Abstract
Complex structures derived from multiple tissue types are challenging to study in vivo, and our knowledge of how cells from different tissues are coordinated is limited. Model organisms have proven invaluable for improving our understanding of how chemical and mechanical cues between cells from two different tissues can govern specific morphogenetic events. Here we used Caenorhabditis elegans as a model system to show how cells from three different tissues are coordinated to give rise to the anterior lumen. While some aspects of pharyngeal morphogenesis have been well-described, it is less clear how cells from the pharynx, epidermis and neuroblasts coordinate to define the location of the anterior lumen and supporting structures. Using various microscopy and software approaches, we define the movements and patterns of these cells during anterior morphogenesis. Projections from the anterior-most pharyngeal cells (arcade cells) provide the first visible markers for the location of the future lumen, and facilitate patterning of the surrounding neuroblasts. These neuroblast patterns control the rate of migration of the anterior epidermal cells, whereas the epidermal cells ultimately reinforce and control the position of the future lumen, as they must join with the pharyngeal cells for their epithelialization. Our studies are the first to characterize anterior morphogenesis in C. elegans in detail and should lay the framework for identifying how these different patterns are controlled at the molecular level.
Keywords: C. elegans, morphogenesis, rosettes, polarity, cell migration, contractility, adhesion
Graphical Abstract
Introduction
Our knowledge of how complex structures form in the developing embryo is limited due to challenges in studying the morphogenesis of multiple tissues simultaneously in vivo. C. elegans is an extremely powerful organism to use for studies of development at the cellular and subcellular level, as they have multiple tissues with a defined number of cells. They are also a powerful genetic model, and the development of transgenic and microscopy tools makes them ideal for studying complex events in vivo. As the mechanisms regulating cell polarity and migration are highly conserved, studies of tissue morphogenesis in C. elegans have provided insight into how tissues develop in more complex organisms (Jacinto et al., 2001; Muller and Bossinger, 2003; Campanale et al., 2017).
Anterior morphogenesis is required for development of the anterior lumen, and involves the coordination of epidermal cells, pharyngeal cells and neuroblasts (neuronal precursor cells). Due to this complexity, it is not known how all three tissues form the anterior lumen, but revealing this could provide fundamental knowledge that is relevant for other complex developmental processes. The timing of anterior morphogenesis coincides with epidermal and pharyngeal morphogenesis, which are outlined below. The anterior-most epidermal cells migrate toward the anterior of the embryo after the epidermal cells meet at the ventral midline. Similarly, a large subset of pharyngeal cells polarizes and forms a cyst to define a lumen that aligns with the intestinal cells (Rasmussen et al., 2012). Anterior to the cyst are the arcade cells, which migrate anteriorly before moving back inward as development progresses (Portereiko and Mango, 2001; Portereiko et al., 2004; Mango 2009). They also polarize and coordinate with the epidermal cells for epithelialization of the anterior pharynx, but it is not clear how this occurs (Von Stetina and Mango, 2015). During anterior morphogenesis, the neuroblasts presumably also undergo specific movements and patterning, but with the exception of the amphid neurons this has not been extensively studied (Fan et al., 2019). Importantly, how all of these cell types are coordinated to give rise to a properly positioned anterior lumen remains poorly understood.
Epidermal morphogenesis has been relatively well-characterized, and requires chemical and mechanical signaling between different cell types. During mid-embryogenesis, the dorsal epidermal cells intercalate, followed by migration of the ventral epidermal cells toward the ventral midline to enclose the embryo through a process called ventral enclosure (Williams-Masson et al., 1997; Chisholm and Hardin, 2005). Ventral epidermal cell migration relies on precisely positioned neuroblasts for chemical and/or mechanical signaling (Bernadskaya et al., 2012; Ikegami et al., 2012; Wernike et al., 2016). EFN-VAB signaling is required for neuroblast positioning and for ventral enclosure, although it is not clear if the ligands and/or receptors are required in the neuroblasts or epidermal cells (George et al., 1998; Chin-Sang et al., 1999; Bernadskaya et al., 2012). Other signaling pathways implicated in controlling ventral epidermal cell migration include semaphorin (MAB-20) and plexin (PLX-2; Roy et al., 2000; Nakao et al., 2007; Ikegami et al., 2012). One model is that the receptors are expressed in the epidermal cells and receive cues from the neuroblasts or neighbouring epidermal cells to regulate branched F-actin assembly for their migration (Withee et al., 2004; Chisholm and Hardin, 2005; Patel et al., 2008; Bernadskaya et al., 2012; Patel and Soto, 2013; Wallace et al., 2018). In addition, subsets of neuroblasts in the middle-posterior of the embryo form rosettes, likely via planar cell polarity, to elongate the tissue in preparation for epidermal elongation (Wernike et al., 2016; Shah et al., 2017). Rosettes are patterns required for the morphogenesis of different metazoan tissues, and form by apical-basal polarity or planar cell polarity, the latter of which are often associated with transient rosettes that facilitate tissue re-organization (Blankenship et al., 2006; Harding et al., 2014). The neuroblast rosettes that form in the ventral pocket may mechanically influence migration of the overlying epidermal cells during ventral enclosure (Wernike et al., 2016). Non-muscle myosin (NMY-2) is required in both the underlying neuroblasts and the epidermal cells for ventral enclosure, suggesting that these two tissues are mechanically connected (Wernike et al., 2016). Mechanical signaling also occurs between the epidermal and muscle cells to drive later stages of elongation, the subsequent step in epidermal morphogenesis (Zhang et al., 2011). A recent study showed that the amphid neuronal precursors form lateral rosettes and maintain their organization as they co-migrate anteriorly with the epidermal cells (Fan et al., 2019). The authors proposed that there is chemical and/or mechanical feedback between these cell-types (Fan et al., 2019). Other neuroblast-epidermal cell interactions presumably also guide the anterior migration of epidermal cells for anterior morphogenesis, although this is not known.
Coincident with ventral epidermal morphogenesis, a large subset of pharyngeal cells polarizes to form a cyst to define the pharyngeal lumen. These cells apically constrict with PAR-3 accumulated apically and laminin basally to form a cyst (rosette) with a central lumen (Rasmussen et al. 2012). Apical rosettes are formed by cells that apically constrict as a result of actomyosin activity, which is coordinated at the intercellular level via adhesion junctions (Sawyer et al., 2010; Martin and Goldstein, 2014). This gives rise to a bulb-like organization of cells with their narrow tips coming together to form a small hole apically (Harding et al., 2014). Apically formed rosettes are typically highly stable and ultimately give rise to lumens (Lecaudey et al., 2008; Nechiporuk and Raible, 2008). Interestingly, the anterior arcade cells remain distinct from the large pharyngeal cyst, yet must polarize to form a contiguous lumen (Portereiko and Mango, 2001; Portereiko et al., 2004; Mango 2009; Von Stetina and Mango, 2015). These cells need to be precisely positioned in line with the more posterior pharyngeal cells and the anterior epidermal cells, but as indicated earlier, it is not clear how they do this.
In metazoans, apicobasal polarity occurs due to the recruitment and maintenance of apical complexes (e.g. Par3, Par6, aPKC; Etemad-Moghadam et al., 1995; Hung and Kemphues, 1999; McMahon et al., 2001). Many factors contribute to the onset and establishment of this asymmetric segregation of proteins, including the trafficking and turnover of active Cdc42, actomyosin organization, asymmetric distribution of specific phospholipids, adhesion junctions and mutual antagonism (Jiang et al., 2015; Campanale et al., 2017; Jewett and Prekeris, 2018; Motegi et al., 2020). In C. elegans, intestinal cells acquire apicobasal polarity, which is crucial for formation of the interior lumen (Leung et al., 1999; Bernadskaya et al., 2012; Shafaq-Zadah et al., 2012; Rasmussen et al., 2012; Bossinger et al. 2015; Asan et al., 2016). Epidermal cells also have apicobasal polarity, which is reinforced by adhesion junction components that connect neighboring cells (Patel et al., 2008; Patel and Soto, 2013). These include the DAC (DLG-1 and AJM-1 Complex) and the CCC (Catenin Cadherin Complex; E-cadherin [HMR-1], α-catenin [HMP-1] and β-catenin [HMP-2]) which are located sub-apically (Costa et al., 1998; Labouesse, 2006; Totong et al., 2007; Armenti and Nance, 2012; Pasti and Labouesse, 2015; Gillard et al., 2015; Sasidharan et al., 2018). Adhesion junctions connect actomyosin filaments intercellularly to coordinate cell movements and patterning. The CCC, but not the DAC, is also expressed in neuroblasts (Wernike et al., 2016) and the function of the CCC in these cells is not clear. The expression of different adhesion junction components in different cell types could influence adhesion and how mechanical forces are transmitted between cells.
Here we provide the first detailed description of C. elegans anterior morphogenesis, which involves the coordination of neuronal precursors and their support cells, epidermal and pharyngeal cells. Using diSPIM (dual-view inverted selective plane illumination microscopy) and confocal imaging, we observed that subsets of neuroblasts form concentric patterns around the site of the future lumen and that some of these neuroblasts ingress as the epidermal cells migrate anteriorly. We also observed that a subset of pharyngeal arcade cells forms a stable rosette, which aligns with the previously described pharyngeal cyst. The projections from these cells initially are enriched in PAR-6, NMY-2 and HMP-1 (CCC), which mark the site of the future lumen. Additionally, subsets of neuroblasts form specific patterns of foci around this location, which contain ANI-1, PAR-6, NMY-2 and CCC components. Other foci are closely associated with both the neuroblasts and the migrating epidermal cells and contain PAR-6, NMY-2, CCC and DAC components. All of the foci mature over time to also include the DAC, coincident with when the epidermal cells reach the anterior. The arcade cells regulate the patterning of neuroblasts, while the neuroblasts are required for epidermal cell migration and the timing of lumen formation, and the number of epidermal cells influences the position of the lumen. This is to our knowledge, the first in-depth description of anterior morphogenesis in C. elegans embryos.
Materials and Methods
Strains
Strains were maintained on nematode growth medium (NGM) agar plates with a lawn of Escherichia coli (OP50) according to standard procedures (Brenner, 1974). The list of C. elegans strains used in this study is presented in Table 1. All strains were maintained at 20°C unless indicated otherwise. Genetic crosses were performed using standard protocols (for review, see Fay, 2005).
Table 1.
The C. elegans strains used in this study are listed below.
| UM463 | cpIs42[mex-5p::mNeonGreen::PLCδ-PH::tbb-2 3’UTR, unc-119(+)] II; ItIs37[unc-119(+), Ppie-1::mCherry::HIS-58] IV |
| SM481 | pxIs10 [pha-4::GFP::CAAX + (pRF4) rol-6(su1006)] |
| UM456 | cpSi20[Pmex-5::TAGRFPT::PH::tbb-2 3’UTR; unc-119 (+)] I; unc-119(ed3) III |
| SU159 | ajm-1(ok160) X; jcEx44 [ajm-1::GFP + rol-6(su1006)] |
| SU265 | jcIs17 [hmp-1p::hmp-1::GFP + dlg-1p::dlg-1::DsRed + rol-6(su1006)] |
| ML916 | mcIs40 [lin-26p::ABDvab-10::mCherry+myo-2p::GFP] |
| FT1197 | unc-119(ed3) III; xnIs449 [lin-26::LifeAct::GFP + unc-119(+)] |
| LP216 | par-6(cp45[par-6::mNeonGreen::3xFlag + LoxP unc-119(+) LoxP]) I; unc-119(ed3) III |
| LP162 | nmy-2(cp13[nmy-2::GFP + LoxP]) I |
| MDX29 | ani-1(mon7[mNeonGreen^3xFlag::ani-1]) III |
| AJP100 | mcIs40 [lin-26p::ABDvab-10::mCherry+myo-2p::GFP]; par-6(cp45[par-6::mNeonGreen::3xFlag + LoxP unc-119(+) LoxP]) I; unc-119(ed3) III |
| AJP101 | nmy-2(cp13[nmy-2::GFP + LoxP]) I; cpIs56 [mex-5p::TagRFP-T::PLC(delta)-PH::tbb-2 3’UTR + unc-119 (+)] II |
| AJP102 | cpIs56 [mex-5p::TagRFP-T::PLC(delta)-PH::tbb-2 3’UTR + unc-119 (+)] II; ani-1(mon7[mNeonGreen^3xFlag::ani-1]) III |
| AJP103 | cpIs56 [mex-5p::TagRFP-T::PLC(delta)-PH::tbb-2 3’UTR + unc-119 (+)] II; par-6(cp45[par-6::mNeonGreen::3xFlag + LoxP unc-119(+) LoxP]) I; unc-119(ed3) III |
| AJP104 | pxIs10 [pha-4::GFP::CAAX + (pRF4) rol-6(su1006)]; unc-119(ed3) III; xnIs449 [lin-26::LifeAct::GFP + unc-119(+)]; cpIs56 [mex-5p::TagRFP-T::PLC(delta)-PH::tbb-2 3’UTR + unc-119 (+)] II |
| TU3335 | uls57[unc-119p::YFP + unc-119p::sid-1+ mec-6p::mec-6] |
| LP244 | par-6(cp60[par-6::mKate2::3xMyc + LoxP unc-119(+) LoxP]) I; unc-119(ed3) III |
| AJP105 | uls57[unc-119p::YFP + unc-119p::sid-1+ mec-6p::mec-6]; par-6(cp60[par-6::mKate2::3xMyc + LoxP unc-119(+) LoxP]) I; unc-119(ed3) III |
| AJP106 | pxIs10 [pha-4::GFP::CAAX + (pRF4) rol-6(su1006)]; par-6(cp60[par-6::mKate2::3xMyc + LoxP unc-119(+) LoxP]) I; unc-119(ed3) III |
| pGR71 | hsls391[Pmir-228::myristoylated-GFP; lin-15(+)] |
| OH9729 | otls302[Plsy-6::GFP; Pelt-2::RFP] |
| lin-15(n744); nsEx4011[Phlh-16::GFP::myristoylated-GFP::UTR (pGR133); lin-15(+)] | |
| AJP107 | lin-15(n744); nsEx4011[Phlh-16::GFP::myristoylated-GFP::UTR (pGR133); lin-15(+)];par-6(cp60[par-6::mKate2::3xMyc + LoxP unc-119(+) LoxP]) I; unc-119(ed3) III |
| AJP108 | hsls391[Pmir-228::myristoylated-GFP; lin-15(+)]; par-6(cp60[par-6::mKate2::3xMyc + LoxP unc-119(+) LoxP]) I; unc-119(ed3) III |
RNAi
RNA-mediated interference (RNAi) by bacterial feeding was performed as described (Kamath et al., 2001; Timmons et al., 2001). Briefly, RNAi plates were made from NGM as above, with 50 mg/ml ampicillin and 1 mM IPTG. After growth to OD 0.6–1.0 (~6–12 hours), the cells were pelleted from their initial 5 mL volume and resuspended in varying amounts of LB to control the RNAi strength (e.g., in 100–400 μL). Third and fourth larval stage (L3/L4) hermaphrodites were transferred to plates with E. coli (HT115) transformed with dsRNA constructs; the animals were incubated for 24 hours, and then transferred to fresh RNAi plates, and progeny from these second plates were assessed for phenotype after 48 hours (Kamath et al., 2001). For the analysis of phenotypes after pha-4 RNAi, worms were incubated for 24 hours, while they were only incubated for 3 hours for the analysis of phenotypes after zen-4 RNAi. The Y49E10.19 (ani-1 RNAi), W09C2.1 (elt-1 RNAi), M03D4.1 (zen-4 RNAi) and F38A6 (pha-4 RNAi) clones used in this study were generously provided by J. C. Labbe, IRIC, Montreal and Michael Glotzer, University of Chicago).
Microscopy
Imaging was performed on embryos collected as described (Sulston et al., 1983; Wernike et al., 2016). Images were acquired using a Nikon Eclipse Ti inverted microscope with a NIDAQ/Piezo stage, a 100× PlanApo lens (NA, 1.4), sweptfield confocal illumination (livescan; Bruker), an Ixon3 EMCCD camera (Andor) and NIS-Elements acquisition software. Fluorophores were excited using 488 nm and 561 nm lasers and a dual bandpass emission filter (520/20 nm + 630/30 nm). Z-stacks were collected every 0.5 μm, and embryos were imaged every 2 minutes for up to 2 hours. To limit phototoxicity and photobleaching, exposure times were kept below 300 milliseconds. For RNAi-treated embryos, z-stacks were captured at 0.5 μm intervals every 4 minutes. For en face views, 0.2 μm z-stacks were collected and acquired at intervals of 2–4 minutes. To image embryos en face, they were manipulated to be positioned vertically (upright) in small holes made in the agarose pads.
Images were also acquired using a fiber-coupled diSPIM (parts list and construction detailed in Kumar et al., 2014) with MicroManager software (Open-Source, Vale Lab UCSF). C. elegans embryos were mounted in an open-well chamber as described (Duncan et al., 2019). Image volumes were acquired in either single- or dual-view mode. Z-stacks were collected at 0.5 μm intervals with single-slice acquisition times of 12.75 milliseconds, leading to total volume acquisition times of 1.25 seconds for single-view volumes and 2.76 seconds for dual-view volumes. For single-view datasets, embryos were exposed simultaneously to 488 nm and 561 nm excitation with the emission optically split using a Hamamtsu W-View Gemini image splitter. Multi-point mode was used to capture multiple, spatially separated embryos per imaging session. For dual-view datasets, images were acquired sequentially, although within each path excitation at 488 nm and 561 nm was simultaneous and was again optically split with the Hamamtsu W-View Gemini image splitter. Dual-view data were also acquired using multi-point mode with multiple embryos captured per imaging run.
HILO, modified total internal reflection fluorescence (TIRF) imaging, was performed using an inverted Nikon Ti-E microscope outfitted with a NI-DAQ piezo Z stage (National Instruments), an Evolve (EMCCD) camera, with Elements 4.0 acquisition software (Nikon), filters for 488 and 561 laser diodes, and a 100x CFI Apo TIRF objective. Images were exported as TIFFs and opened in Image J (NIH Image) to create Z-stack projections, perform image rotation and to crop desired regions.
Image Analysis
Data files obtained by sweptfield confocal imaging were deconvolved using AutoQuant X3 (MediaCybernetics) with adaptive point-spread function (PSF) and blind deconvolution. The total number of iterations was set to 10 and the noise level was set to medium. All measurements were performed in FIJI (Fiji Is Just ImageJ; NIH) using the deconvolved images. The datasets were transferred to FIJI to generate hyperstacks and/or maximum-intensity projections and exported as TIFF files.
Single-view data obtained from diSPIM were exported as TIFFstacks through MicroManager software, and are shown in Fig. 1C. Dual-view data were deconvolved after export from MicroManager using custom fusion and deconvolution software (Guo et al., 2019). All datasets were then screened and subsequently analyzed.
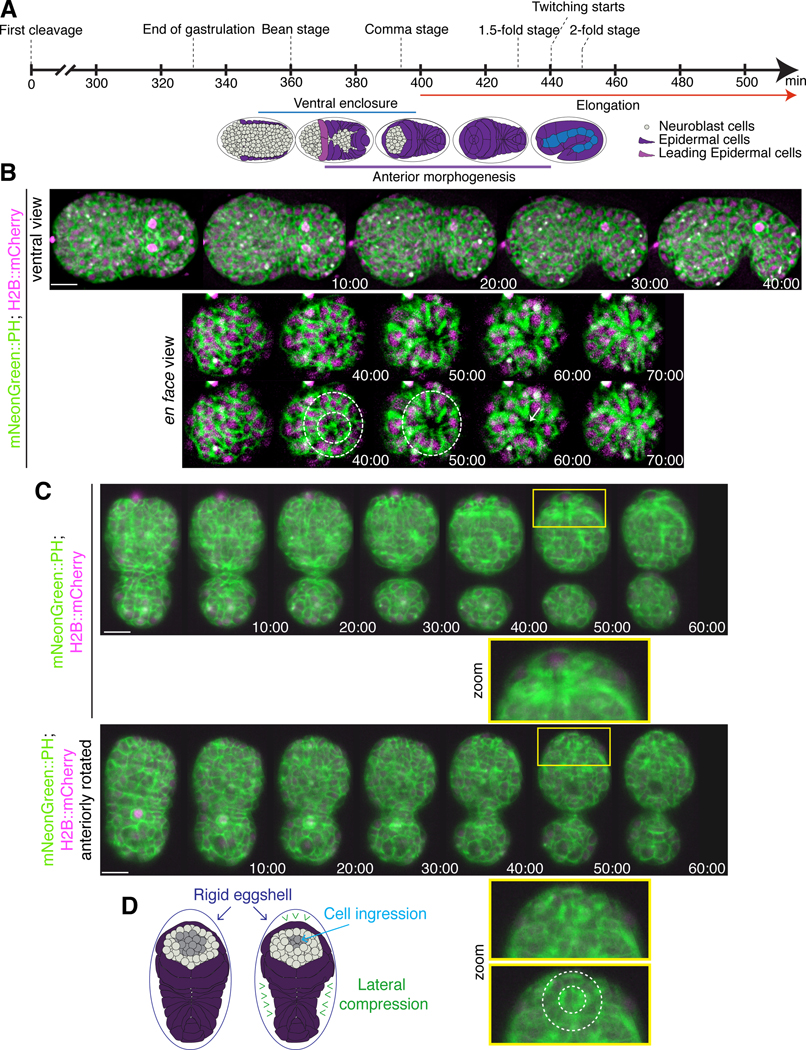
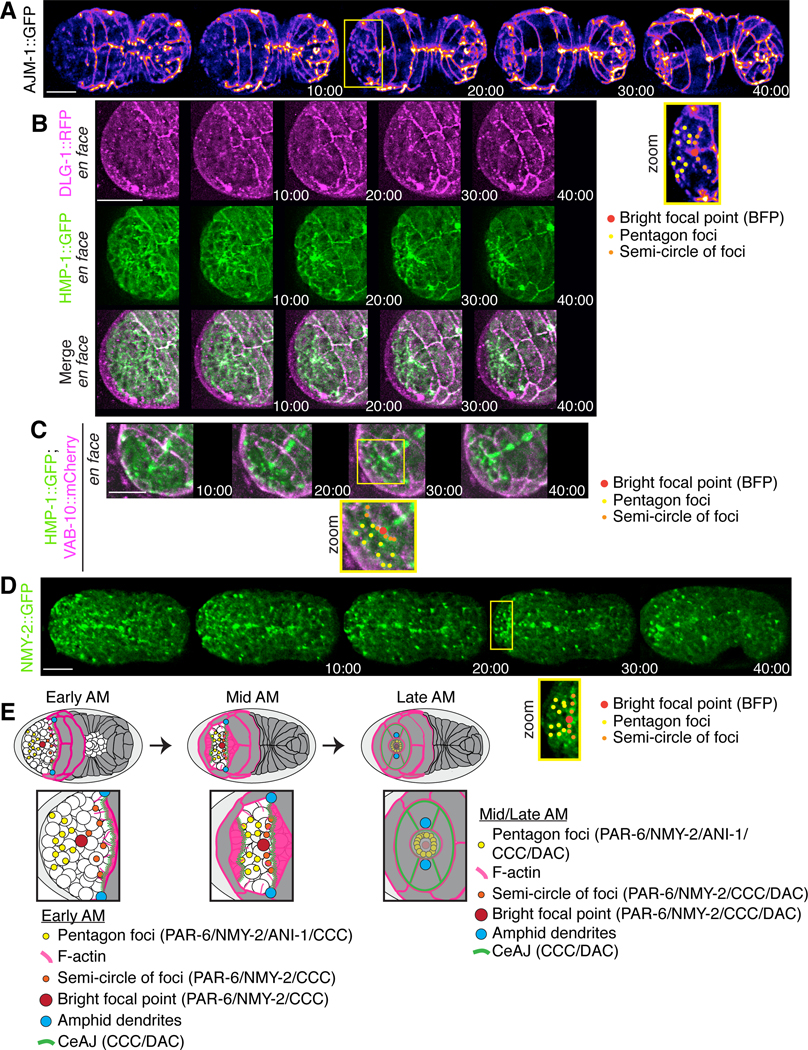
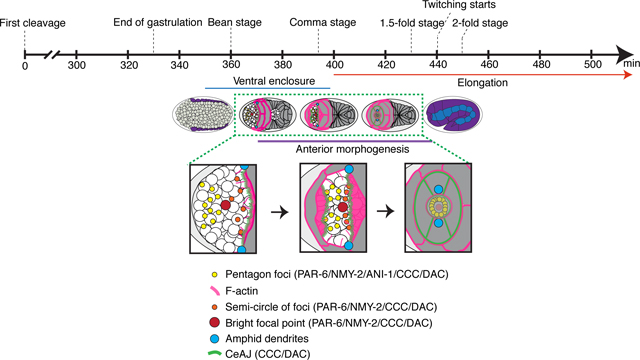
Figure 1. Cells ingress during anterior morphogenesis.
A) The timeline (in minutes) shows the key morphogenetic events that occur during C. elegans embryogenesis. Anterior morphogenesis begins when the leading pair of migrating epidermal cells meet at the ventral midline (t = ~380), and continues throughout ventral enclosure and part of elongation (~440 minutes/twitching stage). B) Time-lapse images obtained by sweptfield microscopy (ventral view; anterior to the left) show an embryo co-expressing mNeonGreen::PH to visualize membranes (green) and H2B::mCherry to visualize nuclei (magenta) through anterior morphogenesis (time in minutes). The en face view shows cell organization at the anterior of the embryo, which results in the formation of a ‘ring in a ring’ (white dashed circles). The star-like pattern in the center marks the site of the future lumen, which forms after cells ingress into the embryo. Times are shown in minutes, and the scale bar is 10 μm. C) Raw time-lapse single-view images (ventral view; anterior pointing up) acquired using diSPIM show embryos as in B during anterior morphogenesis. The top panels show cell ingression at the anterior, whereas the bottom panels are tilted (anterior towards the reader) to show cell patterns (zoom, white dashed circles). Times are shown in minutes, and the scale bar is 10 μm. Zoom images have been magnified by 200%. D) Cartoon schematics show the embryo inside the eggshell. The epidermal cells are shown in purple, the ingressing cells are shown in grey, and arrowheads (green) point to the forces generated laterally to cause ingression anteriorly.
The circumference of the arcade-cell rosette and cell length measurements were performed on Z-stack projections from 1.5-fold embryos expressing pha-4p::GFP::CAAX using FIJI as shown in Figure 2D. To measure the rosette diameter, en-face images were used, and three lines drawn across the diameter were measured for each embryo and averaged together. To measure cell length, lateral-view images were used, and the length of the most in-focus cell was measured from top to bottom. All measurements were converted from pixels to micrometers.
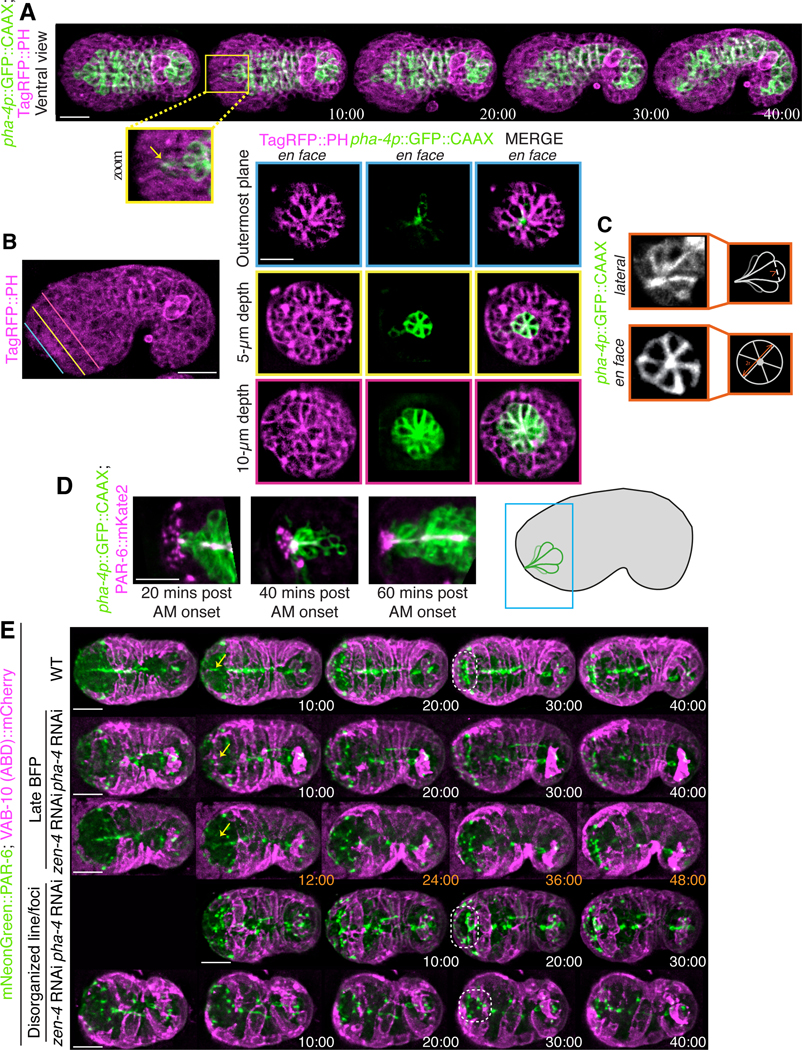
Figure 2. The pharyngeal arcade cells form a rosette.
A) Time-lapse images acquired using sweptfield microscopy show an embryo co-expressing pha-4p::GFP::CAAX to visualize the pharyngeal cells in green and Tag-RFP::PH to visualize membranes in magenta during anterior morphogenesis. The anterior-most pharyngeal cells (arcade cells) form apical constrictions (yellow box) which coalesce (zoom; yellow arrow). Times are shown in minutes. B) The image shows a comma-staged embryo expressing Tag-RFP::PH to visualize membranes (magenta). The colored lines indicate the three depths in the embryo corresponding to the images to the right. Images show separate and merged channels from an embryo co-expressing Tag-RFP::PH (magenta) and pha-4p::GFP::CAAX to visualize pharyngeal cells (green) at ~ 40 min. Three en face views are shown, which correspond to the depths (colored lines). The outermost plane (blue) shows the coalescence of the arcade-cell projections at the anterior of the embryo. At a 5 μm depth into the embryo (yellow) a small multicellular pharyngeal rosette is visible. The larger, previously characterized pharyngeal rosette is visible at a depth of 10 μm (pink). C) Images show en face or lateral views of pha-4p::GFP::CAAX outlining the anterior rosette. The cartoon schematics to the right show the regions used to determine the diameter and length of individual cells. D) Shown are images from excerpts of different time-lapse movies from embryos co-expressing PAR-6::mKate2 (magenta) and pha-4p::GFP::CAAX (green) acquired using sweptfield microscopy during anterior morphogenesis. The embryo in the panel on the left is ~20 minutes after the onset of anterior morphogenesis (when the leading epidermal cells meet at the ventral midline), while the middle is at ~30 minutes, and the right is at ~40 minutes. A cartoon on the right shows the relative position of the images in the embryo. E) Time-lapse images acquired using sweptfield microscopy show control (top panels), pha-4 or zen-4 RNAi embryos expressing mNeonGreen::PAR-6 (green) and lin-26p::VAB-10 (ABD)::mCherry (magenta) during anterior morphogenesis. Yellow arrows point to the position of the bright focal point (BFP), which demarcates the site of the future lumen, while the dotted circles highlight the location of the pentagon and semi-circle of foci. Times are indicated in minutes. For all images, the scale bar is 10 μm.
To determine the effects of ani-1 RNAi on epidermal cell migration, we used NIS-Elements Viewer (Nikon) to measure the time needed for the amphid dendrites to reach their anterior location as a read-out for epidermal cell migration. The ‘start’ of anterior morphogenesis (t = 0 minutes) was when the leading pair of ventral epidermal cells met at the ventral midline (Fig. 1A). Control and ani-1 RNAi embryos were analyzed, and the phenotypes were categorized based on the time required for the amphid dendrites to reach an anterior position (Fig. 6). A chi-square test was used to determine statistical significance (p<0.001) between control and ani-1 RNAi phenotypes, as well as the proportion of phenotypes in each category with pharyngeal epithelialization (p<0.01). Epithelialization was determined based on the accumulation of PAR-6 in the pharyngeal cells.
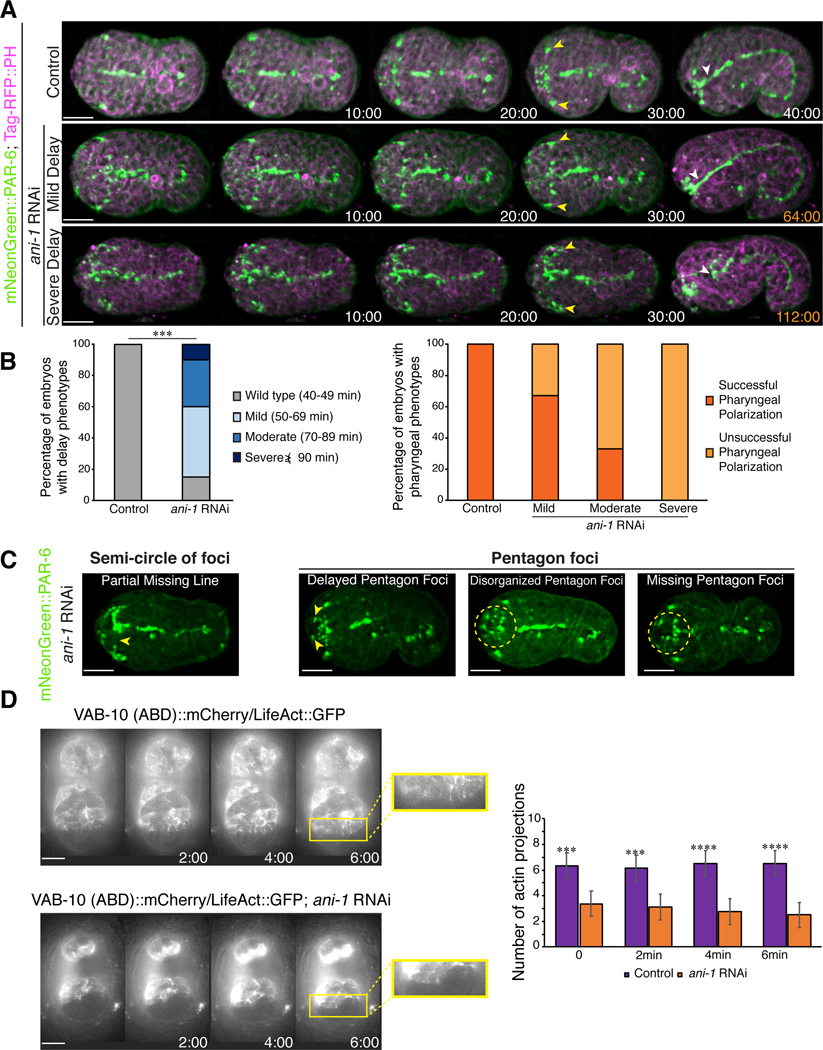
Figure 6. Disrupting neuroblast division causes delays in anterior epidermal cell migration.
A) Time-lapse images acquired using sweptfield microscopy show ventral views of embryos (anterior to the left) expressing mNeonGreen::PAR-6 (green); Tag-RFP::PH (magenta) in control (top panel) and after ani-1 RNAi to disrupt neuroblast cell division (middle and bottom panels). Delays in epidermal cell migration, marked by the amphid dendrites (yellow arrowheads), correlated with changes in the number or position of PAR-6 foci and/or failure of the anterior pharynx to polarize (white arrowheads) in ani-1-depleted embryos compared to control embryos. Times are shown in minutes and the scale bars are 10 μm. B) Epidermal cell migration was monitored by measuring the time it took for the amphid dendrites to reach the anterior region. The bar graph on the left shows the proportion of embryos with different delays in epidermal cell migration after ani-1 RNAi when compared with control embryos. The bar graph on the right shows the proportion of control and ani-1 RNAi embryos with delay phenotypes in which the anterior pharynx failed to polarize. C) Images show changes in the patterns of PAR-6 foci observed in mNeonGreen::PAR-6 (green) embryos with epidermal cell migration delays after ani-1 RNAi. The scale bar for all embryos is 10 μm. D) Time-lapse images acquired by HILO microscopy show ventral views of control or ani-1 RNAi embryos expressing lin-26p::LifeAct::GFP or lin-26::VAB-10 (ABD)::mCherry in greyscale. Zoomed in regions (yellow box) show the projections more clearly. To the right, the bar graph shows the average number of projections for each time point as indicated. The asterisks indicate p<0.0005.
We quantified the number of actin-rich projections using FIJI (NIH). The image files were opened and staged to be 10 minutes after the start of anterior morphogenesis (t = 0). Then the number of projections were counted at this time point, and subsequently at 2-minute intervals for 6 minutes. The average number of projections were measured for each time point and averaged together. Then the control and ani-1 RNAi conditions were compared using a two-tailed t test for significance, which we found was p<0.0005 for each time point.
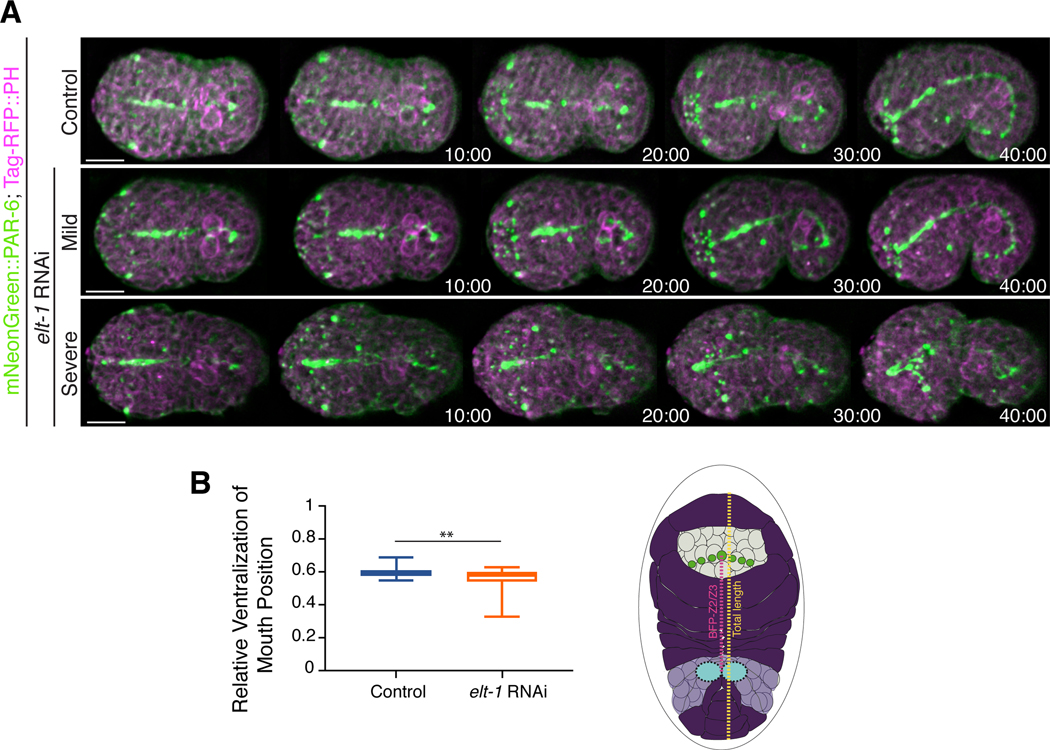
We determined the effects of elt-1 RNAi on lumen position using deconvolved movie files that were analyzed with FIJI. Embryos were measured at t = 20 minutes after the leading pair of ventral epidermal cells met at the midline. The ratio of the distance between the Z2/Z3 cells (germline precursor cells) and the bright focal point (subset of PAR-6 positive cells) and the total length of the embryo was determined for each embryo. Although there was no difference in the average ratio per se, an F-test revealed that the variability was significantly higher in the elt-1 RNAi embryos (p<0.01).
All figures were generated using Adobe Photoshop and Illustrator, after generating 8-bit format images using FIJI. In figures were Fire LUTs were used, which is a pseudocolor that converts the signal into different colors based on intensity levels, where white or red is high, and blue or violet is low.
Results
Dynamic cell movements and patterning occur during anterior morphogenesis
We set out to characterize the cell movements and patterns that occur during anterior morphogenesis of C. elegans embryos. Anterior morphogenesis initiates during ventral enclosure when the leading pair of ventral epidermal cells meet at the ventral midline (Fig. 1A) and continues until the 1.7-fold stage of embryogenesis when the muscles start twitching (Chisholm and Hardin, 2005). Initially the anterior-most part of the embryo is composed primarily of neuroblasts, and the epidermal cells migrate anteriorly to cover the head in epidermal tissue (Chisholm and Hardin, 2005). To visualize the cell patterns and movements during anterior morphogenesis, we imaged embryos co-expressing mNeonGreen-tagged pleckstrin homology (PH) domain, which localizes to membranes and mCherry-tagged histone H2B to visualize nuclei (Fig. 1B-D). We used two types of imaging methods—sweptfield confocal microscopy (n = 37; Fig 1B) and diSPIM (Fig. 1C; n = 8; Movie S1) to visualize changes in cell movements and patterns at different resolutions. diSPIM captures two orthogonal image volumes for two different views of the sample at each time point, allowing computational processing to create a single isotropic volume with better resolution than traditional imaging systems. Single-view images are shown here (Fig. 1C). At 10–15 minutes after the start of anterior morphogenesis, rings of neuroblasts formed around the site of the future lumen (Fig. 1B-C; Fig. S1). As the epidermal cells migrated toward the anterior, a subset of these neuroblasts underwent ingression (Fig. 1B-C). To better visualize the cell movements occurring during anterior morphogenesis, we imaged embryos en face (Fig. 1B; see Methods). Using this approach, we were able to more clearly observe the ring-like patterns formed by the neuroblasts during anterior morphogenesis. The forces generated by cell movements and resistance of the rigid eggshell could facilitate the ingression and/or inward direction of neuroblasts prior to elongation (Fig. 1D). Since a large proportion of the neuroblasts will differentiate into neurons that form the nerve ring, which encircles the pharynx, this inward movement likely helps position these neuronal precursors appropriately (Hobert, 2010). When the epidermal cells reached the anterior region, the membrane marker revealed a star-like pattern. Since this marker contains a domain that binds to phospholipids, this pattern likely corresponds to the growing axons, with the central point corresponding to the site of the future lumen (Fig. 1B-C). Thus, several distinct cell movements occurred during anterior morphogenesis that require further characterization.
The arcade cells of the pharynx form a stable rosette
While we were imaging embryos using the membrane marker, we observed a rosette that formed at a depth of ~ 4–5 μm and aligned with the site of the future lumen (Fig. 2A-C). This rosette formed anteriorly to the larger, previously described cyst formed by pharyngeal cells (Fig. 2B; Rasmussen et al. 2012). This rosette contained six cells and persisted for an extended period of time, which is characteristic of apical polarity-derived rosettes known to give rise to lumens (Fig. 2C; Sawyer et al., 2010; Harding et al., 2014; Martin and Goldstein, 2014). To determine if these cells are pharyngeal and to follow their patterning more specifically, we imaged embryos co-expressing pha-4p::GFP::CAAX (n = 18). This probe localizes to the membranes of pharyngeal cells, as pha-4 encodes a forkhead box A (FOXA) transcription factor required for pharyngeal cell fate, and CAAX is post-translationally farnesylated (farnesyl is a lipid moiety; Horner et al., 1998; Kalb et al., 1998; Gaudet & Mango 2002; Roberts et al., 2008; Manolaridis et al., 2013). At the start of anterior morphogenesis, the most anterior pha-4 positive cells moved toward the anterior of the embryo, but subsequently moved posteriorly within the embryo while retaining a narrow region of signal that aligned with the center of the rosette (Fig. 2A). Approximately 40–50 minutes after the start of anterior morphogenesis, these cells adopted a teardrop shape (Fig. 2A, C). Imaging pha-4p::GFP::CAAX and TagRFP::PH 1.5-fold embryos en face revealed the rosette more clearly. Consistent with what we had observed earlier, the rosette was positioned at a depth of 5 ± 0.40 μm from the anterior tip of the embryo (Fig. 2B, middle panel), whereas the previously characterized, larger pharyngeal rosette was at 11 ± 0.40 μm (n = 13 embryos; Portereiko & Mango, 2001; Fig. 2B, bottom panel). To better characterize the small pha-4 positive rosette, we measured its diameter along three axes per rosette, as well as the length of individual cells in the rosette (Fig. 2C). We found that the average diameter was 6.9 ± 0.13 μm (n = 13 embryos), whereas the average cell length was 4.9 ± 0.04 μm (n = 16 cells), consistent with what we had observed for depth. Given the location and number of these cells, the anterior rosette is likely composed of a subset of arcade cells. The arcade cells formed projections that extended anteriorly and remained in place as the arcade cells moved back inside the embryo.
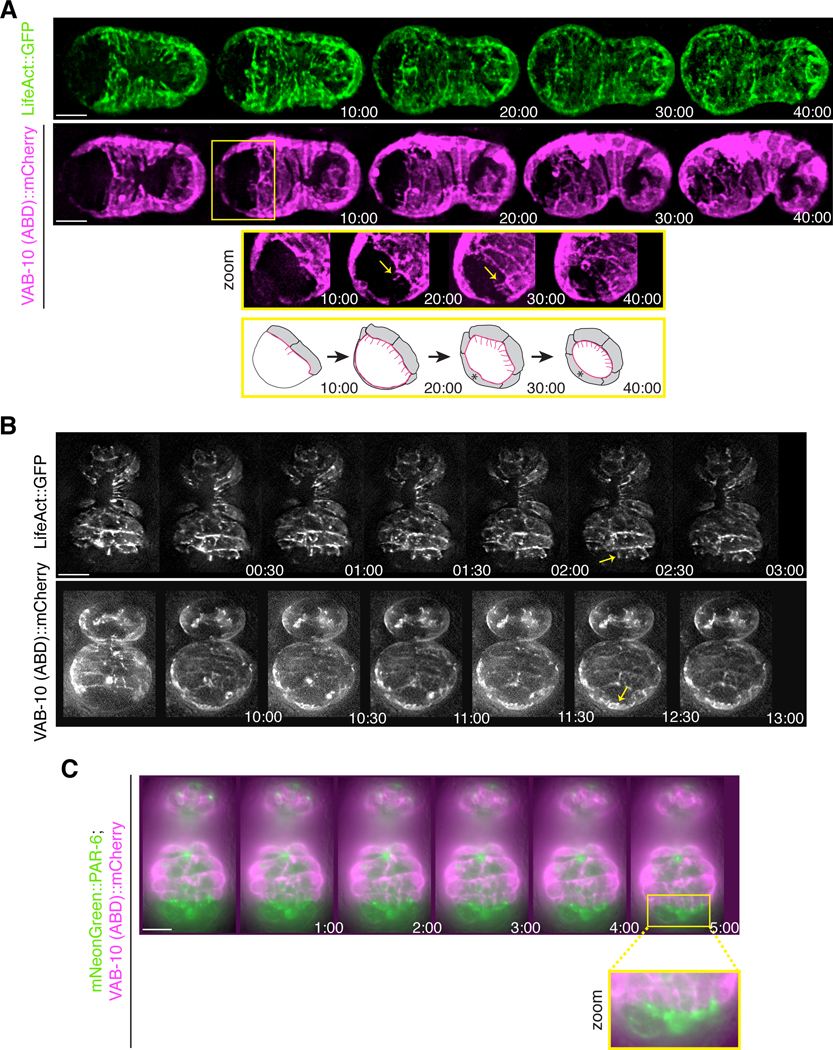
As the arcade cell projections were aligned with the center of the rosette, this polarized structure could be one of the first anterior-positioned markers that defines the site of the future lumen. To determine if a marker of apical polarity is enriched at this location, we co-imaged embryos expressing PAR-6::mKate2 and pha-4p::GFP::CAAX during anterior morphogenesis. After ~10 minutes, we observed the appearance of a bright focal point (BFP) that corresponded to the tip of the arcade cell projections as they grew toward the anterior (n = 18; Fig. 2D; Movie S2). We also observed the emergence of other distinct PAR-6 foci, which likely correspond to other cell types.
Distinct patterns of PAR-6 foci form in the anterior region of the embryo
Next, we characterized the patterns of PAR-6 foci during anterior morphogenesis. We speculated that these correspond to subsets of neuroblasts and/or epidermal cells. We imaged embryos co-expressing mNeonGreen::PAR-6 and mCherry-tagged actin-binding domain (ABD) from VAB-10 driven by the epidermal region of the lin-26 promoter (Landmann et al., 2004; Gally et al., 2009; Zilberman et al., 2017) to visualize epidermal F-actin (n = 15). As reported recently by the Bao lab, we observed that PAR-6 was enriched at the vertex of the amphid dendrite tips (Fig. 3A; Fan et al., 2019). As embryos progressed through morphogenesis, we saw PAR-6 localize to foci that formed two pentagons on either side of the BFP, and to a semi-circle of foci that moved toward the BFP (Fig. 3A, S2A; Movie S3). The BFP first appeared on the ventral side of the embryo ~10 minutes after the start of anterior morphogenesis. At ~20 minutes, we observed a semi-circle of foci that aligned with the leading edge of the migrating epidermal cells (Fig. 3A, zoom; S2A; Movie S3). The two pentagon patterns initiated at a more dorsal position and then moved more ventrally to meet with the semi-circle (Figs 3A; S2A; Movie S3). The most dorsal foci of the pentagons aligned with the leading edge of epidermal cells migrating from more dorsal positions (Figs 3A; S2B). By 40 minutes, the foci formed a rectangular pattern with the semi-circle and the three ventral foci from each pentagon marking one side and the two dorsal foci marking the other, which subsequently resolved into a circular pattern around the lumen (Fig. 3A, zoom; S2A; Movie S3).
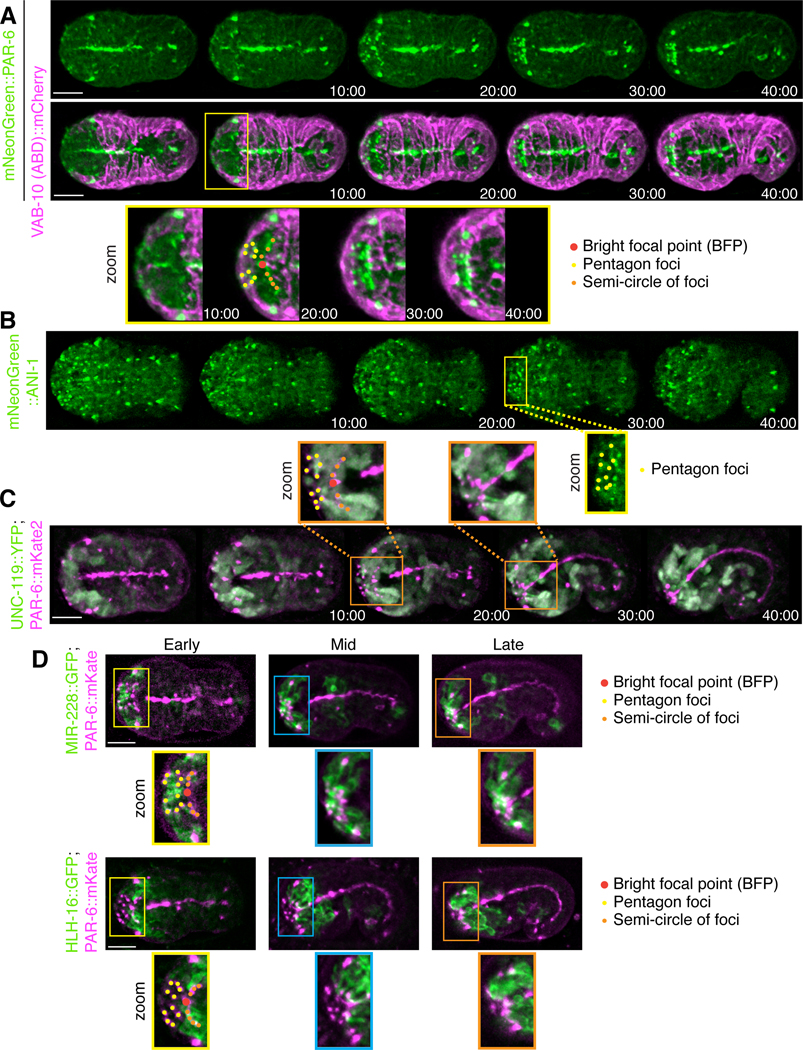
Figure 3. Distinct patterns formed by PAR-6 foci correlate with the migrating anterior epidermal cells.
A) Time-lapse images acquired using sweptfield microscopy show ventral views (anterior to the left) of an embryo expressing mNeonGreen::PAR-6 (green; top panel) and epidermal VAB-10 (ABD)::mCherry (magenta; middle panel) during anterior morphogenesis. Times are indicated in mimutes. Zoomed-in regions (bottom panel; 200%) show the patterns formed by PAR-6 more clearly (yellow box). PAR-6 localizes to the BFP (red), the pentagon foci (yellow), and the semi-circle of foci (orange) that corresponds to the anterior-most boundary of ventral epidermal cells. B) Time-lapse images acquired using sweptfield microscopy show ventral views of embryos expressing mNeonGreen::ANI-1 (green). Times are indicated in minutes. A zoomed-in region (yellow box; 150%) shows the pentagon foci highlighted by yellow dots. C) Time-lapse images acquired using sweptfield microscopy show embryos expressing UNC-119::YFP (green) and PAR-6::mKate2 (magenta). Times are indicated in minutes. Zoomed-in regions (orange boxes; 150%) show the neuronal projections in the anterior region with the pentagons, BFP and semi-circle of foci marked in yellow, red and orange, respectively. D) Images show embryos expressing MIR-228::GFP (green) and PAR-6::mKate2 (magenta), or HLH-16::GFP (green) with PAR-6::mKate2 (magenta). Zoomed-in regions (150%) are indicated by yellow, blue or orange boxes and reflect early (10–20 minutes), mid (30–35 minutes) and later (>40 minutes) stages of anterior morphogenesis. As above, the pentagons, BFP and semi-circle of foci are indicated in yellow, red and orange, respectively. For all images, the scale bar is 10 μm.
To determine if the pentagons or semi-circle of foci correspond to neuroblasts, we imaged embryos expressing mNeonGreen::ANI-1 (anillin) during anterior morphogenesis. We previously found that ANI-1 is specifically enriched in subsets of neuroblasts (Wernike et al., 2016), and is an indicator of polarity during cell division as it binds to actomyosin. ANI-1 localized to the pentagon foci, but not to the BFP as expected, or the (majority of) foci that form the semi-circle (n = 29; Fig. 3B). This suggests that the pentagon foci originate in the neuroblasts (Fig. 3B). To further show this, we imaged embryos co-expressing a pan-neuronal marker, UNC-119::YFP, and PAR-6::mKate2. As embryos progressed through anterior morphogenesis, we observed neuroblast projections aligned with the PAR-6 foci in the pentagons, and to the outermost foci of the semi-circle (n = 35; Fig 3C zoom; Movie S4). Since this expression could reflect neuronal precursors or their support cells, we imaged markers that are known to be expressed in these tissues during embryogenesis (Rapti et al., 2017). Myristoylated (post-translational modification that targets proteins to the membrane) MIR-228::GFP, which is a pan-glial cell marker (CEPsh, labial sh and amphid sh), localized to projections that aligned with the PAR-6 pentagon foci (early; n = 6 MIR-228::GFP; PAR-6::mKate2; Fig. 3D; Movie S5; n = 6 MIR-228::GFP; Fig. S2C; Movie S6; Rapti et al., 2017). Myristoylated HLH-16::GFP, which is expressed in the SMDD/AIY and SIAD/SIBV lineages, localized to some of the semi-circle of foci early, and to most foci by mid stages of anterior morphogenesis (n = 20 HLH-16::GFP; PAR-6::mKate; Fig. 3D; Movie S7; n = 9 HLH-16::GFP; Fig. S2C; Movie S8; Bertrand et al., 2011; Rapti et al., 2017). We also imaged embryos expressing LSY-6::GFP, which is expressed in a small subset of neurons including the ASEL neuron, but these projections did not reach the anterior (n = 7; Fig. S2C; Rapti et al., 2017). Therefore, this data demonstrates that subsets of glial and neuronal precursor cells form polarized projections with distinct patterns during anterior morphogenesis. The proximity of a subset of these foci with the leading edge of the epidermal cells suggests that these cells are in close association during epidermal migration.
Junction proteins are enriched in foci at the anterior of the embryo
To characterize foci in the epidermal cells, we imaged embryos expressing different adhesion components in the DAC (DLG-1 and AJM-1 complex) and CCC (cadherin/HMR-1 and α-catenin/HMP-1 complex; Costa et al., 1998; McMahon et al., 2001; Chisholm and Hardin, 2004; McMahon et al., 2001). AJM-1::GFP is enriched in epidermal cells, and we used Fire LUTs to better visualize changes in AJM-1 intensity to reveal more detailed localization patterns during anterior morphogenesis (Fig. 4A). As anterior morphogenesis progressed, AJM-1 foci appeared at the leading edge of the anterior ventral epidermal cells (n = 15; Fig. 4A). While AJM-1 was not apparent in the BFP and only weakly visible in anterior foci initially (e.g. at 10 minutes), it increased in intensity in these locations as anterior morphogenesis progressed (Fig. 4A). This likely reflects the maturation of adhesion junctions as the epidermal cells contact neighboring cells at the anterior. Ultimately, the epidermal cells arranged themselves to form two bilateral rings, which fused and joined the pharynx, with another ring forming around it (Fig. S3A). Imaging embryos co-expressing DLG-1::RFP and HMP-1::GFP revealed that DLG-1 had a similar pattern of localization to AJM-1, albeit weaker (n = 8; Figs 4B; S3B). We previously showed that the CCC, but not the DAC, is enriched in the mid-posterior neuroblasts that forms transient rosettes for tissue re-organization during ventral enclosure (Wernike et al., 2016). The CCC component HMP-1 localized to the BFP by 10 minutes, and was enriched in the pentagons and semi-circle of foci (n = 8; Fig. 4A). Imaging embryos co-expressing HMP-1::GFP and lin-26p::VAB-10(ABD)::mCherry revealed that the anterior-most epidermal cells were in close contact with the HMP-1-rich semi-circle of foci (n = 8; Fig. 4C). As actomyosin is typically enriched at junctions, we also imaged embryos expressing GFP::NMY-2 (non-muscle myosin) and observed patterns similar to PAR-6 and HMP-1 (n = 23; Fig. 4D). Therefore, the enrichment of PAR-6, HMP-1 and NMY-2 within subsets of cells reflects their polarity and adhesion with neighboring cells.
Figure 4. Adhesion junctions are enriched in subsets of cells during anterior morphogenesis.
A) Time-lapse images acquired using sweptfield microscopy show ventral views (anterior to the left) of an embryo expressing AJM-1::GFP (Fire LUTs). A zoomed-in region (150%) is shown underneath (yellow box; bottom right) with the pentagon foci, BFP and semi-circle of foci labeled in yellow, red and orange as indicated. Times are shown in minutes and the scale bar is 10 μm. B) Time-lapse images show ventral, en face views of an embryo expressing HMP-1::GFP (green) and DLG-1::RFP (magenta) during anterior morphogenesis. Times are shown in minutes, and the scale bar is 10 μm. C) Time-lapse images show ventral, en face views of an embryo expressing HMP-1::GFP (green) and lin-26p::VAB-10(ABD)::mCherry (magenta) during anterior morphogenesis. One of the images is duplicated below and labeled as in A). Times are shown in minutes, and the scale bar is 10 μm. D) Time-lapse images show ventral views of embryos expressing NMY-2::GFP (green). Zoomed-in regions (yellow box; 150%) show the anterior foci as labeled in A). Times are shown in minutes, and the scale bar is 10 μm. E) Cartoon schematics show the patterns of foci and associated proteins during anterior morphogenesis. The pentagon foci are in yellow, the BFP is in red, and the semi-circle of foci are in orange. Also shown are adhesion junctions (green) formed by the DAC and CCC, F-actin projections (pink) and amphid foci (blue). At early stages of anterior morphogenesis, the pentagon foci contain PAR-6, ANI-1, HMP-1 (CCC) and NMY-2, while the BFP contains PAR-6, HMP-1 (CCC) and NMY-2, and the semi-circle of foci contains PAR-6, HMP-1 (CCC), DAC and NMY-2. During mid and late stages of anterior morphogenesis, the pentagon foci and BFP also contain the DAC.
Ventral and dorsal epidermal cells migrate to an anterior-ventral position
Next, we characterized anterior epidermal cell migration. To do this, we monitored the localization of F-actin expressed in epidermal cells during anterior morphogenesis in embryos expressing either lin-26p::VAB-10(ABD)::mCherry (n = 32) or lin-26p::GFP-tagged LifeAct (n = 12; Fig. 5A). As the anterior ventral epidermal cells migrated anteriorly, we observed the formation of actin-rich projections (Fig. 5A, zoom). The dorsal epidermal cells also migrated anteriorly during this time, which need to move a greater distance to reach the site where the ventrally positioned lumen will form (Fig. 5A, zoom).
Figure 5. Projections form at the leading edge of the migrating anterior ventral epidermal cells.
A) Time-lapse images acquired using sweptfield microscopy show ventral views (anterior to the left) of embryos expressing lin-26p::LifeAct::GFP (green) or lin-26p::VAB-10(ABD)::mCherry (magenta) during anterior morphogenesis. Times are shown in minutes, and the scale bar is 10 μm. At the onset of anterior morphogenesis, the anterior of the embryo is devoid of epidermal cells, which subsequently migrate into this space. The ventral cells migrate a shorter distance compared to the dorsal cells. The yellow box highlights a region that has been tilted to better visualize the F-actin projections (yellow arrows). The bottom row shows cartoon schematics that indicate the orientation of the projections in pink. B) Time-lapse images acquired by HILO microscopy show ventral views (anterior pointing down) of embryos expressing epidermal lin-26p::LifeAct::GFP (top panel) or lin-26p::VAB-10(ABD)::mCherry (bottom panel). Times are shown in minutes, and the scale bar is 10 μm. The projections are dynamic and longer at the leading edge of the ventral epidermal cells (top panel; yellow arrow), whereas the dorsal epidermal cells have smaller, more uniform projections (bottom panel; yellow arrow). C) Time-lapse images acquired by HILO microscopy show a ventral view of an embryo co-expressing mNeonGreen::PAR-6 (green) and lin-26p::VAB-10 (ABD)::mCherry (magenta). Times are shown in minutes, and the scale bar is 10 μm. A zoomed in region (yellow box) shows the alignment of projections with the foci.
To follow epidermal cell migration with a higher temporal resolution, we imaged F-actin in embryos using HILO (highly inclined and laminated optical sheet) microscopy (Wernike et al., 2016). With this method, TIRF (total internal reflection fluorescence) objectives are used and the illumination beam is angled to obtain a thicker z plane (e.g., in this case to a depth of ~1 μm) emerging obliquely from the sample. In Fig. 5B, the top panel shows images of LifeAct taken every 30 seconds (anterior is pointing down; n = 8; Movie S9). Projections from the anterior ventral epidermal cells were dynamic, extending anteriorly for several microns. However, they did not extend far past the leading edge of the cell, and aligned with mNeonGreen::PAR-6 foci (Fig. 5C). The bottom panel of Fig. 5B shows the migrating epidermal cells positioned more dorsally in an embryo expressing VAB-10(ABD)::mCherry (see arrow; anterior pointing down; n = 9; Movie S10). The extensions at the leading edge were more uniform and smaller in length compared to the ventral epidermal cells (Fig. 5B). It is striking how epidermal cells positioned ventrally and dorsally coordinated their movements to reach the location of the future lumen at the same time, suggesting that their movements are controlled by cues associated with other cells in this location.
Anterior epidermal cell migration depends on the arcade cells and subsets of anterior neuroblasts
We propose that the patterns formed by the arcade cells and neuroblasts influence epidermal cell migration. Given the timing of the BFP appearance, the arcade cells may provide the first signal to coordinate the surrounding neuroblasts, which in turn provide a substrate and/or cues for the epidermal cells. First, we perturbed the arcade cells to determine the effect on anterior morphogenesis by depleting pha-4, which is required for pharyngeal cell fate (Kalb et al., 1998), or zen-4, which is required for their polarization (Portereiko et a., 2004). Since zen-4 is also required for cytokinesis and ventral enclosure (Raich et al., 1998; Hardin et al., 2008), embryos were only treated with zen-4 RNAi for a short period of time. In pha-4 or zen-4 RNAi embryos co-expressing mNeonGreen::PAR-6 and lin-26p::VAB-10(ABD)::mCherry, we observed that the BFP was delayed in its formation (n = 7/9 for pha-4, and 14/15 for zen-4; Fig. 2E), and the other foci were disorganized (n = 6/14 for pha-4, and 8/16 for zen-4; Fig 2E). The majority of embryos from both treatments showed defects in anterior morphogenesis, ranging from mild to more severe (n = 14/18 for pha-4, and 21/21 for zen-4). We also noticed that a BFP-like signal appeared later in development, although dimmer in appearance, and we speculate that this comes from neuronal or support cell projections (e.g. Fig. 3C). This data suggests that the arcade cells provide early cues to organize the surrounding neuroblasts.
Next, we determined how perturbing the neuroblasts impacts epidermal cell migration. We previously showed that ani-1 is required for neuroblast cell division (Fotopoulos et al., 2013; Wernike et al., 2016). Depleting ani-1 in embryos co-expressing mNeonGreen::PAR-6 and lin-26p::VAB-10(ABD)::mCherry caused changes in PAR-6 localization and delayed anterior epidermal cell migration (n = 34; Fig. S4). We observed a correlation in the severity of anterior morphogenesis phenotypes with higher degrees of perturbed PAR-6 localization and delayed migration, including failed polarization of the anterior pharynx (Fig. S4).
We quantified the different anterior morphogenesis phenotypes caused by ani-1 RNAi in embryos co-expressing mNeonGreen-tagged PAR-6 and a Tag-RFP membrane marker (n = 15 control and n = 28 ani-1 RNAi embryos; PH domain; Fig. 6A). To measure the duration of anterior epidermal cell migration, we determined the time it took for the amphid dendrites to reach the anterior region. The Bao lab recently showed that the amphid dendrites extend with epidermal cells, and we used this as a marker for epidermal migration (Fan et al., 2019). We observed embryos with a range in delay phenotypes that we categorized as mild (50–69 minutes; 45%), moderate (70–89 minutes; 30%) and severe (≥90 minutes; 10%), based on the amphid dendrite extension time relative to that of control embryos (40–49 minutes; 15%; Fig. 6B). We also observed that PAR-6 failed to localize within the anterior pharynx in a subset of the ani-1 RNAi embryos. Of the embryos that displayed a mild delay phenotype, 33% failed to polarize, whereas 67% of embryos with moderate delays failed and 100% of the severely delayed embryos failed (Fig. 6B). We correlated the changes in neuroblast/foci patterns with the observed phenotypes and found that more of the foci that form the semi-circle or pentagons were lost or severely disorganized as the delay worsened from mild to severe (Fig. 6C).
The delays in epidermal migration in ani-1 RNAi embryos likely are due to cytoskeletal changes that impact their ability to migrate. As described earlier, we observed F-actin projections at the leading edges of the anterior ventral epidermal cells. We performed HILO imaging on control and ani-1 RNAi embryos expressing lin-26p::LifeAct::GFP or lin-26p::VAB-10(ABD)::mCherry, and quantified the length and number of projections from the onset of anterior morphogenesis over several time points (n = 6 for control, and 8 for ani-1 RNAi; Fig. 6D). While there was no change in the length of the projections (0.6 μm in both conditions), there was a significant decrease in the number of projections at each time point, which correlated with their delayed migration (Fig. 6D).
It is important to note that we did not observe changes in the position of lumen formation (e.g., in embryos with a less-severe delay) in ani-1 RNAi embryos, nor was the BFP altered in intensity or appearance. Therefore, neuroblasts are required for controlling the speed of epidermal cell migration, but not their direction. Also, the failure to polarize the pharynx in the more severely delayed embryos reflects a threshold requirement for the timing of cell-cell contacts to complete lumen formation.
Epidermal cells determine the position of the lumen
Next, we determined how mild perturbation of epidermal cell fate affects anterior morphogenesis. To accomplish this, we partially depleted elt-1 in embryos co-expressing mNeonGreen::PAR-6 and Tag-RFP::PH (n = 15 control vs. n = 35 elt-1 RNAi). elt-1 encodes a GATA-like transcription factor and is essential for determining epidermal cell fate (Fig. 7A; Page et al. 1997). After partial elt-1 depletion, there was no significant delay in the migration of anterior epidermal cells. However, we observed a ventral shift in the position of the anterior lumen formation (Fig. 7 A, B). To quantify this, we measured the ratio of the distance between the lumen (marked by PAR-6) and the Z2/Z3 cells (germline precursors) to the total length of the embryo. Whereas the average ratio was not significantly different, the variability was significantly higher in mildly perturbed elt-1 RNAi embryos (n = 19) as compared with control embryos (n = 10; Fig. 7B). In a subset of the more severely affected embryos we saw that the PAR-6 foci formed patterns that were initially similar to those of control embryos, but these foci failed to coalesce and shifted more ventrally as compared to control embryos (Fig. 7A). The BFP also underwent a more ventral shift in severely affected embryos, and the pharynx failed to epithelialize. Therefore, the position of the anterior lumen was determined by the epidermal cells. Although it is not clear why the lumen tended to shift more ventrally, this could be because there are two rows of dorsal cells that intercalate and fuse, and perhaps the loss of one/more of these cells has less of an impact vs. the loss of ventrally positioned cells. These cells also migrate over a longer distance and may interpret cues differently as compared with the ventrally positioned cells.
Figure 7. Altering epidermal cell fate causes a change in lumen position.
A) Time-lapse images acquired using sweptfield microscopy show ventral views (anterior to the left) of embryos expressing mNeonGreen::PAR-6 (green); Tag-RFP::PH (magenta) in control (top panel) and elt-1 RNAi embryos (middle and bottom panels). Times are shown in minutes, and the scale bar is 10 μm. A range of phenotypes was observed: 1) mild, in which PAR-6 foci patterns were similar to those of control embryos, although there was a shift toward a more ventrally positioned lumen, and 2) severe, in which foci failed to coalesce into patterns, and the lumen was extremely displaced. B) A graph shows the average ratio of the distance between the lumen and Z2/Z3 cells (magenta in the cartoon schematic; future germline) to the total length of the embryo (yellow in the cartoon schematic). Only mildly perturbed embryos were measured, and the variability of the ratio was significantly increased in elt-1 RNAi (orange) as compared with control (blue) embryos. The asterisks indicate p<0.01.
Discussion
We characterized anterior morphogenesis of the C. elegans embryo. This developmental process is crucial for giving rise to a properly formed anterior lumen, which must align precisely with the pharynx and intestine. Cells from three different tissues are coordinated for anterior morphogenesis: neuroblasts, pharynx and epidermis. But how they did this was not known, largely due to the complexity of studying multiple tissues simultaneously in vivo. We used various types of microscopy and post-acquisition software to define the patterns formed by different cell types during anterior morphogenesis. We observed the ingression of neuroblasts as epidermal cells migrated anteriorly to enclose the anterior surface of the embryo in a layer of epidermis. The neuroblasts arranged into a circle-like pattern that likely facilitates their inward movement. We propose that their movement could in part be due to forces generated from cell movements and patterning during ventral enclosure. We also observed that the arcade cells, the anterior-most pharyngeal cells, formed a rosette positioned at a depth of ~5 μm. These cells formed projections that reached the anterior, marking the location of the future lumen. The ends of the projections accumulated markers of apical polarity including PAR-6, NMY-2 and HMP-1. Since the projections coalesce causing fluorophores detecting these proteins to be bright compared to other cellular locations, we referred to them as the ‘bright focal point’ (BFP; Fig. 4E). While the BFP initially reflected signals associated with the arcade cells, neuronal and/or support cells also could join this location later in anterior morphogenesis. Soon after the appearance of the BFP, foci expressing PAR-6, HMP-1, NMY-2 and ANI-1 arranged themselves into two pentagons that formed on either side of the BFP and aligned with UNC-119 and MIR-228 neuronal and glial cell projections (Fig. 4E). Another set of foci expressing PAR-6, HMP-1, NMY-2 and HLH-16 (in a subset) formed a semi-circle and corresponded to the leading edge of the ventral epidermal cells as they migrated anteriorly (Fig. 4E). The majority of anterior foci also expressed AJM-1, which initially was weak, but strengthened with time. The ventral epidermal cells co-migrated with the semi-circle of foci and remained in a more ventral position, whereas the dorsal epidermal cells co-migrated with the dorsal foci of the pentagons and migrated over a longer distance. We observed F-actin-rich projections in the migrating epidermal cells, which appeared longer and more dynamic in the ventral cells than in the dorsal cells, where they were shorter and more uniform in appearance. We found that the neuroblasts were required for controlling the speed of epidermal migration, as reducing their numbers by ani-1 RNAi caused delays. In contrast, mildly perturbing epidermal cell fate caused a shift in the location of the future lumen but did not affect the timing of its formation.
We propose that the arcade cell projections provide cues that could influence the patterning of the surrounding neuroblasts, which in turn provide a substrate and/or cues to regulate epidermal cell migration. In support of this model, removing the arcade cells or disrupting their polarity caused anterior morphogenesis phenotypes, including the altered organization of neuroblast-associated foci. We also found that using ani-1 RNAi to reduce the number of neuroblasts caused a range of anterior morphogenesis phenotypes, including delayed epidermal cell migration and failure to epithelialize the anterior lumen. During ventral enclosure, where the ventral surface of the embryo is covered by epidermal cells, several signaling pathways have been shown to regulate WSP-1 and WVE-1 to control the formation of branched F-actin in the migrating ventral epidermal cells (Sawa et al., 2003; Withee et al., 2004; Patel et al., 2008; Bernadskaya et al., 2012; Wallace et al., 2018). For ventral enclosure, it is not clear where the ligands for these pathways are located as rescue studies show that they could originate in the neuroblasts or epidermal cells (e.g., Bernadskaya et al., 2012). It is likely that similar pathways influence the migration of anterior epidermal cells, and we propose that they originate from the subset of neuronal precursors and glial support cells that form the pentagons and semi-circle of foci around the arcade cell projections. In support of this hypothesis, we found that fewer actin projections formed in the anterior ventral epidermal cells that were delayed after ani-1 RNAi, however, the length of the projections was not affected as expected if the signal was non-autonomous. The pathways controlling F-actin include those responding to ligands such as UNC-6 (netrin), SLT-1 (slit), EFN-1–4 (ephrins) and MAB-20 (semaphorin) and their receptors UNC-40 (DCC), SAX-3 (Robo), VAB-1 (Eph receptor) and PLX-2 (plexin), respectively (Chin-Sang et al., 1999; George et al., 1998; Wang et al., 1999; Chin-Sang and Chisholm, 2000; Roy et al., 2000; Bernadskaya et al., 2012; Ikegami et al., 2012). We are testing this by perturbing components of these different pathways to determine their impact on anterior epidermal cell migration. As the dorsal cells migrate over a longer distance, they could be influenced by some of the same pathways but with different sensitivities and/or subsets of receptors to account for this distance.
While we saw ventral shifts in the position of the BFP when we perturbed epidermal cell fate, this only occurred at a later timepoint, during polarization of the anterior pharynx. Prior to this, it initiated in the correct position. This suggests that the epidermal cells are crucial for reinforcing lumen position, while the neuroblasts control the timing of lumen formation. We also observed shifts in the position of neuroblast foci just prior to polarization, suggesting that they also associate with and rely on the epidermal cells for late vs. early positioning. This was recently described by the Bao lab for amphid neuroblasts, which form rosettes and piggyback on the migrating epidermal cells for anterior-directed dendrite growth (Fan et al., 2019).
Our studies show how cells from different tissues are coordinated to give rise to the anterior lumen during development. These findings will facilitate more in-depth studies of how chemical cues organize and coordinate the patterning and movements of the different cell types. Given the challenges of studying tissues in more complex metazoans, our studies provide fundamental knowledge of how different cell types can be coordinated for the development of structures.
Supplementary Material
Movie S5. A time-lapse movie (7 fps) of the embryo from Fig. 3D co-expressing myristoylated (post-translational modification that targets proteins to the membrane) MIR-228::GFP (green) and PAR-6::mKate2 (magenta) imaged by sweptfield confocal microscopy through anterior morphogenesis.
Movie S3. A time-lapse movie (7 fps) of the embryo from Fig. 3A co-expressing mNeonGreen::PAR-6 and lin-26p::VAB-10(ABD)::mCherry (magenta) imaged by sweptfield confocal microscopy through anterior morphogenesis.
Movie S1. A time-lapse movie (7 fps) of the embryo from Fig. 1C co-expressing mNeonGreen::PH to visualize membranes (green) and H2B::mCherry to visualize nuclei (magenta), imaged by di-SPIM through anterior morphogenesis.
Movie S7. A time-lapse movie (7 fps) of one of the embryos from Fig. 3D co-expressing myrisotylated HLH-16::GFP (green) and PAR-6::mKate2 (magenta) imaged by sweptfield confocal microscopy through anterior morphogenesis.
Movie S2. A time-lapse movie (7 fps) of an embryo from Fig. 2D co-expressing pha-4p::GFP::CAAX (green) and PAR-6::mKate2 (magenta) imaged by sweptfield confocal microscopy through anterior morphogenesis.
Movie S6. A time-lapse movie (7 fps) of an embryo expressing myristoylated MIR-228::GFP in Fire LUTs imaged by sweptfield confocal microscopy through anterior morphogenesis.
Movie S8. A time-lapse movie (7 fps) of an embryo expressing myristoylated HLH-16::GFP in Fire LUTs imaged by sweptfield confocal microscopy through anterior morphogenesis.
Movie S4. A time-lapse movie (7 fps) of the embryo from Fig. 3C co-expressing UNC-119::YFP (green), and PAR-6::mKate2 (magenta) imaged by sweptfield confocal microscopy through anterior morphogenesis.
Movie S9. A time-lapse movie (7fps) of the embryo from Fig. 5B (top panel) expressing epidermal LifeAct::GFP taken using HILO microscopy (anterior down) to see ventral epidermal cell F-actin projections.
Movie S10. A time-lapse movie (7 fps) of the embryo from Fig. 5B (bottom panel) expressing epidermal VAB-10 (ABD)::mCherry taken using HILO microscopy (anterior down) to see dorsal epidermal cell F-actin projections.
Time-lapse images obtained by sweptfield microscopy show en face views of an embryo co-expressing mNeonGreen::PH to visualize membranes (green) and H2B::mCherry to visualize nuclei (magenta) through anterior morphogenesis. A ‘ring in a ring’ of cells is highlighted (white dashed circles, inner yellow dots). The star-like pattern in the center marks the site of the future lumen, which forms after cells ingress into the embryo. Times are shown in minutes, and the scale bar is 10 μm.
Time-lapse images acquired using sweptfield microscopy show ventral views of an embryo co-expressing mNeonGreen::PAR-6 (green) and lin-26p;VAB-10(ABD)::mCherry (magenta) after ani-1 RNAi. The zoomed insets (yellow box) show the region where the foci would typically be found. While the BFP is still present, some of the foci associated with the pentagons and the semi-circle of foci are not visible. Times are shown in minutes, and the scale bar is 10 μm.
A)Time-lapse images acquired using sweptfield microscopy show en face views of two different embryos expressing AJM-1::GFP (Fire LUTs) during anterior morphogenesis. B) Time-lapse images show ventral views (anterior to the left) of an embryo expressing HMP-1::GFP and DLG-1::RFP during anterior morphogenesis. Times are shown in minutes, and the scale bar is 10 μm.
A) Timelapse images of the embryo from Fig. 3A, co-expressing mNeonGreen::PAR-6 (green) and lin-26p::VAB-10 (ABD)::mCherry (magenta) is shown during anterior morphogenesis without any labels. B) Timelapse images of an embryo as in A), with a more dorsal view to better visualize epidermal cells that are more dorsally positioned. The arrows point to the dorsal foci of the pentagons, and the dotted line indicates the leading edge of epidermal cells migrating from more dorsal positions. C) Time-lapse images show embryos expressing LSY-6::GFP, HLH-16::GFP, and MIR-228::GFP in Fire LUTs as indicated. For all embryos, times are shown in minutes, and the scale bar is 10 μm.
Acknowledgments
We thank Mathew Duguay for his contribution to various aspects of this work. Except where indicated, imaging was performed at the Centre for Microscopy and Cellular Imaging at Concordia University. We thank Hari Shroff for his generosity and use of the diSPIM at the NIH/NIBIB, and Harshad Vishwasrao at the trans-NIH Advanced Imaging and Microscopy Resource (AIM) for his technical assistance and time. We thank Georgia Rapti (EMBL Heidelberg), Michael Glotzer (University of Chicago), Jean-Claude Labbe (IRIC), Susan Mango (Biozentrum of the University of Basel) and Richard Roy (McGill University) for reagents. Also, some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). We acknowledge Wyoming INBRE for editing services, a project that is supported in part by a grant from the National Institute of General Medical Sciences (2P20GM103432) from the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work was funded by a grant from the National Institutes of Health (NIH) to David Fay (R01GM125091). R.C. acknowledges support from the intramural research programs of the National Institute of Biomedical Imaging and Bioengineering, within the National Institutes of Health.
References
- Armenti ST, & Nance J. (2012). Adherens junctions in C. elegans embryonic morphogenesis. Sub-Cellular Biochemistry, 60, 279–299. 10.1007/978-94-007-4186-7_12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asan A, Raiders SA, & Priess JR (2016). Morphogenesis of the C. elegans Intestine Involves Axon Guidance Genes.PLoS Genetics, 12(4). 10.1371/journal.pgen.1005950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernadskaya YY, Wallace A, Nguyen J, Mohler WA, & Soto MC (2012). UNC-40/DCC, SAX-3/Robo, and VAB-1/Eph Polarize F-Actin during Embryonic Morphogenesis by Regulating the WAVE/SCAR Actin Nucleation Complex. PLoS Genetics, 8(8), e1002863. 10.1371/journal.pgen.1002863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand V, Bisso P, Poole R, Hobert O. (2011). Notch-dependent induction of left/right asymmetry in C. elegans interneurons and motorneurons. Current Biology, 21(4), 1225–1231. 10.1016/j.cub.2011.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenship JT, Backovic ST, Sanny JSSP, Weitz O, & Zallen JA (2006). Multicellular Rosette Formation Links Planar Cell Polarity to Tissue Morphogenesis. Developmental Cell, 11(4), 459–470. 10.1016/j.devcel.2006.09.007 [DOI] [PubMed] [Google Scholar]
- Bossinger O, Wiesenfahrt T, & Hoffmann M. (2015). Establishment and maintenance of cell polarity in the C. Elegans Intestine. In Cell Polarity 2: Role in Development and Disease (pp. 33–65). 10.1007/978-3-319-14466-5_2 [DOI] [Google Scholar]
- Brenner S. (1974). The genetics of Caenorhabditis elegans. Genetics, 77(1), 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanale JP, Sun TY, & Montell DJ (2017). Development and dynamics of cell polarity at a glance. Journal of Cell Science, 130(7), 1201–1207. 10.1242/jcs.188599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanale JP, Sun TY, & Montell DJ (2017). Development and dynamics of cell polarity at a glance. Journal of Cell Science, 130(7), 1201–1207. 10.1242/jcs.188599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin-Sang ID, George SE, Ding M, Moseley SL, Lynch AS, & Chisholm AD (1999). The ephrin VAB-2/EFN-1 functions in neuronal signaling to regulate epidermal morphogenesis in C. elegans. Cell, 99(7), 781–790. 10.1016/s0092-8674(00)81675-x [DOI] [PubMed] [Google Scholar]
- Chin-Sang ID, & Chisholm AD (2000, December 1). Form of the worm: Genetics of epidermal morphogenesis in C. elegans. Trends in Genetics, Vol. 16, pp. 544–551. 10.1016/S0168-9525(00)02143-0 [DOI] [PubMed] [Google Scholar]
- Chisholm AD, & Hardin J. (2005). Epidermal morphogenesis. WormBook : The Online Review of C. Elegans Biology, pp. 1–22. 10.1895/wormbook.1.35.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Raich W, Agbunag C, Leung B, Hardin J, & Priess JR (1998). A putative catenin-cadherin system mediates morphogenesis of the caenorhabditis elegans embryo. Journal of Cell Biology, 141(1), 297–308. 10.1083/jcb.141.1.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan LH, Moyle MW, Shao L, Sengupta T, Ikegami R, Kumar A, … Colón-Ramos DA (2019). Isotropic light-sheet microscopy and automated cell lineage analyses to catalogue caenorhabditis elegans embryogenesis with subcellular resolution. Journal of Visualized Experiments, 2019(148), e59533. 10.3791/59533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etemad-Moghadam B, Guo S, & Kemphues KJ (1995). Asymmetrically distributed PAR-3 protein contributes to cell polarity and spindle alignment in early C. elegans embryos. Cell, 83(5), 743–752. 10.1016/0092-8674(95)90187-6 [DOI] [PubMed] [Google Scholar]
- Fan L, Kovacevic I, Heiman MG, & Bao Z. (2019). A multicellular rosette-mediated collective dendrite extension. ELife, 8. 10.7554/eLife.38065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay DS (2005). The cell cycle and development: Lessons from C. elegans. Seminars in Cell and Developmental Biology, Vol. 16, pp. 397–406. 10.1016/j.semcdb.2005.02.002 [DOI] [PubMed] [Google Scholar]
- Fotopoulos N, Wernike D, Chen Y, Makil N, Marte A, & Piekny A. (2013). Caenorhabditis elegans anillin (ani-1) regulates neuroblast cytokinesis and epidermal morphogenesis during embryonic development. Developmental Biology, 383(1), 61–74. 10.1016/j.ydbio.2013.08.024 [DOI] [PubMed] [Google Scholar]
- Gally C, Wissler F, Zahreddine H, Quintin S, Landmann F, & Labouesse M. (2009). Myosin II regulation during C. elegans embryonic elongation: LET-502/ROCK, MRCK-1 and PAK-1, three kinases with different roles. Development (Cambridge, England), 136(18), 3109–3119. 10.1242/dev.039412 [DOI] [PubMed] [Google Scholar]
- Gaudet J, & Mango SE (2002). Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science, 295(5556), 821–825. 10.1126/science.1065175 [DOI] [PubMed] [Google Scholar]
- George SE, Simokat K, Hardin J, & Chisholm AD (1998). The VAB-1 Eph receptor tyrosine kinase functions in neural and epithelial morphogenesis in C. elegans. Cell, 92(5), 633–643. 10.1016/s0092-8674(00)81131-9 [DOI] [PubMed] [Google Scholar]
- Gillard G, Shafaq-Zadah M, Nicolle O, Damaj R, Pećreáux J, & Michaux G. (2015). Control of E-cadherin apical localisation and morphogenesis by a SOAP-1/AP-1/clathrin pathway in C. elegans epidermal cells. Development (Cambridge), 142(9), 1684–1694. 10.1242/dev.118216 [DOI] [PubMed] [Google Scholar]
- Guo H, Xu Y, Li Q, Du S, He D, Wang Q, & Huang Y. (2019). Improved Machine Learning Approach for Wavefront Sensing. Sensors (Basel, Switzerland), 19(16). 10.3390/s19163533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin J, King R, Thomas-Virnig C, & Raich WB (2008). Zygotic loss of ZEN-4/MKLP1 results in disruption of epidermal morphogenesis in the C. elegans embryo. Developmental Dynamics, 237(3), 830–836. 10.1002/dvdy.21455 [DOI] [PubMed] [Google Scholar]
- Harding MJ, McGraw HF, & Nechiporuk A. (2014). The roles and regulation of multicellular rosette structures during morphogenesis. Development (Cambridge, England), 141(13), 2549–2558. 10.1242/dev.101444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. (2010, October 4). Neurogenesis in the nematode Caenorhabditis elegans. WormBook : The Online Review of C. Elegans Biology, pp. 1–24. 10.1895/wormbook.1.12.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner MA, Quintin S, Domeier ME, Kimble J, Labouesse M, & Mango SE (1998). pha-4, an HNF-3 homolog, specifies pharyngeal organ identity in Caenorhabditis elegans. Genes and Development, 12(13), 1947–1952. 10.1101/gad.12.13.1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung TJ, & Kemphues KJ (1999). PAR-6 is a conserved PDZ domain-containing protein that colocalizes with PAR-3 in Caenorhabditis elegans embryos. Development (Cambridge, England), 126(1), 127–135. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/9834192 [DOI] [PubMed] [Google Scholar]
- Ikegami R, Simokat K, Zheng H, Brown L, Garriga G, Hardin J, & Culotti J. (2012). Semaphorin and Eph receptor signaling guide a series of cell movements for ventral enclosure in C. elegans. Current Biology : CB, 22(1), 1–11. 10.1016/j.cub.2011.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto A, Martinez-Arias A, & Martin P. (2001). Mechanisms of epithelial fusion and repair. Nature Cell Biology, Vol. 3, pp. E117–E123. 10.1038/35074643 [DOI] [PubMed] [Google Scholar]
- Jewett CE, & Prekeris R. (2018). Insane in the apical membrane: Trafficking events mediating apicobasal epithelial polarity during tube morphogenesis. Traffic, Vol. 19, pp. 666–678. 10.1111/tra.12579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T, David DJV, & Harris TJC (2015). Epithelial apicobasal polarity in the drosophila embryo. In Cell Polarity 1: Biological Role and Basic Mechanisms (pp. 167–187). 10.1007/978-3-319-14463-4_7 [DOI] [Google Scholar]
- Kalb JM, Lau KK, Goszczynski B, Fukushige T, Moons D, Okkema PG, & McGhee JD (1998). pha-4 is Ce-fkh-1, a fork head/HNF-3alpha, beta, gamma homolog that functions in organogenesis of the C. elegans pharynx. Development, 125(12). [DOI] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, & Ahringer J. (2001). Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biology, 2(1), research0002.1. 10.1186/gb-2000-2-1-research0002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Wu Y, Christensen R, Chandris P, Gandler W, McCreedy E, … Shroff H. (2014). Dual-view plane illumination microscopy for rapid and spatially isotropic imaging. Nature Protocols, 9(11), 2555–2573. 10.1038/nprot.2014.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labouesse M. (2006). Epithelial junctions and attachments. WormBook : The Online Review of C. Elegans Biology, 1–21. 10.1895/wormbook.1.56.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landmann F, Quintin S, & Labouesse M. (2004). Multiple regulatory elements with spatially and temporally distinct activities control the expression of the epithelial differentiation gene lin-26 in C. elegans. Developmental Biology, 265(2), 478–490. 10.1016/j.ydbio.2003.09.009 [DOI] [PubMed] [Google Scholar]
- Lecaudey V, Cakan-Akdogan G, Norton WHJ, & Gilmour D. (2008). Dynamic Fgf signaling couples morphogenesis and migration in the zebrafish lateral line primordium. Development, 135(16), 2695–2705. 10.1242/dev.025981 [DOI] [PubMed] [Google Scholar]
- Leung B, Hermann GJ, & Priess JR (1999). Organogenesis of the Caenorhabditis elegans Intestine. Developmental Biology, 216(1), 114–134. 10.1006/dbio.1999.9471 [DOI] [PubMed] [Google Scholar]
- Mango SE (2009). The molecular basis of organ formation: insights from the C. elegans foregut. Annual Review of Cell and Developmental Biology, 25, 597–628. 10.1146/annurev.cellbio.24.110707.175411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolaridis I, Kulkarni K, Dodd RB, Ogasawara S, Zhang Z, Bineva G, … Barford D. (2013). Mechanism of farnesylated CAAX protein processing by the intramembrane protease Rce1. Nature, 504(7479), 301–305. 10.1038/nature12754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AC, & Goldstein B. (2014). Apical constriction: themes and variations on a cellular mechanism driving morphogenesis. Development, 141(10), 1987–1998. 10.1242/dev.102228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon L, Legouis R, Vonesch JL, & Labouesse M. (2001). Assembly of C. elegans apical junctions involves positioning and compaction by LET-413 and protein aggregation by the MAGUK protein DLG-1. Journal of Cell Science, 114(Pt 12), 2265–2277. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/11493666 [DOI] [PubMed] [Google Scholar]
- Motegi F, Plachta N, & Viasnoff V. (2020). Novel approaches to link apicobasal polarity to cell fate specification. Current Opinion in Cell Biology, Vol. 62, pp. 78–85. 10.1016/j.ceb.2019.09.003 [DOI] [PubMed] [Google Scholar]
- Müller HAJ, & Bossinger O. (2003). Molecular networks controlling epithelial cell polarity in development. Mechanisms of Development, Vol. 120, pp. 1231–1256. 10.1016/j.mod.2003.06.001 [DOI] [PubMed] [Google Scholar]
- Nakao F, Hudson ML, Suzuki M, Peckler Z, Kurokawa R, Liu Z, … Takagi S. (2007). The plexin PLX-2 and the ephrin EFN-4 have distinct roles in MAB-20/semaphorin 2A signaling in Caenorhabditis elegans morphogenesis. Genetics, 176(3), 1591–1607. 10.1534/genetics.106.067116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechiporuk A, & Raible DW (2008). FGF-dependent mechanosensory organ patterning in zebrafish. Science (New York, N.Y.), 320(5884), 1774–1777. 10.1126/science.1156547 [DOI] [PubMed] [Google Scholar]
- Page BD, Zhang W, Steward K, Blumenthal T, & Priess JR (n.d.). ELT-1, a GATA-Iike transcription factor, is required for epidermal cell fates in Caenorhabditis elegans embryos. Pásti G, & Labouesse M. (2015). Epithelial junctions, cytoskeleton, and polarity *. 10.1895/wormbook.1.56.2 [DOI] [PubMed] [Google Scholar]
- Patel FB, Bernadskaya YY, Chen E, Jobanputra A, Pooladi Z, Freeman KL, … Soto MC (2008). The WAVE/SCAR complex promotes polarized cell movements and actin enrichment in epithelia during C. elegans embryogenesis. Developmental Biology, 324(2), 297–309. 10.1016/j.ydbio.2008.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel FB, & Soto MC (2013). WAVE/SCAR promotes endocytosis and early endosome morphology in polarized C. elegans epithelia. Developmental Biology, 377(2), 319–332. 10.1016/j.ydbio.2013.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portereiko MF, & Mango SE (2001). Early Morphogenesis of the Caenorhabditis elegans Pharynx. Developmental Biology, 233(2), 482–494. 10.1006/DBIO.2001.0235 [DOI] [PubMed] [Google Scholar]
- Portereiko MF, Saam J, & Mango SE (2004). ZEN-4/MKLP1 Is Required to Polarize the Foregut Epithelium. Current Biology, 14(11), 932–941. 10.1016/j.cub.2004.05.052 [DOI] [PubMed] [Google Scholar]
- Rapti G, Li C, Shan A, Lu Y, & Shaham S. (2017). Glia initiate brain assembly through noncanonical Chimaerin-Furin axon guidance in C. elegans. Nature Neuroscience, 20(10), 1350–1360. 10.1038/nn.4630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen JP, Reddy SS, & Priess JR (2012). Laminin is required to orient epithelial polarity in the C. elegans pharynx. Development (Cambridge, England), 139(11), 2050–2060. 10.1242/dev.078360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts PJ, Mitin N, Keller PJ, Chenette EJ, Madigan JP, Currin RO, … Der CJ (2008). Rho family GTPase modification and dependence on CAAX motif-signaled posttranslational modification. Journal of Biological Chemistry, 283(37), 25150–25163. 10.1074/jbc.M800882200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy PJ, Zheng H, Warren CE, & Culotti JG (2000). mab-20 encodes Semaphorin-2a and is required to prevent ectopic cell contacts during epidermal morphogenesis in Caenorhabditis elegans. Development, 127(4). [DOI] [PubMed] [Google Scholar]
- Sasidharan S, Borinskaya S, Patel F, Bernadskaya Y, Mandalapu S, Agapito M, & Soto MC (2018). WAVE regulates Cadherin junction assembly and turnover during epithelial polarization. Developmental Biology, 434(1), 133–148. 10.1016/j.ydbio.2017.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M, Suetsugu S, Sugimoto A, Miki H, Yamamoto M, & Takenawa T. (2003). Essential role of the C. elegans Arp2/3 complex in cell migration during ventral enclosure. Journal of Cell Science, Vol. 116, pp. 1505–1518. 10.1242/jcs.00362 [DOI] [PubMed] [Google Scholar]
- Sawyer JM, Harrell JR, Shemer G, Sullivan-Brown J, Roh-Johnson M, & Goldstein B. (2010). Apical constriction: A cell shape change that can drive morphogenesis. Developmental Biology, 341(1), 5–19. 10.1016/j.ydbio.2009.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafaq-Zadah M, Brocard L, Solari F, & Michaux G. (2012). AP-1 is required for the maintenance of apico-basal polarity in the C. elegans intestine. Development, 139(11), 2061–2070. 10.1242/dev.076711 [DOI] [PubMed] [Google Scholar]
- Shah PK, Tanner MR, Kovacevic I, Rankin A, Marshall TE, Noblett N, Tran NN, Roenspies T, Hung J, Chen Z, Slatculescu C, Perkins TJ, Bao Z, & Colavita A. (2017). PCP and SAX-3/Robo Pathways Cooperate to Regulate Convergent Extension-Based Nerve Cord Assembly in C. elegans. Developmental Cell, 41(2), 195–203.e3. 10.1016/j.devcel.2017.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, & Thomson JN (1983, November 1). The embryonic cell lineage of the nematode Caenorhabditis elegans. Developmental Biology, Vol. 100, pp. 64–119. 10.1016/0012-1606(83)90201-4 [DOI] [PubMed] [Google Scholar]
- Timmons L, Court DL, & Fire A. (2001). Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene, 263(1–2), 103–112. 10.1016/S0378-1119(00)00579-5 [DOI] [PubMed] [Google Scholar]
- Totong R, Achilleos A, & Nance J. (2007). PAR-6 is required for junction formation but not apicobasal polarization in C. elegans embryonic epithelial cells. Development, 134(7), 1259–1268. 10.1242/dev.02833 [DOI] [PubMed] [Google Scholar]
- Von Stetina SE, & Mango SE (2015). PAR-6, but not E-cadherin and β-integrin, is necessary for epithelial polarization in C. elegans. Developmental Biology, 403(1), 5–14. 10.1016/j.ydbio.2015.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace AG, Raduwan H, Carlet J, & Soto MC (2018). The RhoGAP HUM-7/Myo9 integrates signals to modulate RHO-1/RhoA during embryonic morphogenesis in Caenorhabditiselegans. Development (Cambridge, England), 145(23). 10.1242/dev.168724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Roy PJ, Holland SJ, Zhang LW, Culotti JG, & Pawson T. (1999). Multiple ephrins control cell organization in C. elegans using kinase-dependent and -independent functions of the VAB-1 Eph receptor. Molecular Cell, 4(6), 903–913. 10.1016/S1097-2765(00)80220-8 [DOI] [PubMed] [Google Scholar]
- Wernike D, Chen Y, Mastronardi K, Makil N, & Piekny A. (2016). Mechanical forces drive neuroblast morphogenesis and are required for epidermal closure. Developmental Biology, 412(2), 261–277. 10.1016/j.ydbio.2016.02.023 [DOI] [PubMed] [Google Scholar]
- Williams-Masson EM, Malik AN, & Hardin J. (1997). An actin-mediated two-step mechanism is required for ventral enclosure of the C. elegans hypodermis. Development, 124(15). [DOI] [PubMed] [Google Scholar]
- Withee J, Galligan B, Hawkins N, & Garriga G. (2004). Caenorhabditis elegans WASP and Ena/VASP proteins play compensatory roles in morphogenesis and neuronal cell migration. Genetics, 167(3), 1165–1176. 10.1534/genetics.103.025676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Landmann F, Zahreddine H, Rodriguez D, Koch M, Labouesse M. (2011). A tension induced mechanotransduction pathway promotes epithelial morphogenesis. Nature, 471(7336), 99–103. 10.1038/nature09765 [DOI] [PubMed] [Google Scholar]
- Zheng G, Cochella L, Liu J, Hobert O, & Li WH (2011). Temporal and spatial regulation of microrna activity with photoactivatable cantimirs. ACS Chemical Biology, 6(12), 1332–1338. 10.1021/cb200290e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman Y, Abrams J, Anderson DC, & Nance J. (2017). Cdc42 regulates junctional actin but not cell polarization in the Caenorhabditis elegans epider. Journal of Cell Biology, 216(11), 3729–3744. 10.1083/jcb.201611061 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S5. A time-lapse movie (7 fps) of the embryo from Fig. 3D co-expressing myristoylated (post-translational modification that targets proteins to the membrane) MIR-228::GFP (green) and PAR-6::mKate2 (magenta) imaged by sweptfield confocal microscopy through anterior morphogenesis.
Movie S3. A time-lapse movie (7 fps) of the embryo from Fig. 3A co-expressing mNeonGreen::PAR-6 and lin-26p::VAB-10(ABD)::mCherry (magenta) imaged by sweptfield confocal microscopy through anterior morphogenesis.
Movie S1. A time-lapse movie (7 fps) of the embryo from Fig. 1C co-expressing mNeonGreen::PH to visualize membranes (green) and H2B::mCherry to visualize nuclei (magenta), imaged by di-SPIM through anterior morphogenesis.
Movie S7. A time-lapse movie (7 fps) of one of the embryos from Fig. 3D co-expressing myrisotylated HLH-16::GFP (green) and PAR-6::mKate2 (magenta) imaged by sweptfield confocal microscopy through anterior morphogenesis.
Movie S2. A time-lapse movie (7 fps) of an embryo from Fig. 2D co-expressing pha-4p::GFP::CAAX (green) and PAR-6::mKate2 (magenta) imaged by sweptfield confocal microscopy through anterior morphogenesis.
Movie S6. A time-lapse movie (7 fps) of an embryo expressing myristoylated MIR-228::GFP in Fire LUTs imaged by sweptfield confocal microscopy through anterior morphogenesis.
Movie S8. A time-lapse movie (7 fps) of an embryo expressing myristoylated HLH-16::GFP in Fire LUTs imaged by sweptfield confocal microscopy through anterior morphogenesis.
Movie S4. A time-lapse movie (7 fps) of the embryo from Fig. 3C co-expressing UNC-119::YFP (green), and PAR-6::mKate2 (magenta) imaged by sweptfield confocal microscopy through anterior morphogenesis.
Movie S9. A time-lapse movie (7fps) of the embryo from Fig. 5B (top panel) expressing epidermal LifeAct::GFP taken using HILO microscopy (anterior down) to see ventral epidermal cell F-actin projections.
Movie S10. A time-lapse movie (7 fps) of the embryo from Fig. 5B (bottom panel) expressing epidermal VAB-10 (ABD)::mCherry taken using HILO microscopy (anterior down) to see dorsal epidermal cell F-actin projections.
Time-lapse images obtained by sweptfield microscopy show en face views of an embryo co-expressing mNeonGreen::PH to visualize membranes (green) and H2B::mCherry to visualize nuclei (magenta) through anterior morphogenesis. A ‘ring in a ring’ of cells is highlighted (white dashed circles, inner yellow dots). The star-like pattern in the center marks the site of the future lumen, which forms after cells ingress into the embryo. Times are shown in minutes, and the scale bar is 10 μm.
Time-lapse images acquired using sweptfield microscopy show ventral views of an embryo co-expressing mNeonGreen::PAR-6 (green) and lin-26p;VAB-10(ABD)::mCherry (magenta) after ani-1 RNAi. The zoomed insets (yellow box) show the region where the foci would typically be found. While the BFP is still present, some of the foci associated with the pentagons and the semi-circle of foci are not visible. Times are shown in minutes, and the scale bar is 10 μm.
A)Time-lapse images acquired using sweptfield microscopy show en face views of two different embryos expressing AJM-1::GFP (Fire LUTs) during anterior morphogenesis. B) Time-lapse images show ventral views (anterior to the left) of an embryo expressing HMP-1::GFP and DLG-1::RFP during anterior morphogenesis. Times are shown in minutes, and the scale bar is 10 μm.
A) Timelapse images of the embryo from Fig. 3A, co-expressing mNeonGreen::PAR-6 (green) and lin-26p::VAB-10 (ABD)::mCherry (magenta) is shown during anterior morphogenesis without any labels. B) Timelapse images of an embryo as in A), with a more dorsal view to better visualize epidermal cells that are more dorsally positioned. The arrows point to the dorsal foci of the pentagons, and the dotted line indicates the leading edge of epidermal cells migrating from more dorsal positions. C) Time-lapse images show embryos expressing LSY-6::GFP, HLH-16::GFP, and MIR-228::GFP in Fire LUTs as indicated. For all embryos, times are shown in minutes, and the scale bar is 10 μm.