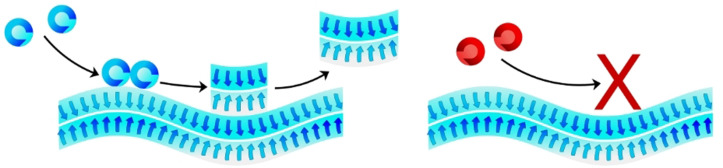
Abstract
Chirality is a fundamental feature of asymmetric molecules and of critical importance for intermolecular interactions. The growth of amyloid fibrils displays a strong enantioselectivity, which is manifested as elongation through the addition of monomers of the same, but not opposite, chirality as the parent aggregate. Here we ask whether also secondary nucleation on the surface of amyloid fibrils, of relevance for toxicity, is governed by the chirality of the nucleating monomers. We use short amyloid peptides (Aβ20‐34 and IAPP20‐29) with all residues as L‐ or all D‐enantiomer in self and cross‐seeding experiments with low enough seed concentration that any acceleration of fibril formation is dominated by secondary nucleation. We find a strong enantio‐specificity of this auto‐catalytic process with secondary nucleation being observed in the self‐seeding experiments only. The results highlight a role of secondary nucleation in strain propagation.
Keywords: aggregation, amyloid-beta peptides, autocatalysis, enantioselectivity, secondary nucleation
Amyloid‐forming peptides are chiral substances. While the growth of amyloid fibrils is known to be enantiospecific, we show here that this applies also to secondary nucleation of monomers in the presence of fibrils. Secondary nucleation is only possible if the monomers in solution and aggregate are of the same chirality.
Surface‐catalyzed secondary nucleation is an autocatalytic process by which new aggregates are formed in a reaction involving both monomers and already existing aggregates.[ 1 , 2 ] Such secondary nucleation is prominent in self‐assembly reactions, including crystallization of small molecules and proteins[ 3 , 4 ] and the formation of fibrous aggregates of proteins and peptides.[ 5 , 6 , 7 , 8 ]
The energy barrier for all forms of nucleation is higher than for growth of existing aggregates; however, the energy barrier for secondary nucleation is much lower than for primary nucleation.[ 9 , 10 ] Thus, as soon as a finite amount of aggregates have formed in an initially monomeric solution, or if a small amount of aggregate is introduced from an external source, secondary nucleation dominates over primary nucleation in the generation of new aggregates. In a system dominated by secondary nucleation, the end point aggregates may be highly monomorphic. [11] This fact is exploited in industrial crystallizers to obtain a homogeneous end product [1] and to generate multiple identical crystals for diffraction analysis; seeding of a supersaturated monomer solution with pre‐formed crystals leads to the generation of new crystals with the same morphology, chirality, crystal packing and space group as the seed due to secondary nucleation of monomers on the original seed.[ 12 , 13 ]
Chiral selectivity of secondary nucleation has been reported for the formation of optically active assemblies from chiral [13] as well as non‐chiral monomers.[ 12 , 13 , 14 ] The formation of optically active aggregates from an initially inactive system was reported in 1954, [15] and a few years later a model was presented for autocatalytic processes leading to the spontaneous formation of product with high enantiomeric excess. [16] In the presence of a chiral product, achiral reactants may preferentially produce crystals of one chirality, leading to a feedback mechanism for the generation and amplification of optical activity. [17] Enantioselectivity of secondary nucleation and growth is thus established in a number of cases.[ 12 , 13 , 14 , 18 , 19 ]
Amyloid‐forming peptides are intensely studied, due to their involvement in neurodegenerative diseases. The formation of amyloid fibrils from monomers is a nucleated, multi‐step process in which unstable reaction intermediates are believed to be the most toxic species. [20] The end product consists of highly ordered fibrils in which every peptide unit is folded in the same way. Replication of the fibril structure by monomer addition contributes to the propagation of fold and morphology in amyloid and prion systems [21] and may contribute to the proliferation of disease‐relevant forms. It has been proposed that structure propagation may originate not only from fragmentation and growth, but also from monomer‐dependent secondary nucleation. [22]
Peptides are chiral substances, naturally only occurring in the L‐form. Studying co‐assembly or self‐sorting in mixtures of L‐ and D‐peptides has yielded important insights into the chiral basis of protein interactions.[ 23 , 24 , 25 , 26 , 27 ] Using this approach with amyloidogenic peptides, it has been observed that L‐ or D‐peptides form similar fibrils but with inverted handedness as reported by circular dichroism (CD) spectroscopy and atomic force microscopy (AFM). [28] In racemic mixtures, monomers have been observed to form heterochiral fibrils with a structure distinct from the homochiral aggregates. [29] If monomers of one enantiomer forms fibrils, it is typically observed that monomers of the other are unable to be incorporated into these mirror‐image seeds. Such enantioselective fibril growth has been established in a number of systems including β2‐microglobulin fragments,[ 30 , 31 , 32 ] amyloid β peptides,[ 31 , 33 , 34 ] and other short amyloidogenic peptides.[ 28 , 35 , 36 ] PolyQ peptides seem to be an exception to this rule. [37]
The aggregation of many different amyloidogenic peptides has been found to be dominated by surface catalyzed secondary nucleation.[ 7 , 8 , 38 ] This process has been linked to massive transient generation of oligomers, [39] which are widely believed to be the toxic species of relevance in disease. [20] Catalysis of nucleation on fibril surfaces is in many cases remarkably specific with regards to peptide sequence,[ 40 , 41 ] implying that, analogously to elongation, secondary nucleation requires a structural compatibility between the parent fibril and nucleating monomers.
Here we investigated the role of chirality in secondary nucleation in amyloid formation. We studied the low seed regime, in which secondary nucleation, if existent, may lead to a significant shortening of the lag phase in a manner dependent on the seed concentration. [42] We used the 20–34 fragment of amyloid β peptide (Aβ(20–34)), for which efficient seeding at low seed concentration has been reported, [43] and global analysis of data at multiple monomer concentrations support secondary nucleation as the dominant route of new aggregate generation (Figure S3, S4). We prepared supersaturated monomeric solutions of the L‐ and D‐peptide, respectively, and monitored their aggregation in the absence and presence of 0.3 % preformed fibrils (seeds) of the same or opposite chirality (Figure 1) using thioflavin T (ThT) fluorescence (Figure 2) and CD spectroscopy (Figure 3 D,E). The end state was investigated using CD spectroscopy and AFM (Figure 3 A–C, Figure S2).
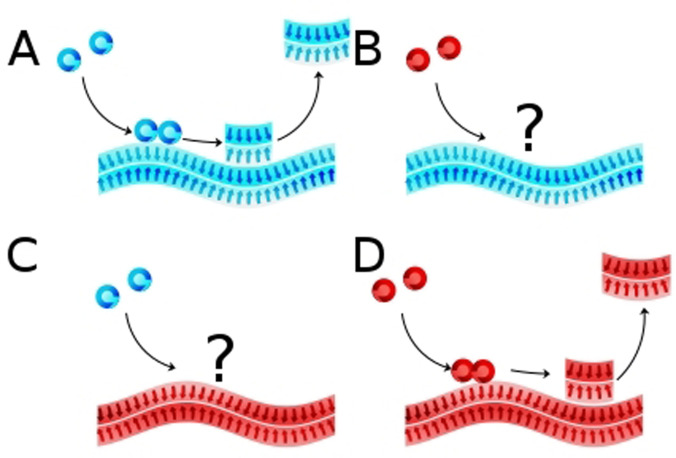
Figure 1.
Secondary nucleation. Cartoons depicting A) L‐fibril + L‐monomer, in which case the seed catalyzes the formation of new aggregates from monomer; B) L‐fibril + D‐monomer, no catalysis; C) D‐fibril + L‐monomer, no catalysis; D) D‐fibril + D‐monomer, seed catalyzes formation of new aggregates. Blue indicates L‐enantiomer and red indicates D‐enantiomer.
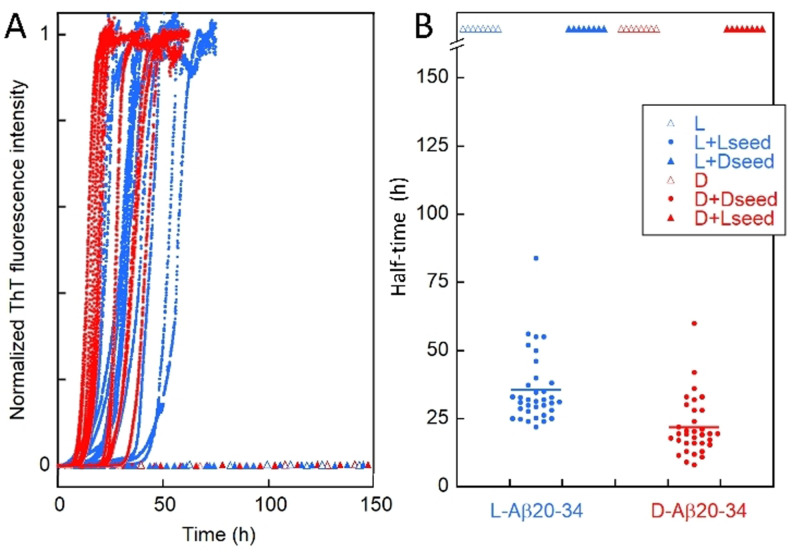
Figure 2.
Self‐ and cross‐seeded aggregation. Aggregation kinetics for 3 mM monomer with or without 10 μM seed of the same or opposite chirality. A) Normalized ThT fluorescence for reactions starting at time zero from L monomer (open blue triangles), L‐monomer plus L‐seed (blue dot), 3 mM L‐monomer plus D‐seed (filled blue triangles), D monomer (open red triangles), D‐monomer plus D‐seed (red dot), D‐monomer plus L‐seed (filled red triangles). Examples of curves are shown. B) Half‐time of aggregation, defined as the point in time where the normalized ThT intensity is 0.5, extracted from all repeats for the self‐seeded cases (L+L‐seed and D+D‐seed). For the non‐seeded (L and D) and cross‐seeded (L+D‐seed and D+L‐seed) cases, no aggregation was observed within one week as indicated by the up‐ward arrowhead symbols. Here, L refers to L‐Aβ20‐34 peptide and D refers to D‐Aβ20‐34 peptide. We note a difference in raw ThT intensity and average half‐time between the L and D peptides (see the Supporting Information).
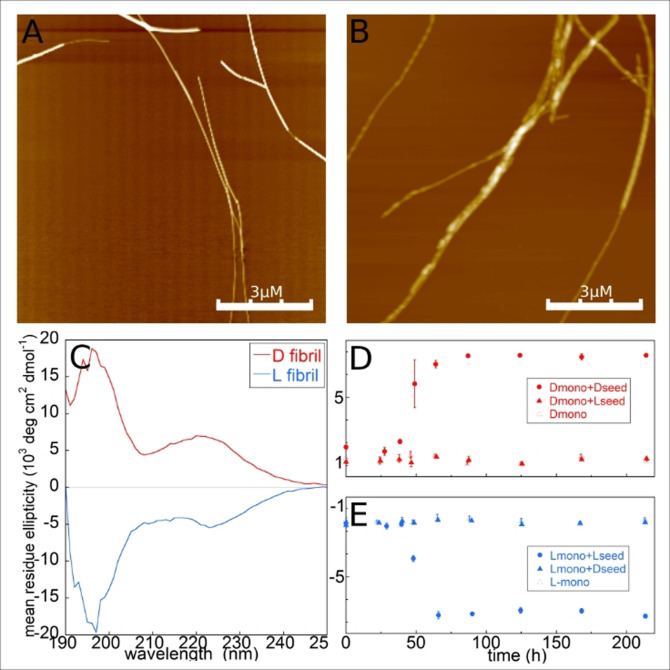
Figure 3.
Fibrillar structure. AFM topography images of fibrils made from A) the L‐peptide and B) the D‐peptide. C) CD spectra of fibril samples made from L‐peptide seeded with L‐seeds (blue trace) and D‐peptide (red trace) seeded with D‐seeds. The spectra reveal in each case the co‐existence of fibrils (β‐sheet signal around 220 nm) and monomers (random coil signal around 198 nm), implying a high solubility (see Supporting Information). D) Mean residue ellipticity at 222 nm recorded at different time points in a sample containing D‐monomer (open red triangles), D‐monomer plus D‐seed (red dot), D‐monomer plus L‐seed (filled red triangles). E) Mean residue ellipticity at 222 nm recorded at different time points in a sample containing L‐monomer (open blue triangles), L‐monomer plus L‐seed (blue dot), L‐monomer plus D‐seed (filled blue triangles).
The results show that the aggregation of L‐monomers is efficiently catalyzed by seed fibrils formed from L‐monomers, but not by D‐fibrils (Figure 1 A, 2). Orthogonal to this, the aggregation of D‐monomers is efficiently catalyzed by seed fibrils formed from D‐monomers, but not by L‐fibrils (Figure 1 B, 2). The cross‐ and self‐ seeding analysis was repeated using L‐ and D‐ versions a peptide comprising of residues 20–29 of human islet amyloid polypeptide (IAPP(20–29)), (SNNFGAILSS), for which secondary nucleation has been reported, [6] and the same lack of cross reactivity was observed (Figure S5). We thus find that surface‐catalyzed secondary nucleation is effective only if the monomer building blocks in the seed and in solution are of the same chirality. This is a strong evidence that in secondary nucleation of amyloid peptides, the structure of the parent seed fibril is replicated by the new fibrillar aggregates that are formed from monomers. The finding of chiral specificity of secondary nucleation underscores the homogeneous nature of this process. Primary nucleation can be either homogeneous, in solution, or heterogeneous, on a foreign surface or the surface of an aggregate of another substance. Surface‐catalyzed secondary nucleation, by definition, occurs on the surface of an aggregate of the same substance as the monomer. The newly formed aggregates will have lowest interfacial tension if they replicate the detailed structure and morphology of the parent aggregate. Detachment and growth by monomer addition serve to extend the new aggregates thereby amplifying the morphology of the original seed. We show here very clearly that the same substance refers not only to the same peptide sequence, but also to the same enantiomer: secondary nucleation is only possible if the nucleating monomers are of the same chirality as those in the parent aggregate.
Our results provide strong evidence for enantioselectivity of secondary nucleation, which implies that catalysis of nucleation on the fibril surface requires that the incoming monomers are able to adopt a structure that is a copy of the template fibril. This highlights a role of secondary nucleation in strain propagation.
Conflict of interest
S.L. is a founder and employee of Wren Therapeutics Ltd
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Supporting Information
Acknowledgements
This work was funded by the Swedish Research council (VR, grant number 2015‐00143).
M. Törnquist, S. Linse, Angew. Chem. Int. Ed. 2021, 60, 24008.
Contributor Information
Dr. Mattias Törnquist, Email: mattias.tornquist@biochemistry.lu.se.
Prof. Dr. Sara Linse, Email: sara.linse@biochemistry.lu.se.
References
- 1. Agrawal S. G., Paterson A. H. J., Chem. Eng. Commun. 2015, 202, 698–706. [Google Scholar]
- 2. Törnquist M., Michaels T. C. T., Sanagavarapu K., Yang X., Meisl G., Cohen S. I. A., Knowles T. P. J., Linse S., Chem. Commun. 2018, 54, 8667–8684. [DOI] [PubMed] [Google Scholar]
- 3. Botsaris G. D. in Industrial Crystallization (Ed.: Mullin J. W.), Springer US, Boston, 1976, pp. 3–22. [Google Scholar]
- 4. Tait S., White E. T., Litster J. D., Cryst. Growth Des. 2009, 9, 2198–2206. [Google Scholar]
- 5. Ferrone F. A., Hofrichter J., Eaton W. A., J. Mol. Biol. 1985, 183, 611–631. [DOI] [PubMed] [Google Scholar]
- 6. Ruschak A. M., Miranker A. D., Proc. Natl. Acad. Sci. USA 2007, 104, 12341–12346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cohen S. I. A., Linse S., Luheshi L. M., Hellstrand E., White D. A., Rajah L., Otzen D. E., Vendruscolo M., Dobson C. M., Knowles T. P. J., Proc. Natl. Acad. Sci. USA 2013, 110, 9758–9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gaspar R., Meisl G., Buell A. K., Young L., Kaminski C. F., Knowles T. P. J., Sparr E., Linse S., Q. Rev. Biophys. 2017, 50, E6. [DOI] [PubMed] [Google Scholar]
- 9. Cartwright J. H. E., García-Ruiz J. M., Piro O., Sainz-Díaz C. I., Tuval I., Phys. Rev. Lett. 2004, 93, 035502. [DOI] [PubMed] [Google Scholar]
- 10. Cohen S. I. A., Cukalevski R., Michaels T. C. T., Šarić A., Törnquist M., Vendruscolo M., Dobson C. M., Buell A. K., Knowles T. P. J., Linse S., Nat. Chem. 2018, 10, 523–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Colvin M. T., Silvers R., Ni Q. Z., Can T. V., Sergeyev I., Rosay M., Donovan K. J., Michael B., Wall J., Linse S., Griffin R. G., J. Am. Chem. Soc. 2016, 138, 9663–9674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kondepudi D. K., Kaufman R. J., Singh N., Science 1990, 250, 975–976. [DOI] [PubMed] [Google Scholar]
- 13. Kondepudi D. K., Asakura K., Acc. Chem. Res. 2001, 34, 946–954. [DOI] [PubMed] [Google Scholar]
- 14. Azeroual S., Surprenant J., Lazzara T. D., Kocun M., Tao Y., Cuccia L. A., Lehn J.-M., Chem. Commun. 2012, 48, 2292. [DOI] [PubMed] [Google Scholar]
- 15. Havinga E., Biochim. Biophys. Acta 1954, 13, 171–174. [DOI] [PubMed] [Google Scholar]
- 16. Calvin M., in Molecular Evolution towards the Origin of Living Systems on the Earth and Elsewhere, Oxford University Press, Oxford, 1969. [Google Scholar]
- 17. Green B. S., Lahav M., J. Mol. Evol. 1975, 6, 99–115. [DOI] [PubMed] [Google Scholar]
- 18. Asakura K., Soga T., Uchida T., Osanai S., Kondepudi D. K., Chirality 2002, 14, 85–89. [DOI] [PubMed] [Google Scholar]
- 19. Asakura K., Hayashi M., Osanai S., Chirality 2003, 15, 238–241. [DOI] [PubMed] [Google Scholar]
- 20. Lambert M. P., Barlow A. K., Chromy B. A., Edwards C., Freed R., Liosatos M., Morgan T. E., Rozovsky I., Trommer B., Viola K. L., Wals P., Zhang C., Finch C. E., Krafft G. A., Klein W. L., Proc. Natl. Acad. Sci. USA 1998, 95, 6448–6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chien P., Weissman J. S., DePace A. H., Annu. Rev. Biochem. 2004, 73, 617–656. [DOI] [PubMed] [Google Scholar]
- 22. Orgel L. E., Chem. Biol. 1996, 3, 413–414. [DOI] [PubMed] [Google Scholar]
- 23. Xu F., Khan I. J., McGuinness K., Parmar A. S., Silva T., Murthy N. S., Nanda V., J. Am. Chem. Soc. 2013, 135, 18762–18765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mortenson D. E., Steinkruger J. D., Kreitler D. F., Perroni D. V., Sorenson G. P., Huang L., Mittal R., Yun H. G., Travis B. R., Mahanthappa M. K., Forest K. T., Gellman S. H., Proc. Natl. Acad. Sci. USA 2015, 112, 13144–13149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kreitler D. F., Yao Z., Steinkruger J. D., Mortenson D. E., Huang L., Mittal R., Travis B. R., Forest K. T., Gellman S. H., J. Am. Chem. Soc. 2019, 141, 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Basak S., Singh I., Ferranco A., Syed J., Kraatz H.-B., Angew. Chem. Int. Ed. 2017, 56, 13288–13292; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2017, 129, 13473–13477. [Google Scholar]
- 27. Guichard G., Benkirane N., Zeder-Lutz G., van Regenmortel M. H., Briand J. P., Muller S., Proc. Natl. Acad. Sci. USA 1994, 91, 9765–9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Koga T., Matsuoka M., Higashi N., J. Am. Chem. Soc. 2005, 127, 17596–17597. [DOI] [PubMed] [Google Scholar]
- 29. Swanekamp R. J., DiMaio J. T. M., Bowerman C. J., Nilsson B. L., J. Am. Chem. Soc. 2012, 134, 5556–5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wadai H., Yamaguchi K., Takahashi S., Kanno T., Kawai T., Naiki H., Goto Y., Biochemistry 2005, 44, 157–164. [DOI] [PubMed] [Google Scholar]
- 31. Ciccotosto G. D., Tew D. J., Drew S. C., Smith D. G., Johanssen T., Lal V., Lau T.-L., Perez K., Curtain C. C., Wade J. D., Separovic F., Masters C. L., Smith J. P., Barnham K. J., Cappai R., Neurobiol. Aging 2011, 32, 235–248. [DOI] [PubMed] [Google Scholar]
- 32. Torbeev V., Grogg M., Ruiz J., Boehringer R., Schirer A., Hellwig P., Jeschke G., Hilvert D., J. Pept. Sci. 2016, 22, 290–304. [DOI] [PubMed] [Google Scholar]
- 33. Esler W. P., Stimson E. R., Fishman J. B., Ghilardi J. R., Vinters H. V., Mantyh P. W., Maggio J. E., Biopolymers 1999, 49, 505–514. [DOI] [PubMed] [Google Scholar]
- 34. Dutta S., Foley A. R., Warner C. J. A., Zhang X., Rolandi M., Abrams B., Raskatov J. A., Angew. Chem. Int. Ed. 2017, 56, 11506–11510; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2017, 129, 11664–11668. [Google Scholar]
- 35. Chung D. M., Nowick J. S., J. Am. Chem. Soc. 2004, 126, 3062–3063. [DOI] [PubMed] [Google Scholar]
- 36. Gupta D., Sasmal R., Singh A., Joseph J. P., Miglani C., Agasti S. S., Pal A., Nanoscale 2020, 12, 18692–18700. [DOI] [PubMed] [Google Scholar]
- 37. Kar K., Arduini I., Drombosky K. W., van der Wel P. C. A., Wetzel R., J. Mol. Biol. 2014, 426, 816–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Padrick S. B., Miranker A. D., Biochemistry 2002, 41, 4694–4703. [DOI] [PubMed] [Google Scholar]
- 39. Michaels T. C. T., Šarić A., Curk S., Bernfur K., Arosio P., Meisl G., Dear A. J., Cohen S. I. A., Dobson C. M., Vendruscolo M., Linse S., Knowles T. P. J., Nat. Chem. 2020, 12, 445—451. [DOI] [PubMed] [Google Scholar]
- 40. Cukalevski R., Yang X., Meisl G., Weininger U., Bernfur K., Frohm B., Knowles T. P. J., Linse S., Chem. Sci. 2015, 6, 4215–4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thacker D., Sanagavarapu K., Frohm B., Meisl G., Knowles T. P. J., Linse S., Proc. Natl. Acad. Sci. USA 2020, 117, 25272–25283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cohen S. I. A., Vendruscolo M., Dobson C. M., Knowles T. P. J., J. Mol. Biol. 2012, 421, 160–171. [DOI] [PubMed] [Google Scholar]
- 43. Warmack R. A., Boyer D. R., Zee C.-T., Richards L. S., Sawaya M. R., Cascio D., Gonen T., Eisenberg D. S., Clarke S. G., Nat. Commun. 2019, 10, 3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Supporting Information