Abstract
Members of the GATA family of transcription factors play important roles in cell fate specification, differentiation, and morphogenesis during mammalian development. GATA5, the only one of the six vertebrate GATA factor genes not yet inactivated in mice, is expressed in a pattern that overlaps with but is distinct from that of other GATA factor genes. During mouse embryogenesis, GATA5 is expressed first in the developing heart and subsequently in the lung, vasculature, and genitourinary system. To investigate the function of GATA5 in vivo, we created mice homozygous for a GATA5 null allele. Homozygous mutants were viable and fertile, but females exhibited pronounced genitourinary abnormalities that included vaginal and uterine defects and hypospadias. In contrast, the genitourinary system was unaffected in male GATA5 mutants. These results reveal a specific role of GATA5 in development of the female genitourinary system and suggest that other GATA factors may have functions overlapping those of GATA5 in other tissues.
There are six members of the GATA family of transcription factors in vertebrates, which play important roles in cell fate specification, differentiation, and morphogenesis. The GATA factors share homology in two evolutionarily conserved zinc fingers that mediate binding to the consensus DNA sequence WGATAR and are generally categorized into two classes based on their expression patterns and amino acid sequence homologies (2, 16, 18).
GATA1, GATA2, and GATA3 are expressed predominantly in hematopoietic cell lineages. GATA1 is required for erythroid and megakaryocyte differentiation (19, 20, 22). GATA2 controls proliferation of hematopoietic precursor cells (24). GATA3 controls T-lymphocyte development and is involved in embryonic liver hematopoiesis and nervous system development (17, 23). In contrast, GATA4, GATA5, and GATA6 are expressed predominantly in the cardiovascular system but also show other sites of expression (1, 4, 8, 12, 13). GATA4 mutant mice die at embryonic day 8.0 (E8) from defects in ventral morphogenesis that prevent formation of the linear heart tube (7, 11), whereas GATA6 mutant mice die before gastrulation, apparently from defects in extraembryonic development (6, 14). The functions of GATA5 in mice have not yet been determined. However, in zebra fish, GATA5 has been shown to be required for expression of myocardial genes and for formation of the heart tube, similar to the role of GATA4 in mice (21).
During mouse embryogenesis, GATA5 expression is detected initially in the precardiac mesoderm between E7 and E8 and continues throughout the heart until E16.5 (13, 15). Beginning at midgestation, GATA5 is also expressed within pulmonary mesenchyme and vascular smooth muscle cells in the developing lung, as well as in the urogenital ridge, in epithelial cells lining the urogenital sinus, in the bladder, and in the gut epithelium. Postnatally, GATA5 expression is restricted to the intestine, stomach, bladder, and lungs. In contrast, GATA4 is expressed predominantly in the adult heart, gut, testes, and ovaries (1, 4, 8), and GATA6 is expressed in the heart, gut, bladder, and vasculature (12).
To investigate the functions of GATA5 in vivo, we generated mice homozygous for a GATA5 null allele. These mutant mice were viable and fertile but showed specific abnormalities in female genitourinary development that included malpositioning of the urogenital sinus, vagina, and urethra, mimicking a condition of proximal hypospadias in human females. These defects reveal an unanticipated role for GATA5 in morphogenesis of the genitourinary tract and suggest that other GATA factors may compensate for the lack of GATA5 in other tissues.
MATERIALS AND METHODS
Targeting of the mouse GATA5 gene.
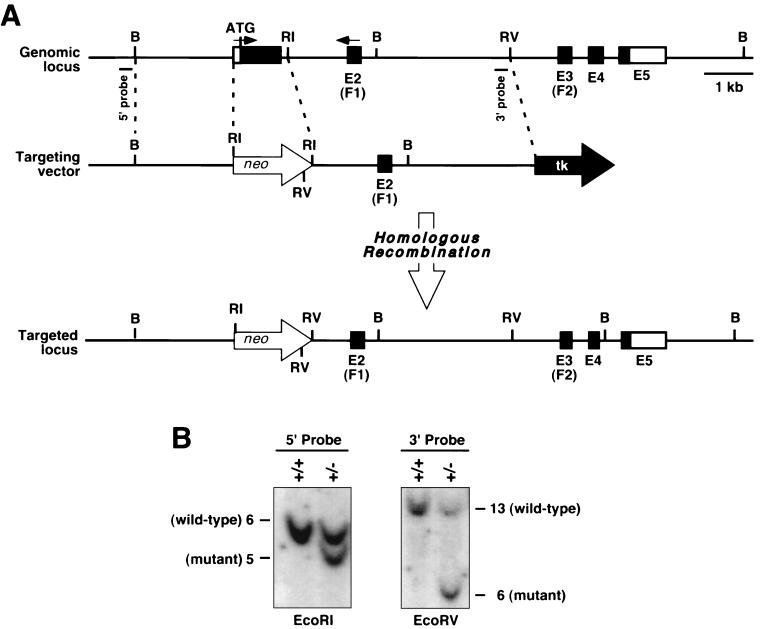
The GATA5-targeting vector was constructed from a 15-kb mouse genomic clone containing the entire coding region (Fig. 1A). As the 5′ region of homology, we used a 1.9-kb genomic fragment immediately upstream of the first coding exon. This was cloned upstream of a neomycin resistance gene linked to the phosphoglycerokinase (PGK) promoter. The 3′ region of homology was created by PCR of genomic DNA to generate a 3.9-kb fragment that extended 3′ from an EcoRI site at the end of the first coding exon of GATA5 to 500 bp 5′ of an EcoRV site (Fig. 1A). This fragment was cloned downstream of PGK-neo, and a thymidine kinase gene under the control of the herpes simplex virus promoter was linked to the 3′ end.
FIG. 1.
Targeting of the GATA5 gene. (A) Structure of the mouse GATA5 gene and strategy for gene targeting. The exons (E) and coding region for zinc fingers (F1 and F2) are shown. The targeting vector contained a neomycin resistance gene and thymidine kinase (tk) gene in the same transcriptional orientation as GATA5. Targeting of the gene resulted in deletion of the ATG and the first 157 amino acids of the protein. Positions of probes used for Southern analysis are shown beneath the genomic map. B, BamHI; RI, EcoRI; RV, EcoRV. (B) Southern blots of tail DNA probed with the 5′ and 3′ probes (shown in panel A) following digestion with EcoRI and EcoRV, respectively.
The targeting vector was linearized by digestion with NotI and electroporated into the KG-1 embryonic stem (ES) cell line. Following positive-negative selection with G-418 (Geneticin, at 180 μg of active concentration per ml; GIBCO BRL) and 0.2 μM fialuridine (FIAU) [1-(2-deoxy-2-fluoro-β-d-arabinofuranosyl)-5-iodouracil], respectively, 400 individual ES cell colonies were isolated and analyzed by Southern blotting for homologous recombination, as described previously (11). Two ES cell clones were found to contain a disrupted GATA5 gene. Both clones were injected into 3.5-day mouse C57BL/6 blastocysts to obtain chimeras. Both ES cell lines gave rise to germ line heterozygous mice, which were intercrossed to obtain homozygous GATA5 mutants.
Genotyping.
Genotypes of mice obtained from GATA5+/− intercrosses were determined by Southern blot analysis of genomic DNA using probes external to the targeted region of the gene (Fig. 1). Hybridization of the 5′ probe to EcoRI-digested DNA yielded bands of 6 and 5 kb for the wild-type and targeted alleles, respectively, and hybridization of the 3′ probe to EcoRV-digested DNA yielded bands of 13 and 6 kb for the wild-type and targeted alleles, respectively.
RT-PCR mRNA quantitation.
Analysis of GATA4 mRNA levels was performed using primers designed to amplify across the first and second exons of GATA5, resulting in a 200-bp product. The primers utilized for GATA5 amplification were 5′ CGACGTAGCCCCTTCGTGG and 5′ GCCACAGTGGTGTAGACAG. Primers used for reverse transcriptase (RT)-PCR of GATA4 and GATA6 were described elsewhere (11). Amplification of mRNA for the ribosome-associated protein L7 was used to control for RNA integrity and loading (11). RT-PCRs were performed with 1 μg of total RNA in the presence of [α-32P]dCTP using 32 cycles of amplification under conditions recommended by the manufacturer (Titan one-tube RT-PCR; Boehringer Mannheim). Products were resolved on a 6% polyacrylamide gel and subjected to PhosphorImager analysis (Molecular Dynamics). RT-PCRs were performed under conditions of linearity with respect to RNA amount.
Histology.
Tissues were harvested from three male and three female 6-week-old mutant mice, fixed in 10% buffered formalin, dehydrated through graded ethanols, embedded in paraffin, sectioned at 10 μm, and stained with hematoxylin and eosin.
RESULTS
Targeting of GATA5 in ES cells and generation of null mice.
To inactivate the mouse GATA5 gene, we created a targeting construct that deleted the first exon, which encodes amino acids 1 to 157 of the protein (Fig. 1A). Although this mutation leaves a portion of the GATA5 protein-coding region intact, this region of the gene does not appear to encode a functional protein, as a deletion mutant lacking the amino-terminal portion does not exhibit either active or dominant negative function in vitro (unpublished results). Moreover, analogous mutations in the GATA4 gene (7) or the GATA6 gene (6; J. Molkentin and E. Olson, unpublished results) result in lethal phenotypes.
The targeting vector was electroporated into KG-1 ES cells that were subjected to positive-negative selection with G-418 and FIAU. Southern blotting analysis of 400 individual ES cell clones (Fig. 1B) identified two targeted clones, representing a targeting frequency of 0.5%. Both ES cell clones heterozygous for the targeted GATA5 gene were injected into blastocysts derived from C57BL/6 mice, and blastocysts were subsequently implanted into pseudopregnant Swiss mouse foster mothers to obtain chimeric mice. Breeding of chimeric mice into a C57BL/6 background resulted in transmission of the mutation through the germ line and generation of GATA5 heterozygous mice of the composite C57BL/6 × Sv 129 genotype. Heterozygotes for the GATA5 mutation showed no apparent phenotype and were intercrossed to generate GATA5 null mice.
Genotyping of offspring from heterozygous intercrosses revealed GATA5 heterozygous and homozygous mutants at approximately the predicted Mendelian frequencies. Specifically, of 78 adult offspring from GATA5+/− intercrosses, 26 (33%) were GATA5+/+, 37 (47%) were GATA5+/−, and 15 (19%) were GATA5−/−. Homozygous GATA5 mutant males appeared to be normal and were fertile. Mutant females were also viable but exhibited obvious abnormalities in the external genitalia (see below). The penetrance of observed defects in female mutants was 100%. Only 1 of more than 20 GATA5 mutant females became pregnant, gave birth, and was able to nurse pups successfully. We conclude that these animals are fertile but have reduced fertility due to obstruction of the vaginal tract.
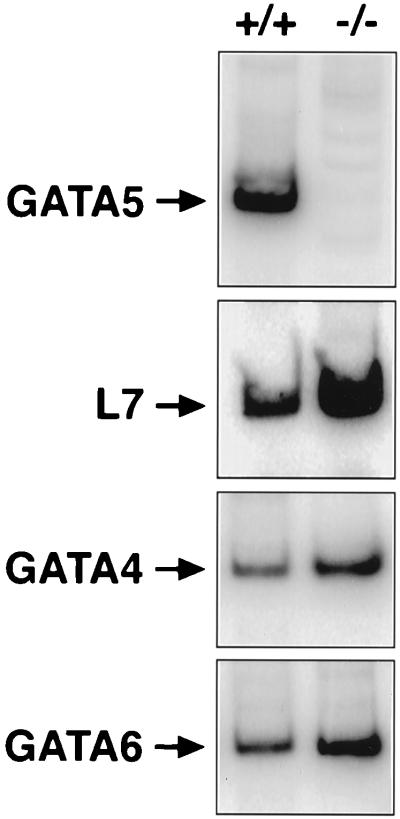
RT-PCR analysis using RNA from the large intestines of wild-type and mutant littermates revealed GATA5 transcripts in wild-type intestine but not in the mutant (Fig. 2). L7 transcripts were measured as an internal control for RNA integrity and were detectable at comparable levels in wild-type and mutant RNA samples (Fig. 2). To determine whether GATA4 or GATA6 might be upregulated in the intestine in response to the absence of GATA5, we also measured these transcripts. We observed no significant difference in abundance of either transcript in intestine from wild-type and mutant littermates (Fig. 2). GATA4 and GATA6 mRNA levels were also unaltered in hearts from GATA5 mutants (data not shown).
FIG. 2.
Detection of GATA5 transcripts by RT-PCR. Total RNA was isolated from large intestines of wild-type and GATA5 mutant mice and analyzed by RT-PCR for expression of GATA5, GATA4, GATA6, and L7 transcripts. No evidence of functional GATA5 transcripts was observed in the mutants. No PCR products were observed in the absence of RT (data not shown).
Gross morphological abnormalities in GATA5 null mutant females.
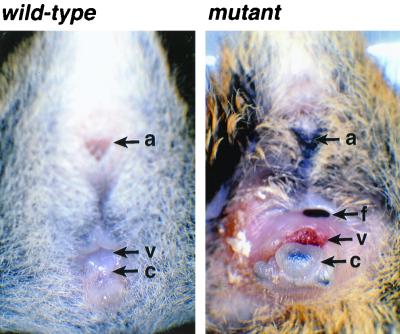
Gross examination of female null mutant mice revealed a consistent abnormality in the perineum (Fig. 3). The anogenital distance was reduced and the clitoris was dramatically enlarged. The vaginal orifice was also abnormal in shape and gaped open, and the vaginal epithelium extended laterally over the perineum, replacing the haired skin. The exteriorized vaginal mucosa was frequently excoriated and inflamed, and fistulas were commonly observed. In contrast, null mutant males were grossly normal.
FIG. 3.
Abnormalities of the external genitalia of GATA5 mutant females. External views of the genital regions of 8-week-old wild-type and GATA5 mutant females. Note the malpositioning and morphological abnormalities of the vagina and clitoris of the mutant. a, anus; c, clitoris; f, fistula; v, vagina.
Histological analysis of genitourinary defects in GATA5 null mutant females.
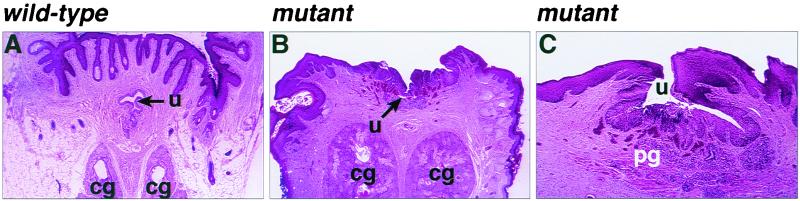
We further examined male and female GATA5 mutant mice by histological analysis of internal organs. No abnormalities were observed in the heart, lung, stomach, small intestine, colon, kidney, uterus, bladder, ovaries, male accessory sex glands, or penis. The ovaries of mutants contained follicles at various stages of maturation, and the vaginal and uterine epithelium presented a morphology characteristic of various periods of the estrus cycle. Lesions were restricted to the distal aspect of the vagina and clitoris. In the normal female, the urethra opens near the tip of the clitoris. In the mutants, the distal urethra did not form a distinct tube but rather formed a trough along the surface of the vaginal wall (Fig. 4). The urethra was lined by hyperplastic transitional epithelium which was often inflamed. Although the urethra was malformed, the periurethral glands and erectile tissue had developed and were evident in the submucosa of the trough-like urethra. The sebaceous clitoral glands were hypoplastic. The normal clitoris contains a small bone comparable to the os penis. This bone was not found in the mutant females.
FIG. 4.
Histological cross sections through the lower genitourinary tracts of wild-type and GATA5 mutant females. Note the absence of the urethra tube in the mutant. The vaginal canal is at the top. cg, clitoral gland; pg, periurethral gland; u, urethra.
DISCUSSION
Analysis of GATA5 null mice revealed a specific role for GATA5 in development of the female genitourinary tract but no apparent defects in male mutant mice. The genitourinary defects in GATA5 mutant mice correlate with sites of GATA5 expression in the genitourinary system during late fetal and postnatal development. GATA5 expression is observed in the urogenital ridge surrounding the lumen of the urogenital sinus as early as E12.5 (13). Expression is also seen at high levels in the wall of the bladder throughout pre- and postnatal development.
The defects observed in GATA5 null mutant females suggest that early morphogenic movements in the lower genitourinary tract are disrupted in the absence of GATA5. The precise embryonic origins of the vagina are unclear. It has been suggested that it has a dual origin, with the cranial portion derived from the Müllerian ducts and the caudal portion derived from the urogenital sinus (3). The urethra develops from the caudal end of the urogenital sinus after its complete separation from the cloaca (10). Abnormalities in GATA5 null mutant females were confined to the distal aspect of the vagina, suggesting that the underlying defect is in the partitioning of the urogenital sinus.
The phenotype of GATA5 null mutant females mimics a rare condition in human females in which the urogenital sinus fails to divide into the vestibule and urethra. Depending on the severity of the defect, the urethra may open into the proximal or distal vagina. In GATA5 null mutant mice, the defect is restricted to the distal urethra, suggesting that the primary defect lies in the urogenital sinus rather than the mesonephric ducts. It is rare for abnormalities of the urogenital sinus to occur independent of other perineal defects or more complex syndromes with ambiguous gentalia or adrenogenital syndromes. However, a very similar case of proximal hypospadias has been described for a female human (5).
Redundant and unique functions of GATA4, GATA5, and GATA6.
In addition to being expressed in the genitourinary system, GATA5 is expressed in the developing heart and lungs during early embryogenesis (9, 13, 15). However, we observed no morphologic or histologic defects in these organs of mutant mice. This suggests that other members of the GATA factor family may substitute for GATA5 in these tissues. Consistent with this notion, GATA4 and GATA6 are coexpressed with GATA5 in the early heart tube. However, in the developing lung, the expression pattern of GATA5 is unique. GATA5 and GATA6 are coexpressed in the developing urogenital ridge, but their functions in that region must not entirely overlap during development of the female genitourinary system.
In an effort to determine whether GATA4 or GATA6 might substitute for GATA5 in tissues that were unaffected in GATA5 mutants, we generated GATA5−/− GATA4+/− and GATA5−/− GATA6+/− mice. Mice of these genotypes were viable, and their phenotypes appeared identical to those of GATA5−/− mutants (J. D. Molkentin and E. N. Olson, unpublished results). Thus, if either GATA4 or GATA6 is able to compensate for the absence of GATA5, a single copy of either gene would appear to be sufficient. We have not generated double null combinations of GATA5 with GATA4 or GATA6 because GATA6−/− mutants die before gastrulation (6, 14) and GATA4−/− mutants die at about E8.0 (7, 11), which is too early to observe functions potentially overlapping with those of GATA5. It is worth noting that GATA4 and GATA6 exhibit an essential threshold of expression for viability, as GATA5+/− and GATA4+/− mice are normal, whereas GATA6+/ GATA4+/− double heterozygotes die during embryogenesis (Molkentin and Olson, unpublished results).
Finally, the phenotype of GATA5 mutant mice raises interesting questions about the functional conservation of GATA genes across species. Recently, the zebra fish mutation faust, which causes cardia bifida and reduced expression of myocardial genes, was shown to represent GATA5 (21). The faust phenotype is similar to the GATA4 mutant phenotype in mice (7, 11), suggesting that the functions of different members of the GATA family may be interchangeable in different species, reflecting the unique expression patterns of the genes.
ACKNOWLEDGMENTS
This research was supported by grants from the NIH to E.N.O. and J.D.M.
We thank A. Tizenor for graphics and J. Page for editorial assistance.
REFERENCES
- 1.Arceci R J, King A A J, Simon M C, Orkin S H, Wilson D B. Mouse GATA-4: a retinoic acid-inducible GATA-binding transcription factor expressed in endodermally derived tissues and heart. Mol Cell Biol. 1993;13:2235–2246. doi: 10.1128/mcb.13.4.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evans T. Regulation of cardiac gene expression by GATA4/5/6. Trends Cardiovasc Med. 1997;7:75–83. doi: 10.1016/S1050-1738(97)00010-8. [DOI] [PubMed] [Google Scholar]
- 3.Julliard M T. Ultrastincture de l'epithelium vaginal de la souris au cours de sa differenciation. Arch Anat Microsc Morphol Exp. 1972;61:33–46. [PubMed] [Google Scholar]
- 4.Kelley C, Blumberg H, Zon L I, Evans T. GATA-4 is a novel transcription factor expressed in endocardium of the developing heart. Development. 1993;118:817–827. doi: 10.1242/dev.118.3.817. [DOI] [PubMed] [Google Scholar]
- 5.Knight H M L, Phillips N J, Mouriquand P D E. Female hypospadias, a case report. J Pediatr Surg. 1995;30:1738–1740. doi: 10.1016/0022-3468(95)90469-7. [DOI] [PubMed] [Google Scholar]
- 6.Koutsourakis M, Langeveld A, Patient R, Beddington R, Grosveld F. The transcription factor GATA6 is essential for early extraembryonic development. Development. 1999;126:723–732. [PubMed] [Google Scholar]
- 7.Kuo C T, Morrisey C C, Anandappa R, Sigrist K, Lu M M, Parmacek M S, Soudais C, Leiden J M. The GATA-4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1998;11:1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- 8.Laverriere A C, MacNeill C, Mueller C, Poelmann R E, Burch J B, Evans T. GATA-4/5/6, a subfamily of three transcription factors transcribed in developing heart and gut. J Biol Chem. 1998;269:23177–23184. [PubMed] [Google Scholar]
- 9.MacNeill C, French R, Evans T, Wessels A, Burch J B. Modular regulation of cGATA-5 gene expression in the developing heart and gut. Dev Biol. 2000;217:62–76. doi: 10.1006/dbio.1999.9539. [DOI] [PubMed] [Google Scholar]
- 10.Maizels M. Normal development of the urinary tract. In: Walsh P C, Retik A B, Stamey T A, Vaughan E D Jr, editors. Campbell's urology. 6th ed. Philadelphia, Pa: W. B. Saunders Co.; 1992. pp. 1317–1329. [Google Scholar]
- 11.Molkentin J D, Lin Q, Duncan S A, Olson E N. Requirement of the transcription factor GATA-4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11:1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- 12.Morrisey E E, Ip H S, Lu M M, Parmacek M S. GATA-6: a zinc finger transcription factor is expressed in multiple cell lineages derived from lateral mesoderm. Dev Biol. 1996;177:309–322. doi: 10.1006/dbio.1996.0165. [DOI] [PubMed] [Google Scholar]
- 13.Morrisey E E, Ip H S, Tang Z, Lu M M, Parmacek M S. GATA-5: a transcriptional activator expressed in a novel temporally and spatial-restricted pattern during embryonic development. Dev Biol. 1997;183:21–36. doi: 10.1006/dbio.1996.8485. [DOI] [PubMed] [Google Scholar]
- 14.Morrisey E E, Tang Z, Sigrist K, Lu M M, Jiang F, Ip H S, Parmacek M S. GATA6 regulates HNF4 and is required for differentiation of visceral endoderm in the mouse embryo. Genes Dev. 1998;12:3579–3590. doi: 10.1101/gad.12.22.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nemer G, Qureshi S T, Malo D, Nemer M. Functional analysis and chromosomal mapping of Gata5, a gene encoding a zinc finger DNA-binding protein. Mamm Genome. 1999;10:993–999. doi: 10.1007/s003359901146. [DOI] [PubMed] [Google Scholar]
- 16.Orkin S H. GATA-binding transcription factors in hematopoietic cells. Blood. 1992;80:575–581. [PubMed] [Google Scholar]
- 17.Pandolfi P P, Roth M E, Karis A, Leonard M W, Dzierzak E, Grosveld F G, Engel J D. Targeted disruption of the GATA3 gene causes severe abnormalities in the nervous system and in fetal liver haematopoiesis. Nat Genet. 1995;11:40–44. doi: 10.1038/ng0995-40. [DOI] [PubMed] [Google Scholar]
- 18.Parmacek M S, Leiden J M. GATA transcription factors and cardiac development. In: Harvey R P, Rosenthal N, editors. Heart development. San Diego, Calif: Academic Press; 1999. pp. 291–307. [Google Scholar]
- 19.Pevny L, Lin C S, D'Agati V, Simon M C, Orkin S H, Constantini F. Development of hematopoietic cells lacking transcription factor GATA-1. Development. 1995;121:163–172. doi: 10.1242/dev.121.1.163. [DOI] [PubMed] [Google Scholar]
- 20.Pevny L, Simon M C, Robertson E, Klein W H, Tsai S F, D'Agati V, Orkin S H, Constantini F. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature. 1991;349:257–260. doi: 10.1038/349257a0. [DOI] [PubMed] [Google Scholar]
- 21.Reiter J F, Alexander J, Rodaway A, Yelon D, Patient R, Holder N, Stainier D Y-R. Gata5 is required for the development of the heart and endoderm in zebrafish. Genes Dev. 1999;13:2983–2995. doi: 10.1101/gad.13.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shivdasani R A, Fujiwara Y, McDevitt M A, Orkin S H. A lineage-selective knockout establishes the critical role of transcription factor GATA-1 in megakaryocyte growth and platelet development. EMBO J. 1997;16:3965–3973. doi: 10.1093/emboj/16.13.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ting C N, Olson M C, Barton K P, Leiden J M. Transcription factor GATA-3 is required for development of the T-cell lineage. Nature. 1996;384:474–478. doi: 10.1038/384474a0. [DOI] [PubMed] [Google Scholar]
- 24.Tsai F Y, Keller G, Kuo F C, Weiss M, Chen J, Rosenblatt M, Alt F W, Orkin S H. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]