Abstract
Opsin‐based transmembrane voltage sensors (OTVSs) are membrane proteins increasingly used in optogenetic applications to measure voltage changes across cellular membranes. In order to better understand the photophysical properties of OTVSs, we used a combination of UV‐Vis absorption, fluorescence and FT‐Raman spectroscopy to characterize QuasAr2 and NovArch, two closely related mutants derived from the proton pump archaerhodopsin‐3 (AR3). We find both QuasAr2 and NovArch can be optically cycled repeatedly between O‐like and M‐like states using 5‐min exposure to red (660 nm) and near‐UV (405 nm) light. Longer red‐light exposure resulted in the formation of a long‐lived photoproduct similar to pink membrane, previously found to be a photoproduct of the BR O intermediate with a 9‐cis retinylidene chromophore configuration. However, unlike QuasAr2 whose O‐like state is stable in the dark, NovArch exhibits an O‐like state which slowly partially decays in the dark to a stable M‐like form with a deprotonated Schiff base and a 13‐cis,15‐anti retinylidene chromophore configuration. These results reveal a previously unknown complexity in the photochemistry of OTVSs including the ability to optically switch between different long‐lived states. The possible molecular basis of these newly discovered properties along with potential optogenetic and biotechnological applications are discussed.
Opsin‐based transmembrane voltage sensors (OTVSs) are NIR fluorescence emitting membrane proteins increasingly used in optogenetic applications to measure voltage changes across cellular membranes for instance in brain imaging studies. A combination of UV‐Vis absorption, fluorescence and FT‐Raman spectroscopy was used to characterize QuasAr2 and NovArch, two closely related OTVSs derived from the proton pump archaerhodopsin‐3. Both QuasAr2 and NovArch can be optically switched repeatedly between long‐lived O‐like and M‐like states revealing a previously unknown complexity in the photochemistry of these OTVSs.
INTRODUCTION
Microbial rhodopsins (type I opsins) are a diverse class of photosensitive 7‐helix transmembrane proteins which have been found in all major domains of life (1). This class of proteins all contain a retinal chromophore covalently linked through a protonated Schiff base (SB) to a highly conserved lysine residue on the G‐helix (Lys‐216 in the case of bacteriorhodopsin (BR)) (2, 3, 4, 5, 6). Besides serving as important models for understanding membrane protein structure and function, type I opsins and variants have been used increasingly in the new field of optogenetics (7, 8).
Optogenetics enables neuroscientists to noninvasively control and monitor neuronal activity using light (8). This is accomplished by transfecting targeted neurons with the genes for photonic proteins which can either modulate neural activity by activating or silencing action potentials or sense key properties such as transmembrane voltage and intracellular Ca++ or c‐AMP levels. In the case of type I opsins, Boyden and Deisseroth (7, 8, 9, 10) first established that channelrhodopsins, a diverse class of type I opsins which function in algae as light‐gated ion channels (11, 12, 13, 14, 15), can be used to photo‐modulate electrical activity in neurons and other cells even in physiologically functional organisms (1, 16).
More recently, it has been found that some type I opsin variants exhibit fluorescence which is sensitive to the transmembrane potential (1, 10). Such fluorescent opsin‐based transmembrane voltage sensors (OTVSs) typically are based on bioengineered mutants of green absorbing proteorhodopsin (GPR) or archaerhodopsin‐3 (AR3) (17, 18), both functioning in their native form as light‐driven proton pumps. Engineering of these proteins using various types of directed evolution methods has resulted in improvement of key properties such as excitation wavelength, voltage sensitivity, brightness and temporal resolution. Examples include QuasAr1 and 2 (19), QuasAr3 and pa‐QuasAr3 (20), NovArch (21) and Archon1 (22). QuasAr2 differs from the native AR3 at 5 positions in the AR3 primary amino acid sequence (see Fig. 1) including substitution of a neutral for a negative residue (Asp to Gln) at position 95, which normally functions as a counterion and proton acceptor in the AR3 proton pump mechanism (as does the homologous residue Asp85 in BR). A similar neutralizing mutation in BR (D85N) and AR3 (D95N) inhibits proton transport and shifts the λ max from 570 to near 600 nm (23, 24). NovArch has an additional 4 mutations compared to QuasAr2 including V59A and I213T in the transmembrane B and G helices, respectively (Fig. 1).
Figure 1.

Predicted 2D folding pattern of native archaerhodopsin‐3 (AR3) along with mutations in QuasAr2 (Q2) and NovArch (NA). Sequence numbering is based on AR3 sequence. The Schiff base counterion at position 95 has the substitution D95Q for both Q2 and NA. Orange circles show AR3 mutations P60S, T80S, D106H and F161V common to both Q2 and NA. Green circles show the additional mutations V59A, K171R, V209I and I213T which transform Q2 into NA (Figure adapted from fig. 1 of (24)).
Despite the great promise of OTVSs in a variety of applications, including understanding of the functioning of intact brain circuitry and the basis for neurodegenerative diseases, there are key limitations which researchers have striven to overcome. These include the following: (I) The relatively weak fluorescence emission of current OTVSs, which requires high excitation intensity compared to other optogenetic sensors such as the optogenetic GCaMP Ca++ sensors (25); (2) desensitization of OTVS after a few seconds, thus limiting their sensitivity (7, 22, 26, 27, 28); (3) scattering and absorption of visible wavelengths of light by brain tissue, thus limiting the utility of OTVSs for deep brain imaging (29) and (4) complex photochemical behavior of OTVSs, in particular at higher excitation levels, which impairs a linear correlation between event and signal intensity.
In order to better understand the molecular basis for some of these limitations and in particular to address limitations 2 and 4 listed above, we used a combination of UV‐Vis absorption, FT‐Raman and fluorescence spectroscopy to characterize the two closely related OTVSs QuasAr2 and NovArch. We already addressed limitations 1 and 3 in a previous study (24). QuasAr2 has already been used as an OTVS to perform all‐optical electrophysiology in mammalian neurons (19). It exhibits a fast time response to voltage changes (~50 µs), although it is less sensitive to voltage changes compared to the slower responding QuasAr1 (19). The more recently reported NovArch and related pa‐QuasAr3 exhibit several interesting properties including enhanced near‐infrared (NIR) fluorescence by irradiation with 1‐photon visible excitation at 488 nm or 2‐photon NIR excitation at 900 nm. These properties enabled optical mapping of action potentials in dendrites in acute mouse brain slices (20, 21).
A key finding of our current study is that both QuasAr2 and NovArch exhibit optical switching between O‐like and M‐like states using relatively low intensity (5 mW cm−2), short duration (5 min) 660 and 405 nm light, similar to optical switching exhibited by some BR mutants (30). Longer red‐light (660 nm) exposure resulted in the formation of long‐lived photoproducts similar to pink membrane (so‐called P490 state) which have been previously observed for acid blue BR and the O intermediate of the BR photocycle and shown to contain 9‐cis retinal chromophores. These properties may explain partially the desensitization of OTVSs in neurons and suggest optical methods for reducing desensitization and increasing sensitivity along with possible new application of opsins such as for transient optoelectronic memories.
MATERIALS AND METHODS
Expression, purification and reconstitution of QuasAr2 and NovArch
Methods for the expression of QuasAr2 and NovArch in E. coli cell cultures, purification and reconstitution using His‐tag Nickel‐chelated nitrilotriacetic acid (Ni‐NTA) agarose beads affinity chromatography and reconstitution into in E. coli polar lipid membrane vesicles or detergent n‐dodecyl‐β‐D‐maltoside (DDM) micelles were previously reported (24). Briefly, pET 28b(+) vectors encoding QuasAr2 and NovArch genes with a C‐terminal His‐tag were synthesized by GenScript (Piscataway, NJ). All‐trans retinal (Millipore‐Sigma) was stored at −80°C in an ethanol stock solution. Octylglucoside (OG) and n‐dodecyl‐β‐D‐maltoside were purchased from Anatrace Products, OH. E. coli (strain BL21 (DH3)), transformed with the pet28b(+) plasmid with the QuasAr2 and NovArch genes were grown in 0.5 L of LB medium with 50 mg L−1 kanamycin, to an O.D. of 0.4 at 600 nm at 35°C. All‐trans retinal (2 µm) and inducer (IPTG, 1 mm) were added, and cells were grown for an additional 4 h in the dark at 35°C. Isolation of plasma membrane and solubilization in 2% OG or 2% DDM was as previously reported (24). Nickel‐chelated nitrilotriacetic acid (Ni‐NTA; Thermo Fisher Scientific) affinity chromatography was used to purify His‐tagged QuasAr or NovArch protein, which was eventually eluted in buffer (50 mm HEPES, 100 mm NaCl, 1% OG, 400 mm imidazole; pH 7.0 at room temperature). The proteins were then reconstituted in E. coli polar lipids (ECPL) (Avanti, Alabaster AL) at 1:10 protein‐to‐lipid (w/w) ratio. The reconstituted proteoliposomes were centrifuged for 3 min at 15 000 rpm (21 000 g) and resuspended in 5 mm K2HPO4, 100 mm NaCl, pH 7.0 buffer, three times consecutively. The resulting QuasAr2 and NovArch membrane samples were stored at 4°C. For more details, see (24).
UV‐VIS spectroscopy
Absorption measurements were made on QuasAr2, NovArch and AR3‐D95Q reconstituted membranes (see above) dissolved in DDM micellar solutions. The samples were prepared in 50 mm bis‐tris‐propane, 150 mm NaCl, 1mM DTT and 2% DDM (w/v), pH 7 (see (24) for details of preparation). The samples were measured at RT in a quartz cuvette (CV10Q700FS, Thorlabs Inc., Newton, NJ) with 2 mm pathlength using a Cary 50 instrument (Agilent Technologies) on typically 200 µL volume. This allowed absorption spectra to be measured both in darkness and while being irradiated. Typically, samples were measured over the range 200–1100 nm at a scanning rate of 600 nm min−1. For illumination of the sample, a fiber optic illuminator was used (either M405FP1 or M660FP1, Thorlabs Inc.) and a DC4104 Four‐Channel LED driver. The tip of the fiber optic was positioned at less than 1 mm from the top of the quartz cuvette that contained the sample. The power density (flux) of illumination was measured using a power meter (Model PM400, Thorlabs Inc.) and was 2.95 mW cm−2 for 660 nm and 3.76 mW cm−2 for 405 nm illumination (100% power density). Sample concentration of the different OTVSs studied was determined from the visible absorption of a sample after 405 nm illumination during the second illumination cycle (see Results below) in order to maximally regenerate the O‐like species and was estimated using the molar extinction coefficient of bacteriorhodopsin at 20.2, 2.8 and 8.3 µm for QuasAr2, NovArch and AR3‐D95Q, respectively.
Fluorescence spectroscopy
Fluorescence measurements were performed on NovArch in DDM micelles prepared as described above. Measurements were made using a Horiba NanoLog 3‐22‐TRIAX spectrofluorometer (HORIBA Jobin Yvon Inc., Edison, NJ) with a 450 W Xenon arc lamp excitation source, CCD detector and double monochromator on both the excitation and emission sides. The 300 μL samples were placed in the quartz cuvette and data collected with excitation wavelength from 500 to 700 nm, where the wavelength increment is 2 nm and integration time is 2 s. The collected data were corrected for lamp power and detector sensitivity. The contour maps emission wavelength is recorded from 588 to 854 nm. Relative brightness of the samples was corrected for absorption of the identical samples measured using a Cary6000i absorption spectrometer equipped with an external diffuse reflectance (DRA) accessory. Illumination of the samples prior to the fluorescence measurement was performed using a 405 nm fiber‐coupled LED (M405FP11, Thorlabs Inc.) with a compact T‐cube driver (LEDD1B, Thorlabs Inc.) and an optical fiber (M35L01, Thorlabs Inc.) with 10.2 mW for 5‐min.
Fourier transform Raman spectroscopy
FT‐Raman measurements of reconstituted QuasAr2 and NovArch in ECPL (proteolipid membrane vesicles) were measured in aqueous buffer at pH 7.3 in glass capillaries as described previously (24). FT‐Raman measurements were made using 1064‐nm laser excitation on a Bruker MultiRam FT‐Raman spectrometer operating at 4 cm−1 resolution and power ranging from 30 to 300 mW. In addition to the 1064‐nm laser, diffuse scattered light from a low‐power (~1 mW) HeNe laser used for calibration of the FT‐interferometer mirror movement also irradiated the sample.
Spectral analysis and structural modeling
Spectral subtractions, baseline corrections, normalization and extraction of peak values were all performed using MATLAB, except where noted.
Baseline correction
Baseline artifacts in the measured absorption and absorption difference spectra arise mainly from Rayleigh scattering and baseline drift due to settling of micellar suspensions over extended periods. In order to correct for this, a multipoint baseline was generated based on a Hermite polynomial function using MathWorks and subtract from the original data. The determination of the optimal set of points used to generate this baseline was made through a manual iterative procedure consisting of inputting points followed by visual inspection of the resulting baseline. After each iteration of a set of user‐input points (typically around 10 points), they are adjusted until the baselines produced display none of peaks visually present in uncorrected spectra.
Exponential time dependence plots
All time‐dependent absorption peak intensities were curve fit using MatLab using a single‐exponential function. Results presented here all had adjusted R 2 > 95% and similarly high values of Pearson’s R coefficient.
Structural modeling
A model for NovArch was constructed using the PyMol 1.3 Molecular Graphics Program from the PDB coordinates of the 1.9 Å crystallographic structure recently reported for the ground state structure of AR3 obtained from LCP crystals using a thin‐film sandwich at room temperature (PDB: 6GUZ) (31). The 9 substitutions in the AR3 sequence present in NovArch are included in this structural model using the Mutagenesis Wizard in PyMol but not energy minimized. The rotamer status for each substitution was chosen as the most common one based on the frequencies of occurrence in proteins.
RESULTS
Optical switching of QuasAr2 between O‐like and M‐like states and formation of a stable O‐like dark state
Absorbance changes of QuasAr2 produced by illumination with two different wavelengths of light (405 and 660 nm) were investigated by measuring the absorption spectrum over the range 250–850 nm. Similar measurements were also made for NovArch and the mutant AR3‐D95Q (see below). These measurements were performed using aqueous solutions of purified proteins incorporated into mixed detergent (DDM)‐lipid micelles to reduce light scattering effects observed using proteoliposome suspensions. Since the Cary 50 instrument can record spectra while the sample is being illuminated, absorption changes could be measured before, during and after illumination.
Both the baseline‐corrected visible absorption spectrum of QuasAr2 (Fig. 2A) and uncorrected baseline spectrum (Figure S1) exhibit a major band near 595 nm and a second smaller band near 409 nm in agreement with previous reports (19, 24). The ~30‐nm red‐shift compared to WT‐AR3 is largely due to substitution of a neutral Gln95 for the negatively charged Asp95 at the Schiff base counterion position in the C helix (Fig. 1) (24, 32). The band in the UV region near 282 nm is mainly due to absorbance of the aromatic tyrosine, tryptophan and phenylalanine residues.
Figure 2.
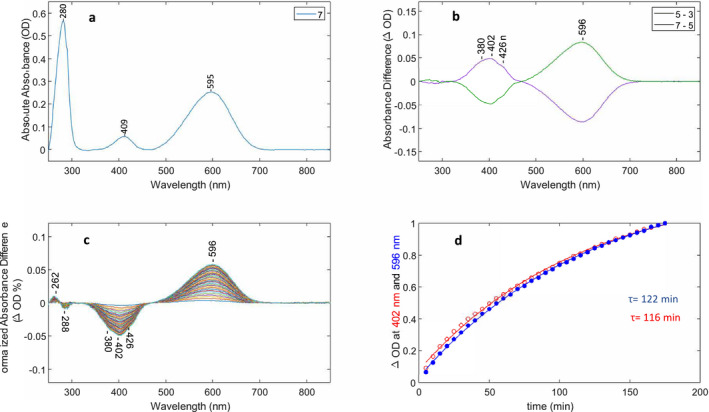
Optical switching of QuasAr2 using 660 and 405 nm light and M decay kinetics. (A) Baseline‐corrected absorption of QuasAr2 from spectrum #7 in cycling sequence (see also Figures S1 and S2). (B) Two baseline‐corrected difference spectra (#5–#3, #7–#5) from cycling sequence. The intensity of the 660 and 405 nm LED illumination was set at 100% (see Materials and Methods). (C) Baseline‐corrected difference spectra recorded after last red‐light illumination in sequence. Each difference spectrum consists of subtraction of spectrum #13 recorded in the dark after the red‐light is turned off from subsequent spectra (e.g. #13……#48). Differences were also normalized to the 280 nm absorption band for each spectrum recorded in dark to correct for sample settling in cuvette. (D) Time course of the difference peak amplitudes in C at 596 and 402 nm reflecting decay and formation of the M‐like and O‐like states, respectively, as a function of time. Changes in the absolute absorbance differences were normalized between 0 and 1 and the decay at 402 nm (red trace) inverted. Time constants were calculated using a single‐exponential function (see Materials and Methods).
In order to test the samples response to different wavelengths of light, the sample is first subjected to three or more cycles of dark–light illumination, with each cycle consisting of 5‐min periods of dark, 405‐nm illumination, dark and 660‐nm illumination (Figure S2). An absorption spectrum is measured at the beginning of each step in the cycle (measurement time is approximately 1.5 min). Following the last cycle, the sample is left in the dark for 3 h and absorption spectra measured every 5 min.
A typical cycle of absorption difference spectra for QuasAr2:A1 is shown in Fig. 2B for the “push” difference (e.g. difference between spectra recorded before and after 660 nm illumination, purple curve) and “reversal” difference (e.g. difference between spectra recorded before and after 405 nm illumination, green curve). These push and reversal differences reflect the transitions between O‐like and M‐like states of QuasAr2 with absorbance maxima near 596 and 402 nm, respectively, in analogy to the BR photocycle O and M intermediates, which absorb maximally at 640 (O640) and 412 nm (M412). As discussed later, these O‐like and M‐like states have a protonated and deprotonated SB, respectively, and a neutral SB counterion analogous to the O640 and M412 states in the BR photocycle (3, 33).
While the O‐like difference band is fairly symmetric and appears to reflect a single species, the M‐like difference band exhibits fine structure with the main peak absorbing near 402 nm and two shoulders appearing near 380 and 426 nm. As discussed previously for the case of GtACR1 and CaChR1,(34, 35), the fine structure could arise from vibronic coupling as commonly seen for other microbial rhodopsins such as NpSRII (36, 37). Interestingly, fine structure bands also appear in the fast and slow decay components of the M intermediate in the BR photocycle at similar wavelengths (380 and 435 nm), and thus, the shoulders may also arise from different M‐like species (38). Note also the main peak in the absorption difference spectrum is downshifted ~7 nm from the M‐like peak appearing at 409 nm in the absolute absorption spectrum which may partially be due to the presence of E. coli cytochromes (414 nm) that tend to be co‐purified with AR3 using Ni‐NTA agarose His‐tag affinity chromatography (39, 40). Importantly, the push and reversal steps in each cycle are almost fully reversible (Fig. 2B).
While only 3 cycles were repeated for the experiment shown in Fig. 2, it was possible to repeat the push‐reversal cycles for at least 10 cycles without significant decay of the cycle difference amplitudes once sample settling in cuvette and possible aggregation was corrected for using the 282 nm band for normalization (Figure S3A and B). Note the initial reversal with 405 nm light (3‐1) (Figure S3A, blue trace) produces much smaller amplitude difference bands most likely because the sample has much less M due to its exposure to room lights prior to insertion in spectrometer compared to subsequent cycles where the sample was exposed first to 660 nm light. The baseline‐corrected absolute absorption also shows the effects of optical switching between the M and O‐like states as reflected by alternating changes in the amplitude of the peaks near 407 and 598 nm (Figure S4A). The downshift of the M band from 412 to 407 nm most likely reflects the presence of a second species absorbing closer to 412 nm (possibly cytochrome) which is not cycling. The cycling fraction of QuasAr2 (CF) defined as the ratio of absorbance of the band at 404 nm or 598 nm in difference spectra (Fig. 3A) to the corresponding band in the absolute absorption spectrum (Figure S4A) prior to illumination was close to 50% for these illumination conditions and did not appreciably change during the measurement of the 10 cycles (Figure S4B). Difference spectra between the sample measured in the dark and the steady‐state mixture formed during illumination with both 405 and 660 nm light are shown in Figure S5A and B. They are similar to the dark–dark difference spectra (e.g. #7‐#5 and #9‐#7; Fig. 2B). This demonstrates that during these short periods of illumination and darkness no formation or decay of transient photoproducts are obvious, in contrast to the longer illumination periods described below.
Figure 3.
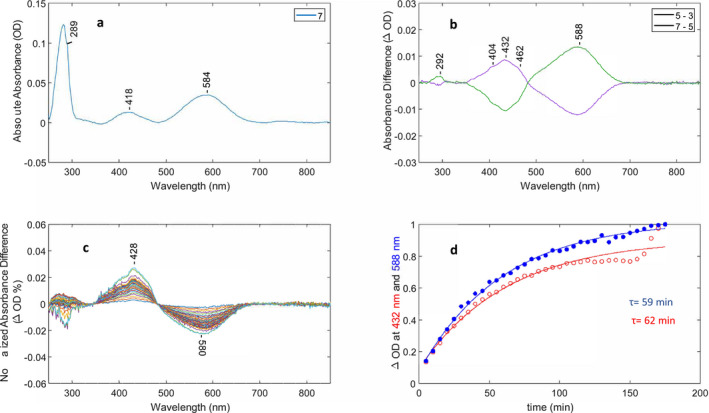
Optical switching of NovArch using 660 and 405 nm light and O decay kinetics. (A) Baseline‐corrected absorption of NovArch from spectrum #7 in cycling sequence (see Figure S2). (B) Two baseline‐corrected difference spectra (#5–#3, #7–#5) from cycling sequence. (C) Baseline‐corrected difference spectra recorded after last red‐light illumination in sequence. Each difference spectrum consists of dark spectrum (N = #13–36) minus spectrum #12. Differences were also normalized to the 280 nm absorption band for each dark spectrum to correct for sample settling in cuvette. (D) Time course of the difference peak amplitudes in C at 588 and 432 nm reflecting decay and formation of the O‐like and M‐like states, respectively, as function of time. Changes in the absolute absorbance differences were normalized between 0 and 1 and the decay at 588 nm (blue trace) inverted. Time constants were calculated using a single‐exponential function (see Materials and Methods).
To determine the lifetime of the M‐like state formed after 660 nm illumination (last illumination after cycling several times), spectra were recorded every 5 min over a 3 h dark period. As shown in Fig. 2C,D, the M‐like 402‐nm state produced by 660 nm illumination slowly decays back to the O‐like state with a single‐exponential decay constant of approximately 120 min (122 and 116 min for the M decay and O formation, respectively). In a proteoliposome environment, the M‐like intermediate of QuasAr2 generated by 660 nm illumination also decayed slowly, but with a slightly faster decay constant (85 min, data not shown). In contrast, as previously reported, dark adaptation of QuasAr2 over a 12‐hour period did not result in any appreciable decay of the O‐like absorbing species (24).
Formation of pink membrane photoproduct
As described above periodic cycling between the O‐ and M‐like states of QuasAr2 using alternating 5‐minute intervals of 405 and 660 nm light (Figure S2) did not lead to any long‐term accumulation of other spectrally distinct states. In contrast, prolonged illumination with red‐light (660 nm) over a 3‐hour period does result in formation of additional photoproducts. For these experiments instead of measuring the sample in dark for 3 h after cycling steps are completed, the 660 nm light was kept on for 3 h (Figure S6A). This resulted in a slow conversion (τ = 27 min) of the O‐like state to a photoproduct with an absorption near 460 nm as indicated by the negative/positive peaks at 600 and 460 nm in the difference spectra recorded after the 660 nm light is turned on (Figure S6B,C). Note differences shown are between the last dark spectrum recorded in the cycle sequence (#39) and successive spectra recorded with red‐light on (#41 to #76). An additional positive band appears initially in these difference spectra at 405 nm, the peak wavelength of the M‐like state of QuasAr2 formed by red‐light in the cycling steps similar to differences shown in Figure S5B. This band gradually decays with a time constant (τ = 36 min) close to the decay constant of the depletion of the O‐species and accumulation of the 460 nm absorbing species (τ = 27 min; Figure S6C). In a similar but extended experiment, the sample was kept and monitored in the dark for 2 h at room temperature after red‐light illumination was turned off. This revealed only minimal decay back to the O‐like state or to a 380 nm decay product of pink membrane (data not shown). Subsequently, the sample was exposed to one hour of 405 nm radiation which led to disappearance of the 460 nm absorbing species and restored the O‐like intermediate to approximately the same level measured at beginning of experiment (data not shown). As discussed later, these results are consistent with formation of a pink membrane photoproduct from the O‐like state similar to previously observed formation of pink membrane from the O intermediate in the BR photocycle (also called P490 and possessing a 9‐cis retinylidene configuration, see discussion) and from acid blue membrane, both having a neutral Schiff base counterion (41, 42, 43).
Optical switching of NovArch and existence of a stable M‐like dark state
As seen in Fig. 3A, the absorption spectrum of NovArch is similar to QuasAr2 with a slightly blue‐shifted λ max of the major band at 584 nm and a red‐shifted λ max of the M‐like band to 418 nm. Cycle difference spectra obtained using the same protocol as for QuasAr2 (Figure S2) reveal reversible optical switching using 405 and 660 nm illumination similar to QuasAr2 (Fig. 3B). The maximum wavelength of the O‐like difference band (588 nm) is similar to the absolute absorption spectrum (584 nm), but the peak of the M‐like difference band appears at a higher wavelength than in the absolute absorption spectrum (432 nm vs 418 nm, respectively) possibly due to cytochrome contributions to the absolute absorption spectrum. As in the case of QuasAr2, shoulders appear on the M difference band at 404 and 462 nm which may arise from additional species that are photoreversible during optical switching. The CF measured for the optical switching was approximately 36% at 588 nm under the illumination conditions of this experiment.
Surprisingly, in contrast to QuasAr2, where dark adaptation of the O‐like intermediate produces little change, prolonged dark adaptation of the NovArch O‐like intermediate results in a partial accumulation of the M‐like species. This was observed after illuminating the sample with 405 nm light which populates the O‐like species and then recording subsequent spectra in dark for 3 h. A slow decay (τ = ~60 min) of over 50% of the O‐like species back to the M‐like species is observed indicating that a significant part of the M‐like species of NovArch is stable in the dark (Fig. 3C,D).
Similar to QuasAr2, prolonged red‐light illumination resulted in the formation of several photoproducts that are likely to be related to pink membrane formation observed for QuasAr2. Figure S7 shows successive difference spectra during 3 h of red‐light illumination using the protocol identical for QuasAr2 (Figure S6A). The O‐like species is initially converted predominantly to the M‐like species absorbing near 435 nm along with shoulders appearing at 460 nm (see green trace in Figure S7). This is similar to the cycling difference produced using only 5 min of red‐light illumination (Fig. 3B). However, later during the 3‐h red‐light illumination both bands decay, while a much weaker band arises near 375 nm which may correspond to the 380 nm absorbing Q380 decay product of P490, which still retains a 9‐cis chromophore configuration but the SB linkage is hydrolyzed (41, 42, 43) (see discussion below “Optical Bistability of Opsins”). Leaving the sample in the dark for an additional 2 h after the extended period of red‐light illumination led to almost no further changes in the NovArch photoproduct population, similar to QuasAr2. Furthermore, illumination for one hour with 405 nm light results in restoration of the O‐like species from the red‐light photoproducts produced over the 3‐h extended illumination.
Optical switching of AR3‐D95Q
Wild‐type AR3 did not exhibit any appreciable optical switching (data not shown) in contrast to QuasAr2 and NovArch. Hence, we investigated the mutant AR3‐D95Q that has a single substitution of a neutral Gln for the native negatively charged Asp at the SB proton counterion position 95, common also to QuasAr2 and NovArch (Fig. 1). This mutation along with substitution of Glu for Asp at the Schiff base donor residue 106 (D106E/D95Q) has previously been shown to have a faster fluorescence response and increased dynamic range compared to AR3‐D95N (44). As shown in Fig. 4A, the native AR3 absorption maximum (~560 nm) shifts to 606 nm, somewhat higher that the 595 nm absorption max of QuasAr2 (Fig. 2A) and significantly higher than NovArch at 584 nm (Fig. 3A). Figure 4B reveals a pattern of reversible cycling between O‐like and M‐like states induced by red (660 nm) and near UV (405 nm) light similar to QuasAr2 and NovArch. The cycling fraction (CF) was found to vary between 27 and 51% for different experiments. In addition, similar to QuasAr2, AR3‐D95Q undergoes a slow decay back to the O‐like state in the dark, with a time constant of 224 min based on the decay of the 404 nm difference band (Fig. 4C,D).
Figure 4.
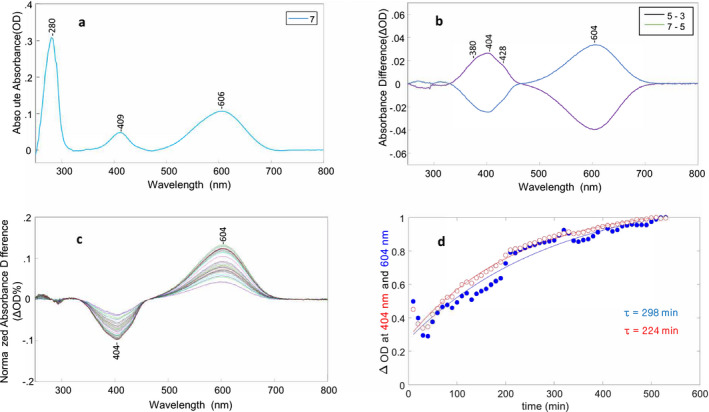
Optical switching of AR3‐D95Q using 660 and 405 nm light and M decay kinetics. (A) Baseline‐corrected absorption of AR3‐D95Q from spectrum #7 in cycling sequence. (B) Two baseline‐corrected difference spectra (#5–#3, #7–#5) from cycling sequence. The intensity of the 660 and 405 nm LED illumination was set as 100% (see Materials and Methods). (C) Baseline‐corrected difference spectra recorded after last red‐light illumination in sequence. Each difference spectrum consists of dark spectrum (N = #22………56) minus first dark spectrum #21. Differences were also normalized to the 280 nm absorption band for each dark spectrum to correct for sample settling in cuvette. (D) Time course of the difference peak amplitudes in C at 604 and 404 nm reflecting decay and formation of the M‐like and O‐like states, respectively, as a function of time. Changes in the absolute absorbance differences were normalized between 0 and 1 and the decay at 404 nm (red trace) inverted. Time constants were calculated using a single‐exponential function (see Materials and Methods).
QuarAr2, NovArch and AR3‐D95Q NIR fluorescence
As previously reported, QuasAr2 exhibits broad excitation/emission peaks with wavelength maxima near 600 and 725 nm, respectively (see figure S3 of (24)). An emission/excitation contour plot of NovArch recorded immediately after 5 min of 405 nm LED illumination to increase the fraction of the O‐like species in the sample reveals similar characteristics, with a broad excitation band near 590 nm and red‐shifted emission maximum near 725 nm (Fig. 5A). Figure 5B shows the fluorescence spectra of QuasAr2, NovArch and AR3‐D95Q using 600 nm excitation. These spectra further confirm that the O‐like species of both QuasAr2 and NovArch are very similar with a broad fluorescence band which may consist of two separate components located near 722 and 759 nm potentially reflecting two different O‐like species. Interestingly, the fluorescence band for AR3‐D95Q is somewhat red‐shifted with a broad maximum extending to 769 nm.
Figure 5.
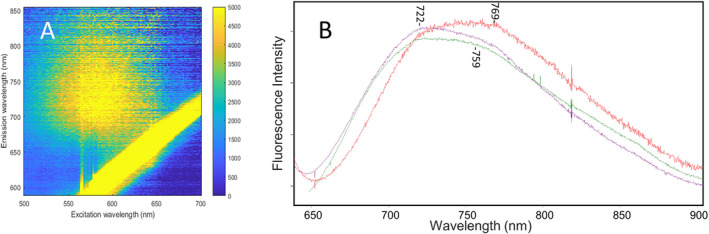
Fluorescence spectra of NovArch, QuasAr2 and AR3‐D95Q: (A) NovArch excitation/fluorescence contour map after 5 min of 405‐nm LED preirradiation. Yellow stripe at lower right quadrant is an artifact due to scattered excitation light. (B) Fluorescence spectra of NovArch (purple), QuasAr2 (green) and AR3‐D95Q (red) recorded using 600 nm excitation. AR3‐D95Q and NovArch samples are measured with 2 s integration time, while the QuasAr2 sample was measured with 1s integration time. NovArch and QuasAr2 spectra are the average of 10 spectra and the AR3‐D95Q spectrum is a single spectrum (not averaged). The Y‐axis tick marks spacing correspond to 5000 fluorescence intensity units measured for NovArch with the two other spectra interactively scaled.
The stable M‐like state of NovArch has a 13‐cis retinylidene chromophore
Preresonance FT‐Raman spectroscopy using 1064‐nm excitation minimizes possible photoreactions of microbial rhodopsins that occur with visible excitation, while still enhancing vibrations of the retinylidene chromophore (45, 46, 47). Furthermore, this approach avoids the production of NIR fluorescence excited by shorter wavelength light which can mask much weaker resonance Raman bands, especially for NIR emitting OTVSs (24).
Similar to earlier FT‐Raman measurements of QuasAr1 and QuasAr2 (24), measurements were performed on NovArch reconstituted into ECPL membrane vesicles (see Materials and Methods). An unusual feature of the NovArch FT‐Raman spectrum compared to QuasAr2 (Fig. 6) and QuasAr1 (24) is the intense 1559 cm−1 band appearing in the 1500–1600 cm−1 C = C stretch ethylenic region. This band is attributed to the ethylenic stretch mode of the M‐like species which is stable in the dark as discussed above. Since the NovArch FT‐Raman spectrum is acquired in the dark over a long period (5.6 h), the O‐like state present originally should have sufficient time to decay partially into the 435 nm M‐like species observed during dark adaptation. In support of this band assignment, a strong 1567 cm−1 band appears in the resonance Raman spectrum of the BR M intermediate (48, 49). The lower frequency of the NovArch M‐like state compared to BR‐M (1559 vs 1567 cm−1) is consistent with the empirical linear correlation between wavelength of absorption (λ max) and ethylenic frequency (νC=C) as described previously (34, 50, 51, 52, 53, 54) (Figure S8). Notably, a band at 1574 cm−1 assigned to the C = C stretching frequency of the UV absorbing state (380 nm) of HKR1 also fits this linear correlation (55). A second weaker band also appears in the ethylenic stretch region of NovArch at 1521 cm−1 (Fig. 6). This band is assigned based on the same linear empirical correlation and similarity to the major ethylenic band in QuasAr2 to the undecayed O‐like species still present in the sample (Figure S8). The appearance of both bands in the Raman spectrum suggests that in the dark the M and O‐like species co‐exist in equilibrium.
Figure 6.
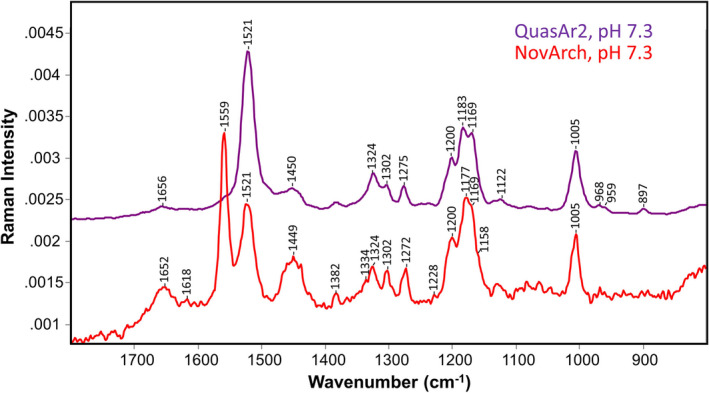
FT‐Raman spectra of QuasAr2 (purple) and NovArch (red) reconstituted in membrane vesicles and recorded at pH 7.3. Spectra were scaled using the band near 1005 cm−1. Y‐axis scale shown is for NovArch. Laser power was 300 mW for both samples, and data acquisition times were 3.5 h for QuasAr2 and 5.6 h for NovArch (see Materials and Methods for additional details). Data for QuasAr2 are reproduced from ref (24).
In order to determine the chromophore configuration of the NovArch dark‐adapted M‐like species, the C‐C stretch fingerprint region (1150–1250 cm−1) which is highly sensitive to the retinylidene configuration was examined (56). The most intense band in this region occurs at 1177 cm−1 with shoulders at 1169, 1158 cm−1 and a second band at 1200 cm−1 (Fig. 6). Significantly, the presence of the 1177 cm−1 band and the overall band structure in this region closely resembles the resonance Raman spectrum of the M intermediate of BR obtained using 413‐nm excitation and dual‐beam flow or spinning cell methods (48, 49). These studies concluded based on the use of BR regenerated with retinal containing stable isotopes at various positions combined with normal mode calculations that the BR M intermediate possesses a predominantly 13‐cis,15‐anti configuration with a deprotonated SB. This conclusion is supported by solid‐state NMR analysis of the M‐state of several microbial rhodopsins (57). The resonance Raman light‐dark difference spectrum using 785‐nm excitation of the BR D96N mutant which displays a slow M decay and the FT‐Raman spectrum of NovArch are also very similar (Figure S9). We thus conclude on the basis of the close similarity of the NovArch FT‐Raman spectrum in the C‐C stretch region that 13‐cis,15‐anti also is the predominant configuration of the chromophore of the NovArch M‐like species which is stable in the dark.
DISCUSSION
We report evidence here that two different variants of AR3: QuasAr2, NovArch which have previously been used for optogenetic transmembrane voltage sensing applications, can also function as optical switches. While optical switching in some forms of animal and microbial and their variants is well known (see discussion below), optical switching between O‐like and M‐like states of OTVSs has previously not been reported. Several major conclusions can be reached from the present studies:
Both QuasAr2 and NovArch optically switch between long‐lived O‐like and M‐like states with alternating red (660 nm) and near UV (405 nm) light.
This optical switching was fully reversible between the two states, when brief cycling periods (5 min) were applied.
The mutant AR3‐D95Q which possesses the same substitution at the SB counterion position as QuasAr2 and NovArch also exhibited similar optical switching properties.
While the QuasAr2 O‐like state is stable in the dark, the NovArch O‐like state partially decays to an M‐like state.
The dark‐adapted M‐like state of NovArch has a 13‐cis, 15‐anti retinylidene chromophore configuration with a deprotonated SB similar to the M intermediate in the BR photocycle.
QuasAr2 and NovArch produce a photoproduct upon extended red‐light (660 nm) illumination with maximal absorbance near 460 nm, which is similar to the pink membrane photoproduct of the O intermediate of BR possessing a 9‐cis chromophore configuration.
The pink membrane‐like photoproduct of QuasAr2 (see #6 above) was stable for several hours in the dark in contrast to NovArch that produces a Q380 decay product, which is also typical of the red‐light induced BR pink membrane.
Optical bistability of opsins
Several types of opsins have previously been found to exhibit optical bistability, defined as the ability to switch between two different stable or long‐lived states with different visible absorption maxima using two different wavelengths of light. For example in the animal rhodopsin family (Type 1 opsins), invertebrate rhodopsins have been found to respond to two different wavelengths of light by cycling between a stable R and Meta state (58, 59). This switching mechanism involves photoisomerization of the retinylidene chromophore between an 11‐cis,15‐anti (R state) and all‐trans, 15‐anti (Meta state) configuration as well as protein conformational changes (60, 61). Examples of bistable animal rhodopsins include squid visual rhodopsin (sRh) (62) and melanopsin (MO) which is found in the animal eye (including human) and many other animal tissues (63, 64, 65).
In the microbial rhodopsin family (Type II opsins), there are also examples of native opsins that exhibit optical bistability such as the photochromic Anabaena Sensory Rhodopsin (ASR) from the freshwater cyanobacterium Anabaena sp. PCC7120. Several studies reveal that ASR can reversibly switch between two stable states containing predominantly all‐trans or 13‐cis retinylidene chromophores (47, 66). Recent striking examples are the histidine kinase rhodopsins and the novel NeoR clade, which probably function as sensory rhodopsin modulators. HKR1 can photocycle between a blue‐shifted state (~485 nm) with a protonated SB and either a 13‐trans/15‐anti or a 13‐cis/15‐syn chromophore configuration and a UV‐shifted state (~380 nm) with an unprotonated SB and a 13‐cis/15‐anti chromophore configuration (67). NeoR can photocycle between a strongly red‐shifted state (ca 690 nm maximum) with a protonated SB and a blue‐shifted M‐like state (ca 360 nm maximum), most likely with a nonprotonated SB (55, 68). The chromophore configuration of these states has not yet been reported.
Optical switching between early K, L and M photocycle intermediates of microbial rhodopsins is also well established, although their physiological lifetimes are typically very short ranging from nanoseconds to milliseconds. However, these photointermediate lifetimes can be significantly extended using low temperatures or dehydrated conditions, where they exhibit true optical bistability. Examples include optical switching between the BR570 and K630 intermediates of BR at 77 K (69) and between BR570 and M412 in dehydrated BR films (70).
Genetic bioengineering can also be used to produce artificial optical bistability of microbial rhodopsins. For example, the D96N mutant of BR exhibits a slowed decay of the M412 intermediate ranging from milliseconds to minutes at pH>7. This occurs due to the absence of the native D96 cytoplasmic proton donor, thereby blocking SB reprotonation and N intermediate formation (71, 72, 73). Because M412 undergoes a back reaction when irradiated with short wavelength light, BR‐D96N can be optically switched back to the light‐adapted BR570 state (74, 75). For example, this property was exploited to produce holographic films of BR‐D96N (76) and all‐optical switching in the near infrared with BR‐D96N‐coated microcavities (77).
Optical switching has also been associated with formation of the P490 species (pink membrane) upon extended red‐light illumination of the BR O intermediate and also in mutants of BR which exhibit extended O intermediate lifetimes. Optical switching is possible since the P490 state can be photoreversed back to the light‐adapted form of BR (BR570) or the O intermediate using blue light (41, 42, 43). For example, films of purple membrane prepared at pH 6.5 in glycerol, exhibit a slowed O intermediate decay (43). Under continuous 647 nm laser illumination, a P490 photoproduct is formed with low quantum yield and long lifetime, which was found to possess a 9‐cis retinylidene configuration similar to earlier studies on acid blue membrane. P490 can be then photoreversed back to the light‐adapted BR or the O intermediate with blue light. The P490 state also thermally decays in the dark to Q380 which possesses a free 9‐cis retinal in the binding site and can be also be converted back to the light‐adapted BR state using near UV wavelength illumination (43). In a second study, BR was suspended in glycerol solutions with low water content (e.g. 85% v/v glycerol/water mixture) which slows O intermediate decay (41). Red‐light produces a P490 photoproduct which was found to consist of separate 445 and 525 component states (41). P490 slowly decays to the Q380 state which can be photoreversed with UV light back to light‐adapted form of BR (BR570). Significantly, Q380 formation appears to depend on the level of hydration of BR, most likely because water in the retinylidene active site near the SB of P490 is required for hydrolysis of the SB to form the free 9‐cis retinal present in Q380 (41). A similar process also appears to occur for the case of acid blue membrane where the SB counterion is protonated at low pH (78).
In an additional example of bioengineered optical bistability, Birge and collaborators used directed evolution to create mutants of BR with long‐lived O intermediates that produce P490 upon red‐light illumination (30). One such mutant involved 4 mutations that did not include the SB counterion Asp85 (V49A, I119T, T121S and A126T). Similar to experiments described above, the 490 nm absorbing species contains a 9‐cis,15‐anti retinylidene chromophore and decayed quickly to a long‐lived “Q” state absorbing at 380 nm (30). Notably, the largest contribution to stabilization of the Q state appears to be the mutation V49A which is homologous to V59A in QuasAr2 and NovArch but not present in AR3‐D95Q.
Similarity between the O intermediate in the BR photocycle and the O‐like state of QuasAr2 and NovArch
It is important to recognize that the long‐lived O‐like states of the various AR3 mutants described here are not identical to the canonical ground state (G) of these mutants which is initially generated during biosynthesis of the mutant by the binding of the retinal chromophore to the opsin apoprotein. In our experiments, the G form is generated during bacterial growth and holoprotein biosynthesis. However, subsequent room light exposure and initial photocycling could result in an altered long‐lived state which is not identical to G. This state has been designated O‐like because of its red‐shifted visible absorption maximum near 600 nm, albeit not as far red‐shifted as the BR O intermediate (640 nm). The O‐like state of BR, QuasAr2 and NovArch all share an uncharged Schiff base counterion. However, in the case of QuasAr2 and NovArch, the SB counterion is neutralized by substitution of a normally charged residue (Asp) with a neutral residue (Gln). In the case of BR, the SB counterion (Asp85) is neutral as early as the formation of the M intermediate due to transfer of a proton from the SB, and Asp85 does not deprotonate until the decay of the O intermediate which leads to formation of the light‐adapted form of BR (79). In both cases, SB counterion neutralization causes a red‐shifted absorption which is predicted on the basis of a simple point charged model first proposed by Honig and coworkers (80). Even in the case of native BR, the “acid blue” form which is generated between pH 2–3 due to protonation of the SB counterion Asp85 exhibits a red‐shifted absorbance maximum near 600 nm (81, 82).
Note that torsion in the alternating single‐bond/double‐bond polyene chain of the BR O intermediate chromophore indicated by an intensified HOOP mode vibrational band near 960 cm−1 is not present in QuasAr2 (24, 79, 83) or NovArch on the basis of the FT‐Raman measurements presented here. This extra torsion in the BR O intermediate most likely contributes to the additional red‐shift to 640 nm compared to QuasAr2 and other OTVSs with neutral counterions such as the mutant D97N of green proteorhodopsin (GPR) (known as PROPS) and the D95N mutant of AR3 (17, 18).
Possible origin of voltage‐sensitive NIR fluorescence
The exact mechanism of voltage‐sensitive fluorescence in QuasAr2 and NovArch is unknown and will require further studies. In this regard, an earlier study of native AR3 based on measuring photogenerated intermediates in E. coli and HEK293 single cells (84) concluded that in this case the fluorescence detected in the NIR involves a 3‐photon absorption process. The first photon absorbed by the ground state of AR3 which has a maximum absorbance wavelength near 570 nm generates the transient N photocycle intermediate which then upon photon absorption forms a previously identified NIR fluorescent Q species (note this is different from Q380 state discussed previously in conjunction with pink membrane) (85). However, it is likely that the O‐like ground states of QuasAr2 and NovArch are distinct from the ground state of native AR3 which has a negatively charged SB counterion. A more appropriate model might be AR3‐D95N which like QuasAR2 and NovArch has a neutral SB counterion. In a study of this mutant based on near‐IR resonance Raman confocal microscopy (32), it was proposed that a slowly decaying 13‐cis retinal containing N‐like species is formed upon photoexcitation of an all‐trans O‐like ground state. Similar to the proposed model for AR3, excitation of the N‐like species produced a NIR fluorescent Q species, which forms an M‐like species upon deprotonation. In this regard, it has yet to be determined whether the photoreversible M‐like state observed here for QuasAr2 and NovArch may also be influenced by the transmembrane potential. Since NovArch as shown here slowly decays in darkness to an M‐like state, its use as an OTVS might be enhanced by first regenerating the O‐like state with UV light. Interestingly, such an approach was reported for NovArch imaging in tissues. In this case, the 1‐photon NIR fluorescence was reversibly enhanced by either blue photons (480 nm) or weak NIR (900 nm) 2‐photon excitation (21).
Optical switching between O‐ and M‐like states does not involve P490 (Pink membrane)
The reversible optical switching we observe for QuasAr2, NovArch and AR3‐D95Q between an O‐like and M‐like state appears to be distinct from optical switching involving pink membrane (P490) and the decay product Q380 discussed above. First, we do not observe any significant accumulation of the P490 or Q380 long‐lived species during or after short 5‐min illumination with 660 nm light (see Figs. 2B, 3B and 4B). In particular, the M‐like photoproduct formed absorbs near 404 nm (QuasAr2, AR3‐D95N) and 434 nm (NovArch) compared to 460 and 380 nm for the P490 photoproduct and Q380 decay product, respectively. In addition, in the case of NovArch, we found that the vibrational spectrum of M‐like state exhibits bands in the fingerprint region which are highly characteristic of a 13‐cis retinylidene configuration (Fig. 6) and not the 9‐cis isomer of P490 or Q380, the later exhibiting characteristic bands at 1212 and 1142 cm−1 (86).
In contrast, longer red‐light illumination (~3 h) of QuasAr2 resulted in photochemical production of a long‐lived state similar to P490 as indicated by the positive band at 460 nm (Figure S6B). In addition, a second positive band at 405 nm which is present after only 5 min of red‐light illumination slowly decays with kinetics similar to the formation of P490 (Figure S6C). The appearance of a P490 photoproduct upon long red‐light illumination was also detected for NovArch along with the band at 435 nm associated with the M‐like photoproduct (Figure S7). Thus, we conclude that the M‐like species which is formed after only short red‐light illumination is distinct from the P490 species which forms after prolonged red‐light illumination.
We also note that unlike QuasAr2 in the case of NovArch a small band appears at 375 nm (Figure S7) indicating that it may form a Q380 decay product. One possible explanation is that the NovArch structure provides a somewhat greater access of water molecules near the SB. This may also explain why NovArch accumulates the M‐like species in the dark (unlike QuasAr2) since greater accessibility to water molecules may lower the pKa of the SB. Note also that the mutant AR3‐D95Q did not exhibit any evidence of formation of a P490 photoproduct or the decay product Q380. However, we cannot eliminate the possibility that P490 is formed and quickly decays into Q380 whose absorption is masked by the more dominant M‐like species near 404 nm, but still this will then be only a minor fraction.
Possible origins of optical switching and M stabilization in QuasAr2 and NovArch
At present, the molecular basis for the reversible O to M optical switching observed here for QuasAr2, NovArch and AR3‐D95Q as well the stability of the M‐like species of NovArch in the dark is unknown and will require further study. However, several possible contributions to these phenomena should be considered:
Effects of the mutation D95Q: The substitution of a Gln for Asp at the SB counterion position, which effectively neutralizes what is normally a negative charge located near the SB, may alone or in part account for the reversible optical switching properties observed for QuasAr2 and NovArch, which also possess this mutation. In particular, the mutant AR3‐D95Q also displayed similar switching properties although with approximately half the efficiency of QuasAr2 and NovArch. Interestingly, the mutant AR3‐D95N which has been previously used as an OTVS (17) displayed only a low level of optical cycling between an O‐like and M‐like state and formed mainly a Q380‐like product upon extended red‐light illumination (Mei et al., to be published). Hence, the extra carbon present in the Gln side‐chain relative to Asn may produce structural changes near the SB relative to AR3‐D95N which facilitates the observed optical switching in this mutant as well as QuasAr2 and NovArch which also share this substitution. This might be facilitated by specific hydrogen bonding between Gln and the SB and/or one or more water molecules located in this region. A similar substitution at the homologous position 85 in BR (e.g. D85Q vs. D85N) produces significantly different absorption shifts (87). Furthermore, a resonance Raman study of the BR mutants comparing D85N and D85A revealed different SB hydrogen bonding strengths (23). It will be important in the future to explore the optical switching properties of additional OTVSs which have different substitutions at the SB counterion position. For example, QuasAr1 and Archon2 have a His substituted for Asp at position 95 in AR3 (22) which are likely to produce significantly different properties.
Effect of the mutation D106H: Both QuasAr2 and NovArch have a His residue instead of the Asp residue at the SB proton donor position (D106) in contrast to AR3 and AR3‐D95N,Q (Fig. 7). Since the BR‐D96N mutant discussed above exhibits a much slower M412 decay due to the absence of a proton donor group for the deprotonated SB, it is not surprising that the mutation BR‐D96H significantly slows M412 decay at pH >7 (~100 fold), although not as dramatically as D96N (88). A similar effect in QuasAr2 and NovArch could account for the accumulation of a M‐like state under red‐light illumination. Furthermore, near UV light photoexcitation is expected to rapidly photoisomerize M back to O as observed in the native BR photocycle and the BR‐D96N mutant.
Effects of the mutation V59A: In the case of NovArch, the V59A substitution, which is not present in QuasAr2 or AR3‐D95Q, may contribute to the stabilization in the dark of the M‐like species we observe. For example, the homologous BR mutation, V49A, appears to interfere with the cytoplasmic uptake of a proton necessary for the decay of the M412 intermediate (89). Furthermore, the high‐resolution X‐ray crystallographic structure of BR‐D96N stabilized in the late M (M2) conformation (90) reveals the proximity of V49 to the SB. One possible explanation for the effect of the mutation V59A in NovArch is that the substitution of the less hydrophobic Ala for Val results in reorganization of water molecules near the SB, thereby lowering the SB pKa.
Effect of the mutation I213T in NovArch: In the case of BR, a proton is ejected to the extracellular medium during M formation. This event is triggered by transfer of a proton from the SB to the SB proton acceptor, D85, and related protonation/ionization changes of two key residues, D194 and D204 along with water molecules located on the extracellular side of the protein below the SB. A variety of studies have shown that this complex, termed the proton release group (PRG), plays a key role at different steps in the BR photocycle including the decay of the O intermediate which requires the PRG to return to its original state prior to absorption of a photon by light‐adapted BR (91, 92). In AR3, I213 is located next to E214, the homologous residue to BR E204 (see Figs. 1 and 7A,B). It is therefore plausible that the mutation I213T, which substitutes a hydrophilic for a hydrophobic residue, would significantly alter the properties of this putative PRG and alter many of the properties of AR3 including its proton transport function, photocycle kinetics and pKa of the SB.
Figure 7.
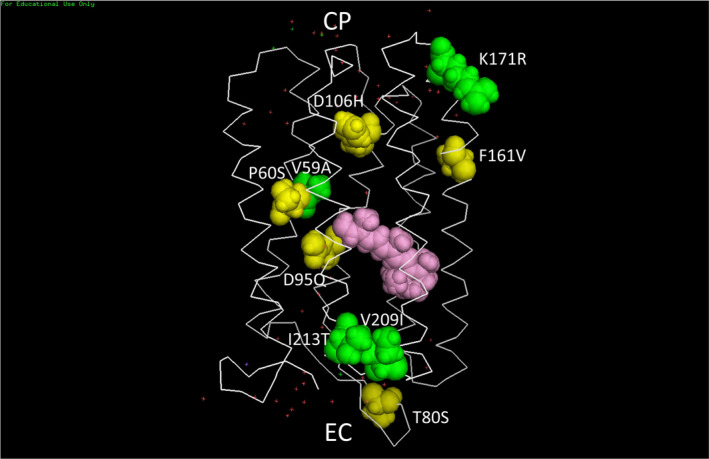
3D Structural Model of NovArch: The model was constructed using the PyMol 1.3 Molecular Graphics Program from the PDB coordinates of the 1.9 Å X‐ray crystallographic determined structure for AR3 (31). Note that a more recent structural model for both light and dark forms of AR3 was recently published (101) but this will not significantly change the relative position of the space‐filling residues shown here. Yellow space‐filling residues are the 5 mutated residues in the AR3 sequence that are common to both QuasAr2 and NovArch. Green space‐filling residues are the 4 residues in the QuasAr2 sequence that are mutated to form NovArch. The all‐trans retinylidene chromophore shown in pink lies inside the 7‐helix transmembrane backbone (white sticks). Red dots show the position of detected water molecules. The protein structure is oriented in the lipid bilayer membrane (not shown) so the extracellular facing side (EC) is oriented at the bottom and the cytoplasmic facing side (CP) is oriented at the top. For additional details, see Materials and Methods.
Structural implications
We conclude that the common principle between type II rhodopsins, that show optical switching behavior, is a blockage of or significant thermal barrier in the SB reprotonation pathway and/or an increased thermal barrier in the chromophore re‐isomerization step. Single mutations such as D96N in BR and D95Q in AR3 probably mainly contribute to the first and second effect, respectively. A more complex pattern of changes like in QuasAr2, NovArch and NeoR probably involves both elements. This optical switching behavior most likely relies on small structural rearrangements in and near the binding pocket and in “soft” structural elements such as rotamer distribution, proton wires and H‐bonded networks including water molecules. When more 3D structures of M‐like states become available, a mechanistic interpretation with help of computational studies should open the way to designing optically switchable rhodopsins with desired properties. That would be very valuable, considering the broad range of biotechnical applications for such switchable systems, like the ones discussed below.
Possible optogenetic and molecular device applications
Engineered forms of opsin proteins or materials which incorporate opsin proteins often display interesting properties such as high temperature thermal stability, altered kinetics, color tuned absorption maxima and light‐activated selective ion gating which can be useful in the field of molecular devices and optogenetics (9, 100). In the field of biomolecular nanotechnology, bioengineering opsins and other photonic proteins provide a powerful strategy for developing molecular devices such as nanosensors, nanopores and nano‐memories.
The discovery of optical switching associated with at least two different forms of OTVSs studied here suggests additional possible optogenetic and molecular device applications. For example, desensitization of opsins such as ChRs for optogenetic control and OTVSs for optogenetic sensing due to continuous or pulsed light excitation imposes limitations on these techniques (7, 22, 26, 27, 28). Our results suggest that at least some of this desensitization may be reversed using properly tuned wavelengths of light to reset the original state(s) of the opsin (e.g. O‐like intermediate). Further research will be needed to explore this possibility.
The ability to optically switch OTVSs between different long‐lived states also introduces the possibility of encoding long‐term stored information in these proteins in 2 and 3D memories similar to earlier work using the BR pink membrane (30). In the case of OTVSs, since optical switching is likely to be strongly affected by transmembrane voltage, opsin‐based memory elements might form the basis for storing and reading out transient electrical neural states that can lead to deeper insight into understanding the complexity of neural brain circuitry.
Supporting information
Figure S1. Absorption spectrum #7 of QuasAr2 measured in the cycle sequence shown in Figure S2 with no base‐line corrections applied.
Figure S2. Schematic showing the sequence of LED illumination used for switching between M‐like and O‐like states of QuasAr2 followed by 3 h in dark. The duration of the time for each measurement is listed in row 2 and illumination condition in row 3. After spectrum #12 was recorded the sample remained in dark for 3 h (from scans #13 to #48). The columns show the number of the measured spectrum in the sequence.
Figure S3. Difference spectra of QuasAr2 cycled 10 times between O‐like and M‐like states. Timing of individual steps in the cycle was similar to those shown in Figure S2 but extended to 10 cycles instead of 3. A) Base‐line corrected differences obtained using 405 nm light for each of 10 cycles. Differences spectra shown are differences between spectra recorded in dark before and after 405 nm illumination (e.g. #3‐#1, #7‐#5 ….. #39‐#37). The blue trace (#3‐#1) reflects the lower level of M‐like intermediate present when the sample was placed in the spectrometer compared to subsequent cycles. B) Same as A showing differences recorded in dark before and after 660 nm illumination (e.g. #5‐3, #9‐#7 ………#37‐#35).
Figure S4. Optical Switching of QuasAr2 during 10 cycles: (A) Base‐line corrected absolute absorption spectra recorded after 405 nm illumination (red traces) and 660 nm illumination (blue traces). (B) Cycling fraction (CF) of QuasAr2 for each individual LED illumination in 10 cycles. Each point is calculated as ratio of change of peak OD (∆OD) between spectrum measured before and after illumination to the absolute OD measured before illumination at wavelength of maximum amplitude of difference spectra (404 and 598 nm (see Figures S3A,B)). Points shown correspond to CF calculated for 405 nm illumination (blue square) or 660 nm illumination (red square) for a particular cycle (x‐axis). Note that in the first and last cycle the CF points that are not part of the equilibrium are not included.
Figure S5. Difference spectra of QuasAr2 cycled 10 times between O‐like and M‐like states measured during illumination. In contrast to Figures S3A and B, differences reflect measurements between dark and illuminated samples. Timing of individual steps in the cycle was similar to those shown in Figure S2 but extended to 10 cycles instead of 3. A) Base‐line corrected differences obtained using 405 nm light for each of 10 cycles. Differences spectra shown are differences between spectra recorded in dark before and during 405 nm illumination (e.g. #2‐#1, #6‐#5 ….. #38‐#37). The blue trace (#2‐#1) reflects the lower level of M‐like intermediate present after sample is place in spectrometer compared to subsequent cycles. B) Same as A showing differences recorded in dark before and during 660 nm illumination (e.g. #4‐#3, #8‐#7 ………#40‐#39).
Figure S6. (A) Schematic showing the sequence of LED illumination used for switching between M‐like and O‐like states of QuasAr2 followed by 3 hours of measurements during red‐light illumination. Note in contrast to 3 cycles of illumination used for data shown in Figure 2B, 10 cycles were used. Each measured spectrum is sequentially numbered. In addition, 4 times (400%) intensity was used for 660 nm illumination compared to the experiment described in Figure S2. The duration of the time for each measurement is listed in row 2 and illumination condition in row 3. The sequence shown was continued up to spectrum #40. After spectrum #40 was recorded the sample remained under the 660 LED illumination for 3 hours (from scans #41 to #76). The columns show the number of the measured spectrum in the sequence. (B) Difference absorption spectra of QuasAr2 during prolonged‐light (660 nm) illumination. Base‐line corrected difference spectra correspond to protocol shown in Figure S6A. Light blue trace with lowest amplitude negative band at 600 nm corresponds to difference (#41‐#39) where red light was on during #40 and #41. Subsequent differences while red light is on [(N‐#42 to #76) minus #39] show increasing loss of the 600 nm band as the O‐like state is depleted and increase of the 460 nm band as the pink membrane species is formed up to difference (#76‐#39). All spectra are scaled to 280 nm band in the absolute absorption spectrum. (C) Kinetics of pink membrane formation: Time course of the difference peak amplitudes in Figure S6B measured at 406 nm (red circles), 460 nm (blue solid dots) and 600 nm, (pink squares) as a percentage of the 280 nm band reflecting decay of the O‐like and M‐like species and formation of a species absorbing near 460 nm (pink membrane). Time constants were calculated using a single‐exponential function (see Materials and Methods).
Figure S7. Base‐line Corrected Difference absorption spectra of NovArch during extended red‐light (660nm) illumination using protocol shown in Figure S6A. Light green trace with lowest amplitude negative band at 581 nm corresponds to difference (#41‐39). Subsequent differences show increasing loss of the 581 nm band up to difference (#76‐39). All spectra are scaled to the 280 nm band in absolute absorption spectrum. Linear 2‐point correction was used (334nmand800nm) for base‐line correction.
Figure S8. Inverse linear correlation plot (purple line) between ethylenic frequency and visible absorption wavelength maximum for various microbial rhodopsins (small blue dots and accompanying blue colored labels along with the FT‐Raman measurement (Figure 5) for assigned ethylenic frequency for NovArch M‐like (amber dot) and O‐like (red dot) species). Adapted from reference (24), supplementary Figure S6 (see accompanying caption for further information).
Figure S9. Comparison of the light‐dark resonance Raman difference spectrum recorded using 785‐nm excitation recorded for the mutant BR‐D96N in intact purple membrane at pH 7.8 (blue trace) and the NovArch FT‐Raman spectrum shown in Figure 6 (red trace). BR D96N exhibits a slowed M decay at a pH above 7. The slowed M decay causes an accumulation of the M intermediate under steady‐state illumination. The difference shown consists of a visual interactive subtraction of the BR‐D96N recorded during white light illumination and under dark conditions such that contributions from the resonance Raman spectrum of the BR‐D96N in the dark (but still light adapted) are neutralized. Y‐scale is for NovArch spectrum.
Acknowledgements
We wish to thank Prof. Xue Han (Boston University) for helpful discussion, Prof. Joel Kralj (UC Boulder) for providing the plasmids for expression of QuasAr2 in E. coli and Dr. Adrian Yi (Boston University) for performing light‐dark resonance Raman difference measurements of the mutant BR‐D95N. This work was supported by the National Science Foundation Division of Chemical, Bioengineering, Environmental and Transport Systems, Grant CBET‐1264434 to KJR. WJdG was financially supported by Leiden University and by the research program of BioSolar Cells (BSC core project Grant C2.9 to WJdG), co‐financed by the Dutch Ministry of Economic Affairs.
REFERENCES
- 1. Zhang, F. , Vierock J., Yizhar O., Fenno L. E., Tsunoda S., Kianianmomeni A., Prigge M., Berndt A., Cushman J., Polle J., Magnuson J., Hegemann P. and Deisseroth K. (2011) The microbial opsin family of optogenetic tools. Cell 147, 1446–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rothschild, K. J. , Argade P. V., Earnest T. N., Huang K. S., London E., Liao M. J., Bayley H., Khorana H. G. and Herzfeld J. (1982) The site of attachment of retinal in bacteriorhodopsin. A resonance Raman study. J. Biol. Chem. 257, 8592–8595. [PubMed] [Google Scholar]
- 3. Rothschild, K. J. and Sonar S. (1995) Bacteriorhodopsin: new biophysical perspectives. In CRC Handbook of Organic Photochemistry and Photobiology (Edited by Horspool W. M. and Song P.‐S.), pp. 1521–1544.CRC Press Inc, Boca Raton. [Google Scholar]
- 4. Lanyi, J. K. (2004) Bacteriorhodopsin. Annu. Rev. Physiol. 66, 665–688. [DOI] [PubMed] [Google Scholar]
- 5. Spudich, J. L. and Kwang‐Hwan J. (2003) Microbial rhodopsins: transport and sensory proteins throughout the three domains of life. In CRC handbook of organic photochemistry and photobiology (Edited by (Horspool W. M.), CRC Press, London. [Google Scholar]
- 6. Spudich, J. L. , Sineshchekov O. A. and Govorunova E. G. (2014) Mechanism divergence in microbial rhodopsins. Biochim. Biophys. Acta 1837, 546–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boyden, E. S. , Zhang F., Bamberg E., Nagel G. and Deisseroth K. (2005) Millisecond‐timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 8, 1263–1268. [DOI] [PubMed] [Google Scholar]
- 8. Boyden, E. S. (2015) Optogenetics and the future of neuroscience. Nat. Neurosci. 18, 1200–1201. [DOI] [PubMed] [Google Scholar]
- 9. Deisseroth, K. (2011) Optogenetics. Nat. Methods 8, 26–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Adamantidis, A. , Arber S., Bains J. S., Bamberg E., Bonci A., Buzsaki G., Cardin J. A., Costa R. M., Dan Y., Goda Y., Graybiel A. M., Hausser M., Hegemann P., Huguenard J. R., Insel T. R., Janak P. H., Johnston D., Josselyn S. A., Koch C., Kreitzer A. C., Luscher C., Malenka R. C., Miesenbock G., Nagel G., Roska B., Schnitzer M. J., Shenoy K. V., Soltesz I., Sternson S. M., Tsien R. W., Tsien R. Y., Turrigiano G. G., Tye K. M. and Wilson R. I. (2015) Optogenetics: 10 years after ChR2 in neurons–views from the community. Nat. Neurosci. 18, 1202–1212. [DOI] [PubMed] [Google Scholar]
- 11. Ernst, O. P. , Sanchez Murcia P. A., Daldrop P., Tsunoda S. P., Kateriya S. and Hegemann P. (2008) Photoactivation of channelrhodopsin. J. Biol. Chem. 283, 1637–1643. [DOI] [PubMed] [Google Scholar]
- 12. Nagel, G. , Brauner M., Liewald J. F., Adeishvili N., Bamberg E. and Gottschalk A. (2005) Light activation of channelrhodopsin‐2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Curr. Biol. 15, 2279–2284. [DOI] [PubMed] [Google Scholar]
- 13. Sineshchekov, O. A. , Govorunova E. G., Li H. and Spudich J. L. (2015) Gating mechanisms of a natural anion channelrhodopsin. Proc. Natl. Acad. Sci. USA 112, 14236–14241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Govorunova, E. G. , Sineshchekov O. A., Rodarte E. M., Janz R., Morelle O., Melkonian M., Wong G. K. and Spudich J. L. (2017) The expanding family of natural anion channelrhodopsins reveals large variations in kinetics, conductance, and spectral sensitivity. Sci. Rep. 7, 43358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yi, A. , Li H., Mamaeva N., Fernandez De Cordoba R. E., Lugtenburg J., DeGrip W. J., Spudich J. L. and Rothschild K. J. (2017) Structural changes in an anion channelrhodopsin: formation of the K and L intermediates at 80 K. Biochemistry 56, 2197–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Deisseroth, K. (2015) Optogenetics: 10 years of microbial opsins in neuroscience. Nat. Neurosci. 18, 1213–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kralj, J. M. , Douglass A. D., Hochbaum D. R., Maclaurin D. and Cohen A. E. (2012) Optical recording of action potentials in mammalian neurons using a microbial rhodopsin. Nat. Methods 9, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kralj, J. M. , Hochbaum D. R., Douglass A. D. and Cohen A. E. (2011) Electrical spiking in Escherichia coli probed with a fluorescent voltage‐indicating protein. Science 333, 345–348. [DOI] [PubMed] [Google Scholar]
- 19. Hochbaum, D. R. , Zhao Y., Farhi S. L., Klapoetke N., Werley C. A., Kapoor V., Zou P., Kralj J. M., Maclaurin D., Smedemark‐Margulies N., Saulnier J. L., Boulting G. L., Straub C., Cho Y. K., Melkonian M., Wong G. K., Harrison D. J., Murthy V. N., Sabatini B. L., Boyden E. S., Campbell R. E. and Cohen A. E. (2014) All‐optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat. Methods 11, 825–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Adam, Y. , Kim J. J., Lou S., Zhao Y., Xie M. E., Brinks D., Wu H., Mostajo‐Radji M. A., Kheifets S., Parot V., Chettih S., Williams K. J., Gmeiner B., Farhi S. L., Madisen L., Buchanan E. K., Kinsella I., Zhou D., Paninski L., Harvey C. D., Zeng H., Arlotta P., Campbell R. E. and Cohen A. E. (2019) Voltage imaging and optogenetics reveal behaviour‐dependent changes in hippocampal dynamics. Nature 569, 413–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chien, M.‐P. , Brinks D., Adam Y., Bloxham W., Kheifets S. and Cohen A. E. (2017) Two‐photon photoactivated voltage imaging in tissue with an Archaerhodopsin‐derived reporter. BioRixiv. https://www.biorxiv.org/content/10.1101/211946v1.full. Cold Spring Harbor Laboratories [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Piatkevich, K. D. , Jung E. E., Straub C., Linghu C., Park D., Suk H. J., Hochbaum D. R., Goodwin D., Pnevmatikakis E., Pak N., Kawashima T., Yang C. T., Rhoades J. L., Shemesh O., Asano S., Yoon Y. G., Freifeld L., Saulnier J. L., Riegler C., Engert F., Hughes T., Drobizhev M., Szabo B., Ahrens M. B., Flavell S. W., Sabatini B. L. and Boyden E. S. (2018) A robotic multidimensional directed evolution approach applied to fluorescent voltage reporters. Nat. Chem. Biol. 14(4), 352–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rath, P. , Marti T., Sonar S., Khorana H. G. and Rothschild K. J. (1993) Hydrogen bonding interactions with the Schiff base of bacteriorhodopsin. Resonance Raman spectroscopy of the mutants D85N and D85A. J. Biol. Chem. 268, 17742–17749. [PubMed] [Google Scholar]
- 24. Mei, G. , Mamaeva N., Ganapathy S., Wang P., DeGrip W. J. and Rothschild K. J. (2020) Analog retinal redshifts visible absorption of QuasAr transmembrane voltage sensors into near‐infrared. Photochem. Photobiol. 96, 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Miesenbock, G. (2009) The optogenetic catechism. Science 326, 395–399. [DOI] [PubMed] [Google Scholar]
- 26. Knopfel, T. , Lin M. Z., Levskaya A., Tian L., Lin J. Y. and Boyden E. S. (2010) Toward the second generation of optogenetic tools. J. Neurosci. 30, 14998–15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Piatkevich, K. D. , Bensussen S., Tseng H. A., Shroff S. N., Lopez‐Huerta V. G., Park D., Jung E. E., Shemesh O. A., Straub C., Gritton H. J., Romano M. F., Costa E., Sabatini B. L., Fu Z., Boyden E. S. and Han X. (2019) Population imaging of neural activity in awake behaving mice. Nature 574, 413–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kiskinis, E. , Kralj J. M., Zou P., Weinstein E. N., Zhang H., Tsioras K., Wiskow O., Ortega J. A., Eggan K. and Cohen A. E. (2018) All‐optical electrophysiology for high‐throughput functional characterization of a human iPSC‐derived motor neuron model of ALS. Stem Cell Rep. 10, 1991–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chernov, K. G. , Redchuk T. A., Omelina E. S. and Verkhusha V. V. (2017) Near‐infrared fluorescent proteins, biosensors, and optogenetic tools engineered from phytochromes. Chem. Rev. 117, 6423–6446. [DOI] [PubMed] [Google Scholar]
- 30. Wagner, N. L. , Greco J. A., Ranaghan M. J. and Birge R. R. (2013) Directed evolution of bacteriorhodopsin for applications in bioelectronics. J. R. Soc. Interface 10, 20130197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vinals Camallonga, J. (2019) Biophysical studies on the structure and function of Archaerhodopsin‐3. DPhil, University of Oxford. [Google Scholar]
- 32. Saint Clair, E. C. , Ogren J. I., Mamaev S., Russano D., Kralj J. M. and Rothschild K. J. (2012) Near‐IR resonance Raman spectroscopy of archaerhodopsin 3: effects of transmembrane potential. J. Phys. Chem. B 116, 14592–14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stoeckenius, W. (1999) Bacterial rhodopsins: evolution of a mechanistic model for the ion pumps. Protein Sci. 8, 447–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ogren, J. I. , Mamaev S., Russano D., Li H., Spudich J. L. and Rothschild K. J. (2014) Retinal chromophore structure and Schiff base interactions in red‐shifted channelrhodopsin‐1 from Chlamydomonas augustae . Biochemistry 53, 3961–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yi, A. , Mamaeva N., Li H., Spudich J. L. and Rothschild K. J. (2016) Resonance Raman study of an anion channelrhodopsin: effects of mutations near the retinylidene Schiff base. Biochemistry 55, 2371–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Takahashi, T. , Yan B., Mazur P., Derguini F., Nakanishi K. and Spudich J. L. (1990) Color regulation in the archaebacterial phototaxis receptor phoborhodopsin (sensory rhodopsin II). Biochemistry 29, 8467–8474. [DOI] [PubMed] [Google Scholar]
- 37. Chizhov, I. , Schmies G., Seidel R., Sydor J. R., Luttenberg B. and Engelhard M. (1998) The photophobic receptor from Natronobacterium pharaonis: temperature and pH dependencies of the photocycle of sensory rhodopsin II. Biophys. J. 75, 999–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dancshazy, Z. , Govindjee R. and Ebrey T. G. (1988) Independent photocycles of the spectrally distinct forms of bacteriorhodopsin. Proc. Natl. Acad. Sci. USA 85, 6358–6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ganapathy, S. , Becheau O., Venselaar H., Frolich S., van der Steen J. B., Chen Q., Radwan S., Lugtenburg J., Hellingwerf K. J., de Groot H. J. and de Grip W. J. (2015) Modulation of spectral properties and pump activity of proteorhodopsins by retinal analogues. Biochem. J. 467, 333–343. [DOI] [PubMed] [Google Scholar]
- 40. Krebs, R. A. , Alexiev U., Partha R., DeVita A. M. and Braiman M. S. (2002) Detection of fast light‐activated H+ release and M intermediate formation from proteorhodopsin. BMC Physiol. 2, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gillespie, N. B. , Wise K. J., Ren L., Stuart J. A., March D. L., Hillebrech J., Li Q., Lavosisier R., Jordan K., Fyvie S. and Birge R. B. (2002) Characterization of the branched‐photocycle intermediates P and Q of bacteriorhodopsin. J. Phys. Chem. B 106, 13352–13361. [Google Scholar]
- 42. Balashov, S. P. (1995) Photoreactions of the photointermediates of bacteriorhodopsin. Israel J. Chem 35, 415–428. [Google Scholar]
- 43. Popp, A. , Wolperdinger M., Hampp N., Bruchle C. and Oesterhelt D. (1993) Photochemical conversion of the O‐intermediate to 9‐cis‐retinal‐containing products in bacteriorhodopsin films. Biophys. J. 65, 1449–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gong, Y. , Li J. Z. and Schnitzer M. J. (2013) Enhanced archaerhodopsin fluorescent protein voltage indicators. PLoS One 8, e66959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sawatzki, G. , Fischer R., Scheer H. and Siebert F. (1990) Fourier‐transform Raman spectroscopy applied to photobiological systems. Proc. Natl. Acad. Sci. USA 87, 5903‐5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rath, P. , DeCaluwe L. L., Bovee‐Geurts P. H., DeGrip W. J. and Rothschild K. J. (1993) Fourier transform infrared difference spectroscopy of rhodopsin mutants: light activation of rhodopsin causes hydrogen‐bonding change in residue aspartic acid‐83 during meta II formation. Biochemistry 32, 10277–10282. [DOI] [PubMed] [Google Scholar]
- 47. Bergo, V. B. , Ntefidou M., Trivedi V. D., Amsden J. J., Kralj J. M., Rothschild K. J. and Spudich J. L. (2006) Conformational changes in the photocycle of Anabaena sensory rhodopsin: absence of the Schiff base counterion protonation signal. J. Biol. Chem. 281, 15208–15214. [DOI] [PubMed] [Google Scholar]
- 48. Braiman, M. and Mathies R. (1980) Resonance Raman evidence for an all‐trans to 13‐cis isomerization in the proton‐pumping cycle of bacteriorhodopsin. Biochemistry 19, 5421–5428. [DOI] [PubMed] [Google Scholar]
- 49. Deng, H. , Pande C., Callender R. H. and Ebrey T. G. (1985) A detailed resonance Raman study of the M412 intermediate in the bacteriorhodopsin photocycle. Photochem. Photobiol. 41, 467–470. [DOI] [PubMed] [Google Scholar]
- 50. Bergo, V. , Amsden J. J., Spudich E. N., Spudich J. L. and Rothschild K. J. (2004) Structural changes in the photoactive site of proteorhodopsin during the primary photoreaction. Biochemistry 43, 9075–9083. [DOI] [PubMed] [Google Scholar]
- 51. Bergo, V. , Spudich E. N., Spudich J. L. and Rothschild K. J. (2002) A Fourier transform infrared study of Neurospora rhodopsin: similarities with archaeal rhodopsins. Photochem. Photobiol. 76, 341–349. [DOI] [PubMed] [Google Scholar]
- 52. Bergo, V. , Spudich E. N., Spudich J. L. and Rothschild K. J. (2003) Conformational changes detected in a sensory rhodopsin II‐transducer complex. J. Biol. Chem. 278, 36556–36562. [DOI] [PubMed] [Google Scholar]
- 53. Kajimoto, K. , Kikukawa T., Nakashima H., Yamaryo H., Saito Y., Fujisawa T., Demura M. and Unno M. (2017) Transient resonance Raman spectroscopy of a light‐driven sodium‐ion‐pump rhodopsin from Indibacter alkaliphilus. J. Phys. Chem. B 121, 4431–4437. [DOI] [PubMed] [Google Scholar]
- 54. Mei, G. , Mamaeva N., Ganapathy S., Wang P., DeGrip W. J. and Rothschild K. J. (2018) Raman spectroscopy of a near infrared absorbing proteorhodopsin: similarities to the bacteriorhodopsin O photointermediate. PLoS One 13, e0209506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hontani, Y. , Broser M., Luck M., Weissenborn J., Kloz M., Hegemann P. and Kennis J. T. M. (2020) Dual photoisomerization on distinct potential energy surfaces in a UV‐absorbing rhodopsin. J. Am. Chem. Soc. 142, 11464–11473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Smith, S. O. , Braiman M. S., Myers A. B., Pardoen J. A., Courtin J. M. L., Winkel C., Lugtenburg J. and Mathies R. A. (1987) Vibrational analysis of the all‐trans‐retinal chromophore in light‐adapted bacteriorhodopsin. J. Am. Chem. Soc. 109, 3108–3125. [Google Scholar]
- 57. Naito, A. , Makino Y., Shigeta A. and Kawamura I. (2019) Photoreaction pathways and photointermediates of retinal‐binding photoreceptor proteins as revealed by in situ photoirradiation solid‐state NMR spectroscopy. Biophys. Rev. 11, 167–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hillman, P. , Hochstein S. and Minke B. (1983) Transduction in invertebrate photoreceptors: role of pigment bistability. Physiol. Rev. 63, 668–772. [DOI] [PubMed] [Google Scholar]
- 59. Gärtner, W. (2000) Molecular mechanisms of visual transduction. In Handbook of Biological Physics, Vol. 3 (Edited by Stavenga D. G., DeGrip W. J. and Pugh E. N.), pp. 298–388.Elseier Press, Amsterdam, The Netherlands. [Google Scholar]
- 60. Suzuki, T. and Callender R. H. (1981) Primary photochemistry and photoisomerization of retinal at 77 K in cattle and squid rhodopsins. Biophys. J. 34, 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sekiya, N. , Kishigami A., Naoki H., Chang C. W., Yoshihara K., Hara R., Hara T. and Tokunaga F. (1991) Fourier transform infrared spectroscopic study on retinochrome and its primary photoproduct, lumiretinochrome. FEBS Lett. 280, 107–111. [DOI] [PubMed] [Google Scholar]
- 62. Murakami, M. and Kouyama T. (2011) Crystallographic analysis of the primary photochemical reaction of squid rhodopsin. J. Mol. Biol. 413, 615–627. [DOI] [PubMed] [Google Scholar]
- 63. Shirzad‐Wasei, N. and DeGrip W. J. (2016) Heterologous expression of melanopsin: present, problems and prospects. Prog. Retin. Eye Res. 52, 1–21. [DOI] [PubMed] [Google Scholar]
- 64. Rodgers, J. , Hughes S., Pothecary C. A., Brown L. A., Hickey D. G., Peirson S. N. and Hankins M. W. (2018) Defining the impact of melanopsin missense polymorphisms using in vivo functional rescue. Hum. Mol. Genet. 27, 2589–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rodgers, J. , Peirson S. N., Hughes S. and Hankins M. W. (2018) Functional characterisation of naturally occurring mutations in human melanopsin. Cell. Mol. Life Sci. 75, 3609–3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kawanabe, A. , Furutani Y., Jung K. H. and Kandori H. (2007) Photochromism of Anabaena sensory rhodopsin. J. Am. Chem. Soc. 129, 8644–8649. [DOI] [PubMed] [Google Scholar]
- 67. Luck, M. , Mathes T., Bruun S., Fudim R., Hagedorn R., Tran Nguyen T. M., Kateriya S., Kennis J. T., Hildebrandt P. and Hegemann P. (2012) A photochromic histidine kinase rhodopsin (HKR1) that is bimodally switched by ultraviolet and blue light. J. Biol. Chem. 287, 40083–40090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Broser, M. , Spreen A., Konold P. E., Peter E., Adam S., Borin V., Schapiro I., Seifert R., Kennis J. T. M., Bernal Sierra Y. A. and Hegemann P. (2020) NeoR, a near‐infrared absorbing rhodopsin. Nat. Commun. 11, 5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rothschild, K. J. and Marrero H. (1982) Infrared evidence that the Schiff base of bacteriorhodopsin is protonated: bR570 and K intermediates. Proc. Natl. Acad. Sci. U S A 79, 4045–4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rothschild, K. J. , Zagaeski M. and Cantore W. A. (1981) Conformational changes of bacteriorhodopsin detected by Fourier transform infrared difference spectroscopy. Biochem. Biophys. Res. Commun. 103, 483–489. [DOI] [PubMed] [Google Scholar]
- 71. Marinetti, T. , Subramaniam S., Mogi T., Marti T. and Khorana H. G. (1989) Replacement of aspartic residues 85, 96, 115, or 212 affects the quantum yield and kinetics of proton release and uptake by bacteriorhodopsin. Proc. Natl. Acad. Sci. U S A 86, 529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Otto, H. , Marti T., Holz M., Mogi T., Lindau M., Khorana H. G. and Heyn M. P. (1989) Aspartic acid‐96 is the internal proton donor in the reprotonation of the Schiff base of bacteriorhodopsin. Proc. Natl. Acad. Sci. U S A 86, 9228–9232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Braiman, M. S. , Mogi T., Marti T., Stern L. J., Khorana H. G. and Rothschild K. J. (1988) Vibrational spectroscopy of bacteriorhodopsin mutants: light‐driven proton transport involves protonation changes of aspartic acid residues 85, 96, and 212. Biochemistry 27, 8516–8520. [DOI] [PubMed] [Google Scholar]
- 74. Karvaly, B. and Dancshazy Z. (1977) Bacteriorhodopsin: a molecular photoelectric regulator. Quenching of photovoltaic effect of bimolecular lipid membranes containing bacteriorhodopsin by blue light. FEBS Lett. 76(1), 36–40. [DOI] [PubMed] [Google Scholar]
- 75. Ormos, P. , Dancshazy Z. and Keszthelyi L. (1980) Electric response of a back photoreaction in the bacteriorhodopsin photocycle. Biophys. J. 31(2), 207–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zheng, Y. , Yao B. L., Wang Y. L., Lei M., Menke N., Cheng G. F. and Hampp N. (2003) Holographic recording properties of BR‐D96N film. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai) 35, 592–595. [PubMed] [Google Scholar]
- 77. Topolancik, J. and Vollmer F. (2007) Photoinduced transformations in bacteriorhodopsin membrane monitored with optical microcavities. Biophys. J. 92, 2223–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Chang, C. H. , Liu S. Y., Jonas R. and Govindjee R. (1987) The pink membrane: the stable photoproduct of deionized purple membrane. Biophys. J. 52, 617–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bousche, O. , Sonar S., Krebs M. P., Khorana H. G. and Rothschild K. J. (1992) Time‐resolved Fourier transform infrared spectroscopy of the bacteriorhodopsin mutant Tyr‐185–>Phe: Asp‐96 reprotonates during O formation; Asp‐85 and Asp‐212 deprotonate during O decay. Photochem. Photobiol. 56, 1085–1095. [DOI] [PubMed] [Google Scholar]
- 80. Honig, B. , Dinur U., Nakanishi K., Balogh‐Nair V., Gawinowicz M. A., Arnaboldi M. and Motto M. G. (1979) An external point‐charge model for wavelength regulation in visual pigments. J. Am. Chem. Soc. 101, 7084–7086. [Google Scholar]
- 81. Smith, S. O. and Mathies R. A. (1985) Resonance Raman spectra of the acidified and deionized forms of bacteriorhodopsin. Biophys. J. 47, 251–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Rath, P. , Marti T., Sonar S., Khorana H. G. and Rothschild K. J. (1993) Hydrogen bonding interactions with the Schiff base of bacteriorhodopsin: resonance raman spectroscopy of the mutants D85N and D85A. J. Biol. Chem. 268, 17742–17749. [PubMed] [Google Scholar]
- 83. Smith, S. O. , Pardoen J. A., Mulder P. P. J., Curry B., Lugtenburg J. and Mathies R. (1983) Chromophore structure in bacteriorhodopsin's O640 photointermediate. Biochemistry 22, 6141–6148. [Google Scholar]
- 84. Maclaurin, D. , Venkatachalam V., Lee H. and Cohen A. E. (2013) Mechanism of voltage‐sensitive fluorescence in a microbial rhodopsin. Proc Natl Acad Sci U S A 110, 5939–5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ohtani, H. , Tsukamoto Y., Sakoda Y. and Hamaguchi H. (1995) Fluorescence spectra of bacteriorhodopsin and the intermediates O and Q at room temperature. FEBS Lett. 359, 65–68. [DOI] [PubMed] [Google Scholar]
- 86. Chang, C. H. , Jonas R., Ebrey T. G. and Hong M. (1987) Protonation changes in the interconversions of the pink membrane, blue membrane, and purple membrane. In Biophysical studies of retinal proteins: Proceedings of a conference in memory of Laura Eisenstein (Edited by Ebrey T., Frauenfelder H., Honig B. and Nakanish K.), pp. 156–165.University of Illinois Press, Urbana, Ill. [Google Scholar]
- 87. Saeedi, P. , Moosaabadi J. M., Sebtahmadi S. S., Behmanesh M. and Mehrabadi J. F. (2012) Site‐directed mutagenesis in bacteriorhodopsin mutants and their characterization for bioelectrical and biotechnological equipment. Biotechnol. Lett. 34, 455–462. [DOI] [PubMed] [Google Scholar]
- 88. Dioumaev, A. K. , Petrovskaya L. E., Wang J. M., Balashov S. P., Dolgikh D. A., Kirpichnikov M. P. and Lanyi J. K. (2013) Photocycle of Exiguobacterium sibiricum rhodopsin characterized by low‐temperature trapping in the IR and time‐resolved studies in the visible. J. Phys. Chem. B 117, 7235–7253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Koyama, K. and Miyasaka T. (2001) The proton uptake channel of bacteriorhodopsin as studied by a photoelectrochemical method. Bioelectrochemistry 53, 111–118. [DOI] [PubMed] [Google Scholar]
- 90. Luecke, H. , Richter H. T. and Lanyi J. K. (1998) Proton transfer pathways in bacteriorhodopsin at 2.3 angstrom resolution. Science 280, 1934–1937. [DOI] [PubMed] [Google Scholar]
- 91. Brown, L. S. , Sasaki J., Kandori H., Maeda A., Needleman R. and Lanyi J. K. (1995) Glutamic acid 204 is the terminal proton release group at the extracellular surface of bacteriorhodopsin. J. Biol. Chem. 270(45), 27122–27126. [DOI] [PubMed] [Google Scholar]
- 92. Balashov, S. P. , Lu M., Imasheva E. S., Govindjee R., Ebrey T. G., Othersen B. 3rd, Chen Y., Crouch R. K. and Menick D. R. (1999) The proton release group of bacteriorhodopsin controls the rate of the final step of its photocycle at low pH. Biochemistry 38, 2026–2039. [DOI] [PubMed] [Google Scholar]
- 93. Oesterhelt, D. , Bräuchle C. and Hampp N. (1991) Bacteriorhodopsin: a biological material for information processing. Q. Rev. Biophys. 4, 425–478. [DOI] [PubMed] [Google Scholar]
- 94. Shen, Y. , Safinya C. R., Liang K. S., Ruppert A. F. and Rothschild K. J. (1993) Stabilization of the membrane protein bacteriorhodopsin to 140 °C in two‐dimensional films. Nature 366, 48–50. [Google Scholar]
- 95. Zeisel, D. and Hampp N. (1994) Bacteriorhodopsin Films Containing The variant D96N as media for dynamic holography and interferometry. Mol. Cryst. Liq. Cryst. 246, 371–374. [Google Scholar]
- 96. Giardi, M. T. and Pace E. (2005) Photosynthetic proteins for technological applications. Trends Biotechnol. 23, 257–263. [DOI] [PubMed] [Google Scholar]
- 97. Kietis, B. P. , Saudargas P., Varo G. and Valkunas L. (2007) External electric control of the proton pumping in bacteriorhodopsin. Eur. Biophys. J. 36, 199–211. [DOI] [PubMed] [Google Scholar]
- 98. Saeedi, P. , Moosaabadi J. M., Sebtahmadi S. S., Mehrabadi J. F., Behmanesh M., Nejad H. R. and Nazaktabar A. (2012) Generation and analysis of bacteriorhodopsin mutants with the potential for biotechnological applications. Bioengineered 3, 275–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Saeedi, P. , Moosaabadi J. M., Sebtahmadi S. S., Mehrabadi J. F., Behmanesh M. and Mekhilef S. (2012) Potential applications of bacteriorhodopsin mutants. Bioengineered 3, 326–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Ranaghan, M. J. , Schwall C. T., Alder N. N. and Birge R. R. (2011) Green proteorhodopsin reconstituted into nanoscale phospholipid bilayers (nanodiscs) as photoactive monomers. J. Am. Chem. Soc. 133, 18318–18327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Bada Juarez, J. F. , Judge P. J., Adam S., Axford D. A., Vinals J., Birch J., Kwan T. O., Hoi K. K., Yen H.‐Y., Vial A., Millhiet P.‐E., Robinson C. V., Schapiro I., Moraes I. and Watts A. (2021) Structures of the archaerhodopsin‐3 transporter reveal that disordering of internal water networks underpins receptor sensitization. Nat. Comm. 12, 629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Absorption spectrum #7 of QuasAr2 measured in the cycle sequence shown in Figure S2 with no base‐line corrections applied.
Figure S2. Schematic showing the sequence of LED illumination used for switching between M‐like and O‐like states of QuasAr2 followed by 3 h in dark. The duration of the time for each measurement is listed in row 2 and illumination condition in row 3. After spectrum #12 was recorded the sample remained in dark for 3 h (from scans #13 to #48). The columns show the number of the measured spectrum in the sequence.
Figure S3. Difference spectra of QuasAr2 cycled 10 times between O‐like and M‐like states. Timing of individual steps in the cycle was similar to those shown in Figure S2 but extended to 10 cycles instead of 3. A) Base‐line corrected differences obtained using 405 nm light for each of 10 cycles. Differences spectra shown are differences between spectra recorded in dark before and after 405 nm illumination (e.g. #3‐#1, #7‐#5 ….. #39‐#37). The blue trace (#3‐#1) reflects the lower level of M‐like intermediate present when the sample was placed in the spectrometer compared to subsequent cycles. B) Same as A showing differences recorded in dark before and after 660 nm illumination (e.g. #5‐3, #9‐#7 ………#37‐#35).
Figure S4. Optical Switching of QuasAr2 during 10 cycles: (A) Base‐line corrected absolute absorption spectra recorded after 405 nm illumination (red traces) and 660 nm illumination (blue traces). (B) Cycling fraction (CF) of QuasAr2 for each individual LED illumination in 10 cycles. Each point is calculated as ratio of change of peak OD (∆OD) between spectrum measured before and after illumination to the absolute OD measured before illumination at wavelength of maximum amplitude of difference spectra (404 and 598 nm (see Figures S3A,B)). Points shown correspond to CF calculated for 405 nm illumination (blue square) or 660 nm illumination (red square) for a particular cycle (x‐axis). Note that in the first and last cycle the CF points that are not part of the equilibrium are not included.
Figure S5. Difference spectra of QuasAr2 cycled 10 times between O‐like and M‐like states measured during illumination. In contrast to Figures S3A and B, differences reflect measurements between dark and illuminated samples. Timing of individual steps in the cycle was similar to those shown in Figure S2 but extended to 10 cycles instead of 3. A) Base‐line corrected differences obtained using 405 nm light for each of 10 cycles. Differences spectra shown are differences between spectra recorded in dark before and during 405 nm illumination (e.g. #2‐#1, #6‐#5 ….. #38‐#37). The blue trace (#2‐#1) reflects the lower level of M‐like intermediate present after sample is place in spectrometer compared to subsequent cycles. B) Same as A showing differences recorded in dark before and during 660 nm illumination (e.g. #4‐#3, #8‐#7 ………#40‐#39).
Figure S6. (A) Schematic showing the sequence of LED illumination used for switching between M‐like and O‐like states of QuasAr2 followed by 3 hours of measurements during red‐light illumination. Note in contrast to 3 cycles of illumination used for data shown in Figure 2B, 10 cycles were used. Each measured spectrum is sequentially numbered. In addition, 4 times (400%) intensity was used for 660 nm illumination compared to the experiment described in Figure S2. The duration of the time for each measurement is listed in row 2 and illumination condition in row 3. The sequence shown was continued up to spectrum #40. After spectrum #40 was recorded the sample remained under the 660 LED illumination for 3 hours (from scans #41 to #76). The columns show the number of the measured spectrum in the sequence. (B) Difference absorption spectra of QuasAr2 during prolonged‐light (660 nm) illumination. Base‐line corrected difference spectra correspond to protocol shown in Figure S6A. Light blue trace with lowest amplitude negative band at 600 nm corresponds to difference (#41‐#39) where red light was on during #40 and #41. Subsequent differences while red light is on [(N‐#42 to #76) minus #39] show increasing loss of the 600 nm band as the O‐like state is depleted and increase of the 460 nm band as the pink membrane species is formed up to difference (#76‐#39). All spectra are scaled to 280 nm band in the absolute absorption spectrum. (C) Kinetics of pink membrane formation: Time course of the difference peak amplitudes in Figure S6B measured at 406 nm (red circles), 460 nm (blue solid dots) and 600 nm, (pink squares) as a percentage of the 280 nm band reflecting decay of the O‐like and M‐like species and formation of a species absorbing near 460 nm (pink membrane). Time constants were calculated using a single‐exponential function (see Materials and Methods).
Figure S7. Base‐line Corrected Difference absorption spectra of NovArch during extended red‐light (660nm) illumination using protocol shown in Figure S6A. Light green trace with lowest amplitude negative band at 581 nm corresponds to difference (#41‐39). Subsequent differences show increasing loss of the 581 nm band up to difference (#76‐39). All spectra are scaled to the 280 nm band in absolute absorption spectrum. Linear 2‐point correction was used (334nmand800nm) for base‐line correction.
Figure S8. Inverse linear correlation plot (purple line) between ethylenic frequency and visible absorption wavelength maximum for various microbial rhodopsins (small blue dots and accompanying blue colored labels along with the FT‐Raman measurement (Figure 5) for assigned ethylenic frequency for NovArch M‐like (amber dot) and O‐like (red dot) species). Adapted from reference (24), supplementary Figure S6 (see accompanying caption for further information).
Figure S9. Comparison of the light‐dark resonance Raman difference spectrum recorded using 785‐nm excitation recorded for the mutant BR‐D96N in intact purple membrane at pH 7.8 (blue trace) and the NovArch FT‐Raman spectrum shown in Figure 6 (red trace). BR D96N exhibits a slowed M decay at a pH above 7. The slowed M decay causes an accumulation of the M intermediate under steady‐state illumination. The difference shown consists of a visual interactive subtraction of the BR‐D96N recorded during white light illumination and under dark conditions such that contributions from the resonance Raman spectrum of the BR‐D96N in the dark (but still light adapted) are neutralized. Y‐scale is for NovArch spectrum.


