Summary
Saprotrophic fungi play an important role in ecosystem functioning and plant performance, but their abundance in intensively managed arable soils is low. Saprotrophic fungal biomass in arable soils can be enhanced with amendments of cellulose‐rich materials. Here, we examined if sawdust‐stimulated saprotrophic fungi extend their activity to the rhizosphere of crop seedlings and influence the composition and activity of other rhizosphere and root inhabitants. After growing carrot seedlings in sawdust‐amended arable soil, we determined fungal and bacterial biomass and community structure in roots, rhizosphere and soil. Utilization of root exudates was assessed by stable isotope probing (SIP) following 13CO2‐pulse‐labelling of seedlings. This was combined with analysis of lipid fatty acids (PLFA/NLFA‐SIP) and nucleic acids (DNA‐SIP). Sawdust‐stimulated Sordariomycetes colonized the seedling's rhizosphere and roots and actively consumed root exudates. This did not reduce the abundance and activity of bacteria, yet higher proportions of α‐Proteobacteria and Bacteroidia were seen. Biomass and activity of mycorrhizal fungi increased with sawdust amendments, whereas exudate consumption and root colonization by functional groups containing plant pathogens did not change. Sawdust amendment of arable soil enhanced abundance and exudate‐consuming activity of saprotrophic fungi in the rhizosphere of crop seedlings and promoted potential beneficial microbial groups in root‐associated microbiomes.
Introduction
In intensively managed arable soils, saprotrophic fungal biomass is low as compared with soils of natural ecosystems (Djajakirana et al., 1996; de Vries and Bardgett, 2012). This is most likely caused by a lack of decomposable plant residues, due to an efficient removal of crop parts and the predominant use of mineral fertilizers (Clocchiatti et al., 2020). Other contributing factors are the destruction of hyphal networks by tillage (Beare et al., 1997; Frey et al., 1999; Jiang et al., 2011) and inhibition of fungal growth by chemical pesticides and fungicides (Duah‐Yentumi and Johnson, 1986; Nettles et al., 2016; Rahman et al., 2017; Shao and Zhang, 2017).
In natural ecosystems and less intensively managed arable ecosystems, it is known that saprotrophic fungi have major positive contributions to ecosystem functioning, for example, by reducing nitrogen‐losses (de Vries et al., 2011) forming soil aggregates (Beare et al., 1997) and suppressing root‐infecting pathogenic fungi (van der Wal et al., 2013). Therefore, agricultural management practises resulting in stimulation of saprotrophic fungi in arable soils can make an important contribution to improve sustainable crop production (de Vries and Bardgett, 2012; Frąc et al., 2018). Fungal biomass stimulation can be achieved by low‐intensity tillage, in the long term (Wang et al., 2017; Chen et al., 2020), whereas the use of cellulose‐rich organic amendments has an immediate effect (Lucas et al., 2014; Clocchiatti et al., 2020).
Strategies that act at multiple scales, such as improving soil functioning and engineering the rhizosphere microbiome, are key for maximizing their benefits on crop performance (Chaparro et al., 2012; Bender et al., 2016). In this light, the influence of fungus‐stimulating organic amendments on the performance of crops could become even more pronounced, when the stimulated fungi in bulk soil can extend their growth and activities into the rhizosphere and roots of seedlings.
Conventional arable soils are bacterial‐dominated and bacteria are also the major consumers of root exudates in the rhizosphere of young plants (Hünninghaus et al., 2019). Only over time, crop plants recruit and modulate bacterial activities so that they are well suited to benefit the plant (Badri et al., 2013; Chaparro et al., 2014). Similarly, saprotrophic fungal biomass increases in the rhizosphere of crops only at later plant developmental stages (Hannula et al., 2012; Pausch et al., 2016), when they become an important microbial group consuming rhizodeposits (Hannula et al., 2012). This can be ascribed to the presence of larger amounts of cellulose‐rich root debris that become available during maturation of crops (Dennis et al., 2010; Eisenhauer et al., 2017; Pausch and Kuzyakov, 2018). Moreover, older roots exude a larger share of complex soluble compounds, such as aromatic acids (Gransee and Wittenmayer, 2000; Chaparro et al., 2013; Zhalnina et al., 2018). The low fungal activity in arable soil provides limited support to the initial growth and health of plants, which, conversely, are highly susceptible to the negative effects of soil‐borne pathogens (Lamichhane et al., 2017) and heavily rely on exogenous chemical inputs.
In a recent study, we showed that incorporation of deciduous wood sawdust in bare arable soils resulted in a rapid and prolonged increase of saprotrophic ascomycetes in bulk soil (Clocchiatti et al., 2020). The stimulation of ascomycetes was ascribed to the accessibility of cellulose polymers in fragmented wood. Therefore, wood amendment could also represent a strategy for increasing the activity of saprotrophic ascomycetes near roots of young crop plants. Increased utilization of exudates by saprotrophic fungi is expected to have impacts on other members of rhizosphere – and root microbiomes (de Boer et al., 2008; de Menezes et al., 2017). Such interactions could be important to establish competitive suppression of root‐infecting pathogens (Fravel et al., 2003; de Boer et al., 2015; Kepler et al., 2017); however, they could also hamper the colonization of roots by beneficial mycorrhizal fungi (de Jaeger et al., 2010).
The aim of the current study was to examine whether fungi stimulated by sawdust in bulk soil can colonize the rhizosphere and root of seedlings and participate in competitive interactions with other microbes for root exudates. In order to test this, we determined abundance and activity of fungi and bacteria in rhizosphere and roots of carrot seedlings that were grown in arable soil amended with beech wood sawdust. For the measurement of fungal and bacterial activity in the rhizosphere, we used whole plant 13CO2 pulse‐labelling and determined 13C accumulation in phospholipid fatty acids (PLFA‐SIP) and microbial DNA markers (DNA‐SIP).
We hypothesized that (i) stimulating fungal biomass in the bulk soil causes an increase in fungal biomass in the rhizosphere, which coincides with (ii) increased activity (i.e. uptake of exudates) of saprotrophic fungi in the rhizosphere of crop seedlings and results in (iii) shifts in the composition of rhizosphere – and root microbiomes. We further investigated the persistence of the effects by sowing seedlings 2 (T1) and 6 weeks (T2) after sawdust amendment.
Results
Plant performance and 13C enrichment
Germination rates and shoot biomass of carrot seedlings were not significantly different between controls and sawdust amended treatments (Supporting Information Table S1). Yet, a slight effect of pre‐incubation time before sowing (T1 = 2 weeks vs. T2 = 6 weeks, Fig. 1) was seen, which consisted of a small decrease in germination rates with longer pre‐incubation time (Supporting Information Table S1). N content of plant parts was constant, with exception of higher N content in plants grown in T1 sawdust‐amended soil.
Fig 1.

Schematic representation of the experimental setup, starting with amendment of sawdust + N or N only (control), followed by sowing and growing carrot seedlings in the greenhouse and completed with 13C‐CO2 labelling in a growth chamber.
Plant seedlings incorporated 13CO2 from the airspace of the air‐tight chamber, as indicated by a decrease in CO2 concentration by the built‐in monitoring system. This was reflected by 13C incorporation in shoot and root parts of carrot seedlings, as well as in rhizosphere soil (Supporting Information Table S1). The 13C values in the control rhizosphere soil and plant parts were within the normal atmospheric value range (δ13C ‐30‰ ± 1.8). The 13C enrichment of the rhizosphere soil was higher than the amount of 13C in unplanted soil (F 1,40 = 33.9, P < 0.05), indicating that pulse‐labelling resulted in 13C‐enriched plant‐derived carbon reaching the rhizosphere (Supporting Information Table S1). The amount of 13C was higher in roots than in shoots (F 1,40 = 322, P < 0.05), suggesting a rapid allocation of new photosynthates to the belowground plant parts. Pre‐incubation time of soil with sawdust before sowing had an effect on the amount of 13C incorporated in carrot seedlings (T1 > T2; F 1,40 = 135, P < 0.05).
Total and labelled biomass of saprotrophic fungi, bacteria and mycorrhizal fungi
An increase in fungal biomass (measured with fungal marker PLFA 18:2ω6,9) was found in sawdust amended treatments for both unplanted and rhizosphere soil (Fig. 2A; Supporting Information Table S2a). This was seen for both pre‐sowing incubation times (F 1,20 = 402.7 for T1, F 1,20 = 57.0 for T2; for both P < 0.001) and the observed pattern was consistent with that observed for ITS2 rRNA copy numbers (Supporting Information Fig. S1b; P < 0.001 for T1, P < 0.01 for T2) and ergosterol (Supporting Information Fig. S1e for Te, P < 0.01). Furthermore, the excess 13C was measured in the PLFA 18:2ω6,9. This was used for quantifying the incorporation of plant‐derived carbon into the biomass of fungi. The excess 13C in the fungal PLFA marker in the rhizosphere was significantly higher (F 1,10 = 7.25, P < 0.05, Tukey HSD P < 0.01; Supporting Information Table S2b) in the sawdust‐amended treatment than in the control for the shortest pre‐sowing incubation (T1). However, for the longer pre‐sowing incubation (T2), the excess 13C in the fungal PLFA was comparable for both treatments (Fig. 2B; Supporting Information Table S2b).
Fig 2.
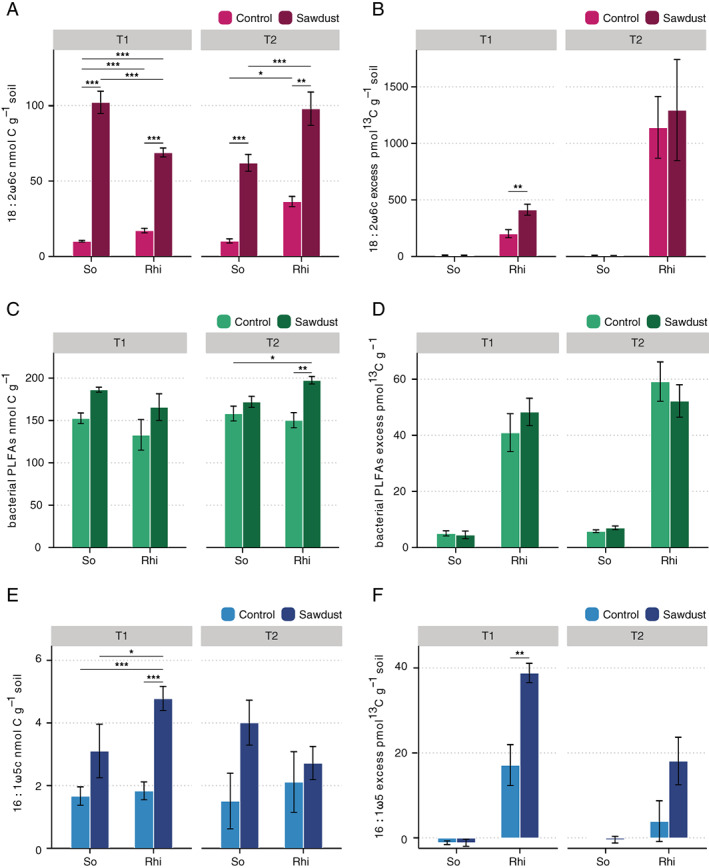
Effect of wood sawdust amendment on biomass and plant‐C incorporation by fungi, bacteria and mycorrhizal fungi. Concentration of total C and 13C excess in PLFA/NLFA markers are shown for fungi (A, B), bacteria (C, D) and arbuscular mycorrhizal fungi (E, F) in unplanted soil (So) and carrot rhizosphere soil (Rhi). T1 and T2 represent 2‐ and 6‐weeks pre‐sowing incubation times of sawdust amendments. The value for bacterial PLFAs is obtained as a sum of PLFAs i15.0, ai15.0, i16.0, 16.1w7c, 16.1w6c, ai17.1w7, i17.0, 17.1w8c, cy‐17.0. 13C excess expresses the net amount of labelled C incorporated in a PLFA/NLFA as a result of pulse‐labelling and is obtained by subtracting the natural 13C abundance found for the same PLFA/NLFA in unlabeled rhizosphere samples.
Based on the cumulative measure of bacterial PLFA markers, bacterial biomass was not affected by sawdust amendment for T1 (Fig. 2C; Supporting Information Table S2c, F 1,20 = 4.4, P = 0.6), however for T2 the PLFA‐based bacterial biomass was higher in the sawdust‐amended rhizosphere as compared with the control rhizosphere (Fig. 2C; P < 0.001). For T1, sawdust amendment was associated with an increase in individual bacterial PLFAs, with higher abundance of PLFAs i16:0, i17:0, ai17:0, 17:1ω8c, cy‐17:0 in the sawdust‐amended soil and rhizosphere (Supporting Information Table S3). The peak of PLFA marker 18:1ω7c/18:1ω9t, which can indicate both bacteria and fungi, was also higher in the sawdust amended soil as compared to the control. We did not, however, detect differences in excess 13C in the rhizosphere for either individual or cumulative bacterial PLFAs between sawdust‐amended and control treatments (Fig. 2D; Supporting Information Tables S2d and S4).
The evaluation of the total bacterial abundance via qPCR‐based quantification of 16S rRNA copy numbers showed a more pronounced increase in bacterial abundance for T1 in both sawdust‐amended soil and rhizosphere, as compared with the control. For T2, qPCR‐based bacterial numbers were higher in the root tissues, but not in the bulk soil or rhizosphere, upon sawdust amendment (Supporting Information Fig. S1a and b).
The marker for arbuscular mycorrhizal fungi NLFA 16:1ω5 was significantly increased in the carrot rhizosphere in the sawdust amended treatment for T1 (F 1,20 = 34, P < 0.001) (Fig. 2E). Moreover, a larger excess of 13C (F 1,10 = 10.8, P < 0.01) was found in NLFA 16:1ω5 for T1, in response to sawdust amendment (Fig. 2F).
Composition of fungal and bacterial communities
Sawdust amendment strongly affected the composition of fungi present in the soil (R 2 = 70.3, P < 0.001; Supporting Information Table S6a), rhizosphere (R 2 = 53.7, P < 0.001; Supporting Information Table S6b), as well as of fungi actively incorporating plant‐derived carbon (R 2 = 17.5, P < 0.001; Supporting Information Table S6c; Figs. 3A and 4A). In addition to this, the effect of sawdust amendment was seen in the root compartment (R 2 = 15.0, P < 0.001; Fig. 3A, Supporting Information Table S6d). The fungal community of unplanted soil without sawdust amendment was dominated by Ascomycota, followed by Basidiomycota and Mortierellomycota (relative abundance > 1.5%, Fig. 4A). Ascomycota and Basidiomycota comprised mostly of saprotrophic guilds, but taxa assigned as plant pathogens were also detected (Fig. 4B). Sawdust amendment led to an increase in the relative abundance of Sordariomycetes in the soil, rhizosphere and 13C‐enriched rhizosphere (Fig. 4A; F 1,80 = 325.7, P < 0.001). We further detected more Sordariomycetes (Fig. 4A; F 1,40 = 13.4, P < 0.001) and Leptosphaeriaceae (Supporting Information Fig. S2a; F 1,40 = 20.3, P < 0.001), as well as arbuscular mycorrhizal fungi (Fig. 4A, Glomeromycota, Archeosporomycetes, F 1,40 = 9.0, P < 0.01) in the fungal community associated with roots in sawdust‐amended soil. Fungal SVs assigned as ‘Plant pathogens’ only or assigned as ‘Plant pathogen or other potential function’ (e.g. ‘Plant pathogen‐Saprotroph’) were grouped in a sub‐community containing potential plant pathogenic fungi. This sub‐community of potential plant pathogens was found in all compartments, but sawdust amendment decreased its abundance, relative to the total fungal community (Fig. 4D, F 1,80 = 43.7, P < 0.001), in favour of a higher relative abundance of fungal SVs assigned as ‘Saprotrophs’ only (Fig. 4C, F 1,80 = 137.7, P < 0.001) in soil, rhizosphere and 13C‐enriched rhizosphere. Functional groups containing potential plant pathogens were found in a higher proportion in roots than in soil compartments, irrespective of the sawdust treatment. Yet, sawdust reduced the relative abundance of these groups in the root fungal community for T1 (F 1,20 = 4.5, P < 0.05), but not for T2. The cumulative relative abundance of all groups potentially containing pathogenic fungi was multiplied by the total fungal biomass, thus estimating the absolute size of this sub‐community. This showed a small, yet significant increase in absolute abundance of groups containing plant pathogenic fungi by sawdust amendment in the soil and rhizosphere (Supporting Information Fig. S3, simple effect of sawdust: P < 0.001 based on PLFA, P < 0.001 based on ITS). However, no change was seen in the absolute abundance of pathogen‐containing groups in the 13C‐enriched rhizosphere and root after sawdust amendment. Conversely, the absolute abundance of the sub‐community of saprotrophic fungi, estimated in the same way, was increased by sawdust in all compartments (Supporting Information Fig. S3, P < 0.001 based on both PLFA and ITS for soil and rhizosphere, P < 0.001 in 13C‐enriched rhizosphere, P < 0.001 in root).
Fig 3.

Effect of sawdust amendment of arable soil on the fungal and bacterial community composition in roots and rhizosphere of carrot seedlings and in bulk soil. Ordination (PCoA based on Bray–Curtis dissimilarity matrix) was performed independently for the whole fungal dataset (A) and for the whole bacterial dataset (B). For both fungi and bacteria, the dissimilarity between communities found in four compartments, with and without sawdust amendment, was displayed in separate plots for each pre‐sowing incubation time (T1 = 2 weeks, T2 = 6 weeks). Communities were obtained from unplanted soil, rhizosphere, 13C‐enriched rhizosphere DNA and roots, after carrot seedlings had grown for 3 weeks.
Fig 4.

Effect of sawdust amendment on fungal community composition in an arable soil, as determined for unplanted soil (So), carrot rhizosphere (Rhi), 13C‐enriched DNA fraction in the carrot rhizosphere (RhiA) and carrot root (Ro). Relative abundance of fungal classes (A) and fungal functional groups (B) are displayed for soil, rhizosphere and root compartments and for each pre‐sowing incubation time (T1 = 2 weeks, T2 = 6 weeks) in sawdust‐amended pots and controls. Fungal taxa and functional groups of relative abundance <1.5% were classified as ‘Other’. (C, D): relative abundance of the functional fungal groups ‘saprotrophs’ and those potentially containing ‘plant pathogens’. The main effect of sawdust amendment is shown alongside the effect of sawdust in each compartment (three‐way ANOVA with planned contrasts, *** P < 0.001, ** P < 0.01, * P < 0.05).
In the unplanted control, the most abundant fungal families containing plant pathogens were Didymellaceae, Plectospherellaceae and Nectriaceae (Fusarium spp.) (Supporting Information Fig. S2b). Decrease in relative abundance of Didymellaceae an Plectospherellaceae contributed to the observed decrease of the relative abundance of pathogen‐containing groups in the soil (F 1,20 = 24.0, P < 0.001), rhizosphere (F 1,40 = 14.0, P < 0.001) and 13C‐enriched rhizosphere (F 1,20 = 10.0, P < 0.01) in response to sawdust amendment. On the other hand, Nectriaceae were not affected by sawdust in any compartment (F 1,120 = 0.9, P = 0.76), whereas Olpidiaceae increased in carrot roots grown in sawdust‐amended soil for the shortest pre‐sowing incubation time (T1: F 1,20 = 5.5, P < 0.05).
The most abundant bacterial classes present in the soil were γ‐ and α‐Proteobacteria, Bacilli and Actinobacteria (Fig. 5A). Overall, sawdust amendment induced a smaller shift in bacterial community composition as compared with that of fungi (Figs. 3B and 5A; Supporting Information Table S7). Yet, significant sawdust‐induced shifts in the bacterial community occurred in unplanted soil (R 2 = 7.9%, P < 0.001; Supporting Information Table S7a), rhizosphere soil (R 2 = 3.4%, P < 0.001; Supporting Information Table S7b) and roots (R 2 = 3.8%, P < 0.01; Supporting Information Table S7d). α‐Proteobacteria increased in relative abundance in response to sawdust amendment in all compartments (F 1,120 = 52.9, P < 0.001) (Fig. 5B). This was also the case for Bacteroidia, although the increase was only significant for unplanted soil (F 1,20 = 6.0, P < 0.05) and roots (F 1,40 = 13.7, P < 0.001, Fig. 5C). Several members of α‐Proteobacteria contributed to the effect observed at class level. Rhizobiaceae increased to a large extent in the root interior after sawdust amendment (F 1,40 = 12.7, P < 0.001), whereas in the soil, the observed increase of α‐Proteobacteria was not only ascribed to Rhizobiaceae but also to several other members of Rhizobiales (F 1,20 = 25.4, P < 0.001, Fig. 5D). With regard to Bacteroidia (Fig. 5E), the relative abundance of Chitinophagaceae was higher after sawdust amendment than in the control (F 1,120 = 5.3, P < 0.05) and this was particularly observed in unplanted soil (F 1,20 = 4.3, P < 0.05). In addition, Sphingobacteriaceae contributed to the higher relative abundance of Bacteroidia (F 1,120 = 9.6, P < 0.01), especially in the root compartment (F 1,40 = 14.1, P < 0.001).
Fig. 5.

Effect of sawdust amendment on bacterial community composition in an arable soil, as determined for unplanted soil (So), carrot rhizosphere (Rhi), 13C‐enriched DNA fraction in the carrot rhizosphere (RhiA) and carrot root (Ro). Results are displayed for each pre‐sowing incubation time (T1 = 2 weeks, T2 = 6 weeks) in sawdust‐amended pots and controls.
A. Relative abundance of bacterial classes (classes of relative abundance <1.5% were classified as ‘Other’).
B,C. Relative abundance of α‐Proteobacteria and Bacteroidia. The main effect of sawdust amendment is shown alongside the effect of sawdust in each compartment (three‐way ANOVA with planned contrasts, ***P < 0.001, **P < 0.01, *P < 0.05).
D,E. Relative abundance of bacterial families belonging to α‐Proteobacteria and Bacteroidia (respectively, families of relative abundance < 1% and < 0.5% were classified as ‘Other’).
Discussion
Effect of sawdust addition on abundance and composition of fungi in soil, rhizosphere and roots
Free‐living, saprotrophic fungi not only occur in soil, they are also common inhabitants of the rhizosphere and are even found inside plant roots (Gkarmiri et al., 2017; Hugoni et al., 2018). However, saprotrophic fungi are negatively affected by intensive agricultural management practises, which tend to favour plant pathogenic fungi (Gao et al., 2019). The addition of organic materials, such as cover crops and straw, can increment the abundance of fungi in soil of arable fields (García‐Orenes et al., 2013; Lucas et al., 2014; Rahman et al., 2017) and fragmented woody material has been shown to be particularly effective (Clocchiatti et al., 2020). The present study not only confirmed the stimulating effect of sawdust on fungi in unplanted arable soil, but also showed that, upon subsequent sowing, fungal stimulation extended to the rhizosphere and even roots of carrot seedlings. Fungal stimulation in rhizosphere and roots was still occurring after 6 weeks of pre‐incubation of sawdust in soil. Saprotrophic Sordariomycetes were the most stimulated fungi in bulk soil, rhizosphere and roots. Stimulation of Sordariomycetes by sawdust is in line with their known ability to efficiently decompose cellulose fractions of organic materials (Koechli et al., 2019). Under conventional agricultural practises, this group tends to associate with older plant roots (Hannula et al., 2010; Han et al., 2017). In order to analyse the impact of sawdust‐amendment on plant‐pathogenic fungi, all functional groups containing 'Pathogen' as the only function or as one of the possible functions were included. This analysis revealed that the relative abundance of the SVs assigned to these functional groups was not increased by sawdust amendment. Yet, the estimation of the absolute abundances of SVs in functional groups containing plant pathogens showed a small increase after sawdust amendment. However, this absolute increase in potential pathogens was limited to the soil and rhizosphere compartments and it was not mirrored by an increase in their uptake of root‐derived 13C or colonization of root tissues. Conversely, saprotrophic fungal stimulation was consistently high in sawdust‐amended soil, based on both estimated absolute and relative abundance. Such promotion of sawdust‐stimulated saprotrophic fungi extended also to the active root‐associated fungi and to the fungi colonizing the interior of roots. Further quantification of pathogens population abundance, combined with bioassays, is required to test the potential of the present saprotroph‐stimulating approach in counteracting the invasion of roots by disease‐causing fungi. Recently, the importance of the mycobiome in reducing the severity of soil‐borne diseases has been suggested (Busby et al., 2016; Poli et al., 2016; Sarrocco, 2016).
An increased abundance of AMF in the rhizosphere (NLFA 16:1ω5) and in roots (ITS2 rRNA amplicon sequences) was found following sawdust amendment. In the current study, the soil received N‐fertilization only, thus the addition of sawdust could have raised the competition for available P in the soil due to increased uptake by saprotrophic fungi (Wu et al., 2007; Zhang et al., 2018). Under conditions of low P availability, AMF increase in abundance and support plant nutrient acquisition, in particular when plant litter is added to an arable soil (Xu et al., 2018). In natural ecosystems, AMF are often associated with plant litter (Joner and Jakobsen, 1995; Gryndler et al., 2002; Bunn et al., 2019), where they are involved in nutrient mining and have a positive feedback on saprotrophic microbes (Quist et al., 2016). To our knowledge, this is the first observation of simultaneous stimulation of AMF and saprotrophic fungi by an organic amendment in arable soil. We suggest that this finding is worth further exploration, given the well‐known beneficial effects of AMF of plant growth and health (Chandanie et al., 2009; Hu et al., 2010).
Effect of sawdust addition on root exudate consumption by fungi
Here, we demonstrate that saprotrophic fungi stimulated by sawdust amendment are actively consuming root‐derived C in the rhizosphere of seedlings, which was measured as increased 13C excess accumulated in the fungal PLFA 18:2ω6,9. Uptake of root‐derived C by saprotrophic fungi was previously observed for mature annual crop plants and perennial crops (Tavi et al., 2013; Pausch et al., 2016), but not for crop seedlings (Hünninghaus et al., 2019). For potato grown in intensively managed soils, it was shown that fungal biomass in the rhizosphere increased in advanced growth stages, probably due to the presence of recalcitrant root deposits (Hannula et al., 2010). Hence, it seems that sawdust amendment creates a situation where the rhizosphere of crops can readily be colonized by root‐exudate consuming saprotrophic fungi at the seedling stage, which normally requires crop maturation. Increased root C uptake after sawdust addition was ascribed mainly to sordariomycetal fungi, as they were prevalent within the 13C‐enriched fungal community. Due to the short duration of the 13CO2 pulse (24 h), followed by immediate harvesting of rhizosphere soil and plants, mainly soluble exudates were probably enriched in 13C (Kaštovská et al., 2017). Thus, the observed 13C labelling in the fungal biomarker can be attributed to the uptake of root exudates like sugars and organic acids. The PLFA marker 18:2w6,9 can be found in plants; hence, care was taken when sampling the rhizosphere soil not to include plant cells. Furthermore, by using qPCR on the ITS2 region and ergosterol as additional measurements of fungal biomass, we showed that these reveal a similar pattern (Supporting Information Fig. S1) and we concluded that the marker 18:2ω6,9 is a useful indicator of fungal biomass in the rhizosphere (Joergensen and Wichern, 2008; Kaiser et al., 2010; Frostegård et al., 2011), especially when combined with 13C pulse‐labelling.
Wood sawdust and exudates greatly differ in biochemical complexity. However, saprotrophic fungi take up soluble sugars released from lignocellulose‐rich organic matter by extra‐cellular enzymes and are, therefore, also able to use labile monomeric exudate compounds (Buée et al., 2009; van der Wal et al., 2013; de Vries and Caruso, 2016). Rapid exudate assimilation by ascomycetal fungi was indeed found in earlier studies (Hannula et al., 2012; Marschner et al., 2012; Gkarmiri et al., 2017; Wang et al., 2019). Our study points out that sordariomycetal fungi are well‐equipped for efficient decomposition of both exudates and of cellulose‐rich organic materials in arable soils. The use of fine wood particles could be important to ensure bridging of fungal hyphae between wood particles and plant roots.
Earlier studies reported that AMF play a dominant role in rapid acquisition of plant‐C, which is transferred only in a later stage to saprotrophic fungi and bacteria (Drigo et al., 2010; Hünninghaus et al., 2019). In the current study, the contributions of saprotrophic and arbuscular mycorrhizal fungi to plant‐C acquisition were not mutually exclusive. On the contrary, a simultaneous stimulation of active incorporation of plant‐C by AMF (13C excess in the NLFA 16:1ω5) and saprotrophic fungi was observed in the rhizosphere of plants sown 2 weeks after sawdust amendment. In line with what is discussed above, this suggest the possibility that after sawdust addition, in absence of P fertilization, no negative interactions occurred between saprotrophic fungi and AMF in the root surroundings.
Effect of sawdust addition on composition and activity of bacteria
PLFA‐ and qPCR‐based quantification of bacteria showed an increase in bacterial abundance in sawdust‐amended soil, but this was less pronounced as compared to fungi. Similarly, sawdust caused a smaller shift in bacterial community composition than in that of fungi. Bacterial groups typically associated with cellulose degradation in arable soils, such as Acidobacteria, β‐, γ‐, δ‐Proteobacteria and Actinobacteria (Kramer et al., 2016) did not increase in relative abundance after sawdust addition. This suggests that fungi were the main primary decomposers of sawdust, while increase of bacteria can be ascribed to the consumption of fungal‐derived carbon or to the consumption of breakdown products derived from fungal sawdust decomposition (Schneider et al., 2010; Ballhausen and de Boer, 2016; de Menezes et al., 2017). Amplicon sequencing revealed that Bacteroidia and α‐Proteobacteria increased after addition of sawdust in the soil. Although Bacteroidia and α‐Proteobacteria could contribute in part to sawdust decomposition (Schellenberger et al., 2009; Haichar et al., 2016; Kramer et al., 2016), these groups are known to engage in fungal‐bacterial interactions. Bacteroidia harbour an array of chitinolytic enzymes, which confers them the ability to utilize fungal debris or predate on fungi (Wieczorek et al., 2019). The utilization of fungal‐derived C by Bacteroidia is in line with the observation that they were more present in the soil, but not in the 13C‐enriched bacterial community, meaning that they potentially received a large share of sawdust‐derived C via fungi. α‐Proteobacteria are often associated with decomposing wood, where they are thought to provide an additional source of N to fungal saprotrophs in exchange of wood breakdown products (Hoppe et al., 2015; Johnston et al., 2016). Hence, the ability of α‐Proteobacteria to respond to wood breakdown products may explain their increase, even though their role of N‐suppliers is unlikely to be important in the current experimental setting where mineral N was supplied together with sawdust. Moreover, the presence of fungal hyphae in the bulk and rhizosphere soil could facilitate the movement of α‐Proteobacteria, and Rhizobiales in particular, towards plant roots, as recently demonstrated in a legume crop (Zhang et al., 2020).
The 13C incorporation in bacterial PLFAs was not affected by sawdust amendment. Thus, the increased consumption of root exudates by saprotrophic fungi did not create the hypothesized constraint for the uptake of plant‐derived carbon by bacteria. This suggests that sawdust‐stimulated fungi utilize an additional pool of plant C than what is used by bacteria in both the control and sawdust‐amended soil. Accordingly, it seems that the total uptake of released root C is more efficient in the rhizosphere when active fungi are present. A similar pattern was reported by Morriën et al. (2017), in a study where an increased activity of fungi led to a better uptake of plant C by the food web of a restored ex‐arable soil.
Even though increased fungal activity did not alter the total bacterial activity, the presence of active fungi in the rhizosphere and roots steered the composition of the plant‐associated bacterial community, especially promoting Bacteroidia and α‐Proteobacteria. In particular, sawdust amendment and fungal stimulation increased the abundance of potentially beneficial bacteria within the roots, namely Chitinophagaceae, Sphingobacteriaceae and Rhizobiales. Chitinophagaceae and Sphingobacteriaceae could contribute to resistance against diseases by antagonistic interactions. Chitinophaga spp. were previously found within sugar beet plants challenged by Rhizoctonia solani and were associated with a higher expression of chitinases (Carrión et al., 2019). Spingobacteriaceae were also found in disease suppressive soils and showed antagonistic activity against plant‐pathogenic fungi, when triggered by interaction with other members of the bacterial community (de Boer et al., 2007; Gómez Expósito, 2017). Although Rhizobiales are mostly known as N‐fixing mutualists of legumes, they can also establish mutualistic interactions with non‐legume plants and contribute to plant growth stimulation and priming of the plant immune system (Garrido‐Oter et al., 2018). Overall, the use of sawdust and the subsequent fungal stimulation steered the soil and rhizosphere bacterial community and influenced the association of seedlings with potentially beneficial bacterial groups.
In this study, changes in size, activity and composition of the fungal and bacterial communities are mainly interpreted as a direct effect of wood particles or sawdust‐stimulated fungi to the resident soil microbiome. In addition to this, it cannot be excluded that wood‐inhabiting microbes were introduced into the soil and contributed to the observed effects (Massart et al., 2015; Mawarda et al., 2020). We argue that their role was minor, as the presence of microbes in commercial sawdust is reduced by drying at high temperatures (Ståhl et al., 2004). Furthermore, we expect that the remaining sawdust‐inhabiting microbes are rapidly outcompeted during wood decomposition in soil (Petrini and Fisher, 1990; van der Wal et al., 2016).
Conclusions and perspectives
Stimulation of saprotrophic fungi by sawdust in arable soil was not restricted to bulk soil, but extended to the rhizosphere and roots of carrot seedlings. This coincided with increased uptake of root exudates by saprotrophic fungi, which was associated with a higher relative abundance of α‐Proteobacteria, Bacteroidia and a higher abundance of active arbuscular mycorrhizal fungi in rhizosphere and roots (Fig. 6). In soil and rhizosphere, sawdust caused a small increase in the estimated abundance of fungal groups containing potential plant pathogens; however, the root exudate consumption and root colonization by these groups did not increase after sawdust amendment. This study shows that an increased biomass and activity of saprotrophic fungi in the rhizosphere and root has a steering effect on other plant‐associated microbes and can potentially promote beneficial microbes. Further research is required for assessing the benefits of such changes on plant growth and health. We highlight the potential of saprotrophic fungi as target group for the design of sustainable agricultural practices, that impact both soil and plant functioning.
Fig 6.

Effect of sawdust on fungi and bacteria in an arable soil and in the root surroundings of carrot seedlings after 2‐ and 6‐weeks (T1, T2) pre‐sowing incubation times. White boxes report the biomass of bacteria, fungi and AMF in the rhizosphere soil, based on PLFA/NLFA markers. The black arrows represent the incorporation of 13C in fungal, bacterial and AMF PLFA/NLFA markers. Pie charts report the proportion of bacterial taxa (top) and fungal functional groups (bottom) that responded to sawdust amendment in the soil and rhizosphere soil (external charts) and in in the 13C‐enriched rhizosphere and root (central charts), based on DNA amplicon sequencing.
Experimental procedures
Experimental setup
In August 2016, soil was sampled at the experimental farm of Wageningen University and Research located at Vredepeel (Clocchiatti et al., 2020). The soil was collected (0–10 cm) from bare patches in a plot (200 m × 18 m) that was conventionally managed and partially covered with triticale. After sieving (4‐mm), soil was stored at 4°C until use. The soil was sandy (92% sand, 6% silt, 2% clay), slightly acidic (pH 6.1) and had a content of 6% organic matter.
For the experiment, the soil was divided in two batches. One batch was amended with beech sawdust (2 mm) at the concentration of 5 g kg−1 soil, (Fagus sylvatica, Sänger Rollenlager GmbH, Waldsolsms, Germany) and ammonium nitrate, 170 mg kg−1 soil. The second batch of soil was mixed with ammonium nitrate only (170 mg kg−1 soil), representing the control. Soil moisture was adjusted to 60% of its water holding capacity and it was maintained as such during the experiment. Portions of 190 g of soil were divided among pots (PET, 10 × 10 × 10 cm) and pre‐incubated for 2 and 6 weeks in a greenhouse under an aluminium cover (Fig. 1). These pre‐incubation times were chosen on basis of sawdust‐induced fungal dynamics in an earlier study (Clocchiatti et al., 2020). After each pre‐incubation period, 10 control pots and 10 sawdust‐amended pots were sown with carrot (40 seeds per pot, Daucus carota subsp. sativus, Bejo Zaden BV, Warmenhuizen, the Netherlands), whereas five control and five sawdust‐amended pots were kept unplanted. Carrot seedlings were grown in the same greenhouse, under a combination of natural and artificial lightning (photoperiod 16:8). Two weeks after sowing, germination of carrot seedlings was scored. One week later, corresponding to the emergence of the first true leaf, 13CO2 pulse‐labelling was performed (Fig. 1).
13CO2 pulse‐labelling and harvesting
Planted and unplanted pots were transferred into a climate chamber and provided with pulses of 99.99 atom‐% 13CO2 (Cambridge Isotope Laboratories, Tewksbury, Massachusetts) for a total of 16 h, under artificially lit (photoperiod 16:8) and air‐tight conditions. The concentration of CO2 was monitored during the labelling period: initially CO2 concentration was allowed to drop from ambient concentration (530 ppm) to 460 ppm, confirming photosynthetic activity of the carrot seedlings. After that, a pulse of 1.5 l of 13CO2 was provided, restoring the initial concentration of CO2 in the chamber. The pulse was repeated five times with 2.5 h intervals making the total amount of 13CO2 added 12.5 l. During the artificial night (8 h darkness), 13CO2 was not provided and air exchange between the climate chamber and the air outside was enabled. The next day, pots were destructively harvested. As controls, the same numbers of sawdust‐amended and un‐amended pots were placed in an identical climate chamber but subjected to pulses of atmospheric CO2 (Fig. 1). We refer to them as 12C controls. The labelling was performed in the same way two times, namely 5 and 9 weeks after the start of the experiment, corresponding to 2 and 6 weeks of soil pre‐incubation, followed by 3 weeks of seedling growth. We refer to the two pre‐incubation time points as T1 and T2, respectively. Bulk soil samples were obtained from unplanted pots. A rhizosphere soil sample was obtained by pooling the rhizosphere soil from all the roots in a pot (26 ± 4 seedlings). Rhizosphere soil was obtained from a soil layer of <1.5 mm attached to the root (Hinsinger et al., 2005; Kuzyakov and Razavi, 2019). The soil was sampled carefully, without damaging or brushing the roots, and the rhizosphere soil sample was further 1‐mm sieved (Butler et al., 2003). In this way, we took care that root parts were excluded from the sample, since plants and fungi do share PLFA markers. Roots and shoots of seedlings harvested per pot were pooled in one sample. The roots were gently washed with sterile demi‐water and used to extract the rhizoplane and endophytic communities together (Li et al., 2019). Root, shoot, rhizosphere and soil samples were rapidly frozen and freeze‐dried. Roots and shoots were weighed and grinded to a fine powder by bead‐beating (<0.1 μm). After that, soil and plant samples were stored at −80°C until use. The determination of 13C content of soil and plant parts is described in the Supporting Information.
Analysis of PLFA and NLFA, combined with stable isotope probing, and ergosterol
PLFA and NLFA were extracted from 3 g soil using the procedure described by Frostegård et al. (1993) PLFA/NLFA concentrations were determined on a GC‐FID (7890 A Agilent; Technologies, Santa Clara, California). δ13 values were measured on a GC‐c‐IRMS (Trace Ultra GC equipped with a Conflo III interface and a Delta V IRMS, Thermo Scientific, Germany). Three C20:0 methyl esters (Schimmelmann, Biogeochemical Laboratories, Indiana University) were used for calibrating the δ value of the in‐house calibration gas. The internal standard methyl nonadecanoate fatty acid (19:0) was used for calculating concentrations. Identification of the compound was based on a BAME mix (Supelco 47080‐ u) and a FAME mix (Supelco 18919‐1AMP). The 13C enrichments for PLFA/NLFA biomarkers (excess 13C pmol g−1 soil) were calculated as described by Boschker and Middelburg (2002). Briefly, δ13C values for each PLFA/NLFA were measured as ratio (R = 13C/12C) by reference to the standard (Vienna Pee Dee Belemnite):
δ13 C values were used to calculate the 13C fraction, F13 = 13C/(13C + 12C), for a PLFA/NLFA in a sample:
Finally, the 13C enrichment of a PLFA/NLFA in a labelled sample was obtained by adjusting the 13C fraction found in a labelled sample () with the average 13C fraction in unlabelled control samples (). Excess ratio was converted into 13C enrichment by using the total PLFA/NLFA concentration in the labelled sample (CP/NLFA):
PLFA 18:2ω6,9 was used as fungal marker, whereas NLFA 16:1ω5 was used as an indicator of AMF (Frostegård et al., 2011; Willers et al., 2015). The following PLFAs were used as bacterial markers: i15:0, ai15:0, i16:0, 16:1ω7c, 16:1ω6c, ai17:1ω7, i17:0, ai17:0, 17:1ω8c, cy‐17:0 (Boschker and Middelburg, 2002; Mauclaire et al., 2003); Actinobacteria were indicated by 10Me‐branched PLFAs (Willers et al., 2015).
As an additional measure for fungal biomass, alkaline extraction of ergosterol was performed starting from 1 g soil, as described by de Ridder‐Duine et al. (2006). Ergosterol was quantified by LC–MSMS (UHPLC 1290 Infinity II, Agilent Technologies and 6460 Triple Quad LC–MS, Agilent Technologies).
DNA extraction and molecular analyses
DNA was extracted form 0.25 g freeze‐dried rhizosphere, soil and root samples by DNeasy PowerSoil Pro Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The DNA concentration was assessed by Qubit dsDNA HS Assay (Termo Fisher Scientific, Waltham, Massachusetts). The DNA was used for quantifying ITS2 and 16S rRNA copy numbers (qPCR). 13C‐enriched DNA was obtained from rhizosphere samples by CsCl gradient fractionation. This, together with soil, rhizosphere and root DNA, was subjected to fungal and bacterial amplicon sequencing. For details on molecular analyses, see the Supporting Information.
Statistical and bioinformatic analysis
The statistical analysis was carried out in R (version 3.6.1). After verifying the assumption of normality and equality of variances, two‐way ANOVA was used for comparing microbial markers among sawdust‐amended/un‐amended soil and between rhizosphere and unplanted soil. Multiple comparisons were obtained by Tukey's post hoc test at family‐wise error rate of 5%. The analysis was carried out independently for each time point and performed for the following markers: the fungal PLFA 18:2ω6,9 (total and 13C excess), the mycorrhizal NLFA 16:1ω5 (total and 13C excess), total and individual bacterial PLFAs listed above (total and 13C excess), ergosterol, ITS2 and 16S rRNA copy number.
Sequencing data were processed in Linux and R. The ITS2 rRNA region was extracted with ITSxpress from fungal sequences (Rivers et al., 2018). After that, the R package dada2 was used for quality filtering (maxEE = 2, truncQ = 2), to join paired‐end reads, to remove chimeric sequences, for modelling sequencing errors and identifying sequence variants (SVs) by the DADA2 algorithm (Callahan et al., 2016). Taxonomy was assigned by using the RDP classifier based on the UNITE v2019 database (Abarenkov et al., 2010). Bacterial sequences were processed in the same way, with exception to the quality filtering parameters (truncLen = 240, maxEE = 2, truncQ = 2) and to the database used for taxonomical assignment (SILVA v132). Overall, the fungal dataset counted 1 646 403 reads and the bacterial dataset had 4 360 774 reads. The datasets were cleaned from singletons and all SVs other than fungi and bacteria, that is, mitochondria and chloroplasts, resulting in 4048 fungal SVs and 16 422 bacterial SVs (R, phyloseq). In the latter dataset, both eubacterial and archeal SVs were retained. Based on the taxonomy, potential functions were assigned to 1871 fungal SVs, using the FUNGuild database v1.0 (Nguyen et al., 2016), followed by manual curation of the functional assignment of the ecological groups of interest in this study: saprotrophic fungi, plant pathogens, fungal parasites, arbuscular mycorrhizal fungi (Supporting Information Table S8).
Permutational multivariate analysis of variance (PERMANOVA) was used to determine the effect of sawdust on fungal and bacterial communities residing in the soil, rhizosphere and root and the rhizosphere communities that actively incorporated ‘heavy’ carbon. PERMANOVA was run independently in each compartment, after testing for homogeneity of multivariate variance (vegan, PERMDISP, Anderson and Walsh, 2013). Bray–Curtis dissimilarities between fungal communities and between bacterial communities found in two treatments (sawdust and control) and four compartments (unplanted soil, rhizosphere, 13C‐enriched rhizosphere and root) were visualized as a result of principal coordinate analysis (PCoA). Differences in relative abundance between sawdust and control were analysed at phylum, class, order and family level and for fungal functional groups. Differences between sawdust and control were analysed for the whole dataset with three‐way ANOVAs, accounting soil treatment, compartment and time point as factors. Normality and homoscedasticity were verified for each model, which also included planned contrasts for comparing sawdust vs control in each compartment, as these comparisons reflect the main research question of this study (effect of sawdust on rhizosphere and root microbial communities). As the variation for fungal taxa and fungal functional groups was larger in the root as compared to the other compartments, the data on the root fungal community were analysed separately from the fungal community data of the three other compartments, by using two‐way ANOVA with soil treatment and time point as factors, combined with planned contrasts for testing the effect of sawdust at each time point.
In order to evaluate the potential effect of sawdust on the size of the sub‐communities of saprotrophic and plant pathogenic fungi, the analysis of functional groups was combined with PLFA and ITS data. The cumulative relative abundance of SVs assigned as plant pathogens (alone or combined with other potential functions) was multiplied, for each sample, for the total fungal abundance, based on both PLFA 18:2ω6,9 (or excess 13C PLFA 18:2ω6,9, for the 13C‐enriched communities) and ITS2. The same estimation was done for the group ‘Saprotroph’. Differences between sawdust‐amended and control soil were compared with ANOVA and Tukey's post hoc test (family‐wise error rate 5%). The data required log‐transformation for meeting the assumptions of homoscedasticity and normality.
Author contributions
A.C., S.E.H. and W.d.B. designed the research, A.C., M.P.J.H. and P.J.A.k.G. carried out the experiment and the measurements, A.C. analysed the data and wrote the manuscript in consultation with S.E.H. and W.d.B.
Conflict of Interest
The authors declare that they have no competing interests.
Supporting information
Fig. S1. Bacterial and fungal abundance in soil, rhizosphere and roots, based on qPCR and ergosterol.
Supplementary Materials and Methods: Measurement of 13C content of soil, rhizosphere, shoot and roots, molecular analyses (qPCR, gradient fractionation and sequencing), bioinformatic processing of raw sequencing data.
Fig. S2. Effect of sawdust on fungal families assigned to saprotrophs and potential pathogens.
Fig. S3. Estimated effect of sawdust on the absolute abundance of fungal functional groups.
Table S1. Biomass, N content and germination rate of seedlings, 13C content of labelled soil, rhizosphere and seedlings.
Table S2. Summary of ANOVA results on PLFAs and NLFA data as a function of organic soil amendment and compartment, as performed independently for each time point.
Table S3. Mean concentration and summary of statistical differences of individual PLFAs and NLFAs (nmol C g−1 soil).
Table S4. Mean labelled fraction and summary of statistical differences of individual PLFAs and NLFAs (pmol excess 13C g−1 soil).
Table S5. Summary of individual fractions pooled into ‘heavy’ and ‘light’ DNA fractions.
Table S6. Summary of permutational multivariate analysis of variance for the fungal community.
Table S7. Summary of permutational multivariate analysis of variance for the bacterial community.
Table S8. Taxa and functional groups identified by ITS2 rDNA sequencing.
Appendix S1 Supporting information.
Acknowledgements
This research is supported by the Dutch Technology Foundation (STW), a branch of the Netherlands Organization for Scientific Research (NWO) (grant number 14012). The authors thank Marlies van den Berg, Ciska Raaijmakers and Nico Helmsing for their technical assistance. The sequencing was done in collaboration with McGill University and Génome Québec Innovation Centre, Montréal, Canada. This is publication number 7208 of the NIOO‐KNAW Netherlands Institute of Ecology.
Data Availability Statement
The data presented in this study is available at doi:10.5061/dryad.2rbnzs7nn. Sequencing data is accessible at the European Nucleotide Archive (accession PRJEB37482(ERP120799)).
References
- Abarenkov, K. , Henrik Nilsson, R. , Larsson, K.‐H. , Alexander, I.J. , Eberhardt, U. , Erland, S. , et al. (2010) The UNITE database for molecular identification of fungi – recent updates and future perspectives. New Phytol 186: 281–285. [DOI] [PubMed] [Google Scholar]
- Anderson, M.J. , and Walsh, D.C.I. (2013) PERMANOVA, ANOSIM, and the mantel test in the face of heterogeneous dispersions: what null hypothesis are you testing? Ecol Monogr 83: 557–574. [Google Scholar]
- Badri, D.V. , Chaparro, J.M. , Zhang, R. , Shen, Q. , and Vivanco, J.M. (2013) Application of natural blends of phytochemicals derived from the root exudates of Arabidopsis to the soil reveal that phenolic‐related compounds predominantly modulate the soil microbiome. J Biol Chem 288: 30503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballhausen, M.‐B. , and de Boer, W. (2016) The sapro‐rhizosphere: carbon flow from saprotrophic fungi into fungus‐feeding bacteria. Soil Biol Biochem 102: 14–17. [Google Scholar]
- Beare, M.H. , Hu, S. , Coleman, D.C. , and Hendrix, P.F. (1997) Influences of mycelial fungi on soil aggregation and organic matter storage in conventional and no‐tillage soils. Appl Soil Ecol 5: 211–219. [Google Scholar]
- Bender, S.F. , Wagg, C. , and van der Heijden, M.G.A. (2016) An underground revolution: biodiversity and soil ecological engineering for agricultural sustainability. Trends Ecol Evol 31: 440–452. [DOI] [PubMed] [Google Scholar]
- de Boer, W. , Hundscheid, M.P.J. , Klein Gunnewiek, P.J.A. , de Ridder‐Duine, A.S. , Thion, C. , van Veen, J.A. , and van der Wal, A. (2015) Antifungal rhizosphere bacteria can increase as response to the presence of saprotrophic fungi. PLoS One 10: e0137988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer, W. , de Ridder‐Duine, A.S. , Klein Gunnewiek, P.J.A. , Smant, W. , and van Veen, J.A. (2008) Rhizosphere bacteria from sites with higher fungal densities exhibit greater levels of potential antifungal properties. Soil Biol Biochem 40: 1542–1544. [Google Scholar]
- de Boer, W. , Wagenaar, A.‐M. , Klein Gunnewiek, P.J.A. , and van Veen, J.A. (2007) In vitro suppression of fungi caused by combinations of apparently non‐antagonistic soil bacteria. FEMS Microbiol Ecol 59: 177–185. [DOI] [PubMed] [Google Scholar]
- Boschker, H.T.S. , and Middelburg, J.J. (2002) Stable isotopes and biomarkers in microbial ecology. FEMS Microbiol Ecol 40: 85–95. [DOI] [PubMed] [Google Scholar]
- Buée, M. , De Boer, W. , Martin, F. , van Overbeek, L. , and Jurkevitch, E. (2009) The rhizosphere zoo: an overview of plant‐associated communities of microorganisms, including phages, bacteria, archaea, and fungi, and of some of their structuring factors. Plant Soil 321: 189–212. [Google Scholar]
- Bunn, R.A. , Simpson, D.T. , Bullington, L.S. , Lekberg, Y. , and Janos, D.P. (2019) Revisiting the ‘direct mineral cycling’ hypothesis: arbuscular mycorrhizal fungi colonize leaf litter, but why? ISME J 13: 1891–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby, P.E. , Ridout, M. , and Newcombe, G. (2016) Fungal endophytes: modifiers of plant disease. Plant Mol Biol 90: 645–655. [DOI] [PubMed] [Google Scholar]
- Butler, J.L. , Williams, M.A. , Bottomley, P.J. , and Myrold, D.D. (2003) Microbial community dynamics associated with rhizosphere carbon flow. Appl Environ Microbiol 69: 6793–6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan, B.J. , McMurdie, P.J. , Rosen, M.J. , Han, A.W. , Johnson, A.J.A. , and Holmes, S.P. (2016) DADA2: high‐resolution sample inference from Illumina amplicon data. Nat Methods 13: 581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrión, V.J. , Perez‐Jaramillo, J. , Cordovez, V. , Tracanna, V. , de Hollander, M. , Ruiz‐Buck, D. , et al. (2019) Pathogen‐induced activation of disease‐suppressive functions in the endophytic root microbiome. Science 366: 606. [DOI] [PubMed] [Google Scholar]
- Chandanie, W.A. , Kubota, M. , and Hyakumachi, M. (2009) Interactions between the arbuscular mycorrhizal fungus Glomus mosseae and plant growth‐promoting fungi and their significance for enhancing plant growth and suppressing damping‐off of cucumber (Cucumis sativus L.). Appl Soil Ecol 41: 336–341. [Google Scholar]
- Chaparro, J.M. , Badri, D.V. , Bakker, M.G. , Sugiyama, A. , Manter, D.K. , and Vivanco, J.M. (2013) Root exudation of phytochemicals in Arabidopsis follows specific patterns that are developmentally programmed and correlate with soil microbial functions. PLoS ONE 8: e55731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaparro, J.M. , Badri, D.V. , and Vivanco, J.M. (2014) Rhizosphere microbiome assemblage is affected by plant development. ISME J 8: 790–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaparro, J.M. , Sheflin, A.M. , Manter, D.K. , and Vivanco, J.M. (2012) Manipulating the soil microbiome to increase soil health and plant fertility. Biol Fertil Soils 48: 489–499. [Google Scholar]
- Chen, H. , Dai, Z. , Veach, A.M. , Zheng, J. , Xu, J. , and Schadt, C.W. (2020) Global meta‐analyses show that conservation tillage practices promote soil fungal and bacterial biomass. Agric Ecosyst Environ 293: 106841. [Google Scholar]
- Clocchiatti, A. , Hannula, S.E. , van den Berg, M. , Korthals, G. , and de Boer, W. (2020) The hidden potential of saprotrophic fungi in arable soil: patterns of short‐term stimulation by organic amendments. Appl Soil Ecol 147: 103434. [Google Scholar]
- Dennis, P.G. , Miller, A.J. , and Hirsch, P.R. (2010) Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol Ecol 72: 313–327. [DOI] [PubMed] [Google Scholar]
- Djajakirana, G. , Joergensen, R.G. , and Meyer, B. (1996) Ergosterol and microbial biomass relationship in soil. Biol Fertil Soils 22: 299–304. [Google Scholar]
- Drigo, B. , Pijl, A.S. , Duyts, H. , Kielak, A.M. , Gamper, H.A. , Houtekamer, M.J. , et al. (2010) Shifting carbon flow from roots into associated microbial communities in response to elevated atmospheric CO2 . Proc Natl Acad Sci U S A 107: 10938–10942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duah‐Yentumi, S. , and Johnson, D.B. (1986) Changes in soil microflora in response to repeated applications of some pesticides. Soil Biol Biochem 18: 629–635. [Google Scholar]
- Eisenhauer, N. , Lanoue, A. , Strecker, T. , Scheu, S. , Steinauer, K. , Thakur, M.P. , and Mommer, L. (2017) Root biomass and exudates link plant diversity with soil bacterial and fungal biomass. Sci Rep 7: 44641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frąc, M. , Hannula, S.E. , Bełka, M. , and Jędryczka, M. (2018) Fungal biodiversity and their role in soil health. Front Microbiol 9: 707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fravel, D. , Olivain, C. , and Alabouvette, C. (2003) Fusarium oxysporum and its biocontrol. New Phytol 157: 493–502. [DOI] [PubMed] [Google Scholar]
- Frey, S.D. , Elliott, E.T. , and Paustian, K. (1999) Bacterial and fungal abundance and biomass in conventional and no‐tillage agroecosystems along two climatic gradients. Soil Biol Biochem 31: 573–585. [Google Scholar]
- Frostegård, Å. , Tunlid, A. , and Bååth, E. (1993) Phospholipid fatty acid composition, biomass, and activity of microbial communities from two soil types experimentally exposed to different heavy metals. Appl Environ Microbiol 59: 3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frostegård, Å. , Tunlid, A. , and Bååth, E. (2011) Use and misuse of PLFA measurements in soils. Soil Biol Biochem 43: 1621–1625. [Google Scholar]
- Gao, Z. , Han, M. , Hu, Y. , Li, Z. , Liu, C. , Wang, X. , et al. (2019) Effects of continuous cropping of sweet potato on the fungal community structure in rhizospheric soil. Front Microbiol 10: 2269–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García‐Orenes, F. , Morugán‐Coronado, A. , Zornoza, R. , and Scow, K. (2013) Changes in soil microbial community structure influenced by agricultural management practices in a Mediterranean agro‐ecosystem. PLoS ONE 8: e80522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido‐Oter, R. , Nakano, R.T. , Dombrowski, N. , Ma, K.‐W. , McHardy, A.C. , and Schulze‐Lefert, P. (2018) Modular traits of the Rhizobiales root microbiota and their evolutionary relationship with symbiotic rhizobia. Cell Host Microbe 24: 155–167.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkarmiri, K. , Mahmood, S. , Ekblad, A. , Alström, S. , Högberg, N. , and Finlay, R. (2017) Identifying the active microbiome associated with roots and rhizosphere soil of oilseed rape. Appl Environ Microbiol 83: e01938–e01917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez Expósito, R. (2017) Microbiome dynamics of disease suppresive soils.
- Gransee, A. , and Wittenmayer, L. (2000) Qualitative and quantitative analysis of water‐soluble root exudates in relation to plant species and development. J Plant Nutr Soil Sci 163: 381–385. [Google Scholar]
- Gryndler, M. , Vosátka, M. , Hršelová, H. , Chvátalová, I. , and Jansa, J. (2002) Interaction between arbuscular mycorrhizal fungi and cellulose in growth substrate. Appl Soil Ecol 19: 279–288. [Google Scholar]
- Haichar, F.E.Z. , Heulin, T. , Guyonnet, J.P. , and Achouak, W. (2016) Stable isotope probing of carbon flow in the plant holobiont. Anal Biotechnol 41: 9–13. [DOI] [PubMed] [Google Scholar]
- Han, L.‐L. , Wang, J.‐T. , Yang, S.‐H. , Chen, W.‐F. , Zhang, L.‐M. , and He, J.‐Z. (2017) Temporal dynamics of fungal communities in soybean rhizosphere. J Soil Sediment 17: 491–498. [Google Scholar]
- Hannula, S.E. , de Boer, W. , and van Veen, J.A. (2010) In situ dynamics of soil fungal communities under different genotypes of potato, including a genetically modified cultivar. Soil Biol Biochem 42: 2211–2223. [Google Scholar]
- Hannula, S.E. , Boschker, H.T.S. , de Boer, W. , and van Veen, J.A. (2012) 13C pulse‐labeling assessment of the community structure of active fungi in the rhizosphere of a genetically starch‐modified potato (Solanum tuberosum) cultivar and its parental isoline. New Phytol 194: 784–799. [DOI] [PubMed] [Google Scholar]
- Hinsinger, P. , Gobran, G.R. , Gregory, P.J. , and Wenzel, W.W. (2005) Rhizosphere geometry and heterogeneity arising from root‐mediated physical and chemical processes. New Phytol 168: 293–303. [DOI] [PubMed] [Google Scholar]
- Hoppe, B. , Krüger, D. , Kahl, T. , Arnstadt, T. , Buscot, F. , Bauhus, J. , and Wubet, T. (2015) A pyrosequencing insight into sprawling bacterial diversity and community dynamics in decaying deadwood logs of Fagus sylvatica and Picea abies . Sci Rep 5: 9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, J.‐L. , Lin, X.‐G. , Wang, J.‐H. , Shen, W.‐S. , Wu, S. , Peng, S.‐P. , and Mao, T.‐T. (2010) Arbuscular mycorrhizal fungal inoculation enhances suppression of cucumber Fusarium wilt in greenhouse soils. Pedosphere 20: 586–593. [Google Scholar]
- Hugoni, M. , Luis, P. , Guyonnet, J. , and Haichar, F.e.Z. (2018) Plant host habitat and root exudates shape fungal diversity. Mycorrhiza 28: 451–463. [DOI] [PubMed] [Google Scholar]
- Hünninghaus, M. , Dibbern, D. , Kramer, S. , Koller, R. , Pausch, J. , Schloter‐Hai, B. , et al. (2019) Disentangling carbon flow across microbial kingdoms in the rhizosphere of maize. Soil Biol Biochem 134: 122–130. [Google Scholar]
- de Jaeger, N. , Declerck, S. , and de la Providencia, I.E. (2010) Mycoparasitism of arbuscular mycorrhizal fungi: a pathway for the entry of saprotrophic fungi into roots. FEMS Microbiol Ecol 73: 312–322. [DOI] [PubMed] [Google Scholar]
- Jiang, X. , Wright, A.L. , Wang, X. , and Liang, F. (2011) Tillage‐induced changes in fungal and bacterial biomass associated with soil aggregates: a long‐term field study in a subtropical rice soil in China. Appl Soil Ecol 48: 168–173. [Google Scholar]
- Joergensen, R.G. , and Wichern, F. (2008) Quantitative assessment of the fungal contribution to microbial tissue in soil. Soil Biol Biochem 40: 2977–2991. [Google Scholar]
- Johnston, S.R. , Boddy, L. , and Weightman, A.J. (2016) Bacteria in decomposing wood and their interactions with wood‐decay fungi. FEMS Microbiol Ecol 92: fiw179. [DOI] [PubMed] [Google Scholar]
- Joner, E.J. , and Jakobsen, I. (1995) Growth and extracellular phosphatase activity of arbuscular mycorrhizal hyphae as influenced by soil organic matter. Soil Biol Biochem 27: 1153–1159. [Google Scholar]
- Kaiser, C. , Frank, A. , Wild, B. , Koranda, M. , and Richter, A. (2010) Negligible contribution from roots to soil‐borne phospholipid fatty acid fungal biomarkers 18:2ω6,9 and 18:1ω9. Soil Biol Biochem 42: 1650–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaštovská, E. , Edwards, K. , and Šantrůčková, H. (2017) Rhizodeposition flux of competitive versus conservative graminoid: contribution of exudates and root lysates as affected by N loading. Plant Soil 412: 331–344. [Google Scholar]
- Kepler, R.M. , Maul, J.E. , and Rehner, S.A. (2017) Managing the plant microbiome for biocontrol fungi: examples from Hypocreales. Curr Opin Microbiol 37: 48–53. [DOI] [PubMed] [Google Scholar]
- Koechli, C. , Campbell, A.N. , Pepe‐Ranney, C. , and Buckley, D.H. (2019) Assessing fungal contributions to cellulose degradation in soil by using high‐throughput stable isotope probing. Soil Biol Biochem 130: 150–158. [Google Scholar]
- Kramer, S. , Dibbern, D. , Moll, J. , Huenninghaus, M. , Koller, R. , Krueger, D. , et al. (2016) Resource partitioning between bacteria, fungi, and protists in the detritusphere of an agricultural soil. Front Microbiol 7: 1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzyakov, Y. , and Razavi, B.S. (2019) Rhizosphere size and shape: temporal dynamics and spatial stationarity. Soil Biol Biochem 135: 343–360. [Google Scholar]
- Lamichhane, J.R. , Dürr, C. , Schwanck, A.A. , Robin, M.‐H. , Sarthou, J.‐P. , Cellier, V. , et al. (2017) Integrated management of damping‐off diseases a review. Agron Sustain Dev 37: 10. [Google Scholar]
- Li, X. , Jousset, A. , de Boer, W. , Carrión, V.J. , Zhang, T. , Wang, X. , and Kuramae, E.E. (2019) Legacy of land use history determines reprogramming of plant physiology by soil microbiome. ISME J 13: 738–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas, S.T. , D'Angelo, E.M. , and Williams, M.A. (2014) Improving soil structure by promoting fungal abundance with organic soil amendments. Appl Soil Ecol 75: 13–23. [Google Scholar]
- Marschner, P. , Marhan, S. , and Kandeler, E. (2012) Microscale distribution and function of soil microorganisms in the interface between rhizosphere and detritusphere. Soil Biol Biochem 49: 174–183. [Google Scholar]
- Massart, S. , Martinez‐Medina, M. , and Jijakli, M.H. (2015) Biological control in the microbiome era: challenges and opportunities. Biol Control 89: 98–108. [Google Scholar]
- Mauclaire, L. , Pelz, O., Thullner, M., Abraham, W.‐R., and Zeyer, J. (2003) Assimilation of toluene carbon along a bacteria‐protist food chain determined by 13C‐enrichment of biomarker fatty acids. J Microbiol Methods 55: 635–649. [DOI] [PubMed] [Google Scholar]
- Mawarda, P.C. , Le Roux, X. , Dirk van Elsas, J. , and Salles, J.F. (2020) Deliberate introduction of invisible invaders: a critical appraisal of the impact of microbial inoculants on soil microbial communities. Soil Biol Biochem 148: 107874. [Google Scholar]
- de Menezes, A.B. , Richardson, A.E. , and Thrall, P.H. (2017) Linking fungal–bacterial co‐occurrences to soil ecosystem function. Curr Opin Microbiol 37: 135–141. [DOI] [PubMed] [Google Scholar]
- Morriën, E. , Hannula, S.E. , Snoek, L.B. , Helmsing, N.R. , Zweers, H. , de Hollander, M. , et al. (2017) Soil networks become more connected and take up more carbon as nature restoration progresses. Nat Commun 8: 14349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, N.H. , Song, Z., Bates, S.T., Branco, S., Tedersoo, L., Menke, J., et al. (2016) FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol 20: 241–248. [Google Scholar]
- Nettles, R. , Watkins, J. , Ricks, K. , Boyer, M. , Licht, M. , Atwood, L.W. , et al. (2016) Influence of pesticide seed treatments on rhizosphere fungal and bacterial communities and leaf fungal endophyte communities in maize and soybean. Appl Soil Ecol 102: 61–69. [Google Scholar]
- Pausch, J. , Kramer, S. , Scharroba, A. , Scheunemann, N. , Butenschoen, O. , Kandeler, E. , et al. (2016) Small but active – pool size does not matter for carbon incorporation in below‐ground food webs. Funct Ecol 30: 479–489. [Google Scholar]
- Pausch, J. , and Kuzyakov, Y. (2018) Carbon input by roots into the soil: quantification of rhizodeposition from root to ecosystem scale. Glob Change Biol 24: 1–12. [DOI] [PubMed] [Google Scholar]
- Petrini, O. , and Fisher, P.J. (1990) Occurrence of fungal endophytes in twigs of Salix fragilis and Quercus robur . Mycol Res 94: 1077–1080. [Google Scholar]
- Poli, A. , Lazzari, A. , Prigione, V. , Voyron, S. , Spadaro, D. , and Varese, G.C. (2016) Influence of plant genotype on the cultivable fungi associated to tomato rhizosphere and roots in different soils. Fungal Biol 120: 862–872. [DOI] [PubMed] [Google Scholar]
- Quist, C.W. , Schrama, M. , de Haan, J.J. , Smant, G. , Bakker, J. , van der Putten, W.H. , and Helder, J. (2016) Organic farming practices result in compositional shifts in nematode communities that exceed crop‐related changes. Appl Soil Ecol 98: 254–260. [Google Scholar]
- Rahman, M.T. , Zhu, Q.H. , Zhang, Z.B. , Zhou, H. , and Peng, X. (2017) The roles of organic amendments and microbial community in the improvement of soil structure of a vertisol. Appl Soil Ecol 111: 84–93. [Google Scholar]
- de Ridder‐Duine, A.S. , Smant, W. , van der Wal, A. , van Veen, J.A. , and de Boer, W. (2006) Evaluation of a simple, non‐alkaline extraction protocol to quantify soil ergosterol. Pedobiologia 50: 293–300. [Google Scholar]
- Rivers, A. , Weber, K., Gardner, T., Liu, S., and Armstrong, S. (2018) ITSxpress: Software to rapidly trim internally transcribed spacer sequences with quality scores for marker gene analysis. F1000Research 7: 1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrocco, S. (2016) Dung‐inhabiting fungi: a potential reservoir of novel secondary metabolites for the control of plant pathogens. Pest Manag Sci 72: 643–652. [DOI] [PubMed] [Google Scholar]
- Schellenberger, S. , Kolb, S. , and Drake, H.L. (2009) Metabolic responses of novel cellulolytic and saccharolytic agricultural soil bacteria to oxygen: metabolic response of soil cellulose degraders. Environ Microbiol 12: 845–861. [DOI] [PubMed] [Google Scholar]
- Schneider, T. , Gerrits, B. , Gassmann, R. , Schmid, E. , Gessner, M.O. , Richter, A. , et al. (2010) Proteome analysis of fungal and bacterial involvement in leaf litter decomposition. Proteomics 10: 1819–1830. [DOI] [PubMed] [Google Scholar]
- Shao, H. , and Zhang, Y. (2017) Non‐target effects on soil microbial parameters of the synthetic pesticide carbendazim with the biopesticides cantharidin and norcantharidin. Sci Rep 7: 5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ståhl, M. , Granström, K. , Berghel, J. , and Renström, R. (2004) Industrial processes for biomass drying and their effects on the quality properties of wood pellets. Pellets 2002 First World Conf Pellets 27: 621–628. [Google Scholar]
- Tavi, N.M. , Martikainen, P.J. , Lokko, K. , Kontro, M. , Wild, B. , Richter, A. , and Biasi, C. (2013) Linking microbial community structure and allocation of plant‐derived carbon in an organic agricultural soil using 13CO2 pulse‐chase labelling combined with 13C‐PLFA profiling. Soil Biol Biochem 58: 207–215. [Google Scholar]
- de Vries, F.T. , and Bardgett, R.D. (2012) Plant–microbial linkages and ecosystem nitrogen retention: lessons for sustainable agriculture. Front Ecol Environ 10: 425–432. [Google Scholar]
- de Vries, F.T. , and Caruso, T. (2016) Eating from the same plate? Revisiting the role of labile carbon inputs in the soil food web. Spec Issue Food Web Interact Root Zone Influ Community Ecosyst Dyn 102: 4–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries, F.T. , van Groenigen, J.W. , Hoffland, E. , and Bloem, J. (2011) Nitrogen losses from two grassland soils with different fungal biomass. Soil Biol Biochem 43: 997–1005. [Google Scholar]
- van der Wal, A. , Geydan, T.D. , Kuyper, T.W. , and de Boer, W. (2013) A thready affair: linking fungal diversity and community dynamics to terrestrial decomposition processes. FEMS Microbiol Rev 37: 477–494. [DOI] [PubMed] [Google Scholar]
- van der Wal, A. , Klein Gunnewiek, P.J.A. , Cornelissen, J.H.C. , Crowther, T.W. , and de Boer, W. (2016) Patterns of natural fungal community assembly during initial decay of coniferous and broadleaf tree logs. Ecosphere 7: e01393. [Google Scholar]
- Wang, J. , Liu, G. , Zhang, C. , Wang, G. , Fang, L. , and Cui, Y. (2019) Higher temporal turnover of soil fungi than bacteria during long‐term secondary succession in a semiarid abandoned farmland. Soil Tillage Res 194: 104305. [Google Scholar]
- Wang, Y. , Li, C. , Tu, C. , Hoyt, G.D. , DeForest, J.L. , and Hu, S. (2017) Long‐term no‐tillage and organic input management enhanced the diversity and stability of soil microbial community. Sci Total Environ 609: 341–347. [DOI] [PubMed] [Google Scholar]
- Wieczorek, A.S. , Schmidt, O. , Chatzinotas, A. , von Bergen, M. , Gorissen, A. , and Kolb, S. (2019) Ecological functions of agricultural soil bacteria and microeukaryotes in chitin degradation: a case study. Front Microbiol 10: 1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willers, C. , Jansen van Rensburg, P.J. , and Claassens, S. (2015) Phospholipid fatty acid profiling of microbial communities–a review of interpretations and recent applications. J Appl Microbiol 119: 1207–1218. [DOI] [PubMed] [Google Scholar]
- Wu, J. , Huang, M. , Xiao, H.‐A. , Su, Y.‐R. , Tong, C.‐L. , Huang, D.‐Y. , and Syers, J.K. (2007) Dynamics in microbial immobilization and transformations of phosphorus in highly weathered subtropical soil following organic amendments. Plant Soil 290: 333–342. [Google Scholar]
- Xu, J. , Liu, S. , Song, S. , Guo, H. , Tang, J. , Yong, J.W.H. , et al. (2018) Arbuscular mycorrhizal fungi influence decomposition and the associated soil microbial community under different soil phosphorus availability. Soil Biol Biochem 120: 181–190. [Google Scholar]
- Zhalnina, K. , Louie, K.B. , Hao, Z. , Mansoori, N. , da Rocha, U.N. , Shi, S. , et al. (2018) Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat Microbiol 3: 470–480. [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Ding, X. , Peng, Y. , George, T.S. , and Feng, G. (2018) Closing the loop on phosphorus loss from intensive agricultural soil: a microbial immobilization solution? Front Microbiol 9: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W. , Li, X.‐G. , Sun, K. , Tang, M.‐J. , Xu, F.‐J. , Zhang, M. , and Dai, C.‐C. (2020) Mycelial network‐mediated rhizobial dispersal enhances legume nodulation. ISME J 14: 1015–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Bacterial and fungal abundance in soil, rhizosphere and roots, based on qPCR and ergosterol.
Supplementary Materials and Methods: Measurement of 13C content of soil, rhizosphere, shoot and roots, molecular analyses (qPCR, gradient fractionation and sequencing), bioinformatic processing of raw sequencing data.
Fig. S2. Effect of sawdust on fungal families assigned to saprotrophs and potential pathogens.
Fig. S3. Estimated effect of sawdust on the absolute abundance of fungal functional groups.
Table S1. Biomass, N content and germination rate of seedlings, 13C content of labelled soil, rhizosphere and seedlings.
Table S2. Summary of ANOVA results on PLFAs and NLFA data as a function of organic soil amendment and compartment, as performed independently for each time point.
Table S3. Mean concentration and summary of statistical differences of individual PLFAs and NLFAs (nmol C g−1 soil).
Table S4. Mean labelled fraction and summary of statistical differences of individual PLFAs and NLFAs (pmol excess 13C g−1 soil).
Table S5. Summary of individual fractions pooled into ‘heavy’ and ‘light’ DNA fractions.
Table S6. Summary of permutational multivariate analysis of variance for the fungal community.
Table S7. Summary of permutational multivariate analysis of variance for the bacterial community.
Table S8. Taxa and functional groups identified by ITS2 rDNA sequencing.
Appendix S1 Supporting information.
Data Availability Statement
The data presented in this study is available at doi:10.5061/dryad.2rbnzs7nn. Sequencing data is accessible at the European Nucleotide Archive (accession PRJEB37482(ERP120799)).


