Abstract
Somatic mutations at Thr-58 of c-Myc have been detected in Burkitt's lymphoma (BL) tumors and have been shown to affect the transforming potential of the Myc oncoprotein. In addition, the N-terminal domain of c-Myc has been shown to interact with microtubules in vivo, and the binding of c-Myc to α-tubulin was localized to amino acids 48 to 135 within the c-Myc protein. We demonstrate that c-Myc proteins harboring a naturally occurring mutation at Thr-58 from BL cell lines have increased stability and are constitutively hyperphosphorylated, which disrupts the in vivo interaction of c-Myc with α-tubulin. In addition, we show that wild-type c-Myc–α-tubulin interactions are also disrupted during a transient mitosis-specific hyperphosphorylation of c-Myc, which resembles the constitutive hyperphosphorylation pattern of Thr-58 in BL cells.
The c-myc gene encodes a nuclear phosphoprotein that has been implicated in the regulation of cell proliferation and the development of human tumors (23, 28). c-Myc is a helix-loop-helix–leucine zipper protein that binds DNA as a heterodimer with Max protein to activate or repress transcription (5, 10, 11). In addition, Myc can complex with Miz to mediate gene repression (33). Miz-1, however, lacks a nuclear localization signal and is observed to accumulate in the cytoplasm in association with microtubules. Overexpression of c-Myc stimulates Miz-1 import to the nucleus, where it is proposed to act as a repressor of transcription (33). Since c-Myc was previously shown to associate with α-tubulin and microtubules in vitro and in vivo (2), it has been proposed that microtubules may act as a cytosolic anchor for both Myc and Miz-1 and to regulate Myc nuclear import (33).
Nuclear import of Myc, however, has been shown to be blocked in both human myeloid leukemia cells and neuronal cells during differentiation, where c-Myc or N-Myc, respectively, was observed to accumulate in the cytoplasm (12, 43). In addition, c-Myc is stored in the cytoplasm in nondividing Xenopus oocytes and is rapidly translocated to the nucleus upon fertilization (19). These findings suggested that cytoplasmic-nuclear exchanges of c-Myc may play an important role in the control of proliferation, differentiation, and development (43) and also implied that microtubules may play a role in sequestration of c-Myc in the cytoplasm (2), although the mechanism of such interaction remains to be determined.
It has been shown that binding of c-Myc to α-tubulin in vitro is mediated through the N-terminal domain of c-Myc (2), which is essential for the transcriptional transactivation and repression as well as transforming activities of the c-Myc protein (27). Several lines of evidence have accumulated indicating that Thr-58 is an important functional residue in the N terminus of the c-Myc protein. For example, mutation of Thr-58 to alanine increases the ability of c-Myc to induce focus formation in embryo fibroblast (17, 34) and enhances the ability of c-Myc-transfected Rat 1A cells to grow in soft agar (22, 24). In addition, Thr-58 is a target for mutations in the majority of v-Myc proteins that are highly transforming relative to v-Myc alleles that retain Thr-58 (8, 32; T. S. Papas and J. A. Lautenberger, Letter, Nature 318:237, 1985), and restoring Thr-58 to the v-Myc protein inhibits its ability to transform cells in culture (41). The observation that Burkitt's lymphoma (BL) tumors frequently contain naturally occurring somatic mutations in Thr-58 (4, 45) further suggests that this is an important functional site within the c-Myc protein. It has also been reported that mutation at Thr-58 leads to hyperphosphorylation of c-Myc at the adjacent Ser-62 site (24, 30). Although the role of Ser-62 remains controversial (22, 34) it has been suggested that phosphorylation at Thr-58 transduces a negative growth signal (22, 34), which may explain the growth-proliferative phenotype of the Thr-58 mutants.
Since the interaction of c-Myc and α-tubulin in vitro has been previously localized to amino acids 48 to 135 in the N-terminal domain of c-Myc (2), we investigated whether mutations at Thr-58 in c-Myc from BL cells affect the binding to α-tubulin. We demonstrated that the Thr-58-to-Ala mutation in c-Myc from BL cells results in constitutive hyperphosphorylation of c-Myc with disruption of Myc–α-tubulin binding in vivo. Since hyperphosphorylation of mutant c-Myc was associated with disruption of Myc–α-tubulin binding and since the N-terminal domain of wild-type (wt) c-Myc is hyperphosphorylated during mitosis (29), we examined binding of wt c-Myc to α-tubulin in HeLa cells arrested at the mitotic stage of the cell cycle. We showed that c-Myc–α-tubulin interaction is also disrupted during mitosis-specific hyperphosphorylation of c-Myc. These data demonstrate that the c-Myc–α-tubulin interaction is regulated by the phosphorylation state of c-Myc and suggest that the loss of c-Myc–α-tubulin interaction at mitosis may be a physiologic requirement for cell division, while disruption of c-Myc–α-tubulin binding due to the constitutive hyperphosphorylation of mutant c-Myc may be associated with the transformed phenotype.
MATERIALS AND METHODS
Cell lines, antibodies, and nocodazole treatment.
Raji, PA682, and KK124 BL cells were kindly supplied by I. Magrath and propagated in RPMI medium containing 10% fetal calf serum (Gibco Biochemicals). HeLa, Cos, and HEK293 cells were grown in Dulbecco's modified Eagle's medium (DMEM) in 10% fetal calf serum (Gibco Biochemicals). Anti-c-Myc clone 9E10 (Ab-1) and Ab-2 were used for immunoblotting, and anti-c-Myc Ab-3 was used for immunoprecipitation as recommended by the manufacturer (Oncogene Research Product). Anti-c-MycFL was used for phosphopeptide mapping and pulse-chase analysis (39). Anti-α-tubulin monoclonal antibody (clone DMIA) was obtained from Amersham. HeLa cells were treated with 100 ng of nocodazole per ml as described previously (29), and the mitotic cells were removed from the monolayer by shake-off after 16 h of incubation.
Expression vectors and transient- and stable-transfection assays.
Myc p64 and Myc p67 expression plasmids were a kind gift from L. Kretzner. Myc p64 and Myc p67 correspond to mutant 3 and mutant 15, which were described previously (5). pSV-MycPA was constructed by subcloning the HindIII/EcoRI 8.1-kb c-myc fragments from the PA682 genomic library into the HindIII/EcoRI 2.1-kb fragment from the pSV2neo vector. The pSV-Myc plasmid was constructed by subcloning the wild-type c-myc genomic clone derived from human liver (13). For transfection to Cos and HEK293 cells, 1.0 × 106 cells were plated in 100-mm-diameter dishes and incubated overnight at 37°C. Subconfluent cells were transfected by calcium phosphate precipitation (37) for 16 h using 10 to 20 μg of plasmid DNA. For transient-transfection experiments, cells were harvested 24 h following the removal of the plasmid precipitate. For stable transfection of PA682 cells, logarithmically growing cells (2 × 107) were transfected with 10 μg of the Myc p64 and Myc p67 expression plasmids, using Lipofectin reagent as recommended by the manufacturer (Gibco BRL).
Cellular extract preparation, immunoprecipitation, immunoblotting, and competition analysis.
Whole-cell extracts used for immunoprecipitation and Western blot analysis were prepared as previously described (2, 26) in lysis buffer (50 mM Tris [pH 8], 200 mM NaCl, 0.5% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, 0.5 mM dithiothreitol, 50 mM NaF) containing leupeptin and aprotinin (10 μg/ml each). Protein lysates were immunoprecipitated with α-tubulin or c-Myc (Ab-3) antibodies by using agarose G-beads as recommended by the manufacturer (Oncogene Research Product). The washed pellets were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose filters, and immunoblotted with anti-c-Myc (clone 9E10) or anti-α-tubulin as described previously (2). Two hundred micrograms of protein extracts was used in immunoblotting, while 2 mg of the same extracts was used for sequential immunoprecipitation and immunoblotting. For HEK293 cells 50 μg of protein extracts was used in immunoblotting, while 500 μg of the same extracts was used for immunoprecipitation with anti-α-tubulin. Blocking of antiserum was performed by incubating 10 μg of a c-Myc peptide for 2 h at 4°C with the c-Myc antiserum (clone 9E10) in a total volume of 200 μl before immunoblotting. The c-Myc peptide, purchased from Oncogene Research Product, was originally used to raise the anti-c-Myc clone 9E10 (15).
PAP, CKII, and MAP kinase treatment.
Protein lysate from PA682 cells was immunoprecipitated with c-Myc (Ab-3) antibody and incubated either with phosphate buffer alone or with potato acid phosphatase (PAP) (Boehringer Mannheim) as described previously (9). The treated and untreated immunoprecipitates were subject to SDS–7.5% PAGE and immunoblotted with anti-c-Myc (Ab-1). For the in vitro kinase assay, glutathione S-transferase (GST)–Myc II fusion protein and GST alone, prepared as described previously (2), were treated with casein kinase II (CKII) and mitogen-activated protein (MAP) kinase (Upstate Biotechnology) as described by manufacturer. CKII- and MAP kinase-treated and untreated proteins were incubated with 1,000 μg of HL60 total cell extract as described previously (2). Precipitated proteins were resolved by SDS–7.5% PAGE and were detected with α-tubulin and c-Myc (Ab-2) antibody.
Pulse-chase experiments.
Cos cells (2.5 × 106 per 100-mm-diameter dish) were plated 24 h before transfection, and 5 μg of the indicated plasmid was used for transfection by the calcium phosphate method. At 6 h after transfection, cells were washed, incubated for additional 24 h, split 1:4 into 100-mm-diameter dishes, and subjected to pulse-chase analysis 24 h later. For the pulse-chase, cells were washed with phosphate-buffered saline and labeled with 300 μCi of [35S]methionine-cysteine (Trans35S-label [ICN]) per plate in methionine- and cysteine-free Dulbecco modified Eagle medium (DMEM) (Gibco BRL) at 37°C for 10 min. After being labeled, the cells were washed with phosphate-buffered saline and chased with complete DMEM containing 10% calf serum at 37°C for the indicated time period. For the BL lines, 5 × 107 cells were pulse-labeled with 1 mCi of Trans35S-label and chased in RPMI 1640 medium containing 10% fetal calf serum for the indicated time period. A total of 107 cells were collected at each time point. Cells were subsequently subjected to immunoprecipitation analysis using anti-MycFL (39). The half-life values were determined by a logarithmic analysis of the data obtained from densitometric scanning of autoradiographs.
In vivo labeling and phosphopeptide analysis.
Cells were labeled with [32P]orthophosphate for 4 h in DMEM–3% dialyzed fetal calf serum. Labeled c-Myc protein was immunoprecipitated from cells with anti-MycFL (39), separated by SDS-PAGE, transferred to nitrocellulose, and digested off the membrane with 10 μg of thermolysin (Worthington Biochemicals). Digestion was followed by performic acid oxidation for 1 h at 0°C. Peptides were then washed twice with H2O and lyophilized. The digested fragments were separated in the first dimension by electrophoresis using a Hunter thin-layer electrophoresis chamber in pH 1.9 buffer (1.5 kV, 30 min) and then separated in the second dimension by ascending chromatography in the phosphochromatography buffer (7).
Synchronization and in vivo labeling of HeLa cells.
HeLa cells were synchronized with a double thymidine block as previously described (40). Cells were labeled with [32P]orthophosphate 2 h before harvest. Labeled c-Myc protein was immunoprecipitated from cells with anti-c-Myc (Ab-3) and anti-α-tubulin, resolved by SDS-PAGE, and visualized by autoradiography. Duplicate cultures were harvested at different time points, and cell cycle analysis was performed (FAST Systems, Inc., Gaithersburg, Md.).
RESULTS
Mutated c-Myc with altered mobility does not bind α-tubulin in vivo.
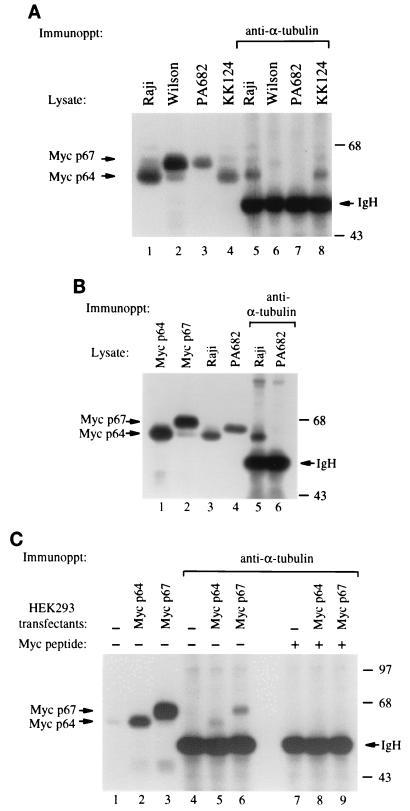
Since the N-terminal domain of c-Myc was shown to interact in vivo and in vitro with α-tubulin and polymerized microtubules (2), we sought to determine whether mutations frequently found in the N-terminal region of c-Myc from BL cells would affect the binding to α-tubulin. We immunoprecipitated protein extracts from BL cells with a monoclonal antibody directed against α-tubulin, followed by immunoblotting with anti-c-Myc. We found that anti-α-tubulin coprecipitated both c-Myc and α-tubulin from Raji and KK124 BL cells, while the binding was defective in the PA682 and Wilson BL cell lines (Fig. 1A). In addition, α-tubulin was coprecipitated with c-Myc from the Ramos and Daudi BL cell lines (data not shown). Immunoblot analysis of the c-Myc protein species from the different lymphoma samples showed that the migration of c-Myc from PA682 and Wilson cells was slightly slower than the migration of p64 c-Myc from Raji, KK124, Ramos, and Daudi BL cells (Fig. 1A, lanes 1 to 4, and data not shown). Since c-myc encodes two related proteins, p64 (Myc 2) and p67 (Myc 1) (21), that can both be phosphorylated, we compared the migration of the endogenous c-Myc species from PA682 cells with the pattern observed in cells transfected with either recombinant p64 or p67 protein (6). The two expression vectors were designed to express exclusively p64 or p67 c-Myc protein by selectively mutating the individual translational start sites (6). We found that c-Myc from PA682 cells migrated at an intermediate position between p64 and p67 (Fig. 1B, lanes 1 to 4). To exclude the possibility that the absence of c-Myc–α-tubulin interaction was due to reduced levels of c-Myc in PA682 cells (Fig. 1A), we repeated the coimmunoprecipitation experiment using a threefold increase in the amount of protein extracts from PA682 cells compared to Raji cells, and we still did not detect binding between c-Myc and α-tubulin in the PA682 cells (Fig. 1B, lanes 5 and 6). In contrast, both recombinant p64 and p67 Myc proteins from the transfected cells bound equally well to α-tubulin (Fig. 1C). To confirm the specificity of c-Myc antibody, we demonstrated that addition of a c-Myc-specific peptide blocked the ability of the c-Myc antibody to react with the immunoprecipitated tubulin complex that contains c-Myc (Fig. 1C, lanes 7 to 9).
FIG. 1.
In vivo interaction of α-tubulin with c-Myc from BL cells. (A) Raji, Wilson, PA682, and KK124 BL protein extracts were resolved by SDS-PAGE and immunoblotted with anti-c-Myc (lanes 1 to 4) or immunoprecipitated (Immunoppt) with anti-α-tubulin and then immunoblotted with anti-c-Myc (lanes 5 to 8) (2). (B) A threefold increase in PA682 extract was tested to equalize for endogenous levels of c-Myc between Raji and PA682 cells. Protein extracts from Raji and PA682 cells were used for immunoblotting with anti-c-Myc (lanes 3 and 4) and for immunoprecipitation with anti-α-tubulin followed by immunoblotting with anti-c-Myc (lanes 5 and 6). Anti-c-Myc immunoblots of HEK293 cells transiently transfected with p64 and p67 c-myc constructs (6) were used as size marker controls for p64 and p67 (lanes 1 and 2). (C) HEK293 cells transiently transfected with p64 and p67 c-myc (lanes 1 to 3) were immunoprecipitated with anti-α-tubulin, followed by immunoblotting with anti-c-Myc (lanes 4 to 9). Blocking of antibody was performed by incubating c-Myc peptide with the c-Myc antibody clone 9E10 as previously described (15). IgH, heavy-chain mmunoglobulin. Numbers on the right are molecular masses in kilodaltons.
Constitutively hyperphosphorylated mutant c-Myc from BL cells is associated with defective Myc-tubulin binding in vivo.
To determine which specific mutations in c-Myc from these cell lines were associated with defective α-tubulin interaction, we obtained nucleotide sequences of c-myc from the BL cells that were not previously published. Nucleotide sequence analysis obtained for the entire coding region of the PA682 c-myc genomic clone revealed the presence of an A-to-G transition that resulted in an threonine-to-alanine substitution at position 58 (Fig. 2). Nucleotide sequence analysis of c-myc from the Wilson cell line also revealed the presence of a mutation at position 58 which resulted in a threonine-to-isoleucine substitution at position 58 (Fig. 2). Although the c-Myc-coding region derived from Raji cells has been reported to contain numerous mutations, the specific mutations differ between independent laboratories (1, 36). We isolated multiple cDNAs from Raji cells using reverse transcription-PCR methodology and found that 50% of the cDNA clones contained wt alleles and the other half contained mutant mRNA which included changes at Arg-10, Glu-39, and Thr-58. Since c-myc in Raji cells migrates as p64, and not as the slower p66 form detected in the PA682 and Wilson cell lines, our results suggest that α-tubulin may coprecipitate with c-Myc translated either from the p64 wt allele (3, 35) or with mutant p64 c-Myc which has not been hyperphosphorylated. The c-myc in KK124 cells consists of wt sequences (4), migrates as p64 Myc, and is coprecipitated by α-tubulin. Moreover, c-Myc–α-tubulin interaction was observed in two additional BL cell lines, Ramos and Daudi, which also express a wt p64 c-Myc protein (36, 44). In summary, (i) the PA682 and Wilson BL cells have the Thr-58 mutation and migrate as p66 c-Myc; (ii) the Ramos, Daudi, and KK224 cells have wt c-Myc sequences; and (iii) the Raji cells express both wt and mutated c-Myc alleles. The nucleotide sequence analysis combined with the c-Myc migration profile on SDS-PAGE suggest that α-tubulin can interact with wt or mutant p64 but not with the mutated p66 c-Myc.
FIG. 2.
Schematic representation of Thr-58 and Ser-62 mutations in c-Myc from PA682 and Wilson BL cells.
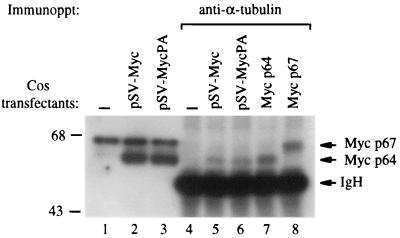
To investigate whether a naturally occurring mutation in Thr-58 was responsible for the disruption of Myc–α-tubulin binding in PA682 cells, we transiently expressed a Thr-58Ala mutated c-myc isolated from PA682 cells (pSV-MycPA) as well as a wt c-myc control (pSV-Myc) in Cos cells and analyzed binding of the transfected c-Myc to α-tubulin. Protein extracts isolated from transfected or control Cos cells were immunoprecipitated with anti-α-tubulin and immunoblotted with anti-c-Myc. We found that α-tubulin antibodies precipitated both wt c-Myc and mutant c-Myc isolated from PA682 cells (PA-Myc), demonstrating that a mutation in Thr-58 alone was not sufficient to abolish c-Myc–α-tubulin interaction in vivo (Fig. 3). However, we observed that the recombinant PA-Myc in Cos cells comigrated with the p64 wt Myc on SDS-PAGE (Fig. 3). The protein band detected just above p67 Myc is a nonspecific signal, since it was not blocked with the specific peptide used to raise the c-Myc antibody (data not shown). Thus, the slightly retarded shift in the migration of c-Myc in PA682 cells (Thr-58 mutant) may be due to posttranslational modification such as hyperphosphorylation that occurs in PA682 but not in Cos cells.
FIG. 3.
c-Myc and α-tubulin binding in transfected Cos cells. A mutated c-myc genomic clone isolated from PA682 cells (pSVMyc-PA) and a wt genomic clone (pSVMyc) were transiently transfected into Cos cells. Protein extracts (500 μg) from transfected and control cells were immunoprecipitated (Immunoppt) with anti-α-tubulin, followed by immunoblotting with anti-c-Myc (lanes 4 to 6). Ten micrograms of control and transfected cells was used for c-Myc immunoblotting (lanes 1 to 3). HEK293 cells transfected with p64 and p67 were used as controls (lanes 7 and 8). IgH, heavy-chain immunoglobulin.
To test if the difference in migration of wt and mutant c-Myc in PA682 cells is due to hyperphosphorylation, we immunoprecipitated c-Myc from PA682 cell extracts and subjected the precipitated protein to phosphatase treatment. We found that following PAP treatment, the mobility of c-Myc from PA682 cells shifted to a single band that comigrates with the p64 c-Myc control (Fig. 4), suggesting that the Thr-58-mutated c-Myc is hyperphosphorylated in BL PA682 cells. In addition, in vitro phosphorylation of a GST-Myc II (2) substrate (containing the N-terminal c-Myc domain [amino acids 1 to 251]) significantly reduced the binding of c-Myc to α-tubulin (Fig. 4B) using CKII or MAP kinase.
FIG. 4.
(A) Mutated c-Myc (Thr-58Ala) is constitutively hyperphosphorylated in PA682 BL cells. c-Myc was precipitated from PA682 protein extracts (500 μg) with anti-c-Myc (Ab-3) (lane 3). Parallel samples (250 μg of immunoprecipitated protein) were treated with increasing concentrations of PAP (lanes 4 and 5). Protein extracts from 293 cells transfected with p64 or p67 c-Myc were directly loaded for SDS-PAGE and used as size marker controls in the immunoblot analysis (lanes 1 and 2). IgH, heavy-chain immunoglobulin. Numbers on the right are molecular masses in kilodaltons. (B) In vitro phosphorylation of c-Myc reduces binding to α-tubulin. The N-terminal portion of c-Myc expressed as a GST fusion protein substrate (GST-Myc II) was treated with CKII and MAP kinase (MAP) and incubated with HL60 cell lysate. Precipitated proteins were resolved by SDS-PAGE and immunoblotted with α-tubulin and c-Myc antibodies.
Mitosis-specific hyperphosphorylation of wt c-Myc disrupts binding to α-tubulin.
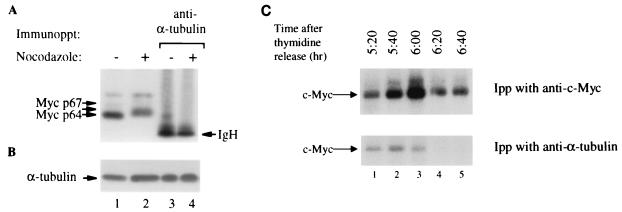
Since the N-terminal domain of c-Myc is reversibly hyperphosphorylated during mitosis (29), we hypothesized that c-Myc–α-tubulin interaction may also be disrupted at the onset of the mitotic stage of the cell cycle. To address this issue, HeLa cells were treated with nocodazole and the protein extracts from both unsynchronized and mitosis-arrested cells were immunoprecipitated with anti-α-tubulin and immunoblotted with anti-c-Myc. We found that hyperphosphorylated c-Myc from synchronized mitotic cells lost the ability to interact with α-tubulin in vivo (Fig. 5A, lanes 2 and 4) compared to the p64 c-Myc control from unsynchronized cultures (Fig. 5A, lanes 1 and 3). The loss of c-Myc–α-tubulin binding during mitosis was not due to lower levels of α-tubulin in the nocodazole-treated cells, since equal levels of α-tubulin were detected in unsynchronized and mitotic cells (Fig. 5B). The hyperphosphorylation of mitotic c-Myc and the lack of its interaction with α-tubulin are not due to nocodazole treatment, since mitotic arrest with a double thymidine block gave similar results (Fig. 5C). HeLa cells were tested for cell cycle progression at 0, 3, and 6 h following thymidine removal. At 6 h following thymidine release, cells were detected at the G2/M phase by flow cytometry (data not shown). To test whether c-Myc interacts with α-tubulin during mitosis, cells were labeled with [32P]orthophosphate for 2 h before harvest and protein lysates were prepared at 20-min intervals (Fig. 5C). Cell lysates were immunoprecipitated with either anti-α-tubulin or anti-c-Myc, and 32P-labeled c-Myc was detected by autoradiography. At 6 h following thymidine release, we observed that the amount of c-Myc bound to α-tubulin decreased, and the binding was abolished when cells entered mitosis (Fig. 5C, lanes 4 and 5).
FIG. 5.
c-Myc from mitotic HeLa cells does not bind α-tubulin. (A) Anti-c-Myc immunoblot analysis of HeLa cells arrested at mitosis with nocodazole treatment (29) (lane 2) and untreated control (lane 1). One milligram of treated and untreated cells was immunoprecipitated with anti-α-tubulin before immunoblotting with anti-c-Myc (lanes 3 and 4). The arrow between p64 and p67 depicts the hyperphosphorylated c-Myc. IgH, heavy-chain immunoglobulin. (B) The membrane from panel A was stripped (2) and immunoblotted with anti-α-tubulin. (C) In vivo 32P-labeled HeLa cells arrested at mitosis using a double thymidine block. 32P-labeled lysates (0.5 mg) were immunoprecipitated (Ipp) with anti-c-Myc or anti-α-tubulin, resolved by SDS-PAGE, and visualized by autoradiography.
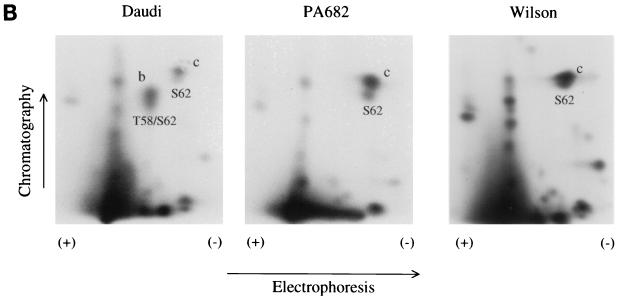
Phosphopeptide mapping of c-Myc from PA682 and Wilson BL cells.
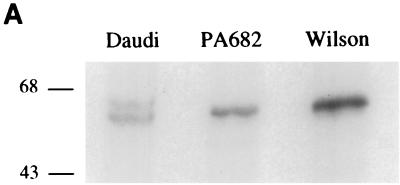
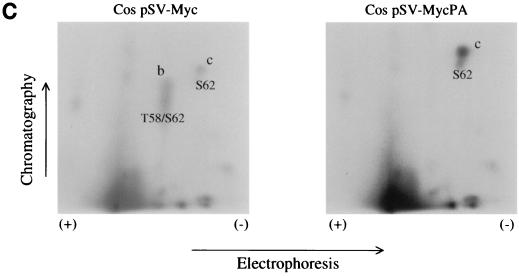
To determine whether the slower-migrating p66 c-Myc from PA682 and Wilson BL cells is phosphorylated differently then wt c-Myc in vivo, the cells were labeled with [32P]orthophosphate and the phosphorylated c-Myc proteins from the PA682 and Wilson cell lines were compared to the p64 wt c-Myc derived from Daudi cells. [32P]orthophosphate-labeled c-Myc from PA682, Wilson, and control Daudi BL cells was immunoprecipitated with anti-c-MycFL (39) and resolved by SDS-PAGE. We found that the slower-migrating c-Myc from PA682 and Wilson cells as well as the p64 c-Myc from Daudi cells were similarly phosphorylated in vivo (Fig. 6A). To determine whether the slower migration pattern of c-Myc from PA682 and Wilson cells was due to a differential phosphorylation at a specific site in the c-Myc protein, we performed phosphopeptide mapping analysis and compared the two-dimensional (2D) maps to the maps of wt c-Myc from Daudi cells (Fig. 6B). Phosphopeptide mapping following thermolysin treatment of wt c-Myc from Daudi cells showed a characteristic pattern (30, 31), where spot b represents a peptide phosphorylated at both Thr-58 and Ser-62 and spot c represents a phosphorylated peptide containing Ser-62 in the c-Myc protein (Fig. 6B). Phosphopeptide mapping of mutated c-Myc from PA682 and Wilson cells demonstrated the absence of spot b in both BL lines, since Thr-58 was replaced by Ala in PA682 cells and by Ile in Wilson cells (Fig. 6B). In addition, we performed phosphopeptide mapping of c-Myc from Cos cells transiently transfected with wt and mutated c-myc expression plasmids. We observed phosphorylation at Thr-58 and Ser-62 in c-Myc from Cos cells transfected with wt c-Myc (spot b) but not in c-Myc from Cos cells transfected with the mutant c-myc gene, which was isolated from PA682 cells (Fig. 6C). In comparison to our wt c-Myc maps, we did not detect a unique phosphorylation site in the endogenous c-Myc from PA682 or Wilson cells, nor did we find qualitative differences in the phosphorylation sites between the endogenous c-Myc from BL cells and exogenous mutant c-Myc (isolated from the PA682 cells) when transfected into Cos cells. We also obtained identical phosphopeptide maps when the c-Myc proteins from PA682 and Wilson were compared to c-Myc in Daudi cells after digestion with trypsin or chymotrypsin (data not shown).
FIG. 6.
Phosphopeptide analysis of wt and mutant c-Myc. (A) c-Myc was immunoprecipitated from [32P]orthophosphate-labeled logarithmically growing Daudi, PA682, and Wilson BL cells and resolved by SDS-PAGE. Numbers on the left are molecular masses in kilodaltons. (B) Thermolytic phosphopeptide analysis of 32P-labeled c-Myc proteins in BL cells. The endogenous c-Myc proteins were immunoprecipitated with anti-c-Myc and processed for 2D phosphopeptide mapping as described in Materials and Methods. (C) Thermolytic phosphopeptide analysis of 32P-labeled exogenous c-Myc proteins in Cos cells. 2D maps of wt c-Myc (pSV-MYC) and Thr-58Ala-mutated c-Myc (pSV-MycPA) are depicted.
Expression of exogenous wt c-Myc in PA682 cells.
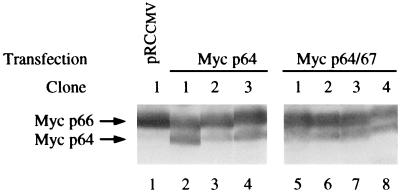
To examine whether the change in the migration of c-Myc observed in PA682 cells is due to the mutation at Thr-58 or whether it is due to a constitutively high kinase activity in the host BL cells, we expressed wt c-Myc in PA682 cells and compared its properties to those of the endogenous mutant c-Myc in these cells. We found that stable transfection of wt c-myc into PA682 cells resulted in a c-Myc product that comigrated with the wt 64-kDa species in SDS-PAGE (Fig. 7). We also transfected c-myc expression plasmids that encoded p64 c-Myc or both p64 and p67 proteins and a control expression vector that did not contain the c-myc gene. Stable G418-resistant clones were derived by serial dilution, and protein extracts were analyzed by immunoblotting with anti-c-Myc. All 10 clones derived after transfection of the PA682 cells with the empty control vector expressed only the endogenous slower-migrating c-Myc protein (representative clone 1) (Fig. 7, lane 1). In contrast, seven clones derived from cells transfected with either the p64- or p64- and p67-encoding plasmids expressed exogenous c-Myc that comigrated at the expected size of 64 kDa (Fig. 7) and could be coprecipitated with α-tubulin (data not shown). Stable expression of wt c-Myc in PA682 cells, therefore, leads to detection of both the endogenous slower-migrating Thr-58Ala-mutated c-Myc and the exogenous p64 c-Myc (Fig. 7). These results suggest that the slower migration of the endogenous Thr-58Ala mutant is not due solely to the intrinsic properties of the Myc kinase system in PA682 cells.
FIG. 7.
Expression of exogenous wt 64-kDa c-Myc in PA682 cells. Immunoblot analysis of protein extracts from stable PA682 transfectants expressing either 64-kDa (Myc p64) or 64- and 67-kDa (Myc p64/67) protein is shown. Arrows depict endogenous p66 Myc and the exogenous transfected p64 c-Myc proteins.
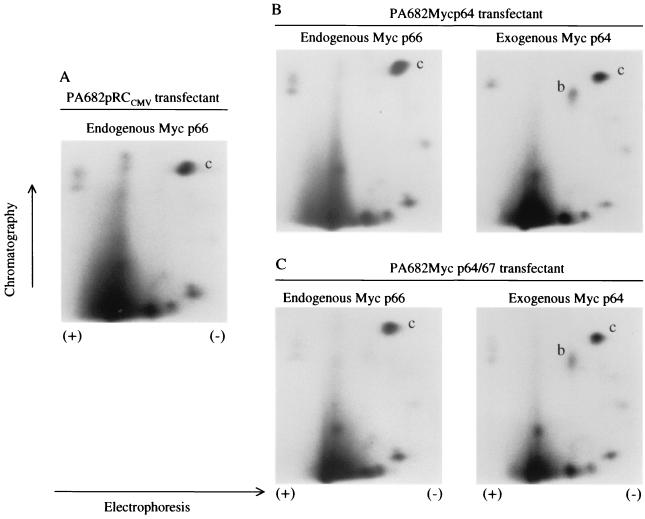
The PA682 transfectants that expressed both wt and mutant c-Myc provided an excellent model system to compare the phosphorylation sites between these two protein species in the same cell background. Stable clones that expressed either the p64 Myc or p64-p67 c-Myc were labeled with [32P]orthophosphate. The exogenous and endogenous c-Myc proteins were then resolved by SDS-PAGE and excised from the gel, and phosphopeptide mapping analysis was performed. We found that the exogenous p64 wt c-Myc gave rise to spot b (Fig. 8B and C, right panels) representing Thr-58 and Ser-62, while this phosphorylation site was missing from the endogenous p66 Myc due to Thr-58Ala mutation (Fig. 8A and B and C, left panels). We did not observe any phosphorylation sites that were different in the endogenous c-Myc from PA682 cells. These data are consistent with our earlier phosphopeptide mapping experiments where we compared the phosphorylation sites between c-Myc proteins from PA682 and Daudi cells (Fig. 6B).
FIG. 8.
Thermolytic phosphopeptide analysis of 32P-labeled endogenous (p66) and exogenous (p64) c-Myc proteins from PA682 transfectants. 2D maps of PA682 stable transformants with empty vector (pRCCMV) (A), pRCMyc p64 (B), and pRCMyc p64/p67 (wt c-myc) (C) vectors are shown.
Increase in stability of c-Myc in PA682 and Wilson cells.
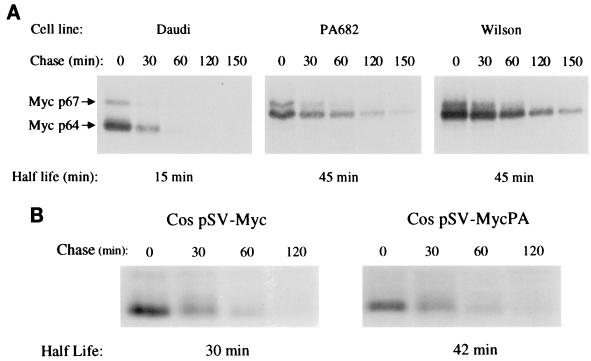
c-Myc is a highly unstable protein, with a half-life of 15 to 30 min (20). It has been recently shown that the c-Myc protein is degraded by ubiquitin-mediated proteolysis (16, 18, 38) and that cancer-associated mutations in c-Myc stabilize the protein (18, 38). Transfection of Thr-58Ala-mutated c-Myc into U2OS or 3T3 cells demonstrated that the mutated protein was more stable than the wt counterpart (18, 38). Moreover, c-Myc was stabilized in BL cells that contain mutations in the vicinity of Thr-58 which are known to abolish Thr-58 phosphorylation (18). Thus, we asked whether the naturally occurring Thr-58 mutations in PA682 and Wilson BL cells affect the stability of c-Myc in vivo. To determine whether c-Myc stability was increased in PA682 and Wilson cells compared to Daudi cells, which contain wt c-Myc, a pulse-chase analysis was performed. We found that the stability of mutant c-Myc from PA682 and Wilson cells was increased threefold as compared to the stability of wt c-Myc from Daudi cells (Fig. 9A). These in vivo results confirm the in vitro results (16, 38) and suggest that a Thr-58 mutation in c-Myc contributes to the increased stability of the c-Myc protein.
FIG. 9.
(A) Stability of c-Myc in BL cells containing wt (Daudi) or mutant (PA682 and Wilson) c-Myc. (B) Stability of wt and mutated c-Myc transfected into Cos cells. Half-lives of c-Myc in BL and Cos cells were measured by in vivo [35S]methionine labeling as described in Materials and Methods.
We also transfected a c-myc genomic clone isolated from PA682 cells (pSV-MycPA), as well as a wt c-myc genomic clone (pSV-Myc), into Cos cells and compared c-Myc protein turnover. We observed again that the hyperphosphorylated Thr-58 mutant c-Myc in PA682 does not undergo hyperphosphorylation in Cos cells (Fig. 3 and 4), and we observed a small difference in the protein half-life between the Thr-58 mutant and the wt c-Myc when transiently expressed in these cells (Fig. 9B). This suggests that while the Thr-58 mutation may contribute to increased stability, the major determinant of stability may be cell dependent.
DISCUSSION
The N-terminal domain of c-Myc, which is required for transformation and transcriptional activity, has been shown to interact with α-tubulin and polymerized microtubules in vitro and in vivo (2). In this study we have demonstrated that the interaction of c-Myc with α-tubulin is regulated by the phosphorylation state of c-Myc in a cell cycle-dependent manner. We observed that c-Myc hyperphosphorylation during mitosis results in a loss of c-Myc–α-tubulin interactions in vivo. Similarly, we demonstrated that a naturally occurring mutated c-Myc from BL cells was associated with constitutive hyperphosphorylation of c-Myc and loss of α-tubulin binding in vivo. We isolated the c-myc gene from PA682 BL cells and found that it contained only one nucleotide substitution, which resulted in a Thr-58Ala mutation in the c-Myc protein. Since we had previously localized the c-Myc–α-tubulin binding domain to amino acid codons 48 to 135 in the c-Myc protein (2), the absence of c-Myc–α-tubulin interactions suggested that Thr-58 was required for Myc–α-tubulin binding in vivo. However, transfection of the mutant Thr-58Ala c-myc gene into Cos cells demonstrated that a mutation at Thr-58 alone was not sufficient to disrupt Myc–α-tubulin binding. The electrophoretic migration pattern of the ectopically expressed PA682 mutated c-Myc in Cos cells was identical to that of the p64 wt c-Myc protein but was distinct from that of the slower-migrating endogenous p66 c-Myc found in PA682 cells. Since this aberrant migration of mutant c-Myc in PA682 cells can be shifted to a wt migration pattern by phosphatase treatment, these results indicated that the mutated Thr-58Ala c-Myc is differentially phosphorylated depending on the cell type and suggest that the phosphorylation state of c-Myc may regulate the interaction with α-tubulin in vivo. The hyperphosphorylation of c-Myc and its altered migration pattern in PA682 cells is associated with the Thr-58Ala mutations, since transfection of wt c-Myc into PA682 cells resulted in c-Myc species that comigrated with wt 64-kDa protein. In addition, we examined a panel of BL cells and found a Thr-58Ile mutant of c-Myc in the Wilson BL cell line that was hyperphosphorylated, similar to c-Myc from PA682, and also did not bind to α-tubulin. wt or mutant c-Myc proteins from 12 other BL cells that were not hyperphosphorylated and that migrated on SDS-PAGE as 64-kDa proteins interacted with α-tubulin.
Mutations at Thr-58 have been observed frequently in BL samples (4, 45) as well as in each of the three different avian acute transforming retroviruses that carry the v-myc oncogene (Papas and Lautenberger, Letter, Nature, 1985). Since it has been shown that Thr-58 mutants result in enhanced transformation activity, the phosphorylation on Thr-58 was suggested to play an inhibitory role in cell growth control (22, 34). Our results suggest that loss of Thr-58 may not solely account for the molecular weight shift of the mutant c-Myc in PA682 cells, since transfected Thr-58Ala-mutated c-Myc migrates similarly to the wt p64 c-Myc protein. In addition, the shift in molecular weight of mutated c-Myc observed in PA682 cells was not associated with a qualitative change in the phosphopeptide mapping pattern, aside from the loss of the Thr-58 site, compared with wt p64 c-Myc protein expressed in PA682 cells.
The shift in molecular weight of the mutant c-Myc protein from PA682 cells resembles the reversible shift of c-Myc observed in HeLa cells during mitosis, and in both cases this shift is collapsed to the faster p64 pattern by phosphatase treatment (Fig. 4) (29). Observations with 32P-labeled mitotic HeLa cells further suggest that phosphorylated c-Myc does not interact with α-tubulin during the mitotic stage of the cell cycle. Phosphopeptide mapping of c-Myc from mitotic or interphase cells in prior studies did not detect the appearance of novel M phase-specific phosphopeptides (29), and it was suggested that either the M phase- and interphase-specific phosphorylation sites may cluster to the same peptides or the increased apparent molecular weight may be the result of a combinatorial effect of several phosphorylation sites (29). This model therefore suggests that a quantitative increase in phosphorylation occurs during mitosis in the wt c-Myc or constitutively in mutant c-Myc from PA682 or Wilson cells (29). However, since there are 10 potential phosphorylation sites that have been described for the c-myc gene, with five of them clustering in the N-terminal portion of the protein (31), it seems unlikely that multiple kinases would be mitosis specific or Thr-58 mutation dependent. Alternatively, a unique site-specific hyperphosphorylation that is Thr-58 mutation-dependent and mitosis-specific may be occurring in PA682 or Wilson cells, but the 2D mapping cannot adequately resolve the unique phosphopeptides.
Since c-Myc is a transcription factor that is located predominantly in the nucleus, the question remains whether the interaction of c-Myc with microtubules occurs in the cytoplasm during interphase or whether this interaction takes place during mitosis following nuclear membrane dissolution. Since c-Myc–α-tubulin interactions were absent during the M phase of the cell cycle and since α-tubulin is not detected in the nucleus during interphase (2), our data suggest that c-Myc–α-tubulin interaction may take place in the cytoplasmic fraction. c-Myc has been shown to accumulate in the cytoplasm in differentiating myeloid and neuronal cells (12, 43), and it was also proposed that subcellular localization of c-Myc may be dependent on the proliferation state of the cell (19, 42). Moreover, it was recently shown that the cytoplasmic protein termed Miz-1, which binds to c-Myc, associates with microtubules and can target c-Myc to the microtubules network before translocation to the nucleus (33). Thus, it was proposed that microtubules might served as a reservoir to sequester c-Myc and to regulate its cellular compartmentalization and transcriptional activity (2). We have previously shown that 5 to 10% of c-Myc was found bound to α-tubulin following two cycles of polymerization and depolymerization of microtubules in vitro. In addition, similar amounts of c-Myc were detected in the cytoplasm of interphase HeLa cells (2). The amount of c-Myc found in the cytoplasm, however, is dependent on the cell type and the proliferation state of the cell. We recently observed predominantly cytoplasmic localization of c-Myc in quiescent normal human foreskin fibroblasts, compared to growing cells where Myc was found predominantly in the nuclear fractions. Treatment of quiescent cells with colchicine, an agent known to disrupt the microtubule network, resulted in translocation of c-Myc into the nuclear fraction (data not shown), suggesting again that Myc may be bound to microtubules in nondividing cells.
c-Myc is a highly unstable protein (half-life of 15 to 30 min) that is regulated by ubiquitin-mediated proteolysis (16, 18, 38). Accordingly, the instability of c-Myc protein is believed to be important in preventing its accumulation in normal cells. It has also been shown that a mutation of Thr-58 stabilizes c-Myc when ectopically expressed in U20S and 3T3 cells (18, 38). We have extended this observation in an in vivo system by showing that the stability of c-Myc was increased in BL cells containing a naturally occurring Thr-58Ala mutation that was associated with constitutive hyperphosphorylation of c-Myc. At the same time we showed that the Thr-58 mutant c-Myc from BL cells does not bind with α-tubulin. In addition, mitotic c-Myc, which migrates slower then interphase c-Myc and behaves similarly to c-Myc from PA682 and Wilson cells, is hyperphosphorylated (29), does not bind to α-tubulin, and is more stable that the interphase c-Myc (18). Thus, binding or sequestration of c-Myc by microtubules occurs in normal cells expressing wt c-Myc and correlates with rapid turnover of the protein. In contrast, in tumor cells expressing mutated c-Myc and in mitotic cells (29), hyperphosphorylation of the protein results in loss of binding to α-tubulin and increased protein stability. Substrate phosphorylation has been shown to be required for protein degradation by the ubiquitin pathway (14, 25), and thus our in vivo results support the in vitro model, which proposes that the phosphorylation status of Thr-58 in c-Myc may play a role in rapid c-Myc degradation (38). However, our results agree with a previous study (18) and suggest that a primary determinant of c-Myc turnover is cell type dependent. Future studies will establish whether enhanced protein stability contributes to oncogenic transformation by mutant c-Myc and define whether microtubules play a direct role in the regulation of c-Myc stability and oncogenic activity.
ACKNOWLEDGMENTS
We thank Shoshana Segal and Greg Otterson for helpful discussions and for critical reading of the manuscript.
REFERENCES
- 1.Albert T, Urlbauer B, Kohlhuber F, Hammersen B, Eick D. Ongoing mutations in the N-terminal domain of c-Myc affect transactivation in Burkitt's lymphoma cell lines. Oncogene. 1994;9:759–763. [PubMed] [Google Scholar]
- 2.Alexandrova N, Niklinski J, Bliskovsky V, Otterson G A, Blake M, Kaye F J, Zajac-Kaye M. The N-terminal domain of c-Myc associates with alpha-tubulin and microtubules in vivo and in vitro. Mol Cell Biol. 1995;15:5188–5195. doi: 10.1128/mcb.15.9.5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.ar-Rushdi A, Nishikura K, Erikson J, Watt R, Rovera G, Croce C M. Differential expression of the translocated and the untranslocated c-myc oncogene in Burkitt lymphoma. Science. 1983;222:390–393. doi: 10.1126/science.6414084. [DOI] [PubMed] [Google Scholar]
- 4.Bhatia K, Huppi K, Spangler G, Siwarski D, Iyer R, Magrath I. Point mutations in the c-Myc transactivation domain are common in Burkitt's lymphoma and mouse plasmacytomas. Nat Genet. 1993;5:56–61. doi: 10.1038/ng0993-56. [DOI] [PubMed] [Google Scholar]
- 5.Blackwood E M, Kretzner L, Eisenman R N. Myc and Max function as a nucleoprotein complex. Curr Opin Genet Dev. 1992;2:227–235. doi: 10.1016/s0959-437x(05)80278-3. [DOI] [PubMed] [Google Scholar]
- 6.Blackwood E M, Lugo T G, Kretzner L, King M W, Street A J, Witte O N, Eisenman R N. Functional analysis of the AUG- and CUG-initiated forms of the c-Myc protein. Mol Biol Cell. 1994;5:597–609. doi: 10.1091/mbc.5.5.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyle W J, van der Geer P, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 1991;201:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- 8.Chen C, Biegalke B J, Eisenman R N, Linial M L. FH3, a v-myc avian retrovirus with limited transforming ability. J Virol. 1989;63:5092–5100. doi: 10.1128/jvi.63.12.5092-5100.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen W-D, Otterson G A, Lipkowitz S, Khleif S N, Coxon A B, Kaye F J. Apoptosis is associated with cleavage of a 5 kDa fragment from RB which mimics dephosphorylation and modulates E2F binding. Oncogene. 1997;14:1243–1248. doi: 10.1038/sj.onc.1201096. [DOI] [PubMed] [Google Scholar]
- 10.Claassen G F, Hann S R. Myc-mediated transformation: the repression connection. Oncogene. 1999;18:2925–2933. doi: 10.1038/sj.onc.1202747. [DOI] [PubMed] [Google Scholar]
- 11.Cole M D, McMahon S B. The Myc oncoprotein: a critical evaluation of transactivation and target gene regulation. Oncogene. 1999;18:2916–2924. doi: 10.1038/sj.onc.1202748. [DOI] [PubMed] [Google Scholar]
- 12.Craig R W, Buchan H L, Civin C I, Kastan M B. Altered cytoplasmic/nuclear distribution of the c-myc protein in differentiating ML-1 human myeloid leukemia cells. Cell Growth Differ. 1993;4:349–357. [PubMed] [Google Scholar]
- 13.Dalla-Favera R, Gelmann E P, Martinotti S, Franchini G, Papas T S, Gallo R C, Wong S F. Cloning and characterization of different human sequences related to the onc gene (v-myc) of avian myelocytomatosis virus (MC29) Proc Natl Acad Sci USA. 1982;79:6497–6501. doi: 10.1073/pnas.79.21.6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elledge S J, Harper J W. The role of protein stability in the cell cycle and cancer. Biochim Biophys Acta. 1998;1377:M61–M70. doi: 10.1016/s0304-419x(98)00005-5. [DOI] [PubMed] [Google Scholar]
- 15.Evan G I, Lewis G K, Ramsay G, Bishop J M. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flinn E M, Busch C M, Wright A P. myc boxes, which are conserved in myc family proteins, are signals for protein degradation via the proteasome. Mol Cell Biol. 1998;18:5961–5969. doi: 10.1128/mcb.18.10.5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frykberg L, Graf T, Vennstrom B. The transforming activity of the chicken c-myc gene can be potentiated by mutations. Oncogene. 1987;1:415–422. [PubMed] [Google Scholar]
- 18.Gregory M A, Hann S R. c-Myc proteolysis by the ubiquitin-proteasome pathway: stabilization of c-Myc in Burkitt's lymphoma cells. Mol Cell Biol. 2000;20:2423–2435. doi: 10.1128/mcb.20.7.2423-2435.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gusse M, Ghysdael J, Evan G, Soussi T, Mechali M. Translocation of a store of maternal cytoplasmic c-Myc protein into nuclei during early development. Mol Cell Biol. 1989;9:5395–5403. doi: 10.1128/mcb.9.12.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hann S R, Eisenman R N. Proteins encoded by the human c-myc oncogene: differential expression in neoplastic cells. Mol Cell Biol. 1984;4:2486–2497. doi: 10.1128/mcb.4.11.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hann S R, Sloan-Brown K, Spotts G D. Translational activation of the non-Aug-initiated c-myc 1 protein at high cell densities due to methionine deprivation. Genes Dev. 1992;6:1229–1240. doi: 10.1101/gad.6.7.1229. [DOI] [PubMed] [Google Scholar]
- 22.Henriksson M, Bakardjiev A, Klein G, Luscher B. Phosphorylation sites mapping in the N-terminal domain of c-myc modulate its transforming potential. Oncogene. 1993;8:3199–3209. [PubMed] [Google Scholar]
- 23.Henriksson M, Luscher B. Proteins of the Myc network: essential regulators of cell growth and differentiation. Adv Cancer Res. 1996;68:109–182. doi: 10.1016/s0065-230x(08)60353-x. [DOI] [PubMed] [Google Scholar]
- 24.Hoang A T, Lutterbach B, Lewis B C, Yano T, Chou T Y, Barrett J F, Raffeld M, Hann S R, Dang C V. A link between increased transforming activity of lymphoma-derived MYC mutant alleles, their defective regulation by p107, and altered phosphorylation of the c-Myc transactivation domain. Mol Cell Biol. 1995;15:4031–4042. doi: 10.1128/mcb.15.8.4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 26.Kaelin W J, Pallas D C, DeCaprio J A, Kaye F J, Livingston D M. Identification of cellular proteins that can interact specifically with the T/E1A-binding region of the retinoblastoma gene product. Cell. 1991;64:521–532. doi: 10.1016/0092-8674(91)90236-r. [DOI] [PubMed] [Google Scholar]
- 27.Kato G J, Barrett J, Villa G M, Dang C V. An amino-terminal c-Myc domain required for neoplastic transformation activates transcription. Mol Cell Biol. 1990;10:5914–5920. doi: 10.1128/mcb.10.11.5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kato G J, Dang C V. Function of the c-Myc oncoprotein. FASEB J. 1992;6:3065–3072. doi: 10.1096/fasebj.6.12.1521738. [DOI] [PubMed] [Google Scholar]
- 29.Luscher B, Eisenman R N. Mitosis-specific phosphorylation of the nuclear oncoproteins Myc and Myb. J Cell Biol. 1992;118:775–784. doi: 10.1083/jcb.118.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lutterbach B, Hann S R. Hierarchical phosphorylation at N-terminal transformation-sensitive sites in c-Myc protein is regulated by mitogens and in mitosis. Mol Cell Biol. 1994;14:5510–5522. doi: 10.1128/mcb.14.8.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lutterbach B, Hann S R. Overexpression of c-Myc and cell immortalization alters c-Myc phosphorylation. Oncogene. 1997;14:967–975. doi: 10.1038/sj.onc.1200920. [DOI] [PubMed] [Google Scholar]
- 32.Palmieri S, Kahn P, Graf T. Quail embryo fibroblasts transformed by four v-myc-containing virus isolates show enhanced proliferation but are non tumorigenic. EMBO J. 1983;2:2385–2389. doi: 10.1002/j.1460-2075.1983.tb01750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peukert K, Staller P, Schneider A, Carmichael G, Hanel F, Eilers M. An alternative pathway for gene regulation by Myc. EMBO J. 1997;16:5672–5686. doi: 10.1093/emboj/16.18.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pulverer B J, Fisher C, Vousden K, Littlewood T, Evan G, Woodgett J R. Site-specific modulation of c-Myc cotransformation by residues phosphorylated in vivo. Oncogene. 1994;9:59–70. [PubMed] [Google Scholar]
- 35.Rabbitts T H, Forster A, Hamlyn P, Baer R. Effect of somatic mutation within translocated c-myc genes in Burkitt's lymphoma. Nature. 1984;309:592–597. doi: 10.1038/309592a0. [DOI] [PubMed] [Google Scholar]
- 36.Rabbitts T H, Hamlyn P H, Baer R. Altered nucleotide sequences of a translocated c-myc gene in Burkitt lymphoma. Nature. 1983;306:760–765. doi: 10.1038/306760a0. [DOI] [PubMed] [Google Scholar]
- 37.Reinhold W, Emens L, Itkes A, Blake M, Ichinose I, Zajac-Kaye M. The myc intron-binding polypeptide associates with RFX1 in vivo and binds to the major histocompatibility complex class II promoter region, to the hepatitis B virus enhancer, and to regulatory regions of several distinct viral genes. Mol Cell Biol. 1995;15:3041–3048. doi: 10.1128/mcb.15.6.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salghetti S E, Kim S Y, Tansey W P. Destruction of Myc by ubiquitin-mediated proteolysis: cancer-associated and transforming mutations stabilize Myc. EMBO J. 1999;18:717–726. doi: 10.1093/emboj/18.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spotts G D, Patel S V, Xiao Q, Hann S R. Identification of downstream-initiated c-Myc proteins which are dominant-negative inhibitors of transactivation by full-length c-Myc proteins. Mol Cell Biol. 1997;17:1459–1468. doi: 10.1128/mcb.17.3.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stein G S, Stein J L. Cell synchronization. In: Baserga R, editor. Cell growth and division: a practical approach. Oxford, United Kingdom: IRL Press; 1990. pp. 133–137. [Google Scholar]
- 41.Symonds G, Hartshorn A, Kennewell A, O'Mara M A, Bruskin A, Bishop J M. Transformation of murine myelomonocytic cells by myc: point mutations in v-myc contribute synergistically to transforming potential. Oncogene. 1989;4:285–294. [PubMed] [Google Scholar]
- 42.Vriz S, Lemaitre J M, Leibovici M, Thierry N, Mechali M. Comparative analysis of the intracellular localization of c-Myc, c-Fos, and replicative proteins during cell cycle progression. Mol Cell Biol. 1992;12:3548–3555. doi: 10.1128/mcb.12.8.3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wakamatsu Y, Watanabe Y, Shimono A, Kondoh H. Transition of localization of the N-Myc protein from nucleus to cytoplasm in differentiating neurons. Neuron. 1993;10:1–9. doi: 10.1016/0896-6273(93)90236-k. [DOI] [PubMed] [Google Scholar]
- 44.Wiman K G, Clarkson B, Hayday A C, Saito H, Tonegawa S, Hayward W S. Activation of a translocated c-myc gene: role of structural alterations in the upstream region. Proc Natl Acad Sci USA. 1984;81:6798–6802. doi: 10.1073/pnas.81.21.6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yano T, Sander C A, Clark H M, Dolezal M V, Jaffe E S, Raffeld M. Clustered mutations in the second exon of the MYC gene in sporadic Burkitt's lymphoma. Oncogene. 1993;8:2741–2748. [PubMed] [Google Scholar]