on fungal attachment and in a noticeable phytotoxic effect, while the complete secretome of T. guizhouense stimulated tomato growth. Interestingly, all recombinantly produced saSSCPs (HFS1, HFB2, HF4, HFB12 and EPL1) showed convergent surface‐modulating activity that favoured the attachment of hyphae indicating that the high diversity of saSSCPs in Trichoderma likely evolved for the efficient conditioning of various surfaces in the hyphosphere to a hydrophilic state, making it suitable for attachment or colonization by the fungus.
Results
Genome mining of T. guizhouense revealed a new family of SSCPs and two new HFBs
We first performed a broad screening of the whole‐genome sequence of T. guizhouense NJAU 4742 (Druzhinina et al., 2018; Kubicek et al., 2019) for saSSCPs such as HFBs and CPs. For this purpose, we expanded the previously available annotations (Kubicek et al., 2019) and collected the small (<30 kDa, <300 a.a.) secreted proteins that contained four to eight single or double cysteine residues (see Experimental procedures for further details). The results, shown in Table 1 and in the Supporting Information 1 Table S1, confirmed the three CPs characterized in this species by Gao et al. (2020). These data also revealed that there are two putative HFBs described by Kubicek et al. (2019) do not belong to this family. The first (GenBank ID: OPB44529) lacks the signal peptide. A comparison with other genomes revealed that it is likely an orphan Trichoderma protein that is present in section Trichoderma (e.g. T. gamsii, T. atroviride and T. asperellum) and Harzianum clade species but absent in the Longibrachiatum clade. The second putative HFB (GenBank ID: OPB38365) has the HFB‐specific cysteine pattern of C‐CC‐C‐C‐CC‐C, but it harbours six more cysteines (14 total) and has a size equivalent to that of three HFBs (28 kDa, 277 a.a.). Although it is probably a member of the SSCPs, it presents as a single copy protein that is relatively conserved in all mined ascomycetes, differing from the HFB family. Thus, we concluded that OPB38365 possibly represents a separate family of SSCPs and excluded it from this investigation. We also found two novel putative HFB‐encoding genes that were previously detected by Kubicek et al. (2019), which we name here as hfb11 (OPB42521, encoding HFB11) and hfb12 (OPB45278, encoding HFB12). Thus, the T. guizhouense genome encodes at least 10 confirmed HFBs (Table 1). Moreover, we found three other proteins with primary structures fulfilling our search criteria (Table 1). These proteins (OPB41482, OPB43804 and OPB40307) had a pattern of eight single cysteine residues and were larger than the HFBs (7–16 kDa) and CPs (12–14 KDa), with molecular weights varying from 21 to 28 kDa and sequences contained 219–280 residues.
Table 1.
Inventory and expression of surface‐active SSCPs of Trichoderma guizhouense. [Colour table can be viewed at wileyonlinelibrary.com]
| saSSCP family | Protein | Protein ID | M w | SP | Gene | Relative expression, folds | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Submerged growth | Aerial hyphae | Conidiation | |||||||||
| Da | a.a. | Av. | Sd | Av. | Sd | Av. | Sd | ||||
| Hydrophobins (HFBs) | HFB2 | OPB38530 | 7044.17 | 16 | hfb2 | 0.083821 | 0.032 | 0.407867 | 0.110 | 0.008979 | 0.004 |
| HFB3 | OPB45549 | 8923.33 | 16 | hfb3 | 0.004052 | 0.002 | 0.168363 | 0.042 | 0.007011 | 0.005 | |
| HFB4 | OPB37525 | 8486.68 | 21 | hfb4 | 0.004519 | 0.001 | 0.641103 | 0.272 | 0.335277 | 0.262 | |
| HFB5 | MF527119 | 8282.49 | 16 | hfb5 | 0.000049 | 0.000 | 0.002428 | 0.001 | 0.015500 | 0.014 | |
| HFB6 | OPB38878 | 15540.76 | 16 | hfb6 | 0.000000 | 0.000 | 0.000000 | 0.000 | 0.000000 | 0.000 | |
| HFB9a | OPB40515 | 14414.52 | 18 | hfb9a | 0.000003 | 0.000 | 0.000004 | 0.000 | 0.000149 | 0.000 | |
| HFB9b | OPB44528 | 12117.36 | 18 | hfb9b | 0.001415 | 0.000 | 0.003648 | 0.001 | 0.001090 | 0.001 | |
| HFB10 | OPB44696 | 11649.88 | 16 | hfb10 | 5.321598 | 2.544 | 7.377791 | 2.857 | 1.071798 | 0.843 | |
| HFB11 | OPB42521 | 7590.57 | 20 | hfb11 | 0.000000 | 0.000 | 0.000000 | 0.000 | 0.000000 | 0.000 | |
| HFB12 | OPB45278 | 7571.42 | 16 | hfb12 | 0.000002 | 0.000 | 0.000010 | 0.000 | 0.000668 | 0.000 | |
| Cerato‐platanins (CPs) | EPL1 | OPB44018 | 12461.81 | 18 | epl1 | 0.380258 | 0.139 | 0.104417 | 0.029 | 0.012128 | 0.008 |
| EPL2 | OPB45524 | 13169.57 | 18 | epl2 | 0.000005 | 0.000 | 0.001414 | 0.000 | 0.003207 | 0.002 | |
| EPL3 | OPB43811 | 14277.99 | 18 | epl3 | 0.000023 | 0.000 | 0.000023 | 0.000 | 0.000113 | 0.000 | |
| Hyphosphere proteins (HFSs) | HFS1 | OPB41482 | 21856.39 | 20 | hfs1 | 0.000043 | 0.000 | 0.000053 | 0.000 | 0.000085 | 0.000 |
| HFS2 | OPB43804 | 21271.61 | 18 | hfs2 | 0.000133 | 0.000 | 0.000185 | 0.000 | 0.000092 | 0.000 | |
| HFS3 | OPB40307 | 27841.06 | 20 | hfs3 | 0.017148 | 0.002 | 0.037480 | 0.007 | 0.025919 | 0.002 | |
M w, molecular weight; SP, signal peptide; Av., average; Sd, standard deviation from four repeats; a.a., amino acid.
As the physiochemical surface‐active properties of the HFBs and CPs in Trichoderma spp. are relatively well studied (Kubicek et al., 2008; Seidl‐Seiboth et al., 2011; Espino‐Rammer et al., 2013; Bonazza et al., 2015; Przylucka et al., 2017b; Gao et al., 2020), we compared their hydropathy profiles to those of the newly detected proteins (Fig. 1A). The principal component analysis (PCA) based on the Kyte and Doolittle hydrophobicity coefficient (Kyte and Doolittle, 1982) and the values of the GRAVY scores, which define a protein sequence's average degree of hydrophobicity, showed that the novel proteins had hydropathy profiles similar to those of the HFBs and CPs (Fig. 1B) and therefore had the potential for being surface‐active or amphiphilic. In particular, the GRAVY scores suggested these novel proteins were comparable to HFB6 and the pseudo‐class I HFB members HFB9a/b in Trichoderma (Seidl‐Seiboth et al., 2011). As they have a unique pattern of eight single cysteine residues (see below) and are considerably larger in size, we assigned them to a putative new family of saSSCPs and named them HFSs what is phonetically similar to ‘hyphosphere’ proteins, see below: HFS1 (OPB41482), HFS2 (OPB43804) and HFS3 (OPB40307).
Fig. 1.
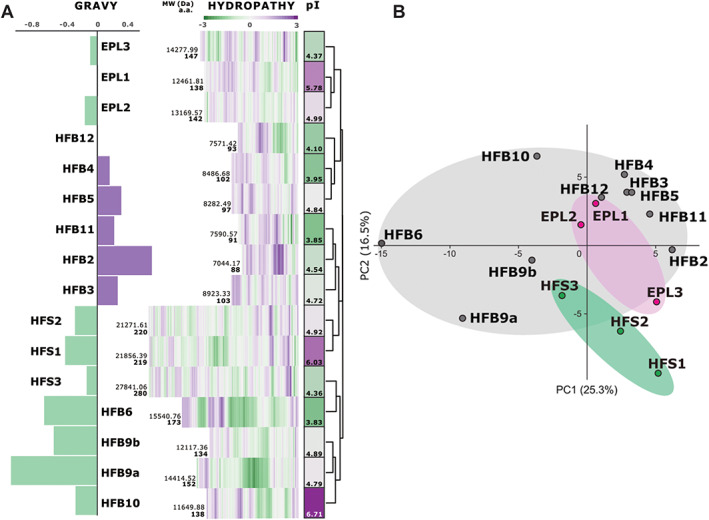
Primary structure analysis of surface‐active small secreted cysteine‐rich proteins (SSCPs) in Trichoderma guizhouense.
A. Deep purple colour highlights relatively high values of either the hydrophobicity based on protein GRAVY scores, the Kyte and Doolittle hydrophobicity coefficient (Kyte and Doolittle, 1982) for individual residues, or the isoelectric point (pI). Green corresponds to the respective low values. The cluster analysis on the right was applied to the hydropathy profile values using complete linkage and Euclidian distances.
B. Principal component analysis (PCA) of the Kyte and Doolittle hydropathy coefficients, which shows a high similarity between proteins from the three families (i.e. both components explain minor variability).
In general, the primary structure analysis indicated that HFB2, HFB3, HFB4, HFB5 and HFB11 are potentially more hydrophobic than HFB6, HFB9a, HFB9b, HFB10, HFS1 and HFS2, which are somewhat hydrophilic. Proteins such as HFB12, CPs (EPL1, EPL2 and EPL3) and HFS3 have intermediate values. The isoelectric point (pI) analysis showed no correlation with a particular family or hydropathy profile, with ranges varying from 3.83 to 6.71 in the HFBs, 4.37 to 5.78 in the CPs and 4.36 to 6.03 in the HFSs. Only a few proteins, namely EPL1, HFS1 and HFB10, had pIs close to the 6–7 range, while the majority of the others could be considered ‘cationic’ (Huber et al., 2020), suggesting they would require unique environmental conditions and potentially precipitate under neutral (pH 7; like water) conditions. All 16 proteins have a relatively short signal peptide (16–21 a.a.), which is characteristic of HFBs (Kottmeier et al., 2011).
To confirm the functionality of the saSSCP‐encoding genes, we tested their expression during the main stages of the asexual life cycle of T. guizhouense when these proteins are known to be active. This included submerged vegetative growth that associated with high CP expression (Gaderer et al., 2014; Gao et al., 2020), and aerial hyphae formation and conidiation which require large amounts of HFBs (Lugones et al., 2004; Winefield et al., 2007; Cai et al., 2020). The qPCR analysis confirmed the expression of all CP‐ and HFB‐encoding genes except hfb6 and hfb11 (Table 1). The expression of hfb12 was low compared with major hfbs such as hfb4, hfb10, hfb2 and hfb3 (Cai et al., 2020), but showed the hfb‐characteristic pattern of a drastic (several hundred folds) increase with the formation of aerial hyphae and conidiation. The three genes encoding HFS proteins showed a relatively low but consistent expression profile that did not change much with the development of the fungus (Table 1). Thus, our data suggest that at least 14 of the detected putative saSSCP‐encoding genes are actively transcribed, while the other two could either be putative pseudogenes or become functional under specific conditions not tested here.
Evolutionary analysis suggests a long evolutionary history of HFSs in filamentous Ascomycota
The newly described HFB11 in T. guizhouense is 91 a.a. long and has a molecular weight of 7.59 kDa. This protein also has the characteristic C‐CC‐C‐C‐CC‐C pattern and a short signal peptide (20 a.a.). The evolutionary analysis revealed it to be an orphan protein present in only a few species in the Harzianum clade of the genus Trichoderma (Fig. 2A), including T. simmonsii, T. afroharzianum, T. harzianum and the putatively new species T. sp. M10 (Cai and Druzhinina, 2021). We did not detect any hfb11 sequences in Harzianum clade species such as T. pleuroti and T. pleuroticola or in the sister clade Virens. The amino acid sequence of HFB11 was ~50% similar to HFB5 (hfb5, MF527119; Fig. 2A), which is in agreement with the similar primary structure of these two proteins (Fig. 1).
Fig. 2.
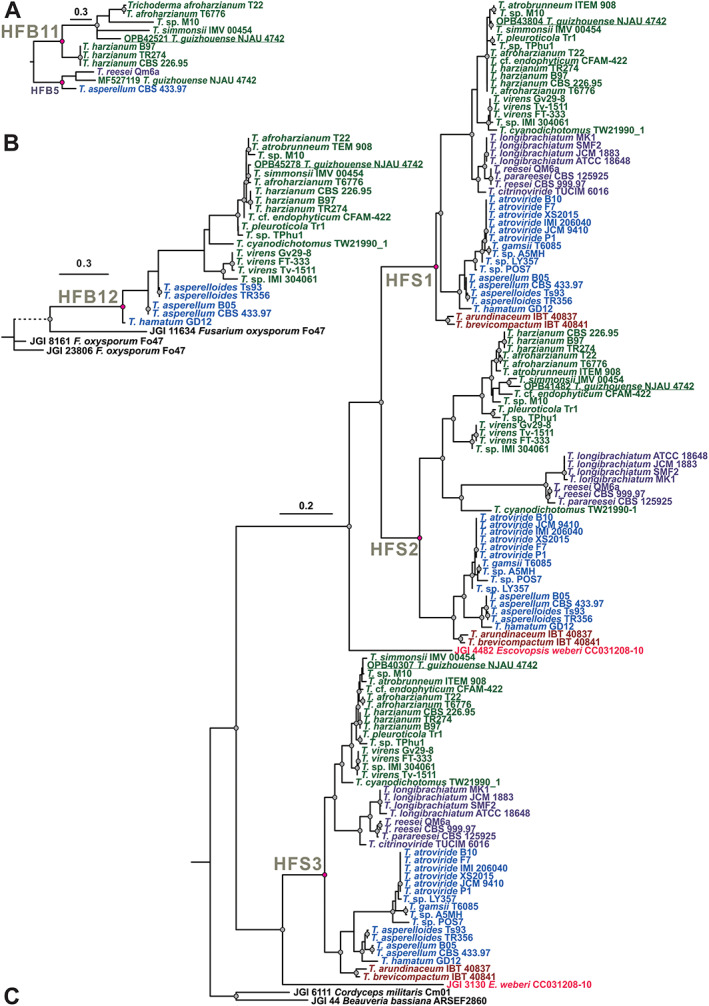
Phylogenetic analysis of newly described surface‐active small secreted cysteine‐rich proteins (saSSCPs) from Trichoderma spp. The evolution of HFB11 (A), HFB12 (B) and HFSs (C) was inferred based on the amino acid alignments. Maximum‐likelihood (ML) phylogenetic trees were constructed using IQ‐TREE 1.6.12 (ultrafast bootstrap, N = 1000). The Bayesian Information Criterion was applied when searching for the best amino acid substitution model with ModelFinder (integrated into the IQ‐TREE program). Statistically supported nodes (bootstrap values >75) are marked by dots. Pink dots indicate the hypothetical ancestor for the particular protein. Trichoderma spp. from the Harzianum and Virens clades, section Longibrachiatum, section Trichoderma, the Brevicompactum clade and Escovopsis weberi are highlighted in dark green, purple, blue, orange and red, respectively, for improved visual assessment. The accession numbers of the individual genomes mined in this study are provided in Supporting Information 1 Table S2.
Another newly described HFB12 has a sequence of 93 a.a. harbouring the typical C‐CC‐C‐C‐CC‐C cysteine pattern of HFBs that provides for an M w of 7.57 kDa with a 16 a.a. signal peptide. The phylogenetic analysis (Fig. 2B) showed that among the 42 Trichoderma spp. (August 2020) whole‐genome sequences, hfb12 can be found in the Harzianum and Virens clades, T. cyanodichotomus (which is phylogenetically close to T. virens), and in some species of section Trichoderma such as T. asperellum, T. asperelloides and T. hamatum, but not in the species T. atroviride or T. gamsii. The gene is also lacking in the Brevicompactum clade and section Longibrachiatum. Interestingly, the phylogeny of HFB12 was not concordant with the phylogeny of the genus (Kubicek et al., 2019), which indicated the likely scenario of a gene loss (GL) event in section Longibrachiatum and a partial loss in the other clades. No homologues to the hfb12 genes were found in other fungi, though the closest neighbour was <50% similar to the Fusarium oxysporum species complex (Hypocreales, Ascomycota). Phylogenetically, hfb12 was highly divergent compared with the other HFBs in Trichoderma. Thus, we assume hfb12 is an orphan Trichoderma gene that is not present in the core genome of the genus or in other fungi.
To investigate the evolution of the T. guizhouense HFSs, we performed a sequence similarity search (BLASTP) against the NCBI and JGI databases and screened all 42 publicly available Trichoderma genomes [August 2020; (Cai and Druzhinina, 2021)] as well as the genome of the closely related hypocrealean fungus Escovopsis weberi (de Man et al., 2016). The multiple sequence alignment revealed that the pattern of eight single cysteine residues of HFSs was highly conserved in all Trichoderma spp. For example, the HFS2 in T. reesei (XP_006967391) had a structure of C49‐C65N66N67L68C69‐C129‐C152‐C174‐C182‐C194. In other fungi (see below), the amino acids and length of the sequence between the C residues slightly varied, except for the space between the second and third cysteines, which were always separated by three amino acids; the first two were most frequently asparagine (N) or aspartic acid (D), while the third was an aliphatic amino acid such as leucine (L) or isoleucine (I). The maximum likelihood (ML) phylogram in Fig. 2C demonstrated that the three HFS proteins were homologous and shared the statistically supported common ancestor. The ancestor of HFS1 and HFS2 was, in turn, monophyletic with HFS3. In Trichoderma, the similarity between the amino acid sequences of the proteins varied from 62% to 100% for HFS1, 78% to 100% for HFS2 and 71% to 100% for HFS3. Three HFS‐encoding genes were harboured in all the screened Trichoderma genomes with the exception of T. citrinoviride, which was missing HFS2. In general, the topologies of the individual HFS subclades were concordant with the topology of the Trichoderma phylogenomic tree (Kubicek et al., 2019), suggesting the vertical evolution and stabilizing selection pressure (Druzhinina et al., 2018; Cai et al., 2020; Gao et al., 2020) that were recorded for the other groups of secreted proteins in Trichoderma.
The topology of the HFS phylogram suggests a GD event in the ancestor of the genus Trichoderma that resulted in paralogous copies of hfs1 and hfs2. To test this, we collected HFS sequences from other fungi and performed NOTUNG and T‐Rex analyses (Chen et al., 2000; Boc et al., 2012; Gao et al., 2020), which allowed us to test for possible GD, GL and LGT events in the evolutionary history of the HFSs in fungi. The results showed that HFSs are common in filamentous Ascomycota, but absent in other Dikarya fungi. The analyses confirmed the evolution of HFS1 and HFS2 by GD (Fig. 3), showing that the event most likely took place in the ancestor of hypocrealean fungi. One duplicated copy of hfs1 was inherited by the fungi from most other families, while hfs2 was only maintained in Trichoderma and likely lost in the other families. Interestingly, our analysis showed that hfs1 was putatively laterally transferred to the genome of Tolypocladium ophioglossoides (Hypocreales, Ascomycota), which has its own copy of hfs1 that was obtained vertically. The third gene, hfs3, also evolved as a result of a GD event but only one copy was maintained in Trichoderma. Similar to the evolution of CPs (Gao et al., 2020), the evolutionary history of the HFSs in filamentous Ascomycota encountered numerous cases of LGTs, GDs and GLs, all indicating the functional significance of these proteins. Our data show that the genes encoding HFSs were present in parasites feeding on any type of live biomass and in polyphages feeding saprotrophically on different organic matter (Fig. 3). Thus, the evolutionary analysis indicates the functional importance of these genes for filamentous Ascomycota, but it does not suggest that the acquisition of these genes was associated with a particular nutritional strategy or lifestyle of the fungi.
Fig. 3.
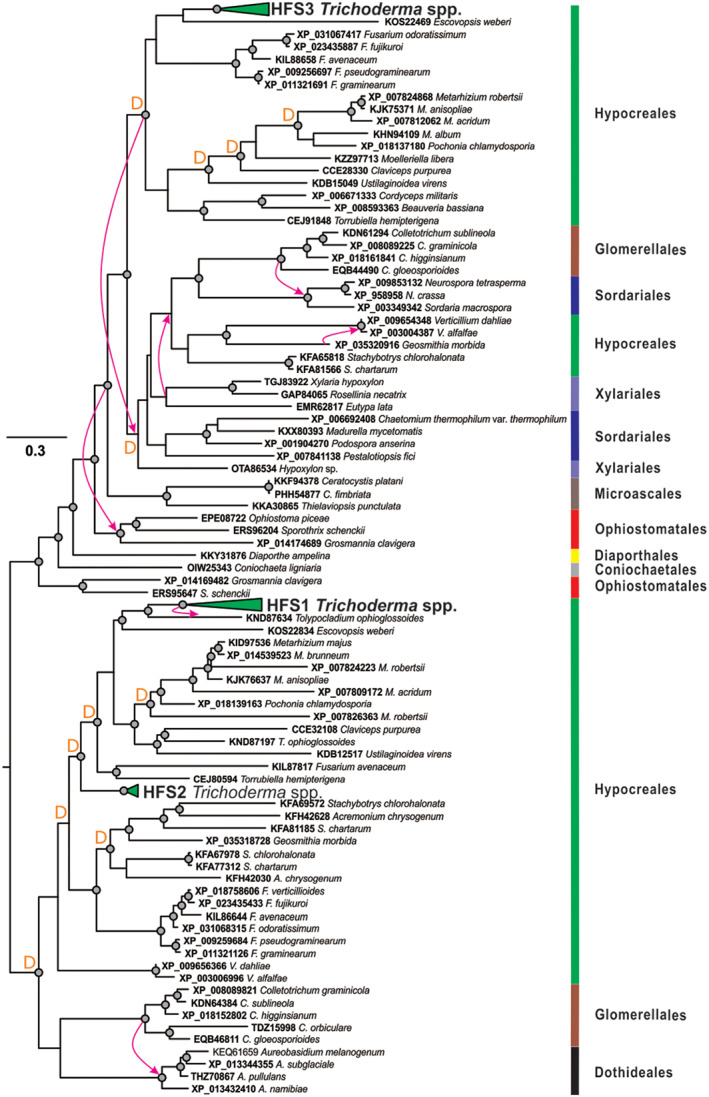
Evolution of the hyphosphere (HFS) proteins in fungi. A maximum‐likelihood phylogenetic tree was constructed using IQ‐TREE 1.6.12 (ultrafast bootstrap, N = 1000). The Bayesian Information Criterion was applied when searching for the best amino acid substitution model with ModelFinder (integrated into the IQ‐TREE program). Statistically supported nodes (bootstrap values >75) are marked by dots. Arrows indicate putative cases of lateral gene transfer as revealed by NOTUNG and T‐Rex; ‘D’ indicates the putative gene duplication events revealed through the same methods. Vertical bars denote the taxonomic positions of the corresponding species.
Heterologous production of saSSCPs in P. pastoris
In previous studies, we explored the use of a gene deletion strategy for the functional characterization of saSSCPs in Trichoderma (Cai et al., 2020; Gao et al., 2020). However, the high number of SSCPs and the low expression profile of the newly revealed proteins indicated that this approach would not be suitable for their characterization. Therefore, we selected HFB12 and HFS1 for recombinant production in a Komagataella phaffii [syn. Pichia pastoris (Kurtzman, 2009)] host. For the comparison, we used the previously characterized saSSCPs of Trichoderma such as the HFBs [HFB2, (Askolin et al., 2005), HFB4 (Cai et al., 2020)] and a CP [EPL1; (Bonazza et al., 2015; Gao et al., 2020)]. For the construction of mutants in P. pastoris, the production of each protein was confirmed by SDS‐PAGE (Fig. 4A) and immunoblotting (Supporting Information 2 Fig. S1). The flask fermentation of the engineered P. pastoris strains yielded at least 0.3–0.5 g L−1 of recombinant protein, varying slightly between the proteins produced and the fermentation batches. The expected size of each recombinant protein, shown in the SDS‐PAGE, excluded the possibility of glycosylation in the recombinants.
Fig. 4.
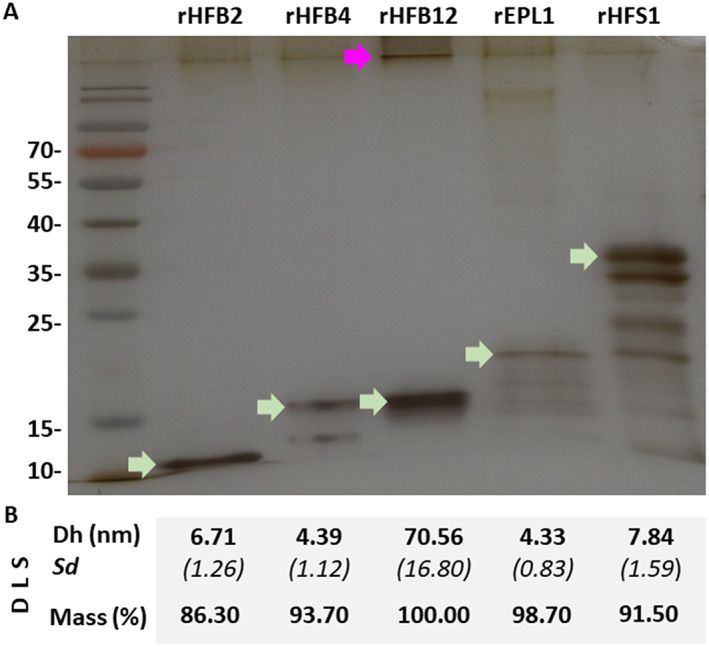
Recombinant production of the five surface‐active small secreted cysteine‐rich proteins (saSSCPs) from Trichoderma guizhouense in Pichia pastoris.
A. The SDS‐PAGE analysis of recombinant saSSCPs (green arrows). The migration of HFB12 through the polyacrylamide gel was impeded (purple arrow).
B. Results of the dynamic light scattering analysis (DLS) applied to the same samples shown in A. Dh corresponds to the mass weight (%) diameter of the proteins obtained from three measurements. Sd, standard deviation.
The behaviour of each recombinant protein (indicated by ‘r’) in water was estimated using dynamic light scattering (DLS) (Przylucka et al., 2017b). The hydrodynamic diameters of the proteins based on the mass weight (%) are shown in Fig. 4B. The majority (mass > 90%) of the rHFB2, rHFB4 and rEPL1 proteins displayed a hydrodynamic size of 4.3–6.7 nm, which corresponded to that of a monomer or dimer of previously reported proteins (Hakanpää et al., 2004; Przylucka et al., 2017b). The rHFS1 protein had a slightly larger particle size of ~7.8 nm. Interestingly, rHFB12 was found mainly (100%) in large aggregates with a dynamic size >70 nm, which could also be seen in the intensity distribution plot (Supporting Information 2 Fig. S2). Therefore, using HFB1 [2–3 nm; (Hakanpää et al., 2004)] from T. reesei as a reference for monomers, rHFB2, rHFB4, rEPL1 and rHFS1 tended to form dimers or oligomers in a water solution, while rHFB12 easily formed aggregates, explaining the retention of this protein in the sample well during the SDS‐PAGE analysis (Fig. 4A).
All saSSCPs convert surfaces to a moderately hydrophilic state
The measurements of the water contact angle (WCA) on initially hydrophobic [poly(ethylene‐terephthalate), PET] and hydrophilic (glass) surfaces coated with the recombinant saSSCPs showed that all five proteins were surface‐active and provided a remarkably consistent result (Fig. 5). Irrespective of the initial hydrophobicity of the material, all proteins converted both surfaces to a moderately hydrophilic state, which corresponds to a WCA between 30° and 45°, excluding rHFB12 on PET. Thus, rHFB4 and rHFB12 increased the surface hydrophobicity of glass (30%–44%, p < 0.05) and decreased that of PET (37%–51%, p < 0.05). The rHFB2, rEPL1 and rHFS1 coatings on glass surfaces did not differ from the control, but the hydrophobicity of PET was reduced by 70% (p < 0.05). As predicted by the primary structure analysis, the newly detected SSCPs (HFS1 and HFB12) have potent surface activity similar to that of HFBs and CPs. The aggregation of HFB12 in water (see above) did not abolish its surface activity, although it was slightly reduced compared with that of the other tested saSSCPs. Considering the shared evolutionary history of the three HFSs and the similarity of their hydropathy profiles, they can be assigned as a family of saSSCPs (Fig. 6). More interestingly, the results indicate that T. guizhouense secretes an arsenal of saSSCPs that are all capable of transforming the surfaces surrounding hyphae to a moderately hydrophilic state. The consistency of the results allows us to hypothesize that hydrophilic surfaces corresponding to a 30°–45° WCA are optimal for the fungus.
Fig. 5.

Surface‐modulating activity of the recombinant surface‐active small secreted cysteine‐rich proteins (rsaSSCPs) from Trichoderma guizhouense revealed by a water contact angle analysis (WCA) on glass and poly(ethylene terephthalate) (PET). Values (mean ± Sd) labelled with the same letter do not statistically significantly differ (ANOVA, p < 0.05).
Fig. 6.
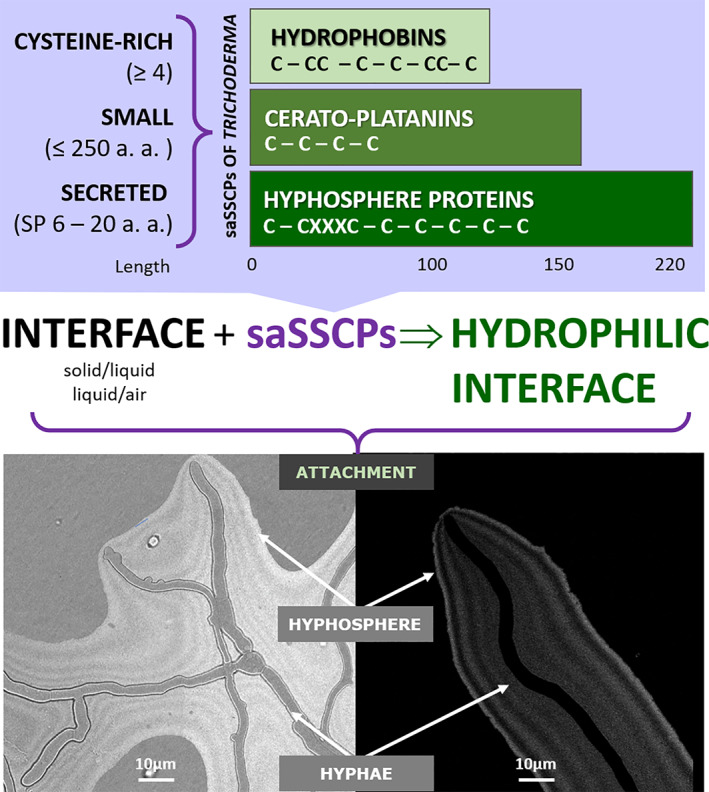
Diagram showing the main distinctive features of the three families of surface‐active small secreted cysteine‐rich proteins (saSSCPs) in Trichoderma guizhouense and their hypothetical function in the hyphosphere. The young hyphae of T. guizhouense NJAU 4742 were imaged on a cellophane surface covering a potato dextrose agar plate 18 h after inoculation (incubated at 25 °C in the dark). The stripes in the hyphosphere correspond to unequal light reflection, likely due to the assembly of different saSSCPs. The right and left images depict a hyphosphere of multiple (left) and a single (right) hyphae imaged using similar setups.
saSSCPs assist Trichoderma hyphal attachment to hydrophilic surfaces
As T. guizhouense has the ecophysiological features of an aero‐aquatic fungus (Cai et al., 2020) and is also rhizosphere competent (Cai et al., 2013, 2015; Zhang et al., 2016), we hypothesized that saSSCPs that are frequently expressed during submerged growth (Table 1) and transform surfaces to a moderately hydrophilic state (similar to that of glass) could also assist mycelial attachment to such modified hydrophilic surfaces in water. (Note that due to the abundance of saSSCPs, all hydrophobic surfaces would ultimately become hydrophilic.) To model such surfaces, we used glass wool (artificial) and the roots of Solanum lycopersicum (tomato) seedlings (natural substrate). The results showed that the T. guizhouense strain had a higher affinity to the glass wool, while attachment to roots was less successful (Fig. 7). The addition of 1 μM of any rsaSSCPs produced a significant (p < 0.05) and multifold increase in hyphae attachment to the glass wool. As for the attachment to roots, three rHFBs produced a similar result, improving colonization, while rEPL1 and rHFS1 had a reverse impact, preventing hyphae attachment. To see whether these functions were species specific, we performed the same experiment with another Trichoderma strain, T. harzianum CBS 226.95, which is related to T. guizhouense (Kubicek et al., 2019) but has a different ecologic profile (Cai et al., 2020). The results for T. harzianum repeatedly showed that rsaSSCPs can assist in the attachment of the fungus to glass wool, but none of the selected rsaSSCPs from T. guizhouense NJAU 4742 could prime the colonization of T. harzianum CBS 226.95 on tomato roots (Fig. 7). This indicates that root attachment is a complex process that is probably regulated by both partners, and the addition of saSSCPs does not necessarily have an impact.
Fig. 7.
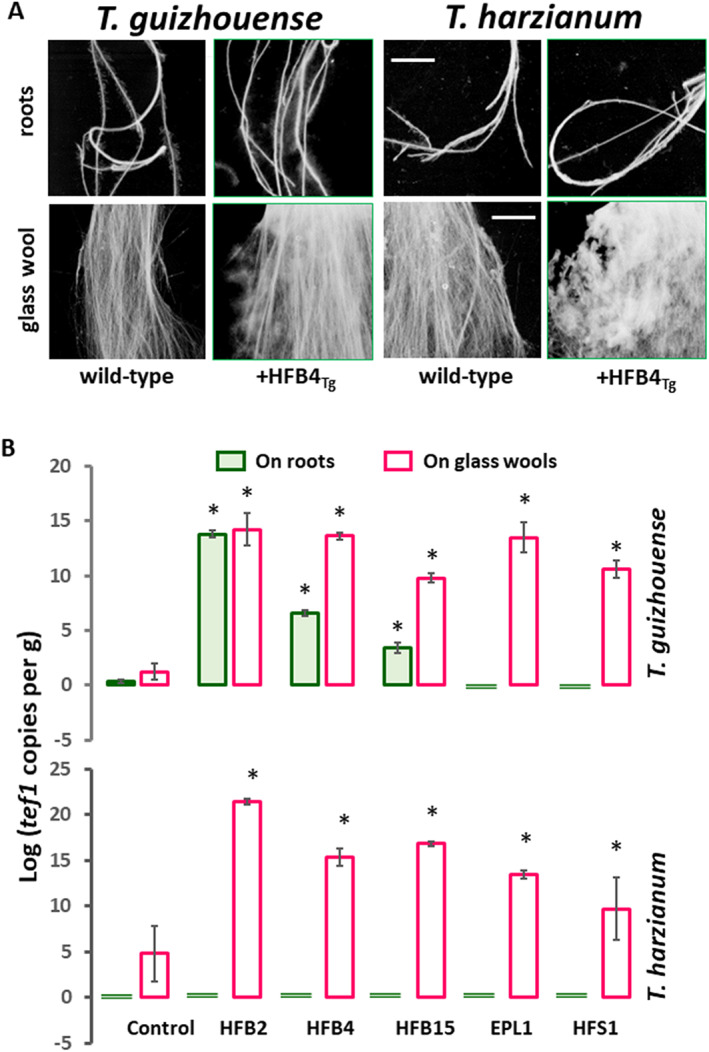
Effect of recombinantly produced surface‐active small secreted cysteine‐rich proteins (saSSCPs) on the ability of Trichoderma hyphae to attach to biotic (tomato roots) and abiotic (glass wool) surfaces.
A. Sample images obtained for T. guizhouense and T. harzianum with or without addition of recombinantly produced HFB4. Images were obtained using a root scanner after 48 h of incubation. Scale, 1 cm.
B. Quantification of fungal attachment to biotic (tomato roots) and abiotic (glass wool) surfaces using a qPCR determination of the single copy constitutively expressed house‐keeping gene tef1 (Cai et al., 2020; Gao et al., 2020).
Some saSSCPs are toxic for plants and can trigger an immune response
As T. guizhouense itself and its secretomes have considerable growth‐promoting effects on plants [Supporting Information 2 Fig. S3 and Cai et al., 2013, Cai et al., 2015], we tested the effect of the rsaSSCPs on the growth of tomato seedlings using 0.1 and 1 μM of a purified protein suspension. Surprisingly, with the exception of HFB12, these treatments had a significantly adverse effect on plant growth that varied in strength and range between the different rsaSSCPs (Fig. 8). One micromole of rHFS1 and rEPL1 significantly (p < 0.05) reduced plant growth to 62% and 64% respectively, of the control biomass without the protein addition. The rHFB2 and rHFB4 proteins had a less severe effect (73%–78%) on plant growth, though it was still statistically significant (p < 0.05) when compared with the control, and the plant height was reduced accordingly. The rHFB12 protein had a neutral effect on the plants, which may be due to its strong aggregation in water (Fig. 4). The growth inhibition effects of the protein application increased when the concentration was raised from 0.1 to 1 μM (Fig. 8).
Fig. 8.
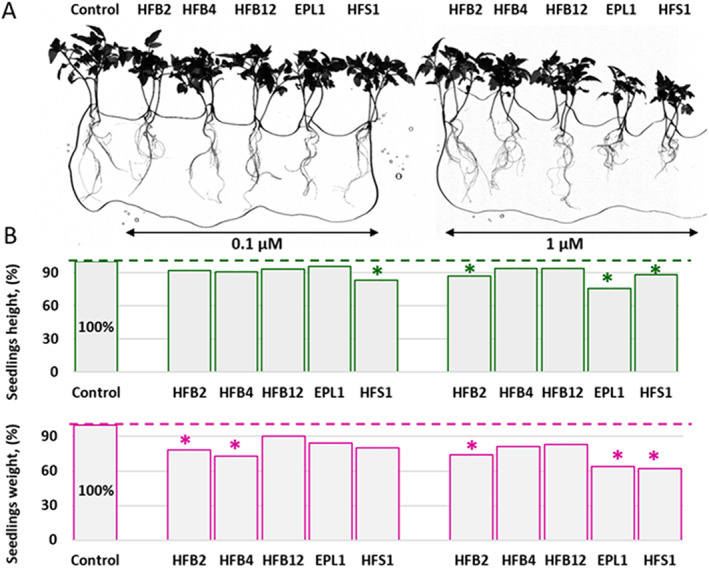
Phytotoxic effect of recombinant surface‐active small secreted cysteine‐rich proteins (rsaSSCPS) on tomato growth.
A. Tomato plants imaged using a root scanner after 21 days of hydroponic growth with or without the addition of two concentrations of rsaSSCPS.
B. Quantification of tomato growth based on seedling height and weight. Values that significantly differ from the control are marked by asterisks (ANOVA, p < 0.05). See A for the concentrations of the rsaSSCPs.
Since saSSCPs are frequently considered effector‐like proteins (Gaderer et al., 2014; Ruocco et al., 2015; Gao et al., 2020), we also tested whether the immune system of the tomato seedlings was activated by the addition of the rsaSSCPs. To this end, we monitored the expression of biomarker genes for the jasmonic acid (JA)‐ and salicylic acid (SA)‐defence pathways in plants. The results (Table 2) show that 0.1 μM of rHFB2 and rHFB12 slightly but significantly (p < 0.05) activated the JA‐ and SA‐mediated defence pathways, as shown by the upregulation of the respective genes LoxA (11–13‐fold) and PAL (~two folds). The rEPL1 protein did likewise. However, the elicitation effect of these proteins was not seen when the protein concentration was increased to 1 μM. In contrast, neither rHFB4 nor rHFS1 was able to trigger the plant systemic defence pathways at the two applied concentrations, likely because under natural conditions, HFB4 is associated with conidiation (Cai et al., 2020) and thus secreted in very low amounts in the rhizosphere, and the expression of HFS1‐encoding gene is generally low (Table 1).
Table 2.
Relative expression of Solanum lycopersicum (tomato) genes involved in induced systemic resistance after application of rsaSSCPs to roots.
| Tomato genes | AOS | LoxA | PAL | PR‐1a | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rsaSSCP conc. (μM) | 0.1 | 1.0 | 0.1 | 1.0 | 0.1 | 1.0 | 0.1 | 1.0 | ||||||||
| Av. | Sd | Av. | Sd | Av. | Sd | Av. | Sd | Av. | Sd | Av. | Sd | Av. | Sd | Av. | Sd | |
| HFB2 | 1.93 | 0.38 | 1.17 | 0.39 | 13.18* | 3.74 | 3.12 | 0.67 | 2.54* | 0.70 | 1.42 | 0.20 | 0.71 | 0.15 | 0.49 | 0.1 |
| HFB4 | 1.37 | 0.66 | 0.96 | 0.73 | 5.37 | 2.25 | 5.51 | 3.17 | 1.76 | 0.54 | 0.86 | 0.59 | 0.29 | 0.11 | 0.38 | 0.27 |
| HFB12 | 1.34 | 0.28 | 1.73 | 0.50 | 11.26* | 3.09 | 4.92 | 1.30 | 2.21 | 0.43 | 1.93 | 0.23 | 0.50 | 0.06 | 0.45 | 0.15 |
| EPL1 | 1.52 | 0.54 | 2.29 | 0.54 | 6.39* | 1.16 | 3.75 | 1.48 | 2.62* | 0.74 | 1.95 | 0.18 | 0.47 | 0.07 | 0.47 | 0.16 |
| HFS1 | 0.93 | 0.37 | 2.01 | 0.79 | 3.98 | 2.46 | 4.03 | 0.84 | 1.37 | 0.34 | 1.76 | 0.71 | 1.08 | 0.36 | 1.43 | 0.81 |
Av., average; Sd, standard deviation from four repeats; Statistically significantly upregulated values compared with no rsaSSCP treatment (control) are marked by asterisks (ANOVA, p <0.05).
Discussion
Studies on HFBs and CPs have frequently focused on the effect of these proteins on other organisms. It is not a surprise that plants and animals recognize these massively produced proteins as indicators of fungal proximity, and they respond by activating their defence mechanisms (Frías et al., 2011; Wang et al., 2019; Gao et al., 2020). In plant‐pathogenic fungi, numerous HFBs and CPs have been reported as effector‐like proteins or elicitors (Rohe et al., 1995; Whiteford et al., 2004; Jeong et al., 2007; Frías et al., 2011). However, our evolutionary analyses and the study of Gao et al. (2020) suggest that their role in plant virulence is likely a secondary function of the saSSCPs, because genes encoding HFBs and CPs are also maintained in the genomes of saprotrophic and animal‐associated fungi.
Besides the detection of a new family of saSSCPs (see below), the results of this study allow us to offer one possible explanation to the overall diversity of HFB‐ and CP‐encoding genes in the genomes of fungi with different lifestyles. We propose that the critical property of HFBs, CPs, the newly detected HFSs and probably other still undetected SSCPs is their ability to modify the interfaces around hyphae (i.e. in hyphosphere). We show that all T. guizhouense saSSCPs can convert the tested abiotic surfaces to a moderately hydrophilic state and facilitate hyphae attachment to these surfaces. We propose that T. guizhouense and a dozen other environmentally opportunistic Trichoderma spp. have a rich reserve of various surface‐active proteins that can secure their attachment to virtually any surface, thus contributing to the nutritional and ecological versatilities of these fungi. This study showed that several HFBs, CPs and HFSs had very similar surface‐modulating abilities. They all transformed the highly hydrophobic surface of PET (which can resemble a plant or insect cuticle) and hydrophilic glass to the same moderately hydrophilic state. However, the 16 tested genes encoding saSSCPs from different families had unequal expression and regulation profiles. This brings us to the hypothesis that for fungi that perform most of their life‐sustaining functions directly through their body surfaces, suitable interface hydrophobicity in hyphosphere is a critical issue. The regulation of saSSCPs is likely fine‐tuned to particular environmental conditions around the hypha, making its surface and the surfaces around it sufficiently hydrophilic for nutrient absorption or coverage in a hydrophobic ‘raincoat’ to protect it from stressors. This hypothesis can be tested in future studies.
This study showed that all saSSCPs improved the attachment of Trichoderma hyphae to a hydrophilic glass surface. Due to the fact that fungi always secrete a diversity of saSSCPs, tests with initially hydrophobic surfaces would not be feasible, as all such surfaces are converted to a hydrophilic state by the activity of such proteins. However, attachment to roots appeared to be a more complicated process. The recombinant HFBs facilitated the attachment of T. guizhouense to tomato roots, while EPL1 and HFS1 prevented attachment. With or without saSSCPs, T. harzianum could not attach to tomato roots. Similar results of improved colonization were obtained when the CP epl1 gene was deleted from T. guizhouense (Gao et al., 2020).
Interestingly, we found several other reports of improved root colonization by fungal mutants lacking one or another CPs (Gao et al., 2020) and cases when the lack of HFBs was associated with a reduced ability to attach to roots (Viterbo and Chet, 2006; Guzmán‐Guzmán et al., 2017). Together, this suggests that it is not EPL1 or HSF1 that prevents the attachment to roots, but rather that plants counteract the attachment process, which we explain by the actions of plant defence mechanisms. The surfaces of roots, in particular root hairs, must be suitable for the efficient absorption of water. The abundant surface‐modulating proteins secreted directly into the rhizosphere by fungi such as Trichoderma can alter the proper state of the root surfaces and impede plant growth. In this study, we demonstrated that the addition of recombinant saSSCPs to hydroponic medium has a significant adverse effect on plant growth. We also showed that those proteins that prevented the attachment of T. guizhouense to roots (EPL1 and HFS1) also triggered an immune response in the tomato plants, while the HFBs went unrecognized by the tomato immune system. The usual silence of these genes during submerged growth may explain the lack of a tomato response to HFBs (Cai et al., 2020). This fits the possibility that roots are usually confronted with CPs and HFSs but not HFBs. As a result, they may lack an efficient system for HFB recognition. Interestingly, the complete secretomes of T. guizhouense and the fungus itself were beneficial for tomato growth, likely due to the presence of such secondary metabolites as plant hormones (Vinale et al., 2009; Cai et al., 2013) and other small molecules (Harman et al., 2004; Li et al., 2015). We assume that under natural conditions, the outcome of fungal–plant interactions for the partners is determined by the trade‐offs between fungal‐beneficial and plant‐beneficial processes and that the balance is determined by the environmental conditions, which in turn, trigger the secretion of a suitable saSSCP cocktail.
We also detected a new family (HFSs) of saSSCPs with an amphiphilic nature similar to that of the HFBs and CPs. Their hydropathy profiles resemble those of relatively hydrophilic HFBs such as HFB6, HFB9a, HFB9b and HFB10. The evolutionary analysis performed for the specific HFS‐encoding genes (Fig. 3) showed that they are commonly present in all Pezizomycotina fungi genomes and have no specificity towards a particular lifestyle. The evolutionary history indicated numerous LGT events and duplications, supporting the functional necessity of these proteins in fungi. Similar results were obtained when the phylogeny of the CPs in Pezizomycotina was studied (Gao et al., 2020).
In conclusion, we would like to highlight that SSCPs in general, and the surface‐active members (saSSCPs) of this group in particular, are unique fungal proteins. Therefore, they should be considered from the perspective of hyphal needs rather than the response of other organisms to their presence. We propose that the saSSCPs studied here constitute a part of a larger group of hyphosphere‐specific proteins that modulate hyphal attachment to surfaces and interactions with other organisms. This work provides initial results in the investigation of a protein‐based surface recognition system in fungi that results in a sophisticated cocktail of surface‐active proteins altering the environment around the hyphae.
Experimental procedures
Microbial strains, plant materials and cultivation conditions
Trichoderma guizhouense strain NJAU 4742 (Zhang et al., 2016; Zhang et al., 2019; Cai et al., 2020; Gao et al., 2020; Pang et al., 2020), which is commercially available as a biofertilizer agent in China, was used as the wild type throughout this work. The sibling species T. harzianum (CBS 226.95) was also included as a reference strain when performing the colonization experiment. To express the Trichoderma genes in yeast, the Pichia pastoris strain KM71H from the EasySelect™ Pichia Expression Kit (Invitrogen) was used, with the plasmid pPICZαA used as the backbone vector. Pichia pastoris strains PpEPL1 (Gao et al., 2020), PpHFB2 and PpHFB4 (Cai et al., 2021) which produce a recombinant EPL1, HFB2 and HFB4 respectively, were generated in our previous works. If not otherwise stated, the Trichoderma strains were maintained on potato dextrose agar (BD Difco, USA) at 25 °C. The Pichia strains were maintained on yeast extract peptone dextrose agar (Sigma‐Aldrich, USA) and fermented in a buffered minimal medium containing glycerol. Protein production was induced by methanol when needed, as described previously (Zhang et al., 2016; Gao et al., 2020). All strains used in this research are listed in Supporting Information 1 Table S3. Tomato seedlings of Solanum lycopersicum cv. HEZUO903 were cultivated in a hydroponic system filled with a 30% Murashige and Skoog basal salt mixture medium (MS; Sigma‐Aldrich), which was maintained under controlled conditions of 25 °C with cycled illumination (light:darkness = 12 h:12 h).
Plant growth promotion assay
The effect on plant growth from inoculating T. guizhouense NJAU 4742 and its metabolites was evaluated by adding 0.4 ml of 106 ml−1 spores or 0.4 ml of 5‐day 30% MS (plus 1% glucose) fermenting broth (without fungal cells) to 40 ml of MS medium supplemented with 1% glucose in the hydroponic system. In this test, 3‐week‐old tomato seedlings showing equal growth were selected and treated as described, and untreated plants were used as the control. The seedlings were allowed to grow for another 3 weeks. Data regarding the plant height and fresh weight were recorded at the end of the experiment. The root growth parameters were determined with a root scanner (Epson Perfection V700 photo, Seiko Epson, Japan).
Standard molecular techniques
The RNeasy Plant Mini Kit (Qiagen, Germany) was used to extract total RNAs from fungal or plant materials according to the manufacturer's instructions. The RevertAid™ First Strand cDNA Kit (Thermo Scientific, USA) was used to synthesize cDNAs with an oligo (dT)18 primer. The qPCR reactions (20 μl) were performed with 0.5 μM of each primer, 100 ng cDNA and 10 μl of iQ™ SYBR Green PCR super mix (BioRad, USA). The thermal program was implemented in a qTOWER real‐time PCR system (Jena Analytics, Germany) with a 3 min initial denaturing at 95 °C followed by 40 cycles of 15 s at 95 °C and 30 s at 60 °C. The specificity of each qPCR reaction was verified by running a melting curve from 55 to 95 °C. PCR reactions (20 μl) contained 0.5 μM of each primer, 100 ng of template DNA, 1× Phanta Max Buffer mix and 2 U Phanta Max Super‐Fidelity DNA Polymerase (Vazyme Biotech, China). The thermal cycling parameters consisted of a 3 min initial denaturing step at 95 °C; 30 cycles of 15 s at 95 °C, 15 s at 59 °C and 30 s at 72 °C; and a 5 min final extension at 72 °C. Supporting Information 1 Table S4 provides the primer information.
Expression of Trichoderma genes in Pichia pastoris
The encoding region of the gene of interest was amplified from T. guizhouense NJAU 4742 cDNA. The signal peptides were predicted by SignalP 5.0 (http://www.cbs.dtu.dk/services/SignalP/) and excluded from the amplicons. Each amplified fragment was cloned into the pPICZαA plasmid between the EcoR I and Xba I sites. The recombinant proteins coded by this construction harbour the yeast α‐factor signal sequence at the N‐terminus and a 6× His epitope at the C‐terminus. The resulting plasmids were transformed into P. pastoris KM71H by standard electroporation transformation, and one positive transformant expressing the designated recombinant protein was chosen for all subsequent experiments. Detailed mutant construction and confirmation are given in Supporting Information 2 Figs S4 and S5. The recombinant proteins were purified as described in Przylucka et al. (2017b), concentrated via Vivaspin ultrafiltration devices (Sartorius, Germany) and re‐suspended in 100 mM potassium phosphate buffer (PB; pH 5.8) or milli‐Q water.
Protein biochemical assays
Protein samples were analysed by 15% SDS‐PAGE followed by silver staining using the SilverQuest™ Silver Staining Kit (Life Technologies, Germany). The proteins separated by SDS‐PAGE were further confirmed by blotting onto an Immobilon®‐PSQ Transfer Membrane (PVDF, 0.2 μm; Merck Millipore, USA) and immunologically visualized by binding with a mouse His‐tag horseradish peroxidase antibody (GenScript, China) as described by Gao et al. (2020). The protein concentration was quantified with the BCA protein assay kit (Thermo Fisher Scientific, USA) according to the manufacturer's instructions.
Protein biophysical assays
The soluble state of each recombinant protein in milli‐Q water was measured by DLS in a Malvern Zetasizer Nano‐ZS (Malvern, UK) at 25 °C. Proteins were prepared at a concentration of 0.6 g L−1 and filtrated through a Whatman® 0.22 μm filter (Sigma‐Aldrich). The mass weight (%) diameters of the proteins were obtained from three measurements. Milli‐Q water without proteins was used as the control.
The surface activity (modification of a material's surface hydrophobicity) of the recombinant proteins was determined by WCA measurements using a Krüss EasyDrop DSA20E (Krüss GmbH, Germany). Four replicates of each surface material (glass or PET) were prepared as described by Espino‐Rammer et al. (2013) with small modifications. Briefly, the surface materials were trimmed to a size of 1.5 × 0.5 cm2 and defatted with 70% (vol./vol.) ethanol. They were then washed in sequence with 5% (wt./vol.) Triton X‐100, 100 mM Na2CO3 and deionized water, with each wash step performed for 30 min at 50 °C. The surfaces were coated with the recombinant proteins by applying a 10 μM (dissolved in PB) concentration of the protein and incubated at 25 °C for 12 h. The coated materials were washed twice with deionized water and dried before the WCA measurements.
In vitro protein bioactivity assays
The in vitro effects of the selected saSSCPs on plant growth and immune response were determined using the hydroponic system as described above. The proteins were applied to 3‐week‐old tomato seedlings at concentrations of 0.1 and 1 μM. In the plant growth promotion test, the seedlings were allowed to grow for an additional 3 weeks, while the seedlings used for testing the immune response were sampled for RNA extraction 48 h after applying the protein. The expression of genes corresponding to different plant defence pathways was monitored with RT‐qPCR as described above. The bioactivity of each SSCP to assist Trichoderma (T. guizhouense NJAU 4742 and T. harzianum CBS 226.95) colonization was investigated by quantifying the tef1 gene copy number of each strain colonized on roots and on an artificial material, i.e. glass wool (see detailed methodology in our previous work; (Gao et al., 2020)).
Genome mining and evolutionary analysis
Genome mining for HFB‐encoding genes and the new SSCP family ‐ HFS‐encoding genes in the 42 Trichoderma whole‐genome sequenced strains (listed in Supporting Information 1 Table S2) ‐ was performed using RapidMiner (version 8.2, USA) with the HFB‐specific cysteine sequence pattern of C‐CC‐C‐C‐CC‐C or the HFS‐specific pattern of C‐CXXX‐C‐C‐C‐C‐C‐C (X represents any possible amino acid). In addition, EffHunter (Carreón‐Anguiano et al., 2020) was used to search for effector proteins that were then manually verified by the presence of a secretory signal peptide (SignalP 5.0). The CP sequences were obtained from our previous report (Gao et al., 2020). The computation of the theoretical isoelectric point (pI) and molecular weight (M w) of the SSCPs from T. guizhouense was performed using the online Compute pI/Mw tool (https://web.expasy.org/compute_pi/). The grand average of hydropathy (GRAVY) value for each protein sequence was calculated using the GRAVY Calculator (http://www.gravy-calculator.de/index.php). Detailed hydropathy profiles were calculated using https://web.expasy.org/protscale/. The possibility of a transmembrane region for each protein was checked using an online prediction program (http://www.detaibio.com/tools/index.php?r=transmembrane%2Findex). The sequences of each protein family were aligned using the MUSCLE algorithm integrated into AliView 1.23 (Edgar, 2004; Larsson, 2014). A ML phylogenetic tree was constructed for the new proteins using IQ‐TREE 1.6.12 (ultrafast bootstrap, N = 1000; (Nguyen et al., 2015)). The Bayesian Information Criterion was applied when searching for the best amino acid substitution model with ModelFinder (integrated into the IQ‐TREE program; (Kalyaanamoorthy et al., 2017)). An evolutionary analysis that included the NOTUNG and T‐Rex programs was performed as described in our previous works (Druzhinina et al., 2018; Gao et al., 2020). In the NOTUNG analysis, the costs of GL, GD and LGT were set at a ratio of 1:3:9, which strictly constrains the possibility of GD and LGT events.
Statistical analysis
The relative expression fold changes of the genes of interest were calculated according to the 2‐ΔΔCt method using tef1 as the housekeeping gene (Cai et al., 2020; Gao et al., 2020). The mean and standard deviation were calculated and statistically analysed by ANOVA and Tukey multiple comparison tests at p < 0.05 using STATISTICA 6 software (StatSoft, USA). The bar plot, heatmap and PCA were performed with R version 3.2.2 (https://cran.r‐project.org/bin/windows/base/old/3.2.2/). Unless otherwise stated, the data regarding plant growth, root colonization and immune response were obtained from at least three biological replicates.
Author Contributions
I.S.D., F.C. and G.B.A. conceived and designed the study. Z.Z., F.C., R.G., M.D., S.J., P.C., G.P. and G.B.A. performed the experiments. I.S.D., F.C. and Z.Z. performed the data analysis and prepared the figures. F.C. and Z.Z. performed the phylogenetic analysis with the assistance of K.C. and I.S.D. I.S.D. and FC wrote the manuscript, with comments from Q.S. and G.B.A. All authors read and approved the manuscript.
Supporting information
Table S1. Small secreted cysteine‐rich protein (SSCP) sequences.
Table S2. Strains used in this study.
Table S3. Primers used in this study.
Table S4. Genomes used in this study.
Fig. S1. Western blot (WB) confirmation of the transformed P. pastoris strains producing recombinant SSCPs (HFB12 or HFS1). Dashed line highlights the aggregated rHFB12 in the sample loading well. The corresponding silver‐stained SDS‐PAGE was shown in Fig. 4. WB visualization was performed by the ONE‐HOUR Western™ Standard Kit (GenScript, China). HRP conjugated Anti‐His Tag Mouse Monoclonal antibody (Invitrogen, USA) was used at a dilution of 1:2000. PageRuler™ Prestained Protein Ladder (Fermentas, USA) was used in the gel electrophoresis.
Fig. S2. The hydrodynamic diameter of each recombinant protein in milli‐Q water. The mass weight (%) diameter was measured by dynamic light scatting (DLS) in a Malvern Zetasizer Nano‐ZS (Malvern, UK) at 25 °C. Proteins were prepared in a concentration of 0.6 g L−1. Milli‐Q water without proteins was used as the control.
Fig. S3. Impact of Trichoderma guizhouense NJAU 4742 inoculation and its secretome on plant growth. To see, whether saSSCPs impact the plant‐growth promoting effect of T. guizhouense NJAU 4742 we collected the culture filtrate (secretome, 1% Sec) corresponding to the submerged stage growth stage when most of saSSCPs were expressed (Table 2). The spore suspension of T. guizhouense NJAU 4742 (104 spores ml−1) (Tri) and the cell‐free Sec to the roots of tomato seedlings cultivated in a hydroponic system. Values (mean ± Sd) labelled with the same letter do not statistically significantly differ (ANOVA, p < 0.05).
Fig. S4. Schematic diagram of expressing SSCP‐encoding genes under a methanol‐inducible promoter (PAOX1) in P. pastoris. α‐Factor, native Saccharomyces cerevisiae α‐factor secretion signal peptide; TAOX1, native transcription termination from AOX1 gene of P. pastoris; Sh ble cassette, ble gene from Streptoalloteichus hindustanus driving resistance to zeocin. Heterologous expression of SSCP‐encoding genes from T. guizhouense NJAU 4742 in P. pastoris strain KM71H was performed as described in Gao et al. (2020). The amplified gene of interest (hfb12 and hfs1) (without signal peptide or intron sequences) was respectively inserted into the position between the restriction site of EcoR I and Xba I of plasmid pPICZαA. The electroporation resulted in numerous positive transformants on zeocin‐contained (100 μg ml−1) YPD plates (shown in Supporting Information 2 Fig. S4 with three randomly selected mutants).
Fig. S5. PCR verification of Pichia mutants expressing hfb12 or hfs1. PpVEC, represents the mutant obtained by transforming the original vector pPICZαA (without a hfb, epl or hfs gene) to P. pastoris cells; PpHFB12, represents mutants harbouring hfb12; PpHFS1, represents mutants harbouring hfs1. DL2000 DNA marker (Vazyme, China) was used in the gel electrophoresis. PCR products were further confirmed by sequencing.
Acknowledgements
The authors wish to thank Dr. Agnes Przylucka (TU WIEN, Vienna, Austria) for her cooperation in the production of recombinant proteins in Pichia pastoris. The authors thank Prof. Erik Reimhult (University of Natural Resources and Life Sciences, Vienna, Austria) for assistance with the DLS analysis and Prof. Hinrich Grothe and Dr. Laura Felgitsch (TU Wien) for providing access to WCA equipment. This work was supported by grants to F.C. from the National Natural Science Foundation of China (31801939), the Ministry of Science & Technology of Jiangsu Province (BK20180533), and China Postdoctoral Science Foundation (2018M630567). The work in Vienna (Austria) was supported (to I.S.D.) by the Austrian Science Fund (FWF) P25613B20 and the Vienna Science and Technology Fund (WWTF), LS13048.
Contributor Information
Feng Cai, Email: fengcai@njau.edu.cn.
Günseli Bayram Akcapinar, Email: gunseli.akcapinar@acibadem.edu.tr.
Irina S. Druzhinina, Email: irina.s.druzhinina@njau.edu.cn.
References
- Aimanianda, V. , Bayry, J. , Bozza, S. , Kniemeyer, O. , Perruccio, K. , Elluru, S.R. , et al. (2009) Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature 460: 1117–1121. [DOI] [PubMed] [Google Scholar]
- Aimanianda, V. , and Latge, J.P. (2010) Fungal hydrophobins form a sheath preventing immune recognition of airborne conidia. Virulence 1: 185–187. [DOI] [PubMed] [Google Scholar]
- Andersson, K.‐M. , Meerupati, T. , Levander, F. , Friman, E. , Ahrén, D. , and Tunlid, A. (2013) Proteome of the nematode‐trapping cells of the fungus Monacrosporium haptotylum . Appl Environ Microbiol 79: 4993–5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askolin, S. , Penttila, M. , Wosten, H.A. , and Nakari‐Setala, T. (2005) The Trichoderma reesei hydrophobin genes hfb1 and hfb2 have diverse functions in fungal development. FEMS Microbiol Lett 253: 281–288. [DOI] [PubMed] [Google Scholar]
- Bayry, J. , Aimanianda, V. , Guijarro, J.I. , Sunde, M. , and Latgé, J.P. (2012) Hydrophobins ‐ unique fungal proteins. PLoS Pathog 8: e1002700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boc, A. , Diallo, A.B. , and Makarenkov, V. (2012) T‐REX: a web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res 40: W573–W579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonazza, K. , Gaderer, R. , Neudl, S. , Przylucka, A. , Allmaier, G. , Druzhinina, I.S. , et al. (2015) The fungal cerato‐platanin protein EPL1 forms highly ordered layers at hydrophobic/hydrophilic interfaces. Soft Matter 11: 1723–1732. [DOI] [PubMed] [Google Scholar]
- Cai, F. , Chen, W. , Wei, Z. , Pang, G. , Li, R. , Ran, W. , and Shen, Q. (2015) Colonization of Trichoderma harzianum strain SQR‐T037 on tomato roots and its relationship to plant growth, nutrient availability and soil microflora. Plant Soil 388: 337–350. [Google Scholar]
- Cai, F. , and Druzhinina, I.S. (2021) In honor of John Bissett: authoritative guidelines on molecular identification of Trichoderma . Fungal Divers 107: 1–69. [Google Scholar]
- Cai, F. , Gao, R. , Zhao, Z. , Ding, M. , Jiang, S. , Yagtu, C. , et al. (2020) Evolutionary compromises in fungal fitness: hydrophobins can hinder the adverse dispersal of conidiospores and challenge their survival. ISME J 14: 2610–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, F. , Yu, G. , Wang, P. , Wei, Z. , Fu, L. , Shen, Q. , and Chen, W. (2013) Harzianolide, a novel plant growth regulator and systemic resistance elicitor from Trichoderma harzianum . Plant Physiol Biochem 73: 106–113. [DOI] [PubMed] [Google Scholar]
- Cai, F. , Zhao, Z. , Gao, R. , Ding, M. , Jiang, S. , and Gao, Q. (2021) Intracellular hydrophobin vesicles modulate the architecture and longevity of fungal colonies. BioRvix, 10.1101/2020.08.18.255406. [DOI] [Google Scholar]
- Carreón‐Anguiano, K.G. , Islas‐Flores, I. , Vega‐Arreguin, J. , Saenz‐Carbonell, L. , and Canto‐Canche, B. (2020) EffHunter: a tool for prediction of effector protein candidates in fungal proteomic databases. Biomolecules 10: 712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. , Kovalchuk, A. , Keriö, S. , and Asiegbu, F.O. (2013) Distribution and bioinformatic analysis of the cerato‐platanin protein family in Dikarya. Mycologia 105: 1479–1488. [DOI] [PubMed] [Google Scholar]
- Chen, K. , Durand, D. , and Farach‐Colton, M. (2000) NOTUNG: a program for dating gene duplications and optimizing gene family trees. J Comput Biol 7: 429–447. [DOI] [PubMed] [Google Scholar]
- Dagvadorj, B. , Ozketen, A.C. , Andac, A. , Duggan, C. , Bozkurt, T.O. , and Akkaya, M.S. (2017) A Puccinia striiformis f. sp. tritici secreted protein activates plant immunity at the cell surface. Sci Rep 7: 1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwiche, R. , Mène‐Saffrané, L. , Gfeller, D. , Asojo, O.A. , and Schneiter, R. (2017) The pathogen‐related yeast protein Pry1, a member of the CAP protein superfamily, is a fatty acid‐binding protein. J Biol Chem 292: 8304–8314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Man, T.J.B. , Stajich, J.E. , Kubicek, C.P. , Teiling, C. , Chenthamara, K. , Atanasova, L. , et al. (2016) Small genome of the fungus Escovopsis weberi, a specialized disease agent of ant agriculture. Proc Natl Acad Sci U S A 113: 3567–3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druzhinina, I.S. , Chenthamara, K. , Zhang, J. , Atanasova, L. , Yang, D.Q. , Miao, Y.Z. , et al. (2018) Massive lateral transfer of genes encoding plant cell wall‐degrading enzymes to the mycoparasitic fungus Trichoderma from its plant‐associated hosts. PLoS Genet 14: e1007322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druzhinina, I.S. , Seidl‐Seiboth, V. , Herrera‐Estrella, A. , Horwitz, B.A. , Kenerley, C.M. , Monte, E. , et al. (2011) Trichoderma: the genomics of opportunistic success. Nat Rev Microbiol 9: 749–759. [DOI] [PubMed] [Google Scholar]
- Edgar, R.C. (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espino‐Rammer, L. , Ribitsch, D. , Przylucka, A. , Marold, A. , Greimel, K.J. , Herrero Acero, E. , et al. (2013) Two novel class II hydrophobins from Trichoderma spp. stimulate enzymatic hydrolysis of poly(ethylene terephthalate) when expressed as fusion proteins. Appl Environ Microbiol 79: 4230–4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, A. , Gao, H. , Zhang, N. , Zheng, X. , Qiu, S. , Li, Y. , et al. (2019) A novel effector gene SCRE2 contributes to full virulence of Ustilaginoidea virens to rice. Front Microbiol 10: 845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman, D. , Amedi, N. , Carmeli, S. , Yarden, O. , and Hadar, Y. (2019) Manipulating the expression of small secreted protein 1 (Ssp1) alters patterns of development and metabolism in the white‐rot fungus Pleurotus ostreatus . Appl Environ Microbiol 85: e00761‐00719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman, D. , Kowbel, D.J. , Glass, N.L. , Yarden, O. , and Hadar, Y. (2017) A role for small secreted proteins (SSPs) in a saprophytic fungal lifestyle: Ligninolytic enzyme regulation in Pleurotus ostreatus . Sci Rep 7: 14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman, D. , Yarden, O. , and Hadar, Y. (2020) Seeking the roles for fungal small‐secreted proteins in affecting saprophytic lifestyles. Front Microbiol 11: 455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frías, M. , González, C. , and Brito, N. (2011) BcSpl1, a cerato‐platanin family protein, contributes to Botrytis cinerea virulence and elicits the hypersensitive response in the host. New Phytol 192: 483–495. [DOI] [PubMed] [Google Scholar]
- Frischmann, A. , Neudl, S. , Gaderer, R. , Bonazza, K. , Zach, S. , Gruber, S. , et al. (2013) Self‐assembly at air/water interfaces and carbohydrate binding properties of the small secreted protein EPL1 from the fungus Trichoderma atroviride . J Biol Chem 288: 4278–4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaderer, R. , Bonazza, K. , and Seidl‐Seiboth, V. (2014) Cerato‐platanins: a fungal protein family with intriguing properties and application potential. Appl Microbiol Biotechnol 98: 4795–4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, R. , Ding, M. , Jiang, S. , Zhao, Z. , Chenthamara, K. , Shen, Q. , et al. (2020) The evolutionary and functional paradox of cerato‐platanins in the mycoparasitic fungus Trichoderma: high diversity, stabilizing selection, and a minor role in biotic interactions. Appl Environ Microbiol 86: e00696‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigues, S. , Gandía, M. , Castillo, L. , Coca, M. , Marx, F. , Marcos, J.F. , and Manzanares, P. (2018) Three antifungal proteins from Penicillium expansum: different patterns of production and antifungal activity. Front Microbiol 9: 2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunbacher, A. , Throm, T. , Seidel, C. , Gutt, B. , Rohrig, J. , Strunk, T. , et al. (2014) Six hydrophobins are involved in hydrophobin rodlet formation in Aspergillus nidulans and contribute to hydrophobicity of the spore surface. PLoS One 9: e94546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzmán‐Guzmán, P. , Alemán‐Duarte, M.I. , Delaye, L. , Herrera‐Estrella, A. , and Olmedo‐Monfil, V. (2017) Identification of effector‐like proteins in Trichoderma spp. and role of a hydrophobin in the plant‐fungus interaction and mycoparasitism. BMC Genet 18: 16–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdu, D. , Huber, A. , Czajlik, A. , Tóth, L. , Kele, Z. , Kocsubé, S. , et al. (2019) Solution structure and novel insights into phylogeny and mode of action of the Neosartorya (Aspergillus) fischeri antifungal protein (NFAP). Int J Biol Macromol 129: 511–522. [DOI] [PubMed] [Google Scholar]
- Hakanpää, J. , Paananen, A. , Askolin, S. , Nakari‐Setälä, T. , Parkkinen, T. , Penttilä, M. , et al. (2004) Atomic resolution structure of the HFBII hydrophobin, a self‐assembling amphiphile. J Biol Chem 279: 534–539. [DOI] [PubMed] [Google Scholar]
- Harman, G.E. , Howell, C.R. , Viterbo, A. , Chet, I. , and Lorito, M. (2004) Trichoderma species ‐ opportunistic, avirulent plant symbionts. Nat Rev Microbiol 2: 43–56. [DOI] [PubMed] [Google Scholar]
- Holzknecht, J. , Kühbacher, A. , Papp, C. , Farkas, A. , Váradi, G. , Marcos, J.F. , et al. (2020) The Penicillium chrysogenum Q176 antimicrobial protein PAFC effectively inhibits the growth of the opportunistic human pathogen Candida albicans . J Fungi (Basel) 6: 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber, A. , Galgóczy, L. , Váradi, G. , Holzknecht, J. , Kakar, A. , Malanovic, N. , et al. (2020) Two small, cysteine‐rich and cationic antifungal proteins from Penicillium chrysogenum: a comparative study of PAF and PAFB. Biochim Biophys Acta Biomembr 1862: 183246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, J.S. , Mitchell, T.K. , and Dean, R.A. (2007) The Magnaporthe grisea snodprot1 homolog, MSP1, is required for virulence. FEMS Microbiol Lett 273: 157–165. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy, S. , Minh, B.Q. , Wong, T.K.F. , von Haeseler, A. , and Jermiin, L.S. (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14: 587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalesi, M. , Jahanbani, R. , Riveros‐Galan, D. , Sheikh‐Hassani, V. , Sheikh‐Zeinoddin, M. , Sahihi, M. , et al. (2016) Antioxidant activity and ACE‐inhibitory of class II hydrophobin from wild strain Trichoderma reesei . Int J Biol Macromol 91: 174–179. [DOI] [PubMed] [Google Scholar]
- Kim, K.‐T. , Jeon, J. , Choi, J. , Cheong, K. , Song, H. , Choi, G. , et al. (2016) Kingdom‐wide analysis of fungal small secreted proteins (SSPs) reveals their potential role in host association. Front Plant Sci 7: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kombrink, A. , Tayyrov, A. , Essig, A. , Stöckli, M. , Micheller, S. , Hintze, J. , et al. (2019) Induction of antibacterial proteins and peptides in the coprophilous mushroom Coprinopsis cinerea in response to bacteria. ISME J 13: 588–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottmeier, K. , Ostermann, K. , Bley, T. , and Rödel, G. (2011) Hydrophobin signal sequence mediates efficient secretion of recombinant proteins in Pichia pastoris . Appl Microbiol Biotechnol 91: 133–141. [DOI] [PubMed] [Google Scholar]
- Krizsán, K. , Almási, É. , Merényi, Z. , Sahu, N. , Virágh, M. , Kószó, T. , et al. (2019) Transcriptomic atlas of mushroom development reveals conserved genes behind complex multicellularity in fungi. Proc Natl Acad Sci U S A 116: 7409–7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicek, C.P. , Baker, S. , Gamauf, C. , Kenerley, C.M. , and Druzhinina, I.S. (2008) Purifying selection and birth‐and‐death evolution in the class II hydrophobin gene families of the ascomycete Trichoderma/Hypocrea . BMC Evol Biol 8: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicek, C.P. , Herrera‐Estrella, A. , Seidl‐Seiboth, V. , Martinez, D.A. , Druzhinina, I.S. , Thon, M. , et al. (2011) Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma . Genome Biol 12: R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicek, C.P. , Steindorff, A.S. , Chenthamara, K. , Manganiello, G. , Henrissat, B. , Zhang, J. , et al. (2019) Evolution and comparative genomics of the most common Trichoderma species. BMC Genomics 20: 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman, C.P. (2009) Biotechnological strains of Komagataella (Pichia) pastoris are Komagataella phaffii as determined from multigene sequence analysis. J Ind Microbiol Biotechnol 36: 1435–1438. [DOI] [PubMed] [Google Scholar]
- Kyte, J. , and Doolittle, R.F. (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157: 105–132. [DOI] [PubMed] [Google Scholar]
- Larsson, A. (2014) AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30: 3276–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, R.X. , Cai, F. , Pang, G. , Shen, Q.R. , Li, R. , and Chen, W. (2015) Solubilisation of phosphate and micronutrients by Trichoderma harzianum and its relationship with the promotion of tomato plant growth. PLoS One 10: e0130081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. , Dong, Y. , Li, L. , Zhang, Y. , Yang, X. , Zeng, H. , et al. (2019) The novel Cerato‐platanin‐like protein foccp1 from Fusarium oxysporum triggers an immune response in plants. Int J Mol Sci 20: 2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder, M.B. , Szilvay, G.R. , Nakari‐Setälä, T. , and Penttilä, M.E. (2005) Hydrophobins: the protein‐amphiphiles of filamentous fungi. FEMS Microbiol Rev 29: 877–896. [DOI] [PubMed] [Google Scholar]
- Lugones, L.G. , de Jong, J.F. , de Vries, O.M.H. , Jalving, R. , Dijksterhuis, J. , and Wosten, H.A.B. (2004) The SC15 protein of Schizophyllum commune mediates formation of aerial hyphae and attachment in the absence of the SC3 hydrophobin. Mol Microbiol 53: 707–716. [DOI] [PubMed] [Google Scholar]
- Lugones, L.G. , Wösten, H.A.B. , Birkenkamp, K.U. , Sjollema, K.A. , Zagers, J. , and Wessels, J.G.H. (1999) Hydrophobins line air channels in fruiting bodies of Schizophyllum commune and Agaricus bisporus . Mycol Res 103: 635–640. [Google Scholar]
- Nguyen, L.T. , Schmidt, H.A. , von Haeseler, A. , and Minh, B.Q. (2015) IQ‐TREE: a fast and effective stochastic algorithm for estimating maximum‐likelihood phylogenies. Mol Biol Evol 32: 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang, G. , Sun, T. , Yu, Z. , Yuan, T. , Liu, W. , Zhu, H. , et al. (2020) Azaphilones biosynthesis complements the defence mechanism of Trichoderma guizhouense against oxidative stress. Environ Microbiol 22: 4808–4824. [DOI] [PubMed] [Google Scholar]
- Pazzagli, L. , Cappugi, G. , Manao, G. , Camici, G. , Santini, A. , and Scala, A. (1999) Purification, characterization, and amino acid sequence of cerato‐platanin, a new phytotoxic protein from Ceratocystis fimbriata f. sp. platani . J Biol Chem 274: 24959–24964. [DOI] [PubMed] [Google Scholar]
- Pellegrin, C. , Morin, E. , Martin, F.M. , and Veneault‐Fourrey, C. (2015) Comparative analysis of secretomes from ectomycorrhizal fungi with an emphasis on small‐secreted proteins. Front Microbiol 6: 1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przylucka, A. , Akcapinar, G.B. , Bonazza, K. , Mello‐de‐Sousa, T.M. , Mach‐Aigner, A.R. , Lobanov, V. , et al. (2017b) Comparative physiochemical analysis of hydrophobins produced in Escherichia coli and Pichia pastoris . Colloids Surf B Biointerfaces 159: 913–923. [DOI] [PubMed] [Google Scholar]
- Przylucka, A. , Akcapinar, G.B. , Chenthamara, K. , Cai, F. , Grujic, M. , Karpenko, J. , et al. (2017a) HFB7 ‐ a novel orphan hydrophobin of the Harzianum and Virens clades of Trichoderma, is involved in response to biotic and abiotic stresses. Fungal Genet Biol 102: 63–76. [DOI] [PubMed] [Google Scholar]
- Quarantin, A. , Castiglioni, C. , Schäfer, W. , Favaron, F. , and Sella, L. (2019) The Fusarium graminearum cerato‐platanins loosen cellulose substrates enhancing fungal cellulase activity as expansin‐like proteins. Plant Physiol Biochem 139: 229–238. [DOI] [PubMed] [Google Scholar]
- Rohe, M. , Gierlich, A. , Hermann, H. , Hahn, M. , Schmidt, B. , Rosahl, S. , and Knogge, W. (1995) The race‐specific elicitor, NIP1, from the barley pathogen, Rhynchosporium secalis, determines avirulence on host plants of the Rrs1 resistance genotype. EMBO J 14: 4168–4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado, I.V. , Rey, M. , Codón, A.C. , Govantes, J. , Moreno‐Mateos, M.A. , and Benítez, T. (2007) QID74 cell wall protein of Trichoderma harzianum is involved in cell protection and adherence to hydrophobic surfaces. Fungal Genet Biol 44: 950–964. [DOI] [PubMed] [Google Scholar]
- Ruocco, M. , Lanzuise, S. , Lombardi, N. , Woo, S.L. , Vinale, F. , Marra, R. , et al. (2015) Multiple roles and effects of a novel Trichoderma hydrophobin. Mol Plant Microbe Interact 28: 167–179. [DOI] [PubMed] [Google Scholar]
- Seidl‐Seiboth, V. , Gruber, S. , Sezerman, U. , Schwecke, T. , Albayrak, A. , Neuhof, T. , et al. (2011) Novel hydrophobins from Trichoderma define a new hydrophobin subclass: protein properties, evolution, regulation and processing. J Mol Evol 72: 339–351. [DOI] [PubMed] [Google Scholar]
- Sun, P. , Li, X. , Yang, M. , Zhao, X. , Zhang, Z. , and Wei, D. (2020) Deletion of a small, secreted and cysteine‐rich protein Cpl1 leads to increased invasive growth of Cryptococcus neoformans into nutrient agar. Microbiol Res 241: 126570. [DOI] [PubMed] [Google Scholar]
- van der Vegt, W. , van der Mei, H.C. , Wösten, H.A. , Wessels, J.G. , and Busscher, H.J. (1996) A comparison of the surface activity of the fungal hydrophobin SC3p with those of other proteins. Biophys Chem 57: 253–260. [DOI] [PubMed] [Google Scholar]
- Vinale, F. , Flematti, G. , Sivasithamparam, K. , Lorito, M. , Marra, R. , Skelton, B.W. , and Ghisalberti, E.L. (2009) Harzianic acid, an antifungal and plant growth promoting metabolite from Trichoderma harzianum . J Nat Prod 72: 2032–2035. [DOI] [PubMed] [Google Scholar]
- Viterbo, A. , and Chet, I. (2006) TasHyd1, a new hydrophobin gene from the biocontrol agent Trichoderma asperellum, is involved in plant root colonization. Mol Plant Pathol 7: 249–258. [DOI] [PubMed] [Google Scholar]
- Wang, W. , Deng, Z. , Wu, H. , Zhao, Q. , Li, T. , Zhu, W. , et al. (2019) A small secreted protein triggers a TLR2/4‐dependent inflammatory response during invasive Candida albicans infection. Nat Commun 10: 1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteford, J.R. , Lacroix, H. , Talbot, N.J. , and Spanu, P.D. (2004) Stage‐specific cellular localisation of two hydrophobins during plant infection by the pathogenic fungus Cladosporium fulvum . Fungal Genet Biol 41: 624–634. [DOI] [PubMed] [Google Scholar]
- Winefield, R.D. , Hilario, E. , Beever, R.E. , Haverkamp, R.G. , and Templeton, M.D. (2007) Hydrophobin genes and their expression in conidial and aconidial Neurospora species. Fungal Genet Biol 44: 250–257. [DOI] [PubMed] [Google Scholar]
- Wösten, H.A. (2001) Hydrophobins: multipurpose proteins. Annu Rev Microbiol 55: 625–646. [DOI] [PubMed] [Google Scholar]
- Yu, C. , Dou, K. , Wang, S. , Wu, Q. , Ni, M. , Zhang, T. , et al. (2020) Elicitor hydrophobin Hyd1 interacts with Ubiquilin1‐like to induce maize systemic resistance. J Integr Plant Biol 62: 509–526. [DOI] [PubMed] [Google Scholar]
- Zajc, J. , Liu, Y. , Dai, W. , Yang, Z. , Hu, J. , Gostinčar, C. , and Gunde‐Cimerman, N. (2013) Genome and transcriptome sequencing of the halophilic fungus Wallemia ichthyophaga: haloadaptations present and absent. BMC Genomics 14: 617–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Bayram Akcapinar, G. , Atanasova, L. , Rahimi, M.J. , Przylucka, A. , Yang, D. , et al. (2016) The neutral metallopeptidase NMP1 of Trichoderma guizhouense is required for mycotrophy and self‐defence. Environ Microbiol 18: 580–597. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Miao, Y. , Rahimi, M.J. , Zhu, H. , Steindorff, A. , Schiessler, S. , et al. (2019) Guttation capsules containing hydrogen peroxide: an evolutionarily conserved NADPH oxidase gains a role in wars between related fungi. Environ Microbiol 21: 2644–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Small secreted cysteine‐rich protein (SSCP) sequences.
Table S2. Strains used in this study.
Table S3. Primers used in this study.
Table S4. Genomes used in this study.
Fig. S1. Western blot (WB) confirmation of the transformed P. pastoris strains producing recombinant SSCPs (HFB12 or HFS1). Dashed line highlights the aggregated rHFB12 in the sample loading well. The corresponding silver‐stained SDS‐PAGE was shown in Fig. 4. WB visualization was performed by the ONE‐HOUR Western™ Standard Kit (GenScript, China). HRP conjugated Anti‐His Tag Mouse Monoclonal antibody (Invitrogen, USA) was used at a dilution of 1:2000. PageRuler™ Prestained Protein Ladder (Fermentas, USA) was used in the gel electrophoresis.
Fig. S2. The hydrodynamic diameter of each recombinant protein in milli‐Q water. The mass weight (%) diameter was measured by dynamic light scatting (DLS) in a Malvern Zetasizer Nano‐ZS (Malvern, UK) at 25 °C. Proteins were prepared in a concentration of 0.6 g L−1. Milli‐Q water without proteins was used as the control.
Fig. S3. Impact of Trichoderma guizhouense NJAU 4742 inoculation and its secretome on plant growth. To see, whether saSSCPs impact the plant‐growth promoting effect of T. guizhouense NJAU 4742 we collected the culture filtrate (secretome, 1% Sec) corresponding to the submerged stage growth stage when most of saSSCPs were expressed (Table 2). The spore suspension of T. guizhouense NJAU 4742 (104 spores ml−1) (Tri) and the cell‐free Sec to the roots of tomato seedlings cultivated in a hydroponic system. Values (mean ± Sd) labelled with the same letter do not statistically significantly differ (ANOVA, p < 0.05).
Fig. S4. Schematic diagram of expressing SSCP‐encoding genes under a methanol‐inducible promoter (PAOX1) in P. pastoris. α‐Factor, native Saccharomyces cerevisiae α‐factor secretion signal peptide; TAOX1, native transcription termination from AOX1 gene of P. pastoris; Sh ble cassette, ble gene from Streptoalloteichus hindustanus driving resistance to zeocin. Heterologous expression of SSCP‐encoding genes from T. guizhouense NJAU 4742 in P. pastoris strain KM71H was performed as described in Gao et al. (2020). The amplified gene of interest (hfb12 and hfs1) (without signal peptide or intron sequences) was respectively inserted into the position between the restriction site of EcoR I and Xba I of plasmid pPICZαA. The electroporation resulted in numerous positive transformants on zeocin‐contained (100 μg ml−1) YPD plates (shown in Supporting Information 2 Fig. S4 with three randomly selected mutants).
Fig. S5. PCR verification of Pichia mutants expressing hfb12 or hfs1. PpVEC, represents the mutant obtained by transforming the original vector pPICZαA (without a hfb, epl or hfs gene) to P. pastoris cells; PpHFB12, represents mutants harbouring hfb12; PpHFS1, represents mutants harbouring hfs1. DL2000 DNA marker (Vazyme, China) was used in the gel electrophoresis. PCR products were further confirmed by sequencing.


