Abstract
Purpose
To extend the double echo steady‐state (DESS) sequence to enable chemical‐shift corrected water‐fat separation.
Methods
This study proposes multiple‐echo steady‐state (MESS), a sequence that modifies the readouts of the DESS sequence to acquire two echoes each with bipolar readout gradients with higher readout bandwidth. This enables water‐fat separation and eliminates the need for water‐selective excitation that is often used in combination with DESS, without increasing scan time. An iterative fitting approach was used to perform joint chemical‐shift corrected water‐fat separation and T2 estimation on all four MESS echoes simultaneously. MESS and water‐selective DESS images were acquired for five volunteers, and were compared qualitatively as well as quantitatively on cartilage T2 and thickness measurements. Signal‐to‐noise ratio (SNR) and T2 quantification were evaluated numerically using pseudo‐replications of the acquisition.
Results
The water‐fat separation provided by MESS was robust and with quality comparable to water‐selective DESS. MESS T2 estimation was similar to DESS, albeit with slightly higher variability. Noise analysis showed that SNR in MESS was comparable to DESS on average, but did exhibit local variations caused by uncertainty in the water‐fat separation.
Conclusion
In the same acquisition time as DESS, MESS provides water‐fat separation with comparable SNR in the reconstructed water and fat images. By providing additional image contrasts in addition to the water‐selective DESS images, MESS provides a promising alternative to DESS.
Keywords: DESS, MR value, multi‐echo, T2 quantification, water‐fat separation
1. INTRODUCTION
The double echo steady‐state (DESS) sequence is a steady‐state free‐precession (SSFP) sequence that acquires two echoes with substantially different contrasts. 1 , 2 , 3 DESS offers high resolution 3D imaging with high scan efficiency, flexibility with respect to the acquired contrasts, and basic T2 estimation. 4 , 5 These properties align well with the recent trend towards high‐value and more quantitative MR imaging. 6
In musculoskeletal imaging research there has been a growing interest in DESS for imaging osteoarthritis progression. Notably, DESS was included in the Osteoarthritis Initiative (OAI) cohort. 7 Examples of usage of DESS include studies of the T2 of cartilage, 4 , 5 , 8 , 9 morphological analysis 10 , 11 , 12 , 13 and segmentation of cartilage. 14 , 15 , 16 , 17 More recently, Chaudhari et al proposed and evaluated a 5‐min DESS sequence for efficient clinical knee examinations. 18 Outside of musculoskeletal imaging, potential applications of DESS include diffusion‐weighted imaging, 8 , 19 , 20 , 21 and T2 mapping in the prostate. 22
Particularly in the knee, DESS is often used in combination with water‐selective radiofrequency (RF) excitation to obtain better contrast on the cartilage. These binomial RF pulses split the excitation into multiple RF pulses, which are timed to coincide with the in‐phase and opposed‐phase effect of water and fat to prevent fat signal from being excited. 23 This increases the duration of the excitation, particularly at lower field strengths, and makes it more specific absorption rate (SAR) intensive.
In this study, we propose the multiple‐echo steady‐state (MESS) sequence, an extension of DESS. For every DESS echo, MESS measures two echoes with higher readout bandwidth and bipolar gradients, that allow Dixon water‐fat separation, 24 without increasing total scan time. Similarly, Chaudhari et al proposed water‐fat separation for ultra‐short echo‐time DESS, 25 which used repeated acquisitions with an echo‐time shift and, therefore, does increase scan time. Water‐fat separation removes the need for water‐selective excitation in applications such as cartilage imaging, which increases scan efficiency and lowers SAR requirements. By preserving and separating the fat signal, anatomical information is preserved that would otherwise need additional scans to obtain. Furthermore, water‐fat separation allows additional contrasts to be reconstructed, such as in‐phase and opposed‐phase images.
We evaluated a 3D MESS acquisition of the knee at 3T, with a 5‐min water‐selective 3D DESS acquisition as a baseline. We implemented an iterative fitting method for joint chemical‐shift corrected water‐fat separation and T2 quantification on the MESS images. The reconstructed images and parameters from MESS were compared qualitatively and quantitatively with the corresponding DESS images. Pseudo‐replications of the acquisitions were used to assess noise sensitivity and signal‐to‐noise ratio (SNR).
2. METHODS
2.1. From DESS to MESS
The DESS sequence is an SSFP acquisition that samples two echoes. The first echo measures an free induction decay (FID) ‐like signal, referred to as S+, which resembles the T1/‐weighted signal from a conventional gradient echo acquisition. The second echo measures an echo‐like signal, referred to as S−, which has a longer effective echo time and therefore has additional T2 weighting. A DESS acquisition typically uses a low readout bandwidth to achieve maximal SNR with a relatively long repetition time.
The MESS sequence replaces each DESS readout by two or more readouts with alternating readout polarity and higher readout bandwidth, which does not increase total scan time. TEs were chosen to be suitable for two‐point Dixon water‐fat separation. 24 Figure 1 shows the extension from DESS to MESS for a four‐echo MESS sequence.
FIGURE 1.
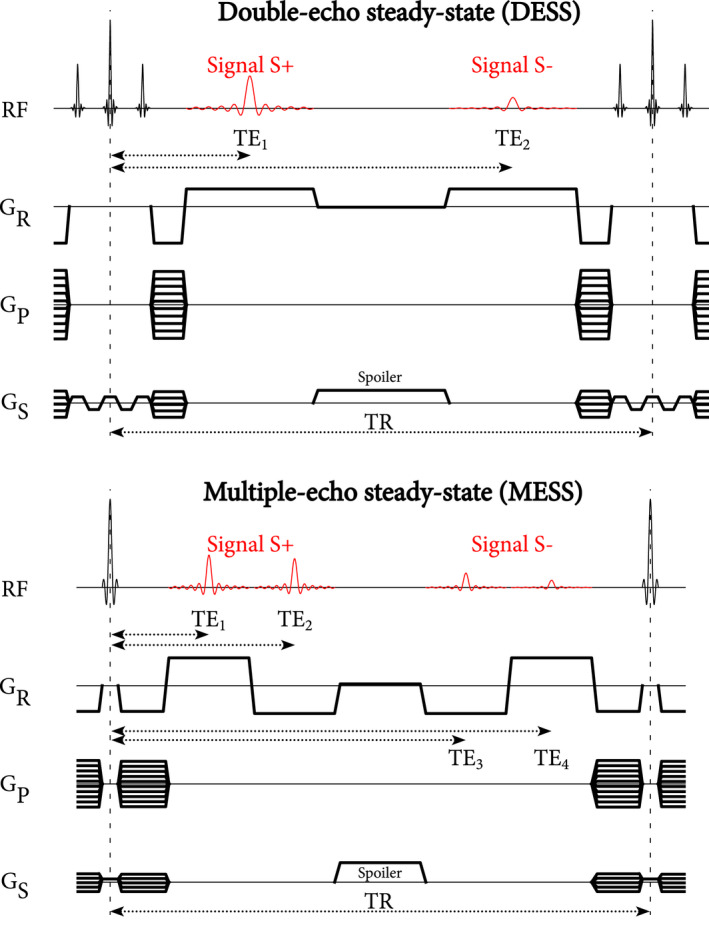
Pulse sequence diagrams for DESS (top) and MESS (bottom). The S+ signal is comparable to an FID signal, whereas the S− is more T2‐weighted and originates from spin echo and stimulated echo pathways. In the same TR, MESS acquires two additional echoes with bipolar readouts. DESS uses a chemically selective binomial RF pulse, whereas MESS uses a standard RF pulse. In both sequences a spoiler gradient is included in the slice direction. During the spoiler, a small gradient is active in the readout direction to compensate for the area of the ramp‐down and ramp‐up of the readouts. Note that, because we implemented DESS and MESS as modified three‐echo and five‐echo GRE sequences with fixed echo spacing, the spoiler duration was 1.56 ms longer in DESS
2.2. Parameter mapping
We used a non‐linear optimization procedure to obtain various parameters from the measured MESS signals. The MESS signal was parametrized based on the flexible two‐point Dixon method, 24 which yields water (), fat (), and () maps, with the additional consideration of T2 decay (expressed as for numerical stability) and possible phase inconsistencies between S+ and S− ( and ).The signal model for the S+ and S− signals is described in Appendix A.
Due to eddy currents in the readout gradients, linear phase errors may occur between the echoes (or equivalently, k‐space shifts), 26 which would prevent a consistent fit using all echoes. To ensure consistent phase behavior, we first performed a linear phase correction along the readout direction, described in Appendix A.
To initialize , we applied the flexible two‐point Dixon method 24 on the S+ echoes to obtain field phasor candidates. The projected power method was used to resolve the field phasor candidates into a smooth phase map . 27 The constant phase was then calculated for the S+ echoes using as per Berglund et al. 24 Similarly, was calculated for the S− echoes.
We first performed a non‐linear least squares fit (Equation 1) with regularization on smoothness of the field inhomogeneity () to refine the ‐parameters using all 4 echoes, fitting . At this stage, water and fat were restricted to real values to provide a stable fit of the ‐parameters. After this initial fit, a second optimization was performed over complex‐valued water and fat, but with the ‐parameters fixed (), which improved separation in voxels affected by partial volume effects.
| (1) |
Here, is the signal model and the measured data for the n’th echo, denotes the fast Fourier transform in the readout dimension, where is the coordinate, and is an phase smoothness term in the readout and phase encoding directions (with ):
| (2) |
In both fits we included regularization on selected frequencies of the water and fat parameters to suppress noise amplification arising from chemical‐shift corrected water‐fat separation in k‐space. 28 The regularized k‐space frequencies () were selected based on the noise amplification factor (NA) as defined by Lu et al 28 with for and for .
The non‐linear fits were implemented in Python 3.8 using the Pytorch 1.8 library for automatic differentiation and graphics processing unit (GPU) acceleration on an NVIDIA RTX 2080 Super GPU. An RMSprop optimizer was used with learning rate 0.01. Each fit was run for 200 iterations, which we empirically established to yield fast and acceptable convergence. The parameter mapping took approximately 218 s/scan.
2.3. Experimental setup
Our baseline acquisition was a 5‐min 3D DESS sequence with a resolution of mm (reconstructed to ) and a field of view of mm. Other scan parameters included: TE1/TE2/TR 4.9/14.3/19.2 ms, flip angle 20°, SENSE, bandwidth 228 Hz/pixel, scan duration 304 s. Here, the TEs are measured from the most recent RF pulse. Water excitation with a 1‐2‐1 binomial RF pulse was used with a duration of ~1.7 ms.
The parameters of the 4‐echo 3D MESS acquisition were matched to the DESS acquisition, except for the following: TE1/TE2/TE3/TE4/TR 3.5/6.5/12.6/15.6/19.1 ms, bandwidth 362 Hz/pixel, scan duration 303 s. The TEs were chosen to provide high scan efficiency and such that water‐fat separation could be applied. A slab‐selective RF pulse was used with a duration of 0.56 ms.
We acquired DESS and MESS images of the knee for five healthy volunteers (mean age 31 y, four male, one female), in accordance with institutional guidelines and with approval of the University Medical Center Utrecht medical ethics committee (NL53099.041.15). All acquisitions were at a field strength of 3T (Philips Ingenia, Best, The Netherlands) using a 16‐channel knee T/R coil. For each volunteer, the femoral and tibial cartilage were manually segmented on both the DESS and MESS images, to measure the mean T2 and thickness of cartilage.
T2 mapping for the DESS sequence was performed as described by Sveinsson et al, 5 which corrects for a reference T1 (articular cartilage, 1200 ms) and the known flip angle.
2.4. Analysis of noise behavior
We numerically assessed the noise behavior in our fitting procedure using the pseudo multiple replica method. 29 In short, additional noise was added to the acquired data with , with muscle tissue intensity normalized to 0.5 (ie, ). On 10 replications of this noisy data, we repeated our fitting procedure and measured the variance of the fitted parameters, which can be attributed to the additional noise.
The same analysis was used for DESS T2 fitting, but with , accounting for the difference in readout bandwidth and with lowered by 16.4% to compensate for suboptimal DESS spoiler gradient duration (Figure 1), ie, using the theoretically achievable SNR for DESS.
We constructed SNR maps for water and fat in MESS and the S+ signal in DESS, with where is the mean and is the SD of the parameter over the 10 replicates. We also calculated the distribution of T2 values in water for the pseudo‐replicates in reference to the T2 fitted to the signal without additional noise.
3. RESULTS
Figure 2A shows the acquired MESS images for one of the subjects. The first set of two echoes (S+) of the MESS sequence display the mixed T1/‐weighted contrast, with in‐phase and opposed‐phase effects in water and fat. The second set of echoes (S−) show increased T2 contrast in the cartilage and synovial fluid. As expected from an acquisition with bipolar readouts, the fat signal showed opposing chemical‐shift displacements in the readout direction (top‐bottom).
FIGURE 2.

A, Acquired 4‐echo MESS images, showing the FID‐like echoes (S+) and echo‐like echoes (S−). Water and fat in echo 1 (TE = 3.5 ms) and echo 4 (TE = 15.6 ms, effective TE = 34.7 ms) are mostly opposed‐phase, while for echo 2 (TE = 6.5 ms) and echo 3 (TE = 12.6 ms, effective TE = 31.7 ms) they are mostly in‐phase. B, Parameter maps reconstructed from the MESS images: water and fat (magnitude and phase), , phase for S+ () and S− (), and magnetic field inhomogeneity (b). Note that the phase parameters wrap around outside the range
The parameters reconstructed from the MESS images are shown in Figure 2B. The reconstructions show a good separation of water and fat with no water‐fat swaps. We observed that the R2 parameter was more affected by artifacts in fat than in water. The field parameters (b, , ) show a well‐defined background field, with smoothness only imposed on the b parameter.
Figure 3A compares water‐selective DESS images with the corresponding MESS water and T2 images. Overall, we observed similar contrasts and resolution in the DESS and MESS images, indicating that the chemical‐shift‐corrected water‐fat separation performed well. The water‐selective RF pulse in DESS provided slightly better fat suppression in the bone marrow, although the DESS S− images showed artifacts not present in MESS, which may have been caused by motion and/or the RF pulse. The zoomed region showed a minor loss in resolution for MESS in some structures perpendicular to the readout, ie, those most affected by chemical‐shift displacement.
FIGURE 3.

A, Comparison of MESS and water‐selective DESS water images and T2 maps in water‐dominant voxels. MESS water images were evaluated at the DESS echo times using the fitted water and parameters. A zoomed region is shown for the S+ images, indicated by a white square in the MESS S+ image. Segmentations of the femoral (red outline) and tibial (green outline) cartilage are shown in the DESS and MESS S+ images. B, Additional contrasts reconstructed from the MESS parameters: in‐phase and opposed‐phase images, and the T2 map in fat‐dominant voxels
Figure 3B shows in‐phase and opposed‐phase images, which are commonly calculated for Dixon sequences. Because these images were calculated after chemical‐shift correction, for example the cortical bone thickness should be more accurately represented than in the source images. Although the T2 of fat regions can be estimated with MESS, our results showed an estimate that was susceptible to noise and artifacts.
Additional water‐fat separated images for all volunteers are available as Supporting Information Figure S1, which is available online. These results show overall robust and high quality water‐fat separation, consistent with Figure 3A. Only two minor water‐fat swaps were visible in the posterior side of the knee for subjects 2 and 3. We verified that the results in the shown slices are representative for all other slices.
The mean cartilage T2 and thickness values for the femoral and tibial cartilage are shown in Table 1. On average, MESS showed a slight overestimation (~9%) of T2 relative to DESS and an increase in variability for the femoral cartilage. The mean cartilage thickness quantification showed marginal differences (<5%) between DESS and MESS quantification.
TABLE 1.
Mean T2 and thickness quantification for the femoral and tibial cartilage for each subject and over the cartilage of all subjects (mean ± SD)
| Subject | Sequence | T2 (ms) | Thickness (mm) | ||
|---|---|---|---|---|---|
| Femoral | Tibial | Femoral | Tibial | ||
| 1 | DESS | 27.9 ± 9.1 | 22.4 ± 12.4 | 1.68 | 2.16 |
| MESS | 29.3 ± 10.2 | 23.1 ± 9.7 | 1.86 | 2.19 | |
| 2 | DESS | 29.4 ± 10.2 | 23.0 ± 10.4 | 1.78 | 1.92 |
| MESS | 33.9 ± 13.3 | 30.5 ± 13.9 | 2.06 | 1.91 | |
| 3 | DESS | 26.9 ± 8.6 | 22.6 ± 11.3 | 1.99 | 2.08 |
| MESS | 28.7 ± 10.6 | 25.7 ± 11.7 | 2.12 | 2.26 | |
| 4 | DESS | 26.1 ± 9.4 | 24.9 ± 12.3 | 2.07 | 2.05 |
| MESS | 28.3 ± 9.8 | 25.7 ± 12.5 | 2.03 | 1.92 | |
| 5 | DESS | 24.7 ± 8.4 | 22.4 ± 10.8 | 1.90 | 2.11 |
| MESS | 25.9 ± 10.3 | 22.6 ± 9.7 | 1.86 | 2.08 | |
| Overall | DESS | 26.8 ± 9.2 | 23.0 ± 11.6 | 1.89 | 2.06 |
| MESS | 29.0 ± 11.1 | 25.3 ± 11.8 | 1.99 | 2.07 | |
The results of the noise analysis are shown in Figure 4. Figure 4A shows the noise levels (relative to DESS) and SNR maps for DESS and MESS. Noise levels and SNR for all subjects are available in Supporting Information Table S1. Qualitatively, Figure 4A shows spatially varying noise behavior in MESS, showing piece‐wise homogeneous regions and some higher variance in regions where the water‐fat separation was uncertain. In bone marrow, we observed a lower noise level for the MESS water image in general, but some uncertain regions showed as spurious higher intensity voxels in the water map (Figure 2). The SNR maps showed very similar patterns for both DESS and the MESS water image, indicating that the noise elevation primarily occurred in low‐intensity regions. The SNR in fat showed more heterogeneity. Quantitatively, the average noise levels in water were ~5% higher in MESS than in DESS, but the average SNR of water was ~2% higher for MESS. The MESS fat images showed substantially higher noise levels (~53%), but simultaneously, SNR in fat was higher than in water (~15%), due to the higher intensity of fat.
FIGURE 4.
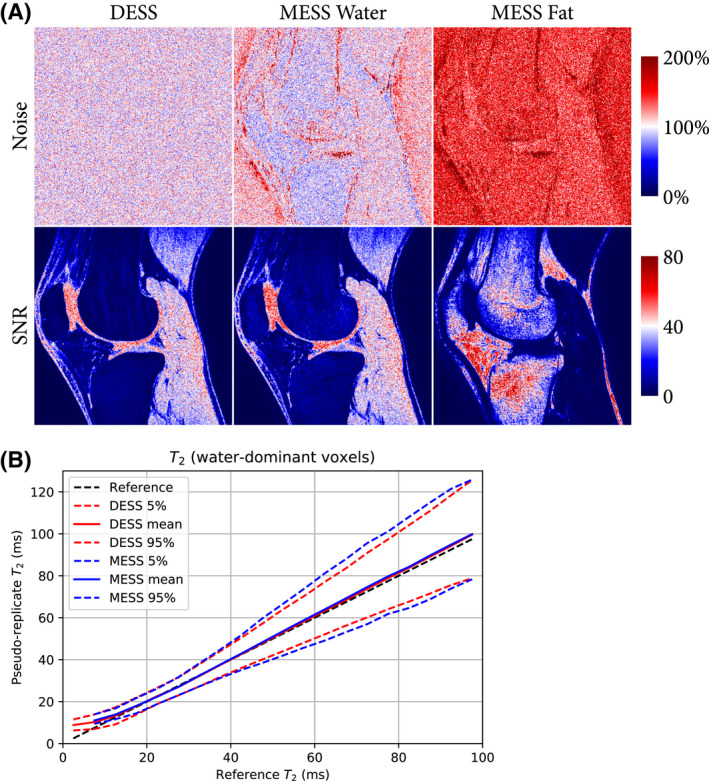
Results of the noise analysis of the pseudo‐replicates. A, Noise levels and SNR maps for DESS and MESS water and fat. The noise was normalized such that the mean noise in DESS equals 100% (ie, showing relative noise levels). Quantification of noise levels and SNR for all subjects is available in Supporting Information Table S1. B, Distribution of the quantification in the pseudo‐replicates, in reference to the fitted to the images without additional noise. The distribution is shown as mean and 5th and 95th percentiles of the values over 10 replications and binned into 20 bins (5 ms range) over the 0‐100 ms range
Figure 4B shows mean values and 5th and 95th percentiles for the T2 in the pseudo‐replicates, in reference to the T2 of the fit without additional noise. This showed negligible noise‐related bias and a slight increase in variability for MESS for ms, consistent with Table 1.
4. DISCUSSION
In this study, we have presented the MESS sequence, a multi‐echo extension of the DESS sequence that includes water‐fat separation in addition to the T2 quantification offered by DESS, without increasing acquisition time. Because of the water‐fat separation, the MESS acquisition does not require water‐selective excitation and, therefore, provides both mixed T1/‐ and T2‐weighted non‐fat‐suppressed images. Through an iterative fitting procedure, water and fat were separated with chemical‐shift correction, and T2 maps and various B0‐related parameters were reconstructed.
The MESS water images reconstructed robustly and with good suppression of fat signal. These images showed good similarity with acquired DESS images. Both the water and fat images produced by MESS showed high resolution details, demonstrating the success and stability of the chemical‐shift correction in the fitting procedure, correcting for a relatively large water‐fat‐shift of 1.3 voxels (ie, a 2.6 voxel shift between two bipolar echoes). Some minor details perpendicular to the readout direction were lost in MESS. Quantitatively, MESS T2 estimation in water was close to the values provided by DESS, showing a minor overestimation and slightly more variance. T2 estimation in fat showed relatively poor results. In both DESS and MESS, T2 quantification is confounded by B1, T1, and diffusion effects. 9 , 25 In MESS, this could influence the water‐fat separation, although we did not observe this happening in this study.
We kept the MESS signal model relatively simple for this proof‐of‐concept study, although fitting of other parameters could be investigated. Notably, T2 could be separately estimated for water and fat, and an estimate of T2 ’ might be possible. Another important limitation of this proof‐of‐concept study is the inclusion of only healthy volunteers. It remains to be investigated whether MESS also performs well at visualizing and quantifying clinically relevant pathological image features.
Assuming identical noise characteristics, the SNR of the individual MESS echoes was a factor lower than the DESS echoes. Note that due to implementation limitations, our DESS SNR was ~8% less than theoretically possible (Figure 1). However, our noise analysis showed that the fitting procedure regained SNR in the reconstructed MESS water and fat images on average. It should be noted that the frequency regularization does affect SNR measurements, where the regularization strength presents a tradeoff between noise amplification, apparent sharpness of the images, and artifacts. In this study, we chose the regularization to achieve fewer artifacts and lower noise amplification at the cost of a slight loss in sharpness, but this should be evaluated per application. Supporting Information Figure S2 illustrates the differences when substantially less regularization is used. Despite regularization, local variations in noise levels did occur in MESS, which could present as artifacts. However, it is worth noting the DESS acquisitions also showed imaging artifacts.
In principle, any DESS sequence with sufficiently long TR can be modified into a MESS sequence without increasing scan time. More flexibility and better SNR would be obtained if MESS sequence parameters are chosen freely (eg, achieving NSA=2 in two‐point Dixon 26 ), rather than mimicking a DESS sequence. The basic principles of MESS can be extended to different echo configurations. For example, a six‐echo MESS acquisition could improve the robustness of water‐fat separation and reduce artifacts from eddy currents. 26 Another alternative is to acquire echoes asymmetrically, eg, three S+ echoes and one S− echo, which could yield additional information on and/or susceptibility effects. MESS could also be used to add water‐fat separation to other multi‐pathway SSFP approaches. 30 , 31
An important benefit of MESS is the absence of spectrally selective RF excitation. First, this saves a modest amount of time compared to water‐selective DESS, around 1.1 ms per TR in this study, and reduces SAR requirements of the RF pulse. Because the duration of the binomial pulse is inversely proportional to the resonance frequency of fat, this time‐benefit would be greater at lower field strengths. Second, RF‐based selective excitation is reported to be more sensitive to field inhomogeneity than Dixon techniques. 32 Finally, conventional RF pulses are less restrictive when acquiring thinner slices.
Both DESS and Dixon imaging have been shown to be useful in musculoskeletal imaging. 11 , 18 , 33 , 34 The combined water, fat, and T2 reconstructions of the MESS sequence could have similar potential. Recent deep learning‐based techniques have shown promising results on both DESS images and Dixon images, for example in cartilage and bone segmentation 16 , 17 , 35 and synthetic CT generation. 36 Such morphological analysis may benefit from the additional contrasts provided by MESS. These benefits could translate outside of musculoskeletal imaging. For example, efficient, co‐registered, multi‐parametric imaging and synthetic CT generation would be well‐suited for MR‐only radiotherapy treatment planning, enabling both organ delineation and dosimetry. 37
With the MESS acquisition, water‐fat separation can be gained from acquiring additional echoes in the same acquisition time as DESS, with relatively minor loss in image quality. MESS efficiently provides additional image contrasts that are lost in a water‐selective DESS sequence. As such, MESS offers a promising alternative in potential applications of DESS.
CONFLICT OF INTEREST
Peter R. Seevinck declares to be a minority shareholder of MRIGuidance BV.
Supporting information
FIGURE S1 MESS water‐fat separation for all 5 subjects (top and middle row), with water‐selective DESS (bottom row) as a reference for the MESS water image. In subjects 2 and 3 a minor water‐fat swap is visible in the posterior side of the knee, indicated by arrows. Subject 3 has two titanium screws (one visible), which showed similar artifacts for the DESS and MESS images
FIGURE S2 The effects of reduced regularization on the water‐fat separation. Reduced regularization () is shown on the left, and the regularization used in this study () on the right. Arrows indicate how the regularization in this study relates to the reduced regularization in terms of sharpness (red arrows), noise amplification (green arrows), artifacts (white arrows)
TABLE S1 A, Noise statistics for DESS and MESS W and F contrasts for all subjects. Noise is shown overall, and for water‐dominant voxels (W>F) and fat‐dominant voxels (F>W) with sufficient signal (W+F>0.2, with intensities normalized to 0.5 for water in muscle). B, SNR statistics for DESS and MESS W and F contrasts for all subjects. SNR is shown overall, and for the region masked for the relevant contrast, water‐dominant voxels for DESS and MESS W, and fat‐dominant voxels for MESS F
ACKNOWLEDGMENTS
This work is part of the research programme Applied and Engineering Sciences (TTW) with project number 15479, which is (partly) financed by the Netherlands Organization for Scientific Research (NWO).
APPENDIX A.
SIGNAL MODEL
We parameterized the signal with the following parameter maps: complex water and fat maps ( and ), R2 map (), and main magnetic field inhomogeneity, expressed as phase maps , and .
The base signal equations for the water () and fat () signals at echo were defined as follows:
| (A1) |
The chemical‐shift displacement of the fat signal is evaluated in k‐space, through the Fast Fourier Transform in the readout dimension (), similar to related work. 28 , 38 , 39 , 40 The echo‐time in k‐space is defined as follows:
| (A2) |
Here, is the k‐space coordinate along the readout axis in the range , and is the readout bandwidth in Hz/pixel.
is the effective echo time for weighting and defined as follows:
| (A3) |
A six‐peak fat model as defined by the 2012 ISMRM challenge 41 was used, where and describe the relative intensity and off‐resonance frequency of the 'th fat component. is the phase induced by main magnetic field inhomogeneity () at echo :
| (A4) |
is the dephasing due to between echo 1 and 2, and, by symmetry of the pulse sequence, echo 3 and 4. is the constant phase at echo 1, and is the constant phase at echo 4.
Using the base water and fat signals, the S+ and S− models at echo are a simple summation:
| (A5) |
The intensity scaling between S+ and S− is defined by , derived from Sveinsson et al, 5 and dependent on the flip angle () and . For water a reference of 1200 ms was used. For fat a reference of 365 ms was used.
PHASE CORRECTION
Linear phase correction terms were fitted to the measured images using a least‐squares solution of the following linear system:
| (A6) |
Per image, the phase correction was described with an offset () and slope () over the readout direction , and applied to the obtain phase‐corrected images :
| (A7) |
Zijlstra F, Seevinck PR. Multiple‐echo steady‐state (MESS): Extending DESS for joint T2 mapping and chemical‐shift corrected water‐fat separation. Magn Reson Med. 2021;86:3156–3165. 10.1002/mrm.28921
DATA AVAILABILITY STATEMENT
The source code and data to reproduce the results in this manuscript are available at: https://github.com/FrankZijlstra/MESS/.
REFERENCES
- 1. Bruder H, Fischer H, Graumann R, Deimling M. A new steady‐state imaging sequence for simultaneous acquisition of two MR images with clearly different contrasts. Magn Reson Med. 1988;7:35‐42. [DOI] [PubMed] [Google Scholar]
- 2. Lee SY, Cho ZH. Fast SSFP gradient echo sequence for simultaneous acquisitions of FID and echo signals. Magn Reson Med. 1988;8:142‐150. [DOI] [PubMed] [Google Scholar]
- 3. Redpath TW, Jones RA. FADE–A new fast imaging sequence. Magn Reson Med. 1988;6:224‐234. [DOI] [PubMed] [Google Scholar]
- 4. Welsch GH, Scheffler K, Mamisch TC, et al. Rapid estimation of cartilage T2 based on double echo at steady state (DESS) with 3 Tesla. Magn Reson Med. 2009;62:544‐549. [DOI] [PubMed] [Google Scholar]
- 5. Sveinsson B, Chaudhari AS, Gold GE, Hargreaves BA. A simple analytic method for estimating T2 in the knee from DESS. Magn Reson Imaging. 2017;38:63‐70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pipe JG. High‐value MRI. J Magn Reson Imaging. 2019;49:e12‐e13. [DOI] [PubMed] [Google Scholar]
- 7. Peterfy CG, Schneider E, Nevitt M. The osteoarthritis initiative: report on the design rationale for the magnetic resonance imaging protocol for the knee. Osteoarthr Cartil OARS Osteoarthr Res Soc. 2008;16:1433‐1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Staroswiecki E, Granlund KL, Alley MT, Gold GE, Hargreaves BA. Simultaneous estimation of T2 and apparent diffusion coefficient in human articular cartilage in vivo with a modified three‐dimensional double echo steady state (DESS) sequence at 3 T. Magn Reson Med. 2012;67:1086‐1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Heule R, Ganter C, Bieri O. Rapid estimation of cartilage T2 with reduced T1 sensitivity using double echo steady state imaging. Magn Reson Med. 2014;71:1137‐1143. [DOI] [PubMed] [Google Scholar]
- 10. Eckstein F, Kunz M, Schutzer M, et al. Two year longitudinal change and test–retest‐precision of knee cartilage morphology in a pilot study for the osteoarthritis initiative. Osteoarthr Cartil. 2007;15:1326‐1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wirth W, Nevitt M, Hellio Le Graverand M‐P, et al. Sensitivity to change of cartilage morphometry using coronal FLASH, sagittal DESS, and coronal MPR DESS protocols – comparative data from the osteoarthritis initiative (OAI). Osteoarthr Cartil. 2010;18:547‐554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Abraham CL, Bangerter NK, McGavin LS, et al. Accuracy of 3D dual echo steady state (DESS) MR arthrography to quantify acetabular cartilage thickness. J Magn Reson Imaging. 2015;42:1329‐1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Duryea J, Cheng C, Schaefer LF, Smith S, Madore B. Integration of accelerated MRI and post‐processing software: a promising method for studies of knee osteoarthritis. Osteoarthr Cartil. 2016;24:1905‐1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dodin P, Pelletier J, Martel‐Pelletier J, Abram F. Automatic human knee cartilage segmentation from 3‐D magnetic resonance images. IEEE Trans Biomed Eng. 2010;57:2699‐2711. [DOI] [PubMed] [Google Scholar]
- 15. Raj A, Vishwanathan S, Ajani B, Krishnan K, Agarwal H. Automatic knee cartilage segmentation using fully volumetric convolutional neural networks for evaluation of osteoarthritis. In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018). 2018, pp. 851‐854.
- 16. Norman B, Pedoia V, Majumdar S. Use of 2D U‐net convolutional neural networks for automated cartilage and meniscus segmentation of knee MR imaging data to determine relaxometry and morphometry. Radiology. 2018;288:177‐185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhou Z, Zhao G, Kijowski R, Liu F. Deep convolutional neural network for segmentation of knee joint anatomy. Magn Reson Med. 2018;80:2759‐2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chaudhari AS, Stevens KJ, Sveinsson B, et al. Combined 5‐minute double‐echo in steady‐state with separated echoes and 2‐minute proton‐density‐weighted 2D FSE sequence for comprehensive whole‐joint knee MRI assessment. J Magn Reson Imaging. 2019;49:e183‐e194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bieri O, Ganter C, Scheffler K. Quantitative in vivo diffusion imaging of cartilage using double echo steady‐state free precession. Magn Reson Med. 2012;68:720‐729. [DOI] [PubMed] [Google Scholar]
- 20. Granlund KL, Staroswiecki E, Alley MT, Daniel BL, Hargreaves BA. High‐resolution, three‐dimensional diffusion‐weighted breast imaging using DESS. Magn Reson Imaging. 2014;32:330‐341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gras V, Farrher E, Grinberg F, Shah NJ. Diffusion‐weighted DESS protocol optimization for simultaneous mapping of the mean diffusivity, proton density and relaxation times at 3 Tesla. Magn Reson Med. 2017;78:130‐141. [DOI] [PubMed] [Google Scholar]
- 22. Dregely I, Margolis DAJ, Sung K, et al. Rapid quantitative T2 mapping of the prostate using three‐dimensional dual echo steady state MRI at 3T. Magn Reson Med. 2016;76:1720‐1729. [DOI] [PubMed] [Google Scholar]
- 23. Hauger O, Dumont E, Chateil J‐F, Moinard M, Diard F. Water excitation as an alternative to fat saturation in MR imaging: preliminary results in musculoskeletal imaging. Radiology. 2002;224:657‐663. [DOI] [PubMed] [Google Scholar]
- 24. Berglund J, Ahlström H, Johansson L, Kullberg J. Two‐point Dixon method with flexible echo times. Magn Reson Med. 2011;65:994‐1004. [DOI] [PubMed] [Google Scholar]
- 25. Chaudhari AS, Sveinsson B, Moran CJ, et al. Imaging and T2 relaxometry of short‐T2 connective tissues in the knee using ultrashort echo‐time double‐echo steady‐state (UTEDESS). Magn Reson Med. 2017;78:2136‐2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Eggers H, Brendel B, Duijndam A, Herigault G. Dual‐echo Dixon imaging with flexible choice of echo times. Magn Reson Med. 2011;65:96‐107. [DOI] [PubMed] [Google Scholar]
- 27. Zhang T, Chen Y, Bao S, et al. Resolving phase ambiguity in dual‐echo Dixon imaging using a projected power method. Magn Reson Med. 2017;77:2066‐2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lu W, Yu H, Shimakawa A, Alley M, Reeder SB, Hargreaves BA. Water–fat separation with bipolar multiecho sequences. Magn Reson Med. 2008;60:198‐209. [DOI] [PubMed] [Google Scholar]
- 29. Robson PM, Grant AK, Madhuranthakam AJ, Lattanzi R, Sodickson DK, McKenzie CA. Comprehensive quantification of signal‐to‐noise ratio and g‐factor for image‐based and k‐space‐based parallel imaging reconstructions. Magn Reson Med. 2008;60:895‐907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Heule R, Ganter C, Bieri O. Triple echo steady‐state (TESS) relaxometry. Magn Reson Med. 2014;71:230‐237. [DOI] [PubMed] [Google Scholar]
- 31. Cheng C‐C, Preiswerk F, Madore B. Multi‐pathway multi‐echo acquisition and neural contrast translation to generate a variety of quantitative and qualitative image contrasts. Magn Reson Med. 2020;83:2310‐2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bley TA, Wieben O, François CJ, Brittain JH, Reeder SB. Fat and water magnetic resonance imaging. J Magn Reson Imaging. 2010;31:4‐18. [DOI] [PubMed] [Google Scholar]
- 33. Pezeshk P, Alian A, Chhabra A. Role of chemical shift and Dixon based techniques in musculoskeletal MR imaging. Eur J Radiol. 2017;94:93‐100. [DOI] [PubMed] [Google Scholar]
- 34. Huijgen WHF, van Rijswijk CSP , Bloem JL. Is fat suppression in T1 and T2 FSE with mDixon superior to the frequency selection‐based SPAIR technique in musculoskeletal tumor imaging? Skeletal Radiol. 2019;48:1905‐1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ambellan F, Tack A, Ehlke M, Zachow S. Automated segmentation of knee bone and cartilage combining statistical shape knowledge and convolutional neural networks: data from the osteoarthritis initiative. Med Image Anal. 2019;52:109‐118. [DOI] [PubMed] [Google Scholar]
- 36. Florkow MC, Zijlstra F, Willemsen K, et al. Deep learning–based MR‐to‐CT synthesis: The influence of varying gradient echo–based MR images as input channels. Magn Reson Med. 2020;83:1429‐1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jonsson J, Nyholm T, Söderkvist K. The rationale for MR‐only treatment planning for external radiotherapy. Clin Transl Radiat Oncol. 2019;18:60‐65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Berglund J, Rydén H, Avventi E, Norbeck O, Sprenger T, Skare S. Fat/water separation in k‐space with real‐valued estimates and its combination with POCS. Magn Reson Med. 2020;83:653‐661. [DOI] [PubMed] [Google Scholar]
- 39. Brodsky EK, Holmes JH, Yu H, Reeder SB. Generalized k‐space decomposition with chemical shift correction for non‐Cartesian water‐fat imaging. Magn Reson Med. 2008;59:1151‐1164. [DOI] [PubMed] [Google Scholar]
- 40. Honorato JL, Parot V, Tejos C, Uribe S, Irarrazaval P. Chemical species separation with simultaneous estimation of field map and T using a k‐space formulation. Magn Reson Med. 2012;68:400‐408. [DOI] [PubMed] [Google Scholar]
- 41. Judging . ISMRM Challenge. http://challenge.ismrm.org/node/17. Published 2012. Accessed December 2, 2020 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 MESS water‐fat separation for all 5 subjects (top and middle row), with water‐selective DESS (bottom row) as a reference for the MESS water image. In subjects 2 and 3 a minor water‐fat swap is visible in the posterior side of the knee, indicated by arrows. Subject 3 has two titanium screws (one visible), which showed similar artifacts for the DESS and MESS images
FIGURE S2 The effects of reduced regularization on the water‐fat separation. Reduced regularization () is shown on the left, and the regularization used in this study () on the right. Arrows indicate how the regularization in this study relates to the reduced regularization in terms of sharpness (red arrows), noise amplification (green arrows), artifacts (white arrows)
TABLE S1 A, Noise statistics for DESS and MESS W and F contrasts for all subjects. Noise is shown overall, and for water‐dominant voxels (W>F) and fat‐dominant voxels (F>W) with sufficient signal (W+F>0.2, with intensities normalized to 0.5 for water in muscle). B, SNR statistics for DESS and MESS W and F contrasts for all subjects. SNR is shown overall, and for the region masked for the relevant contrast, water‐dominant voxels for DESS and MESS W, and fat‐dominant voxels for MESS F
Data Availability Statement
The source code and data to reproduce the results in this manuscript are available at: https://github.com/FrankZijlstra/MESS/.


