Abstract
Background
Current methods fail to accurately predict women at greatest risk of developing fetal growth restriction (FGR) or related adverse outcomes, including stillbirth. Sexual dimorphism in these adverse pregnancy outcomes is well documented as are sex-specific differences in gene and protein expression in the placenta. Circulating maternal serum microRNAs (miRNAs) offer potential as biomarkers that may also be informative of underlying pathology. We hypothesised that FGR would be associated with an altered miRNA profile and would differ depending on fetal sex.
Methods
miRNA expression profiles were assessed in maternal serum (> 36 weeks’ gestation) from women delivering a severely FGR infant (defined as an individualised birthweight centile (IBC) < 3rd) and matched control participants (AGA; IBC = 20–80th), using miRNA arrays. qPCR was performed using specific miRNA primers in an expanded cohort of patients with IBC < 5th (n = 15 males, n = 16 females/group). Maternal serum human placental lactogen (hPL) was used as a proxy to determine if serum miRNAs were related to placental dysfunction. In silico analyses were performed to predict the potential functions of altered miRNAs.
Results
Initial analyses revealed 11 miRNAs were altered in maternal serum from FGR pregnancies. In silico analyses revealed all 11 altered miRNAs were located in a network of genes that regulate placental function. Subsequent analysis demonstrated four miRNAs showed sexually dimorphic patterns. miR-28-5p was reduced in FGR pregnancies (p < 0.01) only when there was a female offspring and miR-301a-3p was only reduced in FGR pregnancies with a male fetus (p < 0.05). miR-454-3p was decreased in FGR pregnancies (p < 0.05) regardless of fetal sex but was only positively correlated to hPL when the fetus was female. Conversely, miR-29c-3p was correlated to maternal hPL only when the fetus was male. Target genes for sexually dimorphic miRNAs reveal potential functional roles in the placenta including angiogenesis, placental growth, nutrient transport and apoptosis.
Conclusions
These studies have identified sexually dimorphic patterns for miRNAs in maternal serum in FGR. These miRNAs may have potential as non-invasive biomarkers for FGR and associated placental dysfunction. Further studies to determine if these miRNAs have potential functional roles in the placenta may provide greater understanding of the pathogenesis of placental dysfunction and the differing susceptibility of male and female fetuses to adverse in utero conditions.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13293-021-00405-z.
Keywords: miRNA, Placenta, Pregnancy, Serum, Biomarker, Placental dysfunction, FGR, Stillbirth, Sexual dimorphism
Highlights
Detection and treatment of pregnancies at high risk of fetal growth restriction (FGR) and stillbirth remains a major obstetric challenge; circulating maternal serum microRNAs (miRNAs) offer potential as novel biomarkers.
Unbiased analysis of serum miRNAs in women in late pregnancy identified a specific profile of circulating miRNAs in women with a growth-restricted infant.
Some altered miRNAs (miR-28-5p, miR-301a-3p) showed sexually dimorphic expression in FGR pregnancies and others a fetal-sex dependent association to a hormonal marker of placental dysfunction (miR-454-3p, miR-29c-3p).
miR-301a-3p and miR-28-5p could potentially be used to predict FGR specifically in pregnancies with a male or female baby, respectively, however larger cohort studies are required.
Further investigations of these miRNAs and their relationship to placental dysfunction will lead to a better understanding of the pathophysiology of FGR and why there is differing susceptibility of male and female fetuses to FGR and stillbirth.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13293-021-00405-z.
Introduction
Fetal growth restriction (FGR) is a pregnancy complication where the fetus fails to reach its genetically determined growth potential and is a significant cause of fetal morbidity and mortality [1]. FGR is associated with a range of placental abnormalities, including impaired growth, villous and vascular structural abnormalities, and impaired nutrient transport and endocrine function [2, 3]. Fetal growth restriction and associated placental dysfunction are linked to increased rates of stillbirth, death of a fetus in utero after 24 weeks gestation [4]. Stillbirth affects 1 in 240 pregnancies in the UK [5], thus identifying pregnancies with placental dysfunction at greatest risk of FGR and stillbirth is an important clinical aim [2].
The only current direct test of placental function in utero in routine clinical use for detection of FGR is Doppler ultrasound assessment of umbilical artery blood flow [2] however, although this screen is sensitive for detecting early-onset FGR (< 34 weeks’ gestation), it is not reliable for late-onset FGR which comprises the majority of all FGR cases [6]. Whilst other tests in late pregnancy such as measurement of placental hormones in maternal blood, e.g. placental growth factor (PlGF), human placental lactogen (hPL) have also been proposed to identify placental dysfunction, a recent systematic review and meta-analysis demonstrated such tests are insufficient to predict FGR pregnancies thus additional biomarkers are required [7].
MicroRNAs are short (~ 20 nucleotide) RNA molecules that post-transcriptionally regulate gene expression by mediating mRNA destabilisation and translational repression [8]. miRNAs are produced in all cells and tissues, including the placenta, where they regulate growth, differentiation, survival and vascular development [9–12]. Vascular maldevelopment and altered trophoblast turnover and function are features of placental dysfunction observed in FGR and pregnancies ending in stillbirth [13] suggesting that the underlying pathology may be a consequence of altered miRNA levels in the placenta. miRNAs have also been shown to be released from tissues, including the placenta, into the circulation either in complexes with proteins such as Argonaute 2 or contained within extracellular vesicles (EVs) [14–16]. In the maternal circulation, specific alterations to miRNA signatures are associated with pregnancy pathologies such as pre-eclampsia (PE), gestational diabetes and pre-term labour [17–20]. Furthermore, in pregnant mice, plasma miRNAs have been proposed to offer a non-hormonal biomarker for direct indication of placental dysfunction [21], suggesting that circulating miRNAs may also offer promise as biomarkers to predict FGR and associated placental dysfunction in human pregnancy.
Indeed, changes in circulating miRNAs in pregnancies complicated by FGR have been reported [22–25], however, none of the miRNAs currently identified have sufficient diagnostic accuracy for use as biomarkers for FGR. This is likely due to key differences between study design, for example the analysis of a limited number of preselected candidate miRNAs rather than global profiling of miRNAs [22, 26]; differences in gestational ages of when serum was drawn between studies [24, 25]; analysing miRNA content in isolated EVs in some studies versus total serum in others [24]; and analysing obstetric populations with mixed FGR subtypes including women with PE and FGR [23]. To date, no published studies have stratified for fetal sex, a variable known to affect both rates of FGR, stillbirth and placental miRNA expression [27–30]. Therefore, we constructed our study to perform unbiased assessment of miRNAs, instead of selecting for specific candidate miRNAs; to profile total serum rather than EV enriched serum; to use a well-defined FGR population with no other known obstetric/medical complications; to stratify for fetal sex; to use in silico analyses to identify miRNAs involved in placental regulatory networks; and to combine analysis of miRNA expression profiles with levels of a known marker of placental dysfunction. We hypothesised that using this methodology, we would identify miRNA profiles that were associated with both FGR and placental dysfunction and that these miRNA profiles may be dependent on the presence of a male or female fetus.
Methods
Participants and clinical samples
Study participants were pregnant women receiving antenatal care at St Mary’s Hospital, Manchester, recruited to the MFHRG Biobank (ethical approval: North West REC (08/H1010/55 + 5)). Written informed consent was obtained and detailed demographic and biophysical data were recorded. Exclusion criteria were multiple pregnancy, pre-term deliveries (< 36 weeks gestation), known fetal anomalies, pre-eclampsia or hypertension, maternal diabetes or any other co-existing obstetric/medical complications. Maternal serum samples were collected in the third trimester (27–42 weeks’ gestation). Briefly maternal venous blood drawn into serum gel tubes (Sarstedt, Numbrecht, Germany) was allowed to clot for 30 min and centrifuged at 3000 g for 10 min. Aliquots were immediately frozen at − 80 °C for analysis. After delivery, obstetric outcome data were collected and the individualised birthweight centile (IBC) calculated using Gestation Related Optimal Weight (GROW) Centile Calculator version 6.7 [31]. Babies were considered FGR when IBC < 5th centile. All FGR samples were matched on a 1:1 basis to appropriately grown (AGA; IBC ≥ 20 ≤ 80th centile) fetuses for the following maternal demographic and biophysical characteristics: infant sex (50:50 female to male), gestation at time of blood sampling, and gestation at delivery. Initial miRNA array profiling was performed on a subset of maternal serum samples drawn between 36–37 weeks’ gestation where primary selector was delivery of a baby with severe FGR (IBC < 3rd centile) (n = 4/group; Table 1). For all other analyses, a larger cohort of 62 participants’ samples (male n = 15, female n = 16; per group; Table 2), were included and FGR was defined as IBC < 5th centile. In both array samples and the larger cohort, only infant birthweight and IBC were significantly different by design between groups (array samples p < 0.05, larger cohort p < 0.0001, Mann–Whitney; Tables 1 and 2).
Table 1.
Demographic and obstetric outcome data for groups used for microRNA array profiling
| Category | AGA n = 4 |
FGR (IBC < 3rd) n = 4 |
|---|---|---|
|
Age (years) |
28 (27–35) |
30.8 (27–35) |
|
Ethnicity Caucasian:other Number (%) |
2:2 (50%:50%) |
2:2 (50%:50%) |
|
Non-smoker Number (%) |
4 (100%) |
4 (100%) |
|
BMI at booking kg/m2 |
24.45 (23–28) |
24.05 (22.3–28.9) |
|
Gestation at delivery (days) |
280 (272–284) |
278.5 (264–284) |
|
Birthweight (gramme) |
3207 (3040–3560) |
2225* (1880–2880) |
| IBC |
32.8 (21.5–46.8) |
0.95* (0.1–3.0) |
|
Male infant Number (%) |
2 (50%) |
2 (50%) |
|
Gestation of blood draw (days) |
252 (252–252) |
252 (252–262) |
Data are median and range unless stated otherwise. Participants were matched 1:1 on all demographic and obstetric factors, except for birthweight and individualised birthweight centile which were significantly different between groups (*p < 0.05 FGR vs AGA, Mann–Whitney test). Data analysed by Mann–Whitney (continuous data) or Fisher’s exact test (categorical data). AGA appropriate for gestational age, BMI body mass index, FGR fetal growth restriction, IBC individualised birthweight centile
Table 2.
Demographic and obstetric outcome data for study participants used for miRNA QPCR analyses stratified by infant sex
| Category | AGA male (n = 15) |
FGR male (n = 15) |
AGA female (n = 16) |
FGR female (n = 16) |
|---|---|---|---|---|
|
Age (years) |
24 (18–40) | 28 (18–40) | 29 (22–39) | 27.5 (20–36) |
|
Ethnicity Caucasian:other Number (%) |
10:5 (67%:33%) |
8:7 (53%:47%) |
7:9 (44%:56%) |
10:6 (63%:37%) |
|
Smoker Number (%) |
4 (27%) | 4 (27%) | 1 (6%) | 1 (6%) |
|
BMI at booking kg/m2 |
25.6 (21.8–29.8) |
24.9 (18.7–30.9) |
23.8 (19.0–28.6) |
25.0 (19.4–30.5) |
|
Gestation at delivery (days) |
282 (257–297) |
280 (260–286) |
280 (265–293) |
274 (256–293) |
|
Birthweight (g) |
3560 (3094–4640) |
2520**** a (1960–3190) |
3390 (2674–3856) |
2370***b (1660–2880) |
| IBC |
41 (20.7–81.3) |
1.54**** a (0.002–5) |
54.1 (20.7–81.1) |
0.69**** b (0.1–3.5) |
|
Gestation of blood draw (days) |
252 (192–287) |
252 (199–281) |
252 (197–291) |
257 (200–282) |
Data are median and range unless stated otherwise. Data analysed by Kruskal–Wallis with Dunn’s post hoc test (continuous data) or Fisher’s exact test (categorical data). AGA appropriate for gestational age, BMI body mass index, FGR fetal growth restriction, IBC individualised birthweight centile. ***p < 0.001, ****p < 0.0001; aFGR male compared to AGA male; bFGR female compared to AGA female; (Kruskal–Wallis with Dunn’s post hoc test)
RNA isolation
Total RNA was extracted from 200 µl of maternal serum using the miRCURY RNA Isolation Kit—Biofluid (Exiqon, Denmark) following the manufacturers’ protocol, with inclusion of an on column DNAse digestion step. For increased reproducibility, a carrier RNA (1 µg MS2 phage RNA; Roche, UK)) was added prior to extraction along with RNA spike-in controls (UniSp2, UniSp4, UniSp5 Exiqon, cat no. 203203) to monitor the technical quality of the RNA isolation, cDNA synthesis and the presence of PCR inhibitors. Samples contaminated by haemolysis where ΔCt miR-23a-3p–miR-451a was > 7 were excluded. Standard methods of RNA yield/purity are inaccurate due to carrier RNA, however we adhered to best practise guidelines by standardising input amounts based on isolating from identical starting volumes and using the same volume of purified RNA for all downstream processes [32].
miRNA array profiling
Isolated maternal serum RNA (19 µl) was reverse transcribed in 95-µl reactions using the miRCURY LNA™ Universal RT microRNA PCR system (Exiqon, Denmark) then diluted 50 × before analysis using the miRCURY LNA™ Universal RT microRNA PCR Human panel I + II Array plates (Exiqon, Denmark) which detects 752 individual miRNAs (list of all miRNAs profiled contained in Additional file 1). Sample quality control was monitored using spike-ins for RNA isolation efficiency (UniSp2, UniSp4, UniSp5) and cDNA synthesis control (UniSp6). Individual miRNA Ct values in each sample, were normalised to the global mean of a panel of 194 miRNA Ct values detected in the same sample (the 194 miRNAs were detected in all samples) as recommended by Mestdagh et al. [33]. The stability of the average of the 194 microRNAs was higher across samples/groups than any single miRNA in the dataset as measured by NormFinder software [34]. The coefficient of variation (CoV) for the array was 0.00024–0.048%. Detection of statistical differences between patient groups was performed using a paired t-test followed by Benjamini and Hochberg multiple testing correction, using the software R/Bioconductor [35]. miRNAs with fold-change increase or decrease of more than 1.2-fold (≤ 0.83 ≥ 1.2) change p < 0.05 were considered significant.
Bioinformatic analysis of altered miRNAs
A list of all 11 miRNAs altered in maternal serum from FGR pregnancies was uploaded to Ingenuity Knowledge Base (Ingenuity Pathway Analysis (IPA), Qiagen, Redwood City, www.qiagen.com/ingenuity) and miRNET version 2.0 [36], a freely available tool that can be accessed at https://www.mirnet.ca., which integrates data from 15 different miRNA databases (including miRbase, TarBase, miRTarBase, miRecords, miRanda, TransmiR2.0 miR2Disease, HMDD, ENCODE and ExoCarta) [36], to allow network-based visualisation of miRNA–target gene interactions coupled with functional analysis.
Networks of interacting genes associated with altered miRNAs were determined using miRNET and functional annotation and enrichment analysis of predicted target genes of altered miRNAs was performed using both miRNET (using Reactome, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses) and IPA. Network connectivity and functions with p < 0.05 were regarded as statistically significant.
Real-time QPCR assessment of serum miRNA levels
Altered miRNAs that had known sexually dimorphic expression in other tissues and/or previous associations with pregnancy pathologies as determined by performing literature searches using PubMed [37] (Table 3), were assessed by QPCR in a larger set of maternal serum samples (Table 2). Reverse transcription of isolated serum RNA (4 µl) was performed using miRCURY™ LNA Universal RT microRNA PCR system (Exiqon, Vedbaek, Denmark), with a UniSp6 RNA spike-in template included as an internal QC to monitor the reaction efficiency. QPCR was performed using ExiLENT SYBR Green master mix (Exiqon) and specific LNA-primer sets for 7 selected candidate miRNAs (miR-28-5p, miR-29c-3p, miR-301a-3p, miR-378a-3p, miR-409-3p, miR-454-3p and miR-526b-5p with target sequences listed in Additional file 2: Table S1) and reference dye 5-carboxy-X-rhodamine (ROX) in a Stratagene MX3000P real-time PCR machine. Thermocycling conditions consisted of polymerase activation for 10 min at 95 °C followed by 40 cycles each one consisting of 10 s at 95 °C then 1 min at 60 °C. The CoV for qPCR assays ranged from 0.2–2.4%. Data were normalised as 2−ΔCT using the geometric mean of two reference miRNAs (miR-23a-3p and miR-191-5p) determined to be most stably expressed between the sample groups as described by Vandesompele et al. [38].
Table 3.
MicroRNAs differentially expressed in maternal serum between FGR and uncomplicated pregnancies identified by microarray
| miRNA ID | SD | ddCq adverse/normal | Fold-change | Unadjusted p value | Sexual dimorphism | Reported association with pregnancy pathology |
|---|---|---|---|---|---|---|
|
miR-28-5p MIMAT0000085 |
0.641 | − 1.258 | − 2.4 | 0.029 | ↑ F non-pregnant plasma [39] | Not reported |
|
miR-200b-3p MIMAT0000318 |
0.169 | − 0.874 | − 1.8 | 0.012 | ↑ M cerebellum + colorectal mucosa [40] |
↑ Placental expression in PE with SGA [41] ↑ Myometrial expression in mouse model of PTB [42] |
|
miR-224-5p MIMAT0000281 |
0.426 | − 0.931 | − 1.9 | 0.022 | Not reported |
↑ Placental expression and maternal serum levels in PE [43] ↑ In trophoblasts exposed to hypoxia [44] |
|
miR-378a-3p MIMAT0000732 |
0.275 | − 0.491 | − 1.4 | 0.038 | ↑ F normal-term placenta [45] |
↑ Placental expression in PTB [46] ↓ Expression in EPL decidua [47] |
|
miR-526b-5p MIMAT0002835 |
0.420 | − 1.463 | − 2.8 | 0.026 | ↑ M cerebellum [40] |
Placental specific C19MC cluster |
|
miR-550a-3p MIMAT0003257 |
0.254 | − 0.429 | − 1.3 | 0.043 | Not reported | Not reported |
|
miR-29c-3p MIMAT0000681 |
0.089 | 0.332 | + 1.3 | 0.0051 |
↑ M plasma [39] ↑ F cerebellum [40] |
↑ Plasma levels in gestationally obese women [51] |
|
miR-301a-3p MIMAT0000688 |
0.364 | 0.588 | + 1.5 | 0.048 | ↑ M peripheral blood [40] | ↑ Maternal serum levels in PE [43] |
|
miR-409-3p MIMAT0001639 |
0.422 | 1.716 | + 3.3 | 0.02 | Plasma concentration + vely correlates with lung function in asthmatic boys [52] |
High placental expression (C14MC cluster) ↓ Maternal serum levels in PE [53] |
|
miR-454-3p MIMAT0003885 |
0.727 | 1.396 | + 2.6 | 0.031 |
↑ M peripheral blood [40] ↑ M FGR fetal heart baboon [54] |
↑ Placental expression in PE [55] |
|
miR-551a MIMAT0003214 |
0.303 | 0.988 | + 2.0 | 0.03 | Not reported | Not reported |
Fold-change in maternal serum expression for FGR < 3rd IBC compared to AGA matched controls profiled by microarray with unadjusted p value. Evidence of sexual dimorphism and/or association with pregnancy pathologies was determined by PubMed literature search accessed 30 May 2021. M male, F female, PE pre-eclampsia, SGA small for gestational age, PTB pre-term birth, EPL early pregnancy loss, FGR fetal growth restriction. ddCq = (dCq miRNA FGR) – (dCq miRNA AGA) as defined in Livak et al. [56]
Measurement of maternal serum human placental lactogen (hPL)
Levels of human placental lactogen (hPL), an established marker of placental endocrine function associated with a poor pregnancy outcome [57, 58], were assessed in matched serum samples which had been analysed for miRNA levels, where available (n = 59 from a total of 62 samples). Maternal serum samples were diluted 1:100 and assessed using a specific hPL ELISA (EIA-1283, DRG Diagnostics, Germany) according to manufacturer’s instructions. Briefly, 10-µl aliquots of samples and standards were applied to ELISA plates in duplicate and absorbance read at 450 nm using a Versamax plate reader and SoftMax Pro software (Molecular Devices, California, USA). The inter- and intra-assay variabilities were 4.3–9.9% and 2.6–5.5%, respectively. To identify altered miRNAs that were associated with placental dysfunction in FGR, maternal hPL concentrations and 2−ΔCT values for altered circulating miRNAs were plotted for each individual patient sample.
In silico analysis to identify validated targets of altered miRNAs
Target genes and previously reported associations between altered miRNAs and pregnancy complications associated with placental dysfunction, were identified using in silico analyses. Briefly, miRNA target genes were selected based on previous experimental validation or that were predicted target genes in more than 1 target prediction program (miRbase v.22 [59] miRTarBase Release 8.0 [60], TargetScan Release 7 [61] and miRDB [62]), based solely on the miRNA seed sequence. To further refine the list a literature search was performed using PubMed [37] to identify published studies supporting a functional role of the gene in the placenta or showing associations between gene target expression and pregnancy pathologies associated with placental dysfunction, including pre-term birth (PTB), FGR, PE, early pregnancy loss (EPL) or gestational diabetes (GDM).
Statistical analysis
Statistical analyses were performed using GraphPad Prism (version 8.4). Demographic and obstetric outcome data were analysed using Mann–Whitney U test for continuous data or Fisher’s exact test for categorical data. Data from QPCR analyses were analysed using Mann–Whitney U test for 2 groups or 2-way ANOVA for multiple groups. Correlations between miRNAs and hPL were analysed using Spearman’s correlation or linear regression. Results were considered significant if p < 0.05. Sample sizes were dictated by power calculations based on previous studies [63] or in the case of validation studies, were determined using a power calculation at 0.95 level of confidence and a p < 0.05 based on the microarray values.
Results
miRNA profiling of maternal serum samples in FGR and uncomplicated pregnancies
Analysis of microarrays revealed 11 miRNAs that were altered between maternal serum samples from FGR (IBC < 3rd) and uncomplicated pregnancies at 36 weeks’ gestation (fold-change of ≤ 0.83 ≥ 1.2; p < 0.05; Fig. 1). Of these, five miRNAs: miR-29c-3p, 409-3p, 551a, 454-3p and 301a-3p, were increased in FGR and six miRNAs: miR-200b-3p, 224-5p, 526b-5p, 28-5p, 378a-3p and 550a-3p, had reduced levels in FGR serum when compared to AGA controls. Eight of the altered miRNAs: miR-28-5p, 200b-3p, 378a-3p, 29c-3p, 526b-5p, 301a-3p, 409-3p and 454-3p, had previously reported sex differences in expression and /or known association with pregnancy pathologies (Table 3).
Fig. 1.
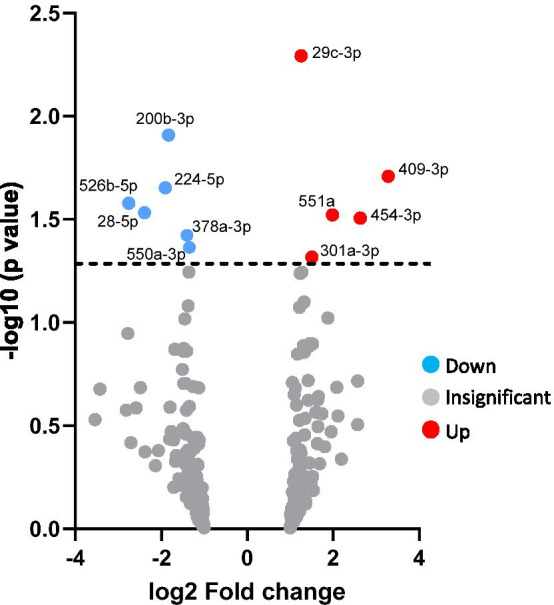
Volcano plot of differentially expressed microRNAs in maternal serum of FGR versus normal pregnancies. Log2 (fold-change) values for detected miRNAs were plotted against − log10 of the unadjusted p-value. Significantly up-regulated genes shown in red, significantly down-regulated genes shown in blue (p < 0.05). Dotted line represents p = 0.05
Functional enrichment analysis of altered miRNAs
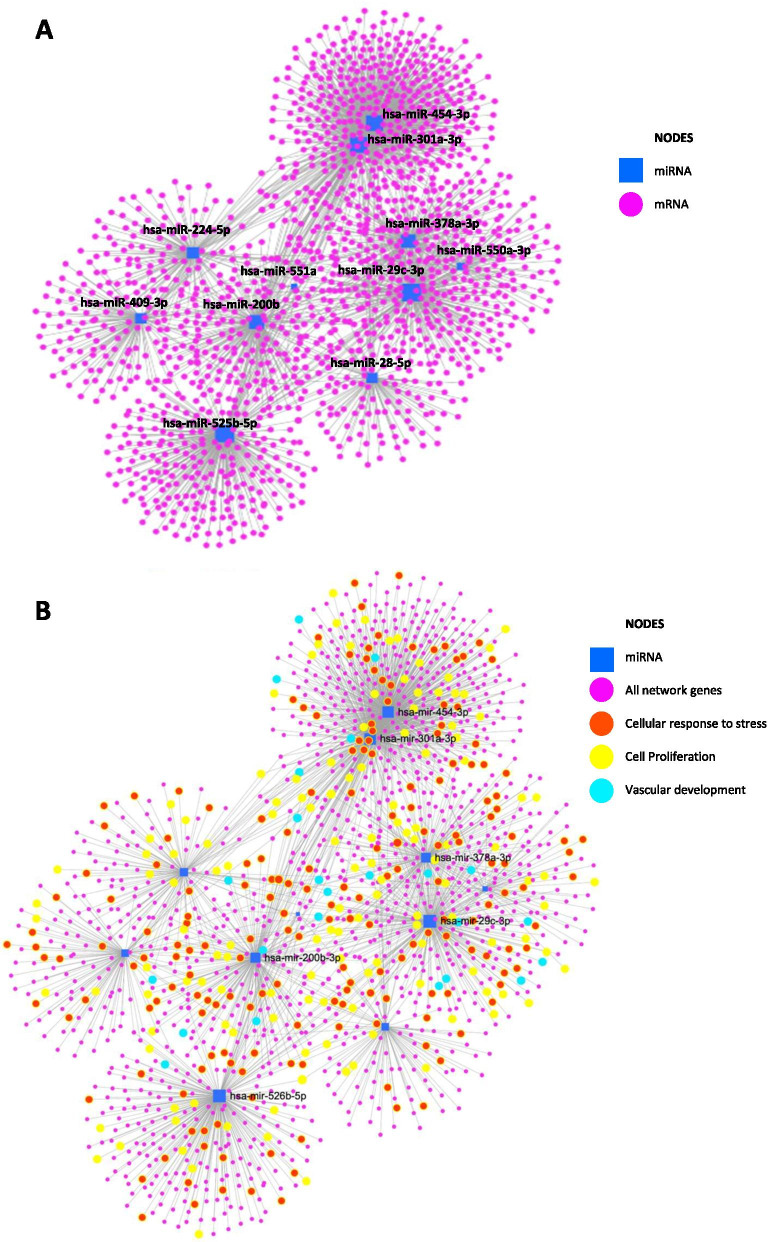
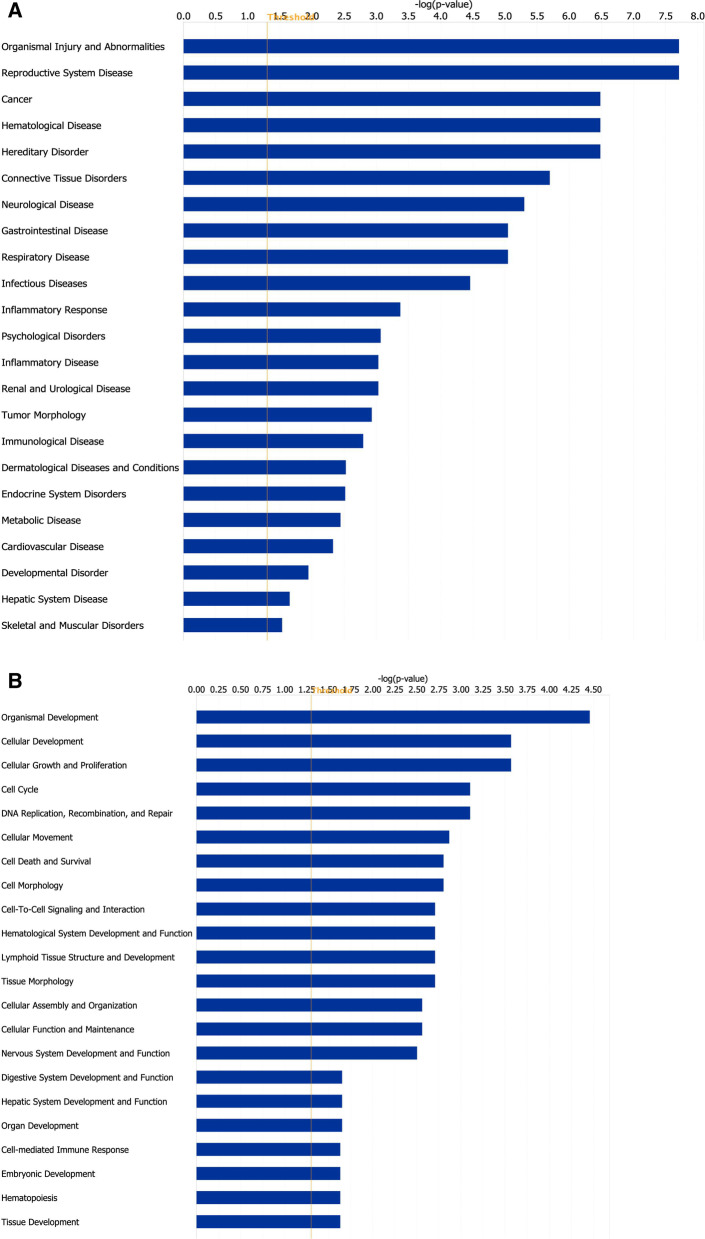
Network analysis, using miRNET, revealed that all 11 miRNAs were present in an interacting gene network (Fig. 2A) enriched in predicted target genes (Fig. 2B) associated with cellular response to stress (77 node genes; Adj p value = 4.9 × 10–6), cell proliferation (111 node genes; Adj p value = 0.00012) and vascular development (97 node genes; Adj p value = 0.00037). Ingenuity pathway analysis (Fig. 3) also revealed a predicted functional effect of altered miRNAs in pathways associated with placental development including cellular growth and proliferation, cellular movement, cell death and survival and cell-to-cell signalling and interaction.
Fig. 2.
Interacting mRNA networks of all altered miRNAs. A, B A list of all 11 miRNAs altered in FGR was uploaded to miRNET and A networks of interacting genes (pink circles) associated with altered miRNAs (blue square) were determined. B Functional enrichment analysis of network genes was performed using Reactome, GO and KEGG. Key functional effects associated with the network were found in pathways associated with cellular response to stress (red; 77 node genes; Adj p = 4.9 × 10–6), cell proliferation (yellow; 111 node genes; Adj p value = 0.00012) and vascular development (turquoise; 97 node genes; Adj p value = 0.00037)
Fig. 3.
Functional enrichment analysis of miRNAs and predicted target genes. A list of the 11 miRNAs altered in FGR was uploaded to Ingenuity Pathway Analysis and A predicted diseases and disorders and B predicted molecular and cellular functions and functions associated with physiological system development were determined from miRNAs and their experimentally validated and predicted targets. Orange line represents significance threshold value of − log p value = 1.5
Sexually dimorphic maternal circulating miRNAs associated with FGR pathology
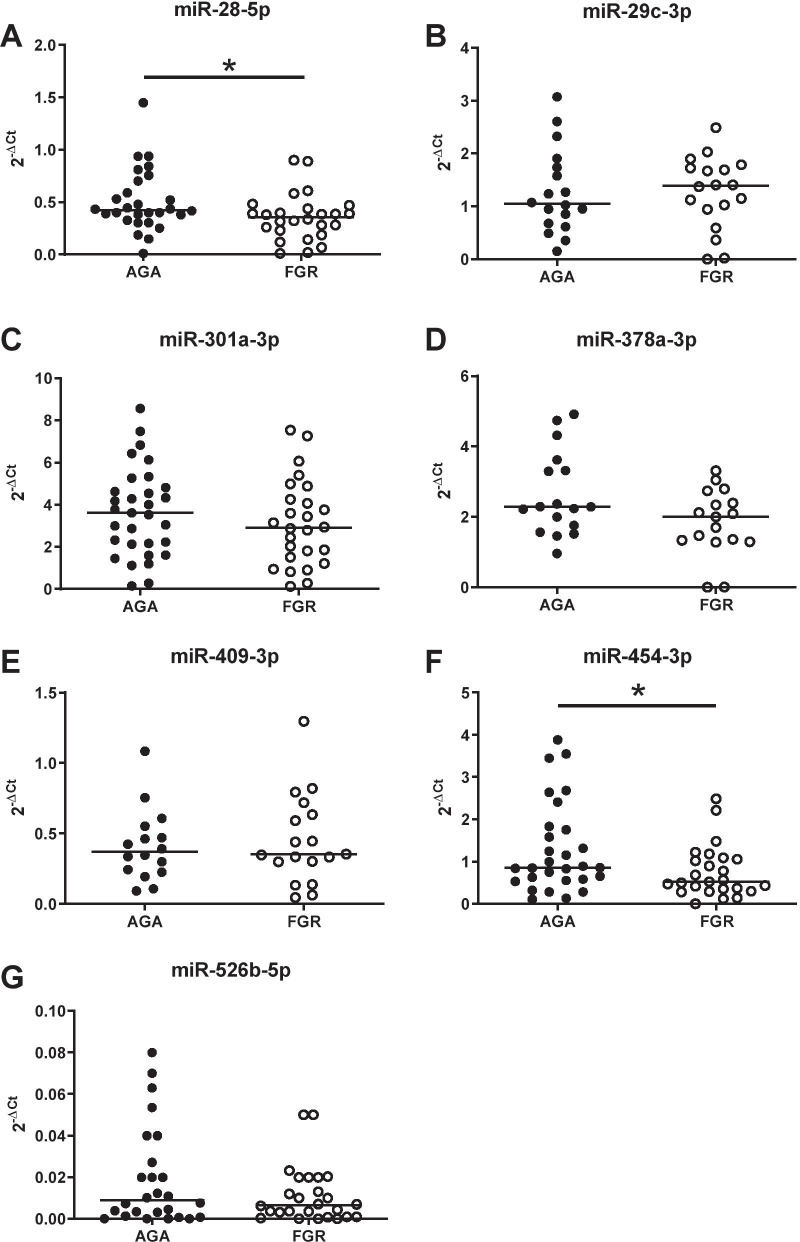
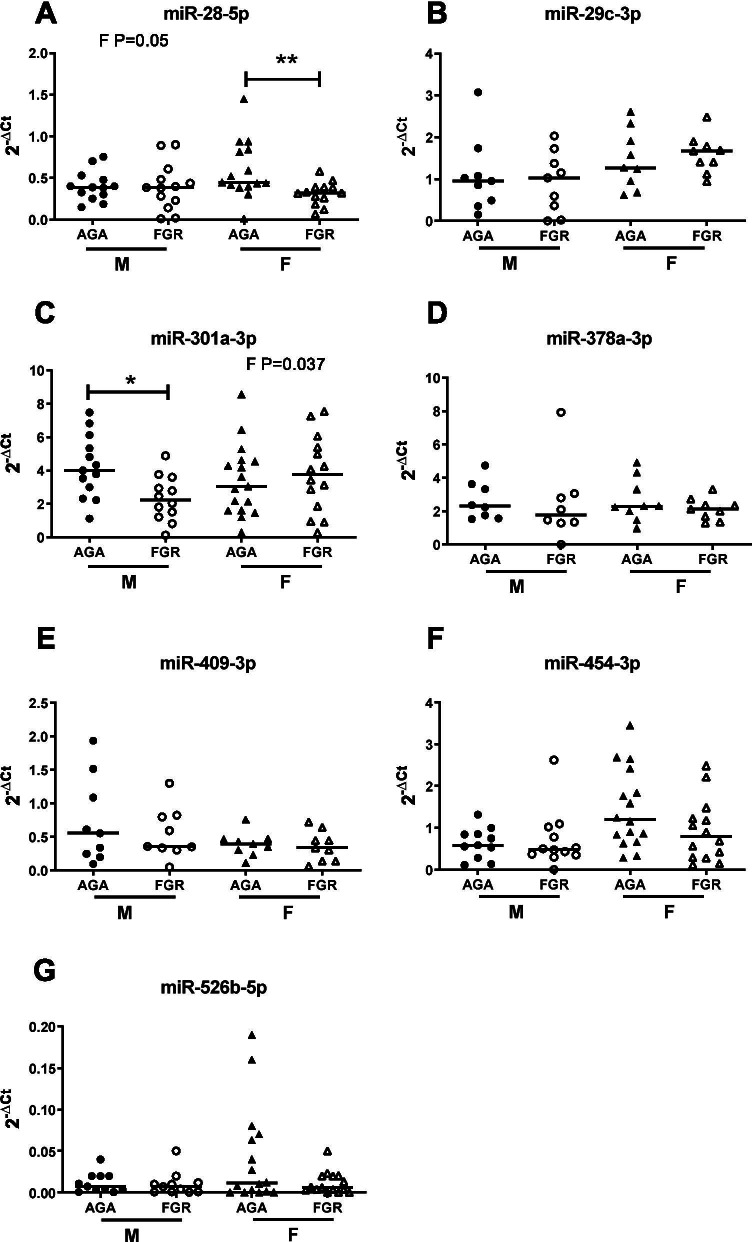
Subsequent analysis in a larger cohort of patients by QPCR revealed that only miRNA-28-5p and miR-454-3p levels were significantly reduced in FGR compared to normal pregnancies (p = 0.028 and p = 0.045; Fig. 4A and F, respectively). miR-378a showed a trend for lower levels in FGR pregnancies, but this did not reach significance (p = 0.08; Fig. 4D). Four remaining miRNAs (miR-29c-3p, miR-301a-3p, miR-409-3p and miR-526b-5p; Fig. 4B, C, E, G), were unaltered between groups. However, male fetuses have poorer outcomes and are at higher risk of FGR and stillbirth [29, 64] than their female counterparts [29, 64]. Whilst mechanisms remain unclear, sex-specific differences in miRNA expression between placentas of male and female fetuses have been reported [27, 30]. When initial miRNA array data were analysed by sex, rather than pathology, sex differences between four miRNAs: miR-29c-3p, miR-32-5p, miR-136-5p (all increased in male samples) and miR-520 h (increased in female), were observed (Table 4). Whilst only one of these miRNAs, miR-29c-3p, was found to also be altered in FGR miRNA array data (Table 3), it suggested that there was potentially sexual dimorphism in miRNA levels in FGR pregnancies. QPCR data obtained for the seven miRNAs assessed in the larger cohort of patient samples was stratified by fetal sex to determine if there were sexual dimorphic patterns for these serum miRNAs in FGR (Fig. 5). Using 2-way ANOVA miR-29c-3p (p = 0.031; Fig. 5B), miR-526b-5p (p = 0.0224; Fig. 5G) and miR-454-3p (p = 0.014; Fig. 5F) were found to have sexually dimorphic expression independent of fetal growth. Additionally, altered miR-301a-3p and miR-28-5p levels in FGR pregnancies were sex-specific; specifically miR-301a-3p was 45% lower in FGR pregnancies when the fetus was male (4.22 ± 1.87 vs 2.33 ± 1.35 Male AGA vs FGR (mean ± standard deviation); interaction (F) p = 0.037; Fig. 5C) but not female, and miR-28-5p was reduced by 28% in FGR pregnancies with a female fetus (0.59 ± 0.34 vs 0.32 ± 0.14 AGA vs FGR (mean ± standard deviation); interaction (F) p = 0.05; Fig. 5A), but not when the fetus was male. No other miRNAs assessed showed sexually dimorphic patterns.
Fig. 4.
Q-PCR validation of candidate microRNAs in maternal serum in uncomplicated and FGR pregnancies. qPCR was performed on individual microRNAs isolated from maternal serum of women with appropriately grown (AGA; IBC 20–80th) or growth-restricted (FGR; IBC < 5th) infants using specific primers for A miR-28-5p, B miR-29c-3p, C miR-301a-3p, D miR-378a-3p, E miR-409-3p, F miR-454-3p and G miR-526b-5p. Data were normalised to 2 reference miRNAs and expressed as 2−ΔCt. Individual data points shown (n = 18–31/group); line represents the median. *p < 0.05 Mann–Whitney U test
Table 4.
Differentially expressed microRNAs between female and male serum samples identified by microarray
| miRNA ID | SD F | SD M | Average dCq F |
Average dCq M |
Fold-change | p-value | Sexual dimorphism | Reported association with pregnancy pathology |
|---|---|---|---|---|---|---|---|---|
|
hsa-miR-520 h MIMAT0002867 |
0.40 | 0.66 | − 2.1 | − 3.5 | 2.6 | 0.018 |
↑ In plasma EVs in GDM pregnancies [19] ↑1st trimester plasma in women who develop PE [65] ↑ Placental expression in PTB [66] ↓ Placental expression in FGR [49] |
|
|
hsa-miR-29c-3p MIMAT0000681 |
0.28 | 0.17 | 2.0 | 2.5 | − 1.4 | 0.038 |
↑ M plasma [39] ↑ F cerebellum [40] |
↑ Plasma levels in gestationally obese women [51] |
|
hsa-miR-32-5p MIMAT0000090 |
0.37 | 0.15 | 1.2 | 1.8 | − 1.5 | 0.039 | ↓Placental expression in PE [67] | |
|
hsa-miR-136-5p MIMAT0000448 |
0.69 | 0.6 | − 2.5 | − 1.4 | − 2.2 | 0.046 |
↑ In plasma EVs in GDM pregnancies [19] ↑Expression in chorionic membranes in PTL [68] |
When comparing the F group to the M group (combined FGR and AGA) using a t-test, 4 microRNAs were found to be differentially expressed using a cutoff of p-value < 0.05. (n = 4/group). Evidence of sexual dimorphism and/or association with pregnancy pathologies was determined by PubMed literature search accessed 30 May 2021. SD standard deviation, F female, M male, dCq = (Cq target miRNA – Cq reference miRNAs), EVs extracellular vesicles, GDM gestational diabetes mellitus, PE pre-eclampsia, PTB pre-term birth, FGR fetal growth restriction, PTL pre-term labour
Fig. 5.
Effect of fetal sex on microRNA levels in maternal serum in uncomplicated and FGR pregnancies. qPCR was performed on individual microRNAs isolated from maternal serum of women with appropriately grown (AGA; IBC 20–80th) or growth-restricted (FGR; IBC < 5th) infants using specific primers for A miR-28-5p, B miR-29c-3p, C miR-301a-3p, D miR-378a-3p, E miR-409-3p, F miR-454-3p and G miR-526b-5p. Data were normalised to 2 reference miRNAs and expressed as 2−ΔCt. Data were stratified into male (n = 15/group) and female (n = 16/group). Individual data points shown, line represents the median. 2-way ANOVA with linear step-up multiple comparison test. Interaction (F) between the groups was considered significant when p < 0.05. Kruskal–Wallis followed by Dunn’s post hoc analysis was used to determine difference between individual groups; *p < 0.05, **p < 0.01
Serum miRNAs correlate with hPL, a marker of placental dysfunction
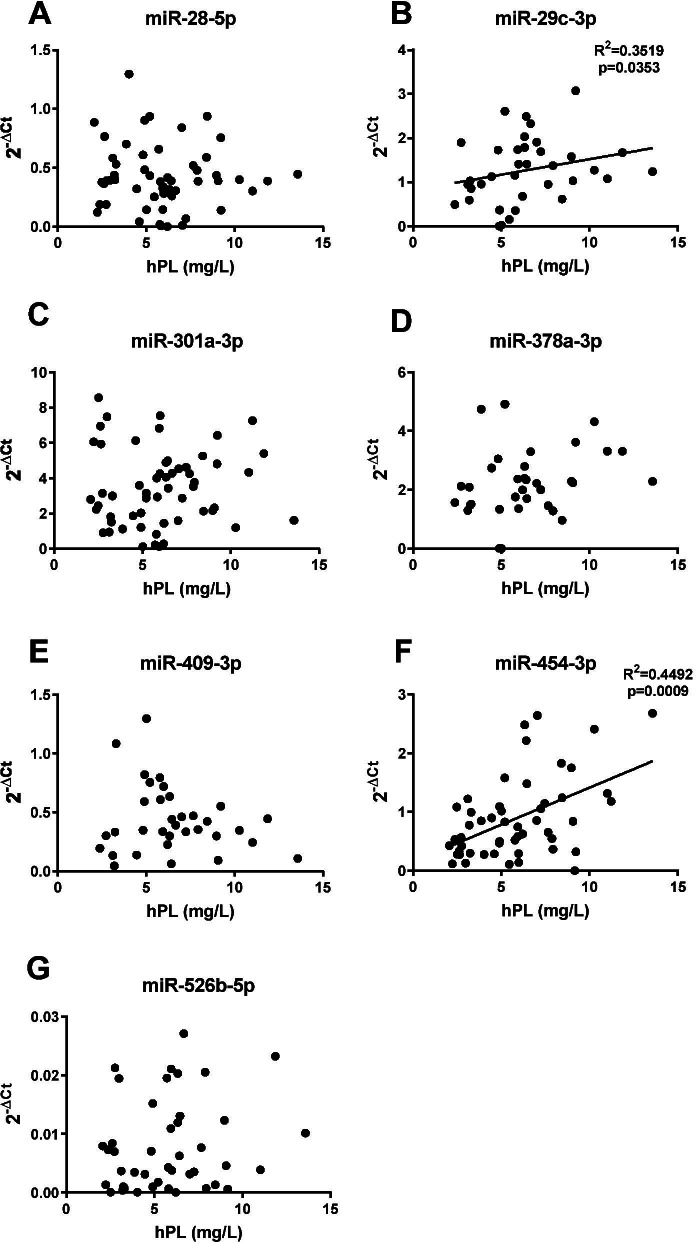
To determine whether levels of serum miRNAs altered in FGR could be indicative of placental dysfunction, the relationship between altered miRNA levels and maternal concentrations of hPL, an established marker of placental endocrine function in late pregnancy which is lower in pregnancies complicated by FGR [57, 58, 69], were assessed (Fig. 6). Positive correlations were found for miR-454-3p (p < 0.001, r = 0.449; Fig. 6F) and miR-29c-3p (p < 0.05, r = 0.352; Fig. 6B), but no associations were found for other miRNAs.
Fig. 6.
Relationship between circulating microRNAs and human placental lactogen (hPL) in maternal serum. Correlation between serum miRNA expression measured by qPCR and hPL a hormone marker of placental dysfunction detected by ELISA for A miR-28-5p, B miR-29c-3p, C miR-301a-3p, D miR-378a-3p, E miR-409-3p, F miR-454-3p and G miR-526b-5p. Positive correlations with hPL were detected for B miR-29c-3p (r2 0.3519, p < 0.05) and F miR-454-3p (r2 0.449, p < 0.001). Individual data points shown (n = 34–56), line represents the median. Spearman rank correlations
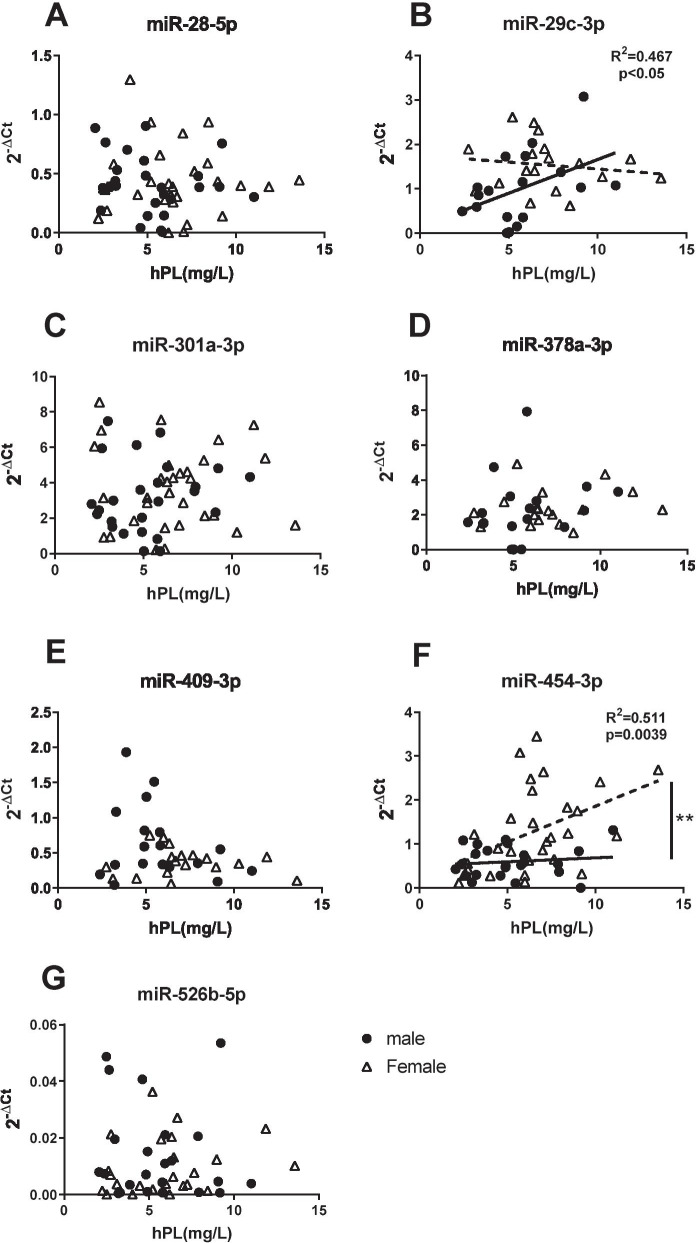
Further stratification of the data, based on fetal sex, revealed that the positive association between miR-454-3p and hPL, was only in pregnancies with a females fetus (r = 0.511, p < 0.01; Fig. 7F), whilst miRNA-29c-3p was only correlated with hPL when there was a male fetus (r = 0.467, p = < 0.05; Fig. 7B). No sex-specific associations with hPL were detected for miRNAs 28-5p, 301a-3p, 378-3p, 409-3p or miR-526b-5p.
Fig. 7.
Effect of fetal sex on correlation between microRNAs and hPL in maternal serum. Correlation between maternal serum miRNA expression from male or female pregnancies measured by qPCR and hPL a hormone marker of placental dysfunction detected by ELISA for A miR-28-5p, B miR-29c-3p, C miR-301a-3p, D miR-378a-3p, E miR-409-3p, F miR-454-3p and G miR-526b-5p. Positive correlations with hPL were detected for B miR-29c-3p (r2 (r = 0.467, p = < 0.05) in males only and F miR-454-3p (r2 r = 0.511, p < 0.010) in females only. Significant difference detected in intercept/elevation for F miR-454-3p (p < 0.01, linear regression). Individual data points shown (n = 17–27 males, n = 18–32 females/group), line represents the median. Spearman rank correlations
Predicted functional roles of altered miRNAs
For a biomarker to be clinically useful, it must reliably be associated with the underlying pathology of a disease [70]. Circulating miRNAs have recently been reported to enter tissues where they interact with target mRNA and affect cellular function [71], thus, in addition to having potential biomarker utility, altered circulating miRNAs may also be causative for underlying pathologies. To assess whether the maternal serum miRNAs have the potential to contribute to placental dysfunction associated with FGR, in silico analysis was performed to determine experimentally validated downstream target genes of the sexually dimorphic miRNAs that were altered in maternal serum and/or correlating with hPL in FGR pregnancies.
Experimentally validated downstream targets for these miRNAs included nutrient transporters, cell cycle or apoptosis regulators and growth/angiogenic factors (Table 5). Many of these are altered in FGR pregnancies, e.g. vascular endothelial growth factor A (VEGFA) [72], pregnancy-associated plasma protein A (PAPPA) [73], insulin-like growth factor 1 (IGF-I) [74], insulin-like growth factor 2 receptor (IGF2R) [75], large neutral amino acid transporter (LAT1) [76], apoptosis regulators Bcl-2-like protein 11 (BCL2L11), tumour protein p53 (TP53), and B-cell lymphoma 2(BCL2) [77, 78] and the taurine transporter SLC6A6 [79].
Table 5.
Bioinformatic analyses of miRNAs altered in maternal serum or correlating with hPL in FGR pregnancies
| miRNA ID | FGR altered miRNA (QPCR data) |
Sexual dimorphism (Current dataset) |
Correlation with biochemical marker of placental dysfunction (hPL) | Selected gene targets Evidence level: -Strong (bold) -Weak (unbolded) -Predicted (italics) |
|---|---|---|---|---|
|
miR-28-5p MIMAT0000085 |
Yes | ↓in FGR females (QPCR) | No |
CDKN1A, MAPK1, IGFI, RAP1B, MAD2L1, SLC7A5, CCND3 PAPPA, CASP3 |
|
miR-29c-3p MIMAT0000681 |
No |
↑ males in microarray + ve correlation [hPL] males |
Yes R2 = 0.467 p < 0.05 |
CDK6, BCL2, MMP2, PTEN, AKT3, LAMC1, LAMC2, DNMT3A, COL1A, COL3A, FBA, FBB, FBG, PDGFRB VEGFA |
|
miR-301a-3p MIMAT0000688 |
Yes | ↓ in FGR males (QPCR) | No | BCL2L11, PAI-1, RUNX3, PTEN, NKRF, TGFβR2, DNMT1, ESR1, XIAP, IGF2R, PUMA, SLC6A6, IGF1, PPARG |
|
miR-454-3p MIMAT0003885 |
Yes | + ve correlation [hPL] females |
Yes R2 = 0.511 p < 0.01 |
SMAD4, TGFβR2, ALK7, CXCL12, TP53, IGF2R, CCND2, MAPK1, XIAP, PARP1, SLC38A2, SLC6A6, IGF1, PPARG, DICER1, H19 |
Potentially relevant gene targets selected from literature-based databases. Bold = experimentally validated as a target using STRONG methods (reporter assay, Western blot, qPCR); unbold = experimentally validated as a target using WEAK methods (microarray, NGS, pSILAC, other); italics = predicted target but not experimentally validated. FGR fetal growth restriction, hPL human placental lactogen. Correlations analysed using Spearman test
Discussion
The current study identified 11 miRNAs that were altered in maternal serum of women that gave birth to a severely FGR infant compared to those delivering AGA babies. In silico analysis of the altered miRNAs revealed that all were present in a gene regulatory network with known roles in placenta dysfunction associated with FGR. Assessment of the altered miRNAs in a larger cohort of patients revealed that the association between levels of some maternal circulating miRNAs and FGR was dependent on fetal sex. Similarly, the association between maternal circulating miRNAs and hPL was fetal sex-dependent.
To date, there have been limited studies of maternal serum miRNA in pregnancies complicated by FGR and existing data are not in agreement [22–25, 49]. In our current investigation, there was no overlap with any of the miRNAs detected in the previous studies even though they were all present in the array used (Fig. 8). However, our approach differed from prior studies in several ways including: methodology [22, 24], classification of FGR with our study focused on later onset FGR and examining the influence of fetal sex. We initially focussed on later onset FGR because the majority of cases of FGR are late-onset and our current ability to detect FGR and fetal compromise is poorer [6, 23, 25, 80]. Although our study was designed to address potential limitations of previous studies, restricting the gestational age window of samples in the initial screen may have excluded some miRNAs seen in earlier studies. Differences between samples and study designs likely account for the lack of overlapping miRNAs found in any published studies.
Fig. 8.
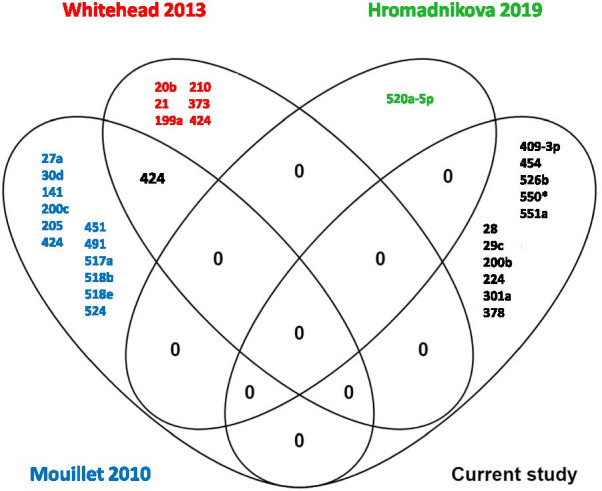
Venn diagram showing the overlapping microRNAs identified from studies comparing maternal serum from uncomplicated and FGR pregnancies. Current study compared with Mouillet et al. [22], Whitehead 2013 [23] and Hromadnikova 2019 [24]
Sex-specific differences in miRNA expression between placentas of male and female fetuses may also contribute to the observed differences between published studies. We found sex differences in miR-526b, miR-454-3p and miR-29c-3p in maternal serum, independent of fetal growth. miR-454-3p and miR-29c-3p are also expressed in a sex-dependent manner in other tissues [39, 40, 54] so accounting for fetal sex when considering the contribution of these miRNAs to disease processes is of obvious importance.
Here, we also report that two miRNAs, miR-28-5p and miR-301a-3p are altered in FGR pregnancies only when there is a female or male baby, respectively. Whilst there are no reports of sex-differences in miR-28-5p during pregnancy, the reduction of miR-301a-3p only in FGR pregnancies with a male fetus, is consistent with previous reports of sexual dimorphic changes in miR-301a-3p in maternal serum in pre-eclampsia (PE) [43]. It is possible therefore, that miR-301a-3p and miR-28-5p could be used to predict FGR specifically in pregnancies with male or female babies. Caution should be taken however, since the heterogeneity of FGR pathophysiology means these miRNAs may only be useful in predicting FGR in the subpopulation of FGR cases in which they were originally identified, that is where the infant is < 3rd centile at birth. Prospective studies in a large independent cohort are required to assess this.
In addition to the potential biomarker utility of these miRNAs, in silico analyses reveal potential functional roles for the altered miRNAs. These miRNAs are enriched in functional networks associated with cellular response to stress, cell proliferation and vascular development. Since these are all hallmarks of placental dysfunction in FGR pregnancies [13, 81, 82], it is possible that they may contribute to the aetiology of FGR by impacting on placental function. Indeed placental sexual dimorphism is well documented [28, 30, 45, 83, 84] with male fetuses having increased susceptibility to adverse perinatal outcomes [64, 85, 86]. It would therefore be interesting to assess whether the sexually dimorphic profiles observed for serum miRNAs are also reflected in placental tissue, and whether the miRNAs have functional roles in the placenta. Indeed, the relationship between some of the altered miRNAs and hPL—a marker of placental dysfunction together with a functional study showing that overexpression of miR-454-3p in trophoblast increases proliferation and invasion whilst reducing apoptosis [55], would support this hypothesis. Roles for the altered miRNAs in other tissues combined with information on their validated targets also provides further support for roles of these miRNAs in regulation of placental function.
IGF-I, a critical regulator of normal human placental development [87, 88], is a validated gene target of miR-28-5p [89] and is strongly associated with FGR in both animal models [90] and human pregnancies [91]. In a study of healthy pregnancies, IGF-I concentrations in cord blood were found to be higher in women carrying a female infant [92]. Target genes of miR-29c-3p and miR-301a-3p in cell death and survival pathways [93–95] may be relevant to FGR as these are known to be altered in FGR pregnancies [77, 96, 97], however further studies are required to assess this.
Perspectives and significance
This manuscript contributes to an increasing understanding of miRNAs in the maternal circulation and how these may be used to predict differing susceptibility of male and female fetuses to FGR and stillbirth. These studies have identified sexually dimorphic miRNA profiles in maternal circulation in pregnancies, specifically those that result in FGR infants. Further studies to establish where these miRNAs originate and investigate potential roles for these miRNAs in the placenta may provide greater understanding of the pathogenesis of placental dysfunction. In addition, it is possible that sex-specific altered circulating miRNAs could contribute to the differences in frequency of pregnancy complications such as increased incidence of term pre-eclampsia and GDM and stillbirth in women carrying a male fetus [86, 98]. At present, fetal sex is not considered in the management of pregnancy conditions, thus the management of cases included here and the observation of increased frequency of complications will not have altered depending upon fetal sex.
Identifying novel predictive biomarkers that can be used in conjunction with existing techniques is important for better predicting late-onset FGR and stillbirth and reducing associated morbidity and mortality [99]. Existing algorithms to detect late-onset placental dysfunction (UAD impedance and PlGF/sFlt1 ratio in maternal serum) [100] may be strengthened by inclusion of one or more of these miRNAs or combining miRNAs with recently identified circulating mRNA markers such as EMP1, which is increased in women who subsequently suffered a stillbirth [101]. Further studies on an extended sample set are necessary to test this hypothesis. Including fetal sex in these algorithms (which would be necessary to interpret the miRNA data) may be challenging as fetal sex is not always accurately determined on ultrasound, parents may not wish to know the sex of their baby and in some states (e.g. India), antenatal determination of fetal sex is prohibited. Nevertheless, the clinical utility of combinations of biomarkers (including fetal sex) should be explored, and if deemed effective in reducing fetal mortality and morbidity, fetal sex could be included.
Conclusions
Our study has identified a distinct profile of circulating miRNAs in women with FGR infants < 3rd centile. Two of these miR-28-5p and miR-454-3p, were also altered in a larger population of pregnancies with infants < 5th centile. Further analyses identified sexually dimorphic changes in some of the altered miRNAs; both miR-28-5p and miR-301a-3p to be altered in maternal serum in FGR pregnancies according to infant sex and miR-29c-3p and miR-454-3p showed sexually dimorphic relationship to placental dysfunction. Furthermore, these altered miRNAs are linked to placental regulatory gene networks. Further investigations to determine the source of these miRNAs and their relationship to placental dysfunction will lead to a better understanding of the relationship between circulating miRNAs and placental dysfunction in FGR and may enable the development of future treatments for placental dysfunction. Ultimately, further studies are required to determine the potential for these altered miRNAs to provide potential biomarkers to predict differing susceptibility of male and female fetuses to FGR and stillbirth.
Supplementary Information
Additional file 1. List of miRNAs in Exiqon QPCR array Human panels I + II.
Additional file 2: Table S1. Target sequences of LNA microRNA primer sets utilised for microRNA Q-PCR.
Acknowledgements
The authors would like to thank the midwives and Biobank technicians of St. Mary’s Hospital, Manchester, UK, for their help with recruitment and sample collection, and the women who donated their tissue for research. This study was facilitated by the Great Manchester Local Clinical Research Network.
Authors' contributions
B.C.B, R.L.J, K.F and A.E.H. designed research; B.C.B, S.L and I.L conducted research; B.C.B, S.L, K.F and R.L.J analysed data; B.C.B, R.L.J. and K.F wrote the paper; B.C.B, R.L.J. and K.F. had primary responsibility for final content. All authors read and approved the final manuscript.
Funding
BCB, SL and AEPH were supported by Tommy’s charity; KF is supported by a Medical Research Council New Investigator Research Grant [REF:MR/R023166/1].
Availability of data and materials
The microarray data generated during the current study has been deposited in NCBI's Gene Expression Omnibus and is accessible through GEO Series accession number GSE188186 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE188186).
Declarations
Ethics approval and consent to participate
Ethical approval was obtained from the local ethical committee North West REC (08/H1010/55 + 5) and all participants gave written informed consent.
Consent for publication
All authors have given their consent for publication of the manuscript in Biology of Sex Differences.
Competing interests
The authors declare no commercial or financial competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Karen Forbes and Rebecca L. Jones are joint last authors
Contributor Information
Bernadette C. Baker, Email: bernadette.baker@manchester.ac.uk
Sylvia Lui, Email: sylvia.lui@manchester.ac.uk.
Isabel Lorne, Email: ilorne17@gmail.com.
Alexander E. P. Heazell, Email: alexander.heazell@manchester.ac.uk
Karen Forbes, Email: K.A.Forbes@leeds.ac.uk.
Rebecca L. Jones, Email: Rebecca.Lee.Jones@manchester.ac.uk
References
- 1.Gordijn SJ, Beune IM, Thilaganathan B, Papageorghiou A, Baschat AA, Baker PN, et al. Consensus definition of fetal growth restriction: a Delphi procedure. Ultrasound Obstet Gynecol. 2016;48(3):333–339. doi: 10.1002/uog.15884. [DOI] [PubMed] [Google Scholar]
- 2.Heazell AE, Worton SA, Higgins LE, Ingram E, Johnstone ED, Jones RL, et al. IFPA Gabor Than Award Lecture: recognition of placental failure is key to saving babies' lives. Placenta. 2015;36(Suppl 1):S20–S28. doi: 10.1016/j.placenta.2014.12.017. [DOI] [PubMed] [Google Scholar]
- 3.Sibley CP, Turner MA, Cetin I, Ayuk P, Boyd CA, D'Souza SW, et al. Placental phenotypes of intrauterine growth. Pediatr Res. 2005;58(5):827–832. doi: 10.1203/01.PDR.0000181381.82856.23. [DOI] [PubMed] [Google Scholar]
- 4.Gardosi J, Kady SM, McGeown P, Francis A, Tonks A. Classification of stillbirth by relevant condition at death (ReCoDe): population based cohort study. BMJ. 2005;331(7525):1113–1117. doi: 10.1136/bmj.38629.587639.7C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Statistics OFN. Births in England and Wales: 2018. 2019.
- 6.Figueras F, Caradeux J, Crispi F, Eixarch E, Peguero A, Gratacos E. Diagnosis and surveillance of late-onset fetal growth restriction. Am J Obstet Gynecol. 2018;218(2S):S790–S802 e1. doi: 10.1016/j.ajog.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Heazell AE, Hayes DJ, Whitworth M, Takwoingi Y, Bayliss SE, Davenport C. Biochemical tests of placental function versus ultrasound assessment of fetal size for stillbirth and small-for-gestational-age infants. Cochrane Database Syst Rev. 2019;5:CD012245. doi: 10.1002/14651858.CD012245.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forbes K, Farrokhnia F, Aplin JD, Westwood M. Dicer-dependent miRNAs provide an endogenous restraint on cytotrophoblast proliferation. Placenta. 2012;33(7):581–585. doi: 10.1016/j.placenta.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 10.Cvitic S, Strutz J, Appel HM, Weiss E, Brandl WT, Thuringer A, et al. Sexual dimorphism of miRNA signatures in feto-placental endothelial cells is associated with altered barrier function and actin organization. Clin Sci (Lond) 2020;134(1):39–51. doi: 10.1042/CS20190379. [DOI] [PubMed] [Google Scholar]
- 11.Ali A, Bouma GJ, Anthony RV, Winger QA. The role of LIN28-let-7-ARID3B pathway in placental development. Int J Mol Sci. 2020;21:10. doi: 10.3390/ijms21103637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayder H, O'Brien J, Nadeem U, Peng C. MicroRNAs: crucial regulators of placental development. Reproduction. 2018;155(6):R259–R271. doi: 10.1530/REP-17-0603. [DOI] [PubMed] [Google Scholar]
- 13.Sun C, Groom KM, Oyston C, Chamley LW, Clark AR, James JL. The placenta in fetal growth restriction: what is going wrong? Placenta. 2020;96:10–18. doi: 10.1016/j.placenta.2020.05.003. [DOI] [PubMed] [Google Scholar]
- 14.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108(12):5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao F, Cheng L, Shao Q, Chen Z, Lv X, Li J, et al. Characterization of serum small extracellular vesicles and their small RNA contents across humans, rats, and mice. Sci Rep. 2020;10(1):4197. doi: 10.1038/s41598-020-61098-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morales-Prieto DM, Chaiwangyen W, Ospina-Prieto S, Schneider U, Herrmann J, Gruhn B, et al. MicroRNA expression profiles of trophoblastic cells. Placenta. 2012;33(9):725–734. doi: 10.1016/j.placenta.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 17.Li H, Ge Q, Guo L, Lu Z. Maternal plasma miRNAs expression in preeclamptic pregnancies. Biomed Res Int. 2013;2013:970265. doi: 10.1155/2013/970265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guarino E, Delli Poggi C, Grieco GE, Cenci V, Ceccarelli E, Crisci I, et al. Circulating microRNAs as biomarkers of gestational diabetes mellitus: updates and perspectives. Int J Endocrinol. 2018;2018:6380463. doi: 10.1155/2018/6380463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gillet V, Ouellet A, Stepanov Y, Rodosthenous RS, Croft EK, Brennan K, et al. miRNA profiles in extracellular vesicles from serum early in pregnancies complicated by gestational diabetes mellitus. J Clin Endocrinol Metab. 2019;104(11):5157–5169. doi: 10.1210/jc.2018-02693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Menon R, Debnath C, Lai A, Guanzon D, Bhatnagar S, Kshetrapal PK, et al. Circulating exosomal miRNA profile during term and preterm birth pregnancies: a longitudinal study. Endocrinology. 2019;160(2):249–275. doi: 10.1210/en.2018-00836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang G, Mouillet JF, Mishima T, Chu T, Sadovsky E, Coyne CB, et al. Expression and trafficking of placental microRNAs at the feto-maternal interface. FASEB J. 2017;31(7):2760–2770. doi: 10.1096/fj.201601146R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mouillet JF, Chu T, Hubel CA, Nelson DM, Parks WT, Sadovsky Y. The levels of hypoxia-regulated microRNAs in plasma of pregnant women with fetal growth restriction. Placenta. 2010;31(9):781–784. doi: 10.1016/j.placenta.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whitehead CL, Teh WT, Walker SP, Leung C, Larmour L, Tong S. Circulating microRNAs in maternal blood as potential biomarkers for fetal hypoxia in-utero. PLoS ONE. 2013;8(11):e78487. doi: 10.1371/journal.pone.0078487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hromadnikova I, Dvorakova L, Kotlabova K, Krofta L. The prediction of gestational hypertension, preeclampsia and fetal growth restriction via the first trimester screening of plasma exosomal C19MC microRNAs. Int J Mol Sci. 2019;20:12. doi: 10.3390/ijms20122972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim SH, MacIntyre DA, Binkhamis R, Cook J, Sykes L, Bennett PR, et al. Maternal plasma miRNAs as potential biomarkers for detecting risk of small-for-gestational-age births. EBioMedicine. 2020;62:103145. doi: 10.1016/j.ebiom.2020.103145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hromadnikova I, Kotlabova K, Ondrackova M, Kestlerova A, Novotna V, Hympanova L, et al. Circulating C19MC microRNAs in preeclampsia, gestational hypertension, and fetal growth restriction. Mediators Inflamm. 2013;2013:186041. doi: 10.1155/2013/186041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muralimanoharan S, Guo C, Myatt L, Maloyan A. Sexual dimorphism in miR-210 expression and mitochondrial dysfunction in the placenta with maternal obesity. Int J Obes (Lond) 2015;39(8):1274–1281. doi: 10.1038/ijo.2015.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clifton VL. Review: sex and the human placenta: mediating differential strategies of fetal growth and survival. Placenta. 2010;31(Suppl):S33–S39. doi: 10.1016/j.placenta.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 29.van der Vlugt ER, Verburg PE, Leemaqz SY, McCowan LME, Poston L, Kenny LC, et al. Sex- and growth-specific characteristics of small for gestational age infants: a prospective cohort study. Biol Sex Differ. 2020;11(1):25. doi: 10.1186/s13293-020-00300-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eaves LA, Phookphan P, Rager JE, Bangma J, Santos HP, Jr, Smeester L, et al. A role for microRNAs in the epigenetic control of sexually dimorphic gene expression in the human placenta. Epigenomics. 2020;12(17):1543–1558. doi: 10.2217/epi-2020-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mongelli M, Gardosi J. Gestation-adjusted projection of estimated fetal weight. Acta Obstet Gynecol Scand. 1996;75(1):28–31. doi: 10.3109/00016349609033279. [DOI] [PubMed] [Google Scholar]
- 32.Blondal T, Jensby Nielsen S, Baker A, Andreasen D, Mouritzen P, Wrang Teilum M, et al. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods. 2013;59(1):S1–6. doi: 10.1016/j.ymeth.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 33.Mestdagh P, Van Vlierberghe P, De Weer A, Muth D, Westermann F, Speleman F, et al. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10(6):R64. doi: 10.1186/gb-2009-10-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64(15):5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 35.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodol) 1995;57(1):289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 36.Chang L, Zhou G, Soufan O, Xia J. miRNet 2.0: network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020;48(W1):W244–W251. doi: 10.1093/nar/gkaa467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.PubMed. https://pubmed.ncbi.nlm.nih.gov/. Accessed May 30, 2021.
- 38.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ameling S, Kacprowski T, Chilukoti RK, Malsch C, Liebscher V, Suhre K, et al. Associations of circulating plasma microRNAs with age, body mass index and sex in a population-based study. BMC Med Genomics. 2015;8:61. doi: 10.1186/s12920-015-0136-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cui C, Yang W, Shi J, Zhou Y, Yang J, Cui Q, et al. Identification and analysis of human sex-biased microRNAs. Genomics Proteomics Bioinform. 2018;16(3):200–211. doi: 10.1016/j.gpb.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pineles BL, Romero R, Montenegro D, Tarca AL, Han YM, Kim YM, et al. Distinct subsets of microRNAs are expressed differentially in the human placentas of patients with preeclampsia. Am J Obstet Gynecol. 2007;196(3):261.e16. doi: 10.1016/j.ajog.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 42.Renthal NE, Chen CC, Williams KC, Gerard RD, Prange-Kiel J, Mendelson CR. miR-200 family and targets, ZEB1 and ZEB2, modulate uterine quiescence and contractility during pregnancy and labor. Proc Natl Acad Sci U S A. 2010;107(48):20828–20833. doi: 10.1073/pnas.1008301107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang S, Li H, Ge Q, Guo L, Chen F. Deregulated microRNA species in the plasma and placenta of patients with preeclampsia. Mol Med Rep. 2015;12(1):527–534. doi: 10.3892/mmr.2015.3414. [DOI] [PubMed] [Google Scholar]
- 44.Mouillet JF, Chu T, Nelson DM, Mishima T, Sadovsky Y. MiR-205 silences MED1 in hypoxic primary human trophoblasts. FASEB J. 2010;24(6):2030–2039. doi: 10.1096/fj.09-149724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsamou M, Vrijens K, Wang C, Winckelmans E, Neven KY, Madhloum N, et al. Genome-wide microRNA expression analysis in human placenta reveals sex-specific patterns: an ENVIRONAGE birth cohort study. Epigenetics. 2020 doi: 10.1080/15592294.2020.1803467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mayor-Lynn K, Toloubeydokhti T, Cruz AC, Chegini N. Expression profile of microRNAs and mRNAs in human placentas from pregnancies complicated by preeclampsia and preterm labor. Reprod Sci. 2011;18(1):46–56. doi: 10.1177/1933719110374115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hong L, Yu T, Xu H, Hou N, Cheng Q, Lai L, et al. Down-regulation of miR-378a-3p induces decidual cell apoptosis: a possible mechanism for early pregnancy loss. Hum Reprod. 2018;33(1):11–22. doi: 10.1093/humrep/dex347. [DOI] [PubMed] [Google Scholar]
- 48.Fu G, Brkic J, Hayder H, Peng C. MicroRNAs in human placental development and pregnancy complications. Int J Mol Sci. 2013;14(3):5519–5544. doi: 10.3390/ijms14035519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Higashijima A, Miura K, Mishima H, Kinoshita A, Jo O, Abe S, et al. Characterization of placenta-specific microRNAs in fetal growth restriction pregnancy. Prenat Diagn. 2013;33(3):214–222. doi: 10.1002/pd.4045. [DOI] [PubMed] [Google Scholar]
- 50.Ishibashi O, Ohkuchi A, Ali MM, Kurashina R, Luo SS, Ishikawa T, et al. Hydroxysteroid (17-beta) dehydrogenase 1 is dysregulated by miR-210 and miR-518c that are aberrantly expressed in preeclamptic placentas: a novel marker for predicting preeclampsia. Hypertension. 2012;59(2):265–273. doi: 10.1161/HYPERTENSIONAHA.111.180232. [DOI] [PubMed] [Google Scholar]
- 51.Carreras-Badosa G, Bonmati A, Ortega FJ, Mercader JM, Guindo-Martinez M, Torrents D, et al. Altered circulating miRNA expression profile in pregestational and gestational obesity. J Clin Endocrinol Metab. 2015;100(11):E1446–E1456. doi: 10.1210/jc.2015-2872. [DOI] [PubMed] [Google Scholar]
- 52.Kho AT, Sharma S, Davis JS, Spina J, Howard D, McEnroy K, et al. Circulating microRNAs: association with lung function in asthma. PLoS ONE. 2016;11(6):e0157998. doi: 10.1371/journal.pone.0157998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stubert J, Koczan D, Richter DU, Dieterich M, Ziems B, Thiesen HJ, et al. miRNA expression profiles determined in maternal sera of patients with HELLP syndrome. Hypertens Pregnancy. 2014;33(2):215–235. doi: 10.3109/10641955.2013.858743. [DOI] [PubMed] [Google Scholar]
- 54.Muralimanoharan S, Li C, Nakayasu ES, Casey CP, Metz TO, Nathanielsz PW, et al. Sexual dimorphism in the fetal cardiac response to maternal nutrient restriction. J Mol Cell Cardiol. 2017;108:181–193. doi: 10.1016/j.yjmcc.2017.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi Z, She K, Li H, Yuan X, Han X, Wang Y. MicroRNA-454 contributes to sustaining the proliferation and invasion of trophoblast cells through inhibiting Nodal/ALK7 signaling in pre-eclampsia. Chem Biol Interact. 2019;298:8–14. doi: 10.1016/j.cbi.2018.10.012. [DOI] [PubMed] [Google Scholar]
- 56.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 57.Bersinger NA, Odegard RA. Second- and third-trimester serum levels of placental proteins in preeclampsia and small-for-gestational age pregnancies. Acta Obstet Gynecol Scand. 2004;83(1):37–45. doi: 10.1111/j.1600-0412.2004.00277.x. [DOI] [PubMed] [Google Scholar]
- 58.Dutton PJ, Warrander LK, Roberts SA, Bernatavicius G, Byrd LM, Gaze D, et al. Predictors of poor perinatal outcome following maternal perception of reduced fetal movements—a prospective cohort study. PLoS ONE. 2012;7(7):e39784. doi: 10.1371/journal.pone.0039784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47(D1):D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chou CH, Shrestha S, Yang CD, Chang NW, Lin YL, Liao KW, et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46(D1):D296–D302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48(D1):D127–D131. doi: 10.1093/nar/gkz757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baker BC, Mackie FL, Lean SC, Greenwood SL, Heazell AE, Forbes K, et al. Placental dysfunction is associated with altered microRNA expression in pregnant women with low folate status. Mol Nutr Food Res. 2017 doi: 10.1002/mnfr.201600646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Verburg PE, Tucker G, Scheil W, Erwich JJ, Dekker GA, Roberts CT. Sexual dimorphism in adverse pregnancy outcomes—a retrospective australian population study 1981–2011. PLoS ONE. 2016;11(7):e0158807. doi: 10.1371/journal.pone.0158807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hromadnikova I, Kotlabova K, Ivankova K, Krofta L. First trimester screening of circulating C19MC microRNAs and the evaluation of their potential to predict the onset of preeclampsia and IUGR. PLoS ONE. 2017;12(2):e0171756. doi: 10.1371/journal.pone.0171756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hromadnikova I, Kotlabova K, Ivankova K, Krofta L. Expression profile of C19MC microRNAs in placental tissue of patients with preterm prelabor rupture of membranes and spontaneous preterm birth. Mol Med Rep. 2017;16(4):3849–3862. doi: 10.3892/mmr.2017.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu XM, Han T, Sargent IL, Yin GW, Yao YQ. Differential expression profile of microRNAs in human placentas from preeclamptic pregnancies vs normal pregnancies. Am J Obstet Gynecol. 2009;200(6):661.e1–7. doi: 10.1016/j.ajog.2008.12.045. [DOI] [PubMed] [Google Scholar]
- 68.Montenegro D, Romero R, Kim SS, Tarca AL, Draghici S, Kusanovic JP, et al. Expression patterns of microRNAs in the chorioamniotic membranes: a role for microRNAs in human pregnancy and parturition. J Pathol. 2009;217(1):113–121. doi: 10.1002/path.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brien ME, Boufaied I, Bernard N, Forest JC, Giguere Y, Girard S. Specific inflammatory profile in each pregnancy complication: a comparative study. Am J Reprod Immunol. 2020 doi: 10.1111/aji.13316. [DOI] [PubMed] [Google Scholar]
- 70.Holland RL. What makes a good biomarker? Adv Precis Med. 2016;1(1):4–11. doi: 10.18063/APM.2016.01.007. [DOI] [Google Scholar]
- 71.Delorme-Axford E, Donker RB, Mouillet JF, Chu T, Bayer A, Ouyang Y, et al. Human placental trophoblasts confer viral resistance to recipient cells. Proc Natl Acad Sci U S A. 2013;110(29):12048–12053. doi: 10.1073/pnas.1304718110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Szentpeteri I, Rab A, Kornya L, Kovacs P, Joo JG. Gene expression patterns of vascular endothelial growth factor (VEGF-A) in human placenta from pregnancies with intrauterine growth restriction. J Matern Fetal Neonatal Med. 2013;26(10):984–989. doi: 10.3109/14767058.2013.766702. [DOI] [PubMed] [Google Scholar]
- 73.Smith GC, Stenhouse EJ, Crossley JA, Aitken DA, Cameron AD, Connor JM. Early pregnancy levels of pregnancy-associated plasma protein a and the risk of intrauterine growth restriction, premature birth, preeclampsia, and stillbirth. J Clin Endocrinol Metab. 2002;87(4):1762–1767. doi: 10.1210/jcem.87.4.8430. [DOI] [PubMed] [Google Scholar]
- 74.Woods KA, Camacho-Hubner C, Barter D, Clark AJ, Savage MO. Insulin-like growth factor I gene deletion causing intrauterine growth retardation and severe short stature. Acta Paediatr Suppl. 1997;423:39–45. doi: 10.1111/j.1651-2227.1997.tb18367.x. [DOI] [PubMed] [Google Scholar]
- 75.Harris LK, Pantham P, Yong HEJ, Pratt A, Borg AJ, Crocker I, et al. The role of insulin-like growth factor 2 receptor-mediated homeobox gene expression in human placental apoptosis, and its implications in idiopathic fetal growth restriction. Mol Hum Reprod. 2019;25(9):572–585. doi: 10.1093/molehr/gaz047. [DOI] [PubMed] [Google Scholar]
- 76.Jansson T, Scholtbach V, Powell TL. Placental transport of leucine and lysine is reduced in intrauterine growth restriction. Pediatr Res. 1998;44(4):532–537. doi: 10.1203/00006450-199810000-00011. [DOI] [PubMed] [Google Scholar]
- 77.Whitehead CL, Walker SP, Lappas M, Tong S. Circulating RNA coding genes regulating apoptosis in maternal blood in severe early onset fetal growth restriction and pre-eclampsia. J Perinatol. 2013;33(8):600–604. doi: 10.1038/jp.2013.16. [DOI] [PubMed] [Google Scholar]
- 78.Heazell AEP, Sharp AN, Baker PN, Crocker IP. Intra-uterine growth restriction is associated with increased apoptosis and altered expression of proteins in the p53 pathway in villous trophoblast. Apoptosis. 2011;16(2):135–144. doi: 10.1007/s10495-010-0551-3. [DOI] [PubMed] [Google Scholar]
- 79.Norberg S, Powell TL, Jansson T. Intrauterine growth restriction is associated with a reduced activity of placental taurine transporters. Pediatr Res. 1998;44(2):233–238. doi: 10.1203/00006450-199808000-00016. [DOI] [PubMed] [Google Scholar]
- 80.Gibbs I, Leavey K, Benton SJ, Grynspan D, Bainbridge SA, Cox BJ. Placental transcriptional and histologic subtypes of normotensive fetal growth restriction are comparable to preeclampsia. Am J Obstet Gynecol. 2019;220(1):110 e1–e21. doi: 10.1016/j.ajog.2018.10.003. [DOI] [PubMed] [Google Scholar]
- 81.Schoots MH, Gordijn SJ, Scherjon SA, van Goor H, Hillebrands JL. Oxidative stress in placental pathology. Placenta. 2018;69:153–161. doi: 10.1016/j.placenta.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 82.Burton GJ, Jauniaux E. Pathophysiology of placental-derived fetal growth restriction. Am J Obstet Gynecol. 2018;218(2S):S745–S761. doi: 10.1016/j.ajog.2017.11.577. [DOI] [PubMed] [Google Scholar]
- 83.Gabory A, Roseboom TJ, Moore T, Moore LG, Junien C. Placental contribution to the origins of sexual dimorphism in health and diseases: sex chromosomes and epigenetics. Biol Sex Differ. 2013;4(1):5. doi: 10.1186/2042-6410-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nugent BM, O'Donnell CM, Epperson CN, Bale TL. Placental H3K27me3 establishes female resilience to prenatal insults. Nat Commun. 2018;9(1):2555. doi: 10.1038/s41467-018-04992-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vatten LJ, Skjaerven R. Offspring sex and pregnancy outcome by length of gestation. Early Hum Dev. 2004;76(1):47–54. doi: 10.1016/j.earlhumdev.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 86.Mondal D, Galloway TS, Bailey TC, Mathews F. Elevated risk of stillbirth in males: systematic review and meta-analysis of more than 30 million births. BMC Med. 2014;12:220. doi: 10.1186/s12916-014-0220-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Forbes K, Westwood M, Baker PN, Aplin JD. Insulin-like growth factor I and II regulate the life cycle of trophoblast in the developing human placenta. Am J Physiol Cell Physiol. 2008;294(6):C1313–C1322. doi: 10.1152/ajpcell.00035.2008. [DOI] [PubMed] [Google Scholar]
- 88.Forbes K, Westwood M. The IGF axis and placental function: a mini review. Horm Res Paediatr. 2008;69(3):129–37. doi: 10.1159/000112585. [DOI] [PubMed] [Google Scholar]
- 89.Shi X, Teng F. Down-regulated miR-28-5p in human hepatocellular carcinoma correlated with tumor proliferation and migration by targeting insulin-like growth factor-1 (IGF-1) Mol Cell Biochem. 2015;408(1–2):283–293. doi: 10.1007/s11010-015-2506-z. [DOI] [PubMed] [Google Scholar]
- 90.Roberts CT, Owens JA, Sferruzzi-Perri AN. Distinct actions of insulin-like growth factors (IGFs) on placental development and fetal growth: lessons from mice and guinea pigs. Placenta. 2008;29(Suppl A):S42–7. doi: 10.1016/j.placenta.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 91.Giudice LC, de Zegher F, Gargosky SE, Dsupin BA, de las Fuentes L, Crystal RA, et al. Insulin-like growth factors and their binding proteins in the term and preterm human fetus and neonate with normal and extremes of intrauterine growth. J Clin Endocrinol Metab. 1995;80(5):1548–55. doi: 10.1210/jcem.80.5.7538146. [DOI] [PubMed] [Google Scholar]
- 92.Geary MP, Pringle PJ, Rodeck CH, Kingdom JC, Hindmarsh PC. Sexual dimorphism in the growth hormone and insulin-like growth factor axis at birth. J Clin Endocrinol Metab. 2003;88(8):3708–3714. doi: 10.1210/jc.2002-022006. [DOI] [PubMed] [Google Scholar]
- 93.Ma F, Zhang J, Zhong L, Wang L, Liu Y, Wang Y, et al. Upregulated microRNA-301a in breast cancer promotes tumor metastasis by targeting PTEN and activating Wnt/beta-catenin signaling. Gene. 2014;535(2):191–197. doi: 10.1016/j.gene.2013.11.035. [DOI] [PubMed] [Google Scholar]
- 94.Zhang L, Zhang Y, Zhu H, Sun X, Wang X, Wu P, et al. Overexpression of miR-301a-3p promotes colorectal cancer cell proliferation and metastasis by targeting deleted in liver cancer-1 and runt-related transcription factor 3. J Cell Biochem. 2019;120(4):6078–6089. doi: 10.1002/jcb.27894. [DOI] [PubMed] [Google Scholar]
- 95.Shi W, Gerster K, Alajez NM, Tsang J, Waldron L, Pintilie M, et al. MicroRNA-301 mediates proliferation and invasion in human breast cancer. Cancer Res. 2011;71(8):2926–2937. doi: 10.1158/0008-5472.CAN-10-3369. [DOI] [PubMed] [Google Scholar]
- 96.Sedlmeier EM, Brunner S, Much D, Pagel P, Ulbrich SE, Meyer HH, et al. Human placental transcriptome shows sexually dimorphic gene expression and responsiveness to maternal dietary n-3 long-chain polyunsaturated fatty acid intervention during pregnancy. BMC Genomics. 2014;15:941. doi: 10.1186/1471-2164-15-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dong Y, Zhang L, Zhang S, Bai Y, Chen H, Sun X, et al. Phosphatase of regenerating liver 2 (PRL2) is essential for placental development by down-regulating PTEN (Phosphatase and Tensin Homologue Deleted on Chromosome 10) and activating Akt protein. J Biol Chem. 2012;287(38):32172–32179. doi: 10.1074/jbc.M112.393462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Broere-Brown ZA, Adank MC, Benschop L, Tielemans M, Muka T, Goncalves R, et al. Fetal sex and maternal pregnancy outcomes: a systematic review and meta-analysis. Biol Sex Differ. 2020;11(1):26. doi: 10.1186/s13293-020-00299-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kingdom JC, Smith GC. Next steps to prevent stillbirth associated with growth restriction. BJOG. 2020;127(9):1081. doi: 10.1111/1471-0528.16237. [DOI] [PubMed] [Google Scholar]
- 100.Higgins LE, Myers JE, Sibley CP, Johnstone ED, Heazell AEP. Antenatal placental assessment in the prediction of adverse pregnancy outcome after reduced fetal movement. PLoS ONE. 2018;13(11):e0206533. doi: 10.1371/journal.pone.0206533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hannan NJ, Stock O, Spencer R, Whitehead C, David AL, Groom K, et al. Circulating mRNAs are differentially expressed in pregnancies with severe placental insufficiency and at high risk of stillbirth. BMC Med. 2020;18(1):145. doi: 10.1186/s12916-020-01605-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. List of miRNAs in Exiqon QPCR array Human panels I + II.
Additional file 2: Table S1. Target sequences of LNA microRNA primer sets utilised for microRNA Q-PCR.
Data Availability Statement
The microarray data generated during the current study has been deposited in NCBI's Gene Expression Omnibus and is accessible through GEO Series accession number GSE188186 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE188186).