Abstract
Objective
To evaluate cell‐free DNA (cfDNA) redraws and pregnancy outcomes following low fetal fraction (FF) cfDNA failures, as it has been suggested that a failed cfDNA screen due to insufficient FF carries increased risk for fetal aneuploidy.
Methods
Here >200,000 consecutive samples were reviewed and >1,100 patients were identified with a failed cfDNA due to low FF using genome‐wide massively parallel sequencing. Redraw results following the initial low FF failure were analyzed, as well as pregnancy outcomes for patients with repeated low FF failure on redraw.
Results
Upon redraw 84.2% of samples yielded a reportable result with no enrichment of aneuploidy observed (p = 0.332). Higher maternal weights and multifetal pregnancy rates were observed in samples with insufficient FF. In patients with repeated low FF failure on redraw, almost all pregnancies resulted in apparently healthy liveborns.
Conclusion
Insufficient FF was not an indicator of aneuploidy risk or adverse pregnancy outcomes in this study. Caution should be taken in generalizing aneuploidy risk to all low FF cfDNA failures. Redrawing may be an appropriate next step, as proceeding directly with diagnostic testing for aneuploidy may be unwarranted for most patients.
Key Points
What's already known about this topic?
Fetal fraction (FF) is affected by a number of factors including maternal weight, gestational age, and some aneuploidies
It has been suggested that cell‐free DNA (cfDNA) screens that fail due to insufficient FF are at increased risk for aneuploidy
What does this study add?
Increased aneuploidy rates were not observed in patients who submitted a new sample after an initial low FF failure
Redrawing cfDNA after an initial low FF failure may be an appropriate next step instead of proceeding directly to diagnostic testing
Caution should be taken in generalizing aneuploidy risk to all low FF failures, as each laboratory methodology and criteria is unique
1. INTRODUCTION
With increased detection and higher positive predictive value compared to conventional screening methods, 1 cell‐free DNA screening (cfDNA) is recommended as a routine aneuploidy screening option by key organizations. 2 , 3 , 4 , 5 , 6 cfDNA screening analyzes cfDNA fragments in maternal plasma, a proportion of which originate from the placental trophoblast and is referred to as the fetal fraction (FF). Occasionally a sample will have an insufficient amount of placental cfDNA fragments (i.e., FF), meaning the pregnancy is inadequately represented in the sample, which will result in a test failure with option for redraw.
Of all the known factors associated with FF, maternal weight is arguably the most well established. Maternal weight has an inverse relationship with FF, which has been thoroughly described in the literature 7 , 8 , 9 and increased cfDNA screening failure rates due to insufficient FF have been observed in patients with high body mass index (BMI). 10 , 11 The increased apoptotic tendencies of adipose tissue are believed to dilute placental cfDNA fragments in maternal plasma. 12 , 13 Maternal medications, namely low molecular weight heparin (LMWH), have also been associated with cfDNA screening failure, though the exact mechanism is speculative. 14 Gestational age, however, has a direct relationship with FF. Generally, as the placenta grows and develops throughout the pregnancy, the FF likewise increases.
There has been concern that aneuploid pregnancies are associated with lower FFs on cfDNA screening. It has been suggested that cfDNA screens with insufficient FF carry increased risk for fetal aneuploidy and is possibly testing platform specific given the different methodology, criteria, and variable failure rates unique to each laboratory. 15 One study found >17% aneuploidy rate in their FF failures, however the high risk arm of this study was already enriched for aneuploidy (26% aneuploidy incidence) as 87% of these patients were already planning to have or already had invasive diagnostic testing. 16 Another study reported a 4.7% aneuploidy rate in FF failures. 1 Following these observations, guidelines have recommended counseling and offering diagnostic testing following a FF failure on cfDNA screening and that a second blood draw may not be appropriate 2 or that a second blood draw may delay diagnosis of fetal aneuploidy. 17 Conversely, a recent study found low FF is not a reliable predictor of aneuploidy risk and repeat noninvasive prenatal test (NIPT) is an appropriate option. 18 Offering diagnostic testing due to a low FF cfDNA failure, regardless of testing platform or contextualization of maternal factors like BMI, is likely unnecessary for most patients and may result in increased procedure related loss. 15
This current study aims to further explore aneuploidy enrichment in low FF failures on a single platform from one laboratory. Here >200,000 consecutive samples were analyzed and >1,100 patients were identified with a cfDNA nonreportable result due to insufficient FF using genome‐wide massively parallel sequencing (MPS) cfDNA technology.
2. METHODS
A retrospective analysis was performed on over 200,000 consecutive cfDNA screening samples submitted to a single clinical laboratory, all subjected to the same assay version. All samples were submitted more than one year ago, making pregnancy outcome information theoretically available for all pregnancies. Demographics were collected when available. Gestational age was determined by LMP or via ultrasound and provided on the test requisition form (TRF). Maternal weight is not required for testing and as such is not available for all samples. For purposes of maternal weight only, <80lbs were excluded as outliers. TRFs with provided weights between 80 and 100lbs and >500lbs were manually reviewed for accuracy.
Maternal blood samples submitted to Sequenom Laboratories for MaterniT®21 PLUS cfDNA screening were subjected to DNA extraction, library preparation, and genome‐wide MPS as previously described. 19 FF calculation (SeqFF) is described in Kim et al. 20 Reportable samples are required to meet both an established minimum FF requirement (≥2.5%) and signal‐to‐noise ratio (SNR) threshold, a method that creates an individualized sample assessment. SNR allows confident reporting of high quality data at lower FFs by separating meaningful information (signal derived from FF) from background information (noise; Figure S1). 21 The threshold of this metric was established to maintain test sensitivity while permitting samples with lower FF to be reported, provided that fetal signal exceeds and is discernable from sample noise. To determine the SNR threshold, simulations were performed using 16,900 samples in which contrived trisomy 21/18/13 events were titrated proportionate to sample FF, and spanning the observed population distribution of FFs. The resulting cumulative T13/18/21 sensitivities at varying SNR cut‐offs were compared to historical clinical validation sensitivity levels 22 , 23 to derive an SNR threshold that optimized test sensitivity. From this large cohort simulation, the cumulative T18/13 sensitivity at the established SNR threshold is 99.98% and >99% sensitivity for T21 (Figure S2).
Failure of a sample to meet the FF and SNR cut offs result in a nonreportable result due to insufficient FF. Multifetal gestations are subjected to higher FF/SNR thresholds, proportional to the number of fetuses.
Two hundred and six thousand one hundred and nine (206,109) total consecutive samples, which included both initial draws and repeat testing (when received), were analyzed and 1,110 patients were identified with a failed cfDNA due to low FF using genome‐wide MPS at their initial draw. De‐identified patient specific IDs were used to determine if a patient submitted a redraw (i.e., repeated NIPT testing) following their initial low FF failure. Collection time between the original and redraw sample was used to determine if both samples represented the same versus a new pregnancy. Redraw sample results representing the same pregnancy were collated and analyzed. For patients with repeated low FF failure at redraw, pregnancy outcomes were elicited via direct communication with the clinical provider.
Additionally, all low FF failure samples were cross‐referenced with cytogenetic and SNP microarray diagnostic results submitted to Laboratory Corporation of America and Integrated Genetics during the corresponding timeframe. For a cfDNA sample to be considered a match to a cytogenetic and/or microarray specimen, the diagnostic and screening results were required to have identical patient identifiers (name and date of birth), and the collection date for the diagnostic test had to be within the term of the pregnancy, given the patient's GA at the cfDNA screening date. Data was analyzed in compliance with IRB approved protocol SCMM‐RND‐402 and Sequenom Laboratories® Notice of Privacy Practices. Identifiable information is not provided. 2‐sided, 2‐sample proportional z‐test and 2‐sample t test were used to calculate significance. p‐values less than 0.05 were considered statistically significant.
3. RESULTS
The low FF failure rate in this time period was 0.6%, which is an improvement from the previously reported rate of 0.9% (p < 0.00001) 24 . Out of the 1,110 patients who had a low FF failure at their initial draw, 704 submitted a repeat cfDNA sample to the laboratory and 406 did not (Figure 1). Out of the 704 patients who sent a redraw sample, 111 received another failed cfDNA, almost all of which was again due to insufficient FF (n = 109). A reportable result was obtained for 593 patients (84.2%) and 98.5% of these patients received a negative screening result upon redraw. Nine patients received a positive screening result upon redraw (T21 = 5, T18 = 1, T13 = 3). For the nine patients who received a positive result, the average gestational age at initial draw was 10 weeks (range 9–12 weeks) and 13 weeks (range 11–16) at redraw. An indication for testing was provided for 6/9 patients and was advanced maternal age in all cases.
FIGURE 1.
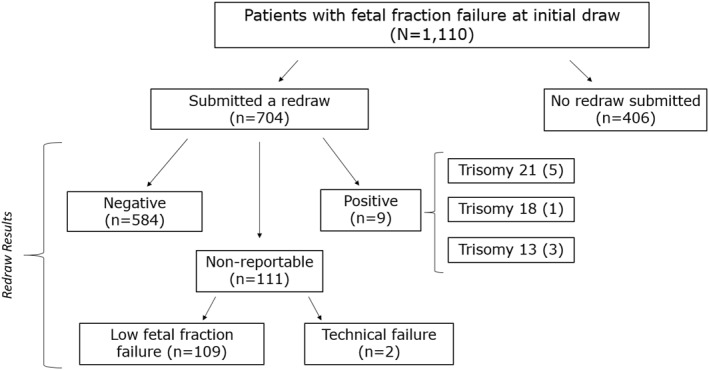
Redraw results following an initial low fetal fraction (FF) failure follows the 1,110 patients who received a failed screen due to low FF at their initial draw and their result upon redraw, if a repeat sample was submitted
The positivity rate in this redraw cohort following an initial low FF failure was 1.5%, which is statistically similar to the overall positivity rate (1.1%) for all samples in this time period (Table 1). These similar positivity rates were consistent when controlling for singletons only versus multifetal pregnancies only. Aneuploidy enrichment was not observed in the patients with successful repeat cfDNA screening following an initial low FF failure.
TABLE 1.
Positivity rates in redraw group versus total tests shows the positivity rates in patients with singleton and/or multifetal pregnancies who had a successful redraw after a low fetal fraction failure versus total testers
| Gestational status | Total sample positivity rate (>200,000) | Redraw sample positivity rate | Significance |
|---|---|---|---|
| Singleton & multifetal | 1.1% | 1.5% (9/593) | Not statistically significant p = 0.332 |
| Singleton only | 1.1% | 1.3% (7/520) | Not statistically significant p = 0.582 |
| Multifetal only | 2.2% a | 2.7% (2/73) | Not statistically significant p = 0.748 |
Multifetal data from Dyr et al. 25 .
Demographics and maternal characteristics are described in Table 2. Gestational age remained similar in all cohorts, with cfDNA typically being drawn towards the end of the first trimester. Notably, maternal weight was higher in all low FF cohorts when compared to the total patient population and highest in the 109 patients receiving low FF failures at both draws (t = 6.241; p < 0.00001). Despite a 95.9% multifetal success rate at initial draw, multifetal pregnancy rate was significantly increased in all low FF failure groups (>12%) versus the multifetal rate observed in the total sample population (2.0%). Of note, there are increased FF requirements for multifetal pregnancies versus singleton gestations. Imposed FF requirements are roughly proportional to fetal number (i.e., triple the requirement in triplets). Chorionicity is not routinely provided nor required on the TRF.
TABLE 2.
Demographics table by cohort provides maternal weight, average gestational age (GA), and percentage of multifetal pregnancies in total testers and various low fetal fraction (FF) failure cohorts
| Cohort | N | Average maternal weight a | Average GA | % Multifetal |
|---|---|---|---|---|
| Total tests | 206,109 | 166 lbs | 13.4 weeks | 2.0% |
| Low FF at initial draw | 1,110 | 215 lbs | 12.1 weeks | 13.5% |
| No redraw submitted | 406 | 216 lbs | 12.9 weeks | 15.5% |
| Redraw submitted | 704 | 214 lbs | 11.7 weeks | 12.4% |
| Repeated low FF at redraw | 109 | 224 lbs | 11.8 weeks | 12.8% |
Maternal weight not provided for all patients.
The indication breakdown was statistically similar between patients who pursued repeat screening and those who did not (Table 3) with both populations having similar proportions of high risk versus average risk patients. This similarity is important from a sample bias consideration, so as to not predispose the 704 patients who submitted a repeat specimen for or against aneuploidy enrichment. Outcome information is limited for the 406 patients who opted not to submit a redraw after low FF on their first draw. Cross‐referencing this patient cohort with a single internal commercial diagnostic testing laboratory (LabCorp and Integrated Genetics), it was found that four patients in this group pursued diagnostic testing via microarray following their low FF failure (two CVS and two amniocenteses). Diagnostic testing was normal in all cases.
TABLE 3.
Indication for testing by cohort
| Cohort | Advanced maternal age | Abnormal serum screening | Ultrasound findings | Family history | Average risk | Multiple indications | No indication provided |
|---|---|---|---|---|---|---|---|
| 206,109 total cohort | 45.0% | 2.1% | 3.6% | 1.5% | 7.5% | 1.9% | 38.4% |
| 1,110 low fetal fraction (FF) at initial draw | 52.3% | 1.3% | 1.4% | 1.5% | 5.9% | 1.2% | 36.3% |
| 704 that sent a redraw | 53.0% | 1.1% | 1.1% | 1.4% | 6.4% | 1.0% | 35.9% |
| 406 that did not redraw | 51.2% | 1.5% | 2.0% | 1.7% | 5.2% | 1.5% | 36.9% |
| 109 low FF at redraw | 62.4% | 0.9% | 0.0% | 0.9% | 5.5% | 1.8% | 28.4% |
3.1. Repeated low fetal fraction cohort
Pregnancy and birth outcome information was elicited for the 109 patients with a repeated low FF cfDNA failure. Birth outcome, defined by the pregnancy resulting in an apparently healthy liveborn or demise, was obtained for 81/109 patients. Eighteen patients were lost to follow up. Partial outcome was obtained for the remaining 10 patients. Partial outcome includes cases where information regarding maternal medications, conditions, and/or ultrasound was available, but information on birth outcome (i.e., status of baby at delivery) was unavailable. Most cases where only partial outcome was available include patients who transferred care at advanced gestational ages.
For the 81 cases in which delivery status was available, 76 were presumably healthy liveborns and five resulted in a fetal demise or termination of pregnancy. Complicating obstetrical factors were present for most fetal demise cases (Table 4). 12/81 cases pursued diagnostic testing (POC, CVS, amniocentesis, or postnatal). An additional 8/81 patients opted for additional screening (NIPT with a different laboratory or serum screening; Table 5). All diagnostic testing/screening was normal except for a twin pregnancy amniocentesis that showed one euploid and one triploid twin. Term information was provided in 61 of the 81 cases with birth outcome (Figure 2). 81% of singleton pregnancies delivered at full term (≥37 weeks gestation). Of the 10 preterm deliveries in singleton pregnancies, the average GA at delivery was 34 weeks (range 28–36.6; median 36). The remainder of preterm deliveries were multifetal pregnancies (n = 5).
TABLE 4.
Birth outcome of 81 patients with repeated low fetal fraction (FF) failures. Birth outcome, defined by the pregnancy resulting in an apparently healthy liveborn or not, was obtained for 81 of the 109 patients with repeated low FF failures on cell‐free DNA and is detailed here
| Birth outcome | N = 81 |
|---|---|
| Healthy liveborn | 76/81 |
| Fetal demise or termination of pregnancy | 5/81 |
| 1. Demise at 19 weeks, normal chromosomes on POC, antiphospholipid syndrome, 56yo patient | |
| 2. TAB, 4.5 mm NT, normal CVS chromosomes & VUS on CVS array | |
| 3. TAB due to MCA at 16 weeks, uncontrolled diabetes with high A1C | |
| 4. 28 weeks delivery due to preeclampsia, uncontrolled diabetes, maternal obesity, normal ultrasounds | |
| 5. 14 weeks miscarriage, no POC |
Abbreviations: MCA, multiple congenital anomalies; POC, products of conception; TAB, therapeutic abortion; wnl, within normal limits.
TABLE 5.
Additional screening & testing in patients with repeated low fetal fraction (FF) cell‐free DNA (cfDNA) details the types of diagnostic or screening pursued in the 81 patients with known birth outcomes
| Additional screening/testing type | Number of patients | Result |
|---|---|---|
| Postnatal | 1 | Normal |
| Products of conception | 1 | Normal |
| CVS | 2 | Normal |
| Amniocentesis | 8 | 7 normal/1 abnormal a |
| Serum biochemical screening | 5 | Negative |
| cfDNA at another laboratory | 3 | Negative |
| Total 20/81 |
Following NIPT, a twin pregnancy showed triploid in one twin and normal chromosomes on the other. Reduced to singleton and healthy fetus delivered at term.
FIGURE 2.
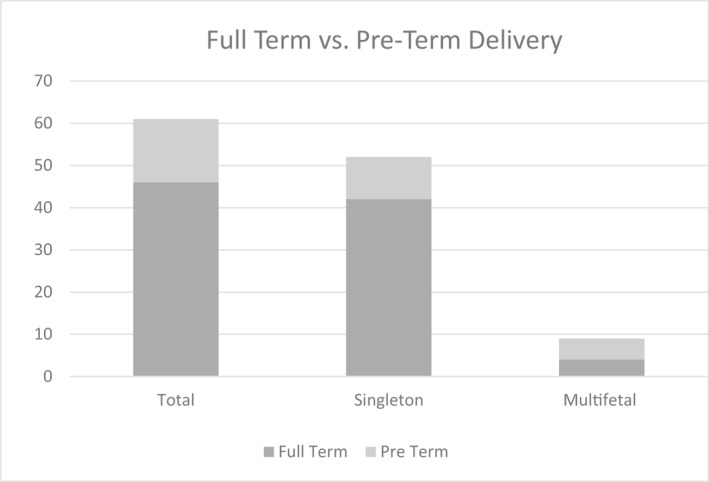
Gestational Age at Delivery (n = 61). Full‐term versus preterm delivery information was provided for 61/81 cases with known birth outcomes. Full‐term is defined as ≥37 weeks gestation
Information regarding pregnancy complications was reported ad hoc when obtaining outcomes for the repeated low FF failure cases (n = 109). Fetal growth restriction (FGR) was reported in five pregnancies (singleton n = 2, multifetal n = 3). Preeclampsia was reported in six pregnancies (singleton n = 4, multifetal n = 2) and both preeclampsia and FGR were reported in one singleton pregnancy. For the two singleton pregnancies with reported FGR, one patient had pre‐existing diabetes and the other had gestational diabetes. Of the four singleton pregnancies with reported preeclampsia, one patient had poorly controlled diabetes, one had pre‐existing hypertension, and one had chorioamnionitis and sickle cell trait. All of these pregnancies were also complicated by maternal obesity. Information regarding maternal medications and conditions was not routinely elicited but was reported ad hoc for 27 patients in the repeated low FF failure cohort (n = 109) (Figure 3). Of these cases, LMWH was reported in 15 patients, making it the most frequently reported medication in patients with low FF cfDNA failures. Maternal diabetes was the most reported maternal condition complicating pregnancy, followed by Lupus/antiphospholipid syndrome.
FIGURE 3.
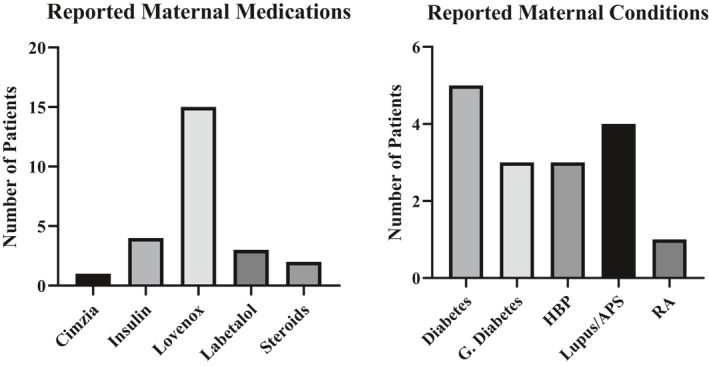
Ad Hoc Reporting of Maternal Medications/Conditions in patients with repeated low fetal fraction (FF) failure G. APS, antiphospholipid syndrome; Diabetes, gestational diabetes; RA, rheumatoid arthritis. *The same patient may be represented multiple times if taking multiple medications and/or both a condition and medication(s) were reported
4. DISCUSSION
4.1. Positivity rate
63% of patients submitted a redraw following an initial low FF failure, consistent with historical reported redraw rates. 24 Increased aneuploidy rates in low FF failures were not observed, similar to other studies. 18 , 23 Of 593 reportable redraw samples, 1.5% yielded a positive result, which is statistically similar to the positivity rate of all samples in this time period (1.1%; p = 0.332) and this statistical similarity holds true when controlling for singleton only, multifetal only, or combined (Figure 1 & Table 1). For the 584 patients receiving a negative result on redraw, average redraw FF was 5.5%. No false negatives in this group have been reported to the laboratory. If following current society guidelines after a low FF failure, these 584 patients may have had diagnostic testing that otherwise might have been avoided.
The trisomy 21/18/13 positivity breakdown after an initial FF failure mirrors the overall breakdown in the total tests, in which trisomy 21 accounts for ∼60% of positive results. There may have been expectation to see an increase in trisomy 13 and 18 positives, as those aneuploidies are associated with smaller placental mass 26 and relatively lower FFs on cfDNA. In the total >200k tests, the average FF in trisomy 21, 18, and 13 cfDNA samples is 9.0%, 8.0% and 8.1% (median 8.6%, 7.5%, 7.3%), respectively and 8.8% average FF (8.3% median) in euploid samples, similar to previously reported FF trends. 18 , 27 , 28 While FF is somewhat lower in the T18 and T13 samples versus T21 and euploid samples, it is not approaching the minimal bounds of FF required for reporting. This is consistent with trends observed in other studies, where the average FF for T18 pregnancies was well above the minimum assay requirement. 18 , 28 While trisomy 13 and 18 pregnancies may have lower FF relative to trisomy 21 and euploid pregnancies, the decrease in FF does not appear to overtly put these pregnancies at increased risk for a low FF failure.
4.2. Maternal characteristics and demographics
Maternal weight was higher in all low FF cohorts when compared to the total patient population and was significantly higher in the 109 patients experiencing repeated failure (t = 6.241; p < 0.00001), supporting maternal habitus as the primary driver of informative cfDNA redraws. 29 While most NIPT samples are collected during the first trimester, slightly earlier gestational ages were observed for samples who received a low FF failure at their first draw (12.1 weeks) and for repeat samples that again resulted in a low FF failures was even earlier (11.8 weeks gestation). Generally, cfDNA in maternal plasma is directly related to increasing gestational age.
FF/SNR reporting requirements are proportionally higher in multifetal gestations (i.e., double for twins). The biological reality however, is that FF is not directly proportionate to the number of fetuses with one study observing a 1.6x FF increase in twins versus singletons. 28 The average FF observed in twins versus singleton pregnancies in this study was 10.2% and 8.7%, respectively. Not surprisingly, given the two fold increased requirement, there were significantly more multifetal gestations (p < 0.00001) in the low FF cohorts as compared to the total study population (Table 2). This should not be confused with the 95.9% multifetal success rate at initial draw.
4.3. Pregnancy outcomes for repeated low FF failures
Birth outcome (i.e., did the pregnancy result in an apparently healthy liveborn or a demise) was obtained for 81/109 patients with repeated low FF failures. Out of the 81 cases with known birth outcome, almost all (76) resulted in apparently healthy liveborns. Aneuploidy was not overtly suspected in most of the fetal demise cases (Table 4), either due to normal prenatal diagnostic results and/or unrelated obstetrical complications. Persistent low FF does not appear to be an indicator for aneuploid pregnancies. Even after repeated low FF failures, when diagnostic testing or additional screening (20/81 patients) was pursued, it was largely reassuring.
Term information was provided for 61 of the 81 birth outcome cases. 60/61 cases with term information resulted in apparently healthy liveborns. In total, two singleton pregnancies delivered <32 weeks gestation, one of which was complicated by poorly controlled maternal diabetes, obesity, hypertension, and preeclampsia (Table 4). The other was complicated by antiphospholipid syndrome, preterm prematrue rupture of membranes, and obesity. The remaining eight singleton preterm births were at ≥32 weeks (median 36 weeks). The remainder of preterm deliveries were multifetal pregnancies (n = 5). Risk factors for premature delivery include but are not limited to, high blood pressure (HBP), obesity, placental previa, diabetes, gestational diabetes, advanced maternal age and multifetal pregnancy. 30 One or more of these factors was observed in all 15 preterm birth cases. Similarly, given the risk factors for FGR and/or preeclampsia, it is challenging to glean whether the persistent low FF itself was an early warning sign or a byproduct of other confounding factors. On the contrary, elevated FF in the second trimester has been associated with prematurity and may be indicative of underlying placental dysfunction. 31 , 32 , 33 Pre‐existing HBP has been associated with lower FF, even when controlling for maternal BMI, and is possibly due to reduced organ perfusion and subsequent “restricted release” of placental cfDNA. 28 HBP was reported in three patients. Maternal medications were reported ad hoc in 27 of the 109 patients receiving repeated low FF failures (Figure 3). Of these cases, LMWH was the medication most often reported. Anticoagulants are a well‐established contributor to low FF cfDNA failures. 13 As medication use and pregnancy complications are not captured on test requisitions, nor routinely ascertained when eliciting outcome, drawing definitive conclusions regarding obstetrical risks and low FF is not possible given ad hoc reporting and the confounding risk factors present. Future studies systematically examining this would be helpful.
5. LIMITATIONS
Outcome information is largely dependent on clinician reporting in this retrospective study. There is limited follow up for the 406 patients who did not pursue repeat cfDNA. Diagnostic testing for this group was cross‐referenced with a single internal laboratory, which does not capture all patients who may have pursued testing elsewhere. Prenatal diagnostic testing results are rarely available following normal screening and birth outcome was not actively elicited for the 584 patients with negative cfDNA upon redraw. No false negatives in this group have been reported to the laboratory. This assay is unable to screen for triploidy, which has been associated with low FF on cfDNA 34 and was observed in one fetus in a twin pregnancy (Table 4). While pregnancy outcomes for patients with repeated FF failures were obtained in 74.3% cases and there were no reports of triploid pregnancies in patients with negative repeat cfDNA results communicated to the laboratory, it is possible that there were additional triploid pregnancies undetected by the assay.
6. CONCLUSION
Data from this retrospective cohort revealed that 84.2% of patients initially receiving a low FF failure achieved a reportable result at redraw with no observable aneuploidy enrichment. Furthermore, pregnancies for patients receiving repeated low FF failures almost always resulted in apparently healthy liveborns. While low FF failures on cfDNA do not rule out aneuploidy in the fetus, taking into account patient factors like weight or medications are important. However, caution should be taken in generalizing redraw success rates and aneuploidy enrichment (or lack thereof) to all low FF nonreportable cfDNA results, as each laboratory reporting criteria and methodology is unique.
CONFLICT OF INTEREST
Authors Caldwell, Almasri, Dyr, and Wardrop are employees and author Cacheris is an independent contractor with Sequenom Laboratories, a wholly‐owned subsidiary of Laboratory Corporation of America Holdings.
Supporting information
Supplementary Material 1
Supplementary Material 2
ACKNOWLEDGEMENTS
Sidra Boshes, MS, LCGC, Michelle Hackbardt, MS, LCGC, Theresa Boomer, MS, LCGC, Juan‐Sebastian Saldivar, MD, FACMG, Hany Magharyous, MD. No external funding was received to support the research in connection with this article.
Caldwell S, Almasri E, Schmidt L, et al. Not all low fetal fraction cell‐free DNA screening failures are at increased risk for aneuploidy. Prenat Diagn. 2021;41(11):1372‐1379. doi: 10.1002/pd.5918
DATA AVAILABILITY STATEMENT
Research data are not shared.
REFERENCES
- 1. Norton M, Jacobsson B, Swamy G, et al. Cell‐free DNA analysis for noninvasive examination of trisomy. N Engl J Med. 2015;372(17):1589‐1597. [DOI] [PubMed] [Google Scholar]
- 2. Gregg A, Skotko B, Benkendorf J, et al. Noninvasive prenatal screening for fetal aneuploidy, 2016 update: a position statement of the American College of Medical Genetics and Genomics. Genet Med. 2016;18:1056‐1065. [DOI] [PubMed] [Google Scholar]
- 3. American College of Obstetricians and Gynecologists . Noninvasive prenatal testing for fetal aneuploidy. Committee Opinion Number 545. Obstet Gynecol. 2012;120:1532‐1534. [DOI] [PubMed] [Google Scholar]
- 4. American College of Obstetricians and Gynecologists . Cell‐free DNA screening for fetal aneuploidy. Committee opinion number 640. Obstet Gynecol. 2015;126:e31‐e37. [DOI] [PubMed] [Google Scholar]
- 5. Wilson K, Czerwinski J, Hoskovec J, et al. NSGC practice guideline: prenatal screening and diagnostic testing options for chromosome aneuploidy. J Genet Counsel. 2013;22:4‐15. 10.1007/s10897-012-9545-3PMID:23179172. [DOI] [PubMed] [Google Scholar]
- 6. Benn P, Borell A, Chiu R, et al. Position statement from the aneuploidy screening committee on behalf of the board of the international society for prenatal diagnosis. Prenat Diagn. 2013;33(7):622‐629. 10.1002/pd.4139PMID:23616385. [DOI] [PubMed] [Google Scholar]
- 7. Livergood M, LeChien K, Trudell A. Obesity and cell‐free DNA “no calls”: is there an optimal gestational age at time of sampling? Am J Obstet Gynecol. 2017;216:413. e1‐9. [DOI] [PubMed] [Google Scholar]
- 8. Wang E, Batey A, Struble C, et al. Gestational Age and maternal weight effects on fetal cell‐free DNA in maternal plasma. Prenat Diagn. 2013;33:662‐666. 10.1002/pd.4119. [DOI] [PubMed] [Google Scholar]
- 9. Rolnik D, Yong Y, Lee T, et al. Influence of BMI on fetal fraction increase with gestation and cfDNA test failure. Obstetrics Gynecol. 2018;132(2):436‐443. [DOI] [PubMed] [Google Scholar]
- 10. Yared E, Dinsmoor M, Endres L, et al. Obesity increases the risk of failure of noninvasive prenatal screening regardless of gestational age. Am J Obstet Gynecol. 2016;215(3). 370. e1–6. [DOI] [PubMed] [Google Scholar]
- 11. Zozzaro‐Smith P, Gray LM, Bacak S, Thornburg L. Limitations of aneuploidy and anomaly detection in the obese patient. J Clin Med Res. 2014;3(3):795‐808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Haghiac M, Vora N, Basu S, et al. Increased death of adipose cells, a path to release cell‐free DNA into systemic circulation of obese women. Obesity. 2012;20(11):2213‐2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hui L. Noninvasive prenatal testing for aneuploidy using cell‐free DNA – new implications for maternal health. Obstet Med. 2016;9(4):148‐152. 10.1177/1753495X16652007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gromminger S, Erkan S, Schock U, et al. The influence of low molecular weight heparin medication on plasma DNA in pregnant women. Prenat Diagn. 2015;35:1155‐1157. [DOI] [PubMed] [Google Scholar]
- 15. Artieri C, Haverty C, Evans E, et al. Noninvasive prenatal screening at low fetal fraction: comparing whole‐genome sequencing and single‐nucleotide polymorphism methods. Prenat Diagn. 2017;37(5):482‐490. [DOI] [PubMed] [Google Scholar]
- 16. Pergament E, Cuckle H, Zimmermann B, et al. Single‐nucleotide polymorphism based noninvasive prenatal screening in a high‐risk and low‐risk cohort. Obstet Gynecol. 2014;124(2 Pt 1):210‐218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. American College of Obstetricians and Gynecologists . Screening for fetal aneuploidy. Bulletin number 163. Obstet Gynecol. 2016;127(5):e123‐e137. [DOI] [PubMed] [Google Scholar]
- 18. Lopes J, Lopes G, Enninga E, et al. Most noninvasive prenatal screen failing due to inadequate fetal cell free DNA are negative for trisomy when repeated. Prenat Diagn. 2020;40(7):1‐7. 10.1002/pd.5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jensen T, Zwiefelhofer T, Tim R, et al. High‐throughput massively parallel sequencing for fetal aneuploidy detection from maternal plasma. PLoS ONE. 2013;8(3):e57381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim S, Hannum G, Geis J, et al. Determination of fetal DNA fraction from the plasma of pregnant women using sequence read counts. Prenat Diagn. 2015;35(8):810‐815. [DOI] [PubMed] [Google Scholar]
- 21. Mazloom M. Sample specific fetal fraction threshold for non‐invasive prenatal testing. Presented at the American College of Medical Genetics Annual Clinical Genetics Meeting. Phoenix; Mar 21‐25, 2017. [Google Scholar]
- 22. Palomaki G, Kloza E, Labert‐Messerlian G, et al. DNA sequencing of maternal plasma to detect Down syndrome: and international clinical validation study. Genet Med. 2011;13(11):913‐920. [DOI] [PubMed] [Google Scholar]
- 23. Palomaki G, Deciu G, Kloza M, et al. DNA sequencing of maternal plasma reliably identifies trisomy 18 and trisomy 13 as well as Down syndrome: and international collaborative study. Genet Med. 2012;14(3):296‐305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mccullough R, Almasri E, Guan X, et al. Non‐Invasive prenatal chromosomal aneuploidy testing – clinical experience: 100,000 clinical samples. PLoS One. 2014;9(10):e109173. 10.1371/journal.pone.0109173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dyr B, Boomer T, Almasri E, et al. A new era in aneuploidy screening: cfDNA testing in >30,000 multifetal gestations: experience at one clinical laboratory. PLoS ONE. 2019;14(8):e0220979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shepard T, FitzSimmons J, Fantel A, Pascoe‐Mason J. Placental weights of normal and aneuploid early human fetuses. Pediatr Pathol. 1989;9(4):425‐431. [DOI] [PubMed] [Google Scholar]
- 27. Palomaki G, Kloza E, Lambert‐Messerlian G, et al. Circulating cell free DNA testing: are some test failures informative? Prenat Diagn. 2015;35(3):289‐293. [DOI] [PubMed] [Google Scholar]
- 28. Zhou Y, Zhu Z, Gao Y, et al. Effects of Maternal and fetal characteristics of cell‐free fetal DNA fraction in maternal plasma. Reprod Sci. 2015;22(11):1429‐1435. [DOI] [PubMed] [Google Scholar]
- 29. Benn P, Valenti E, Shah S, et al. Factors associated with informative redraw after an initial no result in noninvasive prenatal testing. Obstet Gynecol. 2018;132(2):428‐435. [DOI] [PubMed] [Google Scholar]
- 30. National Institutes of Health . What are the risk factors for preterm labor and birth?. https://www.nichd.nih.gov/health/topics/preterm/conditioninfo/who_risk. Accessed June 10, 2019. [Google Scholar]
- 31. Dugoff L, Barberio A, Whittaker P, et al. Cell‐free DNA fetal fraction and preterm birth. Am J Obstet Gynecol. 2016;215(2):231. e1‐7. [DOI] [PubMed] [Google Scholar]
- 32. Levine R, Qian C, Leshane E, et al. Two‐stage elevation of cell‐free fetal DNA in maternal sera before onset of preeclampsia. Am J Obstet Gynecol. 2004;190(3):707‐713. [DOI] [PubMed] [Google Scholar]
- 33. Al Nakib M, Desbriere R, Bonello N, et al. Total and fetal cell‐free DNA analysis in maternal blood as markers of placental insufficiency and intrauterine growth restriction. Fetal Diagn Ther. 2009;26(1):24‐28. [DOI] [PubMed] [Google Scholar]
- 34. Nicolaides K, Syngelaki A, Gil M, et al. Prenatal detection of fetal triploidy from cell‐free DNA testing in maternal blood. Fetal Diagn Ther. 2014;35(3):212‐217. 10.1159/000355655. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1
Supplementary Material 2
Data Availability Statement
Research data are not shared.


