ABSTRACT
Infection by multidrug-resistant (MDR) Acinetobacter baumannii is one of the major causes of hospital-acquired infections worldwide. The ability of A. baumannii to survive in adverse conditions as well as its extensive antimicrobial resistance make it one of the most difficult to treat pathogens associated with high mortality rates. The aim of this study was to investigate MDR A. baumannii that has spread among pediatric cancer patients in the Children’s Cancer Hospital Egypt 57357. Whole-genome sequencing was used to characterize 31 MDR A. baumannii clinical isolates. Phenotypically, the isolates were MDR, with four isolates showing resistance to the last-resort antibiotic colistin. Multilocus sequence typing showed the presence of eight clonal groups, two of which were previously reported to cause outbreaks in Egypt, and one novel sequence type (ST), Oxf-ST2246. Identification of the circulating plasmids showed the presence of two plasmid lineages in the isolates, strongly governed by sequence type. A large number of antimicrobial genes with a range of resistance mechanisms were detected in the isolates, including β-lactamases and antibiotic efflux pumps. Analysis of insertion sequences (ISs) revealed the presence of ISAba1 and ISAba125 in all the samples, which amplify β-lactamase expression, causing extensive carbapenem resistance. Mutation analysis was used to decipher underlying mutations responsible for colistin resistance and revealed novel mutations in several outer membrane proteins, in addition to previously reported mutations in pmrB. Altogether, understanding the transmissibility of A. baumannii as well as its resistance and virulence mechanisms will help develop novel treatment options for better management of hospital-acquired infections.
IMPORTANCE Acinetobacter baumannii represents a major health threat, in particular among immunocompromised cancer patients. The rise in carbapenem-resistant A. baumannii, and the development of resistance to the last-resort antimicrobial agent colistin, complicates the management of A. baumannii outbreaks and increases mortality rates. Here, we investigate 31 multidrug resistant A. baumannii isolates from pediatric cancer patients in Children’s Cancer Hospital Egypt (CCHE) 57357 via whole-genome sequencing. Multilocus sequence typing (MLST) showed the presence of eight clonal groups including a novel sequence type. In silico detection of antimicrobial-resistant genes and virulence factors revealed a strong correlation between certain virulence genes and mortality as well as several point mutations in outer membrane proteins contributing to colistin resistance. Detection of CRISPR/Cas sequences in the majority of the samples was strongly correlated with the presence of prophage sequences and associated with failure of bacteriophage therapy. Altogether, understanding the genetic makeup of circulating A. baumannii is essential for better management of outbreaks.
KEYWORDS: Acinetobacter baumannii, antimicrobial resistance genes, whole-genome sequencing
INTRODUCTION
Acinetobacter baumannii has been recognized as one of the major causes of hospital-acquired infections (1, 2). Its spectrum of illness is wide and includes bloodstream infection, pneumonia, and endocarditis (3). A. baumannii has a remarkable capacity to develop antimicrobial resistance, which is largely related to mobile genetic elements, such as insertion sequences, plasmids, and antibiotic resistance islands (1). The rise in carbapenem-resistant A. baumannii (CRAB) represents a major health threat, as it prohibits the use of the broad-spectrum carbapenem antibiotics that are reserved for the treatment of severe infections in hospitalized patients. The major mechanism of carbapenem resistance in A. baumannii is through the production of class B metallo-β-lactamases such as blaNDM, blaVIM, blaIMP, and blaSIM (4, 5), as well as carbapenem-hydrolyzing class D β-lactamases (CHDLs), in particular subgroups blaOXA-23, blaOXA-24, blaOXA-51, and blaOXA-68 (6–8).
The prevalence of CRAB in Egypt is the highest in the Middle East (9), in particular among immunocompromised and intensive care unit (ICU) patients (10, 11). Several studies have investigated the dissemination of carbapenemases such as blaNDM, blaOXA-51-like, and blaOXA-23-like in A. baumannii and have reported a prevalence of more than 70% among isolates in Egypt (12, 13). The propagation of carbapenemase-encoding genes is in large part due to their association with integrons and plasmids (14, 15), allowing their rapid transmission and accumulation. Insertion sequences (ISs) in the region surrounding these genes cause their overexpression by providing promoter sequences, resulting in higher MICs for carbapenems (16–20).
The cationic peptides polymyxins B and E (colistin) have been available for clinical use since the 1950s due to their effectiveness against Gram-negative bacteria (21). Systemic toxicity due to the administration of polymyxin B, and to a lesser extent colistin, led to their replacement by other antimicrobial drugs (21, 22). The recent prevalence of extensively drug-resistant pathogens such as A. baumannii caused the reemergence of polymyxins as last-resort antibiotic therapy. Polymyxins exert their antimicrobial activity by binding to the lipopolysaccharide (LPS) layer on the outer membrane of Gram-negative bacteria, causing it to swell, which subsequently disrupts the osmotic balance of the cell, causing its death (22). Although still uncommon, the increasing use of colistin caused the emergence of colistin-resistant (ColR) strains, increasing the incidence of pan-drug-resistant pathogens, which are resistant to all available antimicrobials (23). Reports of ColR strains emerging in Egypt have surfaced in the past few years, complicating the management of CRAB infections further and increasing mortality rates (12, 24).
The ability of A. baumannii to develop multidrug resistance, survive on inanimate surfaces, and resist serum killing, as well as its aggressiveness in host epithelial cell invasion makes it one of the most important enemies in intensive care units, being the cause of many outbreaks (1, 25). A. baumannii owes its remarkable aggressiveness and tenacity to an impressive repertoire of virulence genes. Outer membrane porins, such as OmpA, represent a major virulence factor that allows for adherence to and invasion of host epithelial cells (26). Biofilm formation, which depends on AdeFGH efflux pumps, biofilm-associated protein (Bap), Csu fimbriae, poly-β-1,6-N-acetylglucosamine (PNAG) surface polysaccharides, pili, and OmpA, underlies the ability of A. baumannii to survive on abiotic surfaces, allowing it to persist and spread in the hospital environment (27). Capsular and lipopolysaccharides (LPS) in the cell wall of A. baumannii modulate the bacterial resistance to peptide antibiotics (28), evasion of the immune response, and thus, serum resistance (29). Phospholipases, porins, and proteases secreted through outer membrane vesicles (OMVs) aid in the invasion of host epithelial cells (30, 31).
Different methods have been used to investigate outbreaks of A. baumannii within hospitals, such as pulsed-field gel electrophoresis (PFGE) and multilocus sequencing typing (MLST) (32). However, such techniques have important limitations for source tracking of infections in hospitals, as they examine specific sites in the genome, which may result in false clustering of unrelated strains (33). In the past decade, whole-genome sequencing (WGS) has been used to investigate hospital outbreaks using ultrahigh resolution, thus discriminating two isolates differing by just a single mutation (34, 35). Benchtop sequencing instruments now offer a cost-effective approach for bringing bacterial WGS to the clinical environment (36).
The need for new antimicrobials has never been more compelling. Understanding the modes of bacterial resistance and the development of novel antimicrobials is urgently required. In this paper, we utilized next-generation sequencing technology to investigate 31 A. baumannii isolates from clinical samples of 27 hospitalized patients in a tertiary care hospital through whole-genome sequence comparisons and phylogenetic analyses. This was followed by in silico detection of the A. baumannii resistomes and virulomes, as well as CRISPR/Cas and phage sequences. Discerning the genetic elements encompassed in the A. baumannii genome would aid in understanding the transmissibility of the bacteria and the development of prospective treatment options.
RESULTS
Clinical characteristics.
A total of 31 isolates of A. baumannii were obtained from 27 infected pediatric cancer patients with different types of malignancies from Children’s Cancer Hospital Egypt (CCHE) 57357. The recovery rate was 22% (n = 7) and was determined if negative A. baumannii cultures were obtained after treatment. Phenotypic characterization of all 31 isolates by antibiotic susceptibility testing (AST) showed multidrug resistance. Isolates were considered multidrug-resistant (MDR) if they were nonsusceptible to at least one agent in three or more antimicrobial categories and extensively drug-resistant (XDR) if they were nonsusceptible to all but two or fewer antimicrobial classes (23).
Sequence typing.
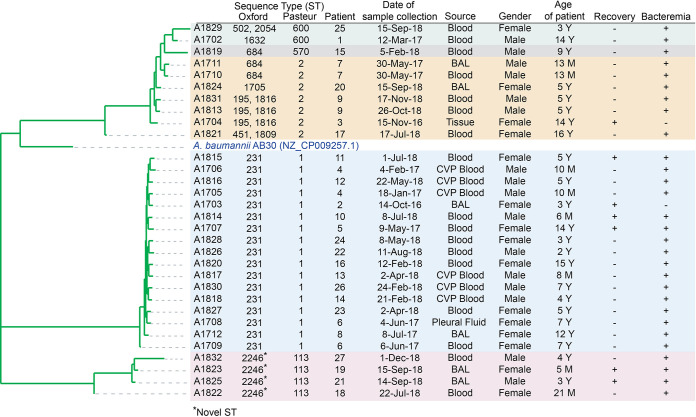
In silico MLST analysis, using the Oxford scheme, revealed the presence of eight groups, Oxf-ST231, Oxf-ST1632, Oxf-ST684, Oxf-ST1705, Oxf-ST195, Oxf-ST451, and Oxf-ST502 and one novel ST, designated Oxf-ST2246. The Oxford scheme detected an alternative gdhB locus (gdhB2) in five isolates, which was classified as gdhB allele 189 in addition to the allele of the proper gdhB locus (37). This led to the detection of two sequence types (STs) in these five isolates, Oxf-ST2054 and Oxf-ST502 in A1829, Oxf-ST1816 and Oxf-ST195 in A1831, A1813, and A1704, and Oxf-ST1809 and to Oxf-ST451 in A1821 (Fig. 1). On the other hand, the Pasteur scheme revealed five groups of STs, Pas-ST1, Pas-ST2, Pas-ST570, Pas-ST600, and Pas-ST113. This is expected, given that the Oxford MLST scheme is more capable at differentiating between closely related isolates (37). However, due to the gdhB paralogy in the Oxford scheme, which complicates ST detection, the Pasteur scheme will be used here. The phylogenetic relationship between different isolates is shown in Fig. 1.
FIG 1.
Phylogenetic tree and demographic data. Merged reads were mapped to the A. baumannii reference (GenBank accession number NZ_CP009257.1) to create a phylogenetic tree. The similarity distance between the isolates was analyzed based on SNP analysis and appended to a table showing the clinical data of the patients, and the sequence type of the isolated A. baumannii is appended. Recovery, + for negative cultures after treatment, – for persistent positive cultures until death of patient; BAL, bronchioalveolar lavage; CVP, central venous port. Isolates are color-shaded based on Pasteur ST.
Plasmid distribution in the different isolates.
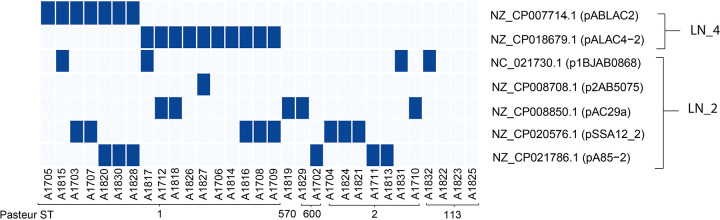
Seven plasmids were detected in the isolates, belonging to two different lineages (LN_2 and LN_4) (38; Fig. 2). Five plasmids (p1BJAB0868, p2AB5075, pAC29a, pSSA12_2, and pA85-2) belong to LN_2, and share 99.8% sequence homology. The other two plasmids (pABLAC2 and pALAC4-2) belong to LN_4 and share 100% sequence identity. The distribution of pABLAC2 and pALAC4-2 is confined to isolates of ST1.
FIG 2.
Heat map showing the presence or absence of each plasmid in each sample. Dark blue shows the presence of the plasmid, while light blue indicates its absence.
Antibiotic resistance profile.
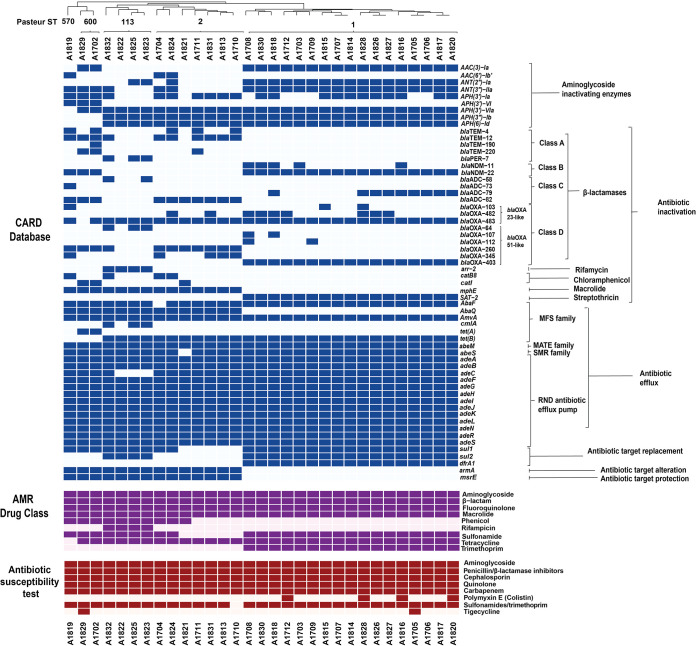
AST was performed on the 31 A. baumannii isolates, and all of them were found to be multidrug-resistant (23; and Fig. 3). Almost all isolates were resistant to the sulfonamide-trimethoprim combination, except for isolate A1710. On the other hand, most isolates were susceptible to colistin (except for isolates A1712, A1816, A1820, and A1828) and tigecycline (except for isolates A1705 and A1829).
FIG 3.
Complex heat maps showing the presence or absence of each antibiotic resistance gene and antibiotic resistance drug class and the observed antibiotic susceptibility. The antibiotic resistance genes detected are shown in blue; dark blue shows the presence of the antibiotic resistance gene, while light blue indicates its absence. The antibiotic classes for which the detected genes confer resistance are shown in purple; dark purple shows expected resistance to a drug class, while light purple shows susceptibility. The phenotypic antibiotic resistance based on AST is shown in maroon; dark maroon shows resistance to the antibiotic, while light maroon shows susceptibility.
Antibiotic resistance genes and mobile genetic elements.
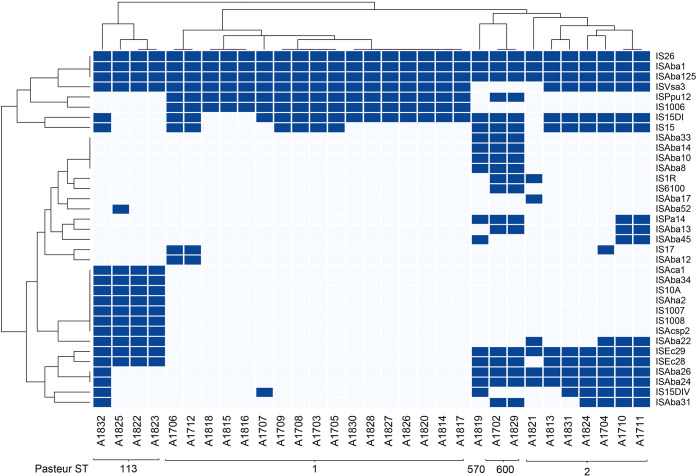
The CARD database, Resfinder, and ISfinder were used to detect different antibiotic resistance genes and ISs in each isolate. The antibiotic resistance genes and ISs detected by WGS are shown in Fig. 3 and 4, respectively, and the resistance mechanisms are shown in Fig. 5. Figure 3 also shows the expected resistance to antibiotic classes based on antibiotic resistance gene presence (purple) and the actual resistance observed by AST (maroon). Clustering based on the prevalence of antibiotic resistance genes and ISs shows significant grouping based on the different sequence types of the isolates. Resfinder (Fig. S1) detected antibiotic resistance genes similar to CARD, with the exception of efflux pumps, which were not detected by Resfinder.
FIG 4.
Heat map showing the presence or absence of each IS. Dark blue shows the presence of the IS, while light blue indicates its absence.
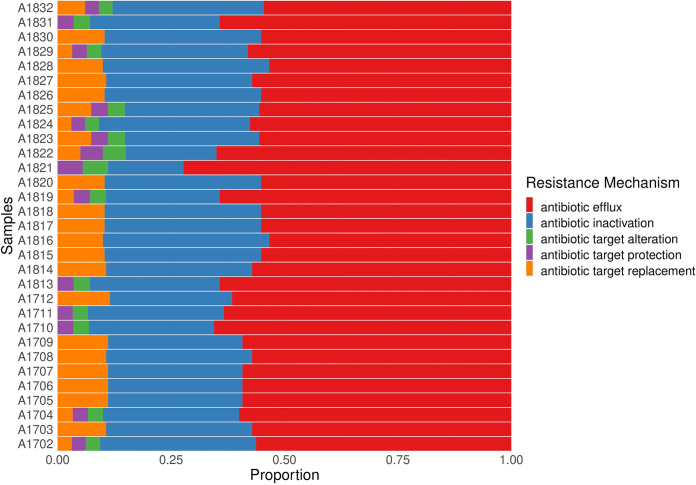
FIG 5.
Stacked bar graph showing the resistance mechanisms represented in each isolate. After grouping of the antibiotic resistance genes by mechanism. Each color represents a mechanism of resistance; red, antibiotic efflux; blue, antibiotic inactivation; green, antibiotic target alteration; purple, antibiotic target protection; orange, antibiotic target replacement.
Heat map showing the presence or absence of each antibiotic resistance gene as detected by the ResFinder database. Dark green shows the presence of the antibiotic resistance gene, while light green indicates its absence. Download FIG S1, TIF file, 1.8 MB (1.8MB, tif) .
Copyright © 2021 Jalal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genes encoding three classes of aminoglycoside-modifying enzymes were detected across the isolates—aminoglycoside adenyltransferases (ANT), aminoglycoside acetyltransferases (AAC), and aminoglycoside phosphotransferases (APH). The plasmid carrying ant-(2″)-1a was predominant in the ST1 group (55% of the isolates, n = 17), whereas the integron-associated ant(3″)-Ia was found across different ST groups (in 74% of the isolates, n = 23). Other aminoglycoside-modifying enzymes were identified in isolates across all ST groups, such as the integron-associated aac(3)-Ia and aac(6′)-1b ′ and five variants of APHs. The most common APH gene was the plasmid-associated aph(3′)-VIa, being detected in 97% (n = 30) of the isolates.
β-lactamases are a class of antibiotic-hydrolyzing enzymes that target the β-lactam ring in penicillins, cephalosporins, carbapenems, and monobactams. Genes encoding several classes of β-lactamases were found in isolates across all ST groups. Extended-spectrum class A β-lactamase (ESBL) genes were only found in a few isolates. This class includes blaPER-7 (3 isolates) and blaTEM variants (12 isolates). Genes encoding class B metallo β-lactamases (MBL) (39), blaNDM, were found in 20 isolates, predominantly belonging to the ST1 group. Genes encoding class C β-lactamases, blaADC, were found distributed across different STs. Finally, genes encoding CHDLs, blaOXA genes, which represent the major carbapenem resistance mechanism in A. baumannii, were found in all of the isolates. Two subgroups of blaOXA genes, intrinsic blaOXA-51-like and acquired blaOXA-23-like, were found in the majority of the isolates. blaOXA-51-like (blaOXA-64, blaOXA-107, blaOXA-112, blaOXA-260, and blaOXA-403) genes were found in 90% of the isolates, whereas blaOXA-23-like (blaOXA-103, blaOXA-482, and blaOXA-483) were found in 97% of the isolates. The presence of ISAba1 upstream of blaOXA genes strongly contributes to antibiotic resistance by overexpressing the blaOXA genes (18, 20, 40, 41). The presence of both ISAba1 and ISAba125 was observed across all isolates, possibly contributing to the carbapenem resistance of these strains (Fig. 3).
Other antibiotic-modifying enzymes were also detected across isolates of different STs. The chloramphenicol acetyltransferase genes, catB8 and catI, were present in 22.6% of the isolates. The rifampin ADP-ribosyl transferase gene arr-2 was only found in isolates belonging to ST113. The macrolide phosphotransferase gene mphE was present in all isolates except those belonging to ST1. The streptothricin acetyltransferase gene (SAT-2) was confined to isolates belonging to ST1.
Genes encoding several classes of efflux pumps that are intrinsic to A. baumannii were detected in the isolates, including resistance-nodulation-division (RND), major facilitator superfamilies (MFS), small multidrug resistance (SMR) family, and multidrug and toxic compound extrusion (MATE) transporter (42). Genes encoding RND family efflux pumps (adeABC, adeFGH, and adeIJK) as well as their regulators (adeL, adeN, and adeRS) were detected in all the isolates. MFS gene abaQ was found in all isolates except those belonging to ST1, whereas cmlA was predominant in ST113. Other members of the MFS were present in almost all the isolates, such as abaF, amvA, and tetAB. abeM and abeS, which belong to the MATE and SMR families, respectively, were found in almost all isolates.
The intrinsic A. baumannii genes sul1 and sul2, which confer resistance to sulfonamides, were found in 84% of the isolates, while the integron-associated dfrA1, which confers resistance to trimethoprim, was only present in ST1 (55% of the isolates). Two genes were absent from ST1 while present in all other STs—the plasmid-associated armA, which provides resistance to aminoglycosides by altering its target 30S ribosomal subunit, and msrE, which provides resistance against erythromycin/streptogramin B by protecting its target 50S ribosomal subunit (Fig. 3 and 5).
Virulence genes in A. baumannii.
The Virulence Factors Database was used to detect virulence factors encoded on both genomic and plasmid DNA in the isolates. As expected for a highly virulent pathogen such as A. baumannii, different classes of virulence genes were detected across all isolates. The gene encoding OmpA, which provides adherence to host cells (43, 44), was detected in all of the isolates. Genes encoding virulence factors that help in biofilm formation, such as the efflux pump AdeFGH (45), the fimbriae CsuABCDE (46), and PNAG polysaccharide biosynthesis gene products PgaABCD (47), were detected in all isolates, except the biofilm-associated protein (bap), which was absent in ST1 isolates and the majority of ST113 isolates. Phospholipases (plc, plcD), which help in host cell lysis (48, 49), were detected in all isolates. Virulence genes responsible for iron uptake through the production of the siderophore acinetobactin (50), such as barAB, basABCFGHI, bauBCDEF, and entE, were found in all isolates except bauA, which was absent in ST1 isolates and the majority of ST113 isolates. Regulatory genes such as those responsible for quorum sensing (abaIR) (51) and the two-component system bfmRS (52) were detected in all isolates (Table 1). We found a significant association between the absence of bauA and bap and recovery in patients with bacteremia (29.4% versus 0%; Table 2), while controlling for colistin sensitivity (22) (r = 0.439, P = 0.032).
TABLE 1.
Virulence factors in each isolate
| Virulence mechanism | Virulence gene | Pasteur ST [no. of isolates (%)] |
All isolates | |||||
|---|---|---|---|---|---|---|---|---|
| ST1 | ST2 | ST600 | ST570 | ST113 | ||||
| Adherence | ompA | 17/17 (100) | 7/7 (100) | 2/2 (100) | 1/1 (100) | 4/4 (100) | 31/31 (100) | |
| Biofilm formation | ||||||||
| Efflux pump | adeF, adeG, adeH | 17/17 (100) | 7/7 (100) | 2/2 (100) | 1/1 (100) | 4/4 (100) | 31/31 (100) | |
| Biofilm-associated protein | bap | 0/17 (0) | 7/7 (100) | 2/2 (100) | 1/1 (100) | 1/4 (25) | 21/31 (67.7) | |
| Csu fimbrae | csuA, csuA/B, csuC, csuD, csuE | 17/17 (100) | 7/7 (100) | 2/2 (100) | 1/1 (100) | 4/4 (100) | 31/31 (100%) | |
| Biofilm formation | pgaA, pgaB, pgaC, pgaD | 17/17 (100) | 7/7 (100) | 2/2 (100) | 1/1 (100) | 4/4 (100) | 31/31 (100%) | |
| Phospholipase | plc, plcD | 17/17 (100) | 7/7 (100) | 2/2 (100) | 1/1 (100) | 4/4 (100) | 31/31 (100%) | |
| Acinetobactin production | barA, barB, basA, basB, basC, basF, basG, basH, basI, basJ, entE | 17/17 (100) | 7/7 (100) | 2/2 (100) | 1/1 (100) | 4/4 (100) | 31/31 (100) | |
| bauA | 0/17 (0) | 7/7 (100) | 2/2 (100) | 1/1 (100) | 1/4 (25) | 21/31 (67.7) | ||
| bauB, bauC, bauD, bauE, bauF | 17/17 (100) | 7/7 (100) | 2/2 (100) | 1/1 (100) | 4/4 (100) | 31/31 (100) | ||
| Regulation | ||||||||
| Quorum sensing | abaI, abaR | 17/17 (100) | 7/7 (100) | 2/2 (100) | 1/1 (100) | 4/4 (100) | 31/31 (100) | |
| Two-component system | bfmR, bfmS | 17/17 (100) | 7/7 (100) | 2/2 (100) | 1/1 (100) | 4/4 (100) | 31/31 (100s) | |
TABLE 2.
Number of recovered patients with bacteremia
| No. of recovered patients with bacteremia |
||
|---|---|---|
| Present (%) | Absent (%) | |
| bauA/bap genes | 0/8 (0) | 5/17 (29.4) |
| ColR | 0/4 (0) | 5/21 (23.8) |
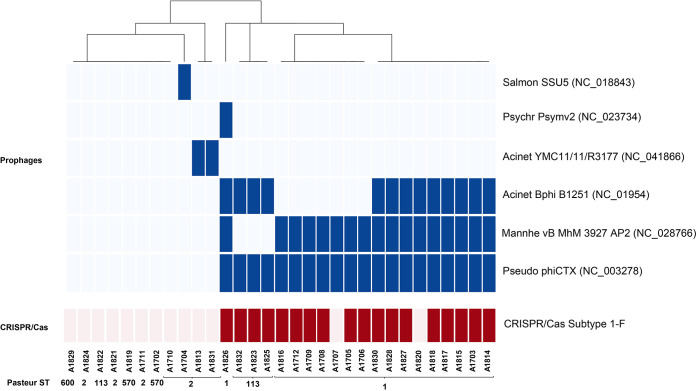
Identification of prophage and CRISPR sequences.
The identification of prophage sequences using the PHASTER tool revealed the presence of intact prophages in 77% (n = 24) of the isolates. The following six intact prophages were detected among all isolates; Pseudo phiCTX (GenBank accession number NC_003278), Mannhe vB MhM 3927AP2 (NC_028766), Acinet Bphi B1251 (NC_019541), Acinet YMC11/11/R3177 (NC_041866), Psychr Psymv2 (NC_023734), and Salmon SSU5 (NC_018843) (Fig. 6).
FIG 6.
Heat map showing the prophage and CRISPR/Cas sequence distribution among isolates.
Identification of the CRISPR regions using the CRISPR-CasFinder tool has revealed the presence of complete CRISPR-Cas subtype I-F in 18 samples, while 3 samples contain CRISPR sequences with no Cas genes. All of the CRISPR sequences identified were 28-nucleotide (nt)-long identical sequences, GTTCATGGCGGCATACGCCATTTAGAAA or TTTCTAAATGGCGTATGCCGCCATGAAC for the forward (+) or reverse (–) direction, respectively. The CRISPR sequences are interspersed among the same 53 spacers in 15 of the isolates all belonging to ST1 or 98 spacers in 3 of the isolates all belonging to ST113.
The presence of intact CRISPR/Cas sequences was predominant in ST1 and ST113 and was found to strongly correlate with the presence of prophages (P = 0.00065), in particular, Pseudo phiCTX (P = 0.00001), Mannhe vB MhM 3927AP2 (P = 0.00026), and Acinet Bphi B1251 (P = 0.0024). The prophages Acinet YMC11/11/R3177, Psychr Psymv2, and Salmon SSU5, on the other hand, were not associated with the presence of CRISPR/Cas.
Emergence of colistin-resistant strains.
Four out of the 31 A. baumannii isolates (A1712, A1816, A1820, and A1828) were found by AST to be colistin resistant. Isolates A1712 and A1816 were obtained from patients that had colistin-susceptible strains prior to colistin administration, suggesting the development of resistance upon colistin exposure. Few mechanisms of colistin resistance have been previously reported, such as the loss of LPS (28) and, more commonly, the modification of lipid A by the addition of phosphoethanolamine (pEtN), by PmrA and PmrB, altering its charge and thus its interaction with colistin (21, 22, 53–57). Horizontal gene transfer of the Escherichia coli pEtN transferase genes mcr1 and mcr4.3 was recently found to contribute to colistin resistance in A. baumannii (58, 59).
The LPS biosynthesis genes, lpsB, lpxA, lpxB, lpxC, lpxD, lpxL, and lpxM, were analyzed using in silico gene analysis and were found in an unmutated intact form in all of the ColR isolates. No mcr genes or associated plasmids were detected during antibiotic resistance gene detection. Mutation analysis was then performed to identify unique mutations that only exist in ColR strains, not in ColS strains (Table 2 and Table S3). Previously reported mutations in pmrA, L20F and M12I (53, 54, 60), were found in isolates A1712 and A1828, respectively (Table 3). Two mutations were identified in lptF, R30H and N82D, a lipopolysaccharide ABC transporter permease previously implicated with colistin resistance (60). Several new mutations in outer membrane proteins were identified, such as the OmpW and OmpA family proteins, the outer membrane porin OprD, the pilin pilA, the multidrug efflux MFS transporter AmvA, TonB-dependent receptor, a LysE family translocator, and a DMT family transporter. Mutations in other proteins, including hypothetical proteins, were also found and are also reported in Table S3.
TABLE 3.
Unique mutations in colistin-resistant strains compared to colistin-susceptible strainsa
| Protein mutation/name | GenBank protein accession no. | Mutations in colistin-resistant strains |
|||
|---|---|---|---|---|---|
| A1712 | A1816 | A1820 | A1828 | ||
| pmrAB mutations | |||||
| DNA-binding response regulator PmrA | WP_000161506.1 | L20F (60) | M12I (53, 54, 60) | ||
| Outer membrane proteins | |||||
| Outer membrane beta-barrel protein, OmpW family | WP_000472952.1 | L81F, N82D | L81F, N82D | ||
| OmpA family protein | WP_000777880.1 | E146fs | |||
| Outer membrane porin, OprD family | WP_171059678.1 | P86T | |||
| TonB-dependent receptor | WP_086242031.1 | V443A | |||
| Pilin pilA | WP_000993718.1 | A29Q | |||
| LPS export ABC transporter permease LptF (60) | WP_000586912.1 | R30H | |||
| ABC transporter permease | WP_000096309.1 | H196Y | A41V | ||
| Multidrug efflux MFS transporter AmvA | WP_042791420.1 | Q294L | |||
| LysE family translocator | WP_024437491.1 | G109D | A108L, G109D | A108L, G109D | |
| DMT family transporter | WP_000340904.1 | V293fs | |||
STOP codon inserted. fs, frameshift.
Additional mutations identified in colistin-resistant strains Table S3, DOCX file, 0.01 MB (15.4KB, docx) .
Copyright © 2021 Jalal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
In this study, 31 hospital-acquired MDR A. baumannii isolates were sequenced to identify the antibiotic resistance mechanisms, virulence genes, and other characteristic sequences, such as ISs, phages, and CRISPR/Cas regions. Our results show distinct patterns of distribution of antibiotic resistance and virulence genes with significant clustering of isolates based on their ST. ST1 and ST2, belonging to international clone I and II (ICI and ICII), respectively, are widely spread throughout the world and have been the cause of many hospital outbreaks in several countries (61–63), including Egypt (10, 13). Even though the prevalence of ST1 and ST2 in our study showed wide temporal distribution (Fig. 1), there was a low rate of acquired mutations across isolates belonging to the same clonal complex (Fig. 3 and 5 and Table 1). The distribution of ST113, on the other hand, was confined to late 2018 and showed a distinct pattern of resistance and virulence. Isolates belonging to ST113 did not belong to a previously identified Oxford ST and were not previously reported in Egypt.
CRAB represents a major global health threat, frequently implicated in MDR outbreaks, and was announced as a global first priority for antibiotic development by the WHO in 2017 (64). The majority of antibiotic resistance genes detected in our study are chromosomally encoded, suggesting transposon-dependent gene transfer. One of the few plasmid-associated antibiotic resistance genes detected in our isolates is the aminoglycoside adenyltransferase ant-(2″)-1a, which is harbored on plasmids pABLAC2 and pALAC4-2 and shows similar patterns of distribution across ST1. Transposon-dependent gene transfer is supported by the large number of ISs detected in all isolates, in particular, ISAba1 and ISAba125, which are frequently associated with β-lactamase genes (16, 17, 41, 65–67). Some insertion sequences were confined to the ST113 group, such as ISAba12, ISAca1, ISAba34, IS10A, ISAha2, IS1007, IS1008, and ISAcsp2. Of these, IS1007, IS1008, ISAba34, and ISAha2 were found to be associated with the sulfonamide resistance gene sul2 (68).
Underlying the wide range of antibiotic resistance exhibited by A. baumannii is the presence of antibiotic efflux pumps. Unlike other antibiotic resistance genes, efflux pumps exhibit substrate promiscuity and thus provide resistance against a large range of antibiotics. Genes encoding RND family efflux pumps are prevalent in A. baumannii (Fig. 3; 69), but their expression is tightly controlled by regulatory genes. The expression of RND antibiotic efflux pumps, adeABC, adeFGH, and adeIJK, is tightly controlled by regulatory genes adeRS, adeL, and adeN, respectively. The most clinically significant efflux pumps are adeABC, as they provide resistance to tigecycline, one of the first-line antibiotics used to treat multidrug-resistant pathogens. All of our isolates were susceptible to tigecycline, with the exception of isolates A1829 and A1705, despite similar RND gene profiles among all isolates (Fig. 3). Interruptions in adeRS by insertion sequence ISAba1 enhance overexpression of the adeABC efflux pump and decrease the susceptibility of A. baumannii to tigecycline (70, 71) and thus may account for the tigecycline resistance observed in these two isolates.
Increasing use of colistin has caused the emergence of colistin-resistant A. baumannii. Colistin resistance mechanisms, while detrimental to patient treatment, reduce the fitness and virulence of A. baumannii (72, 73). This makes colistin-resistant A. baumannii less likely to propagate between patients (74) and more likely to develop following a prior colistin exposure. Here, we report two mutations in pmrA, which have been previously reported to contribute to colistin resistance (53, 54, 60) but not in Egypt (12). We also report novel mutations in several proteins, including outer membrane proteins, such as OmpA, OprD, and AmvA. These mutations need further in vitro studies to understand the effect and mechanism of resistance.
CRAB infections are associated with the highest mortality rates (15 to 35%) among patients with hospital-acquired infections (75–77). Moreover, immune suppression, which is common during cancer therapy, is one of the major factors that negatively affect prognosis, further increasing the fatality from A. baumannii infections (78). The pathogenicity of A. baumannii strongly depends on the arsenal of virulence genes encompassed in its genome. The ability of A. baumannii to survive in vivo principally depends on immune evasion, serum resistance, and iron uptake. A subset of our isolates, belonging to ST1 and ST113, showed the absence of bap and bauA virulence genes, important for biofilm formation and acinetobactin production, respectively. Acinetobactin production, in particular bauA, was previously associated with A. baumannii persistence inside and induction of apoptosis in human alveolar epithelial cells, as well as the infection and killing of mice and Galleria mellonella larvae (50). Consistently, we observed a higher recovery rate in patients with bacteremia among isolates lacking bauA and bap, (29.4% versus 0%), suggesting reduced in vivo survival of A. baumannii. This highlights the importance of iron acquisition for A. baumannii virulence and supports the use of iron chelators/mimics as adjunct antibacterial therapy (79–81).
The worldwide spread of MDR pathogens has renewed interest in the development of novel antimicrobial therapeutic options. Developing resistance to colistin, the last resort in antibiotic therapy, limits the antimicrobial therapy options and poses a huge health care crisis. Reduced susceptibility to antibiotics is frequently overcome by using combination antimicrobial therapy. Immunization against A. baumannii provides an attractive alternative for the management of hospital-acquired outbreaks and could be particularly useful for protecting the highly vulnerable pediatric cancer patients. One of the attractive “old” treatment options is the use of bacteriophages. Susceptibility to phage therapy is largely reduced by the presence CRISPR/Cas sequences in the bacterial genome and increased in the presence of bap and ompA virulence genes (82). Understanding the genetic background of the circulating A. baumannii strains would help decide on the viability of phages as treatment options.
In conclusion, using whole-genome sequencing of MDR A. baumannii from a pediatric cancer hospital, this study revealed the presence of several lineages of A. baumannii circulating in Egypt, including a novel sequence type, ST-2246. Furthermore, through identification of the genetic determinants of multidrug resistance and virulence, we reveal a high rate of horizontal gene transmission of antibiotic resistance genes, in addition to point mutations in outer membrane proteins in individual samples causing colistin resistance. Our results highlight the importance of developing new antimicrobial agents and support the use of iron chelators/mimics as adjunct therapy, as deficient iron acquisition correlates with higher recovery. On the other hand, our results discourage the use of bacteriophage therapy due to the high prevalence of CRISPR/Cas sequences, which negatively affects the response to bacteriophage therapy.
MATERIALS AND METHODS
Bacterial isolates.
A total of 31 A. baumannii isolates were collected between October 2016 and December 2018 from hospitalized patients at CCHE 57357. Bacterial species identification was performed in duplicate, using the Vitek mass spectrometry (MS) in vitro diagnostic (IVD) system (bioMérieux; Marcy l’Étoile, France) according to the manufacturer’s instructions. The bacteria were isolated from different sources—blood (n = 16), central venous port blood (n = 6), bronchioalveolar lavage fluid (n = 6), wound (n = 1), tissue (n = 1), and pleural fluid (n = 1).
Antimicrobial susceptibility testing (AST).
AST was performed using the Vitek 2 AST cards GN222 (bioMérieux SA, Marcy l’Étoile, France) according to the manufacturer’s instructions. Interpretation of results was done according to CLSI guidelines (83).
DNA extraction.
Single colonies were subcultured in 2 ml Luria-Bertani (LB) medium (Oxoid) and incubated overnight at 37°C with shaking (MaxQ 4000 benchtop shaker; Thermo Fisher Scientific). The bacterial cultures were pelleted by centrifugation for 10 min at 5,000 × g. Bacterial DNA extraction was performed using a QIAamp DNA minikit (Qiagen, Germany) following the protocols for bacteria according to the manufacturer’s instructions and stored at −20°C until used for library preparation.
Library preparation and next-generation sequencing.
Library preparation was performed using the Nextera XT DNA library preparation kit (Illumina, USA). The DNA was prepared, fragmented, and tagged using transposome contained in the Nextera XT kit, and unique adapters were added to each sample to label it. A 12-cycle PCR was performed to amplify the tagmented DNA to add the primers and indices for dual-indexed sequencing of pooled libraries. Samples were then normalized, pooled, and subjected to 300-base paired-end read sequencing using an Illumina MiSeqDx system. The sample preparation and sequencing were performed according to the manufacturer’s protocol.
Quality control, genome assembly, and alignment.
The generated read pairs were quality filtered, and adapters were removed using fastp (84). The quality of all samples before and after filtration using fastp are summarized in Table S1. After that, the filtered reads were error corrected and merged using SPAdes BayesHammer (85) and PEAR (86), respectively. The merged reads were de novo assembled using SPAdes (85). The assessment of the assembled files was carried out with QUAST (87) as shown in Table S2.
The quality of all WGS samples before and after filtration and filtering result using fastp Table S1, DOCX file, 0.02 MB (23.7KB, docx) .
Copyright © 2021 Jalal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The evaluation of all assemblies using QUAST Table S2, DOCX file, 0.01 MB (14.4KB, docx) .
Copyright © 2021 Jalal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Variant calling, variant effect prediction, and phylogenetic tree.
Previously merged reads were mapped to the A. baumannii reference (GenBank accession number NZ_CP009257.1) using Bowtie 2 (88). Variant identification and filtration were performed using BCFtools (89), SAMtools (90) and VarScan 2 (91). The resulting variants were used to predict the variant effect using SnpEff (92), as well as consensus sequence generation using BCFtools (89). After that, the multiple sequence alignment was done using MAFFT (93), followed by maximum likelihood phylogenetic tree generation using IQ-TREE (94). The best fit model calculation was performed, and GTR+F+I+G4 was the best model. The phylogenetic tree visualization was done using T-REX (95).
Identification of the resistome, virulome, and mobile genetic elements and multilocus sequence typing (MLST).
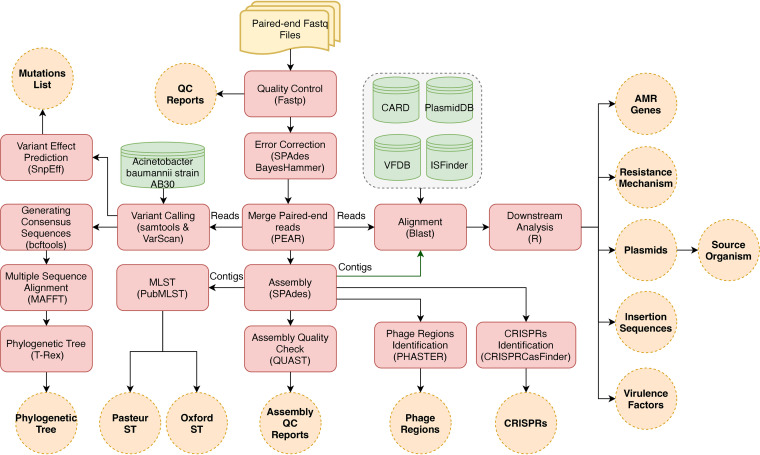
The profiling of antibiotic resistance genes and resistance mechanisms took place by aligning the reads against The Comprehensive Antibiotic Resistance Database (CARD; https://card.mcmaster.ca/) (96). Plasmid-borne virulence factors and insertion sequences were detected by aligning the reads to the Virulence Factors Database (http://www.mgc.ac.cn/VFs/) (97) and ISfinder database (https://github.com/thanhleviet/ISfinder-sequences) (98), respectively. In addition, plasmids were characterized by aligning the contigs of each sample against the database of plasmid sequences (99). All previous alignments were performed using BLAST (100). The bioinformatics pipeline used to analyze and visualize our data is illustrated in Fig. 7.
FIG 7.
Bioinformatics workflow. The workflow shows the bioinformatics pipeline used to analyze the whole-genome sequencing data generated from Illumina MiSeqDx.
The identification and visualization of the phage regions and clustered regularly interspaced short palindromic repeats (CRISPR) were performed using PHASTER (https://phaster.ca/) (101) and CRISPRCasFinder (https://crisprcas.i2bc.paris-saclay.fr/) (102), respectively. PubMLST (https://pubmlst.org/) (103) was used to determine the sequence type of each sample by uploading contigs and determining the allelic variation among seven housekeeping genes in the Oxford (cpn60, gyrB, gltA, gdhB, recA, gpi, and rpoD) and Pasteur (cpn60, fusA, gltA, pyrG, recA, rplB, and rpoB) schemes.
Downstream analysis and visualization.
The results of the alignments were imported to Rstudio (104) for further analysis. Reads with less than 90% identity or a 1e-4 E value were filtered out. The gene coverage was defined as the percentage of covered bases in each gene. Then, the gene-copy-number was calculated by dividing the number of reads aligned to each gene by its length. We set a cutoff coverage of 75%. Hence, only genes with sequencing reads covering over 75% of the length of the gene were included in any downstream analyses. The other results of the analysis were visualized in heat maps, stacked bar plots, and lollipop charts using the ggplot2 package (https://ggplot2.tidyverse.org/) (105).
Statistical analysis.
Fisher’s exact test was used to analyze the association between the presence of prophages and intact CRISPR/Cas sequences. Data with a P value of <0.05 were considered statistically significant.
Ethical approval.
Ethical approval was not required, as the isolates were collected as part of routine clinical care and patient data collection followed patient discharge from the hospital and/or death. No additional isolates were collected beyond those obtained from routine clinical care, and no diagnostic or treatment decisions were affected by the outcomes of this study.
Data availability.
All data generated and analyzed during this study are included in this article and published online on NCBI with the SRA accession number PRJNA688598.
Contributor Information
Ahmed A. Sayed, Email: ahmad.sayed@57357.org.
Paul D. Fey, University of Nebraska Medical Center
REFERENCES
- 1.Peleg AY, Seifert H, Paterson DL. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev 21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roca I, Espinal P, Vila-Farrés X, Vila J. 2012. The Acinetobacter baumannii oxymoron: commensal hospital dweller turned pan-drug-resistant menace. Front Microbiol 3:148. doi: 10.3389/fmicb.2012.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peleg AY, Paterson DL. 2006. Multidrug-resistant Acinetobacter: a threat to the antibiotic era. Intern Med J 36:479–482. doi: 10.1111/j.1445-5994.2006.01130.x. [DOI] [PubMed] [Google Scholar]
- 4.Poirel L, Hombrouck-Alet C, Freneaux C, Bernabeu S, Nordmann P. 2010. Global spread of New Delhi metallo-β-lactamase 1. Lancet Infect Dis 10:832. doi: 10.1016/S1473-3099(10)70279-6. [DOI] [PubMed] [Google Scholar]
- 5.Girija SA, Jayaseelan VP, Arumugam P. 2018. Prevalence of VIM- and GIM-producing Acinetobacter baumannii from patients with severe urinary tract infection. Acta Microbiol Immunol Hung 65:539–550. doi: 10.1556/030.65.2018.038. [DOI] [PubMed] [Google Scholar]
- 6.Brown S, Young HK, Amyes SGB. 2005. Characterisation of OXA-51, a novel class D carbapenemase found in genetically unrelated clinical strains of Acinetobacter baumannii from Argentina. Clin Microbiol Infect 11:15–23. doi: 10.1111/j.1469-0691.2004.01016.x. [DOI] [PubMed] [Google Scholar]
- 7.Brown S, Amyes SGB. 2005. The sequences of seven class D beta-lactamases isolated from carbapenem-resistant Acinetobacter baumannii from four continents. Clin Microbiol Infect 11:326–329. doi: 10.1111/j.1469-0691.2005.01096.x. [DOI] [PubMed] [Google Scholar]
- 8.Héritier C, Poirel L, Fournier P-E, Claverie J-M, Raoult D, Nordmann P. 2005. Characterization of the naturally occurring oxacillinase of Acinetobacter baumannii. Antimicrob Agents Chemother 49:4174–4179. doi: 10.1128/AAC.49.10.4174-4179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moghnieh RA, Kanafani ZA, Tabaja HZ, Sharara SL, Awad LS, Kanj SS. 2018. Epidemiology of common resistant bacterial pathogens in the countries of the Arab League. Lancet Infect Dis 18:e379–e394. doi: 10.1016/S1473-3099(18)30414-6. [DOI] [PubMed] [Google Scholar]
- 10.Al-Hassan L, Zafer MM, El-Mahallawy H. 2019. Multiple sequence types responsible for healthcare-associated Acinetobacter baumannii dissemination in a single centre in Egypt. BMC Infect Dis 19:829. doi: 10.1186/s12879-019-4433-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El Bannah AMS, Nawar NN, Hassan RMM, Salem STB. 2018. Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii in a tertiary care hospital in Egypt: clonal spread of blaOXA-23. Microb Drug Resist 24:269–277. doi: 10.1089/mdr.2017.0057. [DOI] [PubMed] [Google Scholar]
- 12.Fam NS, Gamal D, Mohamed SH, Wasfy RM, Soliman MS, El-Kholy AA, Higgins PG. 2020. Molecular characterization of carbapenem/colistin-resistant Acinetobacter baumannii clinical isolates from Egypt by whole-genome sequencing. Infect Drug Resist 13:4487–4493. doi: 10.2147/IDR.S288865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghaith DM, Zafer MM, Al-Agamy MH, Alyamani EJ, Booq RY, Almoazzamy O. 2017. The emergence of a novel sequence type of MDR Acinetobacter baumannii from the intensive care unit of an Egyptian tertiary care hospital. Ann Clin Microbiol Antimicrob 16:34. doi: 10.1186/s12941-017-0208-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aubert D, Naas T, Héritier C, Poirel L, Nordmann P. 2006. Functional characterization of IS1999, an IS4 family element involved in mobilization and expression of beta-lactam resistance genes. J Bacteriol 188:6506–6514. doi: 10.1128/JB.00375-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin H, Li T-Y, Xie M-H, Zhang Y. 2007. Characterization of the variants, flanking genes, and promoter activity of the Leifsonia xyli subsp. cynodontis insertion sequence IS1237. J Bacteriol 189:3217–3227. doi: 10.1128/JB.01403-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corvec S, Caroff N, Espaze E, Giraudeau C, Drugeon H, Reynaud A. 2003. AmpC cephalosporinase hyperproduction in Acinetobacter baumannii clinical strains. J Antimicrob Chemother 52:629–635. doi: 10.1093/jac/dkg407. [DOI] [PubMed] [Google Scholar]
- 17.Hamidian M, Hall RM. 2013. ISAba1 targets a specific position upstream of the intrinsic ampC gene of Acinetobacter baumannii leading to cephalosporin resistance. J Antimicrob Chemother 68:2682–2683. doi: 10.1093/jac/dkt233. [DOI] [PubMed] [Google Scholar]
- 18.Héritier C, Poirel L, Nordmann P. 2006. Cephalosporinase over-expression resulting from insertion of ISAba1 in Acinetobacter baumannii. Clin Microbiol Infect 12:123–130. doi: 10.1111/j.1469-0691.2005.01320.x. [DOI] [PubMed] [Google Scholar]
- 19.Iacono M, Villa L, Fortini D, Bordoni R, Imperi F, Bonnal RJP, Sicheritz-Ponten T, De Bellis G, Visca P, Cassone A, Carattoli A. 2008. Whole-genome pyrosequencing of an epidemic multidrug-resistant Acinetobacter baumannii strain belonging to the European clone II group. Antimicrob Agents Chemother 52:2616–2625. doi: 10.1128/AAC.01643-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turton JF, Ward ME, Woodford N, Kaufmann ME, Pike R, Livermore DM, Pitt TL. 2006. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol Lett 258:72–77. doi: 10.1111/j.1574-6968.2006.00195.x. [DOI] [PubMed] [Google Scholar]
- 21.Evans ME, Feola DJ, Rapp RP. 1999. Polymyxin B sulfate and colistin: old antibiotics for emerging multiresistant gram-negative bacteria. Ann Pharmacother 33:960–967. doi: 10.1345/aph.18426. [DOI] [PubMed] [Google Scholar]
- 22.Zavascki AP, Goldani LZ, Li J, Nation RL. 2007. Polymyxin B for the treatment of multidrug-resistant pathogens: a critical review. J Antimicrob Chemother 60:1206–1215. doi: 10.1093/jac/dkm357. [DOI] [PubMed] [Google Scholar]
- 23.Magiorakos A-P, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 24.Abdulzahra AT, Khalil MAF, Elkhatib WF. 2018. First report of colistin resistance among carbapenem-resistant Acinetobacter baumannii isolates recovered from hospitalized patients in Egypt. New Microbes New Infect 26:53–58. doi: 10.1016/j.nmni.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jawad A, Seifert H, Snelling AM, Heritage J, Hawkey PM. 1998. Survival of Acinetobacter baumannii on dry surfaces: comparison of outbreak and sporadic isolates. J Clin Microbiol 36:1938–1941. doi: 10.1128/JCM.36.7.1938-1941.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sánchez-Encinales V, Álvarez-Marín R, Pachón-Ibáñez ME, Fernández-Cuenca F, Pascual A, Garnacho-Montero J, Martínez-Martínez L, Vila J, Tomás MM, Cisneros JM, Bou G, Rodríguez-Baño J, Pachón J, Smani Y. 2017. Overproduction of outer membrane protein A by Acinetobacter baumannii as a risk factor for nosocomial pneumonia, bacteremia, and mortality rate increase. J Infect Dis 215:966–974. doi: 10.1093/infdis/jix010. [DOI] [PubMed] [Google Scholar]
- 27.Thummeepak R, Kongthai P, Leungtongkam U, Sitthisak S. 2016. Distribution of virulence genes involved in biofilm formation in multi-drug resistant Acinetobacter baumannii clinical isolates. Int Microbiol 19:121–129. doi: 10.2436/20.1501.01.270. [DOI] [PubMed] [Google Scholar]
- 28.Moffatt JH, Harper M, Harrison P, Hale JDF, Vinogradov E, Seemann T, Henry R, Crane B, St Michael F, Cox AD, Adler B, Nation RL, Li J, Boyce JD. 2010. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob Agents Chemother 54:4971–4977. doi: 10.1128/AAC.00834-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geisinger E, Isberg RR. 2015. Antibiotic modulation of capsular exopolysaccharide and virulence in Acinetobacter baumannii. PLoS Pathog 11:e1004691. doi: 10.1371/journal.ppat.1004691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwon S-O, Gho YS, Lee JC, Kim SI. 2009. Proteome analysis of outer membrane vesicles from a clinical Acinetobacter baumannii isolate. FEMS Microbiol Lett 297:150–156. doi: 10.1111/j.1574-6968.2009.01669.x. [DOI] [PubMed] [Google Scholar]
- 31.Jun SH, Lee JH, Kim BR, Kim S, Park TI, Lee JC, Lee YC. 2013. Acinetobacter baumannii outer membrane vesicles elicit a potent innate immune response via membrane proteins. PLoS One 8:e71751. doi: 10.1371/journal.pone.0071751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA 95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harris SR, Cartwright EJP, Török ME, Holden MTG, Brown NM, Ogilvy-Stuart AL, Ellington MJ, Quail MA, Bentley SD, Parkhill J, Peacock SJ. 2013. Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: a descriptive study. Lancet Infect Dis 13:130–136. doi: 10.1016/S1473-3099(12)70268-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis T, Loman NJ, Bingle L, Jumaa P, Weinstock GM, Mortiboy D, Pallen MJ. 2010. High-throughput whole-genome sequencing to dissect the epidemiology of Acinetobacter baumannii isolates from a hospital outbreak. J Hosp Infect 75:37–41. doi: 10.1016/j.jhin.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 35.Snitkin ES, Zelazny AM, Thomas PJ, Stock F, Henderson DK, Palmore TN, Segre JA, NISC Comparative Sequencing Program Group . 2012. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med 4:148ra116. doi: 10.1126/scitranslmed.3004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J, Pallen MJ. 2012. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol 30:434–439. doi: 10.1038/nbt.2198. [DOI] [PubMed] [Google Scholar]
- 37.Gaiarsa S, Batisti Biffignandi G, Esposito EP, Castelli M, Jolley KA, Brisse S, Sassera D, Zarrilli R. 2019. Comparative analysis of the two Acinetobacter baumannii multilocus sequence typing (MLST) schemes. Front Microbiol 10:930. doi: 10.3389/fmicb.2019.00930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salgado-Camargo AD, Castro-Jaimes S, Gutierrez-Rios R-M, Lozano LF, Altamirano-Pacheco L, Silva-Sanchez J, Pérez-Oseguera Á, Volkow P, Castillo-Ramírez S, Cevallos MA. 2020. Structure and evolution of Acinetobacter baumannii plasmids. Front Microbiol 11:1283. doi: 10.3389/fmicb.2020.01283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jeon JH, Lee JH, Lee JJ, Park KS, Karim AM, Lee C-R, Jeong BC, Lee SH. 2015. Structural basis for carbapenem-hydrolyzing mechanisms of carbapenemases conferring antibiotic resistance. Int J Mol Sci 16:9654–9692. doi: 10.3390/ijms16059654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Figueiredo S, Poirel L, Croize J, Recule C, Nordmann P. 2009. In vivo selection of reduced susceptibility to carbapenems in Acinetobacter baumannii related to ISAba1-mediated overexpression of the natural bla(OXA-66) oxacillinase gene. Antimicrob Agents Chemother 53:2657–2659. doi: 10.1128/AAC.01663-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X, Zong Z, Lü X. 2011. Tn2008 is a major vehicle carrying bla(OXA-23) in Acinetobacter baumannii from China. Diagn Microbiol Infect Dis 69:218–222. doi: 10.1016/j.diagmicrobio.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 42.Li X-Z, Plésiat P, Nikaido H. 2015. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin Microbiol Rev 28:337–418. doi: 10.1128/CMR.00117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smani Y, McConnell MJ, Pachón J. 2012. Role of fibronectin in the adhesion of Acinetobacter baumannii to host cells. PLoS One 7:e33073. doi: 10.1371/journal.pone.0033073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi CH, Lee JS, Lee YC, Park TI, Lee JC. 2008. Acinetobacter baumannii invades epithelial cells and outer membrane protein A mediates interactions with epithelial cells. BMC Microbiol 8:216. doi: 10.1186/1471-2180-8-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alav I, Sutton JM, Rahman KM. 2018. Role of bacterial efflux pumps in biofilm formation. J Antimicrob Chemother 73:2003–2020. doi: 10.1093/jac/dky042. [DOI] [PubMed] [Google Scholar]
- 46.Tomaras AP, Dorsey CW, Edelmann RE, Actis LA. 2003. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology (Reading) 149:3473–3484. doi: 10.1099/mic.0.26541-0. [DOI] [PubMed] [Google Scholar]
- 47.Choi AHK, Slamti L, Avci FY, Pier GB, Maira-Litrán T. 2009. The pgaABCD locus of Acinetobacter baumannii encodes the production of poly-beta-1–6-N-acetylglucosamine, which is critical for biofilm formation. J Bacteriol 191:5953–5963. doi: 10.1128/JB.00647-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Camarena L, Bruno V, Euskirchen G, Poggio S, Snyder M. 2010. Molecular mechanisms of ethanol-induced pathogenesis revealed by RNA-sequencing. PLoS Pathog 6:e1000834. doi: 10.1371/journal.ppat.1000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jacobs AC, Hood I, Boyd KL, Olson PD, Morrison JM, Carson S, Sayood K, Iwen PC, Skaar EP, Dunman PM. 2010. Inactivation of phospholipase D diminishes Acinetobacter baumannii pathogenesis. Infect Immun 78:1952–1962. doi: 10.1128/IAI.00889-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gaddy JA, Arivett BA, McConnell MJ, López-Rojas R, Pachón J, Actis LA. 2012. Role of acinetobactin-mediated iron acquisition functions in the interaction of Acinetobacter baumannii strain ATCC 19606T with human lung epithelial cells, Galleria mellonella caterpillars, and mice. Infect Immun 80:1015–1024. doi: 10.1128/IAI.06279-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Niu C, Clemmer KM, Bonomo RA, Rather PN. 2008. Isolation and characterization of an autoinducer synthase from Acinetobacter baumannii. J Bacteriol 190:3386–3392. doi: 10.1128/JB.01929-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tomaras AP, Flagler MJ, Dorsey CW, Gaddy JA, Actis LA. 2008. Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiology (Reading) 154:3398–3409. doi: 10.1099/mic.0.2008/019471-0. [DOI] [PubMed] [Google Scholar]
- 53.Olaitan AO, Morand S, Rolain J-M. 2014. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol 5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Charretier Y, Diene SM, Baud D, Chatellier S, Santiago-Allexant E, van Belkum A, Guigon G, Schrenzel J. 2018. Colistin heteroresistance and involvement of the PmrAB regulatory system in Acinetobacter baumannii. Antimicrob Agents Chemother 62:e00788-18. doi: 10.1128/AAC.00788-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adams MD, Nickel GC, Bajaksouzian S, Lavender H, Murthy AR, Jacobs MR, Bonomo RA. 2009. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob Agents Chemother 53:3628–3634. doi: 10.1128/AAC.00284-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sepahvand S, Doudi M, Davarpanah MA, Bahador A, Ahmadi M. 2016. Analyzing pmrA and pmrB genes in Acinetobacter baumannii resistant to colistin in Shahid Rajai Shiraz, Iran Hospital by PCR: first report in Iran. Pak J Pharm Sci 29:1401–1406. [PubMed] [Google Scholar]
- 57.Marano V, Marascio N, Pavia G, Lamberti AG, Quirino A, Musarella R, Casalinuovo F, Mazzitelli M, Trecarichi EM, Torti C, Matera G, Liberto MC. 2020. Identification of pmrB mutations as putative mechanism for colistin resistance in A. baumannii strains isolated after in vivo colistin exposure. Microb Pathog 142:104058. doi: 10.1016/j.micpath.2020.104058. [DOI] [PubMed] [Google Scholar]
- 58.Hameed F, Khan MA, Muhammad H, Sarwar T, Bilal H, Rehman TU. 2019. Plasmid-mediated mcr-1 gene in Acinetobacter baumannii and Pseudomonas aeruginosa: first report from Pakistan. Rev Soc Bras Med Trop 52:e20190237. doi: 10.1590/0037-8682-0237-2019. [DOI] [PubMed] [Google Scholar]
- 59.Martins-Sorenson N, Snesrud E, Xavier DE, Cacci LC, Iavarone AT, McGann P, Riley LW, Moreira BM. 2020. A novel plasmid-encoded mcr-4.3 gene in a colistin-resistant Acinetobacter baumannii clinical strain. J Antimicrob Chemother 75:60–64. doi: 10.1093/jac/dkz413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mustapha MM, Li B, Pacey MP, Mettus RT, McElheny CL, Marshall CW, Ernst RK, Cooper VS, Doi Y. 2018. Phylogenomics of colistin-susceptible and resistant XDR Acinetobacter baumannii. J Antimicrob Chemother 73:2952–2959. doi: 10.1093/jac/dky290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karah N, Sundsfjord A, Towner K, Samuelsen Ø. 2012. Insights into the global molecular epidemiology of carbapenem non-susceptible clones of Acinetobacter baumannii. Drug Resist Updat 15:237–247. doi: 10.1016/j.drup.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 62.Matsui M, Suzuki M, Suzuki M, Yatsuyanagi J, Watahiki M, Hiraki Y, Kawano F, Tsutsui A, Shibayama K, Suzuki S. 2018. Distribution and molecular characterization of Acinetobacter baumannii international clone II lineage in Japan. Antimicrob Agents Chemother 62:e02190-17. doi: 10.1128/AAC.02190-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Warner WA, Kuang SN, Hernandez R, Chong MC, Ewing PJ, Fleischer J, Meng J, Chu S, Terashita D, English L, Chen W, Xu HH. 2016. Molecular characterization and antimicrobial susceptibility of Acinetobacter baumannii isolates obtained from two hospital outbreaks in Los Angeles County, California, USA. BMC Infect Dis 16:194. doi: 10.1186/s12879-016-1526-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, WHO Pathogens Priority List Working Group . 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 65.Nigro S, Hall RM. 2015. Distribution of the blaOXA-23-containing transposons Tn2006 and Tn2008 in Australian carbapenem-resistant Acinetobacter baumannii isolates. J Antimicrob Chemother 70:2409–2411. doi: 10.1093/jac/dkv102. [DOI] [PubMed] [Google Scholar]
- 66.Adams-Haduch JM, Paterson DL, Sidjabat HE, Pasculle AW, Potoski BA, Muto CA, Harrison LH, Doi Y. 2008. Genetic basis of multidrug resistance in Acinetobacter baumannii clinical isolates at a tertiary medical center in Pennsylvania. Antimicrob Agents Chemother 52:3837–3843. doi: 10.1128/AAC.00570-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lopes BS, Amyes SGB. 2012. Role of ISAba1 and ISAba125 in governing the expression of blaADC in clinically relevant Acinetobacter baumannii strains resistant to cephalosporins. J Med Microbiol 61:1103–1108. doi: 10.1099/jmm.0.044156-0. [DOI] [PubMed] [Google Scholar]
- 68.Hamidian M, Ambrose SJ, Hall RM. 2016. A large conjugative Acinetobacter baumannii plasmid carrying the sul2 sulphonamide and strAB streptomycin resistance genes. Plasmid 87–88:43–50. doi: 10.1016/j.plasmid.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 69.Magnet S, Courvalin P, Lambert T. 2001. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob Agents Chemother 45:3375–3380. doi: 10.1128/AAC.45.12.3375-3380.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lopes BS, Amyes SGB. 2013. Insertion sequence disruption of adeR and ciprofloxacin resistance caused by efflux pumps and gyrA and parC mutations in Acinetobacter baumannii. Int J Antimicrob Agents 41:117–121. doi: 10.1016/j.ijantimicag.2012.08.012. [DOI] [PubMed] [Google Scholar]
- 71.Ruzin A, Keeney D, Bradford PA. 2007. AdeABC multidrug efflux pump is associated with decreased susceptibility to tigecycline in Acinetobacter calcoaceticus-Acinetobacter baumannii complex. J Antimicrob Chemother 59:1001–1004. doi: 10.1093/jac/dkm058. [DOI] [PubMed] [Google Scholar]
- 72.Da Silva GJ, Domingues S. 2017. Interplay between colistin resistance, virulence and fitness in Acinetobacter baumannii. Antibiot (Basel) 6:28. doi: 10.3390/antibiotics6040028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.López-Rojas R, Domínguez-Herrera J, McConnell MJ, Docobo-Peréz F, Smani Y, Fernández-Reyes M, Rivas L, Pachón J. 2011. Impaired virulence and in vivo fitness of colistin-resistant Acinetobacter baumannii. J Infect Dis 203:545–548. doi: 10.1093/infdis/jiq086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Beceiro A, Moreno A, Fernández N, Vallejo JA, Aranda J, Adler B, Harper M, Boyce JD, Bou G. 2014. Biological cost of different mechanisms of colistin resistance and their impact on virulence in Acinetobacter baumannii. Antimicrob Agents Chemother 58:518–526. doi: 10.1128/AAC.01597-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Le Hello S, Falcot V, Lacassin F, Mikulski M, Baumann F. 2010. Risk factors for carbapenem-resistant Acinetobacter baumannii infections at a tertiary care hospital in New Caledonia, South Pacific. Scand J Infect Dis 42:821–826. doi: 10.3109/00365548.2010.496087. [DOI] [PubMed] [Google Scholar]
- 76.Yang S, Sun J, Wu X, Zhang L. 2018. Determinants of mortality in patients with nosocomial Acinetobacter baumannii bacteremia in Southwest China: a five-year case-control study. Can J Infect Dis Med Microbiol 2018:3150965. doi: 10.1155/2018/3150965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xiao D, Wang L, Zhang D, Xiang D, Liu Q, Xing X. 2017. Prognosis of patients with Acinetobacter baumannii infection in the intensive care unit: a retrospective analysis. Exp Ther Med 13:1630–1633. doi: 10.3892/etm.2017.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Du X, Xu X, Yao J, Deng K, Chen S, Shen Z, Yang L, Feng G. 2019. Predictors of mortality in patients infected with carbapenem-resistant Acinetobacter baumannii: a systematic review and meta-analysis. Am J Infect Control 47:1140–1145. doi: 10.1016/j.ajic.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 79.Arivett BA, Fiester SE, Ohneck EJ, Penwell WF, Kaufman CM, Relich RF, Actis LA. 2015. Antimicrobial activity of gallium protoporphyrin IX against Acinetobacter baumannii strains displaying different antibiotic resistance phenotypes. Antimicrob Agents Chemother 59:7657–7665. doi: 10.1128/AAC.01472-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Antunes LCS, Imperi F, Minandri F, Visca P. 2012. In vitro and in vivo antimicrobial activities of gallium nitrate against multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 56:5961–5970. doi: 10.1128/AAC.01519-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thompson MG, Corey BW, Si Y, Craft DW, Zurawski DV. 2012. Antibacterial activities of iron chelators against common nosocomial pathogens. Antimicrob Agents Chemother 56:5419–5421. doi: 10.1128/AAC.01197-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Leungtongkam U, Thummeepak R, Kitti T, Tasanapak K, Wongwigkarn J, Styles KM, Wellington EMH, Millard AD, Sagona AP, Sitthisak S. 2020. Genomic analysis reveals high virulence and antibiotic resistance amongst phage susceptible Acinetobacter baumannii. Sci Rep 10:16154. doi: 10.1038/s41598-020-73123-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Weinstein MP, Lewis JS, II.. 2020. The Clinical and Laboratory Standards Institute Subcommittee on Antimicrobial Susceptibility Testing: background, organization, functions, and processes. J Clin Microbiol 58:e01864-19. doi: 10.1128/JCM.01864-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang J, Kobert K, Flouri T, Stamatakis A. 2014. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. 2012. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Makarenkov V. 2001. T-REX: reconstructing and visualizing phylogenetic trees and reticulation networks. Bioinformatics 17:664–668. doi: 10.1093/bioinformatics/17.7.664. [DOI] [PubMed] [Google Scholar]
- 96.McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, Bhullar K, Canova MJ, De Pascale G, Ejim L, Kalan L, King AM, Koteva K, Morar M, Mulvey MR, O’Brien JS, Pawlowski AC, Piddock LJV, Spanogiannopoulos P, Sutherland AD, Tang I, Taylor PL, Thaker M, Wang W, Yan M, Yu T, Wright GD. 2013. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother 57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen L, Yang J, Yu J, Yao Z, Sun L, Shen Y, Jin Q. 2005. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res 33:D325–D328. doi: 10.1093/nar/gki008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Brooks L, Kaze M, Sistrom M. 2019. A curated, comprehensive database of plasmid sequences. Microbiol Resour Announc 8:e01325-18. doi: 10.1128/MRA.01325-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 101.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, Wishart DS. 2016. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Couvin D, Bernheim A, Toffano-Nioche C, Touchon M, Michalik J, Néron B, Rocha EPC, Vergnaud G, Gautheret D, Pourcel C. 2018. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res 46:W246–W251. doi: 10.1093/nar/gky425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jolley KA, Bray JE, Maiden MCJ. 2018. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Racine JS. 2012. RStudio: a platform-independent IDE for R and Sweave. J Appl Econ 27:167–172. doi: 10.1002/jae.1278. [DOI] [Google Scholar]
- 105.Wickham H. 2011. ggplot2. WIREs Comp Stat 3:180–185. doi: 10.1002/wics.147. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Heat map showing the presence or absence of each antibiotic resistance gene as detected by the ResFinder database. Dark green shows the presence of the antibiotic resistance gene, while light green indicates its absence. Download FIG S1, TIF file, 1.8 MB (1.8MB, tif) .
Copyright © 2021 Jalal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Additional mutations identified in colistin-resistant strains Table S3, DOCX file, 0.01 MB (15.4KB, docx) .
Copyright © 2021 Jalal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The quality of all WGS samples before and after filtration and filtering result using fastp Table S1, DOCX file, 0.02 MB (23.7KB, docx) .
Copyright © 2021 Jalal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The evaluation of all assemblies using QUAST Table S2, DOCX file, 0.01 MB (14.4KB, docx) .
Copyright © 2021 Jalal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
All data generated and analyzed during this study are included in this article and published online on NCBI with the SRA accession number PRJNA688598.