Summary
The role that common mycorrhizal networks (CMNs) play in plant‐to‐plant transfer of zinc (Zn) has not yet been investigated, despite the proved functions of arbuscular mycorrhizal fungi (AMF) in crop Zn acquisition. Here, two autotrophic Medicago truncatula plants were linked by a CMN formed by Rhizophagus irregularis. Plants were grown in vitro in physically separated compartments (Donor‐C and Receiver‐C) and their connection ensured only by CMN. A symbiosis‐defective mutant of M. truncatula was used as control in Receiver‐C. Plants in both compartments were grown on Zn‐free medium, and only the leaves of the donor plants were Zn fertilized. A direct transfer of Zn was demonstrated from donor leaves to receiver shoots mediated by CMN. Direct transfer of Zn was supported by changes in the expression of fungal genes, RiZRT1 and RiZnT1, and plant gene MtZIP2 in roots and MtNAS1 in roots and shoots of the receiver plants. Moreover, Zn transfer was supported by the change in expression of MtZIP14 gene in AM fungal colonized roots. This work is the first evidence of a direct Zn transfer from a donor to a receiver plant via CMN, and of a triggering of transcriptional regulation of fungal‐plant genes involved in Zn transport‐related processes.
Introduction
In the recent decade, an increasing number of studies have reported the ability of arbuscular mycorrhizal fungi (AMF) to connect plants belonging to the same or different species, genera and families in common mycorrhizal networks (CMNs). The capacity of AMF to develop such CMNs is related to their wide host range, even though a variability in host specificity among AMF has been found (Öpik et al., 2009; Veresoglou and Rillig, 2014; Ciccolini et al., 2016; Davison et al., 2020). These CMNs have attracted the attention of the scientific community due to their presumed roles in the transfer of nutrients and photosynthetic C between interconnected plants (e.g., Newman et al., 1994; He et al., 2003; Simard et al., 2003; Selosse et al., 2006; Voets et al., 2008; Fellbaum et al., 2014), thus playing key roles in the functioning of ecosystems (van der Heijden et al., 1998).
CMNs are complex structures whose density varies with AM fungal isolate and interconnected plant species (Giovannetti et al., 2004; Avio et al., 2006). The high frequency of anastomoses (hyphal fusions; Kirk et al., 2008; 44%–63%) between extraradical hyphae of the same AM fungal isolate, spreading from the roots of different plants, sustains the establishment and functionality of CMNs (Giovannetti et al., 2004; Avio et al., 2006; Croll et al., 2009). Connection between plants can also be established from a mycelium growing from an AM fungal colonized plant to a not inoculated one as reported by Voets and colleagues (2008, 2009) in a bi‐compartmented in vitro cultivation system. In field conditions, a single individual mycelium belonging to a cluster within the Glomus clade was found to link the roots of Hieracium pilosella, covering up to 10 m in length (Rosendahl and Stukenbrock, 2004). Further laboratory studies suggested that ubiquitous AM fungal species, such as Funnneliformis mosseae and Rhizophagus intraradices, interconnected tomato (Solanum lycopersicum) and faba bean (Vicia faba) plants separated by a distance up to 15 cm (Song et al., 2010; Babikova et al., 2013).
Several studies have postulated the transport of nutrients (i.e. phosphorus, P, nitrogen, N and carbon, C) from plant to plant via CMNs in pot and field conditions (e.g., Simard and Durall, 2004; Jakobsen and Hammer, 2015; Wipf et al., 2019). For instance, P absorption by young maize plants was greater in unploughed than in ploughed soil, and this was also confirmed in pots with plants grown in soil cores collected from unploughed and ploughed plots (Read and Birch, 1988). One plausible explanation is that the established AM fungal network provides the way for P inflow to the young plants. In native grasslands, about 20% of the foliar 32P applied to Plantago erecta was transferred to the shoots of interconnected plants (Chiariello et al., 1982). In a microcosm, P concentration in seedlings of Bromus erectum and Brachipodium pinnatum was increased when plants were linked by a CMN (van der Heijden, 2004). Moreover, in a pot experiment with plants interconnected by a CMN, P was preferentially transported to established cucumber (Cucumis sativus) as compared with tomato (Solanum lycopersicum) seedlings (Merrild et al., 2013). Nitrogen transfer was also reported between two interconnected plants of Plantago lanceolata grown in pot conditions (Eissenstat, 1990). In pot studies, it was also shown that inorganic forms of N were transferred from N2‐fixing plants to non‐fixing plants interconnected by AM fungi (e.g., van Kessel et al., 1985; Bethlenfalvay et al., 1991; Frey and Schüepp, 1993; Martin et al., 1995; Johansen and Jensen, 1996). However, although in all cited experiments the transfer of nutrients was proven to occur between plants linked by AMF, it was not possible to firmly discriminate the direct transfer channelled by CMN from the one mediated by, e.g., soil diffusion and uptake by hyphae or roots. Thus, to better understand the importance of CMN in the transfer of nutrients, it is important to exclude or at least to minimize as much as possible indirect transfer caused by diffusion, by transport by unwanted microorganisms or by hyphae release and uptake by roots.
In the last decades, in vitro cultivation systems associating AMF with root organs (e.g., carrots – see review by Fortin et al., 2002) or whole plants (Lalaymia and Declerck, 2020) have been used to investigate transport [e.g., P, N, Caesium (Cs)] from a root‐free compartment to a root compartment or between CMN‐connected plants growing in physically separated compartments. Both systems are adequate to minimize indirect transfer of elements because of the absence of unwanted contaminants and the strict separation of the receiver roots or whole plant from the element to study. For instance, Nielsen and colleagues (2002) using root organ cultures (ROC) of carrot were able to demonstrate the uptake of P by the extraradical mycelium developing in a root‐free hyphal compartment (HC), its translocation to a root compartment (RC) and further transfer within the roots. The same in vitro system was used to investigate the transfer of C and N from HC to RC, but both elements remained in the colonized roots of the receiver plant (Pfeffer et al., 2004; Toussaint et al., 2004; Jin et al., 2005) due to the absence of a true sink, the shoot. Therefore, a system on whole plants was developed by Voets and colleagues (2005) to study the transport of elements from HC to roots and shoots in the RC. This system was used by Dupré de Boulois and colleagues (2006) to investigate the transport of P and Cs from HC to roots and shoots in the RC. For P, ~21% of the initial 33P supplied to the HC was transferred to the shoots, while for Cs, only 1.8% of the initial 134Cs supplied in the HCs was found in the shoots of Medicago truncatula, suggesting the major role of AMF in supplying P to plants, while for Cs, colonized roots were the preferred sink. The system of Voets and colleagues (2005) was further extended to two plants interconnected by a CMN (Voets et al., 2008) for studying plant‐to‐plant transfer of C (Voets et al., 2008) and Cs (Gyuricza et al., 2010). If C was transferred into the receiver roots, it was not transferred from roots to shoot, while for Cs, similar observations as in the study of Dupré de Boulois and colleagues (2006) were made with little accumulation of Cs in shoots. Importantly, in the study of Voets and colleagues (2008) an extremely low amount of C was detected in the non‐colonized control plants suggesting that a tiny amount of C transported by hyphae from RC to HC was released into the medium and further taken up by the non‐AM‐colonized plant. The values were, however, significantly lower than in the AM‐colonized plants suggesting an effective direct transfer between interconnected plants. More recently, the system of Voets and colleagues (2008) was extended to demonstrate the change of expression of defence genes in healthy plants connected to diseased plants (Alaux et al., 2020). Overall, these studies open huge avenues to unravel the full range of functions a web plays in plant‐to‐plant interactions and (agro)‐ecosystems functioning.
The role played by CMNs in interplant transfer of micronutrients, such as copper (Cu), iron (Fe) and zinc (Zn), has not been investigated yet, despite the demonstrated positive roles of AMF in micronutrient plant acquisition (Lehmann et al., 2014; Lehmann, 2015; Ercoli et al., 2017; Coccina et al., 2019; Pellegrino et al., 2020). Among micronutrients, Zn is essential for growth and health of plants and is involved in various physiological functions (Vallee and Auld, 1990; Sasaki et al., 1998; Broadley et al., 2007). Zinc deficiencies reduce plant growth and quality of food products, whereas Zn excess can be toxic in food and reduce plant growth. Thus, plants have developed complex homeostatic mechanisms that ensure optimal cellular Zn concentration when grown in soil with limiting or toxic levels of Zn (Grusak et al., 1999; Sinclair and Krämer, 2012). The active transport of Zn in plants has been largely studied and many membrane transporters belonging to several families have been identified (e.g., Sinclair and Krämer, 2012; Olsen and Palmgren, 2014). One of the most studied families involved in the transport of Zn into the cytoplasm is the Zinc‐Iron‐Regulated Transporter (ZRT‐IRT), called ZIP. The ZIP1, a vacuolar transporter, was found to be upregulated in roots and leaves of M. truncatula deficient in Zn (López‐Millán et al., 2004; Durmaz et al., 2011; Milner et al., 2013). The ZIP2, a plasma membrane transporter into the stele promoting Zn accumulation in the xylem parenchyma, was found to be upregulated in M. truncatula roots and stems, especially at high Zn concentrations (Burleigh et al., 2003). Moreover, other studies found that Zn chelators, such as the non‐proteinogenic amino acid nicotianamine (NA), enabled the intercellular and phloem mobility of Zn in plant (Curie et al., 2008; Deinlein et al., 2012). Recently, other ZIP genes, such as ZIP6, ZIP7 and ZIP14, were reported to be upregulated when plants of M. truncatula were inoculated with AMF under a gradient of Zn concentration in soil (Watts‐Williams et al., 2017, 2020). Specifically, ZIP6 was upregulated under low and medium concentration of Zn in soil (Watts‐Williams et al., 2017), whereas ZIP7 was highly upregulated irrespective to Zn concentration in soil (Watts‐Williams et al., 2020). Finally, ZIP14 was exclusively expressed in AMF‐colonized plants and upregulated irrespective to Zn concentration in soil. MtZIP14 was localized in AMF colonized root cells and specifically in the peri‐arbuscular membrane (PAM). By contrast, the mechanism of Zn homeostasis in AMF was less investigated and only one Zn transporter, belonging to the Cation Diffusion Facilitator (CDF) family, the GintZnT1, was identified in Rhizophagus irregularis (González‐Guerrero et al., 2005, 2016; Tamayo et al., 2014). The GintZnT1 is likely participating in vacuolar sequestration, since it can transport Zn2+ out of the cytosol (González‐Guerrero et al., 2005, 2009). Tamayo and colleagues (2014) identified by an in silico study other putative Zn transporters in R. irregularis.
In the present study, a CMN formed by the AM fungus R. irregularis MUCL 41833 was established in vitro between two M. truncatula plants, one donor plant (AM‐Donor, hereafter) fertilized with Zn on the leaves and one receiver plant (AM‐Receiver, hereafter). Both plants were grown on a Zn‐free medium in a bi‐compartmented system that physically separated the donor compartment (Donor‐C) from the receiver compartment (Receiver‐C; Fig. 1). Zinc availability in soil can be low and is always patchy, and under agricultural systems the homogeneity of application of foliar fertilizers can be variable, from low to medium, according to the application techniques (Ciccolini et al., 2017). Thus, the process of Zn transfer by CMN is ecologically important as it may improve the redistribution of Zn in agro‐ecosystems as well as in natural ecosystems. Therefore, we investigated whether the transfer of Zn was directly associated to the connecting hyphae and postulated that if the fungus transfer Zn from a leaf fertilized plant to an AM‐Receiver plant, a higher Zn concentration and variation in the expression of plant Zn‐transporter genes is observed compared with a non‐mycorrhizal control plant (NM‐Control; Hypothesis 1). The NM‐Control is a mycorrhizal defective mutant isogenic line of M. truncatula that was added to the Receiver‐C to ascertain that transfer was essentially attributable to direct mechanisms. Moreover, to minimize the role of indirect Zn transfer, Zn concentration in the media of Donor‐C and Receiver‐C was analysed. In addition, we hypothesized that after Zn application a larger part of Zn remains in the AM‐Donor, whereas the remaining part is transferred to the AM‐Receiver (Hypothesis 2). This will result in a significant higher Zn concentration and variation in plant gene expression profile in the AM‐Donor as compared with the AM‐Receiver. Moreover, to verify that Zn transfer by CMN does not occur in absence of Zn application, we set up the same in vitro system (Fig. 1), excluding Zn treatment to leaves of the AM‐Donor. In this no‐Zn system, we expected no difference in plant Zn concentration and gene expression profile neither between AM‐Receiver and NM‐Control or between AM‐Donor and AM‐Receiver (Hypothesis 3). Finally, to further support the evidence of Zn transfer by CMN from AM‐Donor to AM‐Receiver, comparisons of Zn‐fertilized and unfertilized plants were performed on plant Zn concentration and gene expression separately for AM‐Donor, AM‐Receiver and NM‐Control, whereas the same comparison was performed on fungal Zn‐transporter genes separately for AM‐Donor and AM‐Receiver.
Fig. 1.
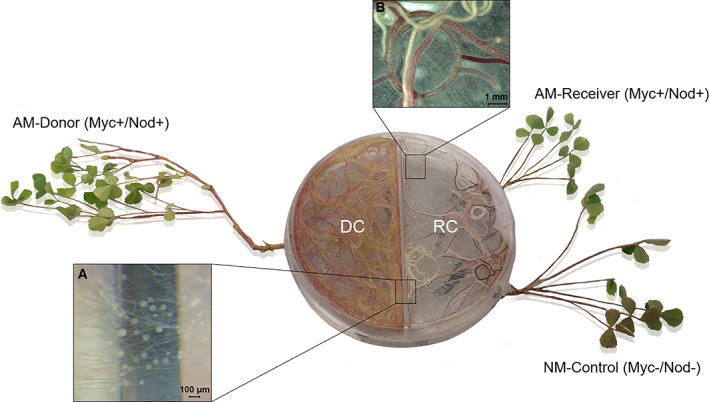
Bi‐compartmented autotrophic in vitro culture system with an arbuscular mycorrhizal (AM) donor plant (AM‐Donor), an AM receiver plant (AM‐Receiver) and non‐mycorrhizal mutant plant (NM‐Control) of Medicago truncatula (Voets et al., 2008). The genotype of the AM‐Donor and AM‐Receiver plants was the wild type J5 and the one of the NM‐Control plants was its isogenic mycorrhiza defective mutant TRV25. A five‐day old AM‐Donor seedling was inoculated with the AM fungus Rhizophagus irregularis (MUCL 41833) 2 weeks after the insertion into the donor compartment (DC). Eight weeks after inoculation, the AM‐Receiver and the NM‐Control seedlings were inserted in the receiver compartment (Receiver‐C). In the Figure, details of the common mycorrhizal network are shown: AM fungal hyphae crossing the partitioning wall (A) and extraradical AM fungal mycelium with roots of the AM‐Receiver plants (B)
In addition, we evaluated the adequacy of the CMN in vitro cultivation system for the plant‐to‐plant Zn transfer study, verifying plant and fungus growth under Zn and no‐Zn application. We expected that, according to differences in plant age (due to the sequence of the operations of the experimental set‐up), the AM‐Donor was larger than the AM‐Receiver, and that the AM‐Receiver grew similarly to the NM‐Control. Moreover, on the basis of literature data on the AM fungus growth in in vitro system, we expected that in an adequate system the fungus extensively grew (e.g., larger mycelium) and produced more spores in Receiver‐C than in the Donor‐C. Finally, to further support the adequacy of the system excluding any toxic effect of Zn, comparisons of plant growth traits under +Zn and −Zn treatments were performed separately for AM‐Donor, AM‐Receiver and NM‐Control, and comparisons of fungal growth traits were performed separately for Donor‐C and Receiver‐C.
Results
Adequacy of the CMN in vitro cultivation system for plant‐to‐plant Zn transfer study
Shoot and root dry weights (SDW and RDW) of AM‐Donor were threefold to fourfold higher as compared with AM‐Receiver in −Zn treatment as well as +Zn treatment (Table S1). By contrast, SDW and RDW of AM‐Receiver and NM‐Control were similar under both +Zn and –Zn treatments. Similarly, stem length and number of leaves were significantly higher in AM‐Donor as compared with AM‐Receiver under both +Zn and −Zn treatments (on average +102% and +157% respectively), and did not vary between AM‐Receiver and NM‐Control. Root length of AM‐Donor was six‐fold higher as compared with AM‐Receiver under both +Zn and −Zn treatments (Table 1). By contrast, root length of AM‐Receiver was similar to NM‐Control under both +Zn and −Zn treatments.
Table 1.
Root length, arbuscular mycorrhizal (AM) fungal root colonization and length of colonized roots of wild type Medicago truncatula donor (AM‐Donor), receiver (AM‐Receiver) and AM‐defective mutant control (NM‐Control) plants, 5 days after application of zinc (0.1 mg plant−1; +Zn) on the leaves of the AM‐Donor plants or in absence of Zn (−Zn). The AM‐Donor and AM‐Receiver plants were linked by the extraradical mycelium of the AM fungus Rhizophagus irregularis (MUCL 41833).
| Plant a | Root length | AM fungal colonization | AM fungal colonized root length |
|---|---|---|---|
| cm | % | cm | |
| AM‐Donor +Zn | 719.4 ± 52.6 | 52.4 ± 2.1 | 372.7 ± 19.1 |
| AM‐Receiver +Zn | 130.3 ± 19.3 | 46.4 ± 1.8 | 59.3 ± 6.6 |
| NM‐Control +Zn | 104.1 ± 18.6 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| AM‐Donor −Zn | 720.7 ± 33.6 | 45.6 ± 3.0 | 325.9 ± 15.9 |
| AM‐Receiver −Zn | 110.7 ± 22.5 | 48.0 ± 2.5 | 54.2 ± 12.9 |
| NM‐Control −Zn | 106.0 ± 17.9 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| Treatments compared (P‐values of linear orthogonal contrasts) | |||
| AM‐Donor versus AM‐Receiver +Zn | <0.001b | 0.020 | <0.001 |
| AM‐Receiver versus NM‐Control +Zn | 0.597 | <0.001 | 0.004 |
| AM‐Donor versus AM‐Receiver −Zn | 0.001 | 0.243 | <0.001 |
| AM‐Receiver versus NM‐Control −Zn | 0.706 | 0.001 | 0.007 |
AM‐Donor: Medicago truncatula Gaertn., cv. ‘Jemalong’ wild type (line J5; Myc+/Nod+); AM‐Receiver: M. truncatula Gaertn., cv. ‘Jemalong’ wild type (line J5; Myc+/Nod+); NM‐Control: M. truncatula Gaertn., cv. ‘Jemalong’ mutant line TRV 25 (Myc−/Nod−). One‐way analysis of variance (ANOVA) was performed to test the effect of plant type and orthogonal contrasts were used to discriminate the differences between AM‐Donor and AM‐Receiver plants and between AM‐Receiver and NM‐Control plants. Values are means ± standard error of five replicates.
b In bold statistically significant values at P ≤ 0.05.
Under Zn application, AM fungal root colonization percentage was significantly higher in AM‐Donor as compared with AM‐Receiver. By contrast, under −Zn treatment, AM fungal root colonization was similar in AM‐Donor and AM‐Receiver. Moreover, under +Zn and −Zn treatments, the AM fungal colonized root length was on average sixfold higher in AM‐Donor as compared with AM‐Receiver. As expected, NM‐Control (i.e., the mycorrhizal defective mutant isogenic line TRV25) did not show any trace of mycorrhizal colonization. The number of spores was significantly higher in the Receiver‐C as compared with the Donor‐C under both +Zn and −Zn treatments (Table 2). Similarly, hyphal length per root length and hyphal length per AM fungal colonized root length were significantly higher in the Receiver‐C as compared with the Donor‐C irrespectively of Zn application. By contrast, total hyphal length and hyphal density were similar in the Donor‐C and the Receiver‐C under both Zn treatments.
Table 2.
Hyphal length, number of spores, hyphal length per root length, hyphal length per arbuscular mycorrhizal (AM) fungal root length and hyphal density in the donor and receiver compartments (Donor‐C and Receiver‐C respectively), 5 days after application of zinc (0.1 mg plant−1; +Zn) on the leaves of the AM‐Donor plants or in absence of Zn (−Zn). The AM‐Donor and AM‐Receiver plants were linked by the extraradical mycelium of the AM fungus Rhizophagus irregularis (MUCL 41833).
| Compartment a | Hyphal length | N° spores | Hyphal length per root length | Hyphal length per AM fungal root length | Hyphal density |
|---|---|---|---|---|---|
| cm | N | cm cm−1 | cm cm−1 | cm cm−3 | |
| Donor‐C +Zn | 1560 ± 102 | 3462 ± 466 ab | 2.19 ± 0.18 a | 4.18 ± 0.22 a | 98.1 ± 6.4 |
| Receiver‐C +Zn | 2047 ± 267 | 9532 ± 1068 b | 17.8 ± 4.3 b | 37.5 ± 7.9 b | 128.7 ± 16.8 |
| Donor‐C −Zn | 1621 ± 297 | 3897 ± 504 a | 2.3 ± 0.5 a | 5.0 ± 1.0 a | 102.0 ± 18.7 |
| Receiver‐C −Zn | 2287 ± 373 | 8048 ± 1144 b | 22.7 ± 3.8 b | 47.8 ± 8.5 b | 143.8 ± 23.4 |
In Donor‐C the Medicago truncatula Gaertn., cv. ‘Jemalong’ wild type (line J5; Myc+/Nod+) was grown. In Receiver‐C the M. truncatula Gaertn. cv. ‘Jemalong’ wild type (line J5; Myc+/Nod+) and the mutant M. truncatula Gaertn., cv. ‘Jemalong’ mutant line TRV 25 (Myc−/Nod−), were grown. For each Zn treatment, t‐test was performed to test the effect of the compartment on the AM fungal traits. Values are means ± standard error of five replicates.
b Values followed by different letters are significantly different at P ≤ 0.05, according the post‐hoc Tuckey‐B test.
Overall, the application of Zn did not modify the phenotype of the plants as evidenced by t‐test conducted on all plant traits for each plant type (AM‐Donor, AM‐Receiver and NM‐Control (Table S2). Similarly, the application of Zn did not impact fungal parameters as evidenced by t‐test (Tables S2 and S3). Finally, the number of active hyphae crossing the partition wall separating Donor‐C from Receiver‐C did not significantly differ between +Zn and −Zn treatments (306.6 ± 42.2 and 244.4 ± 38.4 respectively).
Shoot and root Zn concentrations in plants linked by CMN
Shoot Zn concentration of AM‐Receiver was significantly higher (by 21%) than that of NM‐Control (Fig. 2; Table S4). Moreover, shoot Zn concentration of AM‐Donor was 25‐fold higher as compared with the AM‐Receiver under +Zn treatment. By contrast, under −Zn treatment, no significant differences were noticed in the shoot Zn concentration of AM‐Donor, AM‐Receiver and NM‐Control.
Fig. 2.
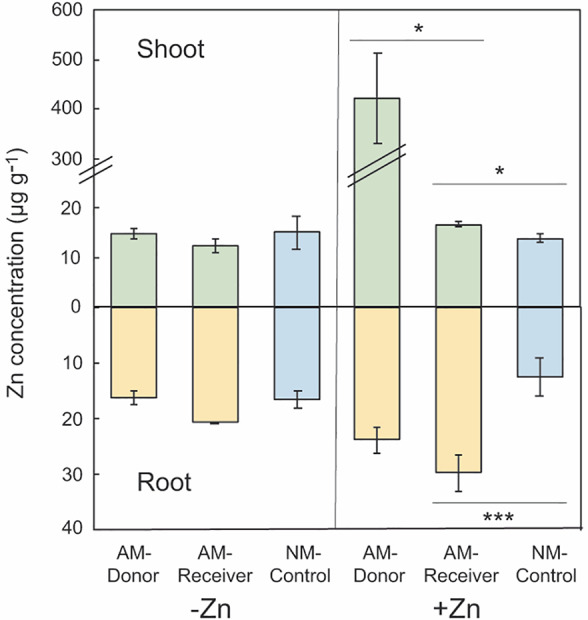
Zinc concentrations (μg g−1) in shoots and roots of 16‐weeks‐old wild type Medicago truncatula donor plants (AM‐Donor; Myc+/Nod+), six‐weeks‐old wild type receiver M. truncatula plants (AM‐Receiver; Myc+/Nod+) and its isogenic mycorrhiza defective mutant control plants (NM‐Control; Myc‐/Nod‐), 5 days after zinc (Zn) application of 0 and 0.1 mg plant−1 (−Zn and + Zn respectively) on the leaves of the AM‐Donor plants. The AM‐Donor and AM‐Receiver plants were linked by the extraradical mycelium of the arbuscular mycorrhizal (AM) fungus Rhizophagus irregularis (MUCL 41833). Values are means ± standard error of three replicates (n = 3). One‐way analysis of variance (ANOVA) was performed to test the effect of plant type and orthogonal contrasts were used to discriminate the differences between AM‐Donor and AM‐Receiver plants and between AM‐Receiver and NM‐Control plants. Different symbols indicate significant differences between plants (*: 0.01 < P ≤ 0.05; **: 0.001 ≤ P ≤ 0.01; ***: P < 0.001).
Under Zn application, root Zn concentration of AM‐Receiver was significantly higher (139%) than that of NM‐Control, while root Zn concentration of AM‐Donor was similar to that of AM‐Receiver (on average 26.8 μg g−1; Fig. 2; Table S4). By contrast, under −Zn treatment no significant differences were noticed among AM‐Donor, AM‐Receiver and NM‐Control.
Shoot Zn concentration was about 28‐fold higher in AM‐Donor in the +Zn treatment as compared with the AM‐Donor in the −Zn treatment, while root Zn concentration was 47% higher (Table S2). Shoot Zn concentration was 35% higher in AM‐Receiver in the +Zn treatment as compared with AM‐Receiver in the −Zn treatment, while root Zn concentration was about 44% higher. By contrast, Zn concentration of shoot and root of NM‐Control was not significantly modified by Zn treatment.
Plant and fungal gene expressions in plants linked by CMN
A schematic representation of the Zn transport‐related proteins for uptake, sequestration, and redistribution of Zn after foliar application to the AM‐Donor linked by the CMN to the AM‐Receiver is reported in Fig. 3. Under Zn application, the gene MtZIP1 was significantly downregulated in AM‐Donor shoots and roots (−78% and −73% respectively) as compared with the AM‐Receiver, which showed expression patterns similar to NM‐Control (Fig. 4A; Table S5). The gene MtZIP2 was not expressed in the shoots of the plants of M. truncatula under both +Zn and −Zn treatments (Table S5). Nevertheless, when Zn was applied to the leaves of AM‐Donor, the gene MtZIP2 was similarly expressed in the roots of AM‐Donor and AM‐Receiver, and MtZIP2 gene expression was about threefold higher in AM‐Receiver than NM‐Control (Fig. 4C; Table S5). Moreover, the gene MtNAS1 was significantly upregulated in the shoots of AM‐Donor (about threefolds) as compared with the AM‐Receiver that also showed a significant upregulation (about 18 folds) as compared with NM‐Control (Fig. 4B; Table S5). However, in roots, the gene MtNAS1 was similarly upregulated in AM‐Donor and AM‐Receiver and MtNAS1 gene expression was about ninefold higher in AM‐Receiver than in NM‐Control. By contrast, under −Zn treatment, the expression of MtZIP1, MtZIP2 and MtNAS1 in shoots and roots did not vary between AM‐Donor and AM‐Receiver and neither between AM‐Receivers and NM‐Control (Fig. 4; Table S5).
Fig. 3.
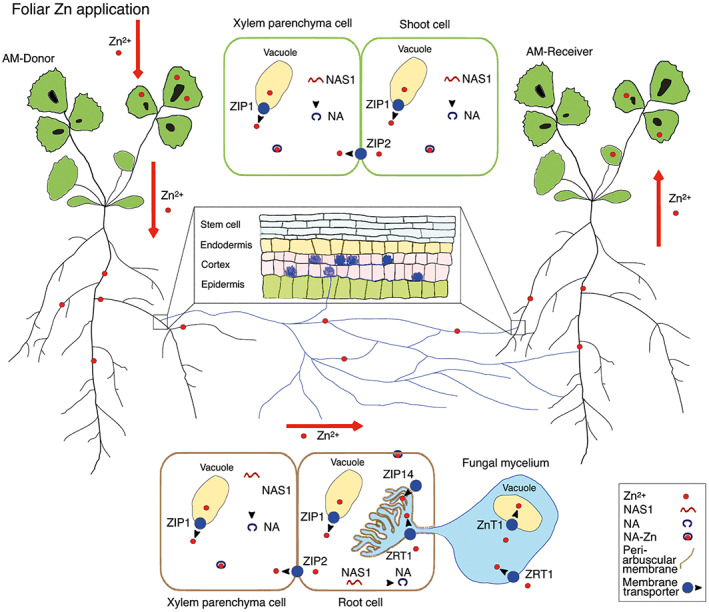
Schematic representation of the studied Zn transport‐related proteins for uptake, sequestration, and redistribution of Zn after foliar application to the arbuscular mycorrhizal (AM) Medicago truncatula donor plant (AM‐Donor; Myc+/Nod+) linked by the common mycorrhizal network to the M. trucatula AM‐Receiver plant (Myc+/Nod+). A putative transport protein (ZIP1) involved in the vacuolar transport of Zn into the cytoplasm (López‐Millán et al., 2004), a putative transport protein (ZIP2) involved in the cellular Zn influx into the xylem parenchyma cells (Burleigh et al., 2003) and an enzyme synthesizing nicotianamine (NAS1; Deinlein et al., 2012) are indicated in the scheme within the cells of shoots and roots. The nicotianamine (NA) and the ligand complex Zn‐nicotianamine (Zn‐NA) are also indicated. The RiZnT1 protein likely participating in Zn vacuolar sequestration (González‐Guerrero et al., 2005), the putative transporter RiZRT1 likely participating in the influx of Zn through the fungal plasma membrane (Tamayo et al., 2014) and the putative Zn transporter ZIP14 localized in AMF colonized root cells and specifically in the peri‐arbuscular membrane (Watts‐Williams et al., 2020) are indicated in the scheme within the cells of the colonized roots.
Fig. 4.
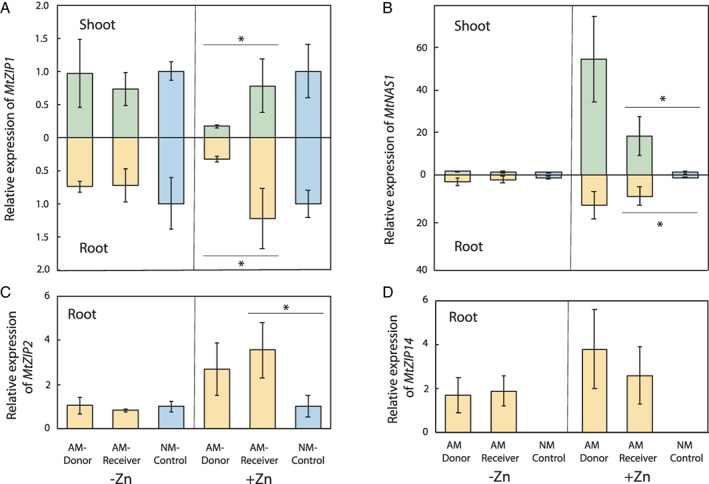
Relative expression of the Zinc‐Iron‐Regulated Transporter ZIP genes, MtZIP1 in shoots and roots (A), MtZIP2 and MtZIP14 in roots (C,D), and of the nicotianamine synthase gene (MtNAS1) (B) of 16‐weeks‐old wild type Medicago truncatula donor plants (AM‐Donor; Myc+/Nod+), six‐weeks‐old wild type receiver M. truncatula plants (AM‐Receiver; Myc+/Nod+) and its isogenic mycorrhiza defective mutant control plants (NM‐Control; Myc−/Nod−), 5 days after zinc (Zn) application of 0 and 0.1 mg plant−1 (−Zn and +Zn respectively) on the leaves of the AM‐Donor plants. The AM‐Donor and AM‐Receiver plants were linked by the extraradical mycelium of the arbuscular mycorrhizal (AM) fungus Rhizophagus irregularis (MUCL 41833). The relative expression analysis of the genes was done by the double standardization method that requires the reference genes and the control treatment (Livak and Schmittgen, 2001; Vandesompele et al., 2002). The transcript levels of actin‐101 (MtACT‐101) and elongation factor 1‐α (MtEF1‐α) were used as reference (Nicot et al., 2005) and the transcript level of the NM‐Control plants was used as control. Values are means ± standard error of five replicates (n = 5). One‐way analysis of variance was performed to test the effect of plant type and orthogonal contrasts were used to discriminate the differences between AM‐Donor and AM‐Receiver plants and between AM‐Receiver and NM‐Control plants. Different symbols indicate significant differences between plants (*: 0.01 < P ≤ 0.05; **: 0.001 ≤ P ≤ 0.01; ***: P < 0.001).
Under Zn foliar application, the AM‐responsive MtZIP14 gene, whose protein was specifically localized in the peri‐arbuscular membrane, was similarly expressed in the AM‐Donor and AM‐Receiver, whereas it was not expressed in NM‐Control (Fig. 4D; Table S5). Moreover, under −Zn treatment, MtZIP14 was upregulated in both AM‐Donor and AM‐Receiver.
Under Zn application, the genes RiZnT1 and RiZRT1 were both expressed in the roots of AM‐Donor and AM‐Receiver (Fig. 5; Table S5). As expected, under Zn application, RiZnT1 and RiZRT1 genes were not expressed in NM‐Control. Conversely, under −Zn treatment, the genes RiZnT1 and RiZRT1 were not expressed in AM‐Donor, very little expressed in AM‐Receiver and not expressed in NM‐Control (Fig. 5; Table S5). The transcript levels of the Ri28S were similar among the roots of AM‐Donor and AM‐Receiver in the +Zn and −Zn treatments (Fig. S1).
Fig. 5.
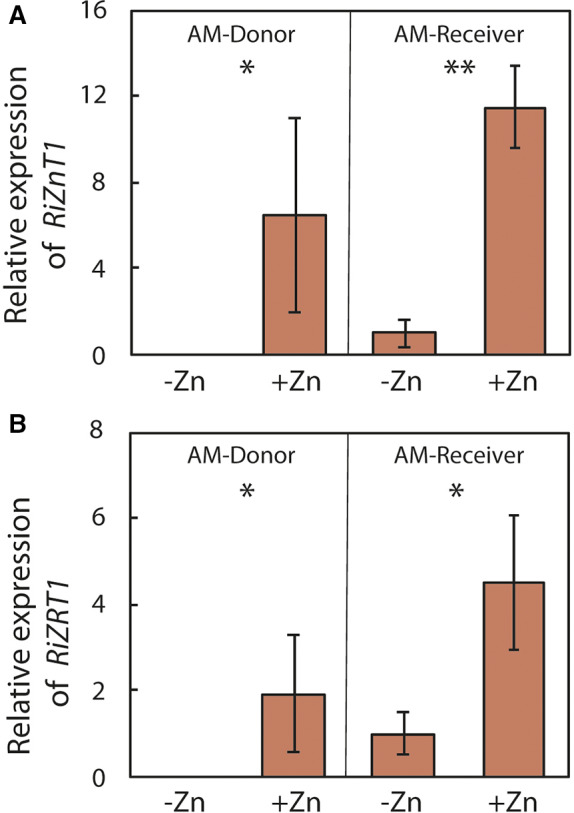
Relative expression of the Rhizophagus irregularis RiZnT1 (A) and RiZRT1 (B) genes in roots of 16‐weeks‐old wild type Medicago truncatula donor plants (AM‐Donor; Myc+/Nod+), and 6‐weeks‐old wild type receiver M. truncatula plants (AM‐Receiver; Myc+/Nod+), 5 days after zinc (Zn) application of 0 and 0.1 mg plant−1 (−Zn and +Zn respectively) on the leaves of the AM‐Donor plants. The AM‐Donor and AM‐Receiver plants were linked by the extraradical mycelium of the arbuscular mycorrhizal (AM) fungus Rhizophagus irregularis (MUCL 41833). The relative expression analysis of the genes was done by the double standardization method that requires the reference genes and the control (−Zn) (Livak and Schmittgen, 2001; Vandesompele et al., 2002). The transcript level of the 28S ribosomal subunit (Ri28S) was used as reference (Nicot et al., 2005) and the transcript level of the AM‐Receiver roots in the −Zn treatment was used as control. Values are means ± standard error of five replicates (n = 5). Pairwise comparisons of Zn‐fertilized and unfertilized plants were performed by t‐test separately for AM‐Donor and AM‐receiver plants. Different symbols indicate significant differences between plants (*: 0.01 < P ≤ 0.05; **: 0.001 ≤ P ≤ 0.01; ***: P < 0.001).
The evidence of Zn transfer from donor to AMF‐receiver plants was further supported by the differences in plant gene expression between +Zn and −Zn treatments in AM‐Donor and AMF‐Receiver (Table S6). Zinc transfer was additionally supported by the differences on fungal gene expression between +Zn and −Zn treatments in AM‐Donor versus AMF‐Receiver.
Discussion
It is only in the recent years that the role of CMNs in the transfer of macronutrients (e.g., N and P) between plants has been shown with AMF (e.g., He et al., 2003; Mikkelsen et al., 2008), while it was not studied for micronutrients, such as Zn. Controversy about whether transfer is direct through CMNs or partly indirect through soil may arise in experiments conducted under pot conditions (He et al., 2003; Bücking et al., 2016). Indeed, the direct contribution of connecting hyphae can only be ascertained in a context where indirect effects, attributed to diffusion in soil, hyphal exudation and decay, impact of microbial communities or biofilm formations, can be excluded. In the present study, a partition wall, physically separating the two compartments of the Petri plate, allowed excluding the diffusion pathway of Zn from the Donor‐C to the Receiver‐C and subsequent uptake by the roots of the receiver plants. Moreover, the introduction of the NM‐Control plant in the Receiver‐C allowed a precise control of the Zn released into the growth medium due to hyphal exudation and decay. The transfer of Zn from AM‐Donor to AM‐receiver plants via the CMN was supported by the significant increase in Zn concentration of the shoots of the AM‐Receiver plants and by the changes in the expression of the fungal genes RiZRT1 and RiZnT1, as well as of MtZIP2, MtZIP14 and MtNAS1 in the roots and of MtNAS1 in the shoots of M. truncatula. However, only the use of radioactive Zn isotopes as tracers would have provided conclusive evidence of the direct Zn transfer between plants connected by CMN (Zhang et al., 2019).
Adequacy of the CMN in vitro cultivation system for Zn plant‐to‐plant transfer studies
In the present study, an in vitro culture system in which two M. truncatula plants were connected by a CMN, was used to demonstrate the plant‐to‐plant transfer of Zn. A Myc− isogenic plant was included in the Receiver‐C as control. Several fungal and plant parameters were first evaluated to ascertain the adequacy of the system. A high number of active hyphae (c. 276 averaged over both −Zn and +Zn treatments) crossed the partition wall separating the Donor‐C from the Receiver‐C. Mycelium developed abundantly in both compartments with hyphal length and density averaging 1878 cm and 118 cm cm−3, irrespective of Zn treatment. These values were almost identical to those reported by Voets and colleagues (2008) and Dupré de Boulois and colleagues (2006) in similar in vitro systems after respectively 10 and 12 weeks of growth. The total number of spores was much higher in the Receiver‐C as compared with the Donor‐C (8790 vs 3679 spores) under both +Zn and −Zn treatments, and these values were similar to those previously found by Voets and colleagues (2008) (6970 vs 2791 spores). These results support the fact that spore production is dependent on extraradical mycelium biomass and on the re‐allocation of resources from intraradical to extraradical hyphal mycelium. Indeed, the hyphal length and hyphal density was higher in the Receiver‐C than in the Donor‐C at both Zn treatments, although no significant differences were detected possibly due to the variability among replicates. Irrespective of Zn treatment, hyphal length per root length and hyphal length per AM fungal colonized root length were much higher in the Receiver‐C than in the Donor‐C. These differences were mainly due the differential root growth in the two compartments, according to the age of the plants and this might be the reason of the differential number of spores in the Donor‐C and Receiver‐C. AM fungal root colonization was confirmed in AM‐Donor and AM‐Receiver plants, both by the classical method of microscope counting and by the quantitative assessment using qPCR methodology. Whatever the treatment, plant root colonization of AM‐Donor and AM‐Receiver was high (in the range 45.6%–52.4%), while it was totally absent in the NM‐Control. While no difference was highlighted in percentage of colonization between AM‐Donor and AM‐Receiver under −Zn, significant difference was detected under +Zn (53% vs 46%) using microscopy. However, this was not confirmed by Ri28S expression that showed a high degree of correlation with the total root colonization quantified by microscopy in all plants, except for the AM‐Receivers under +Zn. It is thus probable that molecular determination is more reliable since less affected by the operator's measurement (Alkan et al., 2004), and consequently AM fungal colonization can be considered similar in all compartments and not affected by Zn treatment. Moreover, according to Parniske (2008), the functionality of the symbiosis was confirmed by the visualization of the arbuscules (data not shown) within the cells of the AM‐Donor and AM‐Receiver roots in both compartments.
No diffusion of medium from Donor‐C to Receiver‐C or root development in the Receiver‐C, and no microbial contamination were noticed in both compartments. Regarding plant growth parameters, Zn limitation was not detrimental to plant growth, as shown by the lack of variation in plant biomass and root length between the plants in the +Zn and –Zn treatments. There were age‐related size differences between the AM‐Donor and the AM‐Receiver, showing that the plants were growing properly, and no differences between the AM‐Receiver and NM‐Control, showing their growth balance.
Zn is transferred from a donor plant to the root and shoot of a receiver plant linked by the CMN
Zinc transfer was demonstrated from donor to receiver plants as earlier demonstrated for 13C from shoot of a donor plant to roots of a receiver plant (Voets et al., 2008) and for radioceasium from shoot of a donor plant to shoot of a receiver plant (Gyuricza et al., 2010) and very recently for warning signals in healthy plants connected to plants attacked by a fungal pathogen (Alaux et al., 2020), extending the transport role of CMNs to semiochemicals. Although in our system indubitable exclusion of the indirect transfer of Zn could not be ascertained, the very low amount of Zn in the Donor‐C medium (about 1 μg g−1 d.w.; data not shown) suggests that if an indirect transfer occur, it would account for negligible quantities. In addition, the very low amount of Zn in NM‐Control was due to the Zn released by hyphal exudation and decay in the Receiver‐C, because no root colonization by the AM fungus was noticed. The Zn released in the receiver growth medium was confirmed by the analysis of the Zn concentration in the medium (about 1 μg g−1 d.w.; data not shown). Quantitatively, the efficiency of Zn application was 19.8%, considering the ratio between the total Zn content in AM‐Donor, AM‐Receiver and media and the amount of Zn applied to the leaves of the AM‐Donor. Thus, about 80% of the applied Zn was not absorbed by leaves and was not released into the MSR medium (see the low amount found in the media of both compartments). However, Zn adhering to the leaf surface was carefully removed before the analytical determination, according to the procedure of sampling preparation (Yilmaz et al., 2017). The total Zn content of the system (19.8 μg Zn plant−1) was partitioned as follows: 89.9% and 8.5% into the shoots and roots of the AM‐Donor, and 1.2% and 0.3% into the roots and shoots of the AM‐Receiver. Thus, considering the percentage of Zn presents in the roots of the donor plant (i.e., 8.5%) and accessible to the AM fungus, 12.2% and 3.4% was transferred to the roots and shoots of the AM‐Receiver respectively, whereas 84.4% remained in the roots of the AM‐Donor. The Zn transfer from roots to shoots in the AM‐Receiver was expected, because it was demonstrated under a same system that P, used as tracer for studying the transport from HC to RC, is transferred from AM‐colonized receiver roots to shoots in a proportion of 20.7% of the initial P supplied in the HC (Dupré de Boulois et al., 2006). Compared with Zn, the larger proportion of P transferred to shoots might be determined by the major role played by P for plant physiology.
Although the proportion of Zn transferred by the CMN from the AM‐Donor to the roots and shoots of the AM‐Receiver was low, the transfer ensures a Zn concentration in the receiver plant tissues above the critical threshold for maintaining cell structure and function (15 μg Zn g−1 dry weight, according to Broadley et al., 2012). Thus, in plant communities or in agricultural systems, the CMN may modulate the transfer and allocation of Zn among plants.
Transcriptional pattern of Zn transport‐related processes in a receiver plant linked by the CMN after Zn application to a donor plant
In the AM‐Receiver an upregulation of the MtZIP2 gene was found in roots as compared with the NM‐Control only in the +Zn treatment. Similarly, after foliar application of Zn, MsZIP2 was significantly upregulated in roots and shoots of Medicago sativa (Cardini et al., 2021). The change of expression found in our +Zn treatment can be interpreted as an indication of Zn storage in the xylem parenchymal cells of the AM‐Receiver roots after transfer of Zn. Indeed, in A. thaliana, the expression of ZIP2 was localized in the root stele, supporting the role of ZIP2 protein in metal transport into the stele that presumably promotes the accumulation of Zn in the xylem parenchyma (Milner et al., 2013). Moreover, our results on the upregulation of MtZIP2 in AM‐Receiver in comparison with NM‐Control cannot be interpreted as an indirect effect of being colonized by AMF, since this gene was significantly affected by Zn and not affected by AMF or the interaction between AMF and Zn (Watts‐Williams et al., 2017). These results are also supported by recent evidences that MtZIP2 is similarly highly upregulated in inoculated and not‐inoculated M. truncatula plants at both low and high Zn soil availabilities (Watts‐Williams et al., 2020). The undetectable MtZIP2 expression in the shoots of AM‐Donor, AM‐Receiver and NM‐Control under +Zn and −Zn treatments, is supported by the pathway of this gene in M. sativa and M. truncatula after Zn application that showed a stronger expression in roots than in shoots/stems and no expression in leaves (Burleigh et al., 2003; Cardini et al., 2021).
As regard MtZIP1, it was only downregulated in the shoots and roots of the AM‐Donor as compared with AM‐Receiver under Zn application, suggesting the success of the Zn application and translocation within the plant. This response is consistent with the expression of ZIP1 when plants are in Zn deficient conditions (Eidie et al., 1996; Ramesh et al., 2003; Ishimaru et al., 2006). Moreover, since the ZIP proteins can transport across membranes not only Zn2+, but also other metals, including Cd2+, Fe3+/Fe2+ , Mn2+, Ni2+, Co2+ and Cu2+ (Grotz et al., 1998; Eckhardt et al., 2001; Mäser et al., 2001), we can hypothesize that CMN can also be involved in the transport of other metals between plants.
Another indication of Zn transfer between plants is represented by the MtNAS1 transcriptional response in roots and shoots of the AM‐Receiver. In the roots and shoots of these plants the upregulation of MtNAS1 as compared with the NM‐Control in the +Zn treatment can support the increased transport of Zn in the form of the ligand complex zinc‐nicotianamine (Zn‐NA). Indeed, NA is considered a fundamental chelator for Zn sequestration in vacuoles and Zn redistribution within the plant (Deinlein et al., 2012). Since the nicotianamine level greatly correlates with NAS transcripts (Talke et al., 2006; Haydon et al., 2012), the NAS expression found in our system can be considered a reliable indicator of NA content and Zn chelation. Moreover, under Zn foliar application, MtNAS1 was upregulated in AM‐Donor shoots and roots. This is similar to the expression observed for NAS1 and NAS2 in M. sativa and Triticum durum plants after Zn foliar application (Deshpande et al., 2018; Cardini et al., 2021). This confirms the strategy of the plants to chelate Zn in the shoot tissues and then to load it into the phloem, enabling its redistribution to roots.
Here, the fact that MtZIP14 gene was similarly expressed in both AM‐Donor and AM Receiver under +Zn and −Zn treatments cannot be explained by the results of Watts‐Williams and colleagues (2020) reporting that MtZIP14 is a membrane protein able to facilitate Zn transport into the root cells. Indeed, given the fact that localization and function of this protein would not change at low and high Zn availabilities, we expected an opposite pattern of the MtZIP14 gene expression in the two compartments. Although in the AM‐Receiver the behaviour of the gene supports Zn transfer from the fungus to the plant, the inconsistency observed in the AM‐Donor plants cannot be explained by the Zn uptake from the donor medium according to recorded low concentration of Zn.
The RiZnT1, belonging to the CDF family, follows this pattern after foliar Zn application: upregulation in the roots of the AM‐Donor and AM‐Receiver and as expected no transcript detection in the roots of the NM‐Control. The transcriptional level of RiZnT1 in the structures of R. irregularis within the roots of the AM‐Receiver supports the increased concentration of Zn in roots mediated by the CMN. The model for metal delivery by AMF to the host plants supports the localization of RiZnT1 in the vacuolar compartments of the extraradical mycelium (González‐Guerrero et al., 2008). However, in our study, the fact that RiZnT1 was expressed in the roots of both AM‐Donor and Receiver under +Zn can support a role of this gene in the vacuolar Zn sequestration also in the intraradical mycelium with vacuoles acting as carrier within the AM‐Donor roots and a possible different role in the intraradical mycelium within the roots of the AM‐Receiver (Ashford, 2002; Javot et al., 2007; González‐Guerrero et al., 2008). The RiZnT1 can play a functional role in addition to other specific transporters identified in the arbuscules (González‐Guerrero et al., 2008), and might have a dual function depending on Zn availability (e.g. change in localization depending on Zn availability as suggested for some yeast Zn transporters and for the ZIP family Zn transporter SIZRT2 of the ectomycorrhizal fungus Suillus luteus; Coninx et al., 2019).
Similarly, under Zn application, the RiZRT1, belonging to the ZIP family, was upregulated in the roots of the AM‐Donor and AM‐Receiver. Thus, in agreement with the results on the expression of RiZnT1, we hypothesize a dual function of the RiZRT Zn transporter. This trait would allow to support the role played by the AM fungus in the trafficking of Zn among plants, and encourage an in‐depth functional characterization of the studied Rhizophagus Zn transporters to confirm our hypothesis.
Finally, the fact that both AM fungal Zn genes are not expressed in the NM‐control under both −Zn and +Zn applications additionally demonstrated that the AM fungus did not colonize the symbiosis‐defective mutant line. Moreover, the different expression profile of the fungal Zn genes in −Zn Donor and Receiver plants is likely to be due to an early fungal response to Zn limiting conditions (i.e., in the Receiver plants), while this response is not detected anymore with plant ageing.
Conclusions
The present work is the first that provides strong evidence for a direct transfer of Zn between two plants only connected by a CMN. It further demonstrates a triggering of the transcription of fungal and plant genes involved in Zn transport‐related processes. Thus, this study increases the range of functions undertaken by CMNs. The in vitro cultivation system allows to study plant‐to‐plant Zn transfer in a context free of any undesirable microorganisms or other unwanted factors (e.g., soil characteristics). However, further studies are needed under pot culture conditions with plants growing in separate compartments only connected by CMN to confirm the results and ascertain the potential applicability of findings in agricultural systems for improving the redistribution of Zn among plants.
Experimental procedures
Biological material
Medicago truncatula Gaertn., cv. ‘Jemalong’ (SARDI, Australia) wild type (line J5; Myc+/Nod+) was used as Zn‐donor plant (AM‐Donor, hereafter) and Zn‐receiver plant (AM‐Receiver, hereafter). Its symbiosis‐defective mutant, line TRV25 (Myc−/Nod−) (Sagan et al., 1995, 1998; Morandi et al., 2005) was used as control plant (NM‐Control, hereafter). More details about this symbiosis‐defective mutant are given in the Method S1. The AM fungus Rhizophagus irregularis (Błaszk., Wubet, Renker, and Buscot) C. Walker and A. Schüßler (2010) strain MUCL 41833, was provided by the Glomeromycota in vitro collection GINCO (http://www.mycorrhiza.be/ginco-bel) on Ri T‐DNA transformed chicory (Cichorium intybus L.) roots and further sub‐cultured following Cranenbrouck and colleagues (2005).
Seeds of M. truncatula were surface‐sterilized and placed in Petri plates (92 mm diameter, 10 seeds per plate) on the Modifed Strullu–Romand (MSR) medium (Declerck et al., 1998), lacking sucrose, vitamins and Zn (MSR −Zn), and solidified with 3 g L−1 of Gellan Gum (Alfa Aesar, Karlsruhe, Germany) (thickness of 0.5 cm). The Petri plates were incubated in the dark at 27°C.
Experimental set‐up
The experimental system consisted in bi‐compartmented Petri plates, with a donor (Donor‐C) and receiver (Receiver‐C) compartment, each containing one autotrophic M. truncatula plant (J5 line) linked by a CMN and, for the Receiver‐C only, a mutant plant (TRV25 line) used as control (see Voets et al., 2008). In the Donor‐C, a hole was made to insert the AM‐Donor J5 plant, while in the Receiver‐C two holes were made to insert the AM‐Receiver J5 and the NM‐Control TRV25 plants (Fig. 4). The Donor‐C and the Receiver‐C were filled with 30 ml of MSR ‐Zn medium.
In each Petri plate, a 5‐day old M. truncatula seedling was inserted into the Donor‐C with the roots plated on the medium and shoot extending outside the plate via the hole. The roots were inoculated after 2 weeks with about 100 spores of R. irregularis following Cranenbrouck and colleagues (2005). Details about how the plants were inoculated and the growth conditions are given in Methods S2. Three weeks after inoculation, 10 ml of sterilized MSR −Zn medium was added in the Donor‐C. This addition was repeated every 2 weeks throughout the experiment. Roots were regularly trimmed to avoid crossing the partition wall. Eight weeks after inoculation, a profuse mycelium was established in the Donor‐C that crossed the partition wall and developed abundantly in the Receiver‐C. At that time, the AM‐Receiver and the NM‐Control plants (5‐day old seedlings) were inserted in the Receiver‐C, following the same procedure as above.
Zinc foliar application and sampling
Zinc was applied to the leaves of the AM‐Donor plant at 0.25 g Zn L−1 as ZnSO4·7H2O (+Zn treatment). A water control (i.e., −Zn treatment) was also included. A total of 34 Petri plates were set up with 17 replicates per treatment (17 for +Zn and 17 for −Zn treatment). The pH of both solutions was adjusted to 6.2. A drop of Tween® 20 (Sigma‐Aldrich, Steinheim, Germany; 10 μl) was added to both solutions to increase the adhesion to the leaves. The solutions were applied to the middle laminae of all leaves of the AM‐Donor plants as 10 droplets (10‐μl each) in four applications at 24 h intervals (at 0, 24, 48 and 72 h) (total volume of 400 μl per plant), corresponding to the doses of 0.1 or 0 mg of Zn per plant.
The adequate time of Zn application/sampling was determined as described in Methods S2. The application of Zn or water solutions was done at week 16 from the plating of the AM‐Donor plants in the Donor‐C (plants were still at vegetative growth stage and any symptoms of nutrient deficiencies was detected), that corresponds to 8 weeks after having insert the AM‐Receiver plants in the Receiver‐C. Five days later, shoots and roots of plants in the DC and RC were sampled for further analysis, and the plates were used to measure the fungal mycelium parameters.
Plant and AM fungus morphological measures
At harvest, the number of leaves, stem and roots lengths of AM‐Donor, AM‐Receiver and NM‐Control plants were assessed on five plates, randomly chosen out of the 17 plates per treatment. Root length was estimated by the gridline intersect method (Newman, 1966). Shoot dry weight was determined after oven drying at 70°C to constant weight. Root fresh weight was measured and root dry weight was determined on a subsample. An additional subsample of roots was soaked in water and checked for the presence on root surface of extraradical hyphae and spores, which were carefully plucked with forceps under a dissecting microscope and successively used for RNA extraction. The roots were cleared in 10% KOH solution at 90°C for 5 min, acidified in HCl 2% for 10 min and stained with 0.05% Trypan blue, using lactic acid instead of phenol (Phillips and Hayman, 1970), at 90°C for 5 min, and percentage of colonization assessed using the gridline intersect method (Giovannetti and Mosse, 1980). The AM fungal colonized root length was calculated multiplying the root length by the percentage of colonization. The total hyphal length was measured in both compartments (after root removal) using the HyLength image analysis tool (Cardini et al., 2020). The hyphal density in the Donor‐C and Receiver‐C was calculated as mean of measures taken from 10 randomly acquired images (20 mm2) in different areas per compartment. Details about images and calculations of hyphal traits are given in Methods S2. The number of active hyphae (i.e., presenting bidirectional flux of cytoplasm/protoplasm) crossing the partition wall was measured under a stereomicroscope (×40). The spore number was assessed in both compartments, following the method of Voets and colleagues (2005).
Plant Zn analysis
The 12 remaining plates of each treatment were used for the Zn concentration analysis. To obtain enough material for Zn analysis four randomly chosen AM‐Donor, AM‐Receiver and NM‐Control plants were merged and thus three replicates were analysed for each treatment. The shoot of each plant was rinsed in a 1 mM CaCl2 solution to remove the Zn adhering to the surface (Yilmaz et al., 2017). Zinc concentration was determined by inductively coupled plasma optical emission spectroscopy (ICP‐OES) on an Optima 8000 spectrometer (Perkin Elmer, Waltham, MA, USA), following the procedure of Nölte (2003). Zinc concentration was also analysed in the MSR ‐Zn medium collected from both compartments of each treatment. However, it was not possible to analyse Zn concentration in the AM fungal mycelium of both compartments due to weight limitation.
Selection of plant–fungal genes encoding Zn transporters and design and validation of RT‐qPCR assays
Three genes of M. truncatula, MtZIP1, MtZIP2 and the gene encoding nicotianamine synthase, Mt‐NAS1, were selected from previous studies (Burleigh et al., 2003; López‐Millán et al., 2004; Curie et al., 2008; Clemens et al., 2013; Cardini et al., 2021) (Fig. 5). Moreover, the Zn transporter gene MtZIP14, localized on the PAM, was selected (Watts‐Williams et al., 2020). Two genes of M. truncatula, actin‐101 (MtACT‐101) and elongation factor 1‐α (MtEF1‐α) were selected as reference genes (Nicot et al., 2005). Two genes of R. irregularis were selected: the RiZnT1 (formerly known as GintZnT1 by González‐Guerrero et al., 2005) and the putative Zn transporter RiZRT1 (Tamayo et al., 2014) (Fig. 5). The R. irregularis 28S ribosomal subunit (Ri28S) was used as reference gene (Alkan et al., 2004).
The designed Medicago sativa qPCR primers, targeting ZIP1, ZIP2 and NAS1 and the two reference genes ACT‐101 and EF1‐α, were validated in M. truncatula, using the procedure described by Cardini and colleagues (2021). Similarly, the primers designed by Watts‐Williams and colleagues (2020) and targeting ZIP14 were validated. Details about qPCR primers and validation are given in Tables S7 and S8, and Methods S3. BLAST of the sequences of amplicons obtained by PCR using the six sets of primers proved the specificity of the assays. Specificity was confirmed by the analysis of melting curves of each qPCR amplicon, which did not show non‐specific amplification products in dissociation (Fig. S2). Very strong precision of the qPCR assays was demonstrated by the coefficients of determination (R2) of the standard curves (R 2: ≥ 0.99; Table S7; Fig. S3), while the high accuracy was proven by the efficiency (E ≥ 99.3%).
Two new RT‐qPCR assays were developed for R. irregularis. Details about primer design and test and PCR and qPCR conditions are given in Tables S5, S7 and Methods S4. Similarly to plant primers, primer specificity was proven by BLAST and confirmed by the analysis of melting curves (Fig. S2). A very strong precision and high accuracy was found (R 2 ≥ 0.99; E ≥ 97.9%; Table S7; Fig. S3).
RNA isolation and Real‐Time RT‐PCR
Total RNA was extracted from 50 mg of representative subsamples of shoot tissues and from 50 mg of representative subsamples of root tissues of AM‐Donor, AM‐Receiver and NM‐Control plants, using the RNeasy Mini Kit (Qiagen, Hilden, Germany). A total of 60 RNA extractions were performed, five replicates for each organ (shoot and root) of three types of plants (AM‐Donor, AM‐Receiver and NM‐Control) per treatment. For additional details see Methods S5. From each RNA extract, 1 μg of total RNA was reverse transcribed to complementary DNA (cDNA) using the iScript cDNA Synthesis Kit (Biorad, Hercules, California) and the RT‐qPCR assays (Table S7) were run in 20 μl reaction volume, as described in Methods S5. The two RT‐qPCR assays (RiZnT1 and RiZRT1; Table S7) were applied only on cDNA of the root samples. Three technical replicates were run for each cDNA sample (the mean of the three technical replicates for each of the five biological replicates were handled in the statistical analyses). The qPCR conditions are given in Methods S5. The transcript levels of MtACT‐101 and MtEF1‐α were determined on all cDNA samples (Nicot et al., 2005) and then used as normalization controls, since they did not vary following Zn application. The transcript levels of the Ri28S of R. irregularis were determined on all cDNA root samples (Alkan et al., 2004) and then used for normalization. The relative expression analysis of all genes was done by the double standardization method (ΔΔCq) that requires the reference genes and the control treatment (Livak and Schmittgen, 2001; Vandesompele et al., 2002). The transcript level of the plant genes in the NM‐Controls (mean of the NM‐Controls in +Zn and −Zn treatment) was used as control, whereas the transcript level of the fungal genes in the AM‐Receiver roots in the −Zn treatment was used as control.
Statistical analyses
To verify the transfer of Zn between Zn treated AM‐Donor plants and AM‐Receiver plants (Hypotheses 1 and 2) we set up a single factor design with plant type (AM‐Donor, AM‐Receiver and NM‐Control) as independent variable and Zn concentration and plant gene expression as dependent variables. Data were analysed by one‐way ANOVA and orthogonal contrasts, such as AM‐Receiver versus NM‐Control (Hypothesis 1) and AM‐Donor versus AM‐Receiver (Hypothesis 2) plants. Additionally, to verify that the transfer mediated by CMN does not occur in absence of Zn application (Hypothesis 3), the same experimental design and statistical approach on Zn concentration and gene expression was applied under no‐Zn application (one‐way ANOVA and orthogonal contrasts). To further support the evidence of Zn transfer from AM‐Donor to AM‐Receiver plants, pairwise comparisons of Zn‐fertilized and unfertilised plants were performed on Zn concentration by t‐test for AM‐Donor, AM‐Receiver and NM‐Control plants, and on fungal gene expression for AM‐Donor and AM‐Receiver.
To verify the adequacy of the system under Zn‐ and no‐Zn application we applied a single factor design with plant type (AM‐Donor, AM‐Receiver and NM‐Control) as independent variable and plant growth traits as dependent variables. Data were analysed by one‐way ANOVA and orthogonal contrasts, such as AM‐Receiver versus NM‐Control and AM‐Donor versus AM‐Receiver plants. Moreover, fungal traits in Donor‐C and Receiver‐C were analysed by t‐tests, under both Zn‐ and no‐Zn application. To verify the adequacy of the system excluding any toxic effect on plants, we analysed the effect of Zn on plant growth traits by t‐test, separately for AM‐Donor, AM‐Receiver and NM‐Control plants. Finally, to verify the adequacy of the system excluding any toxic effect on fungus, we analysed the effect of Zn on fungal traits by t‐test, separately for Donor‐C and Receiver‐C.
Data were transformed when needed to fulfil the assumptions of the ANOVA. All the analyses were performed using the SPSS software package version 21.0 (SPSS, Chicago, IL, USA).
Author contributions
Elisa Pellegrino and Laura Ercoli conceived the ideas and designed the methodology; Stéphane Declerck designed the in vitro system; Alessio Cardini and Maryline Calonne‐Salmon performed in vitro culturing, microscopy and staining analysis; Alessio Cardini and Barbara Mazzolai designed and performed real‐time PCR; Alessio Cardini and Elisa Pellegrino performed chemical analyses; Alessio Cardini and Elisa Pellegrino performed data analysis; Alessio Cardini, Elisa Pellegrino and Laura Ercoli led the writing of the article; all authors contributed critically to the drafts and gave final approval for publication.
Supporting information
Fig. S1. Transcript levels of the Ri28S in the roots of the AM‐Donors and Receivers Medicago truncatula plants colonized by Rhizophagus irregularis under +Zn and −Zn conditions.
Fig. S2. Melting peaks of qPCR products obtained with primers targeting Medicago truncatula genes (MtZIP1, MtZIP2, MtZIP14, MtNAS1, MtACT‐101 and MtEF1‐α) and Rhizophagus irregularis genes (RiZnT1, RiZRT1, Ri28S).
Fig. S3. Standard curves of qPCR products obtained with primers targeting Medicago truncatula genes (MtZIP1, MtZIP2, MtZIP14, MtNAS1, MtACT‐101 and MtEF1‐α) and Rhizophagus irregularis genes (RiZnT1, RiZRT1, Ri28S).
Table S1. Growth parameters of the Medicago truncatula AM‐Donor, AM‐Receiver and NM‐Controls under +Zn and –Zn conditions.
Table S2. P values of the t‐tests on the effect of Zn foliar application on plant growth parameters, AMF root colonization and shoot and root Zn concentrations of AM‐Donor, AM‐Receiver and NM‐Control Medicago truncatula plants.
Table S3. P values of the t‐tests on the effect of Zn foliar application on AMF extraradical mycelium growth parameters and spore number in donor and receiver compartments.
Table S4. Zinc concentrations in shoots and roots of AM‐Donor, AM‐Receiver and NM‐Control Medicago truncatula plants under +Zn and –Zn conditions.
Table S5. P values of the linear orthogonal contrasts on the relative gene expressions of MtZIP1, MtZIP2, MtZIP14, MtNAS1 in shoots and roots, and on the fungal relative gene expressions of RiZnT1 and RiZRT1 in AMF colonized roots under +Zn and –Zn conditions.
Table S6. P values of t‐test on the relative gene expressions of MtZIP1, MtZIP2, and MtNAS1 in shoots and roots, and of MtZIP14, RiZnT1 and RiZRT1 in AM fungal colonized roots under +Zn and –Zn conditions.
Table S7. Sequences of the qPCR primer pairs used in the study and parameters of the validation.
Table S8. The Medicago truncatula and Rhizophagus irregularis sequences used to design the qPCR primers by the Primer‐Blast online tool in NCBI.
Methods S1. Description of the mycorrhizal defective mutant isogenic line TRV25.
Methods S2. Set‐up, such as plant growth conditions and time of Zn application/sampling and details on fungal measures.
Methods S3. Details on RT‐qPCR validation of the MtZIP1, MtZIP2 and MtNAS1 primers.
Methods S4. Details on RT‐qPCR primer design for the Rhizophagus irregularis genes RiZnT1 and RiZRT1 and validation of the new RT‐qPCR assays.
Methods S5. Details on RNA isolation and real‐time RT‐qPCRs.
Acknowledgements
We acknowledge the Italian Institute of Technology (IIT) for funding for PhD Fellowship of AC under the PhD programme in Agrobiosciences at the Scuola Superiore Sant'Anna of Pisa, Italy. We thank the Institut National de la Recherche Agronomique (INRA, Dijon, France) for providing the seeds of both lines of Medicago truncatula. We acknowledge Dr. Hannes A. Gamper for the technical tips for real‐time PCR.
References
- Alaux, P.L. , Naveau, F. , Declerck, S. , and Cranenbrouck, S. (2020) Common mycorrhizal network induced JA/ET genes expression in healthy potato plants connected to potato plants infected by Phytophthora infestans . Front Plant Sci 11: 602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkan, N. , Gadkar, V. , Coburn, J. , Yarden, O. , and Kapulnik, Y. (2004) Quantification of the arbuscular mycorrhizal fungus Glomus intraradices in host tissue using real‐time polymerase chain reaction. New Phytol 161: 877–885. [DOI] [PubMed] [Google Scholar]
- Ashford, A. (2002) Tubular vacuoles in arbuscular mycorrhizas. New Phytol 154: 545–547. [DOI] [PubMed] [Google Scholar]
- Avio, L. , Pellegrino, E. , Bonari, E. , and Giovannetti, M. (2006) Functional diversity of arbuscular mycorrhizal fungal isolates in relation to extraradical mycelial networks. New Phytol 172: 347–357. [DOI] [PubMed] [Google Scholar]
- Babikova, Z. , Gilbert, L. , Bruce, T.J. , Birkett, M. , Caulfield, J.C. , Woodcock, C. , et al. (2013) Underground signals carried through common mycelial networks warn neighbouring plants of aphid attack. Ecol Lett 16: 835–843. [DOI] [PubMed] [Google Scholar]
- Bethlenfalvay, G.J. , Reyes‐Solis, M.G. , Camel, S.B. , and Ferrera‐Cerrato, R. (1991) Nutrient transfer between the root zones of soybean and maize plants connected by a common mycorrhizal mycelium. Physiol Plantarum 82: 423–432. [Google Scholar]
- Broadley, M.R. , Brown, P. , Cakmak, I. , Rengel, Z. , and Zhao, F. (2012) Function of nutrients: micronutrients. In Mineral Nutrition of Higher Plants, Marschner, P. (ed). San Diego, USA: Academic Press, pp. 191–248. [Google Scholar]
- Broadley, M.R. , White, P.J. , Hammond, J.P. , Zelko, I. , and Lux, A. (2007) Zinc in plants. New Phytol 173: 677–702. [DOI] [PubMed] [Google Scholar]
- Bücking, H. , Mensah, J.A. , and Fellbaum, C.R. (2016) Common mycorrhizal networks and their effect on the bargaining power of the fungal partner in the arbuscular mycorrhizal symbiosis. Commun Integr Biol 9: e1107684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burleigh, S.H. , Kristensen, B.K. , and Bechmann, I.E. (2003) A plasma membrane zinc transporter from Medicago truncatula is up‐regulated in roots by Zn fertilization, yet down‐regulated by arbuscular mycorrhizal colonization. Plant Mol Biol 52: 1077–1088. [DOI] [PubMed] [Google Scholar]
- Cardini, A. , Pellegrino, E. , Del Dottore, E. , Gamper, H.A. , Mazzolai, B. , and Ercoli, L. (2020) HyLength: a semi‐automated digital image analysis tool for measuring the length of roots and fungal hyphae of dense mycelia. Mycorrhiza 30: 229–242. [DOI] [PubMed] [Google Scholar]
- Cardini, A. , Pellegrino, E. , White, P. , Mazzolai, B. , Mascherpa, M.C. , and Ercoli, L. (2021) Transcriptional regulation of genes involved in zinc uptake, sequestration and redistribution following foliar zinc application to Medicago sativa . Plan Theory 10: 476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiariello, N. , Hickman, J.C. , and Mooney, H.A. (1982) Endomycorrhizal role for interspecific transfer of phosphorus in a community of annual plants. Science 217: 941–943. [DOI] [PubMed] [Google Scholar]
- Ciccolini, V. , Ercoli, L. , Davison, J. , Vasar, M. , Öpik, M. , and Pellegrino, E. (2016) Land‐use intensity and host plant simultaneously shape the composition of arbuscular mycorrhizal fungal communities in a Mediterranean drained peatland. FEMS Microbiol Ecol 92: fiw186. [DOI] [PubMed] [Google Scholar]
- Ciccolini, V. , Pellegrino, E. , Coccina, A. , Fiaschi, A.I. , Cerretani, D. , Sgherri, C. , et al. (2017) Biofortification with iron and zinc improves nutritional and nutraceutical properties of common wheat flour and bread. J Agr Food Chem 65: 5443–5452. [DOI] [PubMed] [Google Scholar]
- Clemens, S. , Deinlein, U. , Ahmadi, H. , Höreth, S. , and Uraguchi, S. (2013) Nicotianamine is a major player in plant Zn homeostasis. Biometals 26: 623–632. [DOI] [PubMed] [Google Scholar]
- Coccina, A. , Cavagnaro, T.R. , Pellegrino, E. , Ercoli, L. , McLaughlin, M.J. , and Watts‐Williams, S.J. (2019) The mycorrhizal pathway of zinc uptake contributes to zinc accumulation in barley and wheat grain. BMC Plant Biol 19: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coninx, L. , Smisdom, N. , Kohler, A. , Arnauts, N. , Ameloot, M. , Rineau, F. , et al. (2019) SlZRT2 encodes a ZIP family Zn transporter with dual localization in the ectomycorrhizal fungus Suillus luteus . Front Microbiol 10: 2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cranenbrouck, S. , Voets, L. , Bivort, C. , Renard, L. , Strullu, D.G. , and Declerck, S. (2005) Methodologies for in vitro cultivation of arbuscular mycorrhizal fungi with root organs. In In Vitro Culture of Mycorrhizas, Declerck, S. , Fotin, A. , and Strullu, D.G. (eds). Heidelberg, Germany: Springer, pp. 341–375. [Google Scholar]
- Croll, D. , Giovannetti, M. , Koch, A.M. , Sbrana, C. , Ehinger, M. , Lammers, P.J. , and Sanders, I.R. (2009) Nonself vegetative fusion and genetic exchange in the arbuscular mycorrhizal fungus Glomus intraradices . New Phytol 181: 924–937. [DOI] [PubMed] [Google Scholar]
- Curie, C. , Cassin, G. , Couch, D. , Divol, F. , Higuchi, K. , Le Jean, M. , et al. (2008) Metal movement within the plant: contribution of nicotianamine and yellow stripe 1‐like transporters. Ann Bot‐London 103: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison, J. , García de León, D. , Zobel, M. , Moora, M. , Bueno, C.G. , Barceló, M. , et al. (2020) Plant functional groups associate with distinct arbuscular mycorrhizal fungal communities. New Phytol 226: 1117–1128. [DOI] [PubMed] [Google Scholar]
- Declerck, S. , Strullu, D.G. , and Plenchette, C. (1998) Monoxenic culture of the intraradical forms of Glomus sp. isolated from a tropical ecosystem: a proposed methodology for germplasm collection. Mycologia 90: 579–585. [Google Scholar]
- Deinlein, U. , Weber, M. , Schmidt, H. , Rensch, S. , Trampczynska, A. , Hansen, T.H. , et al. (2012) Elevated nicotianamine levels in Arabidopsis halleri roots play a key role in zinc hyperaccumulation. Plant Cell 24: 708–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande, P. , Dapkekar, A. , Oak, M. , Paknikar, K. , and Rajwade, J. (2018) Nanocarrier‐mediated foliar zinc fertilization influences expression of metal homeostasis related genes in flag leaves and enhances gluten content in durum wheat. PloS One 13: e0191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupré de Boulois, H. , Voets, L. , Delvaux, B. , Jakobsen, I. , and Declerck, S. (2006) Transport of radiocaesium by arbuscular mycorrhizal fungi to Medicago truncatula under in vitro conditions. Environ Microbiol 8: 1926–1934. [DOI] [PubMed] [Google Scholar]
- Durmaz, E. , Coruh, C. , Dinler, G. , Grusak, M.A. , Peleg, Z. , Saranga, Y. , et al. (2011) Expression and cellular localization of ZIP1 transporter under zinc deficiency in wild emmer wheat. Plant Mol Biol Rep 29: 582–596. [Google Scholar]
- Eckhardt, U. , Marques, A.M. , and Buckhout, T.J. (2001) Two iron‐regulated cation transporters from tomato complement metal uptake‐deficient yeast mutants. Plant Mol Biol 45: 437–448. [DOI] [PubMed] [Google Scholar]
- Eidie, D. , Broderius, M. , Fett, J. , and Guerinot, M.L. (1996) A novel iron‐regulated metal transporter from plants identified by functional expression in yeast. PNAS USA 93: 5624–5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenstat, D.M. (1990) A comparison of phosphorus and nitrogen transfer between plants of different phosphorus status. Oecologia 82: 342–347. [DOI] [PubMed] [Google Scholar]
- Ercoli, L. , Schüßler, A. , Arduini, I. , and Pellegrino, E. (2017) Strong increase of durum wheat iron and zinc content by field‐inoculation with arbuscular mycorrhizal fungi at different soil nitrogen availabilities. Plant and Soil 419: 153–167. [Google Scholar]
- Fellbaum, C.R. , Mensah, J.A. , Cloos, A.J. , Strahan, G.E. , Pfeffer, P.E. , Kiers, E.T. , and Bücking, H. (2014) Fungal nutrient allocation in common mycorrhizal networks is regulated by the carbon source strength of individual host plants. New Phytol 203: 646–656. [DOI] [PubMed] [Google Scholar]
- Fortin, J.A. , Bécard, G. , Declerck, S. , Dalpé, Y. , St‐Arnaud, M. , Coughlan, A.P. , and Piché, Y. (2002) Arbuscular mycorrhiza on root‐organ cultures. Can J Bot 80: 1–20. [Google Scholar]
- Frey, B. , and Schüepp, H. (1993) Acquisition of nitrogen by external hyphae of arbuscular mycorrhizal fungi associated with Zea mays L. New Phytol 124: 221–230. [DOI] [PubMed] [Google Scholar]
- Giovannetti, M. , and Mosse, B. (1980) An evaluation of techniques for measuring vesicular arbuscular mycorrhizal infection in roots. New Phytol 84: 489–500. [Google Scholar]
- Giovannetti, M. , Sbrana, C. , Avio, L. , and Strani, P. (2004) Patterns of below‐ground plant interconnections established by means of arbuscular mycorrhizal networks. New Phytol 164: 175–181. [DOI] [PubMed] [Google Scholar]
- González‐Guerrero, M. , Azcón‐Aguilar, C. , Mooney, M. , Valderas, A. , MacDiarmid, C.W. , Eide, D.J. , and Ferrol, N. (2005) Characterization of a Glomus intraradices gene encoding a putative Zn transporter of the cation diffusion facilitator family. Fungal Genet Biol 42: 130–140. [DOI] [PubMed] [Google Scholar]
- González‐Guerrero, M. , Benabdellah, K. , Ferrol, N. , and Azcón‐Aguilar, C. (2009) Mechanisms underlying heavy metal tolerance in arbuscular mycorrhizas. In Mycorrhizas—Functional Processes and Ecological Impact, Azcón‐Aguilar, C. , Barea, M. , Gianinazzi, S. , and Gianinazzi‐Pearson, V. (eds). Heidelberg, Germany: Springer‐Verlag, pp. 107–122. [Google Scholar]
- González‐Guerrero, M. , Escudero, V. , Saéz, Á. , and Tejada‐Jiménez, M. (2016) Transition metal transport in plants and associated endosymbionts: arbuscular mycorrhizal fungi and rhizobia. Front Plant Sci 7: 1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González‐Guerrero, M. , Melville, L.H. , Ferrol, N. , Lott, J.N. , Azcon‐Aguilar, C. , and Peterson, R.L. (2008) Ultrastructural localization of heavy metals in the extraradical mycelium and spores of the arbuscular mycorrhizal fungus Glomus intraradices . Can J Microbiol 54: 103–110. [DOI] [PubMed] [Google Scholar]
- Grotz, N. , Fox, T. , Connolly, E. , Park, W. , Guerinot, M.L. , and Eide, D. (1998) Identification of a family of zinc transporter genes from Arabidopsis that respond to zinc deficiency. PNAS USA 95: 7220–7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grusak, M.A. , Pearson, J.N. , and Marentes, E. (1999) The physiology of micronutrient homeostasis in field crops. Field Crops Res 60: 41–56. [Google Scholar]
- Gyuricza, V. , de Boulois, H.D. , and Declerck, S. (2010) Effect of potassium and phosphorus on the transport of radiocesium by arbuscular mycorrhizal fungi. J Environ Radioact 101: 482–487. [DOI] [PubMed] [Google Scholar]
- Haydon, M.J. , Kawachi, M. , Wirtz, M. , Hillmer, S. , Hell, R. , and Krämer, U. (2012) Vacuolar nicotianamine has critical and distinct roles under iron deficiency and for zinc sequestration in Arabidopsis . Plant Cell 24: 724–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, X.H. , Critchley, C. , and Bledsoe, C. (2003) Nitrogen transfer within and between plants through common mycorrhizal networks (CMNs). Crit Rev Plant Sci 22: 531–567. [Google Scholar]
- Ishimaru, Y. , Suzuki, M. , Tsukamoto, T. , Suzuki, K. , Nakazono, M. , Kobayashi, T. , et al. (2006) Rice plants take up iron as an Fe3+‐phytosiderophore and as Fe2+ . Plant J 45: 335–346. [DOI] [PubMed] [Google Scholar]
- Jakobsen, I. , and Hammer, E. (2015) Nutrient dynamics in arbuscular mycorrhizal networks. In Mycorrhizal Networks, Horton, T.R. (ed). Dordrecht, Netherlands: Springer, pp. 91–131. [Google Scholar]
- Javot, H. , Pumplin, N. , and Harrison, M.J. (2007) Phosphate in the arbuscular mycorrhizal symbiosis: transport properties and regulatory roles. Plant Cell Environ 30: 310–322. [DOI] [PubMed] [Google Scholar]
- Jin, H. , Pfeffer, P.E. , Douds, D.D. , Piotrowski, E. , Lammers, P.J. , and Shachar‐Hill, Y. (2005) The uptake, metabolism, transport and transfer of nitrogen in an arbuscular mycorrhizal symbiosis. New Phytol 168: 687–696. [DOI] [PubMed] [Google Scholar]
- Johansen, A. , and Jensen, E.S. (1996) Transfer of N and P from intact or decomposing roots of pea to barley interconnected by an arbuscular mycorrhizal fungus. Soil Biol Biochem 28: 73–81. [Google Scholar]
- Kirk, P.M. , Cannon, P.F. , Minter, D.W. , and Stalpers, J.A. (2008) Ainsworth and Bisby's Dictionary of the Fungi, 10th ed. Wallingford, UK: CABI Publishing. [Google Scholar]
- Lalaymia, I. , and Declerck, S. (2020) The mycorrhizal donor plant (MDP) in vitro culture system for the efficient colonization of whole plants. In Arbuscular Mycorrhizal Fungi. Methods in Molecular Biology, Ferrol, N. , and Lanfranco, L. (eds). Humana: New York, USA, pp. 19–31. [DOI] [PubMed] [Google Scholar]
- Lehmann, A , and Rillig, M.C. (2015) Arbuscular mycorrhizal contribution to copper, manganese and iron nutrient concentrations in crops ‐ a meta‐analysis. Soil Biol Biochem 81: 147–158. [Google Scholar]
- Lehmann, A. , Veresoglou, S.D. , Leifheit, E.F. , and Rillig, M.C. (2014) Arbuscular mycorrhizal influence on zinc nutrition in crop plants—a meta‐analysis. Soil Biol Biochem 69: 123–131. [Google Scholar]
- Livak, K.J. , and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- López‐Millán, A.F. , Ellis, D.R. , and Grusak, M.A. (2004) Identification and characterization of several new members of the ZIP family of metal ion transporters in Medicago truncatula . Plant Mol Biol 54: 583–596. [DOI] [PubMed] [Google Scholar]
- Martin, R.C. , Eaglesham, A.R. , Voldeng, H.D. , and Smith, D.L. (1995) Factors affecting nitrogen benefit from soybean [Glycine max (L.) Merr. cv Lee] to interplanted corn (Zea mays L. cv Co‐op S259). Environ Exp Bot 35: 497–505. [Google Scholar]
- Mäser, P. , Thomine, S. , Schroeder, J.I. , Ward, J.M. , Hirschi, K. , Sze, H. , et al. (2001) Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol 126: 1646–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrild, M.P. , Ambus, P. , Rosendahl, S. , and Jakobsen, I. (2013) Common arbuscular mycorrhizal networks amplify competition for phosphorus between seedlings and established plants. New Phytol 200: 229–240. [DOI] [PubMed] [Google Scholar]
- Mikkelsen, B.L. , Rosendahl, S. , and Jakobsen, I. (2008) Underground resource allocation between individual networks of mycorrhizal fungi. New Phytol 180: 890–898. [DOI] [PubMed] [Google Scholar]
- Milner, M.J. , Seamon, J. , Craft, E. , and Kochian, L.V. (2013) Transport properties of members of the ZIP family in plants and their role in Zn and Mn homeostasis. J Exp Bot 64: 369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morandi, D. , Prado, E. , Sagan, M. , and Duc, G. (2005) Characterisation of new symbiotic Medicago truncatula (Gaertn.) mutants, and phenotypic or genotypic complementary information on previously described mutants. Mycorrhiza 15: 283–289. [DOI] [PubMed] [Google Scholar]
- Newman, E.I. (1966) A method of estimating the total length of root in a sample. J Appl Ecol 3: 139–145. [Google Scholar]
- Newman, E.I. , Devoy, C.L.N. , Easen, N.J. , and Fowles, K.J. (1994) Plant species that can be linked by VA mycorrhizal fungi. New Phytol 126: 691–693. [Google Scholar]
- Nicot, N. , Hausman, J.F. , Hoffmann, L. , and Evers, D. (2005) Housekeeping gene selection for real‐time RT‐PCR normalization in potato during biotic and abiotic stress. J Exp Bot 56: 2907–2914. [DOI] [PubMed] [Google Scholar]
- Nielsen, J.S. , Joner, E.J. , Declerck, S. , Olsson, S. , and Jakobsen, I. (2002) Phospho‐imaging as a tool for visualization and noninvasive measurement of P transport dynamics in arbuscular mycorrhizas. New Phytol 154: 809–819. [DOI] [PubMed] [Google Scholar]
- Nölte, J. (2003) ICP Emission Spectrometry: A Practical Guide, Vol. 1. Owingen: Wiley‐VCH. [Google Scholar]
- Olsen, L.I. , and Palmgren, M.G. (2014) Many rivers to cross: the journey of zinc from soil to seed. Front Plant Sci 5: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öpik, M. , Metsis, M. , Daniell, T.J. , Zobel, M. , and Moora, M. (2009) Large‐scale parallel 454 sequencing reveals host ecological group specificity of arbuscular mycorrhizal fungi in a boreonemoral forest. New Phytol 184: 424–437. [DOI] [PubMed] [Google Scholar]
- Parniske, M. (2008). Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat Rev Microbiol 6: 763–775. [DOI] [PubMed] [Google Scholar]
- Pellegrino, E. , Piazza, G. , Arduini, I. , and Ercoli, L. (2020) Field inoculation of bread wheat with Rhizophagus irregularis under organic farming: variability in growth response and nutritional uptake of eleven old genotypes and a modern variety. Agronomy 10: 333. [Google Scholar]
- Pfeffer, P.E. , Douds, D.D., Jr. , Bücking, H. , Schwartz, D.P. , and Shachar‐Hill, Y. (2004) The fungus does not transfer carbon to or between roots in an arbuscular mycorrhizal symbiosis. New Phytol 163: 617–627. [DOI] [PubMed] [Google Scholar]
- Phillips, J.M. , and Hayman, D.S. (1970) Improved procedures for clearing roots and staining parasitic and vesicular‐arbuscular mycorrhizal fungi for rapid assessment of infection. Trans Brit Mycol Soc 55: 158–161. [Google Scholar]
- Ramesh, S.A. , Shin, R. , Eide, D.J. , and Schachtman, D.P. (2003) Differential metal selectivity and gene expression of two zinc transporters from rice. Plant Physiol 133: 126–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read, D.J. , and Birch, C.P.D. (1988) The effects and implications of disturbance of mycorrhizal mycelial systems. Proc Royal Soc B 94: 13–24. [Google Scholar]
- Rosendahl, S. , and Stukenbrock, E.H. (2004) Community structure of arbuscular mycorrhizal fungi in undisturbed vegetation revealed by analyses of LSU rDNA sequences. Mol Ecol 13: 3179–3186. [DOI] [PubMed] [Google Scholar]
- Sagan, M. , de Larembergue, H. , and Morandi, D. (1998) Genetic analysis of symbiosis mutants in Medicago truncatula . In Biological Nitrogen Fixation for the 21st Century, Elmerich, C. , Kondorosi, A. , and Newton, W.E. (eds). Dordrecht: Kluwer, pp. 317–318. [Google Scholar]
- Sagan, M. , Morandi, D. , Tarenghi, E. , and Duc, G. (1995) Selection of nodulation and mycorrhizal mutants in the model plant Medicago truncatula (Gaertn.) after γ‐ray mutagenesis. Plant Sci 111: 63–71. [Google Scholar]
- Sasaki, H. , Hirose, T. , Watanabe, Y. , and Ohsugi, R. (1998) Carbonic anhydrase activity and CO2‐transfer resistance in Zn‐deficient rice leaves. Plant Physiol 118: 929–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selosse, M.A. , Richard, F. , He, X. , and Simard, S.W. (2006) Mycorrhizal networks: des liaisons dangereuses? Trends Ecol Evol 21: 621–628. [DOI] [PubMed] [Google Scholar]
- Simard, S.W. , and Durall, D.M. (2004) Mycorrhizal networks: a review of their extent, function, and importance. Can J Bot 82: 1140–1165. [Google Scholar]
- Simard, S.W. , Jones, M.D. , and Durall, D.M. (2003) Carbon and nutrient fluxes within and between mycorrhizal plants. In Mycorrhizal Ecology, Van der Heijden, M.G.A. , and Sanders, I.R. (eds). Heidelberg, Germany: Springer, pp. 33–74. [Google Scholar]
- Sinclair, S.A. , and Krämer, U. (2012) The zinc homeostasis network of land plants. BBA‐Mol Cell Res 1823: 1553–1567. [DOI] [PubMed] [Google Scholar]
- Song, Y.Y. , Zeng, R.S. , Xu, J.F. , Li, J. , Shen, X. , and Yihdego, W.G. (2010) Interplant communication of tomato plants through underground common mycorrhizal networks. PloS One 5: e13324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talke, I.N. , Hanikenne, M. , and Krämer, U. (2006) Zinc‐dependent global transcriptional control, transcriptional deregulation, and higher gene copy number for genes in metal homeostasis of the hyperaccumulator Arabidopsis halleri . Plant Physiol 142: 148–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo, E. , Gómez‐Gallego, T. , Azcón‐Aguilar, C. , and Ferrol, N. (2014) Genome‐wide analysis of copper, iron and zinc transporters in the arbuscular mycorrhizal fungus Rhizophagus irregularis . Front Plant Sci 5: 547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint, J.P. , St‐Arnaud, M. , and Charest, C. (2004) Nitrogen transfer and assimilation between the arbuscular mycorrhizal fungus Glomus intraradices Schenck & Smith and Ri T‐DNA roots of Daucus carota L. in an in vitro compartmented system. Can J Microbiol 50: 251–260. [DOI] [PubMed] [Google Scholar]
- Vallee, B.L. , and Auld, D.S. (1990) Zinc coordination, function, and structure of zinc enzymes and other proteins. Biochemistry 29: 5647–5659. [DOI] [PubMed] [Google Scholar]
- van der Heijden, M.G. (2004) Arbuscular mycorrhizal fungi as support systems for seedling establishment in grassland. Ecol Lett 7: 293–303. [Google Scholar]
- van der Heijden, M.G. , Klironomos, J.N. , Ursic, M. , Moutoglis, P. , Streitwolf‐Engel, R. , Boller, T. , et al. (1998) Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 396: 69–72. [Google Scholar]
- van Kessel, C. , Singleton, P.W. , and Hoben, H.J. (1985) Enhanced N‐transfer from a soybean to maize by vesicular arbuscular mycorrhizal (VAM) fungi. Plant Physiol 79: 562–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele, J. , De Preter, K. , Pattyn, F. , Poppe, B. , Van Roy, N. , De Paepe, A. , and Speleman, F. (2002) Accurate normalization of real‐time quantitative RT‐PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veresoglou, S.D. , and Rillig, M.C. (2014) Do closely related plants host similar arbuscular mycorrhizal fungal communities? A meta‐analysis. Plant and Soil 377: 395–406. [Google Scholar]
- Voets, L. , de Boulois, H.D. , Renard, L. , Strullu, D.G. , and Declerck, S. (2005) Development of an autotrophic culture system for the in vitro mycorrhization of potato plantlets. FEMS Microbiol Lett 248: 111–118. [DOI] [PubMed] [Google Scholar]
- Voets, L. , de la Providencia, I.E. , Fernandez, K. , IJdo, M. , Cranenbrouck, S. , and Declerck, S. (2009) Extraradical mycelium network of arbuscular mycorrhizal fungi allows fast colonization of seedlings under in vitro conditions. Mycorrhiza 19: 347–356. [DOI] [PubMed] [Google Scholar]
- Voets, L. , Goubau, I. , Olsson, P.A. , Merckx, R. , and Declerck, S. (2008) Absence of carbon transfer between Medicago truncatula plants linked by a mycorrhizal network, demonstrated in an experimental microcosm. FEMS Microbiol Ecol 65: 350–360. [DOI] [PubMed] [Google Scholar]
- Watts‐Williams, S.J. , Tyerman, S.D. , and Cavagnaro, T.R. (2017) The dual benefit of arbuscular mycorrhizal fungi under soil zinc deficiency and toxicity: linking plant physiology and gene expression. Plant and Soil 420: 375–388. [Google Scholar]
- Watts‐Williams, S.J. , Wege, S. , Ramesh, S.A. , Berkowitz, O. , Gilliham, M. , Whelan, J. , and Tyerman, S.D. (2020) Identification of a unique ZIP transporter involved in zinc uptake via the arbuscular mycorrhizal fungal pathway. BioRxiv. 10.1101/2020.09.28.317669. [DOI] [Google Scholar]
- Wipf, D. , Krajinski, F. , van Tuinen, D. , Recorbet, G. , and Courty, P.E. (2019) Trading on the arbuscular mycorrhiza market: from arbuscules to common mycorrhizal networks. New Phytol 223: 1127–1142. [DOI] [PubMed] [Google Scholar]
- Yilmaz, O. , Kazar, G.A. , Cakmak, I. , and Ozturk, L. (2017) Differences in grain zinc are not correlated with root uptake and grain translocation of zinc in wild emmer and durum wheat genotypes. Plant and Soil 411: 69–79. [Google Scholar]
- Zhang, P. , Misra, S. , Guo, Z. , Rehkämper, M. , and Valsami‐Jones, E. (2019) Stable isotope labeling of metal/metal oxide nanomaterials for environmental and biological tracing. Nat Prot 14: 2878–2899. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Transcript levels of the Ri28S in the roots of the AM‐Donors and Receivers Medicago truncatula plants colonized by Rhizophagus irregularis under +Zn and −Zn conditions.
Fig. S2. Melting peaks of qPCR products obtained with primers targeting Medicago truncatula genes (MtZIP1, MtZIP2, MtZIP14, MtNAS1, MtACT‐101 and MtEF1‐α) and Rhizophagus irregularis genes (RiZnT1, RiZRT1, Ri28S).
Fig. S3. Standard curves of qPCR products obtained with primers targeting Medicago truncatula genes (MtZIP1, MtZIP2, MtZIP14, MtNAS1, MtACT‐101 and MtEF1‐α) and Rhizophagus irregularis genes (RiZnT1, RiZRT1, Ri28S).
Table S1. Growth parameters of the Medicago truncatula AM‐Donor, AM‐Receiver and NM‐Controls under +Zn and –Zn conditions.
Table S2. P values of the t‐tests on the effect of Zn foliar application on plant growth parameters, AMF root colonization and shoot and root Zn concentrations of AM‐Donor, AM‐Receiver and NM‐Control Medicago truncatula plants.
Table S3. P values of the t‐tests on the effect of Zn foliar application on AMF extraradical mycelium growth parameters and spore number in donor and receiver compartments.
Table S4. Zinc concentrations in shoots and roots of AM‐Donor, AM‐Receiver and NM‐Control Medicago truncatula plants under +Zn and –Zn conditions.
Table S5. P values of the linear orthogonal contrasts on the relative gene expressions of MtZIP1, MtZIP2, MtZIP14, MtNAS1 in shoots and roots, and on the fungal relative gene expressions of RiZnT1 and RiZRT1 in AMF colonized roots under +Zn and –Zn conditions.
Table S6. P values of t‐test on the relative gene expressions of MtZIP1, MtZIP2, and MtNAS1 in shoots and roots, and of MtZIP14, RiZnT1 and RiZRT1 in AM fungal colonized roots under +Zn and –Zn conditions.
Table S7. Sequences of the qPCR primer pairs used in the study and parameters of the validation.
Table S8. The Medicago truncatula and Rhizophagus irregularis sequences used to design the qPCR primers by the Primer‐Blast online tool in NCBI.
Methods S1. Description of the mycorrhizal defective mutant isogenic line TRV25.
Methods S2. Set‐up, such as plant growth conditions and time of Zn application/sampling and details on fungal measures.
Methods S3. Details on RT‐qPCR validation of the MtZIP1, MtZIP2 and MtNAS1 primers.
Methods S4. Details on RT‐qPCR primer design for the Rhizophagus irregularis genes RiZnT1 and RiZRT1 and validation of the new RT‐qPCR assays.
Methods S5. Details on RNA isolation and real‐time RT‐qPCRs.


