ABSTRACT
Resistance to the ceftazidime (CAZ)-avibactam (AVI) combination is increasingly being reported. Here, we report a CAZ-AVI-resistant Klebsiella pneumoniae strain belonging to the high-risk sequence type 307 (ST307) clone and producing Klebsiella pneumoniae carbapenemase 39 (KPC-39), a single-amino-acid variant of KPC-3 (A172T). Cloning experiments, steady-state kinetic parameters, and molecular dynamics simulations revealed a loss of carbapenemase activity and increased affinity for CAZ. KPC-39 was identified in a patient without prior exposure to CAZ-AVI, suggesting silent dissemination in European health care settings.
KEYWORDS: avibactam resistance, carbapenemase, KPC
INTRODUCTION
Carbapenemase-producing Enterobacterales (CPE) represent a serious threat to human health (1). Among newly commercialized drugs, avibactam (AVI) is a non-β-lactam inhibitor of β-lactamases that has been successfully used for the treatment of infections caused by Klebsiella pneumoniae carbapenemase (KPC)-producing Klebsiella pneumoniae isolates (2). However, KPC-producing K. pneumoniae isolates that are resistant to ceftazidime (CAZ)-AVI are increasingly being reported. This resistance may be due to either (i) increased expression of KPC after transposition events (3) or increases in the copy number of the KPC-carrying plasmid (4), especially in strains with impaired outer membrane permeability, or (ii) amino acid (AA) substitutions in the KPC protein (5–8). Here, we report the identification of KPC-39, a KPC-3 variant that confers CAZ-AVI resistance to a K. pneumoniae sequence type 307 (ST307) clinical isolate.
K. pneumoniae 181E2 was recovered from a rectal swab sample from a patient upon admission to a French hospital. Because this isolate displayed reduced susceptibility to ertapenem on a disk diffusion antibiogram, it was sent to the French National Reference Center for Carbapenem-Resistant Enterobacterales for further investigations. MICs for β-lactams, determined by Etest (bioMérieux, Marcy-l’Étoile, France), revealed that K. pneumoniae 181E2 was resistant to expanded-spectrum cephalosporins and showed decreased susceptibility to ertapenem but was fully susceptible to imipenem and meropenem, according to EUCAST guidelines (9) (Table 1). Interestingly, K. pneumoniae 181E2 was resistant to CAZ-AVI, with a MIC of 12 μg/ml determined by Etest and a MIC of 16 μg/ml determined using broth microdilution (Sensititre; Thermo Fisher Scientific, Les Ulis, France). In addition, this isolate was resistant to fluoroquinolones but remained susceptible to aminoglycosides, co-trimoxazole, fosfomycin, nitrofurantoin, tigecycline, and colistin, as determined using broth microdilution (Sensititre; Thermo Fisher Scientific) (Table 1). Biochemical tests failed to detect a carbapenem-hydrolyzing enzyme. However, a lateral flow immunoassay (LFIA) detected the production of a KPC-like enzyme, and a PCR assay confirmed the presence of a blaKPC-like allele (Table 1).
TABLE 1.
Antimicrobial susceptibility and carbapenemase confirmation test results for clinical isolate K. pneumoniae 181E2 (KPC-39), E. coli TOP10 transformants, and E. coli TOP10
| Antibiotic or confirmation test | Antibiotic MIC (mg/liter) or confirmation test result |
|||
|---|---|---|---|---|
| K. pneumoniae 181E2 (KPC-39) | E. coli TOP10 (pTOPO-KPC-3) | E. coli TOP10 (pTOPO-KPC-39) | E. coli TOP10 | |
| Antibiotics | ||||
| Amoxicillina | >256 | >256 | >256 | 6 |
| Amoxicillin-clavulanatea | 24 | 48 | 24 | 6 |
| Cefiximea | 6 | 12 | 4 | 0.38 |
| Cefotaximea | 4 | >32 | 2 | 0.064 |
| CAZa | >256 | >256 | >256 | 0.125 |
| CAZ-AVIa | 12 | 0.75 | 4 | 0.125 |
| Cefepimea | 4 | 6 | 2 | 0.0064 |
| Aztreonama | >256 | >256 | >256 | 0.047 |
| Ertapenema | 0.25 | 1 | 0.094 | 0.004 |
| Imipenema | 0.5 | 8 | 1 | 0.25 |
| Meropenema | 0.19 | 3 | 0.094 | 0.032 |
| Imipenem plus relebactamb | 0.12 | |||
| Meropenem plus vaborbactamb | <0.06 | |||
| Cefiderocolb | 0.5 | |||
| Fosfomycinb | <16 | |||
| Tigecyclineb | <0.5 | |||
| Co-trimoxazoleb | 0.125 | |||
| Colistinb | <0.5 | |||
| Amikacinb | <2 | |||
| Tobramycinb | <0.5 | |||
| Ciprofloxacinb | >16 | |||
| Nitrofurantoinb | <8 | |||
| Carbapenemase confirmation tests | ||||
| Biochemicalc | ||||
| CarbaNP test | − | + | − | NDd |
| β CARBA test | − | + | − | ND |
| MBT Star-Carba IVD kit | − | + | − | ND |
| LFIA (NG-Test CARBA-5)e | + | ND | ND | ND |
| Molecular (Xpert Carba-R)f | + | ND | ND | ND |
MIC results obtained using Etests (bioMérieux).
MIC results obtained by broth microdilution (Sensititre; Thermo Fisher Scientific).
CarbaNP test (30), β-CARBA test (Bio-Rad, Marne-la-Coquette, France), and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) MBT Star-Carba IVD kit (Bruker Daltonics, Bremen, Germany).
ND, not determined.
LFIA (NG-Test Carba 5) (NG Biotech, Guipry, France).
Xpert Carba-R PCR assay (Cepheid, Maurens-Scopont, France).
Illumina-based whole-genome sequencing of K. pneumoniae 181E2 was performed as described previously (10). The resistome, as determined using ResFinder v4.1 (11), revealed the presence of four β-lactamase genes, i.e., the chromosomally encoded penicillinase blaSHV-28 and three acquired genes, namely, blaTEM-1, ΔblaOXA-9 (disrupted by a stop codon), and a new blaKPC-3-derived allele named the blaKPC-39 gene. KPC-39 differs from KPC-3 by a single-AA substitution at position 172 (A172T). While OmpK35 is likely not functional due to a premature stop codon, OmpK36 differed from that of K. pneumoniae ATCC 43816 by 25 AA substitutions, 4 AA deletions, and 2 AA insertions (12), which did not have a significant impact on carbapenem susceptibility. Unlike most KPC-producing K. pneumoniae strains, which belong to clonal group 258 (CG258), K. pneumoniae 181E2 belonged to ST307. The blaKPC-39 gene was embedded in Tn4401a and was carried on a multireplicon IncFIIK-IncFIB self-transferable plasmid.
The blaKPC-39 gene was amplified using the primers KPC-RBS (5′-CTCCACCTTCAAACAAGGAAT-3′) and KPC-REV (5′-ATCTGCAGAATTCGCCCTTCGCCATCGTCAGTGCTCTAC-3′) and cloned into the pCR-Blunt II-Topo plasmid (Invitrogen, Villebon-sur-Yvette, France), and the resulting plasmid, pTOPO-KPC-39, was electroporated into Escherichia coli TOP10, as described previously for KPC-3 (13). Similar MICs for amoxicillin, cefixime, and cefepime were obtained for both transformants, but E. coli (pTOPO-KPC-39) was more susceptible to carbapenems, lacked carbapenem hydrolysis, as revealed by biochemical confirmation tests (Table 1), and was 5-fold more resistant to CAZ-AVI, compared to E. coli (pTOPO-KPC-3), suggesting that the A172T substitution is responsible for the resistance to CAZ-AVI and the loss of carbapenemase activity.
KPC-39 and KPC-31 were produced and purified to purity (as revealed by SDS–PAGE analysis) as described previously for KPC-3 (13). Steady-state kinetic parameters of KPC-39 were compared to those of KPC-3 and KPC-31, the most prevalent CAZ-AVI-resistant KPC-3 variant (D179Y). As expected, the catalytic efficiency for imipenem was reduced 13-fold for KPC-39 (Table 2). Globally, kcat values for carbapenems and all tested β-lactams were reduced for KPC-39, suggesting that the A172T substitution led to overall reduced hydrolysis. A kinetic study of KPC-31 confirmed the absence of hydrolysis of selected β-lactams except for CAZ, as had been proposed by Oueslati et al. based on MIC determinations (5). Regarding CAZ hydrolysis, KPC-3, KPC-39, and KPC-31 exhibited high Km values (>1,000 μM), making exact measurement of the catalytic efficiency impossible. AVI 50% inhibitory concentration (IC50) values were 320 nM and 400 nM for KPC-3 and KPC-39, respectively, suggesting that the A172T substitution had no impact on the inhibition properties of AVI, in contrast to KPC-31, for which the IC50 value was about 50-fold higher (20 μM) (5). IC50 values of clavulanate and tazobactam for KPC-39 (46 μM and 31 μM, respectively) were comparable to those of KPC-3 (20 μM and 50 μM, respectively) (6).
TABLE 2.
Steady-state kinetic parameters of KPC-39, KPC-3, and KPC-31 for selected β-lactam substratesa
| Substrate |
Km (μM) |
kcat (s−1) |
kcat/Km (mM−1/s−1) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| KPC-39 | KPC-3 | KPC-31 | KPC-39 | KPC-3 | KPC-31 | KPC-39 | KPC-3 | KPC-31 | |
| Benzylpenicillin | 57 | 40 | NHb | 4 | 16.7 | NDc | 70 | 417 | ND |
| CAZ | >1,000d | >1,000d | >1,000d | >3.7e | >14e | >2.3e | >1.4e | >8e | >2.1e |
| Aztreonam | >1,200 | >1,200 | NH | >20 | >134 | ND | >15 | >57 | ND |
| Imipenem | 137 | 184 | NH | 1.27 | 22.4 | ND | 9.2 | 122 | ND |
| Ertapenem | 6.5 | 22 | NH | 0.12 | 2.5 | ND | 17.7 | 115 | ND |
| Meropenem | 5.4 | 20 | NH | 0.04 | 2.5 | ND | 6.7 | 96 | ND |
Data are the means of three independent experiments. Standard deviations were within 15% of the mean value.
NH, no hydrolysis was observed with 0.7 μM purified enzyme and up to 1,000 μM substrate.
ND, not determined.
Km values were above 1,000 μM and thus could not be determined experimentally.
Because the exact Km values could not be determined, kcat values and catalytic efficiencies were extrapolated.
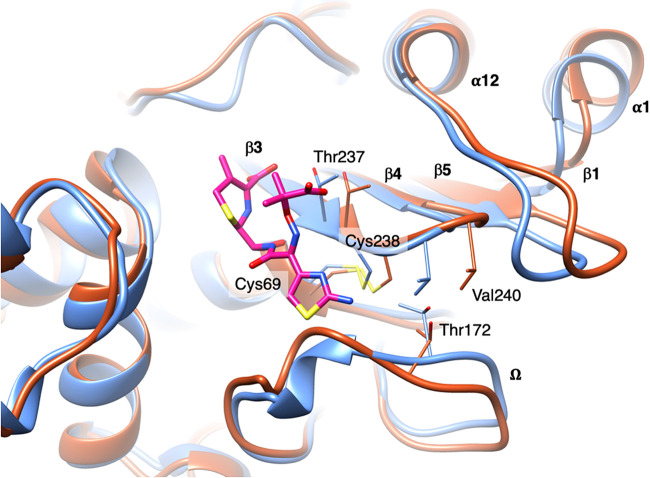
In silico modeling and molecular dynamics of KPC-39 enzyme were studied to understand the mechanism of resistance to CAZ-AVI. The three-dimensional structure of KPC-39 was generated using the swapaa command implemented in UCSF Chimera (14), starting from the structure of KPC-3 (PDB code 6QWD) (15). Molecular dynamics simulations were performed in triplicate with GROMACS v2020.3 (16) using the Amber99SB-ILDN force field (17). The protein was centered in a cubic periodic box, with at least 1.0 nm on each side. The simulation box was then filled with TIP3P water molecules, and the system was neutralized with Na+ and Cl− ions until the physiological ionic strength (150 mM) was reached. Each system was energy minimized to convergence using a steepest-descents algorithm. Molecular dynamics, performed for 100 ns in each case, as described previously (18), showed overall good stability of the three-dimensional structure of KPC-39. Significant conformational changes occurred during the simulation time, compared to KPC-3, for the Ω-loop in the region containing the mutation A172T. Additionally, a correlated movement of β-strands β3, β4, β5, and β1 (with C-α of Val240 shifted by 2.6 Å), which was propagated as far as the α-helix α1, was observed (Fig. 1).
FIG 1.
KPC-39 conformations at the beginning (blue) and at the end (red) of the molecular dynamics simulation. Covalently bound CAZ (PDB code 2ZQD) is represented as magenta-colored sticks.
K. pneumoniae 181E2 was isolated from a patient who had had no prior exposure to CAZ-AVI or to anticancer chemotherapies, which have also been suggested to contribute to the selection of CAZ-AVI-resistant KPC variants (19). It is likely that transmission from a CAZ-AVI-treated patient might have occurred. To date, KPC-39 producers have been reported from Italy (two K. pneumoniae ST512 strains from different patients) (20) and from Spain (a strain of unknown ST from one previously CAZ-AVI-treated patient) (21). These sporadic descriptions of KPC-39 may reflect silent dissemination in European hospitals or several concomitant selections. K. pneumoniae 181E2, described here, belongs to K. pneumoniae ST307, a high-risk clone associated with the spread of blaCTX-M-15 and carbapenemase genes (22) and responsible for hospital outbreaks of CPE in Italy, Germany, Portugal, and France (22, 23).
KPC-39 differs from KPC-3 by one AA substitution at position 172, located in the Ω-loop. Changes in this loop are frequently involved in CAZ-AVI resistance among KPC variants (KPC-31, KPC-33, KPC-35, KPC-40, KPC-48, KPC-51, KPC-52, KPC-53, and KPC-57) or other class A β-lactamases such as SHV or CTX-M (24, 25). Increased hydrolysis of CAZ but loss of hydrolysis of most β-lactams, as observed for KPC-39, has also been reported for CTX-M-93, an L169Q variant of CTX-M-27 (26). Position 169 is conserved in most extended-spectrum β-lactamases (ESBLs), being either a leucine or a methionine. Nine single-AA polymorphisms were identified in the Ω-loop of the 172 CTX-M sequences present in the Beta-Lactamase Database (BLDB) (http://www.bldb.eu) (27). None of the nine substitutions introduced into CTX-M-15 led to CAZ-AVI resistance, but two produced increased enzymatic activity against CAZ, i.e., 4-fold (P167S) and 16-fold (L169Q) increases in the MIC of CAZ (25). However, no substitution at position 172 was evidenced.
CAZ-AVI resistance can be due to better affinity for CAZ (KPC-14, KPC-28 [13], KPC-33 [28], and KPC-41 [6]) or to an impaired capacity of AVI to bind the enzyme (KPC-31 and KPC-33 [5, 13]). Here, the CAZ-AVI resistance mechanism seems more complex. Since the maximum velocity for CAZ could not be reached experimentally, exact Km values could not be determined. Molecular dynamics simulations showed important changes in the shape of the active site induced by the A172T mutation; to avoid the steric hindrance of Thr172 with Val240 and Cys238, the Ω-loop is pushed back and the β-sheet containing β3, β4, β5, and β1 is shifted laterally (while being restrained by the Cys69-Cys238 disulfide bond), with the effect of this movement being propagated as far as the α-helix α1 (Fig. 1). Overall, this larger active site would accommodate CAZ with higher affinity, thus leading to the CAZ-AVI-resistant phenotype determined experimentally.
Accurate identification of KPC variants conferring CAZ-AVI resistance and carbapenem susceptibility to Enterobacterales strains is needed, especially since recent studies showed that carbapenem-based regimens may be used to effectively treat infections (21, 29). Combined use of carbapenemase detection assays (biochemical assays, LFIAs, or molecular assays) is needed to detect these KPC variants, which are increasingly being detected in Europe, even in patients with no prior exposure to CAZ-AVI.
Data availability.
The draft genome of K. pneumoniae 181E2 was deposited in GenBank under accession number JAFFPJ000000000.
ACKNOWLEDGMENTS
We thank Béatrice Gourde (Saint Amand les Eaux), Claire Huart (Valenciennes), and Marie Huyghe (Saint Amand les Eaux) for providing useful information about the patient’s medical history.
This work was partially supported by the Assistance Publique-Hôpitaux de Paris, University Paris-Saclay, Institut National de la Santé et de la Recherche Médicale (INSERM), Laboratory of Excellence in Research on Medication and Innovative Therapeutics (LERMIT), through a grant from the French National Research Agency (grant ANR-10-LABX-33), and by the ANR-BMBF French-German bilateral project Natural-Arsenal (New Antibiotics Tackling mUlti-Resistance by acting on Alternative bacteriaL tARgets in Synergy with mEmbrane-disruptiNg AntimicrobiaL peptides) (grant ANR-19-AMRB-0004).
L.D. is a coinventor of the Carba NP test, the patent for which has been licensed to bioMérieux (La Balmes les Grottes, France).
REFERENCES
- 1.Bonomo RA, Burd EM, Conly J, Limbago BM, Poirel L, Segre JA, Westblade LF. 2018. Carbapenemase-producing organisms: a global scourge. Clin Infect Dis 66:1290–1297. 10.1093/cid/cix893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodríguez-Baño J, Gutiérrez-Gutiérrez B, Machuca I, Pascual A. 2018. Treatment of infections caused by extended-spectrum-beta-lactamase-, AmpC-, and carbapenemase-producing Enterobacteriaceae. Clin Microbiol Rev 31:e00079-17. 10.1128/CMR.00079-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nelson K, Hemarajata P, Sun D, Rubio-Aparicio D, Tsivkovski R, Yang S, Sebra R, Kasarskis A, Nguyen H, Hanson BM, Leopold S, Weinstock G, Lomovskaya O, Humphries RM. 2017. Resistance to ceftazidime-avibactam is due to transposition of KPC in a porin-deficient strain of Klebsiella pneumoniae with increased efflux activity. Antimicrob Agents Chemother 61:e00989-17. 10.1128/AAC.00989-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coppi M, Di Pilato V, Monaco F, Giani T, Conaldi PG, Rossolini GM. 2020. Ceftazidime-avibactam resistance associated with increased blaKPC-3 gene copy number mediated by pKpQIL plasmid derivatives in ST258 Klebsiella pneumoniae. Antimicrob Agents Chemother 64:e01816-19. 10.1128/AAC.01816-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oueslati S, Tlili L, Exilie C, Bernabeu S, Iorga B, Bonnin RA, Dortet L, Naas T. 2020. Different phenotypic expression of KPC β-lactamase variants and challenges in their detection. J Antimicrob Chemother 75:769–771. 10.1093/jac/dkz508. [DOI] [PubMed] [Google Scholar]
- 6.Mueller L, Masseron A, Prod’Hom G, Galperine T, Greub G, Poirel L, Nordmann P. 2019. Phenotypic, biochemical and genetic analysis of KPC-41, a KPC-3 variant conferring resistance to ceftazidime-avibactam and exhibiting reduced carbapenemase activity. Antimicrob Agents Chemother 63:e01111-19. 10.1128/AAC.01111-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antonelli A, Giani T, Di Pilato V, Riccobono E, Perriello G, Mencacci A, Rossolini GM. 2019. KPC-31 expressed in a ceftazidime/avibactam-resistant Klebsiella pneumoniae is associated with relevant detection issues. J Antimicrob Chemother 74:2464–2466. 10.1093/jac/dkz156. [DOI] [PubMed] [Google Scholar]
- 8.Di Pilato V, Aiezza N, Viaggi V, Antonelli A, Principe L, Giani T, Luzzaro F, Rossolini GM. 2020. KPC-53, a KPC-3 variant of clinical origin associated with reduced susceptibility to ceftazidime-avibactam. Antimicrob Agents Chemother 65:e01429-20. 10.1128/AAC.01429-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.European Committee on Antimicrobial Susceptibility Testing. 2020. Clinical breakpoints and dosing of antibiotics. https://www.eucast.org/clinical_breakpoints/.
- 10.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Pontén T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong JLC, Romano M, Kerry LE, Kwong H-S, Low W-W, Brett SJ, Clements A, Beis K, Frankel G. 2019. OmpK36-mediated carbapenem resistance attenuates ST258 Klebsiella pneumoniae in vivo. Nat Commun 10:3957. 10.1038/s41467-019-11756-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oueslati S, Iorga BI, Tlili L, Exilie C, Zavala A, Dortet L, Jousset AB, Bernabeu S, Bonnin RA, Naas T. 2019. Unravelling ceftazidime/avibactam resistance of KPC-28, a KPC-2 variant lacking carbapenemase activity. J Antimicrob Chemother 74:2239–2246. 10.1093/jac/dkz209. [DOI] [PubMed] [Google Scholar]
- 14.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera: a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 15.Tooke CL, Hinchliffe P, Lang PA, Mulholland AJ, Brem J, Schofield CJ, Spencer J. 2019. Molecular basis of class A β-lactamase inhibition by relebactam. Antimicrob Agents Chemother 63:e00564-19. 10.1128/AAC.00564-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pronk S, Páll S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, Shirts MR, Smith JC, Kasson PM, van der Spoel D, Hess B, Lindahl E. 2013. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29:845–854. 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE. 2010. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 78:1950–1958. 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dabos L, Zavala A, Bonnin RA, Beckstein O, Retailleau P, Iorga BI, Naas T. 2020. Substrate specificity of OXA-48 after β5-β6 loop replacement. ACS Infect Dis 6:1032–1043. 10.1021/acsinfecdis.9b00452. [DOI] [PubMed] [Google Scholar]
- 19.Hobson CA, Bonacorsi S, Hocquet D, Baruchel A, Fahd M, Storme T, Tang R, Doit C, Tenaillon O, Birgy A. 2020. Impact of anticancer chemotherapy on the extension of beta-lactamase spectrum: an example with KPC-type carbapenemase activity towards ceftazidime-avibactam. Sci Rep 10:589. 10.1038/s41598-020-57505-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Venditti C, Butera O, Meledandri M, Balice MP, Cocciolillo GC, Fontana C, D’Arezzo S, De Giuli C, Antonini M, Capone A, Messina F, Nisii C, Di Caro A. 2021. Molecular analysis of clinical isolates of ceftazidime-avibactam-resistant Klebsiella pneumoniae. Clin Microbiol Infect 27:1040.e1–1040.e6. 10.1016/j.cmi.2021.03.001. [DOI] [PubMed] [Google Scholar]
- 21.Cano Á, Guzmán-Puche J, García-Gutiérrez M, Castón JJ, Gracia-Ahufinger I, Pérez-Nadales E, Recio M, Natera AM, Marfil-Pérez E, Martínez-Martínez L, Torre-Cisneros J. 2020. Use of carbapenems in the combined treatment of emerging ceftazidime/avibactam-resistant and carbapenem-susceptible KPC-producing Klebsiella pneumoniae infections: report of a case and review of the literature. J Glob Antimicrob Resist 22:9–12. 10.1016/j.jgar.2019.11.007. [DOI] [PubMed] [Google Scholar]
- 22.Peirano G, Chen L, Kreiswirth BN, Pitout JDD. 2020. Emerging antimicrobial resistant high-risk clones among Klebsiella pneumoniae: ST307 and ST147. Antimicrob Agents Chemother 64:e01148-20. 10.1128/AAC.01148-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonnin RA, Jousset AB, Chiarelli A, Emeraud C, Glaser P, Naas T, Dortet L. 2020. Emergence of new non-clonal group 258 high-risk clones among Klebsiella pneumoniae carbapenemase–producing K. pneumoniae isolates, France. Emerg Infect Dis 26:1212–1220. 10.3201/eid2606.191517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Winkler ML, Papp-Wallace KM, Bonomo RA. 2015. Activity of ceftazidime/avibactam against isogenic strains of Escherichia coli containing KPC and SHV β-lactamases with single amino acid substitutions in the Ω-loop. J Antimicrob Chemother 70:2279–2286. 10.1093/jac/dkv094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Compain F, Dorchène D, Arthur M. 2018. Combination of amino acid substitutions leading to CTX-M-15-mediated resistance to the ceftazidime-avibactam combination. Antimicrob Agents Chemother 62:e00357-18. 10.1128/AAC.00357-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Djamdjian L, Naas T, Tandé D, Cuzon G, Hanrotel-Saliou C, Nordmann P. 2011. CTX-M-93, a CTX-M variant lacking penicillin hydrolytic activity. Antimicrob Agents Chemother 55:1861–1866. 10.1128/AAC.01656-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naas T, Oueslati S, Bonnin RA, Dabos ML, Zavala A, Dortet L, Retailleau P, Iorga BI. 2017. Beta-Lactamase Database (BLDB): structure and function. J Enzyme Inhib Med Chem 32:917–919. 10.1080/14756366.2017.1344235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barnes MD, Winkler ML, Taracila MA, Page MG, Desarbre E, Kreiswirth BN, Shields RK, Nguyen M-H, Clancy C, Spellberg B, Papp-Wallace KM, Bonomo RA. 2017. Klebsiella pneumoniae carbapenemase-2 (KPC-2), substitutions at Ambler position Asp179, and resistance to ceftazidime-avibactam: unique antibiotic-resistant phenotypes emerge from β-lactamase protein engineering. mBio 8:e00528-17. 10.1128/mBio.00528-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shields RK, Nguyen MH, Press EG, Chen L, Kreiswirth BN, Clancy CJ. 2017. Emergence of ceftazidime-avibactam resistance and restoration of carbapenem susceptibility in Klebsiella pneumoniae carbapenemase-producing K pneumoniae: a case report and review of literature. Open Forum Infect Dis 4:ofx101. 10.1093/ofid/ofx101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dortet L, Agathine A, Naas T, Cuzon G, Poirel L, Nordmann P. 2015. Evaluation of the RAPIDEC CARBA NP, the Rapid CARB Screen and the Carba NP test for biochemical detection of carbapenemase-producing Enterobacteriaceae. J Antimicrob Chemother 70:3014–3022. 10.1093/jac/dkv213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The draft genome of K. pneumoniae 181E2 was deposited in GenBank under accession number JAFFPJ000000000.