Abstract
Recently described rhizolutin and collinolactone isolated from Streptomyces Gö 40/10 share the same novel carbon scaffold. Analyses by NMR and X‐Ray crystallography verify the structure of collinolactone and propose a revision of rhizolutin's stereochemistry. Isotope‐labeled precursor feeding shows that collinolactone is biosynthesized via type I polyketide synthase with Baeyer–Villiger oxidation. CRISPR‐based genetic strategies led to the identification of the biosynthetic gene cluster and a high‐production strain. Chemical semisyntheses yielded collinolactone analogues with inhibitory effects on L929 cell line. Fluorescence microscopy revealed that only particular analogues induce monopolar spindles impairing cell division in mitosis. Inspired by the Alzheimer‐protective activity of rhizolutin, we investigated the neuroprotective effects of collinolactone and its analogues on glutamate‐sensitive cells (HT22) and indeed, natural collinolactone displays distinct neuroprotection from intracellular oxidative stress.
Keywords: Alzheimer's disease, biosynthesis, natural products, neuroprotection, polyketide
Collinolactone shares the same carbon scaffold with rhizolutin and X‐ray crystallography has provided the absolute configuration. A highly non‐collinear gene cluster was identified. Studies of cell viability using fluorescence microscopy revealed that analogues of collinolactone induce the formation of a monopolar spindle phenotype while collinolactone showed neuroprotective effects by reducing intracellular oxidative stress.
Improved health care by novel treatments and technological innovation has significantly increased life expectancy. Due to the concomitant demographic ageing the proportion of older people has increased and therefore, age‐related diseases such as cancer, diabetes and neurodegenerative diseases are on the rise. [1] In 2020, over 50 million people worldwide have been diagnosed with neurodegenerative diseases, that is, a chronical impairment of memory and altered behavior from declining cognitive abilities. [2] This number is estimated to double every 20 years, leading to more than 131 million cases by 2050. [3] The most common form is Alzheimer's disease (AD) with 50–75 % of the cases. [3] Hallmarks of AD are extracellular deposit of misfolded forms of amyloid‐β (Aβ) and hyperphosphorylated τ‐protein. [4] Although 121 clinical trials are ongoing for AD treatment targeting Aβ or τ‐protein, [5] only one candidate (Aducanumab, 2021) [6] has been approved since 2003. The series of candidates failing in clinical trials is constantly growing [7] and the rate of repurposed agents (43 %) indicates the need of novel structures. [8] Nature has proved to be an incredibly rich source of new and unconventional structural motifs for drug discovery and development. [9]
Recently, Kwon et al. described the discovery of rhizolutin, [10] a 6‐10‐7‐membered tricyclic system with a cyclodecatriene ring flanked by two lactone rings. Investigation of its bioactivity revealed a dissociative effect on Aβ and τ‐tangles in vitro, with significantly reduced apoptosis and inflammation in neural cells. In vivo studies on APP/PS1 double transgenic mice showed substantially dissociated hippocampal plaques. [10]
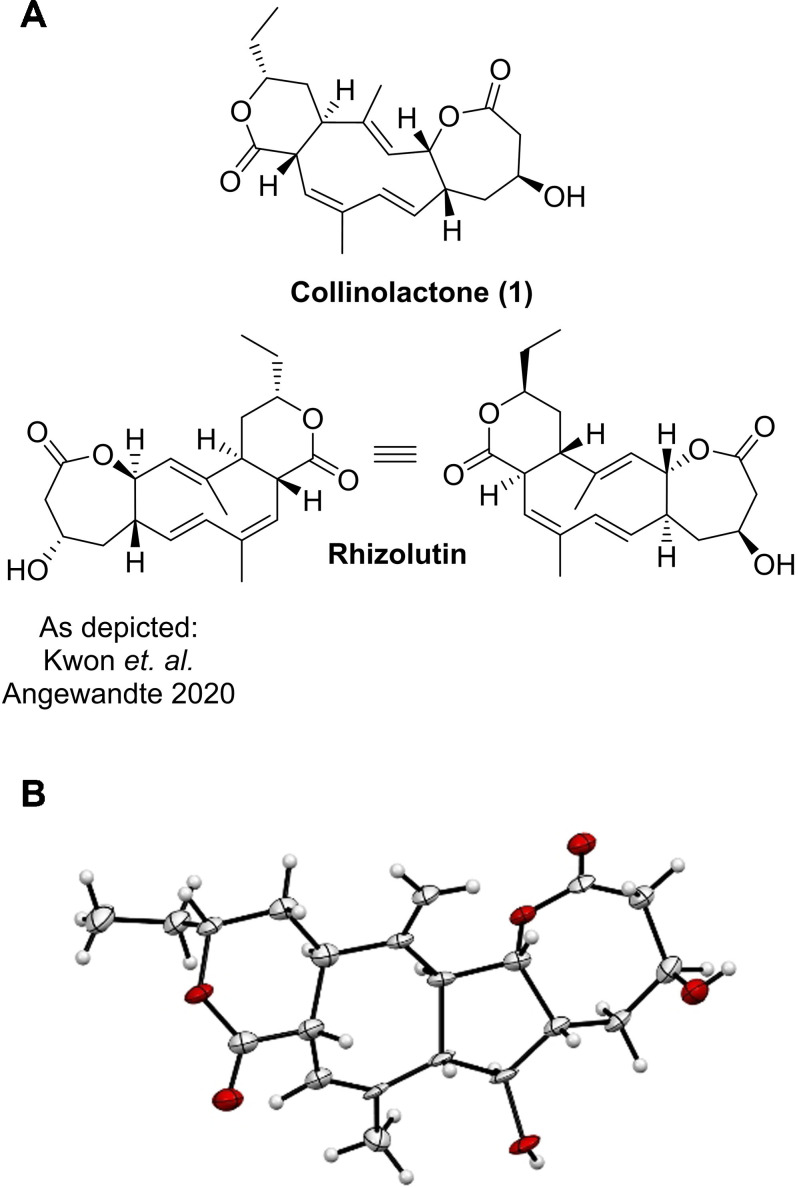
Herein, we present our studies on collinolactone (1), which has originally been described by the Zeeck group [11] and shares the same characteristic core structure elements of its planar structure. After our careful inspection of the structure elucidation carried out for rhizolutin, we have strong evidence to conclude that the stereochemistry of this initial structure has not been correctly assigned, as collinolactone (1) shares NMR spectroscopic data identical to rhizolutin (Figure S1).
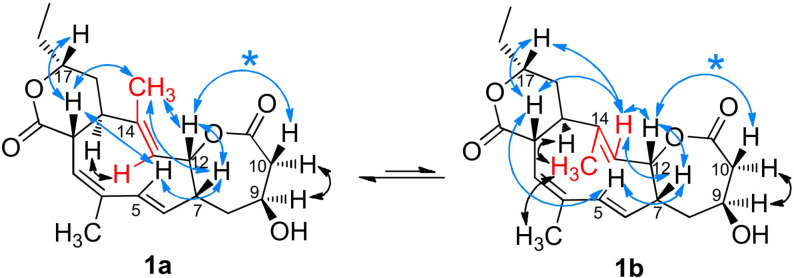
Like rhizolutin, 1 shows two conformers (1 a and 1 b) in NMR spectra due to the rotating double bond at C13−C14. The ratio is dependent on both solvent and temperature: with increasing temperature, the ratio changes in favor of the syn component. The coalescence temperature has not been reached due to solvent and instrument limitations and was interpolated to 398±5.6 K (Figure S3). Reversely, the conversion continued even at 233 K.
2D NMR experiments allowed us to study the conversion in more detail and to state the relative configuration (Scheme 1). For rhizolutin, only one correlation in ROESY was observed to link the left‐hand part of the molecule to the right‐hand part (Scheme 1, correlation of 12‐H/10‐H, marked with an asterisk). Several correlations indicate that 7‐H is in the same plane as 12‐H and 10‐H. In [D5]pyridine, which was used for the newly compiled structure elucidation of rhizolutin by Kwon et al., the signal of 12‐H (δ H=6.12 ppm) overlaps with the signal from 5‐H and can therefore be misinterpreted when attempting to determine the configuration at 7‐H. Notably, using the solvent [D6]benzene resulted in well separated and non‐overlapping signals of hydrogens at 12‐H, 7‐H, and 5‐H, and NOESY spectra showed a strong correlation between 12‐H and 7‐H (Figure S2). In addition, no correlation was found between 9‐H and 7‐H, indicating that these hydrogens are not in the same plane. The comprehensive set of NOE and ROE NMR experiments in [D2]dichloromethane, [D6]benzene, and [D5]pyridine further verifies stereochemical assignments of 1—and due to identical NMR spectra in [D5]pyridine, also of rhizolutin.
Scheme 1.
Important NOE correlations of rotamers (1 a, 1 b). Blue arrows mark NOE above ring plane (* key correlation), black NOE below ring plane. The substructure marked in red is responsible for the dynamic interconversion of the two rotamers (atom numbering, see ref. [10]).
A correlation between 9‐H and 7‐H is displayed in the summary image of ROESY correlations provided by Kwon et al. for rhizolutin. [10] However, we were unable to identify this correlation between hydrogen atoms at δ H=4.67 ppm and δ H=3.31 ppm with reasonable certainty in the provided data as well as in our own recorded ROESY and NOESY spectra of collinolactone (1).
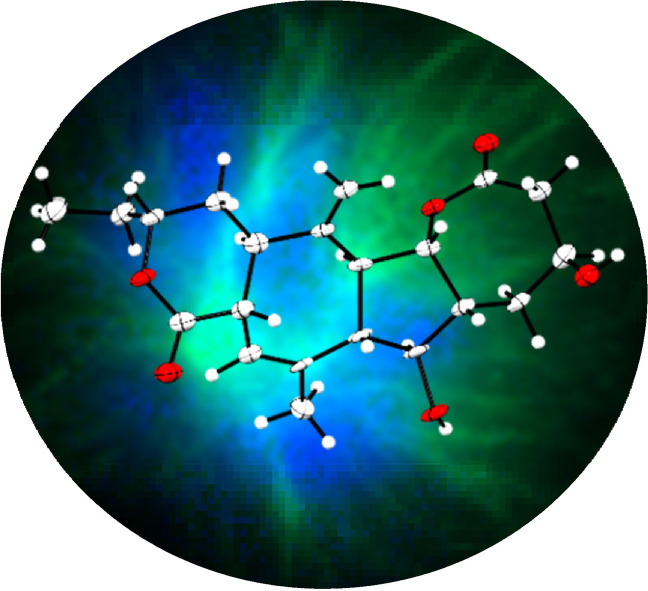
X‐ray crystallography is highly valuable to determine the absolute configuration. [12] All our crystallization attempts of natural product 1 failed due to its flexible scaffold. The fixation of the flexible methyl group at C‐14 using one equivalent of meta‐chloroperoxybenzoic acid (mCPBA) [13] led to 3 as the main product and a crystal suitable for X‐ray crystallography. NMR spectra showed only one distinct set of signals, indicating a rigid scaffold. Structure elucidation [14] revealed that a transannular cyclization has occurred for which we propose a possible mechanism (Scheme S3). Consequently, there is no indication that the cyclization influenced the stereochemistry of the initial scaffold (Figure 1 B). [15] To the best of our knowledge, we conclude that the initial stereochemical structure assignment of rhizolutin was not unambiguous and that collinolactone (1) is the correct structure.
Figure 1.
A) Structure of collinolactone (1) and the proposed structure of rhizolutin by Kwon et al. (left) and flipped by 180° (right). B) X‐Ray structure of semisynthetic tetracyclic collinolactone analogue 3.
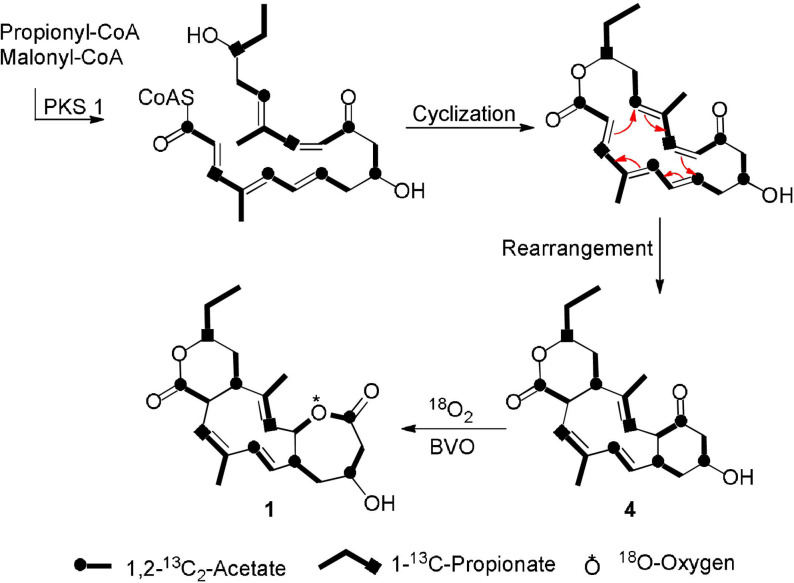
In addition to structure elucidation, the biosynthetic pathway of compound 1 isolated from Streptomyces Gö 40/10 was investigated. From its structure, we assumed a product from a type I polyketide synthase (PKS) assembly line and therefore, we performed feeding experiments using 13C‐labeled common PKS building units. Analyses of the 13C NMR spectra after precursor feeding revealed that three propionate and six acetate units were incorporated. Cultivation of the strain under 18O2 atmosphere led to the incorporation of 18O between C‐11 and C‐12, which was confirmed using the oxygen‐18 isotope shift upon 13C carbon as described before. [16]
The genome sequence of the producer strain was analyzed using antiSMASH 5.1.2. [17] At least 40 biosynthesis gene clusters (BGCs) were identified, supporting the previously described high biosynthetic potential of the strain.[ 11 , 18 ]
Based on the feeding experiments and identified cluster regions, five potential PKS BGCs of 1 were identified and knockouts from each candidate BGC were prepared using CRISPR‐BEST [19] technology or suicide vectors (Tables S1 and S2). In case of the BGC encoded in region 1.23, a stop codon was introduced at the beginning of gene colL (Figure S4), leading to premature termination of translation and thus the inactivation of its gene product. Dysfunctional colL abolished the production of 1 (Figure S5) and indicates that region 1.23 codes for the collinolactone (col) BGC.
A recent study on the biosynthesis of neoabyssomicin proposed a Baeyer–Villiger oxidation type II mechanism involving a flavin reductase and a luciferase‐like monooxygenase (LLM). [20] The genes encoding one flavin reductase and two LLMs were identified in region 1.23 (Figure S4), suggesting a similar oxidation mechanism in the biosynthesis of 1. A detailed analysis of the involved biosynthetic genes is currently ongoing.
Based on the genetic information and feeding experiments, we hypothesize that the PKS‐derived linear nonaketide (Figure S6) undergoes cyclization forming an 18‐membered macrolactone (Scheme 2). A subsequent [6+4] cyclic rearrangement or [4+2] cyclization followed by a [3,3]‐Cope rearrangement, which was studied in detail more recently, [21] leads to the intermediate product 4, which can be isolated in small yields (0.3 mg L−1). The incorporation of molecular oxygen via Baeyer–Villiger‐like oxidation (BVO) leads to the final product 1 (35 mg L−1).
Scheme 2.
Proposed biosynthesis of compound 1 via a type I PKS pathway deduced from isotope‐labeled precursor feeding experiments.
Overexpression of the colA gene encoding a putative luxR‐type positive transcription factor under control of the strong promoter kasOp* [22] led to a 7‐fold increased production compared to the wild‐type strain (Figure S7).
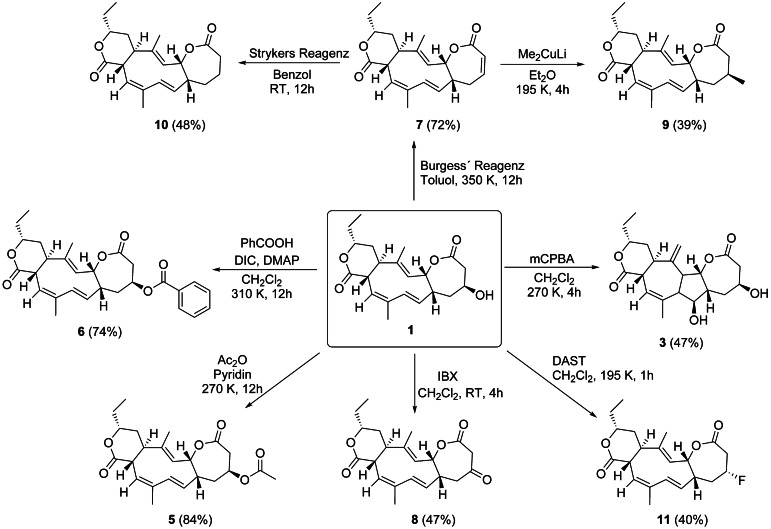
A common approach to investigate the potential mode of action is using (semi)‐synthetic modifications of the original scaffold to tune molecular properties. [23] Collinolactone was acylated with acetic acid anhydride (leading to 5) and by benzoic acid (leading to 6) using a classic Steglich approach for stereochemically hindered secondary alcohols (Scheme 3). Our goal was to improve cellular uptake assuming that acylated compounds cross lipid bilayers more readily and are then unmasked by intracellular esterase activity. During synthesis, the formation of small amounts of collinolactenone (7) was observed as a side product. We propose a six‐membered transition state for the reaction which is heavily influenced by the leaving group ability of the ester‐linked acid.
Scheme 3.
Chemical derivatization of 1. Abbreviations: DIC: N,N′‐Diisopropylcarbodiimide, DMAP: 4‐Dimethylaminopyridine, r.t.: room temperature.
The use of Burgess dehydration gave direct synthetic access to larger amounts (milligram scale) of 7, which features an α,β‐unsaturated Michael system and is therefore favored to be targeted by non‐specific nucleophilic attacks, for example, of free thiol group containing metabolites or proteins. Since small amounts of 7 were always present in purified samples of 5, we were looking for semisynthetic analogues not prone to form 7—presumably resulting in less reactive derivatives.
When oxidizing the hydroxy group of 1 to get 8 using 2‐iodoxybenzoic acid (IBX), formation of 7 was no longer observed as hydrogen bonding is decreased for ketones compared to hydroxy groups.
Organocopper reagents such as Gilmann cuprates and Stryker's reagent tolerate a range of functional groups such as lactones and alkenes making them useful for synthesis of complex natural products. [24] We used Gilmann cuprates to incorporate a methyl group at C‐9 (yielding 9) with the stereoconfiguration determined by NOESY correlations.
Stryker's reagent has been used for selective hydration of the double bond in 1,4 position (to give 10), while all other double bonds in the cyclodecatriene system remained unaffected (Scheme 3). Hydrogenation of the double bond (10) and the insertion of a methyl group (9) successfully suppressed the de novo formation of 7 as anticipated. One commonly used approach to improve pharmacological properties is the replacement of hydroxy groups by fluorine. [25] Compound 11 was synthesized using diethylaminosulfur trifluoride (DAST), which replaces primary and secondary hydroxy groups by fluorine under stereochemical conversion. [26] All synthesized compounds except 3 show two distinct sets of signals in the NMR.
Interestingly, also the biosynthetic intermediate 4 does not show an additional set of signals. This indicates that the scaffold of 4 is more rigid and becomes flexible as soon as the molecular oxygen is incorporated forming ϵ‐lactone in 1.
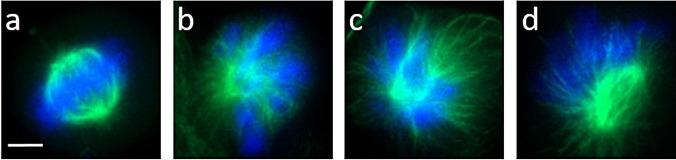
All obtained compounds were tested in distinct sets of bioassays to investigate their biological properties. None of the compounds showed activity against Bacillus subtilis and Escherichia coli or the fungus Rhodotorula rubra. Initial eukaryotic vertebrate cell viability screening revealed that 7 and 5 showed inhibitory effects against L929 cell line (Figure S8). Fluorescence microscopy imaging of PtK2 cell line, as a model for the investigation of mitosis, [27] was used to study the potential mode of action. In cells treated with either 7 or 5 at 30 hours after seeding, the formation of monopolar spindles was observed. This phenotype was first described for the synthetic product monastrol and is assumed to be initiated by the motility inhibition of mitotic kinesin Eg5. [28] No phenotype formation was observed for other derivatives as expected, because formation of 7 was inhibited by chemical modifications (Figure 2). This indicates that the Michael acceptor plays a major role in target binding.
Figure 2.
Fluorescence microscopy images of PtK2 cells during mitosis (scale bar 5 μm). Green: Microtubules form the mitotic spindles; Blue: DNA, condensed to chromosomes. Cells were incubated with concentrations of 25 μm for 12 hours, starting after 30 h of cultivation: a) negative control with 0.2 % DMSO; b) monastrol (positive control for monoastral phenotype); c) collinolactenon (7); d) acetylcollinolactone (5).
The images were further analyzed to investigate effects on cell cycle following a previously published procedure. [29] After treatment with DMSO (30 hours post seeding), an increased number of cells was found in G1 phase compared to non‐treated cells (Figure S9). Note that DMSO itself was reported as an inducer of a reversible arrest in G1 phase. [30] Similar effects were observed for cells treated with 7 and 5 (DMSO concentration as in control) while cells incubated with 1 showed a similar distribution compared to the non‐treated cells. This indicates that 1 might help to overcome the negative effects of DMSO. However, a modulative effect on the cell cycle compared to the non‐treated control could not be observed. Cells influenced by contact inhibition, due to high cell density, did not show these effects (Figure S9).
We have profiled our compounds in an Aβ agregation assay using a previously described procedure. [31] Compound 1 showed similar effects towards Aβ aggregation inhibition as described for rhizolutin. [10] In addition, compound 10 was found to reduce the formation of Aβ aggregates by 25 %. No inhibitory effects could be observed for other derivatives (Figure S11).
Kwon et al. have further investigated the protective effect of rhizolutin against Aβ‐induced cytotoxicity. For their studies, they incubated murine hippocampal HT22 cells with pre‐aggregated Aβ and measured effects on cell viability. [10] We have performed similar experiments with the same cell line in an oxytosis assay for 1 and derivatives 3–11 using the same cell line. The assay is based on the treatment with extracellular glutamate, which causes the production of intracellular stress that ultimately leads to cell death. [32]
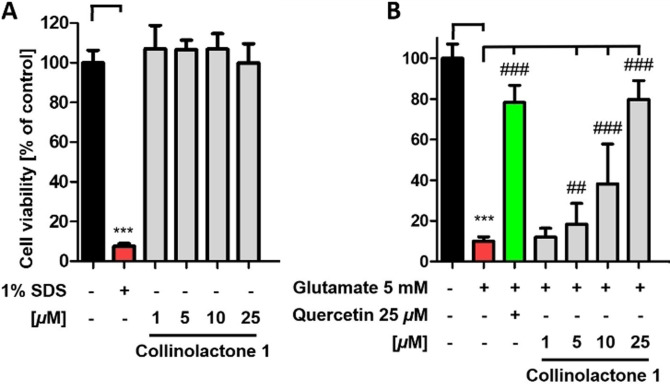
At this point, it should be emphasized that the activity of compounds in this assay is therefore based on an intracellular effect which was not triggered by Aβ. In a first step, compounds were tested for cytotoxic effects on HT22 cells before an oxytosis assay was performed (Figure S10). Besides the already mentioned effects on cell viability for compounds 5 and 7, no additional effects were observed for the derivatives. As expected, the addition of glutamate causes cell death, whereas co‐treatment with quercetin (conc. 25 μm, positive control) protects against this effect (Figure 3). However, 1 also protects against glutamate‐induced oxidative stress in a dose‐dependent manner. Remarkably, no other derivative (3–11) showed any effect of neuroprotection at concentrations up to 25 μm (Figure S10). Therefore, the activity of compound 1 is highly specific and even small structural changes are associated with a loss of activity. It might be linked to other phenomena also, for example, to cell uptake or different metabolism.
Figure 3.
Collinolactone (1) was studied for neurotoxic effects (A) and neuroprotection (B) against glutamate‐induced oxidative stress at 1–25 μm using HT22 cell line. 1 % SDS served as positive control for neurotoxic effects whereas quercetin (25 μm) served as a positive control for neuroprotection. Results of the modified MTT tests are presented as means ± SEM of three independent experiments, each performed in sextuplicate, and refer to untreated control cells which were set as 100 % values. Levels of significance: ***p<0.001; ## p<0.01; ### p<0.001.
In summary, we have compared the stereochemical aspects of rhizolutin and collinolactone by NMR experiments. X‐ray crystallography strongly supports the proposed structure of 1. In addition, we have complemented our study by investigation of the biosynthesis of 1 and identified the corresponding gene cluster. Cell viability was studied for the natural and semisynthetic derivatives of 1; compound 5 and 7 showed effects on L929 cell line in the micromolar range and further fluorescence microscopy studies revealed the formation of monopolar spindles. Finally, the neuroprotective effect of collinolactone and its derivatives against oxidative stress was analyzed using a glutamate‐induced based neurotoxicity assay. Only compound 1 was found to have neuroprotective properties, indicating a highly specific interaction with cellular targets.
Conflict of interest
Eberhard Karls Universität Tübingen and Danmarks Tekniske Universitet have filed a patent application (DE 10 2021 003 944.0) as inventors.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Acknowledgements
J.F.G.G. and T.W. were supported by the Novo Nordisk Foundation [grants NNF16OC0021746, NNF20CC0035580 to T.W.]. R.K., K.F. and S.G. are part of the graduation program IPMB funded by the MWK Baden‐Württemberg. L.S. was funded by SFB 416. The work was further supported by infrastructural funding from Eberhard Karls University of Tuebingen. We especially thank Prof. Dr. Oh and co‐workers from the Natural Products Research Institute, Seoul National University, Korea, who kindly provided NMR data of rhizolutin for a final comparison. Also, we thank Pascal Rath for NMR measurements, José. M. Beltrán‐Beleña Nagel for LC–MS measurements, and Hans‐Jörg Langer for excellent technical assistance. Open Access funding enabled and organized by Projekt DEAL.
J. C. Schmid, K. Frey, M. Scheiner, J. F. G. Garzón, L. Stafforst, J.-N. Fricke, M. Schuppe, H. Schiewe, A. Zeeck, T. Weber, I. Usón, R. Kemkemer, M. Decker, S. Grond, Angew. Chem. Int. Ed. 2021, 60, 23212.
References
- 1. Hou Y., Dan X., Babbar M., Wei Y., Hasselbalch S. G., Croteau D. L., Bohr V. A., Nat. Rev. Neurol. 2019, 15, 565–581. [DOI] [PubMed] [Google Scholar]
- 2. Gouras G. K., in Reference Module in Biomedical Sciences, Elsevier, Amsterdam, 2014. [Google Scholar]
- 3.M. Prince, A. Wimo, M. Guerchet, G.-C. Ali, Y.-T. Wu, M. Prina, World Alzheimer Report 2015, 2015
- 4. Long J. M., Holtzman D. M., Cell 2019, 179, 312–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cummings J., Lee G., Ritter A., Sabbagh M., Zhong K., Alzheimer′s Dementia 2020, 6, e12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Alexander G. C., Emerson S., Kesselheim A. S., J. Am. Med. Assoc. 2021, 325, 1717–1718. [DOI] [PubMed] [Google Scholar]
- 7. Panza F., Lozupone M., Logroscino G., Imbimbo B. P., Nat. Rev. Neurol. 2019, 15, 73–88. [DOI] [PubMed] [Google Scholar]
- 8. Yiannopoulou K. G., Papageorgiou S. G., J. Cent. Nerv. Syst. Dis. 2020, 12, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Newman D. J., Cragg G. M., J. Nat. Prod. 2020, 83, 770–803. [DOI] [PubMed] [Google Scholar]
- 10. Kwon Y., Shin J., Nam K., An J. S., Yang S. H., Hong S. H., Bae M., Moon K., Cho Y., Woo J., Park K., Kim K., Shin J., Kim B. Y., Kim Y., Oh D. C., Angew. Chem. Int. Ed. 2020, 59, 22994–22998; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2020, 132, 23194–23198. [Google Scholar]
- 11. Bode H. B., Bethe B., Höfs R., Zeeck A., ChemBioChem 2002, 3, 619–627. [DOI] [PubMed] [Google Scholar]
- 12. Bijvoet J. M., Peerdeman A. F., van Bommel A. J., Nature 1951, 168, 271–272. [Google Scholar]
- 13. Prileschajew N., Ber. Dtsch. Chem. Ges. 1909, 42, 4811–4815. [Google Scholar]
- 14. Sheldrick G. M., Acta Crystallogr. Sect. A 2008, 64, 112–122. [DOI] [PubMed] [Google Scholar]
- 15.Deposition number 2006674 contains the supplementary crystallographic data for this paper. These data are provided free of charge by the joint Cambridge Crystallographic Data Centre and Fachinformationszentrum Karlsruhe Access Structures service.
- 16. Risley J. M., Van Etten R. L., J. Am. Chem. Soc. 1979, 101, 252–253. [Google Scholar]
- 17. Blin K., Shaw S., Steinke K., Villebro R., Ziemert N., Lee S. Y., Medema M. H., Weber T., Nucleic Acids Res. 2019, 47, W81–W87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schiewe H. J., Zeeck A., J. Antibiot. 1999, 52, 635–642. [DOI] [PubMed] [Google Scholar]
- 19. Tong Y., Whitford C. M., Robertsen H. L., Blin K., Jørgensen T. S., Klitgaard A. K., Gren T., Jiang X., Weber T., Lee S. Y., Proc. Natl. Acad. Sci. USA 2019, 116, 20366–20375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ji X., Tu J., Song Y., Zhang C., Wang L., Li Q., Ju J., ACS Catal. 2020, 10, 2591–2595. [Google Scholar]
- 21. Zhang B., Wang K. B., Wang W., Wang X., Liu F., Zhu J., Shi J., Li L. Y., Han H., Xu K., Qiao H. Y., Zhang X., Jiao R. H., Houk K. N., Liang Y., Tan R. X., Ge H. M., Nature 2019, 568, 122–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang W., Li X., Wang J., Xiang S., Feng X., Yang K., Appl. Environ. Microbiol. 2013, 79, 4484–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Itoh H., Inoue M., Chem. Rev. 2019, 119, 10002–10031. [DOI] [PubMed] [Google Scholar]
- 24. Löffler L. E., Wirtz C., Fürstner A., Angew. Chem. Int. Ed. 2021, 60, 5316–5322; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2021, 133, 5376–5382. [Google Scholar]
- 25. Johnson B. M., Shu Y.-Z., Zhuo X., Meanwell N. A., J. Med. Chem. 2020, 63, 6315–6386. [DOI] [PubMed] [Google Scholar]
- 26. Baumann M., Baxendale I. R., Ley S. V., Synlett 2008, 2008, 2111–2114. [Google Scholar]
- 27. Hamkalo B. A., Elgin S. C. R., Functional Organization of the Nucleus: A Laboratory Guide, Vol. 35, Elsevier Science & Techn., Amsterdam, 1991. [Google Scholar]
- 28. Mayer T. U., Kapoor T. M., Haggarty S. J., King R. W., Schreiber S. L., Mitchison T. J., Science 1999, 286, 971–974. [DOI] [PubMed] [Google Scholar]
- 29. Roukos V., Pegoraro G., Voss T. C., Misteli T., Nat. Protoc. 2015, 10, 334–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fiore M., Zanier R., Degrassi F., Mutagenesis 2002, 17, 419–424. [DOI] [PubMed] [Google Scholar]
- 31. Kim D., Bae G. H., Kim H. Y., Jeon H., Kim K., Shin J., Lee S., Hong S., Kim I., Kim Y., ACS Chem. Neurosci. 2021, 12, 99–108. [DOI] [PubMed] [Google Scholar]
- 32.
- 32a. Tan S., Schubert D., Maher P., Curr. Top. Med. Chem. 2001, 1, 497–506; [DOI] [PubMed] [Google Scholar]
- 32b. Currais A., Maher P., Antioxid. Redox. Sign. 2013, 19, 813–822. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information