Abstract
The microbiome has a fundamental impact on the human host's physiology through the production of highly reactive compounds that can lead to disease development. One class of such compounds are carbonyl‐containing metabolites, which are involved in diverse biochemical processes. Mass spectrometry is the method of choice for analysis of metabolites but carbonyls are analytically challenging. Herein, we have developed a new chemical biology tool using chemoselective modification to overcome analytical limitations. Two isotopic probes allow for the simultaneous and semi‐quantitative analysis at the femtomole level as well as qualitative analysis at attomole quantities that allows for detection of more than 200 metabolites in human fecal, urine and plasma samples. This comprehensive mass spectrometric analysis enhances the scope of metabolomics‐driven biomarker discovery. We anticipate that our chemical biology tool will be of general use in metabolomics analysis to obtain a better understanding of microbial interactions with the human host and disease development.
Keywords: bioorganic chemistry, chemoselectivity, mass spectrometry, metabolites, microbiome
Chemoselective probes based on stable isotope labeling allow for highly sensitive mass spectrometric quantification and detection of metabolites at attomole quantities in three human sample types.
Introduction
Trillions of microbes colonize the surfaces and cavities of the human body and particularly the gastrointestinal tract. [1] These communities are heavily metabolically active and exchange metabolites with other microbes and their host. [2] One of the beneficial relationships of these co‐evolved microbial communities with their host is protection against pathogenic infections. In contrast, abnormal alterations of the composition of the gut microbiota, termed microbiota dysbiosis, has been linked to disease development and other pathologies. [3] While metagenomic sequencing has revealed up‐ and downregulated bacterial species, the molecular mechanisms underpinning communication between the host and the microbiota are largely unknown. [1c] The microbiota genetic pool contains about 400 times more genetic information than the human genome and many of these genes encode metabolic reactions orthogonal to the human metabolism that constitute a major source of xenobiotics.[ 2c , 2d ] Many of these compounds are of unknown bioactivity and more likely remain to be discovered. In order to understand the importance of microbiota‐derived metabolites and their role in human physiology, techniques for the selective discovery and analysis of these metabolites are urgently required so that their bioactivity and impact on the human host can be better studied. [4]
Carbonyl‐containing metabolites are a highly reactive compound class that can form conjugates with DNA and proteins. They are also involved in a wide range of biochemical pathways and their dysregulation has been linked to a host of pathological conditions. [5] Furthermore, six carbonyl‐containing metabolites have been classified as carcinogens by the International Agency for Research of Cancer. [6] Reactive oxygen species (ROS) are one important part of carbonyl metabolites that play a crucial role in cell signaling in many biochemical processes. However, the deregulation of ROS can lead to reaction of carbonyl species with cellular macromolecules resulting in the unfavorable disruption of normal physiology. [7] The pathologies of diseases such as cancer, retinopathy and asthma have been associated with the imbalance between the ROS production and antioxidant defenses. [8]
Deciphering the interactions of reactive microbial metabolites with their host is therefore of critical importance. Liquid chromatography‐coupled mass spectrometry (LC‐MS) is ideally suited for discovery of biomarkers and new metabolites in complex biological matrices due to its sensitivity, dynamic range, and resolution. [9] However, many carbonyl‐containing metabolites possess poor ionization properties making them exceedingly difficult to analyze. [10] It is crucial to develop selective and sensitive analytical methods to obtain a greater overview of this reactive compound class in biological systems. Derivatization methods have been developed for carbonyl‐containing metabolites to improve their analysis and stability.[ 7b , 11 ] However, those methods are limited due to mass spectrometric interferences by complex matrices and to analysis of a low number of compounds only. We have recently developed a chemoselective probe immobilized to magnetic beads for the capture of metabolites in human samples. [12] This method enables increased mass spectrometric sensitivity by up to a factor of one million due to efficient removal of the unreacted matrix background through magnetic separation and improved ionization properties of the captured and tagged metabolites after release from the immobilized beads.
Here, we report the second generation of these probes, which have been redesigned to enable a new chemical biology methodology for advanced mass spectrometric analysis that we have termed Quantitative Sensitive CHEmoselective MetAbolomics (quant‐SCHEMA). The synthesis of 13C/12C isotopically labeled analogues of the probe and separate sample treatment allows for comparative and quantitative analysis of endogenous and microbiota‐derived metabolites in human samples at low concentrations. While isotope‐labeling is a well‐established part of the (chemo)proteomic workflow, it is rarely used in metabolomics due to the higher structural diversity of metabolites and concentration differences. [13] The few reports are based on simple chemistry without separation from the sample matrix, require harsh conditions for coupling, are limited in the validation of metabolites, or have only been applied to a single sample type.[ 11a , 14 ] This second generation of chemoselective probes described in this study now facilitates quantitative analysis that is an important improvement for comparative analysis of two sample sets. The redesign of the probe scaffold that only contains the bioorthogonal cleavage site from the first‐generation probe and now provides several advantages: i) quantitative and comparative analysis between two sample sets by use of a light and a heavy labeled probe; ii) a new coupling chemistry to amino‐activated magnetic beads facilitates a shorter synthetic route; iii) the reactive site is introduced to a primary amine for more efficient activation; iv) precise quantification of selected metabolites at low fmol concentrations without the need of commercially available isotope labeled compounds; v) treatment of three different human sample types with the identical procedure; and vi) biomarker discovery using a combination of isotope‐labeled chemoselective probe methodology and bioinformatic metabolomics analysis.
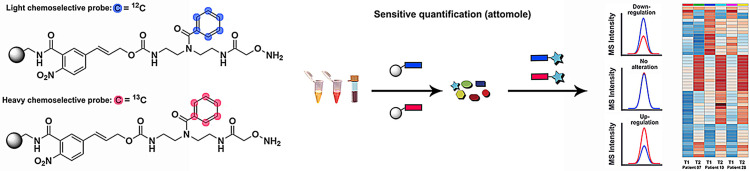
Elimination of undesired mass spectrometric matrix interferences and this new ionizable‐tag technology leads to substantially enhanced detection sensitivity of metabolites at attomole quantities. This method allows for the parallel quantitative analysis of known metabolites and yet unidentified metabolite. This is important for biomarker discovery as only an estimated 2 % of metabolites has been detected in the human metabolome. [15] Analysis of carbonyls in human fecal, plasma and urine samples collected from pancreatic cancer patients led to discovery of a series of unknown metabolites of this reactive compound class as well as a previously inaccessible overview of carbonyl regulation in humans.
Results and Discussion
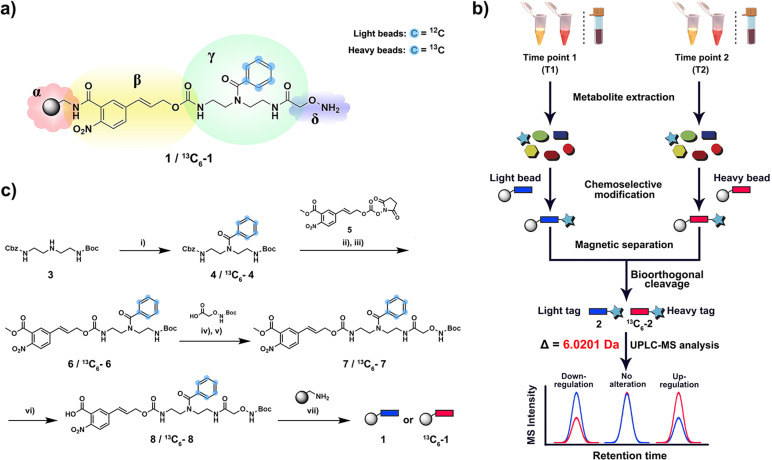
This new quantitative methodology is based on chemoselective probe 1 that is immobilized to magnetic beads and a corresponding stable isotopically labelled analogue 13 C6‐1 (Figure 1 a). The availability of two analogues that only differ in their mass but not in physicochemical properties allows for direct comparative mass spectrometric analysis of two sample sets. Human plasma and urine samples collected at two different time points were treated with the unlabeled (time point 1) and labeled (time point 2) chemoselective probe (Figure 1 b). Fecal samples were treated with the same methodology to investigate microbiome‐derived metabolites. After separate incubation for 16 h to capture carbonyl‐containing metabolites, the beads were isolated from each sample using a magnet, washed, and combined for bioorthogonal cleavage. The resulting tagged metabolites of the general structure 2/ 13 C6‐2 were analyzed using ultra performance liquid chromatography coupled to mass spectrometry (UPLC‐MS). The obtained data was analyzed using the bioinformatic software package XCMS in R. [16] Both conjugated metabolites 2 and 13 C6‐2 can be distinguished by their mass difference of 6.0201 Da without interference from the natural isotope distribution, while both elute at the same retention time due to the same physicochemical properties. These properties increase the accuracy and precision of the analysis and allow for a direct readout of relative changes between each captured metabolite in the two samples. The straightforward synthesis begins with benzoylation of the secondary amine in Boc‐ and Cbz‐protected diethylenetriamine 3 using either natural or 13C6‐labeled benzoic acid (Figure 1 c). After Cbz‐deprotection of intermediate 4, the free amine was coupled to the p‐nitrocinnamyloxycarbonyl (Noc)‐moiety 5 that serves as the bioorthogonal cleavage site. [12c] In a two‐step synthetic procedure, intermediate 6 was functionalized with the Boc‐protected alkoxyamine to yield the full‐length probe 7. This intermediate was saponified to obtain 8 for coupling to amino‐activated magnetic beads under standard peptide‐coupling conditions and TFA treatment furnishes chemoselective probe 1.
Figure 1.
Chemoselective probe for quantitative analysis of metabolites in human samples. a) Chemical structure of chemoselective probe (1 and 13C6−1) immobilized to magnetic beads. α=amino‐activated magnetic beads, β=bioorthogonal cleavage site, γ=metabolite tag containing isotope‐labeled moiety, δ=reactive site. b) Schematic workflow for analysis of human urine, plasma and fecal samples. After metabolite extraction, the metabolome of two different samples is treated with either “light” (1) or “heavy” beads (13C6–1). After separation of captured metabolites, bioorthogonal cleavage of both combined bead types releases tagged metabolites for UPLC‐MS analysis. c) Chemical synthesis of chemoselective probe and immobilization to magnetic beads. Reagents and conditions: i) benzoyl chloride 1.2 equiv, Et3N 3.0 equiv, DCM, room temperature (r.t.) 3 h. ii) Pd/H2, MeOH, r.t., 5 h. iii) N,N‐Diisopropylethylamine (DIPEA) 3.0 equiv, DCM, r.t., 5 h. iv) Trifluoroacetic acid (TFA)/ DCM 1:1, r.t., 2 h. v) 2‐(1H‐Benzotriazol‐1‐yl)‐1,1,3,3‐tetramethyluronium hexafluorophosphate (HBTU) 1.2 equiv, hydroxybenzotriazole (HOBT) 1.1 equiv, DIPEA 3.0 equiv, DCM, r.t., 16 h. vi) 2 M aqueous LiOH, MeOH/H2O 1:1, r.t., 30 min. vii) PyBop 1.5 equiv, HOBT 1.1 equiv, DIPEA 3.0 equiv, DMF/DCM 1:1, r.t., 16 h.
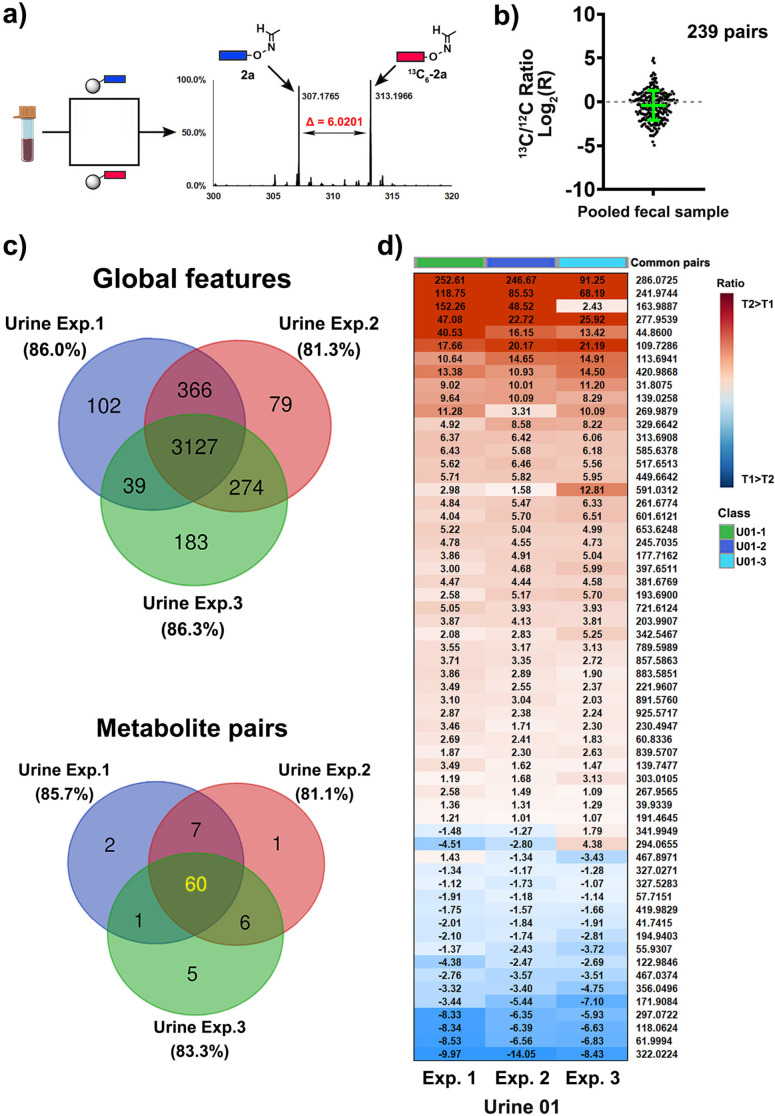
We have conducted a series of experiments to validate this new method for metabolite analysis in biological samples. We initially determined the bead load capacity for the coupling of the chemoselective probe that was applied for all following experiments at 0.8±0.1 μmol g−1 (Figure S1). As the next step, we determined the capture efficiency to be an average of 51.3 % for five selected metabolites (acetaldehyde, acetone, butanone, valeraldehyde, and dihydroxyacetone/ Table S1). An additional analysis of equimolar solutions for each metabolite as well as 1:3 and 3:1 ratios demonstrates the quality of this methodology (Figure S2). We have also determined the recovery rate for three selected carbonyl compounds that are representative for different reactivities of carbonyls in human samples: octanal as an alkylaldehyde, 2,3‐pentanedione as an alkylketone and phenylacetaldehyde as an aromatic aldehyde that is sterically hindered. Comparison of an aqueous solution of these compounds (c=100 μM each) to human urine samples with metabolites spiked in at the same concentration resulted in a recovery rate of 49–67 % with an average RSD of 8.2 % (Table S2). Our method was designed for straightforward identification of the captured carbonyl‐containing metabolites via bioinformatic analysis using XCMS through the difference of Δ=6.0201 Da in the MS chromatogram (Figure 2 a). The efficiency and reproducibility were tested using a pooled fecal sample from ten patients to maximize the number of carbonyl‐containing metabolites. The sample was split in two equal parts and each was either treated with the light (1) or the heavy 13C‐labeled chemoselective probe ( 13 C6‐1), the conjugates were cleaved from the magnetic beads after magnetic separation and analyzed using UPLC‐MS analysis. For the bioinformatic analysis, we used unmodified beads as a control sample that were treated with the same cleavage conditions to resemble the same mass spectrometric background. This experiment led to the identification of 239 metabolite pairs according to these criteria: i) difference of 6.0201 Da; ii) within retention time <0.05 min; iii) mass larger than the free probe (281.1608 Da); and iv) abundance significantly larger than the control sample. This number is greater than identified in fecal samples in any other study and exceeds our first‐generation analysis by a factor of two.[ 9c , 12a ] The reproducibility of this multistep method is illustrated by plotting the ratios of 13C/12C carbonyl‐conjugates, which is commonly used for determination of the reproducibility in related chemoproteomics research (Figure 2 b). This similarity of labeled and unlabeled probes is required for precise sample analysis as it accounts for differences between both synthetic compounds 1/13C6‐1, coupling efficiency to the magnetic beads, metabolite conjugation, bioorthogonal cleavage, and UPLC‐MS analysis.
Figure 2.
Method validation. a) Treatment of pooled fecal sample by both heavy and light chemoselective probe that can be separately analyzed in UPLC‐MS experiments with Δ=6.0201 Da with the example of conjugated acetaldehyde 2a/13C6–2a. b) 13C‐probe/12C‐probe ratios for 239 identified pairs in pooled fecal samples demonstrates the similarity of both treatments. The mean and SD are highlighted in green. A complete list of validated metabolites and m/z values for these carbonyl‐metabolites is reported in Figure S5 and Table S4. c) Reproducibility experiment of the same urine samples collected from one patient at two different time points. Venn diagram illustrates common global features and identified metabolite pairs among three independent experiments. d) Detailed illustration of measured ratios from three independent experiments in urine samples. Data are presented as mean ± SD from independent experimental triplicate (N=3).
This analysis was followed by an additional reproducibility test in urine samples from one patient but collected 6 months apart. Samples T1 and T2 were split in three parts each and each aliquot of T1 was treated with probe 1, while each aliquot of T2 was treated with probe 13 C6‐1. In this reproducibility experiment, 60 out of 82 pairs of labeled and unlabeled tagged metabolites were detected in all independently analyzed samples (81 % of metabolite pairs), which were identified out of 3,127 common features (Figure 2 c). These 60 metabolite pairs were found to be up‐ and down‐regulated in the same direction in repeated analyses, which is demonstrated by the T2/T1 ratios with relative standard deviations of up‐ and down‐regulated features of 21.4 % and 17.9 %, respectively (Figures 2 d and S3).
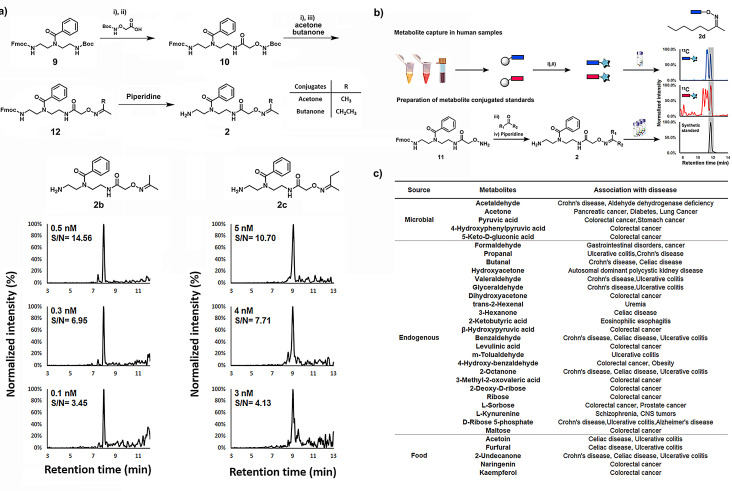
Maximizing the coverage of carbonyls during the mass spectrometric analysis requires the capture of metabolites with high selectivity, optimized ionization properties and solubility of the released tagged metabolite. Conjugates of acetone (2 b) and butanone (2 c) were synthesized from common precursor 10 in a straightforward three‐step synthetic route (Figure 3 a). These purified conjugates were utilized for LOD and LOQ experiments. The acetone‐conjugate 2 b can be detected at a concentration of 100 pM, which corresponds to an LOD of 500 amol. Furthermore, the LOQ for conjugates of acetone (2 b) and butanone (2 c) using this new method is c=0.5‐5 nM. To also include the reaction efficiency, we have also performed LOD measurements with varying concentrations of the carbonyl‐metabolite and a constant concentration of the 13C‐labeled reagent. The LOD values for acetone and butanone were in a similar range and for seven additional carbonyls were determined to be between 500 amol–500 fmol (Table S5).
Figure 3.
Improved mass spectrometric properties and structure validation. a) Synthetic scheme of conjugated acetone 2 b and conjugated butanone 2 c as well as their corresponding LOD/LOQ experiments. Limit of detection (LOD) was determined by signal to noise ratio >3, and limit of quantification (LOQ) was determined by signal to noise ratio >10. Reagent and condition: i) TFA/ DCM 1:1, r.t., 2 h, ii) HBTU 1.2 equiv, HOBT 1.1 equiv, DIPEA 3.0 equiv, DCM, r.t., 16 h. iii) acetone or butanone as solvent, r.t., 16 h. b) Workflow for the metabolite validation using conjugate standard metabolites with 2‐octanone 2 d as an example. c) List of metabolites according to their sources and with association to diseases.
Based on the reproducibility of the relative quantitative method and the highly increased mass spectrometric sensitivity, we constructed a chemical library of unlabeled metabolite conjugates of the general structure 2 (Figure 3 b). These reference compounds are required for structure validation of the captured carbonyl conjugates through comparison of retention times and co‐injection MS experiments (Figure S4 and Table S3). Additionally, the structural E/Z isomers of non‐symmetric aldehydes and ketones after reaction with the alkoxyamine reagent can readily be identified and analyzed in parallel.[ 12a , 17 ] Structure validation is critical as it is the bottleneck of metabolomics analysis and is key to biochemical analysis. [15] We chemically synthesized 107 biologically relevant conjugates 2 using the same straightforward synthetic route as for the LOD experiments for direct UPLC‐MS experiments.
Using this comprehensive library, we validated 60 metabolites in human fecal samples (Figure S5). These metabolites were identified in at least one of the samples and ranged from volatile metabolites acetone or acetaldehyde to dietary compounds, for example, furfural or kaempferol. To our knowledge, this number of validated metabolites has not been exceeded by any previous study, and nine of these metabolites have been detected for the first time in this sample type.[ 11a , 11b ] Carbonyl isomers co‐eluted in three cases and could not be distinguished from each other despite the use of standards. We thus analyzed these regioisomers combined. A series of captured metabolites, which were either of microbial origin, food products or endogenous metabolites, have previously been linked to disease development and can now be investigated in parallel (Figure 3 c). [18]
As fecal samples contain an abundance of microbiota‐derived metabolites generated in the human gut, this sample type represents a readout for microbiome metabolism. Acetaldehyde and acetone are volatile compounds that are common metabolic endpoints of many biochemical pathways in the microbiome. [19] Other examples include pyruvic acid and 4‐hydroxyphenylpyruvic acid that are excreted from E. coli but originate from different metabolic processes. 5‐Keto‐D‐gluconic acid is a known metabolite from the Gluconobacter genus, and is produced from D‐gluconate metabolism through catalysis by D‐gluconate dehydrogenase (GADH). [20] These compounds have been linked to the development of diverse diseases and demonstrate the ability of our procedure to track changes in concentrations of biologically‐relevant molecules in three of the most common sample types.
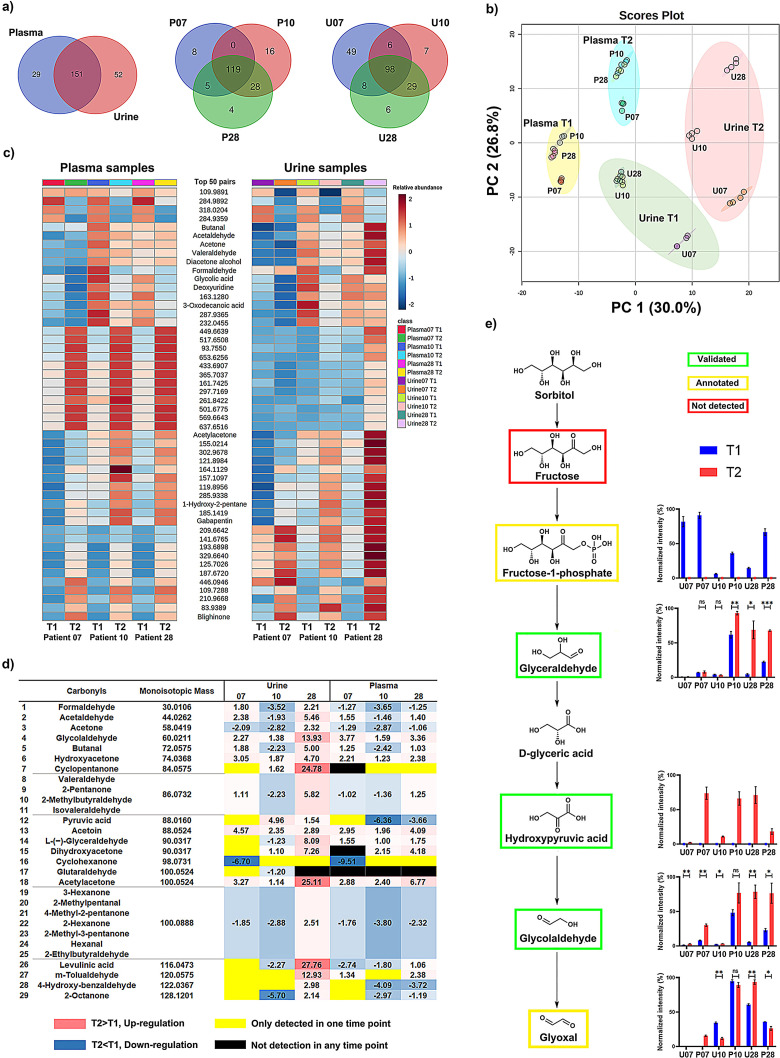
Human urine and plasma samples are the most commonly investigated sample types for metabolomics‐driven biomarker discovery as these ‘liquid biopsies’ are readily accessible. [21] While plasma samples represent a snapshot of ongoing metabolic pathways, urine samples contain the metabolic end‐products of the human clearance process as well as xenobiotics including microbiome‐derived compounds. For this study, we collected urine and plasma samples from the same high‐risk patients for pancreatic cancer at two different time points T1 and T2. We have applied our developed quant‐SCHEMA method for direct comparative analysis of both time points for each of the three individuals. The total number of detected carbonyls in these samples, confirmed by the pairs of the light and heavy tags, are 180 metabolites for plasma and 203 for urine samples (N=6; Figure 4 a). Our analysis revealed distinct pattern of metabolites in all investigated patient samples with 119 and 98 metabolites common to all three investigated plasma and urine samples, respectively. This reliable extraction and isolation of large numbers of carbonyl metabolites enables the large‐scale analysis of this reactive compound class in human samples for biomarker discovery and is enhanced to previous studies due to the increased MS sensitivity and metabolite identification procedure. The treated urine and plasma samples were analyzed in parallel with the same control sample in a randomized sequence allowing for comparison of the samples through principal components analysis (Figure 4 b). This unsupervised multivariate analysis clearly separated plasma and urine samples, but also clearly grouped samples according to collection time, while replicates remained tightly clustered.
Figure 4.
Large‐scale analysis in human patient samples. a) Venn diagrams of the peak pair numbers detected in the plasma (N=6) and urine samples (N=6). b) Principal component analysis (PCA) of plasma and urine samples for three individuals at different time points. c) Heatmap of the top 50 common carbonyl‐pairs in the plasma and urine analysis (p‐value, ANOVA). d) Time‐dependent fold‐change of validated carbonyl‐metabolites in plasma and urine samples. The complete data output from this experiment is shown as a heatmap in Figure S6. e) Investigated metabolites in the sorbitol metabolism pathway in plasma and urine samples.
As samples were collected from the same individuals, the carbonyl‐composition can be directly compared in both sample types (Figures 4 c and S6). Our mass spectrometric investigation into carbonyl‐metabolites revealed distinct individual pattern within the 50 most significant carbonyl‐conjugates that were detected in all six samples. This analysis revealed distinct signatures for individuals as well as collection timepoints. Interestingly, the five metabolites butanal, acetaldehyde, acetone, valeraldehyde, and diacetone alcohol follow the same changes in plasma and urine over time, which are considered volatile organic compounds (VOCs). VOCs are end‐products of different biochemical pathways or can be produced by pathogenic microorganisms. [22] Importantly, some of these compounds have been proposed as potential biomarkers for early‐stage diagnosis of diverse diseases. However, due to their volatile properties, this compound class is difficult to detect using conventional LC‐MS analysis. Our isotope‐labeling‐based methodology of this reactive compound class facilitates measurement over time in urine and plasma samples that is a first step towards monitoring of these important metabolites in humans. By taking a closer look at changes of metabolites with validated structures, we identified individual differences (Figure 4 d). We observed four metabolites upregulated in both sample types for each of the patients at the second timepoint (hydroxyacetone, glycolaldehyde, acetylacetone, and acetoin). It is also intriguing that acetaldehyde and butanal were different for each individual but has the same tendency in urine and plasma samples. This provides the opportunity to monitor changes of these metabolically important metabolites over time in both sample types in parallel. Furthermore, the xenobiotic cyclopentanone was detected in two urine samples, while cyclohexanone was solely detected in the samples from one patient (P10 and U10). Notably, the five microbial metabolites acetaldehyde, acetone, pyruvic acid, 4‐hydroxyphenylpyruvic acid, and 5‐keto‐D‐gluconic acid present in fecal samples were also detected in plasma and urine samples. Important to note that in the latter two sample types, these metabolites can either be products from the host or microbiome metabolism.
The broad applicability of the method has been demonstrated through the investigation of three different human sample types. The analysis of human plasma and urine samples of high‐risk patients at two different timepoints allows for robust mapping and monitoring of metabolic changes in high‐risk patients. Importantly, our method permits identification of metabolic variations in individuals over time that can in the future be utilized to identify disease onset. This has so far been unavailable for broad coverage of carbonyl‐metabolites in clinical human samples. While this method has been developed for metabolite profiling of the highly reactive carbonyl compound class, it can also be extended to analysis of other functionalities through modification of the reactive site δ (Figure 1 a).
Quant‐SCHEMA additionally allows for mapping and quantification of a wide range of metabolic pathways. Examples are the sorbitol and ascorbic acid metabolic pathways that contain carbonyl metabolites. Both are major pathways for endogenous production of glyoxal through autoxidation of glycolaldehyde (Figure 4 e and S7). Glyoxal is an α‐oxoaldehyde that is produced by different organisms including bacteria and humans by glucose oxidation, lipid peroxidation, and DNA oxidation. Additionally, this metabolite is a typical member of the highly reactive 1,2‐dicarbonyl compounds that has been investigated due to its detrimental effect on the gut microbiota. [23] The significantly up‐regulated values of glycolaldehyde in all individuals in both plasma and urine samples might be an indication for increased oxidative damage in these individuals, but requires further investigation.
Metabolomics analyses are being more commonly applied for investigation and prediction of the individual treatment responses such as statins treatment and breast cancer neoadjuvant chemotherapy. [24] In the present study, the precise carbonyl‐containing metabolite profiling of this longitudinal sample cohort has elucidated specific individual metabolic changes that can now be monitored in larger cohorts to identify signatures of disease onset and progression (Figure S6). The current clinical diagnosis and medication of diseases is generally applicable for the majority of the human population. However, symptoms relapse and unexpected side effects are inevitable for some patients owing to the genomic and metabolic differences of individuals. Personalized or precision medicine has been proposed in the past decades for disease management by taking individual's genomic variations, biochemical profiling and lifestyle into account for decisions on disease treatment. [25] Our method with the advantageous sensitivity for mass spectrometric investigation of this reactive compound class has a high potential for profiling this carbonyl‐metabotyping of individuals towards personalized diagnostics and medicine after future validation in large cohorts.
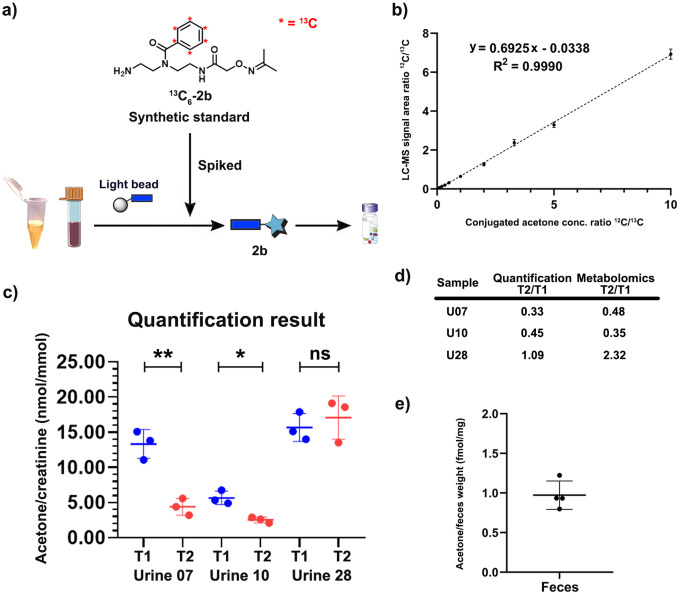
As our quant‐SCHEMA analysis is based on isotope‐labeled metabolite tags, it allows not only for the identification of metabolites but also their precise quantification in complex biological matrices. Additionally, our method provides a new straightforward possibility to synthesize 13C6‐labelled standards for the precise quantification of a metabolite of interest. This is advantageous as not all metabolites are commercially available as stable isotope labelled analogues. We sought to quantify acetone as an important example to further validate our results from the large‐scale carbonyl screening in Figure 4 d. Acetone is a volatile metabolite and difficult to quantify using LC‐MS analysis. It is produced by different metabolic pathways as well as the gut microbiome with importance in diseases such as diabetes. [27] We synthesized light and heavy labelled standards of the acetone conjugate 2 b and 13 C6‐2 b and used these to measure a calibration curve enabling quantification of acetone in urine and fecal samples (Figures 5 a and 5b).[ 9a , 28 ] The three urine samples were treated with the unlabeled probe 1 and the isotope labeled acetone‐standard ( 13 C6‐2 b) was spiked into the sample before the bioorthogonal cleavage. The quantification experiments were highly reproducible and identified significant changes in acetone concentration in urine samples from two individuals (Figure 5 c, average RSD=14.5 %). Importantly, the quantification experiments perfectly mirrored our metabolomics‐based readout from the comparative screening for urine samples (Figures 4 d and 5d). This method was also applied for quantification of acetone in fecal samples, which has never been performed before. We determined the quantity of acetone to be 0.97±0.16 fmol/ mg feces in four independent sample preparation and treatment experiments from the same individual. This demonstrates the high sensitivity and reproducibility of this mass spectrometric method and that the loss of volatile compounds during sample preparation is negligible (Figure 5 e).
Figure 5.
Quantification of acetone. a) Workflow for quantification of acetone in urine and fecal samples. b) Calibration curve of conjugated acetone (2 b). c) Quantification results of acetone in three urine samples at different time points. Data are presented as mean ± SD from experimental triplicates (N=3). ns: not significant, p>0.05; *: p<0.05; **: p<0.01. d) Comparison of quantification and semi‐quantitative metabolomics results. e) Quantification results of acetone in a fecal sample from one individual. Data are presented as mean ± SD from experimental quadruplicates (N=4).
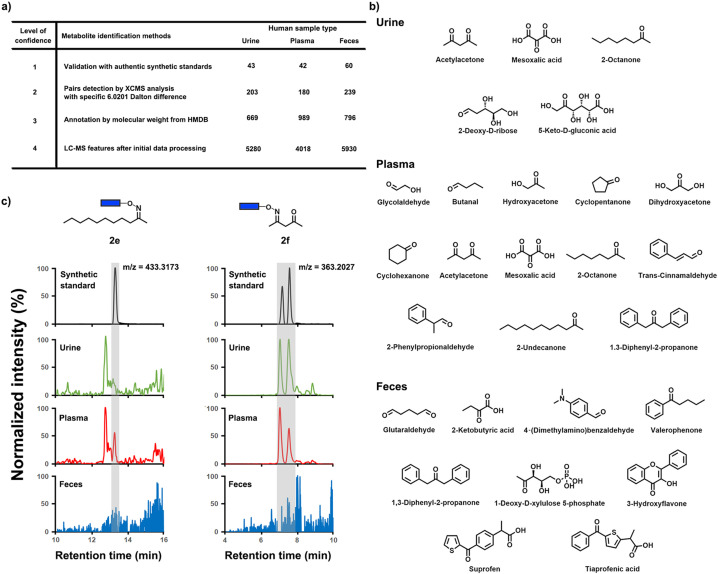
Our sensitive mass spectrometric analysis also led to the identification of previously undetected metabolites in all three investigated sample types. Due to removal of the captured metabolites from the complex sample matrix using magnetic separation, analysis in parallel of metabolites present in all three human sample types is possible. The retention time and peak shape for our analysis is identical for every sample type. This is an important advantage as the magnetic separation removes the common matrix effect that other derivatization methods and standard metabolomics studies need to consider through additional normalization procedures. [26] Validation of carbonyl‐containing metabolites with authentic conjugate standards led to 43 and 42 identified metabolites in urine and plasma, respectively (Figure 6 a and S8). With 60 validated metabolites in fecal samples, this is the highest numbers of validated metabolites in any reported study (Figure 3 c). Five carbonyl metabolites in urine, 13 in plasma and nine in fecal samples were detected for the first time, which belong to diverse compound classes (Figure 6 b). It is important to note that some of these 27 metabolites have previously been detected in another sample type. [18] Our evaluation process to determine the presence or absence of metabolites in the investigated three different sample types, is depicted in Figure 6 c for the conjugates of 2‐undecanone (2 e) and acetylacetone (2 f). The correct conjugate 2 e was identified next to an unknown regioisomer and was only detected in plasma samples, while 2 f was only present in plasma and urine samples.
Figure 6.
Metabolite discovery. a) Overview of identified metabolites in all three investigated sample types based on different levels of confidence. b) 21 newly discovered metabolites in each sample type. c) Representative metabolite detection procedure for conjugated 2‐undecanone (2 e) and acetylacetone (2 f) in urine, plasma and fecal samples compared to the synthetic standard.
Conclusion
In summary, the developed method represents a new and reproducible chemical biology tool that enhances metabolomics and metabolite analysis in complex human samples. This new methodology is based on a chemoselective probe immobilized to magnetic beads with a light and heavy isotope tag that allows for advanced analysis compared to other derivatization methods for metabolite analysis, which lack separation from the sample matrix and must overcome matrix interferences. Due to the combination of natural and isotope‐labeled conjugates before LC‐MS analysis, this new method does not require any normalization procedure including internal standards or normalization of different sample types. This is necessary in general metabolomics analysis to reduce technical errors. The parallel and quantitative analysis of metabolites at attomole quantities and detection of more than 200 metabolites in human fecal, urine and plasma samples has not been reported before. This method also facilitates detection of 21 metabolites that were not previously detected and can now even be quantified. This comprehensive analysis allows for enhanced mass spectrometric approaches to biomarker discovery and the possibility to monitor patients and their disease progression at different timepoints is a new tool towards personalized medicine and diagnostics.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Acknowledgements
We are grateful for funding by the Swedish Research Council (VR 2016–04423/ VR 2020–04707), the Swedish Cancer Foundation (19 0347 Pj), Carl Tryggers Foundation (CTS 2018:820), and a generous start‐up grant from the Science for Life Laboratory to D.G. This study made use of the NMR Uppsala infrastructure, which is funded by the Department of Chemistry—BMC and the Disciplinary Domain of Medicine and Pharmacy.
W. Lin, L. P. Conway, M. Vujasinovic, J.-M. Löhr, D. Globisch, Angew. Chem. Int. Ed. 2021, 60, 23232.
References
- 1.
- 1a. Schmidt T. S. B., Raes J., Bork P., Cell 2018, 172, 1198–1215; [DOI] [PubMed] [Google Scholar]
- 1b. Huttenhower C., et al., Nature 2012, 486, 207–214; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1c. Gill S. R., Pop M., DeBoy R. T., Eckburg P. B., Turnbaugh P. J., Samuel B. S., Gordon J. I., Relman D. A., Fraser-Liggett C. M., Nelson K. E., Science 2006, 312, 1355–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.
- 2a. Visconti A., Le Roy C. I., Rosa F., Rossi N., Martin T. C., Mohney R. P., Li W., de Rinaldis E., Bell J. T., Venter J. C., Nelson K. E., Spector T. D., Falchi M., Nat. Commun. 2019, 10, 4505; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2b. Ballet C., Correia M. S. P., Conway L. P., Locher T. L., Lehmann L. C., Garg N., Vujasinovic M., Deindl S., Lohr J. M., Globisch D., Chem. Sci. 2018, 9, 6233–6239; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2c. Donia M. S., Fischbach M. A., Science 2015, 349, 1254766; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2d. Nicholson J. K., Holmes E., Kinross J., Burcelin R., Gibson G., Jia W., Pettersson S., Science 2012, 336, 1262–1267; [DOI] [PubMed] [Google Scholar]
- 2e. Whidbey C., Sadler N. C., Nair R. N., Volk R. F., DeLeon A. J., Bramer L. M., Fansler S. J., Hansen J. R., Shukla A. K., Jansson J. K., Thrall B. D., Wright A. T., J. Am. Chem. Soc. 2019, 141, 42–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vogtmann E., Goedert J. J., Br. J. Cancer 2016, 114, 237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.
- 4a. Milshteyn A., Colosimo D. A., Brady S. F., Cell Host Microbe 2018, 23, 725–736; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4b. Healy A. R., Herzon S. B., J. Am. Chem. Soc. 2017, 139, 14817–14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.
- 5a. Singh M., Kapoor A., Bhatnagar A., Chem.-Biol. Interact. 2015, 234, 261–273; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5b. Benz C. C., Yau C., Nat. Rev. Cancer 2008, 8, 875–879; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5c. Yan S. D., Chen X., Schmidt A. M., Brett J., Godman G., Zou Y. S., Scott C. W., Caputo C., Frappier T., Smith M. A., Proc. Natl. Acad. Sci. USA 1994, 91, 7787–7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pinsky P., Rabeneck L., Lauby-Secretan B., N. Engl. J. Med. 2018, 379, 301–302. [DOI] [PubMed] [Google Scholar]
- 7.
- 7a. Dickinson B. C., Chang C. J., Nat. Chem. Biol. 2011, 7, 504–511; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7b. Holmquist E. F., Keiding U. B., Kold-Christensen R., Salomon T., Jorgensen K. A., Kristensen P., Poulsen T. B., Johannsen M., Anal. Chem. 2017, 89, 5066–5071. [DOI] [PubMed] [Google Scholar]
- 8. Auten R. L., Davis J. M., Pediatr. Res. 2009, 66, 121–127. [DOI] [PubMed] [Google Scholar]
- 9.
- 9a. Globisch D., Moreno A. Y., Hixon M. S., Nunes A. A. K., Denery J. R., Specht S., Hoerauf A., Janda K. D., Proc. Natl. Acad. Sci. USA 2013, 110, 4218–4223; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9b. Rinschen M. M., Ivanisevic J., Giera M., Siuzdak G., Nat. Rev. Mol. Cell Biol. 2019, 20, 353–367; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9c. Conway L. P., Rendo V., Correia M. S. P., Bergdahl I. A., Sjoblom T., Globisch D., Angew. Chem. Int. Ed. 2020, 59, 14342–14346; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2020, 132, 14448–14452. [Google Scholar]
- 10. Gil A., Siegel D., Permentier H., Reijngoud D. J., Dekker F., Bischoff R., Electrophoresis 2015, 36, 2156–2169. [DOI] [PubMed] [Google Scholar]
- 11.
- 11a. Zhao S., Dawe M., Guo K., Li L., Anal. Chem. 2017, 89, 6758–6765; [DOI] [PubMed] [Google Scholar]
- 11b. Deng P., Higashi R. M., Lane A. N., Bruntz R. C., Sun R. C., Ramakrishnam Raju M. V., Nantz M. H., Qi Z., Fan T. W., Analyst 2017, 143, 311–322; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11c. Kold-Christensen R., Jensen K. K., Smedegard-Holmquist E., Sorensen L. K., Hansen J., Jorgensen K. A., Kristensen P., Johannsen M., Redox Biol. 2019, 26, 101252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.
- 12a. Lin W., Conway L. P., Block A., Sommi G., Vujasinovic M., Lohr J. M., Globisch D., Analyst 2020, 145, 3822–3831; [DOI] [PubMed] [Google Scholar]
- 12b. Conway L. P., Garg N., Lin W., Vujasinovic M., Lohr J. M., Globisch D., Chem. Commun. 2019, 55, 9080–9083; [DOI] [PubMed] [Google Scholar]
- 12c. Garg N., Conway L. P., Ballet C., Correia M. S. P., Olsson F. K. S., Vujasinovic M., Lohr J. M., Globisch D., Angew. Chem. Int. Ed. 2018, 57, 13805–13809; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2018, 130, 14001–14005. [Google Scholar]
- 13.
- 13a. Böttcher T., Pitscheider M., Sieber S. A., Angew. Chem. Int. Ed. 2010, 49, 2680–2698; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2010, 122, 2740–2759; [Google Scholar]
- 13b. Hacker S. M., Backus K. M., Lazear M. R., Forli S., Correia B. E., Cravatt B. F., Nat. Chem. 2017, 9, 1181–1190; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13c. Chokkathukalam A., Kim D. H., Barrett M. P., Breitling R., Creek D. J., Bioanalysis 2014, 6, 511–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.
- 14a. Carlson E. E., Cravatt B. F., J. Am. Chem. Soc. 2007, 129, 15780–15782; [DOI] [PubMed] [Google Scholar]
- 14b. Carlson E. E., Cravatt B. F., Nat. Methods 2007, 4, 429–435. [DOI] [PubMed] [Google Scholar]
- 15.
- 15a. da Silva R. R., Dorrestein P. C., Quinn R. A., Proc. Natl. Acad. Sci. USA 2015, 112, 12549–12550; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15b. Peisl B. Y. L., Schymanski E. L., Wilmes P., Anal. Chim. Acta 2018, 1037, 13–27. [DOI] [PubMed] [Google Scholar]
- 16. Domingo-Almenara X., et al., Nat. Methods 2018, 15, 681–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Byrne P. A., Gilheany D. G., Chem. Soc. Rev. 2013, 42, 6670–6696. [DOI] [PubMed] [Google Scholar]
- 18. Wishart D. S., et al., Nucleic Acids Res. 2018, 46, D608–D617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rondanelli M., Perdoni F., Infantino V., Faliva M. A., Peroni G., Iannello G., Nichetti M., Alalwan T. A., Perna S., Cocuzza C., J. Anal. Methods Chem. 2019, 7247802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Matsushita K., Fujii Y., Ano Y., Toyama H., Shinjoh M., Tomiyama N., Miyazaki T., Sugisawa T., Hoshino T., Adachi O., Appl. Environ. Microbiol. 2003, 69, 1959–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gowda G. A., Zhang S., Gu H., Asiago V., Shanaiah N., Raftery D., Expert Rev. Mol. Diagn. 2008, 8, 617–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Weisskopf L., Schulz S., Garbeva P., Nat. Rev. Microbiol. 2021, 19, 391–404. [DOI] [PubMed] [Google Scholar]
- 23. Lee C., Park C., Int. J. Mol. Sci. 2017, 18, 169. [Google Scholar]
- 24.
- 24a. Trupp M., Zhu H., Wikoff W. R., Baillie R. A., Zeng Z. B., Karp P. D., Fiehn O., Krauss R. M., Kaddurah-Daouk R., PLoS One 2012, 7, e38386; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24b. Wei S., Liu L., Zhang J., Bowers J., Gowda G. A., Seeger H., Fehm T., Neubauer H. J., Vogel U., Clare S. E., Raftery D., Mol. Oncol. 2013, 7, 297–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jacob M., Lopata A. L., Dasouki M., Abdel Rahman A. M., Mass Spectrom. Rev. 2019, 38, 221–238. [DOI] [PubMed] [Google Scholar]
- 26. Wu Y., Li L., J. Chromatogr. A 2016, 1430, 80–95. [DOI] [PubMed] [Google Scholar]
- 27. Bales J. R., Higham D. P., Howe I., Nicholson J. K., Sadler P. J., Clin. Chem. 1984, 30, 426–432. [PubMed] [Google Scholar]
- 28. Globisch D., Pearson D., Hienzsch A., Brückl T., Wagner M., Thoma I., Thumbs P., Reiter V., Kneuttinger A. C., Müller M., Sieber S. A., Carell T., Angew. Chem. Int. Ed. 2011, 50, 9739–9742; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2011, 123, 9913–9916. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information