Abstract
HEF1 (human enhancer of filamentation 1) is a member of a docking protein family that includes p130Cas and Efs. Through assembly of multiple protein interactions at focal adhesion sites, these proteins activate signaling cascades in response to integrin receptor binding of the extracellular matrix. The HEF1 protein is cell cycle regulated, with full-length forms cleaved in mitosis at a caspase consensus site to generate an amino-terminal 55-kDa form that localizes to the mitotic spindle. The identification of a caspase cleavage site in HEF1 led us to investigate whether HEF1 belongs to a select group of caspase substrates cleaved in apoptosis to promote the morphological changes characteristic of programmed cell death. Significantly, inducing expression of HEF1 in MCF-7 or HeLa cells causes extensive apoptosis, as assessed by multiple criteria. Endogenous HEF1 is cleaved into 65- and 55-kDa fragments and a newly detected 28-kDa form in response to the induction of apoptosis, paralleling cleavage of poly(ADP-ribose) polymerase and focal adhesion kinase (FAK); the death-promoting activity of over-expressed HEF1 is associated with production of the 28-kDa form. While the generation of the cleaved HEF1 forms is caspase dependent, the accumulation of HEF1 forms is further regulated by the proteasome, as the proteasome inhibitors N-acetyl-l-leucinyl-l-leucinyl-l-norleucinyl and lactacystin enhance their stability. Finally, the induction of HEF1 expression also increases Jun N-terminal protein kinase (JNK) activation, and activated JNK colocalizes with HEF1, implicating this pathway in HEF1 action. Based on these results, we propose that dysregulation of HEF1 and its family members along with FAK may signal the destruction of focal adhesion sites and regulate the onset of apoptosis.
Apoptosis, or programmed cell death (PCD), is critical in an array of processes as apparently diverse as nervous system development (69), maturation of lymphoid cells (74), and appropriate attachment of epithelial cells (26), but all these processes are linked by the requirement of eliminating defined cellular populations in specific circumstances. Apoptosis can be triggered by extracellular signals such as stimulation of the tumor necrosis factor (TNF) and Fas families of death receptors (3), surface immunoglobulin cross-linking (81), or interference with cellular adhesion to substrate (26), as well as by intracellular perturbations such as changes in mitochondrial permeability (32) or activation of cell damage-sensing pathways (25). A hallmark of apoptosis is the activation of a cascade of cellular proteases, termed caspases, that cleave proteins after aspartic acid residues found in an appropriate amino acid context. Initiator caspases (caspases 8, 10, and others) at the top of the cascade are activated and then sequentially cleave and activate downstream effector caspases (e.g., caspases 3 and 7), which in turn cleave a specific subset of cellular proteins. Cleavage of this subset leads to characteristic changes in cell morphology, including nuclear fragmentation and cytoskeletal rearrangement (55, 80), and ultimately results in production of apoptotic bodies that are engulfed by surrounding cells (41). Because of the central role of apoptosis in many biological processes, it has been of considerable interest to elucidate the steps of these morphological rearrangements and to understand the relation between morphological controls in moribund versus normally growing cells.
It has long been noted that apoptosis and mitosis both require similar programmed changes in cell shape and some conserved elements of signaling (reviewed in reference 42). The first step in both processes requires cell rounding and a reduction in cell attachment to the extracellular matrix (ECM) through modulation of the cell cytoskeleton. Subsequent changes in nuclear morphology, including breakdown of the nuclear envelope and hypercondensation of the chromatin (48), allow the packaging of nuclear fragments into apoptotic bodies or, in the case of mitosis, formation of the mitotic spindle. Although caspases were originally defined as proteins specifically active in apoptosis, recent work has identified additional roles for these proteins in nonapoptotic cells (e.g., see reference 30). For example, caspase 3 activity has been shown to increase upon NIH 3T3 cell spreading; reciprocally, cell spreading is blocked by the general caspase inhibitor z-Asp (86). Two pieces of evidence specifically link caspase activity and cell proliferation. First, a significant transient increase in caspase 3 activity is detected in peripheral T lymphocytes upon T-cell activation, when these cells are actively proliferating but not engaged in apoptosis (60). Second, survivin, a protein which has the ability to inactivate caspases 3 and 7, is upregulated at the G2/M transition and localizes to the mitotic spindle. Expression of survivin can block taxol-induced apoptosis, suggesting that survivin may act to inhibit a constitutive apoptotic signal in mitosis (53). Thus, modulated caspase activity towards specific cellular targets may be important for normal cytoskeletal organization and cell cycle progression, while a dramatic increase or dysregulation of caspase activity is necessary to promote apoptosis.
HEF1 (human enhancer of filamentation 1) was first isolated in a screen for human proteins with the ability to alter Saccharomyces cerevisiae morphology from round to filamentous hyperpolarized cells (45). HEF1 belongs to a larger family of docking adapter proteins including p130Cas and Efs (also known as Sin) (1, 38, 71) termed the Cas family. All members of this family contain multiple protein-protein interaction domains, allowing for the recruitment of additional proteins into a complex that activates signaling cascades following cell adhesion (1, 38, 45, 57, 59, 63, 71, 84). These interaction domains include an amino-terminal SH3 domain that binds polyproline-containing proteins, a substrate domain with multiple tyrosines that when phosphorylated recruit SH2-containing proteins, and a conserved carboxy-terminal domain that may contribute to dimerization of Cas family members (45, 46). In interphase cells, HEF1 and other Cas proteins localize to sites of focal adhesion, bind to focal adhesion kinase (FAK) through the conserved SH3 domain (45), and are phosphorylated by FAK and Src family kinases in response to integrin receptor binding of the ECM. This phosphorylation in turn activates SH2 binding sites to recruit the adapter protein Crk, which then stimulates the Ras/Raf/Jun N-terminal protein kinase (JNK) signaling cascade (4, 6, 24, 45, 57, 59, 66, 73, 77; reviewed in reference 67), contributing to the promotion of cell migration (16, 19, 43, 64; S. J. Fashena, M. B. Einarson, G. M. O'Neill, and E. A. Golemis, unpublished data).
In contrast to p130Cas, the HEF1 protein is regulated at multiple levels in a cell cycle-dependent manner (47), with regulation including changes in steady-state levels, phosphorylation status, and proteolytic processing. As cells traverse through S phase and G2, full-length forms of HEF1 (p105 and p115) accumulate at focal adhesion sites. Strikingly, at the G2/M transition the full-length forms of HEF1 are cleaved at a caspase consensus site to generate an amino-terminal HEF1 form, p55HEF1, which localizes to the mitotic spindle, while carboxy-terminal species are apparently degraded. The cleavage and relocation of HEF1 during mitosis suggest that HEF1 may play a role in coordinating attachment-based signals generated at focal adhesion sites with cell cycle events in the nucleus, thereby promoting the transition from flat substrate-attached interphase cells to rounded mitotic cells. The apparent involvement of a caspase-like activity in the production of p55HEF1 at mitosis therefore led us to investigate whether HEF1 belonged to a select subset of caspase substrates cleaved in apoptosis to promote the cytoskeletal changes characteristic of PCD and whether misregulation of HEF1 might independently contribute to the induction of apoptosis.
The breast adenocarcinoma cell line MCF-7 has been well characterized both for the endogenous expression of HEF1 (47) and its intrinsic ability to undergo apoptosis (7, 13), making these cells suitable for studies of HEF1 biological activities. Here we show that overexpression of the HEF1 protein in MCF-7 cells efficiently induces apoptosis, as assessed by promotion of caspase activation and cleavage of canonical effector caspase targets. HEF1 overexpression results in the activation of JNK kinases and is accompanied by the colocalization of HEF1 and activated JNK at focal adhesions. Induction of apoptosis either by HEF1 overexpression or by treatment with TNF alpha (TNF-α) or other standard proapoptotic agents, leads to cleavage of HEF1 into 65-, 55-, and 28-kDa forms by a caspase 3-like or caspase 7-like activity, in a time period paralleling cleavage of effector caspase targets poly(ADP-ribose) polymerase (PARP) and FAK. p130Cas is also cleaved to produce a 28-kDa species following HEF1 overexpression; comparison of HEF1 and p130Cas sequences facilitated delineation of the likely shared caspase cleavage target site as a DDYD motif that overlaps a previously defined site of phosphorylation by FAK. In transient-transfection assays in which survival rates of cells overexpressing the wild-type, DLVA mutant, p55, or p28 forms of HEF1 were compared, the HEF1 proapoptotic activity was found to depend on the expression of the p28 carboxy-terminal region of the protein. Finally, the various HEF1 cleavage products were found to be subject to degradation via the proteasome; however, the degree of proteasome-dependent cleavage differs in mitosis and apoptosis, likely contributing to the differential abundance of the cleaved HEF1 species during the two processes. These results, together with complementary findings in the literature, suggest a model in which regulation of the forms and abundance of HEF1 family members signals the destruction of focal adhesion sites and regulates onset of mitosis or apoptosis.
MATERIALS AND METHODS
Cell lines and materials.
MCF-7 human breast carcinoma cells and HeLa human cervical carcinoma cells were cultured under standard conditions. The murine B-cell lymphoma line WeHI 231, as well as anti-immunoglobulin M (α-IgM) antibodies, was a generous gift from Kerry Campbell (Fox Chase Cancer Center) and was maintained in a solution containing Dulbecco's modified Eagle medium (DMEM), 10% fetal calf serum, glutamine, penicillin, streptomycin, nonessential amino acids, and β-mercaptoethanol (10 μM). The construction of MCF-7 transfectants carrying either full-length HEF1 or the empty vector under the control of a tetracycline-repressible operator will be described elsewhere (Fashena et al., unpublished data). The cells used in this study are from the HEF1.M1 cell line and the CM1 cell line. Antibodies used in this study include anti-p130Cas, antigelsolin, anti-FAK, and antipaxillin antibodies from Transduction Laboratories (San Diego, Calif.); anti-PARP antibody from Calbiochem (La Jolla, Calif.); anti-phospho-JNK antibody from Promega (Madison, Wis.), anti-JNK antibody from PharMingen (San Diego, Calif.); rhodamine-conjugated goat anti-rabbit antibodies from Molecular Probes (Eugene, Oreg.); and dichlorotriazinyl amino fluorescein-conjugated goat anti-mouse antibodies from Jackson Immunoresearch (West Grove, Pa.). Anti-HEF1-R1 (here denoted HEF1/1) antibodies were previously described (47). Anti-p130Cas antibody has been previously shown to cross-react with the HEF1 C terminus (47) and is therefore referred to here as anti-HEF1/anti-p130Cas antibody. Another HEF1-specific antibody (anti-HEF1/2) was generated by injecting into rabbits the peptide KESSLSASPAQDKR, conjugated to keyhole limpet hemocyanin. TNF-α was purchased from R & D Laboratories. 3-[4,5-Dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) was obtained from Sigma (St. Louis, Mo.). Colorimetric caspase 3 substrate, Ac-DEVD-pNA, was purchased from Calbiochem and dissolved in dimethyl sulfoxide (DMSO) as 5 mM stock solutions and stored at −20°C. The caspase inhibitor peptide z-DEVD-fmk and the specific proteasome inhibitor lactacystin were purchased from Calbiochem, while the proteasome inhibitors N-acetyl-l-leucinyl-l-leucinyl-l-norleucinyl (ALLN) and N-acetyl-l-leucinyl-l-leucinyl-l-methioninal (ALLM) were obtained from Sigma. Puromycin dihydrochloride and tetracycline hydrochloride were from Sigma, while hygromycin B was from Roche (Indianapolis, Ind.). LT2 transfection reagent was purchased from Mirus (Madison, Wis.).
Expression constructs.
Construction of the pCDNA1/HEF1 construct has been previously described (47). C-terminally Myc-His-tagged full-length HEF1 was constructed by PCR amplifying HEF1 with primers containing N-terminal EcoRI and C-terminal KpnI restriction enzyme sites. The fragment was then ligated into EcoRI/KpnI digested pCNA3.Myc/His. The mouse p130Cas cDNA insert was isolated by EcoRI digestion from pCMV5.p130Cas. The resulting fragment was religated into EcoRI-digested pcDNA3, and clones containing inserts in the correct orientation were determined by nucleotide sequencing.
Additional HEF1 cDNAs were cloned in-frame into the pEGFP-C4 vector (Clontech, Palo Alto, Calif.), to create fusion peptides consisting of green fluorescent protein (GFP) fused to the N terminus of the HEF1 peptide. The GFP fusion constructs include (i) pEGFP.HEF1, encoding the complete wild-type HEF1 protein sequence; (ii) pEGFP.55, encoding the N-terminal 363 amino acids (aa) of HEF1 corresponding to the 55-kDa N-terminal peptide produced following caspase cleavage at the conserved DLVD motif (aa 360 to 363); (iii) pEGFP.DLVA, encoding full-length HEF1 carrying a point mutation (D→A) in the caspase cleavage site that has been previously described (47); and (iv) pEGFP.28, encoding the C-terminal 205 aa of HEF1 corresponding to the predicted 28-kDa HEF1 C-terminal peptide including DDYD (aa 627 to 630).
HEF1 expression and assay of cell growth.
Stable cell lines containing the parental tetracycline-regulatable vector with or without the full-length HEF1 cDNA were generated in MCF-7 cells (Fashena et al., unpublished data). These were plated at a density of approximately 106 cells per 100-mm-diameter plate into fresh DMEM plus 10% fetal bovine serum in the absence of tetracycline and other selective antibiotics (induced) or in the presence of 1 μg of tetracycline per ml (uninduced). The viability of cells expressing HEF1 was assayed using the colorimetric MTT assay essentially as described previously (15). Approximately 104 cells were plated in wells of a 96-well plate at the start of each assay, and six replicates were plated per assay. Prior to solubilization of the precipitate with DMSO, plates were centrifuged for 5 min at 800 × g to collect floating cells. The medium was then carefully aspirated and the precipitate was dissolved as described previously (15). Cell numbers were calculated by preparing a standard curve of cell number versus absorbance at 560 nm. Results are expressed as the average cell number of 12 replicates from two independent experiments ± the standard error of the mean.
Assay of caspase activity.
Floating and attached cells were combined and extracted in caspase assay buffer (50 mM HEPES [pH 7.4], 100 mM NaCl, 0.1% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfate [CHAPS], 10 mM dithiothreitol, 1 mM EDTA, and 10% glycerol). The protein content of extracts was determined using the Bio-Rad (Hercules, Calif.) protein assay as per the manufacturer's instructions. Caspase assays were performed as previously described (23). Briefly, 100 μg of total protein extracts were incubated at room temperature in a final volume of 200 μl with Ac-DEVD-pNA (250 μM final concentration) and, where indicated, z-DEVD-fmk (0.5 μM final concentration) in caspase assay buffer. Triplicate samples were incubated at room temperature, and the release of the pNA product was monitored at 405 nm in a microtiter plate reader (Multiskan Plus; Labsystems, Helsinki, Finland). Initial readings were taken at the start of the incubation (T0) and final readings were taken at the completion of the reaction at 24 h (T1). The change in absorbance over time was determined by subtracting T0 from T1 for each sample.
Induction of apoptosis.
MCF-7 cells were plated and grown for 48 h, reaching a cell density of 70% prior to TNF-α addition. Medium was replaced with fresh medium supplemented with TNF-α (100 ng/ml). TNF-α treatment of stable cell lines was carried out by plating cells directly into fresh DMEM plus 10% fetal bovine serum, with or without addition of tetracycline as indicated, containing TNF-α (100 ng/ml). Cells floating in the medium were collected by centrifugation (285 × g for 5 min) and extracted in combination with attached cells at the time points indicated, as described below. WeHI 231 cells were plated at a density of 5 × 104 cells/ml and treated with 1 μg of α-IgM antibodies per ml in complete medium. Cells were collected by centrifugation and lysed as stated below. Blockade of caspase 3 and 7 activation was accomplished by incubation with the cell-permeating caspase inhibitor z-DEVD-fmk at a final concentration of 25 μM, 30 min prior to and continuing after the addition of TNF-α or α-IgM as per the manufacturer's recommendations.
Proteasome inhibition.
The proteasome and calpain were inhibited by the peptide ALLN and its structurally related but less-potent analog ALLM (36). The specific proteasome inhibitor lactacystin was also used for comparison (22). The proteasome inhibitors ALLN and ALLM were added coincident with the addition of TNF-α at a final concentration of 50 μM, while lactacystin was added at a final concentration of 10 μM.
Proteasome inhibition in mitosis was accomplished as follows. MCF-7 cells were blocked in 2 mM thymidine as previously described (47). Cells were washed twice in DMEM and then released in DMEM plus 10% fetal bovine serum for 4 h. Then either a DMSO control or lactacystin at a final concentration of 50 μM was added and the cells were incubated for 5 h more. Mitotic cells were then harvested by gently tapping the dish and collecting cells in the supernatant by centrifugation.
Transient transfections and counting of GFP-positive cells.
HeLa cells were plated 24 h prior to transfection. Transfection was accomplished using the LT2 reagent (Mirus) following the manufacturer's suggested protocol. Cells for Western blot analysis were harvested 24 h following the initiation of the transfection. For counting of GFP-positive cells, HeLa cells were plated 24 h prior to transfection on coverslips. At 24 h posttransfection cells were fixed with 4% paraformaldehyde. Each cDNA construct was transfected in triplicate. Eight random fields were then scored under the 20× objective of a Nikon Eclipse E800 microscope for GFP-positive cells. Each cDNA construct was transfected in triplicate, and the experiment was repeated on three separate occasions. The percentage of GFP-positive cells was determined by dividing the total number of GFP-positive cells in eight fields by the average of three determinations for the GFP vector alone, and the results were multiplied by 100. Data shown consist of the average of nine replicates from three separate experiments for each construct ± the standard error of the mean.
Preparation of cell lysates and Western blot analysis.
Lysates were made as previously described (45, 47) in A-PTY buffer (50 mM HEPES [pH 7.5], 50 mM NaCl, 5 mM EDTA, 1% Triton X-100, 50 mM NaF, 10 mM Na4P2O7), supplemented with 1 mM phenylmethylsulfonyl fluoride, aprotinin (0.01 mg/ml), leupeptin (0.01 mg/ml), and 1 mM Na3VO4. For adherent cultures, floating cells were pooled with the adherent cells prior to lysis. The cell pellet was then lysed in combination with lysates from the attached cells. Western analysis was carried out with the previously described antibody concentrations or those suggested by the manufacturer. Detection was performed via chemiluminescence as described elsewhere (45).
Immunofluorescence.
MCF-7 cells were transfected with the indicated constructs and 24 h posttransfection were fixed with 4% paraformaldehyde and permeabilized with 0.2% Triton X-100. Following incubation with primary antibody for 1 h, bound antibodies were detected with either rhodamine-conjugated goat anti-rabbit or dichlorotriazinyl amino fluorescein-conjugated goat anti-mouse secondary antibodies. Cells were examined and confocal microscopy performed using a Bio-Rad 600 laser scanning confocal microscope.
RESULTS
Induction of HEF1 expression promotes apoptosis, while cleavage of endogenous HEF1 occurs following proapoptotic stimuli.
Endogenous HEF1 exists during interphase as full-length 105-kDa and hyperphosphorylated 115-kDa forms, localized primarily at focal adhesion sites. These endogenous full-length forms are cleaved during mitosis, generating a 55-kDa amino-terminal form that localizes to the mitotic spindle (47), while the carboxy-terminal end of the protein is apparently degraded. We have also shown that HEF1 transiently transfected into HeLa cells is cleaved in a non-cell cycle-regulated manner at a predicted caspase 3-like or caspase 7-like (DLVD) consensus site, producing a 55-kDa amino-terminal peptide comparable in size to the mitotic p55 species (47) and a 65-kDa carboxy-terminal peptide. These results raised the possibility that the overexpressed HEF1 protein might itself be activating the caspase function and/or promoting apoptosis. If so, this suggests that perturbation of HEF1-dependent signaling was a novel initiator of the cell death machinery.
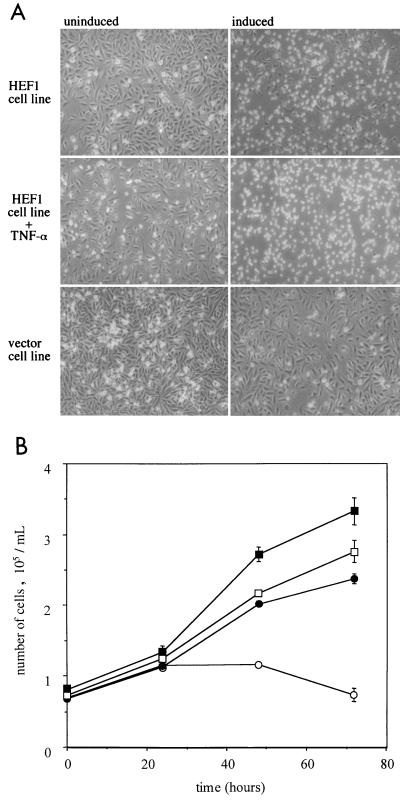
We have recently prepared MCF-7 cell lines stably expressing HEF1 or the parental vector under the control of a tetracycline-regulatable promoter (Fashena et al., unpublished data). To explore the role of HEF1 in apoptosis, we initially assayed whether the stable cell lines also showed signs of apoptosis upon HEF1 induction and whether the cleavage of HEF1 and canonical caspase substrates is concomitantly activated. At 48 h after induction of HEF1 expression, the majority (70%) of cells round up, become refractile, and float off the plate, a behavior consistent with cell death (Fig. 1, top). In contrast, uninduced cells and vector control cells appear normal, with the majority of cells being adherent and proliferating (Fig. 1, top, left-hand panel, and bottom). At this time point, the full-length HEF1 p105 and p115 species, as well as the HEF1 p55-kDa cleavage product, are observed in induced HEF1 stable lines but are not observed in either the uninduced HEF1-containing lines or vector controls, except for endogenous HEF1 (Fig. 2A). To assess the viability of the HEF1-expressing cells we next performed an MTT cell viability assay on cells induced to express HEF1 and compared the results with those for uninduced cells and vector control cells in the presence and absence of tetracycline. At 48 and 72 h after induction of HEF1 expression there is a clear loss of viability of the HEF1-expressing cells compared with the viability of the controls (Fig. 1B). We note that floating cells were collected by centrifugation prior to assaying cleavage of the MTT substrate, and therefore it is expected that the total number of viable cells represents both the floating and adherent populations.
FIG. 1.
HEF1 expression causes cell death. (A) Cells were grown for 48 h in the presence (uninduced) or absence (induced) of tetracycline and, where indicated, TNF-α and were examined by phase-contrast microscopy. HEF1 stable transfectants (HEF1 cell line) were compared with a negative control cell line (vector cell line). (B) Cells were grown in the presence or absence of tetracycline, and cell numbers were determined over a 72-h period using the MTT cell viability assay. Circles indicate the HEF1 cell line and squares indicate the vector control cell line grown in the presence (solid) or absence (open) of tetracycline. Data points represent the average of six replicates from two independent experiments. Error bars indicate the standard errors of the mean.
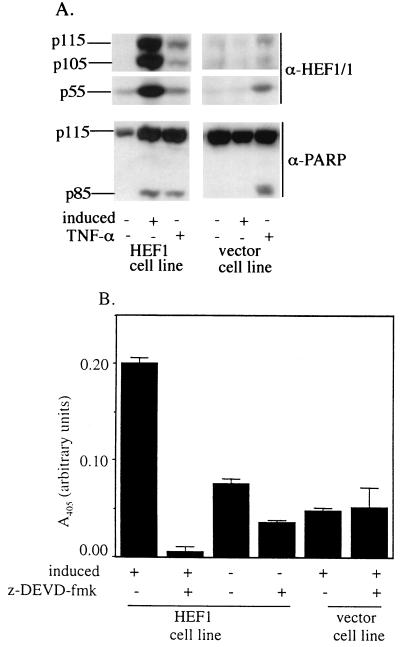
FIG. 2.
HEF1 expression causes protein cleavage and increased caspase activity. (A) Lysates from a HEF1 cell line and a control vector cell line were prepared from cells grown for 48 h under one of the following conditions: induced, noninduced, or noninduced in the presence of TNF-α. Western blots of total cell lysates were probed with antibodies to HEF1 (α-HEF1/1) and to PARP (α-PARP). (B) HEF1-expressing cells and vector control cells were grown for 48 h under inducing or noninducing conditions. Lysates were then incubated with Ac-DEVD-pNA (250 μM) for 24 h. Formation of product was monitored at 405 nm in the absence (−) or presence (+) of the caspase 3 inhibitory peptide z-DEVD-fmk (0.5 μM final concentration) as indicated. Experiments were repeated at least three times, and a representative experiment is shown. Error bars represent the standard errors of triplicate samples.
Caspase induction is a specific indicator of apoptotic cell death. To determine if caspases were being activated by HEF1 expression, we assayed cleavage of the known caspase 3 substrate (8, 49) and apoptosis marker, PARP, following HEF1 induction. As positive controls for PARP cleavage we included uninduced HEF1 cells and vector control cells treated with TNF-α, a treatment previously shown to activate caspases and cause apoptosis in MCF-7 cells (13). PARP cleavage was detected following either TNF-α treatment or HEF1 induction but not in the untreated and uninduced HEF1 and vector control cells (Fig. 2A). Notably, TNF-α treatment resulted in production of the 55-kDa HEF1 cleavage product both in uninduced HEF1-containing and vector control lines, suggesting that endogenous HEF1 was also cleaved. In addition, cells induced to express HEF1 exhibited enhanced cleavage (approximately twofold) of the colorimetric caspase 3 substrate Ac-DEVD-pNA in comparison to control cells (Fig. 2B). Further, activity towards the caspase 3 substrate in the HEF1-expressing cells is potently blocked by the inhibitory peptide z-DEVD-fmk (Fig. 2B), confirming the specificity of the cleavage. These results indicated not only that HEF1 overexpression does induce caspases but that one consequence of their activation is subsequent cleavage of endogenous HEF1. We note that, as the MCF-7 cell line has been described as deficient in caspase 3, the detected activity is likely to correspond to that of caspase 7 or one of the other redundant caspases active in these cells (39).
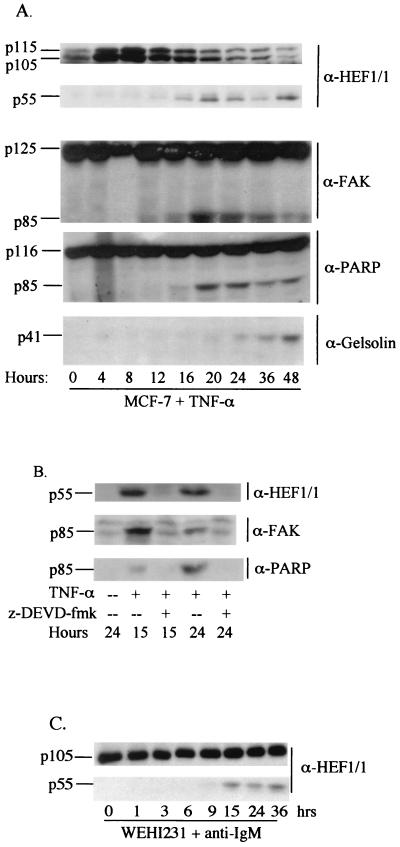
If HEF1 is a significant natural target for cleavage by caspases in apoptosis, then a time course of endogenous HEF1 cleavage should parallel that observed for other defined cytoskeletal and signaling proteins known to be cleaved following apoptotic stimulus. These include the HEF1 interactive partner FAK (87), the morphoregulatory p21-associated kinase (PAK) (70), the actin-severing and/or -capping protein gelsolin (44), and others (e.g., see references 9, 10, 12, and 58). We therefore examined the timing of endogenous HEF1 cleavage in response to TNF-α treatment in parental MCF-7 cells. Approximately 16 h after TNF-α addition, there is a decline in the 105- and 115-kDa full-length forms of HEF1 and an accumulation of the 55-kDa amino-terminal peptide (Fig. 3). For comparison, we also analyzed the cleavage of the caspase substrates FAK, PARP, and gelsolin. FAK and PARP cleavage coincide with the production of the 55-kDa HEF1 form, whereas gelsolin cleavage is detected approximately 4 h later (Fig. 3A). This suggested that HEF1 cleavage, like that of FAK and PARP, is an earlier event in the apoptotic process and may occur prior to dismantling of the cell cytoskeleton. The cell-permeating caspase 3 inhibitory peptide z-DEVD-fmk inhibited generation of p85 FAK, p85 PARP, and the 55-kDa HEF1 form at both 15 and 24 h after TNF-α addition (Fig. 3B), confirming the caspase dependence of the cleavage events. Finally, HEF1 cleavage from full length to p55 was observed in an independent cell type, WeHI 231 B cells, in which apoptosis was induced by an entirely different stimulus, antibody cross-linking of surface IgM. This result indicated that the caspase targeting of HEF1 was not specific to MCF-7 cells and TNF-α treatment (Fig. 3C). In sum, these results indicate that HEF1 expression in the absence of other inducing agents can induce caspase activation and cause apoptosis in MCF-7 cells. Furthermore, HEF1 cleavage is an early downstream event following apoptotic stimulation in multiple cell types.
FIG. 3.
Endogenous MCF-7 HEF1 is cleaved by a caspase 3-like activity. (A) MCF-7 cells treated with TNF-α (100 ng/ml). (B) MCF-7 cells treated with TNF-α (100 ng/ml) in the presence (+) or absence (--) of the caspase 3 inhibitory peptide z-DEVD-fmk at a final concentration of 25 μM. (C) WEHI 231 cells were treated with α-IgM antibodies (1 μg/ml) and extracted at the indicated times. Total proteins were extracted at the indicated time points, and Western blots were probed with the antibodies (α) indicated on the right.
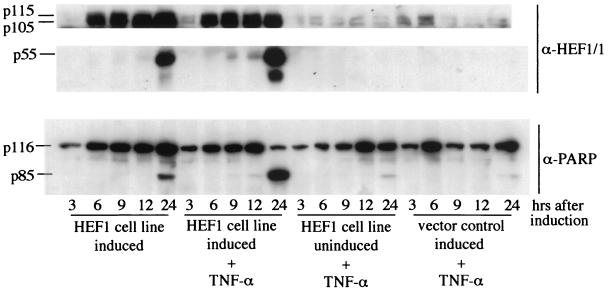
Mechanistically, the fact that TNF-α and HEF1 induction both activate caspases can be explained by hypothesizing that HEF1 overexpression activates a pathway downstream of TNF-α, in which case dual treatment would not have greater effect than treatment with either TNF-α or HEF1 expression alone. Alternatively, HEF1 is activating a separate pathway. To address this point, we determined whether simultaneous overexpression of HEF1 and TNF-α treatment resulted in a more pronounced effect than either stimulus independently. On a gross morphological level, treatment with TNF-α alone resulted in a low level of cell rounding and death; in contrast, HEF1 induction together with TNF-α treatment led to essentially 100% cell death at 48 h (Fig. 1, center panels). Cells were grown under inducing and noninducing conditions with and without the addition of TNF-α as indicated, and cell lysates were prepared and analyzed for HEF1 and PARP cleavage at the indicated time points (Fig. 4). There is an increase in the degree of both HEF1 and PARP cleavage in cells induced to express HEF1 and treated with TNF-α. Control uninduced cells treated with TNF-α displayed significantly less PARP cleavage than induced cells and induced cells receiving TNF-α treatment. This potentiation of cell death indicates that HEF1 expression may either prime the TNF-α death pathway or work synergistically to facilitate apoptosis.
FIG. 4.
Cell death induced by HEF1 expression is potentiated by TNF-α treatment. HEF1 stable transfectants were grown under inducing conditions, inducing conditions plus TNF-α, and noninducing conditions plus TNF-α. Vector control cells were grown under inducing conditions plus TNF-α. Western blots were probed with antibodies to HEF1 (α-HEF1/1) and to PARP (α-PARP).
HEF1 caspase cleavage products are differentially regulated by proteasomal degradation in mitosis and apoptosis.
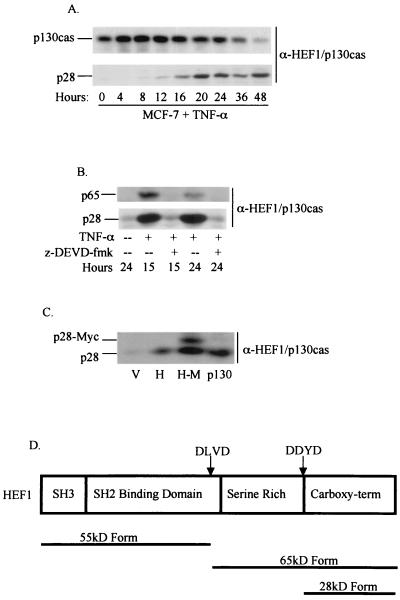
As previously noted, a p65 HEF1 carboxy-terminal cleavage product was readily identified following transient transfection of HEF1, a condition now defined as inducing apoptosis, but was not detectable in mitosis (47). To probe the significance of this difference, we searched for the appearance of lower-molecular-weight HEF1-derived species over the time course for which we had characterized appearance of p55 following TNF-α stimulation of apoptosis (Fig. 5). For this purpose, we used antibodies that were specific for HEF1 at epitopes flanking the previously defined DLVD cleavage site; to scrutinize the extreme carboxy-terminal region of HEF1, it was necessary to use antibodies that cross-reacted with both HEF1 and p130Cas, as antibodies specific for HEF1 are unavailable for this region. Concurrently, we assayed for production of the 65-kDa carboxy-terminal peptide; however, there was very little detectable p65 observed. This is possibly due to the unstable nature of this peptide (47; also see below). Unexpectedly, during this analysis we noted the appearance of an additional immunoreactive protein with an estimated molecular mass of 28 kDa. The generation of this form begins at 12 h following TNF-α treatment of MCF-7 cells (Fig. 5A), paralleling the induction of the 55-kDa species (Fig. 2 and 3), and is inhibited by treatment with z-DEVD-fmk (Fig. 5B).
FIG. 5.
A 28-kDa caspase cleavage product is generated from both HEF1 and p130Cas. (A) MCF-7 cells were treated with TNF-α (100 ng/ml), and total protein lysates were probed with anti-HEF1-p130Cas antibody (α-HEF1/p130Cas). (B) MCF-7 cells were treated with TNF-α (100 ng/ml) in the presence (+) or absence (--) of the caspase 3 inhibitory peptide z-DEVD-fmk at a final concentration of 25 μM. Western blots were probed with α-HEF1/p130Cas. (C) HeLa cells were transfected with the empty vector (V), HEF1 (H), HEF1 carboxy-terminally tagged with the Myc epitope (H-M), or p130Cas (p130). Western blots were probed with α-HEF1/p130Cas. (D) Diagram of the HEF1 caspase cleavage sites and the pieces that are generated.
Bannerman et al. (5) have recently reported that p130Cas is cleaved in response to lipopolysaccharide treatment of epithelial cells (a proapoptotic stimulus), generating a 28-kDa p130Cas peptide. Since MCF-7 cells express both p130Cas and HEF1 and both proteins contain the anti-p130Cas antibody epitope in their C termini, the observed p28 species may have been derived from either protein. To determine the origin of the p28 species, full-length HEF1 was carboxy-terminally tagged with the Myc epitope [HEF1-Myc (H-M)], transfected into HeLa cells and compared with full-length HEF1 (H), p130Cas (p130), or the empty vector (V) similarly transfected. Lysates from HEF1-Myc-transfected cells have two anti-p130Cas immunoreactive bands (Fig. 5C). One band migrates at the same rate as the 28-kDa protein observed in the HEF1- and p130Cas-transfected cells and represents cleaved endogenous HEF1 and/or p130Cas. In contrast, the second band has a comparatively retarded electrophoretic mobility and corresponds to the 28-kDa carboxy-terminal domain of HEF1 fused to the Myc tag. These data indicate that at least some of the 28-kDa form observed in MCF-7 cells is likely to be derived from HEF1. Based on predicted molecular masses, potential caspase sites shared between HEF1 and p130Cas, and the location of the anti-p130Cas antibody epitope, a conserved DDYD (aa 627 to 630) motif is the only apparent candidate cleavage site (shown in Fig. 5D); further, a construct made to express the predicted p28 (HEF1630–834) comigrated with the observed p28 endogenous band (data not shown). We note that, although examination of a DDYD→DDYA mutant would bear on this issue, it is experimentally difficult to evaluate such a mutant, as modulation of the DDYD site destroys the ability of the HEF1/p130Cas antibody to recognize the protein, while appending a carboxy-terminal tag to the expressed p28 form of HEF1 results in other changes that make interpretation difficult (results not shown).
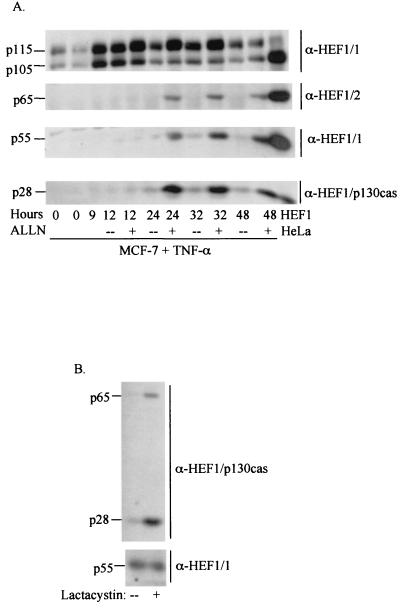
The above results indicate that caspases are required for processing of HEF1 to three distinct forms in apoptosis (p55, p65, and p28) but do not address the differences in abundance of the derivative species in apoptotic and mitotic cells. We therefore investigated the possibility that levels of HEF1-derived peptides might additionally be regulated by proteasomal processing, as activity of the proteasome has been shown to be closely tied to caspase function (reviewed in reference 68). Addition of the calpain and proteasome inhibitor ALLN to MCF-7 cells treated with TNF-α led to a substantial increase in observed levels of the 65-, 55-, and 28-kDa forms of HEF1, indicating that although these forms accumulate to levels higher than those detected in normally dividing MCF-7 cells following TNF-α treatment, they are nevertheless being actively degraded (Fig. 6A). Similar results were obtained with a less-potent analog of ALLN, ALLM, as well as the specific proteasome inhibitor lactacystin (data not shown). In contrast, in a synchronized population of mitotic MCF-7 cells, the addition of lactacystin induced a marked increase in the 65- and 28-kDa HEF1 forms compared to untreated cells, while the amount of the 55-kDa form remained unchanged (Fig. 6B). These results indicate, firstly, that the abundance of the HEF1 forms is controlled not only by caspases but also through degradation via the proteasome and, secondly, that the p55 form is subject to proteasomal degradation during apoptosis, but not mitosis.
FIG. 6.
The stability of HEF1 and its cleavage products (65, 55, and 28 kDa) is regulated by the proteasome. (A) MCF-7 cells were treated with TNF-α in the absence (--) or presence (+) of the proteasome inhibitor ALLN. Lysates were probed with anti-HEF1 antibodies (anti-HEF1/1 antibody [α-HEF1/1] to detect p115, p105, and p55; anti-HEF1/2 antibody to detect p65) or anti-HEF1/p130Cas antibody to detect the 28-kDa species. HeLa cells transfected with the HEF1 cDNA were run in the right lane as a size control for the various HEF1 cleavage products. (B) MCF-7 cells were synchronized by thymidine block. The cells were released for 4 h prior to the addition of the proteasome inhibitor lactacystin (final concentration, 10 μM). Mitotic cells were collected at 9 h by mitotic shake off, and lysates were probed with either anti-HEF1/p130Cas antibodies to detect the 65- and 28-kDa forms or anti-HEF1 antibodies to detect the 55-kDa form.
The C-terminal 28-kDa HEF1 peptide is sufficient to induce cell death.
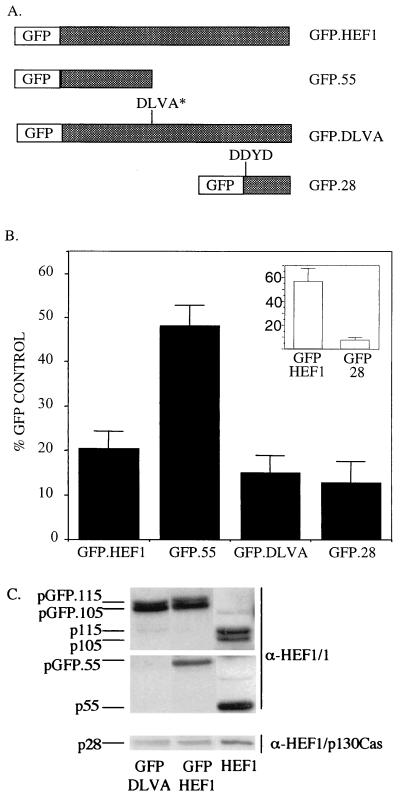
The preceding results indicate that the relative abundance of HEF1 forms is subject to complex regulation, suggesting that processed versus full-length forms of the molecule might possess discrete activities in apoptosis. Therefore, to further assess the role of the full-length versus processed HEF1 versions, we created GFP fusion constructs that correlate to wild-type HEF1, the 55-kDa HEF1 N-terminal fragment, a mutant of HEF1 (DLVD→DLVA) that cannot be cleaved to produce p55 and p65 cleaved forms, and the 28-kDa HEF1 C-terminal fragment (Fig. 7A). Modeling on protocols in which the ability of transiently expressed peptides to induce apoptosis is assessed by counting beta-galactosidase-positive (that is, transfected) cells (e.g., see reference 33), HeLa cells were transiently transfected and GFP-positive cells were scored to determine the effect of expressing HEF1 and the relevant panel of HEF1-derivatives on cell survival. The number of GFP-positive cells for each construct is expressed as a percentage of positive cells transfected with the pEGFP vector control (Fig. 7B). We note that transfected GFP.HEF1 localizes to focal adhesions (see Fig. 8B) and also undergoes cleavage, producing a GFP-fused 55-kDa N-terminal peptide (Fig. 7C), suggesting that the fusion proteins are behaving similarly to the native proteins with respect to at least two important criteria.
FIG. 7.
The 28-kDa HEF1 caspase cleavage product causes cell death. (A) Diagrammatic representation of GFP fusion constructs corresponding to wild-type HEF1 (GFP.HEF1), the 55-kDa N-terminal peptide (GFP.55), the full-length HEF1 molecule containing a mutation in the caspase cleavage site that prevents production of the 55-kDa N-terminal peptide (GFP.DLVA), and the 28-kDa C-terminal peptide (GFP.28). (B) HeLa cells and MCF-7 cells (inset) were transiently transfected with the GFP fusion constructs, and GFP-positive cells were scored as outlined in Materials and Methods. Total numbers of GFP-positive cells are expressed as a percentages of the number of control cells transfected with the GFP vector alone. Data represent the average of nine replicates from three independent experiments. Error bars indicated the standard errors of the mean. (C) Total proteins were extracted from HeLa cells transfected with GFP.DLVA, GFP.HEF1, and pCDNA.HEF1, and Western blots were probed with the indicated antibodies (α prefix).
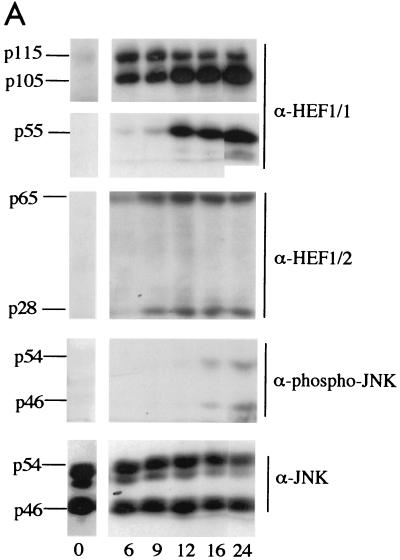
FIG. 8.
HEF1 overexpression causes increased phospho-JNK levels. (A) HEF1 cells were induced for HEF1 expression, and total proteins were extracted at the times indicated. Western blots of total proteins were probed with antibodies to HEF1 (α-HEF1/1 and α-HEF1/p130Cas) or anti-phospho-JNK antibodies (α-phospho-JNK), and to indicate equivalent loadings of total proteins, blots were then probed with antibodies to JNK (α-JNK). (B) Phospho-JNK localizes to focal adhesions and colocalizes with the HEF1. Shown are merged confocal images of MCF-7 cells transfected with the GFP vector alone (panel A) or GFP.HEF1 (panel B). Immunofluorescence was performed with phospho-specific anti-JNK antibodies. GFP is shown in green, and phospho-JNK staining is shown in red; yellow represents merged red and green staining.
As expected from the results described above, transfection of the GFP.HEF1 construct resulted in a reduced yield of GFP-positive cells relative to transfection of the GFP vector (Fig. 7B). The GFP.55 construct produced significantly more transfectants than the GFP.HEF1 construct. Conversely, the percentage of GFP.DLVA-positive cells was at a level similar to that of the GFP.HEF1 transfectants. The DLVA construct does not undergo cleavage to produce the 55-kDa N-terminal peptide (47) (Fig. 7C), although the downstream DDYD cleavage site remains intact, theoretically allowing p28 production. Further analysis demonstrated that a protein band corresponding in size and antibody reactivity to the HEF1 28-kDa C-terminal peptide could be detected in lysates extracted from cells transfected with the GFP.DLVA construct (Fig. 7C), although available antisera do not exclude the possibility that this form results from cleavage of p130Cas (as in Fig. 5C). These results suggest either that the full-length HEF1 protein is proapoptotic or that a proapoptotic moiety residing in the C terminus is produced from the DLVA mutant. In agreement with the latter possibility, transfection with the GFP.28 construct also results in a low percentage of GFP-positive cells. Finally, to demonstrate that this result is not specific to HeLa cells, we repeated assays with the GFP, GFP.HEF1, and GFP.28 constructs in MCF-7 cells. By 24 h after transfection, the frequency of cells expressing GFP.HEF1 is reduced twofold relative to that of cells expressing GFP, while cells expressing GFP.28 are found at less than 10% of control levels, supporting the idea that this domain independently and potently induces cell death (Fig. 7B, inset).
HEF1 induces activation of JNK signaling.
Recent studies have indicated that the HEF1 family member p130Cas associates with the adapter protein Crk to couple extracellular stimuli to activation of the JNK pathway (6, 24). In light of the previously reported association between HEF1 and Crk (57, 59) and the putative role for JNKs in apoptosis (reviewed in reference 37), JNK phosphorylation in HEF1 stable cell lines was examined using antibodies that specifically recognize phosphorylated JNK1 and JNK2, following the observation that activation requires and is reflected by phosphorylation of JNKs on amino acids T183 and Y185 (21). Full-length 105- and 115-kDa HEF1 is abundant by 6 h after induction, with the accumulation of the p65, p55, and p28 cleavage products first noticeable at 9 to 12 h (Fig. 8A). JNK1 and JNK2 are phosphorylated and activated around 16 h after induction of HEF1 expression (Fig. 8A), but not in the uninduced HEF1 cell lines or in vector-expressing control cell lines (data not shown). As a control, Western blots of cell lysates were probed with antibodies to JNK, demonstrating that there are approximately equivalent total JNK levels in all of the cell extracts (Fig. 8A).
Recent reports have noted that activation of regulatory kinases such as MEK kinase 1 and JNK is accompanied by changes in intracellular localization (20, 62). Analysis of untransfected MCF-7 cells or MCF-7 cells transfected with GFP indicates that the population of activated JNK in these cells localizes to focal adhesions, based on colocalization with focal adhesion markers such as paxillin (Fig. 8B and results not shown). Following the observation that the levels of phospho-JNK increase after HEF1 expression, we examined subcellular expression of phospho-JNK in GFP.HEF1-transfected MCF-7 cells. Consistent with the idea that the activation of JNK might be linked with proximity to HEF1, immunofluorescent analysis using the phosphorylation-specific anti-JNK antibody demonstrated extensive colocalization of GFP.HEF1 and phosphorylated JNK (Fig. 8B), suggesting that the observed activation may proceed via the combined function of proteins complexed at focal adhesions. The observation that formation of the HEF1 cleavage products appears to precede JNK phosphorylation and/or activation suggests that these cleavage events may be associated with JNK activation; alternatively, the activation may be due to the sustained overexpression of the full-length form of HEF1.
DISCUSSION
In epithelial cells, the decision to remain quiescent, proliferate, or undergo apoptosis is influenced by positional information such as cell-cell contacts and attachment status. Proteins associated with cell attachment structures may transmit such growth or death stimuli, as they are well positioned to integrate extracellular physical constraints with intracellular signaling cascades. Cas family proteins are important targets of integrin-mediated attachment signals (reviewed in reference 67). While other studies have focused on roles for p130Cas and HEF1 in control of cell motility (16, 19, 43, 64; Fashena et al., unpublished data), we have found that the HEF1 protein is dynamically regulated during the cell cycle, with specific caspase cleavage converting a focal adhesion-associated assembly factor to a truncated form associated with mitotic spindles. This has lead to the hypothesis that changes in HEF1 expression and localization might play important roles in controlling cell attachment and cell cycle progression (45, 47) and potentially cell viability.
Results here provide the first demonstration that HEF1 and p130Cas may additionally play an important role in active induction of apoptosis. The primary findings reported herein supporting this view are first, significantly, that HEF1 overexpression triggers caspase activation and apoptosis in MCF-7 breast carcinoma cells and HeLa cervical carcinoma cells; further, HEF1 overexpression-initiated apoptosis is augmented by TNF-α treatment. Second, we find that the endogenous HEF1 protein is a target of cleavage in apoptosis, with a caspase 3-like activity cleaving the protein into 55-, 65-, and 28-kDa peptides. Third, these cleavages occur relatively early following the initiation of apoptosis, paralleling cleavage of FAK and PARP and preceding that of gelsolin. Fourth, the abundance of the HEF1 cleavage species is additionally controlled by proteasome-mediated degradation. Fifth, structure-function analysis suggests that at least one potent proapoptotic activity resides in the conserved carboxy-terminal region of HEF1. Finally, the conversion of overexpressed HEF1 to caspase-cleaved fragments is followed by the induction of JNK phosphorylation, while HEF1 and activated populations of JNK colocalize at focal adhesions.
Effector caspases have been described as inducing the cleavage of many classes of proteins during apoptosis. These include nuclear proteins involved in DNA surveillance and repair (PARP [49] and DNA-dependent protein kinase [34, 76]), nuclear structure (nuclear lamins [50, 78]), and other diverse functions (U1 snRNP [17, 79], sterol response element binding proteins 1 and 2 [85], huntingtin protein [31], D4 GDP dissociation inhibitor [61], p21 [Cip1/Waf1] [29], and pRb [2, 40]). The majority of cytoplasmic proteins cleaved by caspases are involved in regulating cytoskeletal organization, and include FAK (87), gelsolin (44), PAK (70), Gas2 (9), fodrin (58), and several adherens junction proteins (e.g., β-catenin and adenomatous polyposis coli [10, 12]). Cleavage of target proteins by effector caspases may generate protein fragments that have altered activity which actively promotes apoptosis. PARP, as an example, has a complex role, as its cleavage is thought to prevent depletion of ATP and NAD (important for the later stages of apoptosis) and inhibit the DNA repair mechanism allowing double-stranded breaks in the DNA to accumulate (8, 75). FAK cleavage generates a form that localizes to focal adhesion sites but does not contain the kinase domain and acts as a dominant negative (28). In contrast, gelsolin and PAK become activated by caspase cleavage, with cleavage stimulating gelsolin-dependent cleavage of actin filaments (44) and PAK kinase acquiring enhanced activity toward its substrates (51, 70). In both cases, activation results in changes in the actin cytoskeleton and induction of apoptosis; to date, the degree to which the morphological changes and proapoptotic effect are interdependent remains to be determined.
Our data suggest that the cleavage products of HEF1 are not all simply inactivated forms of the full-length protein but that they possess independent activities. In mitosis, the p55HEF1 species is relatively stable and exhibits a localization pattern distinct from that of the full-length form, suggesting a separate function which may embody action as a sentinel for focal adhesion disassembly and readiness for cytokinesis (47). In contrast, the p28 and p65 forms of HEF1 are virtually undetectable due to proteasomal degradation, suggesting either that their clearance is necessary or that they are functionally mute. In contrast, all forms of the protein are present in apoptotic cells, and hence available to play an active role in the apoptotic process; our results suggest that p28HEF1 function may be particularly relevant in the promotion of apoptosis.
Although Cas family members have been shown to induce motility, no previous report has noted a role for these proteins in apoptotic progression. One possible reason for this difference is that the ability of these proteins to induce apoptosis is restricted by cell type; other studies have focused on Cas protein overexpression in fibroblasts and lymphoid cells. However, we note in this study that the ability of HEF1 to induce apoptosis is not restricted to MCF-7 cells, as comparable results are obtained in HeLa cells (Fig. 7 and results not shown). Alternatively, given that other studies were performed by transient transfection or by creation of stable lines constitutively expressing the Cas family proteins, it may be that the apoptotic effect was more difficult to discern. Our data suggest that p130Cas may have a similar potential to induce apoptosis in epithelial cells in that transfection of a construct expressing p130Cas resulted in the generation of a p28 species (Fig. 5C), most likely due to caspase-mediated cleavage, hence reflecting caspase activation. Finally, it has been noted the HEF1 protein is highly expressed in T and B cells (57, 59, 65). Given the discrete requirements for adherence in cell growth of lymphoid cells, it will be of interest to examine whether variation of HEF1 and/or p130Cas expression affects viability of these populations.
One key question raised by this study is whether the promotion of apoptosis by HEF1 is direct or indirect. Potential indirect mechanisms of HEF1 action in apoptosis might include changes in the cytoskeleton following HEF1 expression that impose physical stress on cells (Fashena et al., unpublished data) and/or inappropriate activation of signaling cascades via HEF1-dependent alteration of focal adhesion complexes. In this context, the similar timings of FAK and HEF1 cleavage are intriguing, given the previously described interactions between the two proteins in which FAK binding and phosphorylation of HEF1 in response to integrin receptor ligation allow the recruitment and activation of various signaling molecules (reviewed in reference 67). FAK has been implicated as a sensor of epithelial cell attachment, based on the observations that constitutively active FAK blocks apoptosis induced by cell detachment (anoikis) (27), whereas inhibition of FAK expression in cultured fibroblasts can promote apoptosis (35). This suggests that one role of endogenous HEF1 and FAK cleavage may be to block survival signals initiated by cell attachment. We note that our observations of HEF1 induction of apoptosis by overexpression, may appear contradictory when compared with cleavage and degradation of the endogenous molecule by proapoptotic stimuli. However, the overexpression results may be explained by posing a model in which levels of HEF1, FAK, and associated proteins at focal adhesions are carefully balanced. Inappropriate expression of individual constituents of focal complexes (such as HEF1) might additionally interfere with adhesion-related survival signaling. Finally, our data suggest that HEF1 and p130Cas are similarly cleaved at a conserved DDYD motif that demarcates the carboxy-terminal region to produce the p28 species. Intriguingly, this site overlaps with the site of FAK phosphorylation and subsequent Src family kinase binding in Cas proteins (77), raising the possibility that the susceptibility of HEF1 and p130Cas to cleavage may be governed by phosphorylation and the degree of association with focal adhesions.
Enhancement of JNK activity has also been implicated as a contributor to apoptosis in a number of studies, although the mechanism of JNK involvement in the process is not yet completely clear and may be cell type and stimulus dependent (18, 52, 54). In our examination of JNK signaling in relation to HEF1 induction, the proximity of HEF1 and phospho-JNK at sites of focal adhesion, along with the observed increases in phospho-JNK expression following HEF1 expression, is particularly interesting given recent reports of the Nsp proteins (Nsp1 to Nsp3 [56]) and the orthologous Cas-HEF1-associated signal transducer (CHAT) (Nsp3) protein. These proteins have been defined as molecules that activate JNK signaling in response to integrin-ECM contact. CHAT has been shown to interact with HEF1 and p130Cas through their carboxy-terminal conserved domains (corresponding to the region encompassed in p28) and is proposed to contribute to the activation of JNK signaling (72). Intriguingly, and indirectly supporting the functional interrelatedness of Nsp and Cas proteins, a retroviral integration screen designed to identify genes whose function was related to estrogen resistance and development of breast cancer turned up a small number of positive candidate loci, of which BCAR1, BCAR2, and BCAR3 have so far been defined. Strikingly, BCAR1 has been shown to be p130Cas (11, 83), while BCAR3 is Nsp2 (82). In sum, these results suggest a model in which the action of the HEF1 carboxy terminus as a docking site for Nsp and CHAT proteins enables activation of JNK kinases. Given that enhanced JNK activation occurs at later stages following HEF1 induction, it may be that the activation event is dependent upon the release of particular truncated forms, such as p28, in signaling dependent on Nsp and CHAT. Notably, the initiator kinase for the JNK signaling pathway, MEK kinase 1, is itself activated by caspase cleavage in anoikis (14). Alternatively, previous work with p130Cas has shown that this protein can activate the JNK pathway through its recruitment of Crk (6, 24, 57, 59), indicating that sequences in the SH2 binding site region (substrate domain) of HEF1 may separately contribute to activation. The importance of HEF1-stimulated JNK activity awaits further experimentation and clarification of the role that JNK activation plays in apoptosis.
In summary, the Cas family members have been implicated in an increasingly diverse set of physiological processes (67), including control of cell attachment, T-cell costimulation, migration, cell cycle, transformation, and now apoptosis. The selective proteolytic processing and relocalization of HEF1 during apoptosis and mitosis detailed here provide an economical and elegant means of extending the function of a single protein to different biological spheres.
ACKNOWLEDGMENTS
S.F.L. and G.M.O. contributed equally to this study.
We are grateful to Maureen Murphy and Kerry Campbell for helpful discussions, critiques, and the gift of reagents. We thank Maureen Murphy and Mary Ann Sells for thorough review of the manuscript.
E.A.G. was supported in this work by NIH grant RO1 CA63366 and core funds CA-06927 (to Fox Chase Cancer Center). S.F.L. was supported by American Cancer Society fellowship PF-4383 and NIH fellowship F32 GM18223. G.M.O. was supported by a W. J. Avery Fellowship. S.J.F. was supported by NIH postdoctoral training grant T32 CA09035.
REFERENCES
- 1.Alexandropoulos K, Baltimore D. Coordinate activation of c-Src by SH3- and SH2-binding sites on a novel, p130Cas-related protein, Sin. Genes Dev. 1996;10:1341–1355. doi: 10.1101/gad.10.11.1341. [DOI] [PubMed] [Google Scholar]
- 2.An B, Dou Q P. Cleavage of retinoblastoma protein during apoptosis: an interleukin 1 beta-converting enzyme-like protease as candidate. Cancer Res. 1996;56:438–442. [PubMed] [Google Scholar]
- 3.Ashkenazi A, Dixit V M. Death receptors: signaling and modulation. Science. 1998;281:1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 4.Astier A, Manie S N, Law S F, Canty T, Hagheyeghi N, Druker B J, Salgia R, Golemis E A, Freedman A S. Association of the Cas-like molecule HEF1 with CrkL following integrin and antigen receptor signaling in B cells. Possible relevance to neoplastic lymphohematopoietic cells. Leuk Lymphoma. 1997;28:65–72. doi: 10.3109/10428199709058332. [DOI] [PubMed] [Google Scholar]
- 5.Bannerman D D, Sathyamoorthy M, Goldblum S E. Bacterial lipopolysaccharide disrupts endothelial monolayer integrity and survival signaling events through caspase cleavage of adherens junction proteins. J Biol Chem. 1998;273:35371–35380. doi: 10.1074/jbc.273.52.35371. [DOI] [PubMed] [Google Scholar]
- 6.Blaukat A, Ivankovic-Dikic I, Gronroos E, Dolfi F, Tokiwa G, Vuori K, Dikic I. Adaptor proteins Grb2 and Crk couple Pyk2 with activation of specific mitogen-activated protein kinase cascades. J Biol Chem. 1999;274:14893–14901. doi: 10.1074/jbc.274.21.14893. [DOI] [PubMed] [Google Scholar]
- 7.Boudreau N, Sympson C J, Werb Z, Bissell M J. Suppression of ICE and apoptosis in mammary epithelial cells by extracellular matrix. Science. 1995;267:891–893. doi: 10.1126/science.7531366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boulares A H, Yakovlev A G, Ivanova V, Stoica B A, Wang G, Iyer S, Smulson M. Role of poly(ADP-ribose) polymerase (PARP) cleavage in apoptosis. Caspase 3-resistant PARP mutant increases rates of apoptosis in transfected cells. J Biol Chem. 1999;274:22932–22940. doi: 10.1074/jbc.274.33.22932. [DOI] [PubMed] [Google Scholar]
- 9.Brancolini C, Benedetti M, Schneider C. Microfilament reorganization during apoptosis: the role of Gas2, a possible substrate for ICE-like proteases. EMBO J. 1995;14:5179–5190. doi: 10.1002/j.1460-2075.1995.tb00202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brancolini C, Lazarevic D, Rodriguez J, Schneider C. Dismantling cell-cell contacts during apoptosis is coupled to a caspase-dependent proteolytic cleavage of beta-catenin. J Cell Biol. 1997;139:759–771. doi: 10.1083/jcb.139.3.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brinkman A, van Der Flier S, Kok E M, Dorssers L C. BCAR1, a human homologue of the adapter protein p130Cas, and antiestrogen resistance in breast cancer cells. J Natl Cancer Inst. 2000;92:112–120. doi: 10.1093/jnci/92.2.112. [DOI] [PubMed] [Google Scholar]
- 12.Browne S J, MacFarlane M, Cohen G M, Paraskeva C. The adenomatous polyposis coli protein and retinoblastoma protein are cleaved early in apoptosis and are potential substrates for caspases. Cell Death Differ. 1998;5:206–213. doi: 10.1038/sj.cdd.4400331. [DOI] [PubMed] [Google Scholar]
- 13.Burow M E, Weldon C B, Tang Y, Navar G L, Krajewski S, Reed J C, Hammond T G, Clejan S, Beckman B S. Differences in susceptibility to tumor necrosis factor alpha-induced apoptosis among MCF-7 breast cancer cell variants. Cancer Res. 1998;58:4940–4946. [PubMed] [Google Scholar]
- 14.Cardone M H, Salvesen G S, Widmann C, Johnson G, Frisch S M. The regulation of anoikis: MEKK-1 activation requires cleavage by caspases. Cell. 1997;90:315–323. doi: 10.1016/s0092-8674(00)80339-6. [DOI] [PubMed] [Google Scholar]
- 15.Carmichael J, DeGraff W G, Gazdar A F, Minna J D, Mitchell J B. Evaluation of a tetrazolium-based semiautomated colorimetric assay: assessment of chemosensitivity testing. Cancer Res. 1987;47:936–942. [PubMed] [Google Scholar]
- 16.Cary L A, Han D C, Polte T R, Hanks S K, Guan J-L. Identification of p130Cas as a mediator of focal adhesion kinase-promoted cell migration. J Cell Biol. 1998;140:211–221. doi: 10.1083/jcb.140.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casciola-Rosen L, Nicholson D W, Chong T, Rowan K R, Thornberry N A, Miller D K, Rosen A. Apopain/CPP32 cleaves proteins that are essential for cellular repair: a fundamental principle of apoptotic death. J Exp Med. 1996;183:1957–1964. doi: 10.1084/jem.183.5.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaudhary P M, Eby M T, Jasmin A, Hood L. Activation of the c-Jun N-terminal kinase/stress-activated protein kinase pathway by overexpression of caspase-8 and its homologs. J Biol Chem. 1999;274:19211–19219. doi: 10.1074/jbc.274.27.19211. [DOI] [PubMed] [Google Scholar]
- 19.Cheresh D A, Leng J, Klemke R L. Regulation of cell contraction and membrane ruffling by distinct signals in migratory cells. J Cell Biol. 1999;146:1107–1116. doi: 10.1083/jcb.146.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christerson L B, Vanderbilt C A, Cobb M H. MEKK1 interacts with alpha-actinin and localizes to stress fibers and focal adhesions. Cell Motil Cytoskel. 1999;43:186–198. doi: 10.1002/(SICI)1097-0169(1999)43:3<186::AID-CM2>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 21.Derijard B, Hibi M, Wu I H, Barrett T, Su B, Deng T, Karin M, Davis R J. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the cJun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 22.Dick L R, Cruikshank A A, Destree A T, Grenier L, McCormack T A, Melandri F D, Nunes S L, Palombella V J, Parent L A, Plamondon L, Stein R L. Mechanistic studies on the inactivation of the proteasome by lactacystin in cultured cells. J Biol Chem. 1997;272:182–188. doi: 10.1074/jbc.272.1.182. [DOI] [PubMed] [Google Scholar]
- 23.DiPietrantonio A M, Hsieh T, Wu J M. Activation of caspase 3 in HL-60 cells exposed to hydrogen peroxide. Biochem Biophys Res Commun. 1999;255:477–482. doi: 10.1006/bbrc.1999.0208. [DOI] [PubMed] [Google Scholar]
- 24.Dolfi F, Garcia-Guzman M, Ojaniemi M, Nakamura H, Matsuda M, Vuori K. The adaptor protein Crk connects multiple cellular stimuli to the JNK signaling pathway. Proc Natl Acad Sci USA. 1998;95:15394–15399. doi: 10.1073/pnas.95.26.15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evan G I, Brown L, Whyte M, Harrington E. Apoptosis and the cell cycle. Curr Opin Cell Biol. 1995;7:825–834. doi: 10.1016/0955-0674(95)80066-2. [DOI] [PubMed] [Google Scholar]
- 26.Frisch S M, Francis H. Disruption of epithelial cell-matrix interactions induces apoptosis. J Cell Biol. 1994;124:619–626. doi: 10.1083/jcb.124.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frisch S M, Vuori K, Ruoslahti E, Chan-Hui P-Y. Control of adhesion-dependent cell survival by focal adhesion kinase. J Cell Biol. 1996;134:793–799. doi: 10.1083/jcb.134.3.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gervais F G, Thornberry N A, Ruffolo S C, Nicholson D W, Roy S. Caspases cleave focal adhesion kinase during apoptosis to generate a FRNK-like polypeptide. J Biol Chem. 1998;273:17102–17108. doi: 10.1074/jbc.273.27.17102. [DOI] [PubMed] [Google Scholar]
- 29.Gervais J L, Seth P, Zhang H. Cleavage of CDK inhibitor p21(Cip1/Waf1) by caspases is an early event during DNA damage-induced apoptosis. J Biol Chem. 1998;273:19207–19212. doi: 10.1074/jbc.273.30.19207. [DOI] [PubMed] [Google Scholar]
- 30.Ghayur T, Banerjee S, Hugunin M, Butler D, Herzog L, Carter A, Quintal L, Sekut L, Talanian R, Paskind M, Wong W, Kamen R, Tracey D, Allen H. Caspase-1 processes IFN-gamma-inducing factor and regulates LPS-induced IFN-gamma production. Nature. 1997;386:619–623. doi: 10.1038/386619a0. [DOI] [PubMed] [Google Scholar]
- 31.Goldberg Y P, Nicholson D W, Rasper D M, Kalchman M A, Koide H B, Graham R K, Bromm M, Kazemi-Esfarjani P, Thornberry N A, Vaillancourt J P, Hayden M R. Cleavage of huntingtin by apopain, a proapoptotic cysteine protease, is modulated by the polyglutamine tract. Nat Genet. 1996;13:442–449. doi: 10.1038/ng0896-442. [DOI] [PubMed] [Google Scholar]
- 32.Green D R, Reed J C. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 33.Gu Z, Flemington C, Chittenden T, Zambetti G P. ei24, a p53 response gene involved in growth suppression and apoptosis. Mol Cell Biol. 2000;20:233–241. doi: 10.1128/mcb.20.1.233-241.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han Z, Malik N, Carter T, Reeves W H, Wyche J H, Hendrickson E A. DNA-dependent protein kinase is a target for a CPP32-like apoptotic protease. J Biol Chem. 1996;271:25035–25040. doi: 10.1074/jbc.271.40.25035. [DOI] [PubMed] [Google Scholar]
- 35.Hungerford J E, Compton M T, Matter M L, Hoffstrom B G, Otey C A. Inhibition of pp125FAK in cultured fibroblasts results in apoptosis. J Cell Biol. 1996;135:1383–1390. doi: 10.1083/jcb.135.5.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ikebe T, Takeuchi H, Jimi E, Beppu M, Shinohara M, Shirasuna K. Involvement of proteasomes in migration and matrix metalloproteinase-9 production of oral squamous cell carcinoma. Int J Cancer. 1998;77:578–585. doi: 10.1002/(sici)1097-0215(19980812)77:4<578::aid-ijc18>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 37.Ip Y T, Davis R J. Signal transduction by the c-Jun N-terminal kinase (JNK)—from inflammation to development. Curr Opin Cell Biol. 1998;10:205–219. doi: 10.1016/s0955-0674(98)80143-9. [DOI] [PubMed] [Google Scholar]
- 38.Ishino M, Ohba T, Sasaki H, Sasaki T. Molecular cloning of a cDNA encoding a phosphoprotein, Efs, which contains a Src homology 3 domain and associates with Fyn. Oncogene. 1995;11:2331–2338. [PubMed] [Google Scholar]
- 39.Janicke R U, Sprengart M L, Wati M R, Porter A G. Caspase-3 is required for DNA fragmentation and morphological changes associated with apoptosis. J Biol Chem. 1998;273:9357–9360. doi: 10.1074/jbc.273.16.9357. [DOI] [PubMed] [Google Scholar]
- 40.Janicke R U, Walker P A, Lin X Y, Porter A G. Specific cleavage of the retinoblastoma protein by an ICE-like protease in apoptosis. EMBO J. 1996;15:6969–6978. [PMC free article] [PubMed] [Google Scholar]
- 41.Kerr J F, Wyllie A H, Currie A R. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.King K L, Cidlowski J A. Cell cycle and apoptosis: common pathways to life and death. J Cell Biochem. 1995;58:175–180. doi: 10.1002/jcb.240580206. [DOI] [PubMed] [Google Scholar]
- 43.Klemke R L, Leng J, Molander R, Brooks P C, Vuori K, Cheresh D A. CAS/Crk coupling serves as a “molecular switch” for induction of cell migration. J Cell Biol. 1998;140:961–972. doi: 10.1083/jcb.140.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kothakota S, Azuma T, Reinhard C, Klippel A, Tang J, Chu K, McGarry T J, Kirschner M W, Koths K, Kwiatkowski D J, Williams L T. Caspase-3-generated fragment of gelsolin: effector of morphological change in apoptosis. Science. 1997;278:294–298. doi: 10.1126/science.278.5336.294. [DOI] [PubMed] [Google Scholar]
- 45.Law S F, Estojak J, Wang B, Mysliwiec T, Kruh G D, Golemis E A. Human enhancer of filamentation 1, a novel p130cas-like docking protein, associates with focal adhesion kinase and induces pseudohyphal growth in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:3327–3337. doi: 10.1128/mcb.16.7.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Law S F, Zhang Y-Z, Fashena S, Toby G, Estojak J, Golemis E A. Dimerization of the docking/adaptor protein HEF1 via a carboxy-terminal helix-loop-helix domain. Exp Cell Res. 1999;252:224–235. doi: 10.1006/excr.1999.4609. [DOI] [PubMed] [Google Scholar]
- 47.Law S F, Zhang Y-Z, Klein-Szanto A, Golemis E A. Cell-cycle regulated processing of HEF1 to multiple protein forms differentially targeted to multiple compartments. Mol Cell Biol. 1998;18:3540–3551. doi: 10.1128/mcb.18.6.3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lazebnik Y A, Cole S, Cooke C A, Nelson W G, Earnshaw W C. Nuclear events of apoptosis in vitro in cell-free mitotic extracts: a model system for analysis of the active phase of apoptosis. J Cell Biol. 1993;123:7–22. doi: 10.1083/jcb.123.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lazebnik Y A, Kaufmann S H, Desnoyers S, Poirier G G, Earnshaw W C. Cleavage of poly(ADP-ribose) polymerase by a proteinase with properties like ICE. Nature. 1994;371:346–347. doi: 10.1038/371346a0. [DOI] [PubMed] [Google Scholar]
- 50.Lazebnik Y A, Takahashi A, Moir R D, Goldman R D, Poirier G G, Kaufmann S H, Earnshaw W C. Studies of the lamin proteinase reveal multiple parallel biochemical pathways during apoptotic execution. Proc Natl Acad Sci USA. 1995;92:9042–9046. doi: 10.1073/pnas.92.20.9042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee N, MacDonald H, Reinhard C, Halenbeck R, Roulston A, Shi T, Williams L T. Activation of hPAK65 by caspase cleavage induces some of the morphological and biochemical changes of apoptosis. Proc Natl Acad Sci USA. 1997;94:13642–13647. doi: 10.1073/pnas.94.25.13642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lenczowski J M, Dominguez L, Eder A M, King L B, Zacharchuk C M, Ashwell J D. Lack of a role for Jun kinase and AP-1 in Fas-induced apoptosis. Mol Cell Biol. 1997;17:170–181. doi: 10.1128/mcb.17.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li F, Ambrosini G, Chu E Y, Plescia J, Tognin S, Marchisio P C, Altieri D C. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396:580–584. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- 54.Liu Z G, Hsu H, Goeddel D V, Karin M. Dissection of TNF receptor 1 effector functions: JNK activation is not linked to apoptosis while NF-kappaB activation prevents cell death. Cell. 1996;87:565–576. doi: 10.1016/s0092-8674(00)81375-6. [DOI] [PubMed] [Google Scholar]
- 55.Los M, Wesselborg S, Schulze-Osthoff K. The role of caspases in development, immunity, and apoptotic signal transduction: lessons from knockout mice. Immunity. 1999;10:629–639. doi: 10.1016/s1074-7613(00)80062-x. [DOI] [PubMed] [Google Scholar]
- 56.Lu Y, Brush J, Stewart T. NSP1 defines a novel family of adaptor proteins linking integrin and tyrosine kinase receptors to the c-Jun N-terminal kinase/stress-activated protein kinase signaling pathway. J Biol Chem. 1999;274:10047–10052. doi: 10.1074/jbc.274.15.10047. [DOI] [PubMed] [Google Scholar]
- 57.Manie S N, Beck A R P, Astier A, Law S F, Canty T, Hirai H, Druker B J, Avraham H, Haghayegi N, Sattler M, Salgia R, Griffin J D, Golemis E A, Freedman A S. Involvement of p130Cas and p105HEF1, a novel Cas-like docking protein, in a cytoskeleton-dependent signaling pathway initiated by ligation of integrin or antigen receptor on human B cells. J Biol Chem. 1997;272:4230–4236. doi: 10.1074/jbc.272.7.4230. [DOI] [PubMed] [Google Scholar]
- 58.Martin S J, O'Brien G A, Nishioka W K, McGahon A J, Mahboubi A, Saido T C, Green D R. Proteolysis of fodrin (non-erythroid spectrin) during apoptosis. J Biol Chem. 1995;270:6425–6428. doi: 10.1074/jbc.270.12.6425. [DOI] [PubMed] [Google Scholar]
- 59.Minegishi M, Tachibana K, Sato T, Iwata S, Nojima Y, Morimoto C. Structure and function of Cas-L, a 105-kD Crk-associated substrate-related protein that is involved in beta-1 integrin-mediated signaling in lymphocytes. J Exp Med. 1996;184:1365–1375. doi: 10.1084/jem.184.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miossec C, Dutilleul V, Fassy F, Diu-Hercend A. Evidence for CPP32 activation in the absence of apoptosis during T lymphocyte stimulation. J Biol Chem. 1997;272:13459–13462. doi: 10.1074/jbc.272.21.13459. [DOI] [PubMed] [Google Scholar]
- 61.Na S, Chuang T H, Cunningham A, Turi T G, Hanke J H, Bokoch G M, Danley D E. D4-GDI, a substrate of CPP32, is proteolyzed during Fas-induced apoptosis. J Biol Chem. 1996;271:11209–11213. doi: 10.1074/jbc.271.19.11209. [DOI] [PubMed] [Google Scholar]
- 62.Nagata K-I, Puls A, Futter C, Aspenstrom P, Schaefer E, Nakata T, Hirokawa N, Hall A. The MAP kinase kinase kinase MLK2 co-localizes with activated JNK along microtubules and associates with kinesin superfamily motor KIF3. EMBO J. 1998;17:149–158. doi: 10.1093/emboj/17.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nojima Y, Morino N, Mimura T, Hamasaki K, Furuya H, Sakai R, Sato T, Tachibana K, Morimoto C, Yazaki Y, Hirai H. Integrin-mediated cell adhesion promotes tyrosine phosphorylation of p130Cas, a Src homology 3-containing molecule having multiple Src homology 2-binding motifs. J Biol Chem. 1995;270:15398–15402. doi: 10.1074/jbc.270.25.15398. [DOI] [PubMed] [Google Scholar]
- 64.Ohashi Y, Iwata S, Kamiguchi K, Morimoto C. Tyrosine phosphorylation of Crk-associated substrate lymphocyte-type is a critical element in TCR- and beta1 integrin-induced T lymphocyte migration. J Immunol. 1999;163:3727–3734. [PubMed] [Google Scholar]
- 65.Ohashi Y, Tachibana K, Kamiguchi K, Fujita H, Morimoto C. T cell receptor-mediated tyrosine phosphorylation of Cas-L, a 105-kDa Crk-associated substrate-related protein, and its association of Crk and C3G. J Biol Chem. 1998;273:6446–6451. doi: 10.1074/jbc.273.11.6446. [DOI] [PubMed] [Google Scholar]
- 66.Ohba T, Ishino M, Aoto H, Sasaki T. Dot far-Western blot analysis of relative binding affinities of the src homology 3 domains of efs and its related proteins. Anal Biochem. 1998;262:185–192. doi: 10.1006/abio.1998.2772. [DOI] [PubMed] [Google Scholar]
- 67.O'Neill G M, Fashena S J, Golemis E A. Integrin signaling: a new Cas(t) of characters enters the stage. Trends Cell Biol. 2000;10:111–119. doi: 10.1016/s0962-8924(99)01714-6. [DOI] [PubMed] [Google Scholar]
- 68.Orlowski R Z. The role of the ubiquitin-proteasome pathway in apoptosis. Cell Death Differ. 1999;6:303–313. doi: 10.1038/sj.cdd.4400505. [DOI] [PubMed] [Google Scholar]
- 69.Pettmann B, Henderson C E. Neuronal cell death. Neuron. 1998;20:633–647. doi: 10.1016/s0896-6273(00)81004-1. [DOI] [PubMed] [Google Scholar]
- 70.Rudel T, Bokoch G M. Membrane and morphological changes in apoptotic cells regulated by caspase-mediated activation of PAK2. Science. 1997;276:1571–1574. doi: 10.1126/science.276.5318.1571. [DOI] [PubMed] [Google Scholar]
- 71.Sakai R, Iwamatsu A, Hirano N, Ogawa S, Tanaka T, Mano H, Yazaki Y, Hirai H. A novel signaling molecule, p130, forms stable complexes in vivo with v-Crk and v-Src in a tyrosine phosphorylation-dependent manner. EMBO J. 1994;13:3748–3756. doi: 10.1002/j.1460-2075.1994.tb06684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sakakibara A, Hattori S. Chat, a Cas/HEF1-associated adaptor protein that integrates multiple signaling pathways. J Biol Chem. 2000;275:6404–6410. doi: 10.1074/jbc.275.9.6404. [DOI] [PubMed] [Google Scholar]
- 73.Sattler M, Salgia R, Shrikhande G, Verma S, Uemura N, Law S F, Golemis E A, Griffin J D. Differential signaling after beta1 integrin ligation is mediated through binding of CRKL to p120CBL and p110HEF1. J Biol Chem. 1997;272:14320–14326. doi: 10.1074/jbc.272.22.14320. [DOI] [PubMed] [Google Scholar]
- 74.Scaffidi C, Kirchhoff S, Krammer P H, Peter M E. Apoptosis signaling in lymphocytes. Curr Opin Immunol. 1999;11:277–285. doi: 10.1016/s0952-7915(99)80045-4. [DOI] [PubMed] [Google Scholar]
- 75.Simbulan-Rosenthal C M, Rosenthal D S, Iyer S, Boulares A H, Smulson M E. Transient poly(ADP-ribosyl)ation of nuclear proteins and role of poly(ADP-ribose) polymerase in the early stages of apoptosis. J Biol Chem. 1998;273:13703–13712. doi: 10.1074/jbc.273.22.13703. [DOI] [PubMed] [Google Scholar]
- 76.Song Q, Lees-Miller S P, Kumar S, Zhang Z, Chan D W, Smith G C, Jackson S P, Alnemri E S, Litwack G, Khanna K K, Lavin M F. DNA-dependent protein kinase catalytic subunit: a target for an ICE-like protease in apoptosis. EMBO J. 1996;15:3238–3246. [PMC free article] [PubMed] [Google Scholar]
- 77.Tachibana K, Urano T, Fujita H, Ohashi Y, Kamiguchi K, Iwata S, Hirai H, Morimoto C. Tyrosine phosphorylation of crk-associated substrates by focal adhesion kinase. A putative mechanism for the integrin-mediated tyrosine phosphorylation of crk-associated substrates. J Biol Chem. 1997;272:29083–29090. doi: 10.1074/jbc.272.46.29083. [DOI] [PubMed] [Google Scholar]
- 78.Takahashi A, Alnemri E S, Lazebnik Y A, Fernandes-Alnemri T, Litwack G, Moir R D, Goldman R D, Poirier G, Kaufmann S H, Earnshaw W C. Cleavage of lamin A by Mch2 alpha but not CPP32: multiple interleukin 1 beta-converting enzyme-related proteases with distinct substrate recognition properties are active in apoptosis. Proc Natl Acad Sci USA. 1996;93:8395–8400. doi: 10.1073/pnas.93.16.8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tewari M, Beidler D R, Dixit V M. CrmA-inhibitable cleavage of the 70-kDa protein component of the U1 small nuclear ribonucleoprotein during Fas- and tumor necrosis factor-induced apoptosis. J Biol Chem. 1995;270:18738–18741. doi: 10.1074/jbc.270.32.18738. [DOI] [PubMed] [Google Scholar]
- 80.Thornberry N A, Lazebnik Y. Caspases: enemies within. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 81.Tsubata T, Wu J, Honjo T. B-cell apoptosis induced by antigen receptor crosslinking is blocked by a T-cell signal through CD40. Nature. 1993;364:645–648. doi: 10.1038/364645a0. [DOI] [PubMed] [Google Scholar]
- 82.van Agthoven T, van Agthoven T L, Dekker A, van der Spek P J, Vreede L, Dorssers L C. Identification of BCAR3 by a random search for genes involved in antiestrogen resistance of human breast cancer cells. EMBO J. 1998;17:2799–2808. doi: 10.1093/emboj/17.10.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.van der Flier S, Brinkman A, Look M P, Kok E M, Meijer-Van Gelder M E, Klijn J G, Dorssers L C, Foekens J A. Bcar1/p130Cas protein and primary breast cancer: prognosis and response to tamoxifen treatment. J Natl Cancer Inst. 2000;92:120–127. doi: 10.1093/jnci/92.2.120. [DOI] [PubMed] [Google Scholar]
- 84.Vuori K, Ruoslahti E. Tyrosine phosphorylation of p130Cas and cortactin accompanies integrin-mediated cell adhesion to extracellular matrix. J Biol Chem. 1995;270:22259–22262. doi: 10.1074/jbc.270.38.22259. [DOI] [PubMed] [Google Scholar]
- 85.Wang X, Zelenski N G, Yang J, Sakai J, Brown M S, Goldstein J L. Cleavage of sterol regulatory element binding proteins (SREBPs) by CPP32 during apoptosis. EMBO J. 1996;15:1012–1020. [PMC free article] [PubMed] [Google Scholar]
- 86.Watanabe Y, Akaike T. Possible involvement of caspase-like family in maintenance of cytoskeleton integrity. J Cell Physiol. 1999;179:45–51. doi: 10.1002/(SICI)1097-4652(199904)179:1<45::AID-JCP6>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 87.Wen L P, Fahrni J A, Troie S, Guan J L, Orth K, Rosen G D. Cleavage of focal adhesion kinase by caspases during apoptosis. J Biol Chem. 1997;272:26056–26061. doi: 10.1074/jbc.272.41.26056. [DOI] [PubMed] [Google Scholar]