Abstract
Zebrafish embryos, with their large size (>0.5 mm) and accessibility, are valuable tools for investigating core cellular processes. Many of those processes, such as cell division, asymmetric inheritance of cellular components, and structural dynamics involved in cell motility and morphology, rely on cytoskeletal rearrangements and associated macromolecules. In addition to the protein-rich cytoskeleton, the early embryo is packed with maternally deposited RNA, which serves essential roles in establishing cell polarity, cell fate, and cell organization. Here, we present methods for visualizing endogenous RNA along with cytoskeletal structures, including microtubules and filamentous actin (F-actin) in the context of an intact vertebrate embryo. Each of the four protocols described herein (embryo fixation, RNA probe design/synthesis, double fluorescent in situ hybridization with tubulin immunofluorescence, and fluorescent in situ hybridization with phalloidin labeling of F-actin) are intended for optimal preservation and visualization of both the cytoskeleton and RNAs of interest. These methods can also be modified and applied to a broad range of other uses.
Keywords: Zebrafish, embryo, cytoskeleton, F-actin, microtubules, in situ hybridization, RNA localization, phalloidin, immunofluorescence
1. Introduction
Interactions between RNA and the cytoskeleton are critical for a number of cellular processes, with notable examples including mRNA transport [1], germ granule aggregation [2], and specific RNA localization necessary for subcellular organization [3]. Simultaneous visualization of both RNA and protein components of cytoskeletal structures, such as tubulin and actin, are therefore useful and necessary experimental tools. The methods described in this chapter are all vital steps for the successful completion of combinatorial RNA and cytoskeletal protein labeling, including: fixation, RNA probe design and synthesis, and in situ hybridization with either microtubule immunofluorescence or F-actin labeling.
These protocols can be readily adapted to a number of RNA-protein combinations. Method 3.1 describes a fixation procedure optimized for preservation of intracellular cytoskeletal structures (Fig. 1a). Several key aspects differentiate this from a typical fixation, including 1) removal of chorions prior to fixation, 2) glutaraldehyde and EGTA as additives in standard 4% PFA, and 3) subsequent reduction of glutaraldehyde with sodium borohydride (NaBH4). These steps may all be omitted for other concurrent RNA and non-cytoskeletal protein labeling purposes. For example, our lab frequently uses beta-catenin protein labeling as a membrane marker while simultaneously visualizing mRNA localization by in situ hybridization, for which the standard fixation procedures are sufficient.
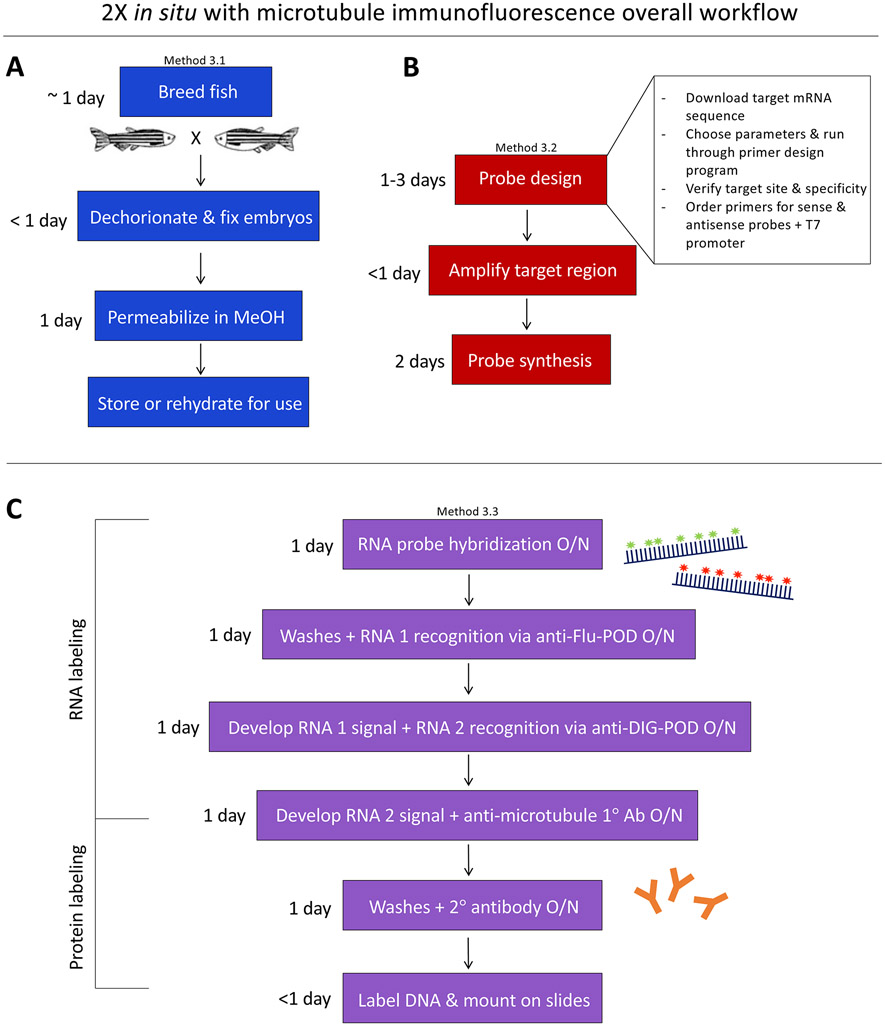
Figure 1.
A generalized diagram and approximate timeline of Methods 3.1 – 3.3, representing the overall workflow required for two-color fluorescence in situ hybridization with microtubule labeling. Note that embryo fixation and RNA probe preparation (Methods 3.1 and 3.2) can be performed well in advance of Method 3.3 (or other downstream uses, such as Method 3.4).
Method 3.2 (Fig. 1b, Fig. 2), which presents a PCR-based strategy for de novo RNA probe design and synthesis using T7-promoter containing primers [4, 5], is similarly adaptable. Although some aspects of probe design are largely a matter of preference, several important considerations, such as probe length and target region [6], are also discussed (see Note 1).
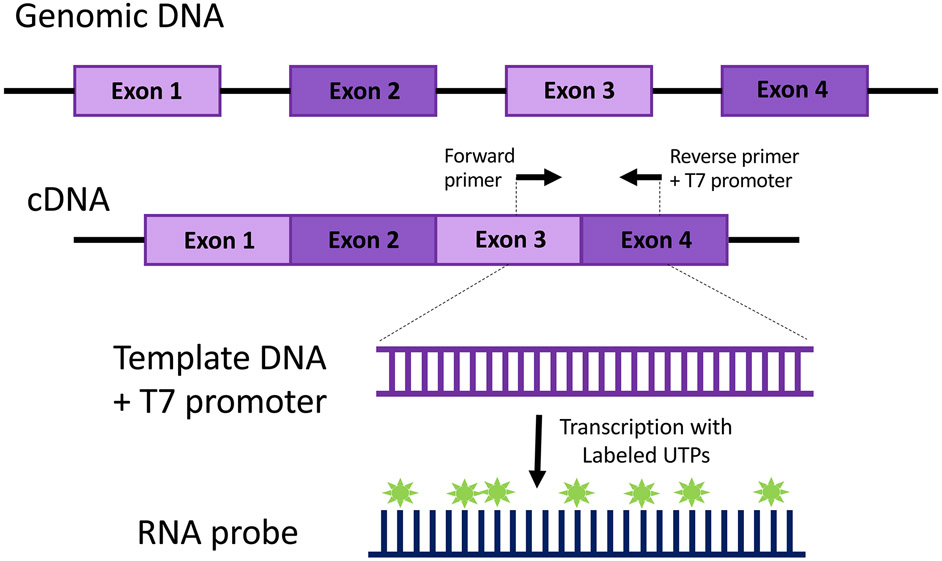
Figure 2.
Generalized schematic of PCR based RNA probe synthesis. This diagram depicts an RNA probe designed to target a region spanning an exon-exon junction, but other design strategies, such as including portions of the 5’ or 3’ UTR, can be employed (see Note 1).
In Method 3.3, we describe the use of Tyramide Signal Amplification (TSA™) [7, 8] with fluorescein (green) and digoxigenin/Cy3 (red) haptens to visualize two different RNA transcripts, and utilize a Cy5 (far-red) secondary antibody to visualize tubulin proteins (Fig. 1c). However, this aspect of the protocol is easily amendable to changes, such as visualizing only one RNA by omitting the second day of in situ labeling and proceeding directly to protein immunofluorescence (Fig. 3) or adding a third day of in situ procedure with another fluorophore to visualize three RNAs of interest (see Note 2). Indeed, the fluorescence in situ portion of the methods described below (Method 3.3, steps 1-32) is largely based on an excellent triple in situ protocol shared by the Talbot lab (Triple Fluorescent In Situ; zfin.org).
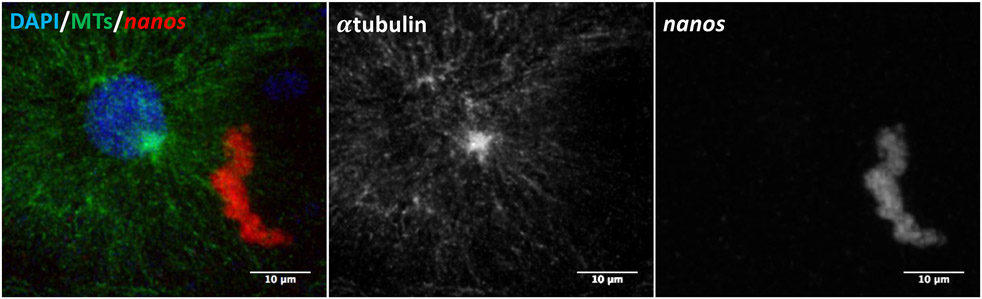
Figure 3.
Micrograph of a germ plasm aggregate (nanos) and microtubules surrounding a nucleus in a fixed 2-hour post-fertilization zebrafish embryo. Using the combined in situ/immunofluorescence protocol described in Method 3.3, digoxigenin-labeled probes targeting nanos RNA and antibodies targeting alpha-tubulin were used to simultaneously visualize an RNA–rich structure (germ plasm) with microtubules and a DAPI-stained nucleus. Image is a 2D projection of a Z-stack acquired on a Zeiss LSM510 confocal microscope.
We have also included a robust, one-step protocol using phalloidin for visualization of filamentous actin (F-actin) in the early embryo. Fluorophore-conjugated versions of phalloidin, a toxic peptide originally isolated from death cap mushrooms [9], is now commonly used to stain actin structures in cells due to its specificity and efficacy in binding F-actin [10, 11]. Here, we present a modified, one-step version of standard phalloidin staining protocols by recommending the addition of fluorophore-conjugated phalloidin to the fixation procedure rather than post-fixation. This optimized procedure provides two major benefits over the traditional method: 1) reduced total time involved as it eliminates the need for a later staining step, and most importantly, 2) improved preservation of F-actin structural details within the sample. The latter is likely achieved due to phalloidin’s ability to stabilize F-actin through its binding site, located in between F-actin subunits and effectively preventing depolymerization [12, 13]. Demonstrating the power of this technique, previously unobserved aspects of F-actin structure in the early embryo, such as micron-scale actin trenches in the cortex and cleavage furrows, were able to be visualized [14].
Other methods of F-actin labeling that do not require phalloidin, such as anti-actin-antibodies or transgenic fluorescent actin reporter lines, provide other potential avenues for simultaneous visualization of actin with RNA, but often have significant additional drawbacks (reviewed nicely in [11]). However, a long-standing limitation of phalloidin staining is its incompatibility with many common in situ hybridization methods, such as the one described here in Method 3.3. There are several aspects of in situ protocols that could lead to this incompatibility, including methanol treatment [15], treatment with high concentrations of formamide, and extended exposure to high temperatures. By addressing each of these key points, we have developed a functional combinatorial phalloidin/in situ protocol (Method 3.4, also see Note 3). This protocol provides a unique opportunity to concurrently visualize both RNAs and F-actin structures at a sub-micron scale in fixed vertebrate embryos (Fig. 4), and we are pleased to share what we hope will be a valuable resource for the research community.
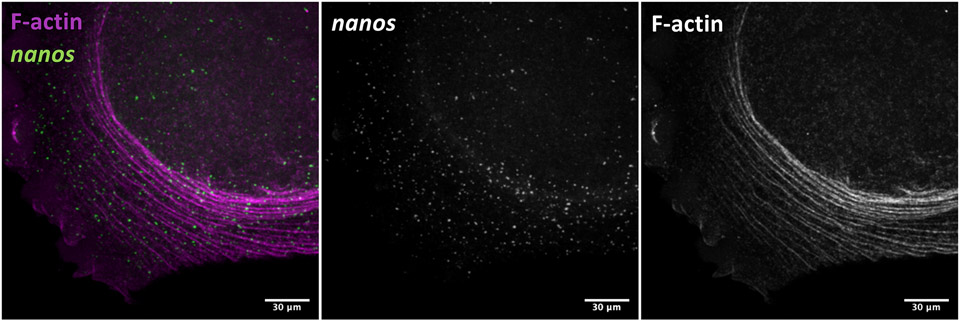
Figure 4.
Micrograph of F-actin arcs and germ plasm RNPs (nanos) in the blastodisc periphery of a fixed 2-cell (45 minutes post-fertilization) zebrafish embryo. As described in Method 3.4, fluorophore-conjugated phalloidin was incorporated with the fixation step to label F-actin (pseudocolored magenta here) followed by in situ hybridization to visualize fluorescein-labeled probes bound to nanos RNA. Image is a 2D projection of a Z-stack acquired on a Zeiss LSM780 confocal microscope.
2.1. Fixation of Zebrafish Embryos
1X PBS-Tween (PBST): 1X PBS, pH 7.0-7.4, 0.1% Tween-20. For 1L stock solution, mix 100 ml 10X PBS, 12.5 ml 20% Tween-20, and add nuclease-free H2O to 1L. Store at room temperature.
Paraformaldehyde (PFA) fix solution: 4% paraformaldehyde (PFA) in PBST. Under a fume hood and in a flask of at least a 250 ml volume capacity, dissolve 4 g paraformaldehyde in 100 ml 1X PBS-Tween by bringing to a boil on a hotplate. Be careful to not overboil. Remove from heat and let cool to room temperature. Aliquot into desired amounts (we typically prepare 1 ml and 10 ml aliquots) and store at −20 °C.
Cytoskeleton Fix: 4% PFA, 0.25% glutaraldehyde, 5 mM EGTA, 0.2% Triton X-100. Per 1 ml of 4% PFA, add 10 μl 25% glutaraldehyde, 10 μl 500 mM EGTA, and 2 μl Triton X-100. Should be prepared freshly each day it is used.
100% Methanol (MeOH).
Pronase: 30 mg/ml stock solution.
1X embryo medium (E3): 5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4, and 10−5 % methylene blue. Prepare 10X stock by dissolving 23.4 g NaCl, 1.0 g KCl, 3.9 g CaCl2·2H2O, 6.5 g MgSO4·7H2O and fill with H2O to a total volume of 8L. For 1X working dilution, dilute 1:10 and add methylene blue to inhibit microbial growth (800 ml 10X stock + 8 ml 0.01% methylene blue and fill with H2O to 8 L total).
Forceps (Dumont #5).
Plastic transfer pipettes (~2 mm opening, for washes and transferring embryos pre-dechorionation).
Glass Pasteur pipettes (for transferring dechorionated embryos).
Glutaraldehyde deactivation solution: 0.5 mg/ml sodium borohydride (NaBH4).
2.2. De Novo RNA Probe Design and Synthesis
Primer design program (e.g. Primer3 http://bioinfo.ut.ee/primer3/ [12]).
Custom DNA oligo primers (we order IDT single-stranded 25 nmole DNA Oligos).
Nuclease-free H2O
Thermocycler
dNTPs and PCR buffer (see Note 4).
cDNA (10-100 ng per reaction) from the embryonic stage of interest (see Note 5).
EconoTaq DNA Polymerase: 5 units/μl.
Materials for diagnostic gel: agarose, Tris/Borate/EDTA (TBE) buffer, gel box, DNA marker size ladder (suggested size range 100-1k bp), loading dye.
Optional: DNA Clean & Concentrator kit (Zymo or equivalent).
Optional: RNase AWAY (ThermoFisher or equivalent).
Incubator or water bath to produce a constant temperature of 37 °C.
RNA labeling mix: DIG (Roche) and/or Fluorescein (Roche).
T7 RNA polymerase (50,000 units/ml) and 10X reaction buffer (NEB or equivalent).
RiboLock RNase inhibitor (40 units/μl) (ThermoFisher or equivalent).
TURBO DNase (2 units/μl) (Ambion or equivalent).
7.5 M Lithium chloride (ThermoFisher or equivalent).
100% Ethanol (also diluted in nuclease-free water to make 70% EtOH).
Centrifuge
100% formamide
Hybridization buffer (hyb): 50% formamide, 5X SSC, 100 μg/ml yeast tRNA, 50 μg/ml heparin, 0.25% Tween-20, 1 M citric acid to pH 6.0 (~0.02 M citric acid final concentration). For a 50 ml stock solution, mix 25 ml formamide, 12.5 ml 20X SSC, 1 ml of 25 mg/ml yeast tRNA, 50 μl of 50 mg/ml heparin, 250 μl 20% Tween-20, ~460 μl 1 M citric acid, 10.74 ml DEPC-treated H2O. Store hyb stock at −20 °C.
PCR strip tubes
1.5 ml Eppendorf tubes
Incubator or water bath to produce a constant temperature of 37 °C.
2.3. Two-Color Fluorescence In Situ Hybridization with Microtubule Immunofluorescence
1X PBS-Tween (PBST): 1X PBS, pH 7.0-7.4, 0.1% Tween-20. For 1 L stock solution, mix 100 ml 10X PBS, 12.5 ml 20% Tween-20, and add nuclease free H2O to 1 L. Store at room temperature.
100% Methanol (MeOH).
Methanol/PBS-Tween series: 2:1, 1:1, and 1:2 mixes of MeOH:PBST can be prepared in advance in 15 ml conical tubes.
Forceps (Dumont #5).
Prehybridization buffer (prehyb): 50% formamide, 5X SSC, 0.1% Tween-20. For a 250 ml stock solution, mix 125 ml formamide, 62.5 ml 20X SSC, 1.25 ml 20% Tween-20, and 62.5 ml nuclease free H2O. Store prehyb stock at −20 °C.
Hybridization buffer (hyb): 50% formamide, 5X SSC, 100 μg/ml yeast tRNA, 50 μg/ml heparin, 0.25% Tween-20, 1 M citric acid to pH 6.0 (~0.02M citric acid final conc.). For a 50 ml stock solution, mix 25 ml formamide, 12.5 ml 20X SSC, 1 ml of 25 mg/ml yeast tRNA, 50 μl of 50 mg/ml heparin, 250 μl 20% Tween-20, ~460 μl 1 M citric acid, 10.74 ml DEPC-treated H2O. Store hyb stock at −20 °C.
RNA probe stock: To be added to hybridization buffer at 150-500 ng/ml (see Method 3.2, step 22). Can be made as described in Method 2.2.
2X SSC: 2X SSC, 0.25% Tween-20. For a 400 ml stock solution, mix 40 ml 20X SSC (sodium citrate buffer), 5 ml 20% Tween-20, and nuclease free H2O to 400 ml. Store at room temperature. 50 ml of 0.2X SSC can be prepared by mixing 5 ml 2X SSC with 45 ml nuclease free H2O.
Prehyb/2X SSC wash solutions: 2:1, 1:1, and 1:2 mixes of Prehyb:2X SSC should be prepared in advance in 15 ml conical tubes and can be stored at room temperature.
Hydrogen peroxide (H2O2): to make 2% dilutions in PBS-Tween or TNT.
TNT: 0.1 M Tris-HCl pH 7.5, 0.15 M NaCl, 0.5% Tween-20. For a 1 L stock solution, mix 100 ml Tris pH 7.5, 30 ml 5M NaCl, 25 ml 20% Tween-20, add nuclease free H2O up to 1 L. Store at room temperature.
Blocking solution/TBSTB: TNT with 0.5% PerkinElmer Blocking Powder. Measure 0.25 g PerkinElmer Blocking Powder or equivalent (see Note 6) into a 50 ml conical or glass bottle and fill to 50 ml with TNT. Invert multiple times to mix and heat to 69 °C for at least one hour (ideally while rocking and/or rotating) to fully dissolve powder into solution. Pipet 1 ml aliquots into Eppendorf tubes and store at −20 °C. Once thawed, never refreeze.
TSA Cyanine 3 and Fluorescein System: PerkinElmer Kit includes 1x Amplification Diluent, Tyr-Fluorescein Amplification Reagent, and Tyr-Cy3 Amplification Reagent. The amplification reagents will need to be resuspended in 150 μl DMSO.
Anti-Fluorescein-POD: Roche or equivalent.
Anti-Digoxigenin-POD: Roche or equivalent.
DAPI: 100 μg/ml stock solution.
Glycerol: used to make 30% and 50% Glycerol in PBS-Tween.
1.5 ml Eppendorf tubes
Plastic transfer pipettes (convenient for washes).
Glass Pasteur pipettes (for transferring embryos).
Incubator or water bath to produce a constant temperature of 69 °C.
2.4. Phalloidin labeling for preservation and visualization of F-actin structures in the early zebrafish embryo (with optional in situ hybridization steps for RNA labeling)
Paraformaldehyde (PFA) fix solution: 4% paraformaldehyde in PBS-Tween. Under a fume hood and in a flask of at least 250 ml capacity, dissolve 4 g paraformaldehyde in 100 ml 1X PBS-Tween by bringing to a boil on a hotplate. Be careful to not overboil. Remove from heat and let cool to room temperature. Aliquot into desired amounts (we typically prepare 1 ml and 10 ml aliquots) and store at −20 °C.
Cytoskeleton Fix: 4% PFA, 0.25% glutaraldehyde, 5 mM EGTA, 0.2 % Triton X-100. Per 1 ml of 4% PFA, add 10 μl 25% glutaraldehyde, 10 μl 500 mM EGTA, and 2 μl Triton X-100. Should be prepared freshly each day it is used.
Conjugated phalloidin [e.g. rhodamine-phalloidin (Cytoskeleton) or Alexa-488-Phalloidin (Thermo Fisher)]; to be added to Cytoskeleton Fix at 0.2 units/ml.
Pronase: 30 mg/ml stock solution.
1X embryo medium (E3): 5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4, and 10−5 % methylene blue. Prepare 10X stock by dissolving 23.4 g NaCl, 1.0 g KCl, 3.9 g CaCl2·2H2O, 6.5 g MgSO4·7H2O and fill with H2O to a total volume of 8L. For 1X working dilution, dilute 1:10 and add methylene blue to inhibit microbial growth (800ml 10X stock + 8ml 0.01% methylene blue and fill with H2O to 8 L total).
Forceps (Dumont #5).
Plastic transfer pipettes (for washes and transferring embryos pre-dechorionation).
Glass Pasteur pipettes (for transferring dechorionated embryos).
0.5 mg/ml sodium borohydride (NaBH4).
DAPI: 100 μg/ml stock solution.
1X PBS-Tween (PBST): 1X PBS, pH 7.0-7.4, 0.1% Tween-20. For 1 L stock solution, mix 100 ml 10X PBS, 12.5 ml 20% Tween-20, and add nuclease free H2O to 1 L. Store at room temperature.
Glycerol: used to make 30% and 50% Glycerol in PBS-Tween.
1.5 ml Eppendorf tubes
Plastic transfer pipettes (convenient for washes).
Glass Pasteur pipettes (for transferring embryos).
Incubator or water bath to produce a constant temperature of 37 °C.
2X SSC: 2X SSC, 0.25% Tween-20. For a 400 ml stock solution, mix 40 ml 20X SSC (sodium citrate buffer), 5 ml 20% Tween-20, and nuclease free H2O to 400 ml. Store at room temperature. 50 ml of 0.2X SSC can be prepared by mixing 5 ml 2X SSC with 45 ml nuclease free H2O.
Minimal Prehybridization buffer (minimal prehyb): 10% formamide, 2X SSC. For a 100 ml stock solution, mix 10 ml formamide and 90 ml 2X SSC. Store minimal prehyb at 4 °C.
Hybridization buffer (hyb): 50% formamide, 5X SSC, 100 μg/ml yeast tRNA, 50 μg/ml heparin, 0.25% Tween-20, 1 M citric acid to pH 6.0 (~0.02 M citric acid final conc.). For a 50 ml stock solution, mix 25 ml formamide, 12.5 ml 20X SSC, 1 ml of 25 mg/ml yeast tRNA, 50 μl of 50 mg/ml heparin, 250 μl 20% Tween-20, ~460 μl 1 M citric acid, 10.74 ml DEPC-treated H2O. Store hyb stock at −20 °C.
RNA probe stock: To be added to hybridization buffer at 150-500 ng/ml (see Method 3.2, step 22). Can be made as described in Method 2.2.
Hydrogen peroxide (H2O2): to make 2% dilutions in PBS-Tween or TNT
TNT: 0.1 M Tris-HCl pH 7.5, 0.15 M NaCl, 0.5% Tween-20. For a 1 L stock solution, mix 100 ml Tris pH 7.5, 30 ml 5M NaCl, 25 ml 20% Tween-20, add nuclease free H2O up to 1 L. Store at room temperature.
Blocking solution/TBSTB: TNT with 0.5% PerkinElmer Blocking Powder. Measure 0.25 g PerkinElmer Blocking Powder or equivalent (see Note 6) into a 50 ml conical or glass bottle and fill to 50 ml with TNT. Invert multiple times to mix and heat to 69 °C for at least one hour (ideally while rocking and/or rotating) to fully dissolve powder into solution. Pipet 1 ml aliquots into Eppendorf tubes and store at −20 °C. Once thawed, never refreeze.
TSA Cyanine 3 and/or Fluorescein System: PerkinElmer Kit includes 1x Amplification Diluent, Tyr-Fluorescein Amplification Reagent, and Tyr-Cy3 Amplification Reagent. The amplification reagents will need to be resuspended in 150 μl DMSO.
Anti-Fluorescein-POD or Anti-Digoxigenin-POD: Roche or equivalent.
3. Methods
3.1. Fixation of Zebrafish Embryos for Cytoskeleton Visualization
Preservation of dynamic networks such as the cytoskeleton requires rapid fixation. Here, chorion removal and the addition of glutaraldehyde to the fixation solution increases the speed of fixation, thus improving the ability to visualize intact cytoskeletal structures in the early embryo.
Collect embryos and let develop in petri dishes containing embryo medium (E3). Shortly prior to desired stage, carefully treat with a pronase solution (2 mg/ml in E3) to facilitate chorion removal. While being treated with pronase, swirl the dish and periodically (every ~30 sec, no longer than 2 minutes) press into chorions with forceps to test their integrity. Once you see chorions start to easily buckle or dimple inwards from gentle pressure, carefully pour off the majority of the pronase/E3 solution, taking care not to expose embryos to the air:water interface (which can cause lysis), and quickly replace with E3. The force of the E3 wash will likely cause the majority of embryos to fall out of their chorions, and remaining chorions can be removed manually with forceps or by carefully pipetting through a glass Pasteur pipette (see Note 7).
Wash the embryos 2 more times by carefully pouring off the E3 (again, taking care not to expose embryos to the air:water interface) and replacing it with fresh, then use a glass Pasteur pipette to transfer embryos into 1.5 ml Eppendorf tubes that have been pre-filled with a small amount of E3 (~300 μl) to reduce risk of direct contact with the tube walls and air:water interface. We recommend fixing no more than 45 embryos at once in a single tube.
Carefully remove as much of the E3 as possible without drying out the embryos, then fill the tube with Cytoskeleton Fix. Lay the tube on its side and let fixation occur at room temperature for at least 4 hours. After 4 hours, you can let the embryos rock gently (see Note 8, Fig. 5) overnight at 4 °C or proceed immediately to next steps.
Wash four times for 5 min each in PBS-Tween at room temperature while rocking gently.
Treat embryos with 500 μl/tube 0.5 mM sodium borohydride (NaBH4) for 30 min at room temperature while rocking gently.
Dehydrate the embryos into methanol through a PBS-Tween/Methanol series (steps 6-9). Important: Omit these steps if planning to visualize actin (see Note 3).
Wash embryos in 2:1 PBS-Tween:MeOH for five minutes while rocking gently at room temperature.
Wash embryos in 1:1 PBS-Tween:MeOH for five minutes while rocking gently at room temperature.
Wash embryos in 1:2 PBS-Tween:MeOH for five minutes while rocking gently at room temperature.
Add 1 ml of 100% methanol to each tube of embryos and leave at least overnight at −20 °C. Embryos can be stored in MeOH at −20 °C for several months.
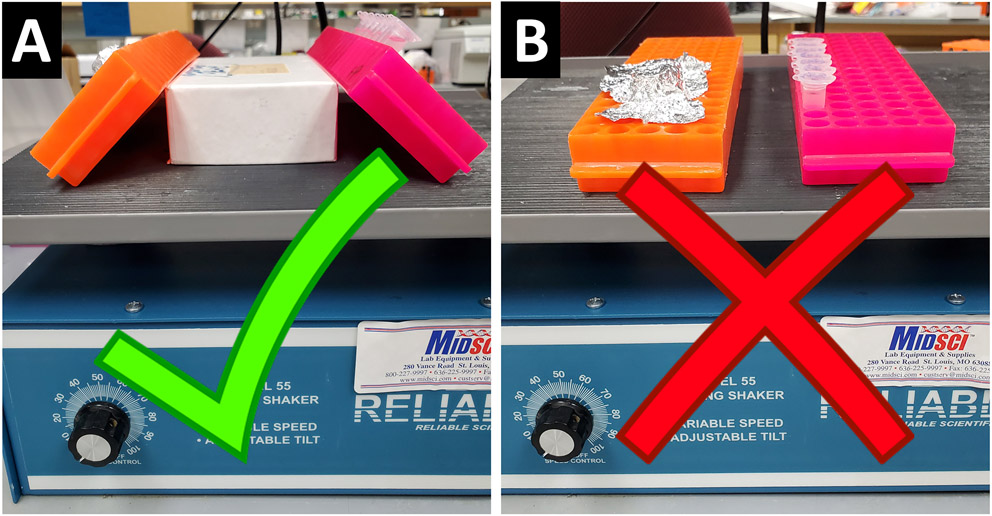
Figure 5.
During washes, we recommend that tube racks should be tilted at an angle (A) rather than sitting flat (B) on the rocker in order to help the wash solutions fully immerse the embryos.
3.2. De Novo RNA Probe Design and Synthesis
This method includes the design and PCR amplification of a probe template (steps 2-7) followed by RNA probe synthesis (steps 8-22). The fluorescence of the RNA probes depends on the RNA labeling mix (conjugated UTPs) used in the transcription reaction mixture (RNA probe synthesis reaction; step 9). If planning to perform two-color fluorescence in situ hybridization (Method 3.3), the probes for each target RNA will need to incorporate a different marker (e.g. fluorescein or digoxigenin).
These initial steps are intended to guide the design of template DNA for RNA probes from scratch using a PCR-based, T7 method [5], but if you already have the DNA template of interest cloned into a plasmid, linearize it (see Note 9) and proceed directly to RNA probe synthesis in step 9.
Upload the exonic sequence of your target gene to a primer design resource, such as Primer3 [16]. Most settings can be left as default, but you will need to select product size ranges. This is up to your discretion, but we typically choose relatively short probes (100 – 300 nt, see Note 1 and Fig. 2).
Select “Pick Primers” and decide which suggested set is most appropriate (see Note 1). For an antisense probe template, add the T7 promoter sequence (see Note 10) to the 5’ end of the reverse primer. For a sense probe template, add the T7 promoter sequence to the 5’ end of the forward primer.
Order primers and resuspend in nuclease-free H2O to a 100 mM stock concentration. For maximum longevity, store at −20 °C and minimize freeze-thaw cycles if possible by aliquoting. However, we find that most primers do well even when stored at 4 °C for several months.
Prepare the following reaction mix for PCR amplification of the template DNA:
| PCR amplification mix (per reaction) | |
|---|---|
| Amount (μl) | |
| Nuclease free H2O | 3.85 |
| dNTP/buffer mix | 17.9 |
| 10mM Fwd primer + 10mM Rev primer mix | 0.15 |
| cDNA (~10-100ng) | 3.00 |
| EconoTaq DNA polymerase | 0.10 |
| Total reaction volume = | 25.0 |
And use the following PCR settings:
| Step | Temperature | Time |
|---|---|---|
| Initial denaturation | 95 °C | 2 min |
| 45 Cycles | 95 °C 45-65 °C (see note 11) 72 °C |
30 sec 15-60 sec 1 min/kb |
| Final extension | 72 °C | 5 min |
| Hold | 4 °C | - |
Run 3 μl of PCR product + 5 μl loading dye vs. a ladder on a 1% agarose gel at 95 V to verify that template was produced and is at the expected size.
Optional: use a Clean & Concentrate kit to increase concentration of PCR product. Only a few microliters of DNA template will be used for the transcription reaction, so we recommend storing the excess at −20 °C for probe synthesis in the future.
From this point forward, be careful about avoiding potential RNase contamination. We find that standard laboratory cleanliness practices and spraying lab bench area, equipment, and gloves with a decontamination reagent such as RNase AWAY is sufficient.
Set up transcription reaction for RNA probe synthesis in RNase-free 1.5 ml Eppendorf tubes as shown below:
| RNA probe synthesis mix (per reaction) | |
|---|---|
| Amount (μl) | |
| Nuclease-free H2O | 12.0 |
| 10X transcription buffer | 2.00 |
| RNA labeling mix (conjugated UTPs) | 2.00 |
| Template DNA (~1-2 μg from PCR or plasmid) | 1.00 |
| Ribolock RNase inhibitor | 1.00 |
| T7 RNA polymerase | 2.00 |
| Total reaction volume = | 20.0 |
Incubate tubes at 37 °C for 2-4 hours.
To remove leftover template DNA at the end of the transcription reaction, add 1 μl of Turbo DNase per tube and incubate at 37 °C for 15 min.
Remove from incubator and precipitate the newly synthesized RNA probes by adding 1.3 μl LiCl and 75 μl chilled 100% ethanol per tube and leave overnight at −20 °C. You will likely observe the solution immediately becomes cloudy as you add the LiCl and EtOH, which is a positive sign that RNA is beginning to precipitate.
Isolate the RNA probes out of solution by centrifugation at 13,000 rpm for 10 min at 4 °C. There should be a visible RNA pellet at the bottom of the tube.
Discard the supernatant without disturbing the RNA pellet and replace with 100 μl 70% EtOH
Centrifuge again at 13,000 rpm for 5 min at 4 °C
Discard as much of the supernatant as possible without disturbing the RNA pellet, then air dry for no more than 5 min at room temperature. One way of doing this is to place several KimWipes on the bench top surface, and prop up the tubes open and upside down on top of the KimWipe. All ethanol and visible liquid should evaporate.
Add 20 μl DEPC-treated H2O and leave at room temperature for 2 min.
Gently vortex at a low setting and/or flick the tube several times to resuspend, then briefly spin down to bottom of tube.
Transfer 2 μl of the resuspension to a PCR tube for later use on a diagnostic gel.
Add 20 μl 100% formamide and store the RNA probe stock, protected from light, at −20 °C to −80 °C.
Run the 2 μl of resuspended RNA that was set aside in step 19 + 5 μl loading dye vs. a ladder on a 1% agarose gel at 120 V to verify that the transcription reaction worked (should visualize a band, not a large smear).
For use in in situ hybridization (colorimetric or fluorescent), prepare a working mix of probes + hybridization buffer, to be stored at −20 °C and which can be reused at least 3 times. If a specific concentration is desired (suggested ranges are 150-500 ng/ml), the RNA concentration can be estimated from the gel or on a Nanodrop and then diluted accordingly. We find that a standard 5 μl of each probe stock per 1 ml of hybridization buffer is sufficient to fully saturate our target transcripts for multiple experiments.
3.3. Two-Color Fluorescence In Situ Hybridization with Microtubule Immunofluorescence
In order to visualize two different target RNAs, this method uses two probes – one made with fluorescein and the other with digoxigenin. Hybridization of both probes is carried out simultaneously, followed by fluorescence development for each color via Tyramide Signal Amplification (TSA™) performed on subsequent days (fluorescein-labeled RNA detection: steps 17-24, digoxigenin-labeled RNA detection: steps 25-31). Next, microtubules are recognized by anti-tubulin antibodies and secondary antibody conjugated to Cy5 (steps 32-38). A suggested DNA stain with DAPI is included in steps 39-40.
Rehydrate the embryos into PBS-Tween through a PBST/Methanol series (steps 2-5).
Wash embryos in 2:1 PBS-Tween:MeOH for five minutes while rocking gently at room temperature (see Note 12, Fig. 5)
Wash embryos in 1:1 PBS-Tween:MeOH for five minutes while rocking gently at room temperature.
Wash embryos in 1:2 PBS-Tween:MeOH for five minutes while rocking gently at room temperature.
Wash at least 2x5 min in PBST. If you are only interested in staining structures in the embryonic blastodisc, you can use this time to remove the yolk using forceps while still in PBST.
Wash once in prehybridization buffer (Prehyb) for 5 min, then add 300 μl of fresh prehyb to each tube of embryos. The embryos can be stored in prehyb at −20 °C or immediately incubated at 69 °C for at least 4 hours. We recommend that each tube used for RNA and/or protein labeling contain only 10-20 embryos so that each embryo is fully saturated by all reagents and wash solutions. Transferring fixed embryos between tubes can be performed with a standard plastic transfer pipette that has an opening wide enough to not damage the embryos as they pass though.
While the embryos are incubating, prepare the desired probe(s)/hyb dilution if it was not already made. We typically use 5 μl of probe stock per 1 ml of hybridization buffer, and aim to have at least 250 μl of probe mix prepared for each tube of embryos. For dual labeling, probes for each target RNA will be added to the same probe hybridization mixture (e.g. 5 μl RNA1-fluorescein probes + 5 μl RNA2-digoxigenin probes in 1 ml hybridization buffer). Once prepared, allow the probe hybridization mix to heat up in the same incubator as the embryos for the remainder of their incubation period.
After the incubation period, remove the prehyb from embryos and replace with at least 250 μl of prepared probe mix. Allow to hybridize in the incubator overnight (see Note 8) at 69 °C.
Preheat the SSC wash solutions to 69 °C for at least 10 min. Remove probe mix and store at −20 °C for reuse, then begin a prehyb:2X SSC wash series (detailed below).
Wash with a 2:1 solution of prehyb: 2X SSC for 5-10 min at 69 °C.
Wash with a 1:1 solution of prehyb: 2X SSC for 5-10 min at 69 °C.
Wash with a 1:2 solution of prehyb: 2X SSC for 5-10 min at 69 °C.
Wash with 2X SSC for 5-10 min at 69 °C.
Wash three times with 0.2X SSC for 15 min each at 69 °C.
Wash with PBS-Tween for 10 min at room temperature while rocking gently (see Note 12).
Freshly prepare a 2% H2O2 in PBS-Tween solution, enough for ~500 μl/tube. Add the 2% peroxide solution to each tube and let gently rock for 60 min. This step is critical to remove potential background from native peroxidase activity but can damage the embryos if left longer than an hour. Be aware that the tubes may pop open due to pressure building up from the peroxide reaction, so be careful not to spill their contents when handling.
Wash four times with TNT at room temperature while rocking gently.
Block for at least 3-4 hours in ~400 μl TBSTB or similar blocking solution (see Note 6) at room temperature while rocking gently. The embryos can be left rocking overnight in blocking solution at 4 °C if needed, but if possible it is best to proceed to the antibody step before leaving overnight.
Remove block and replace with ≥250 μl 1:1000 anti-Fluorescein-POD in blocking solution, and leave rocking gently overnight at 4 °C. If needed, the embryos can be left in antibody solution at 4 °C for multiple nights (3 maximum) before proceeding to next steps.
Discard antibody solution and wash 8 times in TNT while gently rocking at room temperature over the course of 1-2 hours.
Prepare 1:50 Tyr-Fluorescein Amplification Reagent in Amplification Diluent for 25 μl*(n+1) tubes to ensure there will be enough mix for all samples. Add 25 μl of the 1:50 amplification mix to each tube, and cover with foil to protect from light. From this point on, the embryos should be protected from light during all wash steps because the fluorescence has been developed/activated.
Shake upright at room temperature for one hour. If you do not have a shaker, a benchtop rocker will also work.
Wash two times for 5 min each in TNT at while rocking gently at room temperature.
Wash one hour in freshly prepared 2% H2O2 in TNT while rocking gently at room temperature.
Wash three times for 5 min each in TNT at while rocking gently at room temperature.
Block for 1-2 hours in ~400 μl blocking solution at room temperature while rocking gently. The embryos can be left rocking overnight in blocking solution at 4 °C if needed, but if possible it is best to proceed to the antibody step before leaving overnight.
Remove block and replace with ≥250 μl 1:1000 anti-DIG-POD in blocking solution, and leave rocking gently overnight at 4 °C. If needed, the embryos can be left in antibody solution at 4 °C for multiple nights (3 maximum) before proceeding to next steps.
Discard antibody solution and wash 8 times in TNT while gently rocking at room temperature over the course of 1-2 hours.
Prepare 1:50 Tyr-Cy3 in amplification diluent for 25 μl*(n+1) tubes to ensure there will be enough mix for all samples. Add 25 μl of the 1:50 amplification solution to each tube.
Shake upright at room temperature for one hour. If you do not have a shaker, a benchtop rocker will also work.
Wash two times for 5 min each in TNT at while rocking gently at room temperature.
Wash one hour in freshly prepared 2% H2O2 in TNT while rocking gently at room temperature.
Wash 5 min in TNT, then 5 min in 1:1 TNT:PBS-Tween while rocking gently at room temperature.
Wash three times for 10min each in PBS-Tween, then replace with blocking solution and let rock gently at room temperature for at least 2 hours.
Replace blocking solution with at least ≥250 μl /tube of prepared anti-microtubule primary antibody diluted in blocking solution, and leave rocking gently overnight at 4 °C. If needed, the embryos can be left in antibody solution at 4 °C for multiple nights (3 maximum) before proceeding to next steps.
Wash four times for 30 min each in PBS-Tween while rocking gently at room temperature.
Add ~400 μl of blocking solution per tube and let block for at least 1 hour while rocking gently at room temperature.
Replace blocking solution with appropriate secondary antibody dilution in blocking solution and let rock gently overnight at 4 °C. It is possible to reduce the time for this step to 2-4 hours, but we find that leaving the embryos in secondary overnight produces better labeling.
Wash two times for 30 min each in PBS-Tween while rocking gently at room temperature. Wash two additional times if not proceeding to DAPI labeling.
In order to visualize nuclei, add 200 μl per tube of 1:200 DAPI in PBS-Tween and let rock gently at room temperature for 10-20 min.
Wash three times for 5 min each in PBS-Tween while rocking gently at room temperature.
To prepare for mounting, remove the PBS-Tween and replace with ~500 μl of a 30% glycerol in PBS-Tween solution. Leave the tubes on a flat, stationary surface so the embryos can settle to the bottom, then remove the 30% glycerol and replace with 50% glycerol in PBS-Tween. Again, let the embryos settle to the bottom. They can be left in the glycerol solution for multiple nights (maximum 3) at 4 °C before being mounted if necessary.
Mount embryos as desired for imaging (for mounting tips, see Note 13).
3.4. Rapid phalloidin labeling for preservation and visualization of F-actin structures in the early zebrafish embryo (with optional in situ hybridization steps for RNA labeling)
In this method, embryo fixation and F-actin labeling (steps 1-6) take place simultaneously due to the addition of fluorophore-conjugated phalloidin to the fixation solution, which promotes better preservation of F-actin structure. For RNA visualization in combination with the actin cytoskeleton, optional in situ hybridization steps are included (steps 8-25). A suggested DNA stain with DAPI is included in steps 26-27.
Freshly prepare the combined cytoskeleton fixative/phalloidin labeling mix the day of the experiment by mixing the following:
| Cytoskeleton fixative/Phalloidin-labeling mix (per tube of embryos) |
||
|---|---|---|
| Amount (μl) | Final concentration | |
| 4% paraformaldehyde (PFA) | 965.5 | - |
| 25% glutaraldehyde | 10.0 | 0.25% |
| 500 mM EGTA | 10.0 | 5 mM EGTA |
| Triton X-100 | 2.0 | 0.2% |
| 1:200 conjugated phalloidin | 12.5 | 0.2 units/ml |
Important: Whenever possible, protect all phalloidin-containing solutions and phalloidin-labeled samples from light.
Collect embryos and let develop in petri dishes containing embryo medium (E3). Shortly prior to desired stage, carefully treat with a pronase solution (2 mg/ml in E3) to facilitate chorion removal. While being treated with pronase, swirl the dish and periodically (every ~30 sec) press into chorions with forceps to test their integrity. Once you see chorions start to easily buckle or dimple inwards from gentle pressure, carefully pour off the majority of the pronase/E3 solution and quickly replace with E3. The force of the E3 wash will likely cause the majority of embryos to fall out of their chorions, and any remaining chorions can be removed manually with forceps.
Wash the embryos 2 more times by carefully pouring off the E3 and replacing it with fresh, then use a glass Pasteur pipette to transfer embryos into 1.5 ml Eppendorf tubes. We recommend fixing no more than 60 embryos at once in a single tube.
Carefully pipette off as much of the E3 as possible without drying out the embryos, then fill the tube with cytoskeleton fixative/phalloidin-labeling mix. Lay the tube on its side, protect it from light (e.g. covered with foil or left in a drawer) and leave at room temperature for at least 4 hours. After 4 hours, you can let the embryos rock gently overnight at 4 °C or proceed immediately to next steps. We find that phalloidin-labeling is slightly improved if allowed to continue overnight.
Wash four times for 5 min each in PBS-Tween at room temperature while rocking gently (see Note 12, Fig. 5). Continue protecting embryos from light during all of the wash steps.
Treat embryos with 500 μl/tube 0.5 mM sodium borohydride (NaBH4). This step is necessary for deactivating/reducing the glutaraldehyde.
Wash three times for 5 min each in PBS-Tween at room temperature while rocking gently.
If additional protein labeling is desired, the embryos can immediately go into blocking buffer at this point to begin an immunofluorescence assay (e.g. for combined F-actin and microtubule labeling, here proceed to step 34 of Method 3.3 and follow through to end). For RNA labeling via in situ hybridization, proceed to the next steps. Otherwise, skip directly to step 26 for DNA staining and mounting instructions.
Ensure that the embryos are still protected from light whenever possible. Wash twice in a minimal prehybridization buffer (10% formamide in 2X SSC) for 5 min each at room temperature, then add fresh minimal prehyb to each tube of embryos and let incubate at 37 °C for at least 2 hours (see Note 3).
While the embryos are incubating, prepare the desired probe/hyb dilution if it was not already made. We typically use 5 μl of probe stock per 1 ml of hybridization buffer, and aim to have at least 250 μl of probe mix prepared for each tube of embryos. Once prepared, allow the probe mix to heat up to 37 °C for the remainder of their incubation period.
After the incubation period, remove the minimal prehyb from embryos and replace with at least 250 μl of prepared probe mix. Allow to hybridize at 37 °C in the incubator overnight (see Note 8).
Preheat the SSC wash solutions to 37 °C for at least 10 min. Remove probe mix and store at −20 °C for reuse, then begin wash series (detailed below).
Wash with a 1:1 solution of minimal prehyb:2X SSC for 5-10 min at 37 °C.
Wash with 2X SSC for 5-10 min at 37 °C.
Wash three times with 0.2X SSC for 15 min each at 37 °C.
Wash with PBS-Tween for 10 min at room temperature while rocking gently (see Note 12).
Freshly prepare a 2% H2O2 in PBS-Tween solution, enough for ~500 μl/tube. Add the 2% peroxide solution to each tube and let gently rock for 60 min. This step is critical to remove potential background from native peroxidase activity, but can damage the embryos if left longer than an hour. Be aware that the tubes may pop open due to pressure building up from the peroxide reaction, so be careful not to spill their contents when handling.
Wash four times for 5 minutes each with PBS-Tween at room temperature while rocking gently
Block for at least 3-4 hours in ~400 μl TBSTB or similar blocking solution (see Note 6) at room temperature while rocking gently. The embryos can be left rocking overnight in blocking solution at 4 °C if needed, but if possible it is best to proceed to the antibody step before leaving overnight.
Remove block and replace with ≥250 μl 1:1000 anti-Fluorescein-POD in blocking solution, and leave rocking gently overnight at 4 °C.
Discard antibody solution and wash 8 times in PBS-Tween while gently rocking at room temperature over the course of 1-2 hours.
Prepare 1:50 Tyr-Fluorescein Amplification Reagent in Amplification Diluent for 25 μl*(n+1) tubes to ensure there will be enough mix for all samples. Add 25 μl of the 1:50 amplification mix to each tube, and cover with foil to protect from light.
Shake upright at room temperature for one hour. If you do not have a shaker, a benchtop rocker will also work.
Wash two times for 5 min each in TNT at while rocking gently at room temperature.
Wash one hour in freshly prepared 2% H2O2 in TNT while rocking gently at room temperature.
Wash three times for 5 min each in TNT at while rocking gently at room temperature.
In order to visualize nuclei, add 200 μl per tube of 1:200 DAPI in PBS-Tween and let rock gently at room temperature for 10-20 min.
Wash three times for 5 min each in PBS-Tween while rocking gently at room temperature.
To prepare for mounting, remove the PBS-Tween and replace with ~500 μl of a 30% glycerol in PBS-Tween solution. Leave the tubes on a flat, stationary surface so the embryos can settle to the bottom, then remove the 30% glycerol and replace with 50% glycerol in PBS-Tween. Again, let the embryos settle to the bottom. They can be left in the glycerol solution for multiple nights (max 3) at 4 °C before being mounted if necessary.
Mount embryos as desired for imaging (for mounting tips, see Note 13).
4. Notes
There are numerous strategies that can be applied to probe design, and here we will only briefly discuss a few key considerations. Depending on your needs and/or preferences, both sequence length and target region can be extensively varied. Increasing probe length and/or including stretches of the 5’ or 3’ UTR are strategies that increase target specificity. Designing probes that span exon-exon junctions ensures that the probes will only detect spliced transcripts, rather than genomic sequence, although this is less of a concern since RNA:RNA hybrids are much more stable than RNA:DNA hybrids [19]. In older embryos, probes that are longer than ~600 nt may have difficulty fully penetrating the sample; however, we have used full-length probes ~1-1.5 kB in length without difficulty in the early embryo (up to 6 hours post-fertilization). Another potential benefit of longer probes is increased fluorescent signal due to greater numbers of conjugated-UTPs versus the quantity incorporated into short probes. However, for most purposes, short to medium length probes (~100-300 nt) work well because they are short enough to avoid potentially problematic sequence (e.g. repetitive sequence, regions of self-complementarity, extreme G-C content), can easily penetrate whole embryos, typically provide ample fluorescent signal, and the template sequence can be reliably amplified with standard DNA polymerases such as EconoTaq. For additional information, probe design considerations are described in further depth by Aquino de Muro, 2008 [6].
In order to visualize three RNAs in a single sample rather than two, one would need to synthesize an additional RNA probe stock that incorporated UTPs conjugated to a different fluorophore and/or hapten than fluorescein or digoxigenin, such as dinitrophenol (DNP-11-UTP, PerkinElmer), and simply add it to the probe hybridization mixture at the same concentration as the other two probes, as described in Method 3.3, step 7. Further modify Method 3.3 by adding another day of RNA recognition and signal development procedure using the TSA Cyanine 5 System (PerkinElmer) after step 32. For specific directions, see the Triple Fluorescent In Situ protocol shared by the Talbot lab: https://wiki.zfin.org/display/prot/Triple+Fluorescent+In+Situ.
In standard in situ protocols, methanol treatment is intended to increase penetration of RNA probes by permeabilizing the embryo; however, at early stages (cleavage to early blastula), the embryo is naturally quite permeable, so we find that this step (Method 3.1, steps 5-9) can be omitted without negative consequence. For older embryos, other permeabilization agents have been suggested, such as ethanol, glacial acetic acid, or detergents. To address the potential for high formamide concentration negatively impacting phalloidin labeling quality, we found that decreasing the formamide concentration to 10% in 2X SSC, rather than 50% in prehybridization buffer (Materials 2.3, #5), is sufficient to rescue the phalloidin staining quality. This alteration slightly decreases the stringency of post-hybridization washes, so a high-quality/high-specificity RNA probe is necessary. Lastly, simply decreasing the probe hybridization temperature to 37 °C prevents the negative impact of high temperature exposure on phalloidin staining. As with decreasing the formamide concentration of wash buffers, significantly decreasing the hybridization temperature runs the risk of decreasing in situ efficacy, so we recommend first attempting it with well-established, high quality RNA probes.
- Many labs have their own preferred method of adding dNTPs and reaction buffer to their PCR master mix, and we encourage readers to use the method that works best for them. We prepare and freeze ready-to-use 1 ml aliquots of combined reaction buffer and dNTPs in ~200 ml batches by mixing the following materials:
dNTP/buffer mix (aliquots stored at −20C) Amount (mL) Final concentration 1 M MgCl2 0.393 2.05 mM 1 M pH 8.4 Tris-HCl 2.613 13.7 mM 1 M KCl 13.092 68.3 mM BSA (100 mg/ml) 3.468 27.1 mM 100 mM dATP 0.262 137 μM 100 mM dCTP 0.262 137 μM 100 mM dGTP 0.262 137 μM 100 mM dTTP 0.262 137 μM Nuclease-free H2O 171.12 - An in-depth protocol and instructional video for RNA isolation from embryonic zebrafish and cDNA synthesis can be found in Peterson and Freeman, 2009 [17].
The PerkinElmer Blocking Powder Reagent is recommended by the manufacturer when using the PerkinElmer TSA System, and is sometimes conveniently included when purchasing various PerkinElmer kits. Roche Blocking Reagent for nucleic acid hybridization and detection also works well when used at the same concentration as the PerkinElmer powder (0.5% in TNT), and both perform better than 1% BSA. Both the PerkinElmer and Roche powders are primarily comprised of purified casein protein, so we predict that using 0.5% casein powder in TNT as an in-house alternative would perform similarly, although we have not yet tried this formulation.
Embryos can also be manually dechorionated with a pair of fine forceps after a very brief (<1 min) exposure to pronase solution. It may be possible to omit dechorionation via pronase treatment prior to fixation if you do not plan to visualize the cytoskeleton. Pre-fixation dechorionation and the suggested modified 4% PFA solution are used to fix embryos faster and allow better preservation of intracellular structures than traditional methods. Fluorescent in situ hybridization and immunofluorescence of many proteins other than tubulin and/or actin can be performed on early stage embryos (pre-epiboly) that were fixed with 4% PFA without any additional reagents and while still in their chorions. However, especially for proteins that may have a subcellular localization, we recommend always testing whether conditions for fixation of cytoskeletal elements result in improved signal, localization, or resolution. If the cytoskeleton-specific steps are omitted, manually dechorionate post-fixation with forceps prior to permeabilization with 100% MeOH.
We recommend a “lengthy” overnight hybridization timeframe of at least 18 hours. The embryos can also be allowed to hybridize for an additional day and overnight.
A detailed explanation of T7 transcription reactions using plasmid or PCR-amplified target DNA, including a protocol for linearizing plasmids, can be found in Brunelle and Green, 2013 [18].
We include a short spacer sequence at the beginning of the T7 promoter sequence, so that the total sequence added is: 5’-GAAATTAATACGACTCACTATAGG
Determine annealing temperature based on primer pair Tm. For simplicity, our lab finds that a standard annealing setting of 55 °C for 30 sec per cycle works in most cases.
Instead of leaving tube of embryos fully upright in their racks while on a benchtop rocker, place a small object (e.g. box lid or another tube rack) on the rocker and tilt the rack with embryos up against it. This helps spread out the embryos inside the tube and fully immerses them in the wash solutions/reagents (Fig 5).
There are many mounting procedures and we encourage readers to use the one that works best for them and their samples. Our lab typically follows the following methods: To flat mount blastodiscs from cleavage – late blastula stage embryos, we find that stacking two binder hole reinforcement rings on a glass slide provides a conveniently sized “well” for holding the deyolked embryos in place while also serving as a spacer to prevent them from becoming squashed by the cover slip. After the embryos have settled in 50% glycerol, transfer several (≤8) to the well with as little liquid as possible. Add a drop of mounting medium of choice on top of the embryos. There should be a visible “bubble” of liquid slightly raised, but there should not be enough to break surface tension and spill over onto the page reinforcement label surrounding the well. Using forceps, gently move the deyolked embryos so that the blastodiscs are facing up (i.e. so that the animal cortex of the embryo – not the more vegetally located blastodisc edges- will be closest to the coverslip). Blastodiscs mounted facing down typically cannot be visualized properly at high magnifications due to the increased distance to the objective. To better discern the orientation of the blastodiscs, use a dissection needle or forceps to tilt them sideways, then orient them in the desired orientation. Try to avoid placing any directly next to the edge of the well. Carefully place a glass coverslip over the embryos by holding the coverslip on two opposite edges and bringing it as close to the sample as possible before releasing. Very gently press down on the four corners of the coverslip, then paint around all of the edges with clear nail polish (or sealant of choice) to seal it in place and prevent dehydration.
Acknowledgements
This work was supported by NIH grant GM007133-44 to C.L.H. and GM065303 to F.P., with additional support from the College of Agricultural and Life Sciences, the School of Medicine and Public Health, and the Laboratory of Genetics at the University of Wisconsin – Madison.
References
- 1.Jansen RP (1999) RNA-cytoskeletal associations. FASEB J Off Publ Fed Am Soc Exp Biol 13:455–466 [PubMed] [Google Scholar]
- 2.Theusch EV, Brown KJ, Pelegri F (2006) Separate pathways of RNA recruitment lead to the compartmentalization of the zebrafish germ plasm. Dev Biol 292:129–141 . doi: 10.1016/j.ydbio.2005.12.045 [DOI] [PubMed] [Google Scholar]
- 3.Kloc M, Zearfoss NR, Etkin LD (2002) Mechanisms of Subcellular mRNA Localization. Cell 108:533–544 . doi: 10.1016/S0092-8674(02)00651-7 [DOI] [PubMed] [Google Scholar]
- 4.Milligan JF, Uhlenbeck OC (1989) [5] Synthesis of small RNAs using T7 RNA polymerase. In: Methods in Enzymology. Academic Press, pp 51–62 [DOI] [PubMed] [Google Scholar]
- 5.David R, Wedlich D (2001) PCR-based RNA probes: a quick and sensitive method to improve whole mount embryo in situ hybridizations. BioTechniques 30:769–772, 74 [PubMed] [Google Scholar]
- 6.Aquino de Muro M (2008) Probe Design, Production, and Applications. In: Molecular Biomethods Handbook: Second Edition. pp 41–53 [Google Scholar]
- 7.Clay H, Ramakrishnan L (2005) Multiplex Fluorescent In Situ Hybridization in Zebrafish Embryos Using Tyramide Signal Amplification. Zebrafish 2:105–111 . doi: 10.1089/zeb.2005.2.105 [DOI] [PubMed] [Google Scholar]
- 8.Zaidi AU, Enomoto H, Milbrandt J, Roth KA (2000) Dual Fluorescent In Situ Hybridization and Immunohistochemical Detection with Tyramide Signal Amplification. J Histochem Cytochem 48:1369–1375 . doi: 10.1177/002215540004801007 [DOI] [PubMed] [Google Scholar]
- 9.Lynen F, Wieland U (1938) Über die Giftstoffe des Knollenblätterpilzes. IV. Justus Liebigs Ann Chem 533:93–117 . doi: 10.1002/jlac.19385330105 [DOI] [Google Scholar]
- 10.Wulf E, Deboben A, Bautz FA, Faulstich H, Wieland T (1979) Fluorescent phallotoxin, a tool for the visualization of cellular actin. Proc Natl Acad Sci U S A 76:4498–4502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melak M, Plessner M, Grosse R (2017) Actin visualization at a glance. J Cell Sci 130:525–530 . doi: 10.1242/jcs.189068 [DOI] [PubMed] [Google Scholar]
- 12.Dancker P, Löw I, Hasselbach W, Wieland T (1975) Interaction of actin with phalloidin:. Polymerization and stabilization of F-actin. BBA - Protein Struct 400:407–414 [DOI] [PubMed] [Google Scholar]
- 13.Vandekerckhove J, Deboben A, Nassal M, Wieland T (1985) The phalloidin binding site of F-actin. EMBO J 4:2815–2818 . doi: 10.1002/j.1460-2075.1985.tb04008.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eno C, Pelegri F (2018) Modulation of F-actin dynamics by maternal Mid1ip1L controls germ plasm aggregation and furrow recruitment in the zebrafish embryo. Development 145:dev156596 . doi: 10.1242/dev.156596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kellogg DR, Mitchison TJ, Alberts BM (1988) Behaviour of microtubules and actin filaments in living Drosophila embryos. Development 103:675–686 [DOI] [PubMed] [Google Scholar]
- 16.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3—new capabilities and interfaces. Nucleic Acids Res 40:e115 . doi: 10.1093/nar/gks596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peterson SM, Freeman JL (2009) RNA isolation from embryonic zebrafish and cDNA synthesis for gene expression analysis. J Vis Exp JoVE. doi: 10.3791/1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brunelle JL, Green R (2013) Chapter Five - In Vitro Transcription from Plasmid or PCR-amplified DNA. In: Lorsch J (ed) Methods in Enzymology. Academic Press, pp 101–114 [DOI] [PubMed] [Google Scholar]
- 19.Sugimoto N, Nakano S, Katoh M, Matsumura A, Nakamuta H, Ohmichi T, Yoneyama M, Sasaki M (1995) Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry 34:11211–11216 . doi: 10.1021/bi00035a029 [DOI] [PubMed] [Google Scholar]