ABSTRACT
The TonB-dependent transport of scarcely available substrates across the outer membrane is a conserved feature in Gram-negative bacteria. The plasma membrane-embedded TonB-ExbB-ExbD accomplishes complex functions as an energy transducer by physically interacting with TonB-dependent outer membrane transporters (TBDTs). TonB mediates structural rearrangements in the substrate-loaded TBDTs that are required for substrate translocation into the periplasm. In the model heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120, four TonB-like proteins have been identified. Out of these TonB3 accomplishes the transport of ferric schizokinen, the siderophore which is secreted by Anabaena to scavenge iron. In contrast, TonB1 (SjdR) is exceptionally short and not involved in schizokinen transport. The proposed function of SjdR in peptidoglycan structuring eliminates the protein from the list of TonB proteins in Anabaena. Compared with the well-characterized properties of SjdR and TonB3, the functions of TonB2 and TonB4 are yet unknown. Here, we examined tonB2 and tonB4 mutants for siderophore transport capacities and other specific phenotypic features. Both mutants were not or only slightly affected in schizokinen transport, whereas they showed decreased nitrogenase activity in apparently normal heterocysts. Moreover, the cellular metal concentrations and pigment contents were altered in the mutants, most pronouncedly in the tonB2 mutant. This strain showed an altered susceptibility toward antibiotics and SDS and formed cell aggregates when grown in liquid culture, a phenotype associated with an elevated lipopolysaccharide (LPS) production. Thus, the TonB-like proteins in Anabaena appear to take over distinct functions, and the mutation of TonB2 strongly influences outer membrane integrity.
IMPORTANCE The genomes of many organisms encode more than one TonB protein, and their number does not necessarily correlate with that of TonB-dependent outer membrane transporters. Consequently, specific as well as redundant functions of the different TonB proteins have been identified. In addition to a role in uptake of scarcely available nutrients, including iron complexes, TonB proteins are related to virulence, flagellum assembly, pilus localization, or envelope integrity, including antibiotic resistance. The knowledge about the function of TonB proteins in cyanobacteria is limited. Here, we compare the four TonB proteins of Anabaena sp. strain PCC 7120, providing evidence that their functions are in part distinct, since mutants of these proteins exhibit specific features but also show some common impairments.
KEYWORDS: Anabaena, TonB protein, cyanobacteria, metal transport, nitrogenase, outer membrane, siderophores
INTRODUCTION
Cyanobacteria possess a Gram-negative type of cell envelope containing an outer membrane (OM), a peptidoglycan (PG) layer, and a plasma (cytoplasmic or inner) membrane (PM) (1). Macromolecular complexes that reside in the two membranes facilitate the assembly of the cell wall components as well as solute exchange and signaling (2). Among them, the OM-embedded TonB-dependent transport machinery is widely distributed in Gram-negative bacteria (3). The TonB-dependent transport system is important for growth under iron starvation conditions, since iron is an essential but scantily bioavailable nutrient (3–7). Iron-loaded proteins carry out functions in important cellular activities such as electron transport and DNA synthesis (8). This holds particularly true for cyanobacteria, in which iron is required for the synthesis of phycobiliproteins (9) and chlorophyll a (Chl) (10), as well as for photosynthetic complexes that in total require approximately 22 to 23 iron atoms (11). Moreover, in certain cyanobacterial species that are capable of nitrogen fixation, the nitrogenase enzyme is also dependent on iron (12).
Due to its low solubility under oxic conditions at neutral pH, iron rapidly forms insoluble aggregates that are inaccessible to many microorganisms (6, 13). Only a very small amount of dissolved iron exists as inorganic iron, whereas the largest proportion is bound to organic ligands such as siderophores (6). Siderophores are low-molecular-weight compounds that chelate ferric iron with high affinity. The production and secretion of siderophores is a widespread strategy of bacteria, fungi, and plants to cope with iron-limiting conditions (14). Siderophores are divided into three classes depending on the chemical nature of iron coordination, namely, catecholates, hydroxamates, or mixed types that contain another iron complexing group such as hydroxycarboxylate (15).
The TonB-dependent transport system involves a PM-localized energizing TonB-ExbB-ExbD complex and OM-localized TonB-dependent transporters (TBDTs). TBDTs constitute gated channels that facilitate the transport of substrates into the periplasm (16). The translocation process is energy dependent, as the substrates are typically large and rarely abundant (17, 18). Examples besides siderophores are carbohydrates, vitamin B12 (cobalamin), and heme (16, 19). The energy for transport is derived from the proton motive force (pmf) across the PM (20, 21). ExbB and ExbD build up a proton channel that converts the pmf into energy for the translocation process (22). The TonB protein transfers the energy to the TBDT through direct interaction with both, ExbB/ExbD and the TBDT (23–26).
TonB proteins contain a transmembrane α-helix and a conserved C-terminal motif that interacts with the so-called TonB box of the TBDTs (16, 27). Remarkably, more than 40% of the organisms that possess a TonB-dependent system have more than one tonB gene copy (3). For instance, Pseudomonas aeruginosa possesses three TonB proteins (28–30). Here, TonB1 and likely TonB2 facilitate the transport of iron-containing compounds and are required for growth under iron-limiting conditions, while TonB3 is crucial for motility and pilus assembly (28–32). In Pseudomonas putida, one of the two TonB proteins energizes the transport of siderophores, whereas the other TonB protein is important for maintaining the integrity of the cell envelope and flagellum localization (33–35). Also, Vibrio species typically contain multiple tonB copies in the genome. Here, distinct TonB proteins facilitate the transport of both common and individual substrates (36). Thus, multiple TonB proteins in one organism can take over redundant as well as unique functions. They can function in protein complex assembly, cell wall integrity regulation, or global or substrate-specific transport processes.
Little is known about the functionality of TonB proteins in cyanobacteria, which are photoautotrophic organisms that can be found in terrestrial, marine, or freshwater habitats. The number of putative TBDT, TonB, or ExbB/D proteins in the genomes of analyzed cyanobacteria is highly variable (37, 38). For example, in the genome of the filamentous cyanobacterium Anabaena sp. strain PCC 7120 (Anabaena hereafter) 22 different TBDTs were predicted (37). In contrast, only four genes with a tonB signature were assigned by bioinformatics methods (38). TonB1 contains an exceptionally short periplasmic domain that is likely not sufficient in size to reach OM-embedded factors. In contrast, TonB3 is supposed to be a central component of the ferric siderophore transport system (38). The tonB3 mRNA abundance increases under iron-limited conditions, and a single recombination mutant can be generated only in the presence of enhanced iron concentrations (38). The growth of this mutant in the absence of iron is reduced, and siderophore synthesis genes are upregulated in this genetic background (38). Further, we could show recently that the transport of ferric schizokinen, the siderophore secreted by Anabaena, was abolished in a tonB3 mutant (39). In contrast, the tonB1 mutant retained the siderophore transport capacity but was severely impacted in diazotrophic growth. This could be traced back to an abnormal peptidoglycan morphology in the heterocyst septa of the mutant, and therefore, TonB1 was renamed septal junction disc regulator (SjdR) (39).
TonB2 encoded by all3585 or TonB4 encoded by alr5329 has a domain structure comparable to the TonB proteins from Escherichia coli or TonB3 from Anabaena (38). However, the distance between the transmembrane domain and the TonB-box binding domain is smaller than in TonB3 (Fig. 1). Estimation of the size assuming an extended helix suggests a dimension of 12 nm for TonB4 and 22 nm for TonB2, while for TonB3 a 32-nm size is estimated. The latter fits the determined distance between OM and PM in Anabaena, as well as the estimated size of the TolC system (40, 41). Both tonB2 and tonB4 are expressed at highest levels in low-density cultures and lowest levels in the stationary phase (42). Their expression is enhanced at all growth stages in the presence of elevated iron (38, 42), while the expression of tonB4 is also enhanced in the presence of elevated copper concentrations (38, 42).
FIG 1.
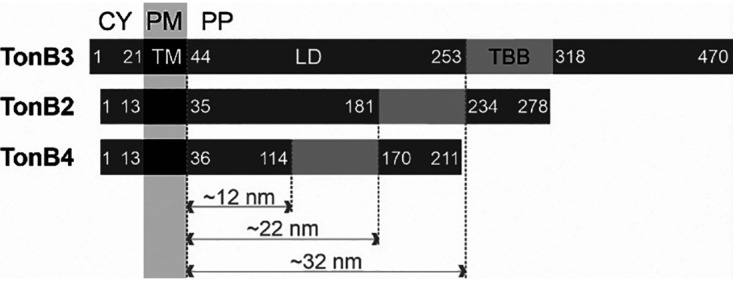
Domain architecture of TonB proteins in Anabaena. TonB3, TonB2, and TonB4 contain an N-terminal cytosolic region (CY), a transmembrane region (TM), a linker domain (LD), the TonB-box binding region (TBB), and a C-terminal extension. Indicated are the amino acids at the border of the cytosolic region, the LD, and the C-terminal extension. The length of the linker domain was calculated assuming an extended helix (3.6 amino acids and 0.54 nm per turn). PM, plasma membrane; PP, periplasm.
Considering the essential role of TonB3 in siderophore transport, opposed to the novel functionality of SjdR, which is not related to TonB-dependent transport, we now aimed to characterize the TonB-like proteins TonB2 and TonB4, since the role of those proteins in Anabaena is still unclear (38, 39). Insertion mutants for tonB2, tonB3, and tonB4 demonstrated alterations in cellular metal levels as well as in carotenoid (Car) or chlorophyll a concentrations compared to the wild type, although to different extents. Moreover, the tonB2 mutant filaments aggregated in liquid cultures, which might be related to an enhanced production of lipopolysaccharide in this strain. Also, the outer membrane integrity as well as the expression of porins was affected in the tonB2 mutant. On the other hand, the tonB2 mutant as well as the tonB4 mutant retains the siderophore transport capacity, which suggests a functional diversity of Anabaena TonB proteins.
RESULTS
The Anabaena tonB mutants bear pigment alterations.
To analyze putative functions of the individual TonB proteins, the growth behavior of mutants of the corresponding genes was examined. The mutant strains, I-sjdR, I-tonB2, I-tonB3, and I-tonB4, were generated through single recombination insertion of a plasmid in the gene of interest (Fig. 2A), as described before (38, 39). In accordance with previous reports, I-sjdR and I-tonB2 were segregated, as no wild-type copy of the respective genes was detectable in the mutants (Fig. 2B) (39). In contrast, I-tonB3 and I-tonB4 could not be segregated, as even after repeated dilution on plates with antibiotics, the wild-type genes were detectable in the corresponding genomic DNA (gDNA) by PCR (Fig. 2A) (38). For I-tonB3 this is consistent with the previous report, where full segregation was obtained only in the presence of enhanced iron (38). This suggests that TonB3 and TonB4 are important for viability under the conditions used in this study.
FIG 2.
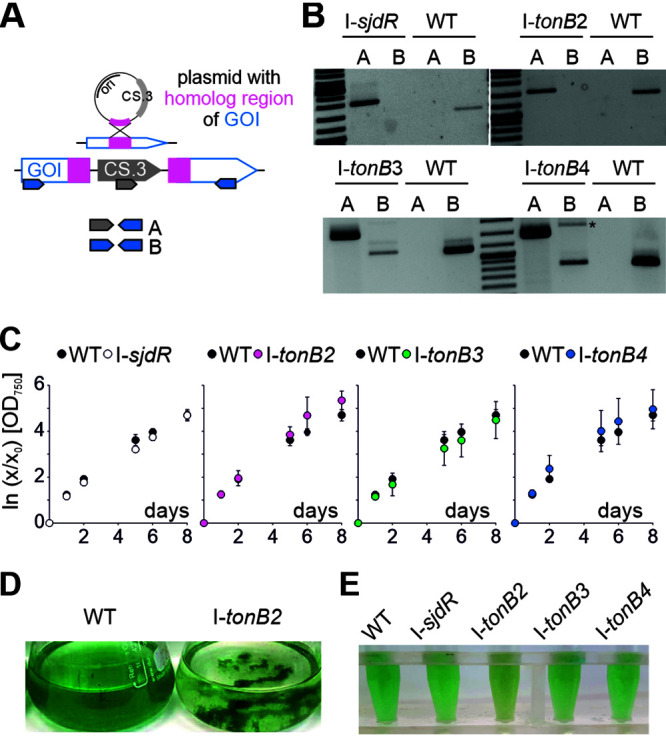
Growth phenotype of the tonB mutants and the sjdR mutant strains. (A) Illustration of the single recombination strategy; a portion of the gene of interest (GOI) is cloned into a plasmid which bears the Spr Smr cassette (CS.3). After the recombination event the GOI in the mutant is interrupted by plasmid insertion. The oligonucleotide pairs utilized for mutant screening are indicated; primer combination A results in a product when the plasmid is integrated into the genome; combination B can generate a product only when the GOI is intact. (B) Segregation analyses of I-tonB3 and I-sjdR were described previously (38, 39). Either an oligonucleotide pair specific for the insertion fragment was used (lanes A), or an oligonucleotide pair specific for the wild-type gene (lanes B). The asterisk marks an unspecific PCR product. (C) The growth of the indicated strains in YBG11 medium was determined by analysis of the OD750; the wild-type experiment is shown in the four diagrams for better comparability to the behavior of each mutant. Values were normalized to initial OD750 of the culture and are expressed as natural logarithm. The mean from at least three biological replicates is shown with the standard deviation as error bar. (D) Wild-type and I-tonB2 cultures photographed after growth for 7 days in YBG11 medium. (E) Wild-type and mutant cultures photographed after growth for 5 days in YBG11 medium; the I-tonB2 sample was homogenized by pipetting up and down prior to the photographing in order to reduce clumping.
None of the tonB mutants exhibited an altered growth behavior compared to wild type under standard conditions (YBG11 medium, Fig. 2C). However, I-tonB2 cells frequently formed aggregates in liquid medium (Fig. 2D). The enhanced tendency of I-tonB2 to aggregate was also verified by sedimentation analysis (see Fig. S1 in the supplemental material). In addition, the color of I-tonB2 was considerably different from that of the wild type (Fig. 2E), suggesting a modification in the cellular pigment content.
Sedimentation of I-tonB2 (white symbols) and wild-type cultures (black symbols). The OD750 was measured in one cuvette after specific time points without further resuspension of the suspension. Average values of four biological replicates per strain with two technical replicates each are given; error bars show the standard deviation. Due to enhanced aggregation, I-tonB2 filaments sediment faster in the cuvette than wild-type samples, which is indicated by a faster decrease of the OD750. Download FIG S1, PDF file, 1.4 MB (1.4MB, pdf) .
Copyright © 2021 Schätzle et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The synthesis of carotenoids and Chl in Anabaena is differentially regulated in response to growth temperature and light intensity (43–46). Therefore, the concentrations of these pigments in the mutants and the wild type were determined after growth of the cultures for 7 days under ambient light (70 μmol photons m−2 s−1) as well as under high-light or low-light conditions (140 and 15 μmol photons m−2 s−1, respectively).
In general, the pigment concentrations under ambient light and low light were comparable. When grown under ambient or low-light conditions, the Chl concentration in the wild type was 9 ± 2 μg ml−1 at an optical density at 750 nm (OD750) of 1, the carotenoid concentration was 2.5 ± 0.3 μg ml−1 at an OD750 of 1, and the phycocyanin (PC) concentration was 26 ± 7 μg ml−1. Compared to that, all mutant strains were diminished in their cellular Chl content under both conditions (Table 1) even though a significant difference was found only for I-sjdR (low light) and I-tonB4 (ambient and low light). TonB3 is involved in siderophore transport (38, 39), and because a reduction in Chl is an indicator of iron starvation in Anabaena, the decrease in Chl possibly mirrors a fast iron starvation (47, 48). SjdR, however, is not involved in TonB-dependent schizokinen transport (39), and therefore, the observed reduction of the Chl content should not be related to iron uptake. Likewise, TonB4 is not involved in iron uptake, and the cause of Chl decrease remains elusive.
TABLE 1.
Chlorophyll a, carotenoid, and phycocyanin concentrations in the wild type and the tonB mutantsa
| Strain | High light |
Ambient light |
Low light |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chl/OD750 (μg ml−1) | Car/OD750 (μg ml−1) | Ratio, Chl/Car | PC/OD750 (μg ml−1) | Chl/OD750 (μg ml−1) | Car/OD750 (μg ml−1) | Ratio, Chl/Car | PC/OD750 (μg ml−1) | Chl/OD750 (μg ml−1) | Car/OD750 (μg ml−1) | Ratio, Chl/Car | PC/OD750 (μg ml−1) | |
| WT | 7 ± 2 | 2.2 ± 0.2 | 3 ± 1 | 27 ± 3 | 9 ± 2 | 2.4 ± 0.3 | 3.6 ± 0.3 | 27 ± 6 | 9 ± 1 | 2.5 ± 0.4 | 3.7 ± 0.3 | 25 ± 3 |
| I-sjdR | 5 ± 2 | 1.6 ± 0.4 | 3 ± 1 | 22 ± 5 | 7 ± 2 | 2.0 ± 0.7 | 3.6 ± 0.5 | 21 ± 7 | 7 ± 2 | 1.8 ± 0.6 | 3.9 ± 0.8 | 24 ± 5 |
| I-tonB2 | 4.4 ± 0.8 | 2 ± 1 | 2 ± 1 | 21 ± 3 | 7 ± 3 | 2 ± 1 | 3.0 ± 0.6 | 12 ± 2 | 7 ± 3 | 3 ± 1 | 3.2 ± 0.8 | 15 ± 4 |
| I-tonB3 | 6 ± 2 | 2.1 ± 0.5 | 2.7 ± 0.9 | 30 ± 3 | 7 ± 2 | 2.6 ± 0.8 | 2.9 ± 0.5 | 24 ± 7 | 8 ± 2 | 2.3 ± 0.4 | 3.4 ± 0.9 | 28 ± 5 |
| I-tonB4 | 5 ± 2 | 2.0 ± 0.3 | 3 ± 1 | 27 ± 9 | 7 ± 1 | 2.0 ± 0.4 | 3.6 ± 0.5 | 24 ± 9 | 7 ± 1 | 1.8 ± 0.5 | 4.2 ± 0.5 | 21 ± 6 |
Given are the concentrations in cultures grown for 7 days under high, ambient, or low light (140, 70, or 15 μmol photons m−2 s−1, respectively) in YBG11 medium. The average from 4 to 10 biological replicates and the standard deviation are given normalized to an OD750 of 1. Values in the mutants that significantly differ from wild-type values are indicated in bold (P < 0.05, Student’s t test with Bonferroni correction). Chl, chlorophyll a; Car, carotenoid; PC, phycocyanin.
Notably, I-tonB3, with 2.6 ± 0.8 μg ml−1, exhibited an elevated carotenoid concentration under ambient light compared to wild type. Under low light, the I-tonB2 carotenoid concentration was enhanced. In contrast to that, I-tonB4 had a significantly lower carotenoid level under both ambient and low-light conditions, and I-sjdR under low light. There were no significant differences of chlorophyll-to-carotenoid ratio under ambient or low-light conditions with the exception of a small decrease in I-tonB3 (ambient light). Under both conditions, I-tonB2 and I-tonB3 had lower ratios than the wild type and the other mutants (Table 1).
The concentrations of phycocyanin were similar in all strains except I-tonB2, in which it was significantly decreased by a factor of 1.5 and 2 (low and ambient light, respectively).
The Chl concentration was higher in all strains under normal (ambient) light or low-light conditions compared to high light (Table 1). For the wild-type strain, an average Chl concentration per OD750 of 7 ± 1 μg ml−1 was determined under high light. Overall, the carotenoid level tended to be lower under high light than under ambient light as well (Table 1), but the difference between these conditions was not as drastic as in the case of Chl. This resulted in a lowered ratio of Chl to Car. These data are consistent with previous observations (49–51). Only in I-sjdR was the carotenoid content significantly decreased compared to the wild type (Table 1). This reduction did not result in compromised growth (Fig. 1). Phycocyanin content did not differ significantly between the strains under high light. While the PC content of I-tonB2 increased relative to ambient light, the PC content of the other strains did not differ significantly between light conditions.
In summary, I-tonB2 contained a strongly lowered level of phycocyanin under ambient and low-light conditions and all mutants showed a mildly lower chlorophyll content compared to wild type under the same condition. Therefore, the color alterations observed for I-tonB2 likely result from the observed alterations in cellular pigmentation.
The cellular metal content is altered in tonB mutants.
The carotenoid concentration in cyanobacteria is affected by metal availability. Elevated Cu, Zn, or Co concentrations result in an elevation of the carotenoid content in Anabaena oryzae (52). Similarly, Ca supplementation enhances the level of pigments in Anabaena (53, 54). Thus, the cellular metal concentrations in I-tonB2, I-tonB3, and I-tonB4 were determined by inductively coupled plasma mass spectrometry (ICP-MS) analyses and compared to the wild-type concentrations that were described before (39) Since SjdR is functionally not related to TonB-dependent transport (39), this strain was excluded from the further studies.
Remarkably, alterations in metal concentrations were observed for all tonB mutants compared to the wild type. (i) I-tonB3 and I-tonB4 exhibited a decrease in cellular Mg and Co concentrations compared to the wild type (Table 2). (ii) In I-tonB2 and I-tonB3 the Mn concentration was decreased. In I-tonB4 the level of Mn showed a large variation, but Mn was always at a lower level than in the wild type. The Mo concentration was (iii) enhanced in I-tonB2 and (iv) reduced in I-tonB4. (v) In I-tonB2 cells an elevated Cu concentration was observed compared to the wild type. Notably, after 7 days of growth in YBG11 medium an alteration in the Fe concentration was not observed, although TonB3 is supposed to be involved in ferric siderophore transport.
TABLE 2.
Metal concentration in wild-type Anabaena and the mutants I-tonB2, I-tonB3, and I-tonB4 expressed as atoms per OD750a
| Metal | 1013 atoms/OD |
Ratio (mutant/WT) |
|||||
|---|---|---|---|---|---|---|---|
| WT | I-tonB2 | I-tonB3 | I-tonB4 | I-tonB2 | I-tonB3 | I-tonB4 | |
| Mg | 500 ± 40 | 410 ± 20 | 350 ± 40 | 280 ± 10 | 0.82 | 0.70 | 0.56 |
| Ca | 61 ± 7 | 70 ± 50 | 60 ± 20 | 40 ± 20 | 1.15 | 0.98 | 0.66 |
| Mn | 51 ± 2 | 17 ± 1 | 11 ± 1 | 30 ± 20 | 0.33 | 0.22 | 0.59 |
| Fe | 34 ± 2 | 35 ± 5 | 29 ± 3 | 30 ± 8 | 1.03 | 0.85 | 0.88 |
| Co | 1.8 ± 0.1 | 1.8 ± 0.1 | 1.2 ± 0.1 | 1.3 ± 0.1 | 1.00 | 0.66 | 0.72 |
| Cu | 5.1 ± 0.5 | 6.7 ± 0.6 | 5 ± 2 | 6 ± 1 | 1.31 | 0.98 | 1.18 |
| Zn | 5.4 ± 0.3 | 6.1 ± 0.4 | 5 ± 4 | 7 ± 3 | 1.13 | 0.93 | 1.29 |
| Mo | 6.4 ± 0.2 | 9.8 ± 0.7 | 7.3 ± 0.6 | 4.8 ± 0.4 | 1.53 | 1.14 | 0.75 |
The ratio of the metal content in wild type and mutants is shown. The values represent averages and standard deviation from three biological measurements. The bold letters indicate significant changes in the ratio column (P < 0.05, Student’s t test).
Ca and Zn levels were not drastically altered in the mutants. In contrast, Cu, which influences the carotenoid content in cyanobacteria (52), was enhanced in I-tonB2, in which the carotenoid level was found to be enhanced as well (Tables 1 and 2). In turn, in I-tonB3 and I-tonB4 the Co levels were reduced, which in the case of I-tonB4 could be related to the reduced carotenoid level (Tables 1 and 2). In summary all tonB mutants exhibited alterations in the cellular metal levels compared to wild type to different extents. Whereas I-tonB3 and I-tonB4 were reduced in Co, Mn (I-tonB3), or Mo (I-tonB4), only I-tonB2 did, besides the observed reduction in Mn, significantly enrich metals, namely, Cu and Mo.
Membrane properties and transcriptional alterations in I-tonB2.
Considering the relative accumulation of Cu and Mo in I-tonB2 and the tendency of this strain to form aggregates in solution, a modification in the cell surface could cause the mentioned effects. Therefore, lipopolysaccharide (LPS) was extracted from the wild-type and I-tonB2 strains and separated by SDS-PAGE. Reproducibly enhanced signals for the O-antigen ladder were observed for the tonB2 mutant compared to wild type, in which the O-chain was barely visible when similar amounts of LPS extracts were loaded (Fig. 3A and Fig. S2). This confirms an increased synthesis of LPS in the mutant strain that could result from an aberrant regulation.
FIG 3.
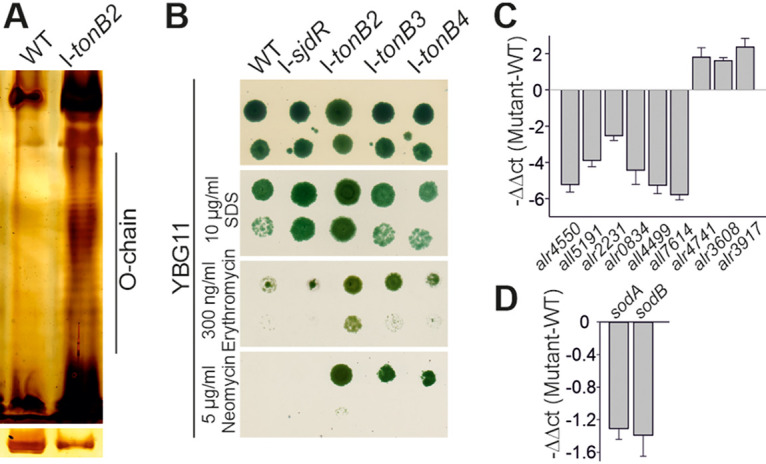
Outer membrane integrity of the tonB mutants and expression patterns in I-tonB2. (A) LPS extracted from the wild type (WT) and I-tonB2 was separated by SDS-PAGE and silver stained. The O-antigen fragments are indicated. The loading control (large subunit of Rubisco) is given in the bottom panel where whole-cell lysate of both strains was separated in amounts proportional to the LPS extract. A representative result is shown; other LPS analyses performed from independently grown cultures are shown in Fig. S2. (B) The wild type (WT) and the tonB mutants were spotted on plates of YBG11 medium supplemented with the indicated compounds. Five microliters of a cell suspension with an OD750 of 1 (upper row in each plate) or an OD750 of 0.1 (bottom row in each plate) was spotted. Images were taken after 10 days of incubation. The experiment was conducted three times with independent cultures, and representative images are shown here. (C and D) The relative expression (I-tonB2 versus WT) in terms of −ΔΔCT is shown for the putative Anabaena porin genes (C) and the superoxide dismutase genes (D). The ΔCT value was normalized to the housekeeping gene rnpB (giving the ΔCT) and the respective ΔCT of the wild type (giving the ΔΔCT). Average values from three independent biological replicates are shown; error bars indicate the standard deviation.
LPS extractions from wild-type Anabaena and I-tonB2 cultures. (1) Cultures grown in tubes in the presence of 1% CO2. (2) Cultures grown without bubbling in shaking flasks; a higher degree of aggregation was observed without CO2 supplementation. The LPS isolated from the cultures was separated by SDS-PAGE and silver stained. The bottom panel shows the loading control (large subunit of Rubisco). Download FIG S2, PDF file, 1.6 MB (1.6MB, pdf) .
Copyright © 2021 Schätzle et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
LPS structure as well as carotenoids are known to influence membrane properties (55–59). In cyanobacteria, carotenoids are found in all membranes including the outer membrane (60–62). Therefore, the integrity of the OM in the tonB mutants was tested by spotting suspensions of the strains on plates containing antibiotics or SDS. Here, I-sjdR was utilized as an unrelated control strain bearing a plasmid insertion.
Interestingly, I-tonB2 exhibited an increased resistance toward SDS, erythromycin, and neomycin compared to the wild type (Fig. 3A). In addition, I-tonB3 and I-tonB4 were more resistant toward the selected antibiotics, but the effect was not as pronounced as for I-tonB2. Moreover, I-sjdR grew in a similar manner as I-tonB2 in the presence of SDS but was as sensitive toward the tested antibiotics as the wild type.
The reduced susceptibility of I-tonB2 suggests a limited uptake of the selected compounds into the cell and confirms an alteration to the cell envelope. Typically, porins mediate the transport of certain antibiotics across the OM, and thus, porin mutants often display hyperresistance toward those compounds (63). Although it has been discussed that lipophilic macrolide antibiotics likely enter the cell through diffusion across the membranes and not through porins (17), for Anabaena a relation to porin function has been proposed (64). Thus, the transcript abundance of nine genes coding for porin-like proteins (1, 65) was examined in I-tonB2.
Remarkably, the transcript abundance of six genes coding for putative porins was reduced in I-tonB2 compared to the wild type after 7 days of growth in YBG11 medium (Fig. 3B). Among those, the transcript abundance of all7614, all4499, and alr4550 was most drastically reduced by 54-, 37-, and 36-fold, respectively. The transcript level of the three genes alr4741, alr3608, and alr3917 was higher in the mutant than in the wild type. The maximum increase was, however, 5-fold (alr3917), which appears to be only moderate compared to the drastic downregulation of other putative porin-encoding genes (Fig. 3B).
In addition to their impact on membranes, carotenoids are involved in the protection of the photosynthetic apparatus from reactive oxygen species (ROS). Thus, an increased carotenoid concentration might result from an elevated level of oxidative stress. To test whether I-tonB2 exhibits a higher oxidative stress level, the expression of superoxide dismutase A (SodA, MnSOD) and B (SodB, FeSOD) was analyzed. Both enzymes confer resistance to oxidative stress under distinct nitrogen regimes (66). The abundance of both transcripts was reduced in I-tonB2 in comparison to the wild type after 7 days of cultivation in YBG11 medium (Fig. 3C). This suggests that the tonB2 mutant does not suffer from increased oxidative stress. The assessment of reactive oxygen (ROS) production in I-tonB2 and wild type by the fluorescent probe 2,7-dichlorodihydrofluorescein diacetate (DCFH-DA) confirmed this finding, as no difference in fluorescence that represents cellular ROS content was detected (Fig. S3).
Estimation of intracellular ROS level. Cultures of wild type and I-tonB2 were grown for 1 week to a density of ∼OD750 of 1, and intracellular ROS levels were determined using the DCHF-AC assay (112) after 20-min pretreatment in the dark, ambient light, or UV-B radiation with or without addition of ascorbic acid. Higher conversion to fluorescent dichlorofluorescein indicates higher ROS levels. I-tonB2 did not behave significantly different from the wild type (Student’s t test). Download FIG S3, PDF file, 0.2 MB (204.4KB, pdf) .
Copyright © 2021 Schätzle et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
I-tonB2 and I-tonB4 are impaired in nitrogenase activity.
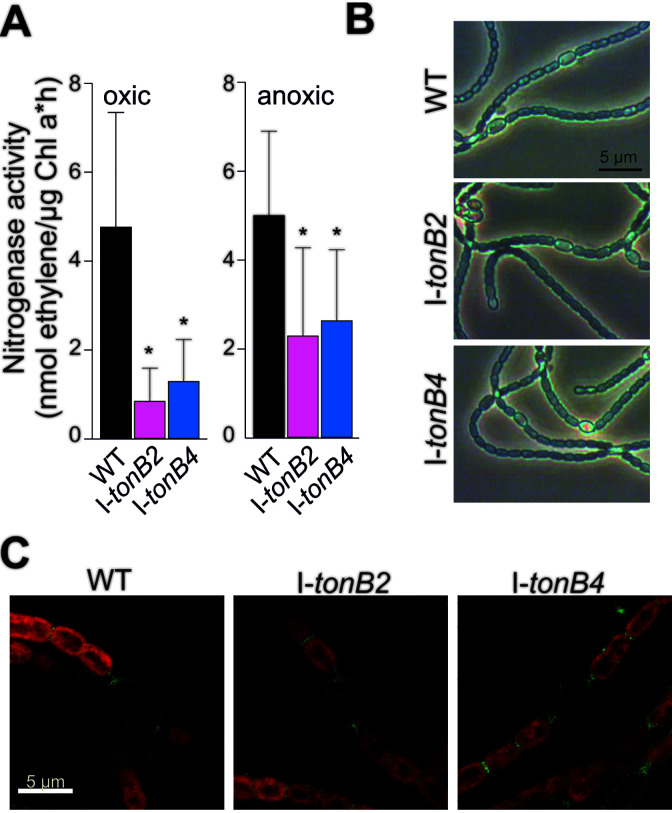
The nitrogenase enzyme in Anabaena has Mo as a cofactor (12). Notably, I-tonB2 and I-tonB4 were altered in the cellular Mo concentration compared to wild type (Table 2), and the abundance of the tonB4 transcript was found to be enhanced in the absence of combined nitrogen (38). Therefore, the nitrogenase activity was determined for I-tonB2 and I-tonB4 by means of the acetylene reduction assay. Under oxic conditions, a nitrogenase activity of 0.8 ± 0.7 or 1.3 ± 0.9 nmol ethylene/μg Chl · h was measured in I-tonB2 and I-tonB4, respectively (Fig. 4A), which was significantly lower than in the wild type (4.7 ± 2.6 nmol ethylene/μg Chl · h). Under anoxic conditions, the nitrogenase activity in both mutants was enhanced compared the oxic conditions. However, the average values of 2 ± 2 and 3 ± 2 nmol ethylene/μg Chl · h for I-tonB2 and I-tonB4, respectively, were again significantly lower than the nitrogenase activity of the wild type. Hence, both strains showed a reduced but not abolished nitrogen fixation capacity.
FIG 4.
Heterocyst formation and nitrogenase activity in I-tonB2 and I-tonB4. (A) Nitrogenase activity was determined under oxic and anoxic conditions in the wild type (WT), I-tonB2, and I-tonB4. Average values of 9 (wild type), 4 (I-tonB2), and 5 (I-tonB4) measurements on independent cultures are given, and error bars represent the standard deviation. Asterisks denote significant differences between each mutant and the wild type (Student’s t test, P < 0.05). (B) Light microscopy images of cultures grown for 3 days on BG110 medium plates; representative images are shown. (C) The peptidoglycan of cells grown for 7 days on BG110 medium plates was labeled with the fluorescent dye Van-FL that specifically binds to the cell wall (see Materials and Methods for details). Fluorescence images were recoded with a confocal laser scanning microscope; a merge of the Van-FL fluorescence (green) and the chlorophyll autofluorescence (red) is shown. For better visibility, the Van-FL signal and the overall intensity were enhanced.
Considering the alterations in nitrogenase activity, the heterocysts of the tonB mutants were analyzed under the light microscope and compared to those of the wild type. I-tonB2 and I-tonB4 mutants differentiated wild-type-like heterocysts as judged from light microscopy (Fig. 4B), and the formation of the constricted heterocyst pole was not altered as determined with fluorescently labeled vancomycin (Fig. 4C). Therefore, TonB2 and TonB4 are not essential for heterocyst differentiation, although tonB4 expression is upregulated under nitrogen starvation (38) and nitrogenase activity in the mutants is somewhat reduced compared to the wild type (Fig. 4A).
Iron starvation and siderophore transport capacities of tonB mutants.
Next, we addressed the question whether TonB2 and TonB4 are involved in the transport of ferric siderophores, as described for TonB3 (39). First, a potential complementation of single tonB insertions through enhanced expression of other tonB genes was analyzed by quantitative reverse transcription-PCR (qRT-PCR). SjdR was excluded from this analysis, as it is no longer considered a TonB candidate (39). The transcript abundance of the nonmutated tonB genes was determined after 7 days of iron starvation and normalized against the expression of the housekeeping gene rnpB. Starvation was applied since the expression of genes involved in TonB-dependent transport in Anabaena is triggered in the absence of iron (38, 42, 48, 67).
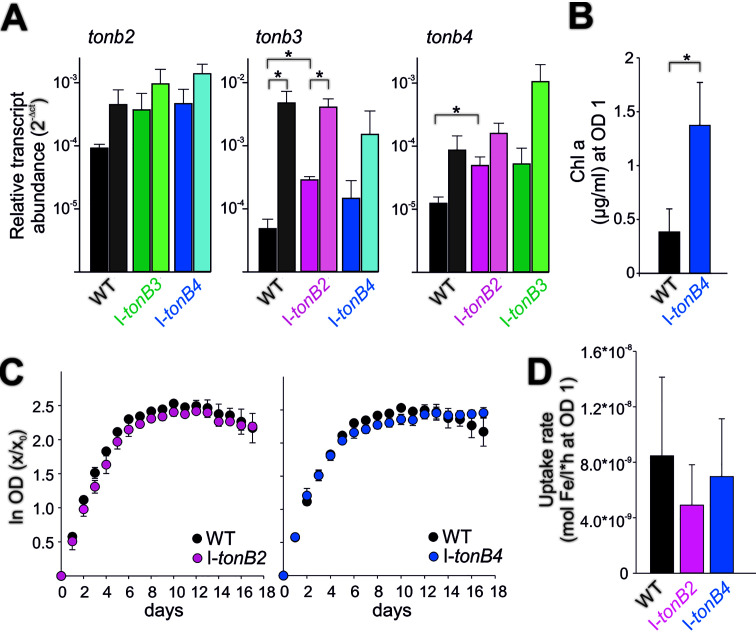
The abundance of tonB2 transcripts in I-tonB3 and I-tonB4 mutants was not significantly different from the wild-type level, both under standard conditions and after iron starvation (Fig. 5A, left). Further, no significant alteration of tonB2 expression was observed in response to iron starvation in any of the strains tested. Similarly, the expression of tonB4 was not significantly affected by iron starvation (Fig. 5A, right) but was enhanced in I-tonB2 compared to the wild type when strains were grown in YBG11 medium.
FIG 5.
Gene expression, growth, and siderophore transport in the tonB mutants. (A) RNA isolated from wild-type and tonB mutant strains after growth in YBG11 medium (left bars in black, dark green, dark blue, and dark pink) and after 7 days of iron starvation (right bars in gray, light green, light blue, and light rose). Gene expression was normalized to rnpB. Significant changes between the mutants and the wild type or the two conditions tested in one strain are marked with an asterisk (Student’s t test, P < 0.05). Three independent biological replicates per strain were analyzed, and the error bars represent the standard deviation. (B) Cultures were grown for 25 days in iron-free medium, and Chl was extracted; averages from three different biological replicates per strain with standard deviation are shown. Asterisk indicates P < 0.05 (Student’s t test). (C) Wild-type, I-tonB2, and I-tonB4 cultures were prestarved and diluted afterward to an OD750 of 0.05, and growth was monitored. The growth of three different cultures per strain was analyzed; for better visualization and comparison, the wild-type values are present in both diagrams. Standard deviations are indicated. (D) Ferric schizokinen transport was measured with the wild type, I-tonB2, and I-tonB4. Average values of 8 (wild type), 7 (I-tonB2), and 7 (I-tonB4) measurements from independent cultures are presented, and standard deviations are given.
The tonB3 transcript was increased under iron starvation in all strains (Fig. 5A, middle), although the change in I-tonB4 was not significant (Fig. 5A, middle). Moreover, tonB3 expression in I-tonB2 was enhanced compared to the wild type when cells were grown in YBG11 medium but not after iron starvation. This may suggest an early level of starvation in I-tonB2, since tonB3 expression is triggered upon iron depletion. However, no changes in Chl (as indicator of iron starvation) and no significant alteration in the cellular iron content were observed in I-tonB2 compared to the wild type (Tables 1 and 2), which does not support iron starvation in I-tonB2.
In contrast, tonB3 expression was not as drastically enhanced after iron starvation in I-tonB4 as observed in the wild type or I-tonB2. This suggests a comparatively lower level of starvation in the tonB4 mutant. To test this, wild-type and mutant cultures were grown for 25 days in iron-free medium and the Chl level was determined as an indicator for the degree of starvation. Indeed, the Chl content was more drastically decreased in the wild type (0.38 μg ml−1 at an OD750 of 1) than in I-tonB4 (1.4 μg ml−1 at an OD750 of 1) (Fig. 5B). Thus, the tonB4 mutant shows an unusually weak iron starvation phenotype, which explains the reduced induction of tonB3 expression during starvation.
In conclusion, no drastic alterations of tonB transcript abundance were observed between the mutants and the wild type after iron starvation. Thus, complementation effects between the tonB genes appear not to take place according to the analysis of the phenotypes under iron starvation. Then, the growth behavior in iron-free medium was analyzed for I-tonB2 and I-tonB4 (Fig. 5C). The cultures were prestarved prior to monitoring growth, since iron starvation in Anabaena which involves the expression of relevant uptake systems requires initiation time (67). We did not observe a compromised growth of the mutants compared to the wild type, which argues against a direct function of TonB2 and TonB4 in iron uptake. To support this conclusion, the transport rates of schizokinen loaded with 55Fe were determined. Although the normalized uptake rates of 4.9 × 10−9 mol Fe/liter · h for I-tonB2 and 7.0 × 10−9 mol Fe/liter · h for I-tonB4 were slightly lower than the wild-type rates (8.5 × 10−9 mol Fe/liter · h), no significant difference could be established (Fig. 5D). Thus, TonB2 and TonB4 do not seem to function in schizokinen transport in Anabaena.
DISCUSSION
The TonB-ExbB-ExbD system is conserved among Gram-negative bacteria, as approximately two-thirds of these bacteria have at least one TonB-encoding gene (3). Notably, many species encode multiple TonB copies in the genome (3), and diverse functions have been assigned to the different genes in one species (30, 31, 36).
In Anabaena four genes have been annotated as encoding possible TonB proteins. For SjdR (formerly annotated as TonB1), a function distinct from TonB-dependent transport was described (39). With respect to the three TonB-like proteins of Anabaena that exhibit a conserved domain architecture (Fig. 1), TonB3 represents the central component of the siderophore-dependent iron uptake system (38, 39). In contrast, TonB2 and TonB4 are likely not related to iron uptake, since schizokinen uptake is not impaired in their mutants (Fig. 5). The abnormal iron starvation behavior of I-tonB4 requires further investigation and cannot be explained at this stage. Notably, Anabaena is capable of transporting other siderophores besides the endogenously synthesized schizokinen, such as aerobactin, ferrioxamine B (48, 67, 68), or ferrichrome (unpublished data). Aerobactin penetrates through the same TBDT as schizokinen does (67); therefore, it is likely that aerobactin transport is TonB3 dependent as well (38). That TonB2 or TonB4 is involved in the TonB-dependent transport of other iron-containing substrates cannot be excluded. However, because the growth of the mutant cultures is not affected in the absence of iron, a relation to ferric siderophore transport seems unlikely. Besides ferric siderophores, also other substrates such as cobalamin, nickel, or sugars are transported in a TonB-dependent manner in some organisms (16, 19). This further broadens the spectrum of possible functions for TonB2 and TonB4 that will need to be investigated, especially considering the high number of 22 TBDTs that are predicted from the Anabaena genome (37).
The phenotypes of I-tonB2 and I-tonB4 were investigated in order to figure out possible functions. Differential characteristics of the strains were unveiled, including alterations of the cellular metal concentrations that were observed, albeit to different extents, for all tonB mutants. Both I-tonB3 and I-tonB4 show decreased cellular contents of Mg and Co, and I-tonB4 also shows a decreased content of Mo (Table 2). Molybdenum is the cofactor of the nitrogenase enzyme (12), which might contribute to the reduced nitrogenase activity in I-tonB4 (Fig. 4). Moreover, in I-tonB4 the Chl level is comparatively decreased (Table 1), which in turn might be a consequence of the reduced Mg content in this strain. Notably, in I-tonB4 the regulation of Chl synthesis seems to be generally affected, considering the remarkably high Chl concentration after iron starvation.
Besides a decrease in Mn in I-tonB2 and I-tonB3, an accumulation of Cu and Mo was observed in I-tonB2 that might be caused by altered outer membrane properties. The anionic LPS surface is involved in metal binding in bacteria (69–72), and it was reported previously that cyanobacterial negatively charged exopolysaccharides are capable of binding metals (73) and might even accumulate trace metals under starvation conditions. Thus, the enhanced LPS production in I-tonB2 might (i) lead to an enhanced adsorption of certain metals that subsequently diffuse into the cell and (ii) cause the formation cell aggregates.
LPS is thought to have an important role in porin trimerization, stability, and conductance (74–76). In the so-called deep rough mutants, strains that are compromised in LPS synthesis and thus produce only truncated LPS, a smaller amount of protein is present in the OM (17). Additionally, those mutants are increasingly susceptible to SDS or hydrophobic antibiotics (77, 78). An opposite effect might take place in I-tonB2, in which the excessive LPS production might result in the monitored decrease in susceptibility toward drugs, possibly reinforced by the reduction of the expression of genes encoding porins observed in this strain. Although it has been speculated that aminoglucoside antibiotics cross the OM through a diffusion-based self-promoted pathway in which the LPS surface is involved (17, 79), for Anabaena a relation of erythromycin uptake to porin function has been established (64). In the I-tonB2 mutant most of the genes coding for porins are downregulated, among them the porins described to be most abundant in Anabaena (40, 65). The decreased porin expression could display a feedback transcriptional response to a putatively enhanced substrate (metal) diffusion into the cell, reflected by a higher metal adsorption in I-tonB2. Hence, considering these characteristics a TolA-like function could be proposed for TonB2 rather than a TonB-like function.
The TolA-TolQ-TolR system embedded in the PM (Tol system here) (80) is structurally related to the TonB system. The C-terminal domains of TonB and TolA are structurally analogous (81), and it is assumed that the Tol system is involved in maintaining OM integrity (82–84). TolA interacts with trimeric porins of E. coli (85, 86), and the Tol-Pal system constitutes a component of the divisome involved in cell constriction and peptidoglycan remodeling (87, 88). Moreover, TolA might modulate the expression of LPS components in E. coli (89, 90), which also might be the case for TonB2.
The lack or low dosage of any TonB-like protein induces pigment alterations in Anabaena (Table 1). The reduction of the phycocyanin level in I-tonB2 is also reflected in the comparatively bright color of its cultures (Fig. 2). Phycocyanin-containing cyanobilins are accessory pigments that harvest light and transfer energy to photosystem II. A possible explanation for the modifications in pigment content is the observed alterations in cellular metal contents because different metal treatments are known to affect cyanobacterial pigment concentrations. For example, the treatment of Anabaena oryzae with 1 to 100 ppm of Cu resulted in an increased carotenoid concentration after 6 to 8 days of incubation (52).
In summary, the four TonB-like proteins found in Anabaena apparently take over distinct functions. Neither I-toB2 nor I-tonB4 is drastically reduced in schizokinen uptake capacity, which suggests that TonB3 exclusively mediates schizokinen transport in Anabaena. On the other hand, both tonB2 and tonB4 mutants are compromised in the production of full nitrogenase activity, especially under oxic conditions, although heterocyst differentiation seems not affected. TonB2 influences OM properties, including LPS synthesis, with an effect on susceptibility toward antibiotics and porin abundance (as deduced from porin gene expression), and its role might be related to that of the Tol system as discussed above. The role of TonB4 is elusive, consistent with its peculiar predicted structure (Fig. 1), although we note that its mutant is affected in the cellular levels of several metals, including Mo, that may result in the observed low nitrogenase levels.
MATERIALS AND METHODS
Anabaena culture conditions.
Anabaena (also known as Nostoc) sp. strain PCC 7120 was stored on plates of BG11 medium with 1% (wt/vol) Bacto agar (BD Biosciences) (91). For liquid culturing either BG11 (91) or YBG11 medium (92) was used. Anabaena cultures were grown under constant shaking at 90 to 100 rpm and constant illumination (ambient light, 70 μmol photons m−2 s−1; Osram L 58 W/954-Lumilux de Luxe, daylight) at 28°C. In the case of mutant strains 5 μg ml−1 of both spectinomycin dihydrochloride pentahydrate (Sp; Duchefa Biochemie) and streptomycin sulfate (Sm; Roth) was added. The growth was monitored spectrophotometrically in terms of optical density at 750 nm (OD750). For growth analysis on plates, 5 μl of cell suspensions with an OD750 of 1 was spotted in a dilution series (1, 1:10, and 1:100), and representative results are shown.
DNA extraction, molecular cloning, and generation of Anabaena mutants.
Transformation of E. coli and isolation and manipulation of plasmid DNA were performed according to standard protocols (93). Anabaena genomic DNA (gDNA) was isolated as described previously (94) with modifications: sodium dodecyl sulfate was not added, and the phenol extraction was done once followed by two washing steps with 400 μl of chloroform.
The Anabaena tonB mutants AFS-I-sjdR, AFS-I-tonB2, AFS-I-tonB3, and AFS-I-tonB4 were utilized in this study and have been introduced previously (38, 39). The annotation stands for Anabaena mutant generated in Frankfurt, Germany by the Schleiff Lab by plasmid insertion; for better readability “AFS-” is omitted throughout the text. In brief, internal fragments of the single genes were ligated into vector pCSV3 (95, 96) in the case of I-tonB2, I-tonB3, and I-tonB4 or pCSEL24 (97) in the case of I-sjdR, both carrying spectinomycin and streptomycin resistance markers. The oligonucleotides and the plasmids used in this study are listed in Tables S1 and S2 in the supplemental material, respectively. Plasmids were transferred to Anabaena with the triparental mating method as previously described (40, 97–99). The Anabaena strains analyzed in this study are listed in Table S3. The genotype of the exconjugants was tested by PCR, in which an oligonucleotide specific for the plasmid in combination with an oligonucleotide specific for the gene region was used.
Oligodeoxynucleotides used in the study. Download Table S1, DOCX file, 0.01 MB (13KB, docx) .
Copyright © 2021 Schätzle et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmids used in this study. Download Table S2, PDF file, 0.09 MB (93.8KB, pdf) .
Copyright © 2021 Schätzle et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Anabaena strains used in this study. Download Table S3, PDF file, 0.09 MB (88.8KB, pdf) .
Copyright © 2021 Schätzle et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Short-term siderophore transport measurements.
The transport rates of ferric schizokinen were determined as described earlier (67, 100). Schizokinen was ordered from EMC Microcollections, and 55FeCl3 was purchased from Perkin-Elmer. A final concentration of 15 nM 55Fe-schizokinen was used in single measurements, and the final cell suspension utilized for measuring was inoculated at an OD750 of 0.05. Cultures were prestarved in iron-free YBG11 before the measurements were conducted, and the degree of starvation was estimated by the chlorophyll a (Chl) concentration at an OD750 of 1, as previous studies indicated that the Chl concentration per cell decreases in Anabaena with ongoing iron starvation (48). The Chl concentrations of experimental cultures are given in Table S4.
55Fe-Schizokinen uptake rates and normalized chlorophyll values. Download Table S4, DOCX file, 0.01 MB (12.7KB, docx) .
Copyright © 2021 Schätzle et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Inductively coupled plasma mass spectrometry (ICP-MS).
The cellular metal concentrations in Anabaena were determined as described earlier (101). In brief, cultures grown for 7 days in YBG11 were harvested by centrifugation and washed twice with 20 mM 2-(N-morpholino)ethanesulfonic acid (pH 5), 10 mM EDTA. Cells were resuspended in 5 ml double-distilled water, and 1 ml was subjected to inductively coupled plasma mass spectrometry (ICP-MS) measurement. The OD750 of the suspension was measured, and cells were counted for normalization. Samples were digested overnight at 120°C in 7 M HNO3 and dissolved in 5% HNO3 for measurement.
Pigment extraction.
The measurements of Chl and carotenoid (Car) concentrations were performed with methanolic extracts as previously described (102, 103). The Chl concentration was determined with the formula Chl (μg/ml) = 12.9447(A665 − A720), and the carotenoid concentration was calculated with the formula Car (μg/ml) = {1,000(A470 − A720) − 2.86 × [Chl (μg/ml)]}/221.
To estimate the Chl concentration for experimental cultures, a simplified equation was utilized, as follows: Chl (mg/ml) = A665/74.5 (104).
Determination of phycocyanin content was carried out as described by Horvath et al. (105). In brief, cultures were harvested by centrifugation (1 min, 4,000 × g) and concentrated to a volume of 0.3 to 2 ml. OD750 of the concentrates was determined in 1/100 dilution, and the remaining material was frozen at −20°C. After thawing at room temperature (RT), a defined volume was diluted to 4 ml with phosphate-buffered saline (PBS) and subjected to sonication with a sonication probe at full power for 90 s. Cell debris was removed by centrifugation (1 min, 20,000 × g). PC concentration was calculated from UV-visible (UV-Vis) absorption spectra as described previously (106): PC (μg/ml) = 154(A618 − A730).
Van-FL staining and microscopy.
For visualization of peptidoglycan, the filaments were stained with BODIPY FL vancomycin (Van-FL) (Invitrogen) as previously described (107). For microscopy a piece of agar was excised and reversely placed onto a coverslip that was utilized as a microscope slide. Images were recorded with a Zeiss LSM 780 using 63× or 40× objectives with immersion oil. Diameter of the pinhole was set to 69.4 μm, and a 488-nm laser source was used for excitation. Specific Van-FL fluorescence was recorded between 500 and 550 nm, Chl autofluorescence was recorded between 630 and 700 nm. Light microscopy images were recorded with a Thorlabs DCC1645C-HQ camera.
RNA extraction, RT-PCR, and qRT-PCR.
RNA was isolated either from strains that had been grown for 7 days in YBG11 medium or, in the case of iron starvation, from samples incubated for 7 days under culture conditions in iron-free YBG11 medium. RNA was extracted with TRIzol (Thermo Fisher Scientific) according to previously described protocols (42). After DNase I digestion the absence of DNA in RNA samples was verified by PCR. Revert Aid reverse transcriptase was used for first strain cDNA synthesis (Thermo Fisher Scientific). qPCR was performed with a StepOnePlus cycler from Thermo Fisher Scientific, and the cycling conditions were set as 50°C, 2 min, and 95°C, 2 min, followed by 40 cycles at 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s. Experiments were performed with PowerUp SYBR green Master Mix from Applied Biosystems. rnpB served as control gene. The threshold cycle (CT) value of the gene of interest was normalized to the rnpB CT value (ΔCT) and, if indicated, further normalized to the corresponding ΔCT of the wild-type (ΔΔCT) (108).
Extraction and analysis of lipopolysaccharide.
Lipopolysaccharide (LPS) was extracted from wild type and I-tonB2, and the cultures were grown in flasks with constant shaking for 8 days or in tubes with supplementation of 1% CO2 in YBG11 medium for 7 days (Fig. S2). Cells corresponding to an OD750 of 3 in 1 ml were harvested for LPS extraction. LPS was extracted as described previously (109) with modifications. In brief, the cells were harvested by centrifugation (6,000 × g, 5 min), washed with 1 ml of phosphate-buffered saline (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4), and resuspended in 50 μl lysing buffer (2% sodium dodecyl sulfate, 4% β-mercaptoethanol, 10% glycerol, 1 M Tris-HCl, pH 6.8, bromphenol blue). After the samples were heated at 100°C for 10 min, 10 μl of lysing buffer containing 100 μg proteinase K was added, followed by 60 min of incubation at 60°C.
The LPS was subjected to denaturing SDS-polyacrylamide electrophoresis utilizing a 12% acrylamide gel; afterward the gel was silver stained (110). For the loading control (large subunit of Rubisco is shown in figures), cells were treated as described for LPS extraction omitting proteinase K.
Nitrogenase activity.
Nitrogenase activity was determined by the acetylene reduction assay (111) carried out under both oxic and anoxic conditions. Cells grown in 25 ml of liquid BG11 medium (supplemented with Sp/Sm when necessary) were harvested by centrifugation, washed with liquid BG110 medium, and incubated at 1 μg Chl ml−1 (without antibiotics) in 25 ml of liquid BG110 medium for 48 h. Cell suspensions of 2 ml at 6 μg Chl ml−1 were transferred to sealed small flasks in which the nitrogenase activity assay was carried out. Under both oxic and anoxic conditions 2 ml of acetylene was injected. For the assays under oxic conditions, the sealed flasks were incubated for 30 min (30°C and shaking) before taking 1-ml samples for determination of ethylene by gas chromatography. For anoxic conditions, the sealed flasks were supplemented with 10 μM 3-(3,4-dichlorophenyl)-1,1-imethylurea (DCMU; from Sigma), bubbled with argon for 4 min, and incubated for 60 min (30°C and shaking) before acetylene injections, and then 1-ml samples were taken for ethylene determination. Under both conditions, samples for ethylene determination were taken for up to 2 h.
Determination of ROS production.
Intracellular conversion of dichlorodihydrofluoresein (DCHF) to dichlorofluorescein (DCF) was measured as established previously (112). Cultures were harvested by centrifugation (1 min, 4,000 × g) and rediluted to an OD750 of 1 with PBS. For comparison, ascorbic acid (100 μM final concentration) was added to some samples. Samples of 10 ml were subjected to the respective light treatment for 20 min. UV treatment was carried out by placing the samples on the transilluminator of a gel electrophoresis documentation system, and ambient light treatment was carried out under cultivation conditions. After addition of 2,7-dichlorodihydrofluorescein (final concentration, 5 μM), samples were incubated for 1 h in the dark. Finally, cells were harvested and transferred to 96-well plates, and fluorescence and OD750 were determined using a microplate reader.
ACKNOWLEDGMENTS
We thank Rafael Pernil for useful discussions and Sotirios Fragkostefanakis for a critical reading of the manuscript.
The work was funded by the Deutsche Forschungsgemeinschaft DFG SCHL585/6-3 and LOEWE/CMMS to E.S. Work in Seville was supported by grant number BFU2017-88202-P from Plan Estatal de Investigación Científica y Técnica y de Innovación, Spain, cofinanced by the European Regional Development Fund, to Enrique Flores. FIERCE is financially supported by the Wilhelm and Else Heraeus Foundation and by the Deutsche Forschungsgemeinschaft (DFG, INST 161/921-1 FUGG and INST 161/923-1 FUGG), which is gratefully acknowledged. This is FIERCE contribution no. 81.
We declare that we have no conflicts of interest.
Contributor Information
Enrico Schleiff, Email: schleiff@bio.uni-frankfurt.de.
Timothy M. LaPara, University of Minnesota
REFERENCES
- 1.Hahn A, Schleiff E. 2014. The cell envelope, p 29–87. In Flores E, Herrero A (ed), The cell biology of cyanobacteria. Caister Academic Press, Poole, United Kingdom. [Google Scholar]
- 2.Mirus O, Hahn A, Schleiff E. 2010. Outer membrane proteins, p 175–228. In König H, Claus H, Varma A (ed), Prokaryotic cell wall compounds: structure and biochemistry. Springer, Berlin, Germany. [Google Scholar]
- 3.Chu BCH, Peacock RS, Vogel HJ. 2007. Bioinformatic analysis of the TonB protein family. Biometals 20:467–483. doi: 10.1007/s10534-006-9049-4. [DOI] [PubMed] [Google Scholar]
- 4.Andrews SC, Robinson AK, Rodríguez-Quiñones F. 2003. Bacterial iron homeostasis. FEMS Microbiol Rev 27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 5.Wandersman C, Delepelaire P. 2004. Bacterial iron sources: from siderophores to hemophores. Annu Rev Microbiol 58:611–658. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- 6.Gledhill M, Buck KN. 2012. The organic complexation of iron in the marine environment: a review. Front Microbiol 3:69. doi: 10.3389/fmicb.2012.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Street JH, Paytan A. 2005. Iron, phytoplankton growth, and the carbon cycle. Met Ions Biol Syst 43:153–193. doi: 10.1201/9780824751999.ch7. [DOI] [PubMed] [Google Scholar]
- 8.Ratledge C, Dover LG. 2000. Iron metabolism in pathogenic bacteria. Annu Rev Microbiol 54:881–941. doi: 10.1146/annurev.micro.54.1.881. [DOI] [PubMed] [Google Scholar]
- 9.Dammeyer T, Frankenberg-Dinkel N. 2008. Function and distribution of bilin biosynthesis enzymes in photosynthetic organisms. Photochem Photobiol Sci 7:1121–1130. doi: 10.1039/b807209b. [DOI] [PubMed] [Google Scholar]
- 10.Pushnik JC, Miller GW, Manwaring JH. 1984. The role of iron in higher plant chlorophyll biosynthesis, maintenance and chloroplast biogenesis. J Plant Nutr 7:733–758. doi: 10.1080/01904168409363238. [DOI] [Google Scholar]
- 11.Raven JA, Evans MCW, Korb RE. 1999. The role of trace metals in photosynthetic electron transport in O2-evolving organisms. Photosynth Res 60:111–150. doi: 10.1023/A:1006282714942. [DOI] [Google Scholar]
- 12.Pernil R, Schleiff E. 2019. Metalloproteins in the biology of heterocysts. Life 9:32. doi: 10.3390/life9020032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Norman L, Cabanesa DJE, Blanco-Ameijeiras S, Moisset SAM, Hassler CS. 2014. Iron biogeochemistry in aquatic systems: from source to bioavailability. Chimia (Aarau) 68:764–771. doi: 10.2533/chimia.2014.764. [DOI] [PubMed] [Google Scholar]
- 14.Khan A, Singh P, Srivastava A. 2018. Synthesis, nature and utility of universal iron chelator – siderophore: a review. Microbiol Res 212–213:103–111. doi: 10.1016/j.micres.2017.10.012. [DOI] [PubMed] [Google Scholar]
- 15.Saha M, Sarkar S, Sarkar B, Sharma BK, Bhattacharjee S, Tribedi P. 2016. Microbial siderophores and their potential applications: a review. Environ Sci Pollut Res Int 23:3984–3999. doi: 10.1007/s11356-015-4294-0. [DOI] [PubMed] [Google Scholar]
- 16.Noinaj N, Guillier M, Barnard TJ, Buchanan SK. 2010. TonB-dependent transporters: regulation, structure, and function. Annu Rev Microbiol 64:43–60. doi: 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vergalli J, Bodrenko I.v, Masi M, Moynié L, Acosta-Gutiérrez S, Naismith JH, Davin-Regli A, Ceccarelli M, van den Berg B, Winterhalter M, Pagès JM. 2020. Porins and small-molecule translocation across the outer membrane of Gram-negative bacteria. Nat Rev Microbiol 18:164–176. doi: 10.1038/s41579-019-0294-2. [DOI] [PubMed] [Google Scholar]
- 19.Schauer K, Rodionov DA, de Reuse H. 2008. New substrates for TonB-dependent transport: do we only see the “tip of the iceberg”? Trends Biochem Sci 33:330–338. doi: 10.1016/j.tibs.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 20.Hancock REW, Braun V. 1976. Nature of the energy requirement for the irreversible adsorption of bacteriophages T1 and Φ80 to Escherichia coli. J Bacteriol 125:409–415. doi: 10.1128/jb.125.2.409-415.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holroyd C, Bradbeer C. 1984. Cobalamin transport in Escherichia coli, p 21–23. In Leive L, Schlessinger D (ed), Microbiology-1984. American Society for Microbiology, Washington, DC. [Google Scholar]
- 22.Celia H, Botos I, Ni X, Fox T, de Val N, Lloubes R, Jiang J, Buchanan SK. 2019. Cryo-EM structure of the bacterial Ton motor subcomplex ExbB–ExbD provides information on structure and stoichiometry. Commun Biol 2:358. doi: 10.1038/s42003-019-0604-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skare JT, Ahmer BMM, Seachord CL, Darveau RP, Postle K. 1993. Energy transduction between membranes. J Biol Chem 268:16302–16308. doi: 10.1016/S0021-9258(19)85421-2. [DOI] [PubMed] [Google Scholar]
- 24.Cadieux N, Kadner RJ. 1999. Site-directed disulfide bonding reveals an interaction site between energy-coupling protein TonB and BtuB, the outer membrane cobalamin transporter. Proc Natl Acad Sci USA 96:10673–10678. doi: 10.1073/pnas.96.19.10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ollis AA, Manning M, Held KG, Postle K. 2009. Cytoplasmic membrane protonmotive force energizes periplasmic interactions between ExbD and TonB. Mol Microbiol 73:466–481. doi: 10.1111/j.1365-2958.2009.06785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ollis AA, Postle K. 2012. Identification of functionally important TonB-ExbD periplasmic domain interactions in vivo. J Bacteriol 194:3078–3087. doi: 10.1128/JB.00018-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferguson AD, Deisenhofer J. 2002. TonB-dependent receptors - structural perspectives. Biochim Biophys Acta 1565:318–332. doi: 10.1016/S0005-2736(02)00578-3. [DOI] [PubMed] [Google Scholar]
- 28.Poole K, Zhao Q, Neshat S, Heinrichs DE, Dean CR. 1996. The Pseudomonas aeruginosa tonB gene encodes a novel TonB protein. Microbiology 142:1449–1458. doi: 10.1099/13500872-142-6-1449. [DOI] [PubMed] [Google Scholar]
- 29.Zhao Q, Poole K. 2000. A second tonB gene in Pseudomonas aeruginosa is linked to the exbB and exbD genes. FEMS Microbiol Lett 184:127–132. doi: 10.1111/j.1574-6968.2000.tb09002.x. [DOI] [PubMed] [Google Scholar]
- 30.Huang B, Ru K, Yuan Z, Whitchurch CB, Mattick JS. 2004. TonB3 is required for normal twitching motility and extracellular assembly of type IV pili. J Bacteriol 186:4387–4389. doi: 10.1128/JB.186.13.4387-4389.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shirley M, Lamont IL. 2009. Role of TonB1 in pyoverdine-mediated signaling in Pseudomonas aeruginosa. J Bacteriol 191:5634–5640. doi: 10.1128/JB.00742-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takase H, Nitanai H, Hoshino K, Otani T. 2000. Requirement of the Pseudomonas aeruginosa tonB gene for high-affinity iron acquisition and infection. Infect Immun 68:4498–4504. doi: 10.1128/IAI.68.8.4498-4504.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ainsaar K, Tamman H, Kasvandik S, Tenson T, Hõrak R. 2019. The TonBm-PocAB system is required for maintenance of membrane integrity and polar position of flagella in Pseudomonas putida. J Bacteriol 201:e00303-19. doi: 10.1128/JB.00303-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bitter W, Tommassen J, Weisbeek PJ. 1993. Identification and characterization of the exbB, exbD and tonB genes of Pseudomonas putida WCS358: their involvement in ferric‐pseudobactin transport. Mol Microbiol 7:117–130. doi: 10.1111/j.1365-2958.1993.tb01103.x. [DOI] [PubMed] [Google Scholar]
- 35.Godoy P, Ramos-González MI, Ramos JL. 2001. Involvement of the TonB system in tolerance to solvents and drugs in Pseudomonas putida DOT-T1E. J Bacteriol 183:5285–5292. doi: 10.1128/JB.183.18.5285-5292.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuehl CJ, Crosa JH. 2010. The TonB energy transduction systems in Vibrio species. Future Microbiol 5:1403–1412. doi: 10.2217/fmb.10.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mirus O, Strauss S, Nicolaisen K, von Haeseler A, Schleiff E. 2009. TonB-dependent transporters and their occurrence in cyanobacteria. BMC Biol 7:68. doi: 10.1186/1741-7007-7-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stevanovic M, Hahn A, Nicolaisen K, Mirus O, Schleiff E. 2012. The components of the putative iron transport system in the cyanobacterium Anabaena sp. PCC 7120. Environ Microbiol 14:1655–1670. doi: 10.1111/j.1462-2920.2011.02619.x. [DOI] [PubMed] [Google Scholar]
- 39.Schätzle H, Arévalo S, Flores E, Schleiff E. 2021. A TonB-like protein, SjdR, is required for the structural definition of intercellular septa in the heterocyst-forming cyanobacterium. mBio 12:e00483-21. doi: 10.1128/mBio.00483-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moslavac S, Reisinger V, Berg M, Mirus O, Vosyka O, Plöscher M, Flores E, Eichacker LA, Schleiff E. 2007. The proteome of the heterocyst cell wall in Anabaena sp. PCC 7120. Biol Chem 388:823–829. doi: 10.1515/BC.2007.079. [DOI] [PubMed] [Google Scholar]
- 41.Wilk L, Strauss M, Rudolf M, Nicolaisen K, Flores E, Kühlbrandt W, Schleiff E. 2011. Outer membrane continuity and septosome formation between vegetative cells in the filaments of Anabaena sp. PCC 7120. Cell Microbiol 13:1744–1754. doi: 10.1111/j.1462-5822.2011.01655.x. [DOI] [PubMed] [Google Scholar]
- 42.Stevanovic M, Lehmann C, Schleiff E. 2013. The response of the TonB-dependent transport network in Anabaena sp. PCC 7120 to cell density and metal availability. Biometals 26:549–560. doi: 10.1007/s10534-013-9644-0. [DOI] [PubMed] [Google Scholar]
- 43.Steiger S, Schäfer L, Sandmann G. 1999. High-light-dependent upregulation of carotenoids and their antioxidative properties in the cyanobacterium Synechocystis PCC 6803. J Photochem Photobiol B 52:14–18. doi: 10.1016/S1011-1344(99)00094-9. [DOI] [Google Scholar]
- 44.Kłodawska K, Bujas A, Turos M, Malec P. 2016. Low temperature induced accumulation of keto-carotenoids canthaxanthin and echinenone in cyanobacterium Anabaena 7120. N Biotechnol 33:S196. doi: 10.1016/j.nbt.2016.06.1397. [DOI] [Google Scholar]
- 45.Várkonyi Z, Masamoto K, Debreczeny M, Zsiros O, Ughy B, Gombos Z, Domonkos I, Farkas T, Wada H, Szalontai B. 2002. Low-temperature-induced accumulation of xanthophylls and its structural consequences in the photosynthetic membranes of the cyanobacterium Cylindrospermopsis raciborskii: an FTIR spectroscopic study. Proc Natl Acad Sci USA 99:2410–2415. doi: 10.1073/pnas.042698799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muramatsu M, Hihara Y. 2012. Acclimation to high-light conditions in cyanobacteria: from gene expression to physiological responses. J Plant Res 125:11–39. doi: 10.1007/s10265-011-0454-6. [DOI] [PubMed] [Google Scholar]
- 47.Guikema JA, Sherman LA. 1983. Organization and function of chlorophyll in membranes of cyanobacteria during iron starvation. Plant Physiol 73:250–256. doi: 10.1104/pp.73.2.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rudolf M, Kranzler C, Lis H, Margulis K, Stevanovic M, Keren N, Schleiff E. 2015. Multiple modes of iron uptake by the filamentous, siderophore-producing cyanobacterium, Anabaena sp. PCC 7120. Mol Microbiol 97:577–588. doi: 10.1111/mmi.13049. [DOI] [PubMed] [Google Scholar]
- 49.Grant CS, Louda JW. 2010. Microalgal pigment ratios in relation to light intensity: implications for chemotaxonomy. Aquat Biol 11:127–138. doi: 10.3354/ab00298. [DOI] [Google Scholar]
- 50.Lakatos M, Bilger W, Büdel B. 2001. Carotenoid composition of terrestrial cyanobacteria: response to natural light conditions in open rock habitats in Venezuela. Eur J Phycol 36:367–375. doi: 10.1080/09670260110001735518. [DOI] [Google Scholar]
- 51.Schagerl M, Müller B. 2006. Acclimation of chlorophyll a and carotenoid levels to different irradiances in four freshwater cyanobacteria. J Plant Physiol 163:709–716. doi: 10.1016/j.jplph.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 52.Chakilam SR. 2012. Metal effects on carotenoid content of Cyanobacteria. Int J Bot 8:192–197. doi: 10.3923/ijb.2012.192.197. [DOI] [Google Scholar]
- 53.Tiwari A, Singh P, Asthana RK. 2016. Role of calcium in the mitigation of heat stress in the cyanobacterium Anabaena PCC 7120. J Plant Physiol 199:67–75. doi: 10.1016/j.jplph.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 54.Singh S, Mishra AK. 2016. Unraveling of cross talk between Ca2+ and ROS regulating enzymes in Anabaena 7120 and ntcA mutant. J Basic Microbiol 56:762–778. doi: 10.1002/jobm.201500326. [DOI] [PubMed] [Google Scholar]
- 55.Ehling-Schulz M, Bilger W, Scherer S. 1997. UV-B-induced synthesis of photoprotective pigments and extracellular polysaccharides in the terrestrial cyanobacterium Nostoc commune. J Bacteriol 179:1940–1945. doi: 10.1128/jb.179.6.1940-1945.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gruszecki WI, Strzałka K. 2005. Carotenoids as modulators of lipid membrane physical properties. Biochim Biophys Acta 1740:108–115. doi: 10.1016/j.bbadis.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 57.Mohamed HE, van de Meene AML, Roberson RW, Vermaas WFJ. 2005. Myxoxanthophyll is required for normal cell wall structure and thylakoid organization in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol 187:6883–6892. doi: 10.1128/JB.187.20.6883-6892.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakao R, Senpuku H, Watanabe H. 2006. Porphyromonas gingivalis galE is involved in lipopolysaccharide O-antigen synthesis and biofilm formation. Infect Immun 74:6145–6153. doi: 10.1128/IAI.00261-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dirienzo JM, Inouye M. 1983. Effect of reduced membrane lipid fluidity on the biosynthesis of lipopolysaccharide of Escherichia coli. Eur J Biochem 135:351–357. doi: 10.1111/j.1432-1033.1983.tb07661.x. [DOI] [PubMed] [Google Scholar]
- 60.Jürgens UJ, Weckesser J. 1985. Carotenoid-containing outer membrane of Synechocystis sp. strain PCC6714. J Bacteriol 164:384–389. doi: 10.1128/jb.164.1.384-389.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Resch CM, Gibson J. 1983. Isolation of the carotenoid-containing cell wall of three unicellular cyanobacteria. J Bacteriol 155:345–350. doi: 10.1128/jb.155.1.345-350.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Masamoto K, Riethman HC, Sherman LA. 1987. Isolation and characterization of a carotenoid-associated thylakoid protein from the cyanobacterium Anacystis nidulans R2. Plant Physiol 84:633–639. doi: 10.1104/pp.84.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fernández L, Hancock REW. 2012. Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin Microbiol Rev 25:661–681. doi: 10.1128/CMR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hahn A, Stevanovic M, Mirus O, Schleiff E. 2012. The TolC-like protein HgdD of the cyanobacterium Anabaena sp. PCC 7120 is involved in secondary metabolite export and antibiotic resistance. J Biol Chem 287:41126–41138. doi: 10.1074/jbc.M112.396010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moslavac S, Bredemeier R, Mirus O, Granvogl B, Eichacker LA, Schleiff E. 2005. Proteomic analysis of the outer membrane of Anabaena sp. strain PCC 7120. J Proteome Res 4:1330–1338. doi: 10.1021/pr050044c. [DOI] [PubMed] [Google Scholar]
- 66.Raghavan PS, Rajaram H, Apte SK. 2011. Nitrogen status dependent oxidative stress tolerance conferred by overexpression of MnSOD and FeSOD proteins in Anabaena sp. strain PCC7120. Plant Mol Biol 77:407–417. doi: 10.1007/s11103-011-9821-x. [DOI] [PubMed] [Google Scholar]
- 67.Rudolf M, Stevanovic M, Kranzler C, Pernil R, Keren N, Schleiff E. 2016. Multiplicity and specificity of siderophore uptake in the cyanobacterium Anabaena sp. PCC 7120. Plant Mol Biol 92:57–69. doi: 10.1007/s11103-016-0495-2. [DOI] [PubMed] [Google Scholar]
- 68.Goldman SJ, Lammers PJ, Berman MS, Sanders-Loehr J. 1983. Siderophore-mediated iron uptake in different strains of Anabaena sp. J Bacteriol 156:1144–1150. doi: 10.1128/jb.156.3.1144-1150.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oh ET, Yun HS, Heo T-R, Koh S-C, Oh K-H, So J-S. 2002. Involvement of lipopolysaccharide of Bradyrhizobium japonicum in metal binding. J Microbiol Biotechnol 12:296–300. [Google Scholar]
- 70.Langley S, Beveridge TJ. 1999. Effect of O-side-chain-lipopolysaccharide chemistry on metal binding. Appl Environ Microbiol 65:489–498. doi: 10.1128/AEM.65.2.489-498.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Strain SM, Fesik SW, Armitage IM. 1983. Structure and metal-binding properties of lipopolysaccharides from heptoseless mutants of Escherichia coli studied by 13C and 31P nuclear magnetic resonance. J Biol Chem 258:13466–13477. doi: 10.1016/S0021-9258(17)43937-8. [DOI] [PubMed] [Google Scholar]
- 72.Ferris FG, Beveridge TJ. 1986. Site specificity of metallic ion binding in Escherichia coli K-12 lipopolysaccharide. Can J Microbiol 32:52–55. doi: 10.1139/m86-010. [DOI] [PubMed] [Google Scholar]
- 73.de Philippis R, Colica G, Micheletti E. 2011. Exopolysaccharide-producing cyanobacteria in heavy metal removal from water: molecular basis and practical applicability of the biosorption process. Appl Microbiol Biotechnol 92:697–708. doi: 10.1007/s00253-011-3601-z. [DOI] [PubMed] [Google Scholar]
- 74.Arunmanee W, Pathania M, Solovyova AS, Le Brun AP, Ridley H, Baslé A, van den Berg B, Lakey JH. 2016. Gram-negative trimeric porins have specific LPS binding sites that are essential for porin biogenesis. Proc Natl Acad Sci USA 113:E5034–E5043. doi: 10.1073/pnas.1602382113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Buehler LK, Kusumoto S, Zhang H, Rosenbusch JP. 1991. Plasticity of Escherichia coli porin channels: dependence of their conductance on strain and lipid environment. J Biol Chem 266:24446–24450. doi: 10.1016/S0021-9258(18)54249-6. [DOI] [PubMed] [Google Scholar]
- 76.Schindler H, Rosenbusch JP. 1981. Matrix protein in planar membranes: clusters of channels in a native environment and their functional reassembly. Proc Natl Acad Sci USA 78:2302–2306. doi: 10.1073/pnas.78.4.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chang V, Chen L-Y, Wang A, Yuan X. 2010. The effect of lipopolysaccharide core structure defects on transformation efficiency in isogenic Escherichia coli BW25113 rfaG, rfaP, and rfaC mutants. J Exp Microbiol Immunol (JEMI) 14:101–107. [Google Scholar]
- 78.Schnaitman CA, Klena JD. 1993. Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiol Rev 57:655–682. doi: 10.1128/mr.57.3.655-682.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hancock REW. 1984. Alterations in outer membrane permeability. Annu Rev Microbiol 38:237–264. doi: 10.1146/annurev.mi.38.100184.001321. [DOI] [PubMed] [Google Scholar]
- 80.Braun V, Herrmann C. 1993. Evolutionary relationship of uptake systems for biopolymers in Escherichia coli: cross‐complementation between the TonB-ExbB‐ExbD and the TolA‐TolQ‐TolR proteins. Mol Microbiol 8:261–268. doi: 10.1111/j.1365-2958.1993.tb01570.x. [DOI] [PubMed] [Google Scholar]
- 81.Witty M, Sanz C, Shah A, Grossmann JG, Mizuguchi K, Perham RN, Luisi B. 2002. Structure of the periplasmic domain of Pseudomonas aeruginosa TolA: evidence for an evolutionary relationship with the TonB transporter protein. EMBO J 21:4207–4218. doi: 10.1093/emboj/cdf417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Davies JK, Reeves P. 1975. Genetics of resistance to colicins in Escherichia coli K-12: cross-resistance among colicins of group A. J Bacteriol 123:102–117. doi: 10.1128/jb.123.1.102-117.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fognini-Lefebvre N, Lazzaroni JC, Portalier R. 1987. tolA, tolB and excC, three cistrons involved in the control of pleiotropic release of periplasmic proteins by Escherichia coli K12. Mol Gen Genet 209:391–395. doi: 10.1007/BF00329670. [DOI] [PubMed] [Google Scholar]
- 84.Bernadac A, Gavioli M, Lazzaroni JC, Raina S, Lloubès R. 1998. Escherichia coli tol-pal mutants form outer membrane vesicles. J Bacteriol 180:4872–4878. doi: 10.1128/JB.180.18.4872-4878.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Derouiche R, Gavioli M, Bénédetti H, Prilipov A, Lazdunski C, Lloubès R. 1996. TolA central domain interacts with Escherichia coli porins. EMBO J 15:6408–6415. doi: 10.1002/j.1460-2075.1996.tb01032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rigal A, Bouveret E, Lloubes R, Lazdunski C, Benedetti H. 1997. The TolB protein interacts with the porins of Escherichia coli. J Bacteriol 179:7274–7279. doi: 10.1128/jb.179.23.7274-7279.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gerding MA, Ogata Y, Pecora ND, Niki H, de Boer PAJ. 2007. The trans-envelope Tol–Pal complex is part of the cell division machinery and required for proper outer-membrane invagination during cell constriction in E. coli. Mol Microbiol 63:1008–1025. doi: 10.1111/j.1365-2958.2006.05571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yakhnina AA, Bernhardt TG. 2020. The Tol-Pal system is required for peptidoglycan-cleaving enzymes to complete bacterial cell division. Proc Natl Acad Sci USA 117:6777–6783. doi: 10.1073/pnas.1919267117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gaspar JA, Thomas JA, Marolda CL, Valvano MA. 2000. Surface expression of O-specific lipopolysaccharide in Escherichia coli requires the function of the TolA protein. Mol Microbiol 38:262–275. doi: 10.1046/j.1365-2958.2000.02094.x. [DOI] [PubMed] [Google Scholar]
- 90.Rivera M, Hancock REW, Sawyer JG, Haug A, McGroarty EJ. 1988. Enhanced binding of polycationic antibiotics to lipopolysaccharide from an aminoglycoside-supersusceptible, tolA mutant strain of Pseudomonas aeruginosa. Antimicrob Agents Chemother 32:649–655. doi: 10.1128/AAC.32.5.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol 111:1–61. [Google Scholar]
- 92.Shcolnick S, Shaked Y, Keren N. 2007. A role for mrgA, a DPS family protein, in the internal transport of Fe in the cyanobacterium Synechocystis sp. PCC6803. Biochim Biophys Acta 1767:814–819. doi: 10.1016/j.bbabio.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 93.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 94.Cai Y, Wolk CP. 1990. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J Bacteriol 172:3138–3145. doi: 10.1128/jb.172.6.3138-3145.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Elhai J, Wolk PC. 1988. A versatile class of positive-selection vectors based on the nonviability of palindrome-containing plasmids that allows cloning into long polylinkers. Gene 68:119–138. doi: 10.1016/0378-1119(88)90605-1. [DOI] [PubMed] [Google Scholar]
- 96.Valladares A, Rodríguez V, Camargo S, Martínez-Noël GMA, Herrero A, Luque I. 2011. Specific role of the cyanobacterial pipX factor in the heterocysts of Anabaena sp. strain PCC 7120. J Bacteriol 193:1172–1182. doi: 10.1128/JB.01202-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Olmedo-Verd E, Muro-Pastor AM, Flores E, Herrero A. 2006. Localized induction of the ntcA regulatory gene in developing heterocysts of Anabaena sp. strain PCC 7120. J Bacteriol 188:6694–6699. doi: 10.1128/JB.00509-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wolk CP, Vonshak A, Kehoe P, Elhai J. 1984. Construction of shuttle vectors capable of conjugative transfer from Escherichia coli to nitrogen-fixing filamentous cyanobacteria. Proc Natl Acad Sci USA 81:1561–1565. doi: 10.1073/pnas.81.5.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Elhai J, Wolk CP. 1988. Conjugal transfer of DNA to cyanobacteria. Methods Enzymol 167:747–754. doi: 10.1016/0076-6879(88)67086-8. [DOI] [PubMed] [Google Scholar]
- 100.Kranzler C, Lis H, Shaked Y, Keren N. 2011. The role of reduction in iron uptake processes in a unicellular, planktonic cyanobacterium. Environ Microbiol 13:2990–2999. doi: 10.1111/j.1462-2920.2011.02572.x. [DOI] [PubMed] [Google Scholar]
- 101.Sharon S, Salomon E, Kranzler C, Lis H, Lehmann R, Georg J, Zer H, Hess WR, Keren N. 2014. The hierarchy of transition metal homeostasis: iron controls manganese accumulation in a unicellular cyanobacterium. Biochim Biophys Acta 1837:1990–1997. doi: 10.1016/j.bbabio.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 102.Sinetova MA, Červený J, Zavřel T, Nedbal L. 2012. On the dynamics and constraints of batch culture growth of the cyanobacterium Cyanothece sp. ATCC 51142. J Biotechnol 162:148–155. doi: 10.1016/j.jbiotec.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 103.Zavrel T, Sinetova M, Cerveny J. 2015. Measurement of chlorophyll a and carotenoids concentration in cyanobacteria. Bio Protoc 5:e1467. doi: 10.21769/BioProtoc.1467. [DOI] [Google Scholar]
- 104.Salomon E, Bar-Eyal L, Sharon S, Keren N. 2013. Balancing photosynthetic electron flow is critical for cyanobacterial acclimation to nitrogen limitation. Biochim Biophys Acta 1827:340–347. doi: 10.1016/j.bbabio.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 105.Horvath H, Kovacs AW, Riddick C, Presing M. 2013. Extraction methods for phycocyanin determination in freshwater filamentous cyanobacteria and their application in a shallow lake. Eur J Phycol 48:278–286. doi: 10.1080/09670262.2013.821525. [DOI] [Google Scholar]
- 106.Sampath-Wiley P, Neefus CD. 2007. An improved method for estimating R-phycoerythrin and R-phycocyanin contents from crude aqueous extracts of Porphyra (Bangiales, Rhodophyta). J Appl Phycol 19:123–129. doi: 10.1007/s10811-006-9118-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lehner J, Berendt S, Dörsam B, Pérez R, Forchhammer K, Maldener I. 2013. Prokaryotic multicellularity: a nanopore array for bacterial cell communication. FASEB J 27:2293–2300. doi: 10.1096/fj.12-225854. [DOI] [PubMed] [Google Scholar]
- 108.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 109.Apicella MA, Griffiss JM, Schneider H. 1994. Isolation and characterization of lipopolysaccharides, lipooligosaccharides, and lipid A. Methods Enzymol 235:242–252. doi: 10.1016/0076-6879(94)35145-7. [DOI] [PubMed] [Google Scholar]
- 110.Kavran JM, Leahy DJ. 2014. Silver staining of SDS-polyacrylamide gel. Methods Enzymol 541:169–176. doi: 10.1016/B978-0-12-420119-4.00014-8. [DOI] [PubMed] [Google Scholar]
- 111.Stewart WD, Fitzgerald GP, Burris RH. 1967. In situ studies on N2 fixation using the acetylene reduction technique. Proc Natl Acad Sci USA 58:2071–2078. doi: 10.1073/pnas.58.5.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.He Y, Häder D. 2002. Involvement of reactive oxygen species in the UV-B damage to the cyanobacterium Anabaena sp. J Photochem Photobiol B 66:73–80. doi: 10.1016/S1011-1344(01)00278-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sedimentation of I-tonB2 (white symbols) and wild-type cultures (black symbols). The OD750 was measured in one cuvette after specific time points without further resuspension of the suspension. Average values of four biological replicates per strain with two technical replicates each are given; error bars show the standard deviation. Due to enhanced aggregation, I-tonB2 filaments sediment faster in the cuvette than wild-type samples, which is indicated by a faster decrease of the OD750. Download FIG S1, PDF file, 1.4 MB (1.4MB, pdf) .
Copyright © 2021 Schätzle et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
LPS extractions from wild-type Anabaena and I-tonB2 cultures. (1) Cultures grown in tubes in the presence of 1% CO2. (2) Cultures grown without bubbling in shaking flasks; a higher degree of aggregation was observed without CO2 supplementation. The LPS isolated from the cultures was separated by SDS-PAGE and silver stained. The bottom panel shows the loading control (large subunit of Rubisco). Download FIG S2, PDF file, 1.6 MB (1.6MB, pdf) .
Copyright © 2021 Schätzle et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Estimation of intracellular ROS level. Cultures of wild type and I-tonB2 were grown for 1 week to a density of ∼OD750 of 1, and intracellular ROS levels were determined using the DCHF-AC assay (112) after 20-min pretreatment in the dark, ambient light, or UV-B radiation with or without addition of ascorbic acid. Higher conversion to fluorescent dichlorofluorescein indicates higher ROS levels. I-tonB2 did not behave significantly different from the wild type (Student’s t test). Download FIG S3, PDF file, 0.2 MB (204.4KB, pdf) .
Copyright © 2021 Schätzle et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Oligodeoxynucleotides used in the study. Download Table S1, DOCX file, 0.01 MB (13KB, docx) .
Copyright © 2021 Schätzle et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmids used in this study. Download Table S2, PDF file, 0.09 MB (93.8KB, pdf) .
Copyright © 2021 Schätzle et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Anabaena strains used in this study. Download Table S3, PDF file, 0.09 MB (88.8KB, pdf) .
Copyright © 2021 Schätzle et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
55Fe-Schizokinen uptake rates and normalized chlorophyll values. Download Table S4, DOCX file, 0.01 MB (12.7KB, docx) .
Copyright © 2021 Schätzle et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.