See “Prolonged impairment of short-chain fatty acid and L-isoleucine biosynthesis in gut microbiome in patients with COVID-19” by Zhang F, Wan Y, Zuo T, et al, on page 548.
The metabolic byproducts and extraordinary coding potential of the dense microbial community inhabiting the gastrointestinal tract have led investigators to describe the gut microbiota as the “forgotten organ,” highlighting the need to consider how its functions are disrupted in disease conditions. Although the ability of intestinal bacteria to competitively exclude enteric pathogens through colonization resistance is now established, numerous studies just in the last decade have uncovered the impact of the gut microbiota on antiviral responses at distal mucosal sites, including the lung. In particular, mouse models demonstrate that efficient activation of both innate and adaptive immune responses during influenza virus infection rely on an intact gut microbiota.1, 2, 3 This long-range protection conferred by the microbiota has been attributed to metabolites secreted by specific bacterial species,4 raising the possibility that interindividual differences in microbial community composition influence the severity of viral lung infections.
At the dawn of the pandemic, it was evident that SARS-CoV-2 caused a devastating respiratory disease in a subset of individuals, with increasing evidence showing that its impacts included significant changes in the gut microbiome composition.5, 6, 7 COVID-19–associated dysbiosis of the microbiota is characterized by loss of commensal bacterial taxa known to have immunomodulatory activity, such as short-chain fatty acid (SCFA) production. However, it is often difficult to distinguish direct effects of viral infection from the impact of hospitalization and treatment on the microbiome, and the functional significance of these findings required additional investigation.
In this issue of Gastroenterology, Zhang et al,8 demonstrate the persistence of functional metabolic consequences of dysbiosis associated with severe COVID-19 infection, thereby solidifying the link between microbiome disruption and disease severity. Their patient cohort included 66 hospitalized patients with COVID-19 representing the spectrum of disease severities—mild, moderate, and severe or critical. Importantly, they addressed changes to the gut microbiome that may occur due to hospitalization by including a control cohort of patients hospitalized with community-acquired pneumonia who were negative for COVID-19. Additionally, all patients were antibiotics-naïve at least 3 months before enrollment, allowing the investigators to decouple the effects of SARS-CoV-2–associated microbial dysbiosis from those precipitated by broad-spectrum antibiotics that are frequently administered to individuals hospitalized for COVID-19.
Using ultradeep metagenomic sequencing of serial stool samples taken from patients with COVID-19 during hospitalization, the authors first defined major changes in gut microbiome composition associated with SARS-CoV-2 infection. In line with previous studies, they found that the gut microbiome of patients with COVID-19 was characterized by a depletion of Bifidobacterium adolescentis, Ruminococcus bromii, and Faecalibacterium prausnitzii, with the latter 2 species showing negative correlations with disease severity. From this sequencing data set, they were able to infer that SCFA and L-isoleucine biosynthesis pathways were depleted in COVID-19 patients with severe disease.
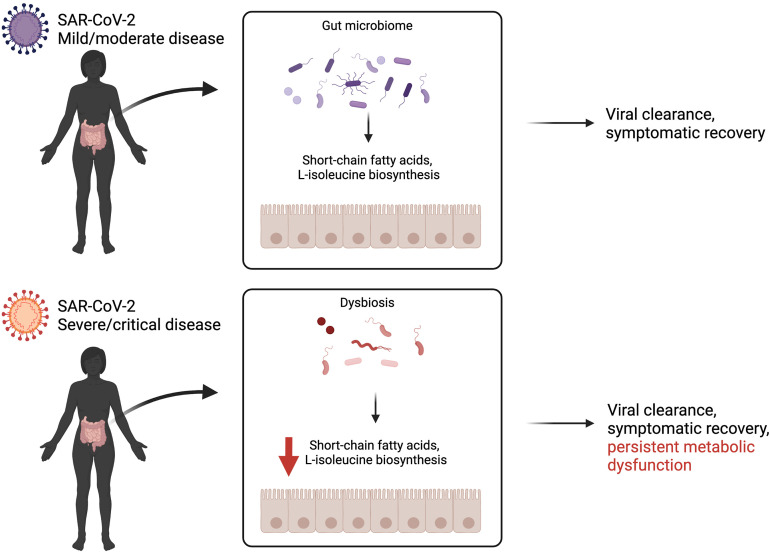
To examine consequences on host immunity downstream of these changes in coding potential of the microbiome, the authors quantified plasma concentrations of inflammatory markers and fecal metabolites. This analysis demonstrated that decreased fecal butyrate levels were associated with increased plasma levels of the cytokines interleukin 10 and C-X-C motif chemokine 10 and the acute-phase reactant C-reactive protein. The authors further validate the reductions in SCFAs and L-isoleucine in longitudinal stool specimens and identify a remarkable persistence of this metabolic dysfunction at time points in which the virus is no longer detectable, up to 41 days after hospital discharge. Together, these findings reveal a direct link between severity of COVID-19 infection and persistent impairment of the metabolic output of the gut microbiota (Figure 1 ).
Figure 1.
Severity of COVID-19 infection is associated with persistent impaired gut microbial metabolism, despite viral clearance and symptomatic recovery. Zhang et al8 find that pathways implicated in short-chain fatty acid and L-isoleucine biosynthesis are significantly depleted in COVID-19 patients with severe disease. Created with BioRender.com.
An important aspect of this study is that it shows that the previously described reduction in diversity of intestinal bacterial communities in patients with COVID-19 leads to consequential changes in the levels of metabolites with known functions in immunity. SCFAs derived from bacteria such as F prausnitzii, a bacterial species reproducibly reduced in patients hospitalized with COVID-19, have received intense scrutiny as mediators of regulatory T-cell differentiation and epithelial gene expression.9 Although less studied in the context of the microbiota-immune axis, numerous effects of L-isoleucine supplementation on immunity have also been documented.10
Therefore, an important research direction would be to determine whether the gut microbiota dysbiosis observed in patients with severe COVID-19 affects the progression and recovery from the disease. It may be particularly worthwhile to examine whether reductions in these metabolites precede certain disease manifestations. Also, it is possible that the prolonged reduction in such microbial metabolites contributes to persistent symptoms, such as respiratory, neurologic, cardiac, and other problems observed months after the initial infection, a condition broadly referred to as post-acute sequelae of SARS-CoV-2 infection (PASC) or “long COVID.”11, 12, 13, 14 Long-term evaluation of microbiome dysbiosis may reveal links between specific taxa and symptoms of PASC, information that can be used to identify patients needing more vigilant clinical follow-up.
Finally, the insightful data from Zhang et al8 suggest the possibility of applying microbiota-based therapies or metabolite supplementation for treating lung infections. Oral probiotics are protective in mouse models of influenza infection15 , 16 and associated with reduced incidence of acute upper respiratory infections in humans.17 Indeed, clinical trials examining the impact of oral probiotics for COVID-19 are currently underway.18 If pharmacologic barriers and safety concerns can be addressed, targeted and personalized approaches that reverse impaired microbial metabolism, informed by findings such as those uncovered by Zhang et al,8 may not be too far on the horizon for treating respiratory viral infections.
Footnotes
Conflicts of interest These authors disclose the following: Ken Cadwell has received research support from or consulted for Pfizer, Takeda, Pacific Biosciences, Genentech, PureTech Health, and AbbVie, and holds U.S. patent 10,722,600 and provisional patent 62/935,035 and 63/157,225. Mericien Venzon discloses no conflicts.
References
- 1.Abt M.C., Osborne L.C., Monticelli L.A., et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37:158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradley K.C., Finsterbusch K., Schnepf D., et al. Microbiota-driven tonic interferon signals in lung stromal cells protect from influenza virus infection. Cell Rep. 2019;28:245–256.e4. doi: 10.1016/j.celrep.2019.05.105. [DOI] [PubMed] [Google Scholar]
- 3.Ichinohe T., Pang I.K., Kumamoto Y., et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci U S A. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steed A.L., Christophi G.P., Kaiko G.E., et al. The microbial metabolite desaminotyrosine protects from influenza through type I interferon. Science. 2017;357:498–502. doi: 10.1126/science.aam5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zuo T., Zhang F., Lui G.C., et al. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology. 2020;159:944–955.e8. doi: 10.1053/j.gastro.2020.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gu S., Chen Y., Wu Z., et al. Alterations of the gut microbiota in patients with coronavirus disease 2019 or H1N1 influenza. Clin Infect Dis. 2020;71:2669–2678. doi: 10.1093/cid/ciaa709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yeoh Y.K., Zuo T., Lui G.C.-Y., et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. 2021;70:698–706. doi: 10.1136/gutjnl-2020-323020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang F., Wan Y., Zuo T., et al. Prolonged impairment of short-chain fatty acid and L-isoleucine biosynthesis in gut microbiome in patients with COVID-19. Gastroenterology. 2022;162:548–561. doi: 10.1053/j.gastro.2021.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hosseinkhani F., Heinken A., Thiele I., et al. The contribution of gut bacterial metabolites in the human immune signaling pathway of non-communicable diseases. Gut Microbes. 2021;13:1–22. doi: 10.1080/19490976.2021.1882927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gu C., Mao X., Chen D., et al. Isoleucine plays an important role for maintaining immune function. Curr Protein Pept Sci. 2019;20:644–651. doi: 10.2174/1389203720666190305163135. [DOI] [PubMed] [Google Scholar]
- 11.Carfì A., Bernabei R., Landi F. Persistent symptoms in patients after acute COVID-19. JAMA. 2020;324:603–605. doi: 10.1001/jama.2020.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carvalho-Schneider C., Laurent E., Lemaignen A., et al. Follow-up of adults with noncritical COVID-19 two months after symptom onset. Clin Microbiol Infect. 2021;27:258–263. doi: 10.1016/j.cmi.2020.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Townsend L., Dyer A.H., Jones K., et al. Persistent fatigue following SARS-CoV-2 infection is common and independent of severity of initial infection. PLoS One. 2020;15 doi: 10.1371/journal.pone.0240784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiong Q., Xu M., Li J., et al. Clinical sequelae of COVID-19 survivors in Wuhan, China: a single-centre longitudinal study. Clin Microbiol Infect. 2021;27:89–95. doi: 10.1016/j.cmi.2020.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahooti M., Abdolalipour E., Salehzadeh A., et al. Immunomodulatory and prophylactic effects of Bifidobacterium bifidum probiotic strain on influenza infection in mice. World J Microbiol Biotechnol. 2019;35:1–9. doi: 10.1007/s11274-019-2667-0. [DOI] [PubMed] [Google Scholar]
- 16.Belkacem N., Serafini N., Wheeler R., et al. Lactobacillus paracasei feeding improves immune control of influenza infection in mice. PLoS One. 2017;12 doi: 10.1371/journal.pone.0184976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shahbazi R., Yasavoli-Sharahi H., Alsadi N., et al. Probiotics in treatment of viral respiratory infections and neuroinflammatory disorders. Molecules. 2020;25:4891. doi: 10.3390/molecules25214891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hung Y.-P., Lee C.-C., Lee J.-C., et al. Gut dysbiosis during COVID-19 and potential effect of probiotics. Microorganisms. 2021;9:1605. doi: 10.3390/microorganisms9081605. [DOI] [PMC free article] [PubMed] [Google Scholar]