Abstract
COVID-19 is a serious infectious disease that has recently swept the world, and research on its causative virus, SARS-CoV-2, remains insufficient. Therefore, this study uses bioinformatics analysis techniques to explore the human digestive tract diseases that may be caused by SARS-CoV-2 infection. The gene expression profile data set, numbered GSE149312, is from the Gene Expression Omnibus (GEO) database and is divided into a 24-h group and a 60-h group. R software is used to analyze and screen out differentially expressed genes (DEGs) and then gene ontology (GO) term and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses are performed. In KEGG, the pathway of non-alcoholic fatty liver disease exists in both the 24-h group and 60-h group. STRING is used to establish a protein–protein interaction (PPI) network, and Cytoscape is then used to visualize the PPI and define the top 12 genes of the node as the hub genes. Through verification, nine statistically significant hub genes are identified: AKT1, TIMP1, NOTCH, CCNA2, RRM2, TTK, BUB1B, KIF20A, and PLK1. In conclusion, the results of this study can provide a certain direction and basis for follow-up studies of SARS-CoV-2 infection of the human digestive tract and provide new insights for the prevention and treatment of diseases caused by SARS-CoV-2.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10528-021-10144-w.
Keywords: SARS-CoV-2, Bioinformatics
Introduction
In December 2019, pneumonia caused by a new type of coronavirus appeared in the world. The World Health Organization (WHO) initially named it COVID-19 (coronavirus disease 2019). Since then, the Coronavirus Research Group of the International Commission on Virology has officially named the virus severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2) (Harapan et al. 2020).
SARS-CoV-2 is a positive-stranded RNA virus with an envelope. It is spherical under a transmission electron microscope, with a diameter ranging from 60 to 140 nm and a unique peak length of 8–12 nm. It belongs to the coronavirus 2b lineage (Zhu et al. 2020; He et al. 2020). Studies have shown that SARS-CoV-2 infects host cells through the combination of spike protein and angiotensin-converting enzyme II (ACE2). It spreads faster and is highly stable, but it is not very lethal (He et al. 2020).
SARS-CoV-2 can spread through the respiratory tract, saliva, contact, and excrement; the possibility of transmission through aerosols is also high (Chan et al. 2020). COVID-19 lurks in the human body for about 6.4 days on average and anyone is susceptible to infection (Wang et al. 2020). Currently, the clinical symptoms of COVID-19 are mainly fever (90% or even higher), cough (about 75%), and breathing difficulties (up to 50%) (Cipriano et al. 2020; Huang et al. 2020; Li et al. 2020). Some patients have symptoms of gastrointestinal diseases, such as nausea and vomiting. Studies have found that SARS-CoV-2 is present in the feces of COVID-19 patients, which suggest the possibility of SARS-CoV-2 infecting the human digestive tract and causing a series of diseases (Han et al. 2020). Furthermore, according to WHO statistics, as of July 27, 2020, there were a total of 15,785,641 cases of COVID-19 and 64,016 deaths worldwide. COVID-19 has become a communicable disease that seriously endangers global public health. Therefore, this study aims to provide new ideas and methods for clinically diagnosing, treating, and limiting the spread of SARS-CoV-2 by studying how it infects the human digestive tract.
Bioinformatics is an essentially interdisciplinary field that uses aspects of computer science, mathematics, and statistics to store, manage, analyze, and interpret biological data (Jin et al. 2020). This study uses bioinformatics analysis to identify and analyze the differential expression of organisms under different conditions and then uses gene ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis to obtain the biological significance of DEGs and establish a visual protein–protein interaction (PPI) network, finally leading to a conclusion (Ostaszewski et al. 2020).
Methods and Materials
Data Collection
We obtained the GSE149312 (Lamers et al. 2020) gene expression profile as microarray data from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). In the original study, the authors treated intestinal organ samples with SARS-CoV or SARS-CoV-2 for 24 or 60 h, respectively. RNA was then extracted under dilation conditions (Exp) or differentiation conditions (DIF). We only took the SARS-COV-2 infection group and blank group from the original study, grouping them according to the infection time. In each group, there were 4 infected samples and 6 control samples.
Differentially Expressed Genes (DEG) Screening
To obtain the DEGs between the experimental group and the control group as well as the corresponding volcano map and heat map, R software (version 4.0.1) and pheatmap package were used in this analysis, and P < 0.05 and | log2 fold change (FC) |> 1 were set as the threshold for judging DEGs. (Xie et al. 2019 Jul) DEGs were considered to have statistical significance within this critical value. The online tool, Venny (version 2.1.0; https://bioinfogp.cnb.csic.es/tools/venny/), was then used to make the Venn diagram of the DEGs shared by the 24-h group and 60-h group. All R scripts have been reported to the GitHub repository. (https://github.com/authentic-zz/mycode.git).
The Function and Pathway Enrichment Analysis of DEGs
R language was used to conduct the GO analysis and KEGG pathway analysis to acquire the biological significance of DEGs. GO is a commonly used approach in bioinformatics analysis. Its function is to annotate genes and their products (Gene Ontology Consortium 2006; Liang et al. 2016). Methods to recognize gene function annotations include the biological process (BP), cell component (CC), and molecular function (MF) categories. KEGG is also a commonly used bioinformatics database for integrating and analyzing a large number of data sets obtained from high-throughput experimental technologies, such as genome sequencing (Liang et al. 2016; Kanehisa and Goto 2000). P < 0.05 was considered significant.
Visualization of the Establishment and Module Selection of PPI Networks and the Identification of the Hub Genes
To evaluate the interrelationship between DEGs and build a visual protein interaction network, the STRING online database (version 11.0; http://string-db.org/) was used. First, the DEGs were entered into the STRING online database and the required minimum cross-evaluation was set to a medium confidence level > 0.4. Second, because the initial PPI obtained in STRING was complicated, Cytoscape was used to build a visual PPI relationship network. The plug-in cytoHubba in Cytoscape was used to analyze the core gene modules of the PPI network complex (the default parameters) and define the top 12 genes of the node as the hub genes.
Verification of the Hub Genes
The 12 defined hub genes were uploaded to GraphPad Prism (version 8.0.2) in the order of high to low. Then GSE150728 was analyzed by bioinformatics, and the data obtained passed t test and non-parametric test and P < 0.05 was used as the standard to screen for statistically significant hub genes.
Results
The Identification of DEGs’
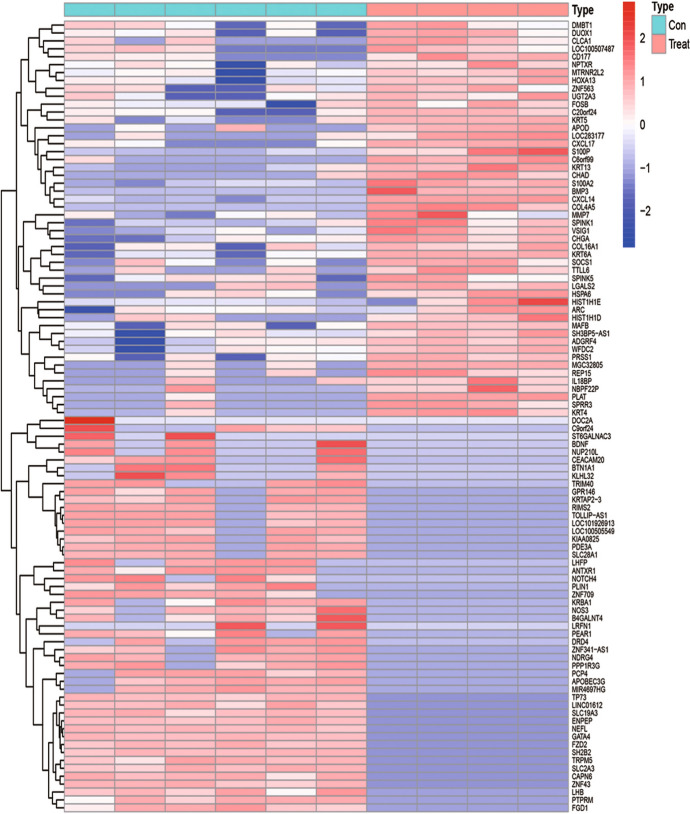
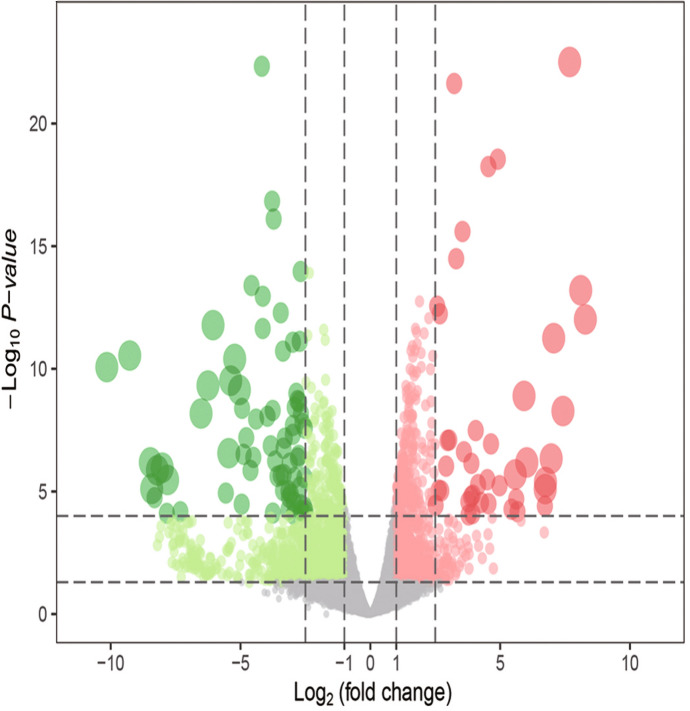
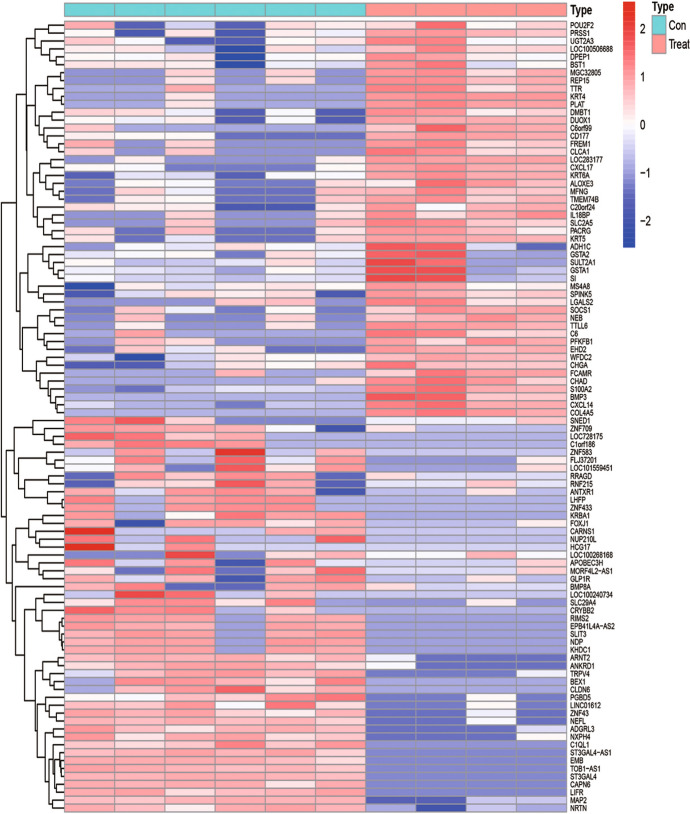
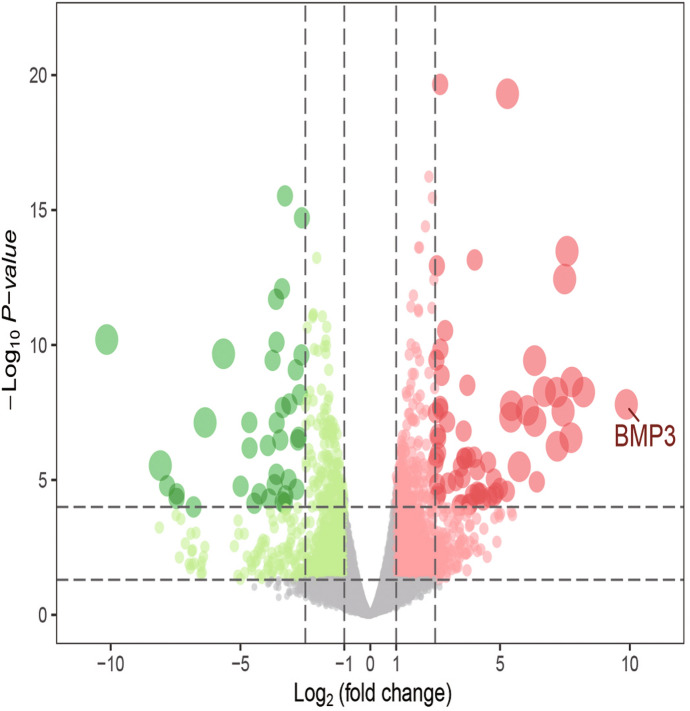
According to the established criteria, 1930 DEGs were screened from the 24-h group, including 748 up-regulated genes and 1182 down-regulated genes. These DEGs are shown in the heat map (Fig. 1) and a volcano plot (Fig. 2) of the 24-h group. From the 60-h group, 1638 DEGs were screened, including 890 up-regulated genes and 748 down-regulated genes. These DEGs are shown in the heat map (Fig. 3) and a volcano plot (Fig. 4) of the 60-h group. Through the analysis of the Venn diagram (Fig. 5), 744 DEGs co-expressed between the 24-h group and the 60-h group were obtained. More detailed data are shown in Table S1.
Fig. 1.
Heat map of the DEGS from the 24-h group
Fig. 2.
Volcano plot of the DEGS from the 24-h group
Fig. 3.
Heat map of the DEGS from the 60-h group
Fig. 4.
Volcano plot of the DEGS from the 60-h group
Fig. 5.
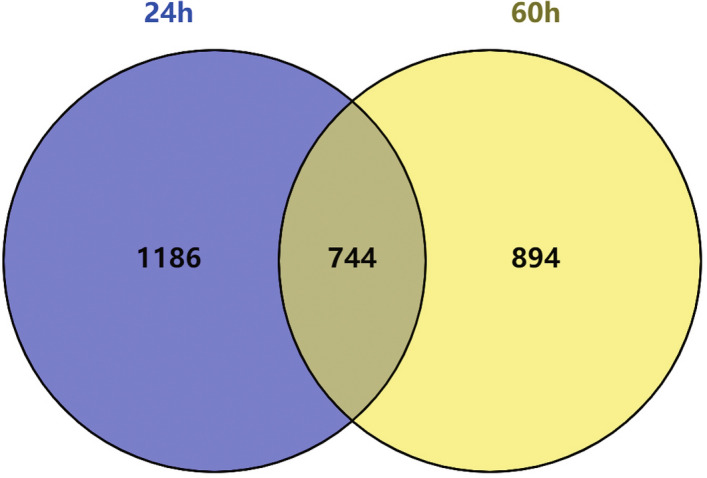
744 DEGs co-expressed between the 24-h group and the 60-h group
GO Term Enrichment Analysis of DEGs
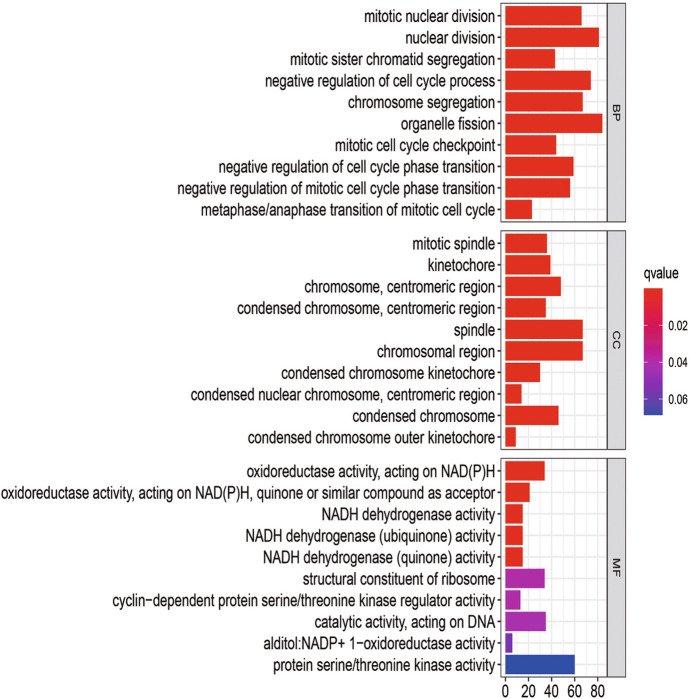
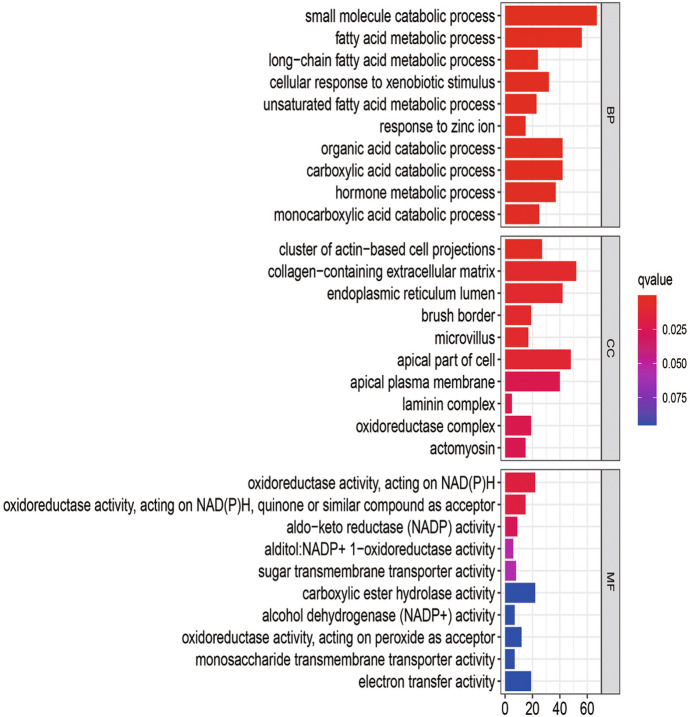
The BP analysis showed that the DEGs of the 24-h group (Fig. 6) were significantly enriched in mitotic nuclear division, nuclear division, mitotic sister chromatid segregation, negative regulation of the cell cycle process, and chromosome segregation. The DEGs of the 60-h group (Fig. 7) were found in small molecule catabolic process and fatty acids and significantly enriched in acid metabolic process, long-chain fatty acid metabolic process, cellular response to xenobiotic stimulus, and unsaturated fatty acid metabolic process. Among them, there was no intersection between the 24-h group and the 60 h group. More detailed data are shown in Table S2.
Fig. 6.
GO analysis of the DEGS from the 24-h group
Fig. 7.
GO analysis of the DEGS from the 60-h group
KEGG Pathway Analysis of DEGs
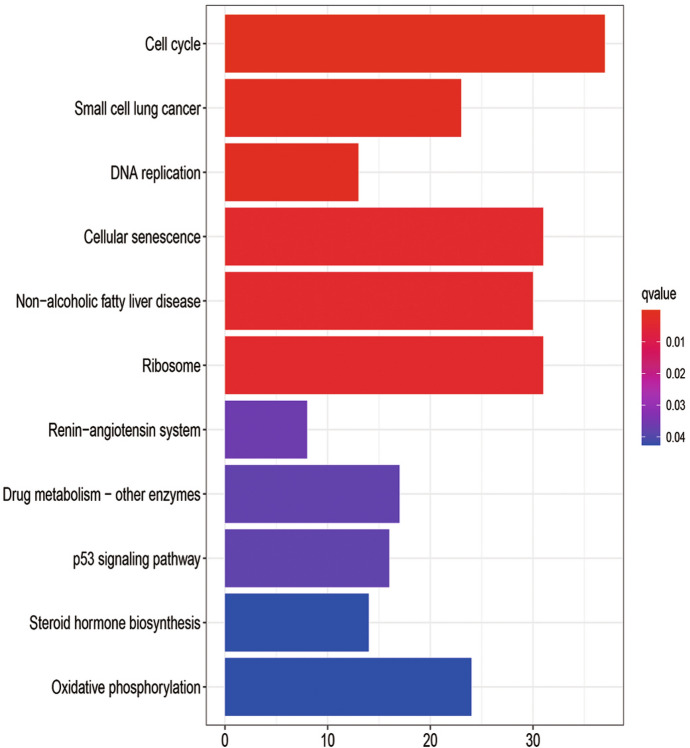
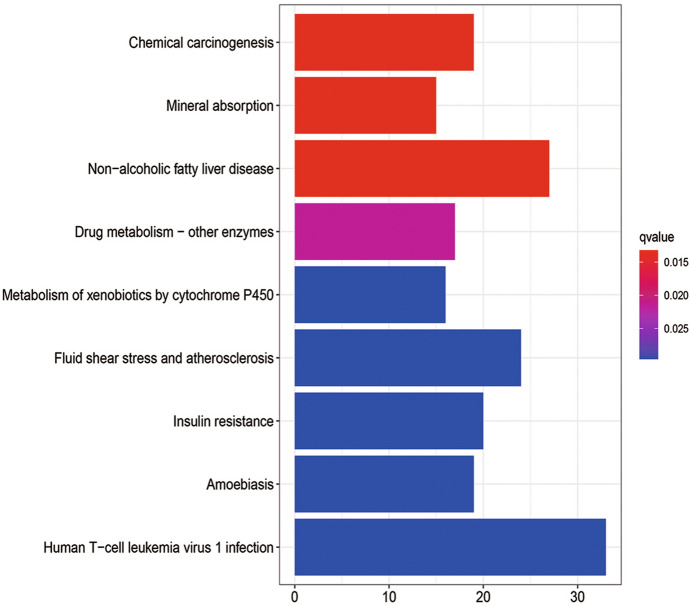
In order to explore the potential mechanism of these DEGs, KEGG pathway analysis was performed using the R software. The results of KEGG analysis showed that the DEGs of 24-h group (Fig. 8) were significantly enriched in cell cycle, small cell lung cancer, DNA replication, cellular senescence, and non-alcoholic fatty liver disease. The DEGs of the 60-h group (Fig. 9) were found in chemical carcinogenesis and mineral absorption and significantly enriched in non-alcoholic fatty liver disease, drug metabolism-other enzymes, and metabolism of xenobiotics by cytochrome P450. Among them, non-alcoholic fatty liver disease was a pathway co-expressed in the 24-h group and 60-h group. More detailed data are shown in Table S3.
Fig. 8.
KEGG pathway enrichment analysis of DEGs from the 24-h group
Fig. 9.
KEGG pathway enrichment analysis of DEGs from the 60-h group
Protein–Protein Interaction Network Analysis of DEGs
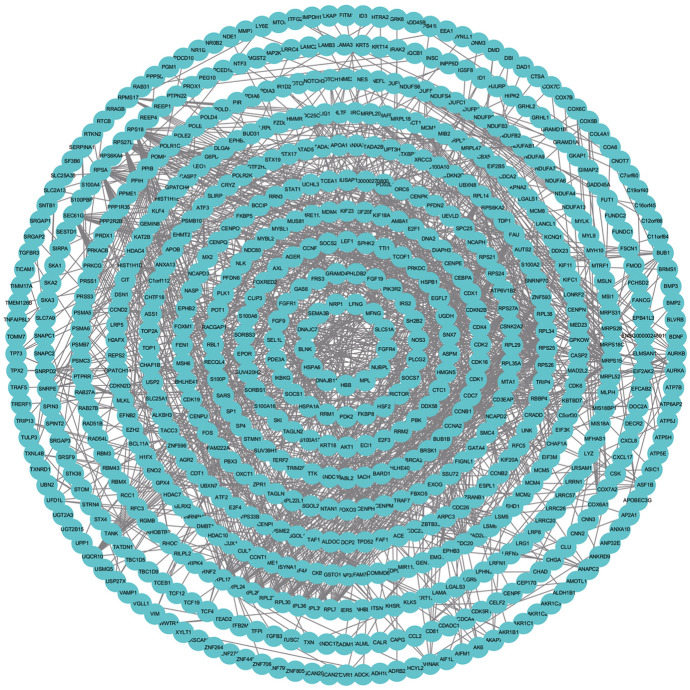
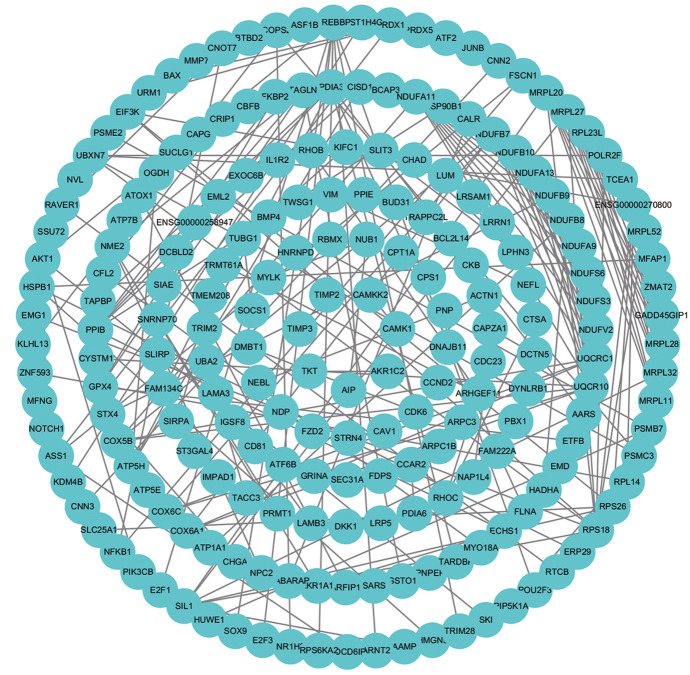
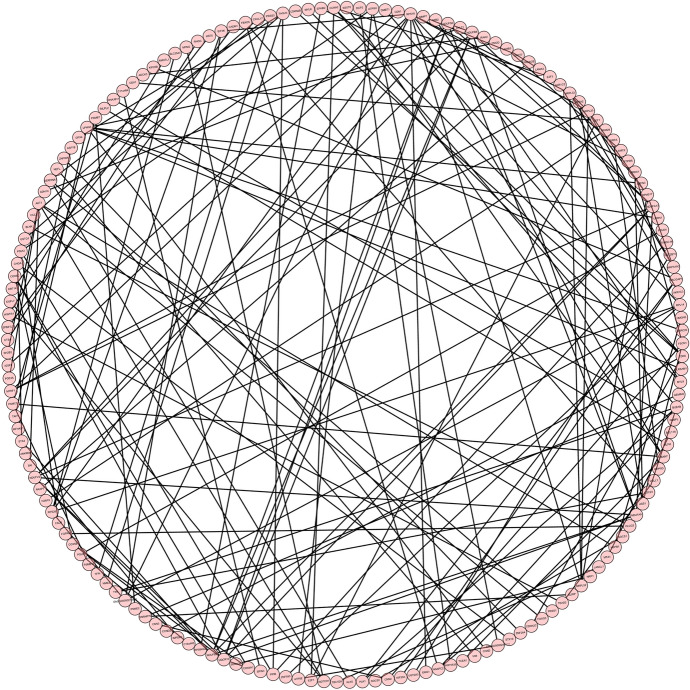
Based on the STRING database, we constructed PPI networks of the DEGs in the 24-h group, 60-h group, and intersection group. The PPI network of the 24-h group (Fig. 10) included 1778 nodes and 1457 edges, the PPI network of the 60-h group (Fig. 11) included 710 nodes and 1457 edges, and the PPI network of the intersection group (Fig. 12) included 692 nodes and 162 edges. The top twelve genes (Fig. 13) were defined as the hub genes, namely AKT1, TIMP1, SERPINA1, NOTCH1, CCNA2, RRM2, TTK, RACGAP1, BUB1B, H2AFX, KIF20A, and PLK1.
Fig. 10.
Protein–protein interaction (PPI) networks of the 24-h group
Fig. 11.
Protein–protein interaction (PPI) networks of the 60-h group
Fig. 12.
PPI network of the intersection between 24- and 60-h groups
Fig. 13.
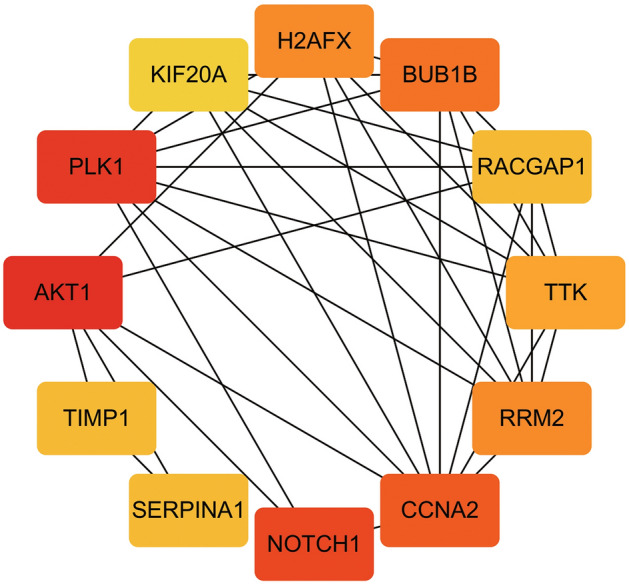
PPI network of the top twelve genes
Verification of the Hub Genes
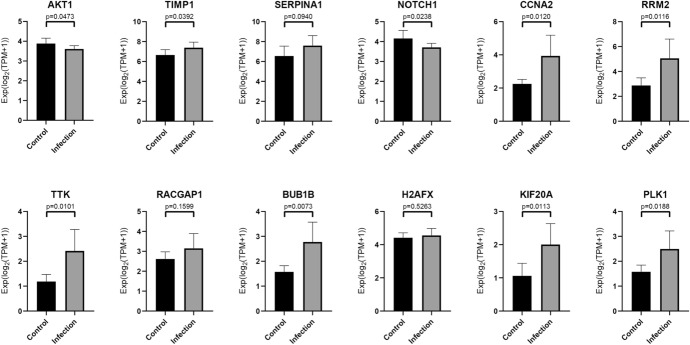
Through t tests and non-parametric tests, with the standard set as P < 0.05, nine statistically significant genes (Fig. 14) were finally obtained, including AKT1, TIMP1, NOTCH1, CCNA2, RRM2, TTK, BUB1B, KIF20A, and PLK1. These genes may play a pivotal role in the impact of SARS-CoV-2 on the human intestine.
Fig. 14.
t tests and non-parametric tests of hub genes
Discussion
Currently, many types of mutations have appeared in SARS-CoV-2, but vaccination has not yet been popularized as vaccines are still in clinical trials. In this study, we identified the DEGs between the SARS-CoV-2 and the normal samples. To understand these DEGs better, we conducted GO function and KEGG pathway analysis on them and constructed a PPI network to determine the hub genes.
Angiotensin-converting enzyme (ACE2) is thought to be the mechanism of SARS‐CoV‐2-infected cells. SARS-CoV-2 activates the intestinal ACE2 receptor, causing inflammation (intestinal inflammation) and eventually diarrhea (Villapol 2020). Existing studies have shown that the high expression of ACE2 is not limited to lung type II alveolar cells (AT2); they are also found in the gastrointestinal tract, especially in the absorptive intestinal epithelial cells of the ileum and colon (Hajifathalian et al. 2020), which provide a scientific basis for the detection of SARS-CoV-2 RNA in the stool samples of some patients, indicating that the digestive system is a potential route of COVID-19 infection (Cipriano et al. 2020). Moreover, in the results of the BP process screening in the 60-h GO functional annotation group, we found that the cells' response to heterologous biological stimuli was significantly enriched. In previous studies, we found that following exposure to the initial stimulus, innate immune cells will experience metabolic, mitochondrial, and epigenetic reprogramming, leading to an immune response with an enhanced memory phenotype (Geller and Yan 2020).
In the analysis of the KEGG pathway, we found that the significant pathways in the 24-h group and 60-h group had a cross-path: non-alcoholic fatty liver disease. Intestinal dysfunction can cause changes in intestinal microbes and increase inflammatory cytokines, leading to the aggravation of symptoms and even more serious complications (Villapol 2020). An increase in the rate of abnormal liver function has been observed in patients with severe COVID-19. The microbiota can aggravate NAFLD through certain mechanisms, including changing the permeability of the intestines and the energy absorbed by the diet (Safari and Gérard 2019), so NAFLD patients may also be more susceptible to the increased cytokine production associated with COVID-19 (Prins and Olinga 2020). Studies have suggested that patients with NAFLD may be particularly vulnerable to SARS-CoV-2 infection and complications resulting from COVID-19 (Portincasa et al. 2020).
In GO analysis and KEGG pathway analysis, DEGs were found to be enriched in cell cycle. Coronavirus N protein is located in the cytoplasm and is involved in virus replication and assembly. Previous reports have indicated that the expression of the coronavirus N protein may affect the cell cycle (Zuwała et al. 2015). Both SARS and COVID-19 are considered to be pandemic infectious diseases caused by coronaviruses, which show that cell cycle research can play a significant role in their prevention and control.
Among the hub genes, the expression of TIMP1, CCNA2, RRM2, TTK, BUB1B, PLK1, and KIF20A were up-regulated, while the expression of ATK1 and NOTCH1 were down-regulated.
AKT1 is one of the subtypes of AKT. The down-regulation of AKT1 expression will promote the M1 polarization of macrophages, and M1 macrophages will secrete high levels of cytokines that can cause inflammation (Arranz et al. 2012). TIMP1 is a member of the tissue inhibitor of the metalloproteinase family (TIMP). As the coronavirus infection progresses, the expression of TIMP will increase correspondingly, and it is clearly expressed in lymphocytes, macrophages, and eosinophils during inflammation (Zhou et al. 2005). It can induce colon cell carcinogenesis through the FAK-PI3K/AKT and MAPK pathways (Song et al. 2016). The ATK1 and TIMP1 genes have a certain stimulating effect on immune cells, such as macrophages, and trigger the immune mechanism of the immune system. This indicates that SARS-CoV-2 may cause severe inflammatory bowel disease, fever, diarrhea, and other symptoms after infecting the human body, eventually causing damage to the human gastrointestinal system. Studies have shown that the overexpression of RRM2 and KIF20A usually accompanies the occurrence of cancer and plays a malignant role in cancer (Kitab et al. 2019; Morikawa et al. 2010; Sheng et al. 2018; Xiong et al. 2019).
In the results of this study, the down-regulation of NOTCH1 expression and the up-regulation of RRM2 and KIF20A provide a feasible direction for subsequent SARS-CoV-2 research. CCNA2 is the cyclin A2 gene, which is the gene encoding cyclin A2 on human chromosome 4 (Pagano et al. 1992). When a virus infects a cell, its genetic information can activate CCNA2 and promote the cell cycle. In addition, the overexpression of CCNA2 can enhance the reproduction, metastasis, and invasive ability of cancer cells and is closely connected to the occurrence and deterioration of ovarian cancer, liver cancer, and esophageal squamous cell carcinoma (ESCC) (Ruan et al. 2017). TTK, also known as MPS1, is a protein kinase. The up-regulation of TTK can activate PKCa/ERK1/2 to promote the division of colon cancer cells and the lack of TTK can lead to apoptosis (Zhang et al. 2017, 2019; Kaistha et al. 2014). The protein expressed by BUB1B is the BUB1 mitotic checkpoint serine/threonine kinase β, which plays an important part in the inspection of the spindle during mitosis. Furthermore, the expression of BUB1B in colon cancer tissues is higher than in normal colon tissues (Burum-Auensen et al. 2007). PLK1 is a member of the Polo-like family of mammals. It is located on the centrosome during mitosis and is usually overexpressed in cancer cells (Malumbres and Barbacid 2007). Moreover, PLK1 is also highly expressed in certain kinds of cancer, such as esophageal cancer and gastric cancer (Takahashi et al. 2003; Song et al. 2018). The TTK, BUB1B, PLK1, and CCNA2 genes and their translation products all perform significant tasks in the normal cell division cycle. During SARS-CoV-2 infection, the up-regulation of these four genes not only provides favorable conditions for the spread of the virus but also hints at the possibility of chromosomal abnormalities and other genetic material damage in the host cells. This result corresponds to the aforementioned expression results of NOTCH1, RRM2, and KIF20A. Therefore, the results of this study can provide a certain direction and basis for subsequent researchers for exploring the relationship between SARS-CoV-2 and cancer.
In conclusion, the nine hub genes obtained through statistical tests in this experiment all play an indispensable role in cell growth, reproduction, and disease. Therefore, this study analyzed their differential expression during the SARS-CoV-2 infection process, aiming to understand the subsequent diseases that SARS-CoV-2 may induce so as to facilitate their timely prevention. However, this study has certain limitations. For instance, the sample size of the experimental data could be further expanded, and the intestinal organs could not perfectly simulate the human environment.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file1 (DOCX 34 kb)
Table S1: List of DGEs considering P < 0.05 and | log2 fold change (FC) | > 1
Supplementary file2 (DOCX 17 kb)
Table S2: Significantly enriched GO terms of DEGs
Supplementary file3 (DOCX 16 kb)
Table S3: Significantly enriched KEGG terms of DEGs
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Arranz A, Doxaki C, Vergadi E, Martinez de la Torre Y, Vaporidi K, Lagoudaki ED, Ieronymaki E, Androulidaki A, Venihaki M, Margioris AN, et al. Akt1 and Akt2 protein kinases differentially contribute to macrophage polarization. Proc Natl Acad Sci USA. 2012;109:9517–9522. doi: 10.1073/pnas.1119038109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burum-Auensen E, Deangelis PM, Schjølberg AR, Røislien J, Andersen SN, Clausen OP. Spindle proteins Aurora A and BUB1B, but not Mad2, are aberrantly expressed in dysplastic mucosa of patients with longstanding ulcerative colitis. J Clin Pathol. 2007;60:1403–1408. doi: 10.1136/jcp.2006.044305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J, Xing F, Liu J, Yip CC, Poon RW, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipriano M, Ruberti E, Giacalone A. Gastrointestinal infection could be new focus for coronavirus diagnosis. Cureus. 2020;12:e7422. doi: 10.7759/cureus.7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller A, Yan J. Could the induction of trained immunity by β-glucan serve as a defense against COVID-19? Front Immunol. 2020;11:1782. doi: 10.3389/fimmu.2020.01782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium The gene ontology (GO) project in 2006. Nucleic Acids Res. 2006;34:D322–326. doi: 10.1093/nar/gkj021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajifathalian K, Mahadev S, Schwartz RE, Shah S, Sampath K, Schnoll-Sussman F, Brown RS, Jr, Carr-Locke D, Cohen DE, Sharaiha RZ. SARS-COV-2 infection (coronavirus disease 2019) for the gastrointestinal consultant. World J Gastroenterol. 2020;26:1546–1553. doi: 10.3748/wjg.v26.i14.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C, Duan C, Zhang S, Spiegel B, Shi H, Wang W, Zhang L, Lin R, Liu J, Ding Z, Hou X. Digestive symptoms in COVID-19 patients with mild disease severity: clinical presentation, stool viral RNA testing, and outcomes. Am J Gastroenterol. 2020;115:916–923. doi: 10.14309/ajg.0000000000000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harapan H, Itoh N, Yufika A, Winardi W, Keam S, Te H, Megawati D, Hayati Z, Wagner AL, Mudatsir M. Coronavirus disease 2019 (COVID-19): a literature review. J Infect Public Health. 2020;13:667–673. doi: 10.1016/j.jiph.2020.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Tao H, Yan Y, Huang SY, Xiao Y. Molecular mechanism of evolution and human infection with SARS-CoV-2. Viruses. 2020;12:428. doi: 10.3390/v12040428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Lian JS, Hu JH, Gao J, Zheng L, Zhang YM, Hao SR, Jia HY, Cai H, Zhang XL, et al. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut. 2020;69:1002–1009. doi: 10.1136/gutjnl-2020-320926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaistha BP, Honstein T, Müller V, Bielak S, Sauer M, Kreider R, Fassan M, Scarpa A, Schmees C, Volkmer H, et al. Key role of dual specificity kinase TTK in proliferation and survival of pancreatic cancer cells. Br J Cancer. 2014;111:1780–1787. doi: 10.1038/bjc.2014.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitab B, Satoh M, Ohmori Y, Munakata T, Sudoh M, Kohara M, Tsukiyama-Kohara K. Ribonucleotide reductase M2 promotes RNA replication of hepatitis C virus by protecting NS5B protein from hPLIC1-dependent proteasomal degradation. J Biol Chem. 2019;294:5759–5773. doi: 10.1074/jbc.RA118.004397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers MM, Beumer J, van der Vaart J, Knoops K, Puschhof J, Breugem TI, Ravelli RBG, Paulvan Schayck J, Mykytyn AZ, Duimel HQ, van Donselaar E, Riesebosch S, Kuijpers HJH, Schipper D, Van de Wetering WJ, Degraaf M, Koopmans M, Cuppen E, Peters PJ, Haagmans BL, Clevers H. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369(6499):50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LQ, Huang T, Wang YQ, Wang ZP, Liang Y, Huang TB, Zhang HY, Sun W, Wang Y. COVID-19 patients' clinical characteristics, discharge rate, and fatality rate of meta-analysis. J Med Virol. 2020;92:577–583. doi: 10.1002/jmv.25757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang B, Li C, Zhao J. Identification of key pathways and genes in colorectal cancer using bioinformatics analysis. Med Oncol. 2016;33:111. doi: 10.1007/s12032-016-0829-6. [DOI] [PubMed] [Google Scholar]
- Malumbres M, Barbacid M. Cell cycle kinases in cancer. Curr Opin Genet Dev. 2007;17:60–65. doi: 10.1016/j.gde.2006.12.008. [DOI] [PubMed] [Google Scholar]
- Morikawa T, Hino R, Uozaki H, Maeda D, Ushiku T, Shinozaki A, Sakatani T, Fukayama M. Expression of ribonucleotide reductase M2 subunit in gastric cancer and effects of RRM2 inhibition in vitro. Hum Pathol. 2010;41:1742–1748. doi: 10.1016/j.humpath.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Ostaszewski M, Mazein A, Gillespie ME, Kuperstein I, Niarakis A, Hermjakob H, Pico AR, Willighagen EL, Evelo CT, Hasenauer J, Schreiber F, Dräger A, Demir E, Wolkenhauer O, Furlong LI, Barillot E, Dopazo J, Orta-Resendiz A, Messina F, Valencia A, Funahashi A, Kitano H, Auffray C, Balling R, Schneider R. COVID-19 Disease Map, building a computational repository of SARS-CoV-2 virus-host interaction mechanisms. Sci Data. 2020;7(1):136. doi: 10.1038/s41597-020-0477-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano M, Pepperkok R, Verde F, Ansorge W, Draetta G. Cyclin A is required at two points in the human cell cycle. EMBO J. 1992;11:961–971. doi: 10.1002/j.1460-2075.1992.tb05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portincasa P, Krawczyk M, Smyk W, Lammert F, Di Ciaula A. COVID-19 and non-alcoholic fatty liver disease: two intersecting pandemics. Eur J Clin Invest. 2020;50:e13338. doi: 10.1111/eci.13338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins GH, Olinga P. Potential implications of COVID-19 in non-alcoholic fatty liver disease. Liver Int. 2020;40(10):2568. doi: 10.1111/liv.14484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan JS, Zhou H, Yang L, Wang L, Jiang ZS, Wang SM. CCNA2 facilitates epithelial-to-mesenchymal transition via the integrin αvβ3 signaling in NSCLC. Int J Clin Exp Pathol. 2017;10:8324–8333. [PMC free article] [PubMed] [Google Scholar]
- Safari Z, Gérard P. The links between the gut microbiome and non-alcoholic fatty liver disease (NAFLD) Cell Mol Life Sci. 2019;76:1541–1558. doi: 10.1007/s00018-019-03011-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Y, Wang W, Hong B, Jiang X, Sun R, Yan Q, Zhang S, Lu M, Wang S, Zhang Z, et al. Upregulation of KIF20A correlates with poor prognosis in gastric cancer. Cancer Manag Res. 2018;10:6205–6216. doi: 10.2147/CMAR.S176147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G, Xu S, Zhang H, Wang Y, Xiao C, Jiang T, Wu L, Zhang T, Sun X, Zhong L, et al. TIMP1 is a prognostic marker for the progression and metastasis of colon cancer through FAK-PI3K/AKT and MAPK pathway. J Exp Clin Cancer Res. 2016;35:148. doi: 10.1186/s13046-016-0427-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song R, Hou G, Yang J, Yuan J, Wang C, Chai T, Liu Z. Effects of PLK1 on proliferation, invasion and metastasis of gastric cancer cells through epithelial-mesenchymal transition. Oncol Lett. 2018;16:5739–5744. doi: 10.3892/ol.2018.9406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Sano B, Nagata T, Kato H, Sugiyama Y, Kunieda K, Kimura M, Okano Y, Saji S. Polo-like kinase 1 (PLK1) is overexpressed in primary colorectal cancers. Cancer Sci. 2003;94:148–152. doi: 10.1111/j.1349-7006.2003.tb01411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villapol S. Gastrointestinal symptoms associated with COVID-19: impact on the gut microbiome. Transl Res. 2020;226:57–69. doi: 10.1016/j.trsl.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang Y, Chen Y, Qin Q. Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID-19) implicate special control measures. J Med Virol. 2020;92:568–576. doi: 10.1002/jmv.25748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S, Jiang X, Zhang J, Xie S, Hua Y, Wang R, Yang Y. Identification of significant gene and pathways involved in HBV-related hepatocellular carcinoma by bioinformatics analysis. PeerJ. 2019;30(7):e7408. doi: 10.7717/peerj.7408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong M, Zhuang K, Luo Y, Lai Q, Luo X, Fang Y, Zhang Y, Li A, Liu S. KIF20A promotes cellular malignant behavior and enhances resistance to chemotherapy in colorectal cancer through regulation of the JAK/STAT3 signaling pathway. Aging (albany NY) 2019;11:11905–11921. doi: 10.18632/aging.102505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XL, Han WX, Ma YM, Baohan HW, Zhao XH, Wang HY. Expression of tyrosine and threonine protein kinase in carcinogenic process of colorectal cancer and its relationship with prognosis. Zhonghua Zhong Liu Za Zhi. 2017;39:172–177. doi: 10.3760/cma.j.issn.0253-3766.2017.03.003. [DOI] [PubMed] [Google Scholar]
- Zhang L, Jiang B, Zhu N, Tao M, Jun Y, Chen X, Wang Q, Luo C. Mitotic checkpoint kinase Mps1/TTK predicts prognosis of colon cancer patients and regulates tumor proliferation and differentiation via PKCα/ERK1/2 and PI3K/Akt pathway. Med Oncol. 2019;37:5. doi: 10.1007/s12032-019-1320-y. [DOI] [PubMed] [Google Scholar]
- Zhou J, Marten NW, Bergmann CC, Macklin WB, Hinton DR, Stohlman SA. Expression of matrix metalloproteinases and their tissue inhibitor during viral encephalitis. J Virol. 2005;79:4764–4773. doi: 10.1128/JVI.79.8.4764-4773.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuwała K, Golda A, Kabala W, Burmistrz M, Zdzalik M, Nowak P, Kedracka-Krok S, Zarebski M, Dobrucki J, Florek D, et al. The nucleocapsid protein of human coronavirus NL63. PLoS ONE. 2015;10:e0117833. doi: 10.1371/journal.pone.0117833. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file1 (DOCX 34 kb)
Table S1: List of DGEs considering P < 0.05 and | log2 fold change (FC) | > 1
Supplementary file2 (DOCX 17 kb)
Table S2: Significantly enriched GO terms of DEGs
Supplementary file3 (DOCX 16 kb)
Table S3: Significantly enriched KEGG terms of DEGs