Abstract
Solute carrier proteins (SLCs) are membrane proteins controlling fluxes across biological membranes and represent an emerging class of drug targets. Here we searched for inhibitors of divalent metal transporters in a library of 1,676 commercially available 3D‐shaped fragment‐like molecules from the generated database GDB‐17, which lists all possible organic molecules up to 17 atoms of C, N, O, S and halogen following simple criteria for chemical stability and synthetic feasibility. While screening against DMT1 (SLC11A2), an iron transporter associated with hemochromatosis and for which only very few inhibitors are known, only yielded two weak inhibitors, our approach led to the discovery of the first inhibitor of ZIP8 (SLC39A8), a zinc transporter associated with manganese homeostasis and osteoarthritis but with no previously reported pharmacology, demonstrating that this target is druggable.
Keywords: membrane transporters, fragments, chemical space, inhibitors, iron transport
Transport restrictions: Screening a library of 1,676 commercially available fragments from the database GDB‐17 revealed two weak inhibitors of DMT1, an iron transporter associated with hemochromatosis, and the first inhibitor of ZIP8, a zinc transporter associated with manganese homeostasis and osteoarthritis.
Introduction
Solute carrier proteins (SLCs) are membrane proteins controlling fluxes of ions and other molecules across biological membranes and represent an emerging class of drug targets.[ 1 , 2 , 3 ] Herein we report the identification of small molecule inhibitors of two divalent metal transporters implicated in a number of diseases, namely DMT1 (SLC11A2)[ 4 , 5 , 6 , 7 , 8 , 9 ] and ZIP8 (SLC39A8),[ 10 , 11 , 12 ] for which few or no modulators are known. DMT1 serves as the key factor for intestinal absorption of dietary inorganic iron across the brush border membrane, it plays a central role in whole body iron homeostasis, and alteration of its activity has been linked to the development of a wide variety of human diseases, including iron deficiency anaemia, hemochromatosis, β‐thalassemia, Parkinson's disease and Alzheimer's disease.[ 7 , 13 , 14 , 15 , 16 ] In contrast, the function of ZIP8 is not yet fully defined. However, it has been shown to be key to maintaining whole body manganese homeostasis, its genetic variants have been linked to a wide variety of human pathologies, including schizophrenia, scoliosis, obesity or Crohn's disease,[ 17 , 18 , 19 ] and lately it has gained great interest as a therapeutic target for the treatment of cartilage and joint diseases.[ 20 , 21 , 22 ]
Although little information is available on their structure, divalent metal transporters possess well defined pockets that should be suitable for inhibition by small molecules, as recently shown by the structural characterization of a competitive inhibitor of DMT1 in complex with a bacterial analog of the transporter.[ 23 , 24 ] In view of our previous difficulties in identifying DMT1 inhibitors caused by interfering metal chelation [25] and the almost complete absence of known inhibitors for such transporters, we set out to identify small molecule inhibitors using a fragment‐based drug discovery approach. Fragment‐based drug discovery consists in testing a small number of structurally diverse fragments, defined as rigid small molecules with few functional groups (Congreve's rule of 3: MW≤300 Da, H‐bond acceptor atoms ≤3, H‐bond donor atoms ≤3, rotatable bonds ≤3, polar surface area ≤60 Å2), for weak target interaction to identify initial hits which can later be optimized for activity.[ 26 , 27 , 28 , 29 ] In our context, this approach had the advantage to be compatible with our activity assays using transfected HEK293T cells, which have a limited throughput.[ 30 , 31 ]
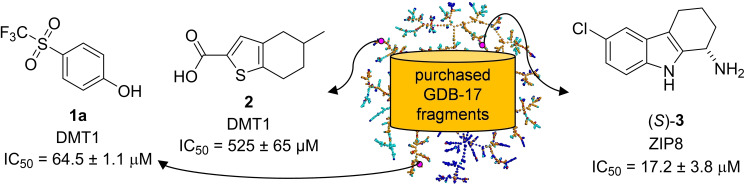
We composed a fragment collection by selecting commercially available molecules with fragment‐like properties appearing in GDB‐17, a database of all possible organic molecules up to 17 atoms following simple rules of chemical stability and synthetic feasibility.[ 32 , 33 ] This fragment‐based discovery project allowed us to experimentally challenge GDB‐17 as a source of diverse fragments for screening, thereby significantly extending our previous drug discovery project with GDB databases,[ 34 , 35 ] which all had been guided by virtual screening [36] on targets with known pharmacology such as NMDA receptors, [37] nicotinic acetylcholine receptors, [38] glutamate transporters [39] and Janus kinases. [40] As detailed below, design and screening of a GDB‐17 derived fragment set resulted in the identification of the trifluoromethylsulfone 1 a and thiophene carboxylic acid 2 as weak inhibitors of DMT1, and of tetrahydrocarbazole (S)‐3 as the first inhibitor of ZIP8 (Figure 1).
Figure 1.
Assembly of GDB‐17 fragment screening set and discovery of divalent metal transporter inhibitors.
Results and Discussion
Design and properties of a GDB‐17 fragment library
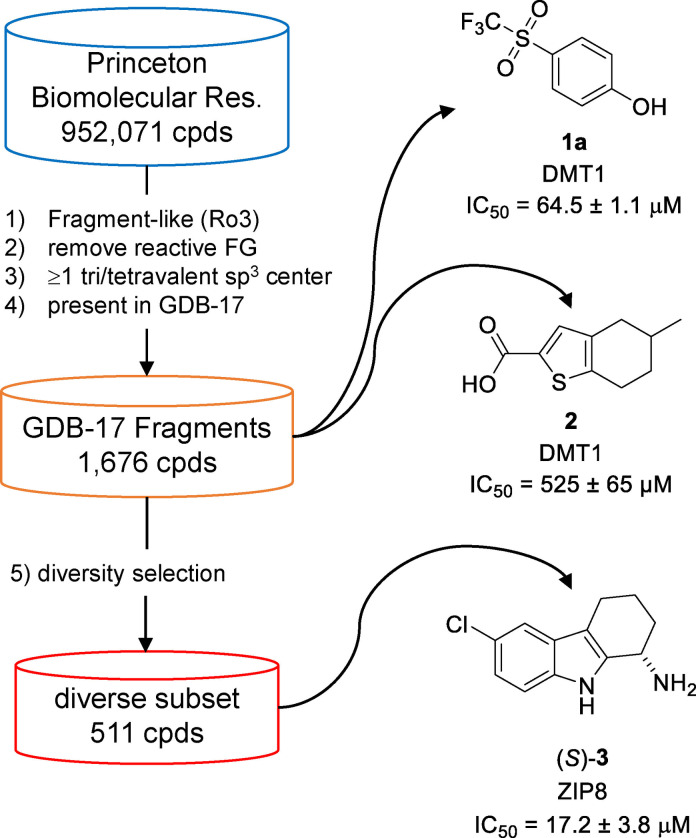
While GDB molecules lie well within the size range required for fragments, [26] most of them are highly functionalized and structurally too complex to be considered as realistic synthetic targets. Therefore, we defined subsets containing molecules of reduced complexity with fragment‐like, [41] drug‐like [42] or ChEMBL‐like features. [43] To assemble a diverse collection of GDB‐17 fragments for experimental screening, we selected molecules as follows (Figure 1): 1) starting from commercially available molecules, retrieve all molecules with up to 17 atoms and following Congreve's rule of 3 (Ro3); [26] 2) eliminate reactive functional groups (FG); [32] 3) select molecules containing at least one saturated trivalent carbon or quaternary center, a structural feature present in most GDB‐17 molecules but underrepresented in commercial compounds; 4) check that every molecule selected is indeed present in GDB‐17 using a previously reported GDB‐17 search tool. [33] We applied this procedure to the catalog of a single commercial provider (Princeton BioMolecular Research, Inc.) and obtained 1,900 fragment‐like molecules from GDB‐17. A total of 1,676 of these molecules were purchased and actually delivered, providing a collection compatible with our screening capacity for DMT1. To adjust for our limited screening capacity concerning ZIP8 (see below), we performed an additional diversity selection of these 1,676 fragments by clustering and minimizing similarities between nearest neighbors to obtain a smaller subset of 511 fragments.
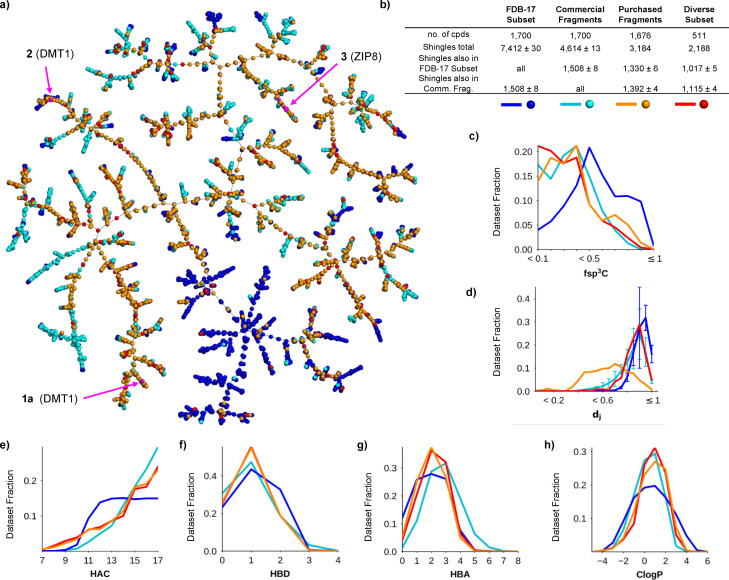
To assess the properties of our purchased fragments, we compared them with a random selection of 1,700 fragments from FDB‐17, a database of 10 million fragments selected from GDB‐17, [41] and a second, equally sized random selection from a cumulated set of 18,151 commercially available fragments up to a size of 17 heavy atoms (Figure 2). The differences between purchased, commercial and FDB‐17 fragments was visible in a TMAP (tree‐map) layout computed using the MAP4 fingerprint combining the three compound sets (Figure 2a).[ 44 , 45 ] In this TMAP, the FDB‐17 subset was in large part separated from purchased and commercial fragments. This difference is caused by the abundance of 3D‐shaped non‐aromatic fragments in the FDB‐17 subset, which are typical of GDB molecules but not well represented among commercially available molecules, including those in our purchased set. Not surprisingly therefore, the purchased fragments or its diverse subset shared a slightly larger fraction of their respective molecular shingles (circular substructures up to a radius of three bonds)[ 43 , 46 ] with the commercial fragment subset (44 % or 51 %) than with the FDB‐17 subsets (42 % or 46 %) (Figure 2b, shingle counts were averaged over 10 different random subsets of FDB‐17 and commercial fragments).
Figure 2.
Comparison of purchased fragments with fragments from FDB‐17 and with commercial fragments. (a) TMAP (tree‐map) layout of the 1,676 purchased fragments (orange+red) including its diversity subset of 511 compounds (red) combined with 1,700 randomly selected fragments from FDB‐17 (blue) and from commercial fragments (cyan). Each point corresponds to a molecule. The active compounds are shown in magenta. An interactive version of this TMAP is accessible at https://tm.gdb.tools/fragment_project/tmap/1.7k_pcc_fdb_mcf.html (b) Analysis of molecular shingles from each subset. The shingle counts are given as the average±standard error across 10 different selections of FDB‐17 subsets and commercial fragments. (c) Histogram of fsp3C (fraction of sp3 carbon atoms). (d) Nearest neighbor similarity analysis in terms of Jaccard distance (dj) according to the molecular fingerprint MAP4. (e–h) Histograms of HAC (heavy atom count), HBD (hydrogen bond donor atom count), HBA (hydrogen bond acceptor atom count), and CLogP (calculated octanol:water partition coefficient). The standard error in histogram values across the ten FDB‐17 subsets (blue lines) and the ten commercial fragment set (cyan lines) were below 1 % for all histograms and are not shown except for the nearest neighbor Jaccard distance histogram.
Furthermore, our selection rule requiring at least one tri‐ or tetrasubstituted tetrahedral atom in each fragment, to avoid the planarity of commercial fragments,[ 47 , 48 ] did not shift significantly the histogram of fsp3C (fraction of sp3 carbon atoms) values towards the high values observed in the FDB‐17 random subset (Figure 2c). Comparing the dissimilarity between nearest neighbors within each set, measured by the Jaccard distance (dj) using the molecular fingerprint MAP4, showed that our purchased subset had many pairs of similar compounds reflecting acquisition from a single commercial provider. However, our diversity subset of 511 compounds showed clearly dissimilar molecules as found in the FDB‐17 and commercial fragment sets (Figure 2d).
The purchased fragments had a similar size distribution to the commercial fragments, while the FDB‐17 subset contained relatively smaller molecules due to its flat size distribution in the range 12≤HAC (heavy atom count)≤17 reflecting the sampling procedure used to assemble FDB‐17 (Figure 2e). Purchased fragments matched commercial fragments in terms of HBD (hydrogen bond donor atom count, Figure 2f), but their HBA (hydrogen bond acceptor atoms count) distribution was closer to the FDB‐17 subset (Figure 2g). This difference in HBA/HBD distribution probably explains the lower polarity of the purchased fragments compared to commercial fragments and the FDB‐17 subset visible in the CLogP histogram (calculated octanol:water partition coefficient, Figure 2h).
In summary, this analysis showed that selecting fragments up to 17 atoms with the conditions that they belong to GDB‐17 produced a fragment set resembling commercial fragments in terms of structural types and molecular property profiles.
Activity screening
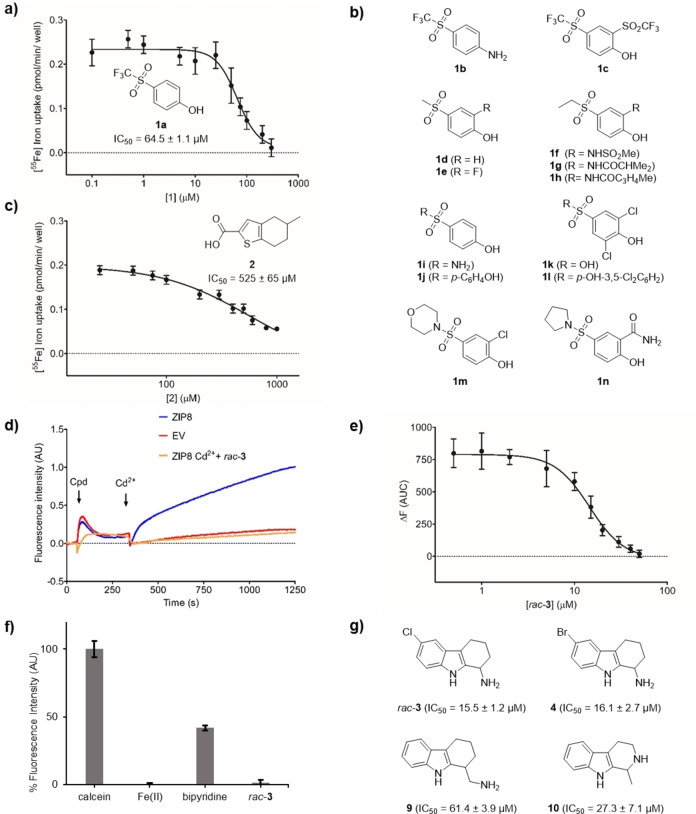
To identify DMT1 inhibitors, we screened the entire library of 1,676 GDB‐17 fragments, conditioned as 10 mM stock solutions in DMSO, by monitoring the uptake of radioactive 55Fe in HEK293 cells stably overexpressing the transporter. We used a concentration of 10 μM screening compound, which we judged sufficiently low to avoid non‐specific effects on cells but high enough to indicate even a weak inhibition, as described earlier. [25] The assay revealed 162 compounds with only weak activity (15–25 %), which were then retested at higher concentration (50 μM) to observe a stronger effect for confirmation. By repurchasing, purification and retesting, we were able to confirm the activity of trifluoromethylsulfone 1 a (IC50=64.5±1.1 μM) corresponding to a ligand efficiency of LE=0.42 kcal.mol−1, [49] an acceptable value for an initial fragment hit (Figure 3a). Unfortunately, testing of 13 purchasable analogs of this hit did not indicate any other active compound, indicating that the relatively weak activity of 1 a was highly sensitive to structural changes (1 b–1 n, Figure 3b). We also characterized a second hit, thiophenecarboxylic acid 2, however this compound showed an even lower potency (IC50=525±65 μM, LE=0.35 kcal.mol−1, Figure 3c). These modest results were in line with previous efforts in our laboratory showing that DMT1 activity screens give a very low hit rate. [31]
Figure 3.
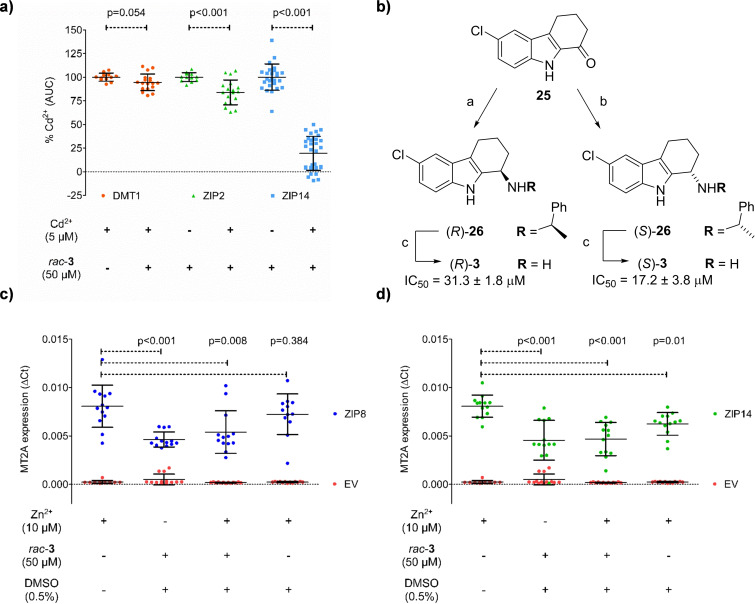
Discovery of DMT1 and ZIP8 inhibitors by GDB fragment screening. IC50 curves determined for compounds 1 a (a) and 2 (c) using HEK293 cells stably overexpressing DMT1. Cells were pre‐incubated for 5 min in the presence of the indicated concentrations of 1 a and 2. Next, radioactive 55Fe2+ was added (1 μM), and cells were incubated for 15 minutes. Each data point represents the Mean±SD (N=6–8) of the 55Fe2+‐uptake determined in the presence of the indicated compound concentrations. (b) Structure of analogs 1 b–1 n found to be inactive against DMT1. (d) Representative trace of the Cd2+‐flux recorded in HEK293T cells transiently transfected with ZIP8 or the empty vector. Cd2+‐flux was monitored using the FLIPR Calcium 5 Assay Kit. A baseline was recorded for 30 sec, then, cells were incubated for 5 minutes in the presence of rac‐3 (50 μM), and finally, Cd2+ (5 μM) was added, and the signal (AU) was recorded for 15 minutes. (e) IC50 curve determined for rac‐3 measuring the Cd2+‐uptake in the presence of the indicated compound concentrations. Cd2+‐uptake was determined as the Area Under the Curve (AUC) of the change in florescence intensity observed upon substrate addition (459–750 s). Each data point represents the Mean±SD (N=6–8) of the AUC determined in the presence of each compound concentration. (f) Calcein quenching assay at pH 7.4. Ligand (20 μM), Fe2+ (4 μM) and ascorbic acid (400 μM) were preincubated for 5 min. Then, an equal volume of calcein (2 μM) in uptake solution was added to give the final concentrations of 1 μM calcein, 10 μM rac‐3 or bipyridine, 2 μM Fe2+ and 200 μM ascorbic acid, which were incubated for 5 min before the fluorescence measurement. Data was obtained from an experiment performed in quintuplicate, and results are presented as the Mean±SD. See Supporting Information for details. (g) Structure of inhibitor rac‐ 3 and analogs 4, 9 and 10. IC50 values in (a), (c) and (e) are the Mean±SD (N=4–6) of the IC50 values calculated from two independent experiments performed in triplicate.
To test inhibition of the human zinc transporter ZIP8 (SLC39A8), we established a cell‐based assay in HEK293T cells transiently transfected with ZIP8, detecting uptake of divalent cadmium with the FLIPR Calcium 5 Assay Kit (Figure 3d). This assay was adapted from a previously published assay for the related ZIP2 using transient transfection because stably expressing the transporter is toxic to the cells.[ 50 , 51 ] Due to the limited throughput of screening using transiently transfected cells, we performed activity assays with the smaller, diversity selected subset of 511 GDB‐17 fragments rather than the entire set and tested directly at a higher concentration (50 μM) to not miss any activity. A first round of screening resulted in 95 compounds with an inhibition of the cadmium flux greater than 50 %. Secondary screening of these initial hits at 25 μM followed by re‐screening of fresh solutions of the 10 best secondary hits resulted in tetrahydrocarbazole rac‐3 as a confirmed inhibitor (IC50=15.5±1.2 μM, Figure 3e).
The relative positioning of the two amines in rac‐3 suggests that the observed iron uptake inhibition might reflect metal chelation as reported recently for pyrazolyl‐pyrimidones. [25] To exclude this possibility, we performed a calcein competition assay, where the quenching of calcein fluorescence indicates the presence of free Fe2+. In contrast to the positive control bipyridine, we could not detect any significant level of iron chelation by rac‐3 (Figure 3f). Additionally, attempts to detect iron complexation by 1H‐NMR were unsuccessful.
A survey of twenty‐one close analogs of rac‐3 (compounds 4–24, Supporting Information Figure S1), which were either retrieved from the complete set of 1,676 fragments or purchased additionally, revealed three analogs with comparable although slightly lower potency than rac‐3, showing that the chloro substituent could be replaced by bromo, and that the amino group was essential for inhibition but could be placed slightly differently in the molecule (4, 9 and 10, Figure 3g and Supporting Information Figure S2a).
Characterization of the ZIP8 inhibitor
To evaluate the specificity of rac‐3, we tested its inhibitory activity with other human divalent metal transporters available in our laboratory, including DMT1, ZIP2 and ZIP14 (Figure 4a). Using the Cd2+‐uptake fluorescence‐based assay, we observed that DMT1 was not inhibited by 50 μM rac‐3. However, while ZIP2 was only weakly inhibited, rac‐3 inhibited ZIP14 as strongly as ZIP8 (IC50=10.57±0.54 μM). This cross‐inhibition probably reflects the fact that ZIP8 and ZIP14 are evolutionary closely related proteins and belong to the same subfamily within the ZIP family.
Figure 4.
Characterization of ZIP inhibition by rac‐3. (a) Average Cd2+‐flux recorded in the absence or presence of rac‐3 in HEK293T cells transiently transfected with DMT1, ZIP2, or ZIP14. Cd2+‐uptake was determined as the Area Under the Curve (AUC) of the change in florescence intensity observed upon substrate addition. Results were expressed as % of the Cd2+‐flux recorded in the absence of rac‐3. Data from three independent experiments performed in triplicate are represented as Mean±SD (N=12–33). (b) Enantioselective synthesis and activity of (R)‐3 and (S)‐3. Conditions: a) i) (S)‐α‐methyl‐4‐methoxybenzylamine, conc. aq. HCl, toluene, reflux overnight, ii) NaBH4, EtOH, −30 °C – RT, overnight, iii) HCl, MeOH, toluene, RT, 60 min. (59 %); b) same as a) using (R)‐α‐methyl‐4‐methoxybenzylamine (66 %); c) BCl3, CH2Cl2, 10 °C, overnight (R: 18 %, S: 47 %). (c–d) detoxification gene expression assay with ZIP8 and ZIP14. MT2A gene expression in HEK293T cells transiently transfected with ZIP8 (blue, left panel), ZIP14 (green, right panel) or the empty vector (EV, red) treated for 2 hours with the indicated solutions. Expression of MT2A mRNA was determined by Real‐Time PCR. Obtained Ct values for MT2A gene were normalized to the housekeeping gene GAPDH following the ΔCt method. Data was obtained from 5 independent experiments performed in triplicate, and results are presented as the Mean±SD (N=10–13). Statistical differences were determined by t‐test or Mann‐Whitney U test ((a) Cd2+ vs Cd2++rac‐ 3; (c–d) Zn2+ (10 μM) vs. each other condition), p values are indicated on top of the corresponding graphs.
We performed an enantioselective synthesis of (R)‐3 and (S)‐3 by reductive amination from the parent ketone 25 with (R)‐ or (S)‐α‐methylbenzylamine to form the corresponding amine (R)‐26 or (S)‐26 stereoselectively followed by hydrogenation using a previously reported procedure. [52] Testing the individual enantiomers showed that (S)‐3 inhibited ZIP8 slightly stronger than (R)‐3 (Figure 4b and Supporting Information Figure S2b). Considering that the chiral center in 3 is adjacent to the primary amino group, the low enantioselectivity was in line with the SAR study discussed above showing that analogs with a differently placed amino group (9 and 10, Figure 3g) retained activity against ZIP8. Both enantiomers of 3 also inhibited ZIP14 to a similar extent.
To test if the effect of rac‐3 on ZIP8 indeed affected the transport of divalent metal ions into cells, we tested the expression of MT2A, a metal detoxification gene that is normally activated if cells are exposed to high levels of divalent metal ions, in the presence or absence of our inhibitor and 10 μM Zn2+. Indeed, expression of the MT2A gene upon exposure to zinc was strongly down‐regulated in the presence of rac‐3, confirming that our inhibitor strongly reduced divalent metal ion uptake into the cells (Figure 4c).
The presence of a primary amine and a tetrahydrocarbazole ring system in rac‐3 suggested that our inhibitor might also interact with neurotransmitter receptors and transporters. To explore the activity profile, we used our polypharmacology browser PPB2 to predict possible off‐targets,[ 53 , 54 ] and tested eight of the suggestions experimentally with the more active enantiomer (S)‐3. The experiment revealed significant activity (>50 % inhibition at 10 μM) for four targets, namely the serotonin transporter, hERG, the dopamine transporter and the serotonin receptor 5‐HT2 A (Table 1). (R)‐3 was also tested for hERG and serotonin transporter and showed similar cross‐reactivity.
Table 1.
Off‐target profiles of ZIP8 inhibitor 3.
|
Target[a] |
Inhib. [%] |
|---|---|
|
5‐HT transporter (h) (antagonist radioligand) |
89.2±0.8 (98.4±0.6)[b] |
|
Potassium Channel hERG (h)‐ [3H] Dofetilide |
72.5±0.07 (54.2±1.8) [b] |
|
5‐HT2A (h) (agonist radioligand) |
55.6±1.7 |
|
Dopamine transporter (h) (antagonist radioligand) |
54.6±1.0 |
|
Ca2+ channel (L, dihydropyridine site) (antagonist radioligand) |
48.9±7.3 |
|
5‐HT2A (h) (antagonist radioligand) |
38.1±5.4 |
|
5‐HT6 (h) (agonist radioligand) |
31.6±5.7 |
|
5‐HT1A (h) (agonist radioligand) |
13.2±0.9 |
|
5‐HT3 (h) (antagonist radioligand) |
8.2±0.4 |
[a] The effect of (S)‐3 was measured at 10 μM in a radiolabelled displacement assay using an agonist or an antagonist specific of each target. Assays were performed by Eurofins Cerep (France). Any inhibition of the control specific binding higher than 50 % was considered significant. Data are shown as a mean of two experiments. h, human. [b] Data for (R)‐3.
Conclusion
In summary, we used a fragment‐based drug discovery approach to identify inhibitors of DMT1 (SLC11A2) and ZIP8 (SLC39A8), two divalent metal transporters implicated in important human diseases but with almost no reported pharmacology. We designed our fragment library by selecting fragment‐like molecules occurring in the generated database GDB‐17 and containing at least one tri‐ or tetravalent tetrahedral center to increase 3D‐shape as a proof‐of‐concept experiment to exploit GDB‐17 for drug discovery.
Screening uncovered compounds 1 a and 2 as two weak inhibitors of DMT1, as well as rac‐3 as the first case of an inhibitor of ZIP8. A structure‐activity relationship study around rac‐ 3 showed that this inhibitor is slightly more active as single enantiomer (S)‐3. The identification of (S)‐3 as the first inhibitor of ZIP8 by a limited fragment‐based screening approach suggests that this target is druggable. While exhibiting excellent selectivity against DMT1 and ZIP2, (S)‐3 showed significant cross‐reactivity with ZIP14, a close analog of ZIP8 which might also represent a therapeutic target,[ 55 , 56 ] as well as significant off‐target effects typical of aliphatic amines with aromatic substituents. Additional screening efforts for analogs of (S)‐3 or from a larger diverse library will be necessary to identify further inhibitors with higher activity and selectivity.
Experimental Section
Cheminformatics
All structures were converted into their major microspecies at pH 7.4 with the ChemAxon tool cxcalc (19.22.0) and successively stored as achiral, canonized SMILES strings using rdkit (2019.03.4).
Selection of purchasable GDB‐17 fragments. Starting from 952,071 compounds in the catalog of Princeton BioMolecular Research, we applied filters for fragment‐likeness (MW≤300, HBD≤3, HBA≤3, CLogP≤3, rotatable bonds ≤3, topological polar surface area ≤60 Å), leaving 30,328 molecules. We then applied filters to remove reactive groups (epoxides, aziridines, maleimide, aldehydes), and only retained molecules with at least one tri‐ or tetravalent sp3 center, leaving 4,366 molecules. Checking for actual presence in GDB‐17 left 1,900 molecules, 1,676 of which were purchased and delivered after ordering.
TMAP generation and nearest neighbor analysis. Commercial fragments of four different libraries were collected and structures with HAC>17 were discarded. (Asinex, Enamine discovery diversity, Enamine sp3‐rich and Life Chemicals ultimate fragments). A random sample of 1,700 compounds was generated comprising equal parts of each library. Another 1,700 molecules were randomly sampled from FDB‐17. Together with the 1,676 acquired compounds, a TMAP was generated based on calculated MAP4 fingerprints (1024 dimensions) with the respective software components that were previously developed in our group.[ 45 , 46 ] A nearest neighbor search was performed using LSH forests for each separate data set. All molecules were queried for their 50 approximate nearest neighbors, followed by a linear scan for the 10 best results among them. The latter were binned by Jaccard distance.
Molecular shingle analysis. Two times ten random samples of 1,700 from commercially available fragments and FDB‐17 were made in the same way as described for the TMAP generation. Unique, circular molecular substructures (shingles) up to distance 3 were extracted using the rdkit and a ten‐fold cross‐validation was performed: At each step, shingles of one of the commercial fragments and FDB‐17 samples were compared with those from the acquired compounds.
Chemistry
The selected GDB‐17 fragments were purchased as 1 mg solid samples from Princeton BioMolecular Research, Inc. and dissolved as ca. 10 mM stock solutions in DMSO for screening. The purity of the active hits was checked by LC‐MS. They were then repurchased in higher amount (usually 25 mg), purified if needed, and analyzed by NMR and HR‐MS. The purity was determined by RP‐UPLC at 214 nm. Characterization of all purchased compounds and the synthesis of (S)‐3 and (R)‐3 are described in the Supporting Information.
Biology
Reagents and chemicals were purchased from Merck unless otherwise stated.
Cell culture. HEK293T cells (ATCC) were used for transient transfection. In previous studies, our laboratory generated a HEK293 cell line (ATCC) stably overexpressing pIRES2 DsRed‐Express2‐DMT1.[ 30 , 31 ] This cell line has been used for the herein described experiments targeting DMT1. Both cell lines were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10 % fetal bovine serum (FBS), 10 mM HEPES, 1 mM sodium pyruvate and 100 μM minimal essential medium (MEM) nonessential amino acids under standard cell culture conditions (37 °C, 95 % humidity and air containing 5 % CO2). Additionally, the media used to culture the HEK293 cells stably overexpressing DMT1 was also supplemented with 500 μg/mL geneticin (Life Technologies).
Cloning human ZIP8 and ZIP14. Human ZIP8 ORF clone (NCBI Reference Sequence: NM_022154.5) with a C‐terminal TurboGFP tag cloned in pCMV6‐AC‐GFP vector was purchased from OriGene (Catalogue #RG204200). After PCR amplification, human ZIP8 was subcloned into the pIRES2 DsRed‐Express2 vector (Takara Clontech) without the TurboGFP tag. C‐terminally eGFP‐tagged human ZIP14 in pEGFP‐N3 vector was purchased from Addgene (Plasmid #104380). Human ZIP14 was PCR amplified without the eGFP tag and subcloned into the pIRES2 DsRed‐Express2 vector (Takara Clontech).
Transient transfection. HEK293T cells were seeded to achieve an 80 % of cell confluency on the day of transfection. Before the seeding, plates were coated with poly‐D‐lysine. When using 96‐well plates, 100 μL of DMEM medium containing 30,000 cells were added into each well. When using 6‐well plates, 2 mL of DMEM medium containing 1 million cells were added into each well. Cells were left in culture for 24 hours, and then, transfected using Lipofectamine 2000 (Life Technologies) as described in the protocol provided with the reagent. DNA constructs used for transient transfection were the following: pIRES2 DsRed‐Express2‐DMT1, pIRES2 DsRed‐Express2‐ZIP2, pIRES2 DsRed‐Express2‐ZIP8, pIRES2 DsRed‐Express2‐ZIP14 or just the backbone pIRES2 DsRed‐Express2 vector, named as the empty vector (EV).[ 30 , 31 , 50 , 51 ]
Radioactive 55 Fe uptake assay. HEK293 cells overexpressing DMT1 were seeded in clear bottom, white‐well, 96‐well plates (Corning) at a density of 50,000 cells/well and kept in culture for 24 hours. Before the experiments, the culture media was removed and the cells were washed 3 times with the uptake buffer (140 mM NaCl, 2.5 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 1.2 mM K2HPO4, 10 mM glucose, 5 mM HEPES, 5 mM MES, pH 7.4). Then, the cells were pre‐incubated for 5 minutes in the presence of 100 μL uptake buffer (pH 5.5, adjusted with 1 M HCl) containing the indicated amounts of the different compounds under study. Next, 10 μL of (10X) uptake solution supplemented with 1 mM ascorbic acid, 10 μM non‐radioactive ferrous iron and 0.5 μCi radioactive 55Fe (American Radiolabeled Chemicals) were added, and the cells were incubated for additional 15 min. Incubation steps were conducted at room temperature. Finally, the solution containing the radioactive 55Fe iron was removed and the plate was washed 3 time with ice‐cold uptake solution (pH 7.4). To determine the iron content, 100 μL of scintillation cocktail Mycrosinth 20 (PerkinElmer) were added to each well, and the plates were incubated for an hour under constant agitation at room temperature. Radioactivity was measured as counts per minute (cpm) with a TopCount Microplate Scintillation Counter (PerkinElmer). Influx rate was determined using the following equation:
Iron uptake was measured in the presence and absence of the tested compounds. To determine the uptake in the absence of compound, cells were pre‐incubated 5 min with uptake buffer containing DMSO at the same % as the tested compounds were dissolved (<0.5 %). Data were expressed as percentage of the average iron uptake measured in the absence of the compound in each individual experiment.
Real‐time fluorescence imaging. HEK293T cells were seeded in clear bottom, black‐well 96‐well plates (Corning) and transfected with the indicated DNA constructs as previously described. 24 hours after the transfection, culture media was removed and cells were loaded with 50 μL of fluorescent dye dissolved in calcium free uptake buffer (117 mM NaCl, 4.8 mM KCl, 1 mM MgCl2, 10 mM glucose, 10 mM HEPES, pH 7.4) according to the manufacturer's protocol for FLIPR Calcium 5 Assay Kit (Molecular Devices). After 1 hour of incubation, real‐time fluorescence measurements were conducted using the FLIPR Tetra fluorescence microplate reader (Molecular Devices) as described previously. [30] Samples were excited using a 470–490 nm LED module, and the emitted fluorescence was detected with a 515–575 nm emission filter. Obtained recordings were monitored and analyzed using the ScreenWorks 3.1.2.002 software (Molecular Devices). Briefly, baseline fluorescence signal was recorded for 50 s, then, 50 μL of the uptake solution containing the indicated amounts (3X) of the different compounds under study were added and the plate was incubated for 5 min. Finally, 50 μL of uptake solution containing 15 μM Cd2+ was added and changes in the fluorescence intensity were recorded during additional 15 minutes. Cells were kept at 37 °C throughout the measurements. Changes in fluoresce intensity were quantified as the Area Under the Curve (AUC) of the fluorescence intensity for a given period of time (459–750 s). Within each experiment, fluorescence intensity was determined in the absence and presence of the different compounds under study, inhibition by each of the compounds was expressed as the percentage of the average fluorescence intensity determined in the absence of the compounds. To determine the fluorescence intensity in the absence of compound, the cells were pre‐incubated for 5 min with uptake solution containing the same % of DMSO as the compounds (<0.5 %). Non‐specific fluorescence was determined in non‐transfected cells, and the obtained average fluorescence value was subtracted from the signal recorded in the transfected cells, prior to the data normalization.
Dose‐response curves. Substrate uptake by the indicated protein transporters was measured in the presence of a range of concentrations of the indicated compounds using either of the previously described functional assays. The obtained curve was analyzed by a 4‐parameter logistic model.
E(D) is the measured parameter, [D] is the drug concentration, IC50 is the concentration of drug at which the half of the maximal inhibition is reached, E0 and E∞ are the top and bottom asymptotes of the response curve.
Real time qRT‐PCR. HEK293T cells were seeded in 6 well plates and transfected with the indicated DNA constructs as previously described. 24 hours after transfection, the medium was removed and the cells were washed with uptake buffer (140 mM NaCl, 2.5 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 1.2 mM K2HPO4, 10 mM glucose, 10 mM HEPES, pH 7.4). Then, cells were incubated in the presence and absence of the indicated 50 μM compound together with 10 μM ZnCl2 dissolved in uptake buffer for 2 hours at 37 °C. After the incubation time, media was removed, cells were washed with PBS and total RNA was extracted using Trizol® reagent as described in the manufacturer's protocol (Invitrogen). Isolated RNA was retro‐transcribed into cDNA using the Taqman® Reverse Transcription kit (Life Technologies). Real‐time PCR was performed using pre‐synthesized Taqman®‐based Assays‐On‐Demand (Life Technologies) on an ABI ViiA 7 System (Thermo Fisher Scientific). The following Taqman® assays were used: MT2 A (Hs02379661_g1) and GAPDH (Hs02786624_g1). Cycle threshold (Ct) values for the different experimental replicates were averaged, and the amounts of mRNA of each gene under study were normalized to the expression of GAPDH by using the ΔCt method.
Statistics. Results are presented as the Mean±Standard Deviation (SD). Normal distribution of the data sets was assessed using the Kolmogorov‐Smirnov (<50 values) and Shapiro‐Wilk tests (>50 values). Data sets showing a normal distribution were statistically compared using the unpaired t‐test, while for those data sets not showing a normal distribution, the statistical comparisons were carried out using the Mann‐Whitney test. Statistical tests were conducted using the IBM SPSS statistics 20 software.
Calcein quenching assay. Calcein (20 mM) was dissolved in 1 M NaOH and diluted to 2 μM with uptake solution (140 mM NaCl, 2.5 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 1.2 mM K2HPO4, 10 mM glucose, 5 mM HEPES, 5 mM MES, pH 7.4). Stock solutions of rac‐3 (10 mM) and bipyridine (10 mM), as positive control, were prepared in MeOH and further diluted to 40 μM with uptake solution. A stock solution of FeSO4 ⋅ 7 H2O (4 mM) with ascorbic acid (400 mM) was prepared and diluted to 8 μM with uptake solution. Solutions of uptake solution with Fe2+ and with or without rac‐3 or bipyridine were prepared in quintuple and incubated at 37 °C for 5 minutes before centrifugation (4000 rpm, 3 minutes). Next, an equal volume of calcein was added to give the final concentrations of 1 μM calcein, 10 μM rac‐3 or bipyridine, 2 μM Fe2+ and 200 μM ascorbic acid. The solutions were transferred into a clear bottom, black‐well 96‐well plate (Corning) and incubated in the dark at 37 °C for 5 minutes. Fluorescence was measured on Tecan M1000 Plate Reader with the excitation wavelength at 495 nm and emission at 515 nm.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Acknowledgements
This work was supported financially by the Swiss National Science Foundation, NCCR TransCure. M. A. H. acknowledges funding by the Swiss National Science Foundation Grant #182272, entitled “Intestinal absorption of transition metals in human health and disease”. The authors thank Matthias Rubin for assistance in screening the fragment library against DMT1, and Dr. Clémence Delalande for help with library setup. Open access funding provided by Universitat Bern.
J. Pujol-Giménez, M. Poirier, S. Bühlmann, C. Schuppisser, R. Bhardwaj, M. Awale, R. Visini, S. Javor, M. A. Hediger, J.-L. Reymond, ChemMedChem 2021, 16, 3306.
Dedicated to the memory of Prof. François Diederich
Contributor Information
Prof. Matthias A. Hediger, Email: matthias.hediger@ibmm.unibe.ch.
Prof. Jean‐Louis Reymond, Email: jean-louis.reymond@unibe.ch.
References
- 1. César-Razquin A., Snijder B., Frappier-Brinton T., Isserlin R., Gyimesi G., Bai X., Reithmeier R. A., Hepworth D., Hediger M. A., Edwards A. M., Superti-Furga G., Cell 2015, 162, 478–487. [DOI] [PubMed] [Google Scholar]
- 2. Lin L., Yee S. W., Kim R. B., Giacomini K. M., Nat. Rev. Drug Discovery 2015, 14, 543–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schumann T., König J., Henke C., Willmes D. M., Bornstein S. R., Jordan J., Fromm M. F., Birkenfeld A. L., Pharmacol. Rev. 2020, 72, 343–379. [DOI] [PubMed] [Google Scholar]
- 4. Olivieri N. F., N. Engl. J. Med. 1999, 341, 99–109. [DOI] [PubMed] [Google Scholar]
- 5. Mariani R., Trombini P., Pozzi M., Piperno A., Mediterr. J. Hematol. Infect. Dis. 2009, 1, e2009006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shawki A., Knight P. B., Maliken B. D., Niespodzany E. J., Mackenzie B., Curr. Top. Membr. 2012, 70, 169–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Montalbetti N., Simonin A., Kovacs G., Hediger M. A., Mol. Asp. Med. 2013, 34, 270–287. [DOI] [PubMed] [Google Scholar]
- 8. Lu L.-N., Qian Z.-M., Wu K.-C., Yung W.-H., Ke Y., Mol. Neurobiol. 2017, 54, 5213–5224. [DOI] [PubMed] [Google Scholar]
- 9. Wang X., Zhang M., Flores S. R. L., Woloshun R. R., Yang C., Yin L., Xiang P., Xu X., Garrick M. D., Vidyasagar S., Merlin D., Collins J. F., Mol. Ther. 2019, 27, 493–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fujishiro H., Himeno S., Biol. Pharm. Bull. 2019, 42, 1076–1082. [DOI] [PubMed] [Google Scholar]
- 11. Jenkitkasemwong S., Wang C.-Y., Mackenzie B., Knutson M. D., BioMetals 2012, 25, 643–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang C.-Y., Jenkitkasemwong S., Duarte S., Sparkman B. K., Shawki A., Mackenzie B., Knutson M. D., J. Biol. Chem. 2012, 287, 34032–34043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Beaumont C., Delaunay J., Hetet G., Grandchamp B., de Montalembert M., Tchernia G., Blood 2006, 107, 4168–4170. [DOI] [PubMed] [Google Scholar]
- 14. Xu H., Jiang H., Xie J., Front. Mol. Neurosci. 2017, 10, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cappellini M. D., Russo R., Andolfo I., Iolascon A., Hematology Am. Soc. Hematol. Educ. Program 2020, 2020, 465–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tybl E., Gunshin H., Gupta S., Barrientos T., Bonadonna M., Celma Nos F., Palais G., Karim Z., Sanchez M., Andrews N. C., Galy B., Hemasphere 2020, 4, DOI 10.1097/HS9.0000000000000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Winslow J. W. W., Limesand K. H., Zhao N., Int. J. Mol. Sci. 2020, 21, DOI 10.3390/ijms21093304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sunuwar L., Frkatović A., Sharapov S., Wang Q., Neu H. M., Wu X., Haritunians T., Wan F., Michel S., Wu S., Donowitz M., McGovern D., Lauc G., Sears C., Melia J., JCI Insight 2020, 5, DOI 10.1172/jci.insight.140978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nebert D. W., Liu Z., Hum. Genomics 2019, 13, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nishimura R., Hata K., Takahata Y., Murakami T., Nakamura E., Ohkawa M., Ruengsinpinya L., Int. J. Mol. Sci. 2020, 21, DOI 10.3390/ijms21041340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vinatier C., Merceron C., Guicheux J., Drug Discovery Today 2016, 21, 1932–1937. [DOI] [PubMed] [Google Scholar]
- 22. Won Y., Shin Y., Chun C.-H., Cho Y., Ha C.-W., Kim J.-H., Chun J.-S., Ann. Rheum. Dis. 2016, 75, 2045–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Manatschal C., Pujol-Giménez J., Poirier M., Reymond J.-L., Hediger M. A., Dutzler R., eLife 2019, 8, e51913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ehrnstorfer I. A., Manatschal C., Arnold F. M., Laederach J., Dutzler R., Nat. Commun. 2017, 8, 14033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Poirier M., Pujol-Giménez J., Manatschal C., Bühlmann S., Embaby A., Javor S., Hediger M. A., Reymond J.-L., RSC Med. Chem. 2020, 11, 1023-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Congreve M., Carr R., Murray C., Jhoti H., Drug Discovery Today 2003, 8, 876–877. [DOI] [PubMed] [Google Scholar]
- 27. Murray C. W., Rees D. C., Nat. Chem. 2009, 1, 187–192. [DOI] [PubMed] [Google Scholar]
- 28. Romasanta A. K. S., van der Sijde P., Hellsten I., Hubbard R. E., Keseru G. M., van Muijlwijk-Koezen J., de Esch I. J. P., Drug Discovery Today 2018, 23, 1596–1609. [DOI] [PubMed] [Google Scholar]
- 29. Mignani S., Rodrigues J., Tomas H., Jalal R., Singh P. P., Majoral J.-P., Vishwakarma R. A., Drug Discovery Today 2018, 23, 605–615. [DOI] [PubMed] [Google Scholar]
- 30. Montalbetti N., Simonin A., Dalghi M. G., Kovacs G., Hediger M. A., J. Biomol. Screening 2014, 19, 900–908. [DOI] [PubMed] [Google Scholar]
- 31. Montalbetti N., Simonin A., Simonin C., Awale M., Reymond J.-L., Hediger M. A., Biochem. Pharmacol. 2015, 96, 216–224. [DOI] [PubMed] [Google Scholar]
- 32. Ruddigkeit L., van Deursen R., Blum L. C., Reymond J. L., J. Chem. Inf. Model. 2012, 52, 2864–2875. [DOI] [PubMed] [Google Scholar]
- 33. Ruddigkeit L., Blum L. C., Reymond J. L., J. Chem. Inf. Model. 2013, 53, 56–65. [DOI] [PubMed] [Google Scholar]
- 34. Reymond J.-L., Ruddigkeit L., Blum L., van Deursen R., WIREs Comput. Mol. Sci. 2012, 2, 717–733. [Google Scholar]
- 35. Meier K., Bühlmann S., Arús-Pous J., Reymond J.-L., Chimia 2020, 74, 241–246. [DOI] [PubMed] [Google Scholar]
- 36. Scior T., Bender A., Tresadern G., Medina-Franco J. L., Martinez-Mayorga K., Langer T., Cuanalo-Contreras K., Agrafiotis D. K., J. Chem. Inf. Model. 2012, 52, 867–881. [DOI] [PubMed] [Google Scholar]
- 37. Nguyen K. T., Syed S., Urwyler S., Bertrand S., Bertrand D., Reymond J. L., ChemMedChem 2008, 3, 1520–1524. [DOI] [PubMed] [Google Scholar]
- 38. Brethous L., Garcia-Delgado N., Schwartz J., Bertrand S., Bertrand D., Reymond J. L., J. Med. Chem. 2012, 55, 4605–4618. [DOI] [PubMed] [Google Scholar]
- 39. Luethi E., Nguyen K. T., Burzle M., Blum L. C., Suzuki Y., Hediger M., Reymond J. L., J. Med. Chem. 2010, 53, 7236–7250. [DOI] [PubMed] [Google Scholar]
- 40. Meier K., Arús-Pous J., Reymond J.-L., Angew. Chem. Int. Ed. Engl. 2021, 60, 2074–2077. [DOI] [PubMed] [Google Scholar]
- 41. Visini R., Awale M., Reymond J. L., J. Chem. Inf. Model. 2017, 57, 700–709. [DOI] [PubMed] [Google Scholar]
- 42. Awale M., Sirockin F., Stiefl N., Reymond J.-L., Mol. Inf. 2019, 38, 1900031. [DOI] [PubMed] [Google Scholar]
- 43. Bühlmann S., Reymond J. L., Front. Chem. 2020, DOI 10.3389/fchem.2020.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Probst D., Reymond J.-L., J. Cheminf. 2020, 12, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Capecchi A., Probst D., Reymond J.-L., J. Cheminf. 2020, 12, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Probst D., Reymond J.-L., J. Cheminf. 2018, 10, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Over B., Wetzel S., Grutter C., Nakai Y., Renner S., Rauh D., Waldmann H., Nat. Chem. 2013, 5, 21–28. [DOI] [PubMed] [Google Scholar]
- 48. Prescher H., Koch G., Schuhmann T., Ertl P., Bussenault A., Glick M., Dix I., Petersen F., Lizos D. E., Bioorg. Med. Chem. 2017, 25, 921–925. [DOI] [PubMed] [Google Scholar]
- 49. Hopkins A. L., Groom C. R., Alex A., Drug Discovery Today 2004, 9, 430–431. [DOI] [PubMed] [Google Scholar]
- 50. Franz M.-C., Simonin A., Graeter S., Hediger M. A., Kovacs G., J. Biomol. Screening 2014, 19, 909–916. [DOI] [PubMed] [Google Scholar]
- 51. Gyimesi G., Albano G., Fuster D. G., Hediger M. A., Pujol-Giménez J., J. Biol. Chem. 2019, 294, 8046–8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Boggs S. D., Cobb J. D., Gudmundsson K. S., Jones L. A., Matsuoka R. T., Millar A., Patterson D. E., Samano V., Trone M. D., Xie S., Zhou X., Org. Process Res. Dev. 2007, 11, 539–545. [Google Scholar]
- 53. Awale M., Reymond J.-L., J. Cheminf. 2017, 9, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Awale M., Reymond J.-L., J. Chem. Inf. Model. 2019, 59, 10–17. [DOI] [PubMed] [Google Scholar]
- 55. Wang G., Biswas A. K., Ma W., Kandpal M., Coker C., Grandgenett P. M., Hollingsworth M. A., Jain R., Tanji K., López-Pintado S., Borczuk A., Hebert D., Jenkitkasemwong S., Hojyo S., Davuluri R. V., Knutson M. D., Fukada T., Acharyya S., Nat. Med. 2018, 24, 770–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shakri A. R., Zhong T. J., Ma W., Coker C., Kim S., Calluori S., Scholze H., Szabolcs M., Caffrey T., Grandgenett P. M., Hollingsworth M. A., Tanji K., Kluger M. D., Miller G., Biswas A. K., Acharyya S., Cancers 2020, 12, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information