ABSTRACT
4′-Ethynyl-2-fluoro-2′-deoxyadenosine (EFdA, MK-8591, islatravir) is a nucleoside reverse transcriptase translocation inhibitor (NRTTI) with exceptional potency against wild-type (WT) and drug-resistant HIV-1 in phase III clinical trials. EFdA resistance is not well characterized. To study EFdA resistance patterns that may emerge in naive or tenofovir (TFV)-, emtricitabine/lamivudine (FTC/3TC)-, or zidovudine (AZT)-treated patients, we performed viral passaging experiments starting with WT, K65R, M184V, or D67N/K70R/T215F/K219Q HIV-1. Regardless of the starting viral sequence, all selected EFdA-resistant variants included the M184V reverse transcriptase (RT) mutation. Using recombinant viruses, we validated the role for M184V as the primary determinant of EFdA resistance; none of the observed connection subdomain (R358K and E399K) or RNase H domain (A502V) mutations significantly contributed to EFdA resistance. A novel EFdA resistance mutational pattern that included A114S was identified in the background of M184V. A114S/M184V exhibited higher EFdA resistance (∼24-fold) than either M184V (∼8-fold) or A114S alone (∼2-fold). Remarkably, A114S/M184V and A114S/M184V/A502V resistance mutations were up to 50-fold more sensitive to tenofovir than was WT HIV-1. These mutants also had significantly lower specific infectivities than did WT. Biochemical experiments confirmed decreases in the enzymatic efficiency (kcat/Km) of WT versus A114S (2.1-fold) and A114S/M184V/A502V (6.5-fold) RTs, with no effect of A502V on enzymatic efficiency or specific infectivity. The rather modest EFdA resistance of M184V or A114S/M184V (8- and 24-fold), their hypersusceptibility to tenofovir, and strong published in vitro and in vivo data suggest that EFdA is an excellent therapeutic candidate for naive, AZT-, FTC/3TC-, and especially tenofovir-treated patients.
KEYWORDS: EFdA, HIV drug resistance, HIV-1 reverse transcriptase, NRTIs, inhibitors, islatravir
INTRODUCTION
As of 2019, 38 million people worldwide are living with HIV/AIDS, with 1.7 million new HIV infections and 690,000 AIDS-related deaths annually (1). However, AIDS-related morbidity and mortality rates have declined in recent years, largely due to the widespread use of highly active antiretroviral therapy (HAART) (2). HAART is typically based on a combination of two nucleoside reverse transcriptase inhibitors (NRTIs) in combination with a nonnucleoside reverse transcriptase inhibitor (NNRTI), a protease inhibitor, or an integrase inhibitor (3). Azidothymidine (AZT), didanosine (ddI), lamivudine (3TC), emtricitabine (FTC), abacavir (ABC), and tenofovir (TFV) are the six NRTIs included in HAART regimens (3–5). However, the prevalence of HIV strains resistant to these compounds is rapidly increasing, both in treatment-experienced and newly infected patients (5–10). High-level resistance to AZT generally requires multiple mutations, including D67N, K70R, T215F, and K219Q (4, 11–13), while an M184I/V mutation grants resistance to both 3TC and FTC (4, 14–16). Meanwhile, the K65R mutation imparts resistance to TFV (17–20). A great concern is the emergence of virus strains with cross-resistance to multiple NRTIs, which can limit treatment options following viral escape from first-line HAART (5). M184V strains show resistance to ddI and abacavir, while mutations selected by AZT are resistant to FTC; also, K65R has reduced sensitivity to ddI, 3TC, and FTC (4, 17, 19, 21, 22). Use of the currently available NRTIs can lead to toxicity and side effects (5, 23–26). In addition, the success of antiretroviral regimens at preventing HIV-1 infection has moved attention to the question of compliance and increased the interest in therapeutic agents that may be suitable for long interval dosing (27, 28). All of the above are reasons for the development of novel therapeutic agents that combine high antiviral efficacy with improved quality-of-life characteristics.
Several studies have focused on a group of novel 4′-substituted NRTIs, the most promising of which is 4′-ethynyl-2-fluoro-2′-deoxyadenosine (EFdA, MK-8591, or islatravir) (reviewed in reference 29). EFdA is a deoxyadenosine analog with an ethynyl group at the 4′ carbon of the ribose and a fluorine at the 2 position of the adenine base (13, 30–32). EFdA also retains a 3′-OH, unlike all other NRTIs currently used in HIV therapeutic regimens. The presence of the 3′-OH improves the recognition of EFdA as a substrate of the cellular deoxycytidine kinase (13, 33, 34) and of the viral HIV reverse transcriptase (RT) at the deoxynucleoside triphosphate (dNTP) binding site (35–37). This may contribute to the efficient production of EFdA-triphosphate (EFdA-TP) and the efficient incorporation of EFdA-monophosphate (EFdA-MP) into nascent HIV DNA during reverse transcription. Deamination of adenosine analogs can reduce intracellular concentrations of the active metabolite, resulting in a decreased efficacy of a drug; notably, deamination of EFdA by cellular enzymes is greatly reduced by the 2-fluorine substitution, thus increasing the intracellular half-life of the molecule compared to similar nonhalogenated compounds (30, 34). The 4′-ethynyl group of EFdA interacts with a hydrophobic pocket in the polymerase active site of RT, affecting translocation and blocking efficient extension of the DNA primer, despite the available 3′-OH (35, 38, 39). EFdA is therefore termed a nucleoside RT translocation inhibitor (NRTTI). EFdA is highly potent against both wild-type (WT) HIV-1, HIV-2, and NRTI-resistant strains (34, 40–42). The NRTI-resistant virus, K65R, shows hypersensitivity to EFdA (41, 43). EFdA has shown in vivo potency against both HIV-1 in humanized mouse (44–46) and SIV in nonhuman primate (47–49) models. EFdA imparts minimal toxicity in the animal models, as well as in multiple in vitro assays, due to minimal inhibition of human DNA polymerases, leading to a high selectivity index (44, 46, 50). Importantly, EFdA is being tested for once-weekly, once-monthly, and once-yearly dosing regimens (48, 49, 51–53). Recently, Merck Co. and Gilead Sciences announced a collaboration that will focus on the development of oral and injectable formulations of two long-acting anti-HIV therapeutics: islatravir and lenacapavir (GS-6207) (54). Taken together, these results suggest that EFdA has great promise as a therapeutic agent.
To our knowledge, there is no information about the capacity of HIV to develop high-level resistance to EFdA. The M184V RT mutation imparts a low decrease in sensitivity to EFdA (13, 34, 41, 42, 55, 56). M184V, along with I142V and T165R, was also selected during passage of WT virus in the presence of EdA, a related compound that has the 4′-ethynyl, but not the 2-fluoro modification (34). The I142V/T165R/M184V mutant virus exhibited a 22-fold increase in EFdA resistance relative to the WT; however, it is uncertain whether these mutations would arise during passage of virus in the clinically relevant EFdA itself or whether novel mutations or combinations of mutations conferring even greater resistance are possible. It is also not well understood how resistance to EFdA develops in virus strains with specific preexisting resistance mutations to other NRTIs, which would be relevant to its potential use as salvage therapy for patients failing HAART. Hence, there is incomplete information on EFdA-resistant strains and their cross-resistance to NRTIs currently used in the clinic.
Here, we explored how preexisting NRTI resistance mutations affect the development of EFdA resistance. We selected viruses resistant to EFdA by serial passage of HIV-1 that was either WT or resistant to TFV (K65R), 3TC/FTC (M184V), or AZT (D67N/K70R/T215F/K219Q). We found that M184V/I was included in all selected EFdA-resistant strains. In some cases, mutations also appeared in the RT connection domain (R358K and E399K) and the RNase H domain (A502V); the latter three changes did not significantly affect resistance when alone, and they also did not appear to compensate for the loss of specific infectivity imparted by primary mutations. We identified a double-mutant virus with a moderate level of resistance, A114S/M184V (∼24-fold), with greater resistance to EFdA than that observed with M184V alone. In addition, these EFdA resistance mutations were found to be remarkably hypersensitive to tenofovir (∼50-fold increase in susceptibility). However, A114S/M184V RT was found to have reduced catalytic efficiency (∼6-fold) and impart reduced virus-specific infectivity.
RESULTS
Virus breakthrough during serial passage of viruses in increasing EFdA concentrations.
Serial passages were initiated by infecting MT-2 cells with WT (xxLAI), K65R, M184V, or D67N/K70R/T215F/K219Q stock virus in the presence of EFdA and monitored as described in Materials and Methods. At the end of the passaging, all EFdA-selected viruses, regardless the initial strain or number of passages, induced ≥75% syncytium formation in untreated MT-2 cells within 7 days of infection, indicating that they had retained replication fitness.
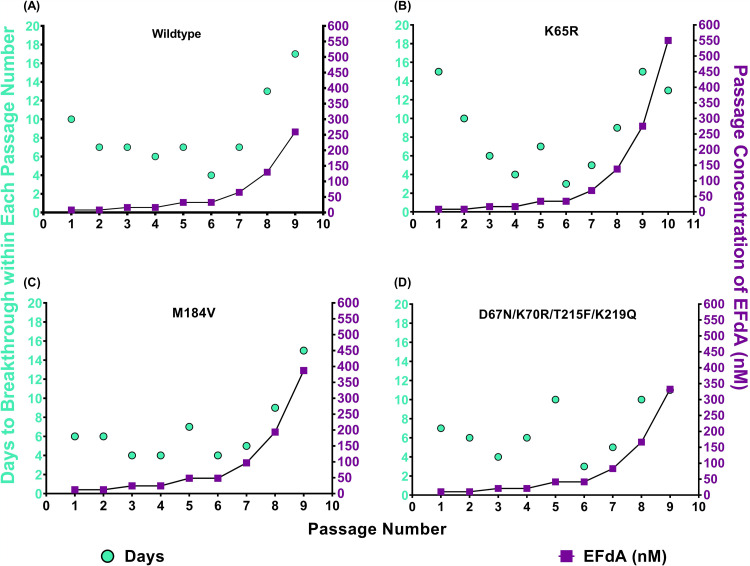
The ability of the viruses to grow in the presence of EFdA was assessed by time to viral breakthrough (defined as 75% syncytium formation) at each passage. This time to breakthrough and the concentration of EFdA in each passage is shown in Fig. 1. For WT, M184V, and D67N/K70R/T215F/K219Q strains, virus breakthrough typically occurred after approximately 7 days. As EFdA concentrations were raised in passages 8 and 9, the time to virus breakthrough increased, until virus replication was no longer observed in passage 10. Passage 10 corresponded to an EFdA concentration of 550 to 800 nM (Fig. 1A, C, and D).
FIG 1.
Selection of resistance to EFdA by serial passage. Time to breakthrough was determined (in MT-2 cells) as the number of days required for ≥75% syncytium formation, at which point the supernatants were harvested, assayed for p24 content, and used to infect the subsequent passage. EFdA concentration was doubled every second passage until P6 and then every passage thereafter. Results represent a single trial for each passage. Passages were initiated with WT (xxLAI) (A), K65R (B), M184V (C), and D67N/K70R/T215F/K219Q (D).
The behavior of K65R deviated from the other mutants studied in two ways. First, there was a reduction in the time to breakthrough from passages 1 to 4, likely because K65R (Fig. 1B), which confers hypersensitivity to EFdA (43), began to be lost as early as P2 and was not detected at passage 6 (Table 1 and Fig. 1B). The time to breakthrough then increased as EFdA concentrations were raised, as observed with other viruses.
TABLE 1.
Amino acid mutations in WT-, K65R-, M184V-, and D67N/K70R/T215F/K219Q-derived viruses during serial passage in progressively increasing concentrations of EFdA
| Virus | Passagea | Sequencingb |
|
|---|---|---|---|
| Amino acid mutation(s)c | Proportion of sequence population (%) | ||
| WT (xxLAI) | 0 | None | 100 |
| 6 | M184I | 100 | |
| 9 | M184I | 36.4 | |
| M184I, E399K | 31.8 | ||
| M184V | 18.2 | ||
| None | 13.6 | ||
| K65R | 0* | None | 100 |
| 6 | R65K, M184I | 90.9 | |
| R65K, M184V | 9.1 | ||
| 9 | R65K, M184V, A502V | 43.5 | |
| R65K, M184I | 34.8 | ||
| R65K, M184V | 17.4 | ||
| R65K, M184I, A502V | 4.3 | ||
| 10 | R65K, A114S, M184V, A502V | 90.9 | |
| R65K, A114S, M184V | 4.5 | ||
| R65K, A114S, A502V | 4.5 | ||
| M184V | 0* | None | 100 |
| 6 | R358K | 45 | |
| None | 55 | ||
| 9 | R358K | 90.5 | |
| None | 9.5 | ||
| 0* | None | 100 | |
| D67N/K70R/T215F/K219Q | 6 | M184V | 66.7 |
| None | 33.3 | ||
| 9 | M184V | 18.2 | |
| M184V, E399K | 81.8 | ||
*, unpassaged stock virus.
Population and clonal sequencing of viral RNA from passage supernatants were performed as described in Materials and Methods.
Relative to unpassaged stock virus of the appropriate strain.
In every case, the passaged viruses were able to replicate in the presence of greater EFdA concentrations than the parental virus, indicating that some degree of EFdA resistance had developed. The most significant resistance appeared in the last and longest passage initiated with K65R virus (Table 1).
Dose response of EFdA-passaged viruses to EFdA.
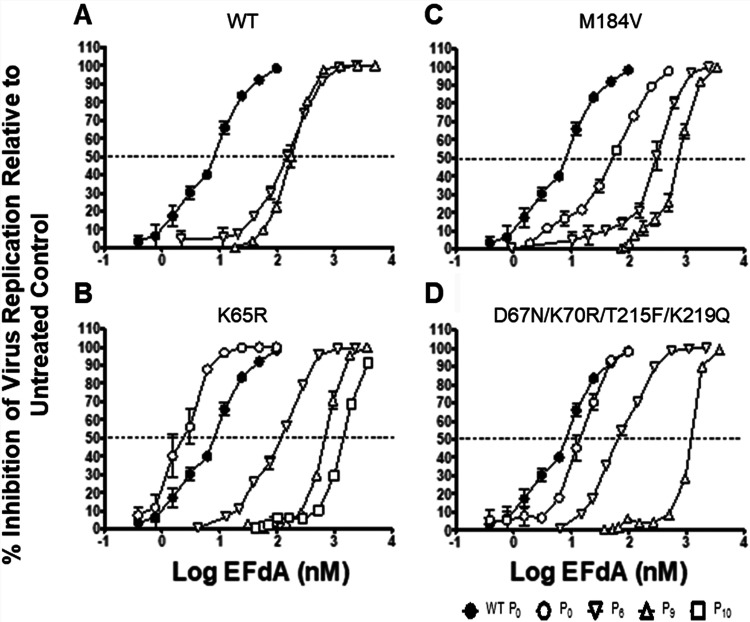
To evaluate the degree of resistance selected in the passaging experiments, sensitivity to EFdA was determined for virus supernatants from passage 6 (P6) onward. Data are presented for the viruses obtained from P6, P9, and, in one case, P10. P6 was the first passage for which extensive sequencing analysis was performed, P9 was the effective endpoint for the WT-, M184V-, and D67N/K70R/T215F/K219Q-derived viruses, and P10 was chosen as the endpoint for the K65R-derived strain. For each of the viruses, the later passages demonstrated increased EC50 values compared to WT and earlier passage viruses, demonstrating that resistance had been acquired as expected (Fig. 2).
FIG 2.
EFdA dose response for viruses selected during serial passage in EFdA. P4-R5 MAGI cells were treated with various concentrations of EFdA and infected with supernatants from the P0, P6, P9, or P10 (where applicable) passages of experiments using WT or mutant-HIV. Untreated control is P0 of WT virus with no EFdA treatment. After incubation for 48 h, the cells were lysed, and the β-galactosidase activity was assessed. (A to D) Inhibition of virus replication relative to infected untreated cells. Dashed line represents 50% inhibition of viral replication. The results represent the means ± the standard deviations (SD) from one experiment with three replicates.
Development of amino acid mutations in reverse transcriptase during serial passage of viruses with EFdA.
Clonal sequencing confirmed the identity of the P0 stock viruses. Similar sequencing was carried out on clones from various passages to identify any changes in RT that may have arisen during replication in the presence of EFdA. Table 1 summarizes the data for P6 and P9 isolates for all parental viruses, as well as viruses from P6, P9, and P10 for K65R.
The WT P6 population sequencing revealed only M184I (Table 1); by P9, approximately 30% of sequences were M184I, 30% were M184I/E399K, and the remainder were divided between M184V and WT. Consistent with the initial passage data, sequencing data showed that all clones of the K65R P6 virus had reverted the K65R mutation back to WT. The reversion occurred rapidly, since K65R was not seen in the P2 population. About 90% of clones in P6 contained M184I, with the remainder containing M184V. Sequencing data also showed that the proportion of M184V increased to approximately one-quarter of the population by P8. The virus harvested following P9 had a more heterogeneous population, with most sequences containing M184V/A502V or M184I. By P10, the diversity decreased dramatically, and A114S/M184V/A502V became the dominant sequence. A114S/M184V and A114S/A502V were found at a much lower frequency.
During the passages that started with M184V, the connection domain mutation R358K appeared and became increasingly dominant as passaging progressed. Passage of the D67N/K70R/T215F/K219Q virus led to the rapid emergence of M184V (dominant in passage 6). This was joined by E399K in late passages (M184V/E399K was dominant in passage 9). None of the starting AZT-resistance-associated mutations were lost during passaging.
Validation of EFdA-resistance associated mutations using molecular clones.
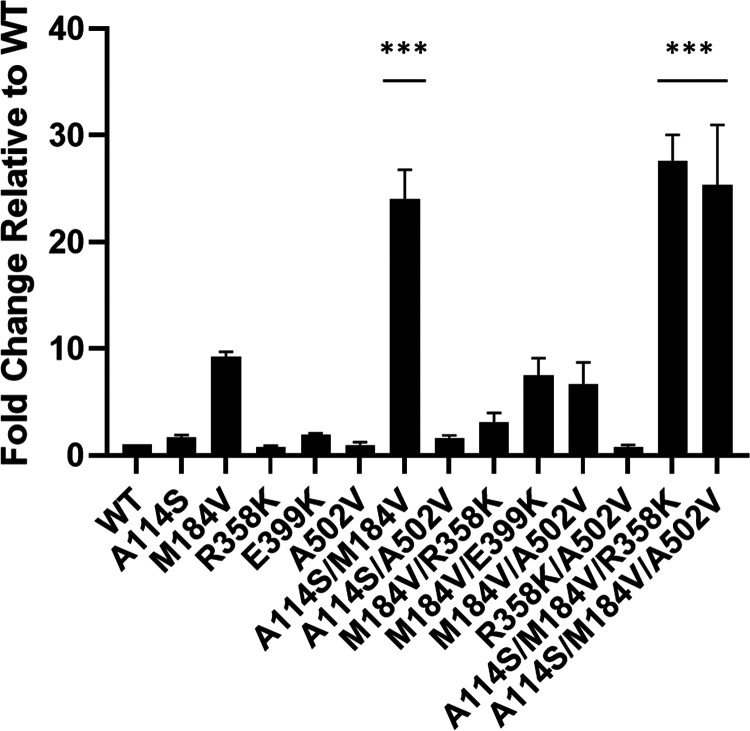
To confirm that the identified mutations conferred EFdA resistance, we constructed relevant molecular clones in a WT (NL4-3) backbone and characterized their infectivity relative to WT (Fig. 3). M184V was the only individual mutation that conferred ∼8-fold EFdA resistance, consistent with previous reports (42, 57). Individual mutations A114S, R358K, E399K, and A502V had no effect on EFdA resistance (Fig. 3). The addition of A114S to M184V further increased the EFdA resistance, as seen in A114S/M184V (24-fold), A114S/M184V/R358K (28-fold), and A114S/M184V/A502V (25-fold). Hence, the highest levels of EFdA resistance were found in the combined presence of A114S and M184V.
FIG 3.
Resistance to EFdA of HIV-1 mutants selected during passaging. Mutations were introduced into NL4-3. TZM-GFP cells were pretreated with EFdA and infected after 24 h. GFP-positive cells (infected cells) were counted in various concentrations of EFdA. Mutants were normalized to WT infection to produce the fold change. EFdA dose-response curves were produced for each mutant and the EC50s calculated. Statistical significance was determined using one-way ANOVA with Tukey’s posttest (***, P < 0.0001). The results represent the means and SD for four independent experiments performed in duplicate.
Susceptibility of select mutants to TDF and FTC.
To determine whether the various EFdA resistance mutations are susceptible to approved drugs such as tenofovir disoproxil fumarate (TDF) and emtricitabine (FTC), we prepared dose-response curves (Table 2). Remarkably, we found that all mutants with the EFdA resistance phenotype were hypersensitized to TDF. Most notably, the A114S mutation by itself and in the presence of M184V or A502V imparted an up to 50-fold decrease in 50% effective dose (EC50; i.e., an increase in EFdA susceptibility). As expected, all M184V mutants were significantly resistant to FTC (>100-fold), which is known resistance mutation and causes therapy failure. There was a 2- to 3-fold decrease in EC50s for FTC with A114S, A502V, and A114S/A502V strains.
TABLE 2.
EC50 fold change compared to WT (NL4-3)
| Parameter | Mean EC50 fold change ± the SD |
|
|---|---|---|
| TDF | FTC | |
| WT | 1 | 1 |
| A114S | 0.04 ± 0.0042 | 0.54 ± 0.043 |
| M184V | 0.57 ± 0.0572 | >100 |
| A502V | 0.12 ± 0.0544 | 0.44 ± 0.071 |
| A114S/A502V | 0.02 ± 0.0014 | 0.39 ± 0.027 |
| M184V/A502V | 0.15 ± 0.054 | >100 |
| A114S/M184V | 0.04 ± 0.0027 | >100 |
| A114S/M184V/A502V | 0.03 ± 0.0052 | >100 |
Replication characteristics of molecular clone viruses.
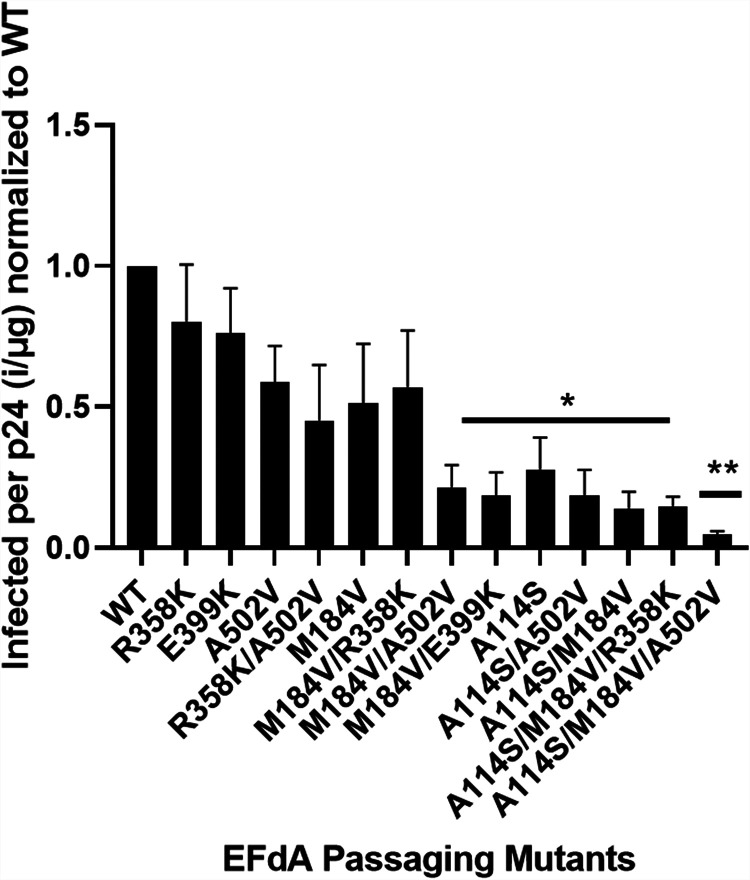
To determine the effect of mutations on viral fitness, we tested viruses in single-cycle replication assays known as specific infectivity. There were no statistically significant differences between the specific infectivities of WT, R358K, or A502V. M184V had a reduced specific infectivity compared to WT; it did not appear to be statistically significant under these experimental conditions. In addition, M184V, in combination with all other mutants besides R358K, showed a significant decrease in specific infectivity. However, there was a decrease in specific infectivity of the A114S-containing mutants, A114S/M184V and A114S/M184V/A502V, compared to WT (Fig. 4). To further validate this result, we examined the biochemical properties of A114S RT, as described below.
FIG 4.
Single-round replication assays using TZM-GFP cells infected with individual mutants. An ELISA was also performed on the virus to determine amount of p24 (total virus in medium). The ratio of infected cells per p24 was then calculated and normalized to the WT. Statistical significance was determined using a one-way ANOVA with Dunnett’s multiple-comparison test (*, P < 0.05; **, P < 0.01). The results represent the means and SD for four independent experiments performed in triplicate.
Steady-state kinetics and EFdA susceptibility of mutant reverse transcriptases.
To better understand the effect of the mutations on the DNA polymerase activity of RT, we performed steady-state kinetics experiments to determine the catalytic efficiency, kcat/Km, for various RT mutants. We cloned, expressed, and purified the mutant RTs listed in Table 3. We found that in the presence of the A114S mutation there was a consistent decrease in the catalytic efficiency kcat/Km compared to WT RT: a 2.1-fold decrease for A114S and a 6.5-fold decrease for A114S/M184V/A502V. The changes in catalytic efficiencies of these mutants appeared to be primarily due to the change in the Km. The Km for A502V was slightly decreased but comparable to that for the WT. Of note, the catalytic efficiency (kcat/Km) for the A502V and M184V single mutants was comparable to that of the WT enzyme (Table 3).
TABLE 3.
Steady-state enzyme kinetics for reverse transcriptase mutants
| Parameter | WT RT | A114S | M184V | R358K | E399K | A502V | 184V/358K/502V | 114S/184V/502V |
|---|---|---|---|---|---|---|---|---|
| Km (nM) | 1,448 | 2,758 | 1,791 | 2,008 | 1,795 | 1,110 | 2,327 | 6,659 |
| Fold change, Km | 1 | 1.9 | 1.2 | 1.4 | 1.2 | 0.8 | 1.6 | 4.6 |
| Fold change, kcat/Km | 1 | 2.1 | 1.2 | 1.4 | 1.2 | 1 | 1.3 | 6.5 |
DISCUSSION
NRTIs are the most widely used therapeutics to treat HIV infection. As such, NRTI-resistant HIV variants are becoming increasingly prevalent in the HIV-1-infected population. EFdA has a strong potential for long-acting first-line therapies for naive HIV-infected individuals or as an agent for preexposure prophylaxis, or salvage therapies. As such, we were interested in identifying and characterizing EFdA resistance mutations that might arise during exposure of WT HIV, and especially NRTI-resistant virus variants, to EFdA. Moreover, there is high interest in discovering novel mutations or combinations of mutations that impart levels of EFdA resistance that may have potential clinical implications.
Regardless the genotype of the starting virus, M184I/V consistently arose during passages through EFdA. M184V confers high-level resistance to both 3TC and FTC (14–16, 58) but only low-level resistance to EFdA (34, 41). Our data confirm the latter, with M184V conferring only an ∼8-fold resistance to EFdA. M184V is also the primary resistance mutation selected during serial passage of WT HIV with the related compounds EdA and Ed4T, suggesting that M184 is critical to the activity of 4′-ethynyl modified nucleoside analogues (34, 59).
D67N/K70R/T215F/K219Q HIV is highly resistant to AZT (11, 12). Similar to another AZT-resistant mutant, M41L/T215Y, which was previously shown to display marginal resistance to EFdA (34), we found that D67N/K70R/T215F/K219Q demonstrated a 1.8-fold increase in EFdA resistance compared to WT. As we have previously shown, the excision unblocking mechanism of resistance is not a major challenge for EFdA; although EFdA can indeed be excised, the efficiency of reincorporation is so high that the net result is not significant overall excision (38). The K65R mutation confers resistance to TFV and is the mutation responsible for virological failure in TFV-based therapies. K65R is also cross-resistant or selected during therapy with ABC, ddI, and 3TC/FTC (5, 17, 19–22). We previously showed that K65R is hypersensitive to EFdA, up to 5-fold lower EC50 compared to the WT (34, 41, 43). The present data are consistent with this finding, since K65R displayed a longer time to breakthrough during initial passage in EFdA, and this breakthrough was associated with the loss of K65R. The rapid reversion of this mutation not only leads to a reduction in sensitivity to EFdA but also confers increased replication fitness to the virus. A114S and A502V were found after the reversion of K65R. Although there is no clear explanation for this observation, it may reflect a stochastic aspect of the drug resistance selection process. In addition, it is possible that the additional passaging steps may have contributed to conditions that led to high concentrations of EFdA, thus increasing the selective pressure on RT and allowing A114S to prevail.
Our results highlight the challenges of selecting for EFdA resistance. Even when starting the passages using a variety of clinically relevant genotypic backgrounds, mutations at residue 184 were consistently the first to emerge. Though in previous studies it was shown that M184V exhibited reduced viral fitness, we observed here a decrease in specific infectivity that was not statistically significant (60–62). This may be related to assay-condition-specific differences. In addition, EFdA was fully effective in maintaining suppression of mutant virus throughout the drug treatment period in WT SIV-infected rhesus macaques (47). EFdA-treated humanized mice infected with HIV remained fully suppressed while on treatment even when M184I emerged after therapy cessation (44). In addition, M184V is known to fail current therapies: 3TC and FTC (reviewed in references 63–67). Here, we have identified a novel EFdA resistance mutation, A114S, which appeared after ∼3 months of EFdA treatment (Fig. 1). The addition of A114S to the M184V background (A114S/M184V, A114S/M184V/R358K, and A114S/M184V/A502V) enhanced EFdA resistance up to ∼28-fold, or 350% higher than M184V by itself. Importantly, both virological and biochemical experiments demonstrated that this mutation, especially in the background of M184V, has a negative impact on specific infectivity (Fig. 4 and Table 3). Similar results were recently presented at a conference reporting that A114S with M184V conferred resistance but had minimal effect on its own (68). Specifically, it seems that the decrease in fitness is likely due to decreased binding of incoming deoxynucleoside triphosphates (dNTPs), reflected by the increased Km, in all cases where A114S mutation was present. The A114S mutation has a longer side chain, which may extend closer to the 3′-OH of an incoming dNTP, thus reducing the available space in the binding pocket, leading to increased selectivity for the canonical dNTP. Consistent with previous data, we found that the A114S mutation decreased the DNA polymerase activity of RT (69, 70). Residues 114 and 184 are located at opposing sides of a hydrophobic pocket, along with Y115, F160, and the aliphatic chain of D185, which accommodates the 4′-ethynyl group of EFdA (35, 71). We speculate that the bulkier side chains of V184 and S114 impinge into the substrate envelope of the 4′-ethynyl binding pocket, thus causing a decrease in dNTP binding that may contribute to the observed decrease in specific infectivity. Nevertheless, A114S-containing mutants may not be a major problem in patients; A114 mutations are rarely seen in clinical samples. According to the Stanford HIV Drug Resistance database, A114S mutants have appeared a total of 7 times in 172,714 sequenced samples (72, 73).
In addition, we have found that TDF inhibits the A114S/M184V EFdA-resistant HIV, significantly more efficiently than the WT (Table 2). We have previously reported that the K65R TDF resistance mutation in RT also hypersensitizes HIV to EFdA (43). Thus, a combination of tenofovir and EFdA could be a very efficient therapy since they complement each other’s resistance profile.
Selected residues R358K and E399K are proximal to G359-A360 and K395-E396, respectively, in the RNase H primer grip region that interacts with the DNA primer strand (74–77). R358K is selected both in NRTI-treated patients and during passage with NRTIs in tissue culture (78–82). E399 is located within a cluster of several conserved tryptophan residues involved in RT dimerization (83, 84). We found no significant variation in EFdA resistance between M184V/E399K or M184V/R358K and M184V alone. Hence, neither R358K nor E399K contribute directly to EFdA resistance. We initially considered that these mutations might provide a fitness benefit. However, we did not observe improved specific infectivity in our single-round replication assays. The A502V late-appearing mutation is located in the primer grip region of the RNase H domain, near residues Y501 and I505 (75). Under the conditions tested, we did not see changes in the biochemical fitness of A502V, but we did see minor decreases in Km. M184V/A502V HIV subsequently acquired the A114S mutation, which coincided with a significant increase in EFdA resistance.
A major concern when introducing any new NRTI to clinical use is the potential for cross-resistance with current NRTIs. Our results confirm that EFdA drives the selection of M184I/V, producing virus with high-level cross-resistance to 3TC and FTC. However, EFdA is a potent inhibitor, even against M184I/V; it retains the ability to inhibit replication of these mutants at therapeutic doses (85). Conversely, M184I/V increases AZT sensitivity in both WT and AZT-resistant backgrounds (10, 16, 86, 87). Similarly, K65R confers resistance to TFV but is hypersensitive to EFdA (43); these data suggest that EFdA may be well suited for combination therapies. We plan to study these mutations in primary cells, as well as various subtypes, in future studies to further confirm our findings and understand EFdA inhibition for implementation on a global scale.
In summary, our data demonstrate that there is a strong barrier to HIV developing high-level EFdA resistance. The development of >10-fold resistance to EFdA required the combination of multiple mutations, including both M184V and A114S, that appear to negatively affect virus-specific infectivity. These RT mutants were also found to be dramatically hypersensitized to tenofovir (∼0.04-fold change compared to WT). Hence, EFdA has the potential to become a highly effective therapeutic and in combination with tenofovir.
MATERIALS AND METHODS
Reagents.
EFdA was synthesized by Life Chemicals (Burlington, Ontario, Canada). Stock solutions (10 mM) of EFdA was prepared in dimethyl sulfoxide and stored in aliquots at −20°C. MT-2 cells (88–90) were cultured in RPMI 1640 medium (Mediatech, Inc., Manassas, VA), supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, UT) and 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin (all from Mediatech, Inc.). P4-R5 MAGI cells were cultured in Dulbecco modified Eagle medium (DMEM; Mediatech, Inc.), supplemented with 10% FBS, 2 mM l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 1 μg/ml puromycin. HEK-293 (91) and HEK-293/17 (92) cells were cultured in DMEM supplemented with 10% FBS, l-glutamine, and penicillin/streptomycin. TZM-GFP cells from Massimo Pizzato laboratory (Trento University) (93–95) were cultured in DMEM (Corning) supplemented with 10% Serum Plus, 2 mM l-glutamine (Thermo Fisher, Waltham, MA), and 100 U/ml penicillin/streptomycin (Thermo Fisher). Jurkat LTR-GFP CCR5+ cells (JLTRG-R5) (96, 97) were cultured in RPMI 1640 medium (Cytiva Life Sciences, Marlborough, MA) supplemented with 10% FBS and 100 U/ml penicillin/streptomycin.
Generation of virus stocks and molecular clones.
Initial K65R, M184V, and D67N/K70R/T215F/K219Q viruses were generated by site-directed mutagenesis on an xxLAI HIV-1 backbone using the QuikChange XL site-directed mutagenesis kit (Agilent Technologies, Inc., Santa Clara, CA) according to the manufacturer’s protocols. Subsequently, 6 × 105 293-T cells were transfected with 10 μg of viral DNA using the PrimeFectimine mammalian transfection reagent (PrimGen, Oak Park, IL). After 72 h of incubation, HEK-293/T cell supernatants were harvested, filtered, and used to infect 1.8 × 106 MT-2 cells. Infected MT-2 cells were incubated at 37°C (5% CO2), inspected daily, and infectious virus was harvested at ≥50% syncytium formation. Emory’s Cloning Core was used to make individual mutants in the backbone of NL4.3. Virus stocks were made using HEK-293/17 cells that were transfected with 6 μg of viral DNA using Xtreme-GENE HP (Roche, Basel, Switzerland) transfection reagent on a 10-cm dish of 60% confluence. After 48 h of incubation, HEK-293/17 cell supernatants were harvested and concentrated overnight with a Lenti-X concentrator (Clontech, Mountain View, CA) according to the manufacturer’s protocol.
Determination of TCID50 values and p24 content.
We determined 50% tissue culture infective doses (TCID50) by infecting 5 × 104 MT-2 cells per well in 96-well flat-bottom plates with 4-fold serial dilutions of virus stock. Three replicates were performed for each virus. Infected plates were inspected daily for syncytium formation; every 3 days, half of the supernatant was replaced with fresh media. The assay was terminated when no additional syncytium formation was noted for 2 days. The TCID50 was then calculated using the Reed-Muench method (98). The p24 content of each virus stock was determined using an HIV-1 p24CA Antigen Capture assay kit (SAIC-Frederick, Frederick, MA).
Serial passage for selection of resistant virus.
MT-2 cells were suspended at 2.5 × 105 cells per ml in 10 ml of medium containing EFdA. Initial EFdA concentrations were chosen based on the EC50 of WT stock virus (8.6 nM), with a similar amount chosen for the K65R virus and slightly higher concentrations for the D67N/K70R/T215F/K219Q (10 nM) and M184V (12 nM) strains. Passages were initiated by immediately adding 200 TCID50 of the appropriate unpassaged (P0) virus stock to the cells, followed by gentle mixing. Untreated cultures were initiated by infecting 2.5 × 105 MT-2 cells per ml with 200 TCID50 of P0 virus stock in 10 ml of drug-free medium. All passages and untreated cultures were grown in T-25 tissue culture flasks. Every 2 to 3 days, the cells were split 1:3 with fresh media containing the appropriate concentration of EFdA. Cultures were visually inspected every 1 to 2 days for the presence of syncytia. At ≥75% syncytium formation, culture supernatants were harvested, concentrated using Amicon Ultra Ultracel 100K centrifugal filters (Millipore, Carrigtwohill, Co. Cork, Ireland) and syringe-filtered through 0.22-μm filters (Millipore). The p24 content of the resulting first passage (P1) supernatant was determined as described above. This procedure was followed for all subsequent passages, with PN initiated by infecting 2.5 × 106 MT-2 cells with PN-1 virus supernatant in media containing the appropriate concentration of EFdA. Untreated cultures were also initiated by infecting 2.5 × 106 MT-2 cells, in drug-free media, with PN-1 virus. Since infectivity differences were expected between the P0 virus and the output strains from each passage, P2 and all subsequent passages were initiated by infecting cells with a p24 amount of PN-1 virus equivalent to the P0 virus p24 content used to initiate P1. The concentration of EFdA was doubled every two passages up to P6, and then the amount of drug was doubled with every passage. Passages lasting more than 60 days without syncytium formation were terminated and repeated, along with the previous passage. If no syncytia were noted in a repeat passage after 30 days, the passage was terminated, and no further attempts were performed.
Sequencing of passaged viruses.
Viral RNA was purified from supernatants using the QIAamp Viral RNA minikit (Qiagen, Valencia, CA), the concentration was determined with a Spectronic BioMate*3 UV spectrophotometer (Thermo Scientific), and 500 ng was used as the template for cDNA synthesis. First-strand PCR was performed using random hexamer primers and the SuperScript III first-strand synthesis system for RT-PCR (Invitrogen, Carlsbad, CA). The resulting cDNA was PCR amplified using the HIV-1 LAI-specific primers ABR-RT-OF (1763 5′-GGAGCCGATAGACAAGGAACTG-3′) and ABR-RT-OR2 (3863 5′-GGCTACTATTTCTTTTGCTACTACAGG-3′). These primers anneal to the 3′ end of gag and the 5′ end of integrase, respectively, and generate a 2,127-bp product spanning the full length of the reverse transcriptase gene. PCR was performed using the Expand High-Fidelity PCR System dNTPack (Roche Diagnostics GmbH, Mannheim, Germany), with 4.5 mM MgCl2 and 3.5 U of enzyme mix used for each reaction. Reactions were run in a PCR Sprint bench-top PCR cycler (Thermo Electron) with an initial denaturation of 3 min at 94°C, followed by 30 amplification cycles, each consisting of 30 s at 94°C, 45 s at 58°C, and 150 s at 72°C. After a 7-min final extension at 72°C, the samples were used immediately or stored at −20°C.
PCR products were separated by electrophoresis on a 1% agarose gel, and the 2,127-bp band was harvested using a QIAquick gel extraction kit (Qiagen). The concentration of DNA was determined by spectrophotometry, and 500 ng of sample was submitted to ACGT, Inc. (Wheeling, IL), for full-length, double-stranded population sequencing. The P0 consensus sequences from each virus were aligned in Clustal X-2 (99) with an independent HIV-1 LAI nucleotide sequence (accession no. NC_001802) to ensure the stock viruses did not contain unexpected mutations. Nucleotide consensus sequences from each passage were aligned to the appropriate P0 consensus. Chromatograms were also inspected visually using Chromatogram Explorer and DNABaser (Heracle BioSoft S.R.L., Pitesti, Romania) for the presence of heterogeneous peaks and minority sequence populations not detectable in the consensus sequence.
Clonal sequencing of full-length reverse transcriptase gene PCR products.
Approximately 105 ng of full-length PCR product was ligated into the pGEM-T vector system (Promega, Madison, WI) at a 3:1 insert/vector molar ratio and incubated overnight at 4°C. Ligations were transformed into MAX Efficiency DH5α competent cells (Invitrogen) by heat shock. Blue-white screening was used to select clones with successful ligations, and plasmids containing the full-length reverse transcriptase gene were isolated with a QIAprep Spin Miniprep kit (Qiagen). A minimum of 20 clones from each sample were sequenced. Primers ABR-RT-OF and ABR-RT-OR2 were used to sequence the 5′ and 3′ ends of reverse transcriptase, while an internal portion of the gene was sequenced with primer ABR-RT-IF (2211 5′-CAGAGATGGAAAAGGAAGGG-3′). Clones were aligned with the appropriate P0 stock virus consensus in Clustal X-2, and the proportion of sequences with the novel substitutions was determined.
Dose response to individual mutants.
All individual mutations were cloned into pNL4.3 using Emory’s Cloning Core. After each virus was generated as described above, TZM-GFP cells were plated at 10,000 cells/well in a 96-well plate and with serially diluted EFdA, TDF, or FTC starting at 100 nM (for EFdA) or 10 μM (TDF and FTC). The cells and drug were incubated for 24 h. Next, the cells were infected with virus and a 1-μg/ml final concentration of DEAE-dextran, followed by incubation for 48 h. The green fluorescent protein (GFP)-positive cells were then counted using Cytation 5 (BioTek, Winooski, VT) with Gen5 v3.03 software. EC50 curves were then determined using Prism 5 (GraphPad) software.
Specific infectivity.
TZM-GFP cells were plated at 10,000 cells/well in a 96-well plate and incubated for 24 h. Then, cells were infected with the various concentrations of the virus and a 1-μg/ml final concentration of DEAE-dextran, followed by incubation for 48 h. The GFP-positive cells were then counted as described above. After 48 h of incubation, the cells were imaged using Cytation 5 to quantify the numbers of GFP-positive cells, which were then split, and the medium was replaced. The cells were subsequently imaged. The p24 content of each virus was also determined using an enzyme-linked immunosorbent assay (ELISA).
Steady-state kinetics and in vitro IC50s.
HIV-1 RT and mutants were expressed and purified as described previously (36, 77, 100–104). RT was expressed in JM109 cells (Invitrogen) and purified by nickel affinity chromatography and Mono-Q anion-exchange chromatography. Steady-state kinetic parameters, Km, for the incorporation of dNTPs were determined using plate-based assays measuring an 18-nucleotide primer annealed to a 100-nucleotide DNA template (primer, 5′-GTCCCTGTTCGGGCGCCA; template, 5′-TAGTGTGTGCCCGTCTGTTGTGTGACTCTGGTAACTAGAGATCCCTCAGACCCTTTTAGTCAGTGTGGAAAATCTCTAGCAGTGGCGCCCGAACAGGGAC-3′) (105–110). The reactions were carried out in RT buffer with 6 mM MgCl2, 40 nM Td100/Pd18, and 10 nM RT in a final volume of 20 μl for 30 min at 37°C and then arrested by 100 mM EDTA. The QuantiFluor dsDNA System (Promega) was used to quantify the amount of formed double-stranded DNA. Reactions were read at ex/em 504/531 nm in a Perkin-Elmer EnSpire Multilabel plate reader. Km values were determined graphically using the Michaelis-Menten equation and Prism 5.
Statistics.
The statistical significance was determined using Prism 5. A one-way analysis of variance (ANOVA) was performed, as well as a Dunnett’s or Tukey’s multiple-comparison test (as mentioned in the figure legends.
Data availability.
Mutation sequences for BankIt2501100 Seq1 to Seq14 have been deposited in GenBank under accession no. OK263149 to OK263162.
ACKNOWLEDGMENTS
Michael Parniak was involved in the early research and writing of this study; as he is now deceased, it has not been possible to obtain his approval of the final version of the manuscript.
This study was supported in part by NIH grants R37 AI076119 to S.G.S. and T32 GM008367 and F31 AI155158-01A1 (provided training funds for M.E.C.). S.G.S. acknowledges funding from the Nahmias-Schinazi Distinguished Chair in Research. This study was supported in part by the Emory Integrated Genomics Core (EIGC), which is subsidized by the Emory University School of Medicine and is one of the Emory Integrated Core Facilities. Additional support was provided by the National Center for Advancing Translational Sciences of the National Institutes of Health under award UL1TR000454. The content is solely the responsibility of the authors and does not necessarily reflect the official views of the National Institutes of Health.
The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: MT-2 cells from Douglas Richman, P4-R5 MAGI cells from Nathaniel Landau, and HEK-293 cells from Andrew Rice. The TZM-GFPs were obtained from Massimo Pizzato (Trento University).
REFERENCES
- 1.UNAIDS. 2020. Fact sheet: World AIDS Day. UNAIDS, New York, NY. [Google Scholar]
- 2.Mahy M, Marsh K, Sabin K, Wanyeki I, Daher J, Ghys PD. 2019. HIV estimates through 2018: data for decision-making. AIDS 33(Suppl 3):S203–S211. 10.1097/QAD.0000000000002321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Merluzzi VJ, Hargrave KD, Labadia M, Grozinger K, Skoog M, Wu JC, Cheng-Kon S, Eckner K, Hattox S, Adams J, Rosehthal AS, Faanes R, Eckner RJ, Koup RA, Sullivan JL, Sarafianos SG, Marchand B, Das K, Himmel D, Michael A, Hughes SH, Arnold E. 1990. Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. Science 250:1411–1413. 10.1126/science.1701568. [DOI] [PubMed] [Google Scholar]
- 4.Menéndez-Arias L. 2008. Mechanisms of resistance to nucleoside analogue inhibitors of HIV-1 reverse transcriptase. Virus Res 134:124–146. 10.1016/j.virusres.2007.12.015. [DOI] [PubMed] [Google Scholar]
- 5.Eggleton JS, Nagalli S. 2020. Highly active antiretroviral therapy (HAART). StatPearls, Treasure Island, FL. [PubMed] [Google Scholar]
- 6.Little SJ, Holte S, Routy J-P, Daar ES, Markowitz M, Collier AC, Koup RA, Mellors JW, Connick E, Conway B, Kilby M, Wang L, Whitcomb JM, Hellmann NS, Richman DD. 2002. Antiretroviral-drug resistance among patients recently infected with HIV. N Engl J Med 347:385–394. 10.1056/NEJMoa013552. [DOI] [PubMed] [Google Scholar]
- 7.Wainberg MA, Zaharatos GJ, Brenner BG. 2011. Development of antiretroviral drug resistance. N Engl J Med 365:637–646. 10.1056/NEJMra1004180. [DOI] [PubMed] [Google Scholar]
- 8.Clutter DS, Jordan MR, Bertagnolio S, Shafer RW. 2016. HIV-1 drug resistance and resistance testing. Infect Genet Evol 46:292–307. 10.1016/j.meegid.2016.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pennings PS. 2013. HIV drug resistance: problems and perspectives. Infect Dis Rep 5:e5. 10.4081/idr.2013.s1.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larder BA, Kemp SD, Harrigan PR. 1995. Potential mechanism for sustained antiretroviral efficacy of AZT-3TC combination therapy. Science 269:696–699. 10.1126/science.7542804. [DOI] [PubMed] [Google Scholar]
- 11.Kellam P, Boucher CA, Larder BA. 1992. Fifth mutation in human immunodeficiency virus type 1 reverse transcriptase contributes to the development of high-level resistance to zidovudine. Proc Natl Acad Sci USA 89:1934–1938. 10.1073/pnas.89.5.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larder BA, Kemp SD. 1989. Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (AZT). Science 246:1155–1158. 10.1126/science.2479983. [DOI] [PubMed] [Google Scholar]
- 13.Nakata H, Amano M, Koh Y, Kodama E, Yang G, Bailey CM, Kohgo S, Hayakawa H, Matsuoka M, Anderson KS, Cheng Y-CY-C, Mitsuya H. 2007. Activity against human immunodeficiency virus type 1, intracellular metabolism, and effects on human DNA polymerases of 4′-ethynyl-2-fluoro-2′-deoxyadenosine. Antimicrob Agents Chemother 51:2701–2708. 10.1128/AAC.00277-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petrella M, Oliveira M, Moisi D, Detorio M, Brenner BG, Wainberg MA. 2004. Differential maintenance of the M184V substitution in the reverse transcriptase of human immunodeficiency virus type 1 by various nucleoside antiretroviral agents in tissue culture. Antimicrob Agents Chemother 48:4189–4194. 10.1128/AAC.48.11.4189-4194.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schinazi RF, Lloyd RM, Nguyen MH, Cannon DL, McMillan A, Ilksoy N, Chu CK, Liotta DC, Bazmi HZ, Mellors JW. 1993. Characterization of human immunodeficiency viruses resistant to oxathiolane-cytosine nucleosides. Antimicrob Agents Chemother 37:875–881. 10.1128/AAC.37.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tisdale M, Kemp SD, Parry NR, Larder BA. 1993. Rapid in vitro selection of human immunodeficiency virus type 1 resistant to 3′-thiacytidine inhibitors due to a mutation in the YMDD region of reverse transcriptase. Proc Natl Acad Sci USA 90:5653–5656. 10.1073/pnas.90.12.5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brenner BG, Coutsinos D. 2009. The K65R mutation in HIV-1 reverse transcriptase: genetic barriers, resistance profile and clinical implications. HIV Ther 3:583–594. 10.2217/hiv.09.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Margot NA, Isaacson E, McGowan I, Cheng AK, Schooley RT, Miller MD. 2002. Genotypic and phenotypic analyses of HIV-1 in antiretroviral-experienced patients treated with tenofovir DF. AIDS 16:1227–1235. 10.1097/00002030-200206140-00004. [DOI] [PubMed] [Google Scholar]
- 19.Miller MD. 2004. K65R, TAMs and tenofovir. AIDS Rev 6:22–33. [PubMed] [Google Scholar]
- 20.Naeger LK, Struble KA. 2006. Effect of baseline protease genotype and phenotype on HIV response to atazanavir/ritonavir in treatment-experienced patients. AIDS 20:847–853. 10.1097/01.aids.0000218548.77457.76. [DOI] [PubMed] [Google Scholar]
- 21.Bazmi HZ, Hammond JL, Cavalcanti SC, Chu CK, Schinazi RF, Mellors JW. 2000. In vitro selection of mutations in the human immunodeficiency virus type 1 reverse transcriptase that decrease susceptibility to (–)-β-d-dioxolane-guanosine and suppress resistance to 3′-azido-3′-deoxythymidine. Antimicrob Agents Chemother 44:1783–1788. 10.1128/AAC.44.7.1783-1788.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang D, Caliendo AM, Eron JJ, DeVore KM, Kaplan JC, Hirsch MS, D’Aquila RT. 1994. Resistance to 2′,3′-dideoxycytidine conferred by a mutation in codon 65 of the human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother 38:282–287. 10.1128/AAC.38.2.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brinkman K, Kakuda TN. 2000. Mitochondrial toxicity of nucleoside analogue reverse transcriptase inhibitors: a looming obstacle for long-term antiretroviral therapy? Curr Opin Infect Dis 13:5–11. 10.1097/00001432-200002000-00002. [DOI] [PubMed] [Google Scholar]
- 24.Brinkman K, Smeitink JA, Romijn JA, Reiss P. 1999. Mitochondrial toxicity induced by nucleoside-analogue reverse-transcriptase inhibitors is a key factor in the pathogenesis of antiretroviral-therapy-related lipodystrophy. Lancet 354:1112–1115. 10.1016/S0140-6736(99)06102-4. [DOI] [PubMed] [Google Scholar]
- 25.Brinkman K, ter Hofstede HJM, Burger DM, Smeitink JAM, Koopmans PP. 1998. Adverse effects of reverse transcriptase inhibitors. AIDS 12:1735–1744. 10.1097/00002030-199814000-00004. [DOI] [PubMed] [Google Scholar]
- 26.Lewis W, Day BJ, Copeland WC. 2003. Mitochondrial toxicity of NRTI antiviral drugs: an integrated cellular perspective. Nat Rev Drug Discov 2:812–822. 10.1038/nrd1201. [DOI] [PubMed] [Google Scholar]
- 27.Margolis DA, Boffito M. 2015. Long-acting antiviral agents for HIV treatment. Curr Opin HIV AIDS 10:246–252. 10.1097/COH.0000000000000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cihlar T, Fordyce M. 2016. Current status and prospects of HIV treatment. Curr Opin Virol 18:50–56. 10.1016/j.coviro.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 29.Markowitz M, Sarafianos SG. 2018. 4′-Ethynyl-2-fluoro-2′-deoxyadenosine, MK-8591: a novel HIV-1 reverse transcriptase translocation inhibitor. Curr Opin HIV AIDS 13:294–299. 10.1097/COH.0000000000000467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirby KA, Michailidis E, Fetterly TL, Steinbach MA, Singh K, Marchand B, Leslie MD, Hagedorn AN, Kodama EN, Marquez VE, Hughes SH, Mitsuya H, Parniak MA, Sarafianos SG. 2013. Effects of substitutions at the 4′ and 2 positions on the bioactivity of 4′-ethynyl-2-fluoro-2′-deoxyadenosine. Antimicrob Agents Chemother 10.1128/AAC.01703-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirby KA, Singh K, Michailidis E, Marchand B, Kodama EN, Ashida N, Mitsuya H, Parniak MA, Sarafianos SG. 2011. The sugar ring conformation of 4′-ethynyl-2-fluoro-2′-deoxyadenosine and its recognition by the polymerase active site of HIV reverse transcriptase. Cell Mol Biol 57:40–46. 10.1170/T900.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kageyama M, Nagasawa T, Yoshida M, Ohrui H, Kuwahara S. 2011. Enantioselective total synthesis of the potent Anti-HIV nucleoside EFdA. Org Lett 13:5264–5266. 10.1021/ol202116k. [DOI] [PubMed] [Google Scholar]
- 33.Gallois-Montbrun S, Schneider B, Chen Y, Giacomoni-Fernandes V, Mulard L, Morera S, Janin J, Deville-Bonne D, Veron M. 2002. Improving nucleoside diphosphate kinase for antiviral nucleotide analogs activation. J Biol Chem 277:39953–39959. 10.1074/jbc.M206360200. [DOI] [PubMed] [Google Scholar]
- 34.Kawamoto A, Kodama E, Sarafianos SG, Sakagami Y, Kohgo S, Kitano K, Ashida N, Iwai Y, Hayakawa H, Nakata H, Mitsuya H, Arnold E, Matsuoka M. 2008. 2′-Deoxy-4′-C-ethynyl-2-halo-adenosines active against drug-resistant human immunodeficiency virus type 1 variants. Int J Biochem Cell Biol 40:2410–2420. 10.1016/j.biocel.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 35.Salie ZL, Kirby KA, Michailidis E, Marchand B, Singh K, Rohan LC, Kodama EN, Mitsuya H, Parniak MA, Sarafianos SG. 2016. Structural basis of HIV inhibition by translocation-defective RT inhibitor 4′-ethynyl-2-fluoro-2′-deoxyadenosine (EFdA). Proc Natl Acad Sci USA 113:9274–9279. 10.1073/pnas.1605223113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michailidis E, Marchand B, Kodama EN, Singh K, Matsuoka M, Kirby KA, Ryan EM, Sawani AM, Nagy E, Ashida N, Mitsuya H, Parniak MA, Sarafianos SG. 2009. Mechanism of inhibition of HIV-1 reverse transcriptase by 4′-ethynyl-2-fluoro-2′-deoxyadenosine triphosphate, a translocation-defective reverse transcriptase inhibitor. J Biol Chem 284:35681–35691. 10.1074/jbc.M109.036616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michailidis E, Huber AD, Ryan EM, Ong YT, Leslie MD, Matzek KB, Singh K, Marchand B, Hagedorn AN, Kirby KA, Rohan LC, Kodama EN, Mitsuya H, Parniak MA, Sarafianosa SG. 2014. 4′-Ethynyl-2-fluoro-2′-deoxyadenosine (EFdA) inhibits HIV-1 reverse transcriptase with multiple mechanisms. J Biol Chem 289:24533–24538. 10.1074/jbc.M114.562694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michailidis E, Huber AD, Ryan EM, Ong YT, Leslie MD, Matzek KB, Singh K, Marchand B, Hagedorn AN, Kirby KA, Rohan LC, Kodama EN, Mitsuya H, Parniak MA, Sarafianos SG. 2014. 4′-Ethynyl-2-fluoro-2′-deoxyadenosine (EFdA) inhibits HIV-1 reverse transcriptase with multiple mechanisms. J Biol Chem 289:24533–24548. 10.1074/jbc.M114.562694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muftuoglu Y, Sohl CD, Mislak AC, Mitsuya H, Sarafianos SG, Anderson KS. 2014. Probing the molecular mechanism of action of the HIV-1 reverse transcriptase inhibitor 4′-ethynyl-2-fluoro-2′-deoxyadenosine (EFdA) using pre-steady-state kinetics. Antiviral Res 106:1–4. 10.1016/j.antiviral.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu VH, Smith RA, Masoum S, Raugi DN, Ba S, Seydi M, Grobler JA, Gottlieb GS, University of Washington–Dakar HIV-2 Study Group. 2017. MK-8591 (4′-ethynyl-2-fluoro-2′-deoxyadenosine) exhibits potent activity against HIV-2 isolates and drug-resistant HIV-2 mutants in culture. Antimicrob Agents Chemother 61:e00744-17. 10.1128/AAC.00744-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maeda K, Desai DV, Aoki M, Nakata H, Kodama EN, Mitsuya H. 2013. Delayed emergence of HIV-1 variants resistant to 4′-ethynyl-2-fluoro-2′-deoxyadenosine: comparative sequential passage study with lamivudine, tenofovir, emtricitabine and BMS-986001. Antivir Ther 19:179–189. 10.3851/IMP2697. [DOI] [PubMed] [Google Scholar]
- 42.Oliveira M, Brenner BG, Xu H, Ibanescu RI, Mesplède T, Wainberg MA. 2017. M184I/V substitutions and E138K/M184I/V double substitutions in HIV reverse transcriptase do not significantly affect the antiviral activity of EFdA. J Antimicrob Chemother 72:3008–3011. 10.1093/jac/dkx280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michailidis E, Ryan EM, Hachiya A, Kirby KA, Marchand B, Leslie MD, Huber AD, Ong YT, Jackson JC, Singh K, Kodama EN, Mitsuya H, Parniak MA, Sarafianos SG. 2013. Hypersusceptibility mechanism of tenofovir-resistant HIV to EFdA. Retrovirology 10:65. 10.1186/1742-4690-10-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stoddart CA, Galkina SA, Joshi P, Kosikova G, Moreno ME, Rivera JM, Sloan B, Reeve AB, Sarafianos SG, Murphey-Corb M, Parniak MA. 2015. Oral administration of the nucleoside EFdA (4′-ethynyl-2-fluoro-2′-deoxyadenosine) provides rapid suppression of HIV viremia in humanized mice and favorable pharmacokinetic properties in mice and the rhesus macaque. Antimicrob Agents Chemother 59:4190–4198. 10.1128/AAC.05036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hattori S, Ide K, Nakata H, Harada H, Suzu S, Ashida N, Kohgo S, Hayakawa H, Mitsuya H, Okada S. 2009. Potent activity of a nucleoside reverse transcriptase inhibitor, 4′-ethynyl-2-fluoro-2′-deoxyadenosine, against human immunodeficiency virus type 1 infection in a model using human peripheral blood mononuclear cell-transplanted NOD/SCID Janus kinase 3 Kno. Antimicrob Agents Chemother 53:3887–3893. 10.1128/AAC.00270-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shanmugasundaram U, Kovarova M, Ho PT, Schramm N, Wahl A, Parniak MA, Garcia JV. 2016. Efficient inhibition of HIV replication in the gastrointestinal and female reproductive tracts of humanized BLT mice by EFdA. PLoS One 11:e0159517. 10.1371/journal.pone.0159517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murphey-Corb M, Rajakumar P, Michael H, Nyaundi J, Didier PJ, Reeve AB, Mitsuya H, Sarafianos SG, Parniak MA. 2012. Response of simian immunodeficiency virus to the novel nucleoside reverse transcriptase inhibitor 4′-ethynyl-2-fluoro-2′-deoxyadenosine in vitro and in vivo. Antimicrob Agents Chemother 56:4707–4712. 10.1128/AAC.00723-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matthews RP, Barrett SE, Patel M, Zhu W, Fillgrove KL, Haspeslagh L, Chen G, Levine V, Zhang S, Goodey A, Forster SP, Vargo R, Grobler JA, Stoch SA, Iwamoto M. 2019. First-in-human trial of MK-8591-eluting implants demonstrates concentrations suitable for HIV prophylaxis for at least one year. J Int AIDS Soc. https://programme.ias2019.org/Abstract/Abstract/4843. [Google Scholar]
- 49.Grobler J, Friedman E, Barrett SE, Wood SL, Ankrom W, Fillgrove KL, Lai M-T, Gindy M, Iwamoto M, Hazuda DJ. 2016. Long-acting oral and parenteral dosing of MK-8591 for HIV treatment or prophylaxis. Top Antivir Med. https://www.croiconference.org/abstract/long-acting-oral-and-parenteral-dosing-mk-8591-hiv-treatment-or-prophylaxis/. [Google Scholar]
- 50.Sohl CD, Singh K, Kasiviswanathan R, Copeland WC, Mitsuya H, Sarafianos SG, Anderson KS. 2012. Mechanism of interaction of human mitochondrial DNA polymerase γ with the novel nucleoside reverse transcriptase inhibitor 4′-ethynyl-2-fluoro- 2′-deoxyadenosine indicates a low potential for host toxicity. Antimicrob Agents Chemother 56:1630–1634. 10.1128/AAC.05729-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grobler J, Friedman E, Barrett SE, Wood SL, Ankrom W, Fillgrove KL, Lai M-T, Gindy M, Iwamoto M, Hazuda DJ. 2017. MK-8591 concentrations at sites of HIV transmission and replication. Top Antivir Med. https://www.natap.org/2017/CROI/croi_92.htm. [Google Scholar]
- 52.Barrett SE, Teller RS, Forster SP, Li L, Mackey MA, Skomski D, Yang Z, Fillgrove KL, Doto GJ, Wood SL, Lebron J, Grobler JA, Sanchez RI, Liu Z, Lu B, Niu T, Sun L, Gindy ME. 2018. Extended-duration MK-8591-eluting implant as a candidate for HIV treatment and prevention. Antimicrob Agents Chemother 62:e01058-18. 10.1128/AAC.01058-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grobler JA, Fillgrove K, Hazuda D, Huang Q, Lai MT, Matthews RP, Rudd DJ, Vargo R. 2019. MK-8591 potency and PK provide high inhibitory quotients at low doses QD and QW. Conference on Retroviruses and Opportunistic Infections (CROI), San Francisco, CA. [Google Scholar]
- 54.Kottle M, Ross J, Danenbaum P, McConnell I. 2021. Gilead and Merck announce agreement to jointly develop and commercialize long-acting, investigational treatment combinations of lenacapavir and islatravir in HIV. Businesswire. [Google Scholar]
- 55.Kodama EI, Kohgo S, Kitano K, Machida H, Gatanaga H, Shigeta S, Matsuoka M, Ohrui H, Mitsuya H. 2001. 4′-Ethynyl nucleoside analogs: potent inhibitors of multidrug-resistant human immunodeficiency virus variants in vitro. Antimicrob Agents Chemother 45:1539–1546. 10.1128/AAC.45.5.1539-1546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang G, Wang J, Cheng Y, Dutschman GE, Tanaka H, Baba M, Cheng YC. 2008. Mechanism of inhibition of human immunodeficiency virus type 1 reverse transcriptase by a stavudine analogue, 4′-ethynyl stavudine triphosphate. Antimicrob Agents Chemother 52:2035–2042. 10.1128/AAC.00083-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kawamoto A, Kodama E, Sarafianos SG, Sakagami Y, Kohgo S, Kitano K, Ashida N, Iwai Y, Hayakawa H, Nakata H, Mitsuya H, Arnold E, Matsuoka M. 2008. 2′-Deoxy-4′-C-ethynyl-2-halo-adenosines active against drug-resistant human immunodeficiency virus type 1 variants. Int J Biochem Cell Biol 40:2410–2420. 10.1016/j.biocel.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 58.Sarafianos SG, Das K, Clark AD, Ding J, Boyer PL, Hughes SH, Arnold E. 1999. Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with β-branched amino acids. Proc Natl Acad Sci USA 96:10027–10032. 10.1073/pnas.96.18.10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nitanda T, Wang X, Kumamoto H, Haraguchi K, Tanaka H, Cheng YC, Baba M. 2005. Anti-human immunodeficiency virus type 1 activity and resistance profile of 2′,3′-didehydro-3′-deoxy-4′-ethynylthymidine in vitro. Antimicrob Agents Chemother 49:3355–3360. 10.1128/AAC.49.8.3355-3360.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deval J, White KL, Miller MD, Parkin NT, Courcambeck J, Halfon P, Selmi B, Boretto J, Canard B. 2004. Mechanistic basis for reduced viral and enzymatic fitness of HIV-1 reverse transcriptase containing both K65R and M184V mutations. J Biol Chem 279:509–516. 10.1074/jbc.M308806200. [DOI] [PubMed] [Google Scholar]
- 61.Hu Z, Kuritzkes DR. 2011. Interaction of reverse transcriptase (RT) mutations conferring resistance to lamivudine and etravirine: effects on fitness and RT activity of human immunodeficiency virus type 1. J Virol 85:11309–11314. 10.1128/JVI.05578-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Andreatta KN, Goodman DD, Miller MD, White KL. 2015. Reduced viral fitness and lack of cross-class resistance with integrase strand transfer inhibitor and nucleoside reverse transcriptase inhibitor resistance mutations. Antimicrob Agents Chemother 59:3441–3449. 10.1128/AAC.00040-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Modrzejewski KA, Herman RA. 2004. Emtricitabine: a once-daily nucleoside reverse transcriptase inhibitor. Ann Pharmacother 38:1006–1014. 10.1345/aph.1D302. [DOI] [PubMed] [Google Scholar]
- 64.Frampton JE, Perry CM. 2005. Emtricitabine: a review of its use in the management of HIV infection. Drugs 65:1427–1448. 10.2165/00003495-200565100-00008. [DOI] [PubMed] [Google Scholar]
- 65.Nelson M, Schiavone M. 2004. Emtricitabine (FTC) for the treatment of HIV infection. Int J Clin Pract 58:504–510. 10.1111/j.1368-5031.2004.00100.x. [DOI] [PubMed] [Google Scholar]
- 66.Das K, Arnold E. 2013. HIV-1 reverse transcriptase and antiviral drug resistance. Part 2. Curr Opin Virol 3:119–128. 10.1016/j.coviro.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cilento ME, Kirby KA, Sarafianos SG. 2021. Avoiding drug resistance in HIV reverse transcriptase. Chem Rev 121:3271–3296. 10.1021/acs.chemrev.0c00967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Diamond T, Ngo W, Xu M, Goh SL, Rodriguez S, Lai M-T, Asante-Appiah E, Grobler J. 2020. Islatravir selects for HIV-1 variants in MT4-GFP cells that profoundly reduce replicative capacity in peripheral blood mononuclear Cells. HIV Glasgow 2020, Glasgow, Scotland. [Google Scholar]
- 69.Van Cor-Hosmer SK, Daddacha W, Kelly Z, Tsurumi A, Kennedy EM, Kim B. 2012. The impact of molecular manipulation in residue 114 of human immunodeficiency virus type-1 reverse transcriptase on dNTP substrate binding and viral replication. Virology 422:393–401. 10.1016/j.virol.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cases-Gonzalez CE, Menéndez-Arias L. 2005. Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114. Biochem J 387:221–229. 10.1042/BJ20041056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Takamatsu Y, Das D, Kohgo S, Hayashi H, Delino NS, Sarafianos SG, Mitsuya H, Maeda K. 2018. The high genetic barrier of EFdA/MK-8591 stems from strong interactions with the active site of drug-resistant HIV-1 reverse transcriptase. Cell Chem Biol 25:1268–1278. 10.1016/j.chembiol.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rhee SY, Kantor R, Katzenstein DA, Camacho R, Morris L, Sirivichayakul S, Jorgensen L, Brigido LF, Schapiro JM, Shafer RW, International Non Subtype B HIV-1 Working Group. 2006. HIV-1 pol mutation frequency by subtype and treatment experience: extension of the HIVseq program to seven non-B subtypes. AIDS 20:643–651. 10.1097/01.aids.0000216363.36786.2b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rhee SY, Gonzales MJ, Kantor R, Betts BJ, Ravela J, Shafer RW. 2003. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res 31:298–303. 10.1093/nar/gkg100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Julias JG, McWilliams MJ, Sarafianos SG, Alvord WG, Arnold E, Hughes SH. 2003. Mutation of amino acids in the connection domain of human immunodeficiency virus type 1 reverse transcriptase that contact the template-primer affects RNase H activity. J Virol 77:8548–8554. 10.1128/jvi.77.15.8548-8554.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sarafianos SG, Das K, Tantillo C, Clark AD, Ding J, Whitcomb JM, Boyer PL, Hughes SH, Arnold E. 2001. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J 20:1449–1461. 10.1093/emboj/20.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ding J, Das K, Hsiou Y, Sarafianos SG, Clark AD, Jacobo-Molina A, Tantillo C, Hughes SH, Arnold E. 1998. Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template-primer and an antibody Fab fragment at 2.8 Å resolution. J Mol Biol 284:1095–1111. 10.1006/jmbi.1998.2208. [DOI] [PubMed] [Google Scholar]
- 77.Schuckmann MM, Marchand B, Hachiya A, Kodama EN, Kirby KA, Singh K, Sarafianos SG. 2010. The N348I mutation at the connection subdomain of HIV-1 reverse transcriptase decreases binding to nevirapine. J Biol Chem 285:38700–38709. 10.1074/jbc.M110.153783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lengruber RB, Delviks-Frankenberry KA, Nikolenko GN, Baumann J, Santos AF, Pathak VK, Soares MA. 2011. Phenotypic characterization of drug resistance-associated mutations in HIV-1 RT connection and RNase H domains and their correlation with thymidine analogue mutations. J Antimicrob Chemother 66:702–708. 10.1093/jac/dkr005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tachedjian G, French M, Mills J. 1998. Coresistance to zidovudine and foscarnet is associated with multiple mutations in the human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother 42:3038–3043. 10.1128/AAC.42.11.3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Delviks-Frankenberry KA, Nikolenko GN, Boyer PL, Hughes SH, Coffin JM, Jere A, Pathak VK. 2008. HIV-1 reverse transcriptase connection subdomain mutations reduce template RNA degradation and enhance AZT excision. Proc Natl Acad Sci USA 105:10943–10948. 10.1073/pnas.0804660105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brehm JH, Koontz D, Meteer JD, Pathak V, Sluis-Cremer N, Mellors JW. 2007. Selection of mutations in the connection and RNase H domains of human immunodeficiency virus type 1 reverse transcriptase that increase resistance to 3′-azido-3′-dideoxythymidine. J Virol 81:7852–7859. 10.1128/JVI.02203-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.von Wyl V, Ehteshami M, Demeter LM, Bürgisser P, Nijhuis M, Symons J, Yerly S, Böni J, Klimkait T, Schuurman R, Ledergerber B, Götte M, Günthard HF. 2010. HIV‐1 reverse transcriptase connection domain mutations: dynamics of emergence and implications for success of combination antiretroviral therapy. Clin Infect Dis 51:620–628. 10.1086/655764. [DOI] [PubMed] [Google Scholar]
- 83.Chiang CC, Tseng YT, Huang KJ, Pan YY, Wang CT. 2012. Mutations in the HIV-1 reverse transcriptase tryptophan repeat motif affect virion maturation and Gag-Pol packaging. Virology 422:278–287. 10.1016/j.virol.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 84.Tachedjian G, Aronson HEG, De Los Santos M, Seehra J, McCoy JM, Goff SP. 2003. Role of residues in the tryptophan repeat motif for HIV-1 reverse transcriptase dimerization. J Mol Biol 326:381–396. 10.1016/S0022-2836(02)01433-X. [DOI] [PubMed] [Google Scholar]
- 85.Murphey-Corb M, Rajakumar P, Michael H, Nyaundi J, Didier PJ, Reeve AB, Mitsuya H, Sarafianos SG, Parniak MA. 2012. Response of simian immunodeficiency virus to the novel nucleoside reverse transcriptase inhibitor 4′-ethynyl-2-fluoro-2′-deoxyadenosine in vitro and in vivo. Antimicrob Agents Chemother 56:4707–4712. 10.1128/AAC.00723-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Boucher CA, Cammack N, Schipper P, Schuurman R, Rouse P, Wainberg MA, Cameron JM. 1993. High-level resistance to (–) enantiomeric 2′-deoxy-3′-thiacytidine in vitro is due to one amino acid substitution in the catalytic site of human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother 37:2231–2234. 10.1128/AAC.37.10.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Boyer PL, Sarafianos SG, Arnold E, Hughes SH. 2002. The M184V mutation reduces the selective excision of zidovudine 5′-monophosphate (AZTMP) by the reverse transcriptase of human immunodeficiency virus type 1. J Virol 76:3248–3256. 10.1128/jvi.76.7.3248-3256.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Charneau P, Mirambeau G, Roux P, Paulous S, Buc H, Clavel F. 1994. HIV-1 reverse transcription: a termination step at the center of the genome. J Mol Biol 241:651–662. 10.1006/jmbi.1994.1542. [DOI] [PubMed] [Google Scholar]
- 89.Haertle T, Carrera CJ, Wasson DB, Sowers LC, Richman DD, Carson DA. 1988. Metabolism and anti-human immunodeficiency virus-1 activity of 2-halo-2′,3′-dideoxyadenosine derivatives. J Biol Chem 263:5870–5875. 10.1016/S0021-9258(18)60646-5. [DOI] [PubMed] [Google Scholar]
- 90.Boufford DE, Spongberg SA. 1983. Eastern Asian-Eastern North American phytogeographical relationships: a history from the time of Linnaeus to the twentieth century. Ann Missouri Bot Gard 70:423–439. 10.2307/2992081. [DOI] [Google Scholar]
- 91.Graham FL, Smiley J, Russell WC, Nairn R. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol 36:59–72. 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 92.Pear WS, Nolan GP, Scott ML, Baltimore D. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA 90:8392–8396. 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rosa A, Chande A, Ziglio S, De Sanctis V, Bertorelli R, Goh SL, McCauley SM, Nowosielska A, Antonarakis SE, Luban J, Santoni FA, Pizzato M. 2015. HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature 526:212–217. 10.1038/nature15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Takeuchi Y, McClure MO, Pizzato M. 2008. Identification of gammaretroviruses constitutively released from cell lines used for human immunodeficiency virus research. J Virol 82:12585–12588. 10.1128/JVI.01726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother 46:1896–1905. 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ochsenbauer-Jambor C, Jones J, Heil M, Zammit KP, Kutsch O. 2006. T-cell line for HIV drug screening using EGFP as a quantitative marker of HIV-1 replication. Biotechniques 40:91–100. 10.2144/000112072. [DOI] [PubMed] [Google Scholar]
- 97.Kutsch O, Levy DN, Bates PJ, Decker J, Kosloff BR, Shaw GM, Priebe W, Benveniste EN. 2004. Bis-anthracycline antibiotics inhibit human immunodeficiency virus type 1 transcription. Antimicrob Agents Chemother 48:1652–1663. 10.1128/AAC.48.5.1652-1663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Reed LJ, Muench H. 1938. A simple method of estimating fifty percent end points. Am J Hyg 27:493–497. 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 99.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and ClustalX version 2. Bioinformatics 23:2947–2948. 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 100.Bauman JD, Das K, Ho WC, Baweja M, Himmel DM, Clark AD, Oren DA, Boyer PL, Hughes SH, Shatkin AJ, Arnold E. 2008. Crystal engineering of HIV-1 reverse transcriptase for structure-based drug design. Nucleic Acids Res 36:5083–5092. 10.1093/nar/gkn464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ndongwe TP, Adedeji AO, Michailidis E, Ong YT, Hachiya A, Marchand B, Ryan EM, Rai DK, Kirby KA, Whatley AS, Burke DH, Johnson M, Ding S, Zheng YM, Liu SL, Kodama EI, Delviks-Frankenberry KA, Pathak VK, Mitsuya H, Parniak MA, Singh K, Sarafianos SG. 2012. Biochemical, inhibition and inhibitor resistance studies of xenotropic murine leukemia virus-related virus reverse transcriptase. Nucleic Acids Res 40:345–359. 10.1093/nar/gkr694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kirby KA, Marchand B, Ong YT, Ndongwe TP, Hachiya A, Michailidis E, Leslie MD, Sietsema DV, Fetterly TL, Dorst CA, Singh K, Wang Z, Parniak MA, Sarafianos SG. 2012. Structural and inhibition studies of the RNase H function of xenotropic murine leukemia virus-related virus reverse transcriptase. Antimicrob Agents Chemother 56:2048–2061. 10.1128/AAC.06000-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sarafianos SG, Clark AD, Tuske S, Squire CJ, Das K, Sheng D, Ilankumaran P, Ramesha AR, Kroth H, Sayer JM, Jerina DM, Boyer PL, Hughes SH, Arnold E. 2003. Trapping HIV-1 reverse transcriptase before and after translocation on DNA. J Biol Chem 278:16280–16288. 10.1074/jbc.M212911200. [DOI] [PubMed] [Google Scholar]
- 104.Le Grice SFJ, Grüninger‐Leitch F. 1990. Rapid purification of homodimer and heterodimer HIV‐1 reverse transcriptase by metal chelate affinity chromatography. Eur J Biochem 187:307–314. 10.1111/j.1432-1033.1990.tb15306.x. [DOI] [PubMed] [Google Scholar]
- 105.Vernekar SV, Tang J, Wu B, Huber AD, Casey MC, Myshakina N, Wilson DJ, Kankanala J, Kirby KA, Parniak MA, Sarafianos SG, Wang Z. 2017. Double-winged 3-hydroxypyrimidine-2,4-diones: potent and selective inhibition against HIV-1 RNase H with significant antiviral activity. J Med Chem 60:5045–5056. 10.1021/acs.jmedchem.7b00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tang J, Do HT, Huber AD, Casey MC, Kirby KA, Wilson DJ, Kankanala J, Parniak MA, Sarafianos SG, Wang Z. 2019. Pharmacophore-based design of novel 3-hydroxypyrimidine-2,4-dione subtypes as inhibitors of HIV reverse transcriptase-associated RNase H: tolerance of a nonflexible linker. Eur J Med Chem 166:390–399. 10.1016/j.ejmech.2019.01.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang L, Tang J, Huber AD, Casey MC, Kirby KA, Wilson DJ, Kankanala J, Parniak MA, Sarafianos SG, Wang Z. 2018. 6-Biphenylmethyl-3-hydroxypyrimidine-2,4-diones potently and selectively inhibited HIV reverse transcriptase-associated RNase H. Eur J Med Chem 156:680–691. 10.1016/j.ejmech.2018.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kankanala J, Kirby KA, Huber AD, Casey MC, Wilson DJ, Sarafianos SG, Wang Z. 2017. Design, synthesis and biological evaluations of N-hydroxy thienopyrimidine-2,4-diones as inhibitors of HIV reverse transcriptase-associated RNase H. Eur J Med Chem 141:149–161. 10.1016/j.ejmech.2017.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tang J, Kirby KA, Huber AD, Casey MC, Ji J, Wilson DJ, Sarafianos SG, Wang Z. 2017. 6-Cyclohexylmethyl-3-hydroxypyrimidine-2,4-dione as an inhibitor scaffold of HIV reverse transcriptase: impacts of the 3-OH on inhibiting RNase H and polymerase. Eur J Med Chem 128:168–179. 10.1016/j.ejmech.2017.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Singh K, Marchand B, Rai DK, Sharma B, Michailidis E, Ryan EM, Matzek KB, Leslie MD, Hagedorn AN, Li Z, Norden PR, Hachiya A, Parniak MA, Xu HT, Wainberg MA, Sarafianos SG. 2012. Biochemical mechanism of HIV-1 resistance to rilpivirine. J Biol Chem 287:38110–38123. 10.1074/jbc.M112.398180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Mutation sequences for BankIt2501100 Seq1 to Seq14 have been deposited in GenBank under accession no. OK263149 to OK263162.