ABSTRACT
Nitrogen fixation, a distinct process incorporating the inactive atmospheric nitrogen into the active biological processes, has been a major topic in biological and geochemical studies. Currently, insights into diversity and distribution of nitrogen-fixing microbes are dependent upon homology-based analyses of nitrogenase genes, especially the nifH gene, which are broadly conserved in nitrogen-fixing microbes. Here, we report the pitfall of using nifH as a marker of microbial nitrogen fixation. We exhaustively analyzed genomes in RefSeq (231,908 genomes) and KEGG (6,509 genomes) and cooccurrence and gene order patterns of nitrogenase genes (including nifH) therein. Up to 20% of nifH-harboring genomes lacked nifD and nifK, which encode essential subunits of nitrogenase, within 10 coding sequences upstream or downstream of nifH or on the same genome. According to a phenotypic database of prokaryotes, no species and strains harboring only nifH possess nitrogen-fixing activities, which shows that these nifH genes are “pseudo”-nifH genes. Pseudo-nifH sequences mainly belong to anaerobic microbes, including members of the class Clostridia and methanogens. We also detected many pseudo-nifH reads from metagenomic sequences of anaerobic environments such as animal guts, wastewater, paddy soils, and sediments. In some samples, pseudo-nifH overwhelmed the number of “true” nifH reads by 50% or 10 times. Because of the high sequence similarity between pseudo- and true-nifH, pronounced amounts of nifH-like reads were not confidently classified. Overall, our results encourage reconsideration of the conventional use of nifH for detecting nitrogen-fixing microbes, while suggesting that nifD or nifK would be a more reliable marker.
IMPORTANCE Nitrogen-fixing microbes affect biogeochemical cycling, agricultural productivity, and microbial ecosystems, and their distributions have been investigated intensively using genomic and metagenomic sequencing. Currently, insights into nitrogen fixers in the environment have been acquired by homology searches against nitrogenase genes, particularly the nifH gene, in public databases. Here, we report that public databases include a significant amount of incorrectly annotated nifH sequences (pseudo-nifH). We exhaustively investigated the genomic structures of nifH-harboring genomes and found hundreds of pseudo-nifH sequences in RefSeq and KEGG. Over half of these pseudo-nifH sequences belonged to members of the class Clostridia, which is supposed to be a prominent nitrogen-fixing clade. We also found that the abundance of nitrogen fixers in metagenomes could be overestimated by 1.5 to >10 times due to pseudo-nifH recorded in public databases. Our results encourage reconsideration of the prevalent use of nifH as a marker of nitrogen-fixing microbes.
KEYWORDS: bioinformatics, computational biology, diazotrophs, genomics, metagenomics, nitrogen fixation
INTRODUCTION
Microbial nitrogen fixation is a prominent process in biogeochemical cycling, and the ecology and evolution of nitrogen-fixing microbes have received extraordinary attention from researchers in various academic fields. While certain clades of bacteria, including cyanobacteria, Clostridium, azotobacter, and legume symbionts, are known for their diazotrophic activities (1), recent genomic and metagenomic surveys have unveiled unexpected diversity among the distributions of diazotrophic communities on Earth (2, 3). Insights into the drivers of nitrogen fixation in the environment are of interest in microbial physiology, ecology, and agriculture, and they are useful in modeling and predicting the dynamics of nitrogen cycling (4, 5). Importantly, nitrogen fixation in gut symbionts has been linked to nitrogen acquisition by the host, which has led to much attention in animal biology studies (6).
A key approach to successful (meta)genomic studies is the use of conserved “core” genes that are essential for nitrogen fixation. Nitrogen fixation is exclusively driven by nitrogenases, and diazotrophic microbes commonly harbor a distinct set of genes, including typical (e.g., nifH, nifD, and nifK) and atypical (e.g., vnfD and anfD) genes, that encode nitrogenase subunits (7). These nitrogenase genes have been regarded as the hallmarks of diazotrophs in genomic and metagenomic analyses.
Particularly popular among these markers is nifH. nifH is a gene encoding an Fe protein named nitrogenase reductase (NifH), which constitutes a subunit of nitrogenase (8). It should be noted that NifH does not directly interact with N2 molecules; rather, it reduces other subunits constituting nitrogenase, namely, NifD/NifK subunits, that catalyze the cleavage of the N–N triple bond. The prevalent use of nifH is presumably attributed to the development of the first degenerative primers for PCR amplification of nifH (9). The use of these primers for fingerprinting (e.g., PCR denaturing gradient gel electrophoresis), quantitative PCR, and amplicon sequencing analyses has substantially expanded scientific knowledge about the diversity of diazotrophic prokaryotes (10–12).
While the straightforward relationship between function and gene presence/absence is useful, it may be not be the case for nifH. Previous studies have suggested that some nifH genes are not involved in nitrogen fixation (13). For example, a group of nifH homologs, named cluster IV (or group IV), belong to nondiazotrophic methanogens, whereas another group of nifH homologs, called cluster V (or group V), include protochlorophyllide reductase or chlorophyllide reductase genes (14). In addition, only nifH homologs have been detected in the genomes of some methanogenic archaea, while nifD and nifK are not (15). These data challenge the long-established conception that nifH is a primary hallmark of diazotrophic potential. In addition, it is speculated that use of nifH as a biomarker would lead to an overestimation of the abundance and diversity of diazotrophic microbes, as well as biased estimation of diazotrophic community structures. Nevertheless, quantitative analysis for such “pseudo-nifH” has been scarcely done, and therefore little is known about how prevalent pseudo-nifH sequences are in public genomic databases and how this affects metagenomic insights into diazotrophic microbiomes.
To quantify distribution of these “pseudo-nifH” genes among prokaryotic genomes and metagenomes, the boundary between “true-nifH” (i.e., contributing to nitrogen fixation) and pseudo-nifH needs to be better clarified. A genome-oriented analysis might provide a way to determine the distribution. For example, a nifH sequence without other genes constituting nitrogenase (e.g., nifD, nifK) in its neighborhood might be a pseudo-nifH. Moreover, if no other nitrogenase gene exists on the genome, that nifH is likely a pseudo-nifH (note that NifH does not directly cleave the N–N triple bond; therefore, NifH alone cannot modulate nitrogen fixation). These kinds of predictions that are based on neighboring genes and coexisting genes on the genomes have been versatile approaches in gene functional annotations (16–18) that complement the conventional homology search.
In this work, we questioned the suitability of nifH as a hallmark of diazotrophs. We aimed to elucidate the distribution of true- and pseudo-nifH among prokaryotic genomes and environmental metagenomes. First, we applied gene coexistence/neighborhood analyses to nifH-harboring genomes stored in highly reputed public databases (i.e., RefSeq and Kyoto Encyclopedia of Genes and Genomes [KEGG]). After confirming the accuracy of our method by checking the consistency with previous isolation-based reports, we further examined the distribution of true- and pseudo-nifH in environmental metagenomes. Finally, we discussed the possible outcomes from prevalent pseudo-nifH stored in public databases and metagenomes.
RESULTS AND DISCUSSION
Cryptic distribution of nifH in publicly available prokaryotic genomes.
To search for the candidates of pseudo-nifH, we first analyzed the distribution of nitrogenase genes in two fundamental, well-annotated, and high-quality genome databases, namely, National Center for Biotechnology Information (NCBI) RefSeq (19) and KEGG (20). Here, we were able to observe a cryptic distribution of nitrogen fixation genes.
Among the 231,908 RefSeq genomes analyzed, 6,529 genomes (excluding ones with the completeness below 95%) harbored one or more coding sequences (CDSs) annotated either as nifH, nifD, nifK, vnfD, vnfK, anfD, or anfK. Here, we accounted for atypical nitrogenase genes, vnf and anf (21), because nifH is homologous to vnfH and anfH and they may be confused in RefSeq (of note, no CDS was annotated as vnfH or anfH). In fact, we observed 66 genomes with nifH neighboring with vnfD or vnfK and 12 genomes with nifH neighboring with anfD or anfK (examples are shown in Table S1 in the supplemental material). While many of the genomes had the same copy numbers of nifH, nifD (including vnfD and anfD), and nifK (including vnfK and anfK), 1,457 genomes (22.3% of the 6,529 genomes) harbored an unequal number of these genes (Fig. 1a). The copy number of nifH was higher than those of nifD and nifK in 972 genomes (66.7% of the 1,457 genomes), and 373 genomes (25.6% of the 1,457 genomes) had only nifH. In contrast, genomes lacking nifH but possessing nifD or nifK were quite rare (96 genomes, 6.59% of the 1,457 genomes). These imbalanced results clearly conflict with the well-established conception that nifH, nifD, and nifK together constitute a gene cluster (nif operon) serving for nitrogen fixation (14). Therefore, the link between nifH and nitrogen fixation might not exist.
FIG 1.
Numbers of RefSeq (a) and KEGG (b) genomes harboring an equal/unequal number of nitrogenase genes. (Left) The line plot shows (im)balances between the copy numbers of nifH, nifD (including vnfD and anfD), and nifK (including vnfK and anfK). The first row indicates genomes harboring an equal copy number of nifH, nifD, and nifK. The four lower rows represent genomes with an unequal copy number of the three genes. Note that genomes with excessive nifH are remarkably abundant, as indicated by the second row (pink bars and line plot).
Examples of syntenies consisting of nifH and vnf or anf genes in RefSeq. Download Table S1, DOCX file, 0.1 MB (88KB, docx) .
Copyright © 2021 Mise et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Using the hidden Markov model (HMM)-based method targeting nifD/vnfD/anfD and nifK/vnfK/anfK (22), we reannotated the CDSs of the 6,529 genomes (see above) to rule out the possibility that nifD and nifK had been overlooked by NCBI’s in-house annotation protocol (PGAP) (19). Note that the annotations in RefSeq bear some inconsistency (variation) even within closely related homologs (for example if the concept of Gene Ontology [23, 24] is recalled), which called for this kind of reannotation. The HMM is suitable for minimizing false-negative results as it is typically more sensitive than BLAST-like algorithms or software (including PGAP) (25). More CDSs were annotated as nifD or nifK by HMM, including those annotated otherwise in RefSeq: of the 373 genomes harboring only nifH according to RefSeq annotations, 136 (36.4% of the 373 genomes) turned out to possess at least one of the nifD (including vnfD and anfD) or nifK (including vnfK and anfK) gene. Nevertheless, nifH remained more prevalent than the other genes in question. The lack of nifD/nifK could be partially attributed to the incompleteness or assembly errors of the genomes; however, their effects would be mostly negligible considering the rigorous quality control procedure of these databases and our in-house filtering of low-completeness (i.e., <95%) genomes.
The KEGG database, where orthologous groups are manually defined based upon a rigorous literature survey, also presented excessive prevalence of nifH (Fig. 1b) in addition to the RefSeq database. Among the 6,509 prokaryotic genomes analyzed, 677 contained one or more of nitrogenase orthologs (excluding ones with the completeness below 95%), namely, nifD/anfD (K02586), nifH (K02588), nifK/anfK (K02591), vnfD (K22896), and vnfK (K22897). nifH (K02588) was distributed in 669 genomes, 72 (10.8%) of which were not concomitant with any of the other nitrogenase orthologs. On the other hand, genomes harboring nifD/vnfD/anfD or nifK/vnfK/anfK but lacking nifH (K02588) were rare (four genomes, 0.6%).
Importantly, we observed three types of nifH CDS on RefSeq/KEGG genomes, which are hereafter called T1-, T2-, and T3-nifH (Fig. 2a to d). T1-nifH is accompanied by at least one of the orthologs encoding nitrogenase subunits (namely, nifD, nifK, vnfD, vnfK, anfD, and anfK) in their neighborhood (not more than 10 CDSs away from nifH). T1-nifH likely constitutes a nitrogen fixation operon that plays a role in nitrogen fixation. T2-nifH is not accompanied by the above-mentioned nitrogenase subunits in their neighborhood, but one or more exist elsewhere on the genome (including plasmids). This type of nifH is somewhat elusive: it appears to be different from the typical structure of the nitrogen fixation operon (14, 26), but it might work in cooperation with other subunits that are encoded distantly (27). T3-nifH is a “stand-alone” type of nifH, meaning no other nitrogenase genes exist in the genome or plasmids. It should not function as nitrogenase reductase (NifH) because of lack of the relevant nitrogenase (NifDK), if no orthologs were overlooked (either by genomic incompleteness or annotation failure). NifH is a member of the ATPase superfamily (28): NifH binds to and hydrolyzes ATP along with transferring electrons to nitrogenase (29). Therefore, T3-NifH may function as some kinds of ATPase by itself. Note that some genomes harbor both T1-nifH and T2-nifH, whereas T3-nifH and the other two are mutually exclusive by definition. The number of RefSeq and KEGG genomes having each type of nifH are summarized in Fig. 2e and g, respectively.
FIG 2.
Illustration of three types of nifH and their distributions in RefSeq and KEGG. (a) nifH accompanied by nifD (including vnfD and anfD) or nifK (including vnfK and anfK) in its neighborhood is called T1-nifH. (b) nifH accompanied by nifD or nifK, not in the neighborhood but somewhere distant on the same genome, is called T2-nifH. (c) T1- and T2-nifH might coexist on one genome. (d) nifH on a genome lacking nifD and nifK is called T3-nifH. (e) Number of RefSeq genomes harboring T1-, T2-, and T3-nifH. The pie chart shows the taxonomic composition of genomes with T3-nifH. (f) Number of RefSeq genomes belonging to the class Clostridia that harbor T1-, T2-, and T3-nifH. (g) Number of KEGG genomes harboring T1-, T2-, and T3-nifH. The pie chart shows the taxonomic composition of genomes with T3-nifH. (h) Number of KEGG genomes belonging to the class Clostridia that harbor T1-, T2-, and T3-nifH.
Genome-based distinction between T1/T2- and T3-nifH is consistent with experimentally validated diazotrophic capability of each species.
Next, we intended to investigate whether our three-class classification of nifH is in line with collective insights into species-level diazotrophic activities reported in numerous previous reports. For this purpose, we referred to FAPROTAX (30), which is a manually curated database that bridges prokaryotic taxonomy names with their functions (including nitrogen fixation). Items in FAPROTAX have been manually propagated from acknowledged and reliable literature sources, such as Bergey’s Manual of Systematic Bacteriology (Bergey’s Manual) and the International Journal of Systematic and Evolutionary Microbiology. A key feature of FAPROTAX is that it is not dependent on genomic sequences. The insights into diazotrophy have not necessarily been coupled with whole-genome sequencing, and therefore, strict correspondence between diazotrophic activity and whole-genome sequences is not available. This warrants the prediction of diazotrophic activity via taxonomic names. Note that FAPROTAX is conceptually much different from PICRUSt, which estimates functional gene profiles from the available genomes of extant prokaryotes, using 16S rRNA gene sequences as the key (31). The phenotype-oriented (rather than genome-oriented) feature of FAPROTAX enabled us to speculate the diazotrophic activities of T1 nifH- and T3 nifH-harboring prokaryotes.
FAPROTAX included approximately 200 records of nitrogen-fixing prokaryotes. Of the 5,749 and 576 prokaryotic strains harboring T1-nifH in RefSeq, 1,600 (27.8%) matched 200 records in FAPROTAX (Table 1). Of the 448 strains harboring T2-nifH but no T1-nifH, 337 (75.2%) were assigned as nitrogen-fixing microbes. Such a high proportion of hits among T2-nifH should be attributed to the taxonomic composition of T2-nifH-harboring genomes. They consisted of long-known and well-characterized diazotrophs, especially Bradyrhizobium, Rhizobium (251 and 39 of 448 strains, respectively). In fact, a previous study provides direct evidence for the diazotrophic activity of T2-nifH-harboring Bradyrhizobium (27). On the other hand, none of the 236 strains harboring T3-nifH overlapped with 200 records of FAPROTAX (Table 1). Genomes in KEGG also showed overall similar trends, although one of the T3-nifH-harboring strains (Methanospirillum hungatei strain JF-1) was exceptionally estimated to be capable of nitrogen fixation (Table 1). This conflict can be explained by within-species diversity of M. hungatei: another strain, GP1, has been shown to fix nitrogen (32), and FAPROTAX has been built upon this knowledge (33). Of note, the genome of strain GP1 (GCF_019263745.1 in RefSeq) bears a T1-nifH accompanied by nifD. On the other hand, the diazotrophic activity of strain JF-1 has not been reported to the best of our knowledge.
TABLE 1.
Numbers of prokaryotic species and strains harboring T1-nifH, T2-nifH but no T1-nifH, and T3-nifH on the genomes from RefSeq and KEGG, with or without a previous report on diazotrophic activity
| Database | Strain or species chararacteristic | No. of prokaryotic species and strains |
|
|---|---|---|---|
| Diazotrophic activity reported | No diazotrophic activity reported/not yet investigated | ||
| RefSeq | Harboring T1-nifH on their genomes | 1,600 | 4,149 |
| Harboring T2-nifH but not T1-nifH on their genomes | 337 | 111 | |
| Harboring T3-nifH on their genomes | 0 | 236 | |
| KEGG | Harboring T1-nifH on their genomes | 132 | 444 |
| Harboring T2-nifH but not T1-nifH on their genomes | 14 | 6 | |
| Harboring T3-nifH on their genomes | 1 | 71 | |
It should be noted that FAPROTAX is not an exhaustive database covering all lineages of prokaryotes. That is, it should be commonplace that strains unlisted in FAPROTAX are capable of nitrogen fixation. In addition, as the aforementioned exception suggests, microdiversity in nitrogen-fixing capabilities, which is beyond the resolution of FAPROTAX, could lead to partially inaccurate estimation. Nevertheless, such microdiversity would not override the stark contrast between T1/T2 (T1/2)- and T3-nifH, which is observed in multiple distinct linages. Overall, the present result is unlikely to contradict our expectation that genome-based distinction between T1- and T3-nifH reflects the presence/absence of strain-level nitrogen fixation capability. In addition, T2-nifH genes are likely to be involved in nitrogen fixation.
Previously, several studies have described the existence of pseudo-nifH or T3-nifH, in line with our results. In particular, some methanogens such as Methanobrevibacter, Methanocaldococcus, and Methanosarcina have been reported to harbor T3-nifH (15) or uncharacterized nifH homologs (13, 14). Part of these genes have been later characterized as coenzyme F430 biosynthesis genes (34). Another example of confusing nifH homologs are protochlorophyllide reductase genes among Cyanobacteria that are serving for biosynthesis of chlorophyll (35). While these gene products are functionally similar to NifH, they have been annotated as such in RefSeq and KEGG, and therefore, they were not included in our analysis.
True- and pseudo-nifH genes are not discernible by short-read sequences or predicted molecular structures.
Among the 236 RefSeq genomes harboring T3-nifH (i.e., genomes without nifDK/vnfDK/anfDK), 136 (57.6%) belonged to Clostridia (Fig. 2e). Notably, Clostridia include long-known diazotrophic bacteria, such as Clostridium spp. In fact, 586 and 351 RefSeq genomes belonging to Clostridia possessed T1-nifH and T2-nifH, respectively (Fig. 2f). Other prokaryotic clades, such as the class Negativicutes and methanogens, also possessed all three types of nifH. The KEGG genomes presented a similar distribution of T3-nifH (Fig. 2g and h).
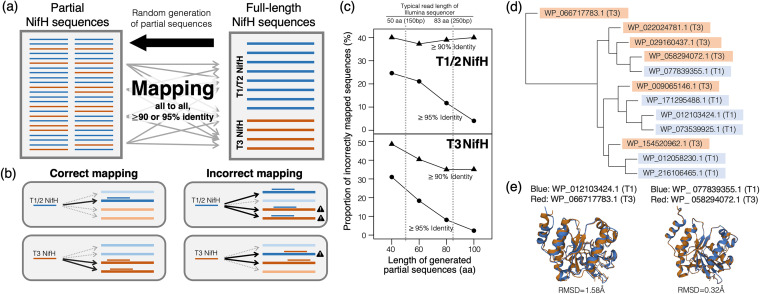
Given this, we questioned whether the biological sequences of T1/2- and T3-nifH can be differentiated (especially based on partial sequences generated by high-throughput sequencers). We randomly generated partial sequences of NifH (40, 60, 80, and 100 amino acids [aa], corresponding to 120 to 300 bases, which cover the range of typical read lengths from Illumina sequencers) (Fig. 3a) and mapped them onto the full-length NifH at a similarity threshold of 90% or 95% (Fig. 3b). A number of query sequences were mapped “incorrectly,” i.e., partial sequences of T1-NifH were mapped to T3-NifH or vice versa (35.0 to 48.6% and 2.4 to 31.0% when the sequence similarity threshold was set at 90% and 95%, respectively). As expected, the proportion of incorrect mapping became larger when the query sequences were shorter or the similarity threshold for mapping was lower (Fig. 3c). This suggests that T1-NifH and T3-NifH are often not distinguishable from their partial sequences that can be generated by high-throughput sequencers. That said, removing T3-NifH sequences from the reference database might not improve the specificity of nifH detection in short-read shotgun metagenomic analyses.
FIG 3.
Proximity between T1/2-NifH and T3-NifH. (a) Schematic diagram showing the generation of partial NifH sequences and their mapping on full-length NifH sequences. (b) Classification between correct mapping and incorrect mapping of partial NifH sequences. If partial T1/2-NifH was mapped only on full-length T1/2-NifH, the mapping was regarded as correct. If it was mapped on T3-NifH (in addition to T1/2-NifH), the mapping was regarded as incorrect. (c) Proportion of incorrect mapping with different query lengths and identity thresholds. Gray dotted lines indicate 50 aa and 83 aa, corresponding to 150 bp and 250 bp on DNA, respectively, which are typical read lengths of Illumina short-read sequencers. (d) Cluster dendrogram showing similarities between the protein structures of T1- and T3-NifH. A RefSeq accession number, as well as a type of NifH (T1 or T3), is indicated for each node. Nodes for T1- and T3-NifH are highlighted in blue and red, respectively. (e) Two examples of protein structural alignments between T1- and T3-NifH. RefSeq accession numbers of subjected NifH sequences, as well as the RMSD between two NifH, are indicated. The visualizations were generated on the PDB’s web server.
We also compared molecular structures of T1-NifH and T3-NifH by using AlphaFold2, a state-of-the-art molecular structure predictor (36). We quantified structural differences between T1-NifH and T3-NifH using root mean square deviations (RMSDs). Structural differences between T1-NifH and T3-NifH were minor compared with those within T1-NifH or within T3-NifH (Fig. 3d). RMSDs were overall less than 2 Å, and pairwise structural alignments presented highly conserved secondary structures (Fig. 3e). These features indicated the close functional and evolutionary relationship between true and pseudo-NifH sequences.
Furthermore, we investigated sequence domains and regions that are highly conserved or divergent between T1/2- and T3-NifH. We constructed a multiple sequence alignment (MSA) of T1/2- and T3-NifH sequences (356 aa) and picked 40 column-long subsequences from the MSA (see Fig. S1b in the supplemental material; each subsequence may include several gaps). Then we used sequence similarity networks to evaluate the distinguishability between the subreads of T1/2- and T3-NifH, where each sequence was classified as either a “distinct” or “confusing” sequence (Fig. S1a). The proportion of confusing sequences were quite different in different regions. More specifically, the subsequences from the N end and middle regions of the MSA (30 to 70 and 151 to 230 aa) were confusing, while the C-end subsequences were mostly distinct. This indicates that the N end and middle regions are highly conserved between T1/2- and T3-NifH. In agreement with this, the middle regions (151 to 230 aa) include the ligand-binding site of the NifH molecules (37). Furthermore, many pairs of nifH universal primers have been designed targeting the upper region of nifH genes (38), which are rather conserved between T1/2- and T3-nifH.
Similarity network analysis of partial sequences of T1/2- and T3-NifH. (a) The conceptual diagram of network analysis employed here. Each node represents one NifH sequence, and any pair of similar sequences is connected by an edge. T1/2-NifH (blue nodes) that are not directly connected to T3-NifH are “distinct T1/2-NifH” (as they are clearly distinct from T3-NifH), whereas T3-NifH (orange nodes) without direct connection to T1-NifH are “distinct T3-NifH.” When a pair of T1/2- and T3-NifH sequences are highly similar and directly connected, these sequences are easily confused with the other type of NifH and therefore regarded as “confusing T1/2-NifH” or “confusing T3-NifH.” (b) The proportions of distinct/confusing T1/2- and T3-NifH sequences. For each of the 40-base regions taken from the multiple sequence alignment (MSA) of NifH, partial T1/2- and T3-NifH sequences were fed into network analysis (described above for panel a) and classified into “distinct” or “confusing” sequences. Note that sparse regions of MSA (regions mostly occupied with gaps) were not used. The two rows of bar plots differ in the sequence similarity threshold used for network analysis: 95% for the middle row and 90% for the bottom row. Download FIG S1, TIF file, 2.1 MB (2.1MB, tif) .
Copyright © 2021 Mise et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Impact of prevalent pseudo-nifH on metagenomic analyses.
To elucidate the impact of pseudo-nifH sequences on shotgun metagenomic analyses, we reanalyzed the publicly available short-read metagenomic sequences. Because many of the genomes harboring T3-nifH are affiliated with anaerobes, we predicted that metagenomic analyses of anaerobic environments were subject to pseudo-nifH errors in the reference database. We obtained and processed shotgun metagenomic data sets from sludge, wastewater, human gut, termite gut, paddy soil, and sediment (Table S2) and then counted the number of nifH, nifD, and nifK sequences contained therein.
Metagenomic data sets used in this study. Download Table S2, DOCX file, 0.09 MB (100.4KB, docx) .
Copyright © 2021 Mise et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
As expected, we found that the number of nifH reads were excessive compared with those of nifD and nifK (Fig. 4a and b). Of note, the lengths of nifH sequences are typically shorter than those of nifD and nifK (Table S3), so the differences in read counts cannot be attributed to differences in gene length (Fig. 4a and b; Fig. S2a and b). On the other hand, the number of reads annotated as nifD and nifK were proportional (Fig. 4c; Fig. S2c). Only two outlier samples, where nifD reads were abundant but nifK reads were absent (Fig. 4c), contained reads similar to nifD of Phascolarctobacterium faecium or Selenomonas spp., which possessed only nifD and no nifK. Overall, considering the extensive prevalence of pseudo-nifH among prokaryotic genomes, our results indicated that nifD and nifK are relatively reliable markers of nitrogen-fixing microbes, whereas nifH is not.
FIG 4.
The outcome of focusing on nifH in shotgun metagenomic studies. (a) The relationship between read counts of nifD and nifH. The x and y axes are displayed in logarithmic scale [log (1 + x)]. The position of each point is slightly jittered to mitigate overlap between points [especially around (0,0)]. The gray dotted line indicates the theoretical relationship between read counts of two genes, where the number of nifD reads and nifH reads are proportional to the whole gene lengths of nifD and nifH (894 and 1,497 bp, respectively; the ratio is 0.597). Points statistically deviating from the theoretical proportion (gray dotted line) are colored red (see also Fig. S2 in the supplemental material). (b) Relationship between read counts of nifK and nifH. (c) Relationship between read counts of nifD and nifK. (d) The left panel shows proportions of true, ambiguous, and pseudo-nifH reads. The right panel shows the reciprocal of the proportion of true-nifH reads. This value represents the degree in which nifH abundance is possibly overestimated owing to pseudo-nifH reads. The horizontal axis is displayed in logarithmic scale.
Lengths of three nitrogenase genes (and their products), namely, nifH (NifH), nifD (NifD), and nifK (NifK), in KEGG. Average and quartile lengths (quartile 1 [Q1] to Q3) are indicated. Nucleotide sequence lengths were calculated by tripling the amino acid sequence length. Download Table S3, DOCX file, 0.1 MB (88.8KB, docx) .
Copyright © 2021 Mise et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The outcome of focusing on nifH in shotgun metagenomic studies. While data presented are the same as Fig. 4a to c, the confidence intervals are indicated in this plot. (a) The relationship between read counts of nifD and nifH. The gray dotted line indicates the theoretical relationship between read counts of two genes, where the number of nifD reads and nifH reads are proportional to the whole gene lengths of nifD and nifH (894 bp and 1,497 bp, respectively; the ratio is 0.597). Each sample is represented by a rectangle, which represents the 99% confidence interval of the number of nifD and nifH reads from the sample. The confidence intervals were calculated assuming that the read count follows Poisson distribution. The x and y axes are displayed in logarithmic scale [log (1 + x)]. Boxes deviating from the gray dotted line are indicated in red. (b) Relationship between read counts of nifK and nifH. (c) Relationship between the read counts of nifD and nifK. Download FIG S2, TIF file, 1.8 MB (1.9MB, tif) .
Copyright © 2021 Mise et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We further mapped the nifH reads within metagenomes onto KEGG database using rigorous (i.e., nonheuristic) Needleman-Wunsch algorithm. We classified nifH reads into true-nifH, pseudo-nifH, and ambiguous nifH reads (see Materials and Methods section for classification criteria). As suggested above, partial metagenomic sequences do not enable clear distinction between T1- and T3-nifH sequences (Fig. 3c). Therefore, sequences that were mapped onto both T1-NifH and T3-NifH were classified as ambiguous nifH reads. Here, we highlight three observations that preclude the use of nifH as the hallmark of diazotrophy (Fig. 4d). First, while true-nifH reads were dominant in some samples, others were critically affected by pseudo-nifH reads. In extreme cases, the abundance of nifH reads were exaggerated by more than 3 or 10 times owing to pseudo-nifH reads. This observation is consistent with a previous report on nifH composition in the human gut microbiome, where 444 of 524 nifH sequences were regarded irrelevant to nitrogen fixation (39). Second, this effect of pseudo-nifH was drastically different among samples, even within an environmental category. This would simply lead to inaccurate knowledge on the distribution of diazotrophs among various samples and geographic locations, which has recently been drawing attention (40, 41). Third, ambiguous nifH reads were dominant in many samples. This is in congruence with the results showing that partial sequences of true- and pseudo-nifH can be confused (Fig. 3c; Fig. S1b), meaning that simply eliminating pseudo-nifH reads would not be a satisfying solution. All these factors suggest that nifH would not be a very reliable marker in terms of specificity.
Conclusion and outlook.
In summary, we exhaustively investigated the distribution of nifH genes among high-quality public genomes in RefSeq and KEGG. Using neighborhood/cooccurrence approaches, we found dozens or hundreds of “pseudo” nifH (i.e., nifH homologs unlikely to contribute to nitrogen fixation) in these databases. We also demonstrated that “pseudo” nifH sequences could substantially affect the metagenomic analyses of diazotrophic communities.
We envision that the prevalent use of nifH as the hallmark of nitrogen-fixing prokaryotes should be reconsidered. A simple and easy solution would be to focus on nifD or nifK (and their counterparts in alternative nitrogenases) instead of nifH, as indicated in our massive reanalysis of public metagenomes (Fig. 4a to c). It is unlikely that “pseudo-nifD” or “pseudo-nifK” sequences are prevalent, considering the proportional distributions of nifD and nifK among prokaryotic genomes (Fig. 1) and metagenomes (Fig. 4c).
Another possible solution is to assemble short-read sequences into longer contigs to enable operon-scale analysis, where pseudo-nifH sequences unaccompanied by nifD or nifK can be discarded. In this case, the quantitative nature of short-read sequences may be compromised: reads from true- and pseudo-nifH sequences might not be distinguishable (Fig. 3; Fig. S1); therefore, mapping unassembled reads onto the contigs should be hampered by nonspecific mapping (42). In this regard, simply using nifD or nifK as the marker would be a more practical choice, as it would avoid many errors that nifH-based analyses may incur.
MATERIALS AND METHODS
We downloaded feature tables (i.e., annotation information of CDSs for each genome) of all genomes in the NCBI RefSeq on 21 July 2021 (19). The functional gene annotations provided in RefSeq are rigorously controlled by NCBI using PGAP, and all genomes are annotated under virtually identical (although not strictly identical) conditions. We selected genomes harboring at least one of the core genes of nitrogenase, namely, nifD (including vnfD and anfD), nifH, or nifK (including vnfK and anfK). We evaluated the completeness of each genome using CheckM v1.1.3 (43) with the options “lineage_wf --genes” and used only genomes with a completeness of 95% or higher. Here, CDSs labeled as pseudogenes by NCBI were discarded. To rule out the possibility that nifD and/or nifK has been overlooked by PGAP, we searched all CDSs in the nifH-harboring genomes for nifD and nifK using KofamScan 1.3.0 with the default parameters (22) and the database version as of April 2021. We further parsed CDS neighboring nifH; genes falling within 10 CDSs upstream or downstream of nifH were regarded as neighboring nifH. Only CDSs on the same strand as nifH were included when determining the range of the neighborhood. We classified nifH CDSs into the following three types: T1, nifH accompanied by nifD or nifK genes in their neighborhood; T2, nifH with nifD or nifK somewhere on the genome but not in the neighborhood; and T3, nifH without nifD or nifK on its genome (Fig. 2a to d). Note that some genomes have both T1 and T2, where one cluster of nifHDK (T1-nifH included) and another copy of stand-alone nifH (i.e., T2) coexist on one genome. We also downloaded the KEGG genomes and Kegg Orthology (KO) annotations from KEGG ftp (paywalled content; downloaded May 2021). On the basis of the KO annotations provided by KEGG, we analyzed the cooccurrences and syntenies of nifH, nifD, and nifK genes and classified nifH into three groups in the same way as we did for RefSeq.
Using FAPROTAX v1.2.4 (30), we assessed the diazotrophic activities of prokaryotic strains harboring T1-nifH (including those owning both T1- and T2-nifH), T2-nifH (excepting those owning both T1- and T2-nifH), and T3-nifH. Because the pipeline of FAPROTAX is designed for community-scale analysis, we generated an identity matrix as an operational taxonomic unit (OTU) table. For each type of nifH, we listed the taxonomic names (genus and species) of prokaryotes harboring the nifH, which were fed into FAPROTAX.
Next, we tested whether true- and pseudo-nifH sequences were distinguishable from each other. To mitigate the effect of phylogenetic bias, here we used only sequences from the members of class Clostridia. Furthermore, we clustered T1/2-NifH sequences at a similarity threshold of 95% using CD-HIT version 4.8.1 (44, 45). T3-NifH sequences were also similarly clustered. Hereafter in this analysis, we used only the representative sequences designated by CD-HIT (analogous to 95% operational taxonomic unit). We randomly picked subsequences of 40, 60, 80, and 100 amino acid length (10 subsequences for each length) from each of the T1/2- and T3-NifH sequences. We mapped these subsequences to the full-length T1/2-NifH and T3-NifH through an all-to-all search using the Needleman-Wunsch algorithm implemented in USEARCH v11.0.668 (with the options -search_global and -fulldp). We preformed the whole analysis with two similarity thresholds: 95% and 90%. Here, we employed global alignment, rather than local alignment (e.g., Smith-Waterman algorithm) to preclude short partial alignments. When a query from T1/2-NifH (i.e., a subsequence of T1/2-NifH) was mapped onto the T3-NifH, or vice versa, this mapping was regarded as an incorrect mapping; otherwise, the mapping was regarded as correct. We calculated the proportion of incorrect mapping for two different thresholds.
We also constructed similarity networks of partial NifH sequences. Here again, we used sequences from Clostridia. T1/2-NifH sequences were clustered at 95% similarity threshold to eliminate excessive redundancy in the sequences. T3-NifH sequences were clustered in the same way. The representative sequences of the clusters were subjected to MSA using the “--auto” mode of MAFFT v7.475 (46). From the constructed MSA (356 aa long), we picked 40 column-long subsequences from the MSA (for example at the position of 31 to 70 aa in the MSA, as shown in Fig. S1a in the supplemental material). The subsequences consisted of 40 aa or less, as some of them included gaps. These subsequences were subjected to all-to-all pairwise homology search using the Needleman-Wunsch algorithm implemented in USEARCH v11.0.668 (with the options -search_global and -fulldp). A sequence similarity network was constructed at a similarity threshold of 95 and 90%. If a pair of T1/2 NifH and T3-NifH were directly connected, then these two sequences were regarded “confusing.” Then we calculated the proportion of “confusing” NifH. We repeated this procedure for seven different subsequence positions in MSA: 31 to 70, 71 to 110, 111 to 150, 151 to 190, 191 to 230, 231 to 270, and 271 to 310 aa (Fig. S1). The terminus regions of MSA were occupied with many gaps and deemed unsuitable for this analysis.
Protein structures of T1-NifH and T3-NifH were predicted using AlphaFold2, a highly reliable predictor of protein structures (36). Six sequences were randomly picked from T1-NifH and from T3-NifH of genus Clostridium in RefSeq (listed in Fig. 4d). Each sequence was fed into the web browser interface of AlphaFold2 named ColabFold (https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/AlphaFold2.ipynb; accessed on 7 August 2021) (47), which was implemented using MMseqs2 (48). The RMSD between each pair of predicted structures was calculated using Mican 2019.11.27 (49). Ward’s method was used to hierarchically cluster the structures based on RMSDs. We visualized pairwise structural alignments using Pairwise Structure Alignment toolkit (https://www.rcsb.org/alignment; accessed on 7 August 2021) hosted by the Protein Data Bank (PDB) (50).
Additionally, we assessed how pseudo-nifH sequences in public databases affect metagenomic analyses of environmental samples. We focused on reusable metagenomic data sets in NCBI SRA/EMBL-EBI ERA/DDBJ DRA (51) under the following environmental categories: “activated sludge metagenome,” “human gut metagenome,” “termite metagenome,” “wastewater metagenome,” and “∗ sediment metagenome” (52–64, 72). We randomly picked SRA/ERA/DRA accession numbers that satisfy the following criteria: (i) sequenced on Illumina MiSeq, HiSeq, MiniSeq, NextSeq, or NovaSeq (i.e., not subject to frameshifting read errors by Roche 454); (ii) labeled as a “WGS” (standing for whole-genome shotgun) project; and (iii) described in a peer-reviewed literature (i.e., likely to be technically sound). Two metagenomic data sets from paddy soils, one of which was labeled as “soil metagenome” on SRA/ERA/DRA (65, 66) were also used. If a project consisted of many samples, we picked 5 to 10 samples from that project.
All of the selected data sets consisted of paired-end sequences; therefore, read1 and read2 were merged using USEARCH (with the options -fastq_maxdiffs 5 -fastq_minovlen 20 -fastq_allowmergestagger). The longest consecutive subsequence with the expected number of errors below 0.5 bases was retrieved from each of the merged sequences. To increase the accuracy of sequence annotation, we retained only sequences with a length of 200 bases or more. We picked the first 2,500,000 reads from each sample and discarded samples with less than 2,500,000 filtered reads. Samples used for subsequent analyses are summarized in Table S2.
The filtered sequences were subjected to a homology search against the KEGG database to find nifH, nifD, and nifK reads (including their counterparts in atypical nitrogenase). First, all filtered sequences were mapped to a small database consisting only of nifD/anfD (K02586), nifH (K02588), nifK/anfK (K02591), vnfD (K22896), and vnfK (K22897). Here, we used DIAMOND v2.0.9.147 (67) for homology search (using blastx command with mode “sensitive”; other parameters were set default). Sequences mapped on these nitrogenase genes were again subjected to a homology search against the whole prokaryotic database of KEGG, and the numbers of queries that were annotated as nifH (K02588), nifD/vnfD/anfD (K02586, K22896), and nifK/vnfK/anfK (K02591, K22897) were counted. Here again we used DIAMOND, with a modification that the E-value threshold was set at 1e–10.
To accurately distinguish true-nifH reads and pseudo-nifH reads, we again mapped the translated sequences of nifH (K02588) reads onto the KEGG gene sequences under nifH (K02588) using the nonheuristic Needleman-Wunsch algorithm implemented in USEARCH. For each query, we retrieved hits with similarities above 95% of the maximum similarity. We classified K02588 reads into three groups: (i) reads mapped onto T1- and/or T2-NifH but no T3-NifH, were regarded as true-nifH reads; (ii) reads mapped onto only T3-NifH were regarded as pseudo-nifH reads; and (iii) all other reads were regarded as ambiguous reads, which could either be a true-nifH or a pseudo-nifH.
Throughout this study, taxonomic names of prokaryotes were managed using the NCBI taxonomy system (68) and TaxonKit v0.8.0 (69), and fasta and fastq files were formatted using SeqKit v0.16.1 (70). R 4.0.5 (71) was used for data visualization.
Data availability.
Genomic and metagenomic data sets used for this study are available from NCBI RefSeq, NCBI SRA, and KEGG. Intermediate files will be made available by the authors upon request, except for the paywalled contents of KEGG, which are handled by Pathway Solutions (Tokyo, Japan).
ACKNOWLEDGMENTS
This work was financially supported by JSPS KAKENHI grants JP20H00409, JP20H05679, and JP20K15423, JST-Mirai Program grant JPMJMI20E5, and JPNP18016 commissioned by the New Energy and Industrial Technology Development Organization (NEDO).
We thank Yoshiaki Yasutake (National Institute of Advanced Industrial Science and Technology) and Wataru Iwasaki (The University of Tokyo) for the helpful discussion and Enago for the English language review. Computations were partially performed on the NIG supercomputer at ROIS National Institute of Genetics and the SHIROKANE supercomputer at Human Genome Center, The Institute of Medical Science, The University of Tokyo. We declare that we have no conflicts of interest.
Contributor Information
Kazumori Mise, Email: mise-33@aist.go.jp.
Susannah Green Tringe, U.S. Department of Energy Joint Genome Institute.
REFERENCES
- 1.Brill WJ. 1980. Biochemical genetics of nitrogen fixation. Microbiol Rev 44:449–467. doi: 10.1128/mr.44.3.449-467.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Masuda Y, Itoh H, Shiratori Y, Isobe K, Otsuka S, Senoo K. 2017. Predominant but previously-overlooked prokaryotic drivers of reductive nitrogen transformation in paddy soils, revealed by metatranscriptomics. Microbes Environ 32:180–183. doi: 10.1264/jsme2.ME16179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delmont TO, Quince C, Shaiber A, Esen ÖC, Lee ST, Rappé MS, McLellan SL, Lücker S, Eren AM. 2018. Nitrogen-fixing populations of Planctomycetes and Proteobacteria are abundant in surface ocean metagenomes. Nat Microbiol 3:804–813. doi: 10.1038/s41564-018-0176-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sohm JA, Webb EA, Capone DG. 2011. Emerging patterns of marine nitrogen fixation. Nat Rev Microbiol 9:499–508. doi: 10.1038/nrmicro2594. [DOI] [PubMed] [Google Scholar]
- 5.Masuda Y, Shiratori Y, Ohba H, Ishida T, Takano R, Satoh S, Shen W, Gao N, Itoh H, Senoo K. 2021. Enhancement of the nitrogen-fixing activity of paddy soils owing to iron application. Soil Sci Plant Nutr 67:243–247. doi: 10.1080/00380768.2021.1888629. [DOI] [Google Scholar]
- 6.Ceja-Navarro JA, Nguyen NH, Karaoz U, Gross SR, Herman DJ, Andersen GL, Bruns TD, Pett-Ridge J, Blackwell M, Brodie EL. 2014. Compartmentalized microbial composition, oxygen gradients and nitrogen fixation in the gut of Odontotaenius disjunctus. ISME J 8:6–18. doi: 10.1038/ismej.2013.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bellenger JP, Darnajoux R, Zhang X, Kraepiel AML. 2020. Biological nitrogen fixation by alternative nitrogenases in terrestrial ecosystems: a review. Biogeochemistry 149:53–73. doi: 10.1007/s10533-020-00666-7. [DOI] [Google Scholar]
- 8.Jones R, Woodley P, Birkmann-Zinoni A, Robson RL. 1993. The nifH gene encoding the Fe protein component of the molybdenum nitrogenase from Azotobacter chroococcum. Gene 123:145–146. doi: 10.1016/0378-1119(93)90555-h. [DOI] [PubMed] [Google Scholar]
- 9.Zehr JP, McReynolds LA. 1989. Use of degenerate oligonucleotides for amplification of the nifH gene from the marine cyanobacterium Trichodesmium thiebautii. Appl Environ Microbiol 55:2522–2526. doi: 10.1128/aem.55.10.2522-2526.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaby JC, Buckley DH. 2012. A comprehensive evaluation of PCR primers to amplify the nifH gene of nitrogenase. PLoS One 7:e42149. doi: 10.1371/journal.pone.0042149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuypers MMM, Marchant HK, Kartal B. 2018. The microbial nitrogen-cycling network. Nat Rev Microbiol 16:263–276. doi: 10.1038/nrmicro.2018.9. [DOI] [PubMed] [Google Scholar]
- 12.Silveira R, Mello TDRBD, Sartori MRS, Alves GSC, Fonseca FCDA, Vizzotto CS, Krüger RH, Bustamante MMDC. 2021. Seasonal and long-term effects of nutrient additions and liming on the nifH gene in cerrado soils under native vegetation. iScience 24:102349. doi: 10.1016/j.isci.2021.102349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaby JC, Buckley DH. 2014. A comprehensive aligned nifH gene database: a multipurpose tool for studies of nitrogen-fixing bacteria. Database 2014:bau001. doi: 10.1093/database/bau001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raymond J, Siefert JL, Staples CR, Blankenship RE. 2004. The natural history of nitrogen fixation. Mol Biol Evol 21:541–554. doi: 10.1093/molbev/msh047. [DOI] [PubMed] [Google Scholar]
- 15.Dos Santos PC, Fang Z, Mason SW, Setubal JC, Dixon R. 2012. Distribution of nitrogen fixation and nitrogenase-like sequences amongst microbial genomes. BMC Genomics 13:162. doi: 10.1186/1471-2164-13-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim P-J, Price ND. 2011. Genetic co-occurrence network across sequenced microbes. PLoS Comput Biol 7:e1002340. doi: 10.1371/journal.pcbi.1002340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Overbeek R, Fonstein M, D’Souza M, Pusch GD, Maltsev N. 1999. The use of gene clusters to infer functional coupling. Proc Natl Acad Sci USA 96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foflonker F, Blaby-Haas CE. 2021. Colocality to cofunctionality: eukaryotic gene neighborhoods as a resource for function discovery. Mol Biol Evol 38:650–662. doi: 10.1093/molbev/msaa221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li W, O’Neill KR, Haft DH, DiCuccio M, Chetvernin V, Badretdin A, Coulouris G, Chitsaz F, Derbyshire MK, Durkin AS, Gonzales NR, Gwadz M, Lanczycki CJ, Song JS, Thanki N, Wang J, Yamashita RA, Yang M, Zheng C, Marchler-Bauer A, Thibaud-Nissen F. 2021. RefSeq: Expanding the Prokaryotic Genome Annotation Pipeline reach with protein family model curation. Nucleic Acids Res 49:D1020–D1028. doi: 10.1093/nar/gkaa1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. 2021. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res 49:D545–D551. doi: 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thiel T. 1993. Characterization of genes for an alternative nitrogenase in the cyanobacterium Anabaena variabilis. J Bacteriol 175:6276–6286. doi: 10.1128/jb.175.19.6276-6286.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aramaki T, Blanc-Mathieu R, Endo H, Ohkubo K, Kanehisa M, Goto S, Ogata H. 2020. KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 36:2251–2252. doi: 10.1093/bioinformatics/btz859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. 2000. Gene Ontology: tool for the unification of biology. Nat Genet 25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gene Ontology Consortium. 2021. The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res 49:D325–D334. doi: 10.1093/nar/gkaa1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soding J. 2005. Protein homology detection by HMM-HMM comparison. Bioinformatics 21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 26.Masuda Y, Yamanaka H, Xu Z-X, Shiratori Y, Aono T, Amachi S, Senoo K, Itoh H. 2020. Diazotrophic Anaeromyxobacter isolates from soils. Appl Environ Microbiol 86:e00956-20. doi: 10.1128/AEM.00956-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matos GF, Rouws LFM, Simões‐Araújo JL, Baldani JI. 2021. Evolution and function of nitrogen fixation gene clusters in sugarcane associated Bradyrhizobium strains. Environ Microbiol 23:6148−6162. doi: 10.1111/1462-2920.15533. [DOI] [PubMed] [Google Scholar]
- 28.Koonin EV. 1993. A superfamily of ATPases with diverse functions containing either classical or deviant ATP-binding motif. J Mol Biol 229:1165–1174. doi: 10.1006/jmbi.1993.1115. [DOI] [PubMed] [Google Scholar]
- 29.Burgess BK, Lowe DJ. 1996. Mechanism of molybdenum nitrogenase. Chem Rev 96:2983–3012. doi: 10.1021/cr950055x. [DOI] [PubMed] [Google Scholar]
- 30.Louca S, Parfrey LW, Doebeli M. 2016. Decoupling function and taxonomy in the global ocean microbiome. Science 353:1272–1277. doi: 10.1126/science.aaf4507. [DOI] [PubMed] [Google Scholar]
- 31.Douglas GM, Maffei VJ, Zaneveld JR, Yurgel SN, Brown JR, Taylor CM, Huttenhower C, Langille MGI. 2020. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol 38:685–688. doi: 10.1038/s41587-020-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Belay N, Sparling R, Choi B-S, Roberts M, Roberts JE, Daniels L. 1988. Physiological and 15N-NMR analysis of molecular nitrogen fixation by Methanococcus thermolithotrophicus, Methanobacterium bryantii and Methanospirillum hungatei. Biochim Biophys Acta 971:233–245. doi: 10.1016/0167-4889(88)90138-3. [DOI] [PubMed] [Google Scholar]
- 33.Boone DR, Whitman WB, Koga Y. 2001. Family III. Methanospirillaceae fam. nov. p 264–268. In Boone DR, Castenholz RW, Garrity RM (ed), Bergey’s manual of systematic bacteriology, 2nd ed, vol 1. Springer, New York, NY. [Google Scholar]
- 34.Zheng K, Ngo PD, Owens VL, Yang X, Mansoorabadi SO. 2016. The biosynthetic pathway of coenzyme F430 in methanogenic and methanotrophic archaea. Science 354:339–342. doi: 10.1126/science.aag2947. [DOI] [PubMed] [Google Scholar]
- 35.Fujita Y, Takahashi Y, Chuganji M, Matsubara H. 1992. The nifH-like (frxC) gene is involved in the biosynthesis of chlorophyll in the filamentous cyanobacterium Plectonema boryanum. Plant Cell Physiol 33:81–92. [PubMed] [Google Scholar]
- 36.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D. 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strop P, Takahara PM, Chiu H-J, Angove HC, Burgess BK, Rees DC. 2001. Crystal structure of the all-ferrous [4Fe-4S]0 form of the nitrogenase iron protein from Azotobacter vinelandii. Biochemistry 40:651–656. doi: 10.1021/bi0016467. [DOI] [PubMed] [Google Scholar]
- 38.Angel R, Nepel M, Panhölzl C, Schmidt H, Herbold CW, Eichorst SA, Woebken D. 2018. Evaluation of primers targeting the diazotroph functional gene and development of NifMAP – a bioinformatics pipeline for analyzing nifH amplicon data. Front Microbiol 9:703. doi: 10.3389/fmicb.2018.00703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Igai K, Itakura M, Nishijima S, Tsurumaru H, Suda W, Tsutaya T, Tomitsuka E, Tadokoro K, Baba J, Odani S, Natsuhara K, Morita A, Yoneda M, Greenhill AR, Horwood PF, Inoue J, Ohkuma M, Hongoh Y, Yamamoto T, Siba PM, Hattori M, Minamisawa K, Umezaki M. 2016. Nitrogen fixation and nifH diversity in human gut microbiota. Sci Rep 6:31942. doi: 10.1038/srep31942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pierella Karlusich JJ, Pelletier E, Lombard F, Carsique M, Dvorak E, Colin S, Picheral M, Cornejo-Castillo FM, Acinas SG, Pepperkok R, Karsenti E, de Vargas C, Wincker P, Bowler C, Foster RA. 2021. Global distribution patterns of marine nitrogen-fixers by imaging and molecular methods. Nat Commun 12:4160. doi: 10.1038/s41467-021-24299-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu L, Zhang B, Wang E, Zhu B, Yao M, Li C, Li X. Soil total organic carbon/total nitrogen ratio as a key driver deterministically shapes diazotrophic community assemblages during the succession of biological soil crusts. Soil Ecol Lett, in press. doi: 10.1007/s42832-020-0075-x. [DOI] [Google Scholar]
- 42.Lee STM, Kahn SA, Delmont TO, Shaiber A, Esen Özcan C, Hubert NA, Morrison HG, Antonopoulos DA, Rubin DT, Eren AM. 2017. Tracking microbial colonization in fecal microbiota transplantation experiments via genome-resolved metagenomics. Microbiome 5:50. doi: 10.1186/s40168-017-0270-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 45.Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mirdita M, Ovchinnikov S, Steinegger M. 2021. ColabFold - making protein folding accessible to all. bioRxiv 10.1101/2021.08.15.456425. [DOI] [PMC free article] [PubMed]
- 48.Mirdita M, Steinegger M, Söding J. 2019. MMseqs2 desktop and local web server app for fast, interactive sequence searches. Bioinformatics 35:2856–2858. doi: 10.1093/bioinformatics/bty1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Minami S, Sawada K, Ota M, Chikenji G. 2018. MICAN-SQ: a sequential protein structure alignment program that is applicable to monomers and all types of oligomers. Bioinformatics 34:3324–3331. doi: 10.1093/bioinformatics/bty369. [DOI] [PubMed] [Google Scholar]
- 50.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. 2000. The Protein Data Bank. Nucleic Acids Res 28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arita M, Karsch-Mizrachi I, Cochrane G. 2021. The international nucleotide sequence database collaboration. Nucleic Acids Res 49:D121–D124. doi: 10.1093/nar/gkaa967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peterson D, Bonham KS, Rowland S, Pattanayak CW, RESONANCE Consortium, Klepac-Ceraj V. 2021. Comparative analysis of 16S rRNA gene and metagenome sequencing in pediatric gut microbiomes. Front Microbiol 12:670336. doi: 10.3389/fmicb.2021.670336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang M, Sun Y, Zeng Z, Wang Z. 2021. Metagenomics of wastewater phageome identifies an extensively cored antibiotic resistome in a swine feedlot water treatment environment. Ecotoxicol Environ Saf 222:112552. doi: 10.1016/j.ecoenv.2021.112552. [DOI] [PubMed] [Google Scholar]
- 54.Rossmassler K, Dietrich C, Thompson C, Mikaelyan A, Nonoh JO, Scheffrahn RH, Sillam-Dussès D, Brune A. 2015. Metagenomic analysis of the microbiota in the highly compartmented hindguts of six wood- or soil-feeding higher termites. Microbiome 3:56. doi: 10.1186/s40168-015-0118-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petrovich M, Chu B, Wright D, Griffin J, Elfeki M, Murphy BT, Poretsky R, Wells G. 2018. Antibiotic resistance genes show enhanced mobilization through suspended growth and biofilm-based wastewater treatment processes. FEMS Microbiol Ecol 94:fiy041. doi: 10.1093/femsec/fiy041. [DOI] [PubMed] [Google Scholar]
- 56.Zhao R, Summers ZM, Christman GD, Yoshimura KM, Biddle JF. 2020. Metagenomic views of microbial dynamics influenced by hydrocarbon seepage in sediments of the Gulf of Mexico. Sci Rep 10:5772. doi: 10.1038/s41598-020-62840-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Muller EEL, Pinel N, Laczny CC, Hoopmann MR, Narayanasamy S, Lebrun LA, Roume H, Lin J, May P, Hicks ND, Heintz-Buschart A, Wampach L, Liu CM, Price LB, Gillece JD, Guignard C, Schupp JM, Vlassis N, Baliga NS, Moritz RL, Keim PS, Wilmes P. 2014. Community-integrated omics links dominance of a microbial generalist to fine-tuned resource usage. Nat Commun 5:5603. doi: 10.1038/ncomms6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Karkman A, Berglund F, Flach C-F, Kristiansson E, Larsson DGJ. 2020. Predicting clinical resistance prevalence using sewage metagenomic data. Commun Biol 3:711. doi: 10.1038/s42003-020-01439-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ng C, Tan B, Jiang X-T, Gu X, Chen H, Schmitz BW, Haller L, Charles FR, Zhang T, Gin K. 2019. Metagenomic and resistome analysis of a full-scale municipal wastewater treatment plant in Singapore containing membrane bioreactors. Front Microbiol 10:172. doi: 10.3389/fmicb.2019.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Singleton CM, Petriglieri F, Kristensen JM, Kirkegaard RH, Michaelsen TY, Andersen MH, Kondrotaite Z, Karst SM, Dueholm MS, Nielsen PH, Albertsen M. 2021. Connecting structure to function with the recovery of over 1000 high-quality metagenome-assembled genomes from activated sludge using long-read sequencing. Nat Commun 12:2009. doi: 10.1038/s41467-021-22203-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pérez MV, Guerrero LD, Orellana E, Figuerola EL, Erijman L. 2019. Time series genome-centric analysis unveils bacterial response to operational disturbance in activated sludge. mSystems 4:e00169-21. doi: 10.1128/mSystems.00169-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hildebrand F, Gossmann TI, Frioux C, Özkurt E, Myers PN, Ferretti P, Kuhn M, Bahram M, Nielsen HB, Bork P. 2021. Dispersal strategies shape persistence and evolution of human gut bacteria. Cell Host Microbe 29:1167–1176.e9. doi: 10.1016/j.chom.2021.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zuo T, Wong SH, Lam K, Lui R, Cheung K, Tang W, Ching JYL, Chan PKS, Chan MCW, Wu JCY, Chan FKL, Yu J, Sung JJY, Ng SC. 2018. Bacteriophage transfer during faecal microbiota transplantation in Clostridium difficile infection is associated with treatment outcome. Gut 67:634–643. doi: 10.1136/gutjnl-2017-313952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fukuyama J, Rumker L, Sankaran K, Jeganathan P, Dethlefsen L, Relman DA, Holmes SP. 2017. Multidomain analyses of a longitudinal human microbiome intestinal cleanout perturbation experiment. PLoS Comput Biol 13:e1005706. doi: 10.1371/journal.pcbi.1005706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hartman WH, Ye R, Horwath WR, Tringe SG. 2017. A genomic perspective on stoichiometric regulation of soil carbon cycling. ISME J 11:2652–2665. doi: 10.1038/ismej.2017.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H-Y, Wang H, Wang H-T, Xin P-Y, Xu X-H, Ma Y, Liu W-P, Teng C-Y, Jiang C-L, Lou L-P, Arnold W, Cralle L, Zhu Y-G, Chu J-F, Gilbert JA, Zhang Z-J. 2018. The chemodiversity of paddy soil dissolved organic matter correlates with microbial community at continental scales. Microbiome 6:187. doi: 10.1186/s40168-018-0561-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Buchfink B, Reuter K, Drost H-G. 2021. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat Methods 18:366–368. doi: 10.1038/s41592-021-01101-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Federhen S. 2012. The NCBI Taxonomy database. Nucleic Acids Res 40:D136–D143. doi: 10.1093/nar/gkr1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shen W, Ren H. 2021. TaxonKit: a practical and efficient NCBI taxonomy toolkit. J Genet Genomics 48:844–850. doi: 10.1016/j.jgg.2021.03.006. [DOI] [PubMed] [Google Scholar]
- 70.Shen W, Le S, Li Y, Hu F. 2016. SeqKit: a cross-platform and ultrafast toolkit for FASTA/Q file manipulation. Plops One 11:e0163962. doi: 10.1371/journal.pone.0163962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.R Core Team. 2021. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 72.Yeoh YK, Chen Z, Wong MCS, Hui M, Yu J, Ng SC, Sung JJY, Chan FKL, Chan PKS. 2020. Southern Chinese populations harbour non-nucleatum Fusobacteria possessing homologues of the colorectal cancer-associated FadA virulence factor. Gut 69:1998–2007. doi: 10.1136/gutjnl-2019-319635. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Examples of syntenies consisting of nifH and vnf or anf genes in RefSeq. Download Table S1, DOCX file, 0.1 MB (88KB, docx) .
Copyright © 2021 Mise et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Similarity network analysis of partial sequences of T1/2- and T3-NifH. (a) The conceptual diagram of network analysis employed here. Each node represents one NifH sequence, and any pair of similar sequences is connected by an edge. T1/2-NifH (blue nodes) that are not directly connected to T3-NifH are “distinct T1/2-NifH” (as they are clearly distinct from T3-NifH), whereas T3-NifH (orange nodes) without direct connection to T1-NifH are “distinct T3-NifH.” When a pair of T1/2- and T3-NifH sequences are highly similar and directly connected, these sequences are easily confused with the other type of NifH and therefore regarded as “confusing T1/2-NifH” or “confusing T3-NifH.” (b) The proportions of distinct/confusing T1/2- and T3-NifH sequences. For each of the 40-base regions taken from the multiple sequence alignment (MSA) of NifH, partial T1/2- and T3-NifH sequences were fed into network analysis (described above for panel a) and classified into “distinct” or “confusing” sequences. Note that sparse regions of MSA (regions mostly occupied with gaps) were not used. The two rows of bar plots differ in the sequence similarity threshold used for network analysis: 95% for the middle row and 90% for the bottom row. Download FIG S1, TIF file, 2.1 MB (2.1MB, tif) .
Copyright © 2021 Mise et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Metagenomic data sets used in this study. Download Table S2, DOCX file, 0.09 MB (100.4KB, docx) .
Copyright © 2021 Mise et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Lengths of three nitrogenase genes (and their products), namely, nifH (NifH), nifD (NifD), and nifK (NifK), in KEGG. Average and quartile lengths (quartile 1 [Q1] to Q3) are indicated. Nucleotide sequence lengths were calculated by tripling the amino acid sequence length. Download Table S3, DOCX file, 0.1 MB (88.8KB, docx) .
Copyright © 2021 Mise et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The outcome of focusing on nifH in shotgun metagenomic studies. While data presented are the same as Fig. 4a to c, the confidence intervals are indicated in this plot. (a) The relationship between read counts of nifD and nifH. The gray dotted line indicates the theoretical relationship between read counts of two genes, where the number of nifD reads and nifH reads are proportional to the whole gene lengths of nifD and nifH (894 bp and 1,497 bp, respectively; the ratio is 0.597). Each sample is represented by a rectangle, which represents the 99% confidence interval of the number of nifD and nifH reads from the sample. The confidence intervals were calculated assuming that the read count follows Poisson distribution. The x and y axes are displayed in logarithmic scale [log (1 + x)]. Boxes deviating from the gray dotted line are indicated in red. (b) Relationship between read counts of nifK and nifH. (c) Relationship between the read counts of nifD and nifK. Download FIG S2, TIF file, 1.8 MB (1.9MB, tif) .
Copyright © 2021 Mise et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Genomic and metagenomic data sets used for this study are available from NCBI RefSeq, NCBI SRA, and KEGG. Intermediate files will be made available by the authors upon request, except for the paywalled contents of KEGG, which are handled by Pathway Solutions (Tokyo, Japan).